Abstract
Stearoyl-CoA desaturase 1 (SCD1) is a membrane-embedded metalloenzyme that catalyzes formation of a double-bond on a saturated acyl-CoA. SCD1 has a diiron center and its proper function requires an electron transport chain composed of NADH (or NADPH), cytochrome b5 reductase (b5R), and cytochrome b5 (cyt b5). Since SCD1 is a key regulator in fat metabolism and is required for survival of cancer cells, there is intense interest in targeting SCD1 for various metabolic diseases and cancers. Crystal structures of human and mouse SCD1 were reported recently, however, both proteins have two zinc ions instead of two iron ions in the catalytic center and as a result, the enzymes are inactive. Here we report a general approach for incorporating iron into heterologously expressed proteins in HEK293 cells. We produced mouse SCD1 that contains a diiron center, and visualized its diiron center by solving its crystal structure to 3.5 Å. We assembled the entire electron transport chain using the purified soluble domains of cyt b5 and b5R, and the purified mouse SCD1, and we showed that three proteins coordinate to produce proper products. These results established an in vitro system that allows precise perturbations of the electron transport chain for the understanding of the catalytic mechanism in SCD1.
Keywords: SCD1, membrane enzyme, diiron center, electron transfer, double bond formation
Introduction
Stearoyl-CoA desaturase (SCD) is embedded in the endoplasmic reticulum (ER) membrane and converts saturated acyl-CoAs to monounsaturated acyl-CoAs which are precursors for the biosynthesis of phospholipids, cholesterol esters, and triglycerides [1, 2]. Humans have two SCD genes encoding SCD1 and SCD5 that are of 53% sequence identity, while mice have four (SCD1–SCD4) that are of 77–86% sequence identity. SCD1 is a key enzyme that helps maintain balance between fat consumption and accumulation [3, 4]. Mice with SCD1 knocked out are resistance to high-fat diet, leading to the perspective that drugs that inhibit SCD1 activity could be used to treat metabolic diseases such as obesity and diabetes [5, 6]. Expression of SCD1 is significantly higher in cancer cells due to a higher demand for membrane synthesis, and preclinical research have shown that inhibiting SCD1 activity stops propagation of cancer cells [7–10]. Early biochemical studies demonstrated that the desaturation reaction is catalyzed by a diiron center in SCD1 in a highly regio- and stereo-specific manner [1, 11, 12]. The double bond is formed at the 9th position (Δ9) of the acyl chain (regiospecificity) and with the cis configuration (stereospecificity). SCD1 can utilize saturated acyl-CoAs with an acyl chain length of 14–19 carbons as substrates but favors stearoyl-CoA (18:0 CoA) over others [13].
Crystal structures of both mouse and human SCD1 were reported previously [14, 15]. The mouse SCD1 has an 84% sequence identity with human SCD1 and the two structures are almost identical with a root mean squared deviation (RMSD) of 0.35 Å. SCD1 has a structural fold that is different from soluble desaturases such as acyl-ACP desaturase, which catalyzes a similar reaction [16–18]. SCD1 is a monomer and has a mushroom-like shape with a dimension of 55 × 53 × 41 Å. Each SCD1 has four transmembrane helices (TM1–4) that form the stem of the mushroom and a cytosolic domain that forms the crown of the mushroom (Figure 1a). TM2 and TM4 are longer than TM1 and TM3 and protrude into the cytosolic domain to provide three of the nine histidine residues that coordinate the two metal ions (Figure 1a). The other histidine residues come from the soluble domain. However, further analysis showed that the two metal ions in the structure are zinc ions [14, 15], which is an artefact likely due to overexpression of SCD1 proteins in a heterologous system. In both the mouse and human SCD1 structures, a large non-protein electron density was present, and a stearoyl-CoA was built into the electron density. The coenzyme A moiety is recognized by the surface of the protein while the acyl chain accommodated by a V-shaped tunnel inside the protein (Figure 1b). The dimension of the tunnel is such that Δ9 and Δ10 carbons are positioned at the inflection point of the V-shape and is close to the dimetal center. The V-shaped tunnel also ensures that only the two hydrogens in cis are positioned close to the metal ions for extraction (Figure 1b). Thus, the conformation of the bound stearoyl-CoA provides a simple explanation for both the regioselectivity and stereospecificity imposed by SCD1s.
Figure 1.
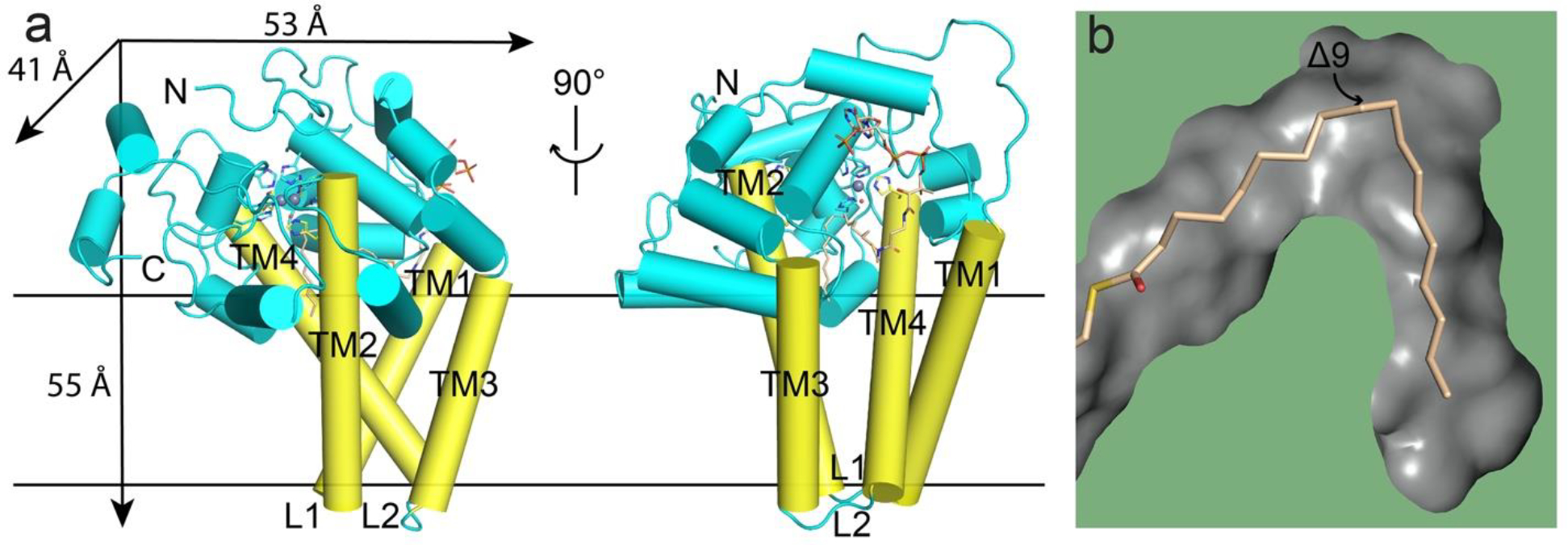
Crystal structure of zinc-containing mouse SCD1 (PDB ID: 4YMK). a. Overall structure of SCD1 shows a mushroom-like architecture with two zinc ions (gray spheres) coordinated by nine histidine residues (side-chain sticks) and one water molecule (red sphere), and one bound stearoyl-CoA (wheat stick). TM, transmembrane helix, in yellow; L, loop region; N and C, N-terminus and C-terminus. b. A long V-shape tunnel (gray surface) buried inside the protein accommodates the acyl chain of the bound acyl-CoA. The Δ9 carbon is in close proximity to the dimetal center located at the turn of the tunnel.
