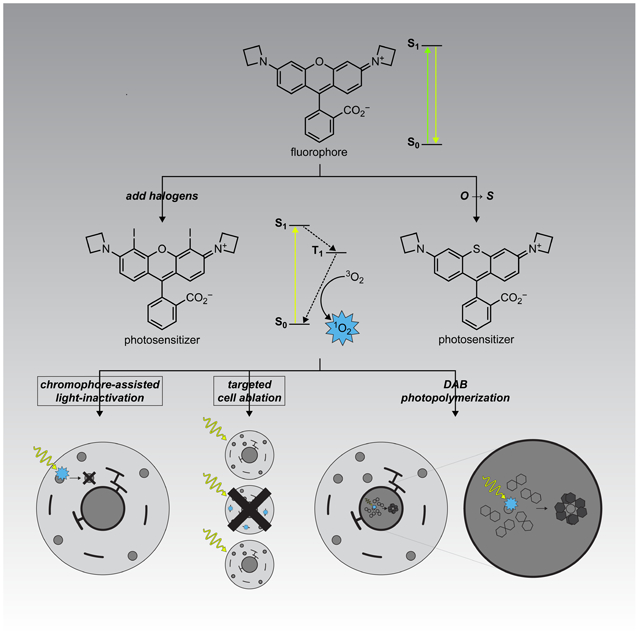
Summary:
Light-mediated chemical reactions are powerful methods for manipulating and interrogating biological systems. Photosensitizers, compounds that generate reactive oxygen species upon excitation with light, can be utilized for numerous biological experiments, but the repertoire of bioavailable photosensitizers is limited. Here, we describe the synthesis, characterization, and utility of two photosensitizers based upon the widely used rhodamine scaffold and demonstrate their efficacy for chromophore-assisted light inactivation, cell ablation in culture and in vivo, and photopolymerization of diaminobenzidine for electron microscopy. These chemical tools will facilitate a broad range of applications spanning from targeted destruction of proteins to high-resolution imaging.
Keywords: photosensitizer, photochemistry, fluorescence, HaloTag, cell ablation, reactive oxygen species, electron microscopy
Graphical Abstract
eTOC Blurb
Binns et. al describe photosensitizers based on rhodamine dyes. Incorporation of halogen or sulfur atoms into the rhodamine structure substantially increases singlet oxygen quantum yield. Combined with the HaloTag labeling system, these photosensitizers allow light-mediated destruction of proteins, ablation of entire cells, or deposition of electron microscopy contrast agents.
Introduction
The ability to measure and manipulate biological systems using light-driven processes has revolutionized biology. The excited state that occurs after a molecule absorbs light can be harnessed in different ways. Fluorescent dyes absorb and then emit light of different wavelengths, allowing sensitive visualization of biological processes inside cells. Photoactivatable (“caged”) compounds use the excited state energy to break chemical bonds and release bioactive molecules with high spatiotemporal precision. Optogenetic reagents use light to drive conformational changes in proteins to affect cellular function. Finally, photosensitizing molecules absorb light energy and then transfer it to adjacent molecules, facilitating specialized chemistries inside cells and tissue.
A key application of photosensitizers is the generation of reactive oxygen species (ROS) through the interaction of the triplet excited state of the photosensitizer with molecular oxygen (3O2) (Baptista et al., 2017; Xiong et al., 2019). The resulting singlet oxygen (1O2) and other ROS can be leveraged to manipulate or interrogate biological systems in a variety of ways: chromophore-assisted light inactivation (CALI) of proteins, targeted ablation of cells, or photooxidation and subsequent polymerization of diaminobenzidine (DAB) to provide contrast in light and electron microscopy (EM). This broad utility has led to the development of different classes of photosensitizers useful in living systems. Small-molecule photosensitizers fall into three main classes: porphyrins and phthalocyanines, transition-metal complexes, and fluorescent dyes appended with heavy atoms (Xiong et al., 2019). Extant compounds possess excellent photophysical properties but often exhibit limited bioavailability and can be difficult to target to specific cells or subcellular regions. Recent work on genetically encoded photosensitizers based on flavoproteins has produced useful reagents such as miniSOG that can be expressed in specific cells or subcellular regions (Bulina et al., 2006; Riani et al., 2018; Ruiz-Gonzalez et al., 2013; Shu et al., 2011), but many protein-based photosensitizers have relatively short excitation wavelengths (Torra et al., 2019). KillerRed and SuperNova are protein-based photosensitizers with longer excitation wavelengths and well-documented utility (de Rosny and Carpentier, 2012; Takemoto et al., 2013), but their potency is lower than small-molecule photosensitizers (Onukwufor et al., 2020).
We set out to develop a hybrid small-molecule:protein photosensitizer system that combines the flexibility and performance of small-molecule dyes with the subcellular specificity of genetically encoded reagents. To complement the recent development of impressive noncovalent hybrid photosensitizer systems (He et al., 2016; Marek and Davis, 2002; Tour et al., 2003) based on labeling systems that require a constant supply of small-molecule ligand (Ayele et al., 2019), we aimed to use the covalent HaloTag labeling system developed by Promega (Los et al., 2008). This “self-labeling” enzyme-derived tag system has been employed in many contexts ranging from single cells (Chong et al., 2018) to animals (Abdelfattah et al., 2019) and provides rapid, specific labeling of protein fusions with synthetic fluorophores. Previous examples of HaloTag-compatible photosensitizers are typically fluorescein derivatives such as Eosin (Fig. 1a) and related derivatives. Such ligands are excited by relatively short excitation wavelengths (Lee et al., 2008; Li et al., 2018; Takemoto et al., 2011), which are phototoxic on their own (Douthwright and Sluder, 2017; Magidson and Khodjakov, 2013) and require ester-masking groups to render the molecules cell-permeable (Takemoto et al., 2011). We now report HaloTag-compatible photosensitizer ligands based on our cell- and tissue-permeable azetidine-containing “Janelia Fluor” rhodamine scaffold (Grimm et al., 2015; Grimm et al., 2017b). These dyes are red-shifted compared to Eosin and do not need ester masking groups for membrane permeability. We demonstrate the utility of these probes for CALI, targeted cell ablation in culture and in vivo, and photoinduced DAB deposition for electron microscopy. This work expands the repertoire of the HaloTag labeling system to ablation of cells in intact animals and generating contrast for electron microscopy.
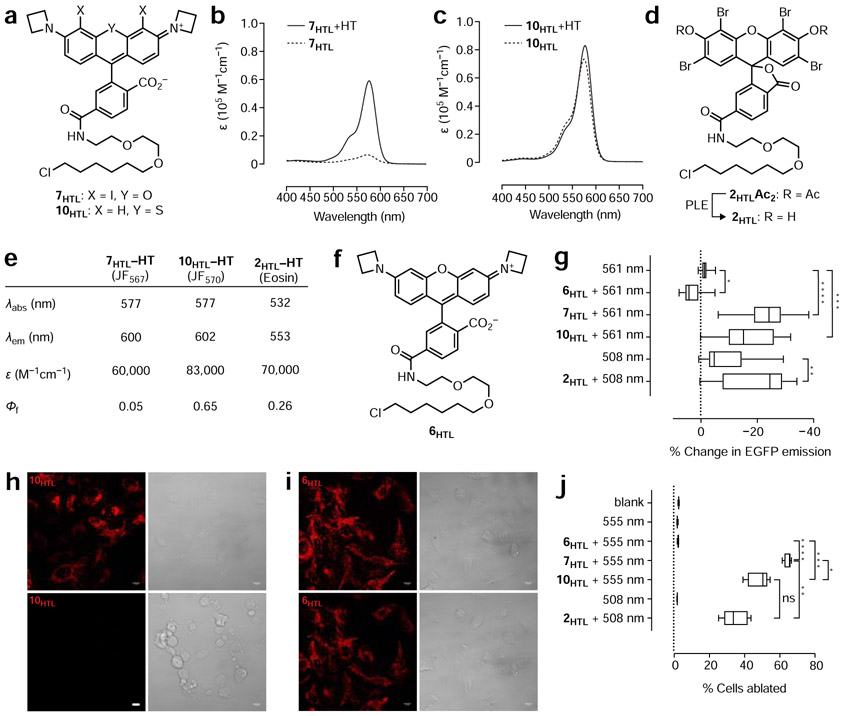
Figure 1. Photosensitizer structures, synthesis, and photophysical characterization.
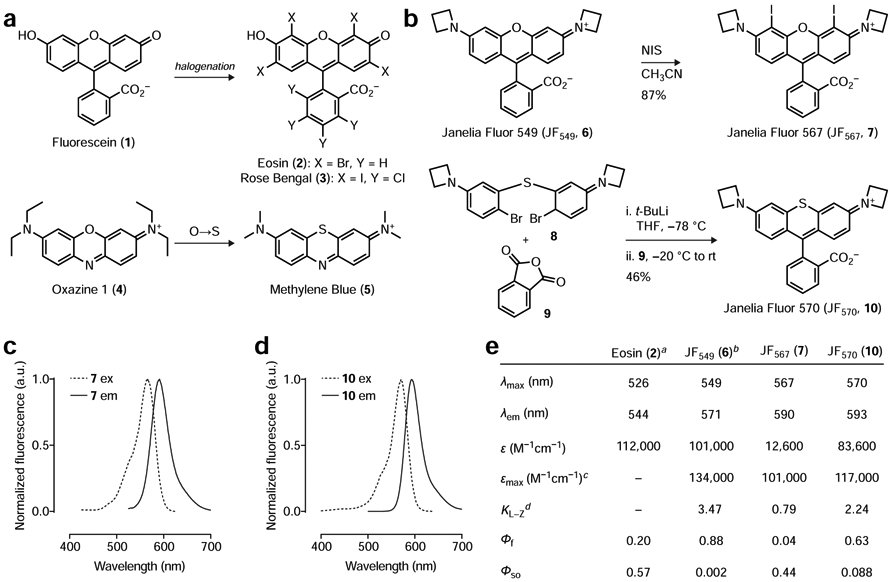
(a) Relationship of classic photosensitizer chemical structures to parent fluorophores. Eosin (2) and Rose Bengal (3) are halogenated derivatives of Fluorescein (1) and Methylene Blue (5) is a sulfur-containing analog of Oxazine 1 (4). (b) Iodination of JF549 (6) to yield JF567 (7) and organolithium-mediated reaction of 8 and 9 to yield JF570 (10). (c–d) Normalized fluorescence excitation (ex) and emission (em) spectra of 7 and 10. (e) Table of photophysical properties for Eosin (2), JF549 (6), JF567 (7), and JF570 (10). Notes: aData for 2 taken from (Fleming et al., 1977; Gandin et al., 1983; Seybold et al., 1969); bData for 6 taken from (Grimm et al., 2015; Grimm et al., 2017b); cεmax measured in trifluoroethanol + 0.1% v/v trifluoroacetic acid; dKL–Z measured in 1:1 v/v dioxane:water.
Results
Rational design and synthesis of Janelia Fluor photosensitizers.
Many classic small-molecule photosensitizers exploit the “heavy atom effect” (Gold, 1987), where the incorporation of an atom with a relatively high atomic number enhances the rate of intersystem crossing from the singlet-excited state to the triplet-excited state. This triplet state then interacts with molecular oxygen (3O2), generating singlet oxygen (1O2) with concomitant relaxation of the dye to the ground state; the efficiency of singlet oxygen generation can be quantified as the singlet oxygen quantum yield (ΦSO).This principle is illustrated by the halogenation of Fluorescein (1, ΦSO = 0.03) to yield classic photosensitizers Eosin (i.e., Eosin Y; 2, ΦSO = 0.57) and Rose Bengal (3, ΦSO = 0.76, Fig. 1a), where the halogen substituents also shift the absorption wavelengths to the red (Fleming et al., 1977; Gandin et al., 1983; Seybold et al., 1969). Likewise, replacement of the oxygen in the phenoxazine dyes such as laser dye Oxazine 1 (4) with sulfur yields well-known photosensitizer Methylene Blue (5, Fig. 1a) (Becker et al., 1990; Francisco et al., 2017). This substitution also elicits a bathochromic shift in wavelength and an increase in ΦSO.
