Abstract
Cell-to-cell communication by exosomes controls normal and pathogenic processes1,2. Viruses can spread in exosomes and thereby avoid immune recognition3. While biogenesis, binding, and uptake of exosomes are well-characterized4,5, delivery of exosome cargo into the cytoplasm is poorly understood3. We report that phosphatidylserine receptor HAVCR16,7 and cholesterol transporter NPC18 participate in cargo delivery from exosomes of hepatitis A virus (HAV)-infected cells (exo-HAV) by clathrin-mediated endocytosis. Using CRISPR/Cas9 knockouts we show that these two lipid receptors, which interact in the late endosome9, are necessary for membrane fusion and delivery of RNA from exo-HAVs into the cytoplasm. The HAVCR1/NPC1 pathway, which Ebola virus exploits to infect cells9, mediates HAV infection by exo-HAV indicating that viral infection by this exosome mimicry mechanism does not require an envelope glycoprotein. The luminal viral RNA but not endosomal uncoating of HAV particles (vpHAV) contained in the exosome is mainly responsible for exo-HAV infectivity as assessed by methylene blue-inactivation of non-encapsidated RNA. In contrast, infectivity of vpHAV is pH-independent and requires HAVCR1 or other yet unidentified receptor(s) but not NPC1. Our findings show that envelope glycoprotein-independent fusion mechanisms are shared by exosomes and viruses, and call for a reassessment of the role of envelope glycoproteins in infection.
Extracellular vesicles (EVs) are heterogeneous cargo-containing vesicles secreted by cells that mediate intercellular communications. EVs include microvesicles, which are approximately 50–1,000 nm in diameter and bud from the plasma membrane, and exosomes, which are approximately 50–150 nm in diameter and produced in endosomal compartments10. Virus-infected cells secrete exosomes containing viral proteins, virus particles, nucleoproteins, and capsid-free genomes that mediate virus spread and pathogenesis while evading immune recognition3. HAV, a non-enveloped positive-sense RNA Picornaviridae that causes acute hepatitis in humans11, presents a unique model to study cargo delivery because establishes persistent infections in cell culture that produce significant amounts of easy-purifiable exosomes containing viral RNA and viral particles in the exosome lumen12, which could be used as markers of cargo delivery. Exosomes from HAV-infected cells have been extensively characterized13, and we have used similar reagents and conditions to produce exosomes in this work. Feng et al.11 termed the exosomes and viral particles purified from HAV-infected cells as enveloped HAV (eHAV) and naked HAV (nHAV), respectively. However, we find this nomenclature misleading and use the terms “exosomes from HAV-infected cells (exo-HAV)” instead of eHAV and “viral particle HAV (vpHAV)” instead of nHAV because HAV is a non-enveloped virus and the exosomes produced in infected cells are bona fide exosomes13, which contain viral particles and genomes as described for a wide variety of other viruses3. After binding to the cell surface, exosomes can trigger cell signaling events, fuse at the cell surface, and/or be internalized via endocytic pathways delivering their cargo into recipient cells via transfer of components such as lipids, membrane-bound proteins, and lumen content including coding and noncoding RNAs4. Binding and uptake of exosomes has been studied extensively but the mechanisms involved in the delivery of lumen cargo into the cytoplasm remain poorly understood. Phagocytic cells uptake exosomes by phagocytosis in a process that is independent of HAVCR1 but requires TIM4, a phosphatidylserine (PS) receptor of the same family, resulting in cargo degradation14. Other cell types use alternative pathways such as clathrin-mediated endocytosis (CME) and micropinocytosis to uptake exosomes and deliver their cargo at the late endosome (LE)15 in a process that avoids degradation in endolysosomes. Some viruses like Ebola virus (EBOV) and Lassa virus uncoat their genomes at LE compartments for productive infection16. However, the mechanisms and host proteins involved in exosome cargo delivery and endosomal uncoating of viruses are far from understood. Here, we studied how exosomes deliver functional mRNA from the lumen cargo into the cytoplasm. Viruses that uncoat their genomes in the LE are likely to share a similar mechanism that we termed exosome mimicry.
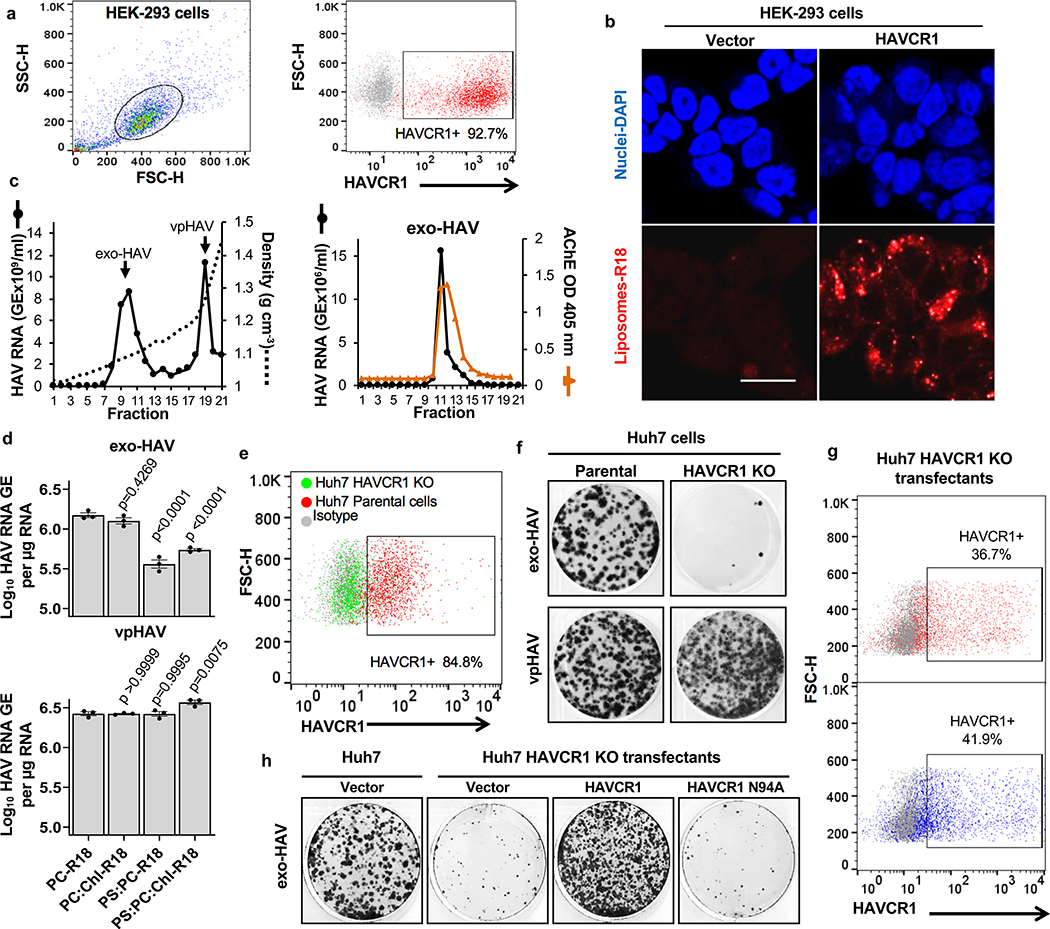
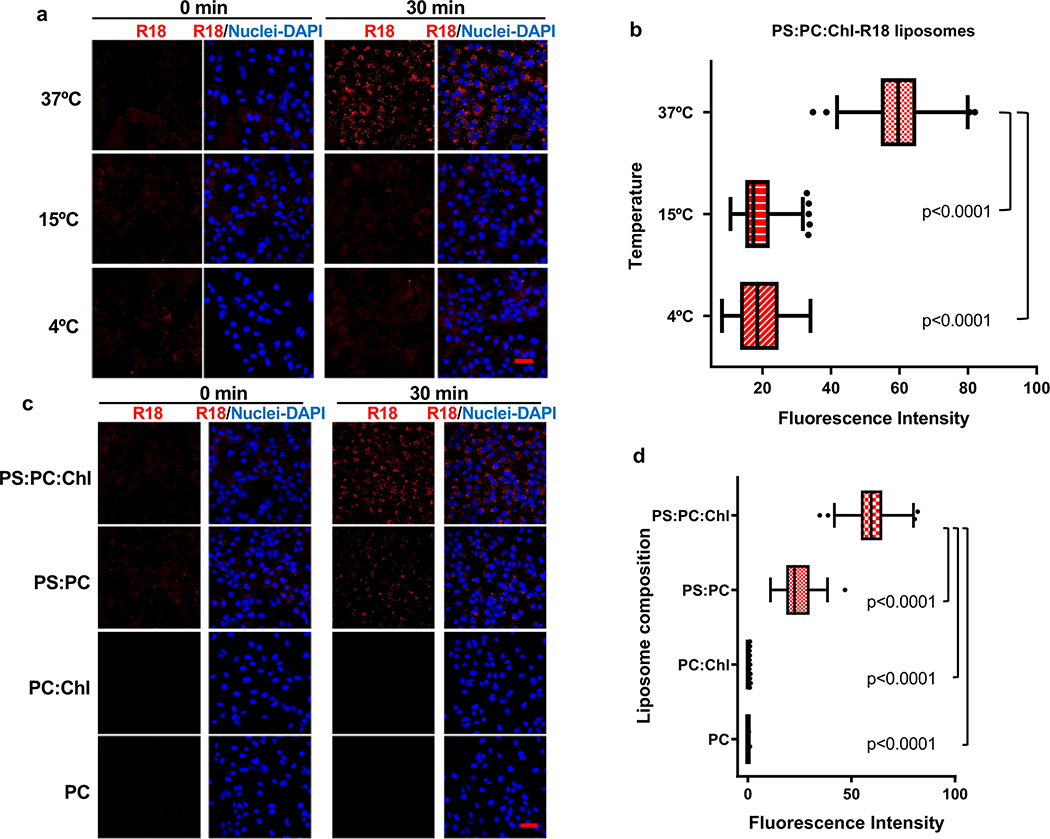
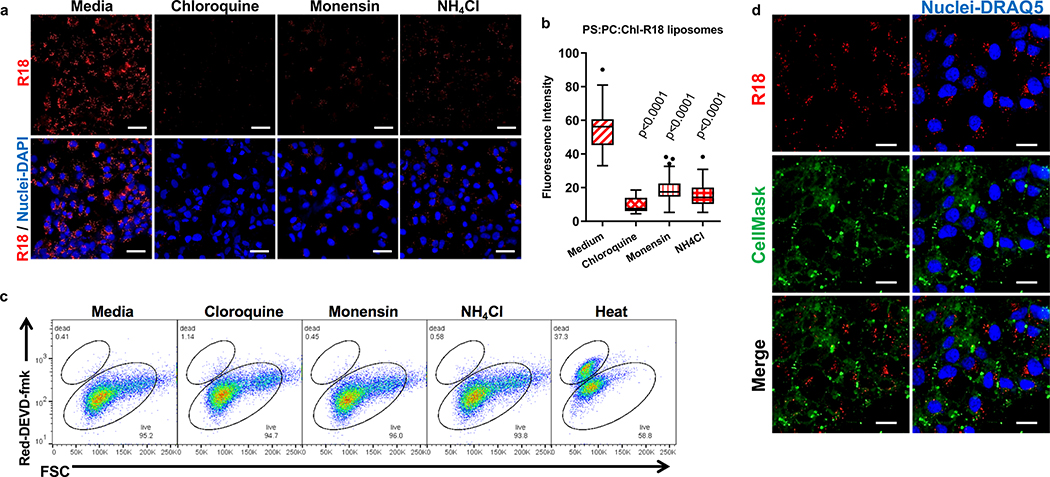
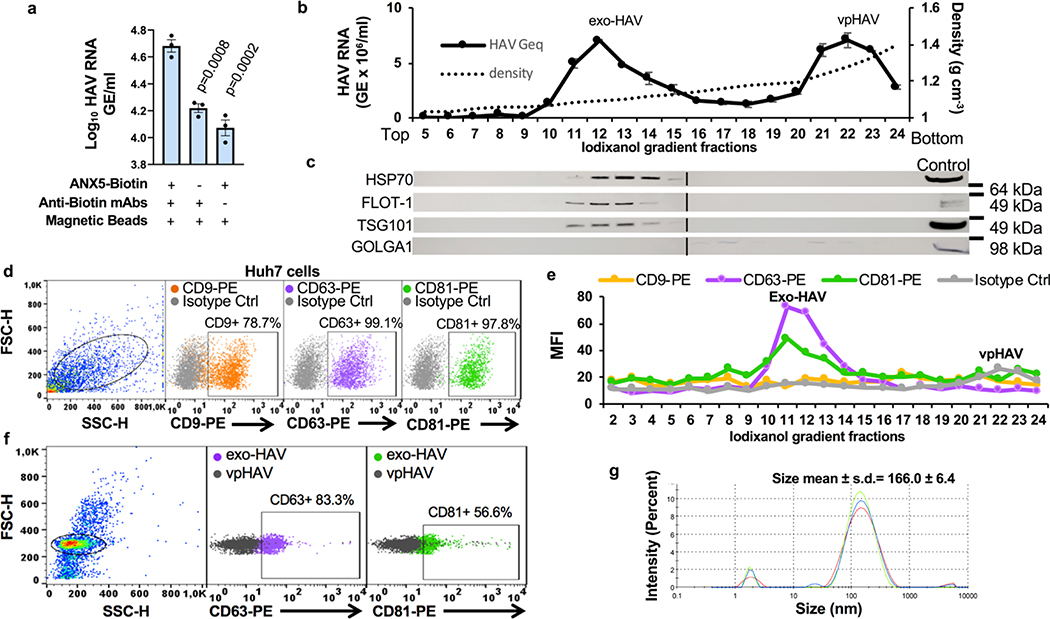
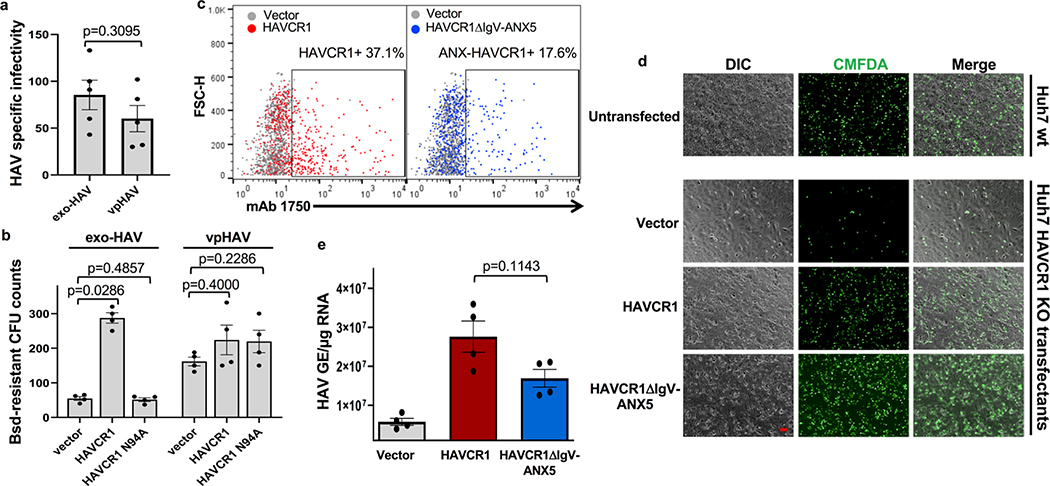
Exosomes lack viral envelope glycoproteins that mediate membrane fusion, therefore, we investigated whether receptors responsible for binding exosomes to the cell surface are also involved in fusion of the exosome and endosomal delimiting membranes. A plethora of receptors including integrins, lectins, PS receptors, and heparan sulfate proteoglycans mediate binding of exosomes to the cell surface3. We focused our attention to HAVCR16,17, a membrane-bound PS receptor that mediates phagocytosis of apoptotic cells7, because PS is enriched on the outer leaflet of the exosome delimiting membranes14,18 and HAVCR1 functions as a virus receptor 6,9,19–21. To study the role of HAVCR1 in fusion, we transfected HAVCR1 cDNA into HEK-293 cells, which resulted in the expression of functional HAVCR1 at the cell surface (Fig. 1a and Supplementary Fig. 1). We analyze fusion using labeled liposomes containing equal amounts of PS, phosphatidylcholine (PC), and cholesterol (Chl) with R18 (PS:PC:Chl-R18 liposomes), an auto-quenched membrane dye that fluoresces upon fusion to cell membranes. The PS:PC:Chl-R18 liposomes rapidly fused with membranes in HAVCR1 transfectants (Fig. 1b) whereas fusion was not detected in vector transfected cells. In human hepatoma Huh7 cells, which naturally express HAVCR1 at the cell surface and are susceptible to HAV infection20, fusion of the PS:PC:Chl-R18 liposomes was temperature dependent occurring mainly at 37°C compared to 4°C and 15°C (Extended Data Fig. 1a,b). Fusion of liposomes was also dependent on i) the presence of Chl and PS on the liposomes (Extended Data Fig. 1c,d), and ii) low intracellular pH since treatment of cells with pH inhibitors cloroquine, monensin, or NH4Cl inhibited fusion (Extended Data Fig. 2a,b,c). Fusion occurred at intracellular membranes and not at the cell surface (Extended Data Fig. 2d) indicating that HAVCR1 forms part of a cellular pathway that mediates endosome fusion, which is in disagreement with previous findings14. To determine whether HAVCR1 can mediate cargo delivery of functional RNAs from the lumen of exosomes to the cytoplasm, we analyzed the mechanism of infection of exo-HAV, which contain PS at the surface (Extended Data Fig. 3a) and HAV free RNA and viral particles in the lumen that serve as markers of cargo delivery. Exo-HAV and vpHAV were purified by isopycnic gradient centrifugation13,20 from supernatants of cells infected with wild type (WT) HAV containing a blasticidin (Bsd) selectable marker (HAV.8Y-Bsd)22 (Fig. 1c, left panel), which confers Bsd-resistance to infected cells. Characterization of exo-HAV according to the recommendations of the International Society for Extracellular Vesicles (ISEV) 23 showed enrichment of i) acetylcholinesterase (Fig. 1c, right panel), ii) chaperone 70-kDa heat shock protein (HSP70), lipid raft marker flotillin-1 (FLOT-1), and biogenesis factor tumor susceptibility gene 101 protein (TSG101) but not golgi-specific golgin subfamily A member 1 (GOLGA1) protein (Extended Data Fig. 3b,c), and iii) tetraspanins CD63 and CD81 but not CD9 at the exo-HAV surface (Extended Data Fig. 3d,e,f), and a particle size of 166 ± 6.4 nm (Extended Data Fig. 3g) that are characteristics