Abstract
Immune cells are known to be critical for successful limb regeneration in the axolotl (Ambystoma mexicanum), but many details regarding their identity, behavior, and function are yet to be resolved. We isolated peripheral leukocytes from the blood of adult axolotls and then created two samples for single-cell sequencing: 1) peripheral leukocytes (N = 7,889) and 2) peripheral leukocytes with presumptive macrophages from the intraperitoneal cavity (N = 4,998). Using k-means clustering, we identified 6 cell populations from each sample that presented gene expression patterns indicative of erythrocyte, thrombocyte, neutrophil, B-cell, T-cell, and myeloid cell populations. A seventh, presumptive macrophage cell population was identified uniquely from sample 2. We then isolated cells from amputated axolotl limbs at 1 and 6 days post-amputation (DPA) and performed single cell sequencing (N = 8,272 and 9,906 cells respectively) to identify immune and non-immune cell populations. Using k-means clustering, we identified 8 cell populations overall, with the majority of cells expressing erythrocyte-specific genes. Even though erythrocytes predominated, we used an unbiased approach to identify infiltrating neutrophil, macrophage, and lymphocyte populations at both time points. Additionally, populations expressing genes for epidermal cells, fibroblast-like cells, and endothelial cells were also identified. Consistent with results from previous experimental studies, neutrophils were more abundant at 1 DPA than 6 DPA, while macrophages and non-immune cells exhibited inverse abundance patterns. Of note, we identified a small population of fibroblast-like cells at 1 DPA that was represented by considerably more cells at 6 DPA. We hypothesize that these are early progenitor cells that give rise to the blastema. The enriched gene sets from our work will aid future single-cell investigations of immune cell diversity and function during axolotl limb regeneration.
Keywords: limb regeneration, axolotl, single-cell, 10X genomics, gene expression
1. Introduction
Axolotls (Ambystoma mexicanum) provide powerful models for studies of tissue regeneration. The regenerative ability of axolotls is often associated with the production of a blastema, a specialized tissue comprised of progenitor cells that reforms damaged tissues and whole organs after amputation (Goss, 1969). However, biological processes that are activated very early after injury and before blastema formation are essential for successful regeneration, including innate responses of immune cells. For example, macrophages are activated within an hour after laser ablation of neurosensory neuromasts in the axolotl tail and their activities are associated with clearance of debris by phagocytosis (Jones and Corwin, 1996). Additionally, macrophage depletion during axolotl limb and heart regeneration yields an atypical extracellular matrix and regenerative failure in both cases (Godwin et al 2013; 2017a). These and other studies have clearly established that immune cells are essential for successful tissue regeneration (reviewed by Godwin et al 2017b; Julier et al 2017; Mescher et al 2017); however, much remains to be learned about their identities, behaviors, and functions.
Two recent studies of limb regeneration using single-cell transcript sequencing identified cells that presented gene expression signatures consistent with immune cells (Gerber et al 2018; Leigh et al 2018). Single-cell transcript sequencing quantifies the abundances of mRNAs for hundreds to thousands of genes and these data can be used to identify and classify cell populations into different cell types. The power of single-cell sequencing to resolve the identity of specific cells within a complex tissue depends upon their numerical abundance and sampling efficiency. To date, there has been no attempt to enrich for immune cells in the highly regenerative axolotl and characterize cell types by single cell sequencing. Gerber et al (2018) profiled cells from intact, uninjured limb and reported the discovery of immune cells but did not attempt to classify these into cell types. Leigh et al (2018) sequenced cells that were isolated from uninjured limbs and at three different stages of limb regeneration: wound healing, early bud, and medium bud. They discovered immune cell populations at all three stages and used differentially expressed genes to classify these into various cell types, including B-cells, T-cells, macrophages, and neutrophils. While this study showed that immune cell types are present in the limb stump after injury, additional studies are needed to detail peripheral leukocyte diversity and investigate the recruitment of various immune cell types to the injury site.
Here we used single-cell sequencing and gene expression signatures to identify immune cell types from the blood, a likely tissue source from which immune cells are recruited. We also enriched for macrophages by collecting cells from the intraperitoneal (IP) cavity, a well-known reservoir of macrophages in most vertebrates that mitigate pathogens and repair damaged internal organs (Wang and Kubes 2016). We also performed single-cell sequencing to document the presence of the identified immune cell types for two regeneration time points (1 and 6 days post-amputation - DPA). Our results provide greater resolution of immune cell types and temporal changes in cell abundance during early regeneration. We also detail non-immunological cell types, including the identity of a fibroblast-like cell population that may contribute progenitor cells to the early blastema.
2. Methods and Materials
2.1. Animal Care and Use
Standard and ethical animal husbandry and use protocols in this study were approved by the Institutional Animal Care and Use Committee (IACUC) at University of Kentucky (Protocol Number 2017–2580). Animals were obtained from the Ambystoma Genetic Stock Center (RRID:SCR_006372).
2.2. Single Cell Preparation from Axolotl Peripheral Leukocytes and Limb Tissues
To create a peripheral leukocyte sample for single cell sequencing, an adult female axolotl (RRID:AGSC_110AF) was anesthetized in a benzocaine solution (0.2 g / L). Approximately 300 μL of blood was collected from the femoral vein with an insulin syringe and then placed into 1 mL of 50 mM EDTA in 0.8X DPBS + 2% BSA. The blood sample was layered on an 85% Percoll solution and centrifuged at 500 g for 5 minutes at room temperature. The interface was collected and washed with 5 mL 0.8X DPBS + 2% BSA. Leukocytes were pelleted at 500 g for 5 minutes at 4° C and resuspended in 200 μL 0.8X DPBS and filtered through a 40 μm cell strainer. Cells were manually counted using a hemocytometer.
Three individually enriched samples of neutrophils, monocytes, and macrophages were isolated and then combined to create a second sample for single-cell sequencing. This was accomplished according to the following method. Peripheral blood was collected as described above from two female adult axolotls. The first blood sample was layered on top of an 80/85% discontinuous Percoll gradient for the enrichment of neutrophils, and the second blood sample was layered on top of a 75/80% discontinuous Percoll gradient for the enrichment of monocytes. These gradients were centrifuged at 500 g for 15 minutes at room temperature. Both interfaces were separately collected and independently washed with 5 mL 0.8X DPBS + 2% BSA. Cells were resuspended in 100–200 μL 0.8X DPBS + 2% BSA and roughly 5000 cells were spun in a Cytospin centrifuge at 600 rpm for 4 minutes onto slides and stained with hematoxylin to identify populations that were enriched for each target cell type. These analyses showed that the 80% interface from the first sample was enriched for neutrophils and the 80% interface from the second sample was enriched for monocytes. For a macrophage-enriched sample, the peritoneal cavity of an adult female axolotl was lavaged with 10 mL of 0.8X PBS. Isolated peritoneal cells were added to 5 mL of 0.8X PBS/50 mM EDTA/2% BSA. The cells were centrifuged at 500 g for 5 minutes at 4° C and resuspended in 1 mL 0.8X DPBS. Equal cell numbers of neutrophil-enriched, monocyte-enriched, and macrophage-enriched cell populations were then pooled to create a sample for single-cell sequencing.
