Abstract
Sickle cell disease (SCD) is a worldwide hematological disorder causing painful episodes, anemia, organ damage, stroke, and even deaths. It is more common in sub-Saharan Africa and other resource-limited countries. Conventional laboratory-based diagnostic methods for SCD are time-consuming, complex, and cannot be performed at point-of-care (POC) and home settings. Optical microscope-based classification and counting demands a significant amount of time, extensive setup, and cost along with the skilled human labor to distinguish the normal red blood cells (RBCs) from sickled cells. There is an unmet need to develop a POC and home-based test to diagnose and monitor SCD and reduce mortality in resource-limited settings. An early-stage and timely diagnosis of SCD can help in the effective management of the disease. In this article, we utilized a smartphone-based image acquisition method for capturing RBC images from the SCD patients in normoxia and hypoxia conditions. A computer algorithm is developed to differentiate RBCs from the patient’s blood before and after cell sickling. Using the developed smartphone-based technique, we obtained similar percentage of sickle cells in blood samples as analyzed by conventional method (standard microscope). The developed method of testing demonstrates the potential utility of the smartphone-based test for reducing the overall cost of screening and management for SCD, thus increasing the practicality of smartphone-based screening technique for SCD in low-resource settings. Our setup does not require any special storage requirements and is particularly useful in assessing the severity of the SCD. This is the characteristic advantage of our technique as compared to other hemoglobin-based POC diagnostic techniques.
Introduction:
Sickle cell disease (SCD) is a common worldwide genetic disorder caused by the single point mutation in the beta-globin gene [1–3]. The β−6 glutamic acid is substituted by Valine, leading to the transformation of normal hemoglobin into HbS [4]. At low levels of oxygen, the HbS polymerizes, and results in sickled shape RBCs [5, 6]. This sickling of cells makes them hard and sticky and as a result, severely affects their oxygen transport efficiency and blood circulation. Patient experiences acute vaso-occlusive pain in children as well as in adults [7, 8]. The children born in resource-limited settings are at a greater risk of SCD [9]. Centers for Disease Control (CDC) has reported about 100,000 cases of individuals with homozygous genotype from a 2008 census in the USA population mainly in African Americans [10]. SCD is the most prevalent disease in sub-Saharan Africa with the highest incidence of deaths in children under 5 years of age [11]. Approximately 700 children in Africa are born with SCD every day [2, 11]. Over half of them die due to lack of diagnosis and treatment of SCD. This disease can damage any part of the body, especially spleen [12]. Children having SCD are liable to the development of systemic infections due to loss of splenic functions. Another major organ affected in SCD is the lung [13]. Patients with SCD are at high risk of pulmonary hypertension at a very young age, which crucially increases mortality rates in children. Cerebrovascular disorders are also responsible for much morbidity and mortality with SCD [14, 15]. The most common risk factors associated with SCD are stroke and silent infarction. The likelihood of a child with SCD having a risk of stroke is 200 times greater than a healthy one with a top incidence of ischemic stroke (caused by a blood clot that blocks a blood vessel in the brain) between 2 to 5 years of age.
An early-stage detection of SCD can be quite effective in managing the disease, especially in home-based, point-of-care (POC), and other resource-limited settings [16]. Hemoglobin electrophoresis and high-performance liquid chromatography (HPLC) are gold standard methods for the detection of SCD [17]. They require heavy laboratory equipment, a continuous supply of electricity, approximately 1mL patient blood sample, and trained staff to operate and interpret the test. Other existing techniques rely heavily on the use of optical microscopes. The morphological changes in the sickle cells are observed during their oxygenated (normoxia) and deoxygenated (hypoxia) states. This use of bulky and expensive microscopes is a time-consuming process that can only be performed at a diagnostic lab. These standards are often impossible to meet in many parts of sub-Saharan Africa and other low-resource countries.
Microfluidic devices have been widely utilized in the detection of several microorganisms such as bacterial cells (Escherichia Coli, Salmonella spp, Vibrio cholerae, and Mycobacterium tuberculosis), viruses (Human Immunodeficiency Virus type 1, Hepatitis B virus, and Zika virus) and other tumor markers (Neuron-specific enolase in small cell lung cancer) [18–21]. Several researchers have developed electrical impedance spectroscopy (EIS) based methods for the detection of SCD in microfluidic chips [22]. This combination of EIS and microfluidic has resulted in a reliable, accurate, and efficient method that can easily distinguish normal and sickled RBC and offers several advantages like label-free and non-invasiveness. The variations in the measured electrical impedance differential of sickle RBCs can work as a new biomarker of SCD. Another electrical impedance-based microflow cytometry technique with oxygen control seems potentially useful for SCD diagnosis [5].
Optical methods for sickle cell detection are based on the measurement of number of cells and different form factors. Electrical methods rely on the cellular dielectric properties, cell size, and difference in cell interior, Hb types and concentrations. They do not need advanced image processing. However, they do not provide information on disease severity. The equipment used for EIS (impedance analyzer) and oxygen control is relatively expensive and large in size, and not suitable for home-based and POC settings. It is vital to make sure that SCD diagnostic device must fulfill the World Health Organization’s criteria of being affordable, sensitive, specific, user-friendly, rapid and robust, equipment free, and delivered to those who need it, leading to the acronym “ASSURED” [23]. The further addition of features like real-time testing, communication of results and ease of sample collection leading to the acronym “REASSURED” can make such SCD devices even more suitable [24]. A POC SCD device developed on REASSURED criteria can diagnose SCD in newborn babies and adults and can significantly reduce the associated pain episodes and mortality [25]. A variety of techniques such as HemeChip and μPADs have been reported in this effort. See reference [26] for a comprehensive review.
The widespread use of smartphones worldwide has opened new avenues for home and POC-based biomedical diagnostics [3]. A myriad of attachments has been developed to integrate with smartphones to enhance their imaging capabilities and observe medical conditions. Smartphone-based techniques have been used for rapid imaging, detection, quantification, and monitoring of infections and diseases [27–32]. Various researchers have developed mobile phone-based fluorescence microscopy devices for POC diagnosis [33, 34]. These devices have demonstrated comparable performance to standard laboratory methods. A magnetic levitation-based platform was developed by researchers to detect sickled cells [3]. The developed setup eliminates the need for expensive centrifuge machines and microscopes for SCD detection. It rather utilizes magnetic levitation and a smartphone to capture the images of RBC levitating in the magnetic field using a smartphone camera. The sample is illuminated by an external LED and a lens is utilized for image enhancement purposes. Sickle cell levitation patterns are inherently different than those of normal RBC and this criterion may be used to distinguish the disease. This technique is limited to SCD detection and may be further developed to detect disease severity or sickle cell trait (SCT).
In this article, we have developed a POC and home-based portable and standalone setup for the diagnosis and treatment monitoring of SCD based on shape change in RBCs under hypoxia. It consists of a custom-designed 3D structure that can be easily attached to the smartphone camera. This setup supports an external lens to enhance the image quality along with the microchip to contain the blood sample. The sample is illuminated with an external LED and images of cells are captured using the smartphone camera. The captured images are further analyzed using the computer algorithm written in MATLAB. The normal RBCs can be automatically distinguished from the sickled cells based on their morphology. The developed setup is cost-effective that significantly reduces the per-test cost and can easily be utilized in any home-based settings. It can diagnose the SCD and can also be utilized for monitoring the treatment. Using our technique, it is possible to determine the percentage of sickled blood cells and may potentially adjust the dose of medication. The whole diagnostic process can be completed within 16 minutes time with minimal user input.
Materials and methods:
Microfluidic device design and fabrication:
The microfluidic device was developed using previously reported method utilizing 1.5 mm thick Poly-(methylmethacrylate) (PMMA) sheets (McMaster-Carr, Atlanta, GA), the double-sided adhesive tape (DSA) (3M, St. Paul, MN, 20 μm thick) and microscope slides (Fisherbrand plain, pre-cleaned glass slides) [35–38]. The design for the device was developed using AutoCAD software. PMMA sheets were machined with VLS 2.30 laser cutter (VersaLaser, Scottsdale, AZ). A 20 mm × 2 mm channel was cut inside DSA sheet to hold the blood sample. The top layer provided an inlet and outlet with diameter 0.4 mm. The schematic of the microchip fabrication is shown in Figure 1 (a). The whole structure containing PMMA and DSA was affixed to a 75 mm × 25 mm glass slide to make a composite microfluidic device (Figure 1 (b)). These dimensions were purposely chosen as per the requirements of smartphone attachment. Approximately 1 μl of blood was injected into the microchannel as shown in Figure 1(c,d).
Figure 1:
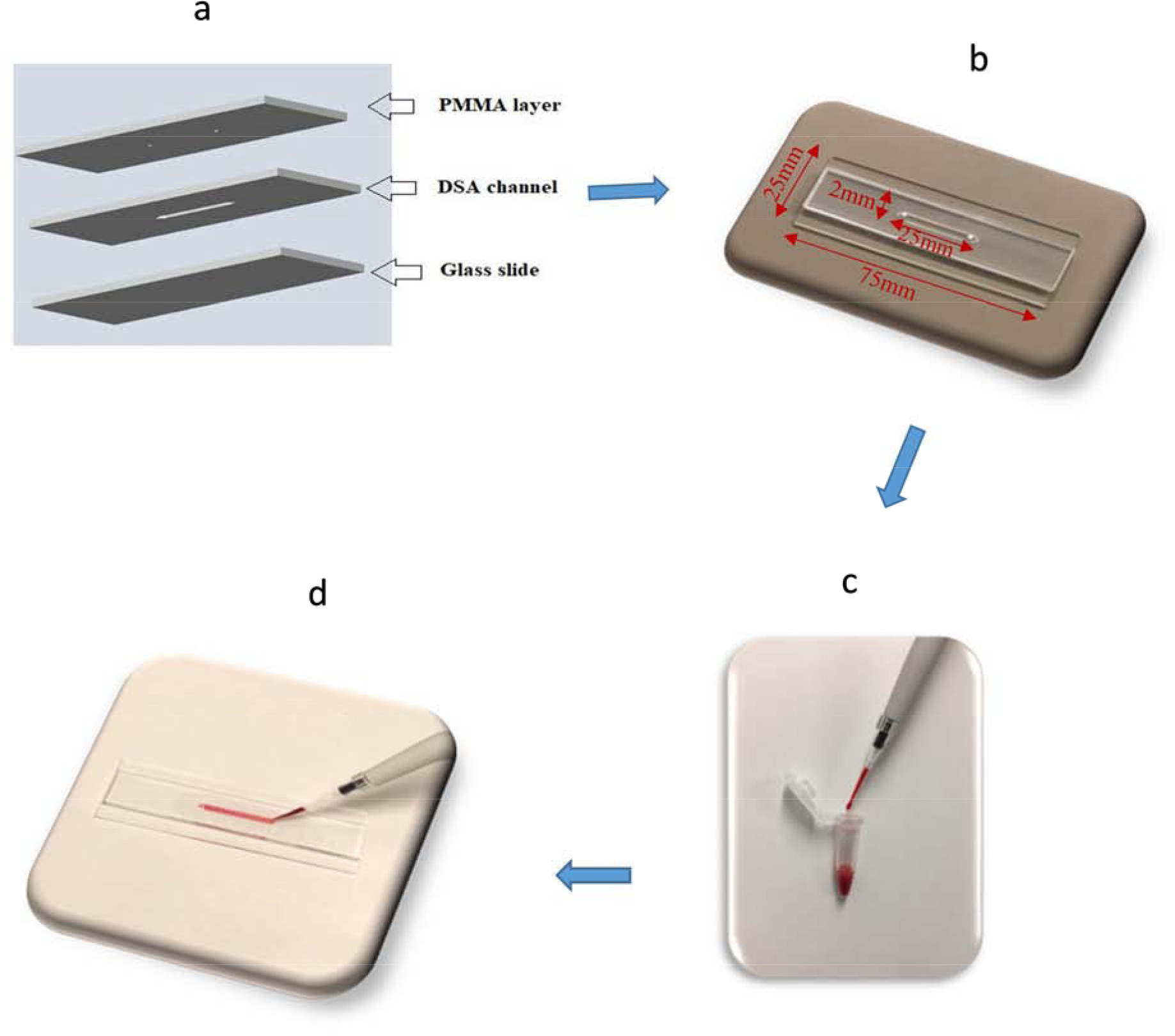
(a) Schematic illustration of microfluidic chip assembly consisting of three layers (PMMA, DSA, and glass slide) (b) Photograph of assembled microfluidic channel (length=25mm, width=2mm, height=0.02mm) (c) Blood sample used ~1 μL (d) Prepared chip containing RBCs.
Optical attachment and smartphone-based setup:
The setup was designed to operate with the rear camera of the smartphone. The optical element is composed of an aspheric lens with 2 mm height and 3.8 mm diameter. This lens is extracted from an internal Blu-ray drive. The lens is pasted on flexible film which cleanly sticks on the glass of any smartphone covering the camera. It was attached directly to the rear camera of the smartphone. The sample was trans-illuminated by a bright light source, powered by 2 Lithium batteries (3V each) with optical intensity of 2259 lux. This value was measured using the C-700R Spectromaster (Skeonic, USA.). The illumination source, microfluidic chip, and stage holding the microchip were placed in a complete dark setting. First of all, dark calibration was performed. After this calibration step, C-700R spectromaster instrument was brought to normal operation mode and the optical intensity of the illumination source was measured. Two consecutive readings were measured, and their average was calculated. It has the correct intensity for supplying good illumination through the samples without saturating the camera sensor. This light source is compatible with the safety standard EN62471:2008. The setup allows to recognize details of about 4.38 microns. We used the USAF 1951 slide and distinguished group 7 element 6 (Supplementary Figure S1). A stage for positioning smartphone and slide support was designed in AutoCAD. It was cut using a VLS 2.30 laser cutter (VersaLaser, Scottsdale, AZ) to support the delicate control of the focal distance and for the alignment of the light source (Figure 2). The effective focal point was established practically through trial and error. The stage was designed for a good alignment of the light source under the lens and a correct setting of the distance between lens and sample slide (1/8 inch or 3mm). The light is placed at about 20mm distance for illuminating the sickle blood sample. This distance is provided by the slide support of the smartphone stage. Figure 2 shows the top and side views for our developed smartphone-based setup detailing the smartphone positioning, slide support and alignment of light source. The dimensions of the stage were selected carefully to avoid manual focusing and errors that arise from focal distance and unstable mechanical support. The image magnification in our optical attachment was achieved using digital zoom of smartphone camera (5x with iPhone 6s). The developed smartphone system was able to image RBCs inside microchannel covering a field-of-view ~ 0.12 mm × 0.1 mm.
Figure 2:
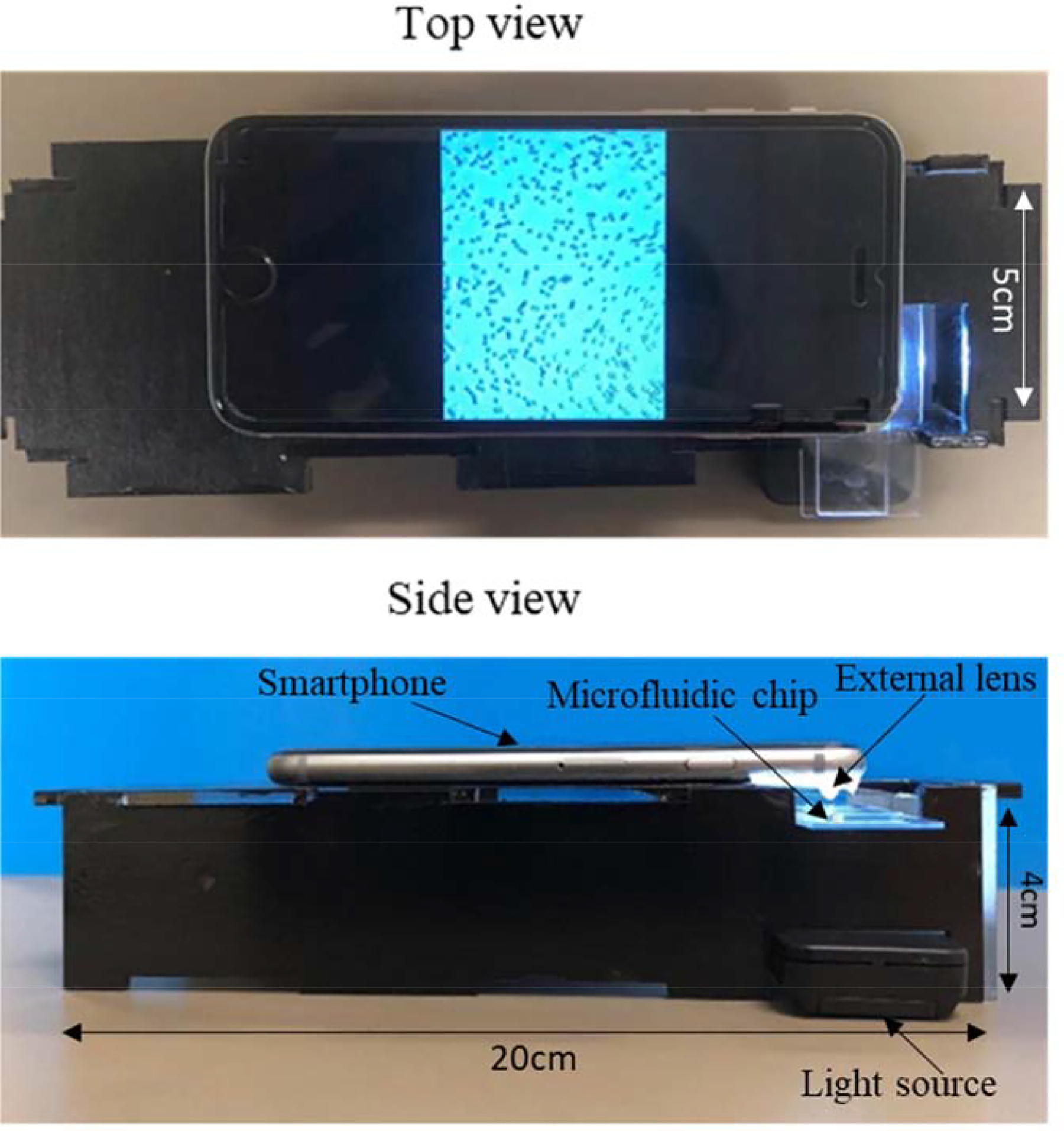
Smartphone-based platform for sickle cell blood imaging with dimensions of stage (length = 20cm, width = 5cm, height = 4cm).
Sample preparation:
De-identified whole blood from healthy donors was obtained from a local blood bank (Continental Services Group, Inc. Miami, USA). K2 anticoagulant EDTA tubes were used to collect these blood samples. De-identified sickle blood samples were obtained following Institutional Review Board (IRB) approvals from Florida Atlantic University and the University of Miami. All blood samples were diluted using phosphate-buffered saline (PBS) at a constant ratio of 1:100. We calculated the percentage of sickled cells in the blood sample obtained from two SCD patients under normal oxygen levels and hypoxia. Normally hypoxia is based on measure of; oxygen saturation in blood (SO2) <95%, partial pressure of oxygen (PaO2) <80mmHg, PH <7.35, partial pressure of carbon dioxide (PaCO2) >45mmHg or concentration of bicarbonate (HCO3) <22meq/L [39]. Less than 1 μl blood was loaded into the microfluidic device. Images were obtained using the microscope (Nikon Eclipse TE2000-S) & smartphone setup and labeled as “Before treatment.” Sodium metabisulfite (2% solution) was used to deoxygenate the RBCs (0.2 g of sodium metabisulfite in 10 ml of nano-pure water) [4, 40]. Sodium metabisulfite is a reducing agent which promotes sickling [41, 42]. The diluted blood from the normal and sickle blood samples was mixed separately into the prepared sodium metabisulfite controlling the dilution factor as 1:100. This sample was incubated at room temperature for 15 minutes and injected into another microfluidic chamber. Sodium metabisulfite reduces the oxygen tension inducing the typical sickle-shape to RBCs. Images were taken for these samples with deoxygenated hemoglobin inside RBCs using the microscope and the developed smartphone setup for comparison.
Image processing and algorithm development:
Computer-based image processing techniques are developed for counting different types of erythrocytes according to their variable morphology [43]. In this work, MATLAB R2019a is used for image analysis and quantification of sickled cells with a single click using an adaptive thresholding algorithm. Visual representation of our developed computer-based program can be seen in Figure 3. The MATLAB codes can be easily ported to the iOS/Android as well. The algorithm developed here can assess the concentration of sickled cells under hypoxia and normoxia conditions. The image processing time to calculate the percentage of sickled cells in a sample using the smartphone system is <15 seconds.
Figure 3:
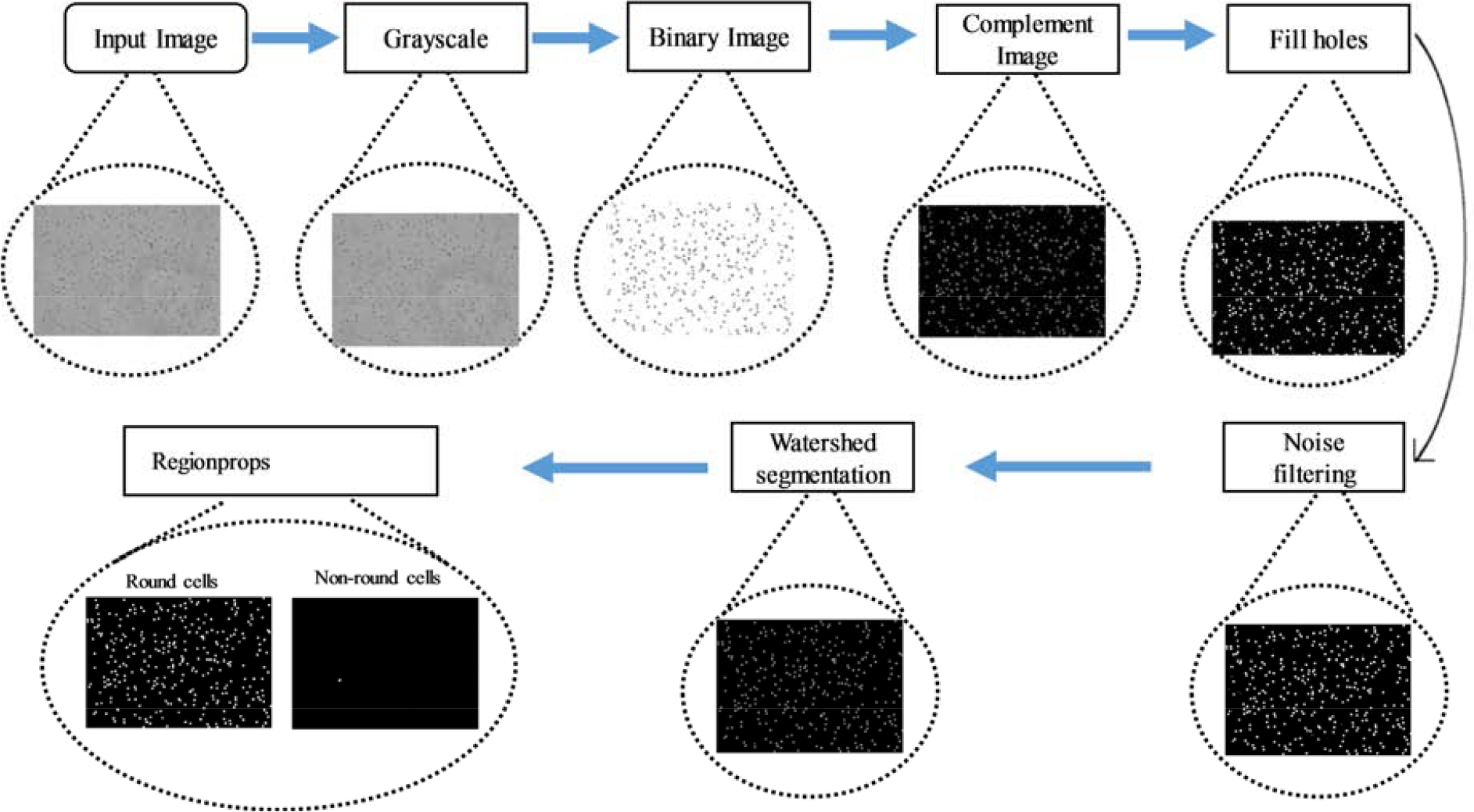
Diagram detailing the MATLAB architecture for diseased cells counting using regionprops function (microscope image).
The algorithm developed in this testing is described below:
Step 1. The human RBCs image captured either with a microscope or smartphone is fed to the program as input and is first converted to gray and then binary image using an adaptive thresholding that separates the foreground from the background with nonuniform illumination.
Step 2. The binary image is complemented, and the small holes inside the objects are filled up using imfill operator to increase the accuracy of the further calculation.
Step 3. For the ease of further processing, cells on the borders are removed using imclearborders.
Step 4. The watershed transform is used to segment adjacent RBCs into separate bodies. The watershed transforms “watershed ridge lines” in an image by treating it as a surface where light pixels represent high elevations, and dark pixels represent low elevations.
Step 5. Convex hull image is generated from the binary image using bwcovhull.
Step 6. Small unnecessary spots are removed from the image using bwareaopen operation. It eliminates all the objects in the diagram containing smaller number than the number of pixels mentioned in the threshold level.
Step 7. Connected components were labeled in a 2-D binary image using bwlabel.
Step 8. Area and perimeter of each of the components is calculated using regionprops on the objects.
Step 9. Discarding the smaller background objects, only the larger RBCs are considered and preprocessed for going through the rest of the steps.
Step 10. Metric [i.e. (perimeter2/4*π*area)] is calculated for each object. A circle, with radius r has an area of π*r2 and a perimeter of 2*π*r. A common place resolution independent measure of roundness is calculated by:
where C is the measure of circularity. If the value of C is equal to one, its circle, and if not, then it has other shapes [44, 45].
Step 11. By this metric value, any deviation or change in the shape of RBC is detected, which gives the number of sickled cells in the sample.
Results:
The smartphone setup used for screening sickled cells is illustrated in Figure 2. The whole platform comprised of an aspheric lens, printed stage, and a light source to capture the images of RBCs inside microfluidic channel. The stage was designed to stabilize the smartphone upright and the sample was input into microfluidic chamber through an inlet. The smartphone camera captured images of the microfluidic chip through the aspheric lens that is pasted on the smartphone camera lens to focus the RBCs. An LED under the channel is used to enhance imaging. It can be easily modified for other smartphones. Blood samples were diluted using PBS and images were captured under microscope and smartphone setup to make a comparison between the results. 2% sodium metabisulfite solution (reducing agent) was prepared and mixed with the samples in separate eppendorfs and imaged under microscope and smartphone likewise.
To evaluate the optical attachment setup and MATLAB-based program, first we performed the experiment using normal blood samples from healthy donors. Validation image results were obtained with two healthy blood samples using 3 different areas (near the inlet, near the outlet, and at the center of the microchannel to obtain consistent results) for field-of-view each covering 0.12 mm × 0.1 mm (~0.036 mm2 total area) for all samples. Images were captured with our mobile setup and laboratory-grade standard microscope for direct comparison. MATLAB algorithm performed the necessary image processing. Our system automatically executed cell classification and calculated the number of total cells, extracted sickled cells, and normal cells. For the normal blood sample 1, a very small average number of healthy blood cells (0.2%±0.08) were calculated as abnormal/sickled cells before treatment using the microscope images and 0.26%±0.09 cells were classified as abnormal cells after treatment (false-positive results). Likewise, for cellphone images, 0.86%±0.12 and 3.6%±1.3 healthy RBCs were misclassified as sickled cells pre and post-treatment, respectively. Similarly, for the second normal blood sample, 0.3%±0.14 cells were classified as sickled before treatment using microscopic images, and 0.45%±0.17 cells were detected as sickled after treatment. In images taken with smartphone 1.65%±0.77 and 2.9%±1.22 cells were counted as sickled before and after treatment, respectively. Our developed system calculated an insignificant number of sickled cells for known healthy blood samples and presented comparable results for both types of images. To avoid false positives, we have set a threshold of 5%. If sickle cells are more than this threshold of 5%, then the blood sample would be classified as SCD positive. The percentage of sickled cells calculated by our platform for each one of these normal blood samples is listed in Table 1, and an example of image transformation through different digital processing steps is also shown in Figure 4.
Table 1:
Result set for the percentage of sickled blood cells.
| Serial # | Percentage of sickled cells | |||||||
|---|---|---|---|---|---|---|---|---|
| Normal blood sample 1 | Normal blood sample 2 | Patient 1 | Patient 2 | |||||
| Smartphone Image | Microscope Image | Smartphone Image | Microscope Image | Smartphone Image | Microscope Image | Smartphone Image | Microscope Image | |
| Before treatment | ||||||||
| Image 1 | 1% | 0.3% | 1.7% | 0.23% | 6.9% | 6% | 2.4% | 2.8% |
| Image 2 | 0.9% | 0.1% | 0.86% | 0.3% | 5.4% | 5.6% | 5.3% | 5.5% |
| Image 3 | 0.7% | 0.2% | 2.4% | 0.5% | 6.7% | 6.8% | 3.5% | 2% |
| Average | 0.86%±0.12 | 0.2%±0.08 | 1.65%±0.77 | 0.3%±0.14 | 6.3%±0.66 | 6.14%±0.49 | 3.7%±1.19 | 3.4%±1.49 |
| After treatment | ||||||||
| Image 1 | 2.9% | 0.2% | 3.5% | 0.26% | 37% | 35.6% | 45.4% | 40.5% |
| Image 2 | 5.7% | 0.4% | 3.7% | 0.5% | 38.1% | 32.6% | 41% | 40.7% |
| Image 3 | 2.9% | 0.2% | 1.5% | 0.6% | 34% | 36.13% | 44% | 40.4% |
| Average | 3.6%±1.3 | 0.26%±0.09 | 2.9%±1.22 | 0.45%±0.17 | 36.3%±1.7 | 32.7%±1.5 | 43.7%±1.4 | 40.5%±0.12 |
Figure 4:
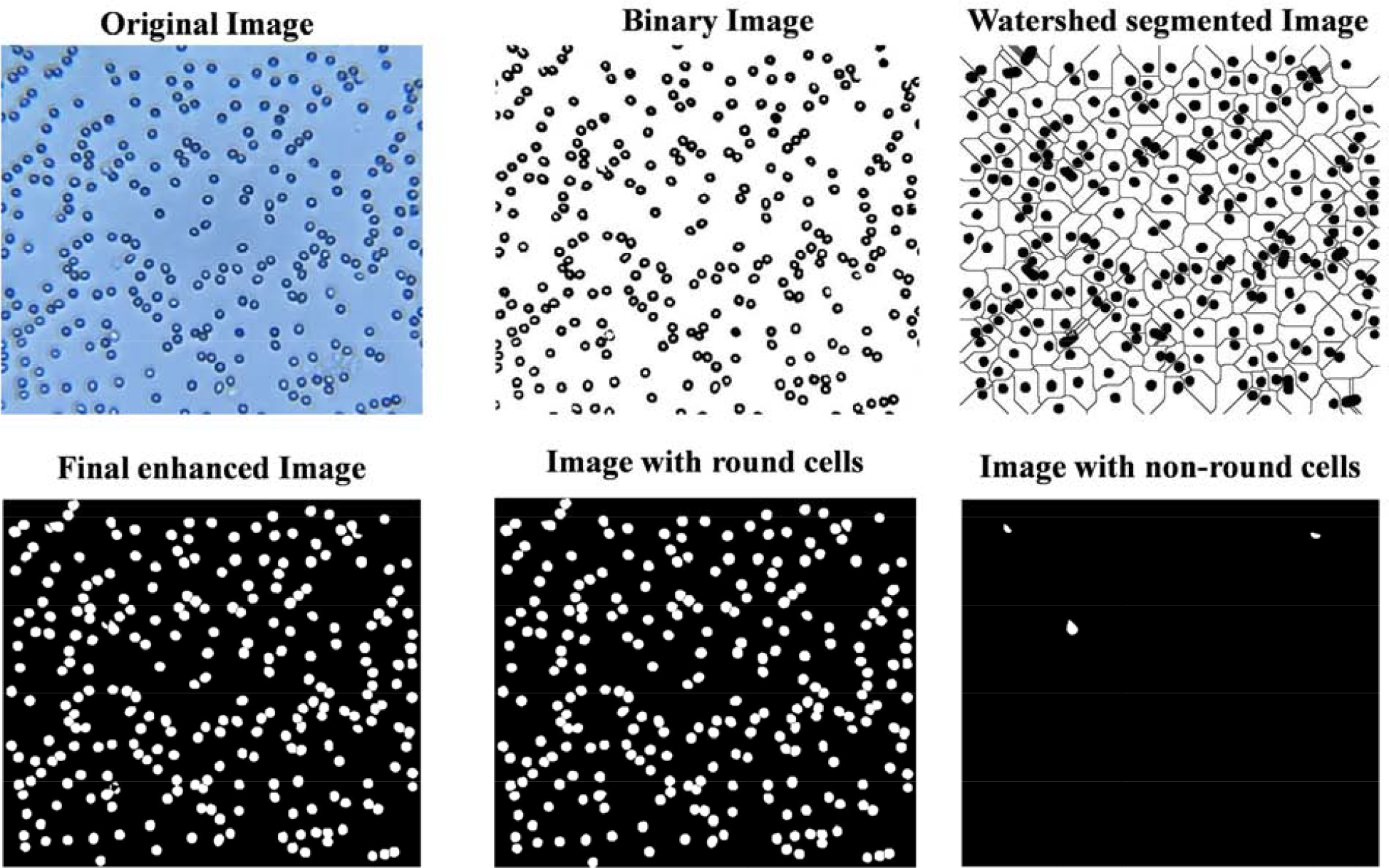
Normal blood cells image captured using smartphone-based setup before treatment.
Following the validation of our developed framework, we performed the same testing technique using blood samples from two SCD patients (patient 1 & patient 2). Three different patches inside the channel were imaged covering a total area of approximately 0.036mm2. The system classified and counted RBCs using the clinical blood samples before deoxygenation and presented a different set of results. An average of 6.1% and 6.3% sickled cells for patient 1 blood sample and an average of 3.4% and 3.7% for patient 2 blood sample indicated the deformation of cells in a very low proportion under normal oxygen levels. A significant number of sickled cells were obtained using post-treatment clinical samples from both the patient 1 & 2. Computing the values of percentages of sickled cells, system gave us an average of 32.7% ± 1.5 and 36.3% ± 1.7 for two types of images (patient 1). The percentage values for patient 2 blood sample were also consistent for microscope and smartphone images (40.5% ± 0.12 and 43.7% ± 1.4 respectively). Using our platform, patient 1 and patient 2 blood samples were classified as SCD positive. Supplementary Figure S2 represents examples of sickle blood sample images using a smartphone-based setup and microscope for the same sample. The percentage of sickled cells after deoxygenation is associated with predictable physiologic and clinical parameters and can be used as a measure of SCD severity and patient’s disease outcome [46]. Our developed system has the potential to be used as a screening tool for SCD and other blood cell disorders in resource-limited settings.
Discussion:
Here, we present the development and evaluation of a smartphone-based optical setup for the detection and quantification of sickled cells using disposable microfluidic chip containing a small volume of blood ~1 μL. The main advantages of using microchip are the ease of utilization, the requirement of only finger prick blood volume and high uniformity of blood cells in microfluidic device. Blood smears are frequently utilized in hematological analysis where the uniformity of the cells may be affected by the expertise of the operator [47]. We imaged the evenly distributed RBCs in the microfluidic chip with our developed smartphone-based setup and compared the results with the optical microscope image results. We have used the blood from normal & SCD patients and captured the images under normal and hypoxic conditions using our smartphone-based setup and microscope. A program written in MATLAB counted number of cells for each kind and classified them based on their shapes to count percentages of sickled cells. The blood samples containing more than a significant number of sickled cells were categorized as being SCD positive. If the average percentage number of sickled cells was insignificant, the sample was classified as normal. Our results are comparable with the results obtained using microscope for the same sample. For example, for patient 1, we obtained a percentage of 6.3% sickled cells under normal conditions and 36.3% sickled cells under hypoxic conditions. Using optical microscope images 6.1% sickled cells before treatment and 32.7% sickled cells after treatment were calculated which are consistent with the results obtained using our developed method. Some other medical health conditions such as hyperthyroidism, abetalipoproteinemia, leukemia, and liver diseases may also induce changes in the shape of RBCs but differential diagnosis based on signs and symptoms rules out inaccuracy of the analysis of results before the laboratory investigations and diagnosis of disease [26, 48–51].
Previous studies have shown a close relationship between sickle red blood cells (SS-RBCs) morphology and hemoglobin polymers aligned inside the cells [52]. The maximum sickled fraction indicated a strong positive correlation with the HbS percentage level during the long-term deoxygenated state [53]. It is proved that there is a clear inverse correlation between the percentage of sickled erythrocytes under hypoxia and fetal hemoglobin (HbF) levels [46]. Percentage of sickled RBCs after deoxygenation are correlated negatively with pH and positively with the presence of long fibers inside the erythrocytes. Environmental factors, such as pH, and patient related factors, such HbF levels are well known to influence the rate of HbS polymer formation [54, 55]. HbS polymer fraction is associated directly with disease severity [56]. Decrease in pH may lead to painful crisis in SCD patients and treatment with alkali administration may be beneficial [57]. Hydroxyurea has been the only Food and Drug Administration (FDA) approved drug to treat SCD in adults from previous two years [58, 59]. It reduces the frequency of painful episodes and enhances the amount of HbF and hemoglobin. Recently, FDA approved Adakveo® (crizanlizumab-tmca) medicine to reduce the frequency of painful crises in adult and pediatric patients with SCD [60–62]. This is an important advancement for people living with this disease. Adakvo binds to a cell adhesion protein called P-selectin that plays a vital role in multicellular interactions that can activate vaso-occlusion [8, 63].There is no useful drug reported to prevent or reverse the polymerization of HbS [64].
This POC technology can potentially be applied to diagnose and improve SCD management in resource-limited settings [26]. Most of the laboratory-based technologies are expensive and limited to testing centers [65]. The developed technology has the potential to reduce the costs significantly for SCD testing and care. The smartphone-based system presented here requires a mass-producible and economical microfluidic device with an optical smartphone attachment with overall cost less than $10. The total material cost to fabricate the microfluidic chip was approximately <$1, which includes $0.1 for PMMA, ~10 cents for the double-sided adhesive (DSA), $0.10 for glass slide. The material cost to fabricate the smartphone attachment was <$7, with ~$1 for the 3D printed smartphone accessory, $5.91 for an LED, $1 for the lenses, and $2 for the battery. Sodium metabisulfite for a single test cost is ~$0.01. This assay is user-friendly and does not require any trained personnel and can potentially be self-performed. A drop of blood can be mixed using disposable transfer pipette with PBS. A 15-minute waiting period is required, after mixing blood with sodium metabisulfite. Loading the sample into microfluidic channel, capturing the image with smartphone, and one click analysis can be performed in less than a minute. The sodium metabisulfite can potentially be dried inside microfluidic device for self-testing purpose if needed.
Early-stage diagnosis and screening of SCD can significantly improve the management of disease, particularly in resource-constrained areas. Critical complications, including stroke, organ damage, blindness, leg ulcers, and others can be prevented if proper management of the disease is done and mortality rate can be drastically reduced. Our system can rapidly quantify the sickled RBCs. Based on its properties, each cell is examined and classified as normal/sickled. This system has potential for screening of SCD and its management.
Conclusion:
We have demonstrated a simple, rapid, and cost-effective smartphone-based SCD detection method. This developed platform utilizes an external lens that can be easily attached to the smartphone camera to record images of various blood samples inside a microchip. The captured images are rapidly processed using a MATLAB program and the total number of sickled cells is automatically counted. To evaluate the performance of our setup, we used normal blood sample as well as the SCD patients’ samples. Our developed image processing algorithm accurately quantified sickled cells in deoxygenated blood. The smartphone-based quantification results were compared and validated with the optical microscope data. With its simple sample preparation combined with small volume, our developed setup is well-suited for resource-limited settings providing remote diagnosis opportunities.
Supplementary Material
Research Highlights:
Simple, reliable, and user-friendly smartphone-based diagnostic platform
Potential utility for sickle cell disease (SCD) detection at POC settings
Enables SCD detection directly from a drop of whole blood
Useful in assessing the SCD severity, and can guide adjusting the dose of medication
Acknowledgments:
We acknowledge research support from NIH R15AI127214, R56AI138659, Institute for Sensing and Embedded Networking Systems Engineering (I-SENSE) Research Initiative Award, FAU Faculty Mentoring Award, Humanity in Science Award, and a start-up research support from College of Engineering and Computer Science, Florida Atlantic University, Boca Raton, FL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest:
The authors declare no competing financial interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References:
- 1.Alapan Y, et al. , Emerging point-of-care technologies for sickle cell disease screening and monitoring. Expert review of medical devices, 2016. 13(12): p. 1073–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Modell B and Darlison M, Global epidemiology of haemoglobin disorders and derived service indicators. Bulletin of the World Health Organization, 2008. 86: p. 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowlton S, et al. , Sickle cell detection using a smartphone. Scientific reports, 2015. 5: p. 15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oyenike MA, et al. , In-vitro anti-sickling and membrane stability potentials of Mishenland polyherbal extract on sickle red blood cells. The Egyptian Journal of Haematology, 2019. 44(1): p. 65. [Google Scholar]
- 5.Liu J, et al. , Electrical impedance microflow cytometry with oxygen control for detection of sickle cells. Sensors and Actuators B: Chemical, 2018. 255: p. 2392–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darrow MC, et al. , Visualizing red blood cell sickling and the effects of inhibition of sphingosine kinase 1 using soft X-ray tomography. J Cell Sci, 2016. 129(18): p. 3511–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, et al. , Electrical impedance characterization of erythrocyte response to cyclic hypoxia in sickle cell disease. ACS sensors, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rees DC, Williams TN, and Gladwin MT, Sickle-cell disease. The Lancet, 2010. 376(9757): p. 2018–2031. [DOI] [PubMed] [Google Scholar]
- 9.Kumar AA, et al. , Evaluation of a density-based rapid diagnostic test for sickle cell disease in a clinical setting in Zambia. PloS one, 2014. 9(12): p. e114540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adenmosun O, Asghar W, and Kumi-Diaka J, Sickle cell sperm selection with Hb-S mab: a future application for intracytoplasmic genotypically selected sperm injection (IGSI). Arch Clin Microbiol, 2017. 8: p. 34–36. [Google Scholar]
- 11.Makani J, et al. , Mortality in sickle cell anemia in Africa: a prospective cohort study in Tanzania. PloS one, 2011. 6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Salem AH, Splenic complications of sickle cell anemia and the role of splenectomy. ISRN hematology, 2010 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gladwin MT and Vichinsky E, Pulmonary complications of sickle cell disease. New England journal of medicine, 2008. 359(21): p. 2254–2265. [DOI] [PubMed] [Google Scholar]
- 14.De Montalembert M and Wang W, Cerebrovascular complications in children with sickle cell disease, in Handbook of clinical neurology. 2013, Elsevier. p. 1937–1943. [DOI] [PubMed] [Google Scholar]
- 15.Lynch JK, Cerebrovascular disorders in children. Current neurology and neuroscience reports, 2004. 4(2): p. 129–138. [DOI] [PubMed] [Google Scholar]
- 16.Vichinsky E, et al. , Newborn screening for sickle cell disease: effect on mortality. Pediatrics, 1988. 81(6): p. 749–755. [PubMed] [Google Scholar]
- 17.Kanter J, et al. , Validation of a novel point of care testing device for sickle cell disease. BMC medicine, 2015. 13(1): p. 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasseri B, et al. , Point-of-care microfluidic devices for pathogen detection. Biosensors and Bioelectronics, 2018. 117: p. 112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y, et al. , A wireless point-of-care testing system for the detection of neuron-specific enolase with microfluidic paper-based analytical devices. Biosensors and Bioelectronics, 2017. 95: p. 60–66. [DOI] [PubMed] [Google Scholar]
- 20.Herrada CA, et al. , Advances in diagnostic methods for Zika virus infection. Journal of medical devices, 2018. 12(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabir MA, et al. , Advances in HIV diagnosis and monitoring. Critical Reviews in Biotechnology, 2020: p. 1–16. [DOI] [PubMed] [Google Scholar]
- 22.Dona G, et al. Electrical Equivalent Circuit Model of Sickle Cell. in ASME 2017International Mechanical Engineering Congress and Exposition. 2018. American Society of Mechanical Engineers Digital Collection. [Google Scholar]
- 23.Sher M, et al. , based analytical devices for clinical diagnosis: recent advances in the fabrication techniques and sensing mechanisms. Expert review of molecular diagnostics, 2017. 17(4): p. 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Land KJ, et al. , REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nature microbiology, 2019. 4(1): p. 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steele C, et al. , Point-of-care screening for sickle cell disease in low-resource settings: A multi-center evaluation of HemoTypeSC, a novel rapid test. American journal of hematology, 2019. 94(1): p. 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilyas S, Simonson AE, and Asghar W, Emerging Point-of-Care Technologies for Sickle Cell Disease Diagnostics. Clinica Chimica Acta, 2019. [DOI] [PubMed] [Google Scholar]
- 27.Natesan M, et al. , A smartphone-based rapid telemonitoring system for Ebola and Marburg disease surveillance. ACS sensors, 2018. 4(1): p. 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neill DL, et al. , A smartphone based device for the detection of sulfane sulfurs in biological systems. Sensors and Actuators B: Chemical, 2019. 292: p. 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calabretta MM, et al. , Nano-lantern on paper for smartphone-based ATP detection. Biosensors and Bioelectronics, 2020. 150: p. 111902. [DOI] [PubMed] [Google Scholar]
