Abstract
Androgen receptor (AR) antagonism increases overall survival in prostate cancer; however, treatment failure leads to tumor progression and patient mortality. The effect of AR modulation on AR+ non-tumor cells that participate in the resistance to AR antagonism is poorly understood. Tumor-infiltrating myeloid cells, including macrophages and myeloid-derived suppressor cells (MDSCs), express AR and promote prostate cancer progression. We investigated how AR antagonism affects myeloid cell function and metabolism in an AR-independent murine colon tumor model. Systemic blockade of AR with enzalutamide resulted in increased MC-38 tumor growth in vivo even when AR was knocked out of MC-38 tumor cells. MC-38 tumor growth was also increased when immunocompetent, but not immuno-deficient, mice were co-injected with tumor cells and MDSCs treated with enzalutamide or lacking AR, suggesting that AR regulated the ability of MDSCs to suppress adaptive immunity. Myeloid AR knockout (MARKO) male mice also displayed increased growth of TRAMP C2 prostate tumors when compared to WT. Inhibition of AR signaling suppressed mitochondrial respiration in myeloid cells via MPC/AMPK signaling pathways; suppression of mitochondrial respiration increased MDSC tumor-promoting functions. Our work showed that AR regulates a tumor-promoting myeloid cell phenotype and influences myeloid cell metabolism. These findings suggest that tumor resistance to AR antagonism is due in part to changes in myeloid cell function and metabolism.
Keywords: Androgen receptor, Enzalutamide, Myeloid Cells, Immunosuppression, Metabolism
Introduction
Prostate cancer (PCa) has the highest malignancy rates and is the second leading cause of cancer mortality in men. Hormone ablation therapy is a commonly used treatment for PCa patients, as early-stage prostate tumors require androgens for growth. Limiting the amount and function of androgen decreases tumor cell proliferation, reduces tumor sizes, and improves overall survival [1, 2]. Androgen deprivation therapy (ADT) reduces testosterone concentrations in the blood, whereas antiandrogen therapy directly inhibits androgen receptor (AR) signaling in prostate cells. Although initially effective, hormone therapy eventually fails in a proportion of patients and leads to progression of castration-resistant prostate cancer (CRPC) [1]. Various tumor cell intrinsic mechanisms are described for treatment resistance, such as genetic alterations of AR (amplifications, mutations, splice variants) and the upregulation of pathways that support AR signaling [1].
Systemic hormone therapy might also affect the tumor microenvironment through modulation of AR+ cells in the tumor niche [3–6]. Myeloid cells in the tumor niche, including tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs), promote tumor development and progression by inducing inflammation, immunosuppression, and angiogenesis, thus supporting treatment resistance [4, 7, 8]. Myeloid cell infiltration is associated with poor cancer prognosis [9, 10]. The role of AR expression by myeloid cells is controversial in murine prostate tumor models. Genetic deletion of AR in macrophages delays initial tumorigenesis [4], but increases progression and metastasis [11]. ADT and AR antagonism with the antiandrogen enzalutamide increases TAM infiltration in murine prostate tumors, leading to PCa progression [12]. Thus, the direct role of AR in myeloid cell function in cancer and their response to antiandrogen therapy remains unclear.
Myeloid cell function is highly dependent on metabolism [13] and upregulation of glycolysis increases tumor-promoting capacity of TAMs and immunosuppression by MDSCs [14–17]. Androgen stimulation of AR in prostate cancer cells induces glycolysis [18]. It is unknown if AR antagonism modulates myeloid cell metabolism, and whether treatment-induced metabolic changes impact myeloid cell function.
In order to understand AR regulation of myeloid cells in tumors, we utilized the AR-independent MC-38 colon tumor model [19], and the TRAMP C2 prostate tumor model [20]. The effect of AR on myeloid cell function was investigated using pharmacologic inhibition and myeloid cell specific genetic deletion of AR. Pharmacological blockade of AR in myeloid cells with enzalutamide increased their tumor-promoting capacity by inhibiting of adaptive immunity. AR pharmacological inhibition also induced VEGF and Arg1 expression, and directly increased the suppressive activity of MDSCs. Blocking AR signaling with enzalutamide altered myeloid cell metabolism by decreasing mitochondrial respiration and increasing glycolysis. The effects on metabolism were mediated in an MPC/AMPK-dependent manner. Our work suggests that resistance to AR antagonism and subsequent relapse of CRPC was due in part to the effects of AR antagonism on myeloid cell function and metabolism.
Material and Methods
Animal studies
Seven- to ten-week old C57BL/6J male and female and B6-severe combined immunodeficiency (B6-SCID) male mice were purchased from Taconic Laboratory (Hudson, NY) and the LysMcre C57BL/6 male mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Male SCID pathogen-free mice aged 7–8 weeks were obtained through the Laboratory Animal Resource of RPCCC. ARfloxed mice were generated by De Gendt Lab at Katholieke Universiteit Leuven [21] and kindly shared by the Agoulnik Lab at Florida International University. GFP+ mice were kindly donated by Dr. Andrei Gudkov Lab at Roswell Park Cancer Institute. For the generation of Myeloid AR KnockOut (MARKO) mice, Lys-Mcre males were crossed with ARfloxed females to generate MARKO males. Mice were housed in microisolator cages in a laminar flow unit under ambient light at 24°C. The RPCCC Institutional Animal Care and Use Committee (IACUC) approved all procedures and experiments for this study.
Genotyping
Tail clips of littermates from Lys-Mcre and ARfloxed breeding pairs were digested in tail lysis buffer (Viagen Biotech 102-T) with 200ug proteinase K (Viagen Biotech 502-PK) following manufacturer’s instructions, and DNA concentrations were determined using a photometer (BioRad). For PCR reactions, Platinum Taq DNA polymerase was used following manufacturer’s instructions for PCR reaction (ThermoFisher 10966018), with AR PCR primers F 5’ AGCCTGTATACTCAGTTGGGG 3’ and R 5’ AATGCATCACATTAAGTTG ATACC 3’. WT AR band was genotyped as 855 bp, ARfloxed as 952 bp and ARKO as 404 bp.
Primary cultures
Bone marrow cells were isolated by flushing out BM from femur and tibia with needle/ syringe. BMDMs were generated by culturing 1×106 unfractioned bone marrow cells from C57BL/6 WT or MARKO male mice with 30ng/mL M-CSF (ThermoFisher 14–8983-80) in 10cm dishes or culturing 0.2×106 bone marrow cells in 6-well plates in phenol red free RPMI 1640 (ThermoFisher 11835030) supplemented with 100ug/ml Penicillin-Streptomycin-Glutamine (ThermoFisher 10378016) and 10% FBS Premium Select (Atlanta S11595) at 37 °C and 5% CO2 for 5 days. BMDMs generated in 10cm dishes were used in flow cytometry, admixture, suppression, seahorse, and western blot experiments, while BMDMs generated in 6-well plates were utilized in RNA experiments. Mature day 5 BMDMs were then stimulated with DMSO or 5uM enzalutamide (Selleckchem S1250) and other pathway inhibitors (see below in this section) for 24h. Murine MDSCs were generated by culturing 2.5×106 unfractioned bone marrow cells from male WT, MARKO or GFP+ C57BL/6 mice with 40ng/mL GM-CSF (R&D Systems 415-ML) and 40ng/mL IL-6 (Preprotech 216–16) in the presence or absence of 5uM enzalutamide (Selleckchem S1250) and other pathway inhibitors (see below in this section) in 10cm dishes in phenol red free media RPMI 1640 (ThermoFisher 11835030) supplemented with 100ug/ml Penicillin-Streptomycin-Glutamine (ThermoFisher 10378016) and 10% FBS Premium Select (Atlanta S11595) at 37 °C and 5% CO2 for 4 days [22]. After 4 days of MDSC culture, MDSCs were harvested for use in assays. For intracellular cytokine detection, day 4 MDSCs were cultured with GolgiStop (BD 554724) following manufacturer’s instructions for 4h. The following inhibitors were utilized utilized and diluted according to manufacturer’s instructions: mTOR (100nM rapamycin, Selleckchem S1039), HIF-1a (10uM YC-1, Selleckchem S7958), AMPK (6.25uM dorsomorphin, Selleckchem S7306), AKT (5uM MK-2206, Selleckchem S1076), MPC (40uM UK5509, Selleckchem S5317), and 2-DG (1.5mM Sigma D6134).
