Abstract
The rapid expansion of machine learning is offering a new wave of opportunities for nuclear medicine. This paper reviews applications of machine learning for the study of attenuation correction (AC) and low-count image reconstruction in quantitative positron emission tomography (PET). Specifically, we present the developments of machine learning methodology, ranging from random forest and dictionary learning to the latest convolutional neural network-based architectures. For application in PET attenuation correction, two general strategies are reviewed: 1) generating synthetic CT from MR or non-AC PET for the purposes of PET AC, and 2) direct conversion from non-AC PET to AC PET. For low-count PET reconstruction, recent deep learning-based studies and the potential advantages over conventional machine learning-based methods are presented and discussed. In each application, the proposed methods, study designs and performance of published studies are listed and compared with a brief discussion. Finally, the overall contributions and remaining challenges are summarized.
1. INTRODUCTION
Positron emission tomography (PET) has been used as a non-invasive functional imaging modality with a wide range of clinical applications. By providing the information of metabolic processes in the human body, it is utilized for various purposes including tumor staging and detection of metastases in oncology [1–9], gross target volume definition in radiation oncology [10–12], myocardial perfusion in cardiology [13, 14], and investigation of neurological disorders [15]. Among these applications, the accuracy of tracer uptake quantification has been less recognized than other characteristics of PET such as sensitivity [16]. Recently, with the focus shifting towards precision medicine, it is of great clinical interest to accurately quantify tracer uptake towards expansion of PET in more demanding applications such as therapeutic response monitoring [17–19], and treatment outcome prediction as a prognostic factor [20].
Long-standing challenges in quantitative PET accuracy remain such as degraded image quality caused by inaccurate photon attenuation and scattering [21, 22], low count statistics [23, 24], and the partial-volume effect [25–27]. Improperly addressing these challenges can result in bias, uncertainty and artifacts in the PET images, which can degrade the utility of both qualitative and quantitative assessments. With the introduction of advanced PET scanners and applications in the recent years, such as Magnetic Resonance (MR)-combined PET (PET/MR), low-count scanning protocol and high-resolution PET, these novel PET technologies aim to incorporate anatomical imaging modality for better soft tissue visualization, reduce administered activity and shorten scan time of standalone PET and Computed tomography (CT)-combined PET (PET/CT), and increase detection capability on radiopharmaceutical accumulation in structures of millimeter size, all of which are highly desirable for clinical practice. On the other hand, unconventional or suboptimal scan schemes are proposed to achieve these clinical benefits, which bring new technical barriers that have not been seen in conventional PET/CT systems. It is important to address these problems without compromising the quantitative capabilities of PET.
Many methods have been proposed to deal with the inherent deficiencies of PET and are still being improved to tackle the difficulties emerging from these novel implementations. For example, before PET/CT was widely implemented, transmission scanning with an external positron emitting source (e.g. Ge-68) rotating around the patient was used to determine noisy but accurate 511keV linear attenuation coefficient maps of the patient body for attenuation correction [28]. With the addition of CT images in PET/CT, 511keV linear attenuation coefficient maps can be computed by a piecewise linear scaling algorithm reducing the noise contribution compared to Ge-68 transmission scans in the reconstruction process [21, 29, 30]. In recent years, the incorporation of MR with PET has become a promising alternative to PET/CT due to the availability of its excellent anatomical soft tissue visualization without ionizing radiation. However, MR voxel intensity is related to proton density rather than electron density, thus it cannot be directly converted to 511 keV linear attenuation coefficient maps for use in the AC process. To overcome this barrier, conventional methods have assigned piecewise constant attenuation coefficients on MR images based on the segmentation of tissues [31, 32]. The segmentation can be done by either manually-drawn contours [33] or automatic classification methods [34–36]. However, these methods are limited by misclassification and inaccurate prediction of bone and air regions due to their ambiguous relationships in MR. Instead of segmentation, other methods have warped atlases of MR images labeled with known attenuation factors to patient-specific MR images by deformable registration or pattern recognition but their efficacy highly depends on the performance of the registration algorithm. Moreover, the atlases are usually created on normal anatomy and cannot represent anatomic abnormalities seen in clinical practice [37, 38].
Low-count PET protocols aim to reduce the administrated activity, which is attractive for it clinical utility [39]. The reduction in radiation dose is desirable in the pediatric population since accumulated imaging dose can be a big concern for younger patients who are more sensitive to radiation [40, 41]. Reducing administered activity can also help patients undergoing radiation therapy who have multiple serial PET scans as part of their pretreatment and radiotherapy response evaluation [42, 43]. Considerations should be given to the need for optimal imaging and minimization of patient dose in order to reduce the potential risk of secondary cancer [44]. Secondly, shortened scan time is beneficial for minimizing patient motion as well as potentially increasing patient throughput. More importantly, in dynamic PET imaging employing a sequence of time frames, counts at each frame are much lower than those in static PET imaging resulting in increased image noise, reduced contrast-to-noise ratio, and measurement bias [16]. Dynamic PET imaging aims to provide a voxel-wise parametric analysis for kinetic modeling and the accuracy highly depends on the image quality [45]. However, the low count statistics would result in increased image noise, reduced contrast-to-noise ratio, and large bias in uptake measurement [16]. The immunoPET introduced recently also encounters low-count issue since the commonly administered activity is about 37 to 75 MBq of 89Zr [46, 47], which is much lower than 370 MBq used for 18F-FDG of a standard PET. Both hardware-based and software-based solutions have been proposed for the low-count PET scanning. Advances in PET instruments such as lutetium-based detectors, silicon photomultipliers and time-of-flight compatible scanners are able to considerably increase the acquisition efficiency thus lower the measurement uncertainty [48, 49]. Meanwhile, software-based post-processing and the usage of noise regularization in reconstruction would penalize the differences among neighboring pixels in order to have a smooth appearance [50–52].
Hardware advances have mainly focusing on the developments in the scintillator and photodetector technologies, which have substantially improved the performance of PET systems [53, 54]. Paired with software improvements, particularly novel image processing and reconstruction algorithms, software advances may complement hardware improvements and be easier to implement at a lower cost.
Inspired by the rapid expansion of artificial intelligence in both industry and academia in recent years, many research groups have attempted to integrate machine learning-based methods into medical imaging and radiation therapy [55–59]. The common machine learning applications include detection, segmentation, characterization, reconstruction, registration and synthesis [60–66]. Before the permeation of artificial intelligence into these sub-fields, conventional image processing methods have been developed for decades. The conventional algorithms are usually very different from each other in many aspects such as workflow, assumptions, complexity, requirements on prior knowledge, and the detailed implementation highly depends on the task [67–71]. Compared with conventional methods, machine learning-based methods share a general framework using a data-driven approach. Supervised learning workflows usually consists of a training stage, i.e. a machine learning model is trained by the training datasets to find the patterns between the input and its training target; and a predication stage, where the trained model maps an input to an output. Unlike the conventional methods which usually require case-by-case parameter tuning for optimal performance, machine-learning based methods are more robust, of which the effectiveness largely depends on the representativeness of the training datasets used, the designed network architecture and hyperparameter settings.
Recently, there are review papers generally summarizing the current studies of machine learning and PET scanning [72, 73]. In this paper, we reviewed the emerging machine learning-based methods for quantitative PET imaging. Specifically, we reviewed the general workflows and summarized the recently published studies of learning-based methods in dealing with PET AC and low-count PET reconstruction in the following sections respectively, and briefly introduced the representative ones among them with a discussion on the trend and future directions at the end of each section. Note that the degrading PET effects such as scattering and partial volume are not reviewed in this study but also play an important role in PET quantification accuracy. The PET scans in these reviewed literatures used glucose analogue 2-18F-fluoro-2-deoxy-D-glucose (FDG) for uptake if not explicitly stated otherwise.
2. PET AC
As mentioned in the introduction, CT is able to provide the attenuation coefficient maps to correct for the loss of annihilation photons due to the attenuation process in the patient body. However, in a PET-only scanner or a PET/MR scanner the absence of CT images causes difficulty in attenuation correction. Many studies have proposed schemes to address this issue and machine learning is involved in different forms with different purposes. The general workflow can be divided into two pathways depending on whether anatomical images were acquired (MR in PET/MR) or not (PET-only scanner). Most proposed methods have been implemented on PET brain scans, with a few on pelvis and whole body studies. The ground truth for training and evaluation was PET with AC by CT images. Relative bias in percentage from ground truth in selected volumes-of-interest (VOIs) were usually reported to evaluate the quantification accuracy. Several studies compared their proposed methods with conventional MR-based PET AC methods such as atlas-based methods and segmentation-based methods.
2.1. Synthetic CT for PET/MR
For a PET/MR scanner, the most common strategy is to generate synthetic CT (sCT) images from MR and use the sCT in the PET AC procedure The synthesis methods among images of different modalities have been developed in recent years [74–87]. Many studies have investigated the feasibility of using sCT for PET AC in brain and whole body imaging, which are summarized in Table I. Examples of AC PET using learning-based methods for brain are shown in Figure 1 which is redrawn based on the method proposed in reference [88]. AC PET images by MR using learning-based methods show a very similar appearance and intensity scale with ground truth. The bias among 11 VOIs as selected in reference [88] ranged from −1.72% to 3.70%, with a global mean absolute error of 2.41%. The AC PET by MR successfully maintained the relative contrast among different brain regions, which can be helpful for diagnostic purposes, although the improvement on diagnostic accuracy is difficult to quantify based on current studies [38, 89].
Table I.
Overview of learning-based PET AC methods.
| Methods and strategy | PET or PET/MR | Site | # of patients in training/testing datasets | Reported bias+ (%) | Authors |
|---|---|---|---|---|---|
| Deep convolutional autoencoder (CAE) network MR->tissue class (bone, soft-tissue, air) | PET/MR (3T Postcontrast T1-weighted) | Brain | 30 train/ 10 test | −0.7±1.1 (−3.2, 0.4) among 23 VOls | Liu et al., 2018 [90] |
| Gaussian mixture regression MR->sCT | PET/MR (T2-weighted+ultrashort echo time) | Brain | N.A.*/9 test | −1.9±4.1 −61, 34) in global | Larsson et al., 2013 [91] |
| Support Vector Regression MR->sCT | PET/MR (UTE and Dixon-VlBE) | Brain | 5 train/ 5 test | 2.16±1.77 (1.32, 3.45) among 13 VOIs |
Navalpakkam et al., 2013 [92] |
| Alternating random forests MR->sCT | PET/MR (T1-weighted MP-RAGE) | Brain | 17 leave-one-out | (−1.61, 3.67) among 11 VOIs | Yang et al., 2019 [88] |
| Unet MR->sCT | PET/MR(ZTE) | Brain | 23 train/ 47 test | −0.2(−1.8, 1.7) among 70 VOIs | Blanc-Durand et al., 2019 [94] |
| Unet MR->sCT | PET/MR 3T ZTE and Dixon |
Pelvis | 10 train/ 26 test | −1.11±2.62 in global | Leynes et al., 2018 [95] |
| Unet MR->sCT | PET/MR Dixon |
Pelvis | N.A./19 patients with 28 scans in test. | −0.95±5.09 in bone −0.03±2.98 in soft tissue 0.27±2.59 in fat |
Torrado-Carvajal et al., 2019 [96] |
| Unet MR->sCT | PET/MR (1.5T T1-weighted) | Brain | 44 train/ 11 validation/ 11 test | −0.49±1.711C-WAY-100635 PET −1.52±0.73 11C-DASB in global |
Spuhler et al., 2019 [98] |
| Unet MR->sCT | PET/MR (ZTE and Dixon) | Brain | 14 leave-two-out | Absolute error (1.5%−2.8%) among 8 VOIs | Gong et al., 2018 [99] |
| Unet MR->sCT | PET/MR (UTE) | Brain | 79 (pediatric) 4-fold cross validation | −0.1(−0.2, 0.5) in 95%CI among all tumor volumes | Ladefoged et al., 2019 [97] |
| Deep learning adversarial semantic structure (DL-AdvSS) MR->sCT |
PET/MR (3T T1 MP-RAGE) | Brain | 40 2-fold cross validation | <4 Among 63 VOIs | Arabi et al., 2019 [100] |
| Hybrid of CAE and Unet NAC PET->sCT->AC PET | PET (18F-FP-CIT) | Brain | 40 5-fold cross validation | About −8, −4) among 4 VOIs | Hwang et al., 2018 [103] |
| Unet NAC PET-> sCT->AC PET | PET | Brain | 100 train/ 28 test | −0.64±1.99 −4.18, 2.22) among 21 regions | Liu et al., 2018 [104] |
| Generative adversarial networks (GAN) NAC PET->sCT->AC PET |
PET | Brain | 50 train/ 40 test | −2.5, 0.6) among 7 VOIs | Armanious et al., 2019 [105] |
| Cycle-consistent GAN NAC PET->sCT->AC PET | PET | Whole body | 80 train/ 39 test | (−1.06,10.72) among 7 VOIs 1.07±9.01 in lesion |
Dong et al., 2019 [106] |
| Unet NAC PET->AC PET | PET | Brain | 25 train/ 10 test | 4.0±15.4 among 116 VOIs | Yang et al., 2019 [108] |
| Unet NAC PET->AC PET | PET | Brain | 91 train/ 18 test | −0.10±2.14 among 83 VOIs | Shiri et al., 2019 [109] |
| Cycle-consistent Generative adversarial networks (GAN) NAC PET->AC PET | PET | Whole body | 25 leave-one-out+ 10 patients*3 sequential scan tests | −17.02,3.02) among 6 VOIs, 2.85±5.21 in lesions | Dong et al., 2019 [107] |
N.A.: not available, i.e. not explicitly indicated in the publication
Numbers in parentheses indicate minimum and maximum values.
