Abstract
Immune dysfunction plays a role in the development of Parkinson’s disease (PD). Natural killer (NK) cells regulate immune functions and are modulated by killer cell immunoglobulin-like receptors (KIR). KIR are expressed on the surface of NK cells and interact with human leukocyte antigen (HLA) class I ligands on the surface of all nucleated cells. We investigated KIR allelic polymorphism to interrogate the role of NK cells in PD. We sequenced KIR genes from 1,314 PD patients and 1,978 controls using next generation methods and identified KIR genotypes using custom bioinformatics. We examined associations of KIR with PD susceptibility and disease features including age at disease onset and clinical symptoms. We identified two KIR3DL1 alleles encoding highly expressed inhibitory receptors associated with protection from PD clinical features in the presence of their cognate ligand: KIR3DL1*015/HLA-Bw4 from rigidity (pc = 0.02, OR = 0.39, 95% CI 0.23 – 0.69) and KIR3DL1*002/HLA-Bw4i from gait difficulties (pc = 0.05, OR = 0.62, 95% CI 0.44 – 0.88), as well as composite symptoms associated with more severe disease. We also developed a KIR3DL1/HLA interaction strength metric and found that weak KIR3DL1/HLA interactions were associated with rigidity (pc = 0.05, OR = 9.73, 95% CI 2.13 – 172.5). Highly expressed KIR3DL1 variants protect against more debilitating symptoms of PD, strongly implying a role of NK cells in PD progression and manifestation.
Keywords: Parkinson’s disease, KIR, expression, natural killer cells, neurological disorders, brain
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disease characterized by multiple symptoms including tremors, gait disturbances, postural instability, and rigidity, as well as numerous symptoms impacting sensory perception, cognition, and psychological function. PD is the second most common neurologic disease after Alzheimer’s disease, affecting roughly 1% of adults over the age of 50 in the United States (1). The etiology of PD is thought to involve complex interactions between genetics and environmental risk factors (2–4), but it also shares many features of autoimmune neurological diseases, such as immune system dysfunction and disease progression over time. Our recent work (5) and that of others (1, 2, 6) have demonstrated association of PD risk and protection with polymorphism in human leukocyte antigen (HLA) genes and with other genes involved in the immune system. However, PD is unique in the environmental factors associated with risk and protection. In marked contrast to most other autoimmune diseases, cigarette smoking, coffee consumption and female sex are all factors associated with protection from PD (4). In addition to overall disease risk, clinical presentations of PD align with differences in disease prognosis. Symptoms impacting the ability to move (gait disturbances, postural instability, rigidity and bradykinesis) are all associated with a more severe PD course (7–10). Conversely, asymmetric onset of symptoms and tremor-dominant disease are associated with milder forms of PD and follow a less dramatic disease course (7, 11). Finally, contrasting with other degenerative brain disorders and dementias such as Alzheimer’s, in PD later age of onset is correlated with more rapidly progressing disease (12–14). The heterogeneous presentation of PD is thus characterized by a varying constellation of disease features and complications (7–10).
The precise nature of immune regulatory dysfunction in PD is not known, although research has demonstrated a role for multiple immune cells in protection from and development of disease. For example, in PD patients, T cells respond to alpha-synuclein peptides and these responses drive T cell cytotoxic activity (15). B cells specific for alpha-synuclein are also found in PD. Some studies observed increased numbers of natural killer (NK) cells in the peripheral blood of PD patients (16, 17). NK cells are critical in protection from viral infection, but also have immunoregulatory functions and are implicated in multiple autoimmune diseases and cancers (18–24). NK cells are negative regulators of B cell somatic hypermutation, linking NK cell activity to regulation of antibody production (25). By contrast with PD, in multiple sclerosis (MS), an autoimmune disease of the central nervous system, NK cell numbers decrease during clinical relapse and increase as the patient’s symptoms improve (26). Differences in NK activity and regulation suggest alternative roles for NK cells in neurological disease development.
Key modulators of NK cell functions are the killer immunoglobulin-like receptors (KIR) (19, 27, 28). KIR are present on the surface of NK cells and engage HLA class I ligands that are present on the surface of all nucleated cells (27). These interactions modulate NK cell activity, including the cytotoxic killing of virus infected cells and tumors, or induction of cytokine secretion (29, 30). KIR are encoded by the fast evolving and highly polymorphic KIR gene family on chromosome region 19 (19q13.4) (31, 32). Individual KIR genes can be present or absent, with individual KIR gene-content haplotypes having 4-14 genes, and exhibiting considerable variation between individuals and populations (33, 34). KIR molecules are structurally and functionally distinct, interacting differently with specific sequence motifs of HLA class I allotypes (29).
Thus KIR3DL1 recognizes a sequence motif at positions 77-83 in the α1 domain and forming the Bw4 epitope present on some HLA-A and HLA-B (35). An isoleucine at position 80 characterizes the HLA-Bw4i ligand, which confers a stronger interaction with KIR than the HLA-Bw4t ligand, which has threonine at position 80 (36, 37). KIR allotypes vary in their inhibitory capacity, which is governed by the strength of their binding to HLA class I and the magnitudes of their cell surface expression (38–41). Several studies report associations between KIR gene-content and the development of autoimmune disease, including neurological autoimmunity (24, 42–45). Given the evidence for disruption of NK cell homeostasis in PD, we investigated the possibility that KIR gene content and allelic variation play a role in PD. This is the first study to analyze KIR allele-level variation in any neurological autoimmune disease.
Using a recently developed sequencing method and custom bioinformatics pipeline (46), we determined and analyzed the association of individual KIR alleles with PD and as predictors of specific clinical symptoms. We found that variation associated with high levels of inhibitory KIR expression was associated with protection from specific disease symptoms and may promote a distinct disease course characterized by fewer symptoms impacting movement
Material and Methods
Study populations:
Cohorts of 1,314 Parkinson’s disease patients and 1,978 healthy controls were analyzed. Patients and controls consisted of the National Institutes of Health (NIH)-GWAS and NIH-NeuroX datasets derived from the National Institute of Neurological Disorders and Stroke-funded Neurogenetics Repository at the Coriell Institute for Medical Research, as well as the UCSF-EPIC dataset (47). All patients were unrelated Euro-descendants from the United States with idiopathic PD. Controls were unrelated and self-reported Euro-descendants and free from neurologic disorders. Ancestral outliers were identified through analysis of genome-wide markers and removed from the cohorts prior to analysis, as described by Hollenbach et al. 2019 (5). Informed consent was obtained for each participant under locally approved protocols.
Clinical phenotype information.
Information pertaining to clinical phenotypes in PD were acquired from Coriell (48). We analyzed presence and absence of the following clinical phenotypes with respect to KIR-HLA combinations: activation tremor, resting tremor, rigidity, bradykinesis, asymmetric onset, gait difficulties and postural instability.
KIR and HLA genotyping.
DNA samples were genotyped for all KIR genes according to Norman 2016 (46). To determine KIR gene content and allelic genotypes from NGS data, we used an extended version of our custom bioinformatics pipeline PING (Pushing Immunogenetics into the Next Generation) (46); which takes short-read sequences as input and through multiple alignment and filtration steps identifies KIR genotypes as previously described, with additional modifications (Marin et. al, in prep). The HLA-A, -B, -C genotypes for the study cohorts were reported previously (5).