Two adult female axolotls were anesthetized in benzocaine (0.2 g / L) and administered amputations through the forearm. Each of these axolotls contributed a single 1 mm distal stump tissue sample, one at 1 DPA and the other at 6 DPA. Tissues were rinsed twice with 500 μL 0.8X DPS (Mg2+/ Ca2+ free) to reduce peripheral blood cell contamination. Tissues were transferred to 500 μL digestion buffer (0.175 mg / mL Liberase in 0.8X DPBS with 0.1U / mL of DNAse I (NEB)) and cut into small pieces then placed on a rocker at room temperature for 15 minutes. Cells and tissue pieces were filtered using a 100 μm cell strainer and a 1 mL syringe plunger was used to further mechanically dissociate tissues through the strainer. The strainer was washed three times with 1 mL of 0.8X DPBS + 2% BSA and the single-cell eluate was filtered through a 40 μm strainer. To prevent clogging of the 10X Genomics chip, extracellular matrix (ECM) debris in the 1 dpa sample required centrifugation at 500 g for 5 minutes over a 40% Percoll solution. This helped remove ECM as well as dead cells from the sample (as detected with propidium iodide staining). The pelleted cells were then washed, centrifuged, and resuspended with 200 μL 0.8X DPBS + 2% BSA. Cells were concentrated through centrifugation at 500 g and resuspended in 50–100 μL of 0.8X DPBS and filtered again through a 40 um cell strainer. Cells were manually counted using a hemocytometer. Cells were kept on ice until performing single cell sequencing.
2.3. Single-cell sequencing analysis
Single cell libraries were generated using the Chromium Single Cell 3’ RNAseq kit (v2 Chemistry) using manufacturer recommended protocols and targeting 10,000 cells per library. The resulting libraries were sequenced on an illumina NovaSeq 6000 (2×150nt reads using S4 chemistry) to generate >1 billion read triplets per library (forward, reverse and index reads) and demultiplexed with the mkfastq option of Cell Ranger 2.1.1. The exact number of read triplets for each study were: peripheral leukocytes (N = 1,041,310,488), peripheral leukocytes and macrophages (N = 1,061,876,582); 1 day post amputation limb stump (N = 1,146,707,096) and 6 day post amputation limb stump (N = 1,135,967,095). Assignment of barcodes to cells and transcript quantification were performed using Cell Ranger 3.0.2 and default alignment and cell assignment parameters. Quality control and sampling metrics indicated that each library sampled several thousand cells and several thousand unique molecules within these cells, with high sequence saturation (Table 1). For consistency with other studies, we employed a previously reported reference transcriptome (Gerber et al 2018) for gene annotation. K-means clustering was used to identify distinct cell populations and Loupe Cell Browser 3.0.1 was used to create t-SNE plots. Genes that were significantly enriched (Benjamani-Hochberg adjusted p < 0.05) within cell populations were used to annotate cell types. The data from this project were submitted to NCBI Gene Expression Omnibus for public release upon publication.
Table 1.
Quality control and sampling metrics for single cell sequencing.
| Sample | Estimated Number of Cells | Mean Reads/Cell | Median Genes/Cell | Median UMI Counts/Cell |
|---|---|---|---|---|
| Peripheral Leukocytes | 7,889 | 131,995 | 1,008 | 3,610 |
| Peripheral Leukocytes + Macrophages | 4,988 | 212,886 | 1,530 | 13,671 |
| One Day Post Amputation | 8,272 | 138,625 | 573 | 4,231 |
| Six Days Post Amputation | 9,906 | 114,674 | 646 | 3,977 |
3. Results
3.1. Characterization of Immune Cells from Axolotl Blood
Peripheral leukocytes were isolated from adult female axolotls to create two samples. Sample 1 contained a mixture of leukocytes and thrombocytes isolated from whole blood using density-gradient centrifugation, and Sample 2 contained enriched fractions of neutrophils (21%) and monocytes (16%) from whole blood, and presumptive macrophages (74%) that were isolated from the IP cavity. Because enrichment for neutrophils and monocytes was low, we observed several different immune cell types in Sample 2, including neutrophils, monocytes, and macrophages (Figure 1)(Supplemental Figure 1).
Figure 1.
