Abstract
Yarrowia lipolytica has emerged as an important non-model host for terpene production. However, three main challenges remain in industrial production using this yeast. First, considerable knowledge gaps exist in metabolic flux across multiple compartments, cofactor generation, and catabolism of non-sugar carbon sources. Second, many enzymatic steps in the complex-terpene synthesis can pose rate-limitations, causing accumulation of toxic intermediates and increased metabolic burdens. Third, metabolic shifts, morphological changes, and genetic mutations are poorly characterized under industrial fermentation conditions. To overcome these challenges, systems metabolic analyses, protein engineering, novel pathway engineering, model-guided strain design, and fermentation optimization, have been attempted with some successes. Further developments that address these challenges are needed to advance the Yarrowia nonmodel yeast platform for industrial-scale production of high-value terpenes, including those with highly complex structures such as the anticancer withanolides and insecticidal limonoids.
Keywords: acetyl-CoA, genetic stability, metabolic burden, metabolic shift, peroxisome
Graphical Abstract
Introduction
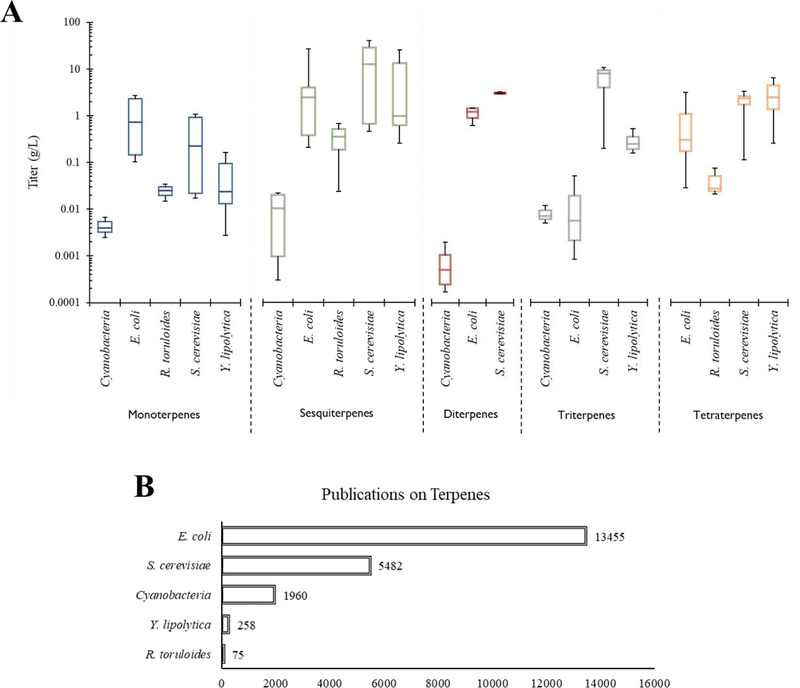
Extensive research has been aimed at producing terpenes (a vast group of high value natural compounds) using microbial hosts. Model organisms like Escherichia coli and Saccharomyces cerevisiae have been widely used for the production of terpenes as nutraceuticals [1], fuels (e.g., bisabolene [2]), and pharmaceuticals such as cannabinoids [3], with encouraging results. The microbial hosts and production titers for terpenes of different molecular weights (monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), triterpenes (C30), and tetraterpenes (C40)) are summarized in Figure 1. However, for commercial applications, E. coli has drawbacks such as bacterial toxin contamination, acetate inhibition, low activity of expressed plant-based enzymes, and an inability to post-translationally modify and compartmentalize complex proteins. S. cerevisiae also has drawbacks such as relatively slow growth rates, complex growth medium requirements, and the Crabtree effect. Recently, new microbial platforms, including non-model yeasts, microalgae, and consortia, have been developed via their unique traits for terpene biosynthesis [1,4,5]. For example, the red yeast Rhodosporidium toruloides is capable of using cheap feedstocks such as aromatics, glycerol, and methane, while cyanobacteria can utilize CO2. Among these non-model hosts, oleaginous yeast Yarrowia lipolytica exhibits high chemical tolerance and robust secretion abilities, and has received strong research interests, facilitating the rapid development of relevant genetic and systems biology tools [6]. These tools include genome editing via CRISPR/Cas9, novel secretion pumps, promoter tuning, pathway assembly, and modular cloning strategies, as well as genome-scale modeling to guide rational metabolic engineering [7]. Thus, Y. lipolytica has emerged as an attractive non-conventional host, and has been demonstrated for its capability of high terpenoid production (e.g., 6 g/L β-carotene [8]).