Molecular Dynamics Simulations of KIR3DL1*015 and KIR3DL1*002.
The structure of KIR3DL1 was obtained from the protein data bank (PDB ID: 3vh8) and residue 47 was replaced by valine to model the 3DL1*015 allotype. For 3DL1*002, residue 238 was also changed to arginine using the mutate plugin in Visual Molecular Dynamics (VMD) package (49). Molecularmodels were solvated in the TIP3P explicit solvent and minimized for 100,000 steps and equilibrated for 10ns prior to the production run. All simulations were carried out using NAMD molecular dynamics package and CHARMM36 forcefield (50). The 3DL1*015 allotype was simulated for 90ns, while the 3DL1*002 simulation continued to 120ns. The NPT ensemble was used in which temperature and pressure were maintained at 310 K and 1 bar using Langevin thermostat and Langevin piston Nose-Hoover, respectively. Timestep of 2fs was used and periodic boundary condition was applied in all directions. Error bars in energy plots were obtained via dividing the standard deviation by where n is the number of frames. Visualization and post-processing analyses were performed using VMD and Bio3D packages (51).
KIR interaction scores.
We developed KIR-HLA interaction scores to distinguish within locus differences in KIR and HLA binding propensity. KIR3DL1 interaction scores were determined based on published data of KIR3DL1 allotype binding to HLA (41). We created three categorical measures of KIR3DL1 binding strength (strong, medium, weak) based on the interquartile ranges for the binding percentages determined by Saunders et. al (41). These categories were consistent with a previous analysis of KIR3DL1/HLA interaction strength (52). Our dataset of KIR and HLA genotypes included alleles that were not measured by Saunders et al 2016 (41). To include KIR3DL1-HLA pairs in our dataset for which there is no published functional data, we imputed the strength of the binding to KIR by taking into account 1) similar inhibitory capacity, 2) cell surface expression level and 3) sequence similarity to published allotypes (53, 54); for HLA, we considered sequence similarity to alleles with experimentally measured KIR binding data (39, 41). We identified the closest KIR protein sequence in regions known to interact with HLA and a composite score based on the full potential pairing possible between the KIR and HLA allotypes was given to each individual. All KIR/HLA pairs and their assigned strengths are given in Supplementary Table 1.
Statistical analyses.
Impact of genotype on disease and clinical outcomes were calculated by multivariate regression models using R version 3.6.0 (55). For discrete variables, we analyzed only KIR alleles with a frequency >1%, which were analyzed by logistic regression (presence/absence clinical phenotype). We adjusted the models for sex as a covariate. All p-values reported are two-tailed and were adjusted for multiple comparisons using the Bonferroni method (56).
Results
High expression KIR3DL1 alleles in combination with HLA-Bw4/Bw4i reduce gait difficulties and rigidity in Parkinson’s disease.
KIR allotypes differ in surface expression and strength of interaction with cognate HLA ligands (39, 52, 53). We analyzed KIR allelic polymorphism for all the KIR genes in combination with known HLA class I ligands for association with PD disease risk and clinical outcomes (57). No statistically significant KIR-HLA associations were observed with respect to disease predisposition in a case/control analysis (Supplemental Figure 1). In contrast, we found evidence that the high expression allele KIR3DL1*015 in combination with HLA-Bw4 was associated with protection from rigidity, an important clinical feature of PD (Figure 1, p=0.002, pc = 0.02, OR = 0.39, 95% CI 0.23 – 0.69). Another highly expressed allele KIR3DL1*002, in combination with Bw4, was also strongly suggestive for protection from gait impairment (p=0.006, pc = 0.06, OR = 0.62, 95% CI 0.44 – 0.88).
Figure 1. KIR3DL1 alleles associated with protection from PD symptoms.
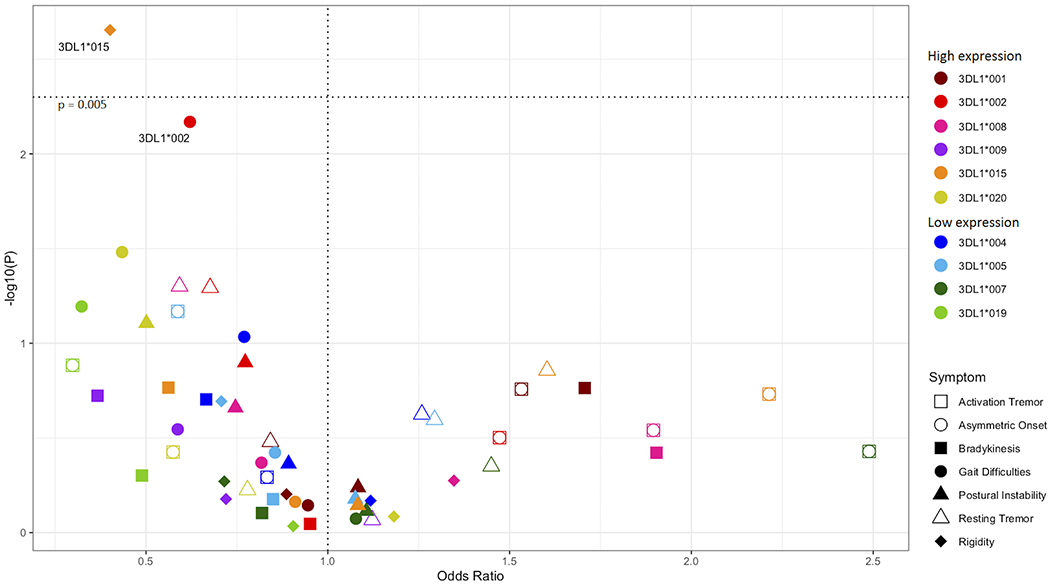
All KIR3DL1 alleles were analyzed in combination with HLA-Bw4 ligands. Odds ratios (x – axis) by -log10(p-value) (y – axis). Each dot represents the odds ratio and corresponding -log10(p-value) for a KIR allele/symptom pair. PD symptoms are indicated by shapes and KIR3DL1 alleles are indicated by color. The horizontal dotted line indicates the significant corrected p-value.
Weak KIR3DL1/HLA interactions associate with higher risk of rigidity and lower risk of resting tremor.
We developed a scoring system to evaluate aggregate KIR-HLA interactions at the allele level and their potential inhibitory impact. Using the percent maximum binding values of Saunders et al. 2016 (41), we computed a range of KIR3DL1-HLA interaction values. Interaction strength is not associated with disease risk overall. On analyzing interaction strength with respect to PD symptoms, we found that strong KIR3DL1-HLA interactions are associated with the presence of resting tremors (Figure 2, pc = 0.04, OR = 1.52, 95% CI 1.07 – 2.20). By contrast, weak KIR3DL1-HLA interactions were negatively associated with resting tremors (Figure 2, pc = 0.04, OR = 0.56, 95% CI 0.35 - 0.92). We also found that weak KIR3DL1-HLA interactions are strongly associated with rigidity (Figure 2, pc = 0.05, OR = 9.73, 95% CI 2.13 – 172.5).