Although Zn2+ serves as a surrogate to maintain the structural integrity because of its similar ionic radius and charge property to a ferrous ion (Fe2+), misincorporation of Zn2+ renders the enzyme inactive and thus hampers further investigation of the enzymatic reaction mechanism. The misincorporation of Zn2+ is not unique to SCD1, and it was also reported in the production of other enzymes such as yeast fatty acid α-hydroxylase [19] and quinol-dependent nitric-oxide reductase [20].
Proper function of SCD1 requires two additional membrane-embedded proteins, cytochrome b5 reductase (b5R) and cytochrome b5 (cyt b5). b5R contains a single transmembrane helix at its N-terminus, and a soluble cytosolic domain composed of an N-terminal and C-terminal halves. A flavin adenine dinucleotide (FAD) binds to the N-terminal half while a NAD(P)H binds to its C-terminal half. The primary redox partner of b5R is cyt b5, which is composed of a soluble cytosolic domain and a single transmembrane domain at the C-terminus. The soluble domain contains a b-type heme, which is ligated to two axial histidine ligands. b5R and cyt b5 are both anchored to the ER membrane by their single transmembrane domain. An electron transport chain is formed sequentially from NADH/NADPH, b5R, and cyt b5, which then interacts with downstream enzymes such as oxygenase and desaturase (Figure 2) [21, 22]. Since it is not known how the three proteins interact, there are two potential ways the electrons can be delivered. Cyt b5 may need to shuttle between its two partners so that binary complexes b5R-cyt b5 and cyt b5-SCD1 form alternately. It’s also possible that a ternary complex of b5R-cyt b5-SCD1 could form and allow for continuous electron transfer.
Figure 2.
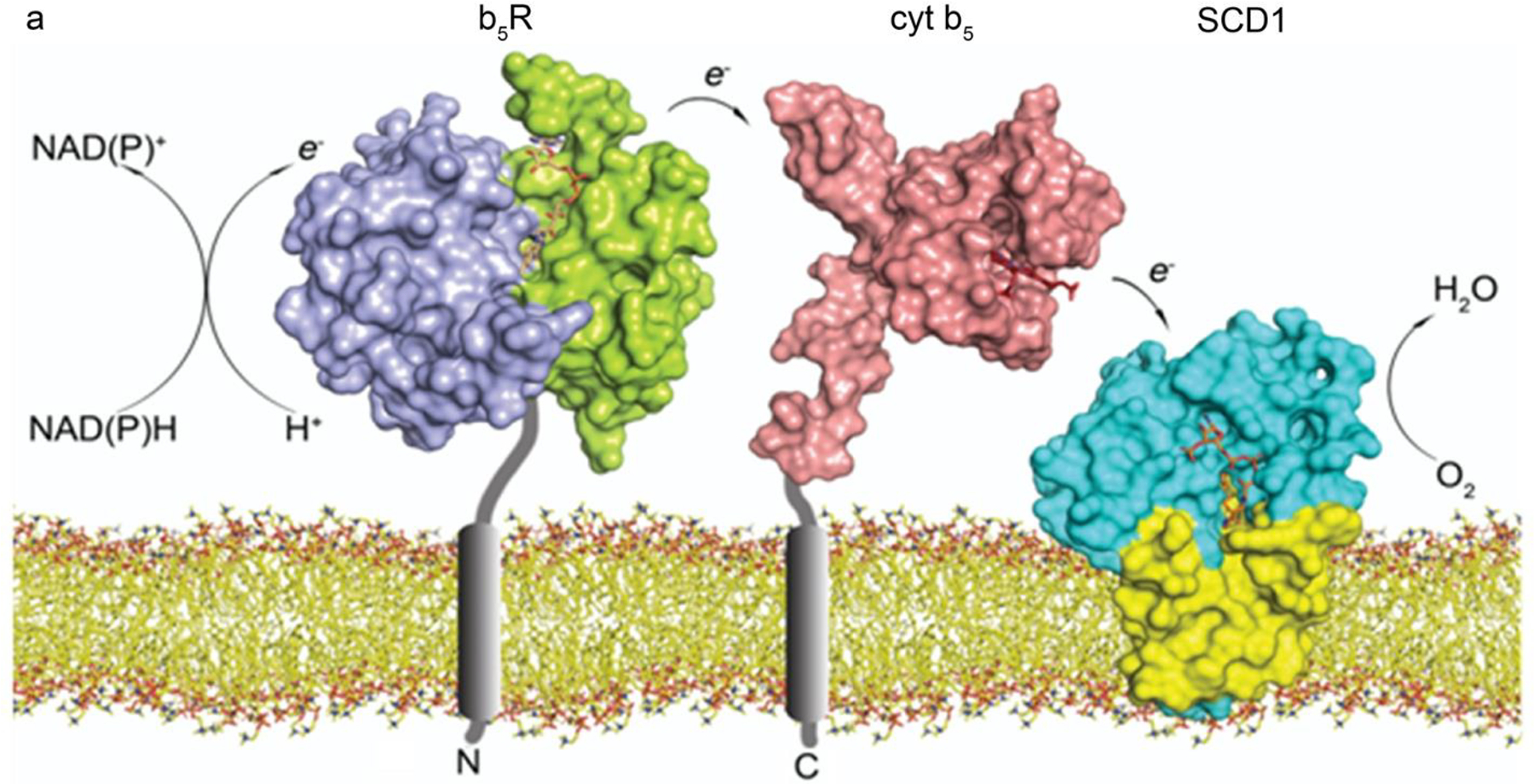
Proteins involved in the desaturation reaction. Electrons transfer sequentially from NADH, to b5R (PDB ID: 1UMK), cyt b5 (PDB ID: 2I96), SCD1 (PDB ID: 4YMK), and molecular oxygen. The single transmembrane domains in b5R and cyt b5 are not in their structures and shown as gray cylinders. Color coding: NADH-binding domain of b5R (purple), FAD-binding domain of b5R (green), FAD (wheat), cyt b5 (pink), heme (red), soluble (cyan) and transmembrane (yellow) domains of SCD1. Lipid bilayer is shown as sticks.
Functional studies on SCD1 have been reported previously [21]. SCD1 activity in isolated rat liver microsomes was monitored by oxidation of NADPH [23, 24]. SCD1 has also been produced in a cell-free translation system and its activity was demonstrated using a radiolabeled substrates [25]. Both systems have provided information on required components for the desaturation reaction. However, these systems are inadequate for structural and spectroscopic studies that require large amount of protein. To further understand the mechanism of SCD1 catalysis, it is imperative that we produce fully functional SCD1 so that the redox state of the bound irons can be tracked by precise spectroscopic studies such as UV-vis spectroscopy and electron paramagnetic resonance and by structural studies via X-ray crystallography. As a first step towards this goal, we developed a protocol to consistently produce iron-containing SCD1 and solved its crystal structure to 3.5 Å. We also purified the soluble domains of cyt b5 and b5R and reconstituted the stearoyl-CoA desaturation reaction in vitro.
Results
Production of iron-containing SCD1 and other proteins
As an initial effort to replace zinc with iron in SCD1s, we added iron in cell culture media but the majority of the purified SCD1 still has zinc ions. This indicates that iron was not efficiently taken up by the insect cells and led us to try the HEK cell expression system. We reasoned that the endogenous transferrin receptor could facilitate iron uptake. We expressed mouse SCD1 in human embryonic kidney (HEK) 293 cells and supplemented the cell culture media with transferrin and ferric ions (Figure 3a). SCD1 produced by this protocol has an iron occupancy of more than 90% as determined by inductively coupled plasma mass spectrometry (ICP-MS) (Figure 3c). After optimizing the expression and purification protocol, we routinely produce ~1 mg pure iron containing SCD1 per liter of cell culture. The iron-loaded SCD1 shows a small and yet significant absorbance peak around 340 nm (A340/A280 ≈ 0.082), which is typical for a non-heme diiron center [26] (Figure 3b).
