Abstract
Recent evidence in humans and mice supports the notion that mitochondrial metabolism is active and necessary for tumor growth. Mitochondrial metabolism supports tumor anabolism by providing key metabolites for macromolecule synthesis and generating oncometabolites to maintain the cancer phenotype. Moreover, there are multiple clinical trials testing the efficacy of inhibiting mitochondrial metabolism as a new cancer therapeutic treatment. In this review, we discuss the rationale of using these anti-cancer agents in clinical trials and highlight how to effectively utilize them in different tumor contexts.
Introduction
Historically, mitochondrial metabolism has been viewed as inconsequential to support the metabolic demands of rapidly proliferating cancer cells (Warburg, 1956). This view is founded upon the seminal observation, made in the 1920s by Otto Warburg, that tumor slices take up glucose and produce excess lactate regardless of oxygen availability (Koppenol et al., 2011). This has been referred to as aerobic glycolysis or the Warburg effect and has shaped the way generations of scientists think about the role of mitochondrial metabolism in cancer. Warburg postulated “injury to respiration” as a prerequisite for malignant transformation. Thus, glycolysis was ascribed to be the primary metabolic pathway necessary for tumor proliferation (Warburg, 1956). Ultimately, mitochondrial dysfunction and aerobic glycolysis have become widely accepted as hallmarks of cancer (Hanahan and Weinberg, 2011).
The long-standing belief that mitochondrial metabolism was dispensable for tumor growth has been challenged in recent decades by both human and mouse studies. In fact, the Warburg effect was shown to be dispensable for B16 melanoma tumor growth due to increased mitochondrial metabolism (Ždralević et al., 2018). Mitochondrial metabolism is required for oncogenic Kras-driven mouse models of lung adenocarcinoma (Guo et al., 2011; Weinberg et al., 2010). Positron emission tomography (PET) imaging using a radiotracer that measures mitochondrial membrane potential (MMP) in autochthonous mouse models of lung cancer demonstrates high MMP in lung adenocarcinoma (Momcilovic et al., 2019). Importantly, intraoperative infusions of [U-13C]glucose in human lung and brain tumors demonstrate high levels of glucose oxidation and tricarboxylic acid (TCA) cycle labeling, exceeding that of adjacent normal tissue (Hensley et al., 2016; Maher et al., 2012). Analysis of the Cancer Genome Atlas (TCGA) revealed that the mitochondrial DNA (mtDNA) content of cancerous tissues varies relative to their normal tissue counterparts. For example, lung adenocarcinomas display elevated mtDNA content relative to adjacent normal lung tissue (Reznik et al., 2016). Contrary to conventional wisdom, analysis of over 30 cancer types revealed that mitochondria with mtDNA mutations that are pathogenic are less likely to be maintained in cancer cells, suggesting that there is a positive selection for functional mitochondria to drive tumor growth (Ju et al., 2014). Furthermore, genetic defects leading to defective mitochondrial respiratory function produce a metabolic checkpoint that prevents malignant transformation (Joshi et al., 2015). These studies indicate that mitochondrial metabolism is an active essential process for tumor growth. More recent work suggests that this metabolic reprogramming is a dynamic process throughout tumorigenesis with metabolic flexibility serving the needs of the tumor at every stage, from tumor initiation to metastasis (Faubert et al., 2020). In this review, we highlight recent advances in our understanding of the essential role of mitochondrial metabolism and its potential as a target for cancer therapy.
Mitochondrial Metabolism-Dependent Macromolecule Synthesis and Oncometabolite Production Support Tumor Growth
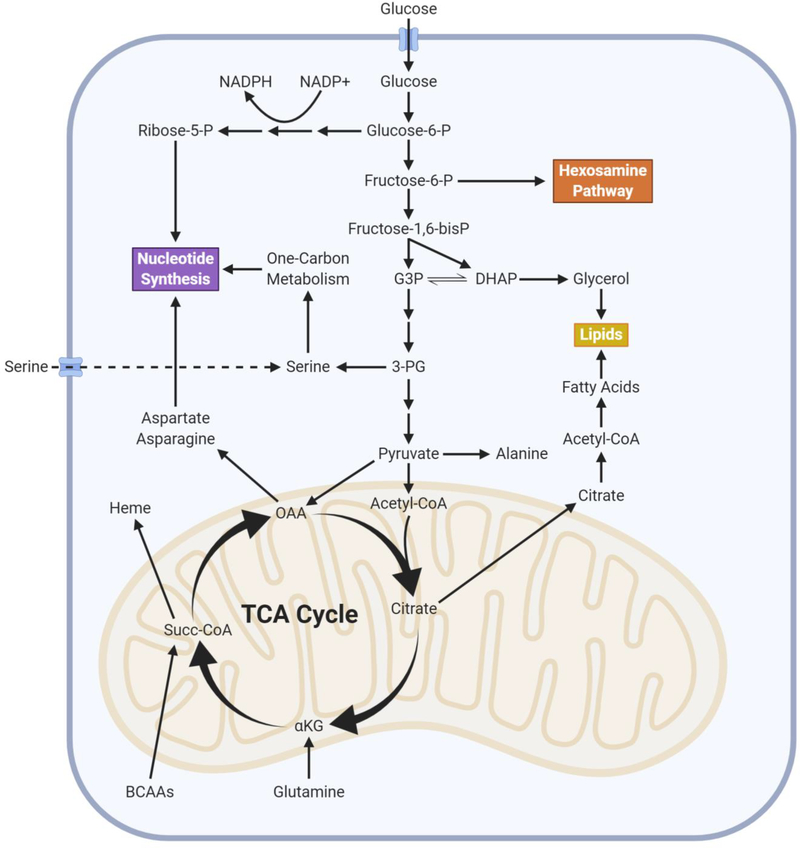
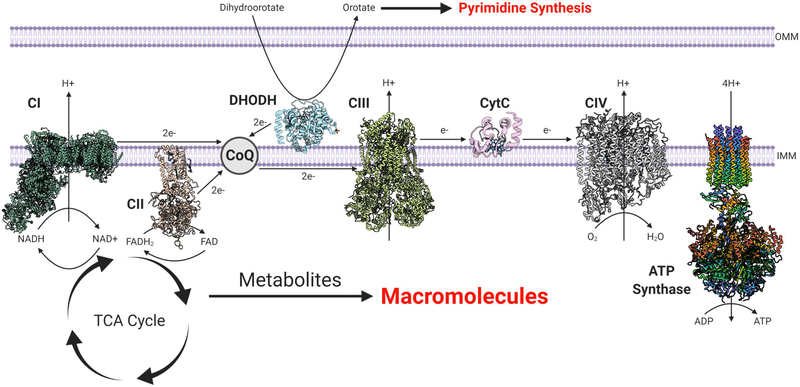
Tumor cells undergo metabolic reprogramming as a consequence of driver mutations, whereby metabolic flux through conventional metabolic pathways utilized by normal cells is increased or decreased in tumor cells relative to their premalignant tissue of origin (DeBerardinis and Chandel, 2016). Tumor cells robustly engage in both glycolysis, and its branching pathways, and TCA cycle metabolism in order to generate ATP, NADPH, and the building blocks necessary for macromolecule (nucleotides, lipids, and amino acids) synthesis, which are all essential for cell proliferation (Figure 1) (DeBerardinis and Chandel, 2020). Activation of major oncogenic drivers, such as Myc and Kras, and deregulation of signaling pathways, including the PI3K pathway, in part account for the elevated rate of glycolysis and TCA cycle flux seen in cancer cells. The elevated glycolytic rate allows for the generation of metabolic intermediates that can be shunted into multiple biosynthetic pathways required for cell proliferation, such as the pentose phosphate pathway (PPP) for ribose and cytosolic NADPH production, to sustain nucleotide synthesis and antioxidant activity, respectively, as well as one-carbon metabolism for mitochondrial NADPH production, nucleotide synthesis, and methylation reactions (DeBerardinis and Chandel, 2016; Vander Heiden and DeBerardinis, 2017). TCA cycle flux allows the generation of metabolites that funnel into nucleotide, lipid, amino acid, and heme synthesis (Zong et al., 2016). For example, oxaloacetate produced in the TCA cycle is exported from the mitochondrial matrix to the cytosol for nucleotide synthesis (Birsoy et al., 2015; Sullivan et al., 2015). Shunting of TCA cycle intermediates for biosynthetic purposes creates a need for replenishment of carbons to allow the TCA cycle to continue functioning, i.e., anaplerosis. There are multiple anaplerotic reactions utilized by cancer cells including the stepwise oxidation of glutamine to generate the TCA cycle intermediate α-ketoglutarate, branched-chain amino acid catabolism into succinyl-CoA, and pyruvate carboxylase generation of oxaloacetate (Cluntun et al., 2017). The TCA cycle also generates NADH and FADH2 that need to be regenerated to NAD+ and FAD by the mitochondrial electron transport chain (ETC) to allow the oxidative TCA cycle to function (Figure 2) (Chandel, 2015). Recent work demonstrates that oxidation of ubiquinol back to ubiquinone is the essential role of the ETC for tumor growth (Martinez-Reyes et al., 2020). Mitochondrial complex I and II donate electrons to ubiquinone generating ubiquinol. Mitochondrial complex III oxidizes ubiquinol back to ubiquinone, which allows complexes I and II to continue functioning and regenerate NAD+ and FAD. Ubiquinone is also used as an electron acceptor by dihydroorotate dehydrogenase (DHODH), an enzyme required for de novo pyrimidine synthesis. Tumor cells with a diminished ability to regenerate mitochondrial ubiquinone have an impaired ability to form tumors in vivo. Moreover, mitochondrial complex III subunits are essential genes for cancer cell proliferation in vitro (Dempster et al., 2019; Meyers et al., 2017). Thus, tumor growth requires a functional ETC for oxidation of ubiquinol, which is necessary to maintain oxidative TCA cycle function and DHODH activity.
