Abstract
The healing capacity of bones after fracture implies the existence of adult regenerative cells. However, information on identification and functional role of fracture-induced progenitors is still lacking. Paired-related homeobox 1 (Prx1) is expressed during skeletogenesis. We hypothesize that fracture recapitulates Prx1’s expression, and Prx1 expressing cells are critical to induce repair. To address our hypothesis, we used a combination of in vivo and in vitro approaches, short and long-term cell tracking analyses of progenies and actively expressing cells, cell ablation studies, and rodent animal models for normal and defective fracture healing. We found that fracture elicits a periosteal and endosteal response of perivascular Prx1+ cells that participate in fracture healing and showed that Prx1-expressing cells have a functional role in the repair process. While Prx1-derived cells contribute to the callus, Prx1’s expression decreases concurrently with differentiation into cartilaginous and bone cells, similarly to when Prx1+ cells are cultured in differentiating conditions. We determined that bone morphogenic protein 2 (BMP2), through C-X-C motif-ligand-12 (CXCL12) signaling, modulates the downregulation of Prx1. We demonstrated that fracture elicits an early increase in BMP2 expression, followed by a decrease in CXCL12 that in turn down-regulates Prx1, allowing cells to commit to osteochondrogenesis. In vivo and in vitro treatment with CXCR4 antagonist AMD3100 restored Prx1 expression by modulating the BMP2-CXCL12 axis. Our studies represent a shift in the current research that has primarily focused on the identification of markers for postnatal skeletal progenitors, and instead we characterized the function of a specific population (Prx1+ cells) and their expression marker (Prx1) as a crossroad in fracture repair. The identification of fracture-induced perivascular Prx1+ cells and regulation of Prx1’s expression by BMP2 and in turn by CXCL12 in the orchestration of fracture repair, highlights a pathway in which to investigate defective mechanisms and therapeutic targets for fracture non-union.
Keywords: Prx1, BMP2, CXCL12, fracture healing, skeletal cells, progenitor
1. Introduction
The combined lifetime incidence of hip, forearm and vertebral fractures is around 40%, which is equivalent to the incidence of cardiovascular diseases. Fractures account for the largest total lifetime cost associated with any one injury type, with over $99 billion in estimated annual medical costs and productivity loss in the United States.(1) Fracture healing is a complex, multistep process that involves multiple cell lineages and is still not fully understood. Although new advances in orthopedic surgery have substantially enhanced fracture healing outcomes, there is a subset of fractures that continue to be deficient in bone repair and culminate in failure of healing (non-unions). Revealing the key mechanisms that orchestrate fracture repair may identify therapeutic targets to promote healing in patients that suffer from non-unions.
The high regenerative capacity of bones after fracture implies the existence of adult progenitor cells capable of contributing to the reparative process.(2–4) However, the nature of these progenitor cells remains elusive. It is evident that the periosteum is critical to fracture repair as indicated by the fact that its removal impedes repair.(5–10) However, the cell population(s) that govern the regenerative ability of the periosteum are largely unknown. Although some markers for periosteal cells have been identified, most of the studies have been based on tracing cells (or their progeny), tagged with an imaging reporter system, during fracture repair.(11,14) A functional role of these specific markers and the contribution of the progenitor cells expressing those markers to the reparative process have not been fully elucidated. Furthermore there is evidence, including from our laboratory, that the endosteum contributes to the repair, either by providing cell population(s) derived from the bone marrow (BM) that are critical in the early stage of fracture repair, or by contributing to the formation of the callus.(8, 10, 15, 16) Characterizing the roles of specific cell population(s) in the fracture repair process can shed light on the defective mechanisms underlying non-unions and lead to the development of cell-based therapies to treat them.
Prrx1 (paired-related homeobox protein 1) is a homeobox gene expressed in several developing tissues, including skeletal elements.(16–19) In humans, mutation of Prrx1 can lead to agnathia-otocephaly complex, a rare condition characterized by several skeletal abnormalities including underdeveloped mandible, club foot, rib, and sacral bone dysplasia.(20) Progeny of cells embryologically tagged with Prx1-Cre mediated imaging reporters, namely Prx1-derived cells, localize in the periosteum and within the callus of fractured bones.(21) Adult cells tagged with Prx1-cre imaging reporters can be identified within the callus and are believed to be derived from the periosteum.(6, 10, 12, 14, 22, 23) Wilk et al. reported that postnatal calvarial skeletal stem cells expressing Prx1 reside in the sutures and contribute to calvaria regeneration.(24) The function of cells actively expressing Prx1 and the role of Prx1 in fracture repair is largely unknown. In vitro studies have shown that Prx1 inhibits Osterix (Osx) and RUNX2 expressions as well as the osteogenic differentiation of MC3T3 cells and mesenchymal stromal cells(25). Prx1 is also reported to maintain the stemness of adult neural cells and to inhibit adipogenesis by activating TGF-β signaling.(26, 27)
If the characterization of fracture-induced progenitors is critical, equally important is the identification of humoral factors that control fracture regeneration. There is compelling evidence that BMP2 has a critical role in fracture repair, including failed fracture repair in mice with BMP2-deficient osteo-chondroprogenitor cells.(28–31) Furthermore, BMP2 levels are reduced in fracture biopsies from patients with non-unions, and exogenous BMPs have some beneficial effects in treating non-unions.(32–34) We previously reported that in BMP2-haploinsufficient mice failure of proper fracture healing is associated with a disarranged increase of chemokine C-X-C motif-ligand-12 (CXCL12) expressed by pericytic cells.(13) CXCL12 is a critical cytokine for cell mobilization and to support hematopoiesis.(35) It is also expressed by osteoprogenitors and perivascular cells.(36–41) CXCL12+ cells are present in bone and cartilage.(42–44) CXCL12 binds to CXCR4 and CXCR7, although this latter receptor lacks an intracellular signaling.(35) AMD3100, an antagonist of CXCR4 and CXCR7, that has an excellent safety profile and has undergone pharmacokinetic characterization in rodents and humans,(45–48) is FDA-approved to induce hematopoietic stem cell mobilization.(49)
In this study, we analyze the regulation of Prx1’s expression in postnatal life and its correlation to fracture healing. We have found that Prx1’s expression is fracture-induced, decreases with callus formation and while Prx1-derived cells contribute to the callus they lose Prx1 expression during differentiation into cartilaginous and bone cells, and when cultured in differentiating conditions. Using in vivo and in vitro approaches, we discovered that Prx1 is downstream of CXCL12, and that BMP2 through CXCL12 signaling, modulates the expression of Prx1. We report that fracture elicits an early increase of BMP2 that leads to a decrease of CXCL12 that in turn down-regulates Prx1, allowing cells to commit to osteochondrogenesis. Further, our findings on AMD3100 being able to regulate Prx1’s expression by modulating the BMP2-CXCL12 axis represent a significant breakthrough for exploring its potential clinical use to treat non-unions, for which we lack a pharmacological treatment.
2. Materials and Methods
2.1. Antibodies and reagents
Primary antibodies and fluorochrome-conjugated secondary antibodies are summarized in Supplemental Table 1. Safranin O (S2255) was purchased from Fluka Chemical (Milwaukee, WI). Hematoxylin (H3136), Orange G (O7252), glacial acetic acid (A6283), aluminum ammonium sulfate (A2140), IdU (I7125), collagenase (C1889), trypsin (T6567) and 4OH-TAM (4-hydroxy-tamoxifen) (H6278) were obtained from Sigma Aldrich (St Louis, MO). Fast green (F7252) and sodium iodate (S4007) are from Fisher Scientific (Waltham, MA). The inhibitor AMD3100 (Plerixafor) was from AdooQ Bioscience (A13074, Irvine, California, USA).
2.2. Animal models
Prx1-CreER-GFP:ROSA26-lox-STOP-lox-ßGal double mutant mice were generated by crossing male Prx1-CreER-GFP mice(6) (provided by Dr. Shunichi Murakami, Case Western Reserve University, Cleveland, Ohio, USA) with female ROSA26-lox-STOP-lox-ßGal reporter mice (Jackson Laboratory, Bar Harbor, Maine, USA). In early postnatal development, Prx1-CreER-GFP:ROSA26-lox-STOP-lox-ßGal male mice received a single intraperitoneal (i.p.) injection of 4OH-tamoxifen (4OH-TAM) (0.25 mg/g body weight) at age 3 days (P3). Mice were sacrificed at 6, 9, 12, 15, 18, and 21 days of age for histologic analysis; CreER-negative littermates that received 4OH-TAM were used as controls. In adult life studies, male Prx1-CreER-GFP:ROSA26-lox-STOP-lox-ßGal double mutant mice, as well as their male littermate controls, received i.p. injections of 4OH-TAM (0.5 mg/g body weight) at the age of 16–18 weeks for 5 days. Mice were sacrificed either 6, 28 or 168 days later. Prx1-CreER-GFP:R26-DTA double mutant mice were generated by crossing male Prx1-CreER-GFP mice with female R26-DTA mice (Jackson Laboratory, Bar Harbor, Maine, USA; Stock #009669)(50), in which a loxP-conditional DTA allele was introduced into the ubiquitously expressed ROSA26 locus, allowing the specific ablation of Prx1-expressing cells. To generate the Prx1CreER;R26-DTA-tdTom mice, R26-tdTomato mice (Jackson Labs, Stock #007905)(51) were crossed with Prx1-CreER-GFP:R26-DTA mice and Prx1-CreER-GFP;R26-tdTomato littermates (control) received 4OH-TAM for 5 days starting 2 days before fracture. In Prx1CreER;R26-DTA-tdTom mice, we genetically ablated Prx1-expressing cells through toxicity of DTA whose expression is activated via Prx1CreER recombination after 4OH-TAM. In studies where animals were injected with 4OH-TAM only male mice were used, because the concern of effects of 4OH-TAM on females that would potentially interfere with data interpretation. BMP2flox/flox mice on a mixed C57B/L6 background were kindly provided by Dr. James Martin (Texas A&M Health Science Center, Houston, TX).(52) To generate BMP2cKO/+mice, BMP2flox/flox mice were crossed with Prx1 limb enhancer driven-Cre transgenic mice (C. Tabin, Harvard Medical School, Boston, MA),(52) which drives Cre-expression in osteochondroprogenitors. BMP2cKO/+ mice were crossed with BMP2flox/flox to produce BMP2cKO/cKO. Cre-negative sibling mice, regardless of flox-status, were almost always used as controls, other controls are otherwise specified. We confirmed the genotypes of the transgenic mice by PCR analyses of genomic DNA isolated from mouse ears. Genotyping was done by PCR analysis using the primers reported in Supplemental Table 2. All mice had been backcrossed on the C57BL/6 strain for at least ten generations.
2.3. Tibia fracture model
Semi-stabilized tibia fractures, in which intramedullary fixation enables relative stability to facilitate endochondral ossification, were produced in C57B/L6 background male mice between 8 and 16 weeks of age as described.(53) The intramedullary pin was carefully removed after dissection. In male Prx1-CreER-GFP:ROSA26-lox-STOP-lox-ßGal mice, 4OH-TAM administrations were performed for 5 days (from 2 days before to 2 days after fracture) (Study 1) or for 5 days starting 33 days before fracture (Study 2). Mice were euthanized 7 days after fracture. In male Prx1-CreER-GFP:R26-DTA and Prx1CreER;R26-DTA-tdTomato mice, 4OH-TAM i.p. injections were performed (0.5 mg/daily), as in Study 1 regimen, and mice were sacrificed at 14 and 21 days later. For treatment with AMD3100, mice were i.p.-injected with 2.5 mg/g of bodyweight of AMD3100-PBS solution twice per day on the 2 days preceding fracture and then twice daily from day 2 to day 7 post-fracture, followed by harvest at 14 days post-fracture. All procedures used were consistent with the guidelines of the National Institutes of Health and approved by the Institutional Animal Care and Use Committees of Rush University Medical Center (Chicago, IL, USA).