Figure 3.
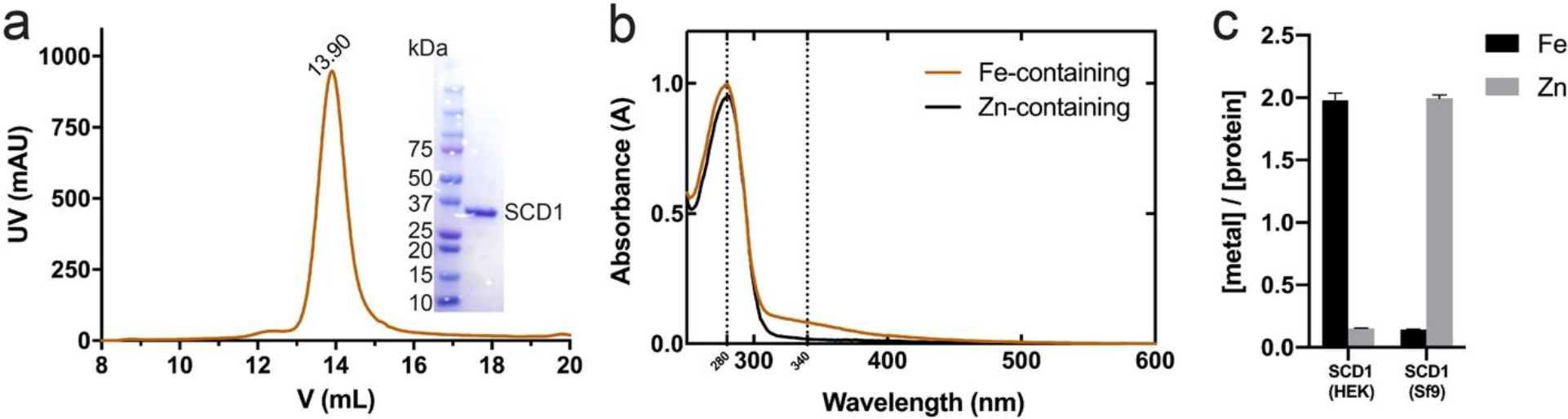
Purification and characterization of HEK-expressed mouse SCD1. a. Size exclusion chromatography (SEC) profile and SDS-PAGE gel of the purified mouse SCD1. b. UV/Vis spectra of iron-containing and zinc-containing SCD1. SCD1 with the diiron center showed a characteristic broad peak around 340 nm. Vertical dash lines mark the absorbance at 280 nm and 340 nm. c. ICP-MS results of SCD1 expressed in HEK or Sf9 cells. The molar ratio of iron to protein in HEK-expressed SCD1 is 1.98 ± 0.06.
Crystal structure of SCD1 with diiron center
We proceeded to crystalize the iron containing mouse SCD1 and obtained crystals using the lipidic cubic phase (LCP) method [27]. These crystals diffracted to ~3.5 Å and the structure was solved by molecular replacement using the previous SCD1 structure as a search model and with both the metal ions and the histidine residues that coordinate the metal ions omitted. Metal ions and histidine residues were then built into electron densities of the “omit” map. A composite omit map covering the entire contents of the unit cell was also calculated to further reduce model bias (Supplementary Figure 1). The final structural model was refined to R/Rfree of 21.9%/ 27.6%; the final structure includes residues 41 to 361, two iron ions, and one oleoyl CoA. The structure of the current iron containing SCD1 is almost identical to the previous zinc-containing SCD1 with a RMSD of only ~0.3 Å for all backbone atoms (Figure 4a). The structures also show that the conserved histidine residues that coordinate the di-metal center have the same coordination geometry (Figure 4a). These results suggest that although Zn2+ cannot catalyze the redox reaction, it is a suitable surrogate for Fe2+ to maintain the three-dimensional structure likely due to its similar ionic radius to Fe2+ (0.88 Å for Zn2+ and 0.92 Å for Fe2+) and charge property.
Figure 4.
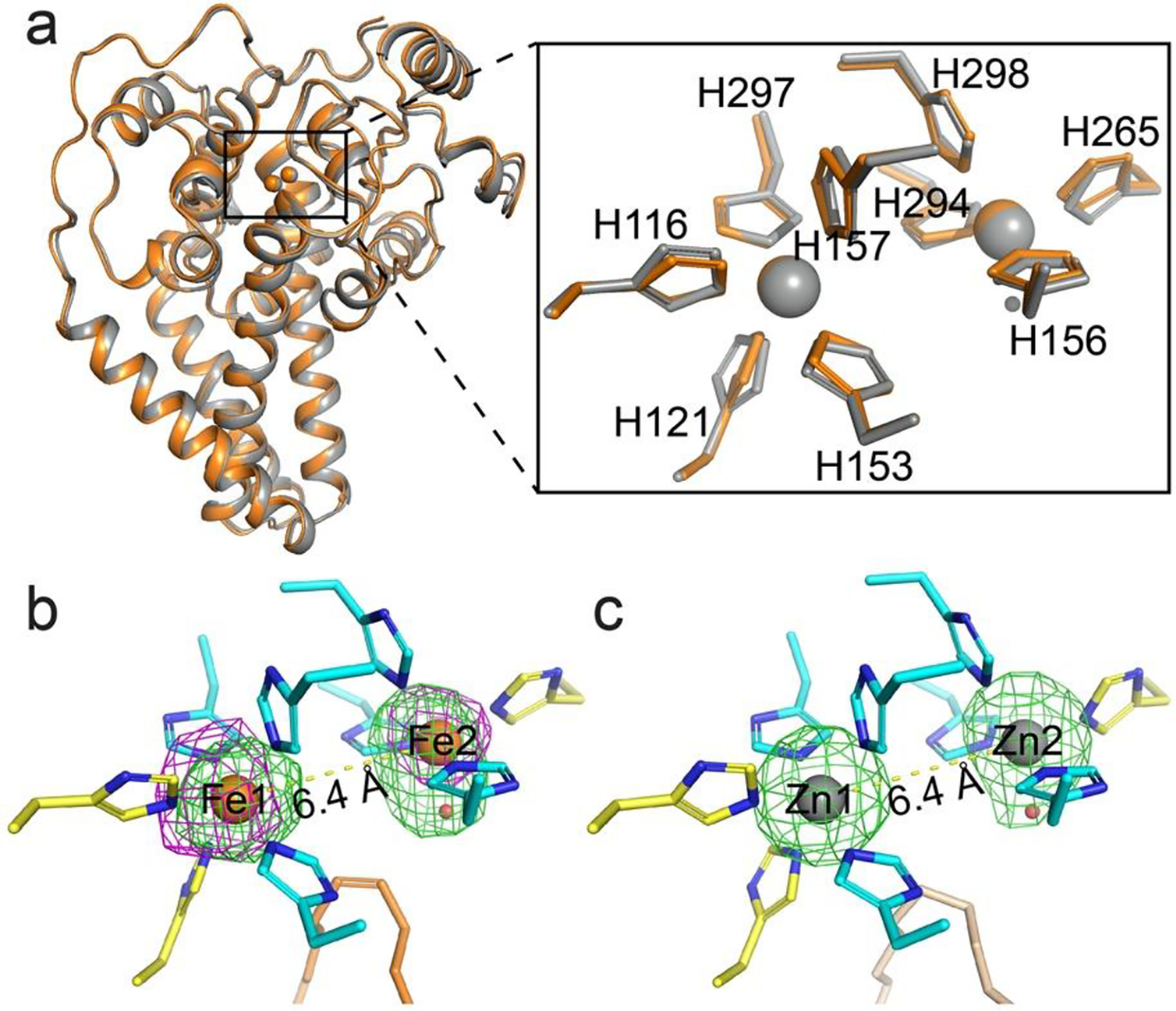
Crystal structure of iron-containing mouse SCD1. a. Structural alignment of iron-containing (orange) and zinc-containing (gray) SCD1. Inset, alignment of the dimetal center and the coordinating histidine residues. b. The reaction center is occupied by a diiron center with a unique coordination scheme that is preserved in the structure. The iron ions (orange) and a water molecule (red) are shown as spheres. Nine histidine ligands (residues from TMs in yellow; from the soluble domain in cyan) and the oleoyl-CoA (orange) are shown as sticks. Green mesh, Fo - Fc map with iron ions omitted contoured at 3 σ; magenta mesh, anomalous difference map at 3.5 σ. c. Metal coordination in the zinc-containing structure (PDB ID: 4YMK). An elongated density is also observed at Zn2. The stearoyl-CoA are shown as wheat stick. Green mesh, Fo - Fc map with zinc ions omitted contoured at 4.0 σ.
