Abstract
Numerous environmental toxicants have been shown to induce the epigenetic transgenerational inheritance of disease and phenotypic variation. Alterations in the germline epigenome are necessary to transmit transgenerational phenotypes. In previous studies, the pesticide DDT (dichlorodiphenyltrichloroethane) and the agricultural fungicide vinclozolin were shown to promote the transgenerational inheritance of sperm differential DNA methylation regions, non-coding RNAs and histone retention, which are termed epimutations. These epimutations are able to mediate this epigenetic inheritance of disease and phenotypic variation. The current study was designed to investigate the developmental origins of the transgenerational differential histone retention sites (called DHRs) during gametogenesis of the sperm. Vinclozolin and DDT were independently used to promote the epigenetic transgenerational inheritance of these DHRs. Male control lineage, DDT lineage and vinclozolin lineage F3 generation rats were used to isolate round spermatids, caput epididymal spermatozoa, and caudal sperm. The DHRs distinguishing the control versus DDT lineage or vinclozolin lineage samples were determined at these three developmental stages. DHRs and a reproducible core of histone H3 retention sites were observed using an H3 chromatin immunoprecipitation-sequencing (ChIP-Seq) analysis in each of the germ cell populations. The chromosomal locations and genomic features of the DHRs were analyzed. A cascade of epigenetic histone retention site alterations was found to be initiated in the round spermatids and then further modified during epididymal sperm maturation. Observations show that in addition to alterations in sperm DNA methylation and ncRNA expression previously identified, the induction of differential histone retention sites (DHRs) in the later stages of spermatogenesis also occurs. This novel component of epigenetic programming during spermatogenesis can be environmentally altered and transmitted to subsequent generations through epigenetic transgenerational inheritance.
Keywords: Epigenetics, Histones, Transgenerational, Inheritance, Vinclozolin, DDT, Sperm, Spermatogenesis
Introduction
Transgenerational epigenetic inheritance refers to the transmission of epigenetic information to the next generation through the germline. The main epigenetic processes previously studied are DNA methylation, functional noncoding RNA, chromatin structure, histones modifications, and RNA methylation [1]. Changes in these epigenetic parameters can have short or long-term effects on genome activity and become transgenerational when the germline is involved [2]. The first epigenetic molecular mark identified was DNA methylation, where a methyl group is chemically attached to a cytosine base at CpG residues in DNA [3, 4]. DNA wraps around an octamer of histones (a family of basic proteins) to form a nucleosome, which is the basic subunit of chromatin. Post-translational histone modifications are essential for gene expression and cells to properly function. For example, it has been shown that histone deacetylases control embryonic stem cell differentiation [5], histone phosphorylation is involved in mitosis [6, 7], and dysregulated function of histone modifying enzymes can be linked to the initiation of cancer development [8]. Proper regulation of epigenetic processes is crucial throughout spermatogenesis to ensure sperm function and subsequent embryonic development following fertilization. Therefore, the sperm epigenetics plays an essential role in establishing epigenetic marks and transcription in the embryo [9].
Numerous studies in humans and in animal models have demonstrated that embryonic and fetal development is sensitive to environmental exposures to adverse conditions which can result in numerous pathologies, such as cardiometabolic, neurobehavioral and reproductive disorders in later life [10]. These observations have led to the concept of “Developmental Origins of Health and Diseases” (DOHaD). Interestingly, these phenotypes have also been shown not only in the first generation (F1) but may be transmitted to a second (F2) or a number of subsequent generations through non-genetic epigenetic mechanisms [11–13]. Epidemiological studies have also observed such effects on birth weight and cardiovascular risk [14–17]. Animal studies have also been undertaken to confirm these findings where prenatal insults appear to influence the health of future generations with effects on reproductive health and behavior [2].
Previous observations have demonstrated that a gestating female exposed to environmental toxicants such as the agricultural fungicide vinclozolin and the pesticide DDT (dichlorodiphenyltrichloroethane) during the fetal gonadal sex determination window promotes an altered developmental programming of the male germline epigenome [2, 18]. The fungicide vinclozolin is an anti-androgenic endocrine disruptor compound used in the fruit and vegetable agricultural industries [19], and was one of the first compounds shown to promote the epigenetic transgenerational inheritance of pathologies [2]. Vinclozolin induced F3 transgenerational pathology which includes testis, prostate, kidney and ovarian disease. The pesticide DDT is an estrogenic endocrine disruptor compound that was used in the agricultural industry in the 1950s and 1960s and banned in the early 1970s in the USA due to human and animal health impacts [20]. DDT was one of the first endocrine disruptors shown to promote transgenerational obesity [18], as well as testis, prostate, kidney and ovarian disease. Caloric restriction, high fat diets, traumatic stress, and a variety of different toxicants have also been linked to the transgenerational epigenetic inheritance phenomenon [21–23]. This non-genetic form of inheritance requires epigenetic modifications of the germline (sperm and egg) to pass altered epigenomes to the early embryo, that can then impact the transcriptomes and epigenetics of all subsequently derived somatic cells [2, 21]. Different epigenetic processes are involved in the transgenerational germline transmission where the environment can alter the health and evolution of the species [22–25].
The male germline is derived from its stem cell population of primordial germ cells (PGCs) that develop and migrate to the genital ridge to colonize the undifferentiated gonad [26]. Upon gonadal sex determination, the PGCs develop into the male or female germline lineage based on the sex of the gonad [27]. The early stage testis germ cells develop into the pro-spermatogonia during the male gonadal sex determination period by embryonic day 16 (E16) in the rat gonad [28, 29]. Following birth, the germline differentiates into spermatogonia by the postnatal day 10 (P10) pre-pubertal period. Following the onset of puberty, the initial wave of spermatogenesis in the testis begins, and the spermatocyte stages develop including the meiotic stage of pachytene spermatocytes. Following meiosis, the round spermatid stage develops. In the final stage, the spermatozoa are released into the lumen of the seminiferous tubules [30]. The spermatozoa then enter the epididymis for further differentiation at the caput stage of the epididymis and mature during transit to the final caudal stage of the epididymis where the sperm have acquired the capacity for motility [31, 32]. The mature caudal epididymal sperm are then stored in the vas deferens prior to ejaculation or degradation.
Male germ cells go through major chromatin remodeling events during the haploid stages of spermiogenesis [33–35]. A multistep replacement of histones by transition proteins and then protamines takes place, resulting in a unique sperm chromatin structure with the major part of the DNA tightly packaged with protamines [36–38]. This chromatin remodeling is required as protamination is important in condensing the DNA into the head of the sperm and protecting the DNA [39–42]. Alterations in the sperm protamine content often leads to DNA damage and male infertility [43–47]. Furthermore, protamine packaging is involved in gene silencing, preventing transcriptional activity during the latter half of spermiogenesis and in sperm. Therefore, the sperm predominantly contain protamines and are transcriptionally silent, but some histone retention sites have been previously observed in sperm and vary in genome content from 5–10% depending on the species [41, 48]. Upon fertilization, the highly compacted sperm chromatin is again remodeled when protamines are removed and replaced by maternally derived histones, except for the paternally derived histone retention sites [41, 49, 50]. The paternally retained histones can associate with imprinted genes and influence subsequent embryonic cell epigenetics and transcriptomes [51, 52]. The sperm paternal histone retention alterations provide a mechanism to promote epigenetic inheritance [52, 53], that has been shown to appear in subsequent offspring sperm [52]. Epigenetic transgenerational inheritance of parent-of-origin allelic transmission of paternally derived sperm epimutations and pathology is proposed to in part be mediated through environmentally altered sperm histone retention [54].
Epigenetic regulation is subject to change based on environmental stimuli. Several of our transgenerational studies using rats have demonstrated that a specific set of histone retention sites are retained in the transgenerational F3 generation control lineage sperm [25, 55, 56]. This same set of histones is retained in F3 generation rats ancestrally exposed to vinclozolin or DDT, but additional histone retention sites (DHRs) are induced in the vinclozolin and DDT lineage sperm [25, 55, 56]. Previous literature supports a role for sperm epigenetic alterations and histone modifications to impact subsequent embryo epigenetics and gene expression in mice [52, 57], humans [58, 59], and worms [60]. One of the most direct demonstrations of histone modifications in the sperm that can regulate embryo gene expression and epigenetics was observed in the frog embryo [61].This suggests that histone retention and modifications may be associated in part with sperm mediated transgenerational inheritance of disease following ancestral DDT or vinclozolin exposure.
The later haploid male germline developmental periods used for the current study are the round spermatids, caput epididymal sperm, and caudal epididymal sperm to investigate the developmental origins of the transgenerational sperm differential histone retention regions (DHRs). The current study was designed to investigate the developmental programming and origins of the vinclozolin and DDT induced epigenetic transgenerational inheritance of sperm DHRs. The hypothesis tested is that, similarly to what was observed with the DMRs (differential DNA methylated regions) [62, 63], a cascade of epigenetic changes occur during development, such that the majority of DHRs arise in later haploid spermatogenic stages.
Results
Germ Cell Isolation
Three different germ cell stages from transgenerational F3 generation great-grand offspring control, DDT and vinclozolin lineage male rats were used in this experimental design for epigenetic analysis. The F0 generation gestating female rats at 90 days of age were transiently exposed to DDT, vinclozolin or a vehicle control during gestational embryonic days E8–14. At this stage of fetal development, the PGCs have migrated to colonize the undifferentiated gonad which then initiates the early stages of gonadal sex determination [1, 64]. At 90 days of age, the F1 generation offspring were bred within the control, DDT and vinclozolin lineages (i.e. not crossed with control) to generate the F2 generation (grand offspring), which were then bred at 90 days of age to obtain the F3 generation for each control, DDT and vinclozolin lineage. An outbred group of unrelated Harlen Sprague-Dawley male and female rats were used with no cousin or sibling breeding to avoid any inbreeding artifacts. Only the F0 generation gestating females were exposed to DDT or vinclozolin, which also directly exposed the F1 generation fetus and the germline within the F1 generation fetuses that will generate the F2 generation. Therefore, the F3 generation is the first transgenerational generation not directly exposed [1, 65], so was used to study the developmental origins of the transgenerational sperm histone retention. The F3 generation control, DDT and vinclozolin lineage male rats were aged to 8–10 months for isolation of round spermatid, caput spermatozoa, and cauda epididymal sperm. The isolation of the round spermatid cell type was done using a gravity sedimentation StaPut protocol, as described in the Methods. The epididymal caput spermatozoa and cauda sperm were isolated from the appropriate section of the epididymis from the same animals, as described in the Methods. Three different pools each with different individual sets of animals from different litters were obtained with 3 adult males per pool. The H3 histones and methylation H3K27me3 histones were isolated from each purified round spermatid stage pool, and from individual animals for epididymal caput spermatozoa and cauda sperm and then three pools generated for subsequent epigenetic analysis.
Differential Histone Retention (DHR) Identification
The analysis of the DHRs was performed with a chromatin immunoprecipitation (ChIP) procedure followed by next generation sequencing for a ChIP-Seq protocol described in the Methods [55, 56]. At each developmental stage studied, round spermatid (RS), caput spermatozoa, and cauda sperm, the F3 generation control versus DDT or control versus vinclozolin lineage pools were compared to identify the environmental toxicant lineage DHRs in the male germline. The DHRs at various p-value thresholds are presented at each developmental stage in Fig. 1C–H. An EdgeR p-value of p<1e-04 was selected at all stages for further analysis of the DHRs. The All Windows is all DHRs with at least one 100 bp region (window) with statistical significance, and the Multiple Windows being ≥2 neighboring significant 100 bp windows (Fig. 1). The number of multiple windows and number of DHRs at p<1e-04 is presented for each data set, Fig. 1. In addition, the F3 generation control lineage developmental stages were compared within the control lineage, including the round spermatid (RS) versus caput spermatozoa (caput) and the caput spermatozoa (caput) versus cauda sperm (cauda), Fig. 1A &B. The number of epigenetic changes (i.e. DHRs) were higher in the control lineage developmental stage comparisons. The highest numbers of DHRs were between the caput spermatozoa and the cauda sperm stages with 188 DHRs (Fig. 1B). The majority of DHRs were single 100 bp window sites. The chromosomal locations of the DHRs at each developmental stage are presented in Fig. 2. The red arrowheads represent the location of a DHR, while the black boxes indicate clusters of DHRs. The DHRs were spread throughout the genome with some DHR clusters. Although mitochondrial DNA (MT) contains primarily nucleoids [66], histones have been observed [67, 68], as seen in Fig. 2A & G. At each stage of development, the genomic features of the DHRs were examined, Supplemental Tables S1–S3. The CpG density of the DHRs at all stages was 1–4 CpG per 100 bp with 1 CpG per 100 bp being predominant, which is characteristic of a low density CpG deserts [69], Supplemental Figure S1. This has been observed with previous transgenerational DHRs and DMRs [55]. The length of the DHRs at all the developmental stages were between 1–4 kb with 1 kb length being predominant, Supplemental Figure S2. Consequently, the DHRs are primarily 1 kb in size with around 10 CpGs, as previously observed [55]. This is anticipated to consist of five nucleosomes with approximately 200 bp each.
Figure 1.
