TO THE EDITOR
The human epidermis is a stratified epithelial tissue that protects the body from the outside environment. The integrity of this tissue relies on the balance of self-renewal and differentiation of the stem and progenitor keratinocyte population of the basal layer (Ge and Fuchs, 2018). Issues affecting various aspects of the differentiation program can result in a compromised skin barrier. A compromised barrier can lead to diseases such as ichthyosis, atopic dermatitis, and psoriasis which can impact up to 20% of the population (Lopez-Pajares et al., 2013).
Work from our lab and others have shown that differentiation gene expression depends on the activity of lineage determining transcription factors (LDTFs) such as KLF4, MAF/MAFB, CEBPA/B, ZNF750, and GRHL3 (Boxer et al., 2014, Hopkin et al., 2012, Lopez et al., 2009, Lopez-Pajares et al., 2015, Segre et al., 1999, Sen et al., 2012). While these factors have been studied extensively, the downstream mechanisms by which they regulate transcriptional control of epidermal differentiation genes remains poorly understood. It has recently been shown in acute myeloid leukemia cells that BRD4 works downstream of hematopoietic LDTFs to promote their transcriptional program (Roe et al., 2015). BRD4 belongs to the bromodomain and extra-terminal domain (BET) family and utilizes its bromodomains to bind to acetylated lysine groups on both histones (i.e. H3K27ac) and transcription factors. It has been shown to bind active promoters and enhancers to promote gene transcription at the initiation and elongation steps (Donati et al., 2018, Lee et al., 2017, Roe et al., 2015).
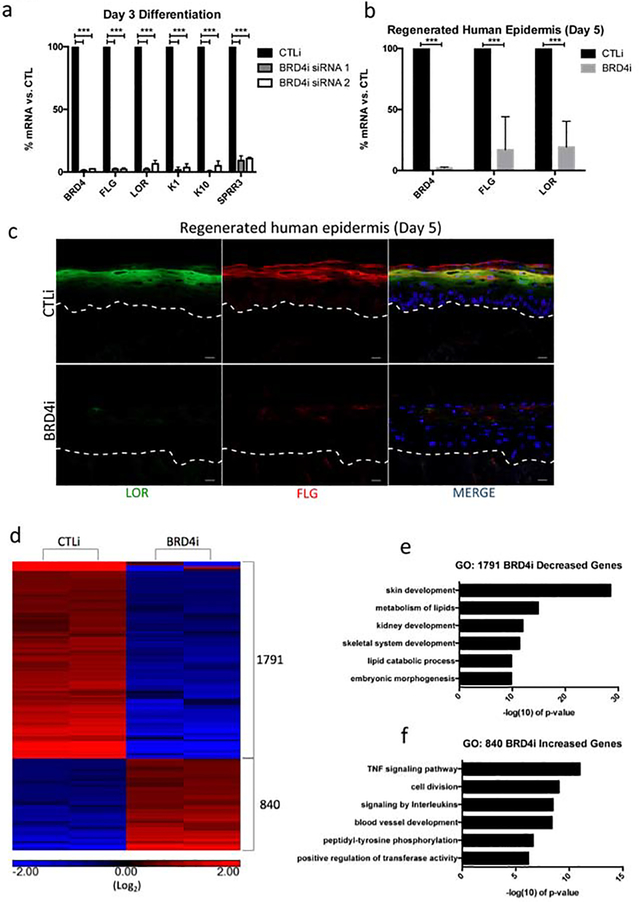
To determine if BRD4 is necessary for differentiation gene expression, keratinocytes were treated with scrambled control (CTLi) or BRD4 targeting (BRD4i) siRNA and cultured in differentiation conditions for three days. Two distinct siRNAs targeting different regions of BRD4 were used to ensure the phenotype seen was not the result of off-target siRNA effects. Knockdown of BRD4 resulted in a significant reduction in critical differentiation genes such as K1, K10, LOR, FLG, and SPRR3 (Figure 1a). In regenerated human epidermis, BRD4 depletion resulted in the loss of K10 protein expression but did not impact K5 levels (Supplementary Figure 1a). Knockdown of BRD4 blocked terminal epidermal differentiation as evidenced by the reduction in stratum corneum formation and loss of structural proteins such as FLG and LOR on the mRNA and protein level (Figure 1b–c, Supplementary Figure 1b). To characterize the BRD4 knockdown phenotype on a genome wide transcriptional level, RNA sequencing (RNA-Seq) was performed on replicate control and BRD4 knockdown samples cultured in differentiation conditions for three days. Analysis of BRD4 knockdown associated differential gene expression (≥2 fold change vs. CTLi and FDR ≤ 0.05) revealed 1,791 genes with decreased expression and 840 genes with increased expression (Figure 1d, Supplementary Table 1). More than twice the number of genes was downregulated upon BRD4 depletion than upregulated which supports the role of BRD4 as a transcriptional activator. Gene ontology (GO) analysis of the 1,791 decreased genes revealed a significant enrichment for differentiation related terms, such as skin development and metabolism of lipids (Figure 1e). Analysis of the 840 increased genes revealed terms related to cell division, as well as immune system functions (Figure 1f). Together, these data suggest that BRD4 is necessary for promoting epidermal differentiation gene expression while suppressing cell cycle and immune response genes.