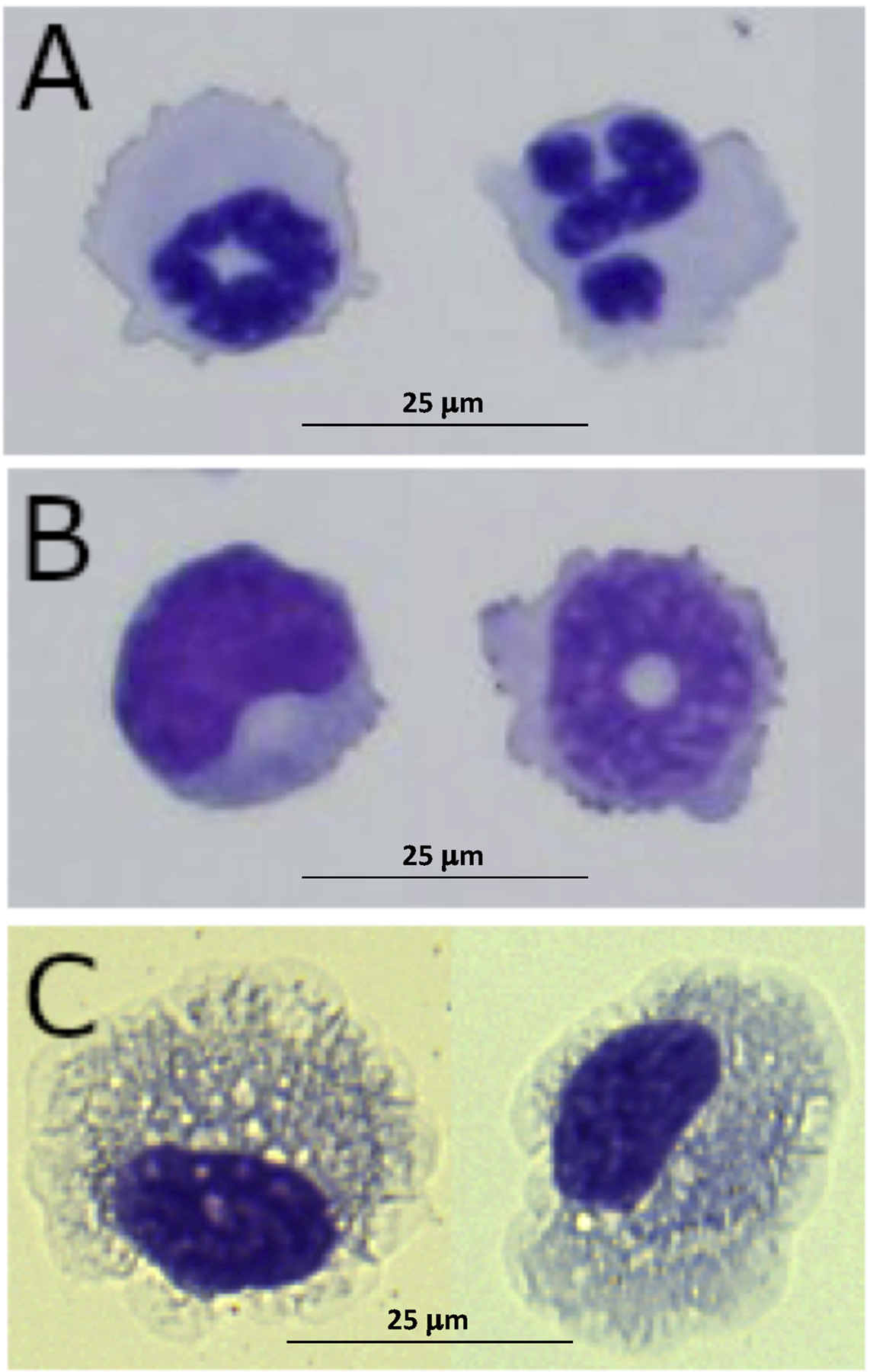
Representative figures of A) neutrophils, B) monocytes, and C) macrophages from enriched fractions that were combined to make Sample 2.
3.2. Single-Cell Sequencing of Peripheral Immune Cells
Cells from Sample 1 (N = 7,889) and Sample 2 (N = 4,988) were isolated and used to profile gene expression using the 10x Genomics platform. Using K-means clustering of genes, 7 cell populations were identified that were supported by 25 or more significantly up-regulated genes (Benjamani-Hochberg adjusted p < 0.05) in comparison to other populations (Supplemental Table 1). Cells from Samples 1 and 2 contributed to all but population 6, which was only comprised of cells from Sample 2. Thus, population 6 likely contained macrophages from the IP cavity (Wang and Kubes 2016) as this cell isolate was only included in Sample 2 and expresses several biomarkers that are considered to be diagnostic of macrophages (below).
We examined significantly enriched genes within populations and then searched NCBI databases including PubMed to identify genes that could be used to assign cell type identities to each population (Figure 2). In particular, we looked for genes that encoded cell-specific structural proteins, enzymes, and transcription factors. We also considered genes that were used by Leigh et al (2018) to classify immune cells in their single-cell study. We note that some of the most highly enriched genes within the populations below are anonymous with respect to gene identity at this stage of axolotl genome resource development, and thus our ability to characterize cellular diversity is conservative.
Figure 2.
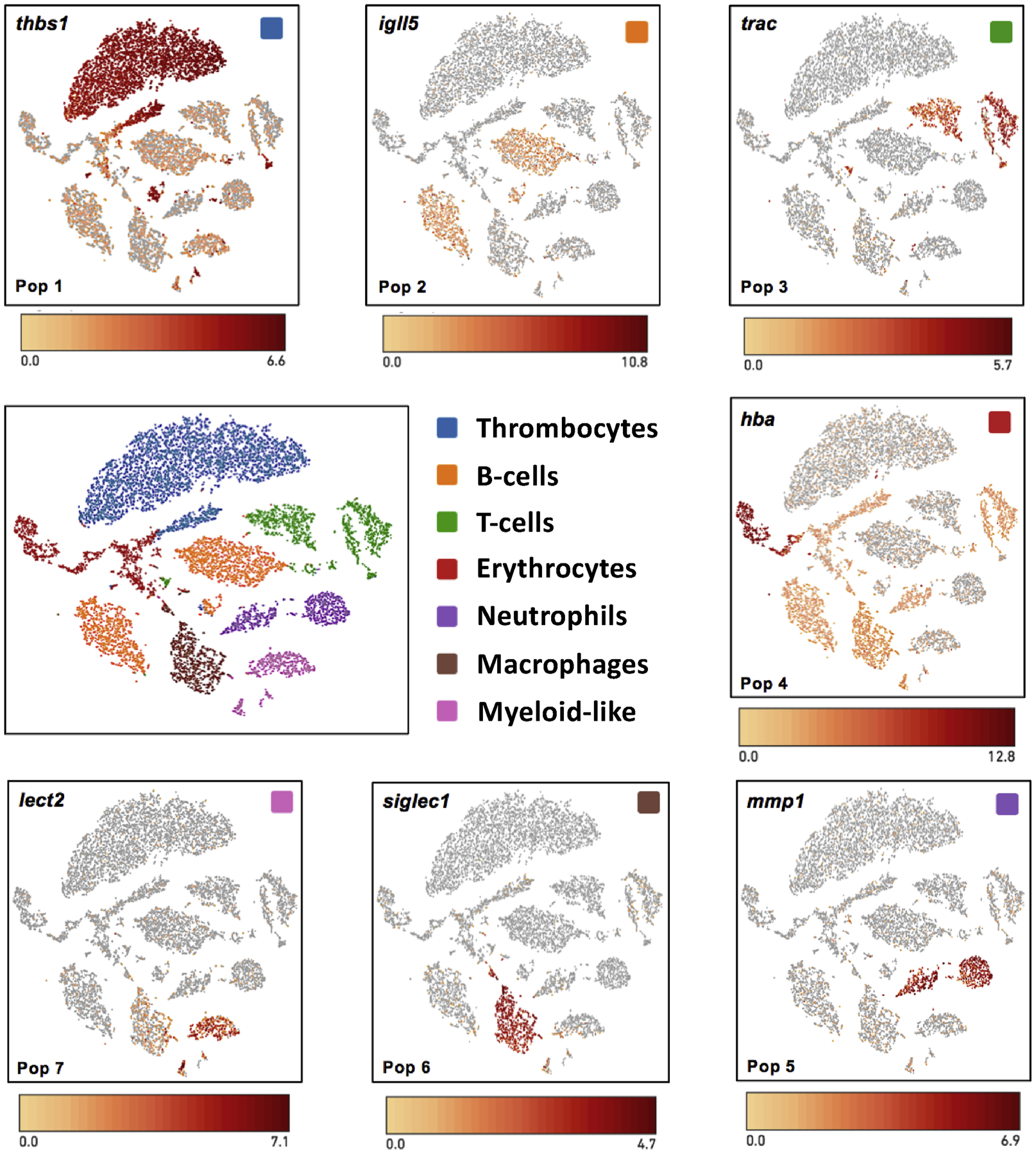
t-SNE plots showing cell populations that were identified by k-means clustering of transcripts. Example gene expression markers are shown for cell types isolated from blood and the intraperitoneal cavity.