Figure 2. Strong interactions between KIR3DL1 and HLA-Bw4 alleles are associated with clinical features of PD.
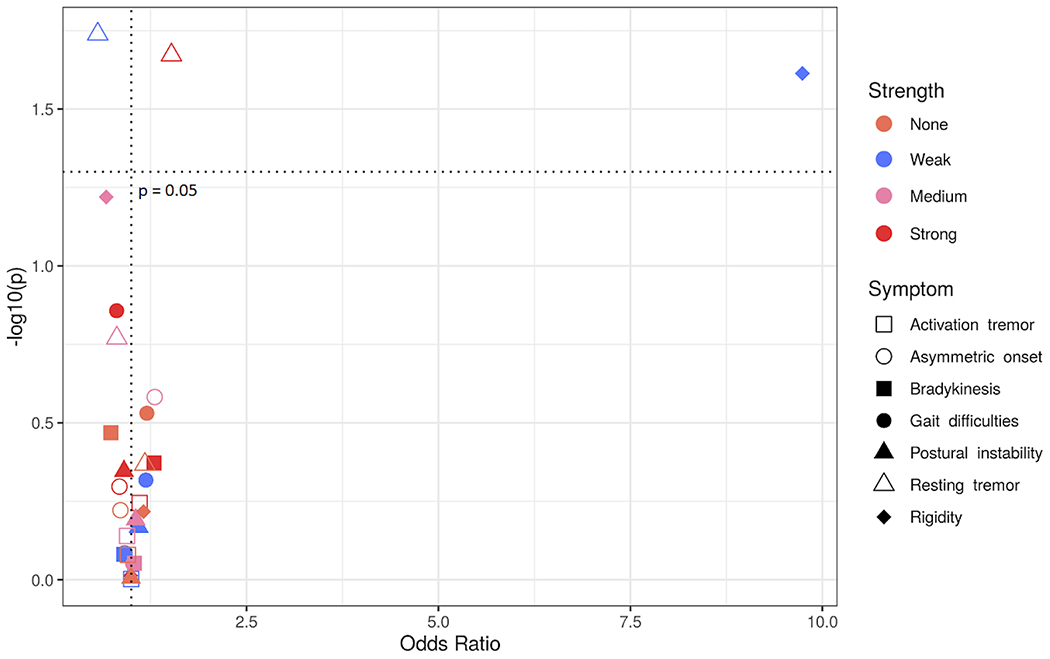
Odds ratios (x – axis) by -log10(p-value) (y – axis). Each dot represents the odds ratio and corresponding -log10(p-value) for a KIR interaction strength/symptom pair. Different PD symptoms are indicated by shapes and KIR3DL1 interaction strengths are indicated by color.
High-expressing KIR3DL1*002 is at higher frequency in patients who present with symptoms related to movement.
Thus far we have identified a pattern in PD, whereby high expressing KIR3DL1 alleles and strong KIR3DL1-HLA interactions are associated with protection from PD symptoms impacting movement. In contrast, low expressing KIR3DL1 and weak KIR3DL1-HLA interactions are associated with risk of more debilitating symptoms. From these observations, we categorized the patients based upon their symptoms of PD.
Among the most debilitating PD symptoms are postural instability and gait difficulty, whereas resting tremors and asymmetric onset appear to correspond to a milder disease course (8, 9). Postural instability and gait difficulty at disease onset and absence of resting tremors predicts poor survival in PD patients (58). Additionally, it has been shown that there are distinct differences in brain matter and morphology between subsets of patients with resting tremors and those with postural instability and gait difficulties (59, 60). Because postural instability and gait difficulty appear to distinguish subgroups of PD patients (8, 9, 60), we assigned the patients into two groups according to their symptoms: one consisting of patients having gait difficulties or postural instability, or both (PIGD+); the other group (PIGD-) comprised patients having neither symptom (Figure 3A). These two groups were then compared for other associations. In the PIGD+ group, the mean age of onset is 59, whereas in the PIGD- group it is 57 (p = 0.005). This difference suggests that this subdivision of PD patients has captured an additional factor important in disease prognosis. Furthermore, the PIGD+ group shows significantly increased symptoms of rigidity and bradykinesis, and decreased symptoms of asymmetric onset and resting tremors compared to the PIGD- group (Supplementary Figure 2, Supplementary Table 2).
Figure 3. Stratification of PD patients by symptom type.
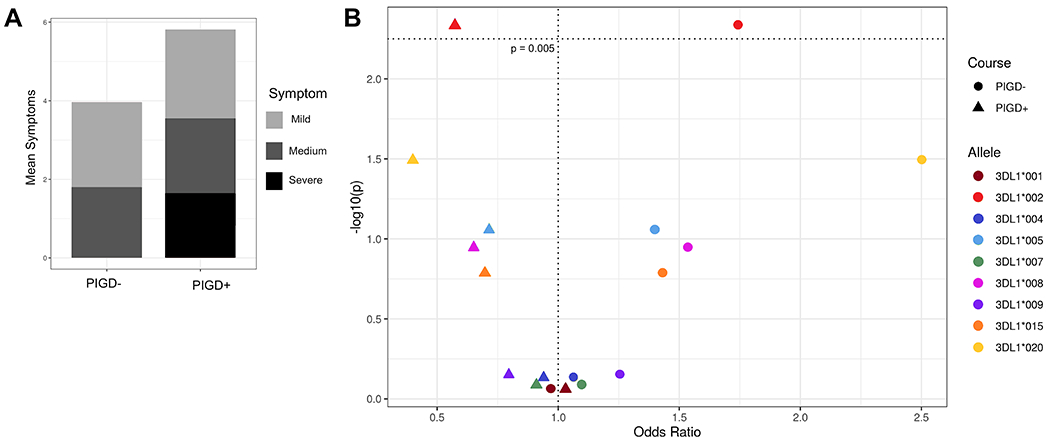
A) Mean number of symptoms (y – axis) per cluster (x – axis). Symptoms are broken down into three categories where red indicates the most debilitating (postural instability and gait difficulty), coral the moderate symptoms (rigidity and bradykinesis), and light pink the mild symptoms associated with less severe disease (resting and activation tremors and asymmetric onset). B) KIR3DL1 allele-HLA-Bw4 associations with disease group. Odds ratio (x – axis) by -log10(p-value) (y – axis). Each dot represents a KIR3DL1 allele-HLA ligand pair, the different shapes indicate PIGD- (circle) or PIGD+ (triangle) disease. The horizontal dotted line indicates the value for a significant corrected p-value.
We also examined KIR3DL1 allelic variation, expression levels and strength of KIR3DL1-HLA interaction in the two groups of PD patients. The combination of KIR3DL1*002 and HLA-Bw4 was associated with protection from the PIGD+ category of disease symptoms (Figure 3B, pc = 0.04, OR = 0.57, 95% CI 0.39 – 0.85). Although no other alleles had significant associations after correction for multiple comparisons, we observed a strong trend for strong KIR3DL1 allotypes providing protection from the PIGD+ category of disease symptoms (Figure 3B).