In addition to the ICP-MS results, the presence of two iron ions was confirmed by anomalous x-ray scattering signal, which unequivocally locates the iron ions in the structure (Figure 4b). The two iron ions are separated by ~6.4 Å, similar to that between the two zinc ions in the previous SCD1 structures (Figure 4c) [14, 15]; This distance exceeds the range allowed for the formation of known reaction intermediates, such as cis-μ−1,2 peroxo or diferryl, proposed in other enzymes with a diiron center. One of the iron ions (Fe1) is coordinated by five histidine residues while another (Fe2) by four and a putative water molecule. These histidine residues are highly conserved and segregate into four histidine-rich motifs that are distant in the primary sequence [14]. Both the coordination by all histidine residues and the distance between the ions are different from all other diiron centers found in soluble redox enzymes, such as acyl-ACP desaturase [28], ribonucleotide reductase [29], and methane monooxygenase [30, 31] (Supplementary Figure 2). There is no obvious electron density between the two iron ions, suggesting that an oxo-bridge, which is a common feature in other diiron centers, is either not present or short-lived. These properties of the diiron center suggest that the activation of molecular oxygen in SCD1 has a novel mechanism.
Oleoyl-CoA is present in both structures
As is observed in the zinc-containing SCD1, one acyl-CoA molecule is present in our structure (Figure 5a). In the current structure, clear and strong electron density is present for the CoA moiety, while the electron density for the acyl chain is less continuous, likely due to the modest resolution and a lower occupancy of the acyl-CoA. In the previous structure which has a higher resolution, the electron density is continuous for the entire acyl-CoA (Figure 5b) [14].
Figure 5.
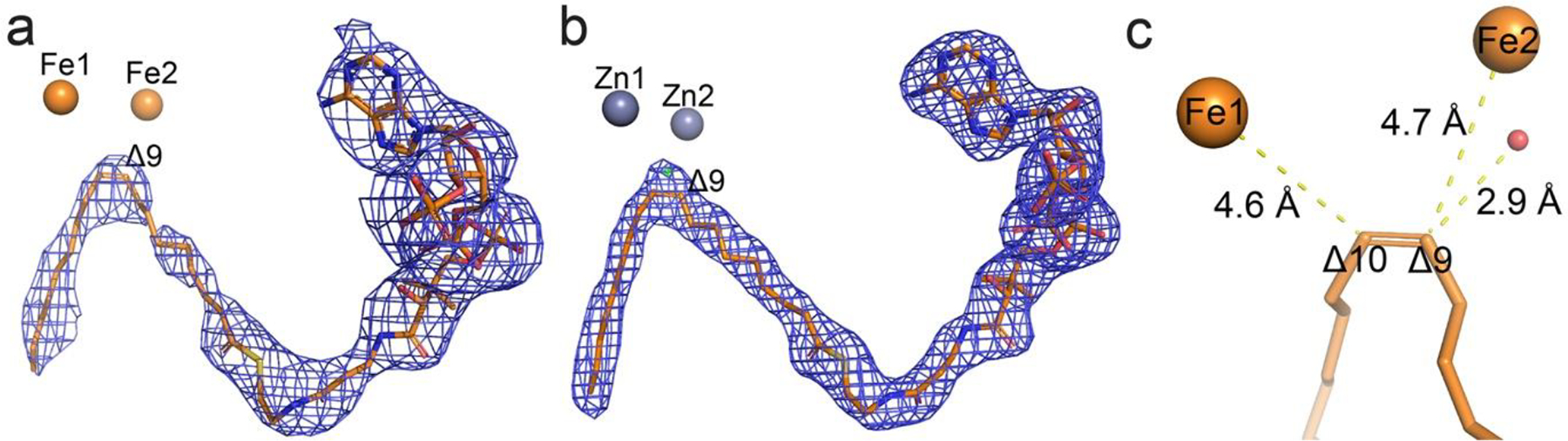
An oleoyl-CoA is bound to the SCD1. a. The oleoyl-CoA molecule (orange stick) fits well in the iron-containing structure. Blue mesh, 2Fo - Fc map at 1.0 σ. b. An oleoyl-CoA (orange stick) was modeled to into the non-protein electron density in the previous zinc-containing structure (PDB ID: 4YMK). Blue mesh, 2Fo - Fc map at 1.5 σ; green mesh, Fo - Fc map at 4.0 σ. The extra density is present whether an oleayl-CoA or a stearoyl-CoA was modeled. c. Distances between the two irons and the Δ9 and Δ10 carbons on the oleoyl-CoA.
We developed a protocol to extract and identify the bound acyl-CoA from the SCD1 and examined the SCD1 produced from the previous insect cell expression system and the current HEK cell expression system. We found that the bound ligand is oleoyl-CoA and its molar ratio to protein is close to 1:1 (Methods). Therefore, we built an oleoyl-CoA into the previous mouse SCD1 structure (PDB ID: 4YMK) and refined the model. It turns out that an 18:1 acyl chain fits better at the Δ9 and Δ10 position than the previously built 18:0 acyl chain (Figure 5b). This geometry restraint imposed by the double bond results in slightly different conformation of the zigzag string of acyl chain between the Δ10 carbon and the thioester sulfur, whose positioning is stabilized by the extensive interactions between CoA group and SCD1. The Δ9 carbon on the oleoyl-CoA is ~3 Å to the water bound to Fe2 (Figure 5c).
Electron transfer from cyt b5 to SCD1
We expressed and produced the soluble domains of cyt b5 and b5R (Supplementary Figure 3a and 3c). Spectroscopic features of the two proteins confirm the presence of appropriate cofactors [32, 33]. In the presence of b5R, addition of NADH can reduce the heme group in cyt b5 as demonstrated by a shift of the Soret peak from 413 nm to 423 nm, which indicates that both proteins function properly (Supplementary Figure 3b and 3d).
To examine the redox communication between cyt b5 and SCD1, we measured the kinetics of cyt b5 oxidation in presence of SCD1. We first prepared ferrous cyt b5 with b5R and NADH. Reduced cyt b5 can be gradually oxidized by molecular oxygen in solution, which interferes with the electron transfer between cyt b5 and SCD1. Therefore, the reactions were carried out anaerobically. The SCD1 accelerates the oxidation of cyt b5, which exhibits a biphasic time course with a k1 = 1.86 min−1 and k2 = 0.246 min−1 as observed by following the decrease of Soret peak at 423 nm (Figure 6). These results indicate that the purified cyt b5 and SCD1 are both functional and form an electron transport pair.