- 30.Coleman B, et al. , Point-of-care colorimetric analysis through smartphone video. Sensors and Actuators B: Chemical, 2019. 282: p. 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman B, Coarsey C, and Asghar W, Cell phone based colorimetric analysis for point-of-care settings. Analyst, 2019. 144(6): p. 1935–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabir MA, et al. , Development of a flow-free automated colorimetric detection assay integrated with smartphone for Zika NS1. Diagnostics, 2020. 10(1): p. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snow JW, et al. , Rapid imaging, detection, and quantification of Nosema ceranae spores in honey bees using mobile phone-based fluorescence microscopy. Lab on a Chip, 2019. 19(5): p. 789–797. [DOI] [PubMed] [Google Scholar]
- 34.Shrivastava S, Lee W-I, and Lee N-E, Culture-free, highly sensitive, quantitative detection of bacteria from minimally processed samples using fluorescence imaging by smartphone. Biosensors and Bioelectronics, 2018. 109: p. 90–97. [DOI] [PubMed] [Google Scholar]
- 35.Asghar W, et al. , Microfluidic Chip for Detection of Fungal Infections. ACS omega, 2019. 4(4): p. 7474–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rappa K, et al. , Quantitative analysis of sperm rheotaxis using a microfluidic device. Microfluidics and Nanofluidics, 2018. 22(9): p. 100. [Google Scholar]
- 37.Sher M and Asghar W, Development of a multiplex fully automated assay for rapid quantification of CD4+ T cells from whole blood. Biosensors and Bioelectronics, 2019. 142: p. 111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coarsey C, et al. , Development of a flow-free magnetic actuation platform for an automated microfluidic ELISA. RSC advances, 2019. 9(15): p. 8159–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castro D and Keenaghan M, Arterial Blood Gas. 2019. [PubMed] [Google Scholar]
- 40.Pauline N, et al. , The in vitro antisickling and antioxidant effects of aqueous extracts Zanthoxyllum heitzii on sickle cell disorder. BMC Complementary and Alternative Medicine, 2013. 13(1): p. 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torabian K, et al. , Substituting sodium hydrosulfite with sodium metabisulfite improves long-term stability of a distributable paper-based test kit for point-of-care screening for sickle cell anemia. Biosensors, 2017. 7(3): p. 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chikezie PC, Sodium metabisulfite–induced polymerization of sickle cell hemoglobin incubated in the extracts of three medicinal plants (Anacardium occidentale, Psidium guajava, and Terminalia catappa). Pharmacognosy magazine, 2011. 7(26): p. 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oleiwi SR, Humaidi AJ, and Fadhel MA, Image Processing-Based Diagnosis of Sickle cell Anemia in Erythrocytes. [Google Scholar]
- 44.Kanthathas K, Willmot D, and Benson P, Differentiation of developmental and post-orthodontic white lesions using image analysis. The European Journal of Orthodontics, 2005. 27(2): p. 167–172. [DOI] [PubMed] [Google Scholar]
- 45.Ritter N and Cooper J, New resolution independent measures of circularity. Journal of Mathematical Imaging and Vision, 2009. 35(2): p. 117–127. [Google Scholar]
- 46.Van Beers EJ, et al. , Imaging flow cytometry for automated detection of hypoxia-induced erythrocyte shape change in sickle cell disease. American journal of hematology, 2014. 89(6): p. 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y-C, et al. Lens-free holographic microscopy for complete blood analysis on a chip. in Optical Diagnostics and Sensing XX: Toward Point-of-Care Diagnostics. 2020. International Society for Optics and Photonics. [Google Scholar]
- 48.Dodge JT, et al. , Peroxidative hemolysis of red blood cells from patients with abetalipoproteinemia (acanthocytosis). The Journal of clinical investigation, 1967. 46(3): p. 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viana MB, Anemia and infection: a complex relationship. Revista brasileira de hematologia e hemoterapia, 2011. 33(2): p. 90–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spaulding SW and Lippes H, Hyperthyroidism. Causes, clinical features, and diagnosis. The Medical clinics of North America, 1985. 69(5): p. 937–951. [DOI] [PubMed] [Google Scholar]
- 51.Smith JA, Lonergan ET, and Sterling K, Spur-cell anemia: hemolytic anemia with red cells resembling acanthocytes in alcoholic cirrhosis. New England Journal of Medicine, 1964. 271(8): p. 396–398. [DOI] [PubMed] [Google Scholar]
- 52.Lei H and Karniadakis GE, Predicting the morphology of sickle red blood cells using coarse-grained models of intracellular aligned hemoglobin polymers. Soft Matter, 2012. 8(16): p. 4507–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du E, et al. , Kinetics of sickle cell biorheology and implications for painful vasoocclusive crisis. Proceedings of the National Academy of Sciences, 2015. 112(5): p. 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afolabi IS, et al. , Solenostemon monostachyus, Ipomoea involucrata and Carica papaya seed oil versus Glutathione, or Vernonia amygdalina: Methanolic extracts of novel plants for the management of sickle cell anemia disease. BMC complementary and alternative medicine, 2012. 12(1): p. 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christoph GW, Hofrichter J, and Eaton WA, Understanding the shape of sickled red cells. Biophysical journal, 2005. 88(2): p. 1371–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brittenham GM, Schechter AN, and Noguchi CT, Hemoglobin S polymerization: primary determinant of the hemolytic and clinical severity of the sickling syndromes. Blood, 1985. 65(1): p. 183–189. [PubMed] [Google Scholar]
- 57.EH K, Studies on the destruction of red blood cells. XIII. Observations on the role of pH in the pathogenesis and tr eatment of painful crisis in sickle-cell disease. [DOI] [PubMed] [Google Scholar]
- 58.Agrawal RK, et al. , Hydroxyurea in sickle cell disease: drug review. Indian Journal of Hematology and Blood Transfusion, 2014. 30(2): p. 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Food U and Administration D, FDA approves hydroxyurea for treatment of pediatric patients with sickle cell anemia. [Google Scholar]
- 60.Prime B, New Novartis medicine Adakveo®(crizanlizumab-tmca) approved by FDA to reduce frequency of pain crises in individuals living with sickle cell disease. [Google Scholar]
- 61.Euronext N, New Novartis medicine Adakveo®(crizanlizumab) approved by FDA to reduce frequency of pain crises in individuals living with sickle cell disease. [Google Scholar]
- 62.Anlage E, Kolumnen E, and Newswires DJ, New Novartis medicine Adakveo®(crizanlizumab) approved by FDA to reduce frequency of pain crises in individuals living with sickle cell disease. [Google Scholar]
- 63.Lawrence MB and Springer TA, Leukocytes roll on a selectin at physiologic flow rates: distinction from and prerequisite for adhesion through integrins. Cell, 1991. 65(5): p. 859–873. [DOI] [PubMed] [Google Scholar]
- 64.Halsey C and Roberts IA, The role of hydroxyurea in sickle cell disease. British journal of haematology, 2003. 120(2): p. 177–186. [DOI] [PubMed] [Google Scholar]
- 65.Bond M, et al. , Towards a point-of-care strip test to diagnose sickle cell anemia. PloS one, 2017. 12(5): p. e0177732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