Figure 1: A survey of microbial terpene production.
A. Terpene production titers relative to hosts and terpenes, with the data mostly derived from a recent review [4]. B. The number of terpene-related publications for each host, according to a PubMed (PMC) search conducted on January 15, 2020, based on the keyword combination “(host) AND (isoprenoid or terpene or terpenoid)”.
Knowledge gaps in Y. lipolytica metabolism
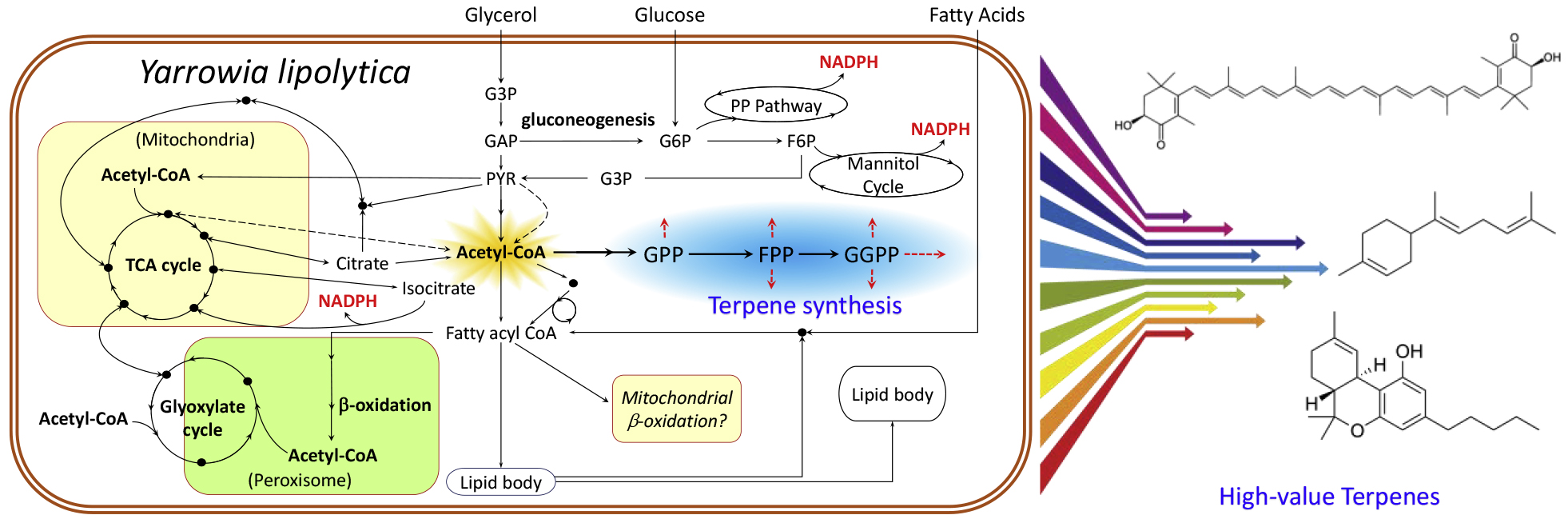
There are still grand challenges in using nonmodel yeasts for biomanufacturing of terpenes. Detailed metabolic understanding is necessary for performing rational strain engineering, but knowledge gaps still exist in Y. lipolytica metabolism [9,10]. First, cytosolic acetyl-CoA is the key terpene pathway precursor, but its generation is not completely elucidated, particularly when Y. lipolytica metabolizes non-sugar based feedstock. ATP-citrate lyase (ACL) is known to cleave cytosolic citrate to form acetyl-CoA and considered the major generation route. However, there are multiple putative acetyl-CoA generation and consumption routes, and acetyl-CoA translocation between compartments (mitochondria or peroxisome) is not yet fully understood. For example, Y. lipolytica can degrade fatty acids via β-oxidation and is able to grow on them as a sole carbon source. Acetyl-CoA is generated in the peroxisomes, but the transportation of peroxisomal acetyl-CoA to the cytosol is unclear. One likely route is shuttling via glyoxylate (Glyox) cycle intermediates, which can enter the mitochondria from the peroxisome. Another potential route is the carnitine shuttle that translocates acetyl-CoA between mitochondria and cytosol [11]. However, the activity of carnitine acetyltransferases in the oleaginous yeast is still not well-characterized compared to S. cerevisiae [12]. Finally, the cytosolic pyruvate bypass can produce acetyl-CoA via the intermediate acetate, but this pathway’s contribution and activity during growth on non-acetate carbon sources are not quantified in Y. lipolytica. Different engineering efforts have been explored to enhance cytosolic acetyl-CoA and push the flux towards mevalonate. The pyruvate bypass and pyruvate dehydrogenase (PDH) overexpression have resulted in the highest increases of product titers; modification of the peroxisomal structural protein may also result in increased acetyl-CoA supplies for biosynthesis. However, enzyme engineering of ACL is less effective to rewire acetyl-CoA flux [11,13–15].