Figure 6.
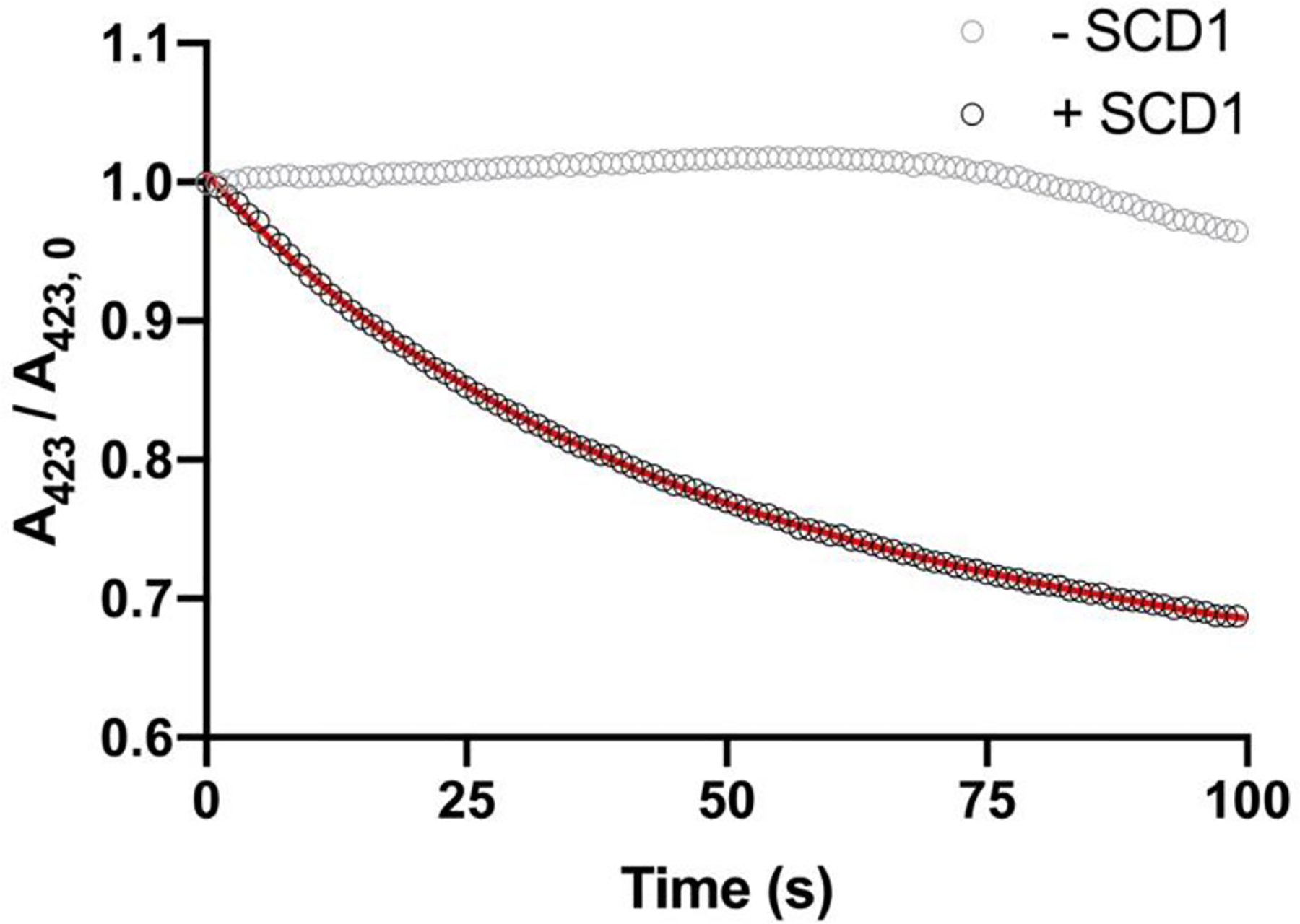
Electron transfer between cyt b5 and SCD1. Ferrous cyt b5 can be oxidized by diferric SCD1 as followed by Soret peak at 423 nm (black circle). A double exponential decay model was fitted to the data as shown in red line. Cyt b5 remained in its reduced state without SCD1 in anaerobic environment (gray circle).
In vitro assembly of the continuous turnover system
We next tested if we can assemble an in vitro enzymatic reaction. Detergent-solubilized SCD1, the cytosolic domains of cyt b5 and b5R, and stearoyl-CoA were included in the reaction mixture, and the reaction was initiated by the addition of NADH. The reactions were stopped at different time points and the amount of oleoyl-CoA was determined (Methods) (Figure 7a). Detection of de novo synthesis of oleoyl-CoA provides unequivocal evidence that we have assembled an in vitro system for steady-state turnover. The initial rate of oleoyl-CoA production at different concentrations of stearoyl-CoA was measured and a Michaelis-Menten curve was fitted to the data with a KM and a kcat of 6.3 ± 1.2 μM and 2.78 ± 0.59 min−1, respectively (Figure 7b).
Figure 7.
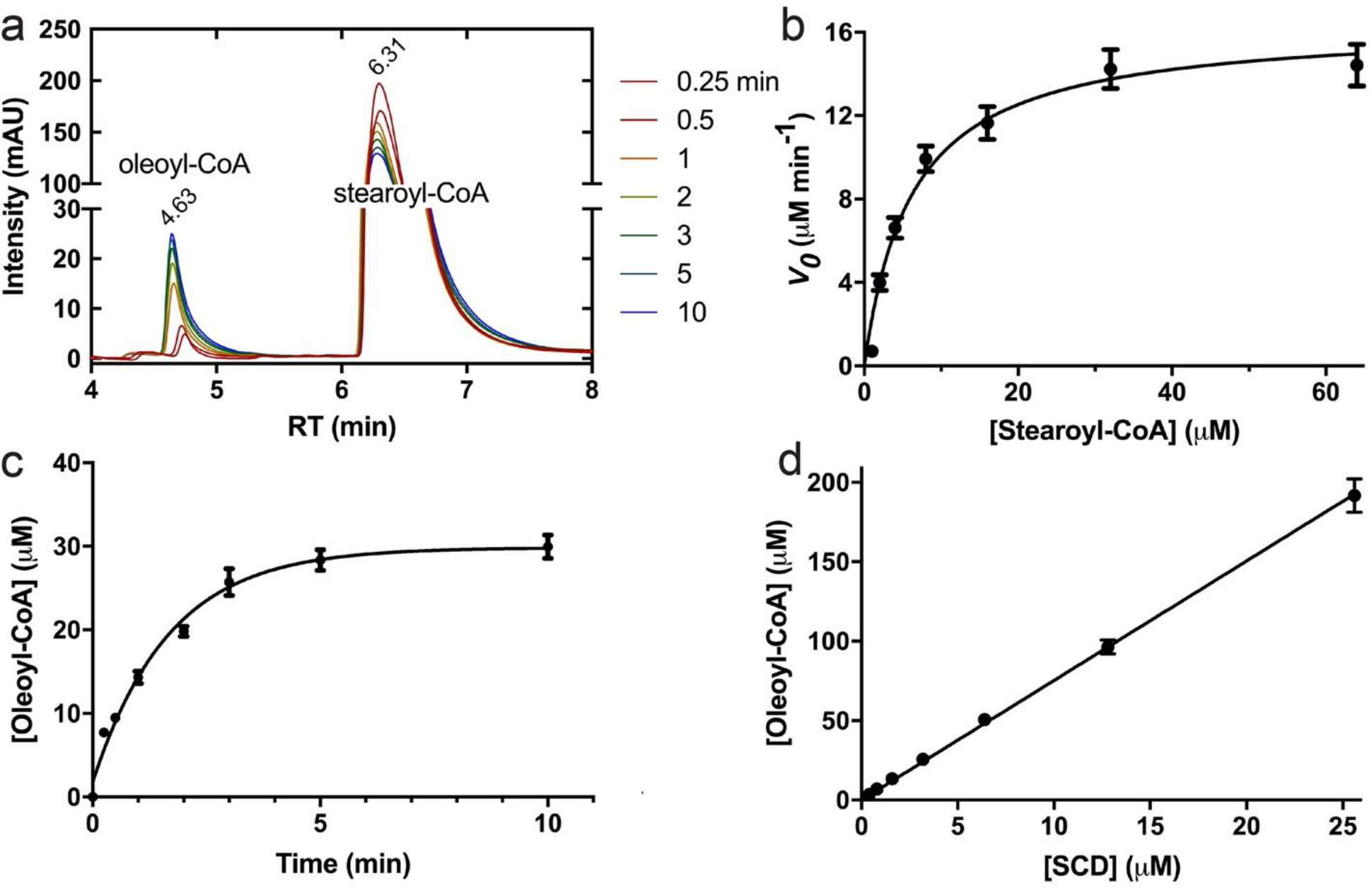
Continuous turnover reaction of SCD1. a. HPLC profiles of reaction products in the assembled electron transfer chain including SCD1, cyt b5 and b5R, quenched at different time points (left). Increasing peak of oleoyl-CoA and simultaneous decreasing peak of stearoyl-CoA confirmed stearoyl-CoA to oleoyl-CoA conversion. b. Michaelis-Menten kinetics of the steady-state enzymatic reaction quantified by initial rates of oleoyl-CoA production before reaching product inhibition. All the error bars stand for experiments of three repeats. c. The time course of oleoyl-CoA production showed a profound product inhibition as most of the excess stearoyl-CoA was not reacted (>90%). d. Linear correlation of final [product] with the [SCD1] in the presence of excess substrate.