Cell culture
Murine MC-38 colon cancer cells (purchased from Kerafast ENH204 in 2018) were cultured in Dulbecco’s modified MEM (DMEM) (ThermoFisher 11965–118) with 10% FBS Optima (Atlanta S12495), 100ug/ml Penicillin-Streptomycin-Glutamine (ThermoFisher 10378016), 0.1 mM nonessential amino acids (ThermoFisher 11140035), 1 mM sodium pyruvate (ThermoFisher 11360070) and 10 mM Hepes (ThermoFisher 15630080) at 37°C and 5% CO2. Human PC3M prostate tumor cells (obtained from Dr. I. Gelman (RPCCC) in 2016) and cultured in RPMI 1640 (ThermoFisher 11875–119) 10% FBS Optima (Atlanta S12495) and 100ug/ml Penicillin-Streptomycin-Glutamine (ThermoFisher 10378016) at 37 °C and 5% CO2. All cell lines were cultured for a maximum of 15 passages (maximum of 2 weeks). Cell lines were not authenticated in the past year and are routinely tested for Mycoplasma; only Mycoplasma negative cells are used for experiments. For in vitro experiments, 0.066×106 MC-38 cells were plated in 6-well plates. On day 1, cells were treated with diluent DMSO or 5uM enzalutamide for 24, 48, 72 and 96h for cell number and viability assessment by trypan blue staining (>90% viability was used). TRAMP C2 prostate tumor cells (obtained from ATCC CRL-2731 in year?) were cultured in the presence of 10−8 M dihydrotestosterone (Sigma D-073) at 37 °C and 10% CO2 and as previously described [20]. MC-38 ARKO cells were generated using CRISPR/Cas9 gene editing. MC-38 cells were transfected with AR-Crispr/Cas9 KO (sc-419181, Santa Cruz Biotechnology) and AR-HDR (sc-419181-HDR) plasmids, which contain sequences encoding green fluorescent protein (GFP) or a puromycin resistance gene respectively for selection of ARKO cells, according to manufacturer’s instruction. MC-38 control cells were transfected with the pGIPZ-GFP plasmid. For transfection, plasmids in equivalent ratios were diluted in Plasmid Transfection Medium (sc-108062) and mixed with UltraCruz Transfection Reagent (sc-395739). Prior to transfection, MC-38 growth medium was replaced with fresh antibiotic-free medium, and the transfection complexes (5 ug of each plasmid, 50 ul of transfection reagent in 1.5 ml of transfection medium) were added dropwise to the fresh antibiotic-free growth medium (10 ml in 100-mm dish). The medium was replaced in 24 hours. MC-38 cells were harvested 72 hours post-transfection and sorted for GFP expression (BD FACSAria II, BD Biosciences) to enrich the target population of transfected cells. GFP expressing cells were plated in growth medium, and cells where Cas9-induced DNA cleavage has occurred were selected with puromycin. The ARKO phenotype of MC-38 cells was confirmed by WB using the AR antibody (06–680, MilliporeSigma; Supplementary Figure 2A).
In vivo tumor experiments and tumor processing
C57BL/6 males were inoculated subcutaneously on the shoulder with 100uL of 105 or 106 MC-38 cells. When tumors inoculated with 106 MC-38 cells reached 100mm3, mice were treated with saline or enzalutamide 20mg/kg daily by oral gavage in less than 5 ml/kg of body weight. For admixture experiments, either 2×105 BMDMs or MDSCs were mixed in a 2:1 ratio with MC-38 cells in PBS and 100uL were implanted subcutaneously on the shoulder of C57BL/6 males. C57BL/6 and MARKO male mice were inoculated subcutaneously on the shoulder with 100uL of 106 TRAMP C2 prostate tumor cells in PBS. SCID males were inoculated subcutaneously on the shoulder with 100uL of 106 PC3M cells in PBS. Tumors were measured with an external caliper and tumor volume was calculated by Volume = Length × (Widtĥ2) × 1/2. Tumor growth was measured until tumors reached endpoint of 2000 mm3. A human prostate cancer xenograft (PCaX) was also studied (sample obtained with written consent and in accordance with the U.S. Common Rule), in collaboration with Dr. Barbara A. Foster (RPCCC)). PCaX derives from one caucasian male diagnosed with PCa at 55 years of age. Tumor staging is 4 Gleason primary/ 5 Gleason secondary, T1c, N0, M1b. tumors from a human prostate cancer xenograft (PCaX). PCaX tumor cells were implanted in NSG males, and when tumors reached 200mm3, mice were left either untreated or were treated with enzalutamide (25mg/kg 5 days a week by oral gavage) until tumors reached the endpoint of 1000mm3. Tumors were digested for 1h with 5mg collagenase (Sigma C6885) and 50ug DNaseI (Sigma D4527–200KU) using gentleMACS octo Dissociator with heaters using gentleMACS C tubes (Miltenyi) and program 37-m-TDK-3
Suppression Assay
Spleens were collected and splenocytes were harvested from C57BL/6 male mice by mashing spleens, centrifuging and lysing RBCs with RBC lysis buffer. Pan T cells were isolated by negative selection following manufacturer’s instructions (Miltenyi Biotec 130–095-130 and 130–042-401) and Pan T cell enrichment was confirmed by flow cytometry (>90% CD3+ T cells). Pan T cells were stained with CTV following manufacturer’s instyructions to allow monitoring of T cell proliferation through dye dilution (ThermoFisher C34557). CTV-stained PanT cells were stimulated with anti-CD3/CD28 beads according to manufacturer’s instructions (ThermoFisher 11452D) in a 1:1 ratio, and MDSCs generated (see above Primary cultures) were cultured with T cells in a ratio of 1:1, 1:2 and 1:4 MDSC:T cell for 4 days in phenol red free RPMI 1640 (ThermoFisher 11835030) supplemented with 100ug/ml Penicillin-Streptomycin-Glutamine (ThermoFisher 10378016) and 10% FBS Premium Select (Atlanta S11595). Cells were then stained with surface antibodies and analyzed for T cell proliferation by flow cytometry (details under Flow cytometry and Imagestream section).
Isolation of CD11b+ myeloid cells
CD11b+ tumor-infiltrating cells were isolated by positive selection using magnetic bead separation following manufacturer’s instructions from single cell suspensions generated from digested tumors (Miltenyi Biotec 130–049-601 and 130–042-401). The purity of the enriched CD11b+ cells was analyzed by flow cytometry (as described in Flow cytometry and Imagestream section), with the percentage of CD11b+ cells being > 80% for each separation.
RNA expression
Total RNA extraction was performed using the Trizol® method according to manufacturer’s instructions (ThermoFisher 15596018). RNA concentrations were determined using a photometer (BioRad). cDNA synthesis was done using iScript cDNA Synthesis Kit (BioRad 1708891) and q-PCR using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad 1725275), both according to the manufacturer’s instructions. RNA expression was normalized to GAPDH levels. PCR primers were purchased from IDT. Primer sequences: Arg1 F 5’ AAGAAAAGGCCGATTC ACCT 3’ R 5’ CATGATATCTAGTCCTGAAAGG 3’, GAPDH F 5’ GGCAAGTTCAACGGCACAGTCAAG 3’ R 5’ GCACATACTCAGCACCAGCATC AC 3’, GLUT1 F 5’ TATCGTCAACACGGCCTTCAC TGT 3’ R 5’ CACAAAGCCAAAGA TGGCCACGAT 3’, IL-1β F 5’ AAGGAGAACCAAGCAACGACAAAA 3’ R 5’TGGGGAA CTCTGCAGACTCAAACT 3’, IL-6 F 5’ GACAAAGCCAGAGTCCTTCAGAGAG 3’ R 5’ CTAGGTTTGCC GAGTAGATCTC 3’, IL-10 F 5’GAGACTTGCTCTTGCACTACC 3’ R 5’ CTCTCTTTTCT GCAAGGCTG 3’, iNOS F 5’ CATCTCCGCAAATGTAGAGG 3’ R 5’ CAAACCCAAGGT CTACGTTCA 3’, MPC-2 F 5’ CCGCTTTACAACCACCCGGCA 3’ R 5’ CAGCACACAC CAATCCCCATTTCA 3’, TGF-β F 5’ CGTCAGACATTCGGGAAGA 3’ R 5’ CGTATCAG TGGGGGTCAGCA 3’, VEGF F 5; GAGGATGTCCTCACTCGGATG 3’. Gene expression was determined by 2^–ΔΔCT.