Peripheral Population 1 (N = 4,513; Sample 1 = 4,126; Sample 2 = 387) Thrombocytes.
Several genes that are specifically expressed by thrombocytes were significantly enriched among cells in population 1. These encode GPIb-V-IX system proteins (gp1ba, gp1bb, gp9) and other proteins (gucy1b3, thbs1) that function in the regulation of platelet adhesion (Dixit et al 1985; Ignarro 1990; Savage et al 1996).
Peripheral Population 2 (N = 2,604; Sample 1 = 1480; Sample 2 = 1,124) B-cells.
This population was characterized by a large number of significantly enriched genes that encode ribosomal proteins and two immunoglobulin loci (igll5 and igj). Leigh et al (2018) annotated a cell population with these expression patterns as early B-cells.
Peripheral Population 3 (N = 1,754; Sample 1 = 888; Sample 2 = 886) T-cells.
The alpha chain component of the T-cell receptor (trac) and a serine peptidase (gzmb) that functions in cytotoxic T-cell mediated apoptosis (Trapani 1996) and wound healing processes (Shen et al 2018) were significantly enriched in this population. Leigh et al (2018) used trac as a biomarker of T-cells.
Peripheral Population 4 (N = 1,339; Sample 1 = 228; Sample 2 =1,111) Erythrocytes.
Multiple hemoglobin genes (hbb, hba, hbg1) were up-regulated in this population, as well as alas2 which encodes an erythroid-specific, mitochondrially located enzyme that catalyzes the first step in the heme biosynthetic pathway (Bishop et al 1990).
Peripheral Population 5 (N = 991; Sample 1 = 580; Sample 2 = 411) Neutrophils.
Several genes expressed in vertebrate neutrophils were significantly enriched in this population, including mmp1 and cebpe, an essential transcription factor for neutrophil differentiation in mammalian cells (Bartels et al 2015). Several other genes (camp, mmp2, chit1) that were highly expressed in this population were identified as significantly enriched in a neutrophil population by Leigh et al (2018). This population may contain other granulocytes as eosinophil peroxidase (epx) was also enriched.
Peripheral Population 6 (N = 983; Sample 1 = 0; Sample 2 = 983) Macrophages.
This population was only observed in Sample 2 and thus presumably corresponded to macrophages isolated from the IP cavity. In support of this idea, two genes (fabp1, siglec1) that are expressed in mammalian macrophages (Schachtrup et al 2004; Roos et al 2019) were significantly enriched in this population. Other highly expressed genes (marco, c1qb, c1qc) were identified as biomarkers of recruited macrophages by Leigh et al (2018).
Peripheral Population 7 (N = 693; Sample 1 =587; Sample 2 = 106) Myeloid-like.
A diversity of immune cell gene expression markers were observed for this population, suggesting that it may contain monocytes that were enriched in Sample 2. Conservatively, we refer to this population of cells as myeloid-like. Two genes (cybb, cyba) associated with the microbicidal oxidase system of phagocytes, including macrophages and neutrophils (Dale et al 2008), were significantly enriched in this population along with gpx1, which was identified from neutrophils, macrophages, and dendritic cells by Leigh et al (2018). The most highly enriched gene (lect2) was identified as a highly abundant protein in zebrafish neutrophils (Singh et al 2013). As was observed for neutrophils in Population 5, epx was also enriched.
Summary
Gene expression data generated in our study suggest that thrombocytes, B-cells, T-cells, erythrocytes, neutrophils, and a myeloid-like cells with characteristics of macrophages and neutrophils were isolated from axolotl blood (Table 2). Proportionally more thrombocytes and neutrophils were sequenced in Sample 1, while proportionally more T-cells, B-cells, erythrocytes, and macrophages were sequenced in Sample 2. These differences likely reflect the enrichment strategy that was used to isolate immune cells for Sample 2, which was created to identify macrophages from the IP cavity.
Table 2.
Populations, cell types, number of significant genes, and corresponding biomarkers identified from single-cell sequencing of peripheral axolotl immune cells. The top 5 sig genes are annotated genes that were the most significantly enriched in each population.
| Population | Cell Type | Sig Genes | Top 5 Sig Genes |
|---|---|---|---|
| Pop1 | Thrombocyte | 286 | thbs1, f2rl3, gp9, cald1, wipf3 |
| Pop2 | B-cell | 76 | cd5l, igll5, atp1b3, rpl22l1, hvcn1 |
| Pop3 | T-cell | 30 | trac, ccl19, psapl1, igll59, gzmb |
| Pop4 | Erythrocyte | 34 | hba, hbb, hbg1, alas2, slc4a1 |
| Pop5 | Neutrophil | 212 | mmp1, chit1, camp, cebpe, btn1a1 |
| Pop6 | Macrophage | 201 | cr2, c4a, siglec1, fabp1, lta4h |
| Pop7 | Myeloid-like | 185 | lect2, epx, lgals1, prtn3, psap |
3.3. Single-Cell Sequencing of Cells Isolated from Regenerating Limbs
To investigate the representation of immune cell populations during limb regeneration, we collected stump tissue from two post-amputation time points and performed single-cell isolation and sequencing. The first sample was collected from the distal limb stump at 1 DPA and the other at 6 DPA. We reasoned from results of a previous study (Godwin et al 2013) that the first sample would yield stump and early infiltrating immune cells, and the second sample would yield stump and either lingering or second-wave immune cells. We note that the 6 DPA sample represents an intermediate time point between limb amputation and the appearance of an early limb bud (Tank et al 1976), a developmental stage that indicates when the blastema has reached a critical size to engender distal limb outgrowth.