The second main knowledge gap involves NADPH generation, which is the reducing power for terpene synthesis. In Y. lipolytica grown using glucose as the carbon source, the oxidative pentose phosphate pathway was shown to be the primary source of NADPH for lipid overproduction [16]. However, it is unclear what is mainly responsible for NADPH generation when other carbon sources such as lipids are used. Most commonly, a cytosolic NADP+-dependent malic enzyme can provide NADPH during oleaginous microbial growth [17]. However, Y. lipolytica lacks a cytosolic version of the malic enzyme, while the mitochondrial version is NAD+-dependent [17]. Expression of a heterologous NADP+-malic enzyme in the cytosol of Y. lipolytica did not lead to an increase in lipid productivity [18]. Therefore, Y. lipolytica may employ multiple cofactor generation routes, including a cytosolic NADP+-dependent isocitrate dehydrogenase (ICDH) for NADPH synthesis. Another possible source of NADPH in many non-model yeasts is the mannitol cycle, which converts NADH and ATP to NADPH [19]. Transcriptional analysis of Y. lipolytica cultures indicated that both the mannitol cycle and ICDH were upregulated to support lipogenic activities [20]. Genome-scale modeling also showed that the ICDH and mannitol cycle could be two key NADPH sources in Y. lipolytica [21].
Technological difficulties in constructing multi-step pathways for terpene synthesis
Engineering terpene synthesis requires multiple heterologous enzymes that may pose rate limitations, flux imbalances, accumulation of toxic intermediates, and metabolic burdens. Some enzymes, such as the cytochrome P450 oxygenase [26,27], are difficult to functionally express in heterologous hosts [28]. Further, many secondary metabolite pathways involve metabolite channeling via membrane or covalent binding of multiple pathway enzymes [29]. The multi-enzyme complexes help to channel the flux of substrates into different branches of the pathways [30]. A better understanding of the native channeling mechanisms will improve the efficacy of the heterologously expressed pathways while also reducing metabolic burdens and metabolite transport challenges [31–33]. Some successes in synthetic multi-enzyme complexes have been met by utilizing protein scaffolds [34,35] and by natural organelle engineering to cluster cascade enzymes. In addition, innate channels may prevent the release of intermediate metabolites to engineered pathways. For example, heterologous enzymes could not effectively hijack the geranyl diphosphate (GPP) precursor for monoterpene synthesis in Yarrowia [36,37]. Thus, a rich area for future research will be compartmentalization of native biosynthesis pathways with downstream heterologous enzymes, as well as the development of membraneless synthetic channels with minimal metabolic disruptions.
Metabolic shifts, morphological changes, and genetic instability under bioreactor conditions
Y. lipolytica can ferment diverse substrates to various organic acids and its overflow metabolism is highly sensitive to growth conditions and fermentation stages. For example, Y. lipolytica ionone fermentation has been observed to secrete mevalonate and other organic acids in the early fermentation stage and reuse these acids during the late production phase [38]. Moreover, Y. lipolytica is dimorphic, i.e., capable of transition between ovoid and filamentous morphologies, which is influenced by pH, dissolved oxygen, temperature, and nutrient conditions, and can alter fermentation broth rheological property and mixing efficiency [39,40]. The mechanism of such morphological changes is not entirely clear, but it is believed to result from the interplays among complex genetic and environmental factors [41]. In general, industrial-scale bioreactor operations tend to intensify cell stress responses due to constant exposure of the cells to highly varying hydrostatic pressures and ununiformed O2 and substrate concentrations [24]. Decoupling the growth and production phase or the use of consortia systems could potentially alleviate stresses on the engineered yeast cells [42].
