SUMMARY:
Hepatocellular carcinoma (HCC) is a complex and deadly disease lacking druggable genetic mutations. The limited efficacy of systemic treatments for advanced HCC implies that predictive biomarkers and drug targets are urgently needed. Most HCC drugs target protein kinases and their kinase-dependent signaling networks to drive HCC progression. To identify HCC signaling networks that determine responses to kinase inhibitors (KIs), we apply a pharmacoproteomics approach integrating kinome activity in 17 HCC cell lines with their responses to 299 KIs, resulting in a comprehensive dataset of pathway-based drug response signatures. By profiling patient HCC samples, we identify signatures of clinical HCC drug responses in individual tumors. Our analyses reveal kinase networks promoting the epithelial-mesenchymal transition (EMT) and drug resistance, including a FZD2-AXL-NUAK1/2 signaling module, whose inhibition reverses the EMT and sensitizes HCC cells to drugs. Our approach identifies cancer drug targets and molecular signatures of drug response for personalized oncology.
Keywords: Hepatocellular Carcinoma, Epithelial-Mesenchymal Transition, Protein Kinase, Kinobeads, Pharmacoproteomics, Kinase Inhibitors, Drug Resistance
Graphical Abstract

In Brief:
We describe our kinome-centric pharmacoproteomics approach to study kinase signaling networks controlling cancer drug sensitivity and resistance to kinase-targeted drugs. Our analyses reveal that the epithelial-mesenchymal transition (EMT) state of hepatocellular carcinoma (HCC) cells affects their sensitivity to hundreds of clinical and pre-clinical kinase inhibitors. We show that FZD2-AXL-NUAK1/2 signaling promotes the HCC cell EMT, and that its inhibition reverses the EMT and sensitizes mesenchymal-like HCC cells to drug treatment.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the fourth most common cause of cancer-related death worldwide (Global Burden of Disease Cancer Collaboration, 2017) and has many etiologies, including viral hepatitis, alcoholic cirrhosis, and nonalcoholic steatohepatitis (NASH) (Garber, 2019). Among solid cancers, HCC has one of the fewest druggable genetic alterations, limiting treatment options for advanced HCC (The Cancer Genome Atlas Research Network, 2017; Zehir et al., 2017). Five of the seven FDA-approved drugs for advanced HCC target protein kinases, including the small molecule drugs sorafenib (Llovet et al., 2018), regorafenib (Bruix et al., 2017), lenvatinib (Kudo et al., 2018) and cabozantinib (Abou-Alfa et al., 2018), as well as the antibody Ramucirumab (Zhu et al., 2019), highlighting the importance of kinase-dependent signaling networks in HCC progression. However, predictive biomarkers that could guide clinical use of these kinase inhibitors (KIs) are lacking, likely contributing to the poor response rates of 10-15% (Villanueva, 2019).
Even in HCCs that initially respond to treatment, drug resistance invariably develops; this has been particularly well-documented for sorafenib and suggests that HCCs activate compensatory signaling pathways to drive rebound growth (Abou-Alfa et al., 2018; Firtina Karagonlar et al., 2016). In carcinomas, many of these compensatory pathways are linked to the interconversion between an epithelial-like to a mesenchymal-like cancer cell phenotype, i.e. the epithelial-mesenchymal transition (EMT) (Kalluri and Weinberg, 2009; Shibue and Weinberg, 2017). Under physiological conditions, the EMT is an integral part of tissue development and repair (Thiery et al., 2009). In cancer, however, cell signaling networks that control the EMT are hijacked to promote tumor metastasis and drug resistance. Protein kinases act as central nodes in most cell signaling networks and are commonly dysregulated in cancer (Fleuren et al., 2016). Despite known roles for certain kinases in phenotypic transition (Gay et al., 2017; Gujral et al., 2014), there are no studies that comprehensively map EMT-associated kinase pathways. Reasoning that analyses of uncharted and dysregulated HCC kinase signaling could reveal new mechanisms of EMT-related drug resistance, molecular markers of drug response, and new drug targets, we took a kinome-centric pharmacoproteomics approach to study HCC responses to kinase inhibitor drugs.
Quantifying the activity of kinase-dependent signaling networks requires measuring kinase expression levels, their post-translational modifications (PTMs), and their association with regulatory proteins. Kinase expression and phospho-activation can be assessed by mass spectrometry (MS)-based proteomics techniques such as global proteomics and phosphoproteomics (Beekhof et al., 2019), targeted MS analyses of kinase activation loop phosphorylation sites (Schmidlin et al., 2019), and by kinobead kinase affinity enrichment coupled to phosphopeptide analyses (kinobead/LC-MS) (Bantscheff et al., 2007; Daub et al., 2008). Kinobead/LC-MS offers deep analytical coverage of kinases, their PTMs and kinase-regulatory proteins, quantifying kinome activity in a comprehensive and unbiased manner (Golkowski et al., 2020).
Consequently, we used kinobead/LC-MS to map features of kinome activity to growth inhibition caused by 299 kinase-targeted drugs across a panel of 17 HCC cell lines. We applied gene set enrichment analysis (GSEA) to score the association of 275 kinase-dependent cancer pathways with drug responses (Fabregat et al., 2018; Subramanian et al., 2005), generating a comprehensive database of KI drug response signatures in HCC. Analyzing human HCC samples with kinobead/LC-MS, we quantified drug response signatures that could act as candidate predictive markers for personalized treatment. Our analyses identified distinct signaling networks and drug response phenotypes closely linked to the EMT, and that the cellular EMT-state broadly impacts kinase expression and activation. Specifically, we identified a FZD2-AXL-NUAK1/2 signaling module that promotes HCC cell EMT. Genetic knockdown or small-molecule inhibition of these proteins reversed the EMT, activated replication stress signaling, and increased sensitivity of HCC cells to drugs.
RESULTS
Kinome-centric pharmacoproteomics analysis of HCC
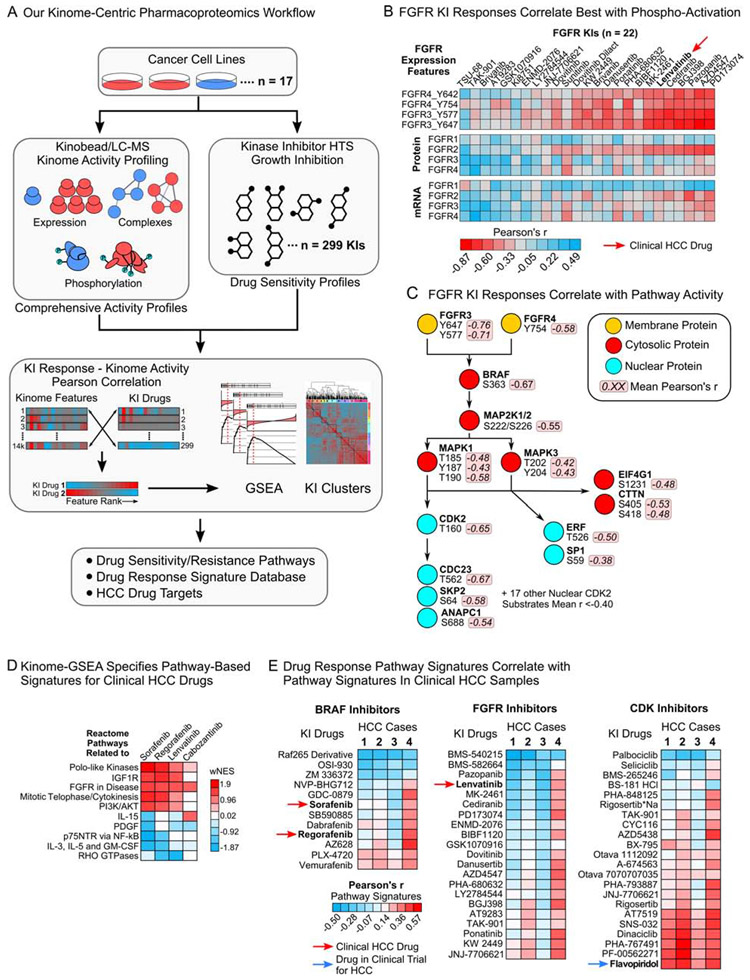
We selected a panel of 17 HCC lines from the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al., 2012) with diverse kinase mRNA expression and profiled these lines using our kinobead/LC-MS platform to measure kinase expression, their phosphorylation states, and kinase-interacting proteins (Figure 1A, S1A and Table S1) (Golkowski et al., 2020); this quantified 2731 proteins and 11204 phosphorylation sites, including 346 kinases, 2821 kinase phosphorylation sites, and 886 kinase-interacting proteins (Figure S1B and C, and Table S2). Functionally characterized phosphosites (Hornbeck et al., 2015), kinase-interacting proteins, and their phosphorylation sites specified the activation states of 284 of the 346 kinases (see ‘STAR Methods’). We used a 299-member diversity library of experimental, preclinical and clinical KIs that together inhibit at least 145 primary kinase targets to obtain seven-point dose-response curves for each KI in all 17 HCC cell lines (see ‘STAR Methods’, Figure 1A, Table S1 and S3). Using the area under the dose-response curve (AUC) as a measure of drug efficacy, we observed diverse responses to inhibitors. For instance, FGFR, EGFR, IGF1R and BRAF inhibitors, blocked cell growth in certain HCC lines, while inhibitors of MEK, cell cycle-related kinases and MTOR were broadly active, efficiently inhibiting cell growth in 10 of the 17 cell lines (Figure S1D, Table S3).
Figure 1. A pharmacoproteomics platform linking kinome features to drug response.
(A) Schematic of our kinome-centric pharmacoproteomics platform.
(B) Pearson’s r-values for FGFR1-4 expression features correlated with the AUCs of all 22 FGFR KIs in our drug screen.