The production of oleoyl-CoA stalled after ~10 minutes well before the exhaustion of substrate stearoyl-CoA, and no more oleoyl-CoA was produced even with addition of more NADH (Figure 7c). The maximum oleoyl-CoA production was proportional to the amount of SCD1 in the system, and the reaction seemed to stall with a turnover number of 7.52 ± 0.047 (Figure 7d). We speculate that product inhibition occurs because of limited capacity of a detergent micelle around SCD1.
Discussion
When the SCD1 was misincorporated with Zn2+, the bound Zn2+ resists extraction by extensive dialysis or high-affinity zinc chelators like TPEN (Methods). The current protocol provides an efficient procedure for large-scale production of iron-containing SCD1. We anticipate that this protocol can be adapted for production of other iron-containing proteins.
The structures of SCD1 show a tightly bound acyl-CoA, which we have identified to be an oleoyl-CoA even in the non-functional Zn-containing SCD1. After refinement, the distance between Δ9 and Fe2 is 4.7 Å, which is slightly less than that in the previous structure (Figure 5c). We speculate that the release of oleoyl-CoA may require larger conformational changes of SCD1 because the acyl chain becomes more rigid when a double bond is present. We suspect that product inhibition could be due to a combination of higher apparent affinity towards oleoyl-CoA and limited diffusion of oleoyl-CoA in detergent micelles surrounding SCD1. Such product inhibition may be lifted when SCD1 are on the ER membrane, where free diffusion and drainage of products by downstream enzymes, such as DGAT and ACAT [21], will reduce product concentration, thus sustaining continuous turnover.
SCD1 is embedded in ER membrane while cyt b5 and b5R are anchored to the ER membrane by their single transmembrane helix (Figure 2). This configuration limits their diffusion to 2-dimension and enhances their interactions. The rate-limiting step in our in vitro assembled chain is the interactions between cyt b5 and SCD1, as the rate of electron transfer from cyt b5 to SCD1 (k1 = 1.86 min−1) is comparable to the steady-state rate of oleoyl-CoA formation (kcat = 2.78 min−1). Since our experiments are performed with the three proteins all in the solution, their communications are likely not as efficient as in the native environment. Further optimization could come from reconstitution of all three proteins into a nanodisc or generating fusion proteins of either two or all three partners. Fusion of a cyt b5-like domain occurs in certain desaturases, such as OLE-1 [34] and FAT5–6 [35], which may reflect a strategy of achieving efficient electron transfer in the evolution of these enzymes.
Methods
Iron incorporation into Sf9-expressed SCD1
Mouse SCD1 was expressed and purified from Sf9 (Spodoptera frugiperda) cells as previously described [14]. We tried several approaches, and found that the most efficient way of replacing zinc ions was to dialyze SCD1 first against two strong zinc chelators, N,N,N’,N’-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine (TPEN) (100 μM) and dipicolinic acid (DPA) (10 mM), and then against high concentrations of ferrous ions (10 mM). However, the maximum iron occupancy was less than 70% as determined by ICP-MS and there was a significant loss of protein during the required prolonged dialysis. Since both zinc ions have to be replaced in order for the protein to be functional, a 70% occupancy translates to less than 50% (0.72) of functional protein.
Construct of mouse SCD1
Mouse SCD1 gene with deletion of residue 2–23 at the N-terminus [14] (mΔSCD1) was codon-optimized for human cell expression and subcloned into a BacMam expression vector [36] with green fluorescent protein (GFP) tag attached to the C-terminus. A tobacco etch virus (TEV) protease recognition site was inserted between the C-terminus of the protein and the GFP tag.
Overexpression and purification of SCD1 from HEK293 cells
The resulting plasmid containing cDNA of mouse SCD1 was used to generate baculoviruses following the manufacturer’s protocol (Invitrogen). HEK293S GnTI- suspension cells at 3×106 cells/mL in Freestyle 293 medium (GIBCO) supplemented with 2% FBS were infected with 10% P3 virus. Iron-saturated human transferrin (Sigma) in 50 mM sodium bicarbonate (pH 8.5) and 10 mM FeCl3 solution were filtered and added to the cell culture to reach a final concentration of 5 mg/L transferrin and 5 μM FeCl3. Infected cells were incubated at 37°C overnight (~16 h). The protein expression was then induced by adding 10 mM sodium butyrate and temperature was decreased to 30 °C for two days (~48 h) before harvesting [36]. Cell pellets were homogenized in buffer A containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM PMSF, 5 mM MgCl2, and DNaseI. 40 mM n-Decyl-β-D-Maltopyranoside (DM, Anatrace) was added to solubilize the membranes at 4 °C for 2 h under gentle agitation. After centrifugation (55,000g, 45 min, 4 °C) to remove the insoluble fraction, the supernatant containing the detergent-solubilized proteins was collected and loaded onto a column of CNBR-activated Sepharose beads (GE Healthcare) coupled to GFP nanobody (GFPnb). The GFPnb was derived from a llama single chain antibody and was expressed and purified as previously reported [37]. The columns were pre-equilibrated with buffer B [20 mM HEPES (pH 7.5), 150 mM NaCl, 4 mM DM]. After 1 h incubation at 4 °C, GFPnb resins were washed with 20 column volume of buffer B. The proteins were cleaved off the column with TEV protease at room temperature for 1 h. The TEV protease with a polyhistidine tag was removed by loading the supernatant to pre-equilibrated cobalt-based affinity resins (Talon, Clontech). The released proteins were collected, concentrated (Amicon 50 kDa cutoff, Millipore), and loaded onto a size-exclusion column (Superdex 200 10/300 GL, GE Health Sciences) equilibrated with buffer B. Peak fractions were collected and analyzed by SDS-PAGE. The predicted molecular mass and extinction coefficient at 280 nm of mΔSCD1 are 39,512 g/mol and 73,800 M−1cm−1 [38], respectively, and were used to calculate protein concentration.