Furthermore, Y. lipolytica strains isolated from different sources are known to contain high genetic and metabolic variabilities [43], and these issues have not been fully studied [44]. The inherently stochastic nature of cellular machinery often leads to DNA mismatch during replication, and the accumulation of these defects can lower productivity [45,46]. In general, the combined effect of growth stresses and long replication generations leads to genetic instability [47]. Particularly, highly transcribed genes tend to accumulate mutations at higher rates [48], and synthetic biological components (such as efflux pumps) may increase the propensity for strain mutations [49]. Overall, overcoming Y. lipolytica genetic instability associated with bioreactor conditions and fermentation scale-up is an emerging area deserves more investigations to succeed in industrial applications.
Future perspective
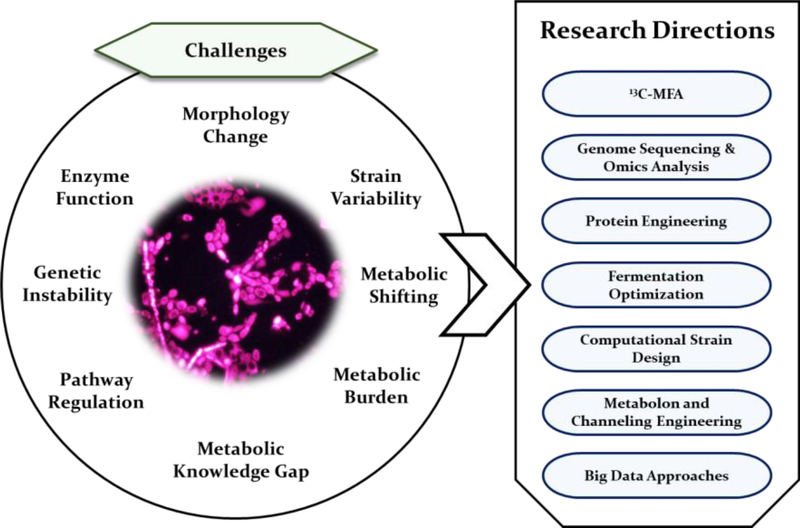
There has been considerable progress in developing non-model yeast platforms. However, challenges are still present, as shown in Y. lipolytica studies (Figure 3). Traditional strain development relies on the design-build-test-learn (DBTL) cycle, and significant time is spent on strain development and testing of desired phenotypes. Computational designs can reduce the duration of DBTL cycles and facilitate strain development. For example, genome-scale modeling can predict flux organization for optimal biosynthesis in oleaginous yeast Y. lipolytica [50]. However, such techniques rely on the completeness of metabolic knowledge. Thus, further characterization of Y. lipolytica using 13C-metabolic flux analysis is necessary to quantify cofactor balances, cellular maintenance costs, acetyl-CoA fluxes, and metabolic burdens. Moreover, machine learning approaches can analyze omics data to decipher dynamic metabolic regulations and to estimate the hidden constraints/limitations on strain engineering outcomes [51]. The data-driven approaches may also assist genome-scale modeling to capture cellular features and predict effective strain engineering targets [52].
Figure 3:
Challenges and future perspective of terpene production from non-model yeasts.
Recently, the biosynthesis of terpenoids with highly complex structures has been elucidated, and engineered into S. cerevisiae, including steroid-derived metabolites such as the anticancer withanolides [53] and insecticidal limonoids [54]. Intertwining long-step terpenoid pathway with the endogenous yeast pathway requires intensive engineering efforts on regulations of enzyme assemblies and temporal control of metabolic reactions. These successful engineering strategies in S. cerevisiae can be potentially applied to the non-model yeasts to promote production yields [55–57]. Finally, with the success already demonstrated for producing the more conventional terpenoid products such as β-carotene that accumulated to a titer over 6 g/L, meeting the challenges including those highlighted in Figure 3 will likely advance the Yarrowia non-model yeast platform to produce even more diverse and complex terpenoid compounds.