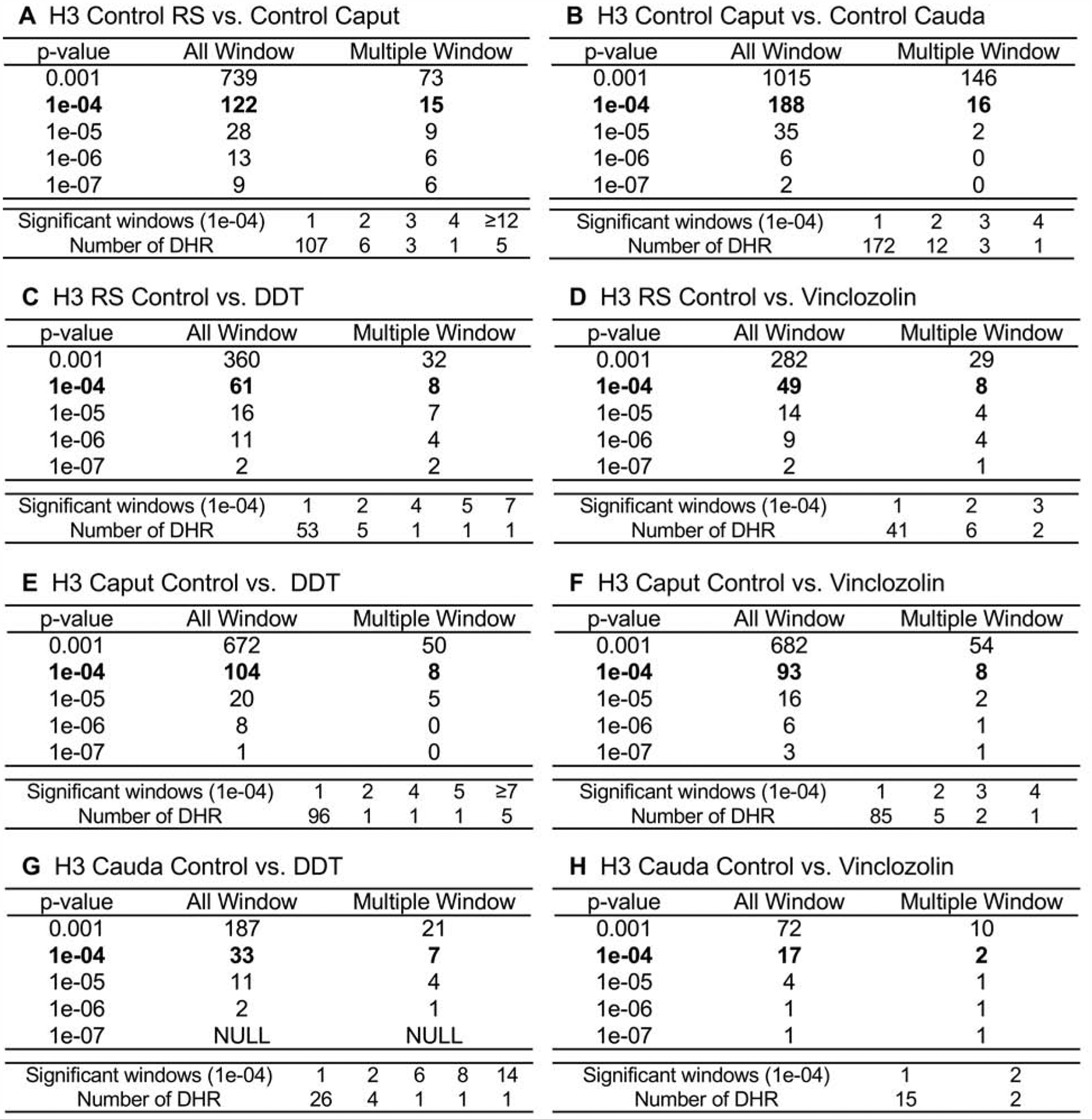
DHR identification. The number of DHRs found using different p-value cutoff thresholds. The All Window column shows all 100 bp windows at the p-value to identify the DHRs. The Multiple Window column shows the number of DHRs containing at least two significant nearby 100 bp windows with the number of DHRs. The specific number of significant windows for the p-value threshold of p<1e-04 is shown. (A) H3 control lineage caput versus round spermatids. (B) H3 control lineage cauda versus caput. (C) H3 round spermatids (RS) control versus DDT. (D) H3 RS control versus vinclozolin. (E) H3 caput control versus DDT. (F) H3 caput control versus vinclozolin. (G) H3 cauda control versus DDT. (H) H3 cauda control versus vinclozolin.
Figure 2.
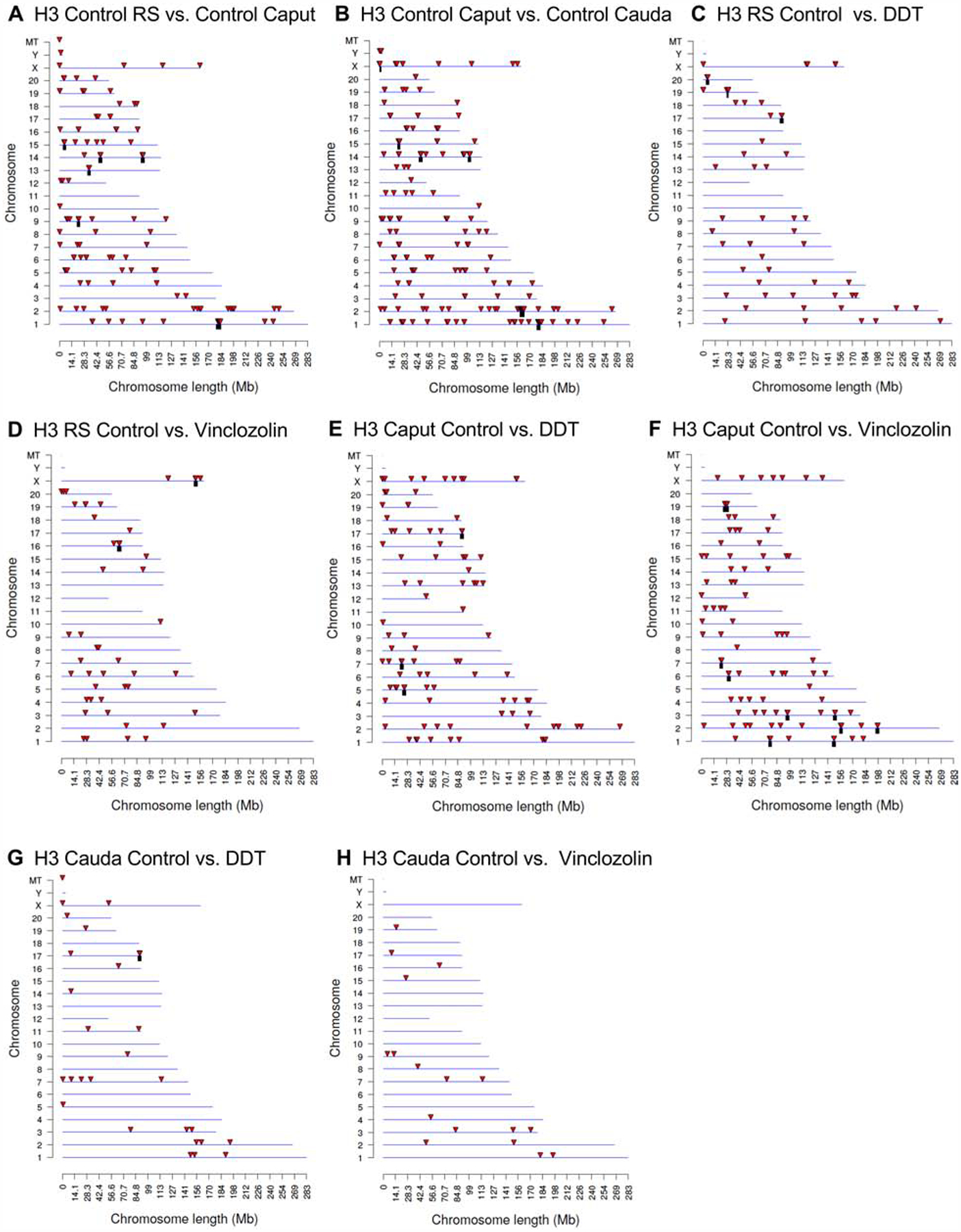
DHR chromosomal locations. The DHR locations on the individual chromosomes. All DHRs at a p-value threshold of p<1e-04 are presented. The chromosome number, X, Y, and mitochondrial (MT) are presented versus the size of the chromosome (megabase). (A) H3 control lineage caput versus control RS. (B) H3 control lineage cauda versus control caput. (C) H3 RS control versus DDT. (D) H3 RS control versus vinclozolin. (E) H3 caput control versus DDT. (F) H3 caput control versus vinclozolin. (G) H3 cauda control versus DDT. (H) H3 cauda control versus vinclozolin.
DHR Comparisons
The overlap between the various developmental stage DHRs was investigated for the control lineage comparisons and is presented in Fig. 3A. The DHRs were found to be primarily stage comparison specific with a very low number of DHRs overlapping between the stage comparisons with a p<1e-04. Comparison between the stages round spermatids, caput epididymal spermatozoa and cauda sperm had limited overlap of DHRs, Fig. 3A. These observations suggest a cascade of epigenetic programming occurs throughout the later haploid stages of male germline development.
Figure 3.
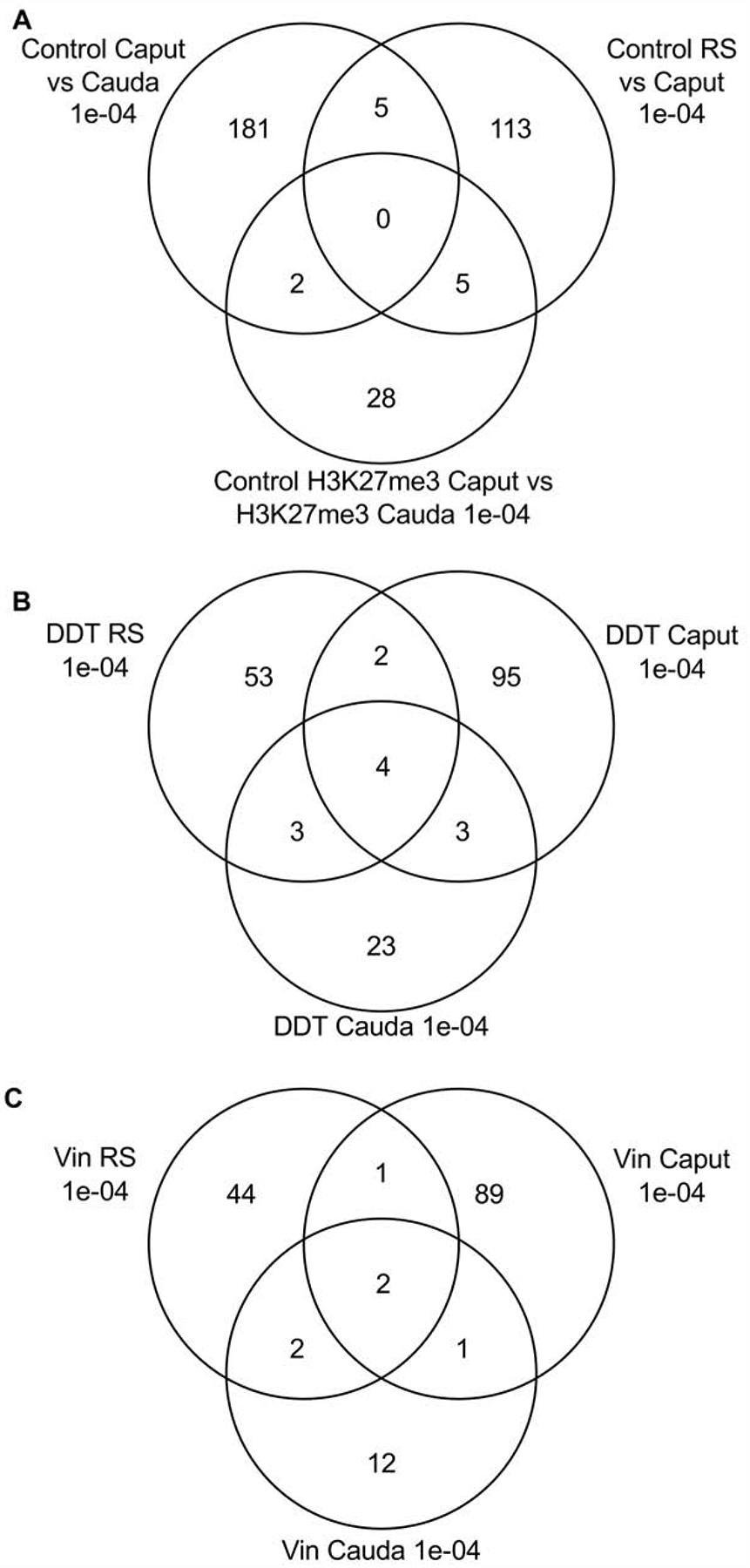
DHR overlap Venn diagram. The overlapping DHR between different data sets at p<1e-04 for (A) Control caput lineage versus cauda, round spermatids (RS) versus caput, and control H3K27me3 caput versus cauda. (B) Control versus DDT for RS, caput and cauda. (C) Control versus vinclozolin for RS, caput and cauda DHRs.