(C) Phosphosites in the FGFR-RAF/MEK/ERK-cell cycle signaling cascade correlated with responses to the seven FGFR KIs that showed the strongest correlation with FGFR3 and FGFR4 tyrosine phosphorylation (see (B)).
(D) Examples for pathways that are associated with sensitivity (positive wNES) or resistance (negative wNES) to clinical HCC drugs. wNES is the FDR-weighted normalized Reactome pathway enrichment score.
(E) Correlation of pathway-based drug response signatures defined in 17 HCC lines with kinome pathway signatures identified in clinical HCC samples, showing that response signatures for specific KI drugs are enriched in human tumors (see Table S5).
Kinase activation states are powerful predictors of drug response
To rank proteomics features by their association with drug responses, we correlated the MS intensities of all 13935 quantified features with each KI’s AUC values across the 17 HCC line panel, where r < 0 indicates drug sensitivity (high MS intensity – low AUC) while r > 0 denotes cell survival and KI resistance (high MS intensity – high AUC, see ‘STAR Methods’, Table S2, S3 and S4). For example, we examined the correlation of FGFR expression features with the response to 22 FGFR inhibitors, including lenvatinib (Figure 1B). Activating FGFR3 phosphorylation sites Y647 and Y577, and the activating Y754 on FGFR4 correlated very well with responses to most FGFR inhibitors (mean r = −0.40), whereas kinase protein and mRNA expression correlated much less (mean r = −0.12 and −0.09, Figure 1B). Similarly, sensitivity to 12 BRAF inhibitors including sorafenib and regorafenib correlated better with kinase phosphorylation and activation than mRNA or protein expression (Figure S2A). Analyzing the correlation of other phosphosites, we observed that sites on numerous FGFR pathway members were tightly linked with FGFR inhibitor sensitivity (Figure 1C). For instance, activating sites on MEK1/2, ERK1/2 and CDK2 (T160), and substrates of these kinases that regulate the cell cycle (e.g. CDC23 and SKP2), transcription (ERF and SP1) and translation (EIF4G1) all correlated with FGFR KI responses (Figure 1C), representing a pathway-based drug response signature for FGFR inhibitors.
Unbiased GSEA of kinome features defines pathway-based drug response signatures
To identify pathway-based drug response signatures using a quantitative and statistical framework, we applied GSEA with 327 cancer-relevant Reactome pathways as the gene sets (Fabregat et al., 2018; Kim et al., 2012; Subramanian et al., 2005) (see ‘STAR Methods’ and Table S1). This yielded normalized enrichment scores (NES) for 275 of the 327 pathways, ranking them for their association with sensitivity (positive NES) or resistance (negative NES) to each drug. Here, the selective FGFR and EGFR inhibitors pazopanib and lapatinib enriched pathways associated with FGFR and cell cycle activation or pathways associated with EGFR, PI3K and NF-κB activation (Figure S2B, S2C and Table S3). This analysis also identified pathways known to promote resistance to FGFR and EGFR inhibitors (i.e. with a negative NES) such as interleukin signaling (JAK-STAT pathway, Figure S2B) (Casas-Selves et al., 2012) and Wnt-signaling (Figure S2C) (Jones et al., 2016); while this analysis highlights the power of unbiased GSEA to identify pathways related to well characterized FGFR and EGFR inhibitors, our dataset should similarly identify signaling pathways related to the activity of other less well-studied KIs. Analyzing drug response pathways of the clinical HCC drugs sorafenib, regorafenib and lenvatinib confirmed that these drugs are effective when FGFR and cell cycle pathways are active, and ineffective when survival pathways such as interleukin and NF-κB signaling are engaged (Figure 1D). In contrast, the clinical AXL and MET inhibitor, cabozantinib, correlated less well with cell cycle pathway activity. To evaluate if our pathway-based drug response signatures can be detected in clinical HCC specimens, we analyzed four tumor-normal adjacent liver (NAL) pairs with kinobead/LC-MS. Gratifyingly, performance of our kinobead protocol in HCC tissue was comparable to our HCC cell line experiments and we quantified 2151 kinase phosphosites on 286 kinases, as well as 680 kinase interactors (Table S5, Figure S3A-C). We applied GSEA to identify Reactome pathways upregulated in tumors over NAL, correlating their pathway NES’ with those from the 17 HCC cell lines to identify drug response signatures for each tumor sample (see ‘STAR Methods’, Table S5). Pathway-based signatures of clinical HCC drugs were highly enriched in specific tumors (Figure 1E). For instance, HCC case 4 showed enrichment of pathways that specify sorafenib and regorafenib sensitivity (r of ~0.4), as well as sensitivity to FGFR inhibitors. In contrast, CDK inhibitor pathway signatures were highly enriched in three out of four tumors (r of ~0.4 to ~0.5, Figure 1E), including flavopiridol that is currently in clinical trials in HCC (Shen et al., 2019).
The HCC cell EMT state broadly impacts responses to kinase inhibitors
To identify the principal pathways that control responses to a broad range of clinical and pre-clinical KI drugs, we explored similarities in response pathways among all 299 drugs tested. We classified drugs into 11 KI clusters with similar pathway signatures and calculated mean NES values for 34 representative Reactome terms from the larger panel of 275 scored pathways, followed by unsupervised hierarchical clustering (see ‘STAR Methods’, Figure 2A, S4A, Table S3); this produced a clear separation into two distinct groups. KIs in clusters 5-7 and 9-11 formed one group with positive NES values for pathways commonly overexpressed in rapidly proliferating cells, including FGFR-, IGF1R-, cell cycle-, and mitosis-related pathways. This group contained 199 drugs, mainly inhibiting BRAF, FGFR isoforms, the IGF1R, and cell cycle-related kinases (PLK1, CDKs, CHEK1/2), and had negative enrichment scores for pathways related to MET, TGF-β, cytokine and NF-kB signaling (Figure 2A) – pathways that are known to regulate the EMT, an important mechanism of cancer cell metastasis and drug resistance (Kalluri and Weinberg, 2009; Shibue and Weinberg, 2017). Conversely, 100 compounds in clusters 1-4 and 8 (e.g. EGFR, MET and SRC KIs) showed positive enrichment scores for these same EMT-associated pathways and negative scores for FGFR- and cell cycle-related terms (Figure 2A). This opposing behavior of EMT pathway activation and KI drug response suggested: 1) the presence of mesenchymal HCC cells in our panel, and that 2) the EMT promotes resistance to two-thirds of the tested KI drugs. To test if our panel contains cell lines in different EMT states, we examined the mRNA expression of 50 important EMT and stem cell markers in the 17 HCC cell lines (Barretina et al., 2012) (Table S1). Indeed, semi-supervised hierarchical clustering of EMT markers classified the panel into seven epithelial and ten mesenchymal lines (Figure 2B and Table S1).
Figure 2. The EMT state defines HCC drug resistance phenotypes and kinase signaling network activity.
(A) Clustering of mean wNES values for 34 representative Reactome pathways across the 299 drugs grouped into 11 classes with similar response pathway signatures (see Figure S4A and Table S3 for complete Reactome pathway terms).
(B) Hierarchical clustering of EMT marker mRNA expression in the 17 CCLE HCC lines (data for 50 EMT markers in Table S1).
(C) Difference in cell motility between epithelial and mesenchymal HCC cells (wound healing assay, Student’s T-test, P = 0.02). We tested 16 of the 17 CCLE HCC lines and the mesenchymal FOCUS line.
(D) Kinases (circles, left) and their phosphorylation sites (right) significantly overexpressed in epithelial or mesenchymal HCC lines (BH-FDR = 0.05). Kinases with a log2 MS ratio >2 are labeled. The largest phosphosite ratio between EMT phenotypes for each kinase is plotted and kinases with activating phosphorylation sites are highlighted.
Because EMT is typically associated with increased cell motility (Lamouille et al., 2014), we also assayed the cell migration of 16 of the 17 HCC lines as well as the mesenchymal FOCUS HCC line, confirming that mesenchymal lines exhibited significantly enhanced wound closure compared to epithelial lines at 24 h (two sample T-test: P = 0.02, Figure 3C). Furthermore, comparing our HCC line classification by drug response (Figure S1D) with our classification by EMT marker expression (Figure 2B), we found that epithelial lines were highly enriched in the drug sensitive cluster (6 out of 7, hypergeometric T-test: P = 0.0037).
Figure 3. FZD2-AXL-NUAK1/2 signaling drives HCC cell EMT.
(A) Kinase protein (left) and phosphosite (right) expression differences in FOCUS AXL RNAi cells over WT cells overlaid on the human kinome dendrogram. The top 20 highest regulated kinases are labeled. The phosphorylation site with the highest absolute MS ratio on each kinase was plotted.
(B) Comparing AXL and FZD2 RNAi-dependent kinase expression changes in FOCUS cells with EMT-dependent kinase expression in the 17 HCC line panel (all kinases with MS ratio >4-fold).
(C) Log2 MS ratios of the nine common RNAi- and EMT-dependent kinases (see (B))
(D) qPCR of AXL mRNA in FOCUS STAT3 RNAi cells.
(E) Western blot of pSTAT3-Y705 in FOCUS AXL RNAi cells.
(F) Wound healing assay in FOCUS AXL RNAi cells compared to control shRNA cells.
(G) Wound healing assay in FOCUS NUAK1 and NUAK2 RNAi cells compared to controls.
(H) Comparing the effect of NUAK1 RNAi, NUAK2 RNAi and AXL RNAi on kinase expression in FOCUS cells (MS ratio >4-fold).
(I) Change of protein expression of AXL, NUAK1, and NUAK2 with three kinases (AKT1, SRC and MERTK) as internal controls upon knockdown of either of these kinases.
(J) qPCR of mRNA changes in EMT markers upon knockdown of either NUAK1 or NUAK2 in FOCUS cells.