2.4. Micro—computed tomography analysis of fracture calluses
Fractured tibias were dissected at 14 and 21 days after fracture, fixed in 4% paraformaldehyde in PBS for 18 hours at 4°C, rinsed, and scanned by micro-computed tomography (μCT) (Scanco Medical μCT 50). Native scans were performed in 70% ethanol at 55 kVp, 145 μA, 300 ms integration time, and at 6-μm isotropic voxel edge along a length of the tibia centered on the fracture line. Volumetric analysis of tissue composition was measured only in the callus by narrowing the analysis from the first proximal to the last distal sign (indicated by periosteal enlargement) of the callus formation when examining the coronal plane of the μCT images as reported.(53, 54) Further narrowing to within the fracture line was done by including only areas with signs of breaking within the cortical bone in the transversal plane, as reported.(53, 54) In this region, we evaluated with a direct-model morphometric measure of bone volume (BV) over total volume (TV), and HA/BV segmentation(55) was performed based on a calibration curve derived from manufacturer-supplied phantoms containing known hydroxyapatite (HA) composition: voxels with a linear attenuation coefficient ≥ 1.76 cm−1 (corresponding to 330 mg HA/cm3) were considered mineralized tissue. A Gaussian segmentation filter with kernel of 2 and standard deviation of 1.2 voxels was uniformly applied to the volume of interest. Within the mineralizing callus, different ranges of thresholds were identified based on a parametric thresholding study combining serial μCT scanning and histological analysis, described previously (Low mineralized tissue = 100–325; High mineralized tissue = 326–425).(53, 54) Results presented are volumes of each threshold range divided by the number of slices encompassing the callus to normalize for varying callus size. Soft tissues were evaluated by using a post-mortem phosphotungstic acid (PTA) contrast-enhanced scanning that allows a better visualization of soft tissue through PTA-collagen binding at basic amino side chains.(56, 57) Samples were stained with 5% PTA in 70% ethanol for 10 days before scanning. The volume of soft tissue in the callus was obtained by subtracting the volume of total mineralized tissue from the total tissue volume measured with contrast-enhanced scanning. Three-dimensional reconstructions were done using 3D software of Scanco Medical μCT 50.
2.5. Biomechanical testing
The mechanical properties of the fractures were measured with a standardized four-bending technique. Fractured tibias were dissected 21 days after fracture and loaded into the ElectroForce 3200 (TA Instruments, New Castle, DE) for four-point bending test. The loading rate was set at a constant speed of 5 mm/minute until failure. Force- bending and stress-strain curves were recorded to identify the ultimate bending load (maximum force at failure) and bending stiffness (slope of the linear part of the curve).(58)
2.6. Staining, immunohistochemistry and immunofluorescence studies
Tibias from at least four individual animals per time point per study were dissected and subjected to staining, immunohistochemistry (IHC) or immunofluorescence (IF) analysis. Tibias were fixed in cold 0.2 % glutaraldehyde for 2 hours (young) or 18 hours (adult and fractured), then decalcified in 14% EDTA solution for 3 days (young) or 7–10 days (adult and fractured) before soaking in 30% sucrose for approximately 24–48 hours. Tissue was embedded in OCT and sectioned on a cryostat at 7–8 μm. Cryo-sectioned tibias were subjected to LacZ histochemical staining, as previously described.(53) Briefly, cryo-sections were fixed in 4% paraformaldehyde for 90 minutes at 4°C, then washed in PBS and finally incubated in staining solution overnight at 37°C; then, the sections were subjected to Safranin-O/Orange G or Safranin-O/Fast Green staining. In fractured tibias, the center of the fracture gap was identified as the largest diameter of the callus in which the fracture line was clearly seen following a serial Safranin O/Orange G staining. All further histological analyses were performed within 500 μm of the center of the fracture line.
For IF, to mitigate autofluorescence, sections were pretreated with 1% NaBH4 for 20 minutes, blocked for endogenous mouse IgG using M.O.M Kit (Mouse on Mouse Kit, Jackson ImmunoResearch) combined with 5% normal donkey serum (NDS) or goat normal serum (0.1 M PBS with 0.01% Triton X) for 1 hour and incubated in primary antibodies in 2% NDS buffer overnight at room temperature; sections were incubated in the appropriate secondary antibody diluted in 2% NDS buffer for 1 hour. For IHC analysis, Vectastain ABC kit (Vector Laboratories) was used according to the manufacturer’s instructions. Briefly, sections were incubated in 3 % H2O2 diluted with 0.01M PBS for 10 min, blocked for 1 hour with appropriate serum and incubated in primary antibody (diluted in blocking solution) overnight at room temperature. Sections were incubated in the appropriate biotinylated secondary antibody diluted in blocking buffer for 1 hour. Sections were then incubated in Avidin-Biotin complex and then visualized with diaminobenzidine. As a control, sections were processed with the omission of primary antibodies which completely abolished specific staining. The list of used primary and secondary antibodies are reported in Supplemental Table 1. Sections for IF analysis were counterstained with DAPI for nuclear staining and mounted with Aquamount (Thermofisher). Images were taken with either an Olympus BX60 Microscope with a DP71 camera or an Olympus FV1000 MPE SIM Laser Scanning Confocal Microscope with a 60x PlanApo oil immersion lens zoomed 2x using sequential scanning. Images were viewed with Olympus FV10-ASW Viewer software and final images merged with Image J or Photoshop.
2.7. Quantification of positive cells
For histological analysis, at least six sections per animal and four animals per genotype, were analyzed and examined. For quantification of genetically-labeled periosteal and endosteal cells in early and late postnatal development studies, tibias of Prx1-CreER-GFP:ROSA26-lox-STOP-lox-βGal mice, as well as their littermate controls, were sectioned by vibrating microtome (7 μm), LacZ/Safranin O stained at every sixth section and imaged using a predetermined area (100 x 150 μm) from the growth plate to the fibula-tibia insertion, hereby called ROI (Region of Interest). LacZ+ cells were counted in each predetermined area and in each section. Supplemental Table 3 shows the relative number of LacZ+ cells of the total number of cells [(% LacZ+ cells/Total number of cells) ± SD]. For quantification of immunohistochemically-labeled 5-iodo-2’-deoxyuridine (IdU) and Ki67, tibias of Prx1-CreER-GFP-ROSA-LacZ mice were sectioned, stained, and imaged as described above. Both IdU+ cells and Ki67+ cells were counted in each predetermined area and in each section and reported as the relative number of positive cells of the total number of βGal+ cells [(% IdU+ cells/βGal+ cells) ± SD; % Ki67+ cells/βGal+ cells) ± SD]. For immunofluorescence analysis, at least four sections per animal and three-five animals per genotype, were analyzed and examined. For quantification of genetically-labeled βGal cells in periosteum of Prx1CreER-GFP-ROSA-LacZ mice, tibias were cryo-sectioned (8 μm), stained at every sixth section, and imaged using a predetermined area (50 x 50 μm). βGal+ cells were counted in each predetermined area and in each section and reported as the relative number of Gal+ cells of the total number of Prx1+ cells [(% βGal+ cells/Prx1+ cells) ± SD] or GFP+ cells [(% βGal+ cells/GFP+ cells) ± SD]. For quantification of genetically-labeled tdTomato cells in fractured Prx1CreER;R26-DTA-tdTomato mice, as well as R26-DTA-tdTomato littermate controls, calluses were cryo-sectioned (8 μm), stained at every sixth section, and imaged using a predetermined area (50 x 50 μm). tdTomato+ cells were counted in each predetermined area and in each section and reported as the relative number of tdTomato+ cells of the total number of cells [(% tdTomato+ cells/DAPI+ cells) ± SD]. For quantification of other labeling (i.e. Sox9, Osx, Ocn, DMP1, PECAM), calluses were cryo-sectioned, stained and imaged as described above. Positive cells were counted in each predetermined area and in each section and reported as the relative number of positive cells of the total number of cells [(% positive cells/DAPI+ cells) ± SD].
2.8. IdU incorporation study
In order to label slow-proliferating cells, 3-month Prx1-CreER-GFP-ROSA-LacZ old mice received IdU during the postnatal life through daily supply of drinking water (1 mg/mL) for one month; after a washout period of 58 days, mice received daily i.p. injections of 4OH-TAM (0.5 mg/each) for 5 days. The mice were sacrificed 3 days after last injection and tibias were subjected to IF analysis.
2.9. Periosteal and endosteal cell isolation
Periosteal and endosteal cells were freshly isolated from long bones of 5 month-old Prx1-CreER-GFP:ROSA26-lox-STOP-lox-βGal mice. Tissues attached to the periosteal surface of tibias and femurs were scraped off and pooled in a Petri dish. Following digestion with Collagenase D (1 mg/ml) for 45min, all periosteal cells were pooled and suspended in aMEM medium containing 1% penicillin and streptomycin, 1% glutamine and 10% fetal bovine serum (Atlanta Biologicals, Norcross, GA, USA). Endosteal cells were isolated from the BM of tibias and femurs using the procedure of Balduino et al.(13, 59, 60) Briefly, tibias and femurs were dissected and then treated with five incubations for 30 min in 0.1% collagenase and 0.125% trypsin in Hank’s balanced salt solution. The BM was removed by flushing and bones were broken into segments and treated twice with 0.1% collagenase for 40 min at 37°C, for isolating endosteal cells. Cells were grown in Dulbecco’s modified Eagle’s medium with antibiotics and 10% fetal bovine serum (Maintenance media) (Atlanta Biologicals) to allow confluence (undifferentiated conditions). Cells were grown at 37°C and 5% CO2 for three passages before being using for experiments. Osteoblastic differentiation was carried out using StemXVivo osteogenic base media plus 0.5x StemXVivo osteogenic supplement (R&D Systems). Media was changed every three days for the length of the experiment. For in vitro AMD3100 treatment of confluent endosteal cells, osteogenic differentiation was initiated as described above; beginning on day 7, AMD3100 (400 μM) was added to the cells every three days until day 14, when cells were harvested.
2.10. FACS sorting, RNA isolation and qRT-PCR
Before cell sorting, periosteal and endosteal cell suspensions were filtered with a cell strainer and kept on ice. GFP-positive and -negative cells were sorted into 15 ml plastic tubes containing aMEM modification medium (10% FCS, 2X P/S, 1X Fungizone) by MoFlo Astrios cell sorter (Beckman Coulter Life sciences, Indianapolis, IN, USA) located in the Flow Cytometry Core at the University of Illinois at Chicago. When indicated, mRNA was harvested from cells using the mMACS mRNA Isolation kit (Miltenyi Biotec, San Diego, CA, USA). Contaminating DNA was removed with DNase I (New England BioLabs, Ipswich, MA, USA) and extracted mRNA was converted to cDNA using mMACS One-Step cDNA Kit (Miltenyi Biotec). To determine the expression of marker genes, we performed qRT-PCR using 2_ Sso Advanced SYBR Green Supermix and StepOnePlus™ Real-Time System and StepOne™ software (Applied Biosciences, Beverly Hills, CA, USA). Analysis of relative gene expression was made using the Pfaffl method with GAPDH and/or 18s as housekeeping gene. Primers are reported in Supplemental Table 2.
2.11. Statistical analysis
Data are expressed as mean ± SD. For cell culture, each experiment included 3–5 samples. Statistical analyses were performed in GraphPad Prism 8, using a two-tailed unpaired Student’s t-test for single comparisons or one-way ANOVA followed by Sidak’s test for multiple comparisons. Statistical significance was set at p ≤ 0.05.