Figure 1. Metabolism Supports Macromolecule Synthesis for Growth.
Cancer cells upregulate both glycolysis and TCA cycle metabolism in order to provide the substrates required for synthesis of macromolecules such as lipids and nucleotides that are required for cell proliferation. Multiple substrates feed into these biosynthetic pathways, thus providing cancer cells with metabolic flexibility to support tumor growth.
Figure 2. Mitochondrial ETC Serves Bioenergetic and Biosynthetic Needs of Cancer Cells.
The five complexes of the ETC serve to produce the majority of ATP utilized by cancer cells as well as oxidize NADH and FADH2 to NAD+ and FAD, respectively. This allows for the TCA cycle to continue functioning, producing metabolites that support macromolecule synthesis. DHODH donates electrons to mitochondrial ubiquinone (CoQ) during the conversion of dihydroorotate to orotate, a key step in de novo pyrimidine synthesis. Atomic structures: mitochondrial complex I (PDB: 6RFR) (Parey et al., 2019), complex II (PDB: 1ZOY) (Sun et al., 2005), DHODH (PDB: 4LS1), complex III (PDB: 6Q9E) (Letts et al., 2019), cytochrome c (PDB: 2B4Z) (Mirkin et al., 2008), complex IV (PDB: 5Z62) (Zong et al., 2018), and ATP synthase (PDB: 5FL7) (Hahn et al., 2016).
Besides the TCA cycle’s essential role in supporting tumor cell anabolism, it can also generate oncometabolites in certain cancer contexts, defined as an accumulation of a metabolite that drives tumor growth. Although the large majority of cancers contain functional mitochondria, there exists a small subset that displays mutations in TCA cycle proteins that leads to an accumulation of oncometabolites (Nowicki and Gottlieb, 2015; Yong et al., 2020). In particular, loss-of-function mutations in the TCA cycle enzymes succinate dehydrogenase (SDH) and fumarate hydratase (FH) result in accumulation of succinate and fumarate, respectively (Linehan et al., 2019). Germline heterozygous genetic mutations in SDH complex subunits are observed in patients with hereditary paragangliomas and pheochromocytomas. The neoplastic transformation occurs when there is the loss of the remaining wild-type allele in the somatic cells, i.e., loss of heterozygosity (LOH), leading to the complete loss of enzymatic function. Similarly, LOH occurs in germline FH mutations in patients with hereditary leiomyomatosis and renal cell cancer (HLRCC). It remains unknown why these tissues can tolerate these mutations, as these genes are essential in most cancer cell lines (Dempster et al., 2019; Meyers et al., 2017). SDH- and FH-deficient tumors are dependent on glycolysis for the generation of ATP necessary for cellular proliferation and survival. It was presumed that these tumors would not be able to generate TCA cycle intermediates; however, FH tumors utilize glutamine-dependent “reductive carboxylation,” a process whereby α-ketoglutarate generated from glutamine takes a reverse path in the TCA cycle, to generate citrate for biosynthetic purposes (Metallo et al., 2011; Mullen et al., 2011; Wise et al., 2011), while SDH tumors have robust pyruvate carboxylase activity to generate oxaloacetate for nucleotide synthesis (Cardaci et al., 2015; Lussey-Lepoutre et al., 2015). Both FH- and SDH-deficient tumors are also dependent on part of the oxidative TCA cycle to generate the TCA cycle intermediates leading up to the deficiency (Cardaci et al., 2015; Sullivan et al., 2013). Importantly, tumors harboring SDH or FH mutations have an accumulation of succinate and/or fumarate, which inhibits α-ketoglutarate-dependent dioxygenases, involved in histone and DNA methylation (Xiao et al., 2012), and the resulting epigenetic modifications are thought to contribute to malignant transformation (Sciacovelli et al., 2016). Aside from direct genetic lesions that disrupt the oxidative TCA cycle, there are tumors with an intact TCA cycle that use reductive carboxylation and oncometabolites to sustain tumor growth. A salient example is that patients with clear cell renal cell carcinoma (ccRCC) given [U-13C]glucose infusions display enhanced glycolytic intermediate labeling, suppressed flux through pyruvate dehydrogenase, and reduced TCA cycle labeling (Courtney et al., 2018). Furthermore, glutamine-dependent reductive carboxylation is observed in vivo in subcutaneous xenografts of RCC cells in nude mice (Gameiro et al., 2013). This is due in part to the los of the von-Hippel Lindau (VHL) tumor suppressor resulting in stabilization of hypoxia-inducible factors, activating a transcriptional program resulting in suppressed pyruvate oxidation (Gameiro et al., 2013). These tumors accumulate L-2-hydroxyglutarate (L-2HG), which like succinate and fumarate inhibits α-ketoglutarate-dependent dioxygenases; increases methylation of histones, RNA, and DNA; and is necessary for tumor growth (Shim et al., 2014). Collectively, these observations indicate that although some tumors rely exclusively on glycolysis to meet their bioenergetic needs, they still rely on residual or reprogrammed aspects of mitochondrial metabolism to maintain pools of TCA cycle intermediates to meet the biosynthetic demands required for cell proliferation and generate oncometabolites that promote tumorigenesis.
Targeting Mitochondrial ETC for Cancer Therapy
The cores of many solid tumors are poorly vascularized and, thus, contain nutrient-poor environments with limited glucosa and oxygen availability (Jain et al., 2002). These tumor cores continue to use respiration (Le et al., 2014) since the ETC is able to function optimally even at oxygen levels as low as 0.5% (Rumsey et al., 1990). Therefore, poorly vascularized tumor cores have limited glucose availability but have enough oxygen to continue generating mitochondrial ATP for survival. Furthermore, as discussed above, decreasing ETC function prevents oxidative TCA cycle from functioning, thus diminishing macromolecule synthesis to support tumor growth. To date, the biguanide metformin as a putative mitochondrial ETC complex I inhibitor has been tried in multiple clinical trials as an anticancer agent in combination with standard of care therapies (Pollak, 2014).
Metformin is best known as a first-line therapy for patients with type 2 diabetes. Metformin’s therapeutic effect in part is due to decreased hepatic gluconeogenesis resulting in improved insulin sensitivity. Initially, an epidemiological retrospective study reported an association between metformin use for controlling blood sugar and reduced cancer incidence (Evans et al., 2005). Patients who began taking metformin for blood sugar control after already developing cancer had an increased survival rate (Dowling et al., 2012). Additionally, multiple laboratory-based studies have also reported that metformin acts as an anticancer agent (Algire et al., 2011; Buzzai et al., 2007; Hirsch et al., 2009; Memmott et al., 2010; Tomimoto et al., 2008). It is important to note that dosing of metformin in mice is comparable to human studies (Chandel et al., 2016; Dowling et al., 2016). The efficacious dose of metformin (1,750 mg/day) in reducing tumor growth in humans is likely to be close to twice the anti-diabetic dosage (1,000 mg/day), but well below the maximum tolerated dose (MTD). There are a handful of clinical trials that have reported some efficacy of metformin in various cancers, while others have not observed robust anti-cancer efficacy. Recently, a phase II clinical trial found that combining metformin with standard EGFR-TKI therapy in patients with advanced lung adenocarcinoma significantly improved both progression-free survival and overall survival (Arrieta et al., 2019). Furthermore, a stage II clinical trial in ovarian cancer demonstrated better-than-expected overall survival in the metformin-treated group (Brown et al., 2020). A multicenter phase III clinical trial at the University of Toronto will report their results in the coming year to establish the potential of metformin (1,750 mg/day) as a viable therapeutic strategy against breast cancer (Goodwin et al., 2015).