Flow cytometry and Imagestream
For flow cytometry and Imagestream staining, single cell suspensions (digested tumors, BMDMs, MDSCs) were treated for 10min with 2:1 diluted mouse IgG: PBS (ThermoFisher 10400C) to block Fc receptors expressed on myeloid cells, followed by incubation with antibodies against cell surface molecules for 15min. If intracellular staining was performed, cells were washed twice with FACs buffer, and cells were fixed/permeabilized (ThermoFisher 00–5523-00) for a minimum of 30min, following manufacturer instructions. Permeabilized cells were treated for with 2:1 diluted mouse IgG: PBS for 15min, followed by incubation with antibodies against intracellular molecules for 30min. Flow cytometry analysis was done by flow cytometer (BD Fortessa, BD LSRII). ImageStream analyses were performed using ImageStreamX Mark II. For p65 nuclear translocation control, MDSCs were stimulated with 100 ng/mL LPS (Sigma L4516) and 2 μM ionomycin (Sigma I0634) for 30min at 37 °C and 5% CO2, stained for surface markers, fixed with 4% formaldehyde, permeabilized and stained for intracellular markers. Flow cytometry and Imagestream analyses were performed using FlowJo™ and IDEAS, respectively. Surface antibodies used for flow cytometry and Imagestream were CD45 (BD 550994), CD11b (BD 553311), F4/80 (ThermoFisher 25–4801-82), CD115 (ThermoFisher 12–1152-82), Gr-1 (BD 553127), Ly6-G (BioLegend 127612), Ly6-C (BioLegend 128033), CD4 (BD 550954), CD8 (BD 553033), CD3e (ThermoFisher MA5–17658), and PD-L1 (Thermo 12–5982-82). Antibodies against intracellular proteins were p65 (Cell signaling 8242), and IL-23p90 (Thermo 50–7023-82). DAPI (Thermo D1306) and LD Aqua (ThermoFisher L34957) were utilized to assess viability.
Protein expression
Protein lysates were made using MT lysis buffer (Sigma C3228) in the presence of protease (Sigma P8340) and phosphatase inhibitors (Santa Cruz sc-45044 and sc-45045) according to manufacturer’s instructions. Protein concentrations were determined using Bradford assays (BioRad 5000006) and 20–40ug of protein were run on 10% bis-tris gels (ThermoFisher NP0303BOX) and then transferred to nitrocellulose membranes (ThermoFisher LC2001). Blots were probed for antibodies specific for ACC (Cell Signaling 3676S), p-ACC (Cell Signaling 11818S), AMPK (Cell Signaling 2532S), p-AMPK (Cell Signaling 2535S), AR (Sigma 06–680), HIF-1α (Cell Signaling 3716) and β-Actin (Sigma A2228). Secondary incubations were performed with HRP-antimouse (Santa Cruz SC-516102) and HRP-anti rabbit (Enzo ADI-SAB-300-J). Primary antibody incubations were done overnight at 4°C and secondary antibody incubations were done for 1h at room temperature. HRP was developed (BioRad 170–5061) and recorded using a Bio-Rad imager (BioRad ChemiDoc XRS+) and quantified using ImageLab software (Bio-Rad, Hercules, California).
Metabolism
Glucose uptake was assessed by culturing BMDMs (section Primary cultures) treated with DMSO or 5uM enzalutamide for 9h, incubating cells with 100uM of fluorescent glucose analog 2-NBDG (ThermoFisher N13195) in serum-free media without glucose for 30min at 37°C and quantifying glucose uptake by flow cytometry. Mitochondria was detected by flow cytometry with Mitotracker (ThermoFisher M7512), mitochondrial superoxide with MitoSOX (ThermoFisher M36008) and cellular ROS with CM-H2DCFDA (ThermoFisher C6827) following manufacturer’s instructions.
105 MDSCs were plated in Cell-Tak coated plates (ThermoFisher CB40241) and 5×104 BMDMs were plated in XF96 Cell Culture Microplates. Glycolytic rates were measured using Seahorse XF Glycolysis Stress Test Kit (Agilent 103020–100), mitochondrial respiration was measured with Seahorse XF Cell Mito Stress Test Kit (Agilent 103015–100) according to manufacturer’s instructions by Roswell Park’s Immune Analysis Facility. The carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) concentration utilized was 2uM. Fatty acid oxidation (FAO) was performed using the XF Palmitate-BSA FAO Substrate (Agilent 102720–100) according to manufacturer’s instructions. FAO was calculated by subtracting values for before and after 40uM etomoxir treatment. Normalization of results was performed for adherent cells using methylene blue. Metabolic analyses were run on a Seahorse Xfe 96 Extracellular Flux Analyzer.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.0 software. When comparing two groups, statistical analyses were performed using two-tailed Mann-Whitney or paired tests. When comparing two groups or more groups, 1-way or 2-way ANOVA were performed. Multiple comparison correction was applied when necessary. Tumor growth rate was analyzed using mixed model analyses for random slope and intercept. For metabolic profile plots, ellipses from were estimated with ggplot2 in R. Differences were considered significant when P values were ≤ 0.05.
Results
AR antagonism increased colon tumor growth
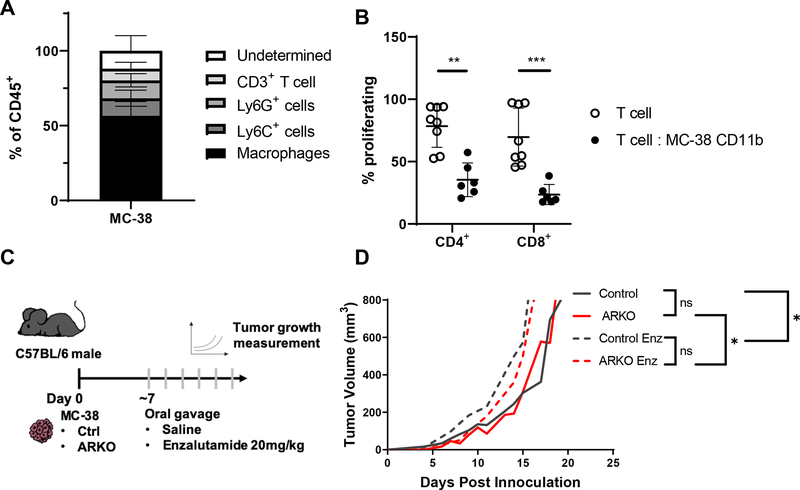
Tumor-infiltrating myeloid cells can promote tumor development and progression by a number of mechanisms [8]. The MC-38 is a murine colon tumor model that is syngeneic to C57BL/6 and exhibits myeloid-biased leukocyte infiltration (Figure 1A, Supplementary Figure 1A; [19]). MC-38 infiltrating CD11b+ myeloid cells, which include tumor-associated macrophages (TAMs) and MDSCs, suppress T cell proliferation ex vivo (Figure 1B; Supplementary Figure 1B). To examine the tumor-independent effects of AR blockade, we first investigated whether tumor cell expression of AR affected tumor growth. MC-38 cells have low expression of AR (Supplementary Figure 2A); to assess the impact of AR on tumor growth we used CRISPR/Cas9 gene editing to eliminate AR expression in MC-38 tumor cells (MC-38 ARKO). MC-38 ARKO cells had significantly lower expression of AR when compared to MC-38 cells (p<0.01; Supplementary Figure 2A). To assess if AR had an indirect effect on tumor growth, we examined the effect of AR antagonist, enzalutamide, treatment on tumor growth. MC-38 and MC-38 ARKO tumor bearing mice were treated daily with enzalutamide or saline through oral gavage (Figure 1C). Tumor growth was not significantly different in male C57BL/6 mice inoculated with MC-38 control or MC-38 ARKO tumor cells (Figure 1D). Enzalutamide treatment had a modest, but significant effect on accelerating both MC-38 control and MC-38 ARKO tumor growth over that of saline treated MC-38 and MC-38 ARKO tumor growth, respectively (Figure 1D). In contrast, enzalutamide treatment of MC-38 cells in vitro reduced cell numbers over time (Supplementary Figure 2B); the effects of enzalutamide on MC-38 ARKO cell proliferation was delayed as compared to the effect on control cells, which is likely a result of the reduced AR expression (Supplementary Figure 2A). In total, these results supported the hypothesis that AR antagonism enhanced the tumor-promoting properties of non-tumor cells. Enzalutamide treatment reduced the percentage of total infiltrating leukocytes, with this difference being attributed to decreased macrophage infiltration in tumors (Supplementary Figure 2C).
Figure 1. AR inhibition accelerated MC-38 colon tumor growth.