Using again the 10x Genomics platform, we generated transcripts for 8,272 and 9,906 cells from 1 DPA and 6 DPA timepoints respectively, and used k-means clustering of genes to identify 8 cell populations (Figure 3)(Supplemental Table 2). To approach the annotation of cell identities in an unbiased way, we identified the 30 most significantly enriched genes within each of the 8 limb cell populations and then compared these to the 30 most significantly enriched genes identified for immune cell populations reported above. We compared 30 genes because only 30 significant genes were identified for the T-cell population and we didn’t want pairwise comparisons to be biased by differences in the number of significant genes identified among populations. These comparisons revealed overlapping genes between peripheral immune cells types and some, but not all of the limb cell populations (Tables 3 and 4). Overlapping genes identified for limb populations 1–4 suggested these corresponded to erythrocytes, macrophages, lymphocytes, and neutrophils, respectively. Absence of overlapping genes for populations 5–7 and only two overlapping genes for population 8 suggested that these corresponded to non-immune cell populations. We investigated this reasoning further by more closely examining genes within each of the limb cell populations. We note that while our methodology for isolating leukocytes in the earlier study effectively filtered erythrocytes, this was not the case for methods used to isolate cells from limb stumps. Approximately 66% (N = 11,390) of the cells that were sequenced were annotated as erythrocytes (Table 3). Clearly, future studies would benefit from a method to specifically remove erythrocytes from regenerating tissues. Still, the 5,788 non-erythrocytic cells that were sequenced revealed immune and non-immune cell types that we review below.
Figure 3.
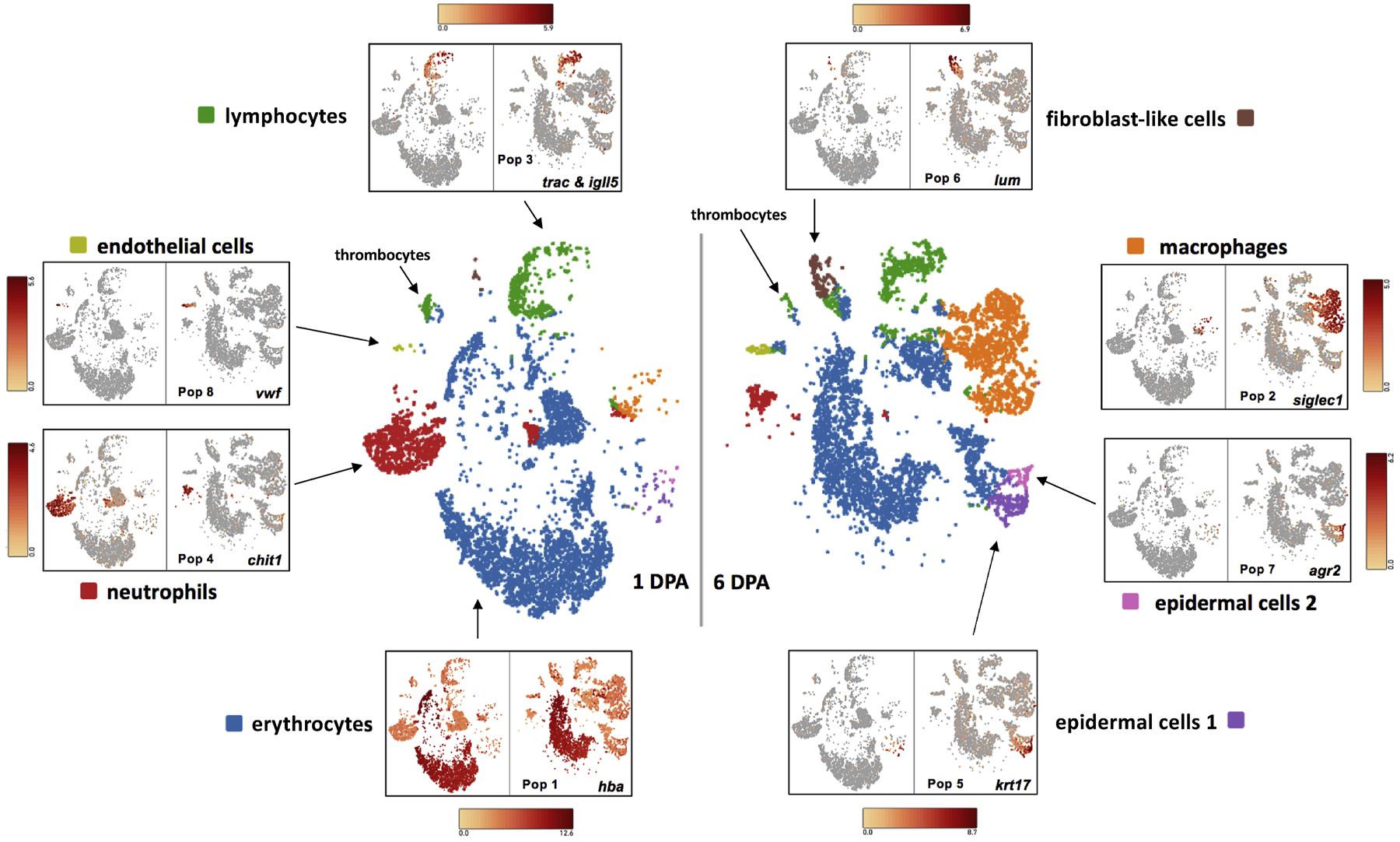
t-SNE plots showing cell populations that were identified by k-means clustering of transcripts. Example gene expression markers are shown for cell types from 1 and 6 DPA limb stumps.
Table 3.
Pairwise comparison of the 30 most significantly enriched genes among peripheral and limb cell populations.
| 2 | 3 | ||||||
| 1 | 9 | ||||||
| 13 | |||||||
| 18 | |||||||
| 14 | |||||||
| 9 | |||||||
| 2 |
Table 4.
Populations, cell types, and number of significantly enriched genes identified from single-cell sequencing of 1 and 6 DPA axolotl limb stump samples. The top 5 sig genes are annotated genes that were the most significantly enriched in each population.
| Population | Cell Type | # Sig Genes | Top 5 Sig Genes |
|---|---|---|---|
| Pop1 | Erythrocyte | 80 | hbb, hba, hbg1, rhag, alas2 |
| Pop2 | Macrophage | 600 | ctsla, gpnmb, marco, trem2, apoe |
| Pop3 | Lymphocyte | 131 | igll5, trac, psapl1, cyr61, igj |
| Pop4 | Neutrophil | 266 | chit1, btn1a1, ifitm2, camp, mmp1 |
| Pop5 | Epidermal cell 1 | 225 | krt5, krt75, krt17, vwa7, serpinb6 |
| Pop6 | Fibroblast-like | 232 | lum, col3a1, dcn, col5a2, ptx3 |
| Pop7 | Epidermal cell 2 | 169 | fcgbp, psca, thdl20, mal, upk1b |
| Pop8 | Endothelial cell | 75 | vwf plvap, mmrn1, egfl7, ahnak |
Limb Population 1 (N = 11,390; 1 DPA = 5,939; 6 DPA = 5,451) Erythrocytes.