of bonafide exosomes10 and further support the notion that exo-HAV are exosomes containing HAV. Exo-HAV and vpHAV showed similar specific infectivity in Huh7 cells (Extended Data Fig. 4a). Infectivity of exo-HAV (Fig. 1d, upper panel) but not vpHAV (lower panel) was blocked by liposomes containing PS:PC:Chl and PS:PC but not PC:Chl and PC indicating that endocytosis of liposomes and exo-HAV share a common pathway. Huh7 HAVCR1 knockout (HAVCR1 KO) cells, which lack HAVCR1 expression (Fig. 1e), were resistant to exo-HAV infection compared to parental Huh7 cells as assessed by the number of Bsd-resistance colony forming units (CFU)20 (Fig. 1f, upper panels). Control vpHAV infected both parental and HAVCR1 KO cells (Fig. 1f, lower panels) indicating that Huh7 cells express an alternative yet unidentified HAV receptor specific for HAV particles but not exosomes. Huh7 HAVCR1 KO cells transfected with HAVCR1 cDNA expressed HAVCR1 at the cell surface (Fig. 1g) and regained susceptibility to exo-HAV (Fig. 1h and Extended Data Fig. 4b). Transfectants expressing HAVCR1 containing an N94A mutation, which abrogates PS-receptor function24, failed to regain exo-HAV cargo delivery function. Taken together, these data further confirmed the role of the HAVCR1 pathway in exosome cargo delivery.
Figure 1.
Exosomes require HAVCR1 for cargo delivery. a, HAVCR1 expression at the cell surface of HEK-293 cells by flow cytometry. Gating strategy (left panel) and HAVCR1 cell surface expression in HAVCR1- (red dots) or vector-transfected (grey dots) cells stained with anti-HAVCR1 mAb 1D12 (right panel). b, HEK-293 transfectants incubated with PS:PC:Chl-R18 liposomes for 45 to 90 min at 37ºC, fixed, and analyzed using a LSM 710 confocal microscope. Micrographs were taken using 40X oil objective showing nuclear DAPI (blue) and R18 endosomal fusion (red) fluorescence. Scale bar represents 20 μm. c, Purification by isopycnic ultracentrifugation of exo-HAV and vpHAV from supernatants of Huh7 cells infected with HAV.8Y-Bsd. HAV RNA genome equivalents (GE) in gradient fractions quantified by RT-qPCR (left panel). Arrows indicate exo-HAV peak (fractions 9–10, density of 1.08–1.10 g cm−3) and vpHAV peak (fraction 19, density of 1.27 g cm−3). Purified exo-HAV was subject to a second iodixanol gradient and fractions tested for acethylcolinesterase (AChE) content (right panel). d, Liposomes containing PS block infectivity of exo-HAV but not vpHAV. Huh7 cells treated with 50 mM PC-R18, PC:Chl-R18, PS:PC-R18, or PS:PC:Chl-R18 liposomes and infected with exo-HAV (upper panel) or vpHAV (lower panel) from (c). HAV RNA quantitated by HAV RT-qPCR at 72 h p.i. Data are mean ± sem, n=3 from 3 independent experiments. P values between PC-R18 and other treatments were determined by one-way ANOVA with Dunnett’s post-test. e, Flow cytometry analysis of HAVCR1 cell surface expression on Huh7 parental (red dots) and HAVCR1 KO cells (green dots) stained with anti-HAVCR1 mAb 1D12 or isotype mAb (grey dots). f, Bsd-resistant CFU assays of Huh7 parental and HAVCR1 KO cells treated with purified exo-HAV or vpHAV from (c). Colonies stained with crystal violet (dark spots). g, Flow cytometry analysis of HAVCR1 cell surface expression on HAVCR1- (red dots), HAVCR1 N94A- (blue dots), or vector-transfected (grey dots) Huh7 HAVCR1 KO cells stained with HAVCR1–1 mAb. h, Bsd-resistant CFU assay of Huh7 parental and HAVCR1 KO cells transfected with HAVCR1, HAVCR1 N194A, or vector and treated with exo-HAV as in (f). Data in all panels are representative of 3 independent experiments.
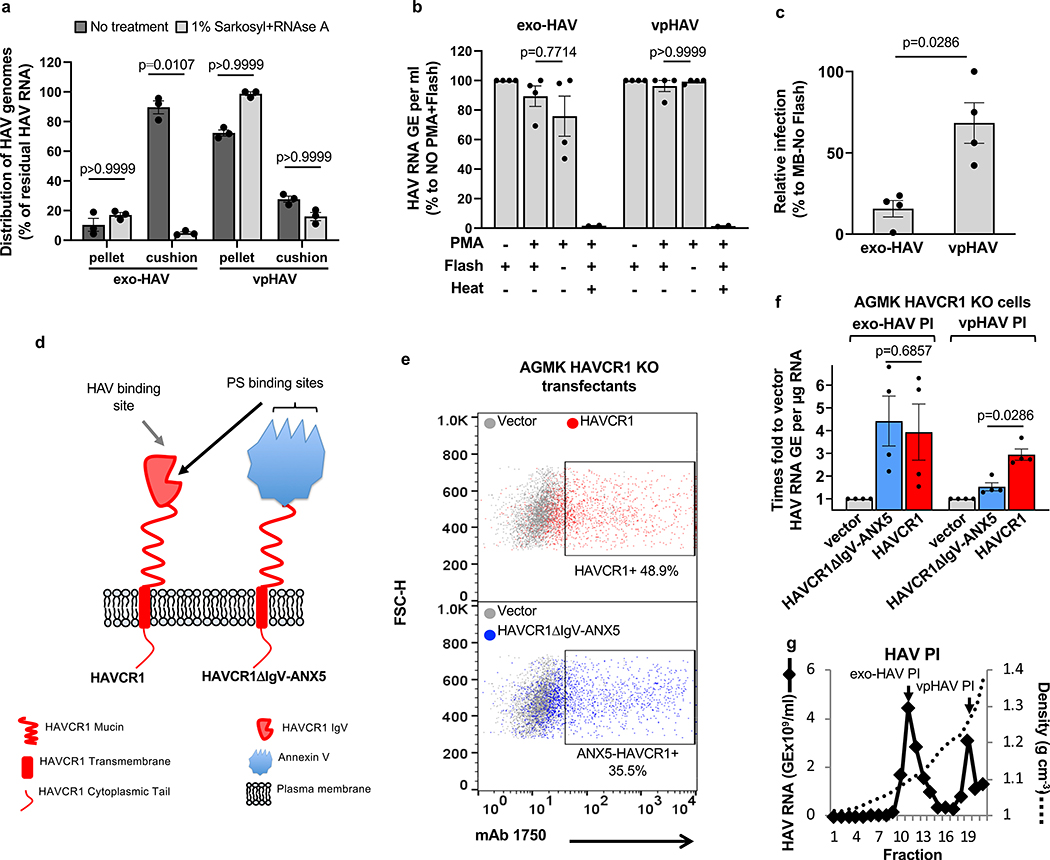
We hypothesized that exo-HAV-mediated infection could be due to two different mechanisms: i) degradation of the exo-HAV delimiting membrane by lipases and endosomal uncoating of vpHAV by intracellular receptors, or ii) fusion of the exo-HAV and endosome limiting membranes and delivery of the capsid-free HAV genomes into the cytoplasm. To test our hypothesis, we analyze the role of the capsid-free HAV genome cargo in the exo-HAV infection. Treatment of exo-HAV with detergent and RNase before sedimentation through a 40% sucrose cushion revealed that the exo-HAV cargo contained approximately 90% capsid-free HAV RNA (RNase-sensitive), which migrated into the cushion, and 10% vpHAV (RNase-resistant), which was pelleted through the sucrose cushion (Fig. 2a). The free RNA was protected within the exosome lumen since treatment with propidium monoazide (PMA), an impermeant nucleic acid-intercalating photoreactive dye that blocks RT-qPCR25, did not affect nucleic acid amplification except after a heat shock (Fig. 2b). Treatment with methylene blue (MB), a photoreactive dye that penetrates membranes but not viral capsids26, significantly reduced the infectivity of exo-HAV but not vpHAV (Fig. 2c). Huh7 HAVCR1 KO transfectants expressing the chimeric receptor HAVCR1ΔIgV/ANX5 (Fig. 2d and Extended Data Fig. 4c) in which the IgV domain of HAVCR1 that interacts with PS and vpHAV24 was replaced with annexin V, a PS- but not a vpHAV-receptor, bound apoptotic cells (Extended Data Fig. 4d) and regained susceptibility to exo-HAV infection (Extended Data Fig. 4e). Furthermore, AGMK HAVCR1 KO cells, which are resistant to both exo-HAV and vpHAV infection20, transfected with HAVCR1ΔIgV/ANX5 cDNA expressed this chimeric receptor at the cell surface (Fig. 2e) and regained susceptibility to exo-HAV but not vpHAV (Fig. 2f) purified from supernatants of AGMK cells infected with HAV PI (Fig. 2g). Taken together, these data showed that the HAVCR1 pathway mediates cargo delivery of free RNA from the lumen of exosomes into the cytoplasm, and that endosomal uncoating of vpHAV is not required for exo-HAV-mediated infection.
Figure 2.
Cargo delivery of free HAV RNA from the lumen of exo-HAV RNA into the cytoplasma. a, Analysis of the HAV free RNA and vpHAV content in the lumen of exo-HAV produced in Huh7 cells infected with HAV-8Y-Bsd. Samples treated or not with detergent and RNAse A were ultracentrifuged through a 40% sucrose cushion, and HAV RNA in cushion and pellet was quantified by RT-qPCR. Data are mean ± sem, n=3 from 3 independent experiments with technical duplicates. P values between untreated and treated samples were determined by unpaired, non-parametric, one-way ANOVA and Dunn’s post-test. b, PMA photoinactivation of exo-HAV and vpHAV produced in Huh7 cells infected with HAV.8Y-Bsd quantified by HAV RT-qPCR. Heat treated exo-HAV and vpHAV as HAV RNA inactivation controls. Data are mean ± sem, n=4 from 2 independent experiments. c, Methylene blue (MB) photoinactivation of exo-HAV and vpHAV produced in Huh7 cells infected with HAV.8Y-Bsd assessed by residual HAV infectivity determined by ARTA. Data are mean ± sem, n=4 from 4 independent experiments. d, Schematic representation of HAVCR1 (red) and chimeric HAVCR1ΔIgV/ANX5 (red and blue), in which the IgV domain of HAVCR1 was replaced by human annexin V (blue). e, Expression of HAVCR1 (upper panel, red dots) or HAVCR1ΔIgV/ANX5 (bottom panel, blue dots) at the cell surface of AGMK HAVCR1 KO cell transfectants by flow cytometry using anti-HAVCR1 mucin 1750 mAb. Data are representative of 3 independent experiments. f, AGMK HAVCR1 KO cells, which lack exo-HAV and vpHAV receptors, transfected with HAVCR1, HAVCR1ΔIgV/ANX5, or vector were treated with exo-HAV or vpHAV from (g) and HAV replication was determined by RT-qPCR. Data are mean ± sem, n=4 from 2 independent experiments. P values between exo-HAV and vpHAV, or between HAVCR1- and HAVCR1ΔIgV/ANX5-transfected cells were analyzed by unpaired, two-sided Mann-Whitney test. g, Exosomes (exo-HAV PI) and viral particles (vpHAV PI) purified from supernatants of AGMK cells infected with HAV PI by isopycnic ultracentrifugation as in Fig. 1c. Data are representative of 3 independent experiments.