Because KIR3DL1*015 and KIR3DL1*002 differ only at position G238R (Figure 4) and are both high expressing allotypes, we hypothesized that their association with protection from different PD symptoms is associated with conformational differences induced by position 238, which may in turn have important functional differences. To test this hypothesis, we performed all-atom molecular dynamics (MD) simulations of both KIR3DL1 allotypes and tracked conformational changes throughout the trajectories. In 3DL1*002, R238 anchors D2 to D1 and allows surface residues of the two domains to interact more effectively as shown in Figure 5A. Simulations indicated that the overall interaction energy between D1 and D2 was 50 kcal/mol higher in 3DL1*002 than 3DL1*015 (Figure 5B). The G to R substitution at position 238 resulted in 10-fold stronger association of this residue with D1 as shown in Figure 5C. We also observed a negative correlation between G238 and HLA binding residues E201, S227, S228, and D230, in 3DL1*015, which was diminished in 3DL1*002 (Figure 5D).
Figure 4. KIR3DL1*015 and KIR3DL1*002 Differ by A Single Amino Acid at Position 238.
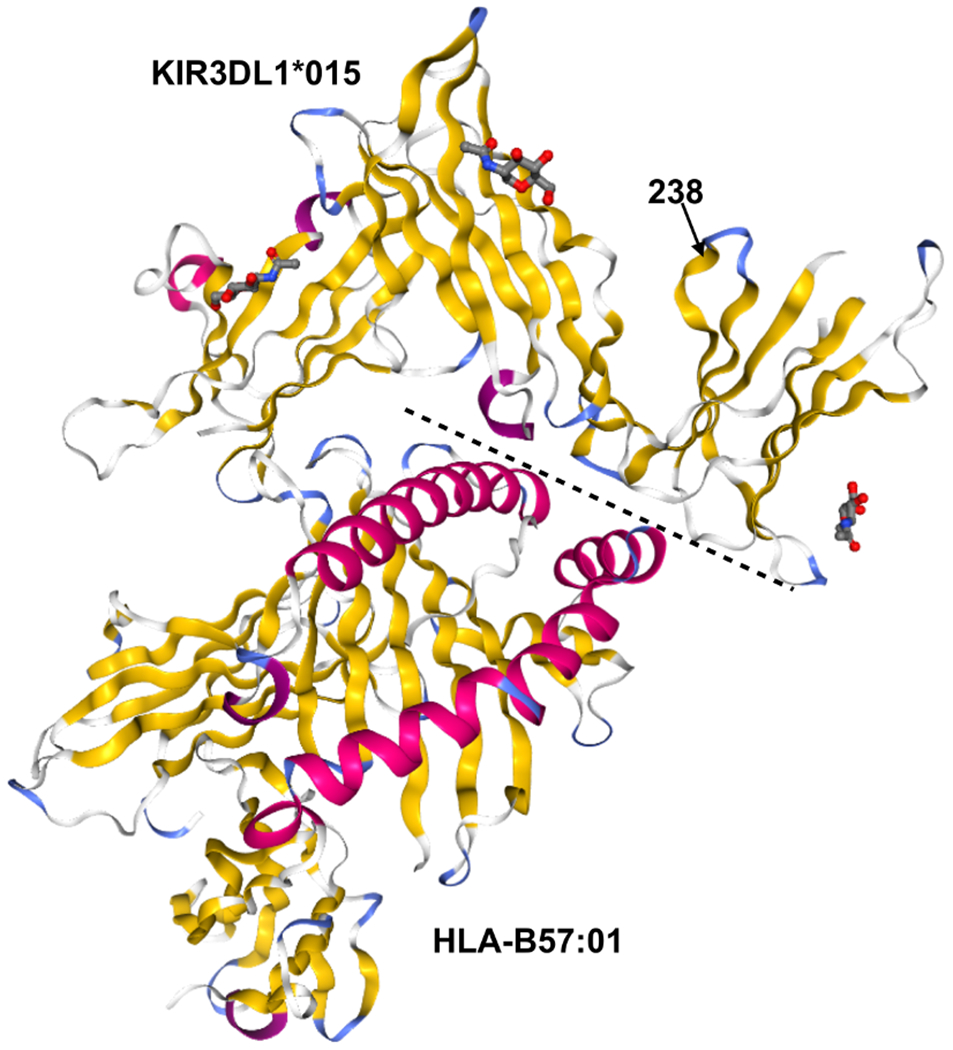
Crystal structures of KIR3DL1*015 (top) complexed with its ligand HLA-B*57:01 (bottom). Secondary structures are shown by color: alpha helices (pink) and beta sheets (yellow). The dashed line indicates the binding interface between KIR3DL1*015 and HLA-B*57:01. Position 238 is indicated by an arrow. Protein Data Bank accession no. 5B39 (41).
Figure 5. Molecular dynamics simulations of KIR3DL1*015 and KIR3DL1*002.
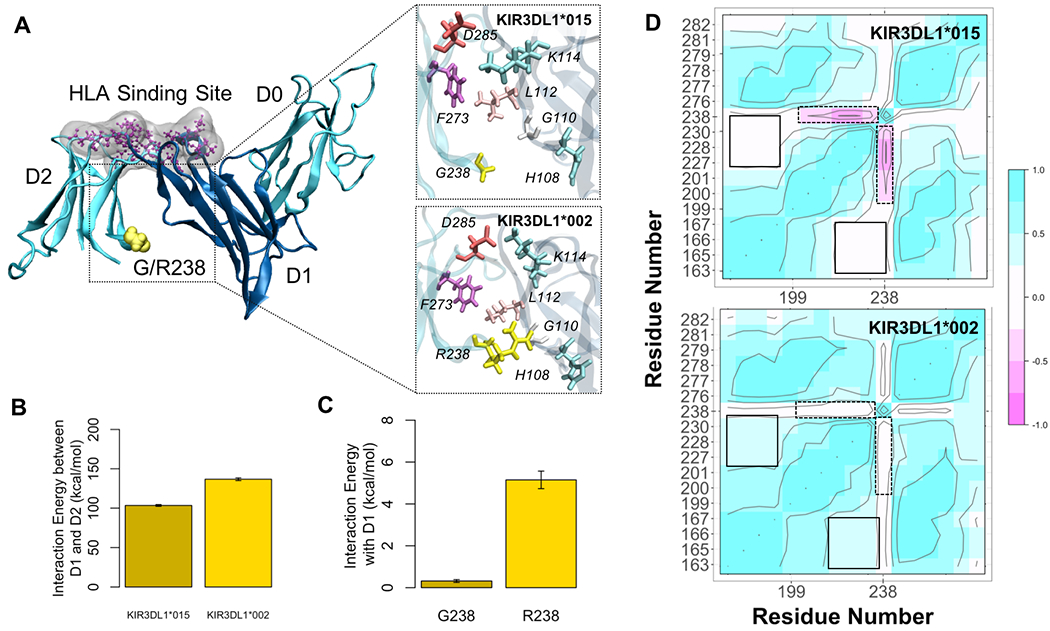
A) KIR3DL1 is composed of three extracellular domains (D0-D2). The HLA binding site (purple) is located at the hinge between D1 and D2. Residue 238 (yellow) is in the D2 domain facing towards D1 serving as an anchorage point. The G to R substitution in 3DL1*002 positions D2 closer to D1 allowing more contacts between surface residues of D1 and D2. B) This led to an increased D1-D2 interaction energy by 50 kcal/mol in 3DL1*002 compared to 3DL1*015. C) The association between residue 238 and D1 was also 10-fold higher in 3DL1*002 as opposed to 3DL1*015. D) Residue cross correlation (RCC) map depicts pairwise correlations between residue 238 and the HLA binding site, mapped on residues P163, M165, L166, A167, P199, Y200, E201, S227, S228, D230, F276, R277, H278, S279, Y281, and E282. The negative correlation between G238 and residues E201, S227, S228, and D230 in 3DL1*015 was eliminated upon G238R substitution in 3DL1*002 as shown by the dashed boxes. On the contrary, the positive correlation between HLA binding residues is enhanced in 3DL1*002 (solid boxes).