(A, B) Eight- to twelve-week old C57BL/6 males were injected subcutaneously on the shoulder with 106 MC-38 colon tumor cells. Analyses were done when tumors reached 1000mm3. Graphs show pooled data of 2 experiments with 3 mice per group per experiment and show mean and standard deviation. (A) Percentage of tumor infiltrating leukocyte populations within CD45+ cells determined by flow cytometry. CD11b+ cells from tumors were isolated and measured for suppressive activity in vitro. Graph depicts (B) proliferation of CD4+ and CD8+ T cells cocultured with tumor-infiltrating CD11b+ cells, analyzed with Man-Whitney test. (C) C57BL/6 males were injected subcutaneously on the shoulder with 106 MC-38 control or MC-38 ARKO colon tumor cells and given saline or enzalutamide 20mg/kg daily by oral gavage when tumors reached 100mm3. (D) Tumor growth curve is depicted with pooled data of 2 experiments with 5 mice per group analyzed with random mixed model. Mean tumor size vs. time is shown. Black lines indicate MC-38 control, red lines indicate MC-38 ARKO, and dashed lines indicate enzalutamide treatment. * p<0.05, **p<0.01 and ***p<0.001.
AR inhibition increased tumor-promoting capacity of myeloid cells
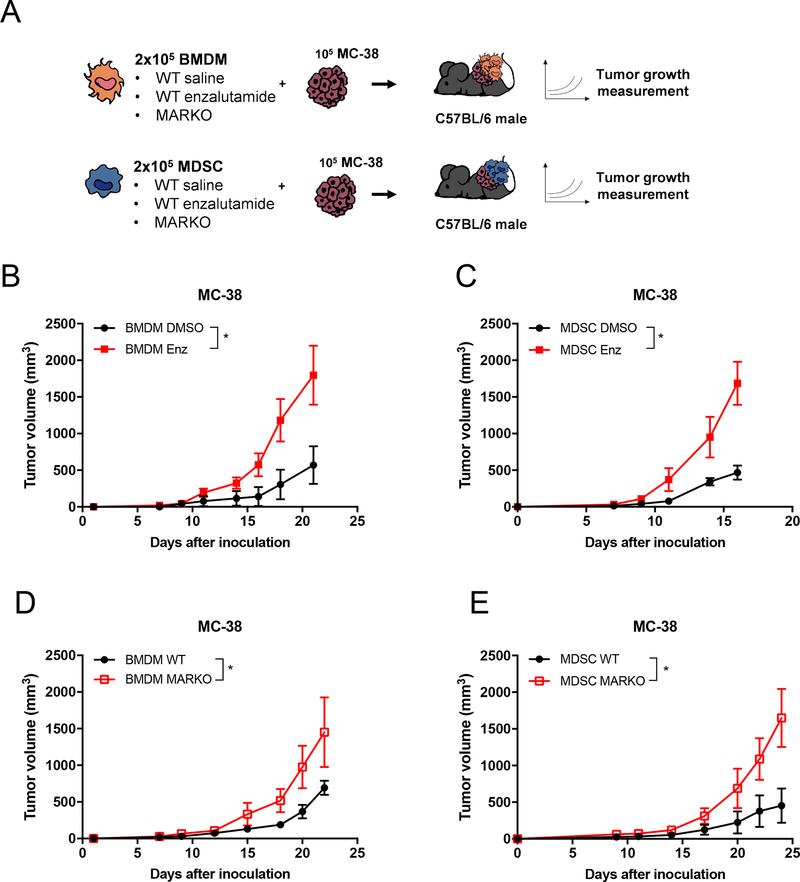
AR antagonism potentially affects a variety of AR+ cell types that compose the tumor microenvironment [4, 23]. Since the majority of tumor infiltrating immune cells in MC-38 tumors were myeloid-biased (Figure 1A, Supplementary Figure 2C) and myeloid cells accelerates in vivo MC-38 tumor growth (Supplementary Figure 3A), we next assessed whether genetic deletion or pharmacologic blockade of AR in macrophages or MDSCs could further enhance colon tumor growth. BMDMs and MDSCs were generated in vitro from Lys-Mcre ARfloxed myeloid-specific AR knockout mice (MARKO) or WT (C57BL/6) bone marrow in the presence of vehicle (DMSO) or enzalutamide. Myeloid cell populations were then mixed with 105 MC-38 tumor cells in a 2:1 myeloid: tumor cell ratio. The mixture was injected subcutaneously into C57BL/6 males and tumor progression was followed (Figure 2A). Enzalutamide treatment of either BMDMs or MDSCs led to enhanced MC-38 tumor growth when compared to DMSO-treated myeloid cells (Figures 2B, C). Myeloid cells from the initial GFP+ MDSC:MC-38 mixture remained in tumors, with about 10% of total tumor infiltrating macrophages and 6% of Ly6G+ cells belonging to initial GFP+ myeloid cells. Nonetheless, the enzalutamide-treated macrophages appeared to be more labile than their DMSO control counterparts (Supplementary Figure 3B). DMSO and enzalutamide-treated MDSC admixed tumors did not display differences in percentage of leukocyte subpopulation infiltration (Supplementary Figure 3C). Mixture of MC-38 cells with MARKO myeloid cells also led to increased MC-38 tumor growth when compared to WT myeloid cells (Figures 2D, E), indicating that inhibition of AR in myeloid cells enhanced their tumor-supporting capacity. The observed effects were AR-dependent, as enzalutamide treatment of MARKO myeloid cells did not further alter MC-38 tumor growth (Supplementary Figures 3D–E).
Figure 2. Tumor-supporting properties of myeloid cells were increased upon AR inhibition.
(A) Experimental design: BMDMs and MDSCs were generated in vitro from WT or MARKO bone marrow of C57BL/6 males in the presence of DMSO or enzalutamide. BMDMs or MDSCs were then mixed with MC-38 tumor cells in a 2:1 myeloid: tumor cell ratio and implanted subcutaneously on the shoulder of C57BL/6 male mice. (B-E) Graphs depict tumor growth curves of mice injected with MC-38 cells mixed with DMSO or enzalutamide-treated WT (B) BMDMs and (C) MDSCs or mixed with WT or MARKO (D) BMDMs or (E) MDSCs. Graphs depict mean and standard deviation of representative experiment of 2–3 experiments with 3 mice/ group. For all graphs, black dots denote DMSO-treated WT MDSCs, red solid squares enzalutamide-treated MDSCs and red empty squares MARKO MDSCs. Statistical analyses were done using two-way ANOVA. *p<0.05.
AR antagonism enhanced MDSC immunosuppression
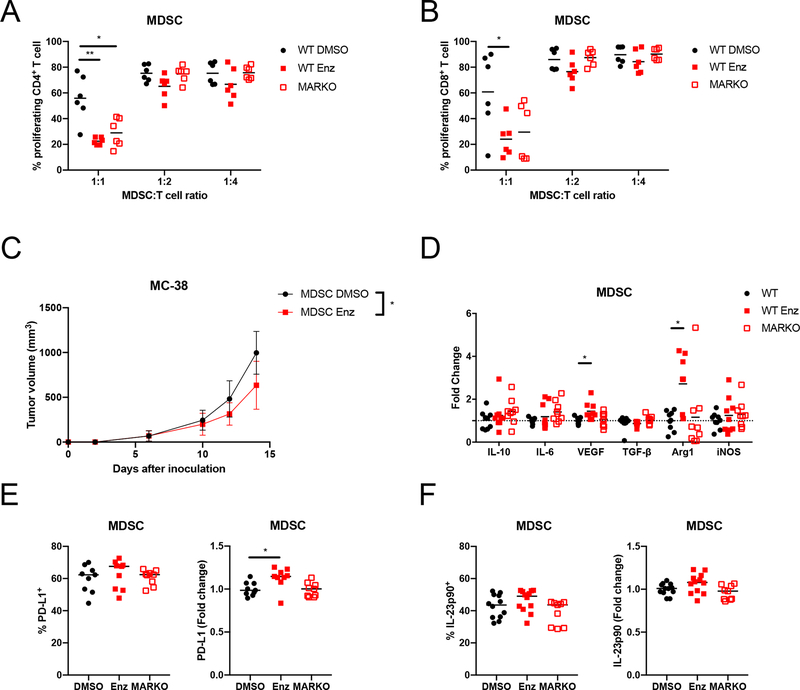
MDSCs can support tumor growth by suppressing anti-tumor immune responses [8]. Both genetic deletion and pharmacological inhibition of MDSCs AR led to increased immunosuppressive activity in suppression assays, as demonstrated by decreased CD4+ and CD8+ T cell proliferation in MDSC co-cultures (Figures 3A and B). MDSCs treated with enzalutamide were not able to enhance MC-38 cancer growth in immunocompromised B6-SCID mice, indicating that enzalutamide enhanced MDSC tumor-supporting capacity through modulation of the adaptive immune system (Figure 3C).
Figure 3. AR inhibition enhanced MDSC immunosuppression.