A comprehensive map of the EMT state-associated kinome
To identify kinases that could be exploited as drug targets to block the EMT and overcome drug resistance, we applied T-test statistics to our dataset of MS intensity values in epithelial (n = 7) vs mesenchymal (n = 10) HCC cells, identifying 101 kinases, 380 kinase phosphosites, and 938 other phosphoproteins that differed significantly in expression between the two EMT phenotypes (Figure 2B, 2D and Table S2). The protein expression of 66 out of 101 EMT kinases was upregulated in mesenchymal cells, including the clinically important cabozantinib targets AXL (>100-fold) and MET (5-fold) (Abou-Alfa et al., 2018), the receptor tyrosine kinase EPHB2 (>100-fold) and the non-receptor kinases FYN, AKT3, CAMK1D, NUAK1 and NUAK2 (all > 4-fold, Figure 2D). The EMT-associated phosphokinome revealed that AXL’s kinase activity, but not MET’s, was increased in mesenchymal HCC cells, suggesting that AXL plays a more important role in EMT than MET. EPHB2, CAMK1D, FYN and 18 other kinases were also highly activated in mesenchymal HCC cells, indicating that these kinases promote the EMT (Figure 2D, S4B and Table S2). Conversely, epithelial HCC cells showed increased expression of 35 kinases, including the lenvatinib targets FGFR3 and 4 (~15-fold), and the sorafenib and regorafenib target BRAF (3-fold, Figure 2D). Other highly upregulated kinases (> 4-fold) included NEK3, CDK3, PLK1 and CHEK1 that have important roles in cell cycle and DNA damage response (DDR) signaling. Additionally, we identified activating phosphosites on kinases and specific kinase substrates enriched in the epithelial cell phosphoproteome that confirmed increased activity of the cell cycle through CDK2 and mitogenic signaling through FGFR3/4, BRAF and MAPK1/3 (Figure 2D, S4C and Table S2).
AXL drives reprogramming of the EMT-associated kinome
Our kinome profiling data revealed that activating phosphorylation sites on AXL and the protein itself are highly enriched in mesenchymal over epithelial HCC cells (Figure 2D and S4B). AXL is an important player in cancer cell EMT (Gay et al., 2017) and the development of tumor metastasis and drug resistance in HCC (Lee et al., 2014; Liu et al., 2016), as highlighted by the recent success of the AXL and MET inhibitor, cabozantinib, as a second-line treatment for sorafenib-resistant HCCs (Abou-Alfa et al., 2018). However, no detailed proteomics studies of AXL signaling have been published to date. Because such studies may reveal important AXL pathway components that can serve as EMT markers and molecular targets to break drug resistance, we used RNAi to knock down AXL in the FOCUS cell line, a widely used mesenchymal HCC cell model (Figure S4D) (Gujral et al., 2014). Western blot analysis of four EMT marker proteins and qPCR quantification of 43 EMT marker mRNAs confirmed that AXL RNAi induced widespread expression changes indicating reversal of the EMT (Figure S4E-G). Next, we compared the FOCUS AXL RNAi line to the wild-type (WT) control using kinobead/LC-MS and found that 159 kinases and 187 phosphosites on 104 kinases significantly changed in expression (T-Test result, BH-FDR = 0.05, n = 6 each, Figure 3A, Table S6). Notably, among kinases that decreased most with AXL RNAi in FOCUS cells (MS ratio >4-fold), 13 overlapped with the EMT state-associated kinome in the 17 HCC lines (Figures 3B, S4H and S4I), including the mesenchymal kinases NUAK1 and 2, FYN, EGFR and MET (Figure 3C). Concurrently, kinases associated with the epithelial state such as FGFR2/3 and CHEK2 increased in expression in AXL RNAi cells (Figure 3A, S4H and Table S6).
FZD2 is a master regulator of AXL expression and EMT-associated kinome rewiring
Having identified various kinases downstream of AXL, we next sought to identify upstream signaling components that could regulate AXL expression and the EMT in HCC cells. We found previously that AXL mRNA tightly co-expresses with FZD2 mRNA in various cancer cell lines and that this G-protein coupled receptor for WNT5A/B regulates HCC cell EMT via a FYN/STAT3-dependent pathway (Gujral et al., 2014). To investigate a possible functional connection between FZD2, AXL, and other downstream pathway components, we profiled our FOCUS FZD2 RNAi cell model with kinobead/LC-MS. Indeed, FZD2 knockdown affected the expression of 118 kinases, including AXL and several of its effector kinases (T-Test result, BH-FDR = 0.05, n = 6 each, Figure S5A and S5B, Table S6). Among kinases most affected by AXL and FZD2 RNAi (MS ratio >4-fold), 61% are transcriptional targets of both AXL and FZD2 signaling (Figure 3B). Importantly, comparing kinases affected by RNAi with those associated with the EMT state in our 17 HCC lines identified nine common kinases, and among those, NUAK1 and NUAK2 were particularly sensitive to AXL and FZD2 RNAi (Figure 3B and C). To investigate if AXL expression is connected to FZD2’s activation of FYN and STAT3 (Gujral et al., 2014), we knocked down STAT3 in FOCUS cells and found that AXL mRNA levels decreased drastically (Figure 3D). We also found that AXL RNAi in FOCUS cells affects the phosphorylation of STAT3 at its activating site, pY705 (Figure 3E).
NUAK1 and NUAK2 reinforce AXL expression and promote the HCC cell EMT
To identify candidate drug targets that can reverse the EMT and overcome drug resistance, we next looked for kinases downstream of the FZD2-FYN/STAT3-AXL signaling module. The nuclear serine/threonine kinases NUAK1 and 2 were tightly associated with the mesenchymal state in the 17 HCC lines and their expression greatly decreased with AXL and FZD2 RNAi (20 to 30-fold, Figure 3C). NUAK1 is implicated in tumor metastasis (Chen et al., 2013) and both NUAK1 and 2 were shown to promote tumor cell survival (Legembre et al., 2004; Port et al., 2018). To test if NUAK1 and 2 are bona fide drivers of the HCC cell EMT, we generated stable FOCUS NUAK1 or NUAK2 RNAi cell lines (Figure S5C); knockdown reduced cell migration by 60-70%, compared to a 25% reduction in FOCUS AXL RNAi cells (Figure 3F and G), suggesting a prominent role of NUAK1 and 2 in HCC cell EMT. Next, we compared FOCUS NUAK1 and 2 RNAi cells to WT cells using kinobead/LC-MS and found that NUAK1 or NUAK2 knockdown affected the expression of 150 and 135 protein kinases, respectively (T-test result, BH-FDR = 0.05, n = 6 each, Figure S5D, Table S6). Surprisingly, the kinome profiles of NUAK1 and 2 RNAi cells were very similar. The expression of 43 out of 52 highly regulated kinases (MS ratio >4-fold) was affected in both knockdown lines and the LFQ-MS ratios of all affected kinases had a Pearson’s r value of 0.91 (Figure 3H and Table S6). When we compared AXL RNAi with NUAK1 and 2 RNAi, 85% of highly affected kinases (MS ratio >4-fold) were common between AXL, NUAK1 or NUAK2 RNAi experiments and the MS ratios of all regulated kinases showed a r-value of 0.93 (Figure 3H). Furthermore, AXL expression levels were greatly decreased by either NUAK1 or NUAK2 RNAi and vice versa (Figure 3I). This hinted at a positive feedback mechanism where NUAK kinases promote AXL expression or protein stability. Indeed, our qPCR analysis of EMT markers confirmed that AXL, CD44 and MMP2 were decreased in FOCUS NUAK RNAi cells (Figure 3J). Additionally, ectopic expression of NUAK1 in epithelial-type C3A and SNU398 cells caused at least partial EMT as indicated by increased expression of MMP2 and MMP9, AXL and SERPINE1 (Figure S5E, S5F and S5G).
AXL-NUAK1/2 signaling promotes resistance to cell cycle checkpoint kinase inhibitors
To identify combinatorial treatment strategies targeting mesenchymal and drug resistant HCC cells, we next analyzed drug sensitivity pathways that become activated upon AXL and NUAK1/2 inhibition and EMT reversal. We found that FZD2, AXL and NUAK1/2 RNAi increased expression of FGFR isoforms and activation of mitogenic signaling and cell cycle cues (Table S6, Figure S6A). We also observed increased cell cycle activity among epithelial lines in the 17 HCC line panel (Table S3, Figure S6B), that was accompanied by elevated activity of kinases promoting DDR signaling and cell survival under conditions of replication stress (Dobbelstein and Sorensen, 2015). For instance, we observed activation of CHEK1 and 2 along with increased phosphorylation of ATR, CHEK1/2 and WEE1 substrates (Figure 4A, S6C). Similarly, AXL RNAi in FOCUS cells activated the DDR kinases CHEK1 and WEE1, increasing phosphorylation of their substrates CDK11B and CDK2 (Figure 4B and S6D). These findings suggest that AXL and NUAK1/2 suppress the cell cycle and DDR signaling and protect HCC cells from replication stress. Concurrently, the efficacy of KI drugs that target cell cycle-related kinases was strongly reduced in the mesenchymal fraction of the 17 HCC lines (Figure 4C). Thus, we found that the effects of the broadly active CDK, PLK1 and CHEK1/2 inhibitors (KI cluster 9) were among the inhibitor classes most dependent on the EMT state (Figure 4C and S1D), indicating that epithelial HCC cell survival may indeed depend on DDR signaling. We reasoned, therefore, that combinatorial inhibition of AXL or NUAK1/2, as well as cell cycle checkpoint kinases could efficiently kill mesenchymal HCC cells (Figure 4D) (Lin et al., 2017). To test this strategy, we cotreated the mesenchymal and drug resistant SNU449 line (Table S1 and S3) with the selective NUAK1/2 inhibitor WZ4003 (Banerjee et al., 2014) and the CHEK1/2 inhibitor AZD7762. Additionally, we used the CDK inhibitor dinaciclib to test if elevated, EMT state-dependent, cell cycle activity translates into increased efficacy of such drugs. Remarkably, cotreatment reproducibly decreased EC50s, i.e. ~3-fold and ~2-fold for both CHEK and CDK inhibitors (Figure 4E and 4F). This effect was dose-dependent for both AZD7762 and WZ4003 in SNU449 cells, and still significant at 1 μM of WZ4003 (Figure 4G). These encouraging results led us to establish SNU449 NUAK1 or NUAK2 RNAi cells (Figure S6E). Gratifyingly, treating SNU449 NUAK RNAi cells and WT controls with varying doses of AZD7762 and dinaciclib recapitulated the drug co-treatment results (~3- to 4-fold decrease in EC50s, Figure 4H). To consolidate our findings, we tested our FOCUS NUAK and AXL RNAi cell lines against AZD7762 and dinaciclib. As in SNU449 cells, NUAK1/2 RNAi FOCUS cells were sensitized to kinase inhibition up to ~5-fold (Figure 4J).