3. Results
3.1. Temporal expression pattern of Prx1 and Prx1-expressing cells during fracture repair
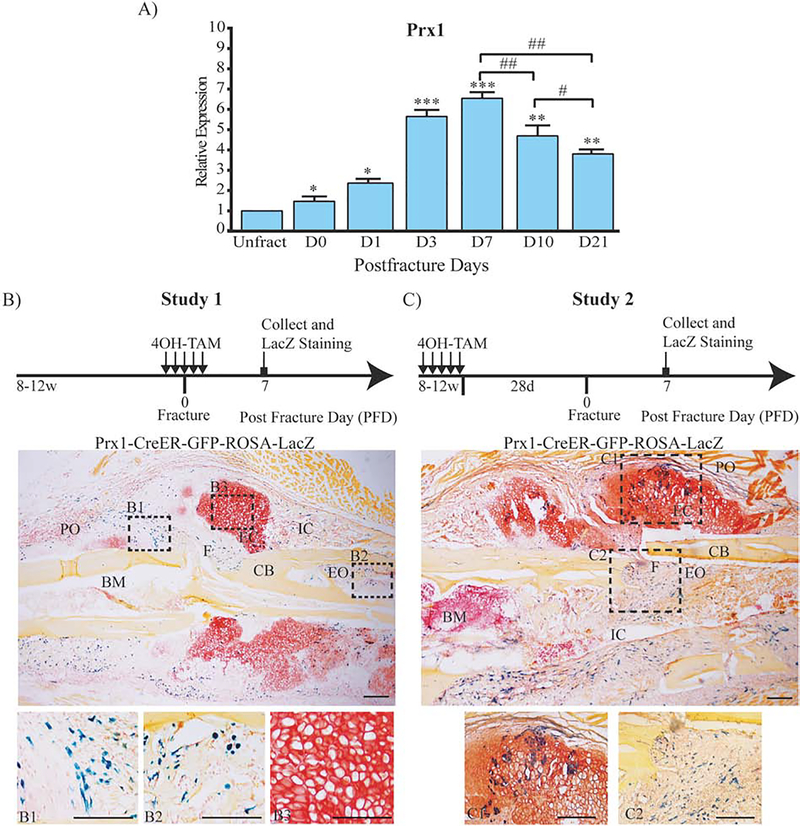
We explored the expression of Prx1 mRNA in fractured bones using a semi-stabilized tibia fracture model in which intramedullary fixation enables relative stability to facilitate endochondral ossification. As shown in Fig. 1A, Prx1 mRNA expression increased rapidly after fracture, peaked by day 7 and decreased by day 10 and day 21. We then explored the expression pattern in fractured tibias using Prx1-CreER-GFP-ROSA-LacZ mice in which 4OH-TAM induces Prx1-Cre-driven recombination that results in LacZ expression. As shown in Fig. 1, B–C, to evaluate Prx1-CreER-βGal cells, we have used two different studies based on the timeline of 4OH-TAM administration. In Study 1 that was aimed at labeling cells that express Prx1 during fracture repair 4OH-TAM (0.5 mg/day i.p.) was given for 5 days, from 2 days before to 2 days after fracture (Fig. 1B). In Study 2 that was aimed at tracing cells that expressed Prx1 before fracture and their progeny contribution to repair, 4OH-TAM was given for 5 days starting 33 days before fracture (Fig. 1C). For both studies, mice were euthanized 7 days after fracture and tibias were subjected to LacZ staining. We found that at this early stage of fracture repair (7 days after fracture), Prx1-CreER-βGal positive cells were detected within the periosteum and endosteum adjacent to the fracture line (Fig. 1, B1–B2), but no Prx1-CreER-βGal cells was detected in the cartilaginous callus (Fig. 1, B3). Differently from Study 1, in Study 2 at the same time point (7 days after fracture), Prx1-CreER-βGal positive cells that expressed Prx1 before fracture (or their progeny) were found within the endochondral callus resembling hypertrophic chondrocytes and within the intramembranous callus (Fig. 1, C1–C2). This difference seemed to indicate that Prx1-CreER-βGal cells differentiate into chondrocytes and bone cells, but they lose Prx1 expression during the differentiation.
Figure 1.
Temporal expression pattern of Prx1 in fracture healing.
(A) mRNA expression levels of Prx1 in fractured Prx1-CreER-GFP mice quantified by qRT-PCR. Data are reported as mean ± SD of triplicate assays, each with n=10 separate samples normalized to unfractured (Unfract). *, p < 0.05, **, p < 0.01, ***, p < 0.001, compared to Unfract by one-way ANOVA and Sidak’s multiple comparison test. #, p < 0.05, ##, p < 0.01, by unpaired two-tail t-test. (B) Representative histological LacZ/Safranin O/Orange G staining for cell lineage tracing Study 1 and Study 2 (C) in Prx1-CreER-GFP-ROSA-LacZ mice. Dotted squares indicate areas magnified in B1, B2, B3, C1 and C2. PO= periosteum; F= fracture line; CB= cortical bone; EO= endosteum; BM= bone marrow; EC= endochondral callus; IC= intramembranous callus. Scale bars = 100μm.
Of note, as shown in Supplemental Figure 1, A–B, we did not detect any βGal positive cells in the CreER-negative littermate controls, supporting the specificity of the LacZ staining for Prx1-CreER-βGal positive cells. To evaluate potential Cre-recombination leakage (in the absence of TAM) we analyzed Prx1-CreER-GFP;R26-LacZ and Prx1-CreER-GFP;R26-tdTomato fractured mice (n=5 for each transgene) that did not receive TAM. We did not detect either any βGal positive staining in Prx1-CreER-GFP;R26-LacZ or positive IF signal for tdTomato in Prx1-CreER-GFP;R26-tdTomato fractured tibias.
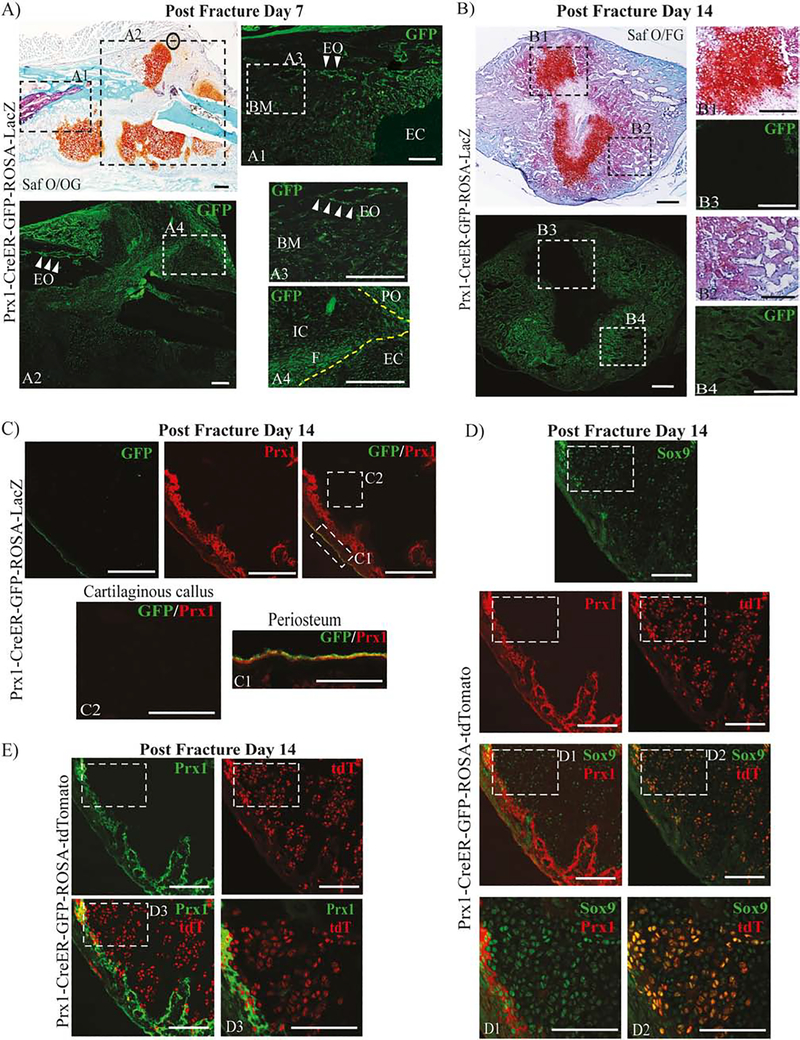
To test the hypothesis that Prx1-CreER-βGal positive cells can differentiate in bone and cartilage cells but lose Prx1’s protein expression with differentiation, we used Prx1CreER-GFP mice in which GFP is driven by the 2.4kb-Prx1 enhancer allowing detection of cells actively expressing Prx1 and performed IF analyses for GFP respectively at 7 and 14 days after fracture. We found that in callus at 7 days after fracture, cells actively expressing Prx1 (GFP+) localized within the fracture line (Fig. 2, A4), within the endosteum (Fig. 2, A3, white arrows) and periosteum (Fig. 2, A4) adjacent to the fracture line but they were almost undetectable within the cartilaginous callus (Fig. 2, A1). In the callus at 14 days after fracture, GFP+ cells were detected in the region surrounding (Fig. 2, B4) but not within the osteo-cartilaginous callus (Fig. 2, B3).
Figure 2.
Expression pattern of Prx1-expressing cells within the repairing fracture sites.
(A) Representative histological Safranin O/Orange G (Saf O/OG) staining of fractured Prx1-CreER-GFP mice at 7 days after fracture. Dotted squares indicate areas magnified of adjacent frozen section stained with GFP antibody (green). White arrows indicate GFP+ cells localized within the endosteum site. See text for more details. (B) Representative histological Saf O/FG staining of fractured Prx1-CreER-GFP mice at 14 days after fracture. See text for more details. Dotted squares indicate areas magnified of adjacent frozen section stained with GFP antibody (green). (C) Representative IF images of fractured Prx1-CreER-GFP mice at 14 days after fracture, immunostained with GFP (green) and Prx1 (red) antibodies. Dotted squares indicate areas magnified in periosteum, C1, and cartilaginous callus, C2. (D) Representative IF images of fractured Prx1-CreER-GFP-ROSA-tdTomato mice at 14 days after fracture, immunostained with Prx1 (red) and Sox9 (green) antibodies. The genetically-labeled tdT cells are shown in red. Dotted squares indicate area magnified in D1, D2 and D3. (E) Representative IF images of fractured Prx1-CreER-GFP-ROSA-tdTomato mice at 14 days after fracture, stained with Prx1 (green) antibody in cartilaginous callus. The genetically-labeled tdT cells are shown in red. PO= periosteum; EO= endosteum; CB= cortical bone; EC= endochondral callus; IC= intramembranous callus, BM=bone marrow. Scale bars = 100μm.
Then, we detected Prx1 expression by IF using Ab against Prx1. We found that at 14 days after fracture, GFP positive cells that were also positive for Prx1, were found within the periosteum (Fig. 2, C1) and endosteum, but we did not detect neither Prx1+ or GFP+ cells within the cartilaginous callus (Fig. 2, C2). In addition to the Prx1-CreER-GFP-ROSA-LacZ mice, to evaluate the pattern of cells tagged by Prx1-CreER induced recombination we used Prx1-CreER-GFP-ROSA-tdTomato mice in which 4OH-TAM induces Prx1-Cre-driven recombination that results in tdTomato (tdT) expression. Mice received 4OH-TAM during fracture (as in Study 1 regimen) but they were sacrificed 14 days later and analyzed for Prx1, tdT, Sox9, Osx, Ocn and DMP1 expressions by IF. As shown in Fig. 2D, we found that within the cartilaginous callus, cells that expressed Sox9 did not express Prx1 (Fig. 2D–D1), while most of the tdT positive cells were positive for Sox9 (63.87%±0.13; n=3) (Fig. 2D–D2). However, Prx1 expressing positive cells did not co-localize with tdT expressing cells (Fig. 2E–D3). Furthermore, within the mineralized callus tdT positive cells co-expressed Osx (72.88%±0.13; n=3), Ocn (69.69%±0.17; n=3) and DMP1 (78.98%±0.09; n=3) (Supplemental Fig. 2). Taken together data indicate that fracture elicits an early increase of Prx1 expression and Prx1-expressing cells (and their progenies) that become differentiated cells within the forming callus, with concurrent loss of Prx1 expression.
In order to determine the nature of Prx1 actively expressing cells, we isolated Prx1-GFP+ cells from endosteum and periosteum of tibias and femurs of Prx1CreER-GFP mice (5 weeks-old). Prx1-GFP+ cells were cultured in osteogenic medium for 21 days. We noted that Prx1 mRNA expression decreased with osteogenic differentiation (Day 21) in periosteal and endosteal cells, while mRNA expressions of osteogenic markers, Osx, Col1a1 and Ocn increased (Supplemental Fig. 3). The in vitro data support the in vivo observation that Prx1-expressing cells are capable of osteogenic differentiation, but differentiation is associated with a decrease in Prx1 expression.
3.2. Phenotypic characterization of Prx1-expressing cells
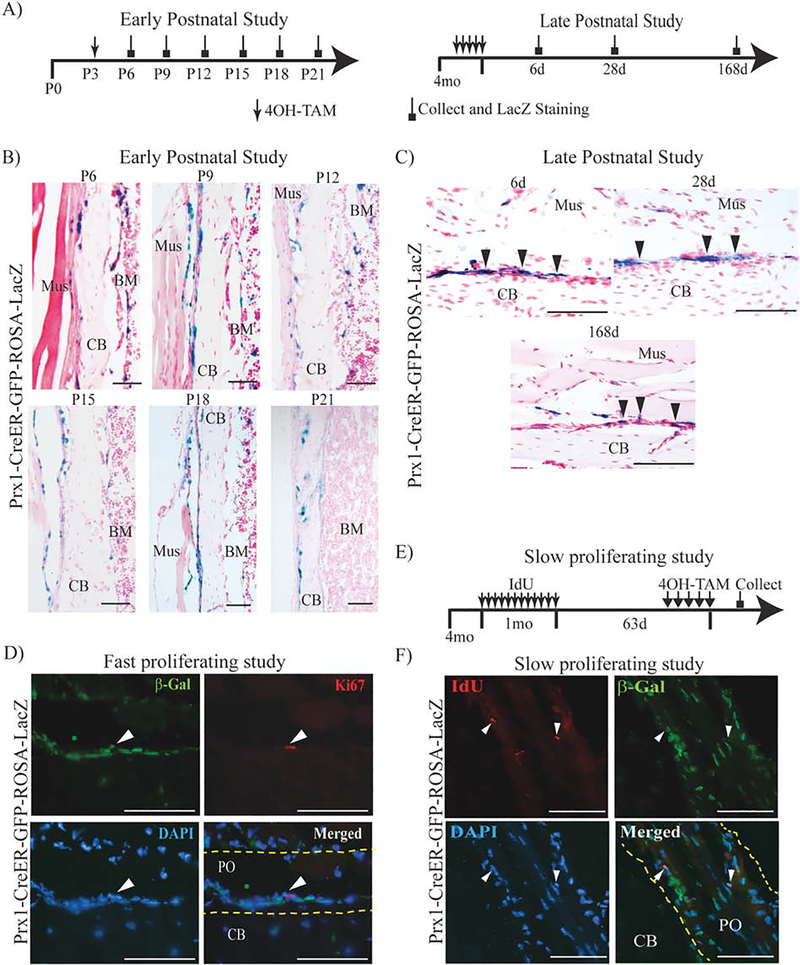
We then explored the expression pattern of Prx1-expressing cells in postnatal intact tibias by using Prx1-CreER-GFP-ROSA-LacZ mice. For the early postnatal study, 4OH-TAM (0.25mg/g body weight) was given at P3 and tibias collected at P6, P9, P12, P15, P18 and P21 were subjected to LacZ/Safranin O staining. As shown in Fig. 3B, we found that as early as P6, Prx1-CreER-βGal+ cells were localized in the endosteum and periosteum, where they persisted throughout the later time points. As showed in Supplemental Table 3, we found that at any given time, more than 10% of cells in the periosteum and endosteum were Prx1-CreER-βGal+ cells and they gradually decreased over time. In older Prx1-CreER-GFP-ROSA-LacZ mice (16–18 weeks), 4OH-TAM was given at 0.5 mg/day for 5 days and mice sacrificed 6, 28, 168 days later. Similarly, to younger mice but in a lower percentage, we found that Prx1-CreER-βGal+ cells were localized in the periosteum (Supplemental Table 3); no Prx1-CreER-βGal+ cells were localized in the endosteum. We also noted that in older mice the percentage of Prx1-CreER-βGal+ cells did not change over the time, and less than 1% of the Prx1-CreER-βGal+ cells were also positive for Ki67 (0.69%±0.11; n=3) (Fig. 3D). To determine whether Prx1-CreER-βGal+ cells were slow-proliferating, we performed a long-term (93 days, Fig. 3E) IdU incorporation study and found that the number of low proliferating IdU+ cells over the total (DAPI+) was low (IdU+/DAPI+ 2.5%±0.11; n=4), but more than 70% of IdU+ were also Prx1-CreER-βGal+ (IdU+/βGal+ 79.8%±0.14; n=4) (Fig. 3E–F). Taken together, our findings indicate that in young adult mice (at the age we perform fracture) Prx1-CreER-βGal+ cells are slow-proliferating cells.
Figure 3.
Lineage tracing studies of Prx1-expressing cells in intact tibias.
(A) Schematic representation of 4OH-TAM injections in postnatal development studies in Prx1-CreER-GFP-ROSA-LacZ mice. (B) Representative histological LacZ/Safranin O staining of early postnatal study. For all pictures, periosteal surface is facing the right side of the images. (C) Representative histological LacZ/Safranin O staining of late postnatal study. For all pictures, periosteal surface is facing the top of the images. (D) Representative IF images in periosteum of Prx1-CreER-GFP-ROSA-LacZ mice, immunostained respectively with β-Gal (green) and Ki67 (red) antibodies, counterstained with DAPI (blue). (E) Schematic representation of slow-proliferating study in Prx1-CreER-GFP-ROSA-LacZ mice. (F) Representative IF images in Prx1-CreER-GFP-ROA-LacZ mice, immunostained respectively with β-Gal (green) and IdU (red) antibody, counterstained with DAPI (blue). White arrows indicate the Prx1-CreER-βGal+/IdU+ cells. Mus=muscle; CB= cortical bone; BM= bone marrow; PO= periosteum. Scale bars = 100μm.
To verify the specificity of β-Gal (tagging CreER-recombination of Prx-1), and GFP (reporting Prx1 expression), we performed a short-term 4OH-TAM study, in which 4OH-TAM was given 0.5 mg/day for 5 days to Prx1-CreER-GFP-ROSA-LacZ and mice were sacrificed at day 6, and sectioned bones were subjected to IF using antibodies against β-Gal, GFP and Prx1. As shown in Supplemental Fig. 4, we found that almost all β-Gal+ positive cells were Prx1+ cells (73.4%±0.17; n=3) and almost all GFP+ cells were β-Gal+ cells (83.2%±0.22; n=3).
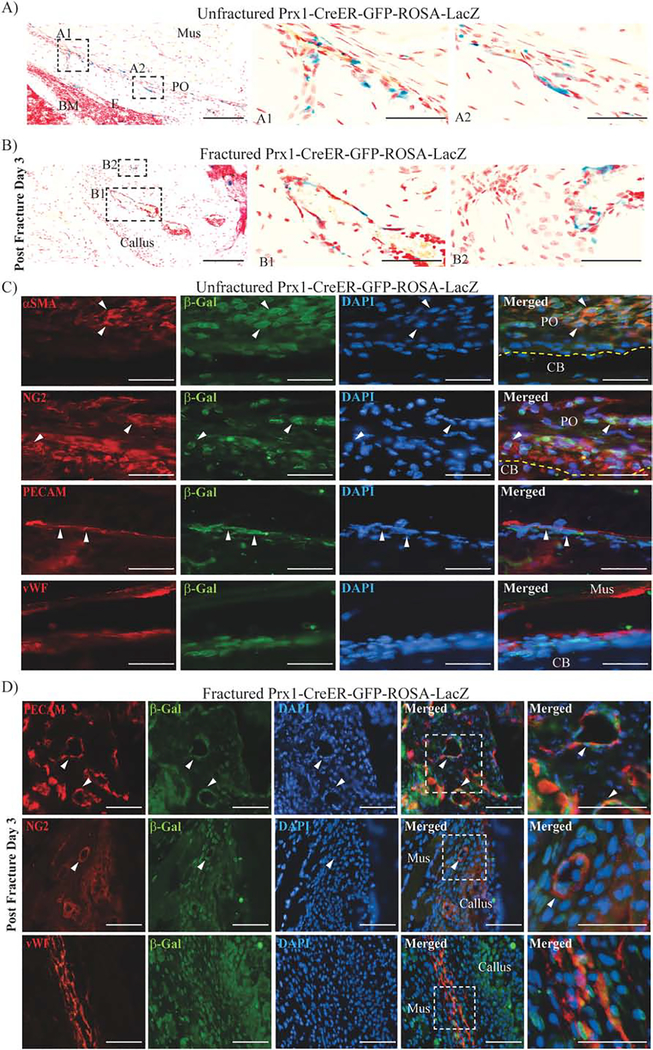
In order to characterize the nature of Prx1-expressing cells we analyzed both Prx1-CreER-GFP-ROSA-LacZ unfractured mice that have received short-term 4OH-TAM (0.5 mg/day for 5 days then sacrificed at day 6) and fractured mice that had received 4OH-TAM (0.5 mg/day i.p.) 2 days before and 2 days after fracture and were sacrificed 3 days after fracture. In unfractured mice (Fig. 4, A1–A2), as well as fractured mice (Fig. 4, B1–B2), βGal+ cells were found to be in the perivasculature. Perivascular cells closely surround endothelial cells and have been termed pericytes in capillaries and microvessels.(61) Beyond their function to support angiogenesis, pericytes are recently emerging as multipotent cells with osteogenic, chondrogenic and adipogenic potentials. (2, 4, 36, 38, 39, 62–67) We found that in unfractured (Fig. 4C) and fractured mice (Fig. 4D),βGal+ cells surround both PECAM+ and vWF+ endothelial cells, and they co-express the pericyte markers αSMA and NG2.
Figure 4.
Prx1-expressing cells co-express pericytes markers, surrounding endothelial cells in unfractured and fractured mice.
(A) Representative histological LacZ/Safranin O staining of unfractured Prx1-CreER-GFP-ROSA-LacZ mice, that have received short-term 4OH-TAM received (0.5 mg/day for 5 days then sacrificed at day 6) and (B) fractured Prx1-CreER-GFP-ROSA-LacZ mice that had received 4OH-TAM (0.5mg/day) 2 days before and 2 days after fracture and were sacrificed 3 days after fracture. Dotted squares indicate areas magnified in A1, A2, B1 and B2. (C) Representative IF images of the periosteum of unfractured Prx1-CreER-GFP-ROSA-LacZ mice that have received short-term 4OH-TAM (0.5 mg/day for 5 days then sacrificed at day 6), immunostained with β-Gal (green) and αSMA, NG2, PECAM or vWF (red) antibodies, counterstained with DAPI (blue). (D) Representative IF images of Prx1-CreER-GFP-ROSA-LacZ fracture tibias obtained 7 days after fracture, stained with β-Gal (green) and PECAM, NG2 or vWF (red) antibodies, counterstained with DAPI (blue). White arrows indicate double-positive cells. Dotted squares indicate magnified regions. PO= periosteum; CB=cortical bone; Mus=muscle. Scale bars = 100μm.
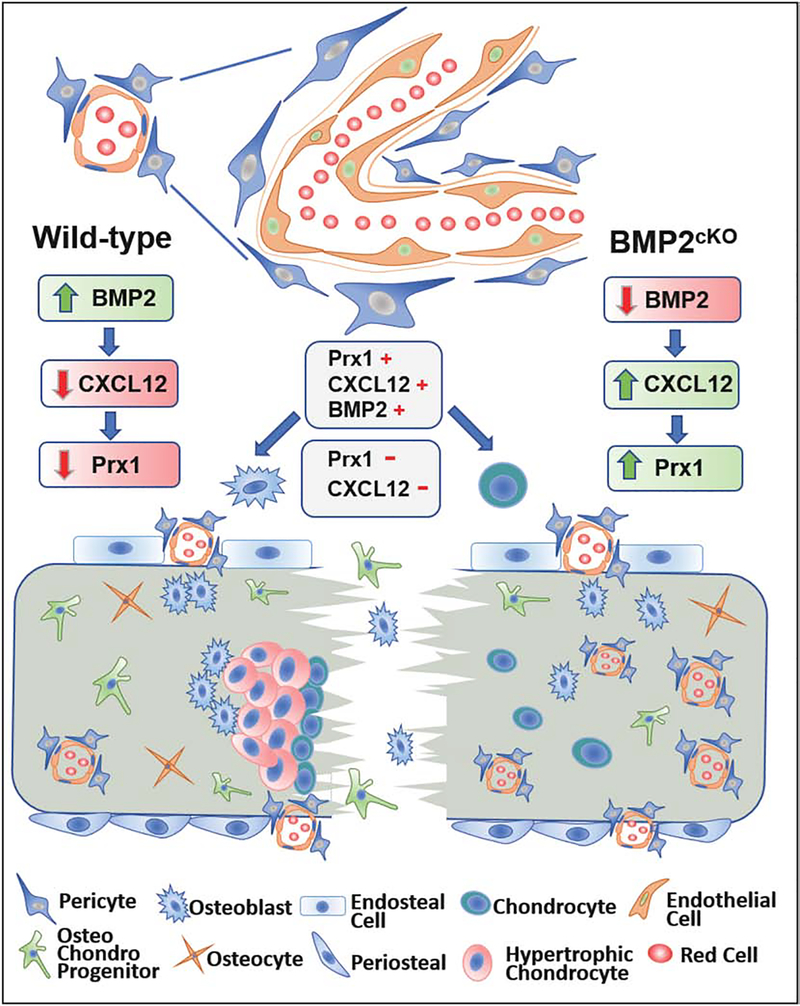
3.3. Prx1 expression pattern during fracture repair is regulated by the interplay between BMP2 and CXCL12
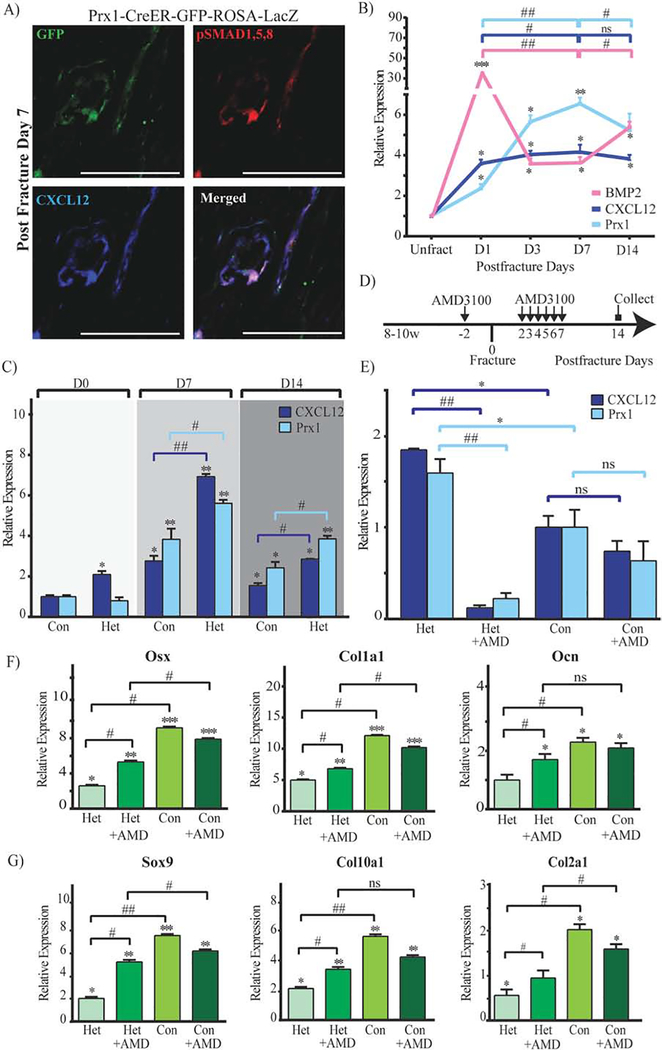
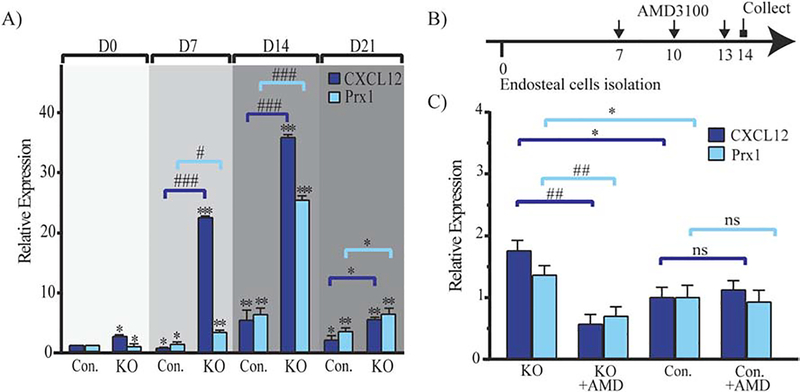
We were intrigued by the expression pattern of Prx1 that peaked at an early stage of fracture repair and then declined with callus formation, and by the fact that Prx1-expressing cells were perivascular. BMP2 has been reported to have a critical role in the initiation of fracture repair through regulation of CXCL12’s expression by perivascular cells.(13) Thus, we investigated if Prx1 expression was related to BMP2 and CXCL12 in fracture repair. As shown in Fig. 5A, we found that fracture-induced Prx1-GFP+ perivascular cells co-expressed CXCL12, and were also positive for pSMAD1,5,8. We then investigated the expression pattern of Prx1 in relationship with expressions of CXCL12 and BMP2. We found that fracture elicited an upsurge of Prx1, BMP2 and CXCL12 as early as 1 day after fracture, with BMP2’s increase being the greatest (Fig. 5B). We also observed, as shown in Fig. 5B, a consistent inverse relationship between BMP2 expression and the expressions of Prx1 and CXCL12: BMP2 had the highest level at day 1 followed by a dramatic decrease by day 3, while Prx1 and CXCL12 peaked later by day 7 when BMP2 had the lowest level. Consistently, at day 14, an increase of BMP2 corresponded with decreasing levels of Prx1 and CXCL12. We have previously reported that mice that carry a Cre-Prx1 mediated conditional inactivation of BMP-2 (either BMP2cKO/cKO or BMP2cKO/+) have abnormally high levels of CXCL12 that is specifically elevated at the endosteal site of fractured and unfractured tibias, and in isolated endosteal cells.(13) We now report that in BMP2cKO/+ mice, Prx1 is elevated compared to control, with the greatest increase noted at 7 days after fracture when CXCL12 also reaches the highest level (Fig. 5C). To investigate whether the deranged expression of Prx1 in BMP2cKO/+ mice could be downstream from CXCL12 signaling, we treated BMP2cKO/+ mice with AMD3100, an antagonist of CXCR4 (the main receptor for CXCL12), 2 days before and from day 2 to day 7 after fracture (Fig. 5D), a regimen that had showed to be able to partially rescue the fracture healing of BMP2cKO/+ mice.(13) As shown in Fig. 5E, AMD3100 dramatically decreased the expression of Prx1 in BMP2cKO/+ mice and had no effect on the Prx1 expression of control fractured mice. At day 14 after fracture, BMP2cKO/+ mice showed reduced levels of cartilage and bone markers that were normalized by AMD3100 treatment (Fig. 5, F–G), confirming our previous findings that AMD3100 is effective in BMP2cKO/+ mice.(13) We next studied the interplay between BMP2, CXCL12, and Prx1 in vitro by using BMP2cKO/cKO endosteal cells. We found that BMP2cKO/cKO endosteal cells, at any given time points during osteogenic differentiation in culture, have increased levels of Prx1 and CXCL12 (Fig. 6A). To further determine that BMP2 and in turn CXCL12 signaling regulates Prx1 expression, BMP2cKO/cKO cells were treated with AMD3100 (400 μM, every 3 days from day 7 to day 14) (Fig. 6B). AMD3100 normalized both Prx1 and CXCL12 expression in cells from BMP2cKO/cKO mice (Fig. 6C) while it had no effect on control cells.
Figure 5.
Prx1 expression is regulated by BMP2/CXCL12 signaling.
(A) Representative IF images of fractured Prx1-CreER-GFP tibia sections obtained 7 days after fracture, immunostained with GFP (green), pSMAD1,5,8 (red) and CXCL12 (blue). Scale bars = 100μm. (B) mRNA expression levels quantified by qRT-PCR of BMP2, CXCL12 and Prx1 in calluses from fractured Prx1-CreER-GFP mice by qRT-PCR. Data are reported as mean ± SD. of triplicate repeats from n=6 (unfractured) or n=10 (D1, D3, D7, D14) samples normalized to unfractured (Unfract) to which it was given a value of 1; *, p < 0.05, **, p < 0.01, ***, p < 0.001, compared to Unfract by one-way ANOVA and Sidak’s multiple comparison test. #, p < 0.05, ##, p < 0.01, by unpaired two-tail t-test. (C) mRNA expression levels quantified by qRT-PCR of CXCL12 and Prx1 in calluses from fractured BMP2cKO/+ (Het) or control (Con) mice obtained at 0 (30 minutes after fracture), 7 and 14 days after fracture. Data are reported as mean ± SD. of triplicate repeats from n=6 (Het) or n=10 (Con) samples normalized to day 0 control. *, p < 0.05, **, p < 0.01, compared to day 0 control by one-way ANOVA and Sidak’s multiple comparison test. #, p < 0.05, ##, p < 0.01, compared to control by unpaired two-tail t-test. (D) Schematic representation of AMD3100 injections in fractured BMP2cKO/+ mice and control mice. See Materials and Methods section for more details. (E) mRNA expression levels quantified by qRT-PCR of CXCL12 and Prx1 in calluses from fractured BMP2cKO/+ (Het) or control (Con) mice, treated with AMD3100 or vehicle (PBS), obtained 14 days after fracture. Data are reported as mean ± SD of triplicate repeats from n=6 (Het) or n=10 (Con) samples normalized to day 14 control. *, p < 0.05, compared to Con day 14 by unpaired two-tail t-test. ##, p < 0.01, compared to Het day 14 by unpaired two-tail t-test. (F) mRNA expression levels of osteogenic (Osx, Col1a1, Ocn) and (G) chondrogenic marker (Sox9, Col10a1, Col2a1) in calluses from BMP2cKO/+ (Het) or control mice (Con), treated with AMD3100 or vehicle (PBS), obtained 14 days after fracture. Data are reported as mean ± SD. of triplicate repeats from n=6 (Het) or n=10 (Con) samples normalized to day 0 control. *, p < 0.05, **, p < 0.01, ***, p < 0.001, by one-way ANOVA and Sidak’s multiple comparison test. #, p < 0.05, ##, p < 0.01, by unpaired two-tail t-test.
Figure 6.
Prx1 expression pattern in isolated BMP2cKO/cKO endosteal cells.
(A) mRNA expression levels quantified by qRT-PCR of CXCL12 and Prx1 in isolated endosteal cells from BMP2cKO/cKO (KO) and control (Con) cultured under undifferentiated conditions (Day 0) or osteogenic differentiation conditions (Day 7, Day 14 and Day 21 of culture). See in the Materials and Methods section for more details. Data are reported as mean ± SD. of triplicate repeats from n=6 samples normalized to day 0 control. *, p < 0.05, **, p < 0.01, ***, p < 0.001, compared to day 0 control by one-way ANOVA and Sidak’s multiple comparison test. #, p < 0.05, ##, p < 0.01, ###, p < 0.001, compared to Con by unpaired two-tail t-test. (B) Schematic representation of AMD3100 treatment in endosteal cells isolated from BMP2cKO/cKO and control mice and cultured under osteogenic differentiation conditions, with or without 400 μM AMD3100 every 3 days beginning on day 7 through day 14. See in the Materials and Methods section for more details. (C) mRNA expression levels quantified by qRT-PCR of CXCL12 and Prx1 in isolated endosteal cells from BMP2cKO/cKO (KO) and control (Con) cultured with or without AMD3100 (as schematically presented in B). Data are reported as mean ± SD. of triplicate repeats from 6 separate samples normalized to day 14 control. *, p < 0.05, compared to control by one-way ANOVA and Sidak’s multiple comparison test. ##, p < 0.01, compared to KO by unpaired two-tail t-test.
3.4. Prx1-expressing cells are required to initiate fracture repair
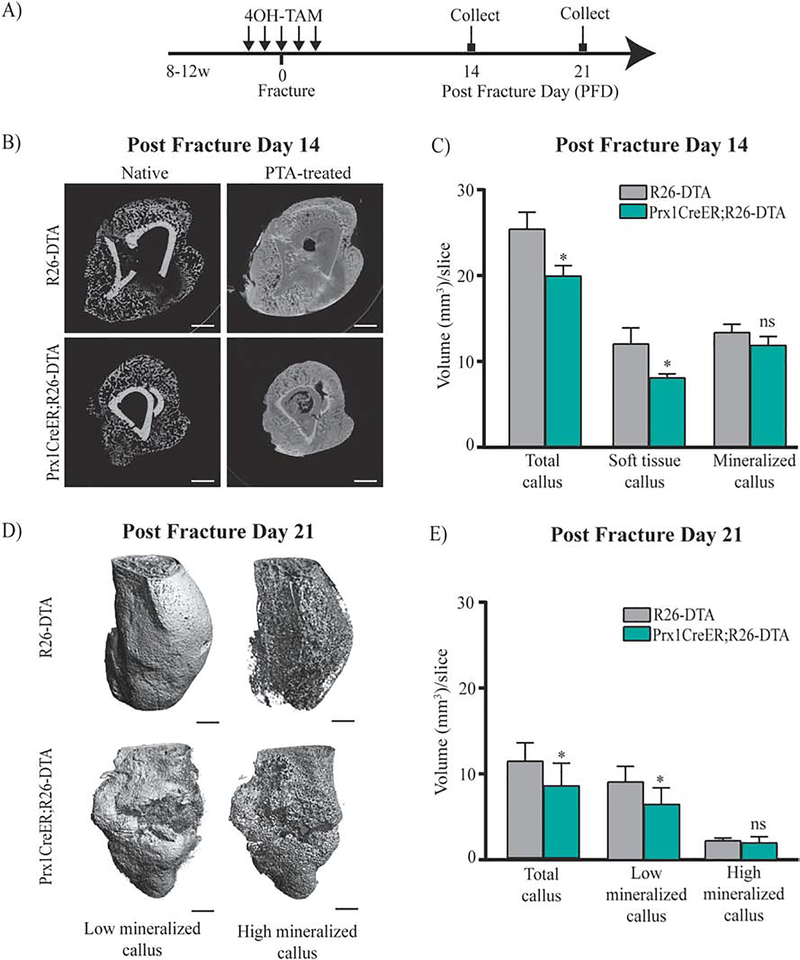
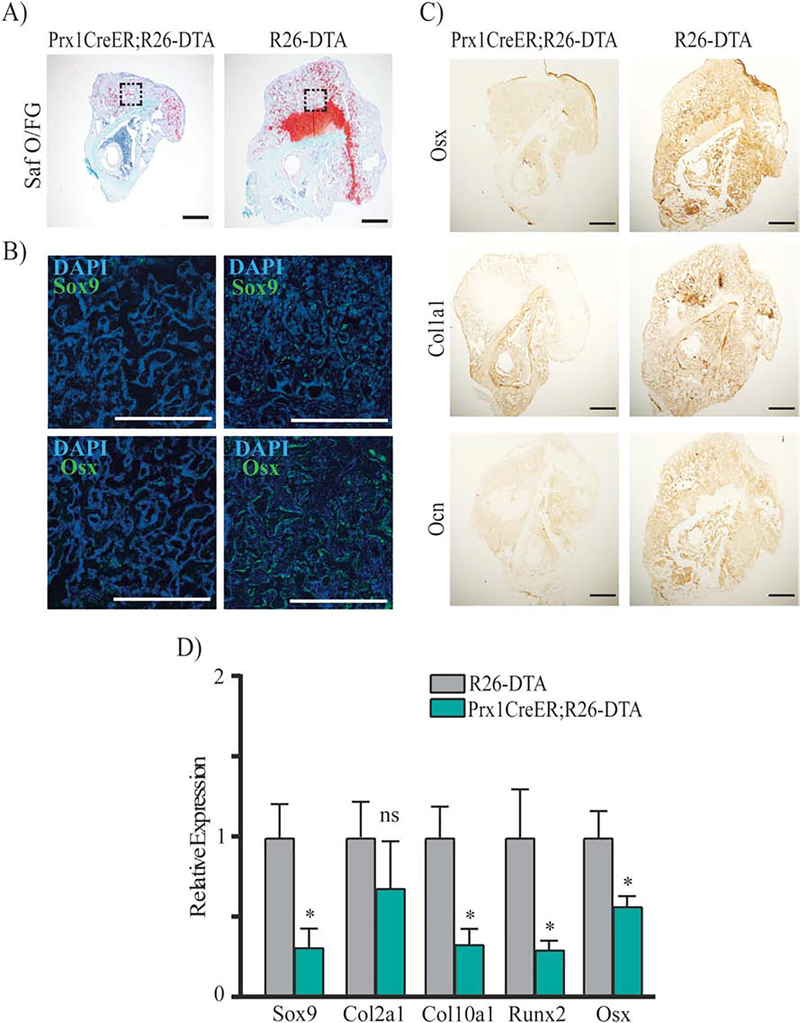
The finding that fracture elicits an early response of Prx1-expressing cells led us to analyze their functional role in initiating the fracture repair process. To this purpose, we have generated the Prx1CreER;R26-DTA mice to genetically ablate Prx1-expressing cells through toxicity of DTA, whose expression is activated via Prx1-CreER recombination after TAM. Prx1CreER;R26-DTA-tdTomato mice and ROSA26-DTA-tdTomato littermate controls received 4OH-TAM for 5 days (2 days before and 2 days after fracture) (Fig. 7A). As shown in Supplemental Fig. 5, we found that within the fracture, ~64% (63.72%±0.36; n=5) of tdTomato+ cells were ablated in Prx1CreER;R26-DTA-tdTomato mice compared to control, indicating incomplete ablation of Prx1-CreER-expressing cells, as reported previously.(50) μCT-PTA and μCT-native analyses showed that 14 days after fracture, the callus of Prx1CreER;R26-DTA compared to R26DTA controls was reduced in total volume and had less soft tissue (Fig. 7, B–C). μCT-PTA and μCT-native analyses at 21 days after fracture showed that compared to R26-DTA controls, Prx1CreER;R26-DTA mice continued to have a smaller and less mineralized callus with some healing reaction surrounding a fracture line center that was still open (Fig. 7, D–E). Histological Safranin O/Fast Green staining (Fig. 8A), IF (Fig. 8B), IHC studies (Fig. 8C), as well as qRT-PCR analyses (Fig. 8D) showed that the calluses of Prx1CreER;R26-DTA mice had less bone and cartilaginous content. Biomechanical testing analyses at 21 days after fracture, showed a significant decrease in stiffness, and a no significant downward trend in ultimate force in Prx1CreER;R26-DTA calluses compared to R26-DTA controls (Supplemental Fig. 6). Interestingly, we found that the small callus formed by Prx1CreER;R26-DTA exhibited an increase in vasculature (Supplemental Fig. 7), as indicated by the increased number of PECAM+ cells. Collectively, our studies in Prx1CreER:R26-DTA mice indicate that Prx1-expressing cells have a key role in initiating the fracture repair process.
Figure 7.
Defective fracture healing in Prx1CreER;R26-DTA mice.
(A) Schematic representation of 4OH-TAM injections in fractured Prx1CreER;R26-DTA and R26-DTA, control. (B) Representative μCT images of control and Prx1CreER;R26-DTA mice at 14 days after fracture, with or without PTA staining (10 days). Scale bars= 1mm. (C) Volumes of total callus, soft tissue and mineralized callus are normalized to the number of slices (mm3/slice) comprising the callus (see Material and Methods section for more details). Data are reported as mean ± SD of duplicate repeats from 5 separate samples. *, p < 0.05, compared to Control by unpaired two-tail t-test. (D) Representative three-dimensional μCT images of control and Prx1CreER;R26-DTA and R26-DTA mice at 21 days after fracture. Scale bars= 1mm. (E) Volumes of total mineralized callus, low mineralized callus and high mineralized callus are normalized to the number of slices (mm3/slice) comprising the callus. Data are reported as mean ± SD of duplicate repeats from 5 separate samples. *, p < 0.05, compared to Control by unpaired two-tail t-test.
Figure 8.
Histological, immunostaining and qRT-PCR analyses of Prx1-CreER;R26-DTA calluses. (A) Representative coronal images of histological Safranin O/Fast Green (Saf O/FG) staining of fractured tibias from Prx1CreER;R26-DTA mice and R26-DTA littermate controls at 14 days after fracture. Adjacent sections were subjected respectively to: B) IF for Osterix [Osx] and Sox9 (Green) and counterstained with DAPI (Blue), areas shown correspond to the dotted squares of mineralized callus indicated in the Saf O/FG images, scale bars = 100μm; C) IHC respectively for Osx, Collagen 1 [Col1a1] and Osteocalcin [Ocn]; scale bars = 100μm. D) mRNA expression levels quantified by qRT-PCR of Sox9, Col2a1, Col10a1, Runx2 and Osx in fractured Prx1CreER;R26-DTA and R26-DTA control calluses obtained 14 days after fracture; data are reported as mean ± SD of duplicate repeats from 5 separate samples normalized to control to which was given value=1; *, p < 0.05, compared to control by unpaired two-tail t-test.
4. Discussion
Despite compelling evidence showing that Prx1 is expressed in the postnatal skeleton,(6, 10, 37, 38) its function and regulation are not fully understood. Our study sought to shift the current research pattern that has primarily focused on the identification of markers for postnatal skeletal progenitors and instead to characterize the function of a specific cell population (Prx1+ cells) and their expressing marker (Prx1) as a crossroad in fracture repair. Using a comprehensive approach that combined cell-tracing, current animal models, and in vitro studies to identify the localization, fate and phenotype of Prx1+ cells before and after fracture, we determined that Prx1-expressing cells are critical in fracture repair and Prx1 expression is regulated by BMP2 through CXCL12 expression. As presented in the graphic model depicted in Fig. 9, our studies determined that: 1) fracture elicits a population of Prx1+ pericytic cells co-expressing CXCL12; 2) Prx1-expressing cells contribute to the cartilaginous and bone callus, but lose Prx1’s expression with differentiation into bone or cartilaginous cells; 3) BMP2’s upregulation after fracture leads to a decrease of CXCL12 that in turn down-regulates Prx1, allowing cells to commit into differentiation; 4) Prx1-expressing cells are essential to initiate a normal fracture healing process; 5) when BMP2 is deficient in osteo-chondroprogenitors (BMP2cKO), CXCL12 and consequently Prx1 remain upregulated leading to a defective fracture healing that can be corrected by blocking the CXCL12 signaling.
Figure 9.
Graphic model describing the functional role of Prx1 and Prx1-expressing cells in fracture repair. Following fracture, a population of Prx1+ periosteal and endosteal pericytic cells, co-expressing BMP2 and CXCL12, are elicited and contribute to the cartilaginous and mineralizing calluses, but Prx1’s expression decreases with callus maturation. BMP2’s upregulation after facture leads to a decrease of CXCL12 that in turn down-regulates Prx1, allowing cells to commit to callus maturation. In absence or insufficiency of BMP2 (BMP2cKO), such regulation of CXCL12 and therefore of Prx1 is lost, leading to abnormal angiogenesis and impaired fracture healing.
The characterization of cells actively expressing Prx1 and the understanding of the mechanisms regulating Prx1’s expression during fracture repair provide the opportunity to better evaluate the regenerative capacity of a skeletal progenitor population and opens the opportunity for cell-based therapies to promote fracture healing. Several sources have been suggested for the skeletal progenitors that participate in the fracture healing process, including the periosteum, endosteum, BM, adipose and muscle tissues adjacent to the fracture, vascular and perivascular cells, and circulating cells recruited at the fracture site.(5, 12, 13, 36, 39, 42, 66, 68–70) Although it has been shown that the periosteum is critical in the repairing process and markers for periosteal cells have been identified, most of the studies have been based on tracing cells after tagging with an imaging reporter system through Cre-recombination.(11–14) Information on a functional role of specific progenitor cells during fracture healing is still partial. Furthermore there is evidence that the endosteum contributes to the repair, either by providing cell population(s) derived from the BM that seem to be critical in the early stage of fracture repair or contributing to the formation of the intramembranous callus.(5, 21, 71, 72) Identifications of specific population(s) of cells, characterization of their role during the fracture repair process, and identification of their natural niches has major clinical relevance. Those findings can shed light on the defective mechanisms underlying non-unions and can lead to the development of cell-based therapies to treat them. Intense research efforts, including from our laboratory, have demonstrated the capacities of skeletal stromal cells to repair injured tissues including fractures.(12–14) Perivascular cells, namely pericytes, are emerging as multipotent cells capable of myogenic, chondrogenic and osteogenic potentials.(21, 54, 62–64, 73–77) Our studies identified that fracture elicits a periosteal and endosteal response of perivascular Prx1+ cells that participate in the fracture healing process and showed that Prx1-expressing cells have a functional role in the repair process.
In embryonic life, Prx1 is highly expressed in developing limbs and in midline skeletal and craniofacial elements.(16, 17) Although Prx1 is expressed in adult tissues, its function in postnatal life still needs to be fully characterized.(23, 24, 73, 78–80) A 2.4 kb Prx1 promoter/enhancer directing the transgene’s expression in osteo-chondrogenic mesenchyme progenitors has been identified and has led to the generation of Prx1-Cre mice that have been useful in directing tissue-specific conditional recombination of numerous genes, and imaging tagging cells (and their differentiated progeny) that expressed Prx1–2.4kb-Cre either during embryonic life or at some point during post-natal life.(6, 81) Progeny of Prx1–2.4 kb-driven-embryologically-derived cells, have been detected in the periosteum and in fractured bones.(71) Interestingly this study noted that 14 days after fracture, no Prx1-actively-expressing cells were found within the callus, but further investigations were not pursued. Importantly, Prx1-Cre tagged adult cells presumably derived from the periosteum were detected within the callus.(6, 10, 12, 14, 22, 23, 79) Postnatally, Prx1-expressing cells have been identified within the skull and contributed to the regeneration of calvaria defects.(24) Moore et al. reported that primary cilia are necessary for cambium layer Prx1-expressing cells to contribute to skeletogenesis.(22) The function of cells actively expressing Prx1 and the role of Prx1 in fracture repair have been largely unknown. Our studies demonstrate that fracture induces a rapid increase of Prx1 expression and cells actively expressing Prx1 are required for fracture regeneration as indicated by the fact that ablation of Prx1-expressing cells lead to fracture healing impairment. We acknowledge that having eliminated Prx1+ cells in other osteochondrogenic sites may affect repair, i.e. by failure to recruit circulating Prx1+ osteochondroprogenitors. However, we estimate that the number of circulating Prx1+ expressing osteochondroprogenitors should be low.(50) Incomplete ablation of Cre-ER-expressing cells by DTA system has been extensively reported, and the percent of Prx1+ ablated cells (~64%) observed in our studies is consistent or even higher than the percentage detected in other studies.(82–84) However, even this incomplete ablation was sufficient to affect the fracture healing, the biomechanical properties and the expressions of chondrogenic and osteogenic markers. In agreement with Comai et al, we speculate that the limited DTA-mediated ablation can be: 1) either because the short window of opportunity for Cre (in our case Prx1-Cre) to be expressed and therefore recombine the loxP sites from the generic Rosa locus as indicated by the fact that its expression levels drop rapidly with fracture repair progression, 2) or an indicator of the differential regulation/processing of the genetically modified (IRES)-Cre allele versus endogenous gene (in our case Prx1) transcripts in vivo. Although in vitro studies using hairpin RNA to knock-down Prx1 might provide some information on the function of Prx1 gene they would not be able to provide information on the role of Prx1 expressing cells. Lastly, we acknowledge that the intramedullary pinning in our fracture model by injuring the BM may have contributed in the response of endosteal osteo-progenitor cells.
The difficulty in identifying adult regenerative cells is multifactorial, due to: population heterogeneity, changes in expression pattern after and depending on the injury, lack of unique markers and poor knowledge of the function of these markers, and the fact that cell identification has largely relied on embryological labeling. This hampers the study of those cells beyond Cre-reporting engineered organisms and the translation of findings to humans. Our approach enabled identification of a novel population of cells that actively express Prx1 in intact and fractured bones. We and others have also identified markers for injury-induced skeletal progenitors.(9, 74–76, 86–88) However, we are still at the early stage of discovery and we are far from having reached a consensus on markers that are either exclusive to skeletal progenitors or expressed by all skeletal progenitors.(89) Our in vitro and in vivo studies showed the phenotypic and changes of Prx1-expressing cells after fracture and in the context of other markers.
Although studies in human subjects are needed to evaluate the role of Prx1 expression and expressing cells in a clinical context, it is interesting to note that expression of Prx1 was found to be differentially expressed in the femurs from subjects with osteoporotic femur neck fractures compared to femurs from subjects without known bone pathology.(90)
This study is the first to demonstrate in vivo postnatal regulation of Prx1 expression, and to relate it to fracture healing. We found that Prx1 expression decreases concurrently with callus formation and the differentiation into cartilaginous and bone cells, similarly to when Prx1+ cells are cultured in differentiating conditions. Using in vivo and in vitro approaches we determined the mechanism by which Prx1 is downregulated. In particular, we found that Prx1 is downstream of CXCL12 and that CXCL12 signaling is the mediator through which BMP2 down-regulates Prx1. We and others have found compelling evidence that BMP2 is essential in fracture repair and that BMP2 exerts its regenerative effects though CXCL12.(13, 31) We now report that fracture elicits an early increase of BMP2 that leads to a decrease of CXCL12 that in turn down-regulates Prx1, allowing cells to commit to osteochondrogenesis. Our results provide in vivo support to previous in vitro studies that reported that Prx1 inhibits osteogenic differentiation, by downregulating Osterix and RUNX2, and adipogenesis by activating TGF-β signaling.(25, 26) They also corroborate the hypothesis that Prx1 plays a role in maintaining stemness, as postulated in neural cells.(27) The effects of AMD3100 in BMP2cKO/cKO cells and BMP2cKO/+ mice provided mechanistic evidence of the interplay between CXCL12 and Prx1 in committing skeletal progenitors to differentiation, suggesting potential therapeutic application to treat fracture non-unions. AMD3100’s clinical use is FDA-approved for patients with BM ablation or failure to induce hematopoietic stem cell mobilization. Its pharmacokinetic properties in rodents and humans are well known, and in the course of many years of clinical use w has shown an excellent safety profile.(46, 47, 49) Our findings on AMD3100 regulation of Prx1 expression by modulating the BMP2-CXCL12 axis might represent a significant breakthrough in the search for a pharmaceutical treatment for non-unions. In future studies, we will further investigate the BMP2 signaling and CXCL12 signaling pathways that regulate Prx1’s expression.
In conclusion, our studies led to the characterization of Prx1-expressing skeletal cells and Prx1 expression in fracture repair. Together, the roles of Prx1 illustrated in this study provide evidence for the regenerative capacity of a skeletal progenitor population and opens the path for more investigations to elucidate the source, the recruitment, and the function of progenitor cell populations. Such findings will help identify targets for cell-based therapies to treat fracture non-unions.
Supplementary Material
Highlights.
The regenerative capacity of bones after fractures implies the existence of adult progenitor cells
In fracture repair, the nature and regulation of progenitor cells remain elusive
Our studies identified the functional role of Prx1expressing cells in fracture
Our studies characterized a novel BMP-CXCL12-Prx1 regulatory cascade in fracture
Acknowledgements
We acknowledge the support of Dr. Ping Ye at University of North Carolina at Chapel Hill in assisting in the cell-tracing studies. We acknowledge the support of M.E. Cruz in assisting in the preparation of the figures and the support of M.R. Sandbulte in proofreading the manuscript. We acknowledge the support of J.D. Temple in assisting in the βCT studies. We acknowledge the Flow Cytometry Core at University of Illinois at Chicago and the Rush Internal Medicine Drug Discovery and Imaging Core for their technical assistance. We are grateful to S. Murakami (Case Western Reserve University) for providing the Prx1-CreER transgenic mice and to Dr. James Martin (Texas A&M Health Science Center, Houston, TX) for providing the BMP2flox/flox transgenic mice.
Funding
This work was supported by a grant from National Institutes of Health-National Institute of Arthritis and Musculoskeletal and Skin Diseases (1R01AR074049-01) to A.S.
Footnotes
Declaration of competing interest
None.
Additional supporting information may be found in the online version of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morshed S, Corrales L, Genant H, Miclau T, 3rd. Outcome assessment in clinical trials of fracture-healing. J Bone Joint Surg Am. 2008;90 Suppl 1:62–7. [DOI] [PubMed] [Google Scholar]
- 2.Bianco P."Mesenchymal" stem cells. Annu Rev Cell Dev Biol.2014;30:677–704. [DOI] [PubMed] [Google Scholar]
- 3.Bianco P, Cao X, Frenette PS, Mao JJ, Robey PG, Simmons PJ, et al. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat Med. 2013;19(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colnot C, Zhang X, Knothe Tate ML. Current insights on the regenerative potential of the periosteum: molecular, cellular, and endogenous engineering approaches. J Orthop Res. 2012;30(12):1869–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawanami A, Matsushita T, Chan YY, Murakami S. Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochemical and biophysical research communications. 2009;386(3):477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19(2):329–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizoguchi T, Pinho S, Ahmed J, Kunisaki Y, Hanoun M, Mendelson A, et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell. 2014;29(3):340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori Y, Adams D, Hagiwara Y, Yoshida R, Kamimura M, Itoi E, et al. Identification of a progenitor cell population destined to form fracture fibrocartilage callus in Dickkopf-related protein 3-green fluorescent protein reporter mice. J Bone Miner Metab. 2016;34(6):606–14. [DOI] [PubMed] [Google Scholar]
- 10.Murao H, Yamamoto K, Matsuda S, Akiyama H. Periosteal cells area major source of soft callus in bone fracture. J Bone Miner Metab. 2013;31(4):390–8. [DOI] [PubMed] [Google Scholar]
- 11.Debnath S, Yallowitz AR, McCormick J, Lalani S, Zhang T, Xu R, et al. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 2018;562(7725):133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duchamp de Lageneste O, Julien A, Abou-Khalil R, Frangi G, Carvalho C, Cagnard N, et al. Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nat Commun. 2018;9(1):773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers TJ,Longobardi L, Willcockson H, Temple JD, Tagliafierro L, Ye P, et al. BMP2 Regulation of CXCL12 Cellular, Temporal, and Spatial Expression is Essential During Fracture Repair. J Bone Miner Res. 2015;30(11):2014–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozaki A, Tsunoda M, Kinoshita S, Saura R. Role of fracture hematoma and periosteum during fracture healing in rats: interaction of fracture hematoma and the periosteum in the initial step of the healing process. J Orthop Sci. 2000;5(1):64–70. [DOI] [PubMed] [Google Scholar]
- 15.Olmedo ML, Landry PS, Sadasivan KK, Albright JA, Meek WD, Routh R, et al. Regulation of osteoblast levels during bone healing. J Orthop Trauma. 1999;13(5):356–62. [DOI] [PubMed] [Google Scholar]
- 16.tenBerge D, Brouwer A, Korving J, Martin JF,Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125(19):3831–42. [DOI] [PubMed] [Google Scholar]
- 17.Balic A, Adams D, Mina M. Prx1 and Prx2 cooperatively regulate the morphogenesis of the medial region of the mandibular process. Dev Dyn. 2009;238(10):2599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MartinJ F, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9(10):1237–49. [DOI] [PubMed] [Google Scholar]
- 19.Peterson RE, Hoffman S, Kern MJ. Opposing roles of two isoforms of the Prx1 homeobox gene in chondrogenesis. Dev Dyn. 2005;233(3):811–21. [DOI] [PubMed] [Google Scholar]
- 20.Schiffer C, Tariverdian G, Schiesser M, Thomas MC, Sergi C. Agnathia-otocephaly complex: report of three cases with involvement of two different Carnegie stages. Am J Med Genet. 2002;112(2):203–8. [DOI] [PubMed] [Google Scholar]
- 21.Colnot C, Huang S, Helms J. Analyzing the cellular contribution of bone marrow to fracture healing using bone marrow transplantation in mice. Biochem Biophys Res Commun. 2006;350(3):557–61. [DOI] [PubMed] [Google Scholar]
- 22.Moore ER, Yang Y, Jacobs CR. Primary cilia are necessary for Prx1-expressingcellsto contribute to postnatal skeletogenesis. J Cell Sci. 2018;131(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang Z, Chen Z, Ishikawa M, Yue X, Kawanami A, Leahy P, et al. Prx1 and 3.2kb Col1a1 promoters target distinct bone cell populations in transgenic mice. Bone. 2014;58:136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilk K, Yeh SA, Mortensen LJ, Ghaffarigarakani S, Lombardo CM, Bassir SH, et al. Postnatal Calvarial Skeletal Stem Cells Expressing PRX1 Reside Exclusively in the Calvarial Sutures and Are Required for Bone Regeneration. Stem Cell Reports. 2017;8(4):933–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu X, Beck GR Jr., Gilbert LC, Camalier CE, Bateman NW, Hood BL, et al. Identification of the homeobox protein Prx1 (MHox, Prrx-1) as a regulator of osterix expression and mediator of tumor necrosis factor a action in osteoblast differentiation. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2011;26(1):209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du B, Cawthorn WP, Su A, Doucette CR, Yao Y, Hemati N, et al. The transcription factor paired-related homeobox 1 (Prrx1) inhibits adipogenesis by activating transforming growth factor-beta (TGFbeta) signaling. J Biol Chem. 2013;288(5):3036–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimozaki K, Clemenson GD Jr., Gage FH. Paired related homeobox protein 1 is a regulator of stemness in adult neural stem/progenitor cells. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33(9):4066–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazard ZW, Heggeness MH, Hipp JA, Sonnet C, Fuentes AS, Nistal RP, et al. Cell-based gene therapy for repair of critical size defects in the rat fibula. J Cell Biochem. 2011;112(6):1563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi M, Jin H, Wang B, Yukata K, Sheu TJ, Ke QH, et al. Chondrocyte BMP2 signaling plays an essential role in bone fracture healing. Gene. 2013;512(2):211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronga M, Fagetti A, Canton G, Paiusco E, Surace MF, Cherubino P. Clinical applications of growth factors in bone injuries: experience with BMPs. Injury. 2013;44 Suppl 1:S34–9. [DOI] [PubMed] [Google Scholar]
- 31.Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, et al. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38(12):1424–9. [DOI] [PubMed] [Google Scholar]
- 32.Kwong FN,Hoyland JA, Evans CH,Freemont AJ.Regionalandcellularlocalisationof BMPs and their inhibitors’ expression in human fractures. Int Orthop. 2009;33(1):281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan A. Treatment of chronic nonunion of a sternal fracture with bone morphogenetic protein. Ann Thorac Surg. 2008;85(2):e12–3. [DOI] [PubMed] [Google Scholar]
- 34.Kwong FN, Hoyland JA, Freemont AJ, Evans CH. Altered relative expression of BMPs and BMP inhibitors in cartilaginous areas of human fractures progressing towards nonunion. J Orthop Res. 2009;27(6):752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagasawa T. CXC chemokine ligand12(CXCL12)anditsreceptorCXCR4.JMol Med (Berl). 2014;92(5):433–9. [DOI] [PubMed] [Google Scholar]
- 36.Chen WC, Saparov A, Corselli M, Crisan M, Zheng B, Peault B, et al. Isolation of blood-vessel-derived multipotent precursors from human skeletal muscle. J Vis Exp. 2014(90):e51195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christopher MJ, Liu F, Hilton MJ, Long F, Link DC. Suppression of CXCL12 production by bone marrow osteoblasts is a common and critical pathway for cytokine-induced mobilization. Blood. 2009;114(7):1331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crisan M, Deasy B, Gavina M, Zheng B, Huard J, Lazzari L, et al. Purification and long-term culture of multipotent progenitor cells affiliated with the walls of human blood vessels: myoendothelial cells and pericytes. Methods Cell Biol. 2008;86:295–309. [DOI] [PubMed] [Google Scholar]
- 39.Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3(3):301–13. [DOI] [PubMed] [Google Scholar]
- 40.Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, et al. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33(3):387–99. [DOI] [PubMed] [Google Scholar]
- 41.Wise JK,Sumne rDR, Virdi AS. Modulatio no fstromal cell-derive dfactor-1/CXC chemokine receptor 4 axis enhances rhBMP-2-induced ectopic bone formation. Tissue Eng Part A. 2012;18(7–8):860–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466(7308):829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shahnazari M, Chu V, Wronski TJ, Nissenson RA, Halloran BP. CXCL12/CXCR4 signaling in the osteoblast regulates the mesenchymal stem cell and osteoclast lineage populations. FASEB J. 2013;27(9):3505–13. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Chen J, Fan W, Zhang J, Hua B, Sun B, et al. Stromal cell-derived factor-1alpha and transforming growth factor-beta1 synergistically facilitate migration and chondrogenesis of synovium-derived stem cells through MAPK pathways. Am J Transl Res. 2017;9(5):2656–67. [PMC free article] [PubMed] [Google Scholar]
- 45.Datema R, Rabin L, Hincenbergs M, Moreno MB, Warren S, Linquist V, et al. Antiviral efficacy in vivo of the anti-human immunodeficiency virus bicyclam SDZ SID 791 (JM 3100), an inhibitor of infectious cell entry. Antimicrob Agents Chemother. 1996;40(3):750–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendrix CW, Flexner C, MacFarland RT, Giandomenico C, Fuchs EJ, Redpath E, et al. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob Agents Chemother. 2000;44(6):1667–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang Y, Chen BJ, Deoliveira D, Mito J, Chao NJ. Selective enhancement of donor hematopoietic cell engraftment by the CXCR4 antagonist AMD3100 in a mouse transplantation model. PLoS One. 2010;5(6):e11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabin L, Hincenbergs M, Moreno MB, Warren S, Linquist V, Datema R, et al. Use of standardized SCID-hu Thy/Liv mouse model for preclinical efficacy testing of anti-human immunodeficiency virus type 1 compounds. Antimicrob Agents Chemother. 1996;40(3):755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pusic I, DiPersio JF. Update on clinical experience with AMD3100, an SDF-1/CXCL12-CXCR4 inhibitor, in mobilization of hematopoietic stem and progenitor cells. Curr Opin Hematol. 2010;17(4):319–26. [DOI] [PubMed] [Google Scholar]
- 50.Voehringer D, Liang HE, Locksley RM. Homeostasis and effector function of lymphopenia-induced "memory-like" T cells in constitutively T cell-depleted mice. J Immunol. 2008;180(7):4742–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA,Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma L, Martin JF. Generation of a Bmp2 conditional null allele.Genesis.2005;42(3):203–6. [DOI] [PubMed] [Google Scholar]
- 53.Granero-Molto F, Weis JA, Miga MI, Landis B, Myers TJ, O’Rear L, et al. Regenerative effects of transplanted mesenchymal stem cells in fracture healing. Stem Cells. 2009;27(8):1887–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Granero-Molto F, Myers TJ, Weis JA, Longobardi L, Li T, Yan Y, et al. Mesenchymal stem cells expressing insulin-like growth factor-I (MSCIGF) promote fracture healing and restore new bone formation in Irs1 knockout mice: analyses of MSCIGF autocrine and paracrine regenerative effects. Stem cells (Dayton, Ohio). 2011;29(10):1537–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25(7):1468–86. [DOI] [PubMed] [Google Scholar]
- 56.Temple JD LL, Li T, Myers TJ, Esposito A,Spagnoli A.. High-Precision Analysis of Subchondral Sclerosis as an Early and Progressive Marker of Post-Traumatic Osteoarthritis Using Micro-Computed Tomography. Annual Meeting American Society for Bone and Mineral Research 2013 2013. [Google Scholar]
- 57.Temple JD LL, Li T, Myers TJ, Spagnoli A. High-Resolution Micro-Computed Tomography of Articular Cartilage and Subchondral Bone Changes in Mouse Models of Osteoarthritis. Osteoarthritits Research Society International (OARSI) Annual Meeting 2013 2013. [Google Scholar]
- 58.Wang X, Nyman JS, Dong X, Leng H, Reyes M. Fundamental Biomechanics in Bone Tissue Engineering Synthesis Lectures on Tissue Engineering Morgan&Claypool Publishers; 2009:55. [Google Scholar]
- 59.Balduino A, Hurtado SP, Frazao P, Takiya CM, Alves LM, Nasciutti LE,et al. Bone marrow subendosteal microenvironment harbours functionally distinct haemosupportive stromal cell populations. Cell Tissue Res. 2005;319(2):255–66. [DOI] [PubMed] [Google Scholar]
- 60.Balduino A, Mello-Coelho V, Wang Z, Taichman RS, Krebsbach PH, Weeraratna AT, et al. Molecular signature and in vivo behavior of bone marrow endosteal and subendosteal stromal cell populations and their relevance to hematopoiesis. Exp Cell Res. 2012;318(19):2427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirschi KK D’Amore PA. Pericytes in the microvasculature. Cardiovasc Res. 1996;32(4):687–98. [PubMed] [Google Scholar]
- 62.Birbrair A, Wang ZM, Messi ML, Enikolopov GN, Delbono O. Nestin-GFP transgene reveals neural precursor cells in adult skeletal muscle. PLoS One. 2011;6(2):e16816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Birbrair A, Zhang T, Wang ZM, Messi ML, Enikolopov GN, Mintz A, et al. Role of pericytes in skeletal muscle regeneration and fat accumulation. Stem Cells Dev. 2013;22(16):2298–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol. 2007;9(3):255–67. [DOI] [PubMed] [Google Scholar]
- 65.Gharaibeh B,Lu A, Tebbets J, Zheng B, Feduska J, Crisan M, et al. Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nat Protoc. 2008;3(9):1501–9. [DOI] [PubMed] [Google Scholar]
- 66.Sacchetti B, Funari A, Michienzi S, DiCesare S, Piersanti S,Saggio I, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324–36. [DOI] [PubMed] [Google Scholar]
- 67.Tavian M, Zheng B, Oberlin E, Crisan M,Sun B, Huard J, et al. The vascular wall as a source of stem cells. Ann N Y Acad Sci. 2005;1044:41–50. [DOI] [PubMed] [Google Scholar]
- 68.Davis KM, Griffin KS, Chu TG, Wenke JC, Corona BT, McKinley TO, et al. Muscle-bone interactions during fracture healing. J Musculoskelet Neuronal Interact. 2015;15(1):1–9. [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, Matthews BG, Yu J, Novak S, Grcevic D, Sanjay A, et al. PDGF Modulates BMP2-Induced Osteogenesis in Periosteal Progenitor Cells. JBMR Plus. 2019;3(5):e10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, Xu J, Chang L, Meyers CA, Zhang L, Broderick K, et al. Relative contributions of adipose-resident CD146(+) pericytes and CD34(+) adventitial progenitor cells in bone tissue engineering. NPJ Regen Med. 2019;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colnot C. Skeletal cell fate decisions within periosteum and bone marrow during bone regeneration. J Bone Miner Res. 2009;24(2):274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colnot C. Cell sources for bone tissue engineering: insights from basic science. Tissue Eng Part B Rev. 2011;17(6):449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bassir SH, Garakani S, Wilk K, Aldawood ZA, Hou J, Yeh SA, et al. Prx1 Expressing Cells Are Required for Periodontal Regeneration of the Mouse Incisor. Front Physiol. 2019;10:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grcevic D, Pejda S, Matthews BG, Repic D, Wang L, Li H, et al. In vivo fate mapping identifies mesenchymal progenitor cells. Stem Cells. 2012;30(2):187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matthews BG, Grcevic D, Wang L, Hagiwara Y, Roguljic H, Joshi P, et al. Analysis of alphaSMA-labeled progenitor cell commitment identifies notch signaling as an important pathway in fracture healing. J Bone Miner Res. 2014;29(5):1283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang C, Inzana JA, Mirando AJ, Ren Y, Liu Z, Shen J, et al. NOTCH signaling in skeletal progenitors is critical for fracture repair. J Clin Invest. 2016;126(4):1471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zilberman Y, Kallai I, Gafni Y, Pelled G, Kossodo S, Yared W, et al. Fluorescence molecular tomography enables in vivo visualization and quantification of nonunion fracture repair induced by genetically engineered mesenchymal stem cells. J Orthop Res. 2008;26(4):522–30. [DOI] [PubMed] [Google Scholar]
- 78.Currie JD, Grosser L, Murawala P, Schuez M, Michel M, Tanaka EM, et al. The Prrx1 limb enhancer marks an adult subpopulation of injury-responsive dermal fibroblasts. Biol Open. 2019;8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu C, Cabahug-Zuckerman P, Stubbs C, Pendola M, Cai C, Mann KA, et al. Mechanical Loading Promotes the Expansion of Primitive Osteoprogenitors and Organizes Matrix and Vascular Morphology in Long Bone Defects. J Bone Miner Res. 2019;34(5):896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang T, Zhang X, Bikle DD. Osteogenic Differentiation of Periosteal Cells During Fracture Healing. J Cell Physiol. 2017;232(5):913–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin JF, Olson EN. Identification of a prx1 limb enhancer. Genesis. 2000;26(4):225–9. [PubMed] [Google Scholar]
- 82.Worthley DL, Churchill M, Compton JT, Tailor Y, Rao M, Si Y, et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160(1–2):269–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao H, Feng J, Ho TV, Grimes W, Urata M, Chai Y. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nature cell biology. 2015;17(4):386–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ivanova A, Signore M, Caro N, Greene ND, Copp AJ, Martinez-Barbera JP. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis (New York, NY: 2000). 2005;43(3):129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Comai G, Sambasivan R, Gopalakrishnan S, Tajbakhsh S. Variations in the efficiency of lineage marking and ablation confound distinctions between myogenic cell populations. Developmental cell. 2014;31(5):654–67. [DOI] [PubMed] [Google Scholar]
- 86.Chan CK, Seo EY, Chen JY, Lo D, McArdle A, Sinha R, et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160(1–2):285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marecic O, Tevlin R, McArdle A, Seo EY, Wearda T, Duldulao C, et al. Identification and characterization of an injury-induced skeletal progenitor. Proc Natl Acad Sci U S A. 2015;112(32):9920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ushiku C, Adams DJ, Jiang X, Wang L, Rowe DW. Long bone fracture repair in mice harboring GFP reporters for cells within the osteoblastic lineage. J Orthop Res. 2010;28(10):1338–47. [DOI] [PubMed] [Google Scholar]
- 89.O’Keefe RJ, Tuan RS, Lane NE, Barry F, Bunnell BA, Colnot C, et al. American Society for Bone and Mineral Research-Orthopaedic Research Society Joint Task Force Report on Cell-Based Therapies. J Bone Miner Res. 2019. [DOI] [PubMed] [Google Scholar]
- 90.Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Gene expression profile of the bone microenvironment in human fragility fracture bone. Bone. 2009;44(1):87–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.