The individual stage comparisons with control versus DDT or control versus vinclozolin DHRs demonstrate primary stage specific DHRs with negligible overlap. The DDT DHRs had four overlapping DHRs with all the stages, Fig. 3B. The vinclozolin DHRs were also primarily stage specific with two DHRs in common with the different stages, Fig. 3C. Observations again suggest a dynamic cascade of DHR development.
Differential Methylated Histone Retention Sites (DMHRs)
At each developmental stage studied, the F3 generation control versus DDT or versus vinclozolin lineage pools were also compared to identify the histone methylation H3K27me3 DMHRs in the caput spermatozoa and the cauda sperm. The H3K27me3 DMHRs at various p-value thresholds are presented at these two developmental stages in Fig. 4. An EdgeR p-value of p<1e-04 was chosen at all stages for further analysis of the DMHRs. This corresponds to a false discovery rate (FDR) of approximately 0.1, except the H3 caput DDT and vinclozolin, which was greater. The epigenetic alterations were similar in the vinclozolin group in the caput spermatozoa with 90 DMHRs and the cauda sperm showing 94 H3K27me3 DMHRs. The lowest number of H3K27me3 DMHRs were in the DDT lineage in the cauda sperm stage. The chromosomal locations of the H3K27me3 DMHRs at each developmental stage are presented in Fig. 5. In most stages, the H3K27me3 DMHRs were found on all the chromosomes in the control lineage caput spermatozoa versus the cauda sperm stage, Fig. 5. At each stage of development, the genomic features of the H3K27me3 DMHRs were examined. The CpG density of the H3K27me3 DMHRs at all stages was 1–4 CpG per 100 bp with 1 CpG per 100 bp being predominant which is characteristic of a low density CpG desert [69], and has been observed with previous transgenerational DHRs [55]. The length of the H3K27me3 DMHRs at all the developmental stages were between 1–5 kb with 1 kb length being predominant, Supplemental Figure S3. Consequently, the H3K27me3 DMHRs are primarily 1 kb in size with around 10 CpGs, as previously observed [55].
Figure 4.
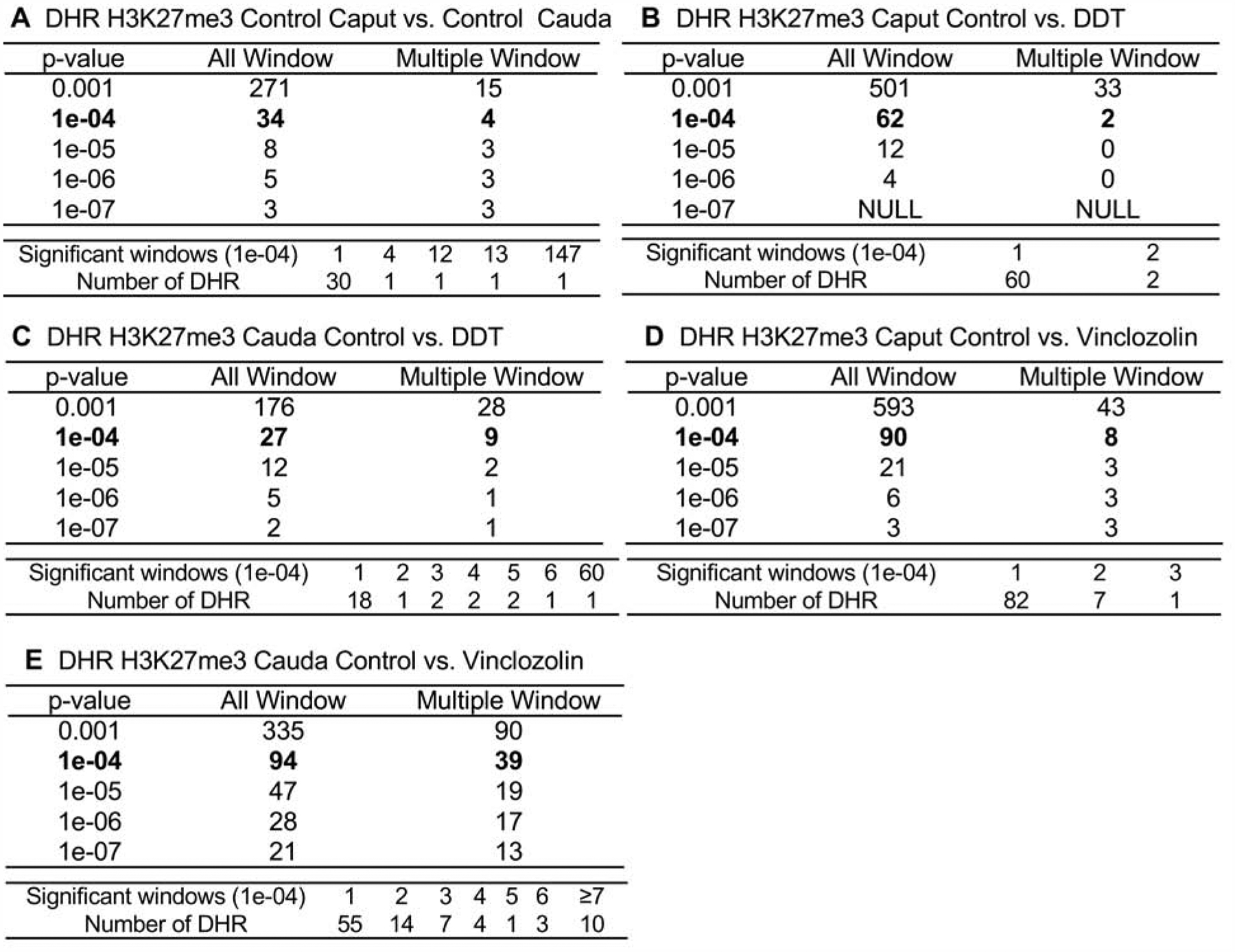
The H3K27me3 differentially methylated histone retention site (DMHRs) identification. The number of differentially methylated histone sites found using different p-value cutoff thresholds. The All Window column shows all 100 bp windows at the p-value to identify the differentially methylated histone sites. The Multiple Window column shows the number of differentially methylated histone sites containing at least two nearby 100 bp significant windows with the number of sites. The lower data set identifies the specific number of significant windows for the p-value threshold of 1e-04. (A) H3K27me3 control lineage cauda versus caput. (B) H3K27me3 caput control versus DDT. (C) H3K27me3 cauda control versus DDT. (D) H3K27me3 caput control versus vinclozolin. (E) H3K27me3 cauda control versus vinclozolin.
Figure 5.
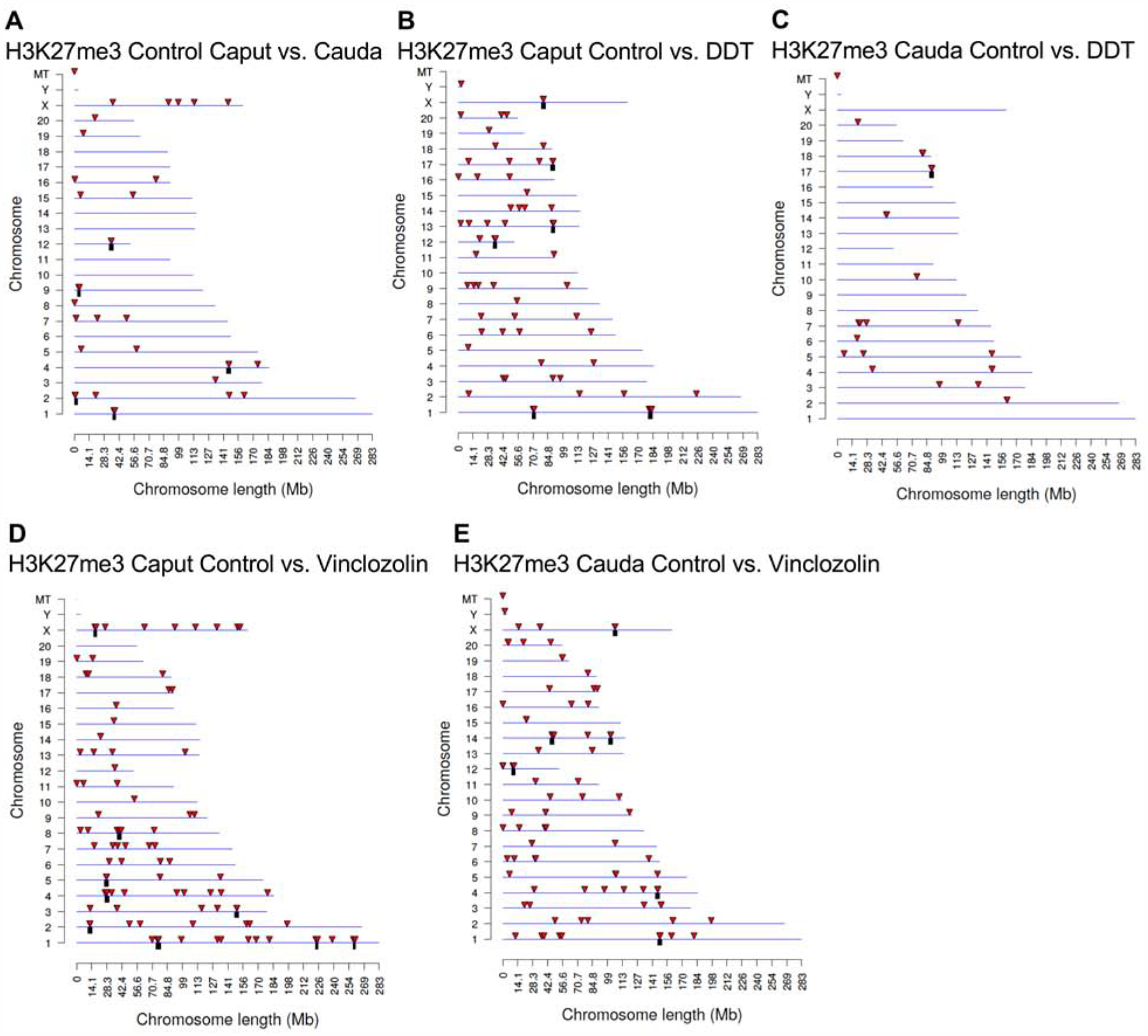
H3K27me3 differentially methylated histone (DMHRs) chromosomal locations. The differentially methylated histone locations on the individual chromosomes. All differentially methylated histone sites at a p-value threshold of 1e-04 are presented. The chromosome number, X, Y, and mitochondrial (MT) are presented versus the size of the chromosome (megabase). (A) H3K27me3 control cauda versus control caput. (B) H3K27me3 caput control versus DDT. (C) H3K27me3 cauda control versus DDT. (D) H3K27me3 caput control versus vinclozolin. (E) H3K27me3 cauda control versus vinclozolin.
The overlap between the various DDT and vinclozolin developmental stage H3K27me3 DMHRs was investigated, and is presented in Fig. 6A. The H3K27me3 DMHRs were found to be primarily stage specific with a very low number of H3K27me3 DMHRs overlapping between the stages with a p<1e-04. The caput epididymal spermatozoa and cauda sperm had 4 H3K27me3 vinclozolin DMHRs overlapping (Fig. 6B) and 2 H3K27me3 DDT DMHR overlapping for the DDT lineage, Fig. 6B, comprising over 4% of the caput epididymal spermatozoa H3K27me3 DMHRs for the vinclozolin lineage and 3% for the DDT lineage.
Figure 6.