Figure 1. BRD4 is necessary for epidermal differentiation gene expression.
(a) RT-qPCR quantifying the relative mRNA expression levels of a panel of epidermal differentiation genes in scrambled control (CTLi) and BRD4 (BRD4i) siRNA treated keratinocytes after three days of differentiation. Two separate siRNAs (siRNA 1 and siRNA 2) targeting different regions of BRD4 mRNA were used (siRNA 1 n=4, siRNA 2 n=3). Statistics: t-test, ***p < 0.001. (b) RT-qPCR quantifying the relative mRNA expression levels of LOR, BRD4, and FLG in CTLi and BRD4i day 5 regenerated human epidermis. n=5 (c) Immunofluorescent staining of late differentiation markers LOR (green) and FLG (red) in CTLi and BRD4i day 5 regenerated human epidermis. Merged image includes Hoechst staining of nuclei. n=3. White Scale bar = 20μm. (d) Heatmap generated for replicate (n=2) RPKM normalized RNASequencing data from CTLi and BRD4i keratinocytes differentiated for 3 days. The expression of genes significantly increased (red) or decreased (blue) is shown. Differential expression was determined with FDR ≤ 0.05 and fold change ≥ 2 vs. CTLi. Graphs are displayed in log2 scale. (e) Gene ontology (GO) term enrichment for the 1,791 genes significantly decreased in BRD4 knockdown cells. (f) Gene ontology (GO) term enrichment for the 840 genes significantly increased in expression in BRD4 knockdown cells.
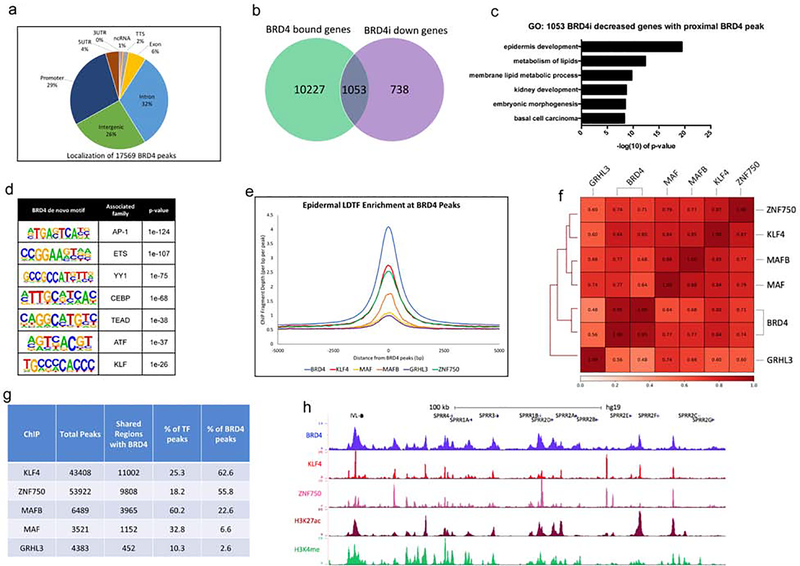
To characterize the genomic localization of BRD4, we performed chromatin immunoprecipitation followed by high throughput sequencing (ChIP-Seq) for BRD4 in differentiated keratinocytes (day 3). ChIP-Seq was performed on replicate BRD4 pulldowns and analysis was done using HOMER (Heinz et al., 2010). 17,569 BRD4 peaks were identified which were primarily located in intronic (32%), intergenic (26%), and promoter (29%) regions (Figure 2a, Supplementary Table 2). Validation of these results by BRD4 ChIP followed by QPCR showed high enrichment for BRD4 at the bound sites whereas no binding was seen with IgG control pulldowns (Supplementary Figure 2a). Since BRD4 is primarily known as a transcriptional activator that binds to enhancers and promoters, we wanted to explore which genes are dependent upon BRD4 for expression(Lee et al., 2017, Roe et al., 2015). Thus, we mapped back the 17,569 BRD4 peaks to its nearest genes which resulted in the identification of 11,280 genes (Figure 2b, Supplementary Table 2). We overlapped the genes that BRD4 are bound with those genes decreased in expression upon BRD4 knockdown (Figure 2b). Importantly, ~70% (1,053/1,791) of the genes downregulated upon BRD4 knockdown were found to have a proximal BRD4 peak, and these genes were enriched for GO terms related to differentiation such as epidermis development and metabolism of lipids (Figure 2b–c). To gain insight into the types of genomic regions BRD4 is binding, we used the ENCODE ChIP-Seq data set as well as previously published data for human keratinocytes for a variety of histone marks (Consortium, 2012, Hopkin et al., 2012, Rubin et al., 2017). These marks included repressive marks H3K27me3 and H3K9me3 (undifferentiated keratinocytes), as well as H3K4me (day 1 of differentiation, representing primed regions of DNA upon differentiation) and H3K27ac (day 3 differentiation, marking open chromatin in the differentiated state) (Consortium, 2012, Hopkin et al., 2012, Rubin et al., 2017). Mapping of these histone marks to the 17,569 BRD4 bound peaks showed high enrichment for H3K27ac, as well as H3K4me (Supplementary Figure 2b). This correlates with the binding affinity of BRD4 to acetylated lysines and its activity as a transcriptional activator. In contrast, the repressive marks of H3K27me3/H3K9me3 had no enrichment at BRD4 regions (Supplementary Figure 2b). This relationship between BRD4 and the various histone marks was confirmed by generating Pearson correlation coefficients between the various sequencing experiments, with H3K27ac having the highest correlation with BRD4 (avg=0.74) (Supplementary Figure 2c). Due to the high enrichment of H3K27ac and H3K4me at BRD4 bound sites, we wanted to characterize how many of these regions were active enhancers or active promoters. We found 11,832 regions of BRD4 co-localization with both H3K27ac and H3K4me (Supplementary Figure 2d). Most of these regions are active enhancers (82%) while a smaller portion are active promoters (18%) (Supplementary Figure 2d). These co-bound regions were mapped to the nearest genes and compared to genes decreased in the BRD4 knockdown. 42.2% (756/1,791) of BRD4i decreased genes were found to have proximal BRD4 bound active enhancer(s) which were highly enriched for the GO term epidermis development (Supplementary Figure 2e). 11.2% (200/1,791) of the genes decreased in the BRD4i cells had an active promoter bound by BRD4 and were enriched for the GO terms metabolism of lipids and regulation of epidermis development (Supplementary Figure 2f). This suggests that BRD4 may be regulating differentiation through both active enhancers and promoters.
Figure 2. BRD4 shares similar genomic binding profiles with epidermal LDTFs.
(a) Genomic localization of the 17,569 BRD4 peaks identified by HOMER. BRD4 ChIP sequencing was performed in replicates (n=2) from day 3 differentiated keratinocytes. (b) Venn diagram showing the number of genes (1,053) decreased in the BRD4 knockdown that also have an associated BRD4 peak. (c) Gene ontology (GO) term enrichment for the 1,053 genes that are decreased in the BRD4 knockdown and have an associated BRD4 peak. (d) de novo motif enrichments and the associated family of transcription factors identified within the 17,569 BRD4 peaks. (e) Mean density profile displaying epidermal LDTF ChIP-Seq profiles (KLF4: red, ZNF750: green, MAFB: orange, MAF: yellow, GRHL3: purple) centered around BRD4 peaks (blue). The 17,569 BRD4 peaks were used as the reference coordinates and the profiles are displayed +/− 5kb from the BRD4 peak centers. (f) Heatmap plot of Pearson correlation coefficients between replicate BRD4 ChIP-Seq data sets and epidermal LDTF ChIP-Seq data sets (RPKM normalized). (g) Table displaying the peak overlap statistics between BRD4 and epidermal LDTFs. Each LDTFs total bound peak numbers identified by HOMER are shown with the number and percentage of overlap with BRD4 peaks. (h) UCSC genome browser track displaying BRD4 (blue), KLF4 (red), ZNF750 (pink), H3K27ac (maroon), and H3K4me (green) ChIP-Seq profiles near IVL and a cluster of SPRR differentiation genes.