Figure 1.
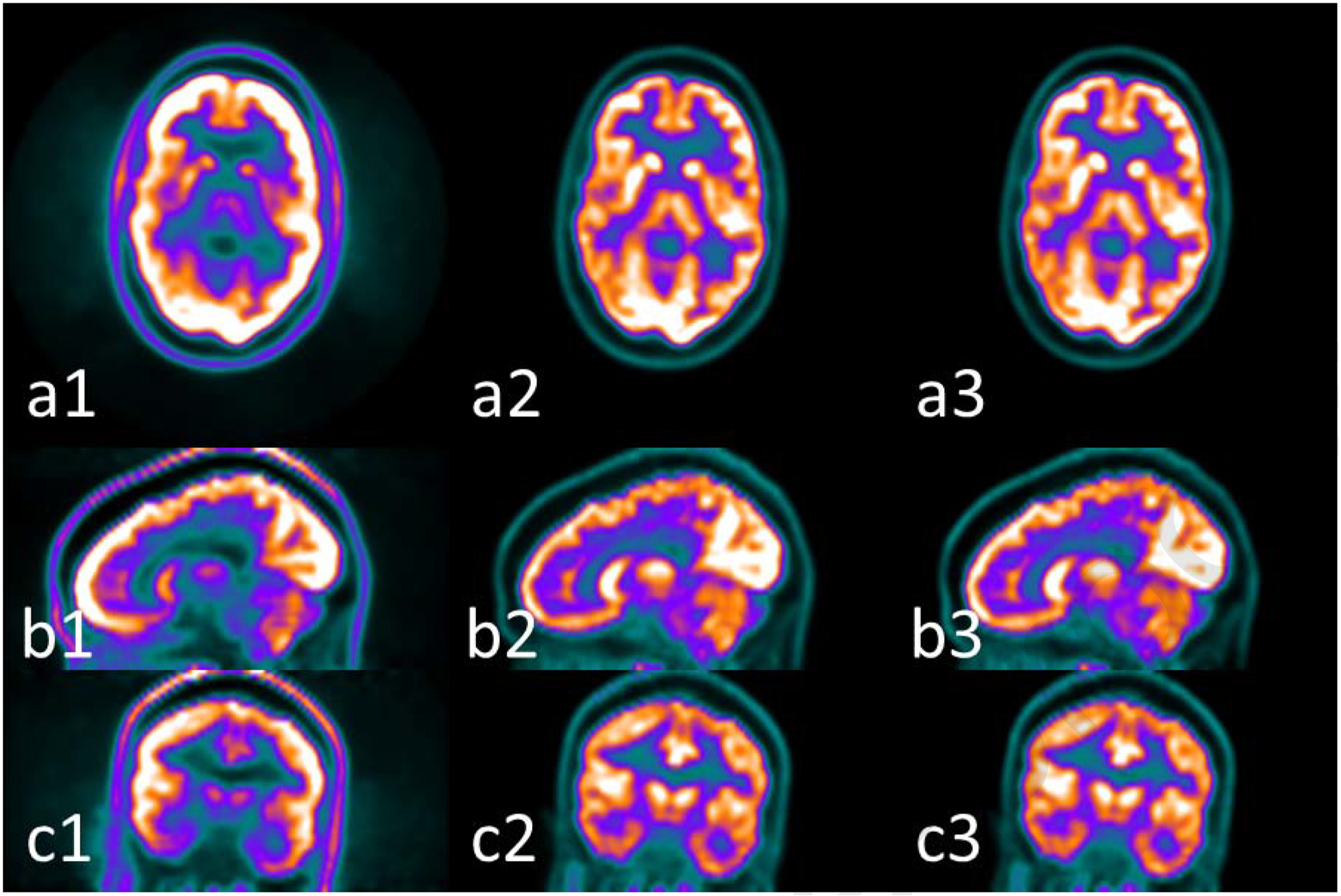
Examples of PET images (1) without AC, (2) with AC by CT and (3) with AC by MRI using the learning-based method presented in reference [88] at (a) axial, (b) sagittal and (c) coronal views. The window width for (1) is [0 5000], and for (2) and (3) is [0 30000] in units of Bq/ml.
Liu et al. [90] proposed to generate the sCT by assigning the CT numbers to the corresponding tissue labels segmented on MR. They used convolutional auto-encoder (CAE), which consisted of a connected encoder and decoder network, to generate CT tissue labels from T1-weighted MR images. The encoder and decoder probe the image features and reconstruct piecewise tissue labels, respectively. They reported a mean bias of −0.7±1.1 % among all selected VOIs in brain iwith PET AC using their sCT, which was significantly better in most of the VOIs compared to the Dixon-based soft-tissue/air segmentation and anatomic CT-based template registration. A limitation of this strategy is its requirement of labelling on training datasets and the small number of classified tissue types.
sCT can also be generated through a direct mapping from MR. Gaussian mixture regression [91], support vector regression [92] and random forest [88] have been used before deep learning was implemented. For example, Yang et al. proposed a random forest-based method to train a set of decision trees. Each decision tree learned the optimal way to separate a set of paired MRI and CT training patches into smaller and smaller subsets to predict CT intensity. When a new MRI patch is put into the model, the sCT intensity can be estimated as the combination of the predicted results of all decision trees. They used a sequence of alternating random forests under the framework of an iterative refinement model to consider both the global loss of the training model and the uncertainty of the training data falling into child nodes with a combination of discriminative feature selection [88]. The authors reported good agreement of brain AC PET with bias ranging from −1.61% to 3.67% among all selected regions. Since deep learning methods have proven to be successful with image style transfer in the computer vision field, sCT from MR can be generated by many off-the-shelf algorithms. One of the most popular networks is Unet and its variants [93]. These convolutional neural network (CNN)-based methods aim to minimize the loss function that includes the pixel-intensity and pixel-gradient difference between training the target CT and generated sCT. The implementation and results of Unet in PET AC have been reported for brain in reference [94], for pelvis [95, 96], for pediatric brain tumor patients [97] and for neuroimaging with 11C-labeled tracers [98]. In order to incorporate multiple MR images from different sequences as input, Gong et al. modified Unet by replacing the traditional convolution module using a group convolutional module to preserve the network capacity while restricting the network complexity [99]. This modification successfully lowered the systematic bias and errors in all regions, which enabled the proposed method to outperform a conventional Unet.
Apart from CNN-based methods, the generative adversarial network (GAN) has also been explored in generating sCT for PET AC. GAN has a generative network and a discriminative network that are trained simultaneously. With MR as input and sCT as output, the discriminative network distinguishes the sCT from training CT images. It can be formulated as solving a minimization problem of the discriminative loss, in addition to pixel-intensity loss and pixel-gradient loss to improve sCT image quality. Arabi et al. proposed a deep learning adversarial semantic structure that combined a synthesis GAN and a segmentation GAN [100]. The segmentation GAN segmented the generated sCT into air cavities, soft tissue, bone and background air to regularize the sCT generation process by back-propagating gradients. The proposed method reported a mean bias of less than 4% in selected VOIs with brain PET, which was slightly worse than atlas-based methods (up to 3%) but better than segmentation-based methods (up to −10%).
2.2. PET AC for PET-only
In a PET-only scanner, anatomical images such as MR are not available. The maximum-likelihood reconstruction of activity and attenuation (MLAA) algorithms are able to simultaneously reconstruct activity and attenuation distributions from the PET emission data by the incorporation of time-of-flight information [101]. However, insufficient timing resolution of clinical PET systems leads to slow convergence, high noise levels the in attenuation maps, and substantial noise propagation between activity and attenuation maps [102]. Recently, it has been shown that the non-attenuation-corrected (NAC) PET could be used to generate sCT by the powerful image style transfer ability of deep learning methods. Similar to the sCT generation from MR, the sCT images are generated from the NAC PET images using a machine learning model trained by pairs of NAC PET and CT images that are acquired from a PET/CT scanner. Again, Unet and GAN are the two common networks adopted for this application [103–106]. Among these studies, learning-based PET AC from whole body PET imaging was investigated by Dong et al. for the first time [106]. A cycle-consistent GAN (CycleGAN) method that combined a self-attention Unet for the generator architecture and a fully convolutional network for the discriminator architecture was employed. The method learned a transformation that minimized the difference between sCT, generated from NAC PET, and the original CT. It also learned an inverse transformation such that the cycle NAC PET images generated from the sCT were close to original NAC PET images. A self-attention strategy was utilized to identify the most informative component and mitigate the disturbance of noise. The average bias of the proposed method on selected organs and lesions ranged from −1.06% to 3.57% except for lungs (10.72%).
The above strategy still requires the step of PET reconstruction using the sCT following the mapping step. Thus, attempts were made to directly map the AC PET from the NAC PET images by exploiting the deep learning methods to bypass the PET reconstruction step. Yang et al. and Shiri et al. both proposed Unet-based methods in brain PET AC and demonstrated the feasibility [108, 109]. Dong et al. again investigated whole body PET direct AC using a supervised 3D patch-based CycleGAN [107]. The CycleGAN had a NAC-to-AC PET mapping and an inverse AC-to-NAC PET mapping strategy in order to constrain the NAC-to-AC mapping to approach a one-to-one mapping. Since NAC PET images have similar anatomical structures to the AC PET images but lack contrast information, residual blocks were integrated into the network to learn the differences between NAC PET and AC PET. They reported the average bias of the proposed method on selected organs and lesions ranging from 2.11% to 3.02% except for lungs (−17.02%). A representative result is shown in Figure 2 which is redrawn based on the method presented in reference [107]. When comparing with the results of Unet and GAN training and testing on the same datasets, the proposed CycleGAN method achieved a superior performance in most metrics evaluated and less bias in lesions.
Figure 2.