Discussion
Few studies have analyzed the impact of KIR allelic variation in immune-mediated diseases (61). To our knowledge this is the first study to examine the role of KIR allotypes and their functional differences in neurological disease. In the context of KIR gene content variation, a protective effect has been observed for the presence of KIR2DS1 in Portuguese MS patients (42). In a study of Spanish MS patients, the activating KIR3DS1 and inhibitory KIR2DL5 were associated with disease (62). Our own work showed that the combination of KIR3DL1 and HLA-Bw4 protects against MS (24). While these studies indicate that KIR polymorphism affects the course of neurological disorders, they also point to the value of comparing the allelic differences within each KIR gene for their functional differences and associations with disease. Importantly, different KIR alleles have known differences in levels of cell surface expression and NK cell inhibition (39, 52, 53, 63). Thus, it is critical to examine KIR associations at the allelic level.
While KIR allelic variation was not associated with overall disease risk in this study, we found that the KIR3DL1*015 and KIR3DL1*002, in combination with HLA-Bw4, were associated with protection from rigidity and gait difficulties in PD patients, respectively, which are each associated with more severe disease course in PD. Further, KIR3DL1*002 was shown to be protective from the more severe PIGD+ disease category. Both highly expressed (64), these two allotypes differ only by the amino acid at position 238 (41). Crystallography mutational analyses indicate that residue 238 does not interact directly with the HLA class I ligand (Figure 4) (41). This fact, however, cannot overrule the possibility that this residue causes differences in binding. Previous results suggested that position 238 may affect conformation of ligand binding loops as well as receptor oligomerization, which could partially explain the difference in strength observed for 3DL1*002 and 3DL1*007 molecules(39). In fact, binding differences have been observed for KIR3DL1*002 and KIR3DL1*015(52). To further explore their functional differences, we studied conformational changes of KIR3DL1*002 and KIR3DL1*015 allotypes in silico. Simulations indicated that residue 238, located on the surface of the D2 domain, is crucial for adjusting the D1-D2 interaction and directly influences the D1-D1 angle, which is a major regulator of HLA binding. In addition, the coordination of movements between residues at the HLA binding site is affected by the type of residue at position 238 implying an allosteric effect of this residue on the KIR3DL1 function. Altogether, these results strongly suggest that differential HLA binding is modulated by residue 238-induced structural differences in KIR3DL1. Therefore, we provide new insights of the role of position 238 in KIR3DL1 and suggest the associations of these two allotypes with different PD symptoms could be the result of functional differences of these two molecules caused by the change G238R. These findings are in accordance with recent experimental work by Saunders et al. (65) that demonstrates that allelic variation in KIR3DL1 results in functional differences impacting the ability of the receptor to interact with HLA.
At the same time, both KIR3DL1*015 and KIR3DL1*002 are highly expressed on the NK surface, which also suggests that high KIR3DL1 expression contributes to protection from some clinical manifestations of disease. Our results suggest that high KIR3DL1 expression has a protective effect in PD. Expression levels of KIR3DL1 have significant functional consequences (63, 66) and NK cells with high-expressing KIR are correlated with greater NK cell reactivity (52). In human immunodeficiency virus infection (HIV), the combination of high expressing KIR3DL1 and HLA-Bw4 results in reduced viral load and slower progression to acquired immunodeficiency syndrome (AIDS) (67). The combination of low-expressing KIR3DL1 and HLA-Bw4 is associated with risk of psoriasis (68), whereas the combination of HLA-Bw4 and KIR3DL1, both high and low, provide protection from ankylosing spondylitis (69).
The overall inhibitory capacity of NK cells significantly impacts NK cell function, aiding to control viral replication and prevent tumor development (70). While KIR expression levels may be a useful measure of inhibitory capacity, quantitative data of HLA and KIR expression and their interaction should allow for more informative characterization of their inhibitory potential. Using a weighted score of inhibitory KIR-HLA interaction strength, higher KIR inhibition overall was shown to be associated with lower loads of HIV, improved spontaneous viral clearance of hepatitis C virus (HCV), and protection from human T lymphocyte virus 1 (HTLV-1) infection (70). Such consideration of specific interactions between KIR and HLA class I allotypes enables interpretation from a functional perspective that fully exploits high-resolution KIR and HLA class I sequencing data. For example, HLA-B*57:01 encodes a KIR3DL1 ligand that binds strongly to most KIR3DL1 allotypes (39, 41), whereas other HLA-B allotypes such as that encoded by HLA-B*44:03 have widely varying interactions that depend upon the particular KIR allotype (41). Based on the interaction scores that we calculated for KIR3DL1 with HLA-Bw4, we find that weak interaction scores are negatively associated with resting tremors, a characteristic symptom of a mild PD subtype (9). Weak interaction also positively associates with risk of rigidity, a symptom of a more severe PD (8, 9, 71). Thus, stronger inhibitory potential of KIR3DL1-expressing NK cells protects against the more severe symptoms of PD.
In conclusion, our finding that KIR3DL1 polymorphism is associated with specific combinations of PD symptoms suggests an impact of these NK cell receptors on disease progression. The high expression allele KIR3DL1*002 combined with the HLA-Bw4 is associated with protection in the PIGD+ patient group, in which movement-associated symptoms predominate. A limitation to our analysis is that it examined disease symptoms at a single time-point, which does not take into account any changes in symptoms over time (72). Further studies examining KIR in PD would benefit from longitudinal analysis to determine whether specific KIR are associated with time to PIGD+ development.
One explanation for the associations observed in this study is related to the immunomodulatory role of NK cells. Key in PD development is the aggregation of alpha-synuclein in Lewy bodies (73). Recently it was shown that antibodies made against alpha-synuclein aid in the clearance of aggregates and prevent their formation, and PD patients have low levels of these protective antibodies (74). An effective B cell response could therefore be essential in preventing PD. NK cells impact B cell antibody responses in several ways: in germinal centers, NK cells may suppress affinity maturation and somatic hypermutation (25), and NK cells may enhance B cell antibody responses through direct interaction with B cells (20, 75). The cause of reduced anti- alpha-synuclein antibody levels in PD patients is unknown but may involve either impaired helper T cells or impaired B cells. In a meta-analysis of lymphocyte subsets in PD, NK cells were more abundant and helper T cells were less abundant than in controls, which could reflect a role for NK cells in regulating helper T cells (17). Thus, NK cell activity may hinder the formation of B cell antibody responses by reducing helper T cell numbers and dampening development of T and B cells.