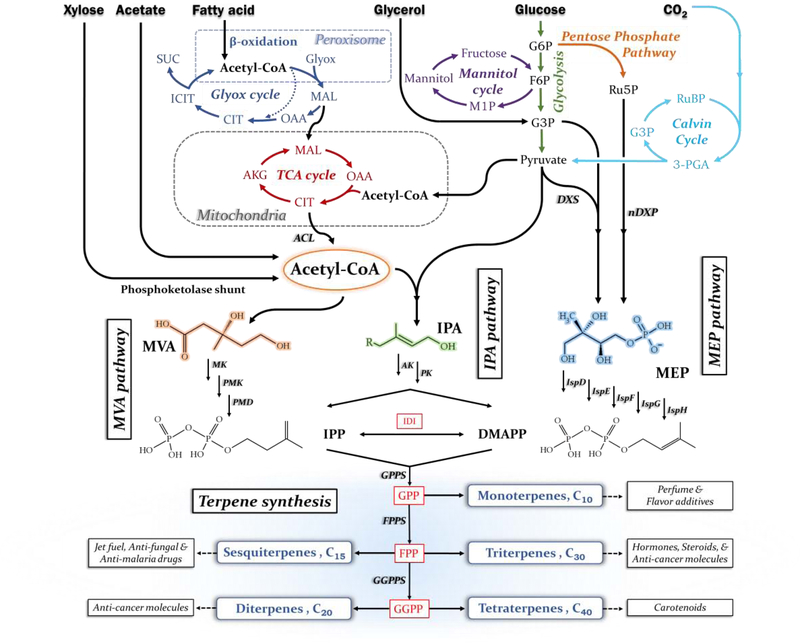
Figure 2: Pathway map for the production of terpenoid compounds.
Using isoprene, a C5 terpene compound, as a building block, larger terpene molecules are created. Bacterial hosts employ the methylerythritol 4-phosphate (MEP) pathway (the non-mevalonate pathway, also called the DXP pathway) to synthesize isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). The Entner-Doudoroff Pathway [22] or nDXP route (synthesis of 1-deoxy-d-xylulose 5-phosphate from pentose) [23] can increase the flux of the MEP pathway. In contrast, eukaryotes employ the mevalonate (MVA) pathway to synthesize IPP. In S. cerevisiae, cytosolic acetyl-CoA generation can be promoted via the pyruvate dehydrogenase bypass or novel phosphoketolase shunt [24]. The newly discovered isoprenoid alcohol (IPA) pathway can convert pyruvate and acetyl-CoA to IPP and DMAPP [25].
Highlights.
Yarrowia has shown the potential for producing complex terpenoids.
Yarrowia may hold multiple cytosolic acetyl-CoA and NADPH generation pathways.
Metabolite channeling is a key consideration in terpene pathway engineering.
Knowledge gaps exist in compartment-specific fluxes for utilizing alternative carbon sources (e.g., lipids).
Metabolic shift, genetic stability, and dimorphism in large-scale Yarrowia fermentation need further studies.
Acknowledgments
Funding: This work was supported by the National Institutes of Health (NIH1R41GM13027701), the National Science Foundation (MCB 1616619), and by the National Institute of Food and Agriculture (hatch project HAW05040-H and multistate project HAW05041-R).
Footnotes
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang C, Liwei M, Park J-B, Jeong S-H, Wei G, Wang Y, Kim S-W: Microbial Platform for Terpenoid Production: Escherichia coli and Yeast. Frontiers in microbiology 2018, 9:2460–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD: Microbial engineering for the production of advanced biofuels. Nature 2012, 488:320. [DOI] [PubMed] [Google Scholar]
- 3.Luo X, Reiter MA, d’Espaux L, Wong J, Denby CM, Lechner A, Zhang Y, Grzybowski AT, Harth S, Lin W, et al. : Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 2019, 567:123–126. [DOI] [PubMed] [Google Scholar]
- 4.Moser S, Pichler H: Identifying and engineering the ideal microbial terpenoid production host. Applied Microbiology and Biotechnology 2019, 103, 5501–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Immethun C, Hoynes-O’Connor A, Balassy A, Moon TS: Microbial Production of Isoprenoids Enabled by Synthetic Biology. Frontiers in Microbiology 2013, 4:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madzak C: Engineering Yarrowia lipolytica for Use in Biotechnological Applications: A Review of Major Achievements and Recent Innovations. Molecular Biotechnology 2018, 60:621–635. [DOI] [PubMed] [Google Scholar]
- 7.Markham KA, Alper HS: Synthetic Biology Expands the Industrial Potential of Yarrowia lipolytica. Trends in Biotechnology 2018, 36:1085–1095. [DOI] [PubMed] [Google Scholar]
- 8.Larroude M, Celinska E, Back A, Thomas S, Nicaud JM, Ledesma-Amaro R: A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of beta-carotene. Biotechnol Bioeng 2018, 115:464–472. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Wu C, Wu Q, Dai J, Song Y: Metabolic Flux Analysis of Lipid Biosynthesis in the Yeast Yarrowia lipolytica Using 13C-Labled Glucose and Gas Chromatography-Mass Spectrometry. PLOS ONE 2016, 11:e0159187.** A compartment specific 13C-MFA model is developed to quantify the metabolism of Y. lipolytica.