(A-B) WT DMSO-treated, WT enzalutamide-treated or MARKO MDSCs were cocultured with CTV-stained anti-CD3/CD28 stimulated CD3+ T cells. Graphs depicts results from 2 pooled experiments of 3 biological replicates each and indicate (A) CD4+ and (B) CD8+ T cell proliferation, which indicate MDSC suppressive potential assessed by flow cytometry. (C) DMSO and enzalutamide-treated MDSCs were generated, mixed in a 2:1 ratio with MC-38 tumor cells and injected subcutaneously into B6-SCID males; plot indicates tumor growth (mean and standard deviation) of 2 pooled experiments with 3–5 mice/group per experiment. (D) RNA was extracted from WT DMSO-treated, WT enzalutamide-treated or MARKO MDSCs and gene expression was assessed by qRT-PCR. Graph depicts gene expression fold change relative to WT MDSCs. (E-F) MDSC protein expression was determined by flow cytometry for PD-L1 (E) and IL-23p90 (F). Dotted line indicates fold change of 1. For all graphs, black dots denote WT MDSCs, red solid squares WT enzalutamide MDSCs and red empty squares MARKO MDSCs. Kruskal-Wallis test were first performed for A, B and D. Lines indicate mean and, when noted, error bars indicate standard deviation. Significant comparisons were then compared by Paired or Mann-Whitney tests with corrections. Two-way ANOVA was performed for data shown in C. *p<0.05, **p<0.01.
To further dissect how AR inhibition enhanced the tumor-supportive capacity of MDSCs, WT MDSCs were treated with enzalutamide and assessed for the expression of tumor-supporting factors. Enzalutamide-treated MDSCs had increased VEGF and Arg1 mRNA expression when compared to DMSO-treated MDSCs (Figure 3D), suggesting that pharmacological inhibition of AR might have enhanced tumor-supporting capability of MDSCs through promotion of angiogenesis (VEGF) and immunosuppression (Arg1). PD-L1 protein expression was increased upon enzalutamide treatment of MDSC (Figures 3E; Supplementary Figure 3F). MDSC IL-23 secretion drives CRPC [7], although enzalutamide treatment did not affect IL-23 protein expression in cultured MDSCs, suggesting an IL-23-independent mechanism (Figure 3F; Supplementary Figure 3F). Genetic deletion of AR in MDSCs did not cause variation in VEGF or Arg1 mRNA expression (Figure 3D), or in PD-L1 and IL-23 protein expression (Figure 3E–F).
AR antagonism increased myeloid cell glycolysis, but decreased mitochondrial respiration
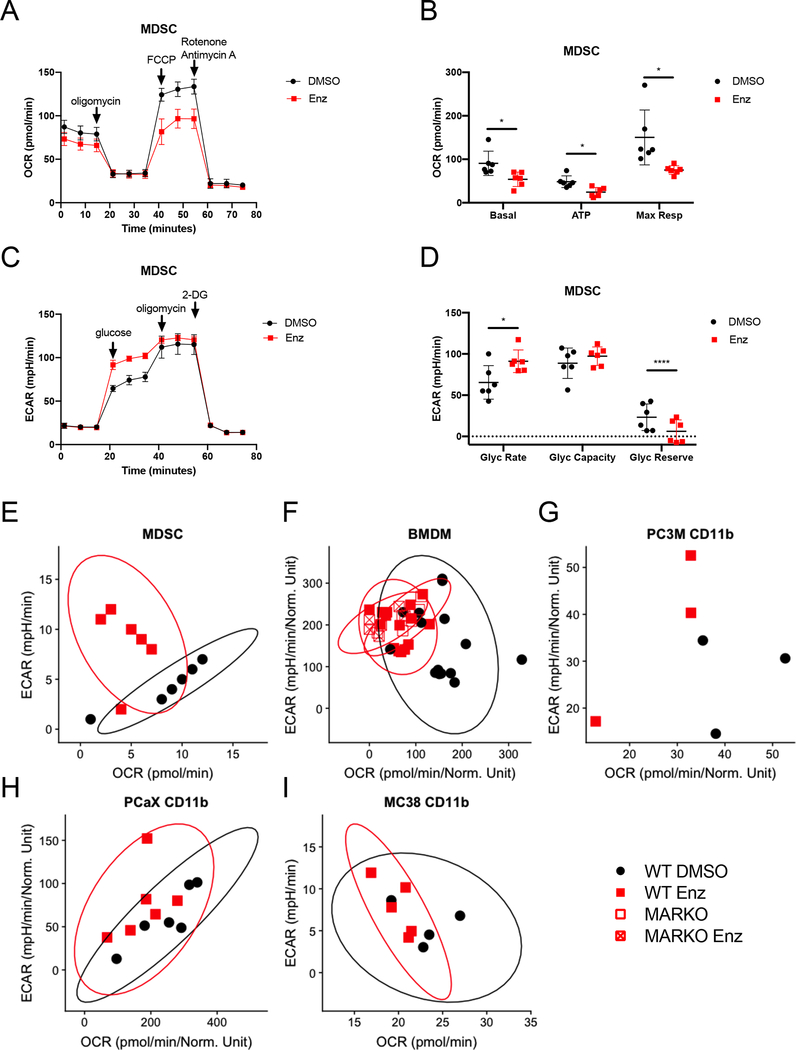
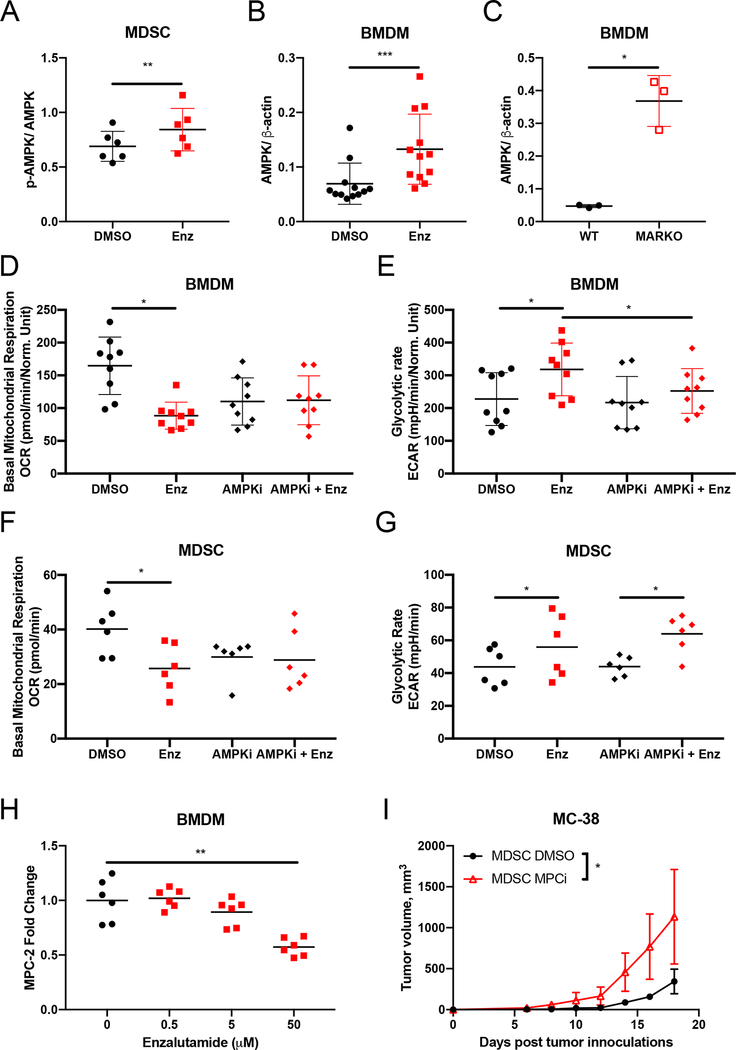
Myeloid cell function is influenced by their metabolic activity [13]. To investigate whether AR blockade altered metabolism in myeloid cells, MDSCs were generated in vitro in the presence of enzalutamide and metabolic changes were assessed using Seahorse technology. Mitochondrial respiration parameters, such as basal respiration, ATP production from oxidative phosphorylation and maximal respiration, were downregulated in MDSCs treated with enzalutamide when compared to DMSO-treated MDSCs (Figures 4A and B). Enzalutamide treatment led to increased glycolytic rate and reduced glycolytic reserve in MDSCs when compared to DMSO-treated controls (Figures 4C and D). Graphing of ECAR vs. OCR showed increase in ECAR in MDSCs treated with enzalutamide (Figure 4E). WT BMDMs treated in vitro with enzalutamide or BMDMs generated from bone marrow of MARKO mice also exhibited decreased mitochondrial respiration and increased glycolysis (Figure 4F). The metabolic changes induced by enzalutamide were dependent on AR, as treatment of MARKO BMDMs with enzalutamide did not alter these metabolic changes (Figure 4F). In addition, in vitro treatment of tumor-associated CD11b+ cells from human prostate tumor xenografts models PC3M and PCaX led to a similar metabolic shift, indicating that tumor-associated myeloid cell metabolism was altered by AR inhibition (Figures 4G, H).