We sought to apply this heavy atom effect to rhodamine dyes, which are widely used fluorescent labels but have received limited attention as photosensitizer scaffolds (Pal et al., 1996). A key advantage of rhodamine dyes over fluorescein-based compounds such as Eosin (2) and Rose Bengal (3) is based on charge. Fluorescein derivatives are anionic at physiological pH and, as highlighted above, require ester masking groups to allow efficient entry into cells. These ester bonds are notoriously unstable in aqueous solution, particularly for halogenated derivatives such as 2 and 3 (Lavis et al., 2006, 2011); their use in vivo typically requires injection of DMSO solutions directly into tissue (Sawinski et al., 2009). In contrast, simple rhodamine dyes are net neutral and exist in equilibrium between a lipophilic lactone and polar zwitterion. This “dynamic amphipathicity” allows rhodamines to rapidly traverse biological membranes without the need for auxiliary groups, allowing facile loading into living cells and even animals (Abdelfattah et al., 2019; Grimm et al., 2017b). We recently developed an improved class of rhodamines, the Janelia Fluor dyes, which incorporate four-membered azetidine rings in place of the common dimethylamino groups found in many classic rhodamines. This simple substitution results in substantial improvements in photophysical properties (Grimm et al., 2015) and facilitates rational fine-tuning of the spectral and chemical features of these molecules (Grimm et al., 2017b). For example, the prototypical Janelia Fluor 549 (JF549, 6, Fig. 1b) exhibits a two-fold improvement in both brightness and photostability in cellular imaging experiments compared to the parent tetramethylrhodamine compound (Grimm et al., 2015). Since increased excited state lifetime and photostability are important for photosensitizer performance, we sought to extend this azetidine-incorporation strategy to photosensitizer dyes. We envisioned either halogenation (à la 3) or sulfur-substitution (à la 5) applied to rhodamine compounds such as 6. For the halogenation strategy, we found that straightforward treatment of JF549 (6) with N-iodosuccinimide (NIS) afforded compound 7 with incorporation of two iodine atoms at the 4- and 5-positions of the xanthene moiety (Fig. 1b). To accomplish the sulfur substitution, we adapted our recent synthetic approach to rhodamine dyes (Grimm et al., 2017a) through lithium–bromide exchange of brominated diarylthioether 8 followed by addition to phthalic anhydride (9) to yield compound 10 (Fig. 1b).
We then examined the spectral and chemical properties of these two dyes comparing to Eosin (2) and JF549 (6; Fig. 1c-e). As in other halogenated xanthene dyes (e.g., 2 and 3), the iodo substituents in rhodamine 7 elicited a bathochromic shift with an absorption maximum (λmax) of 567 nm and an emission maximum (λem) of 590 nm in aqueous buffer; we named this dye “Janelia Fluor 567” (JF567) based on the λmax. Likewise, the replacement of the bridging oxygen with a sulfur in 10 caused a red-shift in its spectra with λmax/λem = 570 nm/593 nm; we gave this dye the moniker “Janelia Fluor 570” (JF570). We then investigated the absorptivity of these dyes in aqueous solution. Although the sulfur-containing JF570 (10) showed high absorptivity with an extinction coefficient (ε) of 83,600 M−1cm−1, the halogenated derivative JF567 (7) gave a substantially lower ε = 12,600 M−1cm−1. As mentioned above, rhodamines exist in equilibrium between a colorless, nonfluorescent lactone form and colored, fluorescent zwitterionic form. We suspected that the electron-withdrawing iodo substituents in compound 7 shifted this equilibrium towards the colorless lactone form. We therefore measured the equilibrium constant (KL–Z) and maximum extinction coefficient (εmax) using trifluoroethanol containing 0.1% v/v trifluoracetic acid, a solvent system that promotes formation of the open form (Grimm et al., 2017b; Zheng et al., 2019) (Fig. 1e). Under these conditions we found that both dyes exhibit high absorptivity (εmax > 100,000 M−1cm−1) demonstrating that the difference in observed ε in aqueous solution is due to a shift in the lactone–zwitterion equilibrium and not significant differences in the inherent absorptivity of the two chromophores.
We then measured the fluorescence quantum yield (Φf) and singlet-oxygen quantum yield (ΦSO) of both potential photosensitizer dyes using Rose Bengal (3; ΦSO = 0.76) (Ezquerra Riega et al., 2017) as a standard. JF567 (7) showed a relatively low Φf = 0.04 and a high ΦSO = 0.44. In contrast, JF570 (10) showed a higher Φf = 0.63 balanced by a more modest ΦSO = 0.088. These ΦSO values were substantially higher than the parent fluorophore JF549 (6), which gave a low ΦSO = 0.002. For comparison, Eosin (2) has a reported ΦSO = 0.57 (Gandin et al., 1983) and the ΦSO for Methylene Blue (5) has been measured at 0.07 in aqueous solution (Francisco et al., 2017). These data show that both halogenation and sulfur substitution substantially increase ΦSO compared to the parent dye and these ΦSO values are comparable to those of established photosensitizers such as Eosin and Methylene Blue.
Synthesis of HaloTag ligands and chromophore-assisted light inactivation (CALI).
To enable attachment of these dyes to specific proteins in living systems, we prepared derivatives of both 7 and 10 containing the chloroalkane HaloTag ligand moiety at the optimal 6-position on the pendant phenyl ring (7HTL and 10HTL; Fig. 2a) (Los et al., 2008). These were synthesized in analogous fashion to the free dyes. The JF567–HaloTag ligand (7HTL; Fig. S1a) involved direct iodination of 6-carboxy-JF549 (11) to give the diiodo 6-carboxy-JF567 (12), which could be amidated with the HaloTag ligand amine (13) to yield 7HTL. The synthesis of 10HTL was more involved (Fig. S1b), starting with the coupling of 3-bromothiophenol (14) and 3-bromoiodobenzene (15) to give diarylthioether 16. Cross-coupling with azetidine gave 17, which was regioselectively brominated with NBS to give 8. Metalation of 8 with t-BuLi and addition to diorthoester methyl benzoate 18 followed by deprotection gave 6-carboxy-JF570 (19). Amidation with the HaloTag ligand amine (13) yielded 10HTL.
Figure 2. Propeties of HaloTag ligands and their utility in CALI and cell ablation.
(a) Chemical structures of JF567–HaloTag ligand (7HTL) and JF570–HaloTag ligand (10HTL). (b–c) Absorbance spectra of 7HTL and 10HTL ± HaloTag protein (HT). (d) Chemical structure of Eosin-Ac2-HaloTag ligand (2HTLAc2) and conversion to 2HTL by incubation with porcine liver esterase (PLE). (e) Table of photophysical properties for 7HTL, 10HTL and 2HTL attached to HaloTag protein. (f) Chemical structure of JF549–HaloTag ligand (6HTL). (g) Comparison of EGFP CALI using 561 nm excitation light (laser; 10 W/cm2) alone (n = 19), 561 nm excitation light (laser; 10 W/cm2) with 6HTL (n = 15), 7HTL (n = 23), and 10HTL (n = 14), 508 nm-centered excitation light (LED; 10 W/cm2) alone (n = 35), or 508 nm-centered excitation light (LED; 10 W/cm2) with 2HTL (n = 30); center line indicates median; box limits indicate upper and lower quartiles; whiskers indicate min–max; data pooled from two experiments and analyzed via Kruskal–Wallis test with Steel–Dwass–Critchlow–Fligner post-hoc test. (h–i) Representative confocal fluorescence (561 nm laser excitation) and brightfield microscopy images of U2OS cells expressing HaloTag–TOMM20 fusion protein incubated with JF570–HaloTag ligand (10HTL; h) or JF549–HaloTag ligand (6HTL; i) before (t = 0 min) and after (t = 30 min) widefield irradiation with 560 nm-centered light; scale bars: 13 μm. (j) Comparison of cell-ablation efficacy using 6HTL, 7HTL, and 10HTL with 555 nm excitation light or 2HTL using 508 nm-centered excitation light in U2OS cells stably expressing HaloTag-TOMM20 fusion protein exposed to 55 mW/cm2 excitation light for 3 min; center line indicates median; box limits indicate upper and lower quartiles; whiskers indicate min–max; data analyzed via Welch’s ANOVA and Games–Howell post-hoc test (n = 5, 10.89 mm2 microplate wells); n = 5 except for blank measurement where n = 10. Statistical significance in g and j reported as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. See also Figure S1, Figure S2, and Figure S3
We evaluated the properties of 7HTL and 10HTL (Fig. 2a) as conjugates with purified HaloTag protein (HT) in vitro. The equilibrium between the open, colored zwitterionic form and the closed, colorless lactone of rhodamine dyes can be leveraged to create chromogenic and fluorogenic ligands—molecules that show large increases in absorption and fluorescence upon binding their cognate biomolecular target. Since JF567 (7) exhibited a relatively low extinction coefficient in aqueous solution and a relatively low KL–Z (Fig. 1e), we predicted the JF567–HaloTag ligand (7HTL) would show increased absorption upon binding the HaloTag protein. Indeed, incubation of this photosensitizer dye with HaloTag protein elicited an 8.6-fold increase in absorption due to binding of the ligand to the HaloTag protein (Fig. 2b), similar in magnitude to the prototypical fluorogenic rhodamine dye tetramethyl-Si-rhodamine (SiR) (Lukinavicius et al., 2013). In contrast, the JF570–HaloTag ligand did not show a substantial increase in absorption (Fig. 2c), as expected from the properties of the free dye (Fig. 1e). We further investigated the properties of the HaloTag conjugates and compared them to Eosin–HaloTag ligand (2HTL); this was prepared by treatment of the commercially available diacetate compound (2HTLAc2) (Takemoto et al., 2011) with porcine liver esterase (PLE; Fig. 2d, Fig. S2a). JF567–HaloTag ligand (7HTL) and JF570–HaloTag ligand (10HTL) showed similar Φf values to the free dyes (7 and 10) with a slight bathochromic shift of 8–12 nm due to attachment of the HaloTag ligand moiety and conjugation to the HaloTag protein (Fig. 1c-e; Fig. 2e; Fig. S2b-c). Both dyes exhibited a substantial bathochromic shift (~45 nm) compared to the conjugate of the Eosin–HaloTag ligand (2HTL, Fig. 2e; Fig. S2d) and therefore constitute the most red-shifted HaloTag-compatible photosensitizers described.
For an initial application, we tested these photosensitizers in chromophore-assisted light inactivation of proteins (CALI) in live cells. CALI allows the targeted inactivation of proteins via excitation of a conjugated photosensitizer which produces damaging oxidants, typically singlet oxygen (1O2) or other radical products such as superoxide (Baptista et al., 2017; Jay, 1988; Liao et al., 1994; Ochsner, 1997). These reactive species act with an effective radius of 4–6 nm (Beck et al., 2002; Linden et al., 1992) via methionine oxidation and protein cross-linking (Yan et al., 2006). Unlike conventional genetic knockout methods, the CALI technique gives temporal control of protein ablation, which may allow for functional knockouts of vital proteins (Vitriol et al., 2007; Wojtovich et al., 2016) with faster time resolution compared to RNAi (Li et al., 2018). CALI has found utility in studying mitochondrial electron transport chain physiology (Wojtovich et al., 2016) and memory maintenance (Takemoto et al., 2017), making bioavailable and targetable photosensitizers of particular interest in various areas of biology.