- 10.Sabra W, Bommareddy RR, Maheshwari G, Papanikolaou S, Zeng A-P: Substrates and oxygen dependent citric acid production by Yarrowia lipolytica: insights through transcriptome and fluxome analyses. Microbial Cell Factories 2017, 16:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu P, Qiao K, Ahn WS, Stephanopoulos G: Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc Natl Acad Sci U S A 2016, 113:10848–10853.** The authors discovered the overexpression of a S. cerevisiae CAT2 gene (encoding carnitine acyltransferase) in Y. lipolytica grown in glucose media enhanced lipid accumulation, presumably through an increased acetyl-CoA supply.
- 12.van Rossum HM, Kozak BU, Niemeijer MS, Dykstra JC, Luttik MAH, Daran J-MG, van Maris AJA, Pronk JT: Requirements for Carnitine Shuttle-Mediated Translocation of Mitochondrial Acetyl Moieties to the Yeast Cytosol. mBio 2016, 7:e00520–00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, Landberg J, Shavarebi F, Bilanchone V, Okerlund A, Wanninayake U, Zhao L, Kraus G, Sandmeyer S: Bioengineering triacetic acid lactone production in Yarrowia lipolytica for pogostone synthesis. Biotechnol Bioeng 2018, 115:2383–2388.* Putative mitochondrial β-oxidation in Y. lipolytica was suggested, based on the identification of Y. lipolytica genes encoding enzymes required for mitochondrial β-oxidation.
- 14.Gao Q, Cao X, Huang YY, Yang JL, Chen J, Wei LJ, Hua Q: Overproduction of Fatty Acid Ethyl Esters by the Oleaginous Yeast Yarrowia lipolytica through Metabolic Engineering and Process Optimization. ACS Synth Biol 2018, 7:1371–1380. [DOI] [PubMed] [Google Scholar]
- 15.Markham KA, Palmer CM, Chwatko M, Wagner JM, Murray C, Vazquez S, Swaminathan A, Chakravarty I, Lynd NA, Alper HS: Rewiring Yarrowia lipolytica toward triacetic acid lactone for materials generation. Proceedings of the National Academy of Sciences 2018, 115:2096.** This paper improves our understanding of Y. lipolytica acetyl-CoA generation routes.
- 16.Wasylenko TM, Ahn WS, Stephanopoulos G: The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab Eng 2015, 30:27–39. [DOI] [PubMed] [Google Scholar]
- 17.Dulermo T, Lazar Z, Dulermo R, Rakicka M, Haddouche R, Nicaud JM: Analysis of ATP-citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acid synthesis. Biochim Biophys Acta 2015, 1851:1107–1117. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Zhang L, Chen H, Chen YQ, Ratledge C, Song Y, Chen W: Regulatory properties of malic enzyme in the oleaginous yeast, Yarrowia lipolytica, and its non-involvement in lipid accumulation. Biotechnol Lett 2013, 35:2091–2098. [DOI] [PubMed] [Google Scholar]
- 19.Hult K, Veide A, Gatenbeck S: The distribution of the NADPH regenerating mannitol cycle among fungal species. Arch Microbiol 1980, 128:253–255. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Marsafari M, Deng L, Xu P: Understanding lipogenesis by dynamically profiling transcriptional activity of lipogenic promoters in Yarrowia lipolytica. Appl Microbiol Biotechnol 2019, 103:3167–3179.* Analysis of transcriptional activities helped identify NADPH upregulated genes, leading to new insights into Yarrowia energy metabolism.