Figure 4. Enzalutamide increased glycolysis and reduced mitochondrial respiration in myeloid cells.
(A-E) MDSCs were generated in DMSO or enzalutamide conditions and were assessed by Seahorse MitoStressTest (A, B) and GlycoStressTest (C, D). Graph indicates mean and standard deviation for mitochondrial respiration test (A) and its calculated parameters (B). Graph indicates mean and standard deviation for glycolytic test (C) and its calculated parameters (D). Metabolic profile plots depict glycolytic rate and basal mitochondrial respiration values from WT MDSCs generated in vitro in the presence of DMSO or enzalutamide (E), in vitro DMSO or enzalutamide treated WT and MARKO BMDMs (F), tumor-associated CD11b+ cells from PCaX (G), and tumor-associated CD11b+ cells from PC3M (H) for 24h. (I) Plot depicts metabolic profile plot from CD11b+ cells sorted from MC-38 tumor-bearing C57BL/6 males that were treated daily with saline or 20mg/kg enzalutamide in vivo through oral gavage. Plots were made from pooled data of 1–3 experiments of 3 biological replicates per group. Ellipses from E, F, G, I and J were estimated with ggplot2 in R. Black filled dots denote DMSO treated, red filled squares indicate enzalutamide-treated, red empty squares indicate MARKO and red crossed squares indicate enzalutamide-treated MARKO myeloid cells. Plots indicate mean and standard deviation. Statistical analyses were performed with paired tests. *p<0.05, ****p<0.0001.
To determine if systemic antiandrogen treatment of tumor-bearing animals affected tumor-associated myeloid cell metabolism, tumor-associated CD11b+ cells were sorted from MC-38 tumor-bearing males treated in vivo with saline or enzalutamide and CD11b+ cell metabolism was assessed. Similar to results observed in vitro, enzalutamide treatment led to decreased mitochondrial respiration and enhancement of glycolysis in tumor-associated CD11b+ cells (Figure 4I), suggesting that in vivo antiandrogen treatment altered the metabolism of tumor-associated myeloid cell directly.
AR antagonism-induced metabolic changes in myeloid cells are MPC-2/AMPK-mediated
Increased glycolysis is frequently associated with a hypoxic environment, particularly in the tumor microenvironment, and linked to an increase in HIF-1α [24]. However, inhibition of HIF-1α did not alter the effects of AR blockade on metabolism (Supplementary Figure 4A). Myeloid cell polarization induces changes in metabolic pathway usage as a consequence of mTOR activation [25]. The effect of AR blockade with enzalutamide on myeloid cellular metabolism was not affected by mTOR inhibition (Supplementary Figure 4B). Increased AKT signaling is associated with increased glucose metabolism [26]; inhibition of AKT did not affect enzalutamide-mediated changes in metabolism (Supplementary Figure 4C). NF-κB signaling positively regulates myeloid cell tumor-promoting function [27], and is involved in regulation of glucose metabolism and resistance to enzalutamide in prostate cancer [28]. AR pharmacological inhibition in MDSCs did not alter NF-κB p65 levels or nuclear translocation (Supplementary Figure 4D).
AMPK regulates energy production through induction of glycolysis and fatty acid oxidation, and is implicated in myeloid cell function [29, 30]. AR pharmacological inhibition enhanced AMPK activation in MDSCs (Figure 5A); AR pharmacological inhibition and genetic deletion of AR upregulated AMPK expression in BMDMs (Figure 5B, C). To investigate whether the metabolic changes associated with enzalutamide were due to AMPK induction in myeloid cells, BMDMs were treated with enzalutamide in the presence or absence of an AMPK inhibitor (dorsomorphin) for 24h. As previously observed (Figure 4), enzalutamide treatment of myeloid cells decreased basal mitochondrial respiration while increasing glycolytic rate. Enzalutamide was unable to downregulate basal mitochondrial respiration or upregulate glycolysis in the presence of AMPK inhibition in macrophages (Figure 5D, E), indicating that enzalutamide-driven metabolic changes in macrophages were dependent on AMPK. AMPK inhibition of enzalutamide-treated MDSCs reversed the downregulation of basal mitochondrial respiration, but not the glycolytic upregulation induced by enzalutamide (Figure 5F, G), suggesting that AR antagonism in MDSCs may have directly affected glycolysis.
Figure 5. Enzalutamide-induced metabolic changes were mediated by AMPK in macrophages.
(A) MDSCs were generated in the presence of DMSO or enzalutamide and quantified for protein expression by western blot. Graphs indicates p-AMPK/AMPK ratio. (B-C) WT or MARKO BMDMs were treated with DMSO or enzalutamide for 24h and protein expression was assessed by western blot. Plots indicate AMPK expression normalized to β-actin for DMSO and enzalutamide-treated BMDMs (B) and WT and MARKO BMDMs (C). (D-E) BMDMs were treated with DMSO or enzalutamide in the presence or absence of the AMPK inhibitor (AMPKi) dorsomorphin for 24h and were assessed by Seahorse MitoStressTest (D) and GlycoStressTest (E). Graph (D) shows basal mitochondrial respiration and graph (E) indicates glycolytic rate for BMDMs. (F-G) MDSCs generated in the presence of DMSO or enzalutamide were treated with AMPKi for the last 24h of MDSC generation and cells were assessed by Seahorse MitoStressTest (F) and GlycoStressTest (G). Graph (F) shows basal mitochondrial respiration and graph (G) indicates glycolytic rate for MDSCs. (H) BMDMs were generated and treated with increasing doses of enzalutamide and RNA expression was assessed by qRT-PCR. Graph depicts MPC-2 fold change in expression relative to 0uM enzalutamide. (I) MDSCs generated in vitro in the presence of DMSO or the MPC inhibitor (MPCi) UK-5509 were mixed with MC-38 tumor cells in a 2:1 myeloid: tumor cell ratio and implanted subcutaneously on the shoulder of C57BL/6 male mice. Graph depicts tumor growth curves. Plots indicate mean and, when noted, error bars indicate standard deviation from pooled data of 2–3 experiments of 3–5 biological replicates per group. Black dots denote DMSO-treated, red squares indicate enzalutamide-treated, black diamonds indicate AMPKi-treated, red diamonds indicate enzalutamide and AMPKi-treated, and red empty triangles indicate MPCi-treated myeloid cells. Statistical analyses were performed with non-parametric one-way ANOVA and paired tests with corrections when necessary. *p<0.05, **p<0.01 and ***p<0.001.
Myeloid cell metabolism supports myeloid cell function [13]. As AR antagonism induced glycolysis and reduced basal mitochondrial respiration (Figure 4), we hypothesized that one of these metabolic pathways was responsible for supporting the increased tumor-promoting ability of enzalutamide-treated myeloid cells (Figure 2). To investigate whether increases in glycolysis were reflective of alterations in glucose uptake, glucose uptake and expression of GLUT1, the major transporter of glucose, were measured. Uptake of glucose was increased in enzalutamide-treated and MARKO BMDMs, and GLUT1 mRNA was induced with enzalutamide in BMDMs, suggesting that AR regulation of glycolysis was due in part to regulation of glucose uptake (Supplementary Figure 5A–C). However, targeting glycolysis with the glucose analog 2-DG upon AR inhibition had a negative impact on MDSC cell number and metabolism (Supplementary Figure 5D–E), further suggesting that the upregulation of glycolysis was compensating for the reduced mitochondrial respiration observed upon enzalutamide treatment.
These results suggested that the enzalutamide-induced increases in glycolysis may have been in response to direct effects of AR blockade on mitochondrial respiration. The reduction of mitochondrial respiration in myeloid cells following enzalutamide treatment may result from inhibition of starting material, i.e. pyruvate or acetyl-CoA, or a decrease in mitochondrial mass or function, as measured by superoxide production [31]. Enzalutamide-treatment did not alter the mitochondrial labeling, superoxide production or fatty acid oxidation needed for acetyl-CoA production (Supplementary Figure 6A–E).