Next, we sought to investigate the relationship between BRD4 and LDTFs of the skin. Motif enrichment analysis of the 17,569 BRD4 peaks revealed an association with families of transcription factors known to regulate epidermal differentiation, such as AP1, ETS, CEBP, and KLF, suggesting that BRD4 may share binding regions with these factors (Figure 2d). Based on this data, we used previously generated ChIP-Seq data sets for LDTFs of the skin including KLF4, ZNF750, MAF, MAFB, and GRHL3 to compare to BRD4 (Boxer et al., 2014, Hopkin et al., 2012, Lopez-Pajares et al., 2015). All of these factors were found to have signal enrichment at BRD4 binding sites, with KLF4, ZNF750, and MAFB having the most enrichment (Figure 2e). This relationship between BRD4 and the LDTFs was confirmed though high Pearson correlation coefficients (Figure 2f). BRD4 had the highest correlation with KLF4 (0.82) followed by ZNF750 (0.725). Analysis of the total peak overlap between BRD4 and the LDTFs also revealed a substantial overlap with KLF4 and ZNF750 (62.6% and 55.8% of BRD4 peaks, respectively) (Figure 2g). Visualization of BRD4 with KLF4 and ZNF750 showed substantial co-localization at active enhancer and promoter regions proximal to IVL, DSG1, DSC1, and the SPRR and keratin family of epidermal differentiation genes (Figure 2h, Supplementary Figure 2g–h).
Since the genomic binding of KLF4 and BRD4 were the most similar to each other, we decided to knockdown KLF4 and perform RNA-Seq. 772 genes were downregulated upon KLF4 depletion which were enriched for epidermal differentiation related GO terms (Supplementary Figure 3a–b, Supplementary Table 3). 1,265 genes were increased upon KLF4 knockdown which were cell cycle and proliferation related (Supplementary Figure 3a,3c, Supplementary Table 3). Comparison of the transcriptome between BRD4 and KLF4 showed positive Pearson correlation (avg=0.70) (Supplementary Figure 3d). ~60% (461/772) of the genes downregulated upon KLF4 depletion was also decreased in BRD4i cells (Supplementary Figure 3e). These genes are enriched for a variety of terms related to skin differentiation (Supplementary Figure 3f). 16.4% (208/1,265) of genes increased in the KLF4i cells were also upregulated in the BRD4i dataset which were enriched for proliferation related GO terms (Supplemental Figure 3g–h). It is also worth noting that the number of genes dependent on BRD4 (1,791) for expression is much greater than KLF4 (772). This could possibly be due to BRD4 being necessary to activate the genes of other epidermal LDTFs such as ZNF750, MAFB, and MAF. Thus, loss of BRD4 may be similar to loss of multiple epidermal LDTFs simultaneously.
Together, these data suggest that BRD4 and epidermal LDTFs functionally overlap and is thus necessary for the epidermal differentiation gene expression program.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (NIH R01AR072590 and R01AR066530) to G.L. Sen.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DATA AVAILABILITY
ChIP-Seq and RNA-Seq data was deposited in GEO with accession number: GSE140992
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev 2014;28(18):2013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer 2018;17(1):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Fuchs E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet 2018;19(5):311–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38(4):576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin AS, Gordon W, Klein RH, Espitia F, Daily K, Zeller M, et al. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS genetics 2012;8(7):e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Park YK, Park S, Jang Y, Waring N, Dey A, et al. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nature communications 2017;8(1):2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez RG, Garcia-Silva S, Moore SJ, Bereshchenko O, Martinez-Cruz AB, Ermakova O, et al. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol 2009;11(10):1181–90. [DOI] [PubMed] [Google Scholar]
- Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, et al. A LncRNA-MAF:MAFB Transcription Factor Network Regulates Epidermal Differentiation. Dev Cell 2015;32(6):693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA. Genetic pathways in disorders of epidermal differentiation. Trends Genet 2013;29(1):31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell 2015;58(6):1028–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AJ, Barajas BC, Furlan-Magaril M, Lopez-Pajares V, Mumbach MR, Howard I, et al. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat Genet 2017;49(10):1522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 1999;22(4):356–60. [DOI] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, et al. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell 2012;22(3):669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.