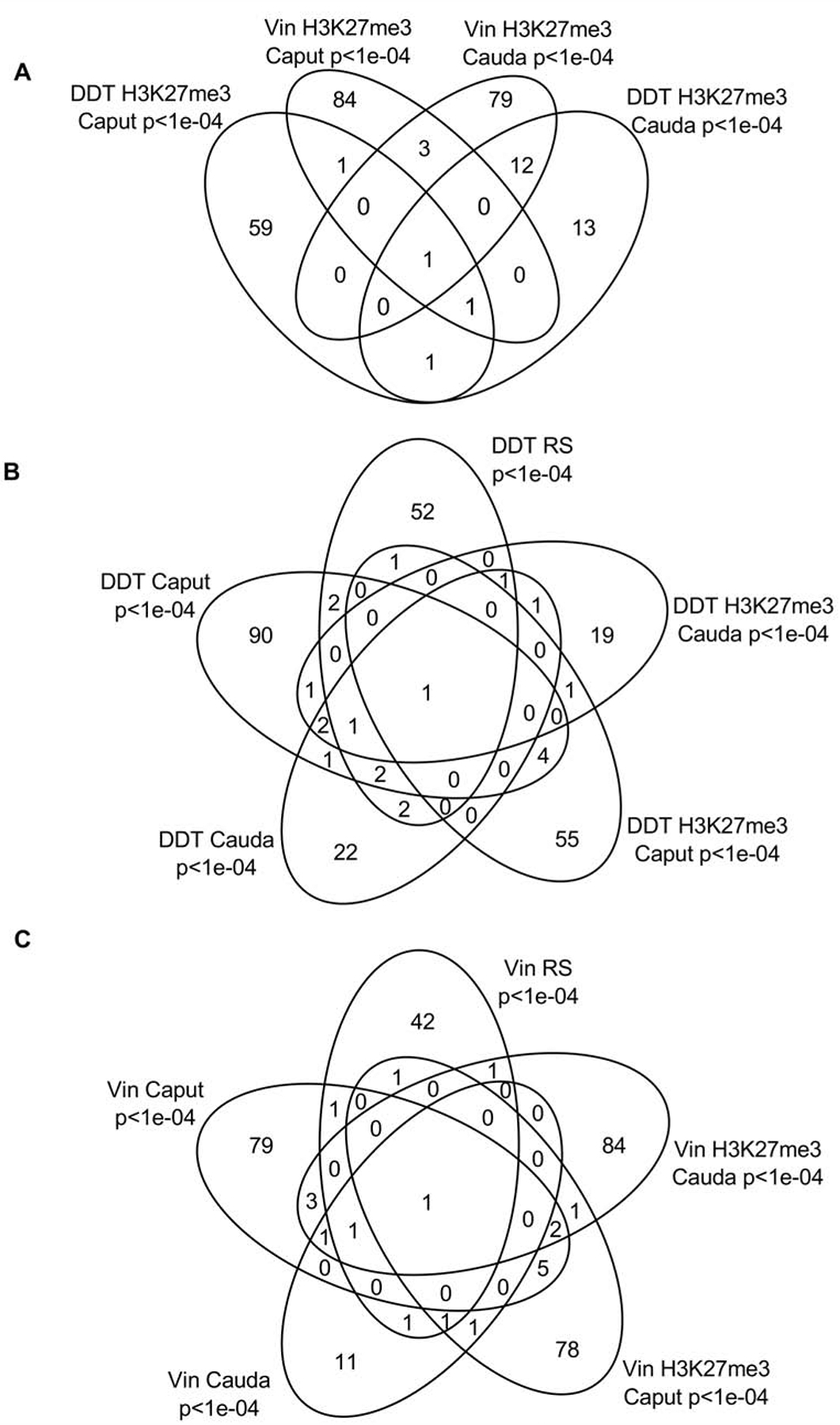
H3K27me3 differentially methylated histone overlap Venn diagram at p<1e-04. (A) The H3K27me3 differentially methylated histone overlap for DDT and vinclozolin (vin) H3K27me3 differentially methylated histone sites. (B) The DDT H3K27me3 differentially methylated histone overlaps with DDT DHRs. (C) The vinclozolin H3K27me3 differentially methylated histone overlaps with vinclozolin DHRs.
Observations suggest a cascade of epigenetic histone retention programming occurs in the male germline epididymal development. An extended overlap was used to compare all control, DDT and vinclozolin DHR sets, Supplemental Figure S4. The DHRs with p<1e-04 were compared to DHRs with p<0.05 statistical threshold to illustrate that there was increased overlap with a comparison at less stringent statistical threshold. In the comparison of all DHR data sets, approximately 25–50% overlap was observed for each comparison, Supplemental Figure S4. Although the more stringent (p<1e-04) statistical threshold overlaps were minimal, the DHRs do have a reasonable overlap with a reduced statistical threshold comparison.
DHR Gene Associations
Genes associated with the DHRs were identified and compared at each of the developmental stages. The increase or decrease in the histone retention in the comparison of the control versus exposure is presented in Supplemental Tables S1–S3 with the log-fold-change (LFC) presented as (+) increase in read depth versus (−) decrease in read depth. The DHRs that had a gene within 10 kb distance (i.e. to include promoters) were identified, and the associated genes and gene functional categories determined, (Supplemental Tables S1–S3). The DHRs with a p<1e-04 were used in this analysis for all the different developmental stages. The number of genes associated with specific gene functional categories is presented in Fig. 7A for control caput versus cauda comparison, Fig. 7B for DDT, and Fig. 7C for vinclozolin. Each of the developmental stages are presented to identify the major gene categories. The signaling, metabolism, transcription and receptor gene categories were the major categories present in the different developmental stages. Additional categories were cytoskeleton, development, transport and protease, that were present in all the developmental stages, Fig. 7A–C.
Figure 7.
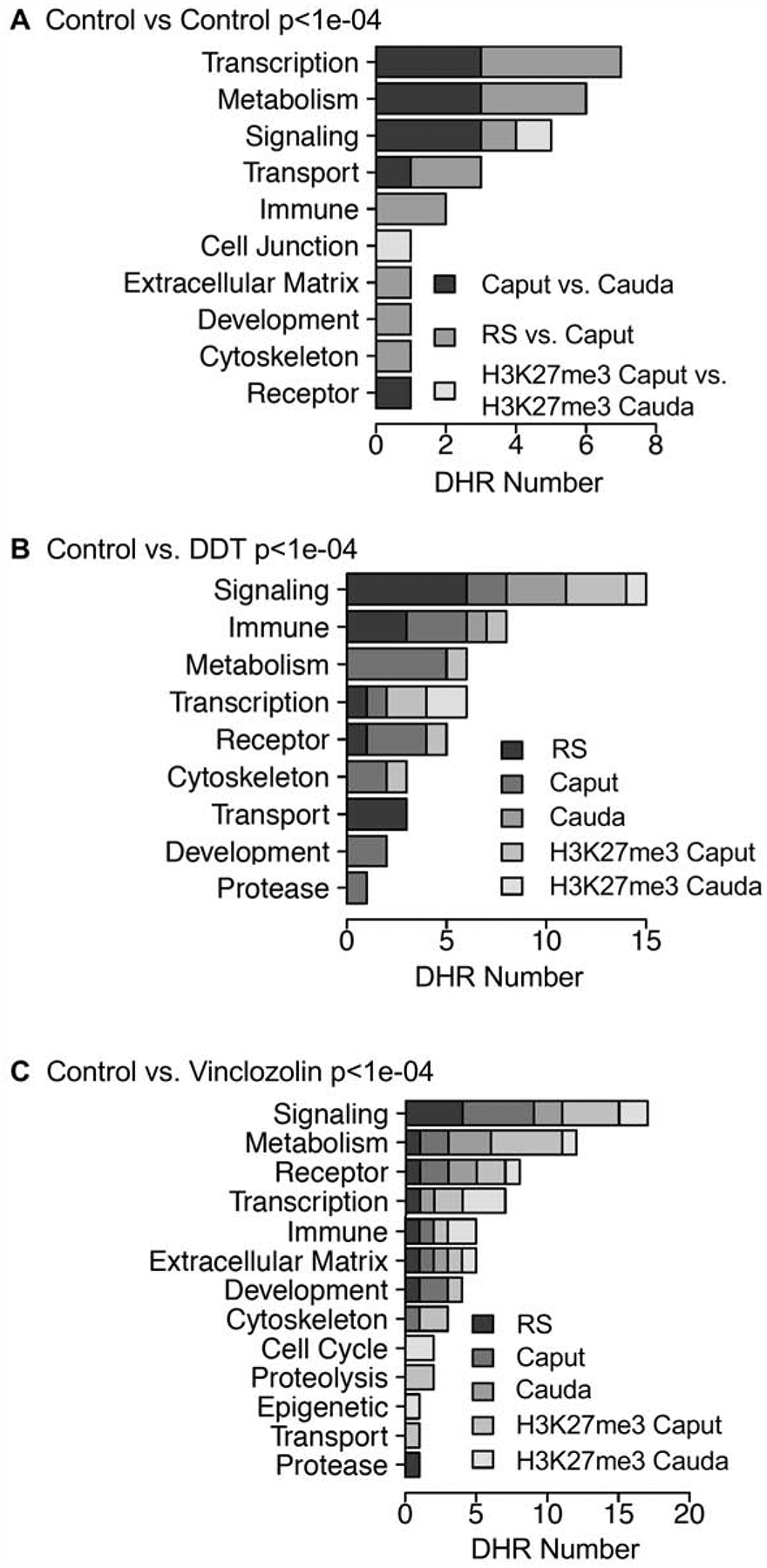
DHR gene association categories. (A) Control vs. control lineage stages at p<1e-04 for different gene categories showing number of DHRs. (B) Control versus DDT p<1e-04 DHRs associated gene categories showing number of DHRs. (C) Control versus vinclozolin p<1e-04 DHRs associated gene categories showing number of DHRs.
The DHR associated genes for each DHR data set were analyzed with a KEGG pathway analysis, as described in the Methods. A number of the DHR data sets with higher DHR numbers had pathways with ≥3 genes, Supplemental Figure S5. The DHR associated gene pathway list shows a number of relevant cellular pathways in later stage germ cell development such as endocytosis, cellular senescence, cell adhesion molecules, allograft rejection, phagocytosis, cellular calcium pathway and axon guidance, Supplemental Figure S5.
DHR Developmental Origins
The developmental origins or alterations of individual DHRs were investigated through comparing the individual stages. The developmental time course of the top 100 most statistically significant DHRs are presented in Fig. 8. The DHRs from a transgenerational exposure lineage greater than control lineage (increase in histone presence) or control lineage greater than exposure lineage (decrease in histone presence) are shown. The scaled sequencing read depth normalization is presented for all the DHRs related to the stage of development of round spermatids (rs), caput spermatozoa (caput), or cauda sperm (cauda). The asterisk indicates a statistical difference for a DHR at a specific stage. For the round spermatid DHRs, the exposure lineage DDT or vinclozolin were high and most declined in subsequent development, Fig. 8B & H. For the round spermatid DHRs, the exposure lineage DHRs generally had a reduction (control > exposure) at later stages of development, Fig. 8E & K, with those which can increase (exposure > control) remaining constant, Fig. 8F & L. For the caput DHRs, there was a mixture of an increase or decrease at the other stages for both control and exposure lineages, Fig. 8C, D, I & J. For the cauda DHRs, the DHRs had similarities between the different stages, Fig. 8E, F, K & L. Overall, the DHRs displayed dynamic patterns during development. Observations support the concept that a complex cascade of DHR change is observed during the developmental process, and that many DHRs are present throughout the developmental process, Fig. 8.
Figure 8.
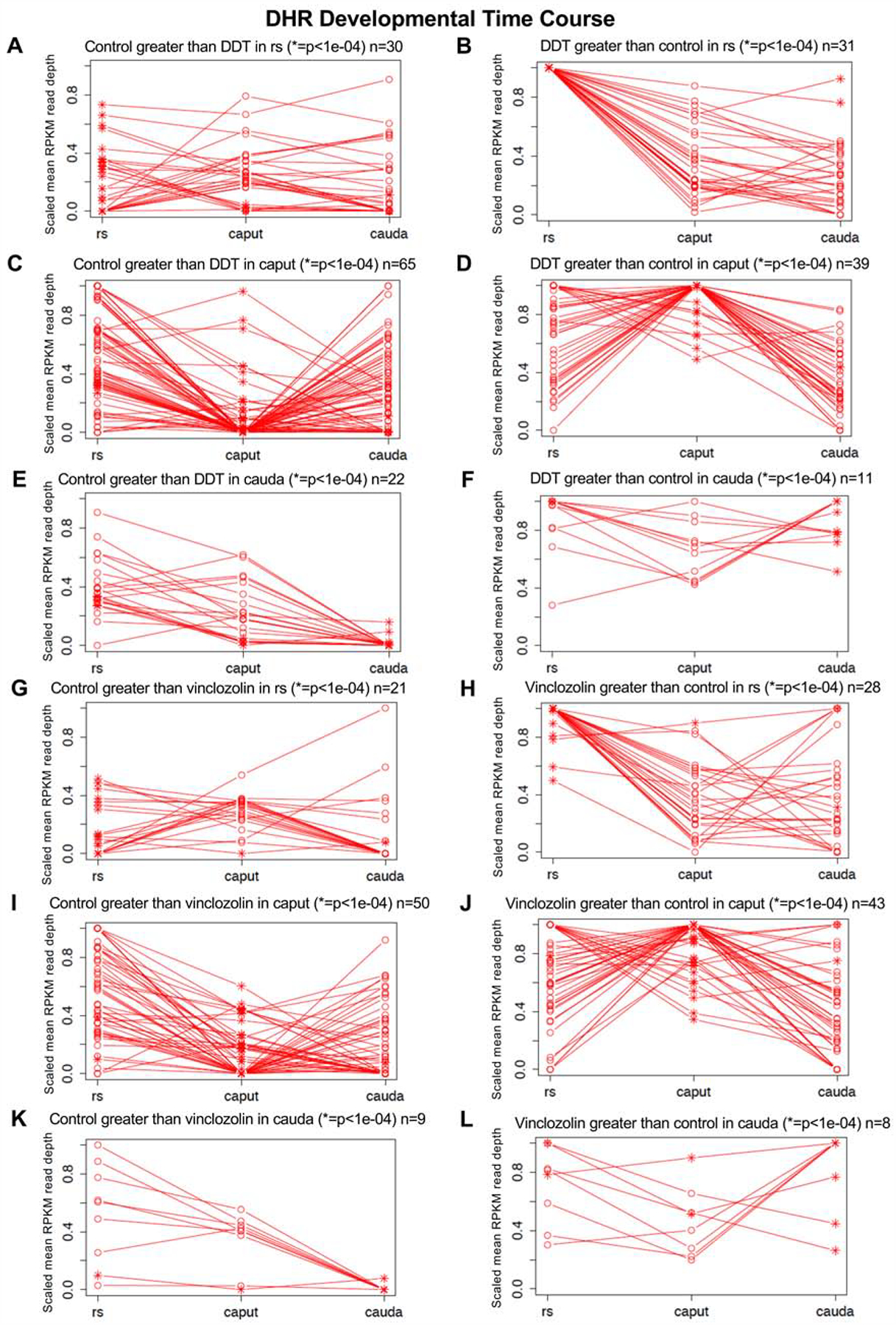
DHR developmental time course for control versus exposure lineage ratio comparisons. The scaled mean RPKM read depth is shown for the exposure lineage samples. (A) Control lineage greater than DDT lineage in rs (* = p < 1e−04) n = 30. (B) DDT lineage greater than control lineage in rs (* = p < 1e−04) n = 31. (C) Control lineage greater than DDT lineage in caput (* = p < 1e−04) n = 65. (D) DDT lineage greater than control lineage in caput (* = p < 1e−04) n = 39. (E) Control lineage greater than DDT lineage in cauda (* = p < 1e−04) n = 22. (F) DDT lineage greater than control lineage in cauda (* = p < 1e−04) n = 11. (G) Control lineage greater than vinclozolin lineage in rs (* = p < 1e−04) n = 21. (H) Vinclozolin lineage greater than control lineage in rs (* = p < 1e−04) n = 28. (I) Control lineage greater than vinclozolin lineage in caput (* = p < 1e−04) n = 50. (J) Vinclozolin lineage greater than control lineage in caput (* = p < 1e−04) n = 43. (K) Control lineage greater than vinclozolin lineage in cauda (* = p < 1e−04) n = 9. (L) Vinclozolin lineage greater than control lineage in cauda (* = p < 1e−04) n = 8.