Alternatively, our findings could point to a role for NK-mediated inflammation in PD. NK cells exert functions as immune regulators through production of cytokines and chemokines that can either augment or activate inflammation (76). We found that strong KIR3DL1-HLA interactions and high KIR3DL1 expression were protective from disease features in PD. This suggests that more inhibition of NK cells results in reduced NK cell mediated inflammation, which would reduce the more severe symptoms of PD. In fact, a recent study clearly demonstrates that KIR3DL1 allotypes associated with higher cell surface expression form an increased number of receptor clusters and transduce more extensive signals (63). We speculate that our findings are consistent with a mechanism in which NK cells function in PD to inhibit B cell production of alpha-synuclein specific antibodies.
In summary, we have performed the first high-resolution analysis of KIR and their functional interactions with HLA class I ligands to identify associations with Parkinson’s disease risk and clinical outcomes. Our results paint a consistent picture whereby compound KIR3DL1-HLA combinations are protective from more acute clinical features of PD and are consistent with a negative impact of NK cell activity in PD (16, 17). We found a distinct pattern of KIR mediated protection from PD whereby highly inhibitory and/or highly expressed alleles were associated with protection from the most severe symptoms of PD including rigidity, gait difficulties and postural instability. Our study is the first to demonstrate evidence of KIR in PD progression and further study could provide support for PD therapies targeting NK cells.
Supplementary Material
Key points:
KIR3DL1 and HLA-Bw4 are associated with different PD clinical features
Highly expressing KIR3DL1 variants protect against more debilitating symptoms of PD
Acknowledgements
We thank all members of the Parkinson Study Group who collected samples used in this study, as well as the patients and their families, whose help and participation made this work possible. The samples used in this work were obtained from the Coriell Biorepository, the Deprenyl and Tocopherol Antioxidative Therapy of Parkinsonism (DATATOP) trial, the UCSF-EPIC project, and the Fox Investigation for New Discovery of Biomarkers (BioFIND; biofind.loni.usc.edu/). BioFIND is sponsored by the Michael J. Fox Foundation for Parkinson’s Research (MJFF) with support from the National Institute for Neurological Disorders and Stroke (NINDS). DATATOP samples were provided for this research by the Indiana University Genetics Biobank with support from the MJFF.
Funding Sources: This study was supported by National Institutes of Health Grant U19NS095774.
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Polymeropoulos MH, Higgins JJ, Golbe LI, Johnson WG, Ide SE, Di Iorio G, Sanges G, Stenroos ES, Pho LT, Schaffer AA, Lazzarini AM, Nussbaum RL, and Duvoisin RC. 1996. Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science 274: 1197–1199. [DOI] [PubMed] [Google Scholar]
- 2.Nussbaum RL, and Polymeropoulos MH. 1997. Genetics of Parkinson’s disease. Hum Mol Genet 6: 1687–1691. [DOI] [PubMed] [Google Scholar]
- 3.Gorell JM, Johnson CC, Rybicki BA, Peterson EL, and Richardson RJ. 1998. The risk of Parkinson’s disease with exposure to pesticides, farming, well water, and rural living. Neurology 50: 1346–1350. [DOI] [PubMed] [Google Scholar]
- 4.Hernan MA, Takkouche B, Caamano-Isorna F, and Gestal-Otero JJ. 2002. A meta-analysis of coffee drinking, cigarette smoking, and the risk of Parkinson’s disease. Ann Neurol 52: 276–284. [DOI] [PubMed] [Google Scholar]
- 5.Hollenbach JA, Norman PJ, Creary LE, Damotte V, Montero-Martin G, Caillier S, Anderson KM, Misra MK, Nemat-Gorgani N, Osoegawa K, Santaniello A, Renschen A, Marin WM, Dandekar R, Parham P, Tanner CM, Hauser SL, Fernandez-Vina M, and Oksenberg JR. 2019. A specific amino acid motif of HLA-DRB1 mediates risk and interacts with smoking history in Parkinson’s disease. Proc Natl Acad Sci U S A 116: 7419–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, Kay DM, Doheny KF, Paschall J, Pugh E, Kusel VI, Collura R, Roberts J, Griffith A, Samii A, Scott WK, Nutt J, Factor SA, and Payami H. 2010. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet 42: 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geraghty JJ, Jankovic J, and Zetusky WJ. 1985. Association between essential tremor and Parkinson’s disease. Ann Neurol 17: 329–333. [DOI] [PubMed] [Google Scholar]
- 8.Zetusky WJ, Jankovic J, and Pirozzolo FJ. 1985. The heterogeneity of Parkinson’s disease: clinical and prognostic implications. Neurology 35: 522–526. [DOI] [PubMed] [Google Scholar]
- 9.Jankovic J, McDermott M, Carter J, Gauthier S, Goetz C, Golbe L, Huber S, Koller W, Olanow C, Shoulson I, and et al. 1990. Variable expression of Parkinson’s disease: a base-line analysis of the DATATOP cohort. The Parkinson Study Group. Neurology 40: 1529–1534. [DOI] [PubMed] [Google Scholar]
- 10.Gasparoli E, Delibori D, Polesello G, Santelli L, Ermani M, Battistin L, and Bracco F. 2002. Clinical predictors in Parkinson’s disease. Neurol Sci 23 Suppl 2: S77–78. [DOI] [PubMed] [Google Scholar]
- 11.Zetusky WJ, and Jankovic J. 1985. Laterality and symptom association in Parkinson’s disease. Arch Neurol 42: 1132–1133. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs D, Sano M, Marder K, Bell K, Bylsma F, Lafleche G, Albert M, Brandt J, and Stern Y. 1994. Age at onset of Alzheimer’s disease: relation to pattern of cognitive dysfunction and rate of decline. Neurology 44: 1215–1220. [DOI] [PubMed] [Google Scholar]
- 13.van der Vlies AE, Koedam EL, Pijnenburg YA, Twisk JW, Scheltens P, and van der Flier WM. 2009. Most rapid cognitive decline in APOE epsilon4 negative Alzheimer’s disease with early onset. Psychol Med 39: 1907–1911. [DOI] [PubMed] [Google Scholar]
- 14.Pagano G, Ferrara N, Brooks DJ, and Pavese N. 2016. Age at onset and Parkinson disease phenotype. Neurology 86: 1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulzer D, Alcalay RN, Garretti F, Cote L, Kanter E, Agin-Liebes J, Liong C, McMurtrey C, Hildebrand WH, Mao X, Dawson VL, Dawson TM, Oseroff C, Pham J, Sidney J, Dillon MB, Carpenter C, Weiskopf D, Phillips E, Mallal S, Peters B, Frazier A, Lindestam Arlehamn CS, and Sette A. 2017. T cells from patients with Parkinson’s disease recognize alpha-synuclein peptides. Nature 546: 656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mihara T, Nakashima M, Kuroiwa A, Akitake Y, Ono K, Hosokawa M, Yamada T, and Takahashi M. 2008. Natural killer cells of Parkinson’s disease patients are set up for activation: a possible role for innate immunity in the pathogenesis of this disease. Parkinsonism Relat Disord 14: 46–51. [DOI] [PubMed] [Google Scholar]
