In most environments, bacteria experience conditions where nutrients may be readily abundant or where nutrients are limited. Under nutrient limitation conditions, even non-spore-forming bacteria may enter a dormant state. Dormancy is accompanied by a variety of cellular physiological changes that are required for the cells to remain viable during dormancy and to resuscitate when nutrients become available. Among the physiological changes that occur in dormant bacteria is the inactivation and preservation of ribosomes by the dormancy protein, hibernation-promoting factor (HPF). In this study, we characterized the activity of HPF of Pseudomonas aeruginosa, an opportunistic pathogen that causes persistent infections, and analyzed the role of HPF in ribosome protection and bacterial survival during dormancy.
KEYWORDS: hibernation-promoting factor, dormancy, Pseudomonas, Pseudomonas aeruginosa, biofilms, ribosome modulation factor, ribosomes
ABSTRACT
Hibernation-promoting factor (HPF) is a ribosomal accessory protein that inactivates ribosomes during bacterial starvation. In Pseudomonas aeruginosa, HPF protects ribosome integrity while the cells are dormant. The sequence of HPF has diverged among bacteria but contains conserved charged amino acids in its two alpha helices that interact with the rRNA. Here, we characterized the function of HPF in P. aeruginosa by performing mutagenesis of the conserved residues and then assaying mutant HPF alleles for their ability to protect ribosome integrity of starved P. aeruginosa cells. The results show that HPF functionally tolerates point mutations in charged residues and in the conserved Y71 residue as well as a C-terminal truncation. Double and triple mutations of charged residues in helix 1 in combination with a Y71F substitution reduce HPF activity. Screening for single point mutations that caused impaired HPF activity identified additional substitutions in the two HPF alpha helices. However, alanine substitutions in equivalent positions restored HPF activity, indicating that HPF is tolerant to mutations that do not disrupt the protein structure. Surprisingly, heterologous HPFs from Gram-positive bacteria that have long C-terminal domains functionally complement the P. aeruginosa Δhpf mutant, suggesting that HPF may play a similar role in ribosome protection in other bacterial species. Collectively, the results show that HPF has diverged among bacteria and is tolerant to most single amino acid substitutions. The Y71 residue in combination with helix 1 is important for the functional role of HPF in ribosome protection during bacterial starvation and resuscitation of the bacteria from dormancy.
IMPORTANCE In most environments, bacteria experience conditions where nutrients may be readily abundant or where nutrients are limited. Under nutrient limitation conditions, even non-spore-forming bacteria may enter a dormant state. Dormancy is accompanied by a variety of cellular physiological changes that are required for the cells to remain viable during dormancy and to resuscitate when nutrients become available. Among the physiological changes that occur in dormant bacteria is the inactivation and preservation of ribosomes by the dormancy protein, hibernation-promoting factor (HPF). In this study, we characterized the activity of HPF of Pseudomonas aeruginosa, an opportunistic pathogen that causes persistent infections, and analyzed the role of HPF in ribosome protection and bacterial survival during dormancy.
INTRODUCTION
The ability of bacteria to maintain viability during prolonged periods of starvation is paramount to cell survival (1). As bacteria undergo changes from nutrient abundance to nutrient deficiency, they enter a dormant state. Dormancy results in a variety of cellular physiological changes, including a reduction in cell size, degradation of macromolecules, and proteolytic reprograming of cellular resources (1). During dormancy, certain cellular functions and macromolecules must be protected, preserving the functions that are required for resuscitation when conditions become favorable (e.g., see reference 2). The best characterized example of a change that occurs as cells enter dormancy is the protection of ribosomes (3–5). Under rapidly growing conditions, ribosome abundance per cell increases, but when the cells enter stationary phase, the ribosomes are degraded and recycled (6), presumably as the need for this energy resource outweighs the need for translation (1). However, bacteria protect and maintain a sufficient supply of ribosomes that are required for de novo protein synthesis during resuscitation (7).
Bacteria within biofilms, defined as cells and their extracellular matrices attached to surfaces (8–10), may experience either nutrient abundance or nutrient limitation, depending on their local environmental position within the biofilms (11, 12). As a result, biofilms induce physiological heterogeneity with some cells having active metabolism, while other cells are dormant. For example, measurement of transcriptional activity and oxygen concentrations (13–15) in vertical strata of Pseudomonas aeruginosa biofilms indicated that cells at the periphery of the biofilms, near the air-biofilm interface, are metabolically active, whereas cells in the interior of the biofilms had very low biosynthetic activity (16–18). In P. aeruginosa biofilms, cells with low metabolic activity, isolated from the interior of the biofilms, were tolerant to the antibiotics ciprofloxacin and tobramycin (18). The dormant antibiotic-tolerant cells in the interior of the biofilms also had reduced ribosome content compared to the number of ribosomes per cell in the periphery of the biofilms (7). However, the cells in the interior of the biofilms maintained a sufficient ribosome supply to enable resuscitation (7).
The mechanism for ribosome maintenance in dormant bacteria is becoming increasingly well characterized (see reviews in references 19 and 20). Several bacterial ribosomal accessory proteins were discovered and characterized from Escherichia coli (3–5, 21–23). These proteins include ribosome modulation factor (RMF), hibernation-promoting factor (HPF), and the HPF paralog, YfiA. Early studies using ribosome purification and sucrose gradient centrifugation showed that these ribosome accessory proteins bind ribosomes and cause conformational changes, inducing the formation of inactive 70S, 90S, and 100S ribosome monomers and dimers (24). The model proposed for inactivating ribosomes in E. coli involves binding of RMF to the ribosome mRNA tunnel between the 30S and 50S subunits, forming an inactive 90S ribosome dimer. HPF binds adjacent to RMF, forming a 100S ribosome dimer (23, 25–27). The HPF paralog, YfiA, competes for the HPF-binding site. YfiA may inhibit RMF binding, resulting in formation of inactive 70S ribosome monomers (3).
The structures of ribosomes with HPF and/or RMF in their active sites have now been solved for several diverse species of bacteria, including E. coli (26, 28), Thermus thermophilus (29), Mycobacterium smegmatis (30, 31), and Staphylococcus aureus (32). Structural studies show that HPF binds within the A and P sites of the 30S ribosomal subunit (26, 33), adjacent to the E site containing a deacylated tRNA that may act as a signal for starvation (28). HPF is positioned between helix 30 and helix 44 of the 16S rRNA and interacts with the rRNA by positively charged residues of its two alpha helices. Although the binding site for HPF is consistent in evolutionarily diverse organisms, variations occur in the mechanisms of ribosome hibernation (28). Most gammaproteobacteria contain a gene for RMF and a gene for HPF that encodes a short C-terminal tail. The Gram-positive bacteria and mycobacteria contain an HPF homolog that has a long C-terminal tail but have no RMF homologs. The long C-terminal tail results in HPF homodimers (34) and 100S ribosome dimer formation in the absence of RMF. The orientation of the 100S ribosome dimers differs for bacteria containing HPF with a long C-terminal tail (long-HPF) versus ribosomes containing the short-HPF/RMF combination (28), with the long-HPF causing ribosome dimerization by protein-protein interactions of HPF, while the short-HPF/RMF induces ribosome dimers by both protein-protein and protein-rRNA interactions (28).
P. aeruginosa PAO1 encodes both an RMF homolog and a short HPF homolog but no YfiA homolog (35). In a previous study characterizing transcriptomics of P. aeruginosa biofilm strata, we identified hpf transcripts as abundant in the dormant antibiotic-tolerant cells deep in biofilms (18). We proposed that HPF is essential for maintenance of viability in the dormant P. aeruginosa subpopulations and tested the effect of an hpf deletion on starved planktonic cultures (2). The results showed that wild-type P. aeruginosa PAO1 survives for many days in nutrient-depleted medium with very little loss of viability. In contrast, an hpf deletion mutant had reduced viability over time during starvation. The reduced number of cells capable of forming a CFU in the starved P. aeruginosa Δhpf strain was accompanied by reduced ribosome content of starved cells as measured by fluorescence in situ hybridization (FISH) and selective loss of the 23S rRNA (2). The starved Δhpf strain had other phenotypes following starvation, including (i) heterogeneity in colony size and (ii) increased lag time for the subpopulation of cells that survived starvation. While these phenotypes were observed in the Δhpf mutant strain, the starved P. aeruginosa Δrmf mutant strain did not show similar phenotypes and behaved similarly to the wild-type strain when starved. Those results indicated that HPF, but not RMF, is required for ribosome preservation and resuscitation of starved P. aeruginosa cells (2). A recent single-particle cryoelectron microscopy study of ribosomes isolated from Bacillus subtilis mutants lacking hpf showed that ribosomes from stationary-phase cells lose two small subunit ribosomal proteins, S2 and S3, and that the purified ribosomes from the hpf mutant were impaired in in vitro translation (36). Those results indicated that the initial stages of ribosome loss in starved cells may proceed by disruption of the integrity and activity of the 30S ribosomal subunit.
In the present study, we used the phenotypic responses of starved wild-type and Δhpf mutant cells to characterize the activity of mutant forms of the HPF protein. We characterized the activity by complementing the P. aeruginosa Δhpf strain with HPF proteins containing defined or randomly generated mutations. We then assayed the complemented strains for impaired resuscitation following starvation, which includes colony heterogeneity, increased lag time required for recovery, and reduction of rRNA integrity. Amino acid substitutions were made in highly conserved regions of HPF, and a C-terminal truncation was made to determine the role of the P. aeruginosa HPF C terminus in ribosome protection. We also used error-prone PCR to identify additional HPF amino acids that are important for HPF function. Finally, we characterized the evolutionary relationship of HPF from distantly related species by complementing the P. aeruginosa Δhpf mutant with heterologous HPF homologs. The results show that HPF tolerates single amino acid changes in highly conserved amino acids but has reduced activity when multiple amino acid changes are made. The activity of HPF is affected by amino acid changes that result in misfolding of its two alpha helices. Even with little primary sequence identity, distantly-related HPF homologs complement the P. aeruginosa Δhpf mutant phenotypes, including ribosome protection during starvation.
