Abstract
Lower urinary tract symptoms (LUTS) are common in aging males. Disease etiology is largely unknown, but likely includes inflammation and age-related changes in steroid hormones. Diagnosis is currently based on subjective symptom scores, and mainstay treatments can be ineffective and bothersome. Biomarker discovery efforts could facilitate objective diagnostic criteria for personalized medicine and new potential druggable pathways. To identify urine metabolite markers specific to hormone-induced bladder outlet obstruction, we applied our custom synthesized multiplex isobaric tags to monitor the development of bladder outlet obstruction across time in an experimental mouse model of LUTS. Mouse urine samples were collected before treatment and after 2, 4, and 8 weeks of steroid hormone treatment and subsequently analyzed by nanoflow ultrahigh-performance liquid chromatography coupled to tandem mass spectrometry. Accurate and high throughput quantification of amine-containing metabolites was achieved by twelve-plex DiLeu isobaric labeling. Metandem, a novel online software tool for large-scale isobaric labeling-based metabolomics, was used for identification and relative quantification of labeled metabolites. A total of 59 amine-containing metabolites were identified and quantified, 9 of which were changed significantly by the hormone treatment. Metabolic pathway analysis showed that three metabolic pathways were potentially disrupted. Among them, the arginine and proline metabolism pathway was significantly dysregulated both in this model and in a prior analysis of LUTS patient samples. Proline and citrulline were significantly changed in both samples and serve as attractive candidate biomarkers. 12-plex DiLeu isobaric labeling with Metandem data processing presents an accessible and efficient workflow for amine-containing metabolome study in biological specimens.
Keywords: urine, metabolomics, lower urinary tract symptoms, mass spectrometry, isobaric labeling, Metandem
Brief Synopsis of the Graphic:
The urine sample is collected from the LUTS mouse model. Extracted metabolites from distinct urine samples are differentially labeled with isobaric DiLeu tags and each metabolite is shown as a single peak at the MS1 level. The subsequent MS2 analysis enables relative quantification based on reporter ion intensities. Metandem software is used for data analysis. Finally, the quantification results can be used for biomarker discovery of LUTS.
Graphical Abstract
INTRODUCTION
Lower urinary tract symptoms (LUTS) is a major public health problem in the aging population. Symptoms include frequency, urgency, nocturia, weak urinary stream, straining to void, and a sense of incomplete emptying (1–3). Recent studies have shown that the etiology of LUTS is multi-factorial (4, 5). In men, prostate enlargement, prostatic inflammation, age-related changes in detrusor function and steroid hormones, and heightened sensitivity to bladder filling may all contribute (4–8). The complex and variegated composition of the LUTS patient population makes it challenging to tease out and validate contributions of different mechanisms. Currently no objective biomarkers exist to inform treatment strategies, so patients are generally treated with drugs to decrease prostate size (5α-reductase inhibitors) or relax smooth muscle (alpha blockers) (9, 10). These treatments are not completely effective, durable in response, nor are they without complication (11). Patients with LUTS that persists after these mainline treatments may undergo invasive transurethral resection of the prostate (12). Overall, better biomarkers and treatment options need to be found. Here, we attempt to characterize the contribution of hormone changes to the urine metabolome under controlled, experimental conditions in a mouse model of bladder outlet obstruction (BOO). Understanding the urine signature of hormone-induced BOO will inform both future efforts to study druggable pathways and validate LUTS biomarkers for patient stratification and personalized treatments.
Our hormone-induced BOO mouse model relies on slow-release, subcutaneous implants of testosterone and 17β-estradiol in adult male C57/BL6 mice to generate marked increases in urinary frequency, bladder volume, bladder mass, and prostate mass as well as decreased urinary volume/void before the 8 week timepoint (13). Additionally, this model, if treated longer (4 months), develops prostatic intraepithelial neoplasia lesions, making this model useful for the study of carcinogenesis in the prostate (14). To characterize the urine metabolome of hormone-induced BOO, we collected samples across the development of this phenotype: before treatment and after 2, 4, and 8 weeks of treatment.
Tandem mass spectrometry (MS/MS)-based relative quantification by isobaric labeling is a useful technique for comparative quantitative metabolomics in biological systems (15–17). N,N-dimethyl leucine (DiLeu) labels are isobaric. This means that analyte precursor m/z between channels is indistinguishable at low MS resolution, but distinct reporter ions are apparent in the low m/z region upon MS2 fragmentation. The intensities of these reporter ions in MS2 spectra reflect the labeled metabolites’ abundance in each sample and, thus, can be compared for relative quantification. Compared to label-free metabolomics, multiplexed isobaric labeling of metabolites greatly reduces run-to-run variation, enhances ionization efficiency, improves chromatographic separation of polar metabolites, and decreases instrument time demand (by 12-fold when using 12-plex DiLeu labeling) (18). DiLeu utilizes a triazine ester to label primary and secondary amines, making this derivatization scheme applicable to many metabolites. Additionally, DiLeu labeling enables polar metabolites to be separated and detected on nanoUPLC systems, improving chromatographic resolution and detection sensitivity over typical standard flow separation techniques (19). Twelve-plex DiLeu isobaric labels, which are designed and synthesized in our lab (20, 21), were employed here for the relative quantification of amine-containing metabolites in urine samples via nanoflow ultrahigh-performance liquid chromatography coupled to tandem mass spectrometry (nanoUPLC-MS/MS).
Isobaric labeling has been widely adopted for quantitative proteomics and peptidomics, but it has only recently been applied to metabolomics analyses (22–24). Thus, data processing workflows for isobaric labeling in proteomics and peptidomics have matured more rapidly than for metabolomics. To date, few metabolomics software tools can process stable isotope label-based metabolomics data, particularly when using reporter ions produced by MS2 for quantification. Therefore, our lab developed a novel online software tool for isobaric labeling-based metabolomics, called Metandem, which integrates metabolite quantification, identification, and statistical analysis in the same software package and is freely available at http://metandem.com/web/ (25). Metandem is also the first omics data analysis software to provide straightforward, online parameter optimization functionality for customization to a particular dataset (25). Here, we employed the Metandem software tool to analyze the 12-plex DiLeu-labeled urinary metabolites at multiple time points after hormone treatment. Metabolite identification, quantification, and statistical analysis were achieved in less than 15 min using our Metandem software tool. It is expected that these powerful tools may identify clinically useful biomarkers of hormone-induced bladder outlet obstruction and new targets for drug treatment.