ICP-MS
HEK-expressed and Sf9-expressed SCD1 were diluted to 15 μM in buffer B. 300 μL of each sample together with blank buffer were subjected to ICP-MS (Agilent 8800 Triple-Q-ICP-MS) at the ICP Analytical Research Lab at University of Houston. Iron and zinc standards were analyzed at the beginning and the end of each run to set up the calibration line. All samples were prepared in 2% HNO3 prior to analysis.
Crystallization and structure determination
Crystallization trials were carried out in LCP methods [27]. Purified SCD1 were concentrated to 30–50 mg/mL. Protein solution was mixed with molten monoolein (Sigma) doped with 10% (w/w) cholesterol (Sigma) [39] at 1:1.5 (v/w) ratio with a coupled syringe device. Gryphon crystallization robot was used to dispense 50 nL protein-lipid mixture into a 96 well glass sandwich plate (Molecular Dimensions) and overlaid with 800 nL precipitant solution in each well. Crystallization conditions were optimized based on an initial crystal hit (100 mM Tris, pH 8.5, PEG400 35%, 50 mM Mg(Ace)2) from a home-made grid screen. The typical condition for good crystal growth is: 100 mM Tris, pH 7.8–8.2, PEG400 37–42%, 20 mM Mg(Ace)2, and 50 mM MgCl2. Crystals were harvested and flash frozen in liquid nitrogen directly without cryoprotection.
Crystals were screened at beamlines 24ID (NE-CAT) at the Advanced Photon Source at the Argonne National Laboratory. A dataset was collected with a resolution of 3.51 Å at a wavelength of 0.9791 Å. The diffraction images were indexed, integrated, and scaled using the XDS [40]. The phases were obtained by the molecular replacement (MR) method in Phenix [41] with mouse SCD1 structure (PDB: 4YMK) [14] as a search model. The obtained solution was subjected to successive rounds of refinement in phenix.refine [41] and manual correction in COOT [42]. Protein geometry was validated by MolProbity [43]. Datasets collected at 1.7389 Å near iron edge were of poorer quality, which diffracted to ~4 Å. X-ray fluorescence emission spectrum confirms the presence of iron in the crystals. The diffraction data were processed similarly by XDS, which indicated that anomalous signals exist in these datasets. Refined model from the datasets of normal wavelength were used as search model. After several rounds of refinement, the resulting anomalous difference map was inspected and four predominant peaks were found in one asymmetric unit, which corresponds to two diiron centers. All structure figures were produced with PyMOL (Schrödinger LLC.).
Overexpression and purification of soluble cyt b5 and b5R
The cDNAs of mouse cyt b5 and b5R were subcloned into a pET vector which appends a polyhistidine tag and a TEV protease site to the C-terminus (for cyt b5) or the N-terminus (for b5R) of the overexpressed protein. High-level expression was achieved by following the previous protocols for soluble cyt b5 [44] and b5R [32]. Cells expressing cyt b5 were supplemented with 0.5 mM δ-aminolevulinic acid, 5 μM FeCl3, and 1 μM hemin chloride, and those expressing b5R with 100 μM FAD in the media. Cell pellets were resuspended in buffer A and then sonicated until cells appeared fully lysed. The following steps are similar to the SCD1 purification protocol. Target protein capture was achieved by cobalt-based affinity resins (Talon, Clontech) and proteins were cleaved off the resins with TEV protease. Buffers without DM and concentrators with 10 kDa cutoff were used.
Electron transfer kinetic measurement
3.1 μM soluble cyt b5-b5R and 3.6 μM SCD1 in buffer B were placed in two tonometers; the solutions were then made anaerobic by 5 cycles of alternate 30 s of degassing followed by 4.5 min of gas displacement with argon. The tonometers and a NADH stock prepared in anaerobic buffer were transferred into an anaerobic chamber. Anaerobic kinetic measurements were conducted with a SX-18MV stopped-flow apparatus (Applied Photophysics, Leatherhead, UK) placed inside the anaerobic chamber. Reduction of cyt b5 domain was achieved by addition of 3.1 μM NADH. The time course of A423 in the reaction between reduced cyt b5 domain and SCD1 was monitored. The decay of A423 was fit with a double exponential function: A = A0 + ΔA1*e−k1*t + ΔA2*e−k2*t, where A and A0 are absorbance and final absorbance, respectively; ΔA1 and ΔA2 are the amplitudes of the two phases; k1 and k2 are the rate constants of the two phases; t is the reaction time.
Continuous turnover reactions and product quantification
Typically, the steady-state reaction system includes detergent-solubilized SCD1 (2 μM), cyt b5 (10 μM), b5R (20 μM), and stearoyl-CoA (300 μM) in buffer B, and the reaction was triggered by the addition of NADH (1 mM). The reaction mixture was quenched at different time points with an acyl-CoA extraction buffer containing 25% iso-propanol and 50% acetonitrile (ACN) in phosphate buffer pH 5.3. We used a previously published protocol [45, 46] with modifications to separate and quantify acyl-CoAs. Briefly, the quenched reaction mixtures were clarified by a brief centrifugation to remove protein precipitates, and the supernatants were loaded in a C18 reversed phase column on a HPLC system. A linear gradient from 50% (v/v) ACN in phosphate buffer pH 5.3 to 70% was applied to elute acyl-CoAs, and the elution of acyl-CoAs was monitored at 254 nm. The identity of oleoyl-CoA and stearoyl-CoA was confirmed by comparing the retention times (RT) to the standard samples and by the mass spectrometry coupled to the HPLC (LC-MS). The amount of acyl-CoAs was calculated based on the integrated area under peak. To generate the Michaelis-Menten curve against [stearoyl-CoA], 1–75 μM of stearoyl-CoA was used in the reaction system. Time courses at different [stearoyl-CoA] were recorded. Straight lines were used to fit time courses of oleoyl-CoA production within 1 min after reaction started. The initial velocities were calculated from the slopes of these straight lines.
Accession Numbers
The coordinate and the structure factor have been deposited in the Worldwide Protein Data Bank (wwPDB) with accession number 6WF2.
Supplementary Material
Highlights.