Figure 4. Perturbation of AXL or NUAK1/2 function reverses the EMT and increases HCC cell sensitivity to cell cycle- and DDR kinase inhibitors.
(A) Activating phosphosites on CHEK1 and 2 and their substrates enriched in seven epithelial vs. ten mesenchymal HCC cell lines.
(B) Phosphorylation sites on CHEK1 and its substrates indicating activation of this kinase in FOCUS AXL RNAi cells over WT that are also associated with EMT in the 17-cell line panel.
(C) Difference of mean AUC values in epithelial vs. mesenchymal HCC cells for the 299 KI panel for the 11 KI pathway clusters (see Figure S4A and Table S3).
(D) Strategy for reversing the EMT and increasing drug sensitivity through NUAK1/2 and AXL inhibition.
(E) and (F) EC50-curves of drug co-treatment experiments in SNU449 cells (n = 4, error bars are S.D.).
(G) Heatmap of drug synergy and titratability of AZD7762 and WZ4003 in SNU449 cells.
(H) and (I) Bar plots of drug treatment experiments with AZD7762 and dinaciclib in the SNU449 NUAK1/2 RNAi cell lines and FOCUS NUAK1/2 and AXL RNAi cell line, (n = 4, error bars are the 95% confidence interval (CI)).
See also Figure S6
DISCUSSION
We introduced a kinome-centric pharmacoproteomics approach integrating kinome activity profiles and drug responses to identify signaling pathways underlying HCC drug sensitivity and resistance. Our data indicate that the phospho-activation states of kinases and their interactors are often better predictors of drug response than mRNA or protein expression, and that phosphorylation events spanning the broader signaling network can predict responses to drugs that target other kinases within these pathways. Our unbiased identification of pathway-based drug response signatures revealed kinase drug targets and suggested rational drug combinations to treat HCC. We conclude that proteome and PTM expression data is an important component currently missing in pharmacogenomics biomarker and drug target discovery (Chambliss and Chan, 2016). We show that our kinobead/LC-MS platform can measure kinome activity in individual patient HCC’s, revealing predictive drug response signatures that might be used to rationally select specific therapies. Our approach can be easily applied to other cell line models, organoids, or organotypic slice cultures (Meijer et al., 2017) to identify markers of drug response, classify drug-sensitive and -resistant disease subtypes, and candidate drug targets. Reproducible and amenable to lab automation, our platform can be easily scaled up to profile hundreds of clinical tumor samples or cell lines. Our study provides a comprehensive map of kinase-dependent signaling networks that define the mesenchymal and epithelial state in cancer; our quantitative measurements of kinome abundance and phosphorylation sites identified 199 kinases associated with EMT phenotypes, providing a clearer picture of the signaling mechanisms underlying the EMT. We identified specific signaling modules that promote the mesenchymal drug-resistant phenotype and demonstrated that our approach can be used to rationally select drug combinations that increase drug sensitivity and cell killing. In addition, we found that FZD2-AXL-NUA1/2 signaling drives HCC cell kinome rewiring and EMT in a FYN-STAT3-dependent manner. Our results establish that NUAK1 and NUAK2 can regulate HCC cell resistance to targeted therapy, highlighting their potential as drug targets to overcome HCC therapy resistance. Our dataset of drug response signatures and EMT-associated signaling pathways is a valuable resource for functional studies of HCC cell signaling and is accessible through a web resource (see ‘STAR Methods’) that allows real-time data interrogation and visualization.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shao-En Ong (shaoen@uw.edu)
Materials Availability:
This study did not generate new materials.
Data and Code Availability:
The MS .raw files and MaxQuant output files generated during this study are available at the MassIVE Repository of the University of California, San Diego (Dataset ID#: MSV000083236). A Shiny app for real time interrogation of the pharmacoproteomics data generated in this study can be accessed at https://quantbiology.org/hcckinome. This study did not generate new code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and tissue culture conditions
C3A, HepG2, SNU398, PLC/PRF/5, Hep3B2.1-7, SKHep1, SNU475, SNU387, SNU423 and SNU449 cell lines were purchased from the American Type Culture Collection (ATCC). NCI-H684, SNU761, SNU886 and SNU878 were purchased from the Korean Cell Line Bank (KCLB). JHH4, JHH6 and HuH-7 cells were purchased form the JRCB Cell Bank. FOCUS cells were obtained from the Laboratory of J. Wands, Brown University (He et al., 1984). All cells were grown at 37°C under 5% CO2, 95% ambient atmosphere. Fifteen cryofrozen cell stocks were generated from the original vial from the cell bank (Passage 3). Experiments were performed with cells at <10 passages from the original vial. All cell media used were supplemented with 100x penicillin-streptomycin-glutamine (Thermo Fisher Scientific, Waltham, MA). FOCUS and HuH-7 cells were grown in Dulbecco’s minimum essential medium (DMEM) supplemented with 10% FBS (VWR Life Science, Seradigm). C3A, HepG2, SNU398, PLC/PRF/5, Hep3B2.1-7, SKHep1, SNU475, SNU387, SNU423 and SNU449 were grown in the ATCC-recommended medium. JHH4 cells were grown in Eagle’s minimum essential medium (MEM), JHH6 cells in William’s E medium and NCI-H684, SNU761, SNU886 and SNU878 in RPMI 1640 medium all supplemented with 10% FBS. Cells were harvested when reaching 90% confluency.
Human HCC and normal adjacent liver specimens
Primary human HCCs with paired non-tumor livers were obtained from patients undergoing liver resection at the University of Washington Medical Center (Seattle, WA, USA). All patients in this study prospectively consented to donate liver tissue for research under the Institutional Review Board protocols #1852. Once the specimens were collected under the direction of Pathology representatives, they were snap-frozen in liquid nitrogen and stored at −80°C until further processing. Patent samples were characterized as follows:
| Case # | Age | Gender | Tumor Description |
Etiology | Histological Grade |
Prior Treatment |
|---|---|---|---|---|---|---|
| 1 | 56 | M | HCC, 2.5 cm | HBV | G2 | None |
| 2 | 77 | F | Mixed ICC/HCC, 3.7 cm | HCV | G2 | None |
| 3 | 61 | M | HCC, 8 cm | HCV | G2 | Recurrent after Resection |
| 4 | 67 | M | HCC, 3 cm | EtOH Cirrhosis | G2 | Recurrent after Resection |
METHOD DETAILS
RNAi knockdown experiments
All lentiviral vectors encoding different shRNAs (STAT3, FYN, FZD2, AXL, NUAK1 and 2) in a pGIPZ vector were purchased from OpenBiosystems (Dharmacon, Lafayette, CO). Cell lines were transfected with shRNA constructs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and 48 h post-transfection selected with 4 μg/ml puromycin (Invitrogen). The clones were sorted by FACS and screened for target mRNA knockdown by Western blot or qPCR analysis (see ‘Western blot analysis and antibodies’ and ‘Quantitative real-time PCR (qPCR) analysis of mRNA expression’ below). Stable cell lines were maintained in DMEM (see ‘Cell lines and cell culture conditions’) supplemented with 2 μg/ml puromycin.
Ectopic expression of NUAK1
The expression construct encoding for full length NUAK1 (NM_ 014840.2) in a lentiviral plasmid (pReceiver-Lv242) was purchased from Genecopoeia (Rockville, MD). Cell lines were transfected with NUAK1 plasmid construct using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and 48 h post-transfection selected in 4 μg/ml puromycin (Invitrogen). Stable cell lines were maintained in DMEM (see ‘Cell lines and tissue culture conditions’) supplemented with 2 μg/ml puromycin.