MATERIALS AND METHODS
Hormone-induced urinary obstruction mouse model
All animal procedures were approved by the University of Wisconsin-Madison Animal Care and Use Committee. Adult male C57BL/6 mice were used for this study (n = 3) (Charles River, Wilmington, MA). Urine was collected from each animal via metabolic cage (26) for a two-hour period. After the pre-treatment urine collection, mice were treated with subcutaneous, slow-release implants containing compressed testosterone (T, 25 mg) and 17β-estradiol (E2, 2.5 mg + 22.5 mg cholesterol binder), as described previously (13). Urine samples from the same mice were then collected at 2, 4, and 8 weeks post-treatment, as above. All samples were stored at −80 °C until further processing.
Mouse urine sample preparation
Urine samples were thawed on ice and mixed well by vortexing. Raw urine (220 μL) was centrifuged at 10,000 g for 10 min to remove particulates and cellular debris. Centrifugal filters (3 kDa, Millipore Amicon Ultra, Burlington, MA) were pre-rinsed 3 times with 500 μL of Milli-Q water at 14,000 g for 20 min. Clarified supernatant (200 μL) of each urine sample was added to the filter unit and centrifuged at 14,000 g for 30 min, followed by 2 rinses with Milli-Q water (200 μL; same centrifugation speed and time) to obtain the metabolite fraction (~600 μL total). Osmolality was determined via a freezing-point depression osmometer (Osmometer Model 3250, Advanced Instruments, Norwood, MA) (27) and all samples were normalized to 50 mOsmoles/kg H2O. Osmolite concentration can represent the total urinary metabolite content, and normalized to osmolality has been proven to be a better method for metabolite normalization prior to instrumental analysis (28). Then all the aliquoted urine samples were lyophilized and stored at −80 °C until labeling.
DiLeu synthesis and labeling procedure
Twelve-plex isobaric DiLeu reagents were synthesized and used for labeling reaction as previously described (29). Briefly, the DiLeu 12-plex reagents were synthesized, aliquoted in inactivated form at 4 °C in a dry box and activated prior to labeling. Lyophilized urine metabolite samples were re-dissolved in 0.5 M triethylammonium bicarbonate solution prior to derivatization of primary and secondary amines by excess activated DiLeu reagent (Fig. 1D). The organic: aqueous ratio was maintained at ~70% via anhydrous dimethylformamide. Reactions were allowed to proceed for 2 hrs at room temperature with vigorous vortexing. Labeling reactions were quenched with 0.25% hydroxylamine (v/v), and labeled samples were combined in equal ratios (v/v) to form pooled 12-plex samples. Excess DiLeu reagents were removed from pooled 12-plex samples via SCX Ziptips (OMIX-SCX, Agilent, Santa Clara, CA) as previously described (19) before lyophilization and storage at −80 °C until analysis.
Figure 1. Details of 12-plex DiLeu labeling.
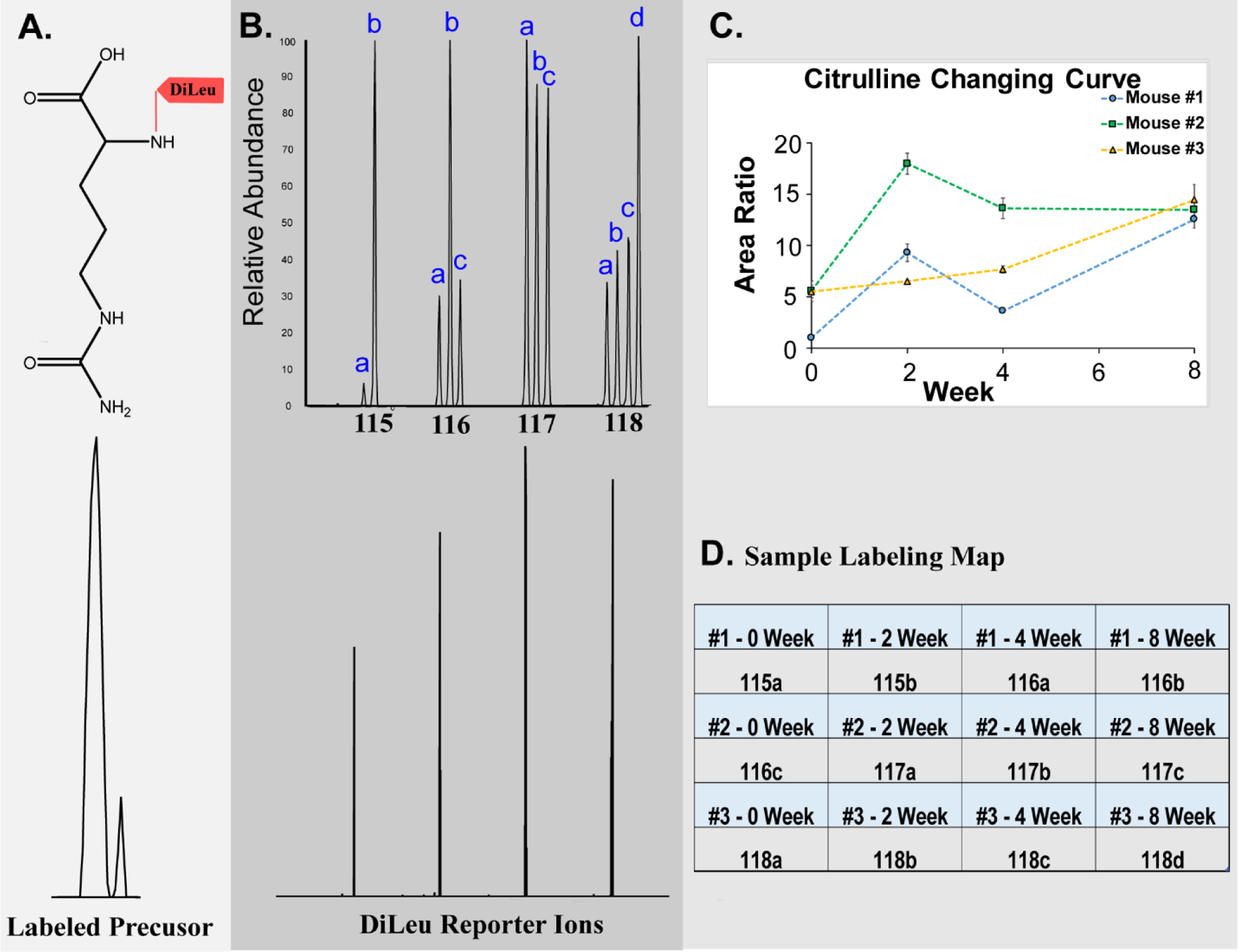
A: Precursor ion of DiLeu-labeled citrulline; B: An MS2 spectrum of the 12-plex DiLeu-labeled citrulline acquired in the Orbitrap at 60 K resolving power. Low m/z region showing distinct DiLeu reporter channels (bottom) and after zooming in, twelve distinct reporter ion peaks are present (bottom); C: Citrulline changing trends from different time points of three biological replicates. D: Sample labeling map showing the 12-plex DiLeu tags and time point (randomized)
LC-ESI-MS analysis
Twelve-plex pooled samples were reconstituted in 3% acetonitrile, 0.1% formic acid (v/v) in water before injection. UPLC-MS/MS analysis was conducted using a Thermo Dionex UltiMate™ 3000 nanoLC system coupled to a Thermo Q Exactive™ HF Orbitrap MS. The analytical column was self-made with an integrated emitter tip and dimensions of 75 μm inner diameter × 15 cm length, packed with 1.7 μm, 150 Å, BEH C18 material (Waters, Milford, MA). Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile (Optima Solvents, Thermo, Waltham, MA). The flow rate was 0.3 μL/min, and the 70-min gradient was as follows: 0–16 min, 3% solvent B; 16–20 min, 3–25% B; 20–30 min, 25–45% B; 30–50 min, 45–70% B; 50–56 min, 70–95% B; 56–60 min 95% B; 60–60.5 min, 95–3% B; 60.5–70 min, 3% B. Positive ionization mode was used for the MS analysis. Full MS scans were acquired from m/z 180 to 1000 at a resolution of 60 K, automatic gain control (AGC) at 1 × 106, and maximum injection time of 50 ms. The top 20 precursors were selected for higher-energy C-trap dissociation tandem mass spectrometry (HCD MS2) analysis with an isolation window of 1 m/z, normalized collision energy (NCE) of 30, resolving power of 60k, AGC target of 1 × 105, maximum injection time of 30 ms, and a lower mass limit of 110 m/z.
Data analysis
Raw data files were acquired using Thermo Scientific Xcalibur™ software and converted into .txt format via the COMPASS software suite (30). Metandem was then used to batch-process three technical replicates of each 12-plex sample for metabolite quantification. Accurate mass of reporter ions was obtained by averaging the mass across several MS/MS spectra. Then, the mass tolerance for metabolite identification was 20 ppm. DiLeu label purity was predetermined, and correction for each channel was performed as previously described (19). Average precursor mass shift due to labeling was 145.1280 Da. Data analysis parameters, such as reporter ion mass tolerance, batch mass tolerance, and retention time tolerance were optimized using the parameter optimization graphs in the Metandem software. Output files with reporter ion information were merged and median-normalized. Molecular weights of detected compounds were calculated based on the charge and mass shift caused by labeling, and then searched against the Human Metabolome Database (HMDB). The unreasonable matches were excluded from the identification results and only the metabolites with primary or secondary amine were kept. Paired t-test was performed comparing metabolite abundance of 2/4/8 weeks to that of 0 week. Any of the three comparisons showing fold change > 1.5 and p-value < 0.05 were considered as significantly changed metabolites. MetaboAnalyte 4.0 software (31) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used for metabolic pathway analysis. Pathways were considered dysregulated when more than two metabolites were identified in the pathway and at least one was significantly changed due to the treatment. Candidate biomarkers and dysregulated pathways were compared with prior patient analyses and other previous reports.
RESULTS AND DISCUSSION
Efficacy of twelve-plex DiLeu isobaric labeling for metabolomics
Twelve-plex DiLeu isobaric labeling allowed multiplexed metabolite identification and quantification in mouse urine samples while also reducing instrumentation time demand, decreasing run-to-run variation, and also improving quantification accuracy. The same labeled metabolite from 12 urine samples showed a single peak in the MS1 spectrum with a mass shift of 145.1280 Da (Fig. 1A). For this peak, twelve distinct reporter ion peaks are present in the MS2 low m/z region (Fig. 1B). The intensity of each reporter ion in MS2 spectra reflects the labeled metabolites’ abundance in each sample and, thus, can be compared for relative quantification (Fig. 1C). The absence of unlabeled metabolite ions in the MS1 (mass difference of 145.1280 compared to unlabeled counterparts) suggested highly efficient labeling of the metabolites by the DiLeu reagent.
Metandem parameter optimization
Metandem is a newly developed custom software platform for large-scale stable isotope labeling-based metabolite identification and quantification. It is also the first omics data analysis software that contains functionality to perform online parameter optimization for customization to a dataset. Results of automated parameter optimization were as follows: optimal reporter ion mass tolerance, 0.5 mDa (Fig. 2A); optimal batch processing mass tolerance, 5 ppm (Fig. 2B); and optimal batch processing retention time tolerance, 0.5 min (Fig. 2C). Metandem also provides the histogram for retention time (Fig. S1A) and detected precursor (Fig. S1B) distribution results.
Figure 2. Metandem parameter optimization results:
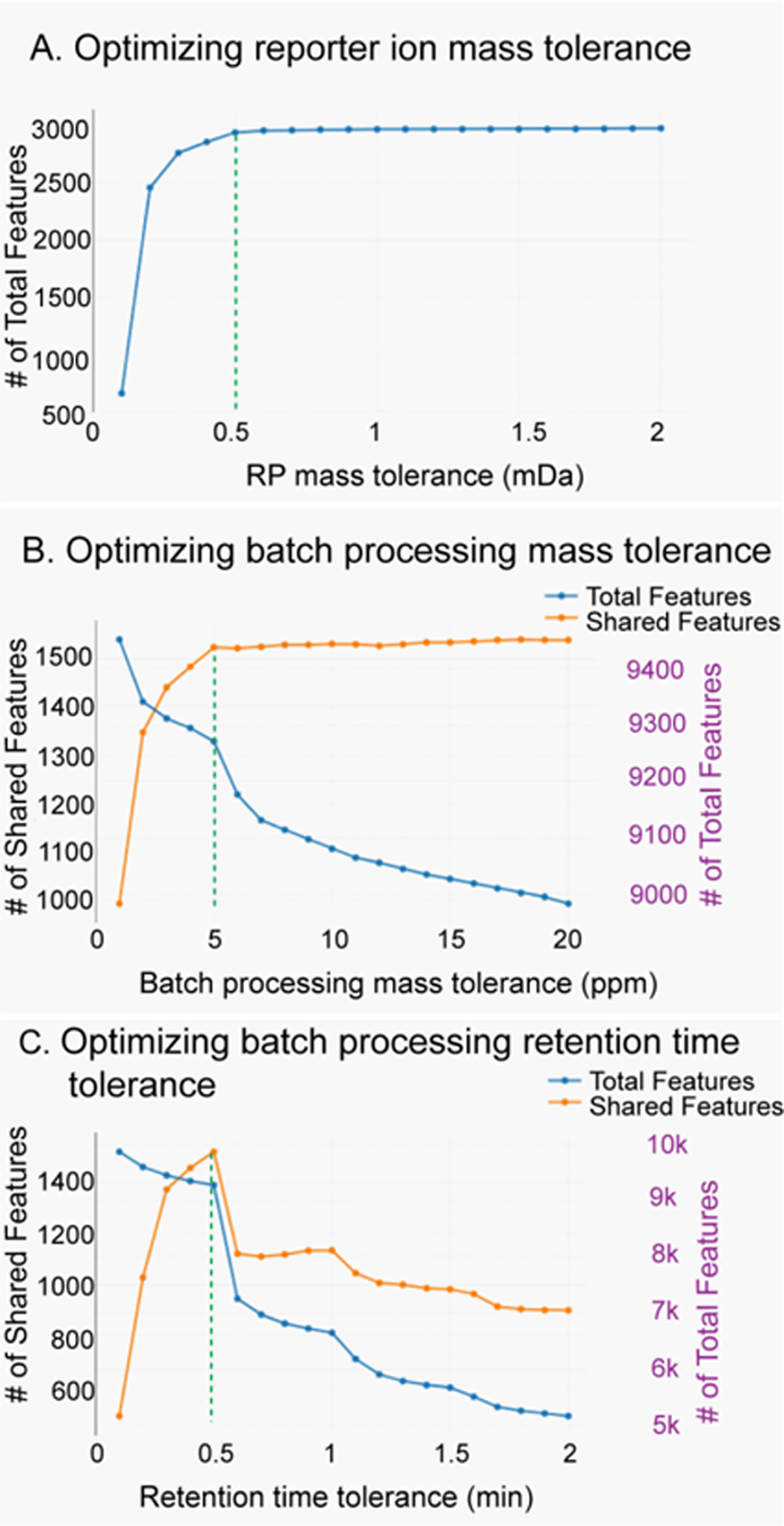
A: Optimizing reporter ion mass tolerance (0.5 mDa); B: Optimizing batch processing retention time tolerance (5 ppm); C: Optimizing batch processing retention time tolerance (0.5 min).
Mouse urine metabolite identification and quantification
Three technical replicates of each 12-plex injection were merged in Metandem. A total of 312 features were identified as putative metabolites after accurate mass matching against the HMDB. After excluding unreasonable matches from the database, 59 were primary or secondary amine-containing metabolites were identified (Table S1). Thirty-seven of these 59 were documented in the urine metabolome database (32), and the twenty-two additional amine-containing metabolites were, to our knowledge, first reported here. After comparing metabolite abundance between timepoints, 9 metabolites were identified as statistically significant biomarker candidates (Paired t-test, fold change > 1.5 and p-value < 0.05. The smallest p-value out of the three was show in Table 1). Among them, eight metabolites were generally increased at all timepoints, while only one metabolite was generally decreased at all timepoints (Fig. 3). For the increased expression patterns, leucine and 5-aminopentanamide have the highest concentration at 2 weeks; N-acetylputrescine at 4 weeks; and proline, citrulline, D-alanyl-D-alanine, O-phosphohomoserine, and 2-Aminobenzoic acid peaked at 8 weeks.
Table 1.
Nine significantly changed urine metabolites after hormone treatment.
| Compound | Molecular Weight | p-value | Expression Pattern | ppm | tr (min) |
HMDB_ID |
|---|---|---|---|---|---|---|
| Proline | 115.0623 | 0.006 | Increase | 9.2 | 13.6 | HMDB00162 |
| 5-Aminopentanamide | 116.0938 | 0.029 | Increase | 10.2 | 14.4 | HMDB12176 |
| N-Acetylputrescine | 130.1108 | 0.042 | Increase | 1.2 | 15.2 | HMDB02064 |
| Citrulline | 175.0936 | 0.005 | Increase | 11.7 | 15.5 | HMDB00904 |
| D-Alanyl-D-alanine | 160.0827 | 0.005 | Increase | 13.1 | 16.5 | HMDB03459 |
| O-Phosphohomoserine | 199.0278 | 0.001 | Increase | 16.1 | 16.7 | HMDB03484 |
| Purine | 120.0450 | 0.001 | Decrease | 11.3 | 8.6 | HMDB01366 |
| 2-Aminobenzoic acid | 137.0464 | 0.023 | Increase | 9.7 | 19.0 | HMDB01123 |
| Leucine | 131.0942 | 0.006 | Increase | 3.0 | 19.0 | HMDB00687 |
Figure 3.
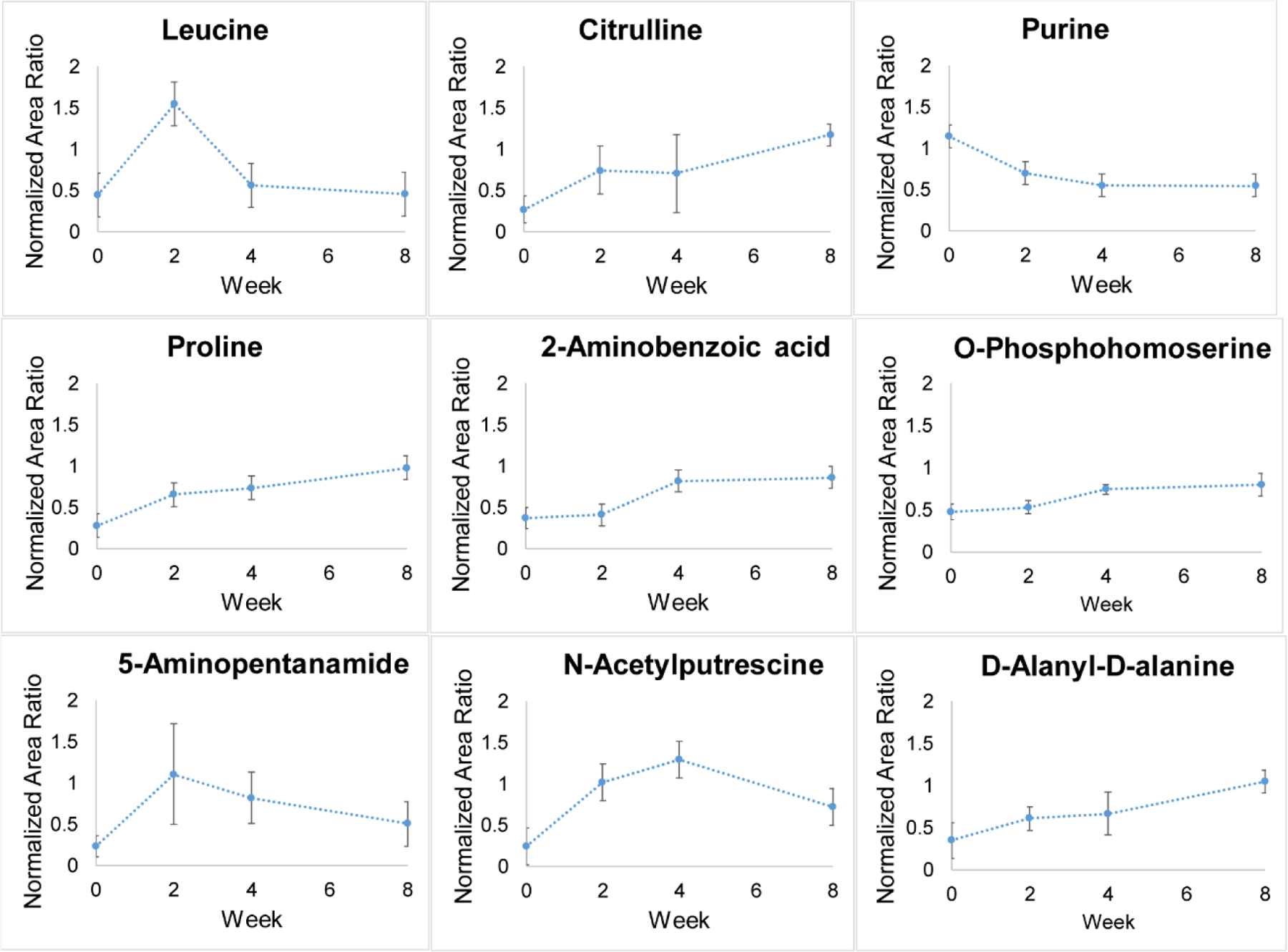
Quantification trends of 9 significantly changed urine metabolites (n = 3; Paired t-test, fold change > 1.5 and p < 0.05).
Metabolic pathway analysis
Metabolic pathway analysis is based on the association between identified metabolites and their related biological processes (33). Herein, all identified metabolites were input into the MetaboAnalyte 4.0 software for metabolic pathway analysis. Three potentially perturbed metabolic pathways were identified: (1) the arginine and proline metabolism pathway; (2) the aminoacyl-tRNA biosynthesis pathway; and (3) the tryptophan metabolism pathway (Table 2. p-value is from Fisher’s exact test). Although tryptophan metabolism pathway has a p-value slightly higher than 0.05, it was also shown here as a candidate disrupted metabolic pathway.
Table 2.
Potentially disrupted metabolic pathways via MetaboAnalyte 4.0 and KEGG pathway analysis, metabolites highlighted with red bold font are significantly changed metabolites (p-value is from Fisher’s exact test).
| Metabolic Pathway | KEGG ID | Matched Metabolites | p-value |
|---|---|---|---|
| Arginine and proline metabolism | Map00330 | citrulline, N-acetylputrescine, proline, glutamic acid, creatine, GABA, 4-aminobutyraldehyde | 4.90E-05 |
| Aminoacyl-tRNA biosynthesis | Map00970 | leucine, proline, cysteine, glycine, alanine, glutamic acid | 4.88E-03 |
| Tryptophan metabolism | Map00380 | 2-aminobenzoic acid, 3-hydroxyanthranilic acid, 5-hydroxyindoleacetic acid | 6.81E-02 |
Comparison with LUTS patient samples and other previous reports
Prior analyses have demonstrated some of the metabolomic and immunohistochemical features of LUTS in patient samples, which represent processes like fibrosis and inflammation (27). Both LUTS patients and this hormone-induced mouse model of BOO show perturbation of the arginine and proline metabolic pathway in the urine metabolome. In particular, the candidate biomarkers citrulline and proline were significantly changed in both LUTS and this mouse model (Fig. 4). For proline, it was increased in both LUTS patients and the hormone-treated mice, represents a strong candidate biomarker for hormone-induced BOO. Urinary proline was also increased in a mouse model of hepatic injury and fibrosis, indicating fibrosis is a potential relevant pathway in the present BOO model (34). Next, citrulline was decreased in LUTS patients but increased in this mouse model. Citrulline is poorly understood in the context of prostate diseases. Citrulline is a non-essential amino acid, a precursor to arginine, and displays antioxidant properties (35). Increased urine citrulline in these mice could be explained by inflammation-induced expression of nitric oxide synthase 2, the enzyme responsible for conversion of arginine to citrulline (36), possibly in response to oxidative stress generated by catechol estrogen metabolites (37).
Figure 4.
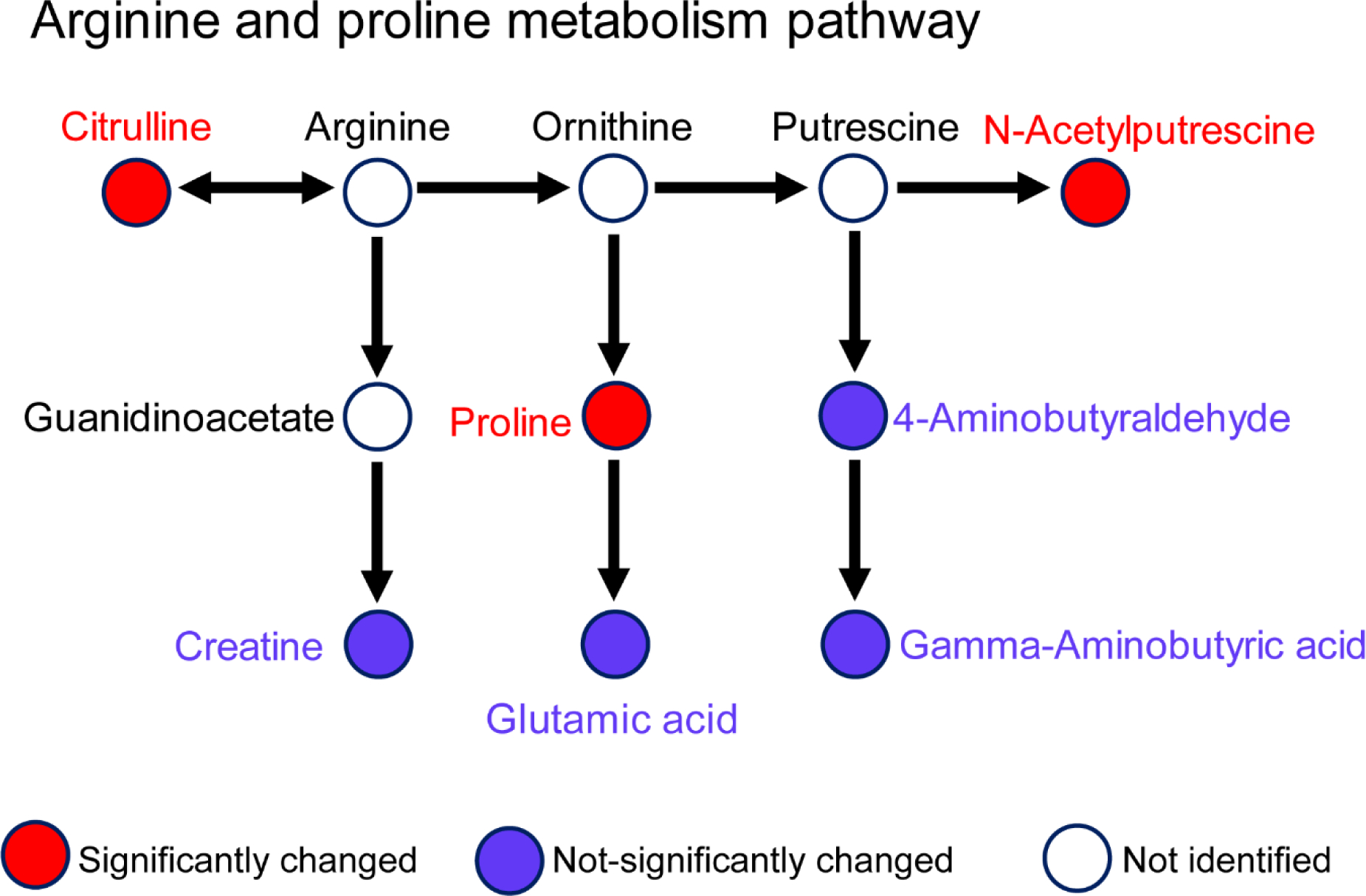
Arginine and proline metabolism pathway is potentially disrupted (MetaboAnalyte, KEGG; Fisher’s exact test, p < 0.07).
Other processes were significantly changed in the present study but not observed in LUTS patients. For example, the aminoacyl-tRNA biosynthesis pathway was disrupted in the urine metabolome of this mouse model but not in LUTS patients. This pathway is pivotal in determining how the genetic code is interpreted as amino acids (38). More specifically, leucine and proline were significantly changed in this metabolic pathway. The tryptophan metabolism pathway, represented by 2-aminobenzoic acid, was also disrupted in this mouse model. Overall, these pathway-level results were similar to a prior report of urine metabolomics in liver injury and fibrosis (39). Interestingly, leucine may play an important role in prostatic proliferation. Leucine is an essential branched-chain amino acid that signals through the mTOR pathway (40). This signaling is pro-proliferative in prostate cancer cells, and as such decreasing leucine transport into tumors is an attractive therapeutic target (41). It is possible the strong initial increases in leucine in this model contribute to the increased prostate mass (benign hyperplasia) or even the development of prostatic intraepithelial neoplasia observed later in this model (14).
CONCLUSIONS
Twelve-plex DiLeu isobaric labeling is an attractive high-throughput strategy for identification and quantification of amine-containing metabolites, and Metandem is a useful tool for large-scale stable isotope labeling-based metabolomics data analysis. Paired together, these tools offer a powerful and accessible method for relative quantification of amine-containing metabolites in disease biomarker research. In this study of urinary amine metabolomics of a hormone-induced LUTS mouse model, we have identified and quantified 59 amine metabolites, and 22 of them were identified in urine for the first time. LUTS patients and this mouse model shared common pathways that are dysregulated compared to control groups, for instance, the arginine and proline metabolism pathway. Proline presents an especially attractive candidate biomarker for hormone-induced BOO, as it was significantly increased in both human LUTS and this mouse model. Future experiments will test the hypothesis that this hormone treatment results in fibrosis of the lower urinary tract, ultimately leading to the pronounced BOO phenotype.
Supplementary Material
Figure S1. Additional statistical graphs from Metandem
Table S1. List of 59 identified metabolites of interest
ACKNOWLEDGEMENTS
The authors would like to thank DiLeu synthesis team in the Li Lab (Dr. Tyler Greer, Dr. Dustin Frost, and Dr. Amanda Rae Buchberger). The authors would also like to thank Seth Hyman for his assistance with urine sample collection and Zeeh Pharmaceutical Experiment Station in School of Pharmacy for instrument access. This work was supported in part by National Institutes of Health through Grants 1P20 DK097826, U54 DK104310, R01DK071801, RF1AG052324, P41GM108538, R01 DK093690, R01 DK099328, DK091193, and DK104310. The Q-Exactive Orbitrap instrument was purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531). L.L. acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
REFERENCES
- 1.Chapple CR, Roehrborn CG. 2006. A Shifted Paradigm for the Further Understanding, Evaluation, and Treatment of Lower Urinary Tract Symptoms in Men: Focus on the Bladder. Eur Urol 49:651–659. [DOI] [PubMed] [Google Scholar]
- 2.Abrams P, Cardozo L, Fall M, Griffiths D, Rosier P, Ulmsten U, Van Kerrebroeck P, Victor A, Wein A. 2003. The standardisation of terminology in lower urinary tract function: report from the standardisation sub-committee of the International Continence Society. Urology 61:37–49. [DOI] [PubMed] [Google Scholar]
- 3.Speakman M, Kirby R, Doyle S, Ioannou C. 2015. Burden of male lower urinary tract symptoms (LUTS) suggestive of benign prostatic hyperplasia (BPH) - focus on the UK. BJU Int 115:508–519. [DOI] [PubMed] [Google Scholar]
- 4.Harley SJD, Wittert G, Brook NR, Secombe P, Campbell J, Lockwood C. 2017. Identifying predictors of change in the severity of untreated lower urinary tract symptoms in men: a systematic review protocol. JBI database Syst Rev Implement reports 15:1585–1592. [DOI] [PubMed] [Google Scholar]
- 5.Edgar AD, Levin R, Constantinou CE, Denis L. 2007. A critical review of the pharmacology of the plant extract of Pygeum africanum in the treatment of LUTS. Neurourol Urodyn 26:458–463. [DOI] [PubMed] [Google Scholar]
- 6.Swerdloff RS, Wang C. 2004. Androgens and the ageing male. Best Pract Res Clin Endocrinol Metab 18:349–362. [DOI] [PubMed] [Google Scholar]
- 7.Nickel JC, Roehrborn CG, Castro-Santamaria R, Freedland SJ, Moreira DM. 2016. Chronic Prostate Inflammation is Associated with Severity and Progression of Benign Prostatic Hyperplasia, Lower Urinary Tract Symptoms and Risk of Acute Urinary Retention. J Urol 196:1493–1498. [DOI] [PubMed] [Google Scholar]
- 8.Noguchi N, Chan L, Cumming RG, Blyth FM, Handelsman DJ, Seibel MJ, Waite LM, Le Couteur DG, Naganathan V. 2016. Lower Urinary Tract Symptoms and Incident Falls in Community Dwelling Older Men: The Concord Health and Ageing in Men Project. J Urol 196:1694–1699. [DOI] [PubMed] [Google Scholar]
- 9.Kim EH, Brockman JA, Andriole GL. 2018. The use of 5-alpha reductase inhibitors in the treatment of benign prostatic hyperplasia. Asian J Urol 5:28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lepor H. 2007. Alpha blockers for the treatment of benign prostatic hyperplasia. Rev Urol 9:181–90. [PMC free article] [PubMed] [Google Scholar]
- 11.Ganzer CA, Jacobs AR, Iqbal F. 2015. Persistent Sexual, Emotional, and Cognitive Impairment Post-Finasteride. Am J Mens Health 9:222–228. [DOI] [PubMed] [Google Scholar]
- 12.Cornu J-N, Ahyai S, Bachmann A, de la Rosette J, Gilling P, Gratzke C, McVary K, Novara G, Woo H, Madersbacher S. 2015. A Systematic Review and Meta-analysis of Functional Outcomes and Complications Following Transurethral Procedures for Lower Urinary Tract Symptoms Resulting from Benign Prostatic Obstruction: An Update. Eur Urol 67:1066–1096. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson TM, Ricke EA, Marker PC, Miano JM, Mayer RD, Timms BG, vom Saal FS, Wood RW, Ricke WA. 2012. Testosterone and 17β-Estradiol Induce Glandular Prostatic Growth, Bladder Outlet Obstruction, and Voiding Dysfunction in Male Mice. Endocrinology 153:5556–5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ricke WA, McPherson SJ, Bianco JJ, Cunha GR, Wang Y, Risbridger GP. 2008. Prostatic hormonal carcinogenesis is mediated by in situ estrogen production and estrogen receptor alpha signaling. FASEB J 22:1512–1520. [DOI] [PubMed] [Google Scholar]