- 17.Jiang S, Gao H, Luo Q, Wang P, and Yang X. 2017. The correlation of lymphocyte subsets, natural killer cell, and Parkinson’s disease: a meta-analysis. Neurol Sci 38: 1373–1380. [DOI] [PubMed] [Google Scholar]
- 18.Bottino C, Moretta L, and Moretta A. 2006. NK cell activating receptors and tumor recognition in humans. Curr Top Microbiol Immunol 298: 175–182. [DOI] [PubMed] [Google Scholar]
- 19.Boudreau JE, and Hsu KC. 2018. Natural killer cell education in human health and disease. Curr Opin Immunol 50: 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao N, Jennings P, Guo Y, and Yuan D. 2011. Regulatory role of natural killer (NK) cells on antibody responses to Brucella abortus. Innate Immun 17: 152–163. [DOI] [PubMed] [Google Scholar]
- 21.James K, and Ritchie AW. 1984. Do natural killer cells regulate B-cell activity? Immunol Today 5: 193–194. [DOI] [PubMed] [Google Scholar]
- 22.Moretta L, Bottino C, Pende D, Castriconi R, Mingari MC, and Moretta A. 2006. Surface NK receptors and their ligands on tumor cells. Semin Immunol 18: 151–158. [DOI] [PubMed] [Google Scholar]
- 23.Hollenbach JA, Ladner MB, Saeteurn K, Taylor KD, Mei L, Haritunians T, McGovern DP, Erlich HA, Rotter JI, and Trachtenberg EA. 2009. Susceptibility to Crohn’s disease is mediated by KIR2DL2/KIR2DL3 heterozygosity and the HLA-C ligand. Immunogenetics 61: 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollenbach JA, Pando MJ, Caillier SJ, Gourraud PA, and Oksenberg JR. 2016. The killer immunoglobulin-like receptor KIR3DL1 in combination with HLA-Bw4 is protective against multiple sclerosis in African Americans. Genes Immun 17: 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rydyznski CE, Cranert SA, Zhou JQ, Xu H, Kleinstein SH, Singh H, and Waggoner SN. 2018. Affinity Maturation Is Impaired by Natural Killer Cell Suppression of Germinal Centers. Cell Rep 24: 3367–3373 e3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kastrukoff LF, Lau A, Wee R, Zecchini D, White R, and Paty DW. 2003. Clinical relapses of multiple sclerosis are associated with ‘novel’ valleys in natural killer cell functional activity. J Neuroimmunol 145: 103–114. [DOI] [PubMed] [Google Scholar]
- 27.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, and Vivier E. 2006. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25: 331–342. [DOI] [PubMed] [Google Scholar]
- 28.Hsu KC, Chida S, Geraghty DE, and Dupont B. 2002. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev 190: 40–52. [DOI] [PubMed] [Google Scholar]
- 29.Parham P, and Moffett A. 2013. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol 13: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper MA, Colonna M, and Yokoyama WM. 2009. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO Rep 10: 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guethlein LA, Norman PJ, Hilton HG, and Parham P. 2015. Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol Rev 267: 259–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wende H, Colonna M, Ziegler A, and Volz A. 1999. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome 10: 154–160. [DOI] [PubMed] [Google Scholar]
- 33.JA Hollenbach IN, MBLadner RM Single, EATrachtenberg 2012. Killer Cell Immunoglobulin-like Receptor (KIR) Gene-Content Variation in the HGDP-CEPH Populations Immunogenetics in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollenbach JA, Augusto DG, Alaez C, Bubnova L, Fae I, Fischer G, Gonzalez-Galarza FF, Gorodezky C, Karabon L, Kusnierczyk P, Noble J, Rickards O, Roberts C, Schaffer M, Shi L, Tavoularis S, Trachtenberg E, Yao Y, and Middleton D. 2013. 16(th) IHIW: Population Global Distribution of Killer Immunoglobulin-like Receptor (KIR) and Ligands. International journal of immunogenetics 40: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gumperz JE, Litwin V, Phillips JH, Lanier LL, and Parham P. 1995. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med 181: 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cella M, Longo A, Ferrara GB, Strominger JL, and Colonna M. 1994. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med 180: 1235–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luque I, Solana R, Galiani MD, Gonzalez R, Garcia F, Lopez de Castro JA, and Pena J. 1996. Threonine 80 on HLA-B27 confers protection against lysis by a group of natural killer clones. Eur J Immunol 26: 1974–1977. [DOI] [PubMed] [Google Scholar]
- 38.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, Holladay M, Rooney B, and Leung W. 2009. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood 114: 5182–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carr WH, Pando MJ, and Parham P. 2005. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol 175: 5222–5229. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, Vlahiotis A, Piccirillo JF, Cella M, Colonna M, Mohanakumar T, Hsu KC, Dupont B, and Yokoyama WM. 2008. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A 105: 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saunders PM, Pymm P, Pietra G, Hughes VA, Hitchen C, O’Connor GM, Loiacono F, Widjaja J, Price DA, Falco M, Mingari MC, Moretta L, McVicar DW, Rossjohn J, Brooks AG, and Vivian JP. 2016. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med 213: 791–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bettencourt A, Silva AM, Carvalho C, Leal B, Santos E, Costa PP, and Silva BM. 2014. The role of KIR2DS1 in multiple sclerosis--KIR in Portuguese MS patients. J Neuroimmunol 269: 52–55. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Xia Q, Fan D, Cai G, Yang X, Wang L, Xin L, Ding N, Hu Y, Liu L, Xu S, Xu J, Wang K, and Pan F. 2015. Association between KIR gene polymorphisms and rheumatoid arthritis susceptibility: A meta-analysis. Hum Immunol 76: 565–570. [DOI] [PubMed] [Google Scholar]
- 44.Rizzo R, Bortolotti D, Gentili V, Rotola A, Bolzani S, Caselli E, Tola MR, and Di Luca D. 2019. KIR2DS2/KIR2DL2/HLA-C1 Haplotype Is Associated with Alzheimer’s Disease: Implication for the Role of Herpesvirus Infections. J Alzheimers Dis 67: 1379–1389. [DOI] [PubMed] [Google Scholar]
- 45.Petrushkin H, Norman PJ, Lougee E, Parham P, Wallace GR, Stanford MR, and Fortune F. 2019. KIR3DL1/S1 Allotypes Contribute Differentially to the Development of Behcet Disease. J Immunol 203: 1629–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, Jayaraman J, Wroblewski EE, Trowsdale J, Rajalingam R, Oksenberg JR, Chiaroni J, Guethlein LA, Traherne JA, Ronaghi M, and Parham P. 2016. Defining KIR and HLA Class I Genotypes at Highest Resolution via High-Throughput Sequencing. Am J Hum Genet 99: 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.University of California, S. F. M SET., Cree BA, Gourraud PA, Oksenberg JR, Bevan C, Crabtree-Hartman E, Gelfand JM, Goodin DS, Graves J, Green AJ, Mowry E, Okuda DT, Pelletier D, von Budingen HC, Zamvil SS, Agrawal A, Caillier S, Ciocca C, Gomez R, Kanner R, Lincoln R, Lizee A, Qualley P, Santaniello A, Suleiman L, Bucci M, Panara V, Papinutto N, Stern WA, Zhu AH, Cutter GR, Baranzini S, Henry RG, and Hauser SL. 2016. Long-term evolution of multiple sclerosis disability in the treatment era. Ann Neurol 80: 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon-Sanchez J, Scholz S, Matarin Mdel M, Fung HC, Hernandez D, Gibbs JR, Britton A, Hardy J, and Singleton A. 2008. Genomewide SNP assay reveals mutations underlying Parkinson disease. Hum Mutat 29: 315–322. [DOI] [PubMed] [Google Scholar]