Western blot analysis and antibodies
Antibodies used were anti-phospho-Stat3 (Tyr705) (Cell signaling Technology, Cat # 9145), anti-E-cadherin (Cell signaling Technology, Cat # 3195), anti-Occludin (BD Transduction Laboratories, Cat # 611091), anti-Vimentin (Millipore, Cat #CS207806), anti-β-Actin (Sigma, Cat #A1978), anti-GAPDH (Santacruz Biotechnology, Cat # Sc-365062), anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Cell signaling Technology, Cat #4370), anti-Axl (C89E7) (Cell signaling Technology, Cat #8661). Briefly, cells were rinsed in phosphate buffered saline (PBS) and lysed in lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100 (v/v), 2 mM EDTA, pH 7.8 supplemented with 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Protein concentrations were determined using the BCA protein assay (Pierce, Rockford, IL) and immunoblotting experiments were performed using standard procedures. For quantitative immunoblots, primary antibodies were detected with IRDye 680-labeled goat-anti-rabbit IgG or IRDye 800-labeled goat-anti-mouse IgG (LI-COR Biosciences, Lincoln, NE) at 1:5000 dilution. Bands were visualized and quantified using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Quantitative real-time PCR (qPCR) analysis of mRNA expression
Cells were seeded in 6-well plates 24 h prior to isolation of total RNA using a RNeasy Mini Kit (QIAGEN, Santa Clara, CA). mRNA levels for EMT-related genes were determined using the validated primer sets (SA Biosciences Corporation, Frederick, MD). Briefly, 1 μg of total RNA was reverse transcribed into first strand cDNA using an RT2 First Strand Kit (SA Biosciences). The resulting cDNA was subjected to qPCR using human gene-specific primers for EMT-associated genes, and two housekeeping genes (GAPDH and ACTB). The qPCR reaction was performed with an initial denaturation step of 2 min at 95°C, followed by 5 s at 95°C and 30 s at 60°C for 40 cycles using a Biorad CFX384 system (Biorad, Hercules, CA). The mRNA levels of each gene were normalized relative to the mean levels of the two housekeeping genes and compared with the data obtained from unstimulated, serum-starved cells using the 2-ΔΔCt method. According to this method, the normalized level of a mRNA, X, is determined using Equation 1:
| (1) |
where Ct is the threshold cycle (the number of the cycle at which an increase in reporter fluorescence above a baseline signal is detected), GOI refers to the gene of interest, and CTL refers to a control housekeeping gene. This method assumes that Ct is inversely proportional to the initial concentration of mRNA and that the amount of product doubles with every cycle.
Kinetic wound healing assay
A wound healing assay was used to study the effect of AXL and NUAK1/2 RNAi knockdown on FOCUS cell migration and to score the cell motility of the 17 wt HCC cell lines. Briefly, cells were plated on 96-well plates (Essen Image Lock, Essen Bioscience, Ann Arbor, MI), and a wound was scratched with a wound scratcher (Essen Instruments). Wound confluence was monitored with Incucyte Live-Cell Imaging System and software (Essen Instruments). Wound closure was observed every 2 hours for 24-72 hours by comparing the mean relative wound density of at least three biological replicates in each experiment.
High throughput growth inhibition assay
The high throughput growth inhibition assay was performed by the QUELLOS high throughput screening facility of the University of Washington (https://iscrm.uw.edu/research/core-resources/quellos-high-throughput-screening-core/) using 299 compounds of a Selleckchem kinase inhibitor Library (http://www.selleckchem.com/screening/kinase-inhibitor-library.html, Selleckchem, Houston, TX). Briefly, the assay was performed in a 384-well plate format in biological duplicate. Compounds were applied at 7 different concentrations ranging from 10 μM to 10 nM and cell viability was measured after 72 h of incubation using the CellTiter-Glo 2.0 assay (Promega, Madison, WI).
Inhibitor co-treatment and AXL/NUAK RNAi growth inhibition
1800 cells/well were seeded onto white flat bottom half area 96-well plates (Greiner Bio-One, Kremsmuenster, AT) in 50 μl of growth medium and allowed to attach in an incubator for 24 h. Then the drugs in DMSO and/or DMSO vehicle controls as 11X solutions in growth medium were added to a total volume of 55 μl and 0.1% DMSO final. The cells were grown in an incubator for another 72 h. Then, 55 μl of CellTiter-Glo 2.0 (Promega, Madison, WI) reagent/well were added according to the manufacturer’s instructions and luminescence was quantified with a SpectraMax 190 plate reader (Molecular Devices, San Jose, CA). The CHEK1/2 inhibitor AZD7762 (Selleckchem, Houston, TX) and the NUAK1/2 inhibitor WZ4003 (Tocris Bioscience, Minneapolis, MN) were applied at 8 different concentrations between 10 μM and 4.6 nM (3-fold dilution steps) and Dinaciclib (Selleckchem) was applied at 8 different concentrations between 1 μM and 0.5 nM (3-fold dilution steps). Experiments were performed in four biological replicates. Growth inhibition curves were fitted using the GraphPad Prism software package (V5.0a) with a least-squares nonlinear regression model for curve fitting (One site - Fit logIC50 function).
Protein extraction from human HCC and normal adjacent liver specimens
Frozen tumor specimens of ca. 100 mg wet weight were ground into a fine powder using the CryoGrinder Kit from OPS Diagnostics (Lebanon, NJ). The powder was then added to ice cold mod. RIPA buffer containing phosphatase and protease inhibitors (see ‘Kinase affinity enrichment and on-bead digestion’ below), vortexed 10 times at max. speed and clarified at 21,000 rcf and 4°C for 20 min. Protein yields from specimens ranged from 5% to 10% depending on the degree of fibrosis.
IMAC phosphopeptide enrichment
IMAC phosphopeptide enrichment was performed according to the published protocol (in-tube batch version) with the following minor modifications (Villen and Gygi, 2008). 20 μl of a 50% IMAC bead slurry composed of 1/3 commercial PHOS-select iron affinity gel (Sigma Aldrich, St Louis, MO), 1/3 in-house made Fe3+-NTA Superflow agarose and 1/3 in-house made Ga3+-NTA Superflow agarose was used for phosphopeptide enrichment (Ficarro et al., 2009). The IMAC slurry was washed three times with 10 bed volumes of 80% aq. ACN containing 0.1% TFA and phosphopeptide enrichment was performed in the same buffer.
Peptide and phosphopeptide desalting with StageTips
Peptides and phosphopeptides were desalted using C18 StageTips according to the published protocol with the following minor modifications for phosphopeptides (Rappsilber et al., 2007). After activation with 50 μl methanol and 50 μl 80% aq. ACN containing 0.1% TFA the StageTips were equilibrated with 50 μl 1% aq. formic acid. Then the peptides that were reconstituted in 50 μl 1% aq. formic acid were loaded and washed with 50 μl 1% aq. formic acid. The use of 1% formic acid instead of 5% aq. ACN containing 0.1% TFA reduces the loss of highly hydrophilic phosphopeptides.
Preparation of optimized kinobead mixture
The seven kinobead affinity reagents used were synthesized in-house as described previously (Golkowski et al., 2014; Golkowski et al., 2017b; Golkowski et al., 2020). For optimal coverage of the human kinome an optimized mixture of the seven kinobead reagents was prepared as previously described (Golkowski et al., 2020). Briefly, 1 ml of reagent 1, 0.5 ml of reagents 2, 3 and 7, respectively, and 0.25 ml of reagents 4, 5 and 6, respectively, were mixed to yield 3.25 ml of the complete kinobead mixture. All reagents were a 50% slurry in 20% aq. ethanol.
Kinase affinity enrichment and on-bead digestion
Kinase affinity enrichment and on-bead digestion was performed as previously described (Golkowski et al., 2020). Briefly, three micro tubes containing 35 μl of a 50% slurry of the in-house-made, optimized kinobead mixture in 20% aq. ethanol were prepared for each pulldown experiment. The beads were washed twice with 350 μl modified RIPA buffer (50 mM Tris, 150 mM NaCl, 0.25% Na-deoxycholate, 1% NP-40, 1 mM EDTA and 10 mM NaF, pH 7.8). 1 mg of protein extract in mod. RIPA buffer containing HALT protease inhibitor cocktail (100x, Thermo Fisher Scientific, Waltham, MA) and phosphatase inhibitor cocktail II and III (100x, Sigma-Aldrich, St Louis, MO) were added to the first tube. The mixture was incubated on a tube rotator for 1h at 4°C and then the beads were spun down rapidly at 2000 rpm on a benchtop centrifuge (5s). The supernatant was pipetted into the next tube with kinobeads for the second round of affinity enrichment. The procedure was repeated once more for a total of three rounds of affinity enrichment. After removal of the supernatant, the beads were rapidly washed twice with 350 μl of ice-cold mod. RIPA buffer and three times with 350 μl ice-cold tris-buffered saline (TBS, 50 mM tris, 150 mM NaCl, pH 7.8) to remove detergents. 100 μl of the denaturing buffer (20% trifluoroethanol (TFE)(Wang et al., 2005), 25 mM Tris containing 5 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP*HCl) and 10 mM chloroacetamide (CAM), pH 7.8), were added and the slurry briefly vortexed at low speed. At this stage, kinobeads from the three tubes are combined and heated at 95°C for 5 min. The mixture was diluted 2-fold with 25 mM triethylamine bicarbonate (TEAB), the pH adjusted to 8-9 by addition 1 N aq. NaOH; 5 μg LysC were added and the mixture agitated on a thermomixer at 700 rpm at 37°C for 2 h. Then 5 μg MS-grade trypsin (Thermo Fisher Scientific, Waltham, MA) were added, and the mixture agitated on a thermomixer at 700 rpm at 37°C overnight. 600 μl of 1% formic acid was added and the mixture acidified by addition of another 6 μl of formic acid to yield 1.2 ml peptide solution in total. An aliquot of 120 μl (10%) of the peptide solution was desalted using StageTips and analyzed in single nanoLC-MS/MS runs for protein quantification. The remaining peptide solution (90%) was dried under vacuum at RT on a SpeedVac. 300 μl of 70% aq. ACN + 0.1 % TFA was added to each tube, the mixture vortexed, and sonicated in a bath sonicator until dried peptide residue dissolved. In case the dried residue could not be fully resuspended, additional 0.1% aq. TFA can be added in 10 μl increments until dissolved. The solution was subjected to IMAC phosphopeptide enrichment protocol and desalted using StageTips (see ‘IMAC phosphopeptide enrichment’ and ‘Peptide and phosphopeptide desalting with StageTips’ above).
nanoLC-MS/MS analyses
The LC-MS/MS analyses were performed as described previously with the following minor modifications (Golkowski et al., 2017b; Golkowski et al., 2020). Peptide samples were separated on a Thermo-Dionex RSLCNano UHPLC instrument (Sunnyvale, CA) using 20 cm long fused silica capillary columns (100 μm ID) packed with 3 μm 120 Å reversed phase C18 beads (Dr. Maisch, Ammerbuch, DE). For whole peptide samples the LC gradient was 120 min long with 10–35% B at 300 nL/min. For phosphopeptide samples the LC gradient was 120 min long with 3–30% B at 300 nL/min. LC solvent A was 0.1% aq. acetic acid and LC solvent B was 0.1% acetic acid, 99.9% acetonitrile. MS data was collected with a Thermo Fisher Scientific Orbitrap Elite or Orbitrap Fusion Lumos Tribrid spectrometer. Data-dependent analysis was applied using Top15 selection with CID fragmentation.