Two k-means clusters were merged to form this large population (Figure 2). While both clusters were characterized by the expression of multiple hemoglobin genes, one cluster was also enriched by genes that Leigh et al (2018) annotated to erythrocytes (e.g. tspo, prdx2, prdx6, nco4, sushd3).
Limb Population 2 (N = 2,384; 1 DPA = 120; 6 DPA = 2,264) Macrophages.
Approximately 69% of the genes that were significantly enriched in macrophages, presumably isolated from the IP cavity, were enriched in this population, including siglec1 (Figure 2). This high gene overlap suggests that macrophages from different tissue niches may express many of the same genes, and thus the IP cavity may provide a useful reservoir to collect macrophages for future functional studies. We note that 18.8x more macrophages were isolated at 6 DPA than 1 DPA. This pattern is consistent with macrophage infiltration and a gradual increase in macrophage number from 1–6 DPA (Godwin et al 2013).
Limb Population 3 (N = 1,729; 1 DPA = 802; 6 DPA = 927) Lymphocytes.
While independent T-cell and B-cell populations were identified from blood, only a single lymphocyte cell population was identified from the limb stump. This population was characterized by the expression of both T-cell (trac) and B-cell (igll5) gene expression markers (Figure 2). Approximately 60% of the genes that were enriched in this population were found in either the T-cell or B-cell population from peripheral blood, with the most significant genes found in common to the T-cell population. We note that a disjunct subgroup of cells within this population (Figure 2) likely corresponds to a group of thrombocytes as these cells are recovered as a distinct population when analyzing the 1 DPA single cell data independently from the 6 DPA data.
Limb Population 4 (N = 1,699; 1 DPA = 1,442; 6 DPA = 257) Neutrophils.
Approximately 65% of the genes that were enriched in the neutrophil population from blood were enriched in this population. Neutrophils were 5.6x more abundant at 1 DPA than 6 DPA, consistent with an early and transient infiltration of the limb stump.
Limb Population 5 (N = 404; 1 DPA = 37; 6 DPA = 367) Epidermal cells 1.
None of the genes that were significantly enriched in peripheral blood or IP cells were identified among the enriched genes in this population. We searched 136 of the annotated genes in this population for enriched Gene Ontology (GO) Biological Process (BP) terms using the ToppGene enrichment tool (Chen et al 2009). This search identified the epithelium development BP term as highly enriched by 35 genes (Supplemental Table 3). Leigh et al (2018) associated several of the genes from this population with epidermal cell populations (e.g. elf3, casp6, sdc1, pkp1, scel, evpl, perp). Thus, it seems likely that this population is comprised of epidermal cells.
Limb Population 6 (N = 309; 1 DPA = 22; 6 DPA = 287) Fibroblast-like cells.
Many fibroblast-like cell markers from Leigh et al (2018) were identified as significantly enriched in this population, including lum, ptx3, fbn1, has2, msmb, sfrp2, apoh, fstl1, ctgf. lum was highlighted as a marker of blastema cells that increased in abundance during limb regeneration. Furthermore, pseudotime analysis performed by Leigh et al (2018) implicated these cells as multipotent progenitors of cartilage and bone in the regenerated limb. Consistent with these results, ToppGene identified 21 (Supplemental Table 3) genes within this population that significantly enriched the skeletal system development BP term. Approximately 13x more fibroblast-like cells were identified at 6 DPA than 1 DPA, consistent with an expanding population.
Limb Population 7 (N = 135; 1 DPA = 9; 6 DPA = 126) Epidermal cells 2.
This population of cells grouped very closely to Population 5 in the t-SNE plot. Many significantlyenriched genes were shared between Populations 5 and 7 and the epithelial development BP term was also enriched by 21 genes from Population 7 (Supplemental Table 3). Leigh et al (2018) identified several different epidermal cell populations and also cellular components of the epidermis (Leydig and secretory cells) in their more comprehensive sampling of later stages of limb regeneration. Our results suggest epidermal cell populations at 1 and 6 DPA are relatively homogeneous.
Limb Population 8 (N = 128; 1 DPA = 21; 6 DPA = 107) Endothelial cells.
Cells in this population significantly expressed several genes (e.g. plvp, mmrn1, egfl7, cd34, kdr) that are known to be expressed by endothelial cells in humans (Thul et al 2017). For example, von Willebrand factor (vwf) functions in platelet aggregation and is expressed in endothelial cells and megakaryocytes (Ruggeri 1999). ToppGene (Supplemental Table 3) identified genes from this population that significantly enriched the cell adhesion (N = 18) and angiogenesis (N = 10) BP terms, consistent with an endothelial cell identity.
In summary, we found very high overlap of significant genes sampled from cells of the same type from blood and limb stump. Consider that 76%, 69%, 60%, and 65% of the genes that were significantly enriched in erythrocytes, macrophages, lymphocytes, and neutrophils in the limb stump respectively, were also significantly enriched in cells of these types that were isolated from blood. Although much of our sequencing effort was consumed by erythrocytes, sufficient numbers of cells were identified to show temporal changes in the abundances of immune and non-immune cells.
4. Conclusions
In this study, we collected enriched fractions of peripheral immune cells and used single-cell transcript sequencing to identify gene expression markers for specific cell types, including erythrocytes, neutrophils, macrophages, and lymphocytes. Because immune cells are known to infiltrate sites of tissue injury, we also sampled cells from regenerating axolotl limbs and searched for immune cells among the different populations of cells that were identified by k-means clustering of transcripts. We identified erythrocytes, thrombocytes, lymphocytes, neutrophils, and macrophages in the limb, and resolved temporal changes in neutrophil and macrophage cell abundances. The enriched gene sets from our work should aid future single-cell investigations of immune cell diversity and function during axolotl limb regeneration. We also identified a fibroblast-like cell population that increased in abundance between 1 and 6 DPA, implicating these cells as early progenitors of the blastema. We discuss these primary results below.
Tissue regeneration is a temporally regulated process that involves many different cell types and molecular functions. A limb for example has many different tissue types, all of which must be reformed during regeneration from different progenitor cell populations. Models of regeneration have focused much attention on the blastema, a tissue unique to appendage regeneration that contains progenitor cells for tissue reformation. However, blastema formation is a relatively late regeneration event. For relatively large juvenile axolotls, a prominent blastema does not form until 9–10 days post-amputation (Tank et al 1976). A number of biological processes must be enacted prior to blastema formation for successful regeneration, including re-epithelialization of the injury site and maturation of the wound epidermis (Goss 1969). More germane to this study are innate immune responses that must be enacted to ensure survival at the whole-organism level. Amputation injury exposes underlying tissues to pathogens, blood loss must be mitigated, and the injury environment must be cleared of debris and dying cells. These non-developmental processes have traditionally received relatively less attention however immune cells that perform essential wound-healing functions are beginning to be appreciated for their roles in orchestrating events that are necessary for blastema formation and successful regeneration (Godwin et al 2017; Julier et al 2017; Mescher et al 2017).