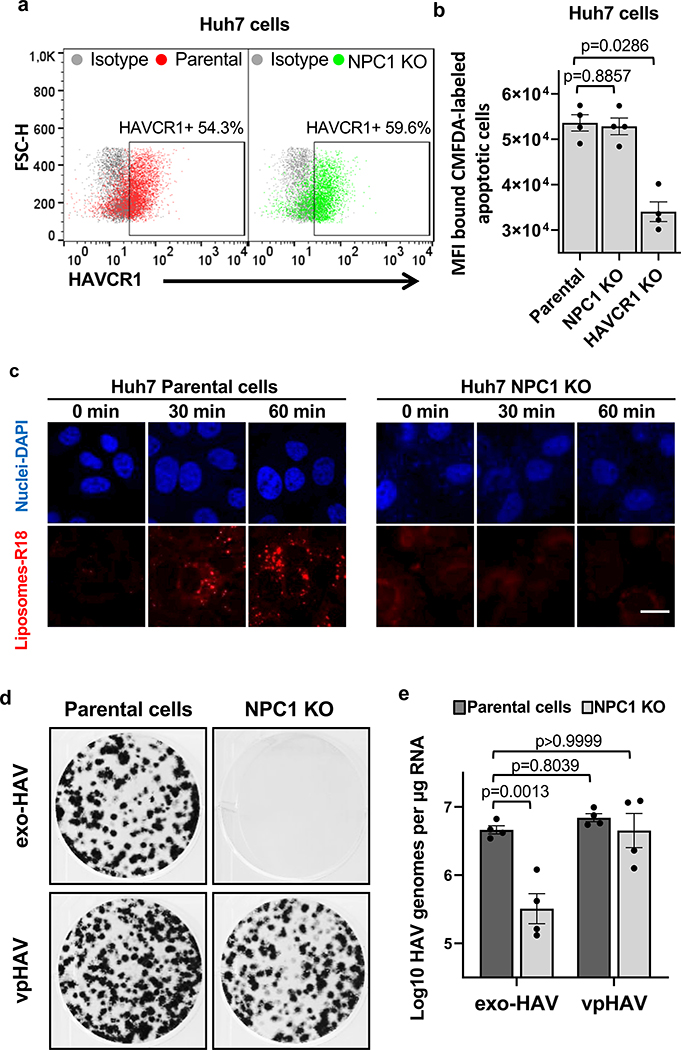
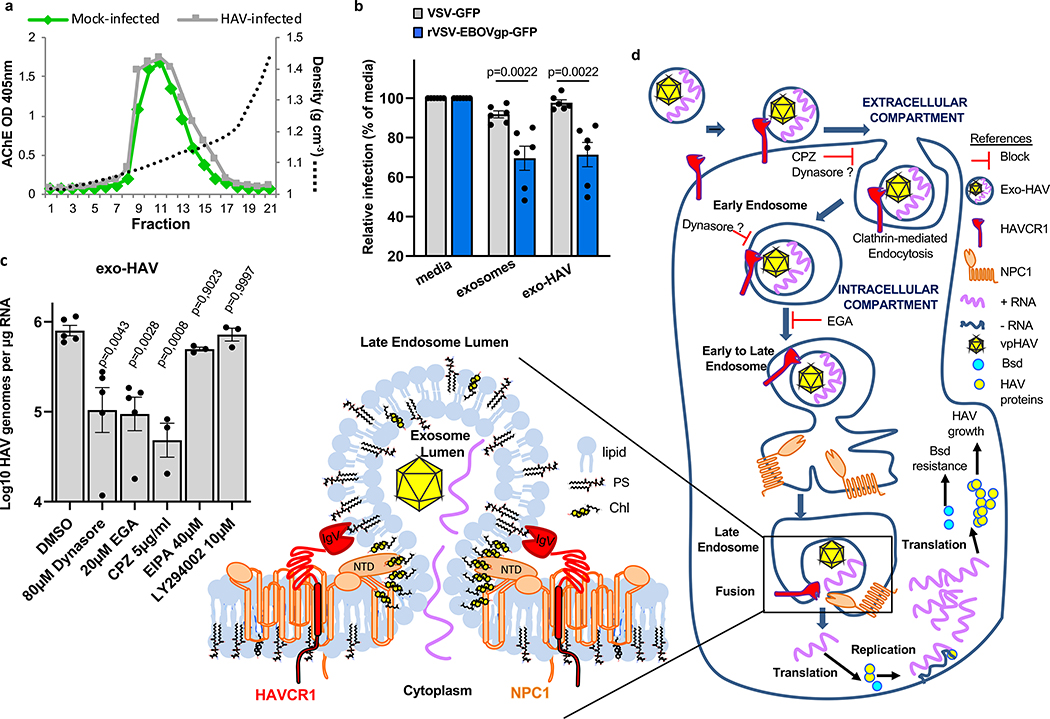
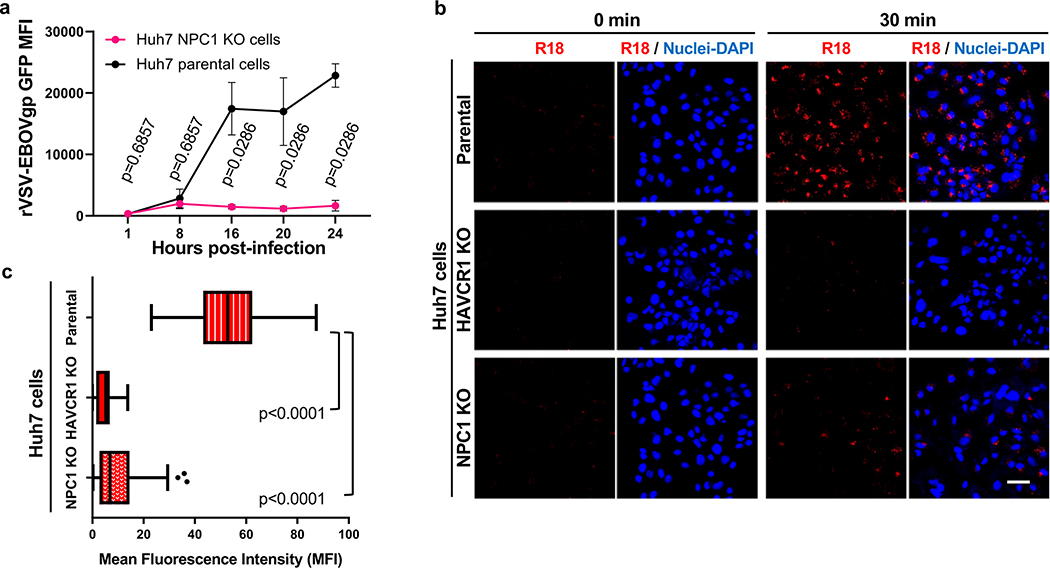
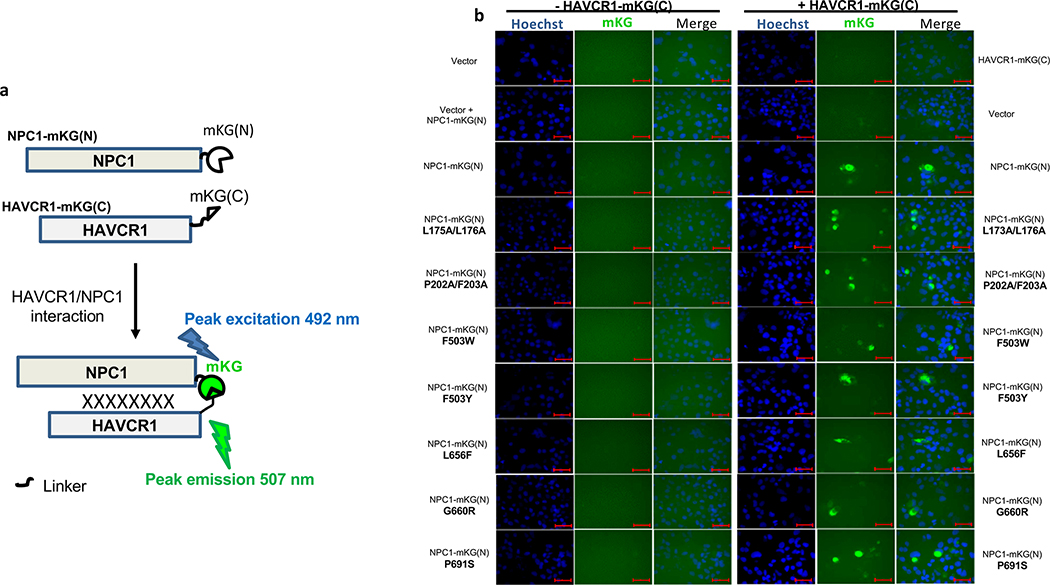
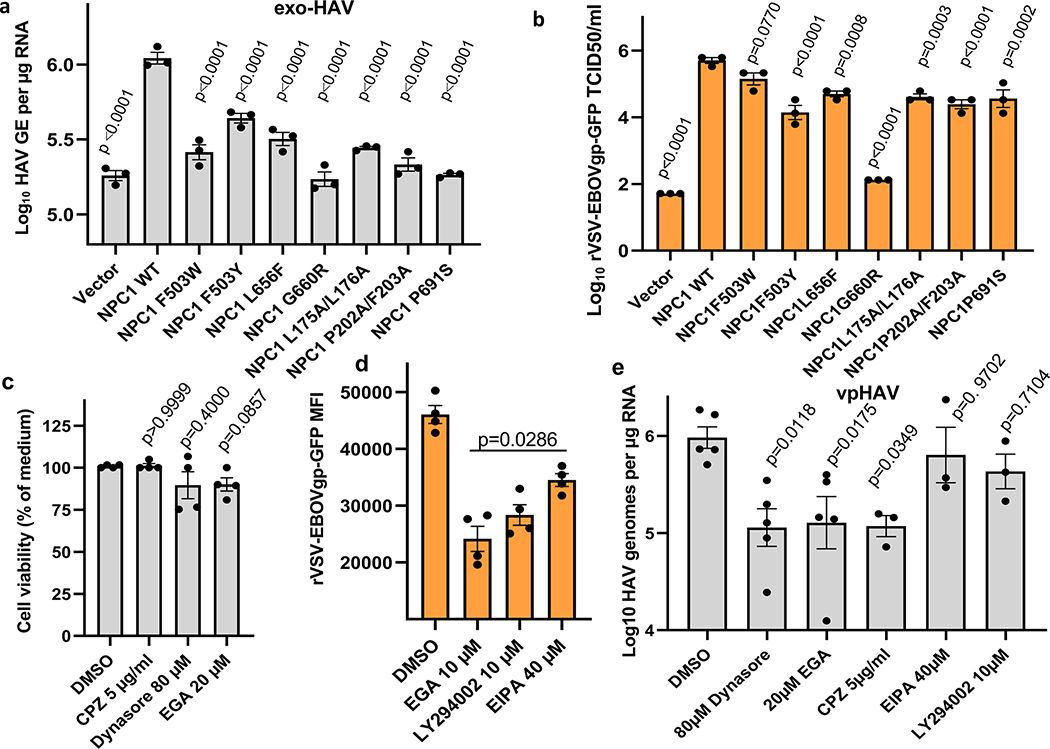
HAVCR1 co-localizes in the LE with NPC19, a cholesterol transporter enriched in the LE27, suggesting that their interaction is important for exosome cargo delivery. To evaluate the role of NPC1 in exosome cargo delivery, we used NPC1 KO cells that are resistant to Ebola virus (EBOV) infection (Extended Data Fig. 5a) but not to other enveloped viruses that also require endosomal acidification for infection28,29. Huh7 NPC1 KO cells expressed HAVCR1 at the cell surface (Fig. 3a) and bound apoptotic cells (Fig. 3b) indicating that the HAVCR1 PS-receptor function was not affected by the NPC1 KO. PS:PC:Chl-R18 liposomes fused at the endosomes of Huh7 parental but not NPC1 KO cells (Fig. 3c and Extended Data Fig. 5b,c) indicating that the NPC1 KO prevented endosomal fusion. Consistent with this finding, Huh7 NPC1 KO cells did not support exo-HAV-mediated infection but were permissive for vpHAV infection (Fig. 3d,e) showing that NPC1 was required for exosome RNA cargo delivery but not HAV replication. Because EBOV requires NPC128,29 to infect cells, we analyze whether exo-HAV and exosomes from uninfected cells (Fig. 4a) shared their cell entry pathway with EBOV. Exo-HAV and exosomes from uninfected cells blocked cell entry of VSV pseudotyped with EBOV glycoprotein (GP) but not VSV infection (Fig. 4b), which does not require NPC1, suggesting that exosomes, exo-HAV, and EBOV share a common cell entry mechanism that requires NPC1. A bimolecular fluorescence complementation (BiFC) assay of the HAVCR1-NPC1 interaction9 based on monomeric Kusabira green reporter fluorescence protein (Extended Data Fig. 6) revealed that double mutations in the Chl-binding pocket at the N-terminal domain (L175A/L176A and P202A/F203A) that prevented NPC1 function30, a mutation in the C domain (F503Y) that reduced EBOV infectivity by 2 logs and a mutation in the same residue (F503W) that enhances EBOV GP binding31, mutations in transmembrane domains (G660R and P691S) that affected NPC1 function30,32,33, and a mutation in the sterol-sensing domain (L656F) that increased Chl binding33 did not affect the expression of NPC1 and its interaction with HAVCR1 at the LE9. However, all these NPC1 mutations affected infection of exo-HAV (Extended Data Fig. 7a) and cell entry of EBOV, except for F503W that did not affect the latter (Extended Data Fig. 7b). Taken together, these results indicate that EBOV and exo-HAV use similar cell entry pathways to infect cells.
Figure 3.