Enzalutamide-mediated decrease in mitochondrial respiration could have also been related to changes in proteins involved in mitochondrial coordination of the TCA cycle and oxidative phosphorylation. Mitochondrial pyruvate carrier 2 (MPC-2) is a member of the MPC complex and a transcriptional target of AR in prostate cancer cells [32]. MPC imports pyruvate formed by the glycolytic pathway into the mitochondria for citric acid cycle (TCA) metabolism [33]. MPC-2 was downregulated in BMDMs upon enzalutamide treatment in a dose-dependent manner (Figure 5H). Inhibition of MPC with the inhibitor UK-5509 (MPCi) lowered mitochondrial respiration in both BMDMs and MDSCs (Supplementary Figure 6F–G). Therefore, to test whether inhibition of mitochondrial respiration increased the tumor-promoting ability of myeloid cells treated with enzalutamide, MDSCs were generated in the presence of DMSO or MPCi, mixed with MC-38 cells and injected in C57BL/6 male mice. Inhibition of MPC in MDSCs resulted in accelerated MC-38 tumor growth (Figure 5I), indicating that blocking mitochondrial respiration phenocopies myeloid AR blockade and enhances myeloid tumor-promoting activity.
Myeloid AR antagonism accelerated prostate tumor growth
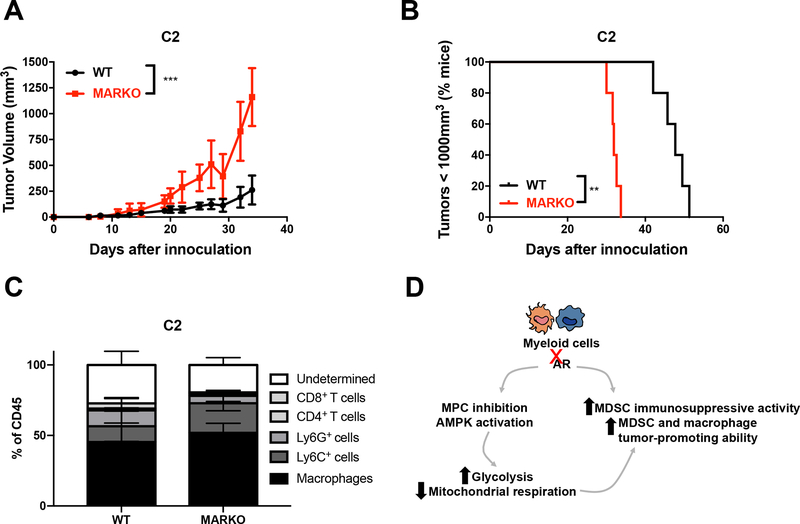
Prostate cancer progression to CRPC involves various mechanisms of treatment resistance, and results in decreased patient survival [1]. To determine whether AR deletion in myeloid cells was a mechanism implicated in CRPC, WT and MARKO C57BL/6 male mice were inoculated with TRAMP C2 prostate tumor cells, which were AR+ (Supplementary Figure 2A), and analyzed tumor growth over time. Mice lacking AR expression in myeloid cells displayed faster tumor growth when compared to mice with intact AR signaling in myeloid cells (Figure 6A–B). Similarly, to myeloid-MC-38 mixture tumors (Supplementary Figure 3C), MARKO mice did not alter tumor leukocyte infiltration when compared to WT mice (Figure 6C), confirming that myeloid cell function, and not infiltration, was linked to accelerated tumor progression (Figure 6D).
Figure 6. Genetic deletion of AR in myeloid cells increased prostate tumor growth and progression.
(A-C) Tramp C2 prostate tumor cells were inoculated in WT and MARKO C56BL/6 male mice and followed for tumor growth. At the endpoint, tumors were accessed for leukocyte infiltration by flow cytometry. Graphs depict mean and standard deviation of tumor growth over time (A), percentage of mice whose PCa progressed (B), and C2 tumor leukocyte infiltration (C). (D) Our proposed model, indicating that enzalutamide treatment of myeloid cells induces functional and metabolic changes, resulting in enhanced tumor progression. Black tumor growth curves denote WT mice, and red empty squares MARKO mice. Statistical analyses were done using two-way ANOVA and Log-rank test. *p<0.05, **p<0.01 and ***p<0.001.
Discussion
Here, we demonstrated that enzalutamide, a commonly used hormone therapy that targets AR+ tumor cells, directly impacted AR+ myeloid cell function and metabolism (summarized in Figure 6D). We showed that blocking myeloid AR enhanced myeloid cell tumor-promoting capacity, increased MDSC immunosuppression of T cell proliferation, and increased tumor progression. We further described MPC-2 and AMPK-dependent changes in metabolism that promoted glycolysis over oxidative phosphorylation and were associated with an enhanced tumor-promoting phenotype.
AR blockade therapy impacts the tumor microenvironment indirectly and directly. In this study, AR antagonism affected myeloid cell function and metabolism similarly when genetic and pharmacological approaches were used, suggesting that enzalutamide modulated myeloid cell phenotype through direct inhibition of AR. TAMs display a predominantly M2-like phenotype [34]. Genetic deletion of AR in macrophages was associated with quicker cutaneous wound healing responses [3], which was a characteristic of M2 macrophage function. However, AR deletion impairs M2 polarization in asthma [35]. These results suggested that the effect of AR signaling in macrophages is context dependent, which may explain the contradictory findings concerning macrophage AR function in prostate tumorigenesis. Inhibition of macrophage AR delays prostate tumor development [4]; however, prostate tumor progression and metastasis is enhanced in mice with AR-deficient macrophages (MARKO mice) [11]. Our findings concur with this study, as AR antagonism of myeloid cells enhanced their immunosuppressive function and enhanced myeloid tumor-promoting capacity and supported tumor progression. One potential explanation for the discrepancy in the effects of AR antagonism is that the frequencies of and functional interactions between tumor, stromal, and immune cells change over the course of tumorigenesis; therefore, the differential effects of inhibition of AR in macrophages on prostate tumor progression might be a consequence of this altered dynamics. Also, the extent to which myeloid AR antagonism affects tumor progression may depend on the aggressiveness of the tumor. In our study and in Izumi et al [11], inhibition of myeloid AR enhanced tumor progression in tumor models that either grew rapidly or had already been established, respectively. Studies comparing slowly and aggressively progressing tumors could potentially clarify this question.
AR antagonism of prostate tumor cell lines and prostate tumor-bearing mice enhances macrophage migration and infiltration [12, 36]. In vivo enzalutamide treatment results in secretion of prostate tumor-derived factors that induce macrophage polarization towards a tumor-promoting phenotype associated with treatment failure [12]. AR antagonism in a murine prostate tumor model mediates immunosuppression by direct inhibition of T cell activation [23]. Our work built on these findings showing that enzalutamide treatment enhanced myeloid cell immunosuppressive function. The combined evidence suggests that PCa treatment with antiandrogens may ultimately contribute to tumor progression by increasing myeloid cell infiltration and by promoting immunosuppression
Enzalutamide can have AR-independent effects such as the ability to inhibit GABA receptors in the brain [37]. We observed divergence between pharmacological and genetic approaches of AR blockade in the alteration of MDSC RNA expression, where enzalutamide treatment, but not AR knockout, induced VEGF and Arg1 mRNA and PD-L1 protein expression in MDSCs. These results implicated either a potential divergence in timing and penetrance of AR genetic knockout vs pharmacological inhibition or, less likely, other unknown AR-independent mechanisms. Nonetheless, we demonstrated an AR-dependent enzalutamide modulation of myeloid cell function and metabolism. However, the specific signaling pathways involved in AR modulation of myeloid cell phenotype remains to be characterized.
Myeloid cell phenotype is tightly regulated by environmental cues and depends on metabolic changes that support effector functions [13]. Tumor-associated macrophages and MDSCs have high glycolytic rates that support tumor-promoting capacity and immunosuppression, respectively [14–17]. We observed that enzalutamide treatment directly inhibited mitochondrial respiration and promoted glycolysis in myeloid cells; it is possible that this metabolic shift facilitated the tumor-promoting capacity of myeloid cells. Reduction of mitochondrial respiration with MPC inhibition phenocopied the enhanced tumor-promoting ability of MDSCs treated with enzalutamide, suggesting a metabolic mechanism by which AR antagonism in myeloid cells enhanced tumor progression. However, a causal link between metabolic changes induced by enzalutamide and the enhanced tumor-supporting capacity of myeloid cells needs to be established.