Thrombocytes play vital roles in early wound hemostatic processes (Nurden 2018) and because salamanders show very little blood loss after amputation, they are likely recruited rapidly to the injury site. While we identified an abundance of thrombocytes in peripheral blood, we identified only a small subgroup of cells that presented thrombocyte gene expression markers within a population of cells that were annotated as lymphocytes. This indicates that thrombocytes and lymphocytes express some of the same transcripts and may therefore perform overlapping functions during early wound healing. This may be generally true of different immune cell types that are sequentially recruited to the injury site. Beyond their classically defined roles in hemostasis and thrombosis, thrombocytes release growth factors, cytokines, metabolites, and extracellular matrix modulators that are required for subsequent immunological events (Etulain 2018). Platelet derived growth factor (PDGF) is thought to be released from thrombocytes as a signal to induce migration of connective tissue fibroblasts in the axolotl, which contribute to the blastema (Currie et al 2016). We did not observe significant upregulation of either PDGF in thrombocytes or its receptor, pdgfra, in fibroblast-like cells. However, we note that pdgfra expression was primarily restricted to the fibroblast-like cell population discussed below.
In mammals and some other vertebrates, neutrophils and macrophages are recruited to sites of injury in a stereotypical manner, with neutrophils appearing before macrophages (Kumar et al 2018). Neutrophils are recruited to injury sites to mitigate pathogens, clear debris, and phagocytize cells. Macrophages also perform these same biological functions, as well as clear apoptotic neutrophils from the injury site. Additionally, macrophages play important roles in adaptive immune and tissue repair processes. Depletion of macrophages in axolotls blocks limb regeneration (Godwin et al 2013) and a requirement for macrophages (and not neutrophils) was also reported for zebrafish tailfin regeneration (Li et al 2012). Godwin et al (2013) observed neutrophils and macrophages as early as 1 DPA during axolotl limb regeneration, suggesting a simultaneous recruitment pattern that may be unique to regeneration-competent axolotls. We also observed neutrophils and macrophages at 1 DPA, however there were many neutrophils at this time and very few macrophages. At 6 DPA, an inverse pattern was observed with more macrophages observed than neutrophils. Thus, while neutrophils and macrophages are present at the same time during limb regeneration, cell numbers reflect infiltration dynamics observed in other vertebrates, specifically an earlier predominance of neutrophils. The significance of this overlapping pattern of immune cell recruitment awaits further study.
While the innate immune response appears to be robust in the axolotl, very little is known about adaptive immune responses during regeneration. We identified a population of lymphocytes in regenerating limb but did not resolve distinct T-cell and B-cell populations. Leigh et al (2018) identified two different B-cell populations during axolotl limb regeneration, one of which was associated with igll5 expression; they also identified a T-cell population that was marked by trac expression. Both of these genes were highly enriched in the lymphocyte population that we identified in our study and co-expression was observed in the same cells. It is possible that at the time we collected cells, T-cells and B-cells express many of the same genes, for example if there is convergence in transcription of ribosomal genes for protein translation. We note that a comprehensive analysis of mouse T-cell and B-cell gene expression datasets found very few transcripts that are specific for either cell type; moreover, many of the same genes are expressed among different immune cell types (Painter et al 2011). It is important to keep in mind that the annotation of cell types from single-cell transcript data is quantitative and threshold dependent, and in some cases may depend upon subtle differences in gene expression. While we noted many examples of overlap between our results and Leigh et al (2018), additional single-cell and lineage tracing studies will be needed to resolve lymphocyte diversity, as well as the diversity of other cell types.
Our analysis of limb also discovered populations of non-immune cells, including epidermal and endothelial cells. Genes that were identified for these populations enriched BP GO terms that are typical of epidermal and endothelial cells and two of these genes (krt5, krt17) were localized previously to the wound epidermis by in situ hybridization (Monaghan et al 2012; Moriyasu et al 2012; Leigh et al 2018). Thus, annotation of these cell types is not controversial. A specialized wound epidermis forms over the limb stump after amputation and over the course of 9–10 days thickens and develops a basal cell layer that secretes factors to underlying mesenchymal cells. Our 1 mm samples of the limb stump contained wound epidermis and epidermis that is typical of an unamputated limb. This might explain why two different epidermal cell populations were identified in our study, although as noted above, many of the same genes were expressed between these populations.
Importantly, we discovered a non-immune cell population that was characterized by the expression of extracellular matrix components and other genes that are typical of connective tissue fibroblasts. Many of these genes were identified from a population of cells that Leigh et al (2018) annotated as fibroblast-like blastema cells. These cells were discovered in non-amputated limbs and at all post-amputation time points that were sampled, and pseudotime analysis suggested these cells to be multipotent progenitors of the blastema. The most highly expressed gene (lum) for fibroblast-like cells in our study was shown by Leigh et al 2018 to be expressed in the medium bud blastema. Our data suggest that this population expands appreciably between 1 and 6 DPA and note that pcna, a marker of cellular proliferation, was significantly enriched only within this population. We hypothesize that these fibroblast-like cells represent a critical, pioneering population of progenitor cells that are activated rapidly following amputation and may initiate the formation of the blastema. In other words, this population creates a niche that supports the subsequent recruitment of non-skeletal system, progenitor cells. Cell-lineage studies of the regenerating limb are needed to test this hypothesis.
Finally, we note a couple of caveats of our study. First, we did not sample all cell types that are expected to be isolated from a regenerating salamander limb. For example, we did not sample muscle cells or Schwann cells that are associated with the peripheral nervous system. We suspect that some large, salamander cells may not be compatible with 10X sequencing and single cell isolation methodologies may need optimization to efficiently capture rare cell populations. Second, given costs in performing single-cell analysis at the sequence/cellular depth required for this study, we only examined single tissue samples for the Day 1 and Day 6 time points. However, we sequenced many more cells than were sequenced by two previous studies of axolotl limb regeneration (Gerber et al 2018; Leigh et al 2018) and we observed co-expression of gene expression biomarkers that were used to annotate cell populations in those studies. Observing the same expression patterns across studies, using different methodologies to construct transcript libraries, speaks to the robustness and repeatability of single cell sequencing technologies and the axolotl limb regeneration model. Data from future single-cell sequencing efforts will further enrich understanding of cells and cellular phenotypes during regeneration.