- 15.Rauniyar N, Yates JR. 2014. Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res. American Chemical Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang F, Ye H, Chen R, Fu Q, Li N. 2010. N,N-Dimethyl leucines as novel Isobaric tandem mass tags for quantitative proteomics and peptidomics. Anal Chem 82:2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gritsenko MA, Xu Z, Liu T, Smith RD. 2016. Large-scale and deep quantitative proteome profi ling using isobaric labeling coupled with two-dimensional lc- ms/ms. Methods Mol Biol 1410:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong S-E, Mann M, Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R, Li J, Steen H, Hansen KC, Schmitt-Ulms G, Chalkley RJ, Hirsch J, Baldwin MA, Burlingame AL. 2005. N,N-Dimethyl Leucines as Novel Isobaric Tandem Mass Tags for Quantitative Proteomics and Peptidomics. Mol Cell Proteomics 1:2817–2825. [Google Scholar]
- 19.Hao L, Zhong X, Greer T, Ye H, Li L. 2015. Relative quantification of amine-containing metabolites using isobaric N,N-dimethyl leucine (DiLeu) reagents via LC-ESI-MS/MS and CE-ESI-MS/MS. Analyst 140:467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frost DC, Li L. 2016. High-throughput quantitative proteomics enabled by mass defect-based 12-plex dileu isobaric tags. Methods Mol Biol 1410:169–194. [DOI] [PubMed] [Google Scholar]
- 21.Frost DC, Rust CJ, Robinson RAS, Li L. 2018. Increased N,N-Dimethyl Leucine Isobaric Tag Multiplexing by a Combined Precursor Isotopic Labeling and Isobaric Tagging Approach. Anal Chem 90:10664–10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao L, Johnson J, Lietz CB, Buchberger A, Frost D, Kao WJ, Li L. 2017. Mass defect-based n,n-dimethyl leucine labels for quantitative proteomics and amine metabolomics of pancreatic cancer cells. Anal Chem 89:1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moulder R, Bhosale SD, Goodlett DR, Lahesmaa R. 2018. Analysis of the plasma proteome using iTRAQ and TMT-based Isobaric labeling. Mass Spectrom Rev 37:583–606. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Elias JE. 2017. Relative protein quantification using tandem mass tag mass spectrometry, p. 185–198. In Methods in Molecular Biology. Humana Press Inc. [DOI] [PubMed] [Google Scholar]
- 25.Hao L, Zhu Y, Wei P, Johnson J, Buchberger A, Frost D, Kao WJ, Li L. 2019. Metandem: An online software tool for mass spectrometry-based isobaric labeling metabolomics. Anal Chim Acta 1088:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Knight J, Fargue S, Buchalski B, Guan Z, Inscho EW, Liebow A, Fitzgerald K, Querbes W, Todd Lowther W, Holmes RP. 2016. Metabolism of13C5-hydroxyproline in mouse models of Primary Hyperoxaluria and its inhibition by RNAi therapeutics targeting liver glycolate oxidase and hydroxyproline dehydrogenase. Biochim Biophys Acta - Mol Basis Dis 1862:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao L, Greer T, Page D, Shi Y, Vezina CM, Macoska JA, Marker PC, Bjorling DE, Bushman W, Ricke WA, Li L. 2016. In-Depth Characterization and Validation of Human Urine Metabolomes Reveal Novel Metabolic Signatures of Lower Urinary Tract Symptoms. Sci Rep 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warrack BM, Hnatyshyn S, Ott KH, Reily MD, Sanders M, Zhang H, Drexler DM. 2009. Normalization strategies for metabonomic analysis of urine samples. J Chromatogr B Anal Technol Biomed Life Sci 877:547–552. [DOI] [PubMed] [Google Scholar]
- 29.Frost DC, Greer T, Li L. 2015. High-Resolution Enabled 12-Plex DiLeu Isobaric Tags for Quantitative Proteomics. Anal Chem 87:1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wenger Craig D., Phanstiel Douglas H., Violet Lee M, Derek J Bailey JJC. 2011. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 92:1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J. 2018. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46:W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. 2013. The Human Urine Metabolome. PLoS One 8:e73076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schilling CH, Schuster S, Palsson BO, Heinrich R. 1999. Metabolic Pathway Analysis: Basic Concepts and Scientific Applications in the Post-genomic Era. Biotechnol Prog 15:296–303. [DOI] [PubMed] [Google Scholar]
- 34.Gou X, Tao Q, Feng Q, Peng J, Zhao Y, Dai J, Wang W, Zhang Y, Hu Y, Liu P. 2013. Urine metabolic profile changes of CCl4-liver fibrosis in rats and intervention effects of Yi Guan Jian Decoction using metabonomic approach. BMC Complement Altern Med 13:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allerton T, Proctor D, Stephens J, Dugas T, Spielmann G, Irving B, Allerton TD, Proctor DN, Stephens JM, Dugas TR, Spielmann G, Irving BA. 2018. l-Citrulline Supplementation: Impact on Cardiometabolic Health. Nutrients 10:921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wijnands K, Castermans T, Hommen M, Meesters D, Poeze M, Wijnands KAP, Castermans TMR, Hommen MPJ, Meesters DM, Poeze M. 2015. Arginine and Citrulline and the Immune Response in Sepsis. Nutrients 7:1426–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhat HK, Calaf G, Hei TK, Loya T, Vadgama JV. 2003. Critical role of oxidative stress in estrogen-induced carcinogenesis. Proc Natl Acad Sci U S A 100:3913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibba M, Söll D. 2000. Aminoacyl-tRNA Synthesis. Annu Rev Biochem 69:617–650. [DOI] [PubMed] [Google Scholar]
- 39.Chang H, Meng H, Liu S, Wang Y, Yang X, Lu F, Wang H. 2017. Identification of key metabolic changes during liver fibrosis progression in rats using a urine and serum metabolomics approach. Sci Rep 7:11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S, Zeng X, Ren M, Mao X, Qiao S. 2017. Novel metabolic and physiological functions of branched chain amino acids: a review. J Anim Sci Biotechnol 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otsuki H, Kimura T, Yamaga T, Kosaka T, Suehiro J, Sakurai H. 2017. Prostate Cancer Cells in Different Androgen Receptor Status Employ Different Leucine Transporters. Prostate 77:222–233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Additional statistical graphs from Metandem
Table S1. List of 59 identified metabolites of interest