There are currently two different widely accepted mechanisms by which metformin may be exerting its antitumor effects that are not necessarily mutually exclusive (Birsoy et al., 2012). First, metformin decreases circulating insulin levels, a known mitogen for tumors. Insulin and insulin-like growth factors (IGFs) can stimulate the pro-tumorigenic PI3K signaling pathway (Pollak, 2012). However, this only applies to those tumors that are positive for insulin and/or insulin growth factor receptor. Since not all cancers are insulin responsive, metformin-mediated reduction of circulating insulin levels would be irrelevant to any potential anticancer effect. The second mechanism by which metformin exerts its anticancer effects is through inhibition of mitochondrial ETC complex I. Two seminal studies at the beginning of the century demonstrated that metformin inhibits mitochondrial complex I in vitro (El-Mir et al., 2000; Owen et al., 2000). Subsequent work in mice demonstrated that metformin inhibits mitochondrial complex I to exert its in vivo anti-tumorigenic effects (Wheaton et al., 2014). An integrative metabolomic analysis of metformin’s mechanism of action in ovarian cancer using patient samples confirmed that the predominant anti-tumorigenic effect is driven by targeting tumor-cell-intrinsic mitochondrial metabolism (Liu et al., 2016). In breast cancer, metformin diminishes TCA cycle intermediate production through inhibition of complex I (Janzer et al., 2014). Integrated pharmacodynamic analysis identified two metabolic adaptation pathways to metformin in breast cancer patients: increased glucose flux and increased transcription of oxidative phosphorylation genes (Lord et al., 2018). Recent studies have shown other mechanisms of resistance including metabolic reprogramming due to activation of BACH1 or HIF-1α, decreasing flux through one-carbon metabolism, and infiltration by tumor-associated macrophages (Khan et al., 2019; Kurelac et al., 2019; Lee et al., 2019; Yang et al., 2020). It will be important to assess the relevance of these resistance mechanisms in future metformin clinical trials as inhibitors of these different pathways may be used in combinatorial therapy.
At first glance, mitochondrial ETC inhibitors like metformin would be toxic. Metformin’s high safety profile is in part due to its mechanism of cellular import. Metformin requires organic cation transporters (OCTs) to enter cells. OCTs are able to transport polyamines, thiamine, carnitine, dopamine, and acetylcholine (Nigam, 2018). Normal kidney, gut, and liver cells express OCTs (Emami Riedmaier et al., 2013). There is considerable heterogeneity within tumors regarding metformin sensitivity that, in part, could be due to OCT expression. Thus, we conducted a CRISPR-based functional genomic screen using a metabolic library to discover genes that confer metformin resistance in a human A549 lung adenocarcinoma cell line, which is sensitive to metformin (Figure S1). Loss of the OCT3 (SLC22A3) gene was the top gene hit that conferred resistance to metformin (Figure S2). Additionally, in both squamous cell carcinomas of the head and neck (HNSCC) and breast cancer, it has been shown that the anti-tumor effect of metformin requires the expression of OCT3 (Cai et al., 2019; Madera et al., 2015). This may in part explain the variability in metformin’s anti-tumor efficacy in clinical trials. Going forward, the identification of OCT protein-expressing tumors similar to Her2-positive tumors for Herceptin should be used to identify tumors that are good candidates for metformin therapy (Rusch et al., 2018). Recently, a group showed that homologous recombination-deficient tumors, such as those with BRCA mutations, are reliant upon mitochondrial metabolism to regenerate ATP for PARP-dependent repair mechanisms, leaving them susceptible to inhibitors such as metformin (Lahiguera et al., 2020). Understanding the interplay between cancer genetics and metabolism will allow for developing rational metabolism-targeted therapies. A robust mitochondrial membrane potential is required for uptake of the positively charged metformin at normal pH into the mitochondrial matrix, where it can inhibit complex I (Bridges et al., 2014; Wheaton et al., 2014). This leads to reversible accumulation within the mitochondrial matrix that contributes to metformin toxicity (Bridges et al., 2014). Recently, a PET radiotracer has been developed that can measure mitochondrial membrane potential and predict therapeutic response to mitochondrial complex I inhibitors such as metformin (Momcilovic et al., 2019). Beyond metformin, there have been other mitochondrial complex I inhibitors and other biguanides, such as phenformin (Birsoy et al., 2014; Shackelford et al., 2013), as well as other inhibitors of ETC complexes, that have shown efficacy in pre-clinical models (Molina et al., 2018; Naguib et al., 2018; Shi et al., 2019; Zhang et al., 2019). In addition to directly inhibiting the mitochondrial ETC, diminishing mitochondrial protein translation and stability has shown promise as another avenue to diminish ETC activity (Kuntz et al., 2017; Siegelin et al., 2011; Skrtić et al., 2011; Zhang et al., 2016).
Targeting Nucleotide Metabolism Linked to Mitochondrial ETC Activity for Cancer Therapy
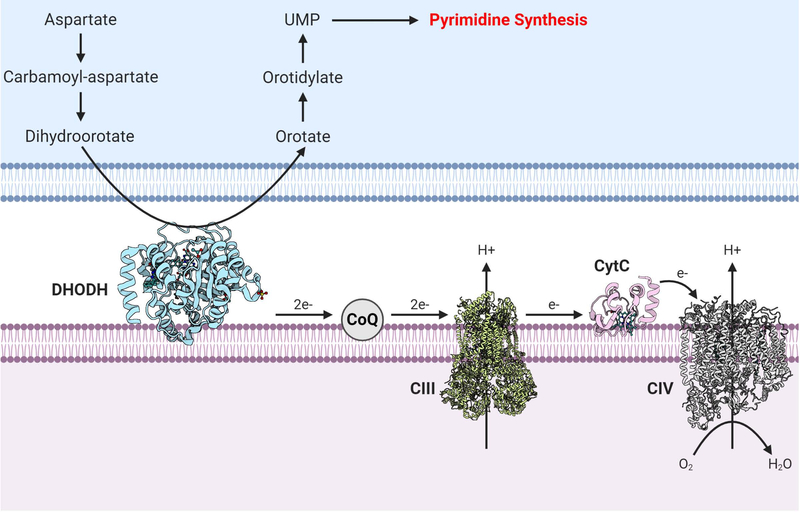
The mitochondrial ETC is intrinsically coupled to pyrimidine nucleotide generation by sustaining DHODH activity (Bajzikova et al., 2019). DHODH catalyzes the fourth enzymatic step, the ubiquinone-mediated oxidation of dihydroorotate to orotate, in de novo pyrimidine biosynthesis (Figure 3). It is found to be located on the outer surface of the inner mitochondrial membrane (Chen and Jones, 1976; Rawls et al., 2000). A recent study demonstrates the availability of ubiquinone to receive electrons from dihydroorotate, which is only compromised when mitochondrial complex III is inhibited, is a key factor for the maintenance of de novo pyrimidine synthesis (Martinez-Reyes et al., 2020). Thus, DHODH activity is dependent on mitochondrial complex III function but does not contribute to the ETC’s role in oxidative phosphorylation or the TCA cycle. DHODH inhibition has demonstrated efficacy in a number of different pre-clinical mouse models of cancer, including highly aggressive small-cell lung cancer (SCLC), acute myeloid leukemia (AML), triple-negative breast cancer, and Kras-driven cancers (Brown et al., 2017; Hosseini et al., 2018; Koundinya et al., 2018; Li et al., 2019; Mathur et al., 2017; Sykes et al., 2016; Wang et al., 2019; White et al., 2011). Although the DHODH inhibitor leflunomide is FDA-approved as an anti-inflammatory drug for rheumatoid arthritis in adults, there are new-generation DHODH inhibitors that show greater potency (Christian et al., 2019; Ladds et al., 2018; Sykes, 2018; Sykes et al., 2016). It remains to be seen whether these new inhibitors will be efficacious as anti-cancer agents in humans.
Figure 3. DHODH Links the Mitochondrial ETC to Pyrimidine Synthesis.
DHODH, a mitochondrial enzyme tethered to the inner mitochondrial membrane, converts dihydroorotate to orotate in the intermembrane space. DHODH donates two electrons to mitochondrial ubiquinone (CoQ) within the ETC. There are currently FDA-approved DHODH inhibitors used for rheumatoid arthritis like leflunomide, as well as other newer DHODH inhibitors. DHODH inhibition has shown promise in preclinical studies of cancer. Atomic structures: DHODH (PDB: 4LS1), complex III (PDB: 6Q9E) (Letts et al., 2019), cytochrome c (PDB: 2B4Z) (Mirkin et al., 2008), and complex IV (PDB: 5Z62) (Zong et al., 2018).