For a proof-of-concept CALI experiment, we expressed EGFP–HaloTag fusion proteins in U2OS cells and then labeled with photosensitizer ligands JF567–HaloTag ligand (7HTL), JF570–HaloTag ligand (10HTL), and Eosin–Ac2–HaloTag ligand (2HTLAc2), with the JF549–HaloTag ligand (6HTL; Fig. 2f) and vehicle-only (DMSO) as controls (Fig. 2g). CALI efficacy was assessed by measuring the decrease in EGFP fluorescence upon illumination of the cells with photosensitizer excitation light; we chose 561 nm for the JF ligands and 508 nm for the Eosin ligand to give comparable absorption cross-sections (Fig. S3a). JF567–HaloTag ligand (7HTL) and JF570–HaloTag ligand (10HTL) both showed effective CALI, showing a decrease in EGFP fluorescence after brief photosensitizer excitation (10 s, 561 nm, 10 W/cm2). Interestingly, cells labeled with JF549–HaloTag ligand (6HTL) showed a small but significant increase in average fluorescence intensity under the same conditions, possibly due to the diminutive amount of generated ROS promoting further maturation of EGFP chromophores. CALI experiments with the Eosin ligand (2HTLAc2) were more complicated due to the overlap of the EGFP and Eosin absorbance spectra. Cells labeled with 2HTLAc2 and excited with 508 nm-centered light elicited a significant decrease in EGFP fluorescence, but the dye-free control experiment also showed significant bleaching of EGFP by the 508 nm-centered light dose. These data demonstrate that both iodination and sulfur substitution yield photosensitizer JF dyes that perform similarly to an established compound in CALI applications. This experiment also showcases the longer absorbance maxima of 7HTL and 10HTL, which could allow concurrent use of these compounds with GFP-based reporters or optogenetic reagents.
Targeted cell ablation in cell culture.
Photosensitizers can also be utilized to ablate entire cells (Miller and Selverston, 1979) presumably via CALI of vital biomolecules or oxidative damage to organelles. Reported targets for effective cell ablation include the nucleus and mitochondria (He et al., 2016; Shirmanova et al., 2015). For nuclear-targeted photosensitization, the hypothesized mechanisms of ablation include DNA damage leading to death of irradiated cells. For mitochondrial-targeted photosensitizers, cell death is presumably caused by mitochondrial membrane damage and subsequent cytochrome c leakage leading to apoptosis or decreased energy output potential leading to necrosis. Photoablation of cells can be utilized for clinical applications such as photodynamic therapy of malignancies (Zhang et al., 2018), photoinactivation of pathogenic microbes (Hu et al., 2018; Wang et al., 2017), or for basic science experiments to study the effect of the ablation of specific cell lineages on animal development or behavior.
We first tested the ablative utility of the JF dyes in cell culture, targeting the HaloTag ligands to the nucleus or mitochondria using HaloTag fusions to either histone H2B (“HaloTag–H2B”) or the mitochondrial outer membrane import protein TOMM20 (“HaloTag–TOMM20”). We found that both JF567–HaloTag ligand (7HTL) and JF570–HaloTag ligand (10HTL) were effective ablative agents of U2OS cells stably expressing HaloTag–H2B when excited with 555 nm-centered light (3 min, 55 mW/cm2; Fig. S3b), ablating 35% and 19% of cells, respectively, when measured 24 h after illumination. This was better or equivalent to the Eosin-Ac2–HaloTag ligand (2HTLAc2; 16% ablation; 3 min, 55 mW/cm2, 508 nm-centered light) and substantially larger than the JF549–HaloTag ligand (9) or DMSO controls, which both showed ~1% ablation under the same illumination conditions (Fig. S3c). Similar trends were observed with HaloTag-TOMM20 expressing cells but at higher overall efficacy with JF567–HaloTag (7HTL), JF570–HaloTag ligand (10HTL), and Eosin-Ac2–HaloTag ligand (2HTLAc2) showing ablation of 65%, 48%, and 35% of cells, respectively, compared to ~2% in the JF549–HaloTag ligand (6HTL) and DMSO-only controls (Fig. 2h-j, Fig. S3d). Increasing illumination intensity (147 mW/cm2) resulted in >80% ablation for both the experiments using the Janelia Fluor photosensitizer HaloTag ligands (Fig. S3e-f). These compounds also showed negligible “dark” toxicity, demonstrating that the cellular ablation is light-mediated (Fig. S3g–h). Ablation was also negligible in the absence of HaloTag fusion proteins, demonstrating that damage due to nonspecifically bound dyes is nominal (Fig. S3i).
Targeted cell ablation in vivo.
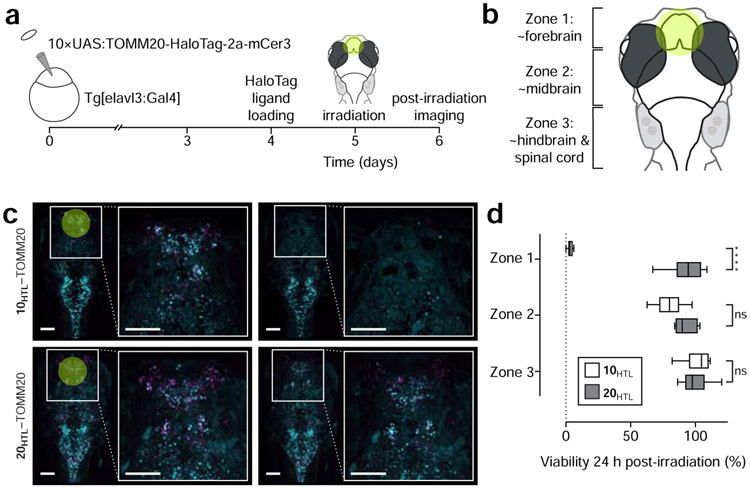
We then tested the utility of this cell ablation strategy in vivo using the zebrafish model system. Transgenic Casper zebrafish (Mifaw2/w2 roya9/a9) embryos expressing a neuron-specific elavl3 GAL4-VP16 driver were injected with a construct resulting in the co-expression of mCerulean3 fluorescent protein and the mitochondria-targeted HaloTag–TOMM20 fusion (Fig. 3a). These neurons were labeled with JF570–HaloTag ligand (10HTL) or the previously described bioavailable JF585–HaloTag ligand (20HTL; Fig. S4a) (Grimm et al., 2017b) by incubation of these transgenic larval fish with these ligands in the system water (33 μM). Attempts to use 7HTL in vivo resulted in poor labeling (data not shown), likely due to the lipophilic iodo substituents combined with the shift in KL–Z towards the lactone form. Both 10HTL and 20HTL dyes labeled neurons throughout the zebrafish brain. We then illuminated the approximate forebrain region (Zone 1, Fig. 3b) of fish labeled with 10HTL with 3 W/cm2 and fish incubated with 20HTL with 12 W/cm2; the higher illumination power used for the JF585 accounts for the differences in absorption coefficients at 560 nm-centered light. We measured cell viability by counting cells using the mCerulean3 fluorescence signal, finding that in Zone 1 (~forebrain) of JF570-labeled fish, only ~3% of cells were viable 24 h post-illumination, whereas use of JF585–HaloTag ligand preserved 93% of the neurons in fish (Fig. 3c-d). Neuronal viability was also quantified in zones roughly corresponding to the midbrain and hindbrain regions (Zones 2 and 3, Fig. 3b, Fig. S4b-c) to assess off-target ablation due to light scattering (Dobrucki et al., 2007) or damage to neuronal processes projecting into the area of illumination. This off-target ablation was modest in Zone 2, with 10HTL zebrafish retaining an 80% of labeled cells, and negligible in Zone 3, which showed no significant ablation (Fig. 3d), indicating that cell death is largely contained to the area of illumination.
Figure 3. Photoablation of neurons in larval zebrafish.
(a) Experimental scheme of larval zebrafish neuronal ablation experiment. (b) Cartoon of larval zebrafish brain “zones” used in this experiment; yellow circle indicates approximate area of irradiation. (c) Representative fluorescence microscopy images of zebrafish pre-irradiation (t = 0 min), and one day post-irradiation (t = 24 h) expressing HaloTag fusions to the mitochondrial protein TOMM20 (magenta) co-expressing mCerulean (cyan) and incubated with either JF570–HaloTag ligand (10HTL, top; 3 W/cm2 excitation) or JF585–HaloTag ligand (20HTL, bottom; 12 W/cm2); scale bars: 100 μm. (d) Quantification of cell-ablation efficacy using JF570–HaloTag ligand (10HTL) with JF585–HaloTag ligand (20HTL) as control; n = 6 for 10HTL experiments and n = 5 for 20HTL experiments; center line indicates median; box limits indicate upper and lower quartiles; whiskers indicate min–max; data analyzed via independent t-tests. Statistical significance reported as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. See also Figure S4.
DAB photopolymerization for electron microscopy.
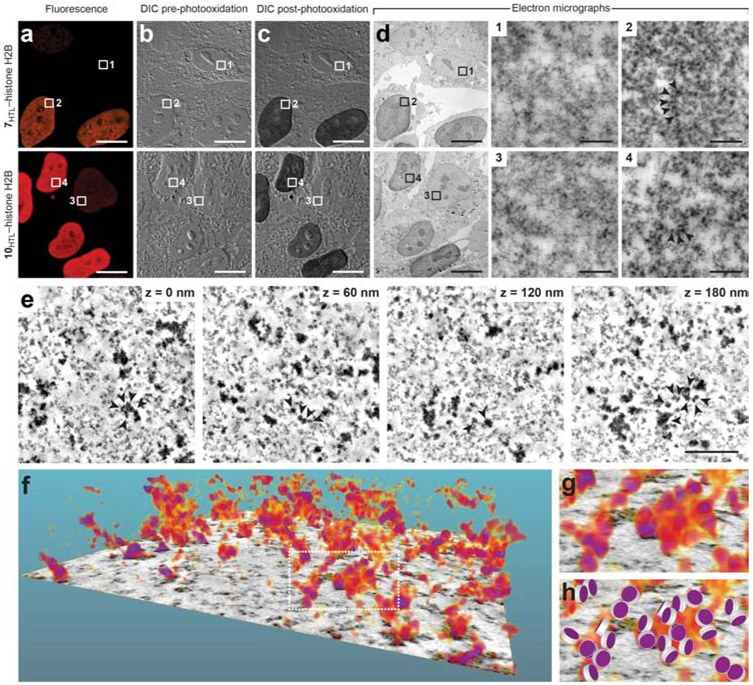
Finally, we tested the JF photosensitizers as agents for diaminobenzidine (DAB) polymerization, which results in the generation of a visibly opaque, osmophilic material. This polymer is an effective contrast agent in both light and electron microscopy (EM). Photosensitizing fluorescent dyes have a rich history in both EM and correlative light and electron microscopy (CLEM) (Deerinck et al., 1994; Gaietta et al., 2002; Grabenbauer et al., 2005; Hoffmann et al., 2010; Liss et al., 2015; Shu et al., 2011), and this photopolymerization complements peroxide-mediated DAB deposition using horseradish peroxidase (Hopkins et al., 2000) and APEX (Lam et al., 2015; Martell et al., 2012). The hybrid system comprised by the photosensitizer Janelia Fluor ligands and the HaloTag combines the modularity and performance of small-molecule photosensitizers such as Eosin (Deerinck et al., 1994) with the genetic specificity of protein-based reagents like miniSOG (Shu et al., 2011) but with longer excitation wavelengths. We expressed HaloTag–H2B fusions in HOS cells, labeled with either JF567–HaloTag ligand (7HTL) or JF570–HaloTag ligand (10HTL), fixed the cells, imaged them by light microscopy, performed DAB photopolymerization and osmium tetroxide treatment, and then imaged the sample using transmission electron microscopy (TEM). Photooxidation of DAB was specific to cells expressing HaloTag–H2B fusion proteins, and this labeling strategy enabled high-resolution electron microscopy in the nucleus (Fig. 4a-d). Histones could be visualized as electron-dense puncta in TEM images and the use of tomographic techniques enabled 3D electron microscopy with resolution approaching the chromatin–nucleosome interface scale (5–24 nm; Fig. 4e-h) (Ou, 2017).