The developmental origins of the DHRs was also determined as shown in Fig. 9A. Approximately 50% of the DHRs developed at the round spermatid stage, and the other half at the cauda sperm stage. Only one DHR developed at the caput spermatozoa stage. The developmental pattern of these DHRs are shown in Fig. 9B & C. The 0 or 1 in the tables represent the absence or presence of the DHRs, respectively, with the total number of DHRs for each developmental pattern listed, Fig. 9. The majority were either present in the round spermatid and in later caput and cauda stages, or present only in the cauda stage. Therefore, as previously observed, DHRs develop at the round spermatid stage, but the novel observation of the current study is that they also appear at the cauda sperm stage of development.
Figure 9.
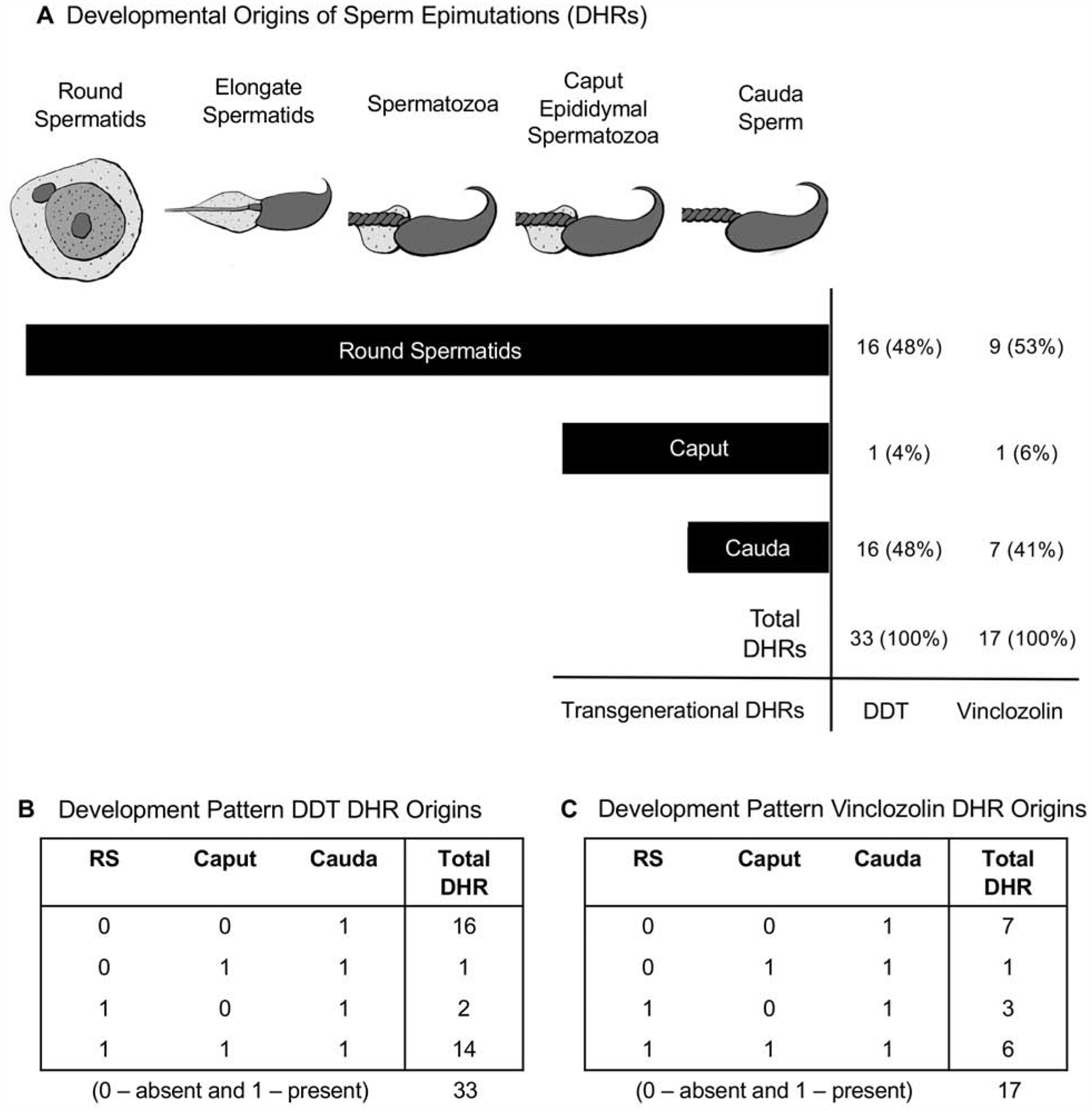
Diagram of the development from spermatid to caput spermatozoa to cauda sperm with the origin of the cauda sperm DHR numbers indicated. (A) Developmental origins of cauda sperm epimutations (DHRs). DHRs are considered to be present in a development stage if they meet a p-value threshold of 0.05. (B) Developmental pattern DDT DHR origins. (C) Developmental pattern vinclozolin DHR origins.
The final analysis examined the potential overlap of the DHRs identified with the previously identified core of highly conserved and consistent histone retention sites found in sperm [55]. A core of 41 histone retention sites previously identified with the significant presence of read depth at the sites [55] was found to also be present in both control and exposure lineages transgenerationally in the current study. These core sites are not differentially present and not altered since they are present both in the control and the exposure lineages, so are not defined as DHRs. A potential overlap with these core histone retention sites was examined and found to be similar in the current study. All the developmental data presented in the current study are unique, however, the cauda sperm DHR data set for DDT and vinclozolin was similar and compared with the previous analysis [25, 55, 56], and found to have a 33% overlap with DDT and 53% overlap with vinclozolin induced DHR with a p<0.05 statistical threshold comparison. Therefore, the cauda sperm DHR had overlap between the current and previous studies [25, 56], but there were fewer DHR in the current study. The previous analysis used postnatal 120-day males and the current study used 8–10 month males. As expected, a 40–50% overlap was observed between the transgenerational DDT DHRs and vinclozolin DHRs. Interestingly, the transgenerational H3K27me3 DMHRs identified in the current study had 3 overlaps with the vinclozolin H3K27me3 DMHRs and the core histone retention sites, and 1 overlap with the DDT H3K27me3 DMHRs and the core histone retention sites [55]. Therefore, the environmentally induced transgenerational DHRs are distinct from the core histone retention sites, but there can be distinct histone methylation of a few of the core histone retention sites.
Discussion
The current study was designed to determine the developmental origins of the cauda sperm transgenerational DHRs, which appear to be involved in the epigenetic transgenerational inheritance phenomenon. In the rat, between day 8–14 of fetal development, the PGCs migrate to and eventually colonize the indifferent fetal gonad. During this developmental period, the F0 generation gestating females were exposed to DDT or vinclozolin. Three different haploid stages of male germ cell development were isolated and studied including round spermatids, caput epididymal spermatozoa and cauda sperm. During spermatid development, one of the dramatic changes is the transition from nucleosome-based histone chromatin to protamine-based chromatin. This event facilitates the uniquely dense condensation of sperm head nuclei, and silences gene expression and protects the paternal DNA from damage and mutagenesis [70]. Despite the fact that the morphological changes during spermatogenesis are well characterized, the molecular basis underlying this chromatin reorganization, especially the histone-to-protamine transition remains largely unknown [70]. Any epigenetic defects during histone-to-protamine transition can not only lead to reduced fertility, but can also be transmitted to subsequent generations [55, 71].
The caput spermatozoa and cauda epididymal sperm were directly isolated, as described in the Methods, then sonicated and washed to remove any contaminating somatic cells, so are pure sperm cell preparations. For the round spermatids, a gravity sedimentation on a StaPut apparatus procedure was used to isolate them as previously described [62, 63, 72]. Among the isolated round spermatids 95% of them were spermatids with 90% of round spermatids and 10% of elongated spermatids and other spermatogenic cell stages [69]. Even though high purity was obtained for the round spermatid population, cell purity should be considered in the data interpretations. DDT and vinclozolin have both been shown to induce the transgenerational inheritance of disease in animals between 6 months to 1 year of age [73]. The postnatal 8–10 month rats were used at a period when disease can be present, such that disease artifacts need to be considered in data interpretation. The focus of the current study was on the transgenerational F3 generation control, DDT and vinclozolin germline samples since the F3 generation is the first to not have any direct exposures, in contrast to the F1 and F2 generations [65]. Observations demonstrated that each developmental stage had DHRs, Fig. 3. Interestingly, these DHRs were found to be primarily distinct between each stage. These stage-specific DHRs suggest a dynamic cascade of epigenetic modifications occurs to program the sperm transgenerational epigenome.
In the round spermatids, the chromatin DNA associated histones are replaced by transition proteins. In elongated spermatids, these transition proteins are then replaced by protamine. These alterations initiate the sperm chromatin condensation. The content of protamine is essential for the final maturation stage of the spermatozoa nucleus. Comparisons between the control (normal) development and the round spermatids, the control and the caput spermatozoa, and the control and the cauda mature sperm were performed. More than a hundred DHRs were observed between the round spermatids and the caput spermatozoa, and almost two hundred between the caput spermatozoa and cauda mature sperm, Fig. 1. This observation suggests that in a normal control setting, the replacement of histones by protamines is a very dynamic phenomenon beginning at the spermatid stage and through epididymal maturation.
Studies have shown that environmental toxicants are able to promote DNA methylation alterations as early as the primordial germ cells stage and prospermatogonia stage [55, 62, 63]. Therefore, in addition to the dynamic cascade of DNA methylation patterns, there is also a cascade of developmental programming histone retention sites. The previous thought in regards to the origins of histone retention was between the round spermatid and elongate spermatid stages when the majority of chromatin remodeling and histone retention was developed [49]. Observations from the current study demonstrate that developmental programming of sperm histone retention can also occur during later stages of epididymal maturation between the caput and cauda epididymal stages. The developmental programming of the cauda sperm DHRs demonstrated a dynamic regulation at all the stages examined. Although approximately 50% develop at the round spermatid stage, Fig. 9, there is also development at the cauda sperm stage promoted by ancestral exposures of environmental factors, such as DDT or vinclozolin. This novel observation suggests a dynamic epigenetic programming of epigenetics throughout gametogenesis, also involving the later stages of epididymal maturation. Similar observations have also been provided for the development of differential DNA methylation regions (DMRs) induced by environmental toxicants such as DDT and vinclozolin [62, 63]. Since DHRs have a similar developmental programming, other epigenetic processes such as ncRNA are also anticipated to be similar.
An overlap of the DHRs between the different stages and exposures at a high statistical threshold demonstrated primarily distinct sets of DHRs between developmental stages, Figs. 3 & 6. However, with an extended overlap at a lower statistical threshold, (p<0.05) a much higher percentage of the DHRs overlapped, Supplemental Figure S4. Therefore, the DDT and vinclozolin exposure transgenerational DHRs were much more common with approximately 50% being similar. The H3K27me methylated DMHRs had a higher level of overlap with the DHRs at nearly 70% overlap for some comparisons, Supplemental Figure S4. Therefore, the different developmental stages and exposure lineages had unique and common DHRs. Although some DHRs appear to be consistent throughout development, others can develop at later stages. This is anticipated to involve the presence of a DHR that becomes differentially present within the exposure lineage, or the developmental transition between stages. The number of DHRs that are not present at early stages and then appear at later stages was investigated and involves a reduction in DHR levels (i.e. DHR site read depth) compared to control or increase in DHR levels (i.e. DHR site read depth) compared to control, as indicated in Supplemental Tables S1–S3 under log-fold-change (LFC) as a (+) or (−) ratio and in Fig. 9. The developmental programming of the DHRs demonstrated nearly half were developed at the round spermatid stage, and the other half at the cauda stage, Fig. 9. A small number originated at the caput epididymal stage. The developmental programming was dynamic for most DHRs between the stages investigated, Fig. 8.