Targeting Mitochondrial TCA Cycle for Cancer Therapy
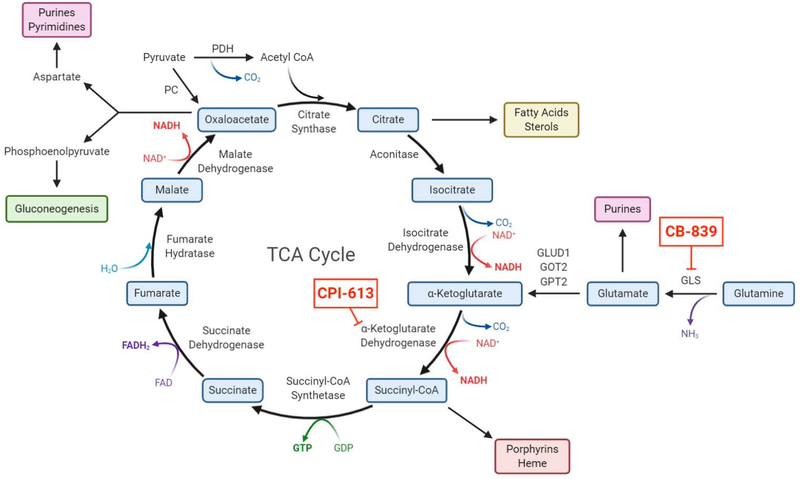
Due to the central role of the TCA cycle in producing the intermediate metabolites for growth, drugs that inhibit the TCA cycle would be predicted to be efficacious. CPI-613 is a first of its kind lipoate analog that can inhibit two major TCA cycle enzyme complexes that require lipoate for their activity, α-ketoglutarate dehydrogenase (α-KGDH) and pyruvate dehydrogenase (PDH) (Figure 4) (Stuart et al., 2014). Although the mechanism by which CPI-613 exerts its anti-cancer activity is not fully understood, it displayed a significant therapeutic index in promising phase I and II results in pancreatic cancer and AML (NCT01835041) (Alistar et al., 2016; Pardee et al., 2014). Currently, CPI-613 is undergoing phase III clinical trials in patients with relapsed/refractory AML or metastatic pancreatic adenocarcinoma (NCT03504410 and NCT03504423).
Figure 4. TCA Cycle Feeds Multiple Biosynthetic Pathways.
Mitochondrial TCA cycle intermediates are utilized as precursors for biosynthetic purposes. This depletion of carbons requires replenishment, i.e., anaplerosis, usually from glutaminolysis and/or pyruvate carboxylase. Multiple inhibitors targeting different steps within the cycle have shown promise in phase I and II clinical trials.
Glutamine is the major carbon source to replenish TCA cycle intermediates and sustain their use for biosynthesis of macromolecules (Altman et al., 2016). Recent work using [U-13C]glutamine infusion in a genetically engineered mouse model of pancreatic cancer demonstrated a large contribution of glutamine into the TCA cycle (Hui et al., 2017). Inhibition of mitochondrial glutaminase (GLS1), which converts glutamine into glutamate, demonstrates efficacy in mouse models of lung adenocarcinoma harboring loss of Keap1, renal cell carcinoma, and MYC-driven hepatocellular carcinoma and lymphoma (Le et al., 2012; Romero et al., 2017; Shroff et al., 2015; Xiang et al., 2015). Glutamate can either be converted into α-ketoglutarate by glutamate dehydrogenase (GLUD) and aminotransferases or be utilized for glutathione synthesis. Human renal cell carcinomas display glutamine carbon incorporation into the TCA cycle (Courtney et al., 2018). Currently, the glutaminase inhibitor CB-839 (Telaglenastat), in combination with the mTOR inhibitor Everolimus or the multi-tyrosine kinase inhibitor Cabozantinib, is in phase II clinical trials for advanced or metastatic renal cell carcinoma (NCT03163667 and NCT03428217). There are also ongoing phase I/II clinical trials using CB-839 in hematological malignancies and solid tumors including NSCLC. Going forward, the use of [U-13C]glutamine infusion in patients to determine whether glutamine contributes carbon into the TCA cycle could identify patients for therapies targeting glutamine metabolism.
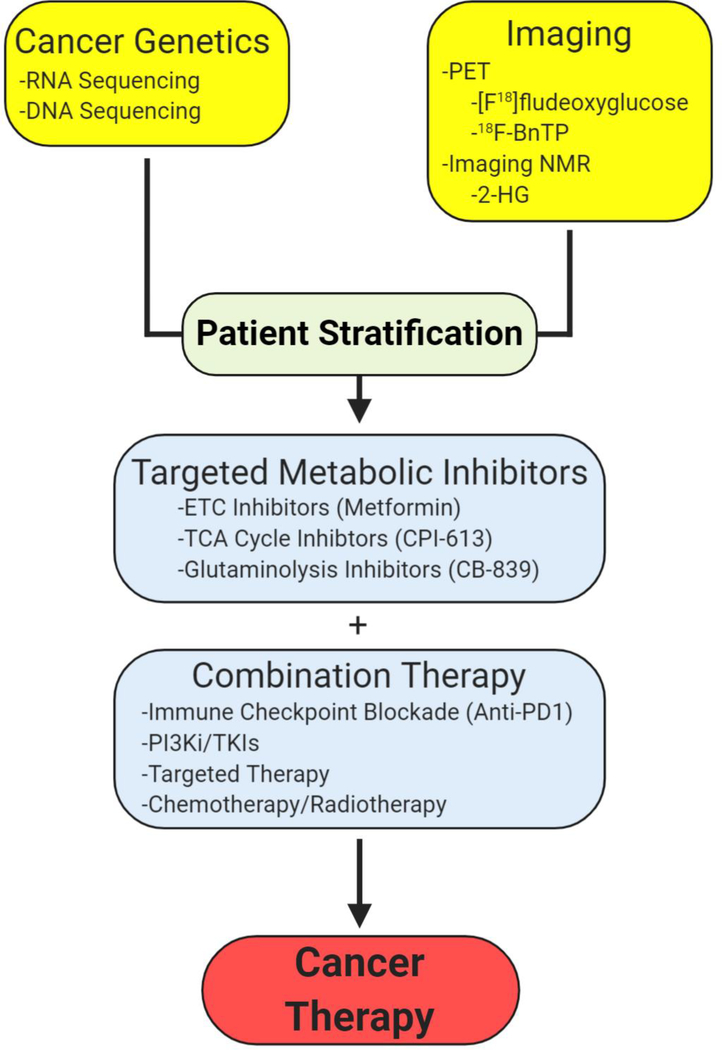
Combining Mitochondrial Metabolism Inhibitors with Other Anti-Cancer Agents
A major advancement in the past two decades is the use of cancer genetics to identify patients that would be best served with a combination of anti-cancer therapies, i.e., personalized medicine. Currently, a major hurdle in using inhibitors targeting mitochondrial metabolism is identifying the right combination of other anti-cancer therapies with appropriate cancer genetics (Figure 5). An emerging theme is that cells that begin to emerge after treatment with chemotherapy, anti-angiogenic therapy, or targeted therapy, e.g., oncogenic Braf or Kras inhibition, are highly dependent on mitochondrial metabolism for survival and proliferation (Caro et al., 2012; Farge et al., 2017; Guièze et al., 2019; Henkenius et al., 2017; Kuntz et al., 2017; Lee et al., 2017; Navarro et al., 2016; Viale et al., 2014). These inhibitors diminish glycolysis and would potentially synergize with agents targeting the mitochondrial ETC or TCA cycle. Mitochondrial respiration within cancer cells, along with low oxygen delivery due to improper tumor vasculature, contributes to intratumoral hypoxia. Thus, inhibiting mitochondrial metabolism would raise tumor oxygen levels, and could significantly improve the tumor cell killing after radiation. Indeed, the FDA-approved drug papaverine inhibits mitochondrial complex I, leading to increased oxygenation and enhanced radiation response in pre-clinical models of cancer (Benej et al., 2018). This pattern of reliance on oxidative phosphorylation is also seen in metastatic lesions. Brain metastases from human melanoma show enrichment for oxidative phosphorylation gene sets (Fischer et al., 2019). It currently remains unclear why there is an increased dependence upon mitochondrial metabolism in advanced disease, but this suggests a potentially shared, targetable metabolic vulnerability across cancers.
Figure 5. Rational Design of Metabolic Cancer Therapy.
Combining Cancer genetics with metabolism-based imaging techniques will allow for patient stratification for targeted metabolic inhibitors. These metabolic inhibitors may be used in combination with chemotherapy, radiotherapy, or even immunotherapy to provide new avenues for cancer therapeutic strategies.