Computation of MS raw files
Data .raw files were analyzed by MaxQuant/Andromeda (Cox et al., 2011) version 1.5.2.8 using protein, peptide and site FDRs of 0.01 and a score minimum of 40 for modified peptides, 0 for unmodified peptides; delta score minimum of 17 for modified peptides, 0 for unmodified peptides. MS/MS spectra were searched against the UniProt human database (updated July 22nd, 2015). MaxQuant search parameters: Variable modifications included Oxidation (M) and Phospho (S/T/Y). Carbamidomethyl (C) was a fixed modification. Max. missed cleavages was 2, enzyme was Trypsin/P and max. charge was 7. The MaxQuant “match between runs” feature was enabled. The initial search tolerance for FTMS scans was 20 ppm and 0.5 Da for ITMS MS/MS scans.
MaxQuant output data processing
MaxQuant output files were processed, statistically analyzed and clustered using the Perseus software package v1.5.6.0 (Tyanova et al., 2016). Human gene ontology (GO) terms (GOBP, GOCC and GOMF) were loaded from the ‘Perseus Annotations’ file downloaded on 01.08.2017. Expression columns (protein and phosphopeptide MS intensities) were log2 transformed and normalized by subtracting the median log2 expression value from each expression value of the corresponding data column. Potential contaminants, reverse hits and proteins only identified by site were removed. Reproducibility between LC-MS/MS experiments were analyzed by column correlation (Pearson’s r) and replicates with a variation of r > 0.25 compared to the mean r-values of all replicates of the same experiment (cell line or knockdown experiment) were considered outliers and excluded from the analyses. Data imputation was performed using a modeled distribution of MS intensity values downshifted by 1.8 and having a width of 0.2. For statistical testing of significant differences in expression, a two-sample Student’s T-test with Benjamini-Hochberg correction for multiple hypothesis testing was applied (FDR = 0.05). For statistical testing of the 17 HCC cell line data (EMT state-association) all biological replicates were used. For MS protein intensities this was n = 42 (epithelial cells) and n = 60 (mesenchymal cells). For MS phosphopeptide intensities this was n = 56 (epithelial cells) and n = 91 (mesenchymal cells).
Pharmacoproteomic AUC – MS intensity correlation
Mean LFQ-MS intensities values for each of the 17 cell lines were calculated using the imputed MS intensity data from Table S2. All proteomics features (n = 13935) were correlated with each compound’s AUC value across the cell line panel (n = 17) using the Pearson’s correlation coefficient resulting in a 13935x299 matrix of r-values (see Table S4). To rank proteomics feature the resulting r-values for each kinase inhibitor were sorted from low to high where negative r-values correspond to drug sensitivity (low AUC - high MS intensity) and positive r-values correspond to drug resistance (high AUC–high MS intensity).
Kinome-GSEA analysis with Reactome pathways
To obtain signaling pathways for GSEA analyses, we used the mapped identifier files ‘NCBI2Reactome_All_Levels.txt’ and ‘ReactomePathwaysRelation.txt’ from Reactome.org (downloaded 22th Oct., 2018). Pathways from the highest hierarchical levels were removed to exclude non-specific pathways. Subsequently, a regular expression match for patterns containing “kinase”, “signal”, “cell cycle”, “migration”, “cancer”, “dna repair”, “mitos” and “mitot” was used to extract 327 cancer relevant pathways (Table S3 ‘Reactome_Pathways’). Member genes of each pathway were mapped to unique identifiers in Table S3 to allow addition of phosphopeptide data. The Pearson correlation coefficients r from correlating drug response with MS intensities of kinome features (see ‘Pharmacoproteomic AUC – MS intensity correlation analyses’ above) were −(x) transformed for GSEA analyses, as the GSEA algorithm ranks features by their correlation with drug response. We used the Bioconductor package, fgsea (Sergushichev, 2016) (https://doi.org/10.1101/060012), with parameters: minSize = 10, maxSize = 500, gseaParam = 2 and nperm = 10000 to compute p-values and enrichment scores, including corrections for multiple hypothesis testing (BH FDR = 0.1).
Classification of KI drugs by correlation-clustering of kinome-GSEA wNES scores
For hierarchical clustering of compounds based on Reactome pathway analysis (Figure S4A), FDR-weighted Reactome pathway normalized enrichment scores (wNES = NES*(1-FDR)) were extracted for each compound (N = 299, Table S3 worksheet ‘299_Compound_NES_FDR’). Pairwise Pearson correlation coefficients (r) for wNES values for all compounds were calculated resulting in a 299X299 matrix of r-values. Clustering of the Pearson’s correlation coefficients using R (gplots::heatmap.2) identified 11 major groups (Figure 4A and Table S3 tab ‘Reactome_Pathway_Clusters’). For these 11 major groups we then calculated mean wNES values for 34 cancer-relevant, non-redundant Reactome pathways representative of the larger panel of Reactome pathways (Figure 2A). These 34 pathways were selected for having the largest difference in mean wNES across all 11 KI drug clusters among Reactome terms in the same overarching pathway theme. The result is a 11X34 matrix of mean wNES values (Figure 2A) that was subjected to hierarchical clustering using R (gplots::heatmap.2).
Kinome-GSEA in clinical HCC specimens and correlation of Pathway signatures
We calculated the difference of the mean imputed MS intensity values for the protein and phosphopeptide expression kinobead LFQ-MS data (Table S5) tumor vs. normal adjacent liver and ranked all proteomics features according to this difference of the mean (positive values was enriched in tumor, negative values was enriched in). We then applied our kinome-GSEA analysis and calculated FDR-weighted Reactome pathway enrichment scores (wNES values, see ‘Kinome-GSEA analysis with Reactome pathways’). We then calculated Pearson correlation coefficients, correlating the wNES values of all 299 KI drugs with the wNES values of the four HCC tumor/NAL pairs. High r-value then indicate enrichment of a KI pathway signature in tumors, and negative values their enrichment in NAL.
Functional phosphosites and kinase-substrate relationships
To determine the biological function of a phosphorylation site and the kinase-substrate relationship of a given phosphorylation site the PhosphoSite Plus datasets ‘Regulatory_Sites’ and ‘Kinase_Substrate_Dataset’ were searched against the 15 amino acid sequence windows centered on the corresponding phosphosite. Human, mouse and rat phosphorylation sites were all considered to assess the biological and biochemical consequences of phosphorylation. The datasets were downloaded from the PhosphoSite Plus webpage on the 31st of July, 2018 (https://www.phosphosite.org/)(Hornbeck et al., 2015).
Identification of protein kinase interactors
Protein kinase interactors were determined using the BioGRID database only considering protein-protein interactions for which two independent lines of evidence exist (Chatr-Aryamontri et al., 2017). To that end, the ‘BIOGRID-MV-Physical-3.5.165.tab2’ file was downloaded on October 6th, 2018 and mined for protein kinase interactions through matching against the gene name in the MaxQuant output files.
Determining kinase activation states
The activation state of a kinase was considered differentially regulated when either 1) regulatory sites on a kinase, 2) corresponding sites on a kinase or kinase interactor with a known kinase-substrate relationship, or 3) a known kinase interactor changed in expression between experiment conditions (T-test significant, see also ‘Functional phosphosites and kinase-substrate relationships’, ‘Identification of protein kinase interactors’, and ‘QUANTITATION AND STATISTICAL ANALYSIS’)
Kinome dendrograms
Kinome dendrograms were prepared using the KinMap web application (http://kinhub.org/kinmap/)(Eid et al., 2017).
Construction of interaction network graphs
Protein-protein interaction network graphs were plotted with the STRING web application (v10.0, https://string-db.org/)(Szklarczyk et al., 2015). Solid edges shown in network figures represent the ‘confidence’ in the existence of a physical interaction
QUANTITATION AND STATISTICAL ANALYSIS
Differences between sample populations were quantified with a two-tailed two sample Student’s T-test. For testing of proteomics data, or where indicated, BH correction for multiple hypothesis testing (FDR = 0.05) was applied.
Supplementary Material
Table S1. Drug Panel, HCC Cell EMT Markers and Kinase mRNA expression; Supplement to Figure 1-4. Identity and primary targets of the 299 kinase inhibitor panel. Log2 mRNA intensity of 50 EMT and stem cell markers as well as 509 protein kinases in the 28 CCLE HCC cell lines.
Table S2. Kinome Profiling 17 HCC Cell Lines; Supplement to Figure 1-4. MaxQuant protein groups and phospho(S/T/Y) output data obtained from kinome profiling of 17 HCC cell lines. Missing values were replaced by data imputation (see ‘STAR Methods’).
Table S3. Kinase Inhibitor HTS Results and kinome-GSEA Analysis; Supplement to Figure 1-4. Results of the HTS of 299 kinase inhibitors in 17 HCC cell lines. Summary table of AUC values. Summary of Reactome pathways used in the kinome-GSEA analysis. Compounds and cumulative NES for clusters from Figure S4A. FDR and NES values for all 299 compounds and 275 Reactome pathways.