A general approach for large-scale production of iron-containing proteins
Crystal structure of mouse stearoyl-CoA desaturase 1 with a diiron center
In vitro assembly of an electron transfer chain composed of NADH, cytochrome b5 reductase, cytochrome b5, and stearoyl-CoA desaturase 1
Acknowledgements
This work was supported by grants from NIH (DK122784 to MZ and AT, HL086392 and GM098878 to MZ), Cancer Prevention and Research Institute of Texas (R1223 to MZ). We acknowledge the access to Northeastern Collaborative Team (NE-CAT) beamlines at the Advanced Photon Source, which is supported by a grant from the National Institute of General Medical Sciences (P30GM124165). Crystals were screened and diffraction data were collected at beamline 24-IDC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
Reference
- [1].Strittmatter P, Spatz L, Corcoran D, Rogers MJ, Setlow B, Redline R. Purification and Properties of Rat Liver Microsomal Stearyl Coenzyme A Desaturase. Proceedings of the National Academy of Sciences. 1974;71:4565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Strittmatter P, Enoch HG. Methods in Enzymology. Section II Microsomal electron transport and cytochrome P-450 systems. 1978;52:188–93. [DOI] [PubMed] [Google Scholar]
- [3].Ntambi JM, Miyazaki M. Regulation of stearoyl-CoA desaturases and role in metabolism. Prog Lipid Res. 2004;43:91–104. [DOI] [PubMed] [Google Scholar]
- [4].Hodson L, Fielding BA. Stearoyl-CoA desaturase: rogue or innocent bystander? Progress in lipid research. 2013;52:15–42. [DOI] [PubMed] [Google Scholar]
- [5].Gutiérrez-Juárez R, Pocai A, Mulas C, Ono H, Bhanot S, Monia BP, et al. Critical role of stearoyl-CoA desaturase-1 (SCD1) in the onset of diet-induced hepatic insulin resistance. Journal of Clinical Investigation. 2006;116:1686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ntambi JM, Miyazaki M, Stoehr JP, Lan H, Kendziorski CM, Yandell BS, et al. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci U S A. 2002;99:11482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends in cell biology. 2014;24:472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Theodoropoulos PC, Gonzales SS, Winterton SE, Rodriguez-Navas C. Discovery of tumor-specific irreversible inhibitors of stearoyl CoA desaturase. 2016;12:218–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tracz-Gaszewska Z, Dobrzyn P. Stearoyl-CoA Desaturase 1 as a Therapeutic Target for the Treatment of Cancer. Cancers (Basel). 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Winterton SE, Capota E, Wang X, Chen H, Mallipeddi PL, Williams NS, et al. Discovery of Cytochrome P450 4F11 Activated Inhibitors of Stearoyl Coenzyme A Desaturase. J Med Chem. 2018;61:5199–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bloomfield DK, Bloch K. The formation of delta 9-unsaturated fatty acids. J Biol Chem. 1960;235:337–45. [PubMed] [Google Scholar]
- [12].Jones PD, Holloway PW, Peluffo RO, Wakil SJ. A requirement for lipids by the microsomal stearyl coenzyme A desaturase. The Journal of biological chemistry. 1969;244:744–54. [PubMed] [Google Scholar]
- [13].Enoch HG, Catalá A, Strittmatter P. Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. The Journal of biological chemistry. 1976;251:5095–103. [PubMed] [Google Scholar]
- [14].Bai Y, McCoy JG, Levin EJ, Sobrado P, Rajashankar KR, Fox BG, et al. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature. 2015;524:252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang H, Klein MG, Zou H, Lane W, Snell G, Levin I, et al. Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate. Nat Struct Mol Biol. 2015;22:581–5. [DOI] [PubMed] [Google Scholar]
- [16].Behrouzian B, Savile CK, Dawson B, Buist PH, Shanklin J. Exploring the Hydroxylation-Dehydrogenation Connection: Novel Catalytic Activity of Castor Stearoyl-ACP Δ9Desaturase. Journal of the American Chemical Society. 2002;124:3277–83. [DOI] [PubMed] [Google Scholar]
- [17].Guy JE, Whittle E, Kumaran D, Lindqvist Y, Shanklin J. The crystal structure of the ivy Delta4–16:0-ACP desaturase reveals structural details of the oxidized active site and potential determinants of regioselectivity. The Journal of biological chemistry. 2007;282:19863–71. [DOI] [PubMed] [Google Scholar]
- [18].Whittle EJ, Tremblay AE, Buist PH, Shanklin J. Revealing the catalytic potential of an acyl-ACP desaturase: Tandem selective oxidation of saturated fatty acids. Proceedings of the National Academy of Sciences. 2008;105:14738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhu G, Koszelak-Rosenblum M, Connelly SM, Dumont ME, Malkowski MG. The Crystal Structure of an Integral Membrane Fatty Acid alpha-Hydroxylase. J Biol Chem. 2015;290:29820–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, et al. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science (New York, NY). 2010;330:1666–70. [DOI] [PubMed] [Google Scholar]
- [21].Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. American Journal of Physiology - Endocrinology and Metabolism. 2009;297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vergeres G, Waskell L. Cytochrome b5, its functions, structure and membrane topology. Biochimie. 1995;77:604–20. [DOI] [PubMed] [Google Scholar]
- [23].Joshi VC, Wilson AC, Wakil SJ. Assay for the terminal enzyme of the stearoyl coenzyme A desaturase system using chick embryo liver microsomes. Journal of lipid research. 1977;18:32–6. [PubMed] [Google Scholar]
- [24].Strittmatter P, Rogers MJ. Apparent dependence of interactions between cytochrome b5 and cytochrome b5 reductase upon translational diffusion in dimyristoyl lecithin liposomes. Proc Natl Acad Sci U S A. 1975;72:2658–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Goren MA, Fox BG. Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expression and Purification. 2008;62:171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jeffcoat R, Brawn PR, Safford R, James AT. Properties of rat liver microsomal stearoyl-coenzyme A desaturase. Biochemical Journal. 1977;161:431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lindqvist Y, Huang W, Schneider G, Shanklin J. Crystal structure of delta9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. Embo j. 1996;15:4081–92. [PMC free article] [PubMed] [Google Scholar]
- [29].Högbom M, Huque Y, Sjöberg B-M, Nordlund P. Crystal Structure of the Di-iron/Radical Protein of Ribonucleotide Reductase fromCorynebacterium ammoniagenes†,‡. Biochemistry. 2002;41:1381–9. [DOI] [PubMed] [Google Scholar]
- [30].Sazinsky MH, Lippard SJ. Correlating Structure with Function in Bacterial Multicomponent Monooxygenases and Related Diiron Proteins. Accounts of Chemical Research. 2006;39:558–66. [DOI] [PubMed] [Google Scholar]
- [31].Rosenzweig AC, Brandstetter H, Whittington DA, Nordlund P, Lippard SJ, Frederick CA. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins. 1997;29:141–52. [PubMed] [Google Scholar]
- [32].Bando S, Takano T, Yubisui T, Shirabe K, Takeshita M, Nakagawa A. Structure of human erythrocyte NADH-cytochrome b5 reductase. Acta Crystallographica Section D: Biological Crystallography. 2004;60:1929–34. [DOI] [PubMed] [Google Scholar]
- [33].Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol Ther. 2003;97:139–52. [DOI] [PubMed] [Google Scholar]
- [34].Stukey JE, McDonough VM, Martin CE. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–9. [PubMed] [Google Scholar]
- [35].Watts JL, Browse J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochemical and biophysical research communications. 2000;272:263–9. [DOI] [PubMed] [Google Scholar]
- [36].Goehring A, Lee C-HH, Wang KH, Michel JC, Claxton DP, Baconguis I, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nature protocols. 2014;9:2574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–8. [DOI] [PubMed] [Google Scholar]
- [38].Gasteiger E, Hoogland C, Gattiker A, Duvaud Se, Wilkins MR, Appel RD, et al. Protein Identification and Analysis Tools on the ExPASy Server In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press; 2005. p. 571–607. [Google Scholar]
- [39].Caffrey M A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr F Struct Biol Commun. 2015;71:3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kabsch W XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. [DOI] [PubMed] [Google Scholar]
- [43].Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mulrooney SB, Waskell L. High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b(5). Protein Expr Purif. 2000;19:173–8. [DOI] [PubMed] [Google Scholar]
- [45].Golovko MY, Murphy EJ. An improved method for tissue long-chain acyl-CoA extraction and analysis. J Lipid Res. 2004;45:1777–82. [DOI] [PubMed] [Google Scholar]
- [46].Su C, Gullberg H, Simko H, Luthman M, Edlund P-OO, Lundbäck T. A novel assay of cellular stearoyl-CoA desaturase activity of primary rat hepatocytes by HPLC. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2010;878:2427–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