NPC1 is required for exosome cargo delivery. a, Cell surface expression of HAVCR1 in Huh7 parental (left panel, red dots) and NPC1 KO (right panel, green dots) cells stained with anti-HAVCR1 1D12 mAb and analyzed by flow cytometry. Data are representative of 3 independent experiments. b, Fluorescence of CMFDA-labeled Jurkat apoptotic cells bound to the cell surface of Huh7 NPC1 KO cells. Data are mean MFI ± sem, n=4 from 4 independent experiments. c, NPC1 is required for membrane fusion at endosomal compartments. Huh7 parental and NPC1 KO cells were incubated with R18-labeled liposomes for 0, 30, and 60 min at 37ºC and fusion (red) was observed by confocal microscopy. Cell nuclei counterstained with DAPI (blue). Data are representative of 3 independent experiments. Scale bar represents 20 μm. d,e, Huh7 parental and NPC1 KO cells treated for 24 h with exo-HAV or vpHAV produced in Huh7 cells infected with HAV.8Y-Bsd. d, Bsd-resistant CFU assay stained 10 days post-treatment with crystal violet (dark spots are Bsd-resistant colonies). Data are representative of 4 independent experiments. e, HAV RT-qPCR of total RNA extracted 48 h post-treatment. Data are mean ± sem, n=4 from 4 independent experiments. P values respect to parental exo-HAV were determined by one-way ANOVA test.
Figure 4.
Exosome cell entry by clathrin-mediated endocytosis (CME) and cargo delivery via the HAVCR1/NPC1 pathway. a, Exosomes purified from cell culture supernatants of mock- or HAV-infected cells by isopycnic ultracentrifugation and peaks identified by acetylcholinesterase assay. Data are representative of 3 independent experiments. b, Blocking of EBOV (VSV-EBOVgp-GFP) and VSV (VSV-GFP) cell entry by treatment with purified exosomes from uninfected cells or exo-HAV from (a). GFP fluorescence determined at 16–20 h post-treatment compared to media-treated cells. Data are mean ± sem, n=6 from 3 independent experiments with 2 biological replicates. P values were determined by unpaired, two-sided Mann-Whitney test. c, Block of exo-HAV cargo delivery by cell entry inhibitors. HAV RNA genomes were quantitated by RT-qPCR at 72h post-treatment. Data are mean ± sem, from left to right n=5, 5, 5, 3, 3, and 3. P values between DMSO and inhibitors were determined by unpaired, one-way ANOVA with Dunnett’s post-test. d, Model of exosome cargo delivery. i) Exosomes bound via PS to the IgV domain of HAVCR1 at the cell surface, are endocytosed by CME. ii) exosomes are translocated to the late endosome (LE) rich in NPC127 where HAVCR1 and NPC1 co-localize9. iii) The N-terminal domain (NTD) of NPC1 binds to Chl30 at the outer leaflet of the exosome membrane. The exosome membrane and the LE delimiting membrane are brought to close proximity by bridges formed by HAVCR1 and NPC1, which are anchored to the LE delimiting membrane by their transmembrane domains and to the outer leaflet of the exosome by the IgV HAVCR1 bound to PS and the NTD NPC1 bound to Chl, resulting in hemifusion. iv) the enrichment of Chl at the hemifusion surface affects the membrane curvature and fluidity inducing complete membrane fusion44, and v) the RNA contained in the lumen of the exosomes passes through the fusion pore into the cytoplasm, and in the case of exo-HAV is translated and transcribed initiating the HAV infection.
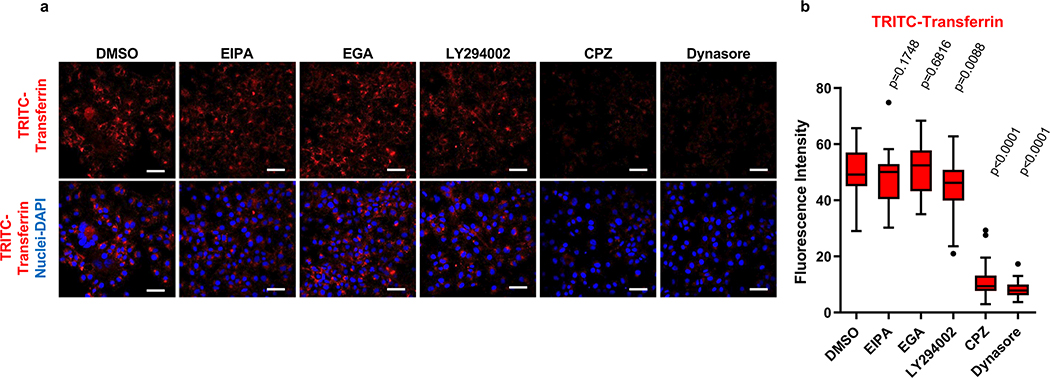
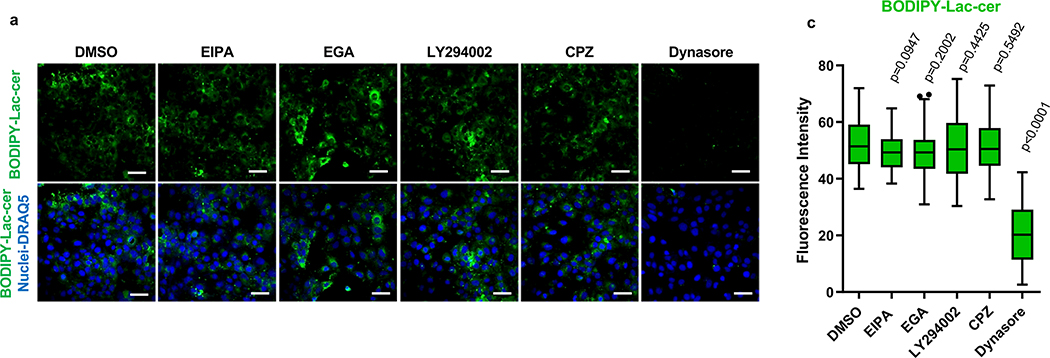
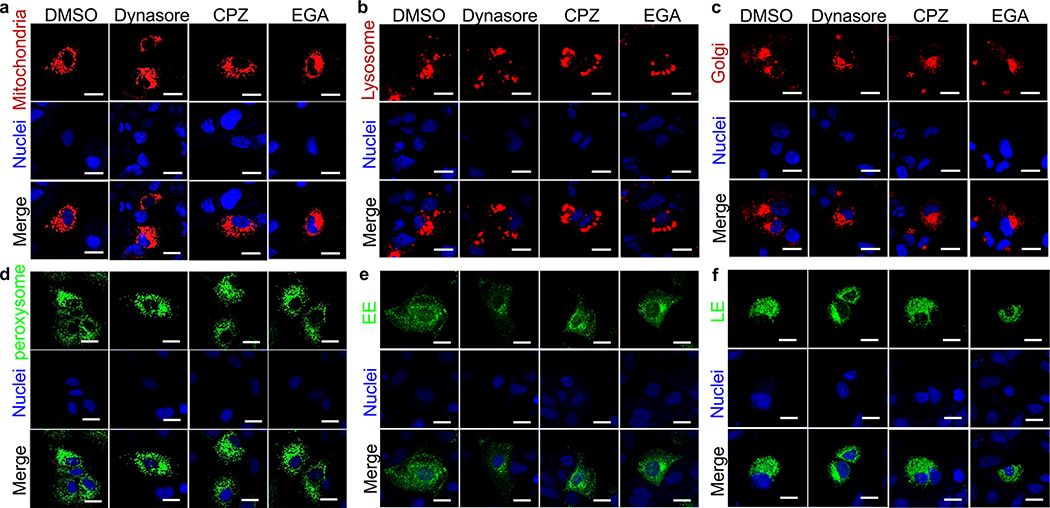
Liposome fusion (Extended Data Fig. 2a,b,c) and cell entry of exo-HAV but not vpHAV12 are pH-dependent endocytic mechanisms. To further understand the exo-HAV cargo-delivery pathway, we used inhibitors of endocytosis34 to block exo-HAV infectivity (Fig. 4c). Treatment with chlorpromazine (CPZ), which specifically blocks CME, or dynasore, which blocks both CME and caveolin-mediated endocytosis and affects the early endosome (EE) compartment35, blocked entry of exo-HAV. However, treatment with 5-(N-ethyl-N-isopropyl) amiloride (EIPA) or LY294002, both of which block micropinocytosis and phagocytosis, did not prevent cell entry of exo-HAV. Interestingly, the EE to LE transport blocker EGA36 also prevented exo-HAV cargo delivery. It should be pointed out that treatment with CPZ, dynasore, and EGA did not affect cell viability (Extended Data Fig. 7c), and that EIPA and LY924002 were active as assessed by their inhibition of EBOV cell entry (Extended Data Fig. 7d). Infection of exo-HAV and vpHAV share the same endocytic pathway (Extended Data Fig. 7e) with the main difference that vpHAV infection is pH independent12. As expected, endocytosis of transferrin, a marker of CME, was inhibited by CPZ and dynasore (Extended Data Fig. 8), whereas lactoceramide, a marker of caveolin-mediated endocytosis, was blocked by dynasore (Extended Data Fig. 9). The endocytosis inhibitors did not affect the localization of specific markers of mitochondria (E1 alpha pyruvate dehydrogenase), lysosomes (Lamp1), Golgi (human Golgi-resident enzyme N-acetylgalactosaminyltransferase 2), peroxisomes (peroxisomal C-terminal targeting sequence), EE (Rab5a), and LE (Rab7a) except for dynasore that, as expected, affected accumulation of Rab5a in EE (Extended Data Fig. 10). Taken together, these data showed that exo-HAV entered the cell by CME and that infectivity was mediated by RNA cargo delivery into the cytoplasm (Fig. 2) via a fusion mechanism (Fig. 3) at the LE or a later compartment, evading degradation of the exo-HAV membrane and the capsid-free HAV RNA in the lumen by lysosomal lipases and nucleases.
The role of exo-HAV in the life cycle of HAV is unclear. HAV is not unique in producing infectious exosomes, which are also produced by enveloped viruses such as HCV and other non-enveloped picornaviruses such as poliovirus and coxsackievirus, which have very different pathogenic processes. Exosomes from virus-infected cells can induce innate immune responses in non-permissive bystander cells that express HAVCR1 such as the activation of plasmacytoid dendritic cells (pDCs) by exosomes from cells infected with HCV 37 or Epstein-Barr virus (EBV) containing a cargo of immunostimulatory small RNAs38. Although pDCs preferentially sense exo-HAV (eHAV) and produce INF-alpha39, the role of type I IFN in HAV infection is unclear since patients with different degrees of disease severity show significant difference in IFN-level responses, and infection of chimps with HAV result in a limited IFN response 40. Clearly, exo-HAV has no role in the fecal-oral route of transmission of HAV because the excreted virus is depleted from lipid membranes in the digestive tract. Therefore, further research is needed to understand the role of exo-HAV in pathogenesis of HAV.
Here, we used a CRISPR/Cas9 gene knockout technology and exosomes from HAV-infected cells to study RNA cargo delivery from the exosome lumen into the cytoplasm, a mechanism mediated by cellular receptors that is poorly understood, paramount to our understanding of the biology of exosomes, and shared by viruses to infect cells. We determined that two lipid receptors, HAVCR1 and NPC1, are required for the delivery of RNA from the lumen of exosomes into the cytoplasm by a fusion mechanism that is independent of viral envelope glycoproteins. EBOV and other filoviruses also use the HAVCR1/NPC1 pathway for the delivery of genetic material into the cytoplasm but in the context of the viral glycoprotein (GP) interaction with NPC1, an endosomal receptor thought to trigger GP conformational changes responsible for the fusion of the viral and LE membranes. However, the endosomal uncoating mechanism of EBOV mediated by the interaction of GP with NPC1 is far from clear16. Our results indicated that fusion of the delimiting membranes of the exosome and LE that results in cargo delivery of RNA and exo-HAV infection is mediated by HAVCR1 and NPC1 in the absence of a viral glycoprotein, which suggested that the GP-NPC1 interaction is not necessary for fusion but may be required for other functions such as stabilizing or enlarging pores for the transfer of a large ribonucleoprotein from the lumen of the viral particle into the cytoplasm leading to productive infection16. Indeed, this exosome mimicry pathway of cargo delivery using multiple lipid receptors may represent a common mechanism in cell entry of other viruses that are difficult to neutralize such as HCV, which uses NPC1-like 1 cholesterol transporter and HAVCR1 as cell entry factors21,41.
We recently showed that the knockout of HAVCR1 in AGMK cells prevented vpHAV and exo-HAV infection, and that transfection of HAVCR1 or its mouse ortholog into AGMK HAVCR1 KO cells restored their infectivity20. Our positive data confirmed that HAVCR1 is a functional HAV receptor in AGMK cells. This differs from the findings of the Lemon and Maury labs42, which showed that truncation of HAVCR1 in Huh7 and Vero E6 cells does not interfere with HAV entry. However, truncation of viral receptors is often not sufficient to abrogate their function43 and both Huh7 and Vero E6 are known to express additional yet unidentified receptors for vpHAV20. Here, we show that Huh-7 cells require HAVCR1 for exo-HAV but not vpHAV infection, which provides further evidence that the HAVCR1/NPC1 pathway is used by exosomes to deliver their luminal cargo into the cytoplasm of the cell.
Our data support an exosome cargo delivery model mediated by two lipid receptors, HAVCR1 and NPC1(Fig. 4d). It is possible that this fusion mechanism could act independently or in conjunction with the SNARE- and viral glycoprotein-mediated mechanisms used by cells and viruses for membrane fusion44 and cargo delivery. Further research will be required to identify other factors involved in the HAVCR1/NPC1 pathway to allow a complete understanding of the exosome cargo delivery mechanism and define therapeutic targets to prevent pathogenesis.
METHODS
Cells and virus
Human hepatoma Huh7 cells (cat# JCRB0403, Japanese Collection of Research Bioresources) selected for the stable growth of wild-type HAV45 were maintained in Dulbecco’s modified Eagle’s (DMEM, Gibco). The clone GL37 of African green monkey kidney (AGMK) cells46 was provided by Dr. Y. Moritsugu (NIH Japan), authenticated by nucleotide sequencing of the monkey HAVCR1 specific allele20 and expression of 190–4 epitope21, and maintained in Eagle’s minimal essential medium (EMEM, Gibco). Human embryonic kidney cells HEK-293 cells (Cat# 11631017, ThermoFisher Scientific) and Vero E6 cells (ATCC, CRL-1586) were mantained DMEM. Media was supplemented with 10% fetal bovine serum (FBS), 2mM L-glutamine, and 1% penicillin-streptomycin. The commercially available Vero E6 and HEK-293 cells were maintained at low passage from the original stock without further authentication. HEK-293 cells were mycoplasma-free as assess by DAPI staining and fluorescence microscope analysis 47. Huh7 cells were authenticated by short tandem repeat (STR) DNA profiling (Genetica Cell Line Testing, LabCorp). Huh7 and AGMK cells tested negative for the presence of HAV, HBV, HCV, HIV1, HIV2, HTLV 1, HTLV 2, and Mycoplasma sp. (h-Impact Profile II test, IDEXX BioResearch). AGMK HAVCR1 KO20, Huh7 HAVCR1 KO, and Huh7 NPC1 KO cells (manuscript in preparation) were developed by Applied StemCell, Inc. using the CRISP/Cas9 gene editing system. Briefly, the selected guide RNAs were cloned into a gRNA/Cas9–2A-Puromycin co-expressing vector and transfected into the cells. The puromycin resistant colonies were further expanded for genotyping. Cas9-mediated-RFLP assay, PCR, and sequencing were used to identify the knockout clones. Huh7 HAVCR1 KO and AGMK HAVCR1 KO cells were authenticated by the lack of expression of HAVCR1 assessed by flow cytometry analysis. Huh7 NPC1 KO cells were authenticated by the resistance to VSV-EBOVgp-GFP but not VSV-GFP infection. These knockout cell lines were further authenticated by nucleotide sequence analysis of the HAVCR1 and NPC1 genes, and were mycoplasma-free as assessed by DAPI staining and fluorescence microscope analysis 47.