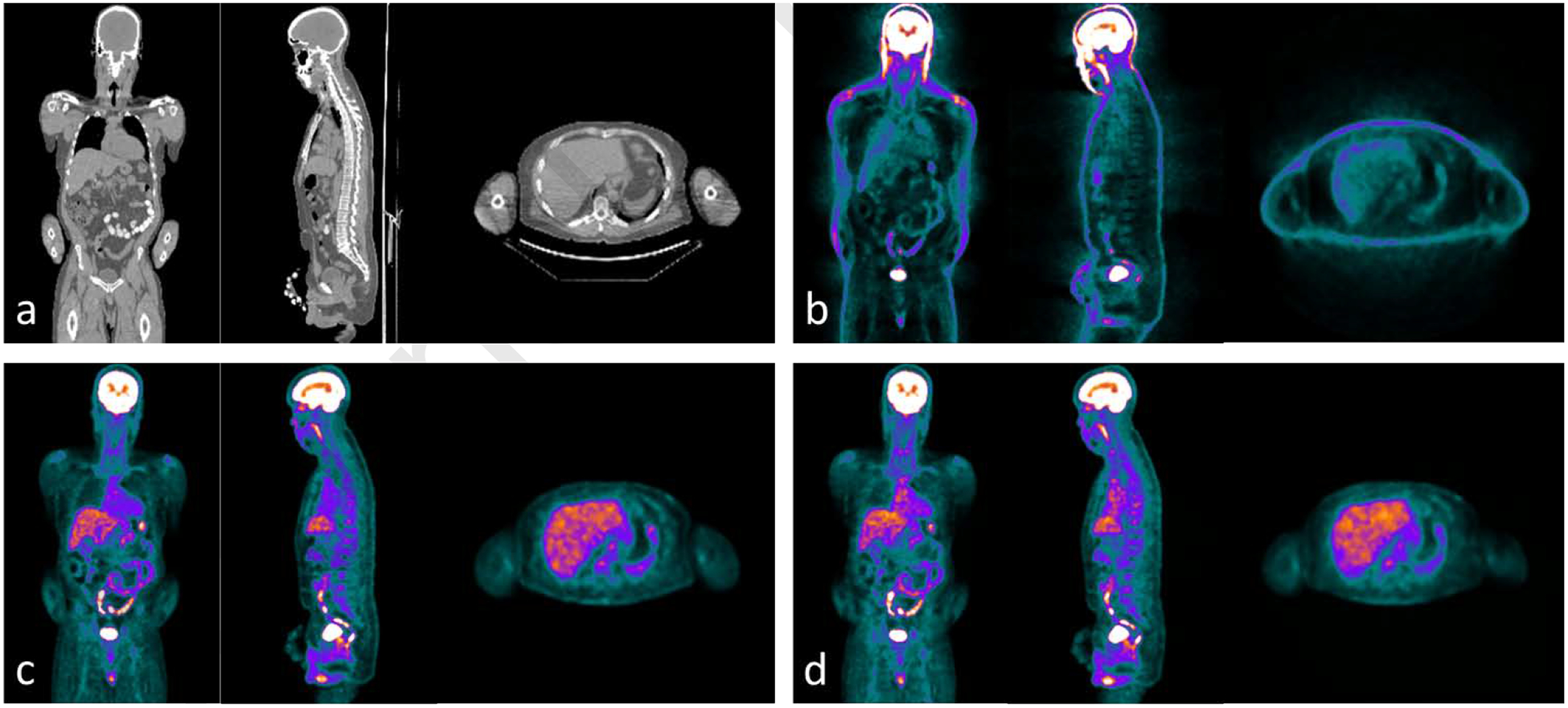
Examples of whole body (a) CT images and PET images (b) without AC, (c) with AC by CT in (a), and (d) synthetic AC PET by learning-based method presented in reference [107] . The window width for (a) are [−300 300] HU, (b) are [0 1000] and for (c) and (d) are [0 10000] with PET units in Bq/ml.
2.3. Discussion
Although it is difficult to specify the tolerance level of quantification errors before they affect clinical judgement, the general consensus is that quantification errors of 10% or less typically do not affect diagnosis [38]. Based on the average relative bias represented by these studies, almost all of the proposed methods in these studies meet this criterion. However, it should be noted that due to the variation among study objects, the bias in some VOIs on a per patient basis may exceed 10% [94, 95, 103, 105–108]. As reported by Armanious et al., under-estimation of about 10% to 20% was observed in small volumes around air and bone structures such as paranasal sinuses and mastoid cells [105]. Hwang et al. reported under-estimation over 10% for a patient around occipital cortex [103] , which may result in a loss of sensitivity for diagnosis of neurodegenerative diseases if cortical hypometabolism occurred in that region. Leynes et al. also reported under-estimation of a soft tissue lesion over 10% for a patient [95]. The under-estimation on the uptake in the lesion may result in misidentification and introduce errors in the definition of shape and position of the lesion for radiation therapy treatment planning, as well as in the evaluation of treatment response. It suggests that special attention should be given to the standard deviation of bias as well as its mean when interpreting results since the proposed methods may have poor local performance that could affect some patients. On the other hand, listing or plotting the results of every data point measure, or at least their range, instead of simply giving a mean and standard deviation would be more informative in demonstrating the performance of the proposed methods. Moreover, compared with static PET, dynamic PET may require higher uptake quantification accuracy since the kinetic parameters derived from it can be more sensitive to uptake reconstruction error than the conventional use of static PET due to the nonlinearity and identifiability of the kinetic models [110].
Various learning-based approaches have been proposed in the reviewed studies. However, the reported errors among these studies cannot be fairly compared because of the use of different datasets, evaluation methodology, and reconstruction parameters. Thus, it is impracticable to choose the best method based on the reported performance in these studies. Some studies compared their proposed methods with others using same datasets, which may reveal the advantages and limitations of the methods selected in comparison. For example, many studies compared their proposed methods with conventional segmentation-based or atlas-based methods, and almost all the learning-based methods gain the upper hand exhibiting less bias on average with less variation among patients and VOIs. The comparison among different learning-based methods is much less common probably because these methods have been just published in the last two years. As mentioned above, Dong et al. compared the CycleGAN, GAN and Unet in direct NAC PET-AC PET mapping on whole body PET images and demonstrated the superiority of CycleGAN over other learning-based approaches owing to the addition of inverse mapping from AC PET to NAC PET that degenerated the ill-posed interconversion between AC PET and NAC PET to be a one-to-one mapping [107].
Among the above PET/MR studies, different types of MR sequences have been attempted for sCT generation. The specific MR sequence used in each study usually depends on the accessibility. The optimal sequence yielding the best PET AC performance has not been studied. T1-weighted and T2-weighted sequences are the two most common MR sequences used in standard of care. Due to their wide availability, sCT model can be trained from a relatively large number of datasets with CT and co-registered T1- or T2-weighted MR images regardless of PET acquisition. Thus, they have been utilized in multiple studies where the MR images in training datasets are not acquired with PET [88, 90, 91, 98, 100]. However, air and bone have little contrast in T1- or T2-weighted MR images, which may impede the extraction of the corresponding features in learning-based methods. The two-point Dixon sequence can separate water and fat, which is suitable for segmentation. It has already been applied in commercial PET/MR scanners integrated with volume-interpolated breath-hold examination (VIBE) for Dixon-based soft-tissue and air segmentation of PET AC attenuation maps [111, 112]. Its drawback is again the poor contrast of bone, which results in the misclassification of bone as fat. Learning-based approaches have been employed on Dixon MR images by Torrado-Carvajal et al.[96] They compared the performance of a Unet-based method with the vendor-provided segmentation based method on the same datasets. The relative bias of the proposed Unet-based method was 0.27±2.59%, −0.03±2.98% and −0.95±5.09% for fat, soft-tissue and bone, respectively, while it was 1.48±6.51%, −0.34±10.00% and −25.1±12.71% for segmentation-based methods. The learning-based methods significantly reduced the bias in bone with a reduced variance in all VOIs among all patients, which indicates its superior capability in PET AC due to improved bone identification. In order to enhance the bone contrast to facilitate the feature extraction in learning-based methods, ultrashort echo time (UTE)– and/or zero echo time (ZTE) MR sequences have been recently highlighted due to their capability to generate positive image contrast from bone [90]. Ladefoged et al. and Blanc-Durand et al. demonstrated the feasibility of UTE and ZTE MR sequences using Unet in PET/MR AC, respectively [94, 97]. The former study reported −0.1% (95%CI: −0.2 to 0.5%) bias on tumors without statistical significance from ground truth using Unet on UTE, which was superior to a significant bias of 2.2% (95%CI: 1.5 to 2.8%) using the vendor-provided segmentation-based method with Dixon. The latter study reported the bias of the Unet AC method on ZTE with a mean of −0.2% and a range from 1.7% in vertex to −1.8% in temporal lobe, and compared with a vendor-provided segmentation-based AC method on ZTE [113] which underestimated in most VOIs with a mean bias of −2.2% that ranged from 0.1% to −4.5%. It should be noted that neither of the studies compared the use of UTE/ZTE or conventional MR sequence under the same deep learning network. Thus, the advantage of this specialized sequence has not been validated. Moreover, compared with conventional T1-/T2-weighted MR images, the UTE/ZTE MR images have little diagnostic value in soft tissue and add to the acquisition time, which may hinder its utility in time-sensitive cases such as whole-body PET/MR scans. Other studies attempted to use multiple MR sequences as inputs in training and sCT generation since it is believed to be superior to a single MR sequence in sCT accuracy [91, 92, 95, 99]. The most common combination is UTE/ZTE and Dixon, which provides contrast in bone against air and fat as well as soft tissue, respectively. However, so far there is no study that has quantitatively investigated the additional gain in PET AC accuracy from a multi-sequence input to that of a single-sequence input. Thus, the necessity of multiple MR sequences for PET AC needs to be further evaluated and balanced with the extra acquisition time.
In the above PET/MR studies, the CT and MR images in the training datasets were acquired on different machines. The reviewed learning-based sCT methods all require image registration between the CT and MR to create CT-MR pairs for training. For the machine learning-based sCT methods such as random forest, the performance is sensitive to the registration error in the training pairs due to its one-to-one mapping strategy. Deep learning methods such as Unet and GAN-based methods are also susceptible to registration error if using a pixel-to-pixel loss. Kazemifar et al. showed that using mutual information as the loss function in the generator of GAN can bypass the registration step in the training [114]. CycleGAN-based methods feature higher robustness to registration error since it introduces cycle consistence loss to enforce the structural consistency between the original and cycle images, (e.g., force the cycle MRI generated images from synthetic CT to be the same as the original MRI) [115–118]. These techniques help reduce the requirement on registration accuracy in MR-sCT generation studies and are worth exploration in the context of PET AC.
Whole-body PET scanning has been an important imaging modality in finding tumor metastasis. Almost all of the reviewed studies developed their proposed methods using brain PET datasets. Although in learning-based methods which are data-driven, the network and architecture are not body part specific, the feasibility accepted for brain images may not translate to whole body due to the high anatomical heterogeneities and inter-subject variability, as can be seen from Figure 2. The only two studies on learning-based whole-body PET AC are from Dong et al. who proposed the CycleGAN-based method and investigated both the sCT generation strategy and direct mapping strategy [106, 107]. They reported average bias within 5% in all selected organs except >10% in lungs in both studies. The authors attributed the poor performance on lung to the tissue inhomogeneity and insufficient representative training datasets. Both studies are performed in PET without the need for CT and so far there is no learning-based methods developed for PET/MR whole body scanners. The integration of whole-body MR into PET AC could be more challenging than brain datasets since the MR acquisition may have a limited field of view (FOV), longer scan times that introduces more motion, and degraded image quality due to a larger inhomogeneous-field region.
3. LOW-COUNT PET RECONSTRUCTION
Low-count PET has important applications in pediatric PET and radiotherapy response evaluation with advantages of better motion control and lower patient dose. However, the low statistics result in increased image noise, reduced contrast-to-noise ratio, and bias in uptake measurements. The reconstruction of diagnostic quality images similar to standard- or full-count PET from the low-count PET cannot be achieved by simple post-processing operations such as denoising since lowering radiation dose changes the noise but also local uptake values [119]. Moreover, even with the same administered dose, the uptake distribution and signal level can vary greatly among patients. Recently, learning-based low-count PET reconstruction methods have been proposed to take advantage of their powerful data-driven feature extraction capabilities between two image datasets. These studies are summarized in Table II. As was described with PET AC methods, the general workflow can be divided into two groups depending on whether anatomical image information was used as an additional input (i.e. both PET and MR images in PET/MR scanner are input) or not (only PET images are input). To the best of our knowledge, there is no study investigating the usage of both PET and CT as inputs. A potential reason could be that CT images do not show high contrast among soft tissues, thus it cannot provide soft tissue structure information to guide PET denoising. Most proposed methods are implemented on PET brain scans, with a few on lung and whole body. Again, the ground truth for training and evaluation is full-count PET. Compared with those evaluations in PET AC, which focus on relative bias, the evaluations in the reviewed studies of low-count PET reconstruction focus on image quality and similarity between the predicted result and its corresponding ground truth. The most common metrics include PSNR (peak signal-to-noise ratio), SSIM (structure similarity index), NMSE (normalized mean square error) and CV (coefficient of variation). Several studies also compared their proposed methods with other competing learning-based methods and conventional denoising methods.
Table II.