RESULTS
Functional analyses for the roles of HPF during bacterial starvation.
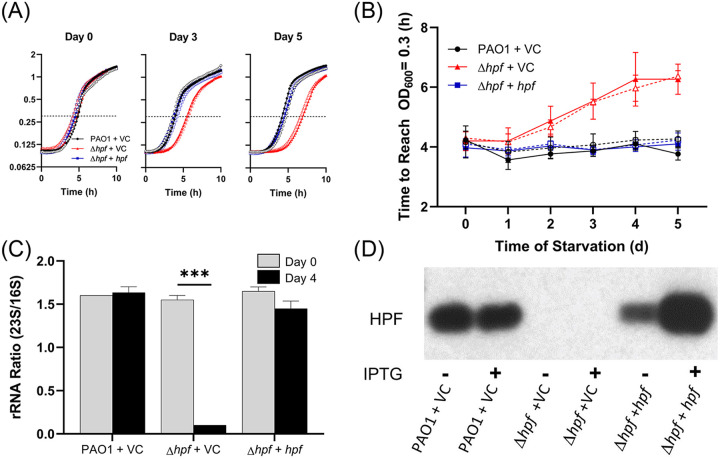
The P. aeruginosa Δhpf strain shows several characteristic phenotypes following starvation that are not observed in the wild-type strain (2) and include the following: (i) a reduction in CFU by 5 days of starvation (see Fig. S1A in the supplemental material); (ii) heterogeneity in colony size for the clones that recover from starvation, including small colony variants that require additional incubation to recover (Fig. S1B); and (iii) loss of ribosome integrity, including selective loss of the 23S rRNA over the course of starvation (see Fig. S2 in the supplemental material). The wild-type phenotypes are restored by complementing the Δhpf mutant with a plasmid copy of hpf (see Fig. S1A and B and Fig. S2). Based on the starvation-induced phenotypes of the P. aeruginosa Δhpf mutant, we developed three HPF functional assays to test the effects of mutant HPF proteins on P. aeruginosa resuscitation and ribosome protection during bacterial starvation. In one assay, we cultured P. aeruginosa Δhpf containing plasmid-carried mutant hpf alleles to stationary phase in tryptic soy broth (TSB) medium (20 h) in the wells of microtiter plates. Aliquots (5 μl) of the cultures were transferred to the wells of microtiter plates containing 225 μl phosphate-buffered saline (PBS) and incubated at 37°C for up to 5 days. Starved cells (5 μl) were removed from the starvation medium each day and allowed to resuscitate in TSB. Regrowth was monitored as an increase in optical density (OD) (Fig. 1A), and the time required for the cells to reach an optical density at 600 nm (OD600) of 0.3 was determined (Fig. 1B). Because of the reduced number of cells capable of resuscitation in the Δhpf mutant (Fig. S1A) and the increased and variable lag time of those capable of resuscitation (2), the time required to reach an OD600 of 0.3 was extended for the Δhpf mutant containing the vector control (VC) (pMF54) compared to that of the wild-type strain carrying the plasmid vector control (Fig. 1B) (P < 0.0001 at days 4 and 5). Complementation of the Δhpf mutant with a plasmid copy of hpf under the Ptrc promoter reduced the time required for starved cells to reach an OD600 of 0.3 to wild-type levels (Fig. 1A and B) (P > 0.99 at days 4 and 5). The second functional assay for the mutant HPF proteins quantified whether the mutant proteins complemented the number of CFU of starved P. aeruginosa Δhpf cells and whether the mutant proteins restored the wild-type colony morphology following starvation (Fig. S1A and B). The third functional assay for HPF characterized ribosome integrity of starved cells by measuring the ratio of 23S/16S rRNA prior to and after 4 days of starvation. The 23S/16S rRNA ratio for the wild-type strain and for the P. aeruginosa Δhpf strain complemented with hpf is approximately 1.6 both prior to and after 4 days of starvation (Fig. 1C). However, for the P. aeruginosa Δhpf strain containing the vector control plasmid, the 23S/16S rRNA ratio is 0.1, indicating selective loss of the 23S rRNA component (Fig. 1C).
FIG 1.
(A) Growth curves of P. aeruginosa PAO1 containing the vector control (VC) plasmid, P. aeruginosa Δhpf with VC, and the P. aeruginosa Δhpf strain complemented with hpf expressed on a plasmid. Open circles indicate that no IPTG was added to the medium, and filled symbols indicate that 1 mM IPTG was added. (B) The time to reach an OD600 of 0.3 over the course of starvation. Dashed lines indicate that no IPTG was added to the medium. Solid lines indicate that 1 mM IPTG was added. The Δhpf strain had increased lag time over the course of starvation compared to that of the wild-type strain. For panels A and B, the error bars show the standard error of the mean for at least three independent biological replicates. (C) Comparison of 23S to 16S rRNA ratios prior to starvation and after 4 days of starvation. The Δhpf strain has 23S/16S rRNA ratios that approach zero after 4 days of starvation, but the ratios are unchanged in the wild-type strain and the hpf complemented strain. Here and throughout the article, the rRNA data show means and standard errors for two biological replicates for the day 0 samples and three biological replicates for the day 4 samples. ***, significant difference at P < 0.0001. (D) Immunoblot analysis of P. aeruginosa strains using anti-HPF antibodies, showing loss of HPF in the Δhpf mutant strain and restoration of HPF in the complemented strain. IPTG induction results in increased HPF amounts compared to the wild-type strain.
Although under the control of the Ptrc promoter, the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to the medium to induce expression of hpf had no effect on the rate of resuscitation in the complemented strain (P > 0.99 at days 4 and 5) (Fig. 1B). Immunoblot analysis using an HPF antibody showed that while IPTG addition increased the amount of HPF produced under the control of the Ptrc promoter, HPF is produced in the absence of IPTG induction (Fig. 1D).
Identification of conserved amino acids in HPF.
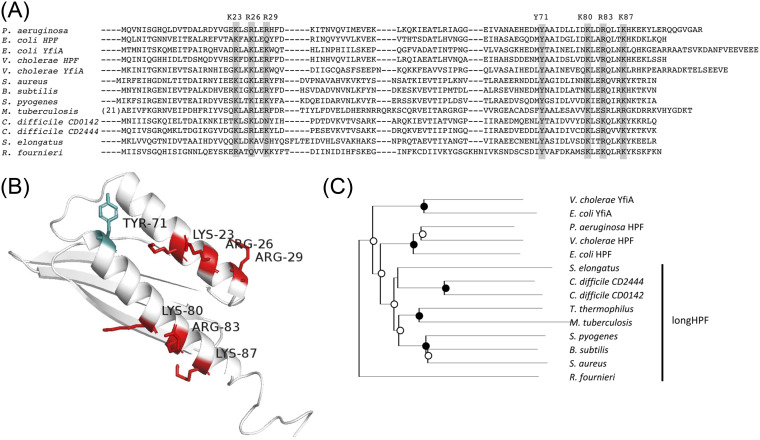
In order to identify targets for HPF mutagenesis studies, we compared the sequence and structures of several HPF homologs from distantly related bacteria. The structures of HPF, including the position of HPF within its ribosome active site, have been solved by X-ray diffraction, solution nuclear magnetic resonance (NMR), and cryoelectron microscopy for a variety of bacteria. Examples are shown in Fig. S3 in the supplemental material, with conserved amino acids highlighted. HPF structures available in the Protein Data Bank (PDB) database (37) include the following: HPF of E. coli (PDB codes 6H58 and 4V8H) (26, 28), YfiA of E. coli (PDB code 4V8I) (26), and Vibrio cholerae HPF (PDB code 4HEI) (38). Gram-positive bacteria contain HPF with a long C-terminal tail (long-HPF). The N-terminal domain of long-HPFs are shown in Fig. S3 and include the following: Bacillus subtilis (PDB code 5NJT) (39), Staphylococcus aureus (PDB codes 5ND9 and 6FXC) (32, 34), and Mycobacterium smegmatis (PDB codes 6DZI and 5ZEP) (30, 31). The C-terminal domain of long-HPF, which plays a role in HPF dimerization, has been characterized for the S. aureus HPF (PDB code 5NKO) (34). We used ClustalX to align the P. aeruginosa HPF homolog with HPF from several other bacteria (Fig. 2A), including the N-terminal region of HPF from Gram-positive bacteria. We also used I-Tasser (40) to model the structure of the P. aeruginosa HPF, showing positions of highly conserved amino acids, including positively charged amino acids that play a role in rRNA interactions (26), and a conserved tyrosine residue (T71) that is found in all known HPF homologs (Fig. 2B). The phylogenetic relationship between the HPF homologs is shown in Fig. 2C. In E. coli, structural analysis of the HPF homolog, YfiA, reveals three positively charged residues in helix I (Arg23, Lys26, and Lys29) and three positively charged residues in helix II (Lys80, Arg83, and Lys87). The E. coli HPF has similar structure but with Lys23 and Gln29 at the equivalent positions to the Arg23 and Lys29 of YfiA (Fig. S3) (26, 33). Multiple sequence alignment of P. aeruginosa HPF with other HPF homologs reveals conservation of the positively charged residues at similar positions within the two alpha helices (Fig. 2A).
FIG 2.
(A) Sequence alignment of the HPF and YfiA from distantly related species of bacteria. The entire HPF or YfiA sequence is shown for the gammaproteobacteria, whereas only the N-terminal domain is shown for the long-HPF of Gram-positive bacteria and mycobacteria. The rectangles show the conserved positively charged amino acids and conserved tyrosine 71 residue that reside within the HPF two alpha helices. The highlighted amino acids of the P. aeruginosa HPF were targets for site-directed mutagenesis studies here. (B) Model of HPF from P. aeruginosa obtained using I-Tasser, highlighting the amino acids targeted for site-directed mutagenesis. (C) Phylogenetic tree of HPF from distantly related bacteria. Black circles indicate bootstrap values of 100%, and open circles indicate bootstrap values of greater than 50%.