- 49.Humphrey W, Dalke A, and Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14: 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- 50.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, and Schulten K. 2005. Scalable molecular dynamics with NAMD. J Comput Chem 26: 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, and Caves LS. 2006. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics (Oxford, England) 22: 2695–2696. [DOI] [PubMed] [Google Scholar]
- 52.Boudreau JE, Mulrooney TJ, Le Luduec JB, Barker E, and Hsu KC. 2016. KIR3DL1 and HLA-B Density and Binding Calibrate NK Education and Response to HIV. J Immunol 196: 3398–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, and Parham P. 2006. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 203: 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas R, Yamada E, Alter G, Martin MP, Bashirova AA, Norman PJ, Altfeld M, Parham P, Anderson SK, McVicar DW, and Carrington M. 2008. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? J Immunol 180: 6743–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Team RC 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 3.6.0 ed, Vienna, Austria. [Google Scholar]
- 56.Hochberg Y, and Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat Med 9: 811–818. [DOI] [PubMed] [Google Scholar]
- 57.Augusto DG, and Petzl-Erler ML. 2015. KIR and HLA under pressure: evidences of coevolution across worldwide populations. Hum Genet 134: 929–940. [DOI] [PubMed] [Google Scholar]
- 58.Pinter B, Diem-Zangerl A, Wenning GK, Scherfler C, Oberaigner W, Seppi K, and Poewe W. 2015. Mortality in Parkinson’s disease: a 38-year follow-up study. Mov Disord 30: 266–269. [DOI] [PubMed] [Google Scholar]
- 59.Benninger DH, Thees S, Kollias SS, Bassetti CL, and Waldvogel D. 2009. Morphological differences in Parkinson’s disease with and without rest tremor. J Neurol 256: 256–263. [DOI] [PubMed] [Google Scholar]
- 60.Rosenberg-Katz K, Herman T, Jacob Y, Giladi N, Hendler T, and Hausdorff JM. 2013. Gray matter atrophy distinguishes between Parkinson disease motor subtypes. Neurology 80: 1476–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeshita LY G-GF, dos Santos EJ, Maia MHv, Rahman MM, Zain SM, Middleton D, Jones AR. 2013. A database for curating the associations between killer-cell immunoglobulin-like receptors and diseases in worldwide populations. Database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia-Leon JA, Pinto-Medel MJ, Garcia-Trujillo L, Lopez-Gomez C, Oliver-Martos B, Prat-Arrojo I, Marin-Banasco C, Suardiaz-Garcia M, Maldonado-Sanchez R, Fernandez-Fernandez O, and Leyva-Fernandez L. 2011. Killer cell immunoglobulin-like receptor genes in Spanish multiple sclerosis patients. Mol Immunol 48: 1896–1902. [DOI] [PubMed] [Google Scholar]
- 63.Kennedy PR, Barthen C, Williamson DJ, Pitkeathly WTE, Hazime KS, Cumming J, Stacey KB, Hilton HG, Carrington M, Parham P, and Davis DM. 2019. Genetic diversity affects the nanoscale membrane organization and signaling of natural killer cell receptors. Sci Signal 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, and Parham P. 2001. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol 166: 2992–3001. [DOI] [PubMed] [Google Scholar]
- 65.Saunders PM, MacLachlan BJ, Pymm P, Illing PT, Deng Y, Wong SC, Oates CVL, Purcell AW, Rossjohn J, Vivian JP, and Brooks AG. 2020. The molecular basis of how buried human leukocyte antigen polymorphism modulates natural killer cell function. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kennedy PR, Barthen C, Williamson DJ, and Davis DM. 2019. HLA-B and HLA-C Differ in Their Nanoscale Organization at Cell Surfaces. Front Immunol 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, and Carrington M. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39: 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahn RS, Moslehi H, Martin MP, Abad-Santos M, Bowcock AM, Carrington M, and Liao W. 2016. Inhibitory KIR3DL1 alleles are associated with psoriasis. Br J Dermatol 174: 449–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez-Larrea C, Blanco-Gelaz MA, Torre-Alonso JC, Bruges Armas J, Suarez-Alvarez B, Pruneda L, Couto AR, Gonzalez S, Lopez-Vazquez A, and Martinez-Borra J. 2006. Contribution of KIR3DL1/3DS1 to ankylosing spondylitis in human leukocyte antigen-B27 Caucasian populations. Arthritis Res Ther 8: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boelen L, Debebe B, Silveira M, Salam A, Makinde J, Roberts CH, Wang ECY, Frater J, Gilmour J, Twigger K, Ladell K, Miners KL, Jayaraman J, Traherne JA, Price DA, Qi Y, Martin MP, Macallan DC, Thio CL, Astemborski J, Kirk G, Donfield SM, Buchbinder S, Khakoo SI, Goedert JJ, Trowsdale J, Carrington M, Kollnberger S, and Asquith B. 2018. Inhibitory killer cell immunoglobulin-like receptors strengthen CD8(+) T cell-mediated control of HIV-1, HCV, and HTLV-1. Sci Immunol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stebbins GT, Goetz CG, Burn DJ, Jankovic J, Khoo TK, and Tilley BC. 2013. How to identify tremor dominant and postural instability/gait difficulty groups with the movement disorder society unified Parkinson’s disease rating scale: comparison with the unified Parkinson’s disease rating scale. Mov Disord 28: 668–670. [DOI] [PubMed] [Google Scholar]
- 72.Kotagal V 2016. Is PIGD a legitimate motor subtype in Parkinson disease? Ann Clin Transl Neurol 3: 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, and Iwatsubo T. 1998. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol 152: 879–884. [PMC free article] [PubMed] [Google Scholar]
- 74.Li X, Koudstaal W, Fletcher L, Costa M, van Winsen M, Siregar B, Inganas H, Kim J, Keogh E, Macedo J, Holland T, Perry S, Bard F, Hoozemans JJ, Goudsmit J, Apetri A, and Pascual G. 2019. Naturally occurring antibodies isolated from PD patients inhibit synuclein seeding in vitro and recognize Lewy pathology. Acta Neuropathol 137: 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strowig T, Brilot F, Arrey F, Bougras G, Thomas D, Muller WA, and Munz C. 2008. Tonsilar NK cells restrict B cell transformation by the Epstein-Barr virus via IFN-gamma. PLoS Pathog 4: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schuster IS, Coudert JD, Andoniou CE, and Degli-Esposti MA. 2016. “Natural Regulators”: NK Cells as Modulators of T Cell Immunity. Front Immunol 7: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