Table S4. Kinome Proteomic Feature-AUC Correlation Results; Supplement to Figure 1. Results from Pearson correlation between mean MS intensity values of all quantified proteomics features (n = 13935) and kinase inhibitor AUCs (n = 299) across the 17 HCC cell line panel.
Table S5. Kinome Profiling of Human HCC Specimens and Kinome-GSEA; Supplement to Figure 1. MaxQuant protein groups and phospho(S/T/Y) output data from LFQ kinobead/LC-MS profiling of 4 paired (tumor/NAL) human HCC specimens. Kinome-GSEA of Reactome Pathways in the 4 paired HCC samples (see ‘STAR Methods’).
Table S6. Kinome Profiling FOCUS RNAi Models; Supplement to Figure 3 and 4. MaxQuant protein groups and phospho(S/T/Y) output data from kinome profiling of the FOCUS cell AXL, FZD2 and NUAK1/2 RNAi models.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-Stat3 (Tyr705) (D3A7) Rabbit mAb | Cell Signaling Technology | Cat# 9145, RRID: AB_2491009 |
| E-Cadherin (24E10) Rabbit mAb | Cell Signaling Technology | Cat# 3195, RRID: AB_2291471 |
| Mouse Anti-Occludin | BD Biosciences | Cat# 611091, RRID: AB_398404 |
| Mouse Anti-Vimentin Antibody, clone V9 | Millipore | Cat# MAB3400, RRID: AB_94843 |
| Mouse Anti-beta-Actin Monoclonal Antibody, Unconjugated, Clone AC-15 | Sigma-Aldrich | Cat# A1978, RRID: AB_476692 |
| GAPDH (G-9) Antibody, Mouse | Santa Cruz Biotechnology | Cat# sc-365062, RRID: AB_10847862 |
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) Rabbit mAb | Cell Signaling Technology | Cat# 4370, RRID: AB_2315112 |
| Axl (C89E7) Rabbit mAb | Cell Signaling Technology | Cat# 8661, RRID: AB_11217435 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| 4 Paired primary HCC and normal adjacent liver specimens, Human | Laboratory of Raymond S. Yeung, University of Washington | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Kinobead affinity capture reagents | Laboratory of Dustin J. Maly, University of Washington | Compounds #1, 2, 3, 4, 5, 6 and 7 (Golkowski et al., 2017a) |
| Dinaciclib | Selleckchem | Cat#: S2768, CAS: 779353-01-4 |
| AZD7762 | Selleckchem | Cat#: S1532, CAS: 860352-01-8 |
| WZ4003 | Tocris Bioscience | Cat#: 5177, CAS: 1214265-58-3 |
| Seradigm Fetal Bovine Serum (FBS) | VWR Life Science | Cat#: 97068-085 |
| Lysyl Endopeptidase, Mass Spectrometry Grade (Lys-C) | Wako | Cat#: 125-05061 |
| Pierce Trypsin Protease, MS Grade | Thermo Fisher Scientific | Cat#: 90058 |
| PHOS-select iron affinity gel | Sigma-Aldrich | Cat#: P9740 |
| Ni-NTA Superflow resin | Quiagen | Cat#: 30410 |
| Critical Commercial Assays | ||
| Kinase Inhibitor High Throughput Screen | QUELLOS HTS Facility, University of Washington | https://iscrm.uw.edu/research/core-resources/quellos-high-throughput-screening-core/ |
| RNeasy Mini Kit | Quiagen | Cat#: 74104 |
| CellTiter-Glo 2.0 Assay | Promega | Cat#: G9241 |
| Deposited Data | ||
| All MS raw files and MaxQuant output files | MassIVE Repository of the University of California, San Diego | Dataset ID #: MSV000083236 |
| Web Application (Shiny) for real time interrogation of our pharmacoproteomics data | Written by Ho-Tak Lau, Shao-En Ong Lab, University of Washington | https://quantbiology.org/hcckinome |
| Experimental Models: Cell Lines | ||
| Human: SNU449, <10 Passages | ATCC | Cat#: CRL-2234 |
| Human: HuH-7, <10 Passages | JRCB Cell Bank | Cat#: JCRB0403 |
| Human: SNU878, <10 Passages | Korean Cell Line Bank (KCLB) | Cat#: 00878 |
| Human: FOCUS, <10 Passages | Laboratory of J. Wands, Brown University, (He et al., 1984) | N/A |
| Human: C3A, <10 Passages | ATCC | Cat#: CRL-10741 |
| Human: HepG2, <10 Passages | ATCC | Cat#: HB-8065 |
| Human: SNU398, <10 Passages | ATCC | Cat#: CRL-2233 |
| Human: PLC/PRF/5, <10 Passages | ATCC | Cat#: CRL-8024 |
| Human: Hep3B2.1-7, <10 Passages | ATCC | Cat#: HB-8064 |
| Human: SKHep1, <10 Passages | ATCC | Cat#: HTB-52 |
| Human: SNU475, <10 Passages | ATCC | Cat#: CRL-2236 |
| Human: SNU387, <10 Passages | ATCC | Cat#: CRL-2237 |
| Human: SNU423, <10 Passages | ATCC | Cat#: CRL-2238 |
| Human: NCI-H684, <10 Passages | Korean Cell Line Bank (KCLB) | Cat#: 90684 |
| Human: SNU761, <10 Passages | Korean Cell Line Bank (KCLB) | Cat#: 00761 |
| Human: SNU886, <10 Passages | Korean Cell Line Bank (KCLB) | Cat#: 00886 |
| Human: JHH4, <10 Passages | JRCB Cell Bank | Cat#: JCRB0435 |
| Human: JHH6, <10 Passages | JRCB Cell Bank | Cat#: JCRB1030 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| GIPZ STAT3 shRNA | Dharmacon | Cat#: RHS4531-EG6774 |
| GIPZ FYN shRNA | Dharmacon | Cat#: RHS4531-EG2534 |
| GIPZ FZD2 shRNA | Dharmacon | Cat#: RHS4531-EG2535 |
| GIPZ AXL shRNA | Dharmacon | Cat#: RHS4531-EG558 |
| GIPZ NUAK1 shRNA | Dharmacon | Cat#: RHS4531-EG9891 |
| GIPZ NUAK2 shRNA | Dharmacon | Cat#: RHS4531-EG81788 |
| Recombinant DNA | ||
| NUAK1 in pReceiver-Lv242 | GeneCopoeia | Cat#: EX-M0778-Lv242, NM_014840.2 |
| Software and Algorithms | ||
| MaxQuant/Andromeda, v1.5.2.8 | https://www.biochem.mpg.de/5111795/maxquant | (Cox et al., 2011) |
| Perseus, v1.5.6.0 | https://www.biochem.mpg.de/5111810/perseus | (Tyanova et al., 2016) |
| R-package, gplots v3.0.1, gplots::heatmap.2 | https://www.rdocumentation.org/packages/gplots/versions/3.0.1 | N/A |
| R-package, Bioconductor: fgsea | http://bioconductor.org/packages/release/bioc/html/fgsea.html | https://www.biorxiv.org/content/early/2016/06/20/060012 |
| GraphPad Prism, V7 | https://www.graphpad.com/ | N/A |
| STRING, database, enrichment analysis | https://string-db.org/ | (Szklarczyk et al., 2015) |
| BioGRID, database | https://thebiogrid.org/ | (Chatr-Aryamontri et al., 2017) |
| PhosphoSite Plus, database | https://www.phosphosite.org/homeAction | (Hornbeck et al., 2015) |
| Reactome, database | https://reactome.org/ | (Fabregat et al., 2018) |
| BioVenn | http://www.biovenn.nl/ | (Hulsen et al., 2008) |
| KinMap | http://kinhub.org/kinmap/ | (Eid et al., 2017) |
| Other | ||
Highlights:
We identify kinases that control hepatocellular carcinoma (HCC) drug responses
Mesenchymal-like HCC cells are resistant to a broad range of kinase inhibitors
FZD2-AXL-NUAK1/2 signaling promotes the epithelial-mesenchymal transition (EMT)
Combinatorial inhibition of EMT kinases enhances HCC cell killing
ACKNOWLEDGMENTS
We thank Dr. Timothy J. Martins and James Annis from the Quellos HTS Core at ISCRM of the University of Washington for providing kinase inhibitor HTS services. We thank John D Scott, Anthony K. Leung, David Shechner, and members of the Ong and Maly Labs for comments on the manuscript. This work was supported by grants from the National Institutes of Health issued under the award numbers R01GM086858, R01GM129090, R01AR065459, R21EB018384, R21CA177402, K22CA201229-01, by the Sidney Kimmel Foundation and by a postdoctoral research fellowship of the German Research Foundation (DFG) awarded to M.G. (GO 2358/1-1). We gratefully acknowledge funding from the Department of Defense under the award number CA150370 as well as the Fibrolamellar Cancer Foundation. This work used an EASY-nLC1200 UHPLC and Thermo Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer purchased with funding from a National Institutes of Health SIG grant S10OD021502 (S-E.O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare that there are no competing financial interests.