Overview of learning-based low-count PET reconstruction methods.
| Methods and strategy | PET or PET+other modalities | Site | # of patients in training/testing datasets | Count fraction (low-count/ full-count) | Reported image quality metrics+: low-count/predicted full-count/full-count | Authors |
|---|---|---|---|---|---|---|
| Random forest | PET+MR(T1-weighted) | Brain | 11, leave-one-out | 1/4 | Coefficient of variation 0.38/0.33/0.31 | Kang et al., 2015 [120] |
| Multi-level canonical correlation analysis | PET, PET+MR(T1-weighted, DTI) | Brain | 11, leave-one-out | 1/4 | PSNR: 19.5/23.9/N.A.* | An et al., 2016 [119] |
| Sparse representation | PET+MR(T1-weighted, DTI) | Brain | 8, leave-one-out | 1/4 | PSNR: 19.02 (15.74, 22.41)/19.98 (16.92, 23.24)/ N.A. | Wang et al., 2016 [121] |
| Dictionary learning | PET+MR(T1-weighted, DTI) | Brain | 18, leave-one-out (only 8 used for test) | 1/4 | PSNR#: (16, 20)/(24, 27)/ N.A. | Wang et al., 2017 [124] |
| CNN | PET or PET+MR(T1-and T2-weighted) | Brain | 40, five-fold crossvalidation 18F-florbetaben | 1/100 | PSNR#: 31/36(PET-only), 38(PET+MR)/ N.A. | Chen et al., 2019 [137] |
| CNN | PET | Brain, lung | Brain: 2 training/1 testing Lung: 5 training/1 testing | Brain: 1/5 Lung: 1/10 | N.A. | Gong et al., 2019 [138] |
| Unet (without training datasets) | PET+MR (T1-weighted) | Brain | 0 training/1 testing | 1/8 | N.A. | Gong et al., 2018 [130] |
| CNN (without training datasets) | PET | Brain | 0 training/ 1 test (monkey) | 1/5 and 1/10 | CNR: N.A./10.74 (1/5 counts), 7.44 (1/10 counts)/11.71 | Hashimoto et al., 2019 [129] |
| Deep autocontext CNN | PET+MR (T1-weighted) | Brain | 16, leave-one-out | 1/4 | PSNR: N.A./24.76/ N.A. | Xiang et al., 2017 [127] |
| Iterative Reconstruction using Denoising CNN | PET | Brain | 27 training (11C-DASB), 1 testing | 1/6 | SSIM: N.A./0.496/ N.A. | Kim et al., 2018 [132] |
| Iterative Reconstruction using CNN | PET | Brain, lung | Brain: 15 training/1 validation/1 testing Lung: 5 training/1 testing | 1/10 | N.A. | Gong et al., 2019 [133] |
| Deep convolutional encoder-decoder | PET (sinogram) | Whole body (simulated) | 245 training, 52 validation, 53 testing | N/A | PSNR: 34.69 | Haggstrom et al., 2019 [134] |
| GAN | PET | Brain | 16, leave-one-out | 1/4 | PSNR#: 20/23/ N.A. (normal) 21/24/ N.A. (mild cognitive impairment) | Wang et al., 2018 [135] |
| GAN | PET+MR(T1, DTI) | Brain | 16, leave-one-out | 1/4 | PSNR: 19.88±2.34/24.61±1.79/ N.A. (normal), 21.33±2.53/25.19±1.98/ N.A. (mild cognitive impairment) | Wang et al., 2019 [131] |
| GAN | PET | Brain | 40, four-fold cross validation | 1/100 | PSNR#: 24/30/ N.A. | Ouyang et al., 2019 [139] |
| Comparison of CAE, Unet and GAN | PET | Lung | 10, five-fold cross validation | 1/10 | N.A. | Lu et al., 2019 [136] |
| CycleGAN | PET | Whole body | 25 leave-one-out + 10 hold-out tests | 1/8 | PSNR: 39.4±3.1/46.0±3.8/ N.A. (leave-one-out) 38.1±3.4/41.5±2.5 (hold-out) | Lei et al., 2019 [117] |
N.A.: not available, i.e. not explicitly indicated in the publication.
Estimated from figures.
Numbers in parentheses indicate minimum and maximum values.
3.1. Machine learning-based methods
Learning-based methods for low dose PET reconstruction have been developed before deep learning-based methods were introduced into this research area. Kang et al. employed a random forest to predict the full dose PET from low dose PET and MR images [120]. Their method first extracted features from patches of low-count PET and MR images based on segmentation to build tissue-specific models for initialization. Then, an iterative refinement strategy was used to further improve the predication accuracy. The method was evaluated on brain PET images with 1/4th of full dose and showed promising results of quantification accuracy and enhanced image quality.
Wang et al. proposed a sparse representation (SR) framework for low dose PET reconstruction [121]. In order to incorporate multi-sequence MR images as inputs with low-count PET, they used a mapping strategy to ensure that the sparse coefficients estimated from the multimodal MR images and low-count PET images could be applied directly to the prediction of full dose PET images. An incremental refinement framework was also added to further improve the performance. A patch selection-based dictionary construction method was used to speed up the prediction process. Their evaluation on brain PET scans with 1/4th of full dose demonstrated significantly improved PSNR and NMSE compared with the original low-count PET. The authors also compared the performance of the proposed mapping-based SR with that of the conventional SR on the same dataset and demonstrated that the proposed method consistently achieved higher PSNR and lower NMSE across all subjects, which indicates that the mapping strategy does help the SR enhance the prediction quality.
An et al. also proposed to formulate full-count estimation as a sparse representation problem using a multi-level canonical correlation analysis-based data-driven scheme [119]. The rationale is that the intra-data relationships between the low-count PET and full-count PET data spaces are different, which results in lower efficacy of directly applying learned coefficients from the low-count PET dictionary to the full-count PET dictionary for estimation. Canonical correlation analysis was then used to learn global mapping with the original coupled low-count PET and full-count PET dictionaries and then mapped both kinds of data into their common space. The multi-level scheme can further improve the learning of the common space by passing on the low-count PET dictionary atoms with nonzero coefficients of patches from the first level to the next level with the corresponding full-count PET dictionary subset, rather than immediately estimating the target full-count PET at the first level. This framework is capable of using PET alone or a combination of PET and multi-sequence MR images as inputs. The evaluation study on low-count brain PET patients showed that the proposed method successfully improved the visual quality and quantification accuracy of the SUV (standardized update value) and significantly outperformed the competing learning-based methods such as SR and random forest as well as the conventional denoising methods such as Block-matching and 3D filtering (BM3D) [122] and Optimized Blockwise Nonlocal Means (OBNM) [123].
Wang et al. pointed out that the performance of the SR approach depends on the completeness of the dictionary, which means that it typically requires coupled samples in the training dataset [124]. However, for low-count PET reconstruction with multi-sequence MR images as additional inputs, it is hard to collect all the MR images of different contrasts for each sample in training. They then proposed to use an efficient semi-supervised tripled dictionary learning method to more effectively utilize all the available training samples including the incomplete ones to predict the full-count PET. The proposed framework enforced node-to-node and edge-to-edge matching between the patches of low-count PET/MR and full-count PET, which can reduce the requirements on their similarity. They validated the proposed method on 18 brain PET patients, only 8 among of which had complete datasets (i.e. having all low-count PET, full-count PET and MR images of three contrasts). It was observed that the proposed method outperformed traditional SR and random forest in PSNR and NMSE. It also compared the performance of the proposed method with and without the incomplete datasets included in the training datasets and the results suggested that the addition of incomplete datasets benefited the performance rather than harmed it and should be used if available.
3.2. Deep learning-based methods
The sparse-learning-based methods reviewed above usually include several steps such as patch extraction, encoding, and reconstruction. It would be time-consuming when testing new cases since it involves a large number of optimization problems, which might not be appropriate for clinical practice. Secondly, these methods tend to over-smooth the image due to its patch-based processing, which may limit the clinical usability in fine structure detection. For example, studies show that over-smoothing on PET images can adversely affect the signal-to-noise ratio on low-contrast lesions, thus degrade lesion detectability [125, 126].
Xiang et al. proposed a deep auto-context CNN to predict full-count PET images based on local patches in low-count PET and MR images [127]. This regression method integrated multiple CNN modules by the auto-context strategy in order to iteratively improve the estimated PET image. A basic four-layer CNN was first used to build a model to estimate the full-count PET from low-count PET and MR images. The estimated full-count PET was treated as the source of the context information and was used as inputs along with the low-count PET and MR images to a new four-layer CNN. Thus, the multiple CNNs were gradually concatenated into a deeper network and were optimized altogether with back-propagation. Validations on brain PET/MR datasets showed the proposed method provided comparative quality to full-count PET, as did the SR-based method proposed by An et al. [119], with much less time in prediction. A potential limitation of this study is that the axial slices extracted from the 3D images were treated as separate 2D images independently for training the deep architecture. This could cause the loss of information in sagittal and coronal directions and discontinuous in estimating results across slices [123]. This patch-based workflow also tends to ignore the global spatial information in the prediction results.
Recent studies have shown that low-noise training datasets are unnecessary for CNNs to produce denoised image since the CNN architecture has an intrinsic ability to solve inverse problems [128]. The proposed deep image prior approach iteratively learns from a pair of random noise and corrupted images, and a denoised image is then obtained as output with moderate iterations [129]. Hashimoto et al. applied the deep image prior approach in a CNN for dynamic brain PET imaging. The static PET images acquired from the start to end were used as the network input and the dynamic PET images were used as the label. They reported the proposed method maintained the CNR (contrast to noise ratio) and outperformed the conventional model-based denoising methods on a dynamic brain PET study in monkeys [129]. Similarly, Gong et al. also used the deep image prior framework with a modified Unet structure for PET/MR imaging [130]. In their method, the MR images served as patient specific prior information and network inputs. The network was then updated during the iterative reconstruction process based on the MR and acquired low-count PET. Results demonstrated that the proposed method was able to recover more cortex details and suppress more noise in white matter than a Gaussian post-filtering method and a pre-trained neural network in the penalty, and provided higher uptake values in tumor than a kernel method.
To fully utilize the 3D information from image volumes, Wang et al. proposed an end-to-end framework based on a 3D conditional GAN to predict full-count PET from low-count PET. They used both convolutional and up-convolutional layers in the generator architecture to ensure the same size between the input and output. The generator network was a 3D Unet-like deep architecture with skip connection strategy, which was more efficient than voxel-wise estimation. Compared with 2D axial-based conditional GAN, the proposed 3D scheme was shown to produce better visual quality with sharper texture on sagittal and coronal views. Compared with the SR-based methods and CNN-based methods proposed in reference [121, 124, 127], the proposed method featured better spatial resolution and quantitative accuracy. To incorporate multi-sequence MR images as inputs for better performance, Wang et al. proposed an auto-context-based “locality adaptive” multi-modality GANs (LA-GANs) model to synthesize the full-count PET image from both the low-count PET and the accompanying multi-sequence MRI [131]. The “locality adaptive” means that the weight of each image contrast depends on the image locations. 1×1×1 kernel was used to learn locality-adaptive fusion mechanisms to minimize the number of parameters selected by the input of multi-sequence images. Compared with their previous method, it can achieve better performance with better PSNR while incurring a smaller number of additional parameters.
Lei et al. proposed a CycleGAN model to estimate diagnostic quality PET images using low count data on whole body PET scans [117]. A representative result of low-count PET reconstruction for whole body is shown in Figure 3 which was redrawn based on the method of Lei et al. CycleGAN learns a transformation to synthesize full-count PET images using low-count PET that would be indistinguishable from a standard clinical protocol. The algorithm also learns an inverse transformation such that cycle low-count PET data (inverse of the synthetic estimate) generated from synthetic full-count PET would be close to the true low-count PET. This approach improves the GAN model’s prediction and uniqueness of the synthetic dataset. Residual blocks were also integrated to the architecture to better capture the difference of low- and full-count images and enhance convergence. Evaluation studies showed that the average bias among all 10 hold-out test patients was less than 5% for all selected organs and the comparison with Unet and GAN-based methods indicated a higher quantitative accuracy and PSNR in all selected organs.
Figure 3.
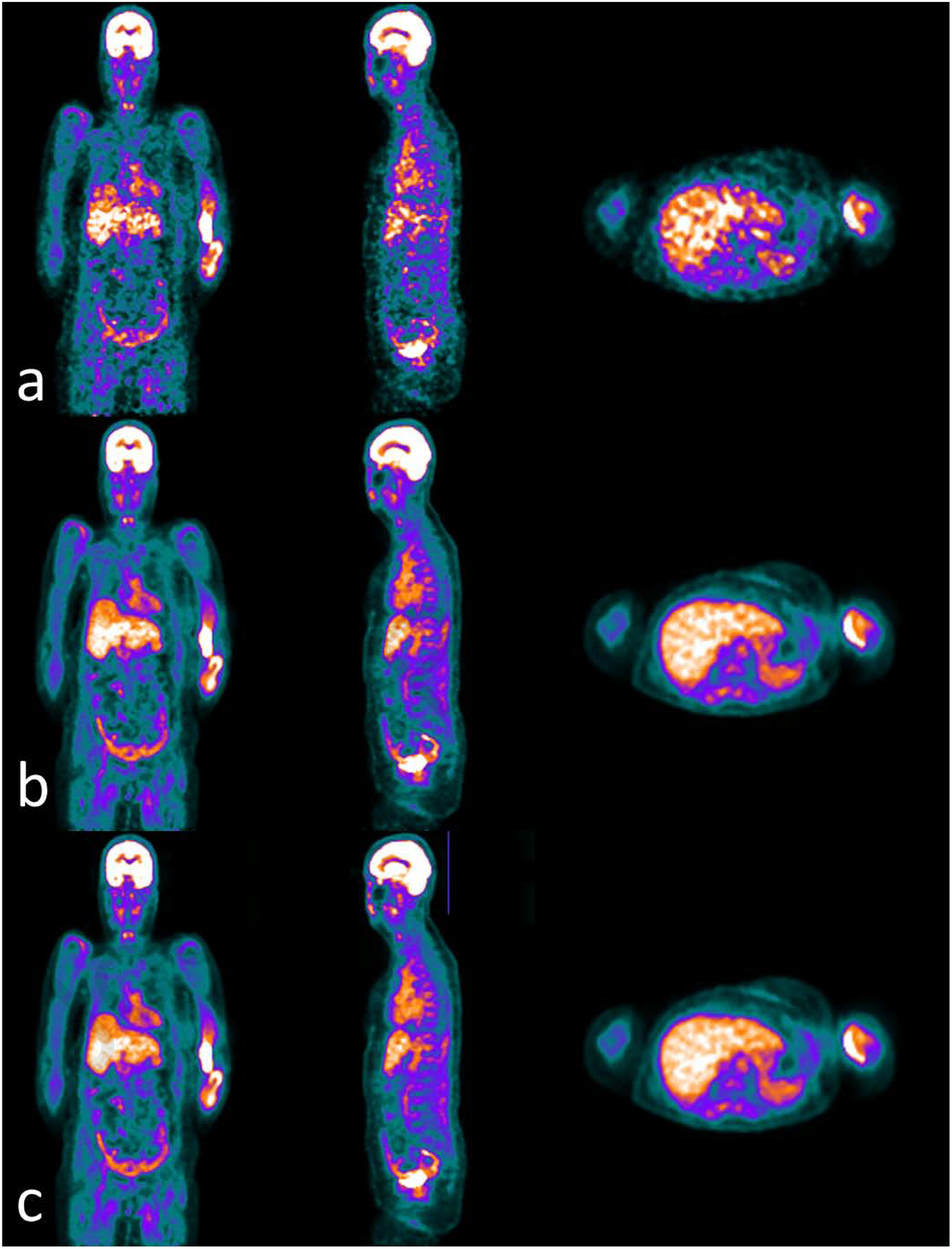
Examples of whole body PET images of (a) low counts, (b) full counts and (c) predicted full counts from (a) low counts using the learning-based method proposed in reference [117]. Window width of [0 10000] with PET units in Bq/ml.
In addition to the direct mapping from low-count PET to full-count PET, some studies investigated the feasibility of embedding the deep learning network into the conventional iterative reconstruction framework. Kim et al. modified the denoising CNN and trained with patient datasets of full-count PET and low-count PET with a preset noise level by simulation [132]. The predicted results from the denoising CNN then served as a prior in the iterative reconstruction setting with a local linear fitting function to correct the unwanted bias caused by the noise level changes in each iteration. Gong et al. used a CNN trained with pairs of high-dose PET and iterative reconstructions of low-count PET [133]. Then, instead of directly feeding a noisy image into the CNN and estimating its output, they used the CNN to define a feasible set of valid PET images as the initial images in the iterative reconstruction. Both methods were compared with conventional denoising and iterative reconstruction algorithms and demonstrated advantages in less noise when quantitative accuracy was kept the same. In contrast to these methods where CNN served as the regularizer in the reconstruction loop, Häggström et al. exploited the deep convolutional encoder-decoder network to take the low-count PET sinogram as input and directly output the high-dose PET images thus the network implicitly learned the inverse problem [134]. On simulated whole-body PET images, the proposed method outperformed the conventional ordered subset expectation maximization (OSEM) method by preserving fine details, reducing noise and reducing reconstruction time.
3.3. Discussion
As categorized above, the reviewed studies can be mainly divided into machine learning-based and deep learning-based approaches. Although the above machine learning-based methods showed good estimation performance, most of them have limitations since they depend on the handcraft features extracted from prior knowledge. Handcrafted features have limited ability in effectively representing images. The voxel-wise estimation strategy usually involves solving a large number of optimization problems online, which is also time-consuming when applied on new test datasets. Moreover, the small-patch-based methods could average the overlapped patches as the last step, thus it tends to generate over-smoothed and blurred images resulting in loss of texture information in PET images. In contrast, the deep learning-based methods can learn representative features directly from the training datasets. The recent advances in GPUs also allow the deep learning-based methods to be efficiently implemented with a large number of parameters and trained with full-sized images. To speed up the convergence and avoid overfitting in training, techniques such as batch normalization, additional bias and ReLU can be integrated into the network. Compared to CNN, which has one generator network, GAN has a generator network and a discriminator network. The purpose of the additional discriminator is to learn to distinguish between the prediction from generator and the real inputs while the generator is optimized to predict samples such that they are indistinguishable from real data by the discriminator. The two networks are trained alternatively to respectively minimize and maximize an objective function.
Comparisons among several reviewed methods have been included in a few studies. For example, Wang et al. [131, 135] compared SR-based, dictionary learning-based and CNN-based methods [119, 121, 124, 127] with their proposed GAN-based method. It was observed that the predicted full-count PET images by SR-based method are more likely to generate over-smoothed images than CNN- and GAN-based methods. GAN-based methods are better than CNN-based methods in preserving the detailed information and it is argued by the authors that it is because CNN-based methods do not consider the varying contributions across image locations. The proposed GAN-based methods also outperforms other competing methods in quantification metrics of image quality and accuracy with statistical significance. The CNN-based and GAN-based methods also feature high processing speed of around several seconds, as compared to minutes and even hours for machine learning-based methods. Note that these comparison studies were from the same group that also proposed the comparison methods. Independent studies in the performance evaluation of different methods are encouraged. Lu et al. [136] compared the performance of CAE, Unet and GAN on lung nodule quantification. They reported that Unet and GAN were slightly better than CAE with less difference between the predicted results and the ground truth, and the prediction of CAE was smoother. Quantitatively, Unet and GAN had significantly lower SUV bias than CAE, especially for small nodules. Unet and GAN had comparable performance while Unet had a lower computational cost.
The effectiveness of a 3D model over a 2D model is confirmed in a few publications [135, 136]. In a 2D model, the whole 2D slices, either at sagittal, coronal or axial view, are used as inputs, while a 3D model uses large 3D image volume patches as inputs. Results show that the advantage of a 3D model is the better performance in all the three views, while a 2D model has obvious discontinuity and blur in the views different from that used in training as well as substantial underestimation in quantification.
The benefit of multi-sequence MR has also been indicated by the reviewed work. Wang et al. compared the performance of their proposed method using T1-weighted and FA (fractional anisotropy) and MD (mean diffusivity) from DTI (diffusion tensor imaging) brain MR images jointly or separately [124]. They found that improvement over low-count PET alone was achieved by including any one MR contrast type as input. Using all MR contrast images with low-count PET jointly can enhance the performance further. It was also observed that the general results by single contrast MR image is similar to each other across all three types of contrast, which suggests that these contrasts contribute similarly in most of the brain areas. Similar conclusions were also drawn in other studies with slight differences in T1-weighting, which may be more helpful than DTI in full-count PET prediction [124, 131]. This is likely because the T1-weighted image can show both white and grey matter more clearly. For the DTI image, it contributes more in the white matter regions than in the grey matter regions.
Lei et al. presented the first study of learning-based low-count PET reconstruction on whole body datasets [117]. Similar to the cases in the PET AC studies, the application of these data-driven approaches to brain data are often less complicated due to less inter-patient anatomical variation in brain images as compared to that on whole body images. Whole body PET images also have higher intra-patient uptake variation, i.e. tracer concentration is much higher in brain and bladder than anywhere else, which may decrease the relative contrast among other organs and introduce difficulty in extracting features. Moreover, considering the challenges in PET/MR with whole body, the benefit from MR images in full-count prediction are worthy of re-evaluation.
SUMMARY AND OUTLOOK
Recent years have witnessed a trend that machine learning, especially deep learning, is increasingly used in the application of PET imaging. Various types of machine learning networks have been borrowed from computer vision field and adapted to specific clinical tasks for PET imaging. As reviewed in this paper, the most common applications are PET AC and low-count PET reconstruction. It is also an emerging field since all of these reviewed studies were published within five years. With the development in both machine learning algorithm and computing hardware, more learning-based methods are expected to facilitate the clinical workflow of PET imaging with more potential in applications of quantification.
In addition to PET AC and low-count reconstruction, there are other topics in PET imaging where machine learning can be exploited. For example, high resolution PET has great potential in visualizing and accurately measuring the radiotracer concentration in structures with dimensions on the order of millimeters, where the partial volume effect substantially limits the activity discriminating of the scanner [140, 141]. In addition to the advancement in state-of-art PET detectors that have achieved sub-nanosecond coincident timing resolution and sub-millimeter coincident spatial resolution [140, 142, 143], the partial volume effect caused by the residual spatial blurring can be further suppressed by image-based correction methods with the aid of a second image which has substantially reduced partial volume effect such as MR or CT [144–148]. Machine learning can be promising since it has excellent performance of direct end-to-end mapping shown in other applications. Encouraging results have been shown by Song et al. [149] in a preliminary study using a CNN. In addition to the improvement on image quality and quantification accuracy, machine learning methods are also attractive to other advanced applications such as segmentation and radiomics, especially when its success in other imaging modalities has been demonstrated [150–163]. Although machine learning has been available for decades, all of these applications are still new to this field. Therefore, it is expected that an increasing number of publications will be coming out in the next few years.
The reviewed studies in the paper are all feasibility studies with small to intermediate number of patients in training/testing. The clinical utility and its potential impact of these learning-based methods cannot be comprehensively evaluated until a large number of clinical datasets are involved in the study due to the data-driven property of machine learning as well as prospective evaluation on a clinical outcome measure. Moreover, the representativeness of training/testing datasets needs special attention in clinical studies where patient cohorts can be heterogeneous. The missing of diverse demographics and pathological abnormalities may reduce the robustness and generality in the performance of these proposed methods. Care has to be taken when the model is trained and applied on data from different center/scanner/protocol, which may lead to unpredictable performance.
Machine learning has been integrated to PET in attenuation correction (AC) and low-count reconstruction in recent years.
The proposed methods, study designs and key results of the current published studies are reviewed in this paper.
Machine learning generates synthetic CT from MR or non-AC PET for PET AC, or directly maps non-AC PET to AC PET.
Deep learning-based methods have advantages over conventional machine learning methods in low-count PET reconstruction.
ACKNOWLEDGEMENT
This research was supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 and Emory Winship Cancer Institute pilot grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- [1].Schrevens L, Lorent N, Dooms C, Vansteenkiste J. The Role of PET Scan in Diagnosis, Staging, and Management of Non-Small Cell Lung Cancer. The Oncologist. 2004;9:633–43. [DOI] [PubMed] [Google Scholar]
- [2].Sugiyama M, Sakahara H, Torizuka T, Kanno T, Nakamura F, Futatsubashi M, et al. 18F-FDG PET in the detection of extrahepatic metastases from hepatocellular carcinoma. Journal of Gastroenterology. 2004;39:961–8. [DOI] [PubMed] [Google Scholar]
- [3].Ma S-Y, See L-C, Lai C-H, Chou H-H, Tsai C-S, Ng K-K, et al. Delayed 18F-FDG PET for Detection of Paraaortic Lymph Node Metastases in Cervical Cancer Patients. Journal of Nuclear Medicine. 2003;44:1775–83. [PubMed] [Google Scholar]
- [4].Strobel K, Dummer R, Husarik DB, Lago MP, Hany TF, Steinert HC. High-Risk Melanoma: Accuracy of FDG PET/CT with Added CT Morphologic Information for Detection of Metastases. Radiology. 2007;244:566–74. [DOI] [PubMed] [Google Scholar]
- [5].Adler LP, Faulhaber PF, Schnur KC, Al-Kasi NL, Shenk RR. Axillary lymph node metastases: screening with [F-18]2-deoxy-2-fluoro-D-glucose (FDG) PET. Radiology. 1997;203:323–7. [DOI] [PubMed] [Google Scholar]
- [6].Abdel-Nabi H, Doerr RJ, Lamonica DM, Cronin VR, Galantowicz PJ, Carbone GM, et al. Staging of primary colorectal carcinomas with fluorine-18 fluorodeoxyglucose whole-body PET: correlation with histopathologic and CT findings. Radiology. 1998;206:755–60. [DOI] [PubMed] [Google Scholar]
- [7].Taira AV, Herfkens RJ, Gambhir SS, Quon A. Detection of Bone Metastases: Assessment of Integrated FDG PET/CT Imaging1. Radiology. 2007;243:204–11. [DOI] [PubMed] [Google Scholar]
- [8].Ohta M, Tokuda Y, Suzuki Y, Kubota M, Makuuchi H, Tajima T, et al. Whole body PET for the evaluation of bony metastases in patients with breast cancer: comparison with 99Tcm-MDP bone scintigraphy. Nuclear Medicine Communications. 2001;22:875–9. [DOI] [PubMed] [Google Scholar]
- [9].Czernin J, Allen-Auerbach M, Schelbert HR. Improvements in Cancer Staging with PET/CT: Literature-Based Evidence as of September 2006. Journal of Nuclear Medicine. 2007;48:78S–88S. [PubMed] [Google Scholar]
- [10].Biehl KJ, Kong FM, Dehdashti F, Jin JY, Mutic S, El Naqa I, et al. 18F-FDG PET definition of gross tumor volume for radiotherapy of non-small cell lung cancer: is a single standardized uptake value threshold approach appropriate? Journal of Nuclear Medicine. 2006;47:1808–12. [PubMed] [Google Scholar]
- [11].Paulino AC, Koshy M, Howell R, Schuster D, Davis LW. Comparison of CT- and FDG-PET-defined gross tumor volume in intensity-modulated radiotherapy for head-and-neck cancer. International Journal of Radiation Oncology • Biology • Physics. 2005;61:1385–92. [DOI] [PubMed] [Google Scholar]
- [12].Nestle U, Kremp S, Schaefer-Schuler A, Sebastian-Welsch C, Hellwig D, Rübe C, et al. Comparison of Different Methods for Delineation of 18F-FDG PET–Positive Tissue for Target Volume Definition in Radiotherapy of Patients with Non–Small Cell Lung Cancer. Journal of Nuclear Medicine. 2005;46:1342–8. [PubMed] [Google Scholar]
- [13].Schwaiger M, Ziegler S, Nekolla SG. PET/CT: Challenge for Nuclear Cardiology. Journal of Nuclear Medicine. 2005;46:1664–78. [PubMed] [Google Scholar]
- [14].Parker MW, Iskandar A, Limone B, Perugini A, Kim H, Jones C, et al. Diagnostic accuracy of cardiac positron emission tomography versus single photon emission computed tomography for coronary artery disease: a bivariate meta-analysis. Circulation Cardiovascular imaging. 2012;5:700–7. [DOI] [PubMed] [Google Scholar]
- [15].Politis M, Piccini P. Positron emission tomography imaging in neurological disorders. Journal of Neurology. 2012;259:1769–80. [DOI] [PubMed] [Google Scholar]
- [16].Boellaard R Standards for PET image acquisition and quantitative data analysis. Journal of Nuclear Medicine. 2009;50 Suppl 1:11s–20s. [DOI] [PubMed] [Google Scholar]
- [17].Ben-Haim S, Ell P. 18F-FDG PET and PET/CT in the Evaluation of Cancer Treatment Response. Journal of Nuclear Medicine. 2009;50:88–99. [DOI] [PubMed] [Google Scholar]
- [18].Shankar LK, Hoffman JM, Bacharach S, Graham MM, Karp J, Lammertsma AA, et al. Consensus Recommendations for the Use of 18F-FDG PET as an Indicator of Therapeutic Response in Patients in National Cancer Institute Trials. Journal of Nuclear Medicine. 2006;47:1059–66. [PubMed] [Google Scholar]
- [19].Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: Evolving Considerations for PET response criteria in solid tumors. Journal of Nuclear Medicine. 2009;50 Suppl 1:122s–50s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Naqa IE. The role of quantitative PET in predicting cancer treatment outcomes. Clinical and Translational Imaging. 2014;2:305–20. [Google Scholar]
- [21].Kinahan PE, Townsend DW, Beyer T, Sashin D. Attenuation correction for a combined 3D PET/CT scanner. Medical physics. 1998;25:2046–53. [DOI] [PubMed] [Google Scholar]
- [22].Watson CC. New, faster, image-based scatter correction for 3D PET. IEEE Transactions on Nuclear Science. 2000;47:1587–94. [Google Scholar]
- [23].Lodge MA, Chaudhry MA, Wahl RL. Noise Considerations for PET Quantification Using Maximum and Peak Standardized Uptake Value. Journal of Nuclear Medicine. 2012;53:1041–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Slifstein M, Laruelle M. Effects of Statistical Noise on Graphic Analysis of PET Neuroreceptor Studies. Journal of Nuclear Medicine. 2000;41:2083–8. [PubMed] [Google Scholar]
- [25].Soret M, Bacharach SL, Buvat I. Partial-volume effect in PET tumor imaging. Journal of Nuclear Medicine. 2007;48:932–45. [DOI] [PubMed] [Google Scholar]
- [26].Sureshbabu W, Mawlawi O. PET/CT Imaging Artifacts. Journal of Nuclear Medicine Technology. 2005;33:156–61. [PubMed] [Google Scholar]
- [27].Blodgett TM, Mehta AS, Mehta AS, Laymon CM, Carney J, Townsend DW. PET/CT artifacts. Clinical Imaging. 2011;35:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Miwa K, Wagatsuma K, Iimori T, Sawada K, Kamiya T, Sakurai M, et al. Multicenter study of quantitative PET system harmonization using NIST-traceable 68Ge/68Ga cross-calibration kit. Physica Medica. 2018;52:98–103. [DOI] [PubMed] [Google Scholar]
- [29].Burger C, Goerres G, Schoenes S, Buck A, Lonn AH, Von Schulthess GK. PET attenuation coefficients from CT images: experimental evaluation of the transformation of CT into PET 511-keV attenuation coefficients. European Journal of Nuclear Medicine and Molecular Imaging. 2002;29:922–7. [DOI] [PubMed] [Google Scholar]
- [30].Witoszynskyj S, Andrzejewski P, Georg D, Hacker M, Nyholm T, Rausch I, et al. Attenuation correction of a flat table top for radiation therapy in hybrid PET/MR using CT- and 68Ge/68Ga transmission scan-based μ-maps. Physica Medica. 2019;65:76–83. [DOI] [PubMed] [Google Scholar]
- [31].Fei B, Yang X, Nye JA, Aarsvold JN, Raghunath N, Cervo M, et al. MR/PET quantification tools: Registration, segmentation, classification, and MR-based attenuation correction. Medical Physics. 2012;39:6443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yang X, Fei B. Multiscale segmentation of the skull in MR images for MRI-based attenuation correction of combined MR/PET. Journal of the American Medical Informatics Association. 2013;20:1037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Goff-Rougetet RL, Frouin V, Mangin J-F, Bendriem B. Segmented MR images for brain attenuation correction in PET. Medical Imaging 1994: SPIE; 1994. 12. [Google Scholar]
- [34].El Fakhri G, Kijewski MF, Johnson KA, Syrkin G, Killiany RJ, Becker JA, et al. MRI-guided SPECT perfusion measures and volumetric MRI in prodromal Alzheimer disease. Archives of Neurology. 2003;60:1066–72. [DOI] [PubMed] [Google Scholar]
- [35].Hofmann M, Pichler B, Scholkopf B, Beyer T. Towards quantitative PET/MRI: a review of MR-based attenuation correction techniques. European Journal of Nuclear Medicine and Molecular Imaging. 2009;36 Suppl 1:S93–104. [DOI] [PubMed] [Google Scholar]
- [36].Zaidi H, Montandon ML, Slosman DO. Magnetic resonance imaging-guided attenuation and scatter corrections in three-dimensional brain positron emission tomography. Medical physics. 2003;30:937–48. [DOI] [PubMed] [Google Scholar]
- [37].Kops ER, Herzog H. Alternative methods for attenuation correction for PET images in MR-PET scanners. IEEE Nuclear Science Symposium Conference Record. 2007;6:4327–30. [Google Scholar]
- [38].Hofmann M, Steinke F, Scheel V, Charpiat G, Farquhar J, Aschoff P, et al. MRI-based attenuation correction for PET/MRI: a novel approach combining pattern recognition and atlas registration. Journal of Nuclear Medicine. 2008;49:1875–83. [DOI] [PubMed] [Google Scholar]
- [39].Catana C The Dawn of a New Era in Low-Dose PET Imaging. Radiology. 2019;290:657–8. [DOI] [PubMed] [Google Scholar]
- [40].Lei Y, Xu D, Zhou Z, Wang T, Dong X, Liu T, et al. A denoising algorithm for CT image using low-rank sparse coding. SPIE Medical Imaging. 2018;10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang T, Lei Y, Tian Z, Dong X, Liu Y, Jiang X, et al. Deep learning-based image quality improvement for low-dose computed tomography simulation in radiation therapy. Journal of Medical Imaging. 2019;6:1–10, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Erdi YE, Macapinlac H, Rosenzweig KE, Humm JL, Larson SM, Erdi AK, et al. Use of PET to monitor the response of lung cancer to radiation treatment. European Journal of Nuclear Medicine. 2000;27:861–6. [DOI] [PubMed] [Google Scholar]
- [43].Cliffe H, Patel C, Prestwich R, Scarsbrook A. Radiotherapy response evaluation using FDG PET-CT-established and emerging applications. The British Journal of Radiology. 2017;90:20160764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Das SK, McGurk R, Miften M, Mutic S, Bowsher J, Bayouth J, et al. Task Group 174 Report: Utilization of [18F]Fluorodeoxyglucose Positron Emission Tomography ([18F]FDG-PET) in Radiation Therapy. Medical Physics. 2019;46:e706–e25. [DOI] [PubMed] [Google Scholar]
- [45].Rahmim A, Lodge MA, Karakatsanis NA, Panin VY, Zhou Y, McMillan A, et al. Dynamic whole-body PET imaging: principles, potentials and applications. European Journal of Nuclear Medicine and Molecular Imaging. 2019;46:501–18. [DOI] [PubMed] [Google Scholar]
- [46].Borjesson PK, Jauw YW, de Bree R, Roos JC, Castelijns JA, Leemans CR, et al. Radiation dosimetry of 89Zr-labeled chimeric monoclonal antibody U36 as used for immuno-PET in head and neck cancer patients. Journal of Nuclear Medicine. 2009;50:1828–36. [DOI] [PubMed] [Google Scholar]
- [47].Jauw YWS, Heijtel DF, Zijlstra JM, Hoekstra OS, de Vet HCW, Vugts DJ, et al. Noise-Induced Variability of Immuno-PET with Zirconium-89-Labeled Antibodies: an Analysis Based on Count-Reduced Clinical Images. Molecular Imaging and Biology. 2018;20:1025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nguyen NC, Vercher-Conejero JL, Sattar A, Miller MA, Maniawski PJ, Jordan DW, et al. Image Quality and Diagnostic Performance of a Digital PET Prototype in Patients with Oncologic Diseases: Initial Experience and Comparison with Analog PET. Journal of Nuclear Medicine. 2015;56:1378–85. [DOI] [PubMed] [Google Scholar]
- [49].Karp JS, Surti S, Daube-Witherspoon ME, Muehllehner G. Benefit of time-of-flight in PET: experimental and clinical results. Journal of Nuclear Medicine. 2008;49:462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Qi J, Leahy RM. A theoretical study of the contrast recovery and variance of MAP reconstructions from PET data. IEEE Transactions on Medical Imaging. 1999;18:293–305. [DOI] [PubMed] [Google Scholar]
- [51].Chan C, Fulton R, Barnett R, Feng DD, Meikle S. Postreconstruction nonlocal means filtering of whole-body PET with an anatomical prior. IEEE Transactions on Medical Imaging. 2014;33:636–50. [DOI] [PubMed] [Google Scholar]
- [52].Christian BT, Vandehey NT, Floberg JM, Mistretta CA. Dynamic PET denoising with HYPR processing. Journal of Nuclear Medicine. 2010;51:1147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Balcerzyk M, Moszynski M, Kapusta M, Wolski D, Pawelke J, Melcher CL. YSO, LSO, GSO and LGSO. A study of energy resolution and nonproportionality. IEEE Transactions on Nuclear Science. 2000;47:1319–23. [Google Scholar]
- [54].Herbert DJ, Moehrs S, D’Ascenzo N, Belcari N, Del Guerra A, Morsani F, et al. The Silicon Photomultiplier for application to high-resolution Positron Emission Tomography. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment. 2007;573:84–7. [Google Scholar]
- [55].Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, et al. Deep learning in medical imaging and radiation therapy. Medical physics. 2019;46:e1–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Giger ML. Machine Learning in Medical Imaging. Journal of the American College of Radiology. 2018;15:512–20. [DOI] [PubMed] [Google Scholar]
- [57].Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. RadioGraphics. 2017;37:505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Feng M, Valdes G, Dixit N, Solberg TD. Machine Learning in Radiation Oncology: Opportunities, Requirements, and Needs. Frontiers in Oncology. 2018;8:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Jarrett D, Stride E, Vallis K, Gooding MJ. Applications and limitations of machine learning in radiation oncology. The British Journal of Radiology. 2019;92:20190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cui G, Jeong JJ, Lei Y, Wang T, Liu T, Curran WJ, et al. Machine-learning-based classification of Glioblastoma using MRI-based radiomic features. SPIE Medical Imaging. 2019;10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X. Deep learning in medical image registration: a review. Physics in Medicine & Biology. 2020;doi: 10.1088/1361-6560/ab843e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lei Y, Liu Y, Wang T, Tian S, Dong X, Jiang X, et al. Brain MRI classification based on machine learning framework with auto-context model. SPIE Medical Imaging. 2019;10953. [Google Scholar]
- [63].Lei Y, Shu HK, Tian S, Wang T, Liu T, Mao H, et al. Pseudo CT Estimation using Patch-based Joint Dictionary Learning. 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). 2018;doi: 10.1109/EMBC.2018.8513475:5150-3. [DOI] [PubMed] [Google Scholar]
- [64].Shafai-Erfani G, Lei Y, Liu Y, Wang Y, Wang T, Zhong J, et al. MRI-Based Proton Treatment Planning for Base of Skull Tumors. International Journal of Particle Therapy. 2019;6:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wang T, Lei Y, Manohar N, Tian S, Jani AB, Shu H-K, et al. Dosimetric study on learning-based cone-beam CT correction in adaptive radiation therapy. Medical Dosimetry. 2019;44:e71–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lei Y, Fu Y, Harms J, Wang T, Curran WJ, Liu T, et al. 4D-CT Deformable Image Registration Using an Unsupervised Deep Convolutional Neural Network. Workshop on Artificial Intelligence in Radiation Therapy. 2019;doi: 10.1007/978-3-030-32486-5_4:26-33. [DOI] [Google Scholar]
- [67].Harms J, Wang T, Petrongolo M, Niu T, Zhu L. Noise suppression for dual-energy CT via penalized weighted least-square optimization with similarity-based regularization. Medical physics. 2016;43:2676–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Harms J, Wang T, Petrongolo M, Zhu L. Noise suppression for energy-resolved CT using similarity-based non-local filtration. SPIE Medical Imaging. 2016;9783:8. [Google Scholar]
- [69].Wang T, Zhu L. Dual energy CT with one full scan and a second sparse-view scan using structure preserving iterative reconstruction (SPIR). Physics in Medicine & Biology. 2016;61:6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang T, Zhu L. Pixel-wise estimation of noise statistics on iterative CT reconstruction from a single scan. Medical physics. 2017;44:3525–33. [DOI] [PubMed] [Google Scholar]
- [71].Wang T, Zhu L. Image-domain non-uniformity correction for cone-beam CT. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). 2017;doi: 10.1109/ISBI.2017.7950611:680-3. [DOI] [Google Scholar]
- [72].Gong K, Berg E, Cherry SR, Qi J. Machine Learning in PET: From Photon Detection to Quantitative Image Reconstruction. Proceedings of the IEEE. 2020;108:51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ravishankar S, Ye JC, Fessler JA. Image Reconstruction: From Sparsity to Data-Adaptive Methods and Machine Learning. Proceedings of the IEEE. 2020;108:86–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lei Y, Harms J, Wang T, Tian S, Zhou J, Shu H-K, et al. MRI-based synthetic CT generation using semantic random forest with iterative refinement. Physics in Medicine & Biology. 2019;64:085001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lei Y, Jeong JJ, Wang T, Shu H-K, Patel P, Tian S, et al. MRI-based pseudo CT synthesis using anatomical signature and alternating random forest with iterative refinement model. Journal of Medical Imaging. 2018;5:1–12, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Lei Y, Shu H-K, Tian S, Jeong JJ, Liu T, Shim H, et al. Magnetic resonance imaging-based pseudo computed tomography using anatomic signature and joint dictionary learning. Journal of Medical Imaging. 2018;5:034001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Liu Y, Lei Y, Wang T, Kayode O, Tian S, Liu T, et al. MRI-based treatment planning for liver stereotactic body radiotherapy: validation of a deep learning-based synthetic CT generation method. The British Journal of Radiology. 2019;92:20190067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Liu Y, Lei Y, Wang Y, Shafai-Erfani G, Wang T, Tian S, et al. Evaluation of a deep learning-based pelvic synthetic CT generation technique for MRI-based prostate proton treatment planning. Physics in Medicine & Biology. 2019;64:205022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Liu Y, Lei Y, Wang Y, Wang T, Ren L, Lin L, et al. MRI-based treatment planning for proton radiotherapy: dosimetric validation of a deep learning-based liver synthetic CT generation method. Physics in Medicine & Biology. 2019;64:145015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Shafai-Erfani G, Wang T, Lei Y, Tian S, Patel P, Jani AB, et al. Dose evaluation of MRI-based synthetic CT generated using a machine learning method for prostate cancer radiotherapy. Medical Dosimetry. 2019;44:e64–e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wang T, Manohar N, Lei Y, Dhabaan A, Shu H-K, Liu T, et al. MRI-based treatment planning for brain stereotactic radiosurgery: Dosimetric validation of a learning-based pseudo-CT generation method. Medical Dosimetry. 2019;44:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lei Y, Tang X, Higgins K, Lin J, Jeong J, Liu T, et al. Learning-based CBCT correction using alternating random forest based on auto-context model. Medical physics. 2019;46:601–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lei Y, Tang X, Higgins K, Wang T, Liu T, Dhabaan A, et al. Improving image quality of cone-beam CT using alternating regression forest. SPIE Medical Imaging. 2018;10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lei Y, Wang T, Harms J, Fu Y, Dong X, Curran WJ, et al. CBCT-Based Synthetic MRI Generation for CBCT-Guided Adaptive Radiotherapy. Workshop on Artificial Intelligence in Radiation Therapy. 2019;doi: 10.1007/9783-030-32486-5_19:154-61. [DOI] [Google Scholar]
- [85].Lei Y, Wang T, Harms J, Shafai-Erfani G, Dong X, Zhou J, et al. Image quality improvement in cone-beam CT using deep learning. SPIE Medical Imaging. 2019;10948. [Google Scholar]
- [86].Lei Y, Wang T, Harms J, Shafai-Erfani G, Tian S, Higgins K, et al. MRI-based pseudo CT generation using classification and regression random forest. SPIE Medical Imaging. 2019;10948. [Google Scholar]
- [87].Lei Y, Wang T, Liu Y, Higgins K, Tian S, Liu T, et al. MRI-based synthetic CT generation using deep convolutional neural network. SPIE Medical Imaging. 2019;10949. [Google Scholar]
- [88].Yang X, Wang T, Lei Y, Higgins K, Liu T, Shim H, et al. MRI-based attenuation correction for brain PET/MRI based on anatomic signature and machine learning. Physics in Medicine & Biology. 2019;64:025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zaidi H, Montandon M-L. Scatter Compensation Techniques in PET. PET clinics. 2007;2:219–34. [DOI] [PubMed] [Google Scholar]
- [90].Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep Learning MR Imaging-based Attenuation Correction for PET/MR Imaging. Radiology. 2018;286:676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Larsson A, Johansson A, Axelsson J, Nyholm T, Asklund T, Riklund K, et al. Evaluation of an attenuation correction method for PET/MR imaging of the head based on substitute CT images. Magma (New York, NY). 2013;26:127–36. [DOI] [PubMed] [Google Scholar]
- [92].Navalpakkam BK, Braun H, Kuwert T, Quick HH. Magnetic resonance-based attenuation correction for PET/MR hybrid imaging using continuous valued attenuation maps. Investigative Radiology. 2013;48:323–32. [DOI] [PubMed] [Google Scholar]
- [93].Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv e-prints 2015. 1505.04597. [Google Scholar]
- [94].Blanc-Durand P, Khalife M, Sgard B, Kaushik S, Soret M, Tiss A, et al. Attenuation correction using 3D deep convolutional neural network for brain 18F-FDG PET/MR: Comparison with Atlas, ZTE and CT based attenuation correction. PloS one. 2019;14:e0223141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, et al. Zero-Echo-Time and Dixon Deep Pseudo-CT (ZeDD CT): Direct Generation of Pseudo-CT Images for Pelvic PET/MRI Attenuation Correction Using Deep Convolutional Neural Networks with Multiparametric MRI. Journal of Nuclear Medicine. 2018;59:852–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Torrado-Carvajal A, Vera-Olmos J, Izquierdo-Garcia D, Catalano OA, Morales MA, Margolin J, et al. Dixon-VIBE Deep Learning (DIVIDE) Pseudo-CT Synthesis for Pelvis PET/MR Attenuation Correction. Journal of Nuclear Medicine. 2019;60:429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ladefoged CN, Marner L, Hindsholm A, Law I, Hojgaard L, Andersen FL. Deep Learning Based Attenuation Correction of PET/MRI in Pediatric Brain Tumor Patients: Evaluation in a Clinical Setting. Frontiers in Neuroscience. 2018;12:1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Spuhler KD, Gardus J 3rd, Gao Y, DeLorenzo C, Parsey R, Huang C. Synthesis of Patient-Specific Transmission Data for PET Attenuation Correction for PET/MRI Neuroimaging Using a Convolutional Neural Network. Journal of Nuclear Medicine. 2019;60:555–60. [DOI] [PubMed] [Google Scholar]
- [99].Gong K, Yang J, Kim K, El Fakhri G, Seo Y, Li Q. Attenuation correction for brain PET imaging using deep neural network based on Dixon and ZTE MR images. Physics in Medicine & Biology. 2018;63:125011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Arabi H, Zeng G, Zheng G, Zaidi H. Novel adversarial semantic structure deep learning for MRI-guided attenuation correction in brain PET/MRI. European Journal of Nuclear Medicine and Molecular Imaging. 2019;46:2746–59. [DOI] [PubMed] [Google Scholar]
- [101].Heußer T, Rank CM, Berker Y, Freitag MT, Kachelrieß M. MLAA-based attenuation correction of flexible hardware components in hybrid PET/MR imaging. EJNMMI physics. 2017;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET Attenuation Map for Whole-Body Time-of-Flight 18F-FDG PET/MRI Using a Deep Neural Network Trained with Simultaneously Reconstructed Activity and Attenuation Maps. Journal of Nuclear Medicine. 2019;60:1183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, et al. Improving the Accuracy of Simultaneously Reconstructed Activity and Attenuation Maps Using Deep Learning. Journal of Nuclear Medicine. 2018;59:1624–9. [DOI] [PubMed] [Google Scholar]
- [104].Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for (18)F-FDG PET attenuation correction. EJNMMI physics. 2018;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Armanious K, Küstner T, Reimold M, Nikolaou K, La Fougère C, Yang B, et al. Independent brain 18F-FDG PET attenuation correction using a deep learning approach with Generative Adversarial Networks. Hellenic Journal of Nuclear Medicine. 2019;22:179–86. [DOI] [PubMed] [Google Scholar]
- [106].Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, et al. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Physics in Medicine & Biology. 2019;64:215016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dong X, Lei Y, Wang T, Higgins K, Liu T, Curran WJ, et al. Deep learning-based attenuation correction in the absence of structural information for whole-body PET imaging. Physics in Medicine & Biology. 2019; 10.1088/1361-6560/ab652c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yang J, Park D, Gullberg GT, Seo Y. Joint correction of attenuation and scatter in image space using deep convolutional neural networks for dedicated brain (18)F-FDG PET. Physics in Medicine & Biology. 2019;64:075019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Shiri I, Ghafarian P, Geramifar P, Leung KH-Y, Ghelichoghli M, Oveisi M, et al. Direct attenuation correction of brain PET images using only emission data via a deep convolutional encoder-decoder (Deep-DAC). European Radiology. 2019;29:6867–79. [DOI] [PubMed] [Google Scholar]
- [110].Wang G, Qi J. Analysis of penalized likelihood image reconstruction for dynamic PET quantification. IEEE Transactions on Medical Imaging. 2009;28:608–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Freitag MT, Fenchel M, Baumer P, Heusser T, Rank CM, Kachelriess M, et al. Improved clinical workflow for simultaneous whole-body PET/MRI using high-resolution CAIPIRINHA-accelerated MR-based attenuation correction. European Radiology. 2017;96:12–20. [DOI] [PubMed] [Google Scholar]
- [112].Izquierdo-Garcia D, Hansen AE, Förster S, Benoit D, Schachoff S, Fürst S, et al. An SPM8-Based Approach for Attenuation Correction Combining Segmentation and Nonrigid Template Formation: Application to Simultaneous PET/MR Brain Imaging. Journal of Nuclear Medicine. 2014;55:1825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wiesinger F, Sacolick LI, Menini A, Kaushik SS, Ahn S, Veit-Haibach P, et al. Zero TE MR bone imaging in the head. Magnetic Resonance in Medicine. 2016;75:107–14. [DOI] [PubMed] [Google Scholar]
- [114].Kazemifar S, McGuire S, Timmerman R, Wardak Z, Nguyen D, Park Y, et al. MRI-only brain radiotherapy: Assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiotherapy and Oncology. 2019;136:56–63. [DOI] [PubMed] [Google Scholar]
- [115].Dong X, Lei Y, Tian S, Wang T, Patel P, Curran WJ, et al. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiotherapy and Oncology. 2019;141:192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Harms J, Lei Y, Wang T, Zhang R, Zhou J, Tang X, et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Medical physics. 2019;46:3998–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lei Y, Dong X, Wang T, Higgins K, Liu T, Curran WJ, et al. Whole-body PET estimation from low count statistics using cycle-consistent generative adversarial networks. Physics in Medicine & Biology. 2019;64:215017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lei Y, Harms J, Wang T, Liu Y, Shu HK, Jani AB, et al. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Medical physics. 2019;46:3565–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].An L, Zhang P, Adeli E, Wang Y, Ma G, Shi F, et al. Multi-Level Canonical Correlation Analysis for Standard-Dose PET Image Estimation. IEEE Transactions on Image Processing. 2016;25:3303–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kang J, Gao Y, Shi F, Lalush DS, Lin W, Shen D. Prediction of standard-dose brain PET image by using MRI and low-dose brain [18F]FDG PET images. Medical physics. 2015;42:5301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Wang Y, Zhang P, An L, Ma G, Kang J, Shi F, et al. Predicting standard-dose PET image from low-dose PET and multimodal MR images using mapping-based sparse representation. Physics in Medicine & Biology. 2016;61:791–812. [DOI] [PubMed] [Google Scholar]
- [122].Dabov K, Foi A, Katkovnik V, Egiazarian K. Image Denoising by Sparse 3-D Transform-Domain Collaborative Filtering. IEEE Transactions on Image Processing. 2007;16:2080–95. [DOI] [PubMed] [Google Scholar]
- [123].Coupe P, Yger P, Prima S, Hellier P, Kervrann C, Barillot C. An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images. IEEE Transactions on Medical Imaging. 2008;27:425–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Wang Y, Ma G, An L, Shi F, Zhang P, Lalush DS, et al. Semisupervised Tripled Dictionary Learning for Standard-Dose PET Image Prediction Using Low-Dose PET and Multimodal MRI. IEEE transactions on bio-medical engineering. 2017;64:569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Wangerin K, Ahn S, Wollenweber S, Ross S, Kinahan P, Manjeshwar R. Evaluation of lesion detectability in positron emission tomography when using a convergent penalized likelihood image reconstruction method. Journal of Medical Imaging. 2016;4:011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Qi J Theoretical evaluation of the detectability of random lesions in Bayesian emission reconstruction. Information Processing in Medical Imaging. 2003;18:354–65. [DOI] [PubMed] [Google Scholar]
- [127].Xiang L, Qiao Y, Nie D, An L, Wang Q, Shen D. Deep Auto-context Convolutional Neural Networks for Standard-Dose PET Image Estimation from Low-Dose PET/MRI. Neurocomputing. 2017;267:406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Lehtinen J, Munkberg J, Hasselgren J, Laine S, Karras T, Aittala M, et al. Noise2Noise: Learning Image Restoration without Clean Data. arXiv e-prints 2018. 1803.04189. [Google Scholar]
- [129].Hashimoto F, Ohba H, Ote K, Teramoto A, Tsukada H. Dynamic PET Image Denoising Using Deep Convolutional Neural Networks Without Prior Training Datasets. IEEE Access. 2019;7:96594–603. [Google Scholar]
- [130].Gong K, Catana C, Qi J, Li Q. PET Image Reconstruction Using Deep Image Prior. IEEE Transactions on Medical Imaging. 2019;38:1655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Wang Y, Zhou L, Yu B, Wang L, Zu C, Lalush DS, et al. 3D Auto-Context-Based Locality Adaptive Multi-Modality GANs for PET Synthesis. IEEE Transactions on Medical Imaging. 2019;38:1328–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Kim K, Wu D, Gong K, Dutta J, Kim JH, Son YD, et al. Penalized PET Reconstruction Using Deep Learning Prior and Local Linear Fitting. IEEE Transactions on Medical Imaging. 2018;37:1478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Gong K, Guan J, Kim K, Zhang X, Yang J, Seo Y, et al. Iterative PET Image Reconstruction Using Convolutional Neural Network Representation. IEEE Transactions on Medical Imaging. 2019;38:675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Häggström I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: A deep encoder–decoder network for directly solving the PET image reconstruction inverse problem. Medical Image Analysis. 2019;54:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 2018;174:550–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, et al. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Physics in Medicine & Biology. 2019;64:165019. [DOI] [PubMed] [Google Scholar]
- [137].Chen KT, Gong E, de Carvalho Macruz FB, Xu J, Boumis A, Khalighi M, et al. Ultra-Low-Dose (18)F-Florbetaben Amyloid PET Imaging Using Deep Learning with Multi-Contrast MRI Inputs. Radiology. 2019;290:649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Gong K, Guan J, Liu C, Qi J. PET Image Denoising Using a Deep Neural Network Through Fine Tuning. IEEE Transactions on Radiation and Plasma Medical Sciences. 2019;3:153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Ouyang J, Chen KT, Gong E, Pauly J, Zaharchuk G. Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Medical physics. 2019;46:3555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Stickel JR, Cherry SR. High-resolution PET detector design: modelling components of intrinsic spatial resolution. Physics in Medicine & Biology. 2005;50:179–95. [DOI] [PubMed] [Google Scholar]
- [141].Funck T, Paquette C, Evans A, Thiel A. Surface-based partial-volume correction for high-resolution PET. Neuroimage. 2014;102 Pt 2:674–87. [DOI] [PubMed] [Google Scholar]
- [142].Vandenbroucke A, Foudray AM, Olcott PD, Levin CS. Performance characterization of a new high resolution PET scintillation detector. Physics in Medicine & Biology. 2010;55:5895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Yang Y, Bec J, Zhou J, Zhang M, Judenhofer MS, Bai X, et al. A Prototype High-Resolution Small-Animal PET Scanner Dedicated to Mouse Brain Imaging. Journal of Nuclear Medicine. 2016;57:1130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Meltzer CC, Kinahan PE, Greer PJ, Nichols TE, Comtat C, Cantwell MN, et al. Comparative evaluation of MR-based partial-volume correction schemes for PET. Journal of Nuclear Medicine. 1999;40:2053–65. [PubMed] [Google Scholar]
- [145].Meltzer CC, Zubieta JK, Links JM, Brakeman P, Stumpf MJ, Frost JJ. MR-based correction of brain PET measurements for heterogeneous gray matter radioactivity distribution. Journal of Cerebral Blood Flow and Metabolism. 1996;16:650–8. [DOI] [PubMed] [Google Scholar]
- [146].Rousset OG, Ma Y, Evans AC. Correction for partial volume effects in PET: Principle and validation. Journal of Nuclear Medicine. 1998;39:904–11. [PubMed] [Google Scholar]
- [147].Sattarivand M, Kusano M, Poon I, Caldwell C. Symmetric geometric transfer matrix partial volume correction for PET imaging: principle, validation and robustness. Physics in Medicine & Biology. 2012;57:7101–16. [DOI] [PubMed] [Google Scholar]
- [148].Erlandsson K, Buvat I, Pretorius PH, Thomas BA, Hutton BF. A review of partial volume correction techniques for emission tomography and their applications in neurology, cardiology and oncology. Physics in Medicine & Biology. 2012;57:R119–R59. [DOI] [PubMed] [Google Scholar]
- [149].Song T-A, Chowdhury SR, Yang F, Dutta J. Super-resolution PET imaging using convolutional neural networks. arXiv e-prints 2019. 1906.03645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Lei Y, Liu Y, Dong X, Tian S, Wang T, Jiang X, et al. Automatic multi-organ segmentation in thorax CT images using U-Net-GAN. SPIE Medical Imaging. 2019;10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Lei Y, Wang T, Wang B, He X, Tian S, Jani AB, et al. Ultrasound prostate segmentation based on 3D V-Net with deep supervision. SPIE Medical Imaging. 2019;10955. [Google Scholar]
- [152].Wang B, Lei Y, Jeong JJ, Wang T, Liu Y, Tian S, et al. Automatic MRI prostate segmentation using 3D deeply supervised FCN with concatenated atrous convolution. SPIE Medical Imaging. 2019;10950. [Google Scholar]
- [153].Wang B, Lei Y, Wang T, Dong X, Tian S, Jiang X, et al. Automated prostate segmentation of volumetric CT images using 3D deeply supervised dilated FCN. SPIE Medical Imaging. 2019;10949. [Google Scholar]
- [154].Wang T, Lei Y, Shafai-Erfani G, Jiang X, Dong X, Zhou J, et al. Learning-based automatic segmentation on arteriovenous malformations from contract-enhanced CT images. SPIE Medical Imaging. 2019;10950. [Google Scholar]
- [155].Wang T, Lei Y, Tang H, Harms J, Wang C, Liu T, et al. A learning-based automatic segmentation method on left ventricle in SPECT imaging. SPIE Medical Imaging. 2019;10953. [Google Scholar]
- [156].Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Medical Physics. 2019;46:2157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Lei Y, Tian S, He X, Wang T, Wang B, Patel P, et al. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net. Medical physics. 2019;46:3194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Wang B, Lei Y, Tian S, Wang T, Liu Y, Patel P, et al. Deeply supervised 3D fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation. Medical physics. 2019;46:1707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Wang T, Lei Y, Tang H, He Z, Castillo R, Wang C, et al. A learning-based automatic segmentation and quantification method on left ventricle in gated myocardial perfusion SPECT imaging: A feasibility study. Journal of Nuclear Cardiology. 2019; 10.1007/s12350-019-01594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Wang T, Lei Y, Tian S, Jiang X, Zhou J, Liu T, et al. Learning-based automatic segmentation of arteriovenous malformations on contrast CT images in brain stereotactic radiosurgery. Medical physics. 2019;46:3133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Hatt M, Laurent B, Ouahabi A, Fayad H, Tan S, Li L, et al. The first MICCAI challenge on PET tumor segmentation. Medical Image Analysis. 2018;44:177–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Lei Y, Dong X, Tian Z, Liu Y, Tian S, Wang T, et al. CT prostate segmentation based on synthetic MRI-aided deep attention fully convolution network. Medical physics. 2020;47:530–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Lei Y, Wang T, Tian S, Dong X, Jani AB, Schuster D, et al. Male pelvic multi-organ segmentation aided by CBCT-based synthetic MRI. Physics in Medicine & Biology. 2020;65:035013. [DOI] [PMC free article] [PubMed] [Google Scholar]