- 21.Kavscek M, Bhutada G, Madl T, Natter K: Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Syst Biol 2015, 9:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Sun Y, Ramos KRM, Nisola GM, Valdehuesa KNG, Lee WK, Park SJ, Chung W-J: Combination of Entner-Doudoroff Pathway with MEP Increases Isoprene Production in Engineered Escherichia coli. PLOS ONE 2013, 8:e83290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirby J, Nishimoto M, Chow RWN, Baidoo EEK, Wang G, Martin J, Schackwitz W, Chan R, Fortman JL, Keasling JD: Enhancing Terpene Yield from Sugars via Novel Routes to 1-Deoxy-d-Xylulose 5-Phosphate. Applied and Environmental Microbiology 2015, 81:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meadows AL, Hawkins KM, Tsegaye Y, Antipov E, Kim Y, Raetz L, Dahl RH, Tai A, Mahatdejkul-Meadows T, Xu L, et al. : Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 2016, 537:694.** Large-scale production of isoprenoid β-farnesene by an engineered S. cerevisiae strain in a 200,000-liter fermenter was demonstrated, and strain stress responses were noticed.
- 25.Clomburg JM, Qian S, Tan Z, Cheong S, Gonzalez R: The isoprenoid alcohol pathway, a synthetic route for isoprenoid biosynthesis. Proceedings of the National Academy of Sciences 2019, 116:12810–12815.** In addition to mevalonate and non-mevalonate pathways, a third terpene pathway has been designed and engineered.
- 26.Renault H, Bassard J-E, Hamberger B, Werck-Reichhart D: Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Current Opinion in Plant Biology 2014, 19:27–34. [DOI] [PubMed] [Google Scholar]
- 27.Duan H, Schuler MA: Heterologous expression and strategies for encapsulation of membrane-localized plant P450s. Phytochemistry Reviews 2006, 5:507–523. [Google Scholar]
- 28.Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, Ajikumar PK: Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proceedings of the National Academy of Sciences 2016, 113:3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jørgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S,Møller BL: Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Current Opinion in Plant Biology 2005, 8:280–291. [DOI] [PubMed] [Google Scholar]
- 30.Conrado RJ, Varner JD, DeLisa MP: Engineering the spatial organization of metabolic enzymes: mimicking nature’s synergy. Current Opinion in Biotechnology 2008, 19:492–499. [DOI] [PubMed] [Google Scholar]
- 31.Abernathy MH, Czajka JJ, Allen DK, Hill NC, Cameron JC, Tang YJ: Cyanobacterial carboxysome mutant analysis reveals the influence of enzyme compartmentalization on cellular metabolism and metabolic network rigidity. Metab Eng 2019, 54:222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abernathy M, Wan N, Shui W, Tang YJ: Dynamic (13)C Labeling of Fast Turnover Metabolites for Analysis of Metabolic Fluxes and Metabolite Channeling. Methods Mol Biol 2019, 1859:301–316. [DOI] [PubMed] [Google Scholar]
- 33.Abernathy MH, He L, Tang YJ: Channeling in native microbial pathways: Implications and challenges for metabolic engineering. Biotechnol Adv 2017, 35:805–814. [DOI] [PubMed] [Google Scholar]
- 34.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL,Keasling JD: Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 2009, 27:753–759. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Nambou K, Wei L, Hua Q: Heterologous production of α-farnesene in metabolically engineered strains of Yarrowia lipolytica. Bioresource Technology 2016, 216:1040–1048. [DOI] [PubMed] [Google Scholar]
- 36.Cao X, Lv Y-B, Chen J, Imanaka T, Wei L-J, Hua Q: Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction. Biotechnology for Biofuels 2016, 9:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Cheng S, Cao J, Qiao J, Zhao G-R: Systematic Optimization of Limonene Production in Engineered Escherichia coli. Journal of Agricultural and Food Chemistry 2019, 67, 7087–7097. [DOI] [PubMed] [Google Scholar]
- 38.Czajka JJ, Kambhampati S, Tang YJ, Wang Y, Allen DK, Application of stable isotope tracing to elucidate metabolic dynamics during Yarrowia lipolytica α-ionone fermentation. iScience 23, 2020, 100854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawasse FM, Amaral PF, Rocha-Leao MH, Amaral AL, Ferreira EC, Coelho MA: Morphological analysis of Yarrowia lipolytica under stress conditions through image processing. Bioprocess Biosyst Eng 2003, 25:371–375. [DOI] [PubMed] [Google Scholar]