The cell culture-adapted HAV derived from infectious cDNA as previously described48 was passaged 100 times in cell culture (HAV PI) and grown in AGMK cells. The recombinant cell culture-adapted HAV containing a blasticidin antibiotic (Bsd) selectable marker22, HAV/Bsd, was derived from infectious cDNA as described and grown in AGMK cells in the presence of 5 μg/ml Bsd (ThermoFisher Scientific). The recombinant wild type HAV containing a Bsd selectable marker45, HAV.8Y-Bsd, was also derived from infectious cDNA as described and grown in Huh7 cells in the presence of 5 μg/ml Bsd. Infectious viral titers were determined by antibiotic resistance titration assay (ARTA)49.
Vesicular Stomatitis Virus (VSV) pseudotypes containing the green fluorescence protein (GFP) gene and the VSV-G envelope (VSV-GFP)50 or the Zaire Ebola virus glycoprotein (VSV-EBOVgp-GFP)51 were produced in Vero E6 cells. Viral titers were determined using the online ID50 program (https://www.ncbi.nlm.nih.gov/CBBresearch/Spouge/html_ncbi/html/id50/id50.cgi) developed by J.L. Spouge52.
Flow cytometry analysis
Mouse anti-human HAVCR1 (CD365) 1D12 mAb (Clone 1D12, BioLegend, Inc.) reacts against an IgV domain epitope conserved between human and monkeys20. HAVCR1–1 mAb was raised against a HAVCR1-Fc fusion protein, reacted against the IgV domain of HAVCR1, protected cells against HAV infection, and blocked the binding of apoptotic cells. Biotin-labeled goat anti-human HAVCR1 1750 Ab (R&D Systems) reacts against the IgV and mucin-like domains of HAVCR1. Parental cells and vector- or HAVCR1-transfected cells were stained with 1D12, HAVCR1–1, or mouse IgG1 isotype control (clone 15H6, Southern Biotech) mAbs as primary antibodies and PE-labeled anti-mouse polyclonal IgG (Southern Biotech) as secondary antibody. Vector-, HAVCR1-, or HAVCR1ΔIgV/ANX5-transfected cells were stained with biotin-conjugated 1750 Ab as a primary antibody and Alexa Fluor 488-labeled Streptavidin (ThermoFisher Scientific). Alternatively, cells were stained with PE-labeled anti-HAVCR1 mAbs 1D12 (Clone 1D12, ThermoFisher Scientific).
To determine tetraspanin expression at the cells surface of Huh7 cells, monolayers of Huh7 cells were dislodged with 0.5 mM EDTA, washed with 5% FBS-PBS, and stained with murine PE-labeled anti-human CD9 (Clone M-L13, BD Biosciences), CD63 (Clone H5C6, BD Biosciences), or CD81 (Clone JS-81, BD Biosciences) mAbs as recommended by the manufacturer. PE-labeled anti-human CD3 (Clone UCHT1, BD Biosciences) was used as isotype control following the manufacturer’s recommendation.
Latex bead-assisted flow cytometry analysis of exosomes was performed as described with minor modifications53. Aliquotes (1/10 of each fraction, 20 μl) of iodixanol density gradient fractions were absorbed to 2μl of 4 μm Aldehyde/Sulfate Latex Beads, 4% w/v (ThermoFisher Scientific), and stained with PE-labeled anti-CD3, -CD9, CD63, or -CD81 mAbs. Median fluorescence intensity (MFI) was determined for each fraction.
Antibody stained cells and exosome-bound latex-beads were analyzed by flow cytometry in a Guava EasyCyte instrument using CytoSoft version 5.3 software (EMD Millipore Corp.). Caspase-3 staining was analyzed by flow cytometry in a FACSCanto II using FACSDiva version 8.0.1 software (BD Biosciences). Data analysis was performed using the FlowJo software (Becton, Dickinson and Company) version 10 for antibody stains and version 8.5 for caspase-3 stains.
Liposome fusion experiments
For fusion studies in HEK-293 cells (Fig. 1b) and fusion kinetics in Huh7 cells (Fig. 3c), liposomes were prepared by the lipid extrusion method using an Avanti Mini-Extruder kit (AvantiPolar Lipids, Inc) with a 30 nm membrane and a combination of phosphatidylserine (PS) (AvantiPolar Lipids, Inc), phosphatidylcholine (PC) (AvantiPolar Lipids, Inc), cholesterol (Chl) (AvantiPolar Lipids, Inc), and Octadecyl Rhodamine B Chloride (R18) (ThermoFisher Scientific) at a molar ratio of 12:12:12:1 as recommended by the manufacturers. Quality control of R18-labeled PS:PC:Chl liposomes was performed by in vitro fluorescence dequenching54 in liposomes treated with 1% Triton X-100 for 15 min at room temperature (r.t.) in a 96-well plate. An approximately 5-fold increase in fluorescence was determined in a Synergy HT fluorescence plate reader (BioTek Instruments) using 530 nm/590 nm excitation/emission filters and Gen5 software, which indicated that the R18 bound to the liposomes was adequately quenched. Cells grown in glass coverslips were incubated with R18-labeled liposomes for 10 min at r.t., washed, incubated at 37ºC under 5% CO2 for different times (0, 15, 30, 45, or 60 min), and fixed with 3% paraformaldehyde (PFA). Cell were mounted on slides using ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI, ThermoFisher Scientific) as nuclear counterstain. Slides were analyzed in a LSM 710 Confocal Microscope using ZEN 2011 software (Carl Zeiss). Micrographs were taken with a 40× oil immersion objective.
To assess phospholipid content, temperature, pH dependency, endosome inhibitors, or HAVCR1 and NPC1 requirement for liposome fusion, Huh7 parental or knockout cells were grown in 8-well glass chamber slides (Nunc, Inc.) for 24h at 37ºC and treated with commercially available R18-labeled liposomes (Encapsula NanoSciences LLC). For the phospholipid content dependency studies, 50 μM of R-18 liposomes containing PS, PC, and Chl (PS:PC:Chl-R18), PS and PC (PS:PC-R18), PC and Chl (PC:Chl-R18), or PC alone (PC-R18), were added to the cells, incubated at 15ºC for 10 min, washed with PBS, and incubated for 30 min at 37ºC. For the temperature dependency studies, 50 μM of PS:PC:Chl-R18 were added to the cells, incubated at 15ºC for 10 min, washed, and incubated for 30 min at 37ºC, 15ºC, or 4ºC. For the pH dependency studies, cells were treated for 6h at 37ºC with 2 μM monensin (ThermoFisher Scientific) or overnight at 37ºC with 30 μM Chloroquine diphosphate (ThermoFisher Scientific), or 5 mM Amonium chloride, treated 50 μM PS:PC:Chl-R18 at 15ºC for 10 min, washed, and incubated for 30 min at 37ºC. After incubation, cells were washed thrice with PBS. To determine whether liposome fusion occurs at the cell surface or intracellularly and to quantitate fusion of liposomes in Huh7 HAVCR1 or NPC1 KO cells, cells were treated with 50 μM PS:PC:Chl-R18 for 10 min at 15ºC, washed, incubated or not for 30 min at 37ºC, washed, stained with plasma membrane stain CellMask (ThermoFisher Scientific) for 15 min at 37ºC and washed extensively. For these liposome fusion studies, cell nuclei were counterstained with Deep Red Anthraquinone 5 (DRAQ5, ThermoFisher Scientific) or DAPI, cells were fixed with 4% PFA, coverslips were mounted with ProLong Gold antifade reagent (Life Technologies), and analyzed with a LSM 700 confocal microscope using ZEN 2011 software (Carl Zeiss). Micrographs were taken with a 40× oil objective. R18 fluorescence due to liposome fusion was quantitated using the ImageJ version 1.8.0 software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, https://imagej.nih.gov/ij/1997-2018).
Cytotoxicity of 2 μM Monensin, 30 μM Chloroquine diphosphate, and 5 mM amonium chloride (NH4Cl) in Huh7 cells was determined by flow cytometry using a Caspase-3 apoptosis assay. Briefly, cells were trypsinized after treatment with the compounds or heat treated for 7 min at 55ºC (apoptosis/dead induction control), stained with 1μl of cell-permeable caspase-3 inhibitor DEVD-FMK conjugated to sulfo-rhodamine (Red-DEVD-FMK) that binds irreversibly to activated caspase-3 (CaspGLOW™ Red Active Caspase-3 Staining Kit, BioVision Inc.), incubated at 37°C for 45 min, washed thrice, and analyzed in a FACSCanto II (BD Biosciences) using the FlowJo v8.5 software (Becton, Dickinson and Company).
HAV-specific quantitative RT-PCR assay
RNA from gradient fractions was extracted with the High Pure Virus RNA Kit (Roche Diagnostics). Alternatively, samples from gradient fractions were diluted 1:5 in RNase-free water and heated at 95°C for 5 min before quantitative RT-PCR (RT-qPCR) analysis. Total RNA from cell lysates was isolated with the RNeasy kit (Qiagen). HAV RNA content was determined by RT-qPCR using HAV-specific primers and FAM/MGB probe55 and the RNA UltraSense one-step quantitative RT-PCR kit (ThermoFisher Scientific). Intracellular HAV RNA was normalized to total extracted RNA. Alternatively, samples were spiked with a known amount of poliovirus, which was used to correct HAV values according to the extraction efficiency. The RT-qPCR analysis was performed in a QuantStudio 6 Flex Real-Time PCR System (ThermoFisher Scientific).
Purification and characterization of Exosomes and viral particles from HAV-infected cells
Exosomes from HAV-infected cells have been extensively characterized13, and we have used the same cells, virus strains, and purification procedures to produce exosomes for this study12,13,20. These purified exosomes had the same density, content of acetylcholinesterase (AchE, an exosome marker56), content of HAV RNA, and infectivity characteristics of the published exosomes. Exosomes and viral particles from HAV-infected cell culture supernatants were produced as described12,13,20. Exosomes and HAV particles were purified from HAV/Bsd-infected AGMK cells (exo-HAV/Bsd and vpHAV/Bsd, repectively), HAV.8Y/Bsd-infected Huh7 cells (exo-HAV and vpHAV, respectively), or HAV PI-infected AGMK cells (exo-HAV PI and vpHAV PI, respectively). Briefly, cell supernatants collected at 8 days post-infection (p.i.) were clarified by centrifugation at 1,000 × g and twice at 10,000 × g, and ultracentrifuged at 100,000 × g for 4h at 4°C. Pellets containing exosomes and viral particles were resuspended in PBS, loaded onto 8–40% iodixanol (OptiPrep Density Gradient Medium, Sigma-Aldrich) step gradients and separated by isopycnic ultracentrifugation at 141,000 × g for 18h at 4°C. Approximately 20–24 fractions were collected from each gradient and analyzed by HAV RT-qPCR and AChE content using a colorimetric Acethylcholinesterase Assay Kit (Abcam Plc.). The density of each gradient fraction was determined using a refractometer (Portable Lab, Mettler Toledo), and the peaks of exosomes (density of 1.08–1.10 g cm−3), which contained HAV RNA and AChE, and HAV particles (density of 1.27 g cm−3), which contained HAV RNA but not AChE, were collected and stored at 4°C for further experimentation.
Presence of PS at the surface of exosomes purified from HAV-infected cells was analyzed using the annexin V (ANX5)-based kit EasySep Dead Cell Removal kit (StemCell Technologies), an immunomagnetic separation based on 3 components: biotinylated ANX5 that binds PS, a cocktail of anti-biotin monoclonal antibodies (mAbs) that bind to the biotinylated ANX5, and magnetic beads that bind the mAbs. Purified exo-HAV/Bsd (0.020 ml per sample) in 0.1 ml of ANX5 binding buffer (Biolegend, inc.) was incubated with or without biotinylated ANX5 for 5 min at r.t. in the presence or not of the anti-biotin mAbs. Magnetic beads were added to all samples, incubated for 5 min at r.t., beads were separated using a magnet and washed twice with ANX5 buffer. RNA from exo-HAV/Bsd bound to the magnetic beads was purified using the RNeasy Mini Kit (Qiagen) as recommended by the manufacturer, and quantitated by HAV RT-qPCR.
Tetraspanin enrichment at the surface of exosomes was determined using latex bead-assisted flow cytometry staining with anti-CD9, -CD63, and CD81 mAbs as described above (see Flow Cytometry Analysis section).
Western blot analysis of iodixanol density gradient fractions was performed using 1/5 to 1/10 of each fraction resolved by denaturing SDS-PAGE in Bolt 4–12% Bis-Tris minigels, transferred to PVDF membranes using the iBolt 2 Dry Blotting System, and probed with antibodies using the iBind Flex Western Device as suggested by the manufacturer (ThermoFisher Scientific). Huh7 cell extracts were prepared in RIPA buffer (Sigma-Aldrich) as suggested by the manufacturer. Rabbit anti-human 70-kDa heat shock protein (HSP70) polyclonal antibody EXOAB-Hsp70A-1 (System Bioscience) at 1:1,000 dilution, and murine anti-human Flotillin-1 (FLOT-1) mAb sc-74566 (Santa Cruz Biotechnology, Inc.), tumor susceptibility gene 101 protein (TSG101) mAb clone 4A10 (Abcam), and golgin subfamily A member 1 (GOLGA1) protein mAb clone CDF4 (ThermoFisher Scientific) at 0.5 μg/ml were used as primary antibodies. Affinity purified peroxidase-labeled goat anti-mouse IgG (H+L) human serum adsorbed (KPL SeraCare) at 1:20,000 dilution or purified peroxidase-labebled goat anti-rabbit IgG antibody EXOAB-HRP (System Bioscience) at 1:20,000 dilution were used as secondary antibodies. Western blots were developed by chemiluminescence using the SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific) and visualized using a KwikQuant Imager (Kindle Biosciences, LLC) containing a Digial Camera X-A2 (Fujifilm Corp.).
Dynamic Light Scattering (DLS) measurements of the density gradient peak of exo-HAV were performed with a ZetaSizer nano ZS90 (Malvern Instruments). Samples were diluted 1:10 in PBS. Three runs of 10 measurement each were performed using standard settings (Refractive Index = 1.33, viscosity = 0.88, temperature = 25°C), the size distribution by intensity was plotted for each run, and the exosome size expressed as mean ± standard deviation.
Purification and characterization of Exosome from uninfected cells
Exosomes from uninfected Huh7 cells were produced and purified in iodixanol gradients as described above for the purification of exosomes from HAV-infected cells. Gradient fractions were analyzed for AChE content using the colorimetric kit described above, and the peak of exosomes at a density of 1.08–1.10 g cm−3 was collected and stored at 4°C for further experimentation.
Inhibition of HAV infection with liposomes
Sub-confluent monolayers of Huh7 cells in 12-well plates containing 0.5 ml of serum-free OptiMEM media (ThermoFisher Scientific) per well were treated with 50 μM PS:PC:Chl-R18, PS:PC-R18, PC:Chl-R18, or PC-R18 liposomes for 30 min at r.t., monolayers were washed 3 times, and infected with vpHAV or exo-HAV at a multiplicity of infection (m.o.i.) of 0.1–0.5 in 0.5 ml of DMEM-10% FBS (complete media) for 2 h at 37°C. Monolayers were washed extensively, complete media was added, and incubated 72 h at 37°C. Monolayers were washed, total RNA was purified using the RNeasy Mini kit, and HAV RNA was quantitated by HAV RT-qPCR.
Blasticidin (Bsd)-resistant colony forming units (CFU) assay
The Bsd-resistant CFU assay was performed as described20 with minor modifications. For AGMK cells, cells were infected at 48 h post-transfection (p.t.), virus was absorbed for 12 h, and incubated at 37°C for 72–96 h before splitting. For Huh7 parental cells, Huh7 HAVCR1 KO or Huh7 NPC1 KO cells in 12-well plates were treated with purified exo-HAV or vpHAV for 2 h, washed, incubated at 37ºC under 5% CO2 for 24 h, trypsinized, seeded at different densities (2×105, 1×105, and 5×104 cells/well) in 6-well plates, and grown in cell culture media containing 5 μg/ml Bsd for 10–12 days. Cells were fixed with 80% methanol, stained with crystal violet, and CFU from duplicate wells were counted manually and imaged using a flatbed scanner.
Constructs and Transfections
Plasmids encoding HAVCR1 and the HAVCR1 N94A mutant were described before21. To construct the chimeric receptor HAVCR1ΔIgV/ANX5, the cDNA of the IgV domain coding for amino acids (aa) 1–125 of HAVCR1 (GenBank accession number AF_043724) was replaced with a synthetic cDNA coding for the preprotrypsin signal peptide (MSALLILALVGAAVA) followed by the full-length human annexin V (aa 1–320, GenBank accession number NM_001154), which lacks a peptide signal for secretion57. The cDNA coding for the chimeric receptor was subcloned into the XbaI/NotI restriction sites of expression vector pEAK12 (Edge Biosystems, Inc.) in which the puromycin selectable marker was changed with a hygromycin resistance gene (pEAK12-Hyg).
A synthetic cDNA construct containing the complete coding sequence of human NPC1 (GenBank accession No. NM_000271.4) coding for aa 1–1,278 but lacking the termination codon was fuse at the C-terminus via a serine and glycine-rich linker to the 168 aa N-terminal fragment of the monomeric Kusabira green fluorescent protein (mKG)58, termed NPC1-mKG(N), and cloned into expression vector pEAK12-Hyg. Single aa mutations L173A/L176A, P202A/F203A, F503W, F503Y, L656F, G660R, or P691S, and double aa mutations L173A/L176A and P202A/F203A were introduced into the NPC1 cDNA in pEAK12-Hyg NPC1mKG(N).
A synthetic construct containing the complete coding sequence of HAVCR1 but lacking the termination codon was fused at the C-terminus via a serine and glycine-rich linker to the 51 aa C-terminal fragment of mKG58, termed HAVCR1-mKG(C), and cloned into expression vector pEAK12-Hyg.
All plasmids were purified by column chromatography (Qiagen) as recommended by the manufacturer.
Cells in 12-well plates were transiently transfected with plasmids using the Lipofectamine 3000 Transfection Reagent (ThermoFisher Scientific) according to the manufactureŕs recommendation. Expression vector pEAK12-Hyg was used as an empty vector transfection control. The expression of HAVCR1 and HAVCR1ΔIgV/ANX5 at the cell surface of transfected cells was determined by flow cytometry at 48h post-transfection staining cells with PE-labeled anti-HAVCR1 IgV mAb 1D12. Transfectants were also stained with unlabeled anti-HAVCR1 1D12 and primary antibody and PE-labeled anti-mouse antibody as secondary antibody or biotin-conjugated 1750 Ab as primary antibody followed by Alexa Fluor 488-labeled Streptavidin.
Detergent/RNase-treatment of exosomes
Exo-HAV and control vpHAV were treated with 1% Sarkosyl (Sigma-Aldrich) for 1 h at r.t. followed by treatment with 50 μg/ml of RNAse A (Sigma-Aldrich) for 30 min at 37°C. Samples were separated by ultracentrifugation through a 40% sucrose (Sigma-Aldrich) cushion at 120,000rpm for 15 min at 4°C in a Sorvall MX 120 Plus Micro-Ultracentrifuge (ThermoFisher Scientific). The cushion and pellet were collected, RNA was extracted, and HAV RNA was quantified by RT-qPCR.
Methylene blue (MB) inactivation
Exo-HAV and vpHAV were incubated with 2 μM MB (Sigma-Aldrich, Inc.) for 20 min at r.t. in the dark, and photoinactivated or not (flash negative controls) for 20 min with fluorescent white light at r.t. as described26. Remaining HAV infectivity in Huh7 cells was determined by ARTA49.
Propidium monoazide (PMA) RT-qPCR assay
Exo-HAV and vpHAV were treated with 200 μM PMA (Biotium, Inc.) for 15 min at r.t. in the dark with constant agitation, and exposed or not (flash negative controls) to a 500W light for 15 min on ice. Samples treated at 85°C for 5 min to expose HAV RNA before incubation with PMA were used as positive inactivation controls.
Inhibition of cell entry
Cell entry inhibitors Chlorpromazine hydrochloride (Sigma-Aldrich), Dynasore hydrate (Sigma-Aldrich), LY294002 (Calbiochem), 5-(N-Ethyl-N-isopropyl)amiloride (EIPA, Sigma-Aldrich), or 4-bromobenzaldehyde N-(2,6-dimethylphenyl) semicarbazone (EGA, Calbiochem) were prepared as suggested by the manufacturers. As negative control, cells were treated with dimethyl sulfoxide (DMSO, Sigma- Aldrich) at the same concentration used with the compounds. Huh7 cells seeded in 24-well plates for 48 h were pretreated with different concentrations of cell entry inhibitors for 1h at 37°C. Exo-HAV or vpHAV were added and incubated for 2 h at 37ºC. Monolayers were extensively washed, fed with DMEM supplemented with 10% FBS and the corresponding inhibitor, and incubated at 37°C for 3 days under 5% CO2. Total RNA was extracted from cells and HAV RNA was quantified by RT-qPCR.
Cytotoxicity of the compounds in Huh7 cells was determined by the Dual-fluorescence Acridine Orange/Propidium Iodide assay (Nexcelom Bioscience) as recommended by the manufacturer.
To confirm the activity of the compounds, Huh7 cells were treated with inhibitors as described above, infected with rVSV-EBOVgp-GFP (see below, Infection with VSV pseudotypes), and GFP fluorescence quantitated in a Synergy HT fluorescence plate reader using Gen5 software.
Specificity of the inhibition was tested by endocytosis of human transferrin and lactosylceramide as markers of clathrin-mediated endocytosis and caveolae-mediated endocytosis, respectively (59 and references therein). For tansferrin uptake assays, Huh7 cells grown in 8-well glass chamber slides were treated with pre-warmed transferrin staining medium (25 mM HEPES pH7.4, 0.5% BSA) for 1 h at 37ºC prior to the addition of 5 μg/ml CPZ, 80 μM Dynasore, 10 μM EGA, 40 μM EIPA, 10 μM LY294002, or DMSO as control. After incubation for 15 min at 37ºC in the presence of inhibitors, 25 μg/ml of Tetramethylrhodamine Conjugate-Transferrin (Tritc-Transferrin) (ThermoFisher Scientific) in transferrin staining medium was added to the cells and incubated for 15 min at 37ºC. Cells were washed with cold 0.2 M sodium acetate pH 4.5 and twice with cold PBS, stained with DRAQ5 as nuclear counterstain, fixed with 4% PFA, and mounted with ProLong Gold antifade reagent. For lactosylceramide uptake assays, Huh7 cells grown in 8-well glass chamber slides were incubated with BODIPY FL C5-Lactosylceramide complexed to BSA (LAC-cer) (ThermoFisher Scientific) for 30 min at 4ºC, washed with cold PBS, treated with 5 μg/ml CPZ, 80 μM Dynasore, 10 μM EGA, 40 μM EIPA, 10 μM LY294002, or DMSO as control for 45 min at 37ºC. After incubation, cells were washed with PBS, stained with DRAQ5 as nuclear counterstain, fixed with 4% PFA, and mounted with ProLong Gold antifade reagent. Micrographs were taken with an LSM 700 confocal microscope using a 40× oil objective.
The effect of the inhibitors in different cell compartments was tested using CellLight Bacmam 2.0 Reagents (Thermo Fisher Scientific), which are recombinant Baculoviruses containing cell compartment-specific markers fused to fluorescent proteins under the control of an eukaryote promoter. Infection with the CellLight Bacmam 2.0 reagents result in the expression of the fluorescent fusion proteins while the Baculovirus genes remain silent. CellLight, BacMam 2.0 reagents used in this study were CellLight™ Early Endosomes-GFP Rab5a fused to GFP; Cat. No. C10586), Peroxisome-GFP BacMam 2.0 (peroxisomal C-terminal targeting sequence fused to GFP; Cat. No. C10604), Golgi-RFP, BacMam 2.0 (human Golgi-resident enzyme N-acetylgalactosaminyltransferase 2 fused to RFP; Cat. No. C10593), Mitochondria-RFP, BacMam 2.0 (leader sequence of E1 alpha pyruvate dehydrogenase fused to RFP; Cat. No. C10601), Lysosomes-RFP BacMam 2.0 (Lamp1 fused to RFP; Cat. No. C10597), and Late Endosomes-GFP, BacMam 2.0 (Rab 7a fused to GFP; Cat. No. C10588). Huh7 cells in 8-wells chamber slides were infected with the CellLight Bacman 2.0 reagents at a multiplicity of infection of 30 particles per cell, incubated at 37°C for 10–12 h, treated with 80 μM dynasore hydrate, 5μg/ml Chlorpromazine hydrochloride solution, 10 μM EGA, or a similar volume of DMSO vehicle as negative control, and incubated for additional 12–14 h at 37°C. Cells were washed with PBS, stained with nuclear counterstain DRAQ5, fixed with 4% PFA, and washed. Coverslips were mounted with ProLong Gold antifade reagent, and cells were analyzed in a LSM 700 confocal microscope. Micrographs were taken using a 63X oil objective. Tritc-Transferrin and Lac-cer fluorescence was analyzed using ImageJ version 1.8.0 software.
Binding of apoptotic cells
Binding of apoptotic cells was assessed as previously described24. In brief, apoptotic Jurkat cells were produced by treatment with 50 μM etoposide for 8 h and stained with 5 μM 5-chloromethylfluorescein diacetate (CMFDA, Thermo Fisher Scientific). Sub-confluent monolayers of Huh7 parental, HAVCR1 KO, or NPC1 KO cells in 12-well plates were incubated with 106 apoptotic cells for 30 min at r.t., monolayers were washed, and the bound apoptotic cells were quantitated in a Synergy HT fluorescence plate reader using 485 nm/528 nm excitation/emission filters. Monolayers were also analyzed under an Axiovert 200 fluorescent microscope (Carl Zeiss) and micrographs were taken with a 20× objective using AxioVision software (Carl Zeiss).
Bimolecular fluorescence complementation (BiFC) assay
The interaction of HAVCR1 and NPC1 using BiFC based on the complementation of monomeric Kusabira green N-terminus [mKG(N)] and C-terminus [mKG(C)] fragments58 has been shown previously9 and performed as described with minor modifications. Huh7 NPC1 KO cells were transfected with pEAK-Hyg NPC1-mKG(N), a plasmid coding for a fusion of NPC1 with mKG(N), pEAK12-Hyg HAVCR1-mKG(C), a plasmid coding for a fusion of HAVCR1 with mKG(C), or vector, or co-transfections of the above mentioned plasmids. After 48 h p.t., cells were stained with Hoechst 33342 as nuclear counterstain and analyzed in an Axiovert 200 fluorescence microscope using filters for Hoeschst 33342 and green fluorescence. Micrographs were taken with a 40× objective using AxioVision software. Co-expression of NPC1-mKG(N) and HAVCR1-mKG(C) resulted in their interaction shown as cytoplasmic green fluorescence. Plasmids coding for NPC1-mKG(N) containing mutations in NPC1 were co-transfected with pEAK-Hyg HAVCR1-mKG(C) to determine expression of NPC1 and the effect of the mutations in the HAVCR1-NPC1 interaction.
Infection with VSV pseudotypes
To assess susceptibility to EBOV infection, Huh7 and Huh7 NPC1 KO cells in 6-well plates were infected with rVSV-EBOVgp-GFP at a m.o.i. of 5 TCID50/cell and incubated at 37°C under 5% CO2. GFP fluorescence was determined at different times p.i. in a Synergy HT fluorescence plate reader and expressed as MFI averages.
For flooding studies, Huh7 cells grown in 12-well plates were treated with exo-HAV at a m.o.i. of 10 TCID50/cell, an equivalent amount of exosomes from uninfected Huh7 cells, or media for 1h at r.t. and then infected with rVSV-EBOVgp-GFP or VSV-GFP at a m.o.i. of 5 TCID50/cell. After incubation at 37°C under 5% CO2 for 16–20 h, GFP fluorescence was determined as above in duplicate wells and expressed as a percentage of the MFI considering media-treated wells as 100% florescence.
To assess the effect of NPC1 mutants to support EBOV infection, Huh7 NPC1 KO cells were transfected with pEAK12-Hyg NPC1-mKG(N) and the derived NPC1 mutants and infected with rVSV-EBOVgp-GFP 48 h p.t, as described above. After 1 h absorption, cells were washed extensively and incubated at 37°C for 24 h. Supernatant samples of the infected cells were taken at 24 h p.i. and titrated by an endpoint dilution assay in 96-well plates containing sub-confluent monolayers of Huh7 cells using 10-fold dilutions in triplicate wells. After 72 h incubation at 37°C, 96-well plates were analyzed under an Axiovert 200 fluorescence microscope to determine green fluorescence and CPE. Viral titers were determined using the online ID50 program 52 (https://www.ncbi.nlm.nih.gov/CBBresearch/Spouge/html_ncbi/html/id50/id50.cgi)
Effect of NPC1 mutations in exo-HAV infection
To analyzed the effect of NPC1 mutations in exo-HAV infection, Huh7 NPC1 KO cells in 12-well plates were transfected with pEAK12-Hyg NPC1-mKG(N), plasmids containing single or double mutations in NPC1, or vector. At 48 h p.t., cells were washed extensively, infected with exo-HAV at a m.o.i. 0.5–1 for 24 h, split 1:4 into 6-well plates, and incubated for 96 h at 37°C. Total RNA was extracted from cells using RNeasy Mini kit, and HAV infection quantitated by HAV RT-qPCR.
Statistical analysis
Statistical methods were not used to predetermine sample size and experiments were not blinded to allocation or accessing outcome.
All experiments were conducted at least three times and the mean values and the standard error of the mean (sem) were calculated using Prism 8 (GraphPad Software) or Excel (Microsoft corp.) software. Statistical significance of log transformed normalized data was calculated using one-way ANOVA with Dunnett’s post-test. For non-parametric data, statistical significance was assessed by two-sided Mann-Whitney test or Kruskal-Wallis test, a non-parametric equivalent of one-way ANOVA, with Dunn’s post-test. Box and whiskers plots were done using the Tukey method to graph whiskers and outliers. Statistical analyses were performed using the Prism 8 software.
Data Availability Statement
The authors declare that the data supporting the findings of this study are available from the corresponding author on reasonable request. Numerical and statistical source data that underlie the graphs in figures, extended data, and supplemental data are provided with the paper.
Extended Data
Extended Data Fig. 1. Fusion of R18-labeled liposomes is temperature dependent and requires phosphatidylserine and cholesterol.
a, Huh7 cells were treated with PS:PC:Chl-R18 liposomes for 30 min at 37ºC, 15ºC, or 4ºC to test for liposome fusion (red). Nuclei were stained with DAPI (blue). Cells were analyzed in a LSM 700 confocal microscope. Micrographs were taken using a 40× oil objective. b, Quantitative analysis of the fusion of R18-labeled liposomes in endosomes from (a). The R18 fluorescence intensity (red) of 20 cells from (a) was measured using ImagJ software and the experiment was repeated 3 times (n=60). c, Huh7 cells were treated with PS:PC:Chl-R18, PS:PC-R18, PC:Chl-R18, or PC-R18 during 30 min at 37ºC. Nuclei were stained with DAPI (blue). Cells were analyzed in a LSM 700 confocal microscope. Micrographs were taken with a 40× oil objective. d, Quantitative analysis of the fusion of R18-labeled liposomes in endosomes from (c). R18 fluorescence intensity (red) of 20 cells from (c) was measured using ImagJ software, and the experiment was repeated 3 times (n=60). Scale bars in (a) and (c) represent 50 mm. In (b) and (d), box and whiskers plots were done using the Tukey method. Box limits are upper and lower quartiles, center lines are the medians, whiskers are 1.5× interquartile range, and points represent outliers. P values were determined by two-sided Mann-Whitney test.
Extended Data Fig. 2. Intracellular fusion of R18-labeled liposomes is pH-dependent.
a, Liposome fusion (red) in Huh7 cells treated with 2 μM Monensin, 30 μM Chloroquine diphosphate, or 5mM Amonium chloride (NH4Cl) prior to the addition of PS:PC:Chl-R18 liposomes. Nuclei were stained with DAPI (blue). Cells were analyzed in a LSM 700 confocal microscope. Micrographs were taken using a 40× oil objective. Scale bars represent 50μm. b, Quantitative analysis of the fusion of R18-labeled liposomes in endosomes from (a). The R18 fluorescence intensity (red) of 20 cells from (a) was measured using ImagJ software and the experiment was repeated 3 times (n=60). Box and whiskers plot was done using the Tukey method. Box limits, upper and lower quartiles; Center line, median; whiskers, 1.5× interquartile range; points, outliers; P values between untreated and treated cells were determined by two-sided Mann-Whitney test. c, Lack of cytotoxicity of the compounds was confirmed by flow cytometry using a caspase-3 apoptosis assay based on the irreversible binding of cell-permeable inhibitor DEVD-fmk conjugated to sulfo-rhodamine (Red-DEVD-fmk) to activated caspase-3. Stained cells were analyzed with a FACSCanto II (BD Biosciences) using FlowJo v8.5 software. Apoptotic/dead Huh7 control cells were prepared by heat treatment at 55ºC for 7 min. d, Liposome fusion with intracellular membranes. Huh7 cells were treated with PS:PC:Chl-R18 liposomes (R18 liposome fusion events, red) for 15 min at 37ºC, the plasma membrane was stained with CellMask (green) for 15 min at 37ºC, and nuclei were stained with DRAQ5 (blue). Cells were analyzed in a LSM 700 confocal microscope. Micrographs were taken with 40× oil objective. Scale Bars represent 25μm. Liposome fusion events detected by R18 fluorescence (red) that did not co-localize with plasma membrane stained with Cell Mask (green). Results are representative of 3 independent experiments.
Extended Data Fig. 3. Characterization of exosomes purified from HAV-infected cells.
a, Presence of PS at the surface of exosomes from HAV-infected cells. Purified exo-HAV/Bsd treated with (+) or without (−) biotinylated ANX5 (ANX5 –Biotin) and anti-biotin mAbs was bound to magnetic beads, washed extensively, and extracted HAV RNA was quantitated by RT-qPCR. Data are mean ± sem, n=3. P values between complete and incomplete immunomagnetic sandwiches were analyzed by one-way ANOVA with Dunnett’s post-test. b, Purification by isopycnic ultracentrifugation of supernatants of HAV.8Y-Bsd-infected Huh7 cells. Gradient collected in 25 fractions from the top. HAV RNA in fractions 5–24 was quantified by RT-qPCR showing peaks of exo-HAV (fractions 11–13, 1.08–1.10 g cm-3) and vpHAV (fraction 21–23, 1.23–1.33 g cm-3). Data are mean ± sd of RT-qPCR duplicates. c, Enrichment of exosomal markers HSP70, FLOT-1, and TSG101 but not Golgi marker GOLGA1 in exo-HAV assessed by Western blot analysis of gradient fractions 5–25 from (b). Fractions aligned with (b). Cell extracts from uninfected Huh7 cells were used as controls. Migration of molecular weight markers is shown in kDa. d, Flow cytometry analysis of cell surface expression of CD9, CD63, and CD81 on Huh7 cells using PE-labeled anti-human CD9, CD63, or CD81 mAs compared to a PE-labeled isotype control. Gating strategy in left panel. e, Flow cytometry analysis of gradient fractions 2–24 from (b) absorbed to latex-beads and stained with mAbs as in (d). f, Content of CD63 and CD81 on exo-HAV (fraction 11 from b) compared to control vpHAV (fraction 22 from b) by flow cytometry as described in (e). Gating strategy in left panel. g, Sizing of exo-HAV (fractions 11–13 from (b) by DLS analysis in a Zetasizer nano ZS90 instrument performed in 3 runs (red, blue, and green lines) of 10 measurements each. Data are representative of 3 independent experiments.
Extended Data Fig. 4. HAVCR1 is an exo-HAV cellular receptor.
a, Specific infectivity of purified exo-HAV and vpHAV from Fig. 1c in Huh7 cells was determined as the ratio between HAV RNA genome equivalents (GE) assessed by RT-qPCR and infectious particles assessed by ARTA. Data are mean ± sem, n=5 from 5 different titrations. b, Expression of HAVCR1 but not HAVCR1 N94A restored exo-HAV cell entry in Huh7 HAVCR1 KO cells. HAVCR1, HAVCR1 N94A, or vector transfectants were infected with exo-HAV or vpHAV from Fig. 1c, and tested for Bsd-resistant CFU. Data are mean ± sem, n=4 from two independent experiments with biological duplicates. c, Flow cytometry analysis of the cell surface expression of HAVCR1 (red dots) or HAVCR1DIgV/ANX5 (blue dots) compared to vector-transfected cells (grey dots) on Huh7 HAVCR1 KO cell transfectants stained with PE-labeled anti-HAVCR1 mucin mAb1750. Gates contain cells expressing the HAVCR1 mucin 1750 epitope. Data representative of 4 independent experiments. d, Huh7 HAVCR1 KO cell transfectants expressing HAVCR1 or HAVCR1DIgV/ANX5 bind apoptotic cells. Monolayers of vector, HAVCR1, or HAVCR1DIgV/ANX5 cell transfectants or control Huh7 parental cells were incubated with CMFDA-labeled apoptotic Jurkat cells and washed extensively. Phase contrast (DIC) and green fluorescence (CMFDA) micrographs were taken with an Axiovert 200 fluorescence microscope using a 20× objective. Scale bar represents 50 mm. Data representative of 3 independent experiments. e, Expression of an annexin V fusion protein at the cell surface restores infectivity of exo-HAV. Huh7 HAVCR1 KO cell transfectants from (c) were infected with purified exo-HAV from Fig. 1c using a m.o.i of 0.1–0.5 for 48 h at 37°C. Total RNA was extracted from the cells and HAV RNA was quantitated by HAV RT-qPCR. Data are mean ± sem, n=4 from 2 independent experiments with 2 biological replicates. P values were determined by two sided Mann-Whitney test.
Extended Data Fig. 5.
Knock out of NPC1 in Huh7 cells blocks EBOV cell entry and endosomal liposome fusion.
a, Growth of rVSV-EBOVgp-GFP in Huh7 parental and NPC1 KO cells. Huh7 parental and NPC1 KO cells were infected with rVSV-EBOVgp-GFP for 24 h. At different times p.i., GFP fluorescence was assessed using a fluorescence plate reader. Data are mean ± sem, n=4 from 4 independent experiments. P values between Huh7 parental and NPC1 KO cells were determined by two-sided Mann-Whitney test. b, Quantitative analysis of the fusion of R18-labeled liposomes in endosomes. Huh7 parental, HAVCR1 KO and NPC1 KO cells were treated with PS:PC:Chl-R18 liposomes 10 min at 15ºC and incubated or not for 30 min at 37ºC to test for liposome fusion (red). Nuclei were stained with DAPI (blue). Cells were analyzed using a LSM 700 confocal microscope (Carl Zeiss), and micrographs were taken using a 40× oil objective. Scale bar represents 50 mm. c, Quantitative analysis of the fusion of R18-labeled liposomes in endosomes from (b). The R18 fluorescence intensity (red) of 25 cells was measured using ImagJ software, and the experiment was repeated 3 times (n=75). Box and whiskers plots were done using the Tukey method. Box limits, upper and lower quartiles; Center line, median; whiskers, 1.5× interquartile range; points, outliers; P values between mean fluorescence intensity (MFI) of Huh7 HAVCR1 KO or Huh7 NPC1 KO cells compared to Huh7 parental cells were determined by two-sided Mann-Whitney test.
Extended Data Fig. 6.
Interaction of NPC1 with HAVCR1 in Huh7 NPC1 KO cell transfectants.
a, Schematic representation of bimolecular fluorescence complementation (BiFC) assay based on the complementation of the monomeric Kusabira green (mKG) protein57 168 amino acid (aa) N-terminus fragment (mKG(N)) fused to the C-terminus of NPC1 (NPC1-mKG(N)) and 51 aa C-terminus fragment (mKG(C)) fused to the C-terminus of HAVCR1 (HAVCR1-mKG(C)). The interaction of NPC1 and HAVCR1 results in BiFC of mKG9 that emits peak fluorescence at 507 nm upon excitation at 492 nm. b, Huh7 NPC1 KO cells transfected with vector, plasmid coding for NPC1-mKG(N) wild type or NPC1 mutants L175A/L176A (double mutation in NTD cholesterol binding pocket prevents function of NPC130), P202A/F203A (double mutation in NTD in the rim of the cholesterol binding pocket prevents function of NPC130), F503W (mutation in domain C increases binding of GP31), F503Y (mutation in domain C reduces infectivity of rVSV-ZEBOV GP by 2 logs31), L656F (mutation in sterol-sensing domain increases cholesterol binding, Millard 200533), G660R (Mutation transmembrane helix 3 prevents function of NPC130, 32, or P691S (mutation in transmembrane domain results in defect in cholesterol uptake and trafficking33) and co-transfected or not with a plasmid coding for HAVCR1-mKG(C) for 48 h to determine HAVCR1-NPC1 interaction (green). Nuclei were stained with Hoechst 33342 (blue). Cells were analyzed in an Axiovert 200 fluorescence microscope, and micrographs were taken using a 40× objective. Scale bars represent 50 mm. Results are representative of 3 independent experiments.
Extended Data Fig. 7.
Effect of NPC1 mutations and cell entry inhibitors in exo-HAV infection.
a, Huh7 NPC1 KO cells were transfected with cDNA of NPC1 WT, NPC1 mutants L173A/L176A, P202A/F203A, F503W, F503Y, L656F, G660R, or P691S, or vector and infected at 48 h p.t. with purified exo-HAV from Fig. 1c. Total RNA was extracted 4 days p.i. and analyzed by HAV RT-qPCR. Data are mean ± sem, n=3 from 3 independent experiments. P values between NPC1 WT and NPC1 mutants or vector were determined by one-way ANOVA with Dunnett’s post-test. b, Cell transfectants as in (a) were infected with rVSV-EBOVgp-GFP for 24 h, and virus in the supernatant was titrated by a fluorescence endpoint dilution assay in 96-well plates containing Huh7 monolayers. Viral titers were determined at 72 h p.i. using the ID50 program. Data are log10 TCID50/ml ± s.d., n=3. P values were determined as in (a). c, Viability of Huh7 cells treated with cell entry inhibitors for 72 h by dual-fluorescence Acridine Orange / Propidium Iodide assay. Data are mean ± sem, n=4 from two independent experiments with biological duplicates. d, Huh7 cells were treated with inhibitors for 1 h, infected with rVSV-EBOVgp-GFP, and GFP fluorescence was quantified at 16–20 h p.i. in a fluorescence plate reader. Data are mean ± sem, n=4 from two independent experiments with biological duplicates. e, Huh7 cells were pretreated with inhibitors for 1 h, infected with purified vpHAV from Fig. 1c. HAV replication was quantitated at 72 h p.i. by RT-qPCR. Data are mean ± sem, from left to right n=5, 5, 5, 3, 3, and 3 biological replicates. P values between DMSO and inhibitors were determined as in (a). In (c) and (d), P values between DMSO and inhibitors were determined by two-sided Mann-Whitney test.
Extended Data Fig. 8.
Chlorpromazine and Dynasore inhibit clathrin-mediated endocytosis.
a, Huh7 cells were treated with inhibitors of endocytic pathways (5 μg/ml CPZ, 80 μM Dynasore, 10 μM EGA, 40 μM EIPA, 10 μM LY294002) or DMSO prior to the addition of tetramethylrhodamine conjugate-transferrin (Tritc-Transferrin), a marker of clathrin-mediated endocytosis. Tritc-Transferrin (red fluorescence) uptake in the presence of inhibitors was analyzed using an LSM 700 confocal fluorescence microscope. Nuclei were stained with DAPI (blue fluorescence). Micrographs were taken with a 40× oil objective. Scale bars represent 50μm. b, Quantitative analysis of the endocytosis of Tritc-Transferrin from (a). The Tritc-Transferrin fluorescence intensity (red) of 16 cells was measured using ImagJ software, and the experiment was repeated 3 times (n=48). Box and whiskers plot was done using the Tukey method. Box limits, upper and lower quartiles; Center line, median; whiskers, 1.5× interquartile range; points, outliers; P values between DMSO and inhibitors were determined by two-sided Mann-Whitney test.
Extended Data Fig. 9.
Dynasore but not chlorpromazine inhibit caveolae-mediated endocytosis.
a, Huh7 cells were treated with BODIPY FL C5-Lactoceramide complexed to BSA (LAC-cer), a marker of caveolae-mediated endocytosis, and treated with inhibitors of endocytosis (5 μg/ml CPZ, 80 μM Dynasore, 10 μM EGA, 40 μM EIPA, 10 μM LY294002) or DMSO as control. LAC-cer (green) uptake was analyzed using a LSM 700 confocal microscope. Nuclei were stained with DRAQ5 (blue). Micrographs were taken with a 40× oil objective. Scale bars represent 50μm. b, Quantitative analysis of the endocytosis of LAC-cer from (a). The LAC-cer fluorescence intensity (green) of 20 cells was measured using ImagJ software, and the experiment was repeated 3 times (n=60). Box and whiskers plot was done using the Tukey method. Box limits, upper and lower quartiles; Center line, median; whiskers, 1.5× interquartile range; points, outliers; P values between DMSO and inhibitors were determined by two-sided Mann-Whitney test.
Extended Data Fig. 10.
Effect of endocytosis inhibitors in different cellular compartments.
Huh7 cells in 8-wells chamber slides were infected with the CellLight Bacman 2.0 reagents fused to GFP (green) or RFP (red) at a multiplicity of infection of 30 particles per cell, incubated at 37°C for 10–12 h, treated with 80 mM Dynasore hydrate, 5mg/ml Chlorpromazine hydrochloride solution, 10 mM EGA, or a similar volume of DMSO vehicle as negative control, and incubated for additional 12–14 h at 37°C. Nuclei were stained with DRAQ5 (blue), cells were fixed with 4% PFA, coverslips mounted with ProLong Gold antifade reagent, and slides analyzed in a LSM 700 confocal microscope. Micrographs were taken using a 63X oil objective. Cells were infected with CellLight Bacmam 2.0 driving the expression of markers of: a, Mitochondria (leader sequence of E1 alpha pyruvate dehydrogenase fused to RFP); b, Lysosomes (Lamp1 fused to RFP); c, Golgi (human Golgi-resident enzyme N-acetylgalactosaminyltransferase 2 fused to RFP); d, Peroxisomes (peroxisomal C-terminal targeting sequence fused to GFP); e, Early endosomes (EE, Rab5a fused to GFP); or f, Late Endosomes (LE, Rab 7a fused to GFP). Scale Bars represent 25μm. Results are representative of 3 independent experiments.
Supplementary Material
Acknowledgements
This work was supported by funding from the U.S. Food and Drug Administration Intramural Program to G.K. and Medical Countermeasures Initiative to G.K. This project was supported in part by appointments to the Research Fellowship Program at the Office of Blood Research and Review, Center for Biologics Evaluation and Research, U.S. Food and Drug Administration, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and FDA (M.I.C., A.A., and H.L.).
Footnotes
Author Competing interests The authors declare no competing interest.
REFERENCES
- 1.Raposo G & Stoorvogel W Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of cell biology 200, 373–383, doi: 10.1083/jcb.201211138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pitt JM, Kroemer G & Zitvogel L Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest 126, 1139–1143, doi: 10.1172/jci87316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Dongen HM, Masoumi N, Witwer KW & Pegtel DM Extracellular Vesicles Exploit Viral Entry Routes for Cargo Delivery. Microbiology and molecular biology reviews : MMBR 80, 369–386, doi: 10.1128/mmbr.00063-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.French KC, Antonyak MA & Cerione RA Extracellular vesicle docking at the cellular port: Extracellular vesicle binding and uptake. Seminars in cell & developmental biology, doi: 10.1016/j.semcdb.2017.01.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hessvik NP & Llorente A Current knowledge on exosome biogenesis and release. Cell Mol Life Sci, doi: 10.1007/s00018-017-2595-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan G et al. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J 15, 4282–4296 (1996). [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi N et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 27, 927–940, doi: 10.1016/j.immuni.2007.11.011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carstea ED et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277, 228–231 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Kuroda M et al. Interaction between TIM-1 and NPC1 Is Important for Cellular Entry of Ebola Virus. J Virol 89, 6481–6493, doi: 10.1128/jvi.03156-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Niel G, D’Angelo G & Raposo G Shedding light on the cell biology of extracellular vesicles. Nature reviews. Molecular cell biology 19, 213–228, doi: 10.1038/nrm.2017.12510.1038/nrm.2017.125. Epub 2018 Jan 17 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Kaplan GG et al. in Viral Hepatitis (eds Thomas Howard C., Zuckerman Arie J., Lok Anna S. F., & Locarnini Stephen A.) 29–42 (Wiley-Blackwell, 2013). [Google Scholar]
- 12.Feng Z et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371, doi: 10.1038/nature12029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKnight KL et al. Protein composition of the hepatitis A virus quasi-envelope. Proc Natl Acad Sci U S A 114, 6587–6592, doi: 10.1073/pnas.1619519114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng D et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic (Copenhagen, Denmark) 11, 675–687, doi: 10.1111/j.1600-0854.2010.01041.x (2010). [DOI] [PubMed] [Google Scholar]
- 15.Tian T et al. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem 289, 22258–22267, doi: 10.1074/jbc.M114.588046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White JM & Whittaker GR Fusion of Enveloped Viruses in Endosomes. Traffic (Copenhagen, Denmark) 17, 593–614, doi: 10.1111/tra.12389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feigelstock D, Thompson P, Mattoo P, Zhang Y & Kaplan GG The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J Virol 72, 6621–6628 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei X et al. Surface Phosphatidylserine Is Responsible for the Internalization on Microvesicles Derived from Hypoxia-Induced Human Bone Marrow Mesenchymal Stem Cells into Human Endothelial Cells. PLoS One 11, e0147360, doi: 10.1371/journal.pone.0147360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feigelstock D, Thompson P, Mattoo P & Kaplan GG Polymorphisms of the hepatitis A virus cellular receptor 1 in African green monkey kidney cells result in antigenic variants that do not react with protective monoclonal antibody 190/4. J Virol 72, 6218–6222 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costafreda MI & Kaplan G HAVCR1 (CD365) and its mouse ortholog are functional hepatitis A virus (HAV) cellular receptors that mediate HAV infection. J Virol 92, e02065–02017, doi: 10.1128/JVI.02065-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kachko A et al. Determinants in the Ig Variable Domain of Human HAVCR1 (TIM-1) Are Required To Enhance Hepatitis C Virus Entry. J Virol 92, e01742–01717, doi: 10.1128/JVI.01742-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konduru K & Kaplan GG Determinants in 3Dpol modulate the rate of growth of hepatitis A virus. J Virol 84, 8342–8347, doi: 10.1128/jvi.01470-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lötvall J et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 3, 26913-26913, doi: 10.3402/jev.v3.26913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manangeeswaran M et al. Binding of hepatitis A virus to its cellular receptor 1 inhibits T-regulatory cell functions in humans. Gastroenterology 142, 1516–1525 e1513, doi: 10.1053/j.gastro.2012.02.039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quijada NM, Fongaro G, Barardi CRM, Hernández M & Rodríguez-Lázaro D Propidium Monoazide Integrated with qPCR Enables the Detection and Enumeration of Infectious Enteric RNA and DNA Viruses in Clam and Fermented Sausages. Frontiers in microbiology 7, 2008, doi: 10.3389/fmicb.2016.02008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Squillace DM, Zhao Z, Call GM, Gao J & Yao JQ Viral Inactivation of Human Osteochondral Grafts with Methylene Blue and Light. Cartilage 5, 28–36, doi: 10.1177/1947603513509650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins ME, Davies JP, Chen FW & Ioannou YA Niemann-Pick C1 is a late endosome-resident protein that transiently associates with lysosomes and the trans-Golgi network. Molecular genetics and metabolism 68, 1–13, doi: 10.1006/mgme.1999.2882 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Carette JE et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343, doi: 10.1038/nature10348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cote M et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477, 344–348, doi: 10.1038/nature10380 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon HJ et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137, 1213–1224, doi: 10.1016/j.cell.2009.03.049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ndungo E et al. A Single Residue in Ebola Virus Receptor NPC1 Influences Cellular Host Range in Reptiles. mSphere 1, doi: 10.1128/mSphere.00007-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahl NK, Reed KL, Daunais MA, Faust JR & Liscum L Isolation and characterization of Chinese hamster ovary cells defective in the intracellular metabolism of low density lipoprotein-derived cholesterol. J Biol Chem 267, 4889–4896 (1992). [PubMed] [Google Scholar]
- 33.Millard EE et al. The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J Biol Chem 280, 28581–28590, doi: 10.1074/jbc.M414024200 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Ivanov AI Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods in molecular biology (Clifton, N.J.) 440, 15–33, doi: 10.1007/978-1-59745-178-9_2 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Mesaki K, Tanabe K, Obayashi M, Oe N & Takei K Fission of tubular endosomes triggers endosomal acidification and movement. PloS one 6, e19764–e19764, doi: 10.1371/journal.pone.0019764 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie EJ et al. Selective inhibitor of endosomal trafficking pathways exploited by multiple toxins and viruses. Proc Natl Acad Sci U S A 110, E4904–4912, doi: 10.1073/pnas.1302334110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dreux M et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell host & microbe 12, 558–570, doi: 10.1016/j.chom.2012.08.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baglio SR et al. Sensing of latent EBV infection through exosomal transfer of 5′pppRNA. Proceedings of the National Academy of Sciences 113, E587–E596, doi: 10.1073/pnas.1518130113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng Z et al. Human pDCs preferentially sense enveloped hepatitis A virions. J Clin Invest 125, 169–176, doi: 10.1172/jci77527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto R, Bosch A & Kaplan G in Liver immunology : principles and practice (eds Gershwin M. Eric, Vierling John M., & Manns Michael P.) 173–189 (Springer, 2013). [Google Scholar]
- 41.Sainz B Jr. et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med 18, 281–285, doi: 10.1038/nm.2581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das A et al. TIM1 (HAVCR1) Is Not Essential for Cellular Entry of Either Quasi-enveloped or Naked Hepatitis A Virions. mBio 8, e00969–00917, doi: 10.1128/mBio.00969-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dveksler GS, Gagneten SE, Scanga CA, Cardellichio CB & Holmes KV Expression of the recombinant anchorless N-terminal domain of mouse hepatitis virus (MHV) receptor makes hamster of human cells susceptible to MHV infection. J Virol 70, 4142–4145 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang ST, Kreutzberger AJB, Lee J, Kiessling V & Tamm LK The role of cholesterol in membrane fusion. Chemistry and physics of lipids 199, 136–143, doi: 10.1016/j.chemphyslip.2016.05.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konduru K & Kaplan GG Stable growth of wild-type hepatitis A virus in cell culture. J Virol 80, 1352–1360, doi: 10.1128/jvi.80.3.1352-1360.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Totsuka A & Moritsugu Y in International Symposium on Viral Hepatitis and Liver Disease (ed Nishioka H Suzuki K., Mishiro S, Oda T) 509–513 (Spring-Verlag, Tokyo, 1994). [Google Scholar]
- 47.Uphoff CC, Gignac SM & Drexler HG Mycoplasma contamination in human leukemia cell lines. I. Comparison of various detection methods. Journal of immunological methods 149, 43–53, doi: 10.1016/s0022-1759(12)80047-0 (1992). [DOI] [PubMed] [Google Scholar]
- 48.Cohen JI, Ticehurst JR, Feinstone SM, Rosenblum B & Purcell RH Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. Journal of Virology 61, 3035–3039 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konduru K, Virata-Theimer ML, Yu MY & Kaplan GG A simple and rapid Hepatitis A Virus (HAV) titration assay based on antibiotic resistance of infected cells: evaluation of the HAV neutralization potency of human immune globulin preparations. Virol J 5, 155, doi: 10.1186/1743-422x-5-155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitt MA Generation of VSV pseudotypes using recombinant DeltaG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods 169, 365–374, doi: 10.1016/j.jviromet.2010.08.006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konduru K et al. Ebola virus glycoprotein Fc fusion protein confers protection against lethal challenge in vaccinated mice. Vaccine 29, 2968–2977, doi: 10.1016/j.vaccine.2011.01.113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spouge JL Statistical analysis of sparse infection data and its implications for retroviral treatment trials in primates. Proc Natl Acad Sci U S A 89, 7581–7585 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Théry C, Amigorena S, Raposo G & Clayton A Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Current Protocols in Cell Biology 30, 3.22.21–23.22.29, doi: 10.1002/0471143030.cb0322s30 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Nunes-Correia I et al. Fluorescent probes for monitoring virus fusion kinetics: comparative evaluation of reliability. Biochimica et Biophysica Acta (BBA) - Biomembranes 1561, 65–75, doi: 10.1016/S0005-2736(01)00457-6 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Costafreda MI, Bosch A & Pinto RM Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Applied and environmental microbiology 72, 3846–3855, doi: 10.1128/aem.02660-05 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cantin R, Diou J, Bélanger D, Tremblay AM & Gilbert C Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. Journal of immunological methods 338, 21–30, doi: 10.1016/j.jim.2008.07.007 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Campos B, Kaetzel MA & Dedman JR Secretion of annexin V from cultured cells requires a signal peptide. Placenta 22, 837–845, doi: 10.1053/plac.2001.0724 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Ueyama T et al. Sequential binding of cytosolic Phox complex to phagosomes through regulated adaptor proteins: evaluation using the novel monomeric Kusabira-Green System and live imaging of phagocytosis. J Immunol 181, 629–640, doi: 10.4049/jimmunol.181.1.629 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Vercauteren D et al. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol Ther 18, 561–569, doi: 10.1038/mt.2009.281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available from the corresponding author on reasonable request. Numerical and statistical source data that underlie the graphs in figures, extended data, and supplemental data are provided with the paper.