Figure 4. Photooxidation of diaminobenzidine (DAB) and electron microscopy.
(a–d) Representative images of HOS cells expressing HaloTag–H2B fusion protein labeled with JF567–HaloTag ligand (7HTL; top) or JF570–HaloTag ligand (10HTL, bottom). (a) Pre-photooxidation fluorescence microscopy images. (b) Pre-photooxidation differential interference contrast (DIC) light microscopy images. (c) Post-photooxidation DIC light microscopy images. (d) Transmission electron microscopy (TEM) images post-photooxidation and OsO4 treatment of the entire field of view and regions of interest (ROIs) 1–4. ROIs 1 and 3 are from cells with low HaloTag–H2B expression and show low staining from DAB photooxidation and OsO4 treatment. ROIs 2 and 4 show increased electron density of the chromatin resulting from specific labeling of 7HTL or 10HTL and subsequent photooxidation of DAB and osmification; arrows highlight representative HaloTag–H2B containing nucleosomes; scale bars: 10 μm (a–d) and 200 nm (ROIs 1–4). (e) Representative electron tomogram slices in the nucleus of a HOS cell expressing HaloTag–H2B fusion proteins labeled with 10HTL, followed by photooxidization of DAB and osmification (cf. ROI 4). Arrows denote a cluster of nucleosomes in the volume. (f) 3D volumetric rendering extracted from the electron tomogram in e (g) Magnified view at the segmented chromatin denoted in f. (h) Manual matching of stained nucleosomes in g with a simplified nucleosome structure consisting of a disc with 11 nm diameter and 5.5 nm height. These imaging experiments were duplicated with similar results.
Discussion
Light-driven chemical reactions allow the manipulation of biological systems with high spatiotemporal control. Photosensitizers generate ROS, which can be harnessed in a variety of applications ranging from inactivation of individual proteins to the synthesis of imaging contrast agents in situ. We transformed the cell-permeable Janelia Fluor scaffold into a photosensitizer system through rational design. The resulting dyes JF567 (7) and JF570 (10) show comparable ΦSO to classic photosensitizers (Fig. 1). Their HaloTag ligand derivatives 7HTL and 10HTL are as effective as the established Eosin ligand (2HTL) in CALI and cell culture ablation experiments but are red-shifted and can be introduced into biological systems without the use of esterase-sensitive masking groups (Fig. 2). This bioavailability of 10 allows extension of this compound’s utility to in vivo experiments (Fig. 3). Both dyes can also be utilized to photopolymerize DAB and show utility in high resolution electron microscopy (Fig. 4). Although JF567 (7) exhibits a substantially higher ΦSO than JF570 (10, Fig. 1e) in vitro, the performance of 7HTL in CALI (Fig. 2g) and cell ablation in culture (Fig. 2j) was only modestly better than 10HTL; both dyes gave comparable DAB photodeposition (Fig. 4a-d). These results, combined with the utility in vivo (Fig. 3), recommend JF570 (10) as a general-purpose photosensitizer with JF567 (7) reserved for applications that require high amounts of 1O2. Having established these photosensitizing modifications to the core Janelia Fluor rhodamine scaffold, we can combine them into a single compound, apply them to other rhodamine variants, and then further fine-tune these dyes to improve bioavailability and modulate their spectral properties (Grimm et al., 2015; Grimm et al., 2017b). Such chemical tools will enable sophisticated cell-ablation experiments in vivo and advanced imaging experiments such as correlated light and electron microscopy (CLEM).
STAR Methods
Resource Availability
Lead Contact.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Luke D. Lavis: lavisl@janelia.hhmi.org.
Materials availability.
Aliquots of JF567–HaloTag ligand (7HTL) and JF570–HaloTag ligand (10HTL) are available by request from the Lead Contact.
Data and code Availability.
This study did not generate new datasets or code.
Experimental Model and Subject Details
Cells utilized in CALI and ablation experiments were U2OS cells (human female, osteosarcoma), some with genetic alterations for the expression of various HaloTag fusion proteins: transiently transfected EGFP; stably expressed HaloTag–histone H2B fusions; stably expressed HaloTag–TOMM20 fusions. U2OS cells for CALI experiments were maintained in Dulbecco’s Modified Eagle Medium (DMEM; 4.5 g/L glucose and sodium pyruvate, −l-glutamine, −phenol red; Corning) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco), 2 mM l-glutamine (Gibco), 100 U/mL penicillin and 100 U/mL streptomycin (Gibco), and 5% CO2 at 37 °C. U2OS cells for ablation experiments were maintained in DMEM (high glucose, −glutamine, −phenol red; Gibco) containing 1× GlutaMax (Gibco), 10% v/v FBS (Gibco) and 5% CO2 at 37 °C. 1× Antibiotic–Antimycotic (Gibco) was used for maintenance only (excluded from imaging/treatment formulations) and cells were passaged at approximately 80% confluence with 0.25% Trypsin–EDTA with phenol red (Gibco). Human osteosarcoma (HOS) cells (female) expressing HaloTag–histone H2B fusions utilized for electron microscopy experiments were cultured on 35 mm MatTek dishes (MatTek Corp) in DMEM (Gibco) containing 10% v/v FBS (Gibco) without antibiotic.
Embryonic Casper (Mitfaw2/w2 roya9/a9) zebrafish (Danio rerio) were utilized from the 1–2 cell stage through day 6 post-fertilization as detailed below. All zebrafish experiments presented in this study were conducted in accordance with the animal research guidelines from the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee and Institutional Biosafety Committee of Janelia Research Campus.
HaloTag protein expressed by and subsequently purified from BL21 chemically competent E. coli.
Method Details
General chemical synthesis details.
Commercial reagents were obtained from reputable suppliers and used as received. All solvents were purchased in septum-sealed bottles stored under an inert atmosphere. All reactions were sealed with septa through which a nitrogen atmosphere was introduced unless otherwise noted. Reactions were conducted in round-bottomed flasks or septum-capped crimp-top vials containing Teflon-coated magnetic stir bars. Heating of reactions was accomplished with a silicon oil bath or an aluminum reaction block on top of a stirring hotplate equipped with an electronic contact thermometer to maintain the indicated temperatures.
Reactions were monitored by thin layer chromatography (TLC) on precoated TLC glass plates (silica gel 60 F254, 250 μm thickness) or by LC/MS (Phenomenex Kinetex 2.1 mm × 30 mm 2.6 μm C18 column; 5 μL injection; 5–98% MeCN/H2O, linear gradient, with constant 0.1% v/v HCO2H additive; 6 min run; 0.5 mL/min flow; ESI; positive ion mode). TLC chromatograms were visualized by UV illumination or developed with p-anisaldehyde, ceric ammonium molybdate, or KMnO4 stain. Reaction products were purified by flash chromatography on an automated purification system using pre-packed silica gel columns or by preparative HPLC (Phenomenex Gemini–NX 30 × 150 mm 5 μm C18 column). Analytical HPLC analysis was performed with an Agilent Eclipse XDB 4.6 × 150 mm 5 μm C18 column under the indicated conditions. High-resolution mass spectrometry was performed by the High Resolution Mass Spectrometry Facility at the University of Iowa.
NMR spectra were recorded on a 400 MHz spectrometer. 1H and 13C chemical shifts were referenced to TMS or residual solvent peaks. Data for 1H NMR spectra are reported as follows: chemical shift (δ ppm), multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublets, m = multiplet), coupling constant (Hz), integration. Data for 13C NMR spectra are reported by chemical shift (δ ppm) with hydrogen multiplicity (C, CH, CH2, CH3) information obtained from DEPT spectra.
JF567 (7):
JF549 (6; 500 mg, 1.22 mmol) was taken up in CH3CN (40 mL), and N-iodosuccinimide (1.10 g, 4.87 mmol, 4 eq) was added portionwise over 10 min. After stirring the reaction at room temperature for 3 h, it was concentrated to half volume, diluted with water, and extracted with CH2Cl2 (2×). The combined organic extracts were washed with brine, dried over anhydrous MgSO4, filtered, and evaporated. Flash chromatography on silica gel (0–30% EtOAc/toluene) afforded 699 mg (87%) of JF567 (7) as a purple solid. 1H NMR (CDCl3, 400 MHz) δ 8.00 (dt, J = 7.3, 1.1 Hz, 1H), 7.65 (td, J = 7.4, 1.3 Hz, 1H), 7.60 (td, J = 7.4, 1.2 Hz, 1H), 7.19 – 7.11 (m, 1H), 6.58 (d, J = 8.7 Hz, 2H), 6.21 (d, J = 8.7 Hz, 2H), 4.24 (q, J = 7.4 Hz, 4H), 4.17 (q, J = 7.4 Hz, 4H), 2.25 (p, J = 7.4 Hz, 4H); 13C NMR (CDCl3, 101 MHz) δ 169.4 (C), 155.2 (C), 153.0 (C), 152.4 (C), 135.0 (CH), 129.8 (CH), 128.2 (CH), 127.2 (C), 125.1 (CH), 124.1 (CH), 110.32 (CH), 110.27 (C), 84.3 (C), 69.1 (C), 54.8 (CH2), 16.3 (CH2); Analytical HPLC: tR = 12.5 min, >99% purity (10–95% MeCN/H2O, linear gradient, with constant 0.1% v/v TFA additive; 20 min run; 1 mL/min flow; ESI; positive ion mode; detection at 575 nm); HRMS (ESI) calcd for C26H21I2N2O3 [M+H]+ 662.9636, found 662.9654.
JF567–HaloTag ligand (7HTL):
6-Carboxy-JF549 (11; TFA salt; 100 mg, 1.22 mmol) was taken up in CH3CN (8 mL), and N-iodosuccinimide (317 mg, 1.41 mmol, 8 eq) was added. After stirring the reaction at room temperature for 18 h, 1 M NaOH (5 mL) was added. Following 5 min of vigorous stirring, the mixture was acidified with 1 M HCl (5.5 mL), diluted with water, and extracted with 15% i-PrOH/CHCl3 (3×). The combined organic extracts were dried over anhydrous MgSO4, filtered, and evaporated. The crude material was purified by reverse phase HPLC (10–75% MeCN/H2O, linear gradient, with constant 0.1% v/v TFA additive). The pooled HPLC product fractions were partially concentrated to remove MeCN and extracted with 10% MeOH/CH2Cl2 (3×). The organic extracts were dried over anhydrous MgSO4, filtered, and evaporated to afford 6-carboxy-JF567 (12) as a dark purple solid (30 mg, 21%, TFA salt).