Supplementary Material
Supplemental Figure 1. Representative figures of A) lymphocytes, B) thrombocytes, and C) eosinophils from Sample 2.
Acknowledgements
This work was supported by the National Institutes of Health (P40OD019794, R24OD010435).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartels M, Govers AM, Fleskens V, Lourenço AR, Pals CE, Vervoort SJ, van Gent R, Brenkman AB, Bierings MB, Ackerman SJ, van Loosdregt J, Coffer PJ. 2015. Acetylation of C/EBPε is a prerequisite for terminal neutrophil differentiation. Blood 125:1782–92. [DOI] [PubMed] [Google Scholar]
- Bishop DF, Henderson AS, Astrin KH. 1990. Human delta-aminolevulinate synthase: assignment of the housekeeping gene to 3p21 and the erythroid-specific gene to the X chromosome. Genomics 7:207–14. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37:W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie JD, Kawaguchi A, Traspas RM, Schuez M, Chara O, Tanaka EM. 2016. Live Imaging of Axolotl Digit Regeneration Reveals Spatiotemporal Choreography of Diverse Connective Tissue Progenitor Pools. Dev Cell 39:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale DC, Boxer L, Liles WC. 2008. The phagocytes: neutrophils and monocytes. Blood 112:935–945; [DOI] [PubMed] [Google Scholar]
- Dixit VM, Haverstick DM, O’Rourke KM, Hennessy SW, Grant GA, Santoro SA, Frazier WA. 1985. A monoclonal antibody against human thrombospondin inhibits platelet aggregation. Proc Natl Acad Sci USA, 82:3472–34766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etulain J 2018. Platelets in wound healing and regenerative medicine. Platelets 29:556–568. [DOI] [PubMed] [Google Scholar]
- Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, Gac-Santel M, Nowoshilow S, Kageyama J, Khattak S, et al. 2018. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362:6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin JW, Debuque R, Salimova E, Rosenthal NA. 2017. Heart regeneration in the salamander relies on macrophage-mediated control of fibroblast activation and the extracellular landscape. NPJ Regen Med 2017:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin JW, Pinto AR, Rosenthal NA. 2013. Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci USA, 110:9415–9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin JW, Pinto AR, Rosenthal NA. 2017. Chasing the recipe for a pro-regenerative immune system. Semin Cell Dev Biol 61:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss RJ. 1969. Principles of Regeneration. Academic Press, New York: IX: 287 pp. [Google Scholar]
- Ignarro LJ. 1990. Haem-dependent activation of guanylate cyclase and cyclic GMP formation by endogenous nitric oxide: a unique transduction mechanism for transcellular signaling. Pharmacol Toxicol 67:1–7. [DOI] [PubMed] [Google Scholar]
- Julier Z, Park AJ, Briquez PS, Martino MM. 2017. Promoting tissue regeneration by modulating the immune system. Acta Biomater 53:13–28. [DOI] [PubMed] [Google Scholar]
- Prame Kumar K, Nicholls AJ, Wong CHY. 2018. Partners in crime: neutrophils and monocytes/macrophages in inflammation and disease. Cell Tissue Res 371:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh ND, Dunlap GS, Johnson K, Mariano R, Oshiro R, Wong AY, Bryant DM, Miller BM, Ratner A, Chen A, et al. 2018. Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat Commun 9: 5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yan B, Shi YQ, Zhang WQ, Wen ZL. 2012. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J Biol Chem 287:25353–25360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher AL, Neff AW, King MW. 2017. Inflammation and immunity in organ regeneration. Dev Comp Immunol 66:98–110. [DOI] [PubMed] [Google Scholar]
- Moriyasu M, Makanae A, Satoh A. 2012. Spatiotemporal regulation of keratin 5 and 17 in the axolotl limb. Dev Dyn 241:1616–1624. [DOI] [PubMed] [Google Scholar]
- Nowoshilow S, Schloissnig S, Fei JF, Dahl A, Pang AWC, Pippel M, Winkler S, Hastie AR, Young G, Roscito JG, et al. 2018. The axolotl genome and the evolution of key tissue formation regulators. Nature 554:50–55. [DOI] [PubMed] [Google Scholar]
- Nurden AT. 2018. The biology of the platelet with special reference to inflammation, wound healing and immunity. Front Biosci 23:726–751. [DOI] [PubMed] [Google Scholar]
- Painter MW, Davis S, Hardy RR, Mathis D, Benoist C, Immunological Genome Project Consortium. 2011. Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J Immunol 186:3047–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos A, Preusse C, Hathazi D, Goebel HH, Stenzel W. 2019. Proteomic profiling nravels a key role of specific macrophage subtypes in sporadic inclusion body myositis. Front Immunol 10:1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri ZM. 1999. Structure and function of von Willebrand factor. Thromb Haemost 82:576–84. [PubMed] [Google Scholar]
- Savage B, Saldivar E, Ruggeri ZM. 1996. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell 84:289–297. [DOI] [PubMed] [Google Scholar]
- Schachtrup C, Scholzen TE, Grau V, Luger TA, Sorg C, Spener F, Kerkhoff C. 2004. L-FABP is exclusively expressed in alveolar macrophages within the myeloid lineage: evidence for a PPARalpha-independent expression. Int J Biochem Cell Biol 36:2042–53 [DOI] [PubMed] [Google Scholar]
- Singh SK, Sethi S, Aravamudhan S, Krüger M, Grabher C. 2013. Proteome mapping of adult zebrafish marrow neutrophils reveals partial cross species conservation to human peripheral neutrophils. PLoS One 8:e73998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zeglinski MR, Turner CT, Raithatha SA, Wu Z, Russo V, Oram C, Hiroyasu S, Nabai L, Zhao H, et al. 2018. Topical small molecule granzyme B inhibitor improves remodeling in a murine model of impaired burn wound healing. Exp Mol Med 50:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank PW, Carlson BM, Connelly TG. 1976. A staging system for forelimb regeneration in the axolotl, Ambystoma mexicanum. J Morph 150:117–128. [DOI] [PubMed] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. 2017. A subcellular map of the human proteome. Science 356:6340. [DOI] [PubMed] [Google Scholar]
- Trapani JA1, Browne KA, Smyth MJ, Jans DA. 1996. Localization of granzyme B in the nucleus. A putative role in the mechanism of cytotoxic lymphocyte-mediated apoptosis. J Biol Chem 271:4127–33. [DOI] [PubMed] [Google Scholar]
- Wang J, Kubes P. 2016. A Reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell 165:668–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Representative figures of A) lymphocytes, B) thrombocytes, and C) eosinophils from Sample 2.