Two metabolic pathways were affected by AR inhibition of myeloid cells in our study: oxidative phosphorylation and glycolysis. AR antagonism reduced oxidative phosphorylation through the inhibition of the AR target MPC-2, thus potentially limiting pyruvate entry into the mitochondria. The increase in glycolysis following AR antagonism was likely an adaptation to the reduction in mitochondrial respiration and was needed to maintain cellular ATP, as inhibition of glycolysis significantly reduced MDSC cell number and metabolism. This hypothesis is in agreement with work in prostate cells where MPC-2 associates with subsequent induction of AMPK pathway and glycolysis [32]. The role of AR in AMPK-mediated metabolism appears to be different between myeloid and prostate cancer cells. Androgen receptor stimulation of PCa cells, rather than inhibition, leads to induction of AMPK signaling to enhance glycolysis [18]. Here, we identified that AR antagonism increased glycolytic and decreases oxidative phosphorylation metabolism through AMPK in macrophages. This divergent effect of AR activation/ inhibition on AMPK-mediated downstream metabolic effects may be potentially explained by an AMPK-AR negative feedback loop [38]; AR induces AMPK activation, which in turn inhibits AR transcriptional activity. By blocking AR in macrophages, this feedback loop could be lost and may potentially lead to increased AMPK activation, which in turn mediated the metabolic changes observed.
Whereas the effects of AR pharmacological inhibition on metabolism were dependent on AMPK signaling in macrophages, enzalutamide only affected mitochondrial respiration through AMPK in MDSCs. It is still possible that AMPK was involved in enzalutamide-mediated upregulation of the glycolytic pathway in MDSCs, as timing of AMPK inhibition was designed to avoid cell death, but may not have been enough to inhibit enzalutamide-induced early AMPK activation. Indeed, AMPK activation of monocytic MDSCs with metformin induces glycolysis and, in turn, glycolysis is important for MDSC immunosuppressive ability [17].
Hormone receptors cross-regulate each other; AR increases and decreases estrogen receptor activity [39]. ERα can increase MDSC mobilization and suppressive activity in a breast tumor model [40]. Therefore, it is possible that the increased MDSC suppressive activity induced by AR blockade is due, in part, to the release of AR mediated inhibition of ER signaling.
Our study highlighted the need to understand how sex hormone modulation therapies affect hormone receptor positive non-tumor cells within the tumor microenvironment. It is apparent from our studies and others that sex hormone antagonism affects multiple cell types within the tumor microenvironment. Tumor resistance to hormone blockade therapies may be associated with the varied responses coming from the complex tumor microenvironment. Here, we identified a mechanism of AR antagonism resistance that affected antitumor immunity directly through myeloid cells. Dissecting this complexity may indicate pathways that can be targeted to increase cancer patient survival. Alternatively, sex hormone receptor targeting can also be used as a strategy to boost antitumor immune responses. Estrogen receptor (ER) antagonism is standard of care in ER+, but not in ER− breast tumor patients. However, MDSC accumulation and immunosuppressive functions are increased with estrogen stimulation, and targeting MDSC ERα reverses these effects in various ER-independent tumor models [40]. This suggests that targeting estrogen receptors in ER− tumors induces a beneficial immunological rewiring towards enhanced antitumor immune responses. Alternatively, even though sex hormone receptor inhibition may have detrimental immunomodulatory effects, as shown here and by Pu et al [23], it may be possible to ameliorate these effects by combination with novel therapies that release this immunomodulatory constraint. One avenue to achieve this is to stimulate antitumor immune responses by metabolic rewiring. Macrophage phagocytic function is enhanced by CpG-mediated induction of fatty acid oxidation, resulting in more efficient cancer cell clearance and delayed tumor progression [41]. Future studies will be needed to investigate whether reversal of enzalutamide-induced metabolic changes in myeloid cells can be targeted to enhance antitumor immunity.
In summary, our work demonstrated that AR signaling affects myeloid cell function and metabolism leading to enhanced tumor-promoting capability in both colon and prostate cancer models. These findings suggest that although AR blockade could inhibit prostate tumor growth, it also promoted tumor resistance by enhancing the immunosuppressive activity of myeloid cells.
Supplementary Material
Acknowledgements
Research reported in this publication was supported in part by the National Cancer Institute of the National Institute of Health under Award 5P01CA98156 (S.O.G.) and the Roswell Park Alliance Foundation. The study used shared resources supported by Roswell Park Cancer Institute Cancer Center Support Grant (P30CA016056). C.R.C. was supported by the Mark Diamond Research Fund from the Graduate Student Association at University at Buffalo and Chateaubriand Fellowship. We would like to acknowledge Roswell Park’s Flow Cytometry core, Kieran O’Loughlin, and Dr. Subhamoy Dasgupta for the technical support and insightful discussions.
Financial Support: Research reported in this publication was supported in part by the National Cancer Institute of the National Institute of Health under Award 5P01CA98156 (S.O.G.) and the Roswell Park Alliance Foundation. The study used shared resources supported by Roswell Park Cancer Institute Cancer Center Support Grant (P30CA016056). C.R.C. was supported by the Mark Diamond Research Fund from the Graduate Student Association at University at Buffalo and Chateaubriand Fellowship. C. B. was supported by the Department of Defense under award PC131811.
Footnotes
Conflict of Interest
The authors declare no potential conflicts of interest.
References
- 1.Mitsiades N, A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer Res, 2013. 73(15): p. 4599–605. [DOI] [PubMed] [Google Scholar]
- 2.Cuzick J and Howell A, A brief review of the breast cancer prevention trials. Eur J Cancer, 2000. 36 Suppl 4: p. S51–3. [DOI] [PubMed] [Google Scholar]
- 3.Lai JJ, et al. , Monocyte/macrophage androgen receptor suppresses cutaneous wound healing in mice by enhancing local TNF-alpha expression. J Clin Invest, 2009. 119(12): p. 3739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang LY, et al. , Infiltrating macrophages promote prostate tumorigenesis via modulating androgen receptor-mediated CCL4-STAT3 signaling. Cancer Res, 2013. 73(18): p. 5633–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leach DA, et al. , Stromal androgen receptor regulates the composition of the microenvironment to influence prostate cancer outcome. Oncotarget, 2015. 6(18): p. 16135–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walecki M, et al. , Androgen receptor modulates Foxp3 expression in CD4+CD25+Foxp3+ regulatory T-cells. Mol Biol Cell, 2015. 26(15): p. 2845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calcinotto A, et al. , IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature, 2018. 559(7714): p. 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian B-Z and Pollard JW, Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell, 2010. 141(1): p. 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lissbrant IF, et al. , Tumor associated macrophages in human prostate cancer: relation to clinicopathological variables and survival. International journal of oncology, 2000. 17(3): p. 445–451. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, et al. , Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget, 2017. 8(18): p. 30576–30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izumi K, et al. , Targeting the androgen receptor with siRNA promotes prostate cancer metastasis through enhanced macrophage recruitment via CCL2/CCR2-induced STAT3 activation. EMBO Mol Med, 2013. 5(9): p. 1383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escamilla J, et al. , CSF1 receptor targeting in prostate cancer reverses macrophage-mediated resistance to androgen blockade therapy. Cancer Res, 2015. 75(6): p. 950–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Neill LA and Pearce EJ, Immunometabolism governs dendritic cell and macrophage function. J Exp Med, 2016. 213(1): p. 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penny HL, et al. , Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology, 2016. 5(8): p. e1191731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hossain F, et al. , Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol Res, 2015. 3(11): p. 1236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D, et al. , Comprehensive Proteomics Analysis Reveals Metabolic Reprogramming of Tumor-Associated Macrophages Stimulated by the Tumor Microenvironment. J Proteome Res, 2017. 16(1): p. 288–297. [DOI] [PubMed] [Google Scholar]
- 17.Wu T, et al. , mTOR masters monocytic myeloid-derived suppressor cells in mice with allografts or tumors. Sci Rep, 2016. 6: p. 20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massie CE, et al. , The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J, 2011. 30(13): p. 2719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar MP, et al. , Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep, 2018. 25(6): p. 1458–1468 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster BA, et al. , Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res, 1997. 57(16): p. 3325–30. [PubMed] [Google Scholar]
- 21.De Gendt K, et al. , A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci U S A, 2004. 101(5): p. 1327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marigo I, et al. , Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity, 2010. 32(6): p. 790–802. [DOI] [PubMed] [Google Scholar]
- 23.Pu Y, et al. , Androgen receptor antagonists compromise T cell response against prostate cancer leading to early tumor relapse. Sci Transl Med, 2016. 8(333): p. 333ra47. [DOI] [PubMed] [Google Scholar]
- 24.Lin N and Simon MC, Hypoxia-inducible factors: key regulators of myeloid cells during inflammation. J Clin Invest, 2016. 126(10): p. 3661–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byles V, et al. , The TSC-mTOR pathway regulates macrophage polarization. Nat Commun, 2013. 4: p. 2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everts B, et al. , TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat Immunol, 2014. 15(4): p. 323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Achyut BR, et al. , Canonical NFkappaB signaling in myeloