Complete genome sequences of two novel torque teno viruses (TTVs) were identified in human brain tissue. These sequences are 3,245 nucleotides (nt) and 2,900 nt long and share 68% and 72% open reading frame 1 (ORF1) identity, respectively, with other human TTVs. This report extends the identification of TTV sequences in the brain.
ABSTRACT
Complete genome sequences of two novel torque teno viruses (TTVs) were identified in human brain tissue. These sequences are 3,245 nucleotides (nt) and 2,900 nt long and share 68% and 72% open reading frame 1 (ORF1) identity, respectively, with other human TTVs. This report extends the identification of TTV sequences in the brain.
ANNOUNCEMENT
Since the discovery of the first torque teno virus (TTV) (family Anelloviridae) from a human sample, a plethora of anelloviruses have been identified in various mammals (including primates) and avian species. Anelloviridae is a family of host-specific diverse viruses (1–5). In primates, a number of TTV genome sequences have been identified from a broad range of tissue and sample types (5–8). Studies in humans have investigated associations between TTV and outcomes of diseases such as cancer and hepatitis (8–10). Previously, two TTV genome sequences were identified in a glioblastoma sample (GenBank accession no. KX810063 and KX810064) (11), and Tisza et al. (12) reported the identification of anelloviruses (partial genomes) in the brain tissue of deceased individuals with Alzheimer’s disease and other forms of dementia.
Given that only two genome sequences of TTV have been identified from brain tissue samples, we undertook a viral metagenomic analysis of human brain tissue to identify additional TTVs. Written informed consent for autopsy was obtained in compliance with institutional guidelines of Banner Sun Health Research Institute. The Banner Sun Health Research Institute Review Board approved this study, including recruitment, enrollment, and autopsy procedures. Individual person(s) and their respective next of kin consented to brain autopsy for the purpose of research analysis as participants in the Banner Sun Health Research Institute Brain and Body Donation Program. The human brain tissue used in this study was from routine existing autopsies, which fully qualifies for 4C exemption by NIH guidelines.
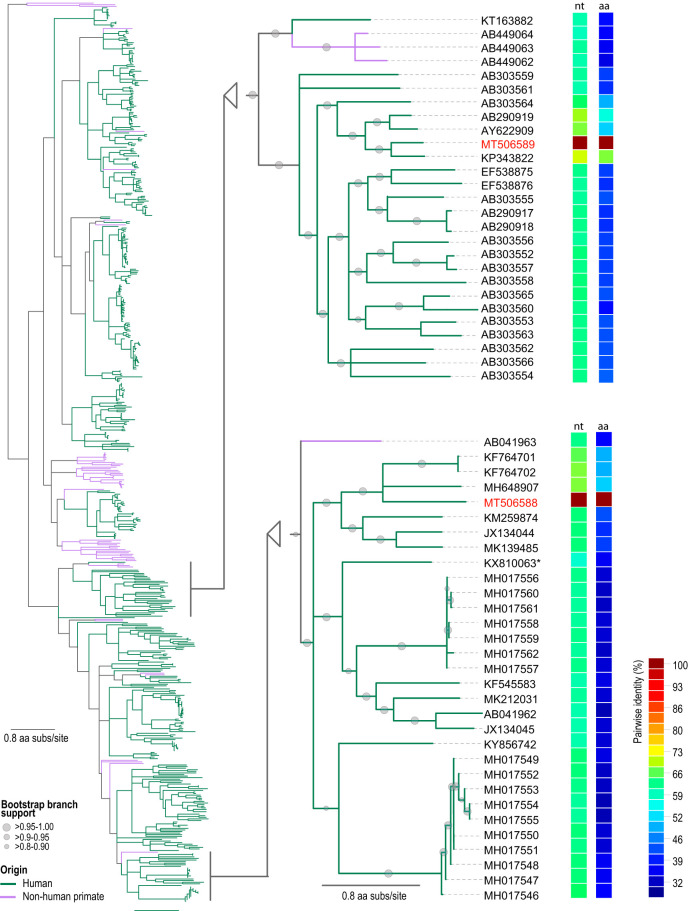
Brain tissue of the middle temporal gyrus (0.1 g) from an 82-year-old deceased individual (identifier [ID] 04-05) with no abnormal brain pathology was homogenized in 300 μl of SM buffer (0.1 M NaCl, 50 mM Tris-HCl [pH 7.4], 10 mM MgSO4), and viral DNA was extracted from the supernatant using the High Pure viral nucleic acid kit (Roche Diagnostics, USA). Circular DNA was preferentially amplified by rolling-circle replication amplification (RCA) with the TempliPhi kit (GE Healthcare, USA) and random hexamers. The resulting RCA DNA was used to generate 2 × 100-bp libraries using the Nextera DNA flex kit (Illumina, USA) and was sequenced at Macrogen, Inc. (South Korea), on the Illumina HiSeq 4000 platform. Raw reads were de novo assembled using metaSPAdes v3.12.0 (13), and the resulting contigs were analyzed using BLASTx against a viral database for similarities to viral proteins (14). Two contigs (1,964 and 3,079 nucleotides [nt]) with similarity to anelloviruses were identified. We designed specific abutting primers (Table 1) which were used to recover the complete genome sequences by PCR with the HiFi HotStart DNA polymerase (Kapa Biosystems, USA). Genomes were cloned and Sanger sequenced at Macrogen, Inc. The two recovered viral genome sequences (GenBank accession no. MT506588 and MT506589) are 2,900 nt (37.2% GC content) and 3,245 nt (42.5% GC content) long and have an organization typically exhibited by anelloviruses. Since the largest open reading frame 1 (ORF1) is relatively conserved, we assembled a data set of primate anellovirus ORF1 sequences available in GenBank (downloaded on 1 April 2020). A maximum likelihood phylogeny of the MUSCLE v3.8.31 (15)-aligned sequences of the ORF1 proteins was constructed using PHYML v3.0 (16) (substitution model rtREV+G+F) (17). Nucleotide and amino acid pairwise identities were determined using SDT v1.2 (18). The analysis (Fig. 1) shows that the two TTVs identified from the brain sample fall into two distinct clades and share the highest similarity with other human TTVs. The sequence under accession no. MT506588 is most closely related to three anelloviruses derived from human blood, under accession no. KF764701, KF764702 (19), and MH648907 (12), shares ∼68% ORF1 gene pairwise identity, and is a member of the Betatorquevirus genus (Fig. 1). The sequence under accession no. MT506589 is most closely related to a sequence from sera taken from a first-trimester mother (KP343822) (20), shares ∼72% ORF1 gene pairwise identity, and is member of the Gammatorquevirus genus (Fig. 1). The sequences under accession no. MT506588 and MT506589 extend our knowledge of TTV diversity in humans and complement the two genome sequences previously identified in the brain samples (11).
TABLE 1.
Details of primers used to amplify the TTV genome
FIG 1.
Maximum likelihood phylogenetic tree of primate anellovirus ORF1 protein sequences. The clades that have the two anelloviruses (GenBank accession no. MT506588 and MT506589; red font) reported here from the human brain sample are shown in detail. Branches with <0.8 approximate likelihood ratio test (aLRT) support have been collapsed using TreeGraph2 (21). Pairwise identity of the ORF1 gene (nt) and protein sequences (aa) in relation to MT506588 and MT506589 in the two separate clades are shown to the right of the accession numbers. *, anellovirus sequence from glioblastoma sample (11).
Data availability.
The TTV sequences have been deposited in GenBank under the accession no. MT506588 and MT506589. Mapped raw reads have been deposited in the SRA database under SRR12450126.
ACKNOWLEDGMENTS
We thank the brain bank at Banner Sun Health Research Institute (USA) for the tissue samples. The Brain and Body Donation Program has been supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026, National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610, Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901, and 1001 to the Arizona Parkinson's Disease Consortium), and the Michael J. Fox Foundation for Parkinson’s Research.
Research reported here was funded by Alzheimer’s Association grant AARGD-17-529197 awarded to D.M.
REFERENCES
- 1.Biagini P. 2009. Classification of TTV and related viruses (anelloviruses). Curr Top Microbiol Immunol 331:21–33. doi: 10.1007/978-3-540-70972-5_2. [DOI] [PubMed] [Google Scholar]
- 2.Bigarré L, Beven V, de Boisséson C, Grasland B, Rose N, Biagini P, Jestin A. 2005. Pig anelloviruses are highly prevalent in swine herds in France. J Gen Virol 86:631–635. doi: 10.1099/vir.0.80573-0. [DOI] [PubMed] [Google Scholar]
- 3.de Souza WM, Fumagalli MJ, de Araujo J, Sabino-Santos G Jr, Maia FGM, Romeiro MF, Modha S, Nardi MS, Queiroz LH, Durigon EL, Nunes MRT, Murcia PR, Figueiredo LTM. 2018. Discovery of novel anelloviruses in small mammals expands the host range and diversity of the Anelloviridae. Virology 514:9–17. doi: 10.1016/j.virol.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Fahsbender E, Burns JM, Kim S, Kraberger S, Frankfurter G, Eilers AA, Shero MR, Beltran R, Kirkham A, McCorkell R, Berngartt RK, Male MF, Ballard G, Ainley DG, Breitbart M, Varsani A. 2017. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol 3:vex017. doi: 10.1093/ve/vex017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hrazdilova K, Slaninkova E, Brozova K, Modry D, Vodicka R, Celer V. 2016. New species of Torque Teno miniviruses infecting gorillas and chimpanzees. Virology 487:207–214. doi: 10.1016/j.virol.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 6.McElvania TeKippe E, Wylie KM, Deych E, Sodergren E, Weinstock G, Storch GA. 2012. Increased prevalence of anellovirus in pediatric patients with fever. PLoS One 7:e50937. doi: 10.1371/journal.pone.0050937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ninomiya M, Takahashi M, Nishizawa T, Shimosegawa T, Okamoto H. 2008. Development of PCR assays with nested primers specific for differential detection of three human anelloviruses and early acquisition of dual or triple infection during infancy. J Clin Microbiol 46:507–514. doi: 10.1128/JCM.01703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spandole S, Cimponeriu D, Berca LM, Mihăescu G. 2015. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol 160:893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- 9.Maggi F, Bendinelli M. 2009. Immunobiology of the Torque teno viruses and other anelloviruses, p 65–90. In de Villiers E-M, zur Hausen H (ed), TT viruses. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 10.Hsu H-Y, Ni Y-H, Chen H-L, Kao J-H, Chang M-H. 2003. TT virus infection in healthy children, children after blood transfusion, and children with non-A to E hepatitis or other liver diseases in Taiwan. J Med Virol 69:66–71. doi: 10.1002/jmv.10249. [DOI] [PubMed] [Google Scholar]
- 11.Ng TFF, Dill JA, Camus AC, Delwart E, Van Meir EG. 2017. Two new species of betatorqueviruses identified in a human melanoma that metastasized to the brain. Oncotarget 8:105800–105808. doi: 10.18632/oncotarget.22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tisza MJ, Pastrana DV, Welch NL, Stewart B, Peretti A, Starrett GJ, Pang YS, Krishnamurthy SR, Pesavento PA, McDermott DH, Murphy PM, Whited JL, Miller B, Brenchley J, Rosshart SP, Rehermann B, Doorbar J, Ta'ala BA, Pletnikova O, Troncoso JC, Resnick SM, Bolduc B, Sullivan MB, Varsani A, Segall AM, Buck CB. 2020. Discovery of several thousand highly diverse circular DNA viruses. Elife 9:e51971. doi: 10.7554/eLife.51971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 17.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jazaeri Farsani SM, Jebbink MF, Deijs M, Canuti M, van Dort KA, Bakker M, Grady BP, Prins M, van Hemert FJ, Kootstra NA, van der Hoek L. 2013. Identification of a new genotype of Torque Teno Mini virus. Virol J 10:323. doi: 10.1186/1743-422X-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bzhalava D, Hultin E, Arroyo Muhr LS, Ekstrom J, Lehtinen M, de Villiers EM, Dillner J. 2016. Viremia during pregnancy and risk of childhood leukemia and lymphomas in the offspring: nested case-control study. Int J Cancer 138:2212–2220. doi: 10.1002/ijc.29666. [DOI] [PubMed] [Google Scholar]
- 21.Stover BC, Müller KF. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The TTV sequences have been deposited in GenBank under the accession no. MT506588 and MT506589. Mapped raw reads have been deposited in the SRA database under SRR12450126.