6-Carboxy-JF567 (12; TFA salt; 30 mg, 36.6 μmol) was combined with DSC (22.5 mg, 87.8 μmol, 2.4 eq) in DMF (2 mL). After adding Et3N (30.6 μL, 219 μmol, 6 eq) and DMAP (0.4 mg, 3.7 μmol, 0.1 eq), the reaction was stirred at room temperature for 1 h. HaloTag(O2)amine (HTL-NH2, 13; TFA salt; 37.1 mg, 110 μmol, 3 eq) in DMF (250 μL) was added, and the reaction was stirred an additional 2 h at room temperature. It was subsequently diluted with saturated NaHCO3 and extracted with EtOAc (2×). The combined organic extracts were washed with brine, dried over anhydrous MgSO4, filtered, and concentrated in vacuo. The residue was purified by reverse phase HPLC (20–70% MeCN/H2O, linear gradient, with constant 0.1% v/v TFA additive); pooled HPLC product fractions were partially concentrated to remove MeCN, diluted with saturated NaHCO3, and extracted with CH2Cl2 (2×). The organics were dried over anhydrous MgSO4, filtered, and evaporated to afford 18.5 mg (55%) of JF567–HaloTag ligand (7HTL) as a purple solid. 1H NMR (CDCl3, 400 MHz) δ 8.07 – 7.97 (m, 2H), 7.53 – 7.48 (m, 1H), 6.75 (bs, 1H), 6.57 (d, J = 8.7 Hz, 2H), 6.21 (d, J = 8.7 Hz, 2H), 4.25 (q, J = 7.4 Hz, 4H), 4.19 (q, J = 7.4 Hz, 4H), 3.66 – 3.57 (m, 6H), 3.55 – 3.48 (m, 4H), 3.37 (t, J = 6.6 Hz, 2H), 2.25 (p, J = 7.4 Hz, 4H), 1.78 – 1.69 (m, 2H), 1.53 – 1.45 (m, 2H), 1.45 – 1.36 (m, 2H), 1.35 – 1.27 (m, 2H); Analytical HPLC: tR = 13.1 min, >99% purity (10–95% MeCN/H2O, linear gradient, with constant 0.1% v/v TFA additive; 20 min run; 1 mL/min flow; ESI; positive ion mode; detection at 575 nm); HRMS (ESI) calcd for C37H41ClI2N3O6 [M+H]+ 912.0768, found 912.0782.
Bis(3-bromophenyl)sulfane (16):
An oven-dried round-bottom flask was charged with CuI (604 mg, 3.17 mmol, 0.1 eq) and K2CO3 (8.77 g, 63.5 mmol, 2 eq). The flask was sealed and evacuated/backfilled with nitrogen (3×). Isopropanol (125 mL) was added, followed by ethylene glycol (3.54 mL, 63.5 mmol, 2 eq), 3-bromothiophenol (14; 3.28 mL, 31.7 mmol), and 3-bromoiodobenzene (15; 4.45 mL, 34.9 mmol, 1.1 eq). The reaction mixture was stirred at 80 °C for 18 h. It was then diluted with saturated NH4Cl (200 mL) and EtOAc (200 mL), vigorously stirred for 30 min, and filtered through Celite. The filtrate was separated, and the aqueous layer was extracted again with EtOAc. The combined organics were washed with brine, dried over anhydrous MgSO4, filtered, and evaporated. Flash chromatography (100% hexanes, linear gradient) afforded 8.93 g (82%) of dibromide 16 as a colorless oil. 1H NMR (CDCl3, 400 MHz) δ 7.48 (t, J = 1.8 Hz, 2H), 7.40 (ddd, J = 7.8, 1.9, 1.2 Hz, 2H), 7.28 – 7.23 (m, 2H), 7.18 (t, J = 7.8 Hz, 2H); 13C NMR (CDCl3, 101 MHz) δ 137.3 (C), 133.8 (CH), 130.79 (CH), 130.75 (CH), 129.8 (CH), 123.3 (C); HRMS (EI) calcd for C12H8Br2S [M]+• 341.8708, found 341.8732.
Bis(3-(azetidin-1-yl)phenyl)sulfane (17):
An oven-dried round-bottom flask was charged with CuI (985 mg, 5.17 mmol, 0.2 eq), L-proline (1.19 g, 10.4 mmol, 0.4 eq), and K2CO3 (14.30 g, 103.5 mmol, 4 eq). The flask was sealed and evacuated/backfilled with nitrogen (3×). A solution of dibromide 16 (8.90 g, 25.9 mmol) in DMSO (100 mL) was added, and the reaction was flushed again with nitrogen (3×). Following the addition of azetidine (10.46 mL, 155.2 mmol, 6 eq), the reaction was stirred at 100 °C for 18 h. It was then cooled to room temperature, diluted with saturated NH4Cl, and extracted with EtOAc (2×). The combined organic extracts were washed with water and brine, dried over anhydrous MgSO4, filtered, and concentrated in vacuo. Purification by flash chromatography on silica gel (0–30% EtOAc/hexanes, linear gradient) afforded 17 (6.08 g, 79%) as a white solid. 1H NMR (CDCl3, 400 MHz) δ 7.11 (t, J = 7.9 Hz, 2H), 6.68 (ddd, J = 7.7, 1.7, 1.0 Hz, 2H), 6.45 (t, J = 2.0 Hz, 2H), 6.30 (ddd, J = 8.1, 2.3, 0.9 Hz, 2H), 3.83 (t, J = 7.2 Hz, 8H), 2.33 (p, J = 7.3 Hz, 4H); 13C NMR (CDCl3, 101 MHz) δ 152.8 (C), 136.3 (C), 129.5 (CH), 119.9 (CH), 113.6 (CH), 110.1 (CH), 52.5 (CH2), 17.1 (CH2); HRMS (ESI) calcd for C18H21N2S [M+H]+ 297.1420, found 297.1428.
Bis(5-(azetidin-1-yl)-2-bromophenyl)sulfane (8):
Sulfide 17 (6.00 g, 20.2 mmol) was taken up in DMF (100 mL). N-Bromosuccinimide (7.20 g, 40.5 mmol, 2 eq) was added portion-wise over 5 min, and the reaction was then stirred at room temperature for 2 h. The reaction mixture was concentrated in vacuo; the resulting residue was diluted with water and extracted with EtOAc (2×). The combined organic extracts were washed with water and brine, dried over anhydrous MgSO4, filtered, and concentrated in vacuo. The crude product was triturated with Et2O, sonicated, and filtered. The filter cake was washed with Et2O and dried to provide the title compound as a white solid. The filtrate was concentrated, chromatographed on silica gel (0–50% Et2O/hexanes, linear gradient), and triturated as before to yield additional dibromide product. The two crops of white powder were combined, affording 6.51 g (71%) of dibromide 8. 1H NMR (CDCl3, 400 MHz) δ 7.40 – 7.34 (m, 2H), 6.23 – 6.18 (m, 4H), 3.76 (t, J = 7.3 Hz, 8H), 2.31 (p, J = 7.2 Hz, 4H); 13C NMR (CDCl3, 101 MHz) δ 151.9 (C), 135.6 (C), 133.3 (CH), 115.0 (CH), 112.3 (C), 112.1 (CH), 52.4 (CH2), 16.9 (CH2); HRMS (ESI) calcd for C18H19Br2N2S [M+H]+ 452.9630, found 452.9632.
JF570 (10):
A solution of dibromide 8 (200 mg, 0.440 mmol) in THF (10 mL) was cooled to −78 °C under nitrogen. tert-Butyllithium (1.7 M in pentane, 1.14 mL, 1.94 mmol, 4.4 eq) was added, and the reaction was stirred at −78 °C for 30 min. It was then warmed to −20 °C, and a solution of phthalic anhydride (9; 143 mg, 0.969 mmol, 2.2 eq) in THF (10 mL) was added dropwise over 30 min via addition funnel. The reaction was allowed to warm to room temperature overnight (18 h). Following the addition of AcOH (100 μL), the mixture was diluted with MeOH, deposited onto Celite, and concentrated to dryness. Silica gel chromatography (0–10% MeOH (2 M NH3)/CH2Cl2, linear gradient; dry load with Celite) afforded 87 mg (46%) of JF570 (10) as a dark purple solid. 1H NMR (CD3OD, 400 MHz) δ 8.14 – 8.08 (m, 1H), 7.68 – 7.59 (m, 2H), 7.25 (d, J = 9.3 Hz, 2H), 7.21 – 7.17 (m, 1H), 6.79 (d, J = 2.3 Hz, 2H), 6.55 (dd, J = 9.3, 2.3 Hz, 2H), 4.22 (t, J = 7.6 Hz, 8H), 2.51 (p, J = 7.6 Hz, 4H); 13C NMR (CD3OD, 101 MHz) δ 172.6 (C), 158.9 (C), 154.0 (C), 144.0 (C), 139.14 (C), 139.10 (C), 136.7 (CH), 131.3 (CH), 130.8 (CH), 130.4 (CH), 130.3 (CH), 120.7 (C), 113.9 (CH), 104.6 (CH), 52.6 (CH2), 16.9 (CH2); Analytical HPLC: tR = 11.8 min, >99% purity (10–95% MeCN/H2O, linear gradient, with constant 0.1% v/v TFA additive; 20 min run; 1 mL/min flow; ESI; positive ion mode; detection at 550 nm); HRMS (ESI) calcd for C26H23N2O2S [M+H]+ 427.1475, found 427.1489.
6-Carboxy-JF570 (19):
A solution of dibromide 8 (600 mg, 1.32 mmol, 1.5 eq) in THF (30 mL) was cooled to −78 °C under nitrogen. tert-Butyllithium (1.7 M in pentane, 3.11 mL, 5.28 mmol, 6 eq) was added, and the reaction was stirred at −78 °C for 30 min. It was then warmed to −20 °C, and a solution of ester 18 (346 mg, 0.880 mmol, 1 eq) in THF (15 mL) was added dropwise over 30 min via addition funnel. The reaction was allowed to warm to room temperature overnight (18 h). It was subsequently diluted with saturated NH4Cl and water and extracted with EtOAc (2×). The combined organic extracts were dried over anhydrous MgSO4, filtered, and concentrated in vacuo. The resulting residue was redissolved in MeOH (10 mL), and 1 M HCl (1 mL) was added. After stirring the solution at room temperature for 30 min, it was diluted with toluene (10 mL), deposited onto Celite, concentrated to dryness, and purified by flash chromatography (0–20% MeOH/CH2Cl2, linear gradient, with constant 1% v/v AcOH additive; dry load with Celite) to provide the bis(2,2-bis(hydroxymethyl)propyl) diester intermediate (608 mg, 94%, acetate salt).
The diester (608 mg, 0.827 mmol) was taken up in 2,2,2-trifluoroethanol (15 mL), and 25% w/w NaOH (5 mL) was added. The reaction was stirred at room temperature for 7 days. It was then acidified with AcOH (5 mL), diluted with water, and extracted with 15% i-PrOH/CHCl3 (3×). The combined organic extracts were dried over anhydrous MgSO4, filtered, and evaporated. Flash chromatography on silica gel (0–20% MeOH/CH2Cl2, linear gradient, with constant 1% v/v AcOH additive) afforded 342 mg (78%, TFA salt) of 6-carboxy-JF570 (19) as a dark purple solid. 1H NMR (CD3OD, 400 MHz) δ 8.41 (d, J = 8.2 Hz, 1H), 8.36 (dd, J = 8.2, 1.6 Hz, 1H), 7.87 (d, J = 1.6 Hz, 1H), 7.15 (d, J = 9.4 Hz, 2H), 6.88 (d, J = 2.3 Hz, 2H), 6.62 (dd, J = 9.4, 2.3 Hz, 2H), 4.27 (t, J = 7.7 Hz, 8H), 2.53 (p, J = 7.6 Hz, 4H); 13C NMR (CD3OD, 101 MHz) δ 167.8 (C), 167.3 (C), 161.8 (C), 154.3 (C), 144.9 (C), 139.1 (C), 136.5 (CH), 136.0 (C), 135.9 (C), 132.9 (CH), 132.5 (CH), 131.8 (CH), 120.1 (C), 114.8 (CH), 104.6 (CH), 52.7 (CH2), 16.9 (CH2); Analytical HPLC: tR = 9.8 min, >99% purity (10–95% MeCN/H2O, linear gradient, with constant 0.1% v/v TFA additive; 20 min run; 1 mL/min flow; ESI; positive ion mode; detection at 575 nm); HRMS (ESI) calcd for C27H23N2O4S [M+H]+ 471.1373, found 471.1376.
Spectroscopy.
Spectroscopy of various dyes, photosensitizers and associated HaloTag Ligand variants was performed using 1 cm path length, 3.5 mL or 1 mL or 300 μL quartz cuvettes (Starna Cells). Absorption spectra were recorded on a Cary Model 100 spectrometer (Agilent). Fluorescence emission spectra were recorded on a Varian Cary Eclipse (Agilent). Absorbance and fluorescence spectroscopy were also carried out using a Flexstation 3 microplate reader (Molecular Devices) when noted. Fluorescence quantum yields were recorded on a Quantaurus-QY (Hamamatsu). Spectroscopy measurements of free dyes were performed 10 mM HEPES, pH 7.3 at ambient temperature (22 ± 2 °C); the reported values of maximum absorption wavelength (λabs), extinction coefficient at λabs (ε), maximum fluorescence emission wavelength (λem), and fluorescence quantum yield Φf) are averages (n = 3). Spectroscopy of HaloTag conjugates were performed in Dulbecco’s phosphate-buffered saline (DPBS, Gibco).
Solutions prepared for spectroscopy contained 3–5 μM dye and 10 μM HaloTag protein when applicable. Spectra of compounds bound to HaloTag protein were allowed a 30 min binding incubation period in the dark at ambient temperature prior to measurement. Masked compound Eosin-Ac2–HaloTag ligand (2HTLAc2) was incubated with 23.75 units/mL porcine liver esterase in the dark at ambient temperature for 6 h prior to incubation with HaloTag protein and subsequent spectroscopy. The reported values of maximum absorption wavelength (λabs), extinction coefficient at λabs (ε), maximum fluorescence emission wavelength (λem), and fluorescence quantum yield Φf) are averages (n = 2) KL–Z was calculated using the following formula: KL–Z = (εdw/εmax)/(1 – εdw/εmax), where εdw is the extinction coefficient in a 1:1 v/v dioxane:water solvent mixture, and εmax is the extinction coefficient in 0.1% v/v trifluoroacetic acid (TFA) in 2,2,2-trifluoroethanol (TFE). The reported values are averages (n = 2).
HaloTag protein purification.
Purified HaloTag protein was obtained via pRSET vector expression in BL21 E. coli and subsequent purification via immobilized metal affinity chromatography (IMAC) using an ÄKTA Avant (GE Healthcare Life Sciences). Protein was dialyzed in DPBS for 48 h prior to quantification and use.
Singlet oxygen quantum yield.
Singlet oxygen quantum yields (ΦSO) of free dyes were measured using the commercially available Singlet Oxygen Sensor Green (SOSG; Thermo Fisher). Solutions of 5 μM dye + 30 μM SOSG in DPBS + 0.1% DMSO dye cosolvent + 0.6% MeOH SOSG cosolvent at a volume of 30 μL were excited in 384-well microplate wells with light doses of increasing exposure times and constant power on a Nikon Ti-Eclipse microscope outfitted with a Lumencor LED excitation system. SOSG emission was measured using a FlexStation 3 microplate reader and corrected for baseline emission and DMSO negative control contributions to emission. Values were first normalized to the estimated photons absorbed by each dye to correct for differential excitation efficiency based upon light source output spectra and dye molar absorptivity spectra; absorption cross-section measurements of dyes for ΦSO determination were measured in DPBS. Well averages were then calculated (n = 3), and data were normalized to the standard dye Rose Bengal (3), which has a ΦSO = 0.76 (Ezquerra Riega et al., 2017).
Chromophore assisted light inactivation (CALI) of EGFP in live cells.
U2OS cells were transfected with pHaloTag-EGFP (Ebner et al., 2017) via the Amaxa Nucleofector II system from Lonza to transiently express a HaloTag-EGFP fusion protein. These cells were labeled with 1 μM ligand in media + 0.1% DMSO cosolvent for 1 h followed by 2 media washes. EGFP fluorescence was imaged via 488 nm laser or 440 nm-centered excitation preceding and immediately following a 10 s exposure to 10 W/cm2 561 nm monochromatic (laser) or 508 nm-centered LED-based light; the 440 nm/508 nm combination was utilized for the Eosin-Ac2–HaloTag ligand (2HTLAC2). Change in mean EGFP signal due to CALI was calculated per manually segmented cell using the FIJI 2.0.0-rc-69/1.52i distribution of ImageJ (Schindelin et al., 2012).
Photoablation of cells in culture.
U2OS cells expressing HaloTag–H2B (Chen et al., 2014), HaloTag–TOMM20, or no exogenous proteins were seeded in a 384-well, tissue-culture treated microplate (Perkin Elmer) at a density of 2K cells/well and allowed to grow for 24 h. Cells were then labeled with 1 μM ligand in media + 0.1% DMSO cosolvent for 1 h followed by 2 media washes and exposed to various light of various power and wavelength for 3 min, one well at a time, through the 4× objective of a Nikon Ti-Eclipse microscope at 5% CO2, 37 °C. After an additional incubation for 24 hr, cells were costained with NucBlue Live ReadyProbes Reagent and NucRed Dead 647 ReadyProbes Reagent (Thermo Fisher) and imaged. The images were analyzed using the FIJI 2.0.0-rc-69/1.52i distribution of ImageJ (Schindelin et al., 2012) as follows. Image segmentation was carried out by autothresholding each image via the Triangle algorithm ± 10 manual value adjustments until image noise was optimally suppressed followed by the binary watershed algorithm. Segmented cells were counted via the Analyze Particles function (5-inf pixel size, 0.15–1.00 roundness). The percentage of ablated cells was determined by measuring the number of dead cells stained in the NucRed Dead 647 ReadyProbe image and the total number of cells stained in the NucBlue Live ReadyProbe image. For the representative confocal and brightfield images (Fig. 2h-i) U2OS cells expressing HaloTag–TOMM20 were labeled with 500 nM ligand in media, washed, and imaged using a Zeiss 880 inverted confocal microscope with an EC Plan-Neofluar 40–/1.30 oil immersion objective and 561 nm laser excitation. Cells were imaged before and 30 min after a 3 min irradiation dose delivered with an X-Cite 120PC Series illuminator using Zeiss filter set 45 (excitation: 560 nm with a 40 nm bandpass; beamsplitter: 585 nm; emission: 630 nm with a 75 nm bandpass).
Cloning of zebrafish ablation construct.
The zebrafish construct 10×UAS:TOMM20-HaloTag-p2a-mCerulean3, myl7:EGFP was assembled using the Tol2kit system with Gateway LR Clonase II Enzyme Mix (Invitrogen). It was composed of the following components: p5E 10×UAS, pME TOMM20-HaloTagv7-p2a-mCerulean3, p3E PolyA (Tol2#302) and the destination vector, pDestTol2pCG2 (Tol2#395) (Grzegorski et al., 2014; Kwan et al., 2007; Lo et al., 2015; Los et al., 2008; Markwardt et al., 2011).
In vivo ablation of zebrafish neurons.
Mifaw2/w2 roya9/a9 (Casper) zebrafish (Kimura et al., 2008) were maintained under standard conditions at 28.5 □ and a 14:10 h light:dark cycle. Embryos (1–2 cell stage) of Tg(elavl3:Gal4-VP16) (Kimura et al., 2008) were injected with 25 ng/μL DNA plasmid encoding the co-expression cassette of mitochondria-localized HaloTag and mCerulean3 under the control of the 10×UAS promoter (10×UAS:TOMM20-HaloTag-p2a-mCerulean3, myl7:EGFP), and 25 ng/×L Tol2 transposase mRNA diluted in E3 medium. Subsequently, the injected embryos with broad expression of mCerulean3 were incubated in system water containing HaloTag dye ligands: JF570–HaloTag ligand (10HTL) or JF585–HaloTag ligand (20HTL) at 30 μM at 4-day post-fertilization (dpf) for 12 h and then washed in dye-free system water. Larvae at 5 dpf were then anesthetized in tricaine methanesulfonate (MilliporeSigma, E10521, MO) dissolved in system water at 160 mg/L and then embedded in 1.6% low melting point agar (MilliporeSigma, 2070-OP, MO). Volumetric images of the entire brain were first acquired with a confocal microscope (Zeiss, LSM880) with a 20× 1.0 NA objective lens (Zeiss, W Plan-Apochromat 20×/1.0) to confirm the distribution of the cells labeled with mCerulean3 and HaloTag ligand prior to ablation. Then yellow, 560 nm-centered ablation light was shone on the forebrain region (Fig. 3a-b; Zone 1) for 5 minutes through the widefield illumination path equipped with a fluorescent filter (Zeiss, Filter Set 45) and a mercury lamp (Leistungselektronik JENA GmbH, HXP 120 V) with the field diaphragm adjusted just to illuminate the target region. The power densities used for 10HTL and 20HTL 3 W/cm2 and 12 W/cm2 respectively. Post-ablation confocal stacks were acquired right after the ablation and 1 day after the ablation (6 dpf) with the imaging parameters identical to those used prior to ablation. Images were analyzed using the FIJI 2.0.0-rc-69/1.52i distribution of ImageJ (Schindelin et al., 2012) as follows. Zone ROIs were manually segmented using brightfield maximum intensity z-projections and kept consistent across image stack channels using FIJI’s ROI manager and crop function. FIJI’s 3D Object Counter module was utilized to count cells in the mCer fluorescence channel image stacks obtained pre-irradiation and 24 h post-irradiation using an 8-bit minimum threshold pixel value of 11 and default size settings. The percentage of remaining cells was determined by measuring the number of cells in pre-irradiation samples and the number of cells in 24 h post-irradiation samples.
DAB photopolymerization for electron microscopy.
HOS cells were incubated for 30 min with 5 μM dye in culture medium at 37 °C. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in 5 mM CaCl2, 0.1 M sodium cacodylate buffer, pH 7.4, incubating at room temperature for 5 min and 0 °C (ice) for 2 h. Cells were washed 5×, 2 min each wash, in 0.1 M sodium cacodylate buffer and then blocked for 15 min in blocking buffer (10 mM glycine, 10 mM potassium cyanide in 0.1 M sodium cacodylate buffer). After 3× washes with 0.1 M sodium cacodylate buffer, cells were bathed in 2.5 mM DAB. (HCl)4 (Sigma) in 0.1 M sodium cacodylate buffer. Cells were placed on a cold-stage Leica SPE-II confocal microscope and photooxidized by continuous epi-fluorescence illumination (150 W Xenon Lamp) through an mCherry filter cube (excitation: 542–582 nm) for 7 min. Cells were washed 5×, 2 min each wash, with 0.1 M sodium cacodylate buffer and fixed for 30 min with 2% glutaraldehyde at 4 °C. After 3× additional washes with 0.1 M sodium cacodylate buffer, the sample was stained for 30 min with 1% osmium tetroxide and 1.5% potassium ferrocyanide in 0.1 M sodium cacodylate buffer. Cells were washed with doubly distilled water 5×, 2 min each wash.
EM sample preparation and imaging.
After DAB polymerization and osmification, HOS cells were dehydrated in ethanol and embedded in Durcupan epoxy resin as described previously (Ou et al., 2015). Briefly, the samples were incubated for 2 min in water containing increasing concentrations of ethanol (20% v/v, 50% v/v, 70% v/v, and 90% v/v) followed by three successive 2 min incubations in 100% ethanol. The samples were then infiltrated with durcupan resin (Sigma, 44610) with three successive 2 h treatments. Finally, the cells were incubated with freshly prepared durcupan resin and cured for 2 d at 60 °C. Epoxy embedded cells were cut into either 70–80 nm sections and mounted onto 200 hexagonal mesh copper grid for TEM or 250 nm sections mounted onto slot grid covered with a luxel film for electron tomography. For conventional transmission electron microscopy (TEM), ultrathin sections were imaged at 80 kV using an FEI Tecnai microscope (FEI Company). For EM tomography, a mixture of 5 nm and 10 nm colloidal gold particles serving as fiducial markers were deposited on both sides of the semi-thin (~250 nm) specimen sections after the sample grids were glow discharged. The tomography data was collected on an FEI Titan Halo electron microscope operating at 300 kV with a DE64 direct electron detector. The grids were loaded into a rotation holder specifically designed for multi-tilt tomography manufactured by Fischione Instruments (Model 2040). During a tilt series, images were acquired tilting the goniometer holding the sample holder from −60° to +60° with 0.5° increments. SerialEM was used to control the FEI Titan for the data acquisition (Mastronarde, 1997). The sample was rotated along the z-axis by 45° for the second tilt series, 90° (relative to origin) for the third tilt series, and 135° (relative to origin) for the final tilt series. Adjustments were made to the stage/sample height to ensure an identical eucentric height and magnification for each tilt series. The sequence of tilt series followed the multilevel access scheme previously described (Phan et al., 2017), which minimizes errors associated with sample shrinkage evenly across all tilt series. EM tomography reconstruction: The entire set of EMT images were aligned and reconstructed using Transform-based Tracking, Bundle adjustment, and Reconstruction (TxBR) package (Lawrence et al., 2006; Phan et al., 2017; Phan et al., 2012). 3D volume rendering was made with Amira 6.3.
Quantification and statistical analyses
For spectroscopy measurements, n values are given in the Methods Details section and represent the number of measurements of different samples prepared from the same dye DMSO stock solution or HaloTag conjugate stock solution. For ΦSO determination, n values are given in the Method Details section and represent the number of separate photochemical experiments using the same dye DMSO stock solutions of dye and SOSG sensor. For CALI experiments, n values are given in the legend of Figure 2 and represent the number of cells. For ablation experiments in culture, n values are given in the legends of Figure 2 and Figure S3 and represent the number of separate experiments counting all and dead cells using NucBlue Live and NucRed Dead. For ablation experiments in vivo, n values are given in the legend of Figure 3 and indicate the number of fish. For electron microscopy experiments, these were duplicated with similar results as indicated in the legend of Figure 4. For all whisker plots, the center line indicates median; box limits indicate upper and lower quartiles; whiskers indicate min–max. Graphs were rendered using GraphPad Prism 7.04. Statictical tests were perfomed using the Addinsoft XLSTAT v2020.1.1 add-in to Microsoft Excel 2019 v1908 build 11929.20838. All data analyzed statistically were assessed for normality and scedasticity assumptions when appropriate using the Shapiro-Wilk test or Levene’s test. Parametric or non-parametric tests are indicated in the figure legends and include the Student’s T-test, Mann–Whitney U test, ANOVA with Tukey’s post-hoc, Welch’s ANOVA with Games–Howell post-hoc, and Kruskal–Wallis H test with Steel–Dwass–Critchlow–Fligner procedure.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Singlet Oxygen Sensor Green | Invitrogen | Thermo Fisher Scientific Catalog #S36002 |
| JF567 (7) | Janelia Research Campus; this paper | N/A |
| JF567–HaloTag ligand (7HTL) | Janelia Research Campus; this paper | N/A |
| JF570 (10) | Janelia Research Campus; this paper | N/A |
| JF570–HaloTag ligand (10HTL) | Janelia Research Campus; this paper | N/A |
| Eosin-Ac2–HaloTag ligand (2HTLAc2) | Promega | N/A |
| Eosin Y | Sigma Aldrich | Sigma Aldrich Catalog #E4009 |
| JF549 (6) | Janelia Research Campus; (Grimm et al., 2015) | N/A |
| JF585–HaloTag ligand (20HTL) | Janelia Research Campus; (Grimm et al., 2017b) | N/A |
| Rose Bengal | Sigma Aldrich | Sigma Aldrich Catalog #330000 |
| JF549–HaloTag ligand (6HTL) | Janelia Research Campus; (Grimm et al., 2015) | N/A |
| Porcine liver esterase (PLE) | Sigma Aldrich | Sigma Aldrich Catalog #E3019 |
| NucBlue™ Live ReadyProbes™ Reagent | Invitrogen | Thermo Fisher Scientific Catalog #R37605 |
| NucRed™ Dead 647 ReadyProbes™ Reagent | Invitrogen | Thermo Fisher Scientific Catalog #R37113 |
| Gateway LR Clonase II Enzyme Mix | Invitrogen | Thermo Fisher Scientific Catalog #11791020 |
| Durcupan™ ACM | Sigma Aldrich | Sigma Aldrich Catalog #44610 |
| Experimental Models: Cell Lines | ||
| Human: U2OS cell line | ATCC | RRID: CVCL_0042 |
| Human: U2OS cell line expressing HaloTag-H2B | Janelia Research Campus; ATTC; HT-H2B sequence; (Chen et al., 2014) | Modified cells of RRID: CVCL_0042 |
| Human: U2OS cell line expressing HaloTag-TOMM20 | Janelia Research Campus; ATCC; TOMM20 sequence from Michael Davidson, FSU | Modified cells of RRID: CVCL_0042 |
| Human: HOS cell line: HaloTag-Histone H2B (HT-H2B) | UCSD; ATTC | Modified cells of RRID:CVCL_0312 |
| Experimental Models: Organisms/Strains | ||
| Mitfaw2/w2 roya9/a9 (Casper) zebrafish (Danio rerio), Tg(elavl3:Gal4-VP16) | Janelia Research Campus; (Kimura et al., 2008) | N/A |
| Recombinant DNA | ||
| pHaloTag-EGFP | Thomas Leonard & Ivan Yudushkin; (Ebner, 2017). | http://n2t.net/addgene:86629; RRID:Addgene_86629 |
| pRSET.HaloTag | Janelia Research Campus; pRSET: Invitrogen; HaloTag: Promega | pRSET - V35120; HaloTag - N/A |
| 10xUAS:TOMM20-HaloTagv7-p2a-mCerulean3, myl7:EGFP | Janelia Research Campus; Tol2 destination vector:(Kwan et al., 2007); Kozak sequence: (Grzegorski et al., 2014); Native TOMM20: Accession NM_001002698; NP_001002698); HaloTagv7: Promega; P2A sequence – (Lo, 2015); mCerulean3 sequence: (Markwardt et al., 2011). | N/A |
| Software and Algorithms | ||
| XLSTAT v2020.1.1 add-in to Microsoft Excel 2019 v1908 build 11929.20838 | Addinsoft | http://www.xlstat.com/en/ |
| GraphPad Prism 7.04 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Fiji ImageJ Distribution 2.0.0-rc-69/1.52i | (Schindelin et al., 2012) | https://fiji.sc/ |
| Amira 6.3 | Thermo Fisher Scientific | N/A |
Highlights.
Incorporation of iodine or sulfur into rhodamine dyes increases 1O2 quantum yield
Rhodamine photosensitizers are compatible with live cell labeling strategies
Generating 1O2 allows photoablation of entire cells in culture or in vivo
Photopolymerization of 3,3'-diaminobenzidine gives contrast in electron microscopy
Significance.
We report the rational design of photosensitizers derived from the azetidine-containing Janelia Fluor rhodamine scaffold. For use in biological systems, we synthesize their HaloTag ligand variants and demonstrate the utility of these compounds in disparate applications. Compared to commercially available photosensitizer HaloTag ligands, these compounds exhibit a bathochromic shift in absorption spectra, allowing illumination with less phototoxic wavelengths of light or multiplexing with fluorescent proteins or optogenetic reagents. These compounds also diffuse in and out of cells and tissue without the need for chemical modification, making use of these chemical tools straightforward. These molecules can be used in a variety of biological experiments, such as destroying individual proteins with high spatiotemporal control, ablating genetically defined cells in a developing or learning animal, or providing multimodal contrast in light and electron microscopy.
Acknowledgements
Paul Tillberg, Claire Deo, Fadi Jradi, Maria Ioannou, and Charles Kim (all at Janelia) for contributive discussions. Xiaorong Zhang, Jordan Towne, Melissa Ramirez, and Kevin McGowan of the Janelia Research Campus Molecular Biology shared resource for assistance with plasmid preparation. Melanie Freeman, Jenny Hagemeier, Kathy Schaefer, Deepika Walpita, and Kevin McGowan of the Janelia Research Campus Cell and Tissue Culture shared resource for assistance with culture work. Margaret Jefferies and Anastasia Osowski for coordinative work. This work was supported by the Howard Hughes Medical Institute (HHMI) and the National Institutes of Health (NIH) through award R01GM086197 to S.R.A. and awards P41GM103412, R24GM137200, and S10OD021784 to M.H.E. T. C. Binns was a HHMI Medical Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interests
US Patent 9,933,417 and patent applications US 2019/0367736 and US 2019/0106573 describing azetidine-containing fluorophores and variant compositions (with inventors J.B.G. and L.D.L.) are assigned to HHMI. L.D.L. serves on the editorial board of Cell Chemical Biology. The authors declare no other competing interests.