REFERENCES
- Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY, Cicin I, Merle P, Chen Y, Park JW, et al. (2018). Cabozantinib in Patients with Advanced and Progressing Hepatocellular Carcinoma. N Engl J Med 379, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Buhrlage SJ, Huang HT, Deng X, Zhou W, Wang J, Traynor R, Prescott AR, Alessi DR, and Gray NS (2014). Characterization of WZ4003 and HTH-01-015 as selective inhibitors of the LKB1-tumour-suppressor-activated NUAK kinases. Biochem J 457, 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, et al. (2007). Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol 25, 1035–1044. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. (2012). The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekhof R, van Alphen C, Henneman AA, Knol JC, Pham TV, Rolfs F, Labots M, Henneberry E, Le Large TY, de Haas RR, et al. (2019). INKA, an integrative data analysis pipeline for phosphoproteomic inference of active kinases. Mol Syst Biol 15, e8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al. (2017). Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 389, 56–66. [DOI] [PubMed] [Google Scholar]
- Casas-Selves M, Kim J, Zhang Z, Helfrich BA, Gao D, Porter CC, Scarborough HA, Bunn PA Jr., Chan DC, Tan AC, et al. (2012). Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res 72, 4154–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambliss AB, and Chan DW (2016). Precision medicine: from pharmacogenomics to pharmacoproteomics. Clin Proteomics 13, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O'Donnell L, Oster S, Theesfeld C, Sellam A, et al. (2017). The BioGRID interaction database: 2017 update. Nucleic acids research 45, D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Li K, Liang Y, Li L, and Zhu X (2013). High NUAK1 expression correlates with poor prognosis and involved in NSCLC cells migration and invasion. Exp Lung Res 39, 9–17. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of proteome research 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, and Mann M (2008). Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Molecular cell 31, 438–448. [DOI] [PubMed] [Google Scholar]
- Dobbelstein M, and Sorensen CS (2015). Exploiting replicative stress to treat cancer. Nat Rev Drug Discov 14, 405–423. [DOI] [PubMed] [Google Scholar]
- Eid S, Turk S, Volkamer A, Rippmann F, and Fulle S (2017). KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinformatics 18, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. (2018). The Reactome Pathway Knowledgebase. Nucleic acids research 46, D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, and Marto JA (2009). Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Analytical chemistry 81, 4566–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firtina Karagonlar Z, Koc D, Iscan E, Erdal E, and Atabey N (2016). Elevated hepatocyte growth factor expression as an autocrine c-Met activation mechanism in acquired resistance to sorafenib in hepatocellular carcinoma cells. Cancer Sci 107, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuren ED, Zhang L, Wu J, and Daly RJ (2016). The kinome 'at large' in cancer. Nat Rev Cancer 16, 83–98. [DOI] [PubMed] [Google Scholar]
- Garber K (2019). The new liver epidemic. Nat Biotechnol 37, 209–214. [DOI] [PubMed] [Google Scholar]
- Gay CM, Balaji K, and Byers LA (2017). Giving AXL the axe: targeting AXL in human malignancy. Br J Cancer 116, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Burden of Disease Cancer Collaboration (2017). Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 3, 524–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golkowski M, Brigham JL, Perera GK, Romano GE, Maly DJ, and Ong SE (2014). Rapid profiling of protein kinase inhibitors by quantitative proteomics. MedChemComm 5, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golkowski M, Maly DJ, and Ong SE (2017a). Proteomic Profiling of Protein Kinase Inhibitor Targets by Mass Spectrometry. Methods Mol Biol 1636, 105–117. [DOI] [PubMed] [Google Scholar]
- Golkowski M, Vidadala RS, Lombard CK, Suh HW, Maly DJ, and Ong SE (2017b). Kinobead and Single-Shot LC-MS Profiling Identifies Selective PKD Inhibitors. Journal of proteome research 16, 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golkowski M, Vidadala VN, Lau HT, Shoemaker A, Shimizu-Albergine M, Beavo J, Maly DJ, and Ong SE (2020). Kinobead/LC-MS Phosphokinome Profiling Enables Rapid Analyses of Kinase-Dependent Cell Signaling Networks. Journal of proteome research 19, 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujral TS, Chan M, Peshkin L, Sorger PK, Kirschner MW, and MacBeath G (2014). A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 159, 844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Isselbacher KJ, Wands JR, Goodman HM, Shih C, and Quaroni A (1984). Establishment and characterization of a new human hepatocellular carcinoma cell line. In Vitro 20, 493–504. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, and Skrzypek E (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic acids research 43, D512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, and Alkema W (2008). BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones VS, Huang RY, Chen LP, Chen ZS, Fu L, and Huang RP (2016). Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochim Biophys Acta 1865, 255–265. [DOI] [PubMed] [Google Scholar]
- Kalluri R, and Weinberg RA (2009). The basics of epithelial-mesenchymal transition. J Clin Invest 119, 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kon M, and DeLisi C (2012). Pathway-based classification of cancer subtypes. Biol Direct 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, Baron A, Park JW, Han G, Jassem J, et al. (2018). Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 391, 1163–1173. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, and Derynck R (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15, 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H-T, Golkowski M, and Ong SE (2019). Kinome features, signaling pathways, and drug response in HCC (https://quantbiology.org/hcckinome) (Ong Lab).
- Lee HJ, Jeng YM, Chen YL, Chung L, and Yuan RH (2014). Gas6/Axl pathway promotes tumor invasion through the transcriptional activation of Slug in hepatocellular carcinoma. Carcinogenesis 35, 769–775. [DOI] [PubMed] [Google Scholar]
- Legembre P, Schickel R, Barnhart BC, and Peter ME (2004). Identification of SNF1/AMP kinase-related kinase as an NF-kappaB-regulated anti-apoptotic kinase involved in CD95-induced motility and invasiveness. J Biol Chem 279, 46742–46747. [DOI] [PubMed] [Google Scholar]
- Lin AB, McNeely SC, and Beckmann RP (2017). Achieving Precision Death with Cell-Cycle Inhibitors that Target DNA Replication and Repair. Clin Cancer Res 23, 3232–3240. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang K, Yan Z, Xia Y, Li J, Shi L, Zou Q, Wan X, Jiao B, Wang H, et al. (2016). Axl Expression Stratifies Patients with Poor Prognosis after Hepatectomy for Hepatocellular Carcinoma. PLoS One 11, e0154767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Montal R, Sia D, and Finn RS (2018). Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 15, 599–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer TG, Naipal KA, Jager A, and van Gent DC (2017). Ex vivo tumor culture systems for functional drug testing and therapy response prediction. Future Sci OA 3, FSO190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port J, Muthalagu N, Raja M, Ceteci F, Monteverde T, Kruspig B, Hedley A, Kalna G, Lilla S, Neilson L, et al. (2018). Colorectal Tumors Require NUAK1 for Protection from Oxidative Stress. Cancer Discov 8, 632–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- Schmidlin T, Debets DO, van Gelder C, Stecker KE, Rontogianni S, van den Eshof BL, Kemper K, Lips EH, van den Biggelaar M, Peeper DS, et al. (2019). High-Throughput Assessment of Kinome-wide Activation States. Cell Syst 9, 366–374 e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev A (2016). An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation bioRxiv, https://doiorg/101101/060012. [Google Scholar]
- Shen S, Dean DC, Yu Z, and Duan Z (2019). Role of cyclin-dependent kinases (CDKs) in hepatocellular carcinoma: Therapeutic potential of targeting the CDK signaling pathway. Hepatol Res. [DOI] [PubMed] [Google Scholar]
- Shibue T, and Weinberg RA (2017). EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 14, 611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research 43, D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network (2017). Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 169, 1327–1341 e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, and Nieto MA (2009). Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, and Nieto MA (2004). Snail blocks the cell cycle and confers resistance to cell death. Genes Dev 18, 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A (2019). Hepatocellular Carcinoma. N Engl J Med 380, 1450–1462. [DOI] [PubMed] [Google Scholar]
- Villen J, and Gygi SP (2008). The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature protocols 3, 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qian WJ, Mottaz HM, Clauss TR, Anderson DJ, Moore RJ, Camp DG 2nd, Khan AH, Sforza DM, Pallavicini M, et al. (2005). Development and evaluation of a micro- and nanoscale proteomic sample preparation method. Journal of proteome research 4, 2397–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM, Assenat E, Brandi G, Pracht M, Lim HY, et al. (2019). Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased alpha-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 20, 282–296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Drug Panel, HCC Cell EMT Markers and Kinase mRNA expression; Supplement to Figure 1-4. Identity and primary targets of the 299 kinase inhibitor panel. Log2 mRNA intensity of 50 EMT and stem cell markers as well as 509 protein kinases in the 28 CCLE HCC cell lines.
Table S2. Kinome Profiling 17 HCC Cell Lines; Supplement to Figure 1-4. MaxQuant protein groups and phospho(S/T/Y) output data obtained from kinome profiling of 17 HCC cell lines. Missing values were replaced by data imputation (see ‘STAR Methods’).
Table S3. Kinase Inhibitor HTS Results and kinome-GSEA Analysis; Supplement to Figure 1-4. Results of the HTS of 299 kinase inhibitors in 17 HCC cell lines. Summary table of AUC values. Summary of Reactome pathways used in the kinome-GSEA analysis. Compounds and cumulative NES for clusters from Figure S4A. FDR and NES values for all 299 compounds and 275 Reactome pathways.
Table S4. Kinome Proteomic Feature-AUC Correlation Results; Supplement to Figure 1. Results from Pearson correlation between mean MS intensity values of all quantified proteomics features (n = 13935) and kinase inhibitor AUCs (n = 299) across the 17 HCC cell line panel.
Table S5. Kinome Profiling of Human HCC Specimens and Kinome-GSEA; Supplement to Figure 1. MaxQuant protein groups and phospho(S/T/Y) output data from LFQ kinobead/LC-MS profiling of 4 paired (tumor/NAL) human HCC specimens. Kinome-GSEA of Reactome Pathways in the 4 paired HCC samples (see ‘STAR Methods’).
Table S6. Kinome Profiling FOCUS RNAi Models; Supplement to Figure 3 and 4. MaxQuant protein groups and phospho(S/T/Y) output data from kinome profiling of the FOCUS cell AXL, FZD2 and NUAK1/2 RNAi models.
Data Availability Statement
The MS .raw files and MaxQuant output files generated during this study are available at the MassIVE Repository of the University of California, San Diego (Dataset ID#: MSV000083236). A Shiny app for real time interrogation of the pharmacoproteomics data generated in this study can be accessed at https://quantbiology.org/hcckinome. This study did not generate new code.