The H3K27me3 methylated DMHRs generally had a higher overlap in the extended overlap approach with a less stringent threshold, Supplemental Figure S4. This suggests the DHRs may have differential methylation at various stages including the later stages of development. Therefore, while the DHR may be consistent and present between the stages of development, the histone modifications may be altered. The observations that several of the core histone retention sites absent in the DHRs had altered histone methylation also supports a role for histone modifications. This adds a level of complexity to the potential role of histone retention in that it may be not only the retention, but also the alterations in histone epigenetic modifications. This supports the conclusion that a more dynamic cascade of developmental programming occurs during the later stages of epididymal sperm maturation.
The DHRs had positional associations with genes and the major functional categories were signaling, metabolism and transcription. This is expected since these functional categories are the larger gene families in the genome. A variety of gene pathways were associated with several DHR data sets and developmental stages, Supplemental Figure S5. The potential functional role of the specific DHR sites and associated genes needs to be considered in future analyses. However, a clear functional impact in the spermatogenic stages can be speculated, as well as following fertilization in the early developing embryo. As previously discussed, observations in several different organisms have demonstrated epigenetic alterations in sperm can influence early embryo epigenetics and gene expression [52, 57–61].
The potential role of epigenetic alterations in the sperm in epigenetic transgenerational inheritance involves potential modifications in the zygote and early embryo following fertilization. The alterations in differential DNA methylation regions (DMRs) sites are retained if the DNA methylation erasure is protected like an imprinted gene site, which has the ability to alter the stem cell epigenetics and transcriptome [1, 73]. The DHRs may also impact the early embryo following fertilization by altering early gene expression events [52, 57–61]. The current study supports the existence of transgenerational DHR alterations, but the functional role following fertilization remains to be established. In the event the embryo stem cell population has a modified epigenetics and corresponding transcriptome, then all somatic cells derived from the stem cell population will have an altered cascade of epigenetic and gene expression programing to result in adult differentiated cells with altered epigenetics and transcriptomes, as previously described [74, 75]. These alterations in somatic cell genome activity and epigenetics are anticipated to alter disease susceptibility. Although the potential for such germline epigenetic alterations to promote such events exist [52, 57–61], many of the specific molecular aspects involved remain to be established. The current study on alterations in DHRs does support a potential role for histone retention in epigenetic transgenerational inheritance, but further studies are required to elucidate the specific molecular events and causal functional significance involved.
Our previous studies focused on cauda epididymal sperm DHRs only and identified transgenerationally altered DHRs in postnatal 120 day young adult males [25, 55, 56]. The current study examined previous developmental stages to identify the developmental programming of the DHRs. An extended overlap at p<0.05 statistical threshold demonstrated 33% of DDT DHRs and 53% of vinclozolin DHRs for the cauda epididymal sperm of the current study were also present in the original studies [25, 56]. The original studies were performed at postnatal 120-day young adult age, while the current study was at an 8–10 month older adult male age. Previous observations have demonstrated in older adult human males alterations in histone retention develop and are associated with infertility [76], and in the boar increased histone retention is observed in younger males than older males [48]. The potential for an age impact on histone retention is suggested, such that the comparison of 120-day old males [25, 55, 56] and 8–10 month old male DHRs in the current study may have differences. The DHR numbers were higher in the previous 120-day old male studies [25, 55, 56] than the current study. Although there is reasonable overlap between the studies, observations suggest the impact of age on DHR development in the male needs to be considered and investigated in the future.
Conclusions
Epigenetic alterations (i.e. epimutations) such as DNA methylation, histone modifications, and non-coding RNA can affect genome activity and become transgenerational when the germline is involved. The current study investigated the developmental origins of transgenerational histone retention alterations in the sperm following ancestral exposure to DDT and vinclozolin. The DHRs were found to primarily originate from the round spermatids and cauda sperm. The vinclozolin and DDT induced transgenerational sperm DHRs occur throughout development with the majority appearing in the round spermatids and cauda sperm, but very little in the caput spermatozoa. Although previous studies have suggested that histone retention primarily occurs between the round spermatid stage and the elongated spermatid stage, the current study suggests an equal number of DHRs develop between the caput spermatozoa and cauda sperm stages. This suggests the replacement of histones by protamines is a much more dynamic phenomenon beginning at the spermatid stage and continuing through epididymal maturation. Alterations in this histone retention appears to be in part involved in the environmental induction of the epigenetic transgenerational inheritance.
Methods
Animal studies and breeding
Female and male rats of an outbred strain Hsd:Sprague Dawley SD obtained from Harlan/Envigo (Indianapolis, IN) at about 70 to 100 days of age were maintained in ventilated (up to 50 air exchanges/hour) isolator cages (cages with dimensions of 10 ¾″ W×19 ¼″ D×10 ¾″ H, 143 square inch floor space, fitted in Micro-vent 36-cage rat racks; Allentown Inc., Allentown, NJ) containing Aspen Sani chips (pinewood shavings) as bedding, and a 14 h light: 10 h dark regimen, at a temperature of 70 F and humidity of 25% to 35%. The mean light intensity in the animal rooms ranged from 22 to 26 ft-candles. Rats were fed ad lib with standard rat diet (8640 Teklad 22/5 Rodent Diet) and ad lib tap water for drinking. The inbreeding coefficient of the non-related males and females was approximately 1% indicating the lack of a common ancestor (i.e. great-great-grandparent) To obtain time-pregnant females, the female rats in proestrus were pair-mated with male rats. The sperm-positive (i.e. sperm plug present) (day 0) rats were monitored for diestrus and body weight. On days 8 through 14 of gestation [73], the females received daily intraperitoneal injections of DDT (25 mg/kg body weight/day), vinclozolin (100 mg/kg body weight/day), or dimethyl sulfoxide (DMSO). The DDT (dichlorodiphenyltrichloroethane) and vinclozolin were obtained from Chem Service Inc., (West Chester, PA). DDT and vinclozolin were dissolved and injected in DMSO vehicle, as previously described [18, 56, 77]. Treatment lineages are designated ‘control’, ‘vinclozolin’, or ‘DDT’ lineages. The treated gestating female rats were designated as the F0 generation. The offspring of the F0 generation rats were the F1 generation. Non-littermate females and males aged 70–90 days from the F1 generation of control, vinclozolin, or DDT lineages were bred to obtain F2 generation offspring. The F2 generation rats were bred to obtain F3 generation offspring. Individuals were maintained for 8–10 months and euthanized for sperm collection. The F1-F3 generation offspring were not themselves treated directly with DDT or vinclozolin. All experimental protocols for the procedures with rats were pre-approved by the Washington State University Animal Care and Use Committee (IACUC approval # 6252).
Epididymal sperm collection
The epididymis was dissected free of connective tissue, a small cut made to the cauda and tissue placed in 5 ml of 1x PBS solution for up to 2 hours at 4°C. The epididymal tissue was minced and the released sperm was centrifuged at 6,000xg, then the supernatant was removed, and the pellet was resuspended in NIM buffer, to be stored at −80 °C until further use. One hundred μl of sperm suspension was sonicated to destroy somatic cells and tissue, spun down at 6,000xg, the sperm pellet was washed with 1x PBS once.
Developing germ cell stage isolation
Harlan Sprague-Dawley rats (Harlan Inc., Indianapolis IN) were used for all experiments. The rats were kept in a temperature-controlled environment and given food and water ad libitum. Estrous cycles of female rats were monitored by cellular morphology from vaginal smears. Rats in early estrus were paired with males overnight and mating confirmed by sperm-positive smears, denoted day 0 of pregnancy. Germ cells were isolated exclusively from males. To isolate round spermatids, testes were collected from 8–10 month old rats suspended in F-12 culture medium (Gibco-Life Technologies, USA. Ref 11765–054) and shipped overnight on ice to Dr. John McCarrey. A StaPut gradient method was used to isolate the developing germ cell stages as previously described [78, 79]. Three pools of cells of each cell type were prepared for each treatment group, with each pool derived from testes of three rats from different litters. DNA was isolated from round spermatids using the same procedure as was used for sperm, with the omission of sonication and DTT treatments.
Histone chromatin immunoprecipitation
Histone chromatin immunoprecipitation with genomic DNA was performed with a procedure previously described [55]. Individual rat sperm collections were generated, and the sperm counts were determined for each individual. Equal numbers of sperm were added from each individual for a total of 1.5 million sperm per pool, and 3 pools with different individual rats for each pool for the control, vinclozolin and DDT lineage animals. The control pools contained equal numbers of sperm for each of 5–6 individuals for a total of n = 17 rats, and the vinclozolin and DDT pools contained equal numbers of sperm for each of 4 individuals in each pool for a total of n = 12 rats per exposure group. To remove any somatic cell contamination sperm samples from each animal were sonicated for 10 seconds using a Sonic Dismembrator Model 300 (Thermo Scientific Fisher, USA) then centrifuged 1800xg for 5 min at 4 °C then resuspended and counted individually on a Neubauer counting chamber (Propper manufacturing Co., Inc., New York, USA) prior to pooling. The sperm pools were reconstituted up to 1 ml with PBS (phosphate buffered saline). To reduce disulfide bonds, 50 μl of 1 M DTT was added to each pool and the pools were then incubated for 2 hours at room temperature under constant rotation. To quench any residual DTT (dithiothreitol, Fisher Scientific, NY USA) in the reaction, 120 μl of fresh 1 M NEM (N-Ethylmaleimide, Thermo Scientific, Rockford, USA) was then added and the samples were incubated for 30 min at room temperature under constant rotation. The sperm cells were pelleted at 450xg for 5 min at room temperature and the supernatant was discarded. Pellets were resuspended in PBS and then spun again at 450xg for 5 min at room temperature. The supernatant was discarded and resuspended in 130 μl of complete buffer supplemented with tergitol 0.5% and DOC 1%. The samples were then sonicated using the Covaris M220. Covaris was set to a 10 min “Chromatin shearing” program and the program was run for each tube in the experiment.
After the Covaris sonication, 10 μl of each sample was run on a 1.5% agarose gel to verify fragment size. Samples were then centrifuged at 12,500xg for 10 min at room temperature. The supernatant was transferred to a fresh microfuge tube. 65 μl of protease inhibitor cocktail (1 tablet dissolved in 500 μl, 20X concentrated) (Roche, cat. no. 11 873 580 001) were added in each sample as well as 3 μl of antibody (anti-histone H3 pan-monoclonal antibody, cat no. 05–928, or anti-trimethyl-histone H3 (Lys27) polyclonal antibody, cat no. 07–449, both with broad spectrum species specifically form Millipore Corp, Temecula CA USA). The DNA-antibody mixture was incubated overnight on a rotator at 4 °C. The following day, magnetic beads (ChIP-Grade protein G magnetic beads, Cell Signaling 9006) were pre-washed as follows: the beads were resuspended in the vial, then 30 μl per sample was transferred to a microfuge tube. The same volume of Washing Buffer (at least 1 ml) was added and the bead sample was resuspended. The tube was then placed into a magnetic rack for 1–2 min and the supernatant was discarded. The tube was removed from the magnetic rack and the beads were washed once. The washed beads were resuspended in the same volume of IP buffer as the initial volume of beads. 30 μl of beads were added to each DNA-antibody mixture from the overnight incubation, then incubated for 2 h on a rotator at 4 °C. After the incubation, the bead-antibody-DNA complex was washed three times with IP buffer as follows: the tube was placed into a magnetic rack for 1–2 min and the supernatant was discarded, then washed with IP buffer 3 times. The washed bead-antibody-DNA solution was then resuspended in 300 μl of digestion buffer (1 M Tris HCI, pH 8.0, 0.5 M EDTA, 10% SDS) and 3 μl proteinase K (20 mg/ml). The sample was incubated for 3 h on a rotator at 56 °C. After incubation the samples were extracted with Phenol-Chloroform-Isoamylalcohol and precipitated with 2 μl of Glycoblue (20 mg/ml), a one-tenth volume of 3 M sodium acetate and two volumes of ethanol overnight at −20 °C.
The precipitate was centrifuged at 18,000xg for 30 min at 4 °C and the supernatant was removed, while not disturbing the pellet. The pellet was washed with 500 μl cold 70% ethanol, then centrifuged again at 18,000xg for 10 min at 4 °C and the supernatant was discarded. The tube was spun briefly to collect residual ethanol to bottom of tube and as much liquid as possible was removed with a gel loading tip. Pellet was air-dried at RT until it looked dry (about 5 min), then resuspended in 20 μl H20. DNA concentration was measured in the Qubit (Life Technologies) with the BR dsDNA kit (Molecular Probes Q32853).
ChIP-Seq analysis
The ChIP pools were used to create libraries for next generation sequencing (NGS) using the NEBNext UltraTM II DNA Library Prep Kit for Illumina (NEB, Ipswich, MA). The manufacturer protocol was followed. Each pool received a separate index primer. NGS was performed at the WSU Spokane Genomics Core using Ilumina HiSeq. 2500 with a read size of approximately 50 bp PE and approximately 20 million reads per pool. Eleven or twelve libraries were run in one lane.