Mitochondrial metabolism inhibitors could also be combined with therapies that diminish glucose metabolism. The PI3K signaling pathway is a major activator of glucose metabolism. Thus, in certain settings the combination of PI3K inhibitors with mitochondrial metabolism inhibitors could be efficacious. Furthermore, certain cancer cells like early-stage lung adenocarcinoma display high levels of the sodium-dependent glucose transporter 2 (SGLT2) (Scafoglio et al., 2018). Targeting SGLT2 with FDA-approved inhibitors, the gliflozins, markedly reduced lung adenocarcinoma growth and prolonged survival in pre-clinical autochthonous mouse models and patient-derived xenografts (Scafoglio et al., 2018). To date, directly inhibiting enzymes in glycolysis has proved to be difficult. Lactate dehydrogenase (LDH) and hexokinase 2 (HK2) inhibition, two key enzymes within glycolysis, has shown efficacy in pre-clinical models (Fantin et al., 2006; Patra et al., 2013). Recent work demonstrated that LDH inhibition in glycolytic tumors leads to redirection of pyruvate to support mitochondrial metabolism, creating a vulnerability to combination therapy with a mitochondrial ETC inhibitor such as metformin (Oshima et al., 2020). HK2 loss in adult mice is well tolerated and, importantly, its inhibition does not affect T cell function (Mehta et al., 2018). Nevertheless, clinical inhibitors that distinguish between HK2 and the widespread isoform hexokinase 1 (HK1) are not currently available.
Multiple mitochondrial inhibitors could be combined since they have distinct targets. For example, metformin could be combined with TCA cycle inhibitor CPI-613 or DHODH inhibitors. In pre-clinical models of prostate cancer, metformin decreases glucose oxidation but increases glutamine-dependent anaplerosis through reductive carboxylation (Fendt et al., 2013; Griss et al., 2015). Interfering with glutamine metabolism may synergize with metformin to improve outcomes. A major limiting factor would be whether these combinations would have a favorable therapeutic index.
The recent successes of immune checkpoint blockade and adoptive cellular therapy (ACT) have revolutionized cancer treatment strategies and have become an established treatment modality moving forward. Similar to cancer, mitochondrial metabolism has been demonstrated to play a critical role in the survival and function of immune cells. As such, when using metabolically targeted therapies for cancer, it is important to consider the potential detrimental effects it may have upon the immune system, as it has been shown that activated immune cells utilize many of the same metabolic pathways attributed to cancer cells (Andrejeva and Rathmell, 2017).
T cells are a key immune effector cell population for a robust and effective anti-tumor immune response. When naive T cells recognize their cognate antigen in the context of co-stimulatory signaling, they increase flux through glycolysis and the TCA cycle to meet the biosynthetic and bioenergetic demands of growth and proliferation (Frauwirth et al., 2002; Ma et al., 2019; Menk et al., 2018). Inhibiting mitochondrial ETC diminishes effector T cell proliferation (Bailis et al., 2019; Sena et al., 2013; Tarasenko et al., 2017) as well as regulatory T cell (Treg) function (Chapman et al., 2018; Fu et al., 2019; Weinberg et al., 2019). Although effector T cells are essential for an anti-tumor response, durable long-lasting immunotherapeutic responses require the establishment of memory T cells. Memory CD8+ T cells preferentially rely on TCA cycle metabolites for function (Geltink et al., 2018). Furthermore, the tumor microenvironment can limit nutrients that can diminish CD8 T cell-dependent tumor killing (Chang et al., 2015). Many studies have observed mitochondrial dysfunction in CD8 T cells within the tumor microenvironment (Scharping et al., 2016). Importantly, enhancing mitochondrial function within these CD8 T cells improved anti-tumor responses (Chamoto et al., 2017; Siska et al., 2017).
A key combinatorial regimen with immune checkpoint blockade is inhibiting glutamine metabolism. After activation, effector T cells can utilize glutamine anaplerosis, similar to cancer cells, due to upregulation of Myc in response to TCR stimulation (Wang et al., 2011). This leads to significant upregulation of SLC1A5, the glutamine transporter, leading to glutamine addiction (Nakaya et al., 2014). Genetic inhibition of GLS diminishes T cell activation and impairs TH17 differentiation in vitro and in vivo. However, transient pharmacologic GLS inhibition leads to increased Th1 and cytotoxic T lymphocyte (CTL) numbers with enhanced anti-tumor immune responses (Johnson et al., 2018). An exciting new study demonstrated that a prodrug for (JHU083) of the glutamine antagonist 6-diazo-5-oxo-L-norlecuine (DON) becomes activated in the tumor microenvironment and enhances T cell mitochondrial metabolism to drive anti-tumor immune responses (Leone et al., 2019). Going forward, it will be interesting to see whether inhibiting glutamine metabolism will be efficacious in patients that have poor responses to immune checkpoint blockade. Furthermore, how other mitochondrial metabolism inhibitors, like CPI-613, perturb immune responses remains to be determined.
Conclusion
The field of cancer metabolism is rooted in the observation that cancer cells exhibit the Warburg effect in vitro. This has misled many to believe that mitochondrial metabolism is either dispensable or only a minor metabolic pathway in tumor growth. Recent advances in our understanding and appreciation of mitochondrial metabolism as a key metabolic driver of cancer and the success of clinical trials targeting mitochondrial metabolism have brought mitochondria to the forefront of both cancer metabolism and immunometabolism fields. Over the next few years, phase III clinical trials of metformin and CPI-613 will be available, and those of us working in cancer metabolism eagerly await these results. The advances in PET imaging and metabolomics and their coupling to cancer genomics can help identify patients that would benefit from use of these inhibitors. We are beginning to understand that the metabolic needs and vulnerabilities of cancer change throughout tumorigenesis, from tumor initiation and growth to metastasis and therapy resistance. In the coming years, elucidating these different vulnerabilities will allow for stage-specific metabolism-targeted therapies. The identification of rational combinations of mitochondrial inhibitors with standard of care treatment including chemotherapy, radiotherapy, and immunotherapy will hopefully bring new and efficacious anti-cancer treatments.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH (5R35CA197532) to N.S.C. and the NIH (5T32CA9560-33) and Northwestern University Pulmonary and Critical Care Department’s Cugell Fellowship to K.V. We thank Hyewon Kong and Colleen Reczek for their input in the CRISPR screen. Figures created with https://biorender.com/.
Footnotes
DECLARATION OF INTERESTS
N.S.C. is an SAB member of Raphael Pharmaceuticals (Devimistat - CPI-613).
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.Org/10.1016/j.cmet.2020.06.019.
REFERENCES
- Algire C, Amrein L, Bazile M, David S, Zakikhani M, and Pollak M (2011). Diet and tumor LKB1 expression interact to determine sensitivity to anti-neoplastic effects of metformin in vivo. Oncogene 30, 1174–1182. [DOI] [PubMed] [Google Scholar]
- Alistar AT, Desnoyers R, D’Agostino RJ, and Pasche B (2016). CPI-613 enhances FOLFIRINoX response rate in stage IV pancreatic cancer. Ann. Oncol. 27, VI228. [Google Scholar]
- Altman BJ, Stine ZE, and Dang CV (2016). From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrejeva G, and Rathmell JC (2017). Similarities and distinctions of cancer and immune metabolism in inflammation and tumors. Cell Metab. 26, 49–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta O, Barrón F, Padilla MS, Avilés-Salas A, Ramírez-Tirado LA, Arguelles Jiménez MJ, Vergara E, Zatarain-Barrón ZL, Hernández-Pedro N, Cardona AF, et al. (2019). Effect of metformin plus tyrosine kinase inhibitors compared with tyrosine kinase inhibitors alone in patients with epidermal growth factor receptor-mutated lung adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol. 5, e192553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis W, Shyer JA, Zhao J, Canaveras JCG, Al Khazal FJ, Qu R, Steach HR, Bielecki P, Khan O, Jackson R, et al. (2019). Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature 571, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajzikova M, Kovarova J, Coelho AR, Boukalova S, Oh S, Rohlenova K, Svec D, Hubackova S, Endaya B, Judasova K, et al. (2019). Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab. 29, 399–416.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benej M, Hong X, Vibhute S, Scott S, Wu J, Graves E, Le Q-T, Koong AC, Giaccia AJ, Yu B, et al. (2018). Papaverine and its derivatives radiosensitize solid tumors by inhibiting mitochondrial metabolism. Proc. Natl. Acad. Sci. USA 115, 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Sabatini DM, and Possemato R (2012). Untuning the tumor metabolic machine: targeting cancer metabolism: a bedside lesson. Nat. Med. 18, 1022–1023. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, Wang T, Chen WW, Clish CB, and Sabatini DM (2014). Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 508, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, and Sabatini DM (2015). An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges HR, Jones AJY, Pollak MN, and Hirst J (2014). Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochem. J. 462, 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KK, Spinelli JB, Asara JM, and Toker A (2017). Adaptive reprogramming of de novo pyrimidine synthesis is a metabolic vulnerability in triple-negative breast cancer. Cancer Discov. 7, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Chan DK, Shank JJ, Griffith KA, Fan H, Szulawski R, Yang K, Reynolds RK, Johnston C, McLean K, et al. (2020). Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI Insight 5, e133247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, and Thompson CB (2007). Systemic treatment with the anti-diabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 67, 6745–6752. [DOI] [PubMed] [Google Scholar]
- Cai H, Everett RS, and Thakker DR (2019). Efficacious dose of metformin for breast cancer therapy is determined by cation transporter expression in tumours. Br. J. Pharmacol. 176, 2724–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardaci S, Zheng L, MacKay G, van den Broek NJF, MacKenzie ED, Nixon C, Stevenson D, Tumanov S, Bulusu V, Kamphorst JJ, et al. (2015). Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat. Cell Biol. 17, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, Polak K, Tondera D, Gounarides J, Yin H, et al. (2012). Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 22, 547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamoto K, Chowdhury PS, Kumar A, Sonomura K, Matsuda F, Fagarasan S, and Honjo T (2017). Mitochondrial activation chemicals synergize with surface receptor PD-1 blockade for T cell-dependent antitumor activity. Proc. Natl. Acad. Sci. USA 114, E761–E770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS (2015). Navigating Metabolism (Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Chandel NS, Avizonis D, Reczek CR, Weinberg SE, Menz S, Neuhaus R, Christian S, Haegebarth A, Algire C, and Pollak M (2016). Are metformin doses used in murine cancer models clinically relevant? Cell Metab. 23, 569–570. [DOI] [PubMed] [Google Scholar]
- Chang C-H, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJW, et al. (2015). Metabolic competition in the tumor microenvironment is a driver of cancer progression.Cell 162, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman NM, Zeng H, Nguyen TM, Wang Y, Vogel P, Dhungana Y, Liu X, Neale G, Locasale JW, and Chi H (2018). mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat. Commun. 9, 2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, and Jones ME (1976). The cellular location of dihydroorotate dehydrogenase: relation to de novo biosynthesis of pyrimidines. Arch. Biochem. Biophys. 176, 82–90. [DOI] [PubMed] [Google Scholar]
- Christian S, Merz C, Evans L, Gradl S, Seidel H, Friberg A, Eheim A, Lejeune P, Brzezinka K, Zimmermann K, et al. (2019). The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies. Leukemia 33, 2403–2415. [DOI] [PubMed] [Google Scholar]
- Cluntun AA, Lukey MJ, Cerione RA, and Locasale JW (2017). Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer 3, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney KD, Bezwada D, Mashimo T, Pichumani K, Vemireddy V, Funk AM, Wimberly J, McNeil SS, Kapur P, Lotan Y, et al. (2018). Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 28, 793–800.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, and Chandel NS (2016). Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, and Chandel NS (2020). We need to talk about the Warburg effect. Nat. Metab. 2, 127–129. [DOI] [PubMed] [Google Scholar]
- Dempster JM, Rossen J, Kazachkova M, Pan J, Kugener G, Root DE, and Tsherniak A (2019). Extracting biological insights from the Project Achilles genome-scale CRISPR screens in cancer cell lines. bioRxiv. 10.1101/720243. [DOI] [Google Scholar]
- Dowling RJO, Niraula S, Stambolic V, and Goodwin PJ (2012). Metformin in cancer: translational challenges. J. Mol. Endocrinol. 48, R31–R43. [DOI] [PubMed] [Google Scholar]
- Dowling RJO, Lam S, Bassi C, Mouaaz S, Aman A, Kiyota T, Al-Awar R, Goodwin PJ, and Stambolic V (2016). Metformin pharmacokinetics in mouse tumors: implications for human therapy. Cell Metab. 23, 567–568. [DOI] [PubMed] [Google Scholar]
- El-Mir MY, Nogueira V, Fontaine E, Avéret N, Rigoulet M, and Leverve X (2000). Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J. Biol. Chem. 275, 223–228. [DOI] [PubMed] [Google Scholar]
- Emami Riedmaier A, Fisel P, Nies AT, Schaeffeler E, and Schwab M (2013). Metformin and cancer: from the old medicine cabinet to pharmacological pitfalls and prospects. Trends Pharmacol. Sci. 34, 126–135. [DOI] [PubMed] [Google Scholar]
- Evans JMM, Donnelly LA, Emslie-Smith AM, Alessi DR, and Morris AD (2005). Metformin and reduced risk of cancer in diabetic patients. BMJ 330, 1304–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, and Leder P (2006). Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434. [DOI] [PubMed] [Google Scholar]
- Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, Bosc C, Sugita M, Stuani L, Fraisse M, et al. (2017). Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 7, 716–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Solmonson A, and DeBerardinis RJ (2020). Metabolic reprogramming and cancer progression. Science 368, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt S-M, Bell EL, Keibler MA, Davidson SM, Wirth GJ, Fiske B, Mayers JR, Schwab M, Bellinger G, Csibi A, et al. (2013). Metformin decreases glucose oxidation and increases the dependency of prostate cancer cells on reductive glutamine metabolism. Cancer Res. 73, 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer GM, Jalali A, Kircher DA, Lee W-C, McQuade JL, Haydu LE, Joon AY, Reuben A, de Macedo MP, Carapeto FCL, et al. (2019). Molecular profiling reveals unique immune and metabolic features of melanoma brain metastases. Cancer Discov. 9, 628–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, and Thompson CB (2002). The CD28 signaling pathway regulates glucose metabolism. Immunity 16, 769–777. [DOI] [PubMed] [Google Scholar]
- Fu Z, Ye J, Dean JW, Bostick JW, Weinberg SE, Xiong L, Oliff KN, Chen ZE, Avram D, Chandel NS, and Zhou L (2019). Requirement of mitochondrial transcription factor A in tissue-resident regulatory T cell maintenance and function. Cell Rep. 28, 159–171.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro PA, Yang J, Metelo AM, Pérez-Carro R, Baker R, Wang Z, Arreola A, Rathmell WK, Olumi A, López-Larrubia P, et al. (2013). In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 17, 372–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geltink RIK, Kyle RL, and Pearce EL (2018). Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36, 461–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin PJ, Parulekar WR, Gelmon KA, Shepherd LE, Ligibel JA, Hershman DL, Rastogi P, Mayer IA, Hobday TJ, Lemieux J, et al. (2015). Effect of metformin vs placebo on and metabolic factors in NCIC CTG MA.32. J. Natl. Cancer Inst. 107, djv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss T, Vincent EE, Egnatchik R, Chen J, Ma EH, Faubert B, Viollet B, DeBerardinis RJ, and Jones RG (2015). Metformin antagonizes cancer cell proliferation by suppressing mitochondrial-dependent biosynthesis. PLoS Biol. 13, e1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guièze R, Liu VM, Rosebrock D, Jourdain AA, Hernández-Sánchez M, Martinez Zurita A, Sun J, Ten Hacken E, Baranowski K, Thompson PA, et al. (2019). Mitochondrial reprogramming underlies resistance to BCL-2 inhibition in lymphoid malignancies. Cancer Cell 36, 369–384.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen H-Y, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JMS, Karantza V, et al. (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Parey K, Bublitz M, Mills DJ, Zickermann V, Vonck J, Kühlbrandt W, and Meier T (2016). Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol. Cell 63, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Henkenius K, Greene BH, Barckhausen C, Hartmann R, Märken M, Kaiser T, Rehberger M, Metzelder SK, Parak WJ, Neubauer A, et al. (2017). Maintenance of cellular respiration indicates drug resistance in acute myeloid leukemia. Leuk. Res. 62, 56–63. [DOI] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. (2016). Metabolic heterogeneity in human lung tumors. Cell 164, 681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Tsichlis PN, and Struhl K (2009). Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 69, 7507–7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini M, Dousset L, Mahfouf W, Serrano-Sanchez M, Redonnet-Vernhet I, Mesli S, Kasraian Z, Obre E, Bonneu M, Claverol S, et al. (2018). Energy metabolism rewiring precedes UVB-induced primary skin tumor formation. Cell Rep. 23, 3621–3634. [DOI] [PubMed] [Google Scholar]
- Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Zhan, Yanxiang Guo, J., et al. (2017). Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, Munn LL, and Fukumura D (2002). Dissecting tumour pathophysiology using intravital microscopy. Nat. Rev. Cancer 2, 266–276. [DOI] [PubMed] [Google Scholar]
- Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, and Struhl K (2014). Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl. Acad. Sci. USA 111, 10574–10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Maseda D, Liberti MV, Paz K, Kishton RJ, et al. (2018). Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Tolkunov D, Aviv H, Hakimi AA, Yao M, Hsieh JJ, Ganesan S, Chan CS, and White E (2015). The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Rep. 13, 1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, Davies HR, Papaemmanuil E, Gundem G, Shlien A, et al. ; ICGC Breast Cancer Group; ICGC Chronic Myeloid Disorders Group; ICGC Prostate Cancer Group (2014). Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife 3, e02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Anshu A, Prasad A, Roy S, Jeffery J, Kittipongdaja W, Yang DT, and Schieke SM (2019). Metabolic rewiring in response to biguanides is mediated by mROS/HIF-1a in malignant lymphocytes. Cell Rep. 29, 3009–3018.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, and Dang CV (2011). Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337. [DOI] [PubMed] [Google Scholar]
- Koundinya M, Sudhalter J, Courjaud A, Lionne B, Touyer G, Bonnet L, Menguy I, Schreiber I, Perrault C, Vougier S, et al. (2018). Dependence on the pyrimidine biosynthetic enzyme DHODH is a synthetic lethal vulnerability in mutant KRAS-driven cancers. Cell Chem. Biol. 25, 705–717.e11. [DOI] [PubMed] [Google Scholar]
- Kuntz EM, Baquero P, Michie AM, Dunn K, Tardito S, Holyoake TL, Helgason GV, and Gottlieb E (2017). Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med. 23, 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurelac I, Iommarini L, Vatrinet R, Amato LB, De Luise M, Leone G, Girolimetti G, Umesh Ganesh N, Bridgeman VL, Ombrato L, et al. (2019). Inducing cancer indolence by targeting mitochondrial Complex I is potentiated by blocking macrophage-mediated adaptive responses. Nat. Commun. 10, 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladds MJGW, van Leeuwen IMM, Drummond CJ, Chu S, Healy AR, Popova G, Pastor Fernández A, Mollick T, Darekar S, Sedimbi SK, et al. (2018). A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage. Nat. Commun. 9, 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiguera Á, Hyroššová P, Figueras A, Garzón D, Moreno R, Soto-Cerrato V, McNeish I, Serra V, Lazaro C, Barretina P, et al. (2020). Tumors defective in homologous recombination rely on oxidative metabolism: relevance to treatments with PARP inhibitors. EMBO Mol. Med. 12, e11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. (2012). Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Stine ZE, Nguyen C, Afzal J, Sun P, Hamaker M, Siegel NM, Gouw AM, Kang B-H, Yu S-H, et al. (2014). Tumorigenicity of hypoxic respiring cancer cells revealed by a hypoxia-cell cycle dual reporter. Proc. Nati. Acad. Sci. USA 111, 12486–12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-M, Giltnane JM, Balko JM, Schwarz LJ, Guerrero-Zotano AL, Hutchinson KE, Nixon MJ, Estrada MV, Sánchez V, Sanders ME, et al. (2017). MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 26, 633–647.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yesilkanal AE, Wynne JP, Frankenberger C, Liu J, Yan J, Elbaz M, Rabe DC, Rustandy FD, Tiwari P, et al. (2019). Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature 568, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone RD, Zhao L, Englert JM, Sun I-M, Oh M-H, Sun I-H, Arwood ML, Bettencourt IA, Patel CH, Wen J, et al. (2019). Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts JA, Fiedorczuk K, Degliesposti G, Skehel M, and Sazanov LA (2019). Structures of respiratory supercomplex I+III2 reveal functional and conformational crosstalk. Mol. Cell 75, 1131–1146.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ng SR, Colón CI, Drapkin BJ, Hsu PP, Li Z, Nabel CS, Lewis CA, Romero R, Mercer KL, et al. (2019). Identification of DHODH as a therapeutic target in small cell lung cancer. Sci. Transl. Med. 11, eaaw7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan WM, Schmidt LS, Crooks DR, Wei D, Srinivasan R, Lang M, and Ricketts CJ (2019). The metabolic basis of kidney cancer. Cancer Discov. 9, 1006–1021. [DOI] [PubMed] [Google Scholar]
- Liu X, Romero IL, Litchfield LM, Lengyel E, and Locasale JW (2016). Metformin targets central carbon metabolism and reveals mitochondrial requirements in human cancers. Cell Metab. 24, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord SR, Cheng W-C, Liu D, Gaude E, Haider S, Metcalf T, Patel N, Teoh EJ, Gleeson F, Bradley K, et al. (2018). Integrated pharmacodynamic analysis identifies two metabolic adaption pathways to metformin in breast cancer. Cell Metab. 28, 679–688.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussey-Lepoutre C, Hollinshead KER, Ludwig C, Menara M, Morin A, Castro-Vega L-J, Parker SJ, Janin M, Martinelli C, Ottolenghi C, et al. (2015). Loss of succinate dehydrogenase activity results in dependency on pyruvate carboxylation for cellular anabolism. Nat. Commun. 6, 8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma EH, Verway MJ, Johnson RM, Roy DG, Steadman M, Hayes S, Williams KS, Sheldon RD, Samborska B, Kosinski PA, et al. (2019). Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity 51, 856–870.e5. [DOI] [PubMed] [Google Scholar]
- Madera D, Vitale-Cross L, Martin D, Schneider A, Molinolo AA, Gangane N, Carey TE, McHugh JB, Komarck CM, Walline HM, et al. (2015). Prevention of tumor growth driven by PIK3CA and HPV oncogenes by targeting mTOR signaling with metformin in oral squamous carcinomas expressing OCT3. Cancer Prev. Res. (Phila.) 8, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EA, Marin-Valencia I, Bachoo RM, Mashimo T, Raisanen J, Hatanpaa KJ, Jindal A, Jeffrey FM, Choi C, Madden C, et al. (2012). Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR Biomed. 25, 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes I, Cardona LR, Kong H, Vasan K, McElroy GS, Werner M, Kihshen H, Reczek CR, Weinberg SE, et al. (2020). Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Stratikopoulos E, Ozturk S, Steinbach N, Pegno S, Schoenfeld S, Yong R, Murty VV, Asara JM, Cantley LC, and Parsons R (2017). PTEN regulates glutamine flux to pyrimidine synthesis and sensitivity to dihydroorotate dehydrogenase inhibition. Cancer Discov. 7, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta MM, Weinberg SE, Steinert EM, Chhiba K, Martinez CA, Gao P, Perlman HR, Bryce P, Hay N, and Chandel NS (2018). Hexokinase 2 is dispensable for T cell-dependent immunity. Cancer Metab. 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memmott RM, Mercado JR, Maier CR, Kawabata S, Fox SD, and Dennis PA (2010). Metformin prevents tobacco carcinogen-induced lung tumorigenesis. Cancer Prev. Res. (Phila.) 3, 1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menk AV, Scharping NE, Moreci RS, Zeng X, Guy C, Salvatore S, Bae H, Xie J, Young HA, Wendell SG, and Delgoffe GM (2018). Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 22, 1509–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. (2011). Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. (2017). Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin N, Jaconcic J, Stojanoff V, and Moreno A (2008). High resolution X-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor. Proteins 70, 83–92. [DOI] [PubMed] [Google Scholar]
- Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, McA-foos T, Morlacchi P, Ackroyd J, Agip AA, et al. (2018). An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 24, 1036–1046. [DOI] [PubMed] [Google Scholar]
- Momcilovic M, Jones A, Bailey ST, Waldmann CM, Li R, Lee JT, Abdelhady G, Gomez A, Holloway T, Schmid E, et al. (2019). In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer. Nature 575, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen P-H, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, and DeBerardinis RJ (2011). Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib A, Mathew G, Reczek CR, Watrud K, Ambrico A, Herzka T, Salas IC, Lee MF, El-Amine N, Zheng W, et al. (2018). Mitochondrial complex I inhibitors expose a vulnerability for selective killing of Pten-null cells. Cell Rep. 23, 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M, Xiao Y, Zhou X, Chang J-H, Chang M, Cheng X, Blonska M, Lin X, and Sun S-C (2014). Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 40, 692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Bueno MJ, Zagorac I, Mondejar T, Sanchez J, Mourón S, Muñoz J, Gómez-López G, Jimenez-Renard V, Mulero F, et al. (2016). Targeting tumor mitochondrial metabolism overcomes resistance to antiangiogenics. Cell Rep. 15, 2705–2718. [DOI] [PubMed] [Google Scholar]
- Nigam SK (2018). The SLC22 transporter family: a paradigm for the impact of drug transporters on metabolic pathways, signaling, and disease. Annu. Rev. Pharmacol. Toxicol. 58, 663–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki S, and Gottlieb E (2015). Oncometabolites: tailoring our genes. FEBS J. 282, 2796–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima N, Ishida R, Kishimoto S, Beebe K, Brender JR, Yamamoto K, Urban D, Rai G, Johnson MS, Benavides G, et al. (2020). Dynamic imaging of LDH inhibition in tumors reveals rapid in vivo metabolic rewiring and vulnerability to combination therapy. Cell Rep. 30, 1798–1810.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MR, Doran E, and Halestrap AP (2000). Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 348, 607–614. [PMC free article] [PubMed] [Google Scholar]
- Pardee TS, Lee K, Luddy J, Maturo C, Rodriguez R, Isom S, Miller LD, Stadelman KM, Levitan D, Hurd D, et al. (2014). A phase I study of the first-in-class antimitochondrial metabolism agent, CPI-613, in patients with advanced hematologic malignancies. Clin. Cancer Res. 20, 5255–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parey K, Haapanen O, Sharma V, Köfeler H, Züllig T, Prinz S, Siegmund K, Wittig I, Mills DJ, Vonck J, et al. (2019). High-resolution cryo-EM structures of respiratory complex I: mechanism, assembly, and disease. Sci. Adv. 5, x9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W, Chandel N, Laakso M, Muller WJ, Allen EL, et al. (2013). Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 24, 213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M (2012). The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat. Rev. Cancer 12, 159–169 [DOI] [PubMed] [Google Scholar]
- Pollak M (2014). Overcoming drug development bottlenecks with repurposing: repurposing biguanides to target energy metabolism for cancer treatment. Nat. Med. 20, 591–593. [DOI] [PubMed] [Google Scholar]
- Rawls J, Knecht W, Diekert K, Lill R, and Löffler M (2000). Requirements for the mitochondrial import and localization of dihydroorotate dehydrogenase. Eur. J. Biochem. 267, 2079–2087. [DOI] [PubMed] [Google Scholar]
- Reznik E, Miller ML, Şenbabaoğlu Y, Riaz N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi AA, and Sander C (2016). Mitochondrial DNA copy number variation across human cancers. eLife 5, e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A, Sánchez-Rivera FJ, et al. (2017). Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumsey WL, Schlosser C, Nuutinen EM, Robiolio M, and Wilson DF (1990). Cellular energetics and the oxygen dependence of respiration in cardiac myocytes isolated from adult rat. J. Biol. Chem. 265, 15392–15402. [PubMed] [Google Scholar]
- Rusch M, Nakitandwe J, Shurtleff S, Newman S, Zhang Z, Edmonson MN, Parker M, Jiao Y, Ma X, Liu Y, et al. (2018). Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat. Commun. 9, 3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafoglio CR, Villegas B, Abdelhady G, Bailey ST, Liu J, Shirali AS, Wallace WD, Magyar CE, Grogan TR, Elashoff D, et al. (2018). Sodium-glucose transporter 2 is a diagnostic and therapeutic target for early-stage lung adenocarcinoma. Sci. Transl. Med. 10, eaat5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, Ferris RL, and Delgoffe GM (2016). The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacovelli M, Gonçalves E, Johnson TI, Zecchini VR, da Costa ASH, Gaude E, Drubbel AV, Theobald SJ, Abbo SR, Tran MGB, et al. (2016). Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 537, 544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang C-R, Schumacker PT, Licht JD, Perlman H, et al. (2013). Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, and Shaw RJ (2013). LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell 23, 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lim SK, Liang Q, Iyer SV, Wang H-Y, Wang Z, Xie X, Sun D, Chen Y-J, Tabar V, et al. (2019). Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature 567, 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E-H, Livi CB, Rakheja D, Tan J, Benson D, Parekh V, Kho E-Y, Ghosh AP, Kirkman R, Velu S, et al. (2014). L-2-Hydroxyglutarate: an epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov. 4, 1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff EH, Eberlin LS, Dang VM, Gouw AM, Gabay M, Adam SJ, Bellovin DI, Tran PT, Philbrick WM, Garcia-Ocana A, et al. (2015). MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc. Natl. Acad. Sci. USA 112, 6539–6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegelin MD, Dohi T, Raskett CM, Orlowski GM, Powers CM, Gilbert CA, Ross AH, Plescia J, and Altieri DC (2011). Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J. Clin. Invest. 121, 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siska PJ, Beckermann KE, Mason FM, Andrejeva G, Greenplate AR, Sendor AB, Chiang YJ, Corona AL, Gemta LF, Vincent BG, et al. (2017). Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2, e93411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrtić M, Sriskanthadevan S, Jhas B, Gebbia M,Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, et al. (2011). Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 20, 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart SD, Schauble A, Gupta S, Kennedy AD, Keppler BR, Bingham PM, and Zachar Z (2014). A strategically designed small molecule attacks alpha-ketoglutarate dehydrogenase in tumor cells through a redox process. Cancer Metab. 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Martinez-Garcia E, Nguyen H, Mullen AR, Dufour E, Sudarshan S, Licht JD, Deberardinis RJ, and Chandel NS (2013). The protooncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol. Cell 51, 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, and Vander Heiden MG (2015). Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, and Rao Z (2005). Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121, 1043–1057. [DOI] [PubMed] [Google Scholar]
- Sykes DB (2018). The emergence of dihydroorotate dehydrogenase (DHODH) as a therapeutic target in acute myeloid leukemia. Expert Opin. Ther. Targets 22, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes DB, Kfoury YS, Mercier FE, Wawer MJ, Law JM, Haynes MK, Lewis TA, Schajnovitz A, Jain E, Lee D, et al. (2016). Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell 167, 171–186.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasenko TN, Pacheco SE, Koenig MK, Gomez-Rodriguez J, Kapnick SM, Diaz F, Zerfas PM, Barca E, Sudderth J, DeBerardinis RJ, et al. (2017). Cytochrome c oxidase activity is a metabolic checkpoint that regulates cell fate decisions during T cell activation and differentiation. Cell Metab. 25, 1254–1268.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimoto A, Endo H, Sugiyama M, Fujisawa T, Hosono K, Takahashi H, Nakajima N, Nagashima Y, Wada K, Nakagama H, and Nakajima A (2008). Metformin suppresses intestinal polyp growth in ApcMin/+ mice. Cancer Sci. 99, 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, and DeBerardinis RJ (2017). Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. (2014). Oncogene ablationresistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, and Green DR (2011). The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang K, Wu Q, Kim LJY, Morton AR, Gimple RC, Prager BC, Shi Y, Zhou W, Bhargava S, et al. (2019). Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Sci. Transl. Med. 11, eaau4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O (1956). On the origin of cancer cells. Science 123, 309–314. [DOI] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GRS, and Chandel NS (2010). Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA 107, 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martínez-Reyes I, Gao P, Helmin KA, Abdala-Valencia H, Sena LA, et al. (2019). Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 565, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB,Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, and Chandel NS (2014). Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife 3, e02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. (2011). DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JES, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, and Thompson CB (2011). Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA 108, 19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Stine ZE, Xia J, Lu Y, O’Connor RS, Altman BJ, Hsieh AL, Gouw AM, Thomas AG, Gao P, et al. (2015). Targeted inhibition of tumorspecific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Invest. 125, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. (2012). Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Garcia Canaveras JC, Chen Z, Wang L, Liang L, Jang C, Mayr JA, Zhang Z, Ghergurovich JM, Zhan L, et al. (2020). Serine catabolism feeds NADH when respiration is impaired. Cell Metab. 31, 809–821.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong C, Stewart GD, and Frezza C (2020). Oncometabolites in renal cancer. Nat. Rev. Nephrol. 16, 156–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ždralević M, Brand A, Di Ianni L, Dettmer K, Reinders J, Singer K, Peter K, Schnell A, Bruss C, Decking S-M, et al. (2018). Double genetic disruption of lactate dehydrogenases A and B is required to ablate the “Warburg effect” restricting tumor growth to oxidative metabolism. J. Biol. Chem. 293, 15947–15961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Frederick DT, Wu L, Wei Z, Krepler C, Srinivasan S, Chae YC, Xu X, Choi H, Dimwamwa E, et al. (2016). Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Invest. 126, 1834–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yao Y, Zhang S, Liu Y, Guo H, Ahmed M, Bell T, Zhang H, Han G, Lorence E, et al. (2019). Metabolic reprogramming toward oxidative phosphorylation identifies a therapeutic target for mantle cell lymphoma. Sci. Transl. Med. 11, eaau1167. [DOI] [PubMed] [Google Scholar]
- Zong W-X, Rabinowitz JD, and White E (2016). Mitochondria and cancer. Mol. Cell 61, 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong S, Wu M, Gu J, Liu T, Guo R, and Yang M (2018). Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28, 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.