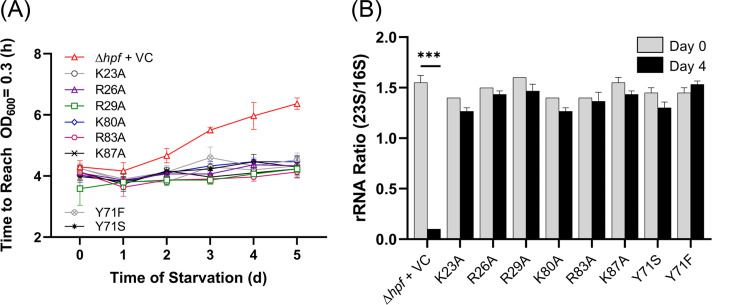
Substitutions of HPF positively charged residues have little effect on HPF function.
We used the functional assays described above to test the effect of changing conserved amino acid residues, which were reported to be required for interactions of the E. coli YfiA or HPF to the rRNA (26). We substituted the conserved charged residues of the P. aeruginosa HPF (K23, R26, R29, K80, R83, and K87) with alanine. The conserved Tyr71 residue was substituted with either a phenylalanine or a serine. All strains containing plasmids with hpf point mutations were compared to the P. aeruginosa Δhpf + hpf control strain and to the wild-type strain carrying the empty vector in the assay testing recovery from starvation as determined by the time required to reach an OD600 of 0.3. Assays were performed on strains cultured both in the presence and absence of IPTG induction of the hpf alleles. All eight mutant proteins complemented the time to reach an OD of 0.3 similar to wild-type P. aeruginosa and to the P. aeruginosa Δhpf + hpf strain (P > 0.99 for all strains at days 0 to 5), and all were statistically different from the P. aeruginosa Δhpf strain with the empty vector control (P < 0.001 on day 4; P < 0.0001 on day 5) (Fig. 3A). IPTG addition to the growth medium (prior to starvation) did not affect the recovery of the mutant strains from starvation, as there was no significant difference from the P. aeruginosa Δhpf strain carrying the wild-type hpf allele, either in the presence or absence of IPTG addition (see Fig. S4A in the supplemental material) (P > 0.99 on days 0 to 5).
FIG 3.
Effect of site-directed mutations on the function of HPF. (A) Resuscitation of the P. aeruginosa Δhpf strain with plasmids containing mutant forms of HPF. Regrowth following starvation curves were used to determine the time to reach an OD600 of 0.3. The P. aeruginosa Δhpf strain containing the vector control is shown as red triangles. The P. aeruginosa Δhpf strain complemented with plasmids containing hpf with point mutations in conserved residues are shown with the colors indicated. Strains were precultured in medium without IPTG. Results of regrowth when strains were precultured with 1 mM IPTG are shown in Fig. S4 in the supplemental material. Error bars here and throughout the manuscript indicate the standard error of the mean for at least three biological replicates for the regrowth data. (B) 23S/16S rRNA ratios of the P. aeruginosa Δhpf strain containing hpf with point mutations prior to starvation and following 4 days of starvation. ***, significant difference at P < 0.0001.
For the CFU assay, the P. aeruginosa Δhpf strain with the vector control had an approximate 5-fold loss in recovery by 5 days of starvation (Fig. S4B), whereas the wild-type strain and the deletion mutant complemented with the wild-type hpf allele had little loss of viability over 5 days of starvation. The P. aeruginosa Δhpf strain containing plasmids with the eight mutant HPF alleles had CFU recovery profiles similar to those of P. aeruginosa Δhpf with the wild-type hpf allele (P > 0.99) (Fig. S4B). To ensure that the plasmids were maintained in all cultures, after 5 days of starvation, the CFU assays were performed by plating on both tryptic soy agar (TSA) medium and on TSA containing carbenicillin. The plasmid maintenance results showed that all strains maintained the plasmids for the 5 days of starvation (see Fig. S5 in the supplemental material) (P > 0.99 for all strains).
We next determined the effect of substitutions of the conserved HPF amino acids on ribosome integrity during P. aeruginosa starvation. For this assay, we compared the 23S/16S rRNA ratio prior to starvation and after 4 days of starvation. The P. aeruginosa Δhpf strain containing wild-type hpf maintained a 23S/16S rRNA ratio of approximately 1.6 after 4 days of starvation (Fig. 1C), whereas the P. aeruginosa Δhpf strain with the vector control plasmid had selective loss of the 23S rRNA, resulting in a 23S/16S ratio that approached zero and was significantly different from that of the wild type (P < 0.0001) (Fig. 3B). Complementing the P. aeruginosa Δhpf strain with a wild-type hpf allele restored the 23S/16S rRNA ratio to wild-type levels following starvation (P > 0.99). Following 4 days of starvation, the 23S/16S rRNA ratios were not significantly different from those of the P. aeruginosa Δhpf + hpf strain for any of these six mutants (P > 0.99 for all strains). In addition, the Y71F and Y71S substitutions had 23S/16S rRNA ratios that did not change significantly following 4 days of starvation and were not different from those of the wild-type allele (P > 0.99) (Fig. 3B).
Overall, these results indicate that single mutations in highly conserved positively charged amino acids and in the conserved Y71 residue of HPF have little effect on the ability of P. aeruginosa to recover from starvation or on ribosome protection by HPF during starvation.
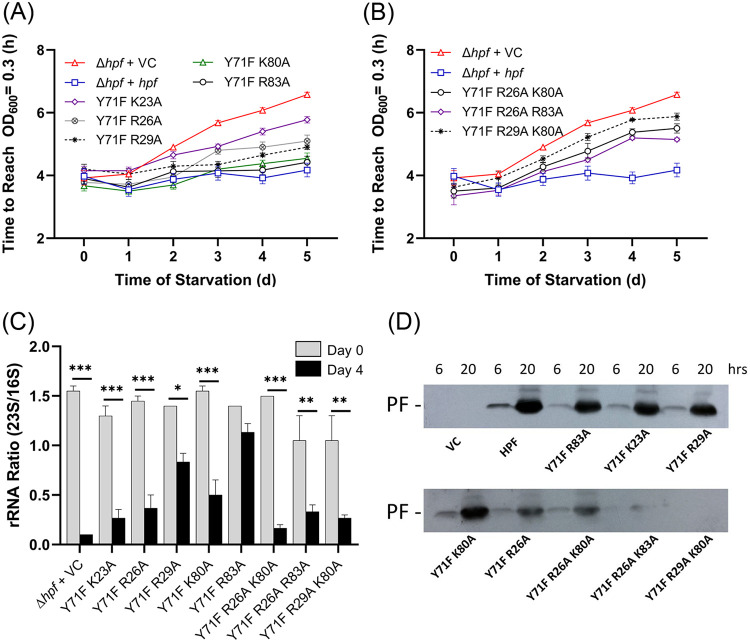
Effect of multiple HPF point mutations on HPF activity.
Since single point mutations in the conserved HPF amino acids had little effect on HPF function, we determined whether multiple point mutations in conserved residues would affect HPF activity. Since the Y71 residue is conserved among all HPF homologs, we constructed double and triple point mutations that included Y71F as one of the mutations and a positively charged amino acid in alpha helix I (K23, R26, or R29) and/or alpha helix II (K80 or R83) as the other mutation(s). Point mutants containing Y71F plus an alanine substitution of a positively charged residue in helix II (Y71F/K80A and Y71F/R83A) had little effect on regrowth kinetics following starvation compared to that of the P. aeruginosa Δhpf strain containing the wild-type allele (P > 0.98 at day 5 of starvation in the absence or in the presence of 1 mM IPTG) (Fig. 4A; see also Fig. S6A in the supplemental material). The Y71F mutation plus a charged residue in helix I (Y71F/K23A, Y71F/R26A, or Y71F/R29A) affected regrowth kinetics following starvation (Fig. 4A; see also Fig. S6), with the Y71F/K23A mutations having the greatest effect on regrowth (P < 0.0001 at day 5) (Fig. 4A). Overexpression of the HPFs with the Y71F/K23A, Y71F/R26A, and Y71F/R29A mutations with 1 mM IPTG partially restored the regrowth kinetics following starvation to near wild-type levels (Fig. S6A). However, the Y71F/K23A and Y71F/R26A alleles were still statistically different from the P. aeruginosa Δhpf strain complemented with the wild-type hpf allele at 5 days of starvation (P < 0.001) (Fig. S6A).
FIG 4.
Effect of double and triple point mutations on the function of HPF. (A) Resuscitation of the P. aeruginosa Δhpf strain with plasmids containing mutant forms of HPF, measured as time to reach an OD600 of 0.3 over the course of starvation. Strains were precultured in the absence of IPTG. Results of regrowth in the presence of 1 mM IPTG are shown in Fig. S6A in the supplemental material. (B) Effect of triple point mutations in HPF on regrowth of the P. aeruginosa Δhpf strain over the course of starvation. (C) 23S/16S rRNA ratios of the P. aeruginosa Δhpf strain containing hpf with double and triple point mutations prior to starvation and after 4 days of starvation. Asterisks indicate significant differences at P < 0.01 (*), P < 0.001 (**), and P < 0.0001 (***). (D) Immunoblot analysis of HPF from strains cultured for 6 h and 20 h.
Triple mutants containing Y71F plus an alanine substitution in both helix I and helix II (Y71F/R26A/K80A, Y71F/R26A/R83A, and Y71F/R29A/K80A) had reduced regrowth kinetics compared to that of the wild-type allele (P < 0.0001 at 5 days) (Fig. 4B). Addition of IPTG to the medium to overexpress the mutant alleles prior to starvation did not restore recovery of these strains to wild-type levels (P < 0.0001) (Fig. S6B).
The CFU assay for regrowth following starvation also showed that the P. aeruginosa Δhpf strain containing HPFs with double point mutations of Y71F plus a mutation helix II (Y71F/K80A or Y71F/R83A) had little effect on recovery of the mutant strains from starvation compared to that of the wild-type allele (P = 0.64 and 0.24) (Fig. S6C). Also similar to the regrowth assay, the greatest effect on CFU following starvation was for the Y71F/K23A mutant (P < 0.01) and for two of the triple mutants (Y71F/R26A/K80A [P < 0.04] and Y71F/R29A/K80A [P = 0.02]) (Fig. S6D). However, in the CFU assay, the Y71A/R26A and Y71A/R29A double mutants and the Y71F/R26A/R83A triple mutant were not significantly different from the wild-type complemented strain (P = 0.39, P = 0.08, and P = 0.32 at 5 days) (Fig. S6D).
The assay for ribosome integrity of the P. aeruginosa Δhpf strain containing double and triple mutations in HPF gave results that reflected the regrowth assays. All double and triple mutant strains had reduced 23S/16S rRNA ratios compared to that of the P. aeruginosa Δhpf strain containing the wild-type allele. However, the least effect on 23S/16S rRNA ratios following 4 days of starvation was for the strain containing HPF with a Y71F mutation paired with a mutation in helix II (Y71F/K80A and Y71F/R83A) in addition to the Y71/R29A strain (Fig. 4C). The greatest effect on ribosome protection was observed when Y71F was paired with a mutation in helix I (Y71F/K23A and Y71F/R26A). The P. aeruginosa Δhpf strain with HPF containing triple point mutants had reduced 23S/16S rRNA following starvation (Fig. 4C), indicative of partial ribosome loss during starvation.
In order to ensure that the mutant HPF proteins were expressed and maintained in the P. aeruginosa Δhpf strain cells, we performed immunoblot analysis of the HPF proteins using HPF antibodies. The results showed that most of the mutant proteins were expressed at levels similar to those encoded by the wild-type allele at 6 and 20 h of growth (Fig. 4D). Exceptions were for two of the triple mutants, Y71F/R26A/K83A and Y71F/R29A/K80A, where there was loss of HPF at the 20-h time point, possibly due to degradation of the mutant protein.
The results of double and triple mutant studies show that HPF with a Y71F substitution coupled with a second mutation in helix II have the least effect on HPF activity, while a Y71F mutation coupled with a mutation in helix I has the greatest effect on HPF activity in vivo. Triple mutations of Y71F plus mutations of charged residues in both helix I and helix II have an additive effect on HPF impairment. However, certain triple mutants may be disrupted to the extent that the mutant proteins are targeted for degradation.
HPF amino acid substitutions that affect HPF activity by disrupting protein stability.
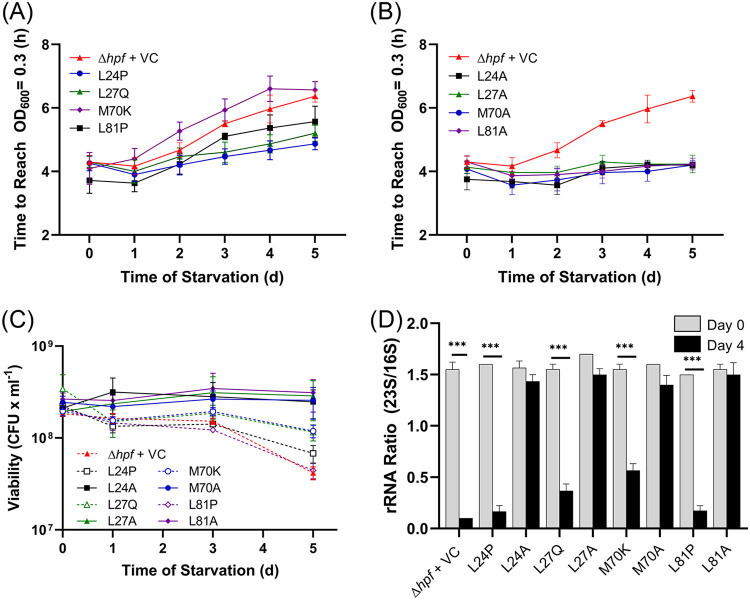
In order to characterize HPF further, we screened for additional amino acid substitutions that disrupted HPF activity. For these studies, we performed random mutagenesis of hpf using error-prone PCR and screened for mutant strains that had delayed regrowth following starvation. A library of mutant hpf alleles was introduced into the P. aeruginosa Δhpf strain, and the resulting strains were assayed for increased lag time on recovery following starvation. Of the 116 mutants that had increased lag time following starvation, many had nonsense mutations in hpf. Sixty-eight mutants contained from one to five amino acid substitutions, with 20 having a single point mutation. Of these 20, many were determined to be false positives upon rescreening. The remaining single point mutants were assayed again for recovery over 5 days using the time to reach an OD600 of 0.3 following starvation. Substitutions with the largest deficit in recovery were M70K and leucine residues adjacent to the conserved positively charged residues described above (L24P, L27Q, and L81P) (Fig. 5A). Overexpression of these mutant alleles by the addition of IPTG to the medium did not influence the ability of these strains to recover from starvation compared to the P. aeruginosa Δhpf strain with the empty vector control (see Fig. S7A in the supplemental material) (P > 0.49 for all strains at days 0 to 5). Included in the HPF mutant alleles that impaired P. aeruginosa recovery from starvation were substitutions that may have affected protein folding (e.g., proline substitutions). Therefore, we used alanine substitutions in equivalent positions (L24A, L27A, L81A, and M70A) and tested the mutant HPF proteins for in vivo activity as described above. When the residues were replaced with alanine, the regrowth kinetics of the P. aeruginosa Δhpf strain containing these alleles were restored to wild-type levels (P > 0.99 for both 0 and 1.0 mM IPTG) (Fig. 5B; see also Fig. S7B).
FIG 5.
(A) Regrowth following starvation of the P. aeruginosa Δhpf strain containing hpf with random point mutations obtained by error-prone PCR and screening for a resuscitation defect and the P. aeruginosa Δhpf strain containing the vector control. (B) Regrowth following starvation of the P. aeruginosa Δhpf strain complemented with hpf where the mutations shown in panel A were replaced by alanine. (C) Resuscitation of P. aeruginosa Δhpf strains containing point mutations, enumerated as CFU over 5 days of starvation. Shown are the responses of hpf with mutations obtained by random mutagenesis and screening for resuscitation defect (dashed lines) and mutations with alanine in equivalent positions as the random mutations (solid lines). (D) The 23S to 16S rRNA ratios of the P. aeruginosa Δhpf strain with mutant hpf alleles prior to starvation and following 4 days of starvation. ***, significant difference at P < 0.0001.
We next tested the P. aeruginosa Δhpf strain containing the hpf mutant alleles identified in the random screening and the alanine substitutions in the equivalent positions for their effect on CFU, colony morphology, and ribosome integrity of the starved cells. The P. aeruginosa Δhpf strain containing HPF with mutations L24P and L81P had reduced CFU during recovery compared to that of the wild-type strain with the empty vector (P = 0.0018 for L24P and P < 0.0001 for L81P) (Fig. 5C). These mutant strains also showed colony heterogeneity upon regrowth, including small colony variants (see Fig. S8 in the supplemental material). However, alanine substitutions in equivalent positions restored the number of CFU to near wild-type levels (P = 0.34 for L24A and P = 0.094 for L81A) (Fig. 5C) and also restored the colony morphologies to those similar to the wild-type strain (Fig. S8). The ribosome integrity assay correlated with the regrowth assays, where the L24P, L27Q, L81P, and M70K mutants resulted in decreased 23S/16S rRNA ratios that were significantly different from those of the wild type (P < 0.0001, analysis of variance [ANOVA]) (Fig. 5D). When L24, L27, L81, and M70 were converted to alanine, the 23S/16S rRNA ratio of starved cells was restored to near wild-type levels (P > 0.99) (Fig. 5D).
The L24P, L27Q, and L81P mutations occurred in helix 1 and helix II of HPF. Since these substitutions may have impacted the HPF alpha helices, we determined whether these proteins were targeted for degradation due to misfolding. Immunoblot analysis of P. aeruginosa PAO1 cells over the course of growth showed that HPF is present 6 h postinoculation and has maximum abundance at 10 h (Fig. 6). The P. aeruginosa Δhpf strain complemented with hpf under the control of the Ptrc promoter also had HPF that was initially detected at 6 h (Fig. 6). Strains with mutations L24P, L27Q, and L81P did not show increasing amounts of HPF over growth. Rather, the abundance of the proteins decreased, with little detectable HPF after 20 h (Fig. 6). When the mutations that caused impairments were converted to alanine (L24A, L27A, and L81A), HPF abundance was restored to levels similar to those of the wild-type HPF (Fig. 6). An exception was the M70K mutation, which had reduced ribosome protection (P < 0.0001, ANOVA) (Fig. 5) but maintained HPF protein levels throughout growth (Fig. 6). The results of the random screening assay indicate that HPF function can be affected by single point mutations that disrupt either of the alpha helices. Disruption of the alpha helices likely targets the misfolded HPF for degradation since alanine substitutions for these amino acids do not cause an impairment in HPF function or stability.
FIG 6.
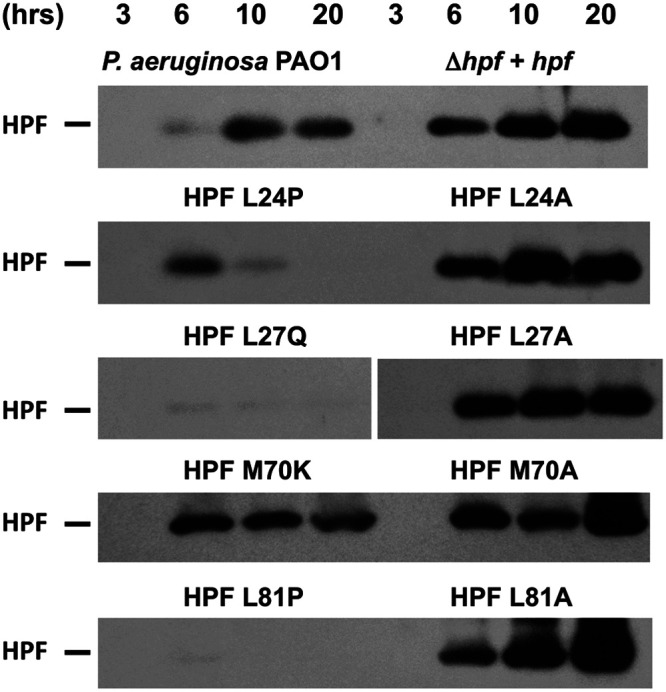
Immunoblot of P. aeruginosa PAO1 and the P. aeruginosa Δhpf strain containing hpf mutant alleles using anti-HPF antibodies. Shown are immunoblots for hpf alleles obtained by random mutagenesis and screening for impaired regrowth and for alanine substitutions in equivalent positions within HPF. Cells were cultured in the presence of 1 mM IPTG, and samples were taken at the times indicated. Representative images from at least three replicates are shown.
The P. aeruginosa HPF C-terminal tail does not affect RMF functional activity.
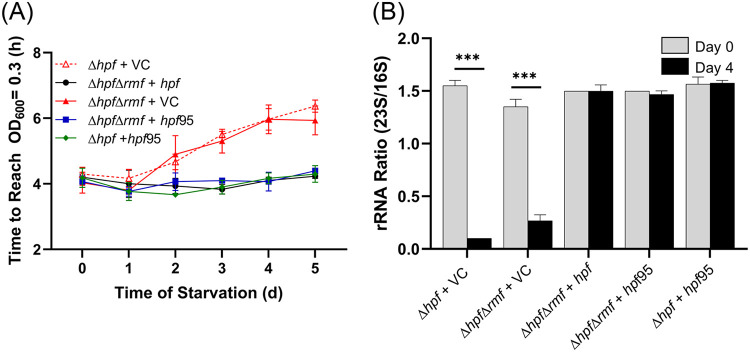
E. coli encodes two hpf paralogs (HPF and YfiA). In E. coli, YfiA has a longer C-terminal tail than HPF (YfiA, 108 amino acids [aa]; HPF, 95 aa), and the extended C-terminal tail of YfiA inhibits RMF binding resulting in formation of inactive 70S ribosome monomers rather than 100S dimers, which contain both HPF and RMF (3). The P. aeruginosa HPF protein has an intermediate length C-terminal tail (HPF, 102 aa) (Fig. 2A). To determine if the P. aeruginosa HPF affects the activity of RMF, we truncated the C-terminal tail of the P. aeruginosa HPF to 95 aa and determined whether the truncated HPF complements the P. aeruginosa Δhpf strain and the P. aeruginosa Δhpf Δrmf double mutant. The Δhpf Δrmf double mutant strain is similar to the Δhpf strain both for resuscitation rate (P > 0.99) (Fig. 7A; see also Fig. S9A in the supplemental material) and for the number of CFU recovered (P > 0.99, ANOVA) (Fig. S9B). The truncated P. aeruginosa HPF protein complemented the P. aeruginosa Δhpf mutant similar to the full-length HPF (Fig. 7A; see also Fig. S9A and B) (P > 0.99 for the regrowth assay; P = 0.95 for the CFU assay). In addition, the truncated HPF protein also complemented the P. aeruginosa Δhpf Δrmf double mutant in both assays similar to full-length HPF (P > 0.99 for the resuscitation assay; P = 0.74 for the CFU assay) (Fig. 7A; see also Fig. S9A and B). Finally, the truncated form of HPF restored the ribosome integrity phenotype of both the Δhpf mutant and the Δhpf Δrmf double mutant strains (P > 0.99 for both strains) (Fig. 7B). The results demonstrate that the C terminus of the P. aeruginosa HPF does not influence the starvation or ribosome protection phenotypes in the Δhpf mutant or in the Δhpf Δrmf double mutant.
FIG 7.
Resuscitation and ribosome protection of the P. aeruginosa Δhpf strain and the Δhpf Δrmf double mutant containing hpf truncated to 95 amino acids. (A) Resuscitation over the course of starvation determined as time to reach an OD of 0.3 for the Δhpf mutant with vector control, the Δhpf Δrmf double mutant with vector control, and complemented strains. The hpf95 truncated gene complemented both the Δhpf mutant and the Δhpf Δrmf double mutant. (B) Ribosome protection of truncated hpf95 in the Δhpf mutant and in Δhpf Δrmf double mutant, measured as 23S/16S rRNA ratio prior to starvation and after 4 days of starvation. ***, significant difference at P < 0.0001.
Distantly related HPF homologs are functional in P. aeruginosa.
While RMF is restricted to the gammaproteobacteria, HPF is broadly distributed in bacteria (Fig. 2). However, HPF from Gram-positive bacteria differs from HPF from gammaproteobacteria both in sequence identity and because the Gram-positive HPFs have a long C-terminal domain (long-HPF), making those proteins approximately twice the size of HPF from P. aeruginosa. The C-terminal domain of long-HPF is a dimerization domain that facilitates ribosome dimerization in the absence of RMF (34, 39).
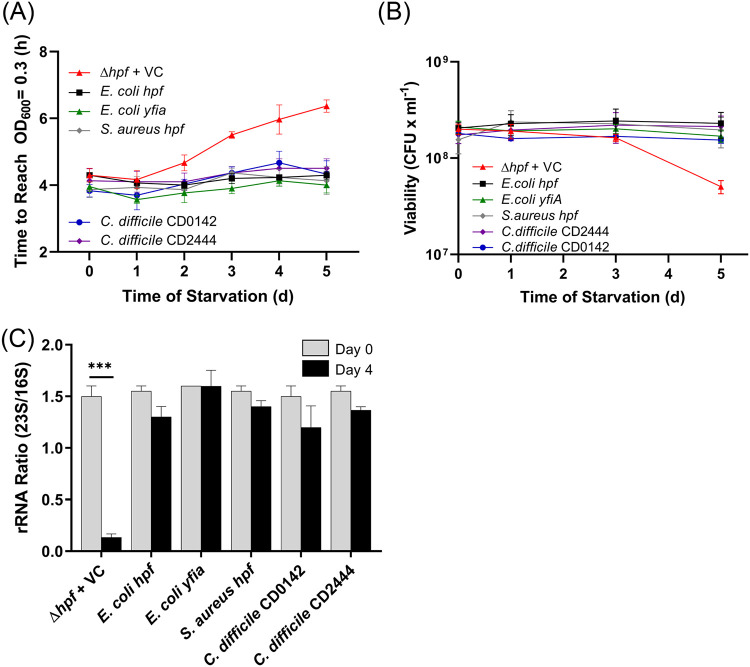
To begin to determine if HPFs from distantly related bacteria play a similar role in ribosome protection during starvation, we asked if heterologous HPF homologs could protect ribosome integrity of P. aeruginosa. Of the heterologous HPFs tested, the E. coli HPF shares the greatest sequence identity to P. aeruginosa HPF (80% identity and 55% similarity) (Fig. 2C). The E. coli hpf complemented the P. aeruginosa Δhpf strain for regrowth (P > 0.99), CFU (P > 0.99), and for ribosome integrity (P = 0.90) following starvation, both in the presence and absence of induction with IPTG (Fig. 8; see also Fig. S10 and S11 in the supplemental material). Similarly, the E. coli YfiA, which has lower sequence identity to the P. aeruginosa HPF (61% identity and 35% similarity) also restored all three phenotypes of the starved P. aeruginosa Δhpf mutant. Overexpression of yfiA with 1 mM IPTG resulted in slightly impaired growth of the P. aeruginosa Δhpf strain that was not observed when the P. aeruginosa hpf or the E. coli hpf was overexpressed in P. aeruginosa (see Fig. S12 in the supplemental material).
FIG 8.
Resuscitation and ribosome protection of the P. aeruginosa Δhpf strain containing heterologous hpf homologs. (A) Time required to reach an OD of 0.3 following starvation for the P. aeruginosa Δhpf strain containing hpf genes from E. coli, S. aureus, and C. difficile. (B) Regrowth of the P. aeruginosa Δhpf strain containing heterologous hpf genes measured as CFU following starvation. (C) Ratio of 23S/16S rRNA of the P. aeruginosa Δhpf strain containing heterologous hpf homologs before starvation and after 4 days of starvation. No IPTG was added to the medium prior to starvation; results with pregrowth with IPTG are shown in Fig. S10 in the supplemental material. ***, significant difference at P < 0.0001.
The long-HPF from Staphylococcus aureus USA300 (KEGG accession number SAUSA300_0736; 190 aa) has 35% identity and 63% similarity to the P. aeruginosa HPF over the first 95-aa N-terminal domain and no similarity to P. aeruginosa HPF or to RMF in its C-terminal dimerization domain. However, the long-HPF homolog from S. aureus USA300 functionally complemented the P. aeruginosa Δhpf mutant in regrowth from starvation assays (P > 0.99 in the presence and absence of IPTG) (Fig. 8A and B; see also Fig. S10). The S. aureus long-HPF also complemented the P. aeruginosa Δhpf mutant for ribosome integrity, in both the presence and absence of IPTG induction (P > 0.99) (Fig. 8C; see also Fig. S10C and S11). Clostridium difficile VPI 10463 (GenBank accession number NZ_CM000604) has two long-HPF paralogs. CD0142 (183 aa) has 38% identity and 66% similarity to the P. aeruginosa HPF over the first 95 amino acids, and CD2444 (187 aa) has 40% identity and 70% similarity to the P. aeruginosa HPF over the first 95 amino acids and no similarity to P. aeruginosa HPF in the C-terminal domain. C. difficile CD0142 and CD2444 complemented the P. aeruginosa Δhpf deletion mutant with respect to regrowth delay following starvation in the presence and absence of IPTG (P > 0.99) (Fig. 8A and B; see also Fig. S10). Without IPTG induction, CD0142 and CD2444 had average 23S/16S rRNA ratios that did not significantly change after 4 days of starvation (P > 0.99) (Fig. 8C). When overexpressed with the addition of 1 mM IPTG, the 23S/16S rRNA ratio of the strain containing CD02444 remained unchanged following starvation (P > 0.99), while the strain containing CD0124 had a small, but statistically significant, reduction in the relative abundance of 23S rRNA (P = 0.0074) (Fig. S10C). All of the heterologous HPF homologs restored the colony morphologies of the P. aeruginosa Δhpf strain to phenotypes similar to those of wild-type P. aeruginosa PAO1. Overall, the results demonstrate that the three long-HPFs functionally complement the ribosome integrity defect associated with the starved P. aeruginosa Δhpf mutant, allowing the strain to resuscitate from starvation in a manner similar to the native P. aeruginosa HPF.
DISCUSSION
Bacteria experience environmental conditions that vary between nutrient abundance and starvation. During periods of starvation, metabolic reprograming includes reduction in cell size and degradation of excess macromolecules that can be used for energy, carbon, nitrogen, or phosphorus sources (1). Ribosome abundances are reduced in stationary-phase cells (6) by ribosome recycling, which includes nuclease cleavage of the rRNAs, followed by degradation of the 23S and 16S rRNA subunits. The ribosomal accessory proteins, HPF and RMF, likely evolved not only to inhibit translation during stationary phase but also to prevent complete degradation of ribosomes during extended periods of starvation (2). HPF allows cells to maintain a minimum reservoir of ribosomes that are needed for protein synthesis when conditions become favorable for resuscitation from dormancy. HPF and RMF bind within the mRNA channel of the ribosome adjacent to a deacylated tRNA in the E site (26, 28), causing conformational changes that result in the formation of ribosome dimers (21). Formation of inactive 100S ribosomes inhibits the initial endonuclease cleavage of the 23S rRNA, preventing ribosome degradation (2). In addition, as shown in B. subtilis, HPF and ribosome dimerization protects the ribosome during stationary phase by preventing loss of the small subunit proteins S2 and S3 (36).
HPF is broadly distributed among bacteria, and homologs are even found in the chloroplasts of plants (41). Therefore, HPF is likely an evolutionarily ancient protein that has been maintained in bacteria, particularly those that experience changes in environmental conditions, from nutrient abundance to nutrient starvation. In contrast, RMF is restricted to the gammaproteobacteria. RMF plays a role in blocking translation (28) but does not appear to be necessary for ribosome maintenance during starvation conditions in P. aeruginosa (2). Although the primary sequence of HPF has diverged through evolution, the HPF structure has been maintained, and certain amino acids are conserved in the HPF homologs, suggesting that these amino acids are important for HPF function. These amino acids include the positively charged amino acids within the two HPF alpha helices that contact rRNA (in P. aeruginosa as follows: K23, R26, R29, K80, R83, and K87) as well as the highly conserved Y71 residue (Fig. 2). In this study, we developed HPF functional assays in a P. aeruginosa Δhpf mutant background to test the role of these conserved amino acids on HPF activity in vivo. The assays included recovery from starvation conditions (regrowth in liquid culture, viability assayed by the number of CFU, and colony morphology of resuscitating cells) as well as loss of ribosome integrity following starvation, measured as the ratio of the 23S to 16S rRNA. Surprisingly, conversion of any of the conserved positively charged amino acids (K23, R26, R29, K80, R83, and K87) to alanine did not affect the function of HPF. Mutant strains containing amino acid substitutions in these conserved residues all recovered from starvation similarly to the wild-type strain, and all mutant strains had protected ribosomes during starvation conditions (Fig. 3). Similarly, mutations in the conserved Y71 residue, when changed to either a phenylalanine or serine, did not affect P. aeruginosa recovery from starvation or the ability of HPF to protect ribosome integrity. These results indicate that HPF is tolerant to amino acid changes in conserved amino acids and likely reflects the role of HPF as a structural protein rather than a typical enzyme with a catalytic domain. The role of HPF in protection of ribosomes during starvation was only disrupted when multiple amino acid changes were made (Fig. 4; see also Fig. S6 in the supplemental material). In particular, when the conserved Y71 residue was paired with a substitution of a charged residue in helix I, HPF activity was reduced. However, HPF activity in the double and triple mutants, while reduced, was not completely eliminated. The results indicate that HPF with multiple mutations still maintains partial activity, particularly when overexpressed.
HPF proteins with impaired in vivo activity were identified by using a random screening method. The mutant proteins, when expressed in the P. aeruginosa Δhpf strain, caused impaired ribosome protection, and therefore, the strains had reduced ability to resuscitate from starvation (Fig. 5). The results further support the role of HPF as a dormancy factor that is required for ribosome maintenance during dormancy. The mutations that caused the greatest impairment in both resuscitation and ribosome protection were in amino acids that likely disrupted HPF structure. In particular, amino acids that caused disruption of the two HPF alpha helices (L24P, L28Q, and L81P) had the greatest influence on the ability of the P. aeruginosa Δhpf strain to resuscitate from dormancy. Since the leucine residues were adjacent to the positively charged amino acids known to be important for HPF interaction with the rRNA, we suspected that the environment of the charged residue was important for activity. However, when the L24P, L28Q, and L81P mutations were converted to L24A, L28A, and L81A, wild-type phenotypes were restored both for ribosome protection during dormancy and resuscitation from dormancy. Therefore, the most likely effect of the L24P, L28Q, and L81P mutations was to disrupt proper folding of the HPF alpha helices. Immunoblot analyses indicated that the HPF L24P, L27Q, and L81P mutant proteins were targeted for degradation, while the HPF L24A, L27A, and L81A mutants were maintained in the cell throughout growth and were functional (Fig. 6). Very few HPF L24P, L28Q, and L81P proteins were observed after 20 h of growth compared to those of the wild-type control strain (Fig. 6). In contrast, the HPF L24A, L28A, and L81A mutant proteins were abundant in the cells at 20 h and would be available for ribosome protection during the starvation period. An exception was the M70K mutation, which was not degraded during growth but did not fully complement the functional role of HPF in ribosome protection. Since the M70 residue is adjacent to Y71, which is highly conserved and important for HPF activity, the change in charge caused by the M70K mutation likely affected the Y71 environment and inhibited HPF activity since nonpolar amino acids adjacent to Y71 are found in all of the HPF homologs (Fig. 2). M70 is located at the N terminus of helix 2 and the M70K mutation likely did not affect protein folding since the mutant protein was not targeted for degradation. When M70 was replaced with a nonpolar alanine residue, activity of HPF was restored. Overall, the results support the conclusion that HPF is capable of tolerating individual amino acid changes as long as they do not induce protein degradation. The results also implicate the importance of Y71 and its environment in HPF activity. It is interesting that while many amino acids within HPF are evolutionarily conserved, they are not essential for HPF activity.
In E. coli, RMF binds ribosomes in the mRNA channel near the anti-Shine-Dalgarno sequence of the 16S rRNA (28). Interactions of RMF with ribosomal protein S1 inhibit translation during the stationary phase of E. coli (28). In our previous study (2), we did not detect a role for RMF in ribosome protection during starvation conditions. P. aeruginosa PAO1 does not have a YfiA homolog. The function of the E. coli HPF and the paralog YfiA differ. Maki et al. (21) demonstrated that YfiA and HPF compete for ribosome binding and that YfiA induces formation of inactive 70S ribosome monomers, while HPF works in concert with RMF to form inactive 100S ribosome dimers. In E. coli, the longer C-terminal tail of YfiA compared to the C-terminal tail of HPF inhibits RMF binding, thereby preventing 100S ribosome dimer formation (21). Since P. aeruginosa HPF has a C-terminal tail with intermediate length between the E. coli HPF and E. coli YfiA, we tested whether HPF behaves more like YfiA with the C-terminal tail that competes for ribosome binding with RMF. We performed assays where the P. aeruginosa HPF was truncated to 95 amino acids (equivalent to the E. coli HPF) and introduced that into both the P. aeruginosa Δhpf mutant strain and a P. aeruginosa Δhpf Δrmf double mutant background to determine if the truncated hpf could functionally complement the resuscitation and ribosome protection phenotypes in the presence and absence of RMF. The truncated form of the HPF protein (HPF-95) fully complemented the Δhpf mutant and Δhpf Δrmf double mutant phenotypes with respect to rRNA protection and cell recovery from starvation (Fig. 7). Therefore, a functional role for the P. aeruginosa RMF in ribosome protection during dormancy is still not apparent.
Interestingly, the yfiA homolog from E. coli caused increased lag time when overexpressed in the P. aeruginosa Δhpf strain, whereas the E. coli hpf (or the P. aeruginosa hpf) did not affect P. aeruginosa growth (see Fig. S12 in the supplemental material). In P. aeruginosa, hpf is expressed as cells enter stationary phase under the control of the Phpf promoter (42). In the experiments here, when IPTG is added to the medium, hpf is expressed earlier in growth. YfiA may be more efficient at blocking translation than HPF, and if expressed early in growth, as described here, it may inhibit the regrowth of P. aeruginosa (Fig. S12).
The HPF primary sequence has diverged over the course of evolution, having about 30% primary sequence identity over the first 95 amino acids of the N-terminal domain. The long-HPFs from Gram-positive bacteria have a C-terminal domain, making the HPF from Gram-positive bacteria approximately twice as long as the HPF from proteobacteria. The C-terminal domain from Gram-positive bacteria is a dimerization domain that results in the formation of HPF homodimers that cause the formation of inactive 100S ribosome dimers (34). Beckert et al. (28) recently demonstrated that the arrangement of 100S ribosomes formed by the RMF/HPF combination in proteobacteria differs from the arrangement formed by the long-HPF of Gram-positive bacteria. Therefore, it is surprising that the long-HPF from Gram-positive bacteria is able to functionally complement the Δhpf deletion in the Gram-negative bacterium P. aeruginosa. However, long-HPFs from S. aureus and from C. difficile are able to complement the P. aeruginosa Δhpf mutant in the resuscitation from starvation assays, the ribosome integrity during starvation assay, and the colony morphology assay (Fig. 8; see also Fig. S10 in the supplemental material). These results indicate that long-HPF in Gram-positive bacteria may be functionally equivalent to HPF in P. aeruginosa for ribosome maintenance when cells are dormant and likely plays similar roles in their native organisms.
The results here show that HPF is a ribosomal accessory protein that is widely distributed among bacteria. While the primary sequence of the N-terminal domain (95 amino acids) has diverged, certain amino acids, particularly positively charged amino acids and Y71, are conserved among the HPF proteins from different bacterial taxa. However, point mutations in individual conserved amino acids do not affect the functional activity of HPF with respect to ribosome protection during starvation or for recovery of P. aeruginosa cells from starvation. The C-terminal tail of the P. aeruginosa HPF does not appear essential for activity. The HPF proteins from diverse species, although inducing differing confirmations in the 100S ribosomes (28), are functionally equivalent in protection of ribosomes from degradation during starvation and in ensuring sufficient ribosome supply during starvation for the bacteria to resuscitate.
MATERIALS AND METHODS
Strains.
Strains used in this study are listed in Table S1 in the supplemental material. All HPF mutant proteins and HPF homologs were carried on a pMF54 vector backbone (43) and expressed in the P. aeruginosa PAO1 Δhpf strain or the P. aeruginosa PAO1 Δhpf Δrmf strain (2). Wild-type P. aeruginosa PAO1 carrying pMF54 and the P. aeruginosa Δhpf strain carrying a wild-type copy of hpf in pMF54 served as positive controls. The P. aeruginosa Δhpf strain carrying pMF54 was used as a negative control.
Media and reagents.
Strains were grown in TSB (BD Bacto tryptic soy broth) with 1.0 mM IPTG (Teknova) with 150 μg/ml carbenicillin (Teknova) when indicated. For starvation studies, strains were incubated in phosphate-buffered saline (PBS)—1.4 g/liter Na2HPO4, 8 g/liter NaCl, 0.2 g/liter KCl, 0.2 g/liter KH2PO4, pH 7.0—for up to 5 days. Restriction enzymes (NcoI and HindIII), high-fidelity Phusion PCR kit, Taq polymerase, and Quick ligase were obtained from New England BioLabs. Primers used for PCR amplification and site-directed mutagenesis were obtained from integrated DNA technology (IDT), and sequences are shown in Table S2 in the supplemental material.
Generation of site-directed HPF mutants.
Site-directed mutations were generated using overlap extension PCR (44) using P. aeruginosa PAO1 genomic DNA as a template and Phusion polymerase (New England BioLabs). For each mutation, two internal primers were designed containing the desired substitution and an overlapping region corresponding to the adjacent strand. The region upstream of the desired mutation was amplified with 5′_NcoI_hpf_inframe and the site-specific UP_REV internal primer (see Table S2). The region downstream of the mutation was amplified with the site-specific Down_FW internal primer and HPF_3'_HindIII primer (Table S2). For overlap PCR, 50 ng of the gel-purified upstream and downstream PCR products were ligated and amplified in a second PCR with 5′_NcoI_hpf_inframe and HPF_3'_HindIII. The sequences of overlapping mutagenic oligonucleotides are shown in Table S2. To generate double and triple point mutants, overlap extension PCR was used with the Y71A internal primers and the plasmid DNA containing the single point mutation as the template.
Generation of heterologous HPF homologs.
HPF homologs were PCR amplified from templates of E. coli TOP10 cells, genomic DNA from Clostridium difficile VPI 10463 (provided by Seth Walk), or genomic DNA from Staphylococcus aureus LAC (USA-300) (provided by Jovanka Voyich-Kane). Gene-specific primers containing flanking NcoI and HindIII sites were used so that heterologous hpf genes could be cloned into pMF54, which contained an IPTG-inducible Ptrc promoter. Plasmids were extracted using the Qiagen MiniPrep kit and confirmed by sequencing (GenScript). Site-directed hpf mutant and heterologous hpf genes were introduced into the P. aeruginosa Δhpf strain by electroporation.
Error-prone PCR for random mutagenesis.
Random mutations of hpf were generated with error-prone PCR. Error-prone PCR was performed in a 100-μl reaction mixture with Taq polymerase in ThermoPol buffer with unbalanced deoxynucleoside triphosphates (dNTPs) (0.2 mM dGTP, 0.2 mM dATP, 1 mM dCTP, 1 mM dTTP), 5.5 mM MgCl2, 0.1 mM MnCl2, and 0.3 mM each primer (5′_NcoI_HindIII_inframe and HPF_3'_HindIII) for 14 cycles. The length of the PCR product was confirmed with gel electrophoresis, and the product was gel excised and purified with a Qiagen MinElute purification kit. The PCR product was digested with NcoI and HindIII, ligated into the NcoI and HindIII sites of pMF54, and transformed into E. coli TOP10 cells. Following overnight incubation, 10 transformation plates with isolated colonies were pooled by resuspension in 10 ml of LB containing 100 μg/ml ampicillin. The pooled culture was incubated for 30 min at 37°C with shaking at 200 rpm in a baffled flask, after which the culture was pelleted into 3-ml pools of E. coli carrying mutant plasmids. Mixed plasmid libraries were extracted from these E. coli pellets with a Qiagen MiniPrep kit. To generate the P. aeruginosa mutant pool, 1.0 μg of the plasmid library was transformed into the electrocompetent P. aeruginosa Δhpf strain with a 15-min recovery in super optimal broth with catabolite repression (SOC) at 37°C prior to plating on LB agar containing 150 μg/ml carbenicillin using methods described by Choi et al. (45). Colonies from these transformation plates were inoculated into the wells of microtiter plates to assay mutant cell recovery following starvation.
Regrowth assay for recovery following starvation.
We adapted an assay described previously (2) to characterize recovery of the P. aeruginosa Δhpf strain containing mutant hpf genes from starvation. P. aeruginosa strains (5 μl of overnight culture) were inoculated into individual wells of a 96-well flat-bottom microtiter plate containing 225 μl/well of TSB with 50 μl carbenicillin ± 1 mM IPTG as indicated. The strains were incubated with shaking at 200 rpm at 37°C for 20 h. For starvation, 5 μl from each well was transferred to 225 μl of PBS in a 96-well microtiter plate. The starvation plates were sealed with parafilm to prevent evaporation and incubated statically at 37°C for up to 5 days. The cultures were sampled daily by inoculating 5 μl of the starved cultures into a flat-bottom microtiter plate containing 195 μl TSB. Regrowth of the starved cultures was monitored over 16 h at 37°C in a BioTek Epoch 2 plate reader with continuous shaking. Resuscitation of the mutant and control strains was determined as the time required for the starved cultures to reach an OD600 of 0.3. Control cultures included wild-type P. aeruginosa PAO1 carrying the empty vector control (pMF54), the P. aeruginosa Δhpf strain with vector control plasmid pMF54, and the P. aeruginosa Δhpf strain complemented with a wild-type copy of hpf. At least three independent biological replicates were performed per mutant strain. The time to reach an OD600 of 0.3 at each day of starvation was plotted in GraphPad Prism v. 8.2 as the mean ± standard error of the mean (SEM). GraphPad Prism was also used to perform 2-way ANOVA with Bonferroni correction for multiple testing to determine significant differences from the control strains at each time point at an α of <0.01.
Viability assay for recovery following nutrient starvation.
Strains were incubated overnight in 3 ml TSB with 150 μg/ml carbenicillin at 37°C. Inoculum (10 μl) of each culture was then transferred to 4 ml TSB with 50 μg/ml carbenicillin ± 1 mM IPTG and incubated for 20 h at 37°C on a test tube roller. Following incubation, 1 ml of culture was washed twice with 1 ml PBS and resuspended in 1.0 ml PBS. Washed cells (350 μl) were added to 4 ml of PBS, pH 7.0, for starvation studies. Starvation cultures were then incubated at 37°C on a roller for up to 5 days. Starvation cultures were sampled for CFU at 30 min, 1 day, 3 days, and 5 days. Aliquots (25 μl) of the starved cultures were removed, serially diluted, and plated for CFU on TSA medium. A minimum of three independent biological replicates was performed for each strain. The mean CFU per milliliter and standard error of the mean (SEM) of the starved cultures were calculated using GraphPad Prism v. 8.2. Log reduction was calculated using the following formula: log reduction = log10 (CFU × ml−1Day5/CFU × ml−1Day1). GraphPad Prism was used to perform ANOVA to determine if the log reduction following starvation in mutant strains was significantly different from the control strains at an α of <0.01. To determine if plasmids were retained in the starved cultures, the CFU of 5 days starved cultures were determined by plating on TSA containing carbenicillin and comparing this to CFU on TSA without carbenicillin. The mean CFU per milliliter ± SEM for the recovering colonies on both types of media were plotted using GraphPad Prism v. 8.2, and a 2-way ANOVA was performed with Bonferroni correction to determine if there was a significant difference in recovery with or without carbenicillin at an α of <0.01. The CFU assay was also used as an indicator of colony morphology following starvation (2). Photographs of colonies were taken after 20 h of regrowth for each starvation sampling time.
rRNA analysis.
Starvation cultures were prepared in 25-ml flasks as in Akiyama et al. (2). After 30 min and after 4 days of starvation, a 1-ml aliquot was removed from each flask, pelleted by centrifugation (9,500 rpm, 2 min, 4°C), and frozen at −80°C. Total RNA was extracted with the hot phenol method as previously described (2). The RNA Clean and Concentrator-5 kit (Zymo Research) was used according to the manufacturer’s instructions. RNA was measured on the NanoDrop 1000 (Thermo Fisher Scientific) and visualized on the Bioanalyzer 2100 (Agilent Technologies) with the Prokaryotic Total RNA 6000 nanoassay (Agilent Technologies). The ratio of 23S rRNA to 16S rRNA was plotted using GraphPad Prism v. 8.2 as the mean and standard error of the mean for two independent biological replicates for nonstarved cultures and three or more independent biological replicates for 4-day starved cultures. We chose to perform only two replicates for the nonstarved cultures because the 23S/16S ratios of nonstarved samples rarely varied from 1.5. GraphPad Prism was also used to perform a two-way ANOVA with Bonferroni correction to determine if the rRNA ratio in each strain was significantly different following 4 days of starvation at an α of <0.01.
Immunoblot analysis.
Anti-HPF antibodies were raised in rabbits as described previously (46). Briefly, hpf was cloned into expression vector pET28A, in frame for an N-terminal fusion to the 6× polyhistidine tag. The 6×His-tagged HPF was purified from E. coli extracts using a nickel affinity column as described previously (46). Purified HPF was used to generate anti-HPF antibodies by Lampire Biological Laboratories (Pipersville, PA). Immunoblots were performed as described previously (46). P. aeruginosa PAO1 and the P. aeruginosa Δhpf strain carrying mutant hpf genes on the pMF54 expression vector were incubated with shaking in microtiter plates in TSB at 37°C. Samples were assayed at 3 h, 6 h, 10 h, or 20 h postinoculation. Aliquots (200 μl) of cultures were removed and centrifuged (10,000 rpm for 3 min), and cell pellets were stored at −80°C. Samples were resuspended in 50 μl of radioimmunoprecipitation assay (RIPA) buffer, and cells were lysed by incubation at 85°C for 5 min. To reduce viscosity, universal nuclease (Thermo Fisher) (1.0 μl diluted 1:20) was added, and incubation was carried out at room temperature for 5 min. Protein sample buffer (50 μl) was added to the samples, and 10-μl aliquots were run on 12% SDS-PAGE gels (47). Gels were electroblotted onto nitrocellulose membranes and probed with anti-HPF antibodies. Detection of antibody binding was determined using secondary antibodies (goat anti-rabbit IgG labeled with horseradish peroxidase) and chemiluminescent imaging.
Bioinformatics.
Sequence alignments for the HPF from P. aeruginosa and from other bacterial species was performed using ClustalX (48). Coordinates for HPF protein structures were obtained from the Protein Data Bank (https://www.rcsb.org/) (37). The structure of HPF from P. aeruginosa was predicted using I-Tasser (40).
Supplementary Material
ACKNOWLEDGMENTS
We thank Seth Walk and Jovanka Voyich-Kane of Montana State University for providing Clostridium difficile and Staphylococcus aureus genomic DNA. We thank Steve Hamner for his support on this work.
This work was funded by a grant from the National Institute of Allergy and Infectious Diseases (NIAID) to Michael J. Franklin under grant number AI113330.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bergkessel M, Basta DW, Newman DK. 2016. The physiology of growth arrest: uniting molecular and environmental microbiology. Nat Rev Microbiol 14:549–562. doi: 10.1038/nrmicro.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama T, Williamson KS, Schaefer R, Pratt S, Chang CB, Franklin MJ. 2017. Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc Natl Acad Sci U S A 114:3204–3209. doi: 10.1073/pnas.1700695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueta M, Yoshida H, Wada C, Baba T, Mori H, Wada A. 2005. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 4.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. 1995. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun 214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 5.Wada A, Yamazaki Y, Fujita N, Ishihama A. 1990. Structure and probable genetic location of a "ribosome modulation factor" associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci U S A 87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutscher MP. 2003. Degradation of stable RNA in bacteria. J Biol Chem 278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 7.Pérez-Osorio AC, Williamson KS, Franklin MJ. 2010. Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J Bacteriol 192:2991–3000. doi: 10.1128/JB.01598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 9.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 10.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 11.Franklin MJ, Chang C, Akiyama T, Bothner B. 2015. New technologies for studying biofilms. Microbiol Spectr 3:MB-0016-2014. doi: 10.1128/microbiolspec.MB-0016-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 13.James GA, Ge Zhao A, Usui M, Underwood RA, Nguyen H, Beyenal H, deLancey Pulcini E, Agostinho Hunt A, Bernstein HC, Fleckman P, Olerud J, Williamson KS, Franklin MJ, Stewart PS. 2016. Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound Repair Regen 24:373–383. doi: 10.1111/wrr.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewandowski Z, Beyenal H. 2001. Limiting-current-type microelectrodes for quantifying mass transport dynamics in biofilms. Methods Enzymol 337:339–359. doi: 10.1016/s0076-6879(01)37024-6. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen K, Lewandowski Z. 1998. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol Bioeng 59:302–309. doi:. [DOI] [PubMed] [Google Scholar]
- 16.Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, Buckingham-Meyer K, Stewart PS. 2007. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol 189:4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenz AP, Williamson KS, Pitts B, Stewart PS, Franklin MJ. 2008. Localized gene expression in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 74:4463–4471. doi: 10.1128/AEM.00710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, Franklin MJ. 2012. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol 194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gohara DW, Yap MF. 2018. Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr Genet 64:753–760. doi: 10.1007/s00294-017-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prossliner T, Skovbo Winther K, Sorensen MA, Gerdes K. 2018. Ribosome hibernation. Annu Rev Genet 52:321–348. doi: 10.1146/annurev-genet-120215-035130. [DOI] [PubMed] [Google Scholar]
- 21.Maki Y, Yoshida H, Wada A. 2000. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 22.Ueta M, Ohniwa RL, Yoshida H, Maki Y, Wada C, Wada A. 2008. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem 143:425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida H, Ueta M, Maki Y, Sakai A, Wada A. 2009. Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase. Genes Cells 14:271–280. doi: 10.1111/j.1365-2443.2008.01272.x. [DOI] [PubMed] [Google Scholar]
- 24.Wada A, Mikkola R, Kurland CG, Ishihama A. 2000. Growth phase-coupled changes of the ribosome profile in natural isolates and laboratory strains of Escherichia coli. J Bacteriol 182:2893–2899. doi: 10.1128/jb.182.10.2893-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato T, Yoshida H, Miyata T, Maki Y, Wada A, Namba K. 2010. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18:719–724. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Polikanov YS, Blaha GM, Steitz TA. 2012. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato A, Watanabe T, Maki Y, Ueta M, Yoshida H, Ito Y, Wada A, Mishima M. 2009. Solution structure of the E. coli ribosome hibernation promoting factor HPF: implications for the relationship between structure and function. Biochem Biophys Res Commun 389:580–585. doi: 10.1016/j.bbrc.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 28.Beckert B, Turk M, Czech A, Berninghausen O, Beckmann R, Ignatova Z, Plitzko JM, Wilson DN. 2018. Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat Microbiol 3:1115–1121. doi: 10.1038/s41564-018-0237-0. [DOI] [PubMed] [Google Scholar]
- 29.Flygaard RK, Boegholm N, Yusupov M, Jenner LB. 2018. Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism. Nat Commun 9:4179. doi: 10.1038/s41467-018-06724-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Sharma MR, Koripella RK, Yang Y, Kaushal PS, Lin Q, Wade JT, Gray TA, Derbyshire KM, Agrawal RK, Ojha AK. 2018. Zinc depletion induces ribosome hibernation in mycobacteria. Proc Natl Acad Sci U S A 115:8191–8196. doi: 10.1073/pnas.1804555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra S, Ahmed T, Tyagi A, Shi J, Bhushan S. 2018. Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA. Sci Rep 8:13587. doi: 10.1038/s41598-018-31850-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matzov D, Aibara S, Basu A, Zimmerman E, Bashan A, Yap MF, Amunts A, Yonath AE. 2017. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat Commun 8:723. doi: 10.1038/s41467-017-00753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH. 2004. Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol 11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 34.Khusainov I, Vicens Q, Ayupov R, Usachev K, Myasnikov A, Simonetti A, Validov S, Kieffer B, Yusupova G, Yusupov M, Hashem Y. 2017. Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EMBO J 36:2073–2087. doi: 10.15252/embj.201696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 36.Feaga HA, Kopylov M, Kim JK, Jovanovic M, Dworkin J. 2020. Ribosome dimerization protects the small subunit. J Bacteriol 202:e00009-20. doi: 10.1128/JB.00009-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berman H, Henrick K, Nakamura H. 2003. Announcing the worldwide Protein Data Bank. Nat Struct Biol 10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 38.De Bari H, Berry EA. 2013. Structure of Vibrio cholerae ribosome hibernation promoting factor. Acta Cryst 69:228–236. doi: 10.1107/S1744309113000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beckert B, Abdelshahid M, Schafer H, Steinchen W, Arenz S, Berninghausen O, Beckmann R, Bange G, Turgay K, Wilson DN. 2017. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J 36:2061–2072. doi: 10.15252/embj.201696189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy A, Kucukural A, Zhang Y. 2010. I-Tasser: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trosch R, Willmund F. 2019. The conserved theme of ribosome hibernation: from bacteria to chloroplasts of plants. Biol Chem 400:879–893. doi: 10.1515/hsz-2018-0436. [DOI] [PubMed] [Google Scholar]
- 42.Akiyama T, Williamson KS, Franklin MJ. 2018. Expression and regulation of the Pseudomonas aeruginosa hibernation promoting factor. Mol Microbiol 110:161–175. doi: 10.1111/mmi.14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franklin MJ, Chitnis CE, Gacesa P, Sonesson A, White DC, Ohman DE. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol 176:1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 45.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Franklin MJ, Ohman DE. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J Bacteriol 184:3000–3007. doi: 10.1128/jb.184.11.3000-3007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 48.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. 1998. Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.