Bioinformatics and statistics
The basic read quality was verified using summaries produced by the FastQC program http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. The raw reads were trimmed and filtered using Trimmomatic [80]. The reads for each ChIP sample were mapped to the Rnor 6.0 rat genome using Bowtie2 [81] with default parameter options. The mapped read files were then converted to sorted BAM files using SAMtools [82]. To identify DHRs, the reference genome was broken into 100 bp windows. The MEDIPS R package [83] was used to calculate differential coverage between control and exposure sample groups. The edgeR p-value [84] was used to determine the relative difference between the two groups for each genomic window. Windows with an edgeR p-value less than 10−4 were considered DHRs. The DHR edges were extended until no genomic window with a p-value less than 0.1 remained within 1000 bp of the DHR. Multiple testing correlation, false discovery rate (FDR), was also performed.
DHRs were annotated using the biomaRt R package [85] to access the Ensembl database [86]. The genes that overlapped with DHRs, and those genes that occurred within 10 kbp of the DHR edge, were then input into the KEGG pathway search [87, 88] to identify associated pathways. The DHR associated genes were then sorted into functional groups by consulting information provided by the DAVID [89] and Panther [90] databases incorporated into an internal curated database www.skinner.wsu.edu/genomic-data-and-r-code-files).
An expanded overlap analysis was performed. For this analysis, DHR identified using a p<1e-4 threshold in one analysis were considered present in a second analysis if they met a relaxed p<0.05 threshold. This analysis was used for Figs. 9 and S4.
For the timeplots shown in Fig. 8, the DHR are separated into two groups based on whether the mean RPKM read depth is elevated in the control or exposure samples. The RPKM read depth is averaged across all sample replicates and genomic windows within the DHR. The mean RPKM is then scaled such that the maximum mean RPKM read depth (in both compared treatment groups) across all development stage pairs is 1 and the minimum is 0.
All molecular data has been deposited into the public database at NCBI (GEO # GSE137963). The specific scripts used to perform the analysis can be accessed at github.com/skinnerlab and at www.skinner.wsu.edu/genomic-data-and-r-code-files.
Supplementary Material
Supplemental Figure S1. The number of DHRs at different CpG densities. All DHRs at a p-value threshold of 1e-04 are presented. (A) H3 control caput versus RS. (B) H3 control cauda versus caput. (C) H3 RS DDT. (D) H3 RS vinclozolin. (E) H3 caput DDT. (F) H3 caput vinclozolin. (G) H3 cauda DDT. (H) H3 cauda vinclozolin.
Supplemental Figure S2. The number of DHR at different lengths. All DHRs at a p-value threshold of 1e-04 are presented. (A) H3 control Caput versus RS. (B) H3 control cauda versus caput. (C) H3 RS DDT. (D) H3 RS vinclozolin. (E) H3 caput DDT. (F) H3 caput vinclozolin. (G) H3 cauda DDT. (H) H3 cauda vinclozolin.
Supplemental Figure S3.The number of DHRs at different CpG densities. All DHRs at a p-value threshold of 1e-04 are presented. (A) H3K27me3 control cauda versus caput. (C) H3K27me3 caput DDT. (E) H3K27me3 cauda DDT. (G) H3K27me3 caput vinclozolin. (I) H3K27me3 cauda vinclozolin. DHR lengths (kilobase) with all DHRs at a p-value threshold of 1e-04 are presented. (B) H3K27me3 control cauda versus caput. (D) H3K27me3 caput DDT. (F) H3K27me3 cauda DDT. (H) H3K27me3 caput vinclozolin. (J) H3K27me3 cauda vinclozolin.
Supplemental Figure S4. Expanded overlap with p<1e-04 on left margin and p<0.05 on top margin. The number of DHR overlaps and percentage of the total presented.
Supplemental Figure S5. DHR gene association Kegg pathways. The pathways associated with each DHR data set presented with the number of DHR in brackets. (https://www.genome.jp/kegg/pathway.html)
Supplemental Table S1. Control DHR lists. (A) Control lineage round spermatid versus caput spermatozoa. (B) Control lineage caput versus cauda. (C) Control lineage H3K27me3 caput versus cauda. The DHR identification, chromosome location, the chromosome nucleotide start and stop site, length (bp), number significant 1 kb windows, p-value, maximum log fold change (max LFC) (positive is increase in presence and negative is decrease in presence), CpG number and density, gene association, and gene functional category.
Supplemental Table S2. DDT DHR lists. (A) DDT round spermatid DHR list. (B) DDT caput DHR list. (C) DDT cauda DHR list. (D) DDT H3k27me caput differentially methylated histone list. (E) DDT H3k27me cauda differentially methylated histone list. The DHR identification, chromosome location, the chromosome nucleotide start and stop site, length (bp), number significant 1 kb windows, p-value, maximum log fold change (max LFC) (positive is increase in presence and negative is decrease in presence), CpG number and density, gene association, and gene functional category.
Supplemental Table S3. Vinclozolin DHR lists. (A) Vinclozolin round spermatid DHR list. (B) Vinclozolin caput DHR list. (C) Vinclozolin cauda DHR list. (D) Vinclozolin H3K27me caput differentially methylated histone list. (E) Vinclozolin H3K27me cauda differentially methylated histone list. The DHR identification, chromosome location, the chromosome nucleotide start and stop site, length (bp), number significant 1 kb windows, p-value, maximum log fold change (max LFC) (positive is increase in presence and negative is decrease in presence), CpG number and density, gene association, and gene functional category.
Highlights.
Environmental induction of new transgenerational sperm histone retention sites.
Identification of a developmental cascade of histone retention.
Potential role of novel sperm histone retention in epigenetic inheritance.
ACKNOWLEDGMENTS
We acknowledge Ms. Michelle Pappalardo, and Mr. Ryan Thompson for technical assistance. We acknowledge Ms. Amanda Quilty for editing and Ms. Heather Johnson for assistance in preparation of the manuscript. We thank the Genomics Core laboratory at WSU Spokane for sequencing data. This study was supported by John Templeton Foundation (50183 and 61174) (https://templeton.org/) grants to MKS and NIH (ES012974) (https://www.nih.gov/) grant to MKS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTERESTS
The authors declare no competing interests.
DATA ARCHIVING
All molecular data has been deposited into the public database at NCBI (GEO # GSE137963), and R code computational tools are available at GitHub (https://github.com/skinnerlab/MeDIP-seq) and www.skinner.wsu.edu.
References
- 1.Jirtle RL and Skinner MK, Environmental epigenomics and disease susceptibility. Nature Reviews Genetics, 2007. 8(4): p. 253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anway MD, Cupp AS, Uzumcu M, and Skinner MK, Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science, 2005. 308(5727): p. 1466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holliday R and Pugh JE, DNA modification mechanisms and gene activity during development. Science, 1975. 187(4173): p. 226–32. [PubMed] [Google Scholar]
- 4.Singer J, Roberts-Ems J, and Riggs AD, Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science, 1979. 203(4384): p. 1019–21. [DOI] [PubMed] [Google Scholar]
- 5.Dovey OM, Foster CT, and Cowley SM, Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci U S A, 2010. 107(18): p. 8242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugiyama K, Sugiura K, Hara T, Sugimoto K, Shima H, Honda K, Furukawa K, Yamashita S, and Urano T, Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene, 2002. 21(20): p. 3103–11. [DOI] [PubMed] [Google Scholar]
- 7.Goto H, Yasui Y, Nigg EA, and Inagaki M, Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells, 2002. 7(1): p. 11–7. [DOI] [PubMed] [Google Scholar]
- 8.Bannister AJ and Kouzarides T, Regulation of chromatin by histone modifications. Cell Res, 2011. 21(3): p. 381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishigami S, Van Thuan N, Hikichi T, Ohta H, Wakayama S, Mizutani E, and Wakayama T, Epigenetic abnormalities of the mouse paternal zygotic genome associated with microinsemination of round spermatids. Dev Biol, 2006. 289(1): p. 195–205. [DOI] [PubMed] [Google Scholar]
- 10.Barker DJ, Osmond C, Golding J, Kuh D, and Wadsworth ME, Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ, 1989. 298(6673): p. 564–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drake AJ and Walker BR, The intergenerational effects of fetal programming: non-genomic mechanisms for the inheritance of low birth weight and cardiovascular risk. J Endocrinol, 2004. 180(1): p. 1–16. [DOI] [PubMed] [Google Scholar]
- 12.Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, Seisenberger S, Hore TA, Reik W, Erkek S, Peters A, Patti ME, and Ferguson-Smith AC, In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science, 2014. 345(6198): p. 1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn GA and Bale TL, Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology, 2011. 152(6): p. 2228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baird D, Changing problems and priorities in obstetrics. Br J Obstet Gynaecol, 1985. 92(2): p. 115–21. [DOI] [PubMed] [Google Scholar]
- 15.Emanuel I, Filakti H, Alberman E, and Evans SJ, Intergenerational studies of human birthweight from the 1958 birth cohort. 1. Evidence for a multigenerational effect. Br J Obstet Gynaecol, 1992. 99(1): p. 67–74. [DOI] [PubMed] [Google Scholar]
- 16.Kaati G, Bygren LO, and Edvinsson S, Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur J Hum Genet, 2002. 10(11): p. 682–8. [DOI] [PubMed] [Google Scholar]
- 17.Kaati G, Bygren LO, Pembrey M, and Sjostrom M, Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet, 2007. 15(7): p. 784–90. [DOI] [PubMed] [Google Scholar]
- 18.Skinner MK, Manikkam M, Tracey R, Guerrero-Bosagna C, Haque MM, and Nilsson E, Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Medicine, 2013. 11: p. 228, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelce WR and Wilson EM, Environmental antiandrogens: developmental effects, molecular mechanisms, and clinical implications. J Mol Med (Berl), 1997. 75(3): p. 198–207. [DOI] [PubMed] [Google Scholar]
- 20.Mansouri A, Cregut M, Abbes C, Durand MJ, Landoulsi A, and Thouand G, The Environmental Issues of DDT Pollution and Bioremediation: a Multidisciplinary Review. Appl Biochem Biotechnol, 2017. 181(1): p. 309–339. [DOI] [PubMed] [Google Scholar]
- 21.Skinner MK, Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol Cell Endocrinol, 2014. 398(1–2): p. 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soubry A, Epigenetic inheritance and evolution: A paternal perspective on dietary influences. Prog Biophys Mol Biol, 2015. 118(1–2): p. 79–85. [DOI] [PubMed] [Google Scholar]
- 23.Vaiserman AM, Koliada AK, and Jirtle RL, Non-genomic transmission of longevity between generations: potential mechanisms and evidence across species. Epigenetics Chromatin, 2017. 10(1): p. 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skinner MK, Environmental Epigenetics and a Unified Theory of the Molecular Aspects of Evolution: A Neo-Lamarckian Concept that Facilitates Neo-Darwinian Evolution. Genome Biol Evol, 2015. 7(5): p. 1296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, and Yan W, Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics & Chromatin 2018. 11(1): p. 8, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, and Reik W, The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell, 2012. 48(6): p. 849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jost A, Vigier B, Prepin J, and Perchellet JP, Studies on sex differentiation in mammals. Recent Prog Horm Res, 1973. 29: p. 1–41. [DOI] [PubMed] [Google Scholar]
- 28.McCarrey JR, Toward a more precise and informative nomenclature describing fetal and neonatal male germ cells in rodents. Biol Reprod, 2013. 89(2): p. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarrey JR, The epigenome as a target for heritable environmental disruptions of cellular function. Mol Cell Endocrinol, 2012. 354(1–2): p. 9–15. [DOI] [PubMed] [Google Scholar]
- 30.Dym M and Fawcett DW, Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol Reprod, 1971. 4(2): p. 195–215. [DOI] [PubMed] [Google Scholar]
- 31.Cornwall GA, New insights into epididymal biology and function. Hum Reprod Update, 2009. 15(2): p. 213–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjorkgren I and Sipila P, The impact of epididymal proteins on sperm function. Reproduction, 2019. [DOI] [PubMed] [Google Scholar]
- 33.Oliva R and Castillo J, Proteomics and the genetics of sperm chromatin condensation. Asian J Androl, 2011. 13(1): p. 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliva and Castillo, Sperm Nucleoproteins, in Sperm Chromatin: Biological and Clinical Applications in Male Infertility and Assisted Reproduction Agarwal AZA, Editor. 2011, Springer: New York: p. 45–60. [Google Scholar]
- 35.Oliva R and de Mateo S, Medical implications of sperm nuclear quality, in Epigenetics and Human Reproduction, Epigenetics and Human Health, Khochbin R, Editor. 2011, Springer-Verlag: Berlin, Heidelberg: p. 45–83. [Google Scholar]
- 36.Oliva R and Dixon GH, Vertebrate protamine genes and the histone-toprotamine replacement reaction. Prog Nucleic Acid Res Mol Biol, 1991. 40: p. 25–94. [DOI] [PubMed] [Google Scholar]
- 37.Balhorn R, The protamine family of sperm nuclear proteins. Genome Biol, 2007. 8(9): p. 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roque A, Ponte I, and Suau P, Secondary structure of protamine in sperm nuclei: an infrared spectroscopy study. BMC Struct Biol, 2011. 11: p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliva R, Protamines and male infertility. Hum Reprod Update, 2006. 12(4): p. 417–35. [DOI] [PubMed] [Google Scholar]
- 40.Torregrosa N, Dominguez-Fandos D, Camejo MI, Shirley CR, Meistrich ML, Ballesca JL, and Oliva R, Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum Reprod, 2006. 21(8): p. 2084–9. [DOI] [PubMed] [Google Scholar]
- 41.Carrell DT, Emery BR, and Hammoud S, The aetiology of sperm protamine abnormalities and their potential impact on the sperm epigenome. Int J Androl, 2008. 31(6): p. 537–45. [DOI] [PubMed] [Google Scholar]
- 42.de Mateo S, Ramos L, de Boer P, Meistrich M, and Oliva R, Protamine 2 precursors and processing. Protein Pept Lett, 2011. 18(8): p. 778–85. [DOI] [PubMed] [Google Scholar]
- 43.Lewis SE and Aitken RJ, DNA damage to spermatozoa has impacts on fertilization and pregnancy. Cell Tissue Res, 2005. 322(1): p. 33–41. [DOI] [PubMed] [Google Scholar]
- 44.de Mateo S, Gazquez C, Guimera M, Balasch J, Meistrich ML, Ballesca JL, and Oliva R, Protamine 2 precursors (Pre-P2), protamine 1 to protamine 2 ratio (P1/P2), and assisted reproduction outcome. Fertil Steril, 2009. 91(3): p. 715–22. [DOI] [PubMed] [Google Scholar]
- 45.Castillo J, Simon L, de Mateo S, Lewis S, and Oliva R, Protamine/DNA ratios and DNA damage in native and density gradient centrifuged sperm from infertile patients. J Androl, 2011. 32(3): p. 324–32. [DOI] [PubMed] [Google Scholar]
- 46.Simon L, Castillo J, Oliva R, and Lewis SE, Relationships between human sperm protamines, DNA damage and assisted reproduction outcomes. Reprod Biomed Online, 2011. 23(6): p. 724–34. [DOI] [PubMed] [Google Scholar]
- 47.Kumar K, Deka D, Singh A, Mitra DK, Vanitha BR, and Dada R, Predictive value of DNA integrity analysis in idiopathic recurrent pregnancy loss following spontaneous conception. J Assist Reprod Genet, 2012. 29(9): p. 861–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czubaszek M, Andraszek K, and Banaszewska D, Influence of the age of the individual on the stability of boar sperm genetic material. Theriogenology, 2020. 147: p. 176–182. [DOI] [PubMed] [Google Scholar]
- 49.Ooi SL and Henikoff S, Germline histone dynamics and epigenetics. Curr Opin Cell Biol, 2007. 19(3): p. 257–65. [DOI] [PubMed] [Google Scholar]
- 50.Aitken RJ and De Iuliis GN, On the possible origins of DNA damage in human spermatozoa. Mol Hum Reprod, 2010. 16(1): p. 3–13. [DOI] [PubMed] [Google Scholar]
- 51.Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, and Carrell DT, Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod, 2011. 26(9): p. 2558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ihara M, Meyer-Ficca ML, Leu NA, Rao S, Li F, Gregory BD, Zalenskaya IA, Schultz RM, and Meyer RG, Paternal poly (ADP-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS Genet, 2014. 10(5): p. e1004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puri D, Dhawan J, and Mishra RK, The paternal hidden agenda: Epigenetic inheritance through sperm chromatin. Epigenetics, 2010. 5(5): p. 386–91. [DOI] [PubMed] [Google Scholar]
- 54.Ben Maamar M, King SE, Nilsson E, Beck D, and Skinner MK, Epigenetic Transgenerational Inheritance of Parent-of-Origin Allelic Transmission of Outcross Pathology and Sperm Epimutations. Developmental Biology, 2020. 458(1): p. 106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben Maamar M, Sadler-Riggleman I, Beck D, and Skinner MK, Epigenetic Transgenerational Inheritance of Altered Sperm Histone Retention Sites. Scientific Reports, 2018(8): p. 5308, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ben Maamar M, Sadler-Riggleman I, Beck D, McBirney M, Nilsson E, Klukovich R, Xie Y, Tang C, Yan W, and Skinner MK, Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate vinclozolin-induced epigenetic transgenerational inheritance of disease. Environmental Epigenetics, 2018. 4(2): p. 1–19, dvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okada Y and Yamaguchi K, Epigenetic modifications and reprogramming in paternal pronucleus: sperm, preimplantation embryo, and beyond. Cell Mol Life Sci, 2017. 74(11): p. 1957–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jenkins TG and Carrell DT, The sperm epigenome and potential implications for the developing embryo. Reproduction, 2012. 143(6): p. 727–34. [DOI] [PubMed] [Google Scholar]
- 59.Castillo J, Jodar M, and Oliva R, The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum Reprod Update, 2018. 24(5): p. 535–555. [DOI] [PubMed] [Google Scholar]
- 60.Samson M, Jow MM, Wong CC, Fitzpatrick C, Aslanian A, Saucedo I, Estrada R, Ito T, Park SK, Yates JR 3rd, and Chu DS, The specification and global reprogramming of histone epigenetic marks during gamete formation and early embryo development in C. elegans. PLoS Genet, 2014. 10(10): p. e1004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, Kwon T, Marcotte EM, Zegerman P, Bradshaw CR, Peters AH, Gurdon JB, and Jullien J, Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res, 2016. 26(8): p. 1034–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ben Maamar M, Nilsson E, Sadler-Riggleman I, Beck D, McCarrey JR, and Skinner MK, Developmental origins of transgenerational sperm DNA methylation epimutations following ancestral DDT exposure. Dev Biol, 2019. 445(2): p. 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skinner MK, Nilsson E, Sadler-Riggleman I, Beck D, and McCarrey JR, Transgenerational Sperm DNA Methylation Epimutation Developmental Origins Following Ancestral Vinclozolin Exposure. Epigenetics, 2019. 14(7): p. 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harikae K, Miura K, and Kanai Y, Early gonadogenesis in mammals: significance of long and narrow gonadal structure. Dev Dyn, 2013. 242(4): p. 330–8. [DOI] [PubMed] [Google Scholar]
- 65.Skinner MK, What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol, 2008. 25(1): p. 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilkerson R, Bravo L, Garcia I, Gaytan N, Herrera A, Maldonado A, and Quintanilla B, The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb Perspect Biol, 2013. 5(5): p. a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matilainen O, Quiros PM, and Auwerx J, Mitochondria and Epigenetics - Crosstalk in Homeostasis and Stress. Trends Cell Biol, 2017. 27(6): p. 453–463. [DOI] [PubMed] [Google Scholar]
- 68.Weyemi U, Paul BD, Bhattacharya D, Malla AP, Boufraqech M, Harraz MM, Bonner WM, and Snyder SH, Histone H2AX promotes neuronal health by controlling mitochondrial homeostasis. Proc Natl Acad Sci U S A, 2019. 116(15): p. 7471–7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skinner MK and Guerrero-Bosagna C, Role of CpG Deserts in the Epigenetic Transgenerational Inheritance of Differential DNA Methylation Regions. BMC Genomics 2014. 15(1): p. 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rathke C, Baarends WM, Awe S, and Renkawitz-Pohl R, Chromatin dynamics during spermiogenesis. Biochim Biophys Acta, 2014. 1839(3): p. 155–68. [DOI] [PubMed] [Google Scholar]
- 71.Bao J and Bedford MT, Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction, 2016. 151(5): p. R55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hermann BP, Mutoji KN, Velte EK, Ko D, Oatley JM, Geyer CB, and McCarrey JR, Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol Reprod, 2015. 92(2): p. 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nilsson E, Sadler-Riggleman I, and Skinner MK, Environmentally Induced Epigenetic Transgenerational Inheritance of Disease. Environmental Epigenetics, 2018. 4(2): p. 1–13, dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guerrero-Bosagna C, Savenkova M, Haque MM, Nilsson E, and Skinner MK, Environmentally Induced Epigenetic Transgenerational Inheritance of Altered Sertoli Cell Transcriptome and Epigenome: Molecular Etiology of Male Infertility. PLoS ONE, 2013. 8(3): p. 1–12, e59922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klukovich R, Nilsson E, Sadler-Riggleman I, Beck D, Xie Y, Yan W, and Skinner MK, Environmental Toxicant Induced Epigenetic Transgenerational Inheritance of Prostate Pathology and Stromal-Epithelial Cell Epigenome and Transcriptome Alterations: Ancestral Origins of Prostate Disease. Scientific Reports, 2019. 9(2209): p. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simon L, Liu L, Murphy K, Ge S, Hotaling J, Aston KI, Emery B, and Carrell DT, Comparative analysis of three sperm DNA damage assays and sperm nuclear protein content in couples undergoing assisted reproduction treatment. Hum Reprod, 2014. 29(5): p. 904–17. [DOI] [PubMed] [Google Scholar]
- 77.Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, and Skinner MK, Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS ONE 2012. 7(2): p. 1–12, e31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCarrey JR, Berg WM, Paragioudakis SJ, Zhang PL, Dilworth DD, Arnold BL, and Rossi JJ, Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev Biol, 1992. 154(1): p. 160–8. [DOI] [PubMed] [Google Scholar]
- 79.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, and Razin A, Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev, 1992. 6(5): p. 705–14. [DOI] [PubMed] [Google Scholar]
- 80.Bolger AM, Lohse M, and Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014. 30(15): p. 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langmead B and Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nature Methods, 2012. 9(4): p. 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome S Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics, 2009. 25(16): p. 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lienhard M, Grimm C, Morkel M, Herwig R, and Chavez L, MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics, 2014. 30(2): p. 284–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson MD, McCarthy DJ, and Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010. 26(1): p. 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Durinck S, Spellman PT, Birney E, and Huber W, Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature Protocols, 2009. 4(8): p. 1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kahari AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, and Flicek P, Ensembl 2015. Nucleic Acids Research, 2015. 43(Database issue): p. D662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kanehisa M and Goto S, KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 2000. 28(1): p. 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, and Tanabe M, Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Research, 2014. 42(Database issue): p. D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang da W, Sherman BT, and Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 2009. 4(1): p. 44–57. [DOI] [PubMed] [Google Scholar]
- 90.Mi H, Muruganujan A, Casagrande JT, and Thomas PD, Large-scale gene function analysis with the PANTHER classification system. Nature Protocols, 2013. 8(8): p. 1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. The number of DHRs at different CpG densities. All DHRs at a p-value threshold of 1e-04 are presented. (A) H3 control caput versus RS. (B) H3 control cauda versus caput. (C) H3 RS DDT. (D) H3 RS vinclozolin. (E) H3 caput DDT. (F) H3 caput vinclozolin. (G) H3 cauda DDT. (H) H3 cauda vinclozolin.
Supplemental Figure S2. The number of DHR at different lengths. All DHRs at a p-value threshold of 1e-04 are presented. (A) H3 control Caput versus RS. (B) H3 control cauda versus caput. (C) H3 RS DDT. (D) H3 RS vinclozolin. (E) H3 caput DDT. (F) H3 caput vinclozolin. (G) H3 cauda DDT. (H) H3 cauda vinclozolin.
Supplemental Figure S3.The number of DHRs at different CpG densities. All DHRs at a p-value threshold of 1e-04 are presented. (A) H3K27me3 control cauda versus caput. (C) H3K27me3 caput DDT. (E) H3K27me3 cauda DDT. (G) H3K27me3 caput vinclozolin. (I) H3K27me3 cauda vinclozolin. DHR lengths (kilobase) with all DHRs at a p-value threshold of 1e-04 are presented. (B) H3K27me3 control cauda versus caput. (D) H3K27me3 caput DDT. (F) H3K27me3 cauda DDT. (H) H3K27me3 caput vinclozolin. (J) H3K27me3 cauda vinclozolin.
Supplemental Figure S4. Expanded overlap with p<1e-04 on left margin and p<0.05 on top margin. The number of DHR overlaps and percentage of the total presented.
Supplemental Figure S5. DHR gene association Kegg pathways. The pathways associated with each DHR data set presented with the number of DHR in brackets. (https://www.genome.jp/kegg/pathway.html)
Supplemental Table S1. Control DHR lists. (A) Control lineage round spermatid versus caput spermatozoa. (B) Control lineage caput versus cauda. (C) Control lineage H3K27me3 caput versus cauda. The DHR identification, chromosome location, the chromosome nucleotide start and stop site, length (bp), number significant 1 kb windows, p-value, maximum log fold change (max LFC) (positive is increase in presence and negative is decrease in presence), CpG number and density, gene association, and gene functional category.
Supplemental Table S2. DDT DHR lists. (A) DDT round spermatid DHR list. (B) DDT caput DHR list. (C) DDT cauda DHR list. (D) DDT H3k27me caput differentially methylated histone list. (E) DDT H3k27me cauda differentially methylated histone list. The DHR identification, chromosome location, the chromosome nucleotide start and stop site, length (bp), number significant 1 kb windows, p-value, maximum log fold change (max LFC) (positive is increase in presence and negative is decrease in presence), CpG number and density, gene association, and gene functional category.
Supplemental Table S3. Vinclozolin DHR lists. (A) Vinclozolin round spermatid DHR list. (B) Vinclozolin caput DHR list. (C) Vinclozolin cauda DHR list. (D) Vinclozolin H3K27me caput differentially methylated histone list. (E) Vinclozolin H3K27me cauda differentially methylated histone list. The DHR identification, chromosome location, the chromosome nucleotide start and stop site, length (bp), number significant 1 kb windows, p-value, maximum log fold change (max LFC) (positive is increase in presence and negative is decrease in presence), CpG number and density, gene association, and gene functional category.