- 40.Pomraning KR, Bredeweg EL, Kerkhoven EJ, Barry K, Haridas S, Hundley H, LaButti K,Lipzen A, Yan M, Magnuson JK, et al. : Regulation of Yeast-to-Hyphae Transition in Yarrowia lipolytica. mSphere 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timoumi A, Guillouet SE, Molina-Jouve C, Fillaudeau L, Gorret N: Impacts of environmental conditions on product formation and morphology of Yarrowia lipolytica. Appl Microbiol Biotechnol 2018, 102:3831–3848. [DOI] [PubMed] [Google Scholar]
- 42.Roell GW, Zha J, Carr RR, Koffas MA, Fong SS, Tang YJ: Engineering microbial consortia by division of labor. Microbial Cell Factories 2019, 18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egermeier M, Russmayer H, Sauer M, Marx H: Metabolic Flexibility of Yarrowia lipolytica Growing on Glycerol. Front Microbiol 2017, 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czajka JJ, Nathenson JA, Benites VT, Baidoo EEK, Cheng Q, Wang Y, Tang YJ: Engineering the oleaginous yeast Yarrowia lipolytica to produce the aroma compound beta-ionone. Microb Cell Fact 2018, 17:136.* This paper investigated Y. lipolytica strain stability in shaking flasks and used machine learning to optimize cultivations. It has shown that high producing strains are much less stable than low producing strains.
- 45.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF: Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 2009, 461:1243. [DOI] [PubMed] [Google Scholar]
- 46.Voordeckers K, Kominek J, Das A, Espinosa-Cantu A, De Maeyer D, Arslan A, Van Pee M, van der Zande E, Meert W, Yang Y, et al. : Adaptation to High Ethanol Reveals Complex Evolutionary Pathways. PLoS Genet 2015, 11:e1005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elena SF, de Visser JAGM: Environmental stress and the effects of mutation. Journal of biology 2003, 2:12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu YO, Siegal ML, Hall DW, Petrov DA: Precise estimates of mutation rate and spectrum in yeast. Proceedings of the National Academy of Sciences 2014, 111:E2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El Meouche I, Dunlop MJ: Heterogeneity in efflux pump expression predisposes antibiotic-resistant cells to mutation. Science 2018, 362:686–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mishra P, Lee NR, Lakshmanan M, Kim M, Kim BG, Lee DY: Genome-scale model-driven strain design for dicarboxylic acid production in Yarrowia lipolytica. BMC Syst Biol 2018, 12:12.* Genome-scale modeling and computer strain design can facilitate Y. lipolytica pathway engineering.
- 51.Costello Z, Martin HG: A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. NPJ Syst Biol Appl 2018, 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Opgenorth P, Costello Z, Okada T, Goyal G, Chen Y, Gin J, Benites V, de Raad M, Northen TR, Deng K, et al. : Lessons from Two Design-Build-Test-Learn Cycles of Dodecanol Production in Escherichia coli Aided by Machine Learning. ACS Synth Biol 2019, 8, 1337–1351.** Machine learning is demonstrated to assist in rational strain design and improve DBTL efficiency.
- 53.Knoch E, Sugawara S, Mori T, Poulsen C, Fukushima A, Harholt J, Fujimoto Y, Umemoto N, Saito K: Third DWF1 paralog in Solanaceae, sterol Δ(24)-isomerase, branches withanolide biosynthesis from the general phytosterol pathway. Proceedings of the National Academy of Sciences of the United States of America 2018, 115:E8096–E8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodgson H, De La Peña R, Stephenson MJ, Thimmappa R, Vincent JL, Sattely ES, Osbourn A: Identification of key enzymes responsible for protolimonoid biosynthesis in plants: Opening the door to azadirachtin production. Proceedings of the National Academy of Sciences 2019, 116:17096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD: Complete biosynthesis of opioids in yeast. Science (New York, N.Y.) 2015, 349:1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown S, Clastre M, Courdavault V, O’Connor SE: De novo production of the plant-derived alkaloid strictosidine in yeast. Proceedings of the National Academy of Sciences 2015, 112:3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Li S, Thodey K, Trenchard I, Cravens A, Smolke CD: Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proceedings of the National Academy of Sciences 2018, 115:E3922. [DOI] [PMC free article] [PubMed] [Google Scholar]