References
- Abdelfattah AS, Kawashima T, Singh A, Novak O, Liu H, Shuai Y, Huang YC, Campagnola L, Seeman SC, Yu J, et al. (2019). Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704. [DOI] [PubMed] [Google Scholar]
- Ayele TM, Knutson SD, Ellipilli S, Hwang H, and Heemstra JM (2019). Fluorogenic photoaffinity labeling of proteins in living cells. Bioconjug Chem 30, 1309–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista MS, Cadet J, Di Mascio P, Ghogare AA, Greer A, Hamblin MR, Lorente C, Nunez SC, Ribeiro MS, Thomas AH, et al. (2017). Type I and Type II photosensitized oxidation reactions: Guidelines and mechanistic pathways. Photochem Photobiol 93, 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S, Sakurai T, Eustace BK, Beste G, Schier R, Rudert F, and Jay DG (2002). Fluorophore-assisted light inactivation: A high-throughput tool for direct target validation of proteins. Proteomics 2, 247–255. [DOI] [PubMed] [Google Scholar]
- Becker RS, Chakravorti S, and Das S (1990). The photosensitizers benzophenoxazine and thiazines: Comprehensive investigation of photophysical and photochemical properties. Photochem Photobiol 51, 533–538. [DOI] [PubMed] [Google Scholar]
- Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, and Lukyanov KA (2006). A genetically encoded photosensitizer. Nat Biotechnol 24, 95–99. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. (2014). Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. (2018). Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosny E, and Carpentier P (2012). GFP-like phototransformation mechanisms in the cytotoxic fluorescent protein KillerRed unraveled by structural and spectroscopic investigations. J Am Chem Soc 134, 18015–18021. [DOI] [PubMed] [Google Scholar]
- Deerinck TJ, Martone ME, Lev-Ram V, Green DP, Tsien RY, Spector DL, Huang S, and Ellisman MH (1994). Fluorescence photooxidation with eosin: A method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J Cell Biol 126, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrucki JW, Feret D, and Noatynska A (2007). Scattering of exciting light by live cells in fluorescence confocal imaging: phototoxic effects and relevance for FRAP studies. Biophys J 93, 1778–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwright S, and Sluder G (2017). Live cell imaging: Assessing the phototoxicity of 488 and 546 nm light and methods to alleviate it. J Cell Physiol 232, 2461–2468. [DOI] [PubMed] [Google Scholar]
- Ebner M, Lučić I, Leonard TA, and Yudushkin I (2017). PI(3,4,5)P(3) engagement restricts Akt activity to cellular membranes. Mol Cell 65, 416–431.e416. [DOI] [PubMed] [Google Scholar]
- Ezquerra Riega SD, Rodriguez HB, and San Roman E (2017). Rose bengal in poly(2-hydroxyethyl methacrylate) thin films: Self-quenching by photoactive energy traps. Methods Appl Fluoresc 5, 014010. [DOI] [PubMed] [Google Scholar]
- Fleming GR, Knight AWE, Morris JM, Morrison RJS, and Robinson GW (1977). Picosecond fluorescence studies of xanthene dyes. J Am Chem Soc 99, 4306–4311. [Google Scholar]
- Francisco CML, Gonçalves JMLA, Brum BS, Santos TPC, Lino-dos-Santos-Franco A, Silva DFT, and Pavani C (2017). The photodynamic efficiency of phenothiazinium dyes is aggregation dependent. New J Chem 41, 14438–14443. [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, and Ellisman MH (2002). Multicolor and electron microscopic imaging of connexin trafficking. Science 296, 503–507. [DOI] [PubMed] [Google Scholar]
- Gandin E, Lion Y, and Van de Vorst A (1983). Quantum yield of singlet oxygen production by xanthene derivatives. Photochem Photobiol 37, 271–278. [Google Scholar]
- Gold V (1987). Compendium of chemical terminology: IUPAC recommendations (Oxford Oxfordshire; Boston: Blackwell Scientific Publications; ). [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, and Nilsson T (2005). Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods 2, 857–862. [DOI] [PubMed] [Google Scholar]
- Grimm JB, Brown TA, Tkachuk AN, and Lavis LD (2017a). General synthetic method for Si-fluoresceins and Si-rhodamines. ACS Cent Sci 3, 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, et al. (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods 12, 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N, et al. (2017b). A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods 14, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorski SJ, Chiari EF, Robbins A, Kish PE, and Kahana A (2014). Natural variability of Kozak sequences correlates with function in a zebrafish model. PLoS One 9, e108475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Wang Y, Missinato MA, Onuoha E, Perkins LA, Watkins SC, St Croix CM, Tsang M, and Bruchez MP (2016). A genetically targetable near-infrared photosensitizer. Nat Methods 13, 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Gaietta G, Zurn A, Adams SR, Terrillon S, Ellisman MH, Tsien RY, and Lohse MJ (2010). Fluorescent labeling of tetracysteine-tagged proteins in intact cells. Nat Protoc 5, 1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C, Gibson A, Stinchcombe J, and Futter C (2000). Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol 327, 35–45. [DOI] [PubMed] [Google Scholar]
- Hu X, Huang YY, Wang Y, Wang X, and Hamblin MR (2018). Antimicrobial photodynamic therapy to control clinically relevant biofilm infections. Front Microbiol 9, 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay DG (1988). Selective destruction of protein function by chromophore-assisted laser inactivation. Proc Natl Acad Sci U S A 85, 5454–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Satou C, and Higashijima S (2008). V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord. Development 135, 3001–3005. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, and Chien CB (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, and Ting AY (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis LD, Chao TY, and Raines RT (2006). Fluorogenic label for biomolecular imaging. ACS Chem Biol 1, 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis LD, Chao TY, and Raines RT (2011). Synthesis and utility of fluorogenic acetoxymethyl ethers. Chem Sci 2, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence A, Bouwer JC, Perkins G, and Ellisman MH (2006). Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. J Struct Biol 154, 144–167. [DOI] [PubMed] [Google Scholar]
- Lee J, Yu P, Xiao X, and Kodadek T (2008). A general system for evaluating the efficiency of chromophore-assisted light inactivation (CALI) of proteins reveals Ru(II) tris-bipyridyl as an unusually efficient "warhead". Mol Biosyst 4, 59–65. [DOI] [PubMed] [Google Scholar]
- Li Y, Aggarwal MB, Ke K, Nguyen K, and Spitale RC (2018). Improved analysis of RNA localization by spatially restricted oxidation of RNA-protein complexes. Biochemistry 57, 1577–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Roider J, and Jay DG (1994). Chromophore-assisted laser inactivation of proteins is mediated by the photogeneration of free radicals. Proc Natl Acad Sci U S A 91, 2659–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden KG, Liao JC, and Jay DG (1992). Spatial specificity of chromophore assisted laser inactivation of protein function. Biophys J 61, 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liss V, Barlag B, Nietschke M, and Hensel M (2015). Self-labelling enzymes as universal tags for fluorescence microscopy, super-resolution microscopy and electron microscopy. Sci Rep 5, 17740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CA, Kays I, Emran F, Lin TJ, Cvetkovska V, and Chen BE (2015). Quantification of protein levels in single living cells. Cell Rep 13, 2634–2644. [DOI] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. (2008). HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 3, 373–382. [DOI] [PubMed] [Google Scholar]
- Lukinavicius G, Umezawa K, Olivier N, Honigmann A, Yang G, Plass T, Mueller V, Reymond L, Correa IR Jr., Luo ZG, et al. (2013). A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat Chem 5, 132–139. [DOI] [PubMed] [Google Scholar]
- Magidson V, and Khodjakov A (2013). Circumventing photodamage in live-cell microscopy. Methods Cell Biol 114, 545–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek KW, and Davis GW (2002). Transgenically encoded protein photoinactivation (FlAsH-FALI): Acute inactivation of synaptotagmin I. Neuron 36, 805–813. [DOI] [PubMed] [Google Scholar]
- Markwardt ML, Kremers GJ, Kraft CA, Ray K, Cranfill PJ, Wilson KA, Day RN, Wachter RM, Davidson MW, and Rizzo MA (2011). An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One 6, e17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, and Ting AY (2012). Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol 30, 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (1997). Dual-axis tomography: An approach with alignment methods that preserve resolution. J Struct Biol 120, 343–352. [DOI] [PubMed] [Google Scholar]
- Miller JP, and Selverston A (1979). Rapid killing of single neurons by irradiation of intracellularly injected dye. Science 206, 702–704. [DOI] [PubMed] [Google Scholar]
- Ochsner M (1997). Photophysical and photobiological processes in the photodynamic therapy of tumours. J Photochem Photobiol B 39, 1–18. [DOI] [PubMed] [Google Scholar]
- Onukwufor JO, Trewin AJ, Baran TM, Almast A, Foster TH, and Wojtovich AP (2020). Quantification of reactive oxygen species production by the red fluorescent proteins KillerRed, SuperNova and mCherry. Free Radic Biol Med 147, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Deerinck TJ, Bushong E, Ellisman MH, and O'Shea CC (2015). Visualizing viral protein structures in cells using genetic probes for correlated light and electron microscopy. Methods 90, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal P, Zeng H, Durocher G, Girard D, Li T, Gupta AK, Giasson R, Blanchard L, Gaboury L, Balassy A, et al. (1996). Phototoxicity of some bromine-substituted rhodamine dyes: Synthesis, photophysical properties and application as photosensitizers. Photochem Photobiol 63, 161–168. [DOI] [PubMed] [Google Scholar]
- Phan S, Boassa D, Nguyen P, Wan X, Lanman J, Lawrence A, and Ellisman MH (2017). 3D reconstruction of biological structures: automated procedures for alignment and reconstruction of multiple tilt series in electron tomography. Adv Struct Chem Imaging 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan S, Lawrence A, Molina T, Lanman J, Berlanga M, Terada M, Kulungowski A, Obayashi J, and Ellisman M (2012). TxBR montage reconstruction for large field electron tomography. J Struct Biol 180, 154–164. [DOI] [PubMed] [Google Scholar]
- Riani YD, Matsuda T, Takemoto K, and Nagai T (2018). Green monomeric photosensitizing fluorescent protein for photo-inducible protein inactivation and cell ablation. BMC Biol 16, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gonzalez R, Cortajarena AL, Mejias SH, Agut M, Nonell S, and Flors C (2013). Singlet oxygen generation by the genetically encoded tag miniSOG. J Am Chem Soc 135, 9564–9567. [DOI] [PubMed] [Google Scholar]
- Sawinski J, Wallace DJ, Greenberg DS, Grossmann S, Denk W, and Kerr JN (2009). Visually evoked activity in cortical cells imaged in freely moving animals. Proc Natl Acad Sci U S A 106, 19557–19562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold PG, Gouterman M, and Callis J (1969). Calorimetric, photometric and lifetime determinations of fluorescence yields of fluorescein dyes. Photochem Photobiol 9, 229–242. [DOI] [PubMed] [Google Scholar]
- Shirmanova M, Yuzhakova D, Snopova L, Perelman G, Serebrovskaya E, Lukyanov K, Turchin I, Subochev P, Lukyanov S, Kamensky V, et al. (2015). Towards PDT with genetically encoded photosensitizer KillerRed: A comparison of continuous and pulsed laser regimens in an animal tumor model. PLoS One 10, e0144617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, and Tsien RY (2011). A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol 9, e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K, Iwanari H, Tada H, Suyama K, Sano A, Nagai T, Hamakubo T, and Takahashi T (2017). Optical inactivation of synaptic AMPA receptors erases fear memory. Nat Biotechnol 35, 38–47. [DOI] [PubMed] [Google Scholar]
- Takemoto K, Matsuda T, McDougall M, Klaubert DH, Hasegawa A, Los GV, Wood KV, Miyawaki A, and Nagai T (2011). Chromophore-assisted light inactivation of HaloTag fusion proteins labeled with eosin in living cells. ACS Chem Biol 6, 401–406. [DOI] [PubMed] [Google Scholar]
- Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, Kotera I, Arai Y, Horiuchi M, Fukui K, et al. (2013). SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci Rep 3, 2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torra J, Lafaye C, Signor L, Aumonier S, Flors C, Shu X, Nonell S, Gotthard G, and Royant A (2019). Tailing miniSOG: Structural bases of the complex photophysics of a flavin-binding singlet oxygen photosensitizing protein. Sci Rep 9, 2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tour O, Meijer RM, Zacharias DA, Adams SR, and Tsien RY (2003). Genetically targeted chromophore-assisted light inactivation. Nat Biotechnol 21, 1505–1508. [DOI] [PubMed] [Google Scholar]
- Vitriol EA, Uetrecht AC, Shen F, Jacobson K, and Bear JE (2007). Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proc Natl Acad Sci U S A 104, 6702–6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Wang Y, Murray CK, Hamblin MR, Hooper DC, and Dai T (2017). Antimicrobial blue light inactivation of pathogenic microbes: State of the art. Drug Resist Updat 33-35, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtovich AP, Wei AY, Sherman TA, Foster TH, and Nehrke K (2016). Chromophore-assisted light inactivation of mitochondrial electron transport chain Complex II in Caenorhabditis elegans. Sci Rep 6, 29695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Tian X, and Ai HW (2019). Molecular tools to generate reactive oxygen species in biological systems. Bioconjug Chem 30, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Xiong Y, Chen B, Negash S, Squier TC, and Mayer MU (2006). Fluorophore-assisted light inactivation of calmodulin involves singlet-oxygen mediated cross-linking and methionine oxidation. Biochemistry 45, 4736–4748. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jiang C, Figueiro Longo JP, Azevedo RB, Zhang H, and Muehlmann LA (2018). An updated overview on the development of new photosensitizers for anticancer photodynamic therapy. Acta Pharm Sin B 8, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Ayala AX, Chung I, Weigel AV, Ranjan A, Falco N, Grimm JB, Tkachuk AN, Wu C, Lippincott-Schwartz J, et al. (2019). Rational design of fluorogenic and spontaneously blinking labels for super-resolution imaging. ACS Cent Sci 5, 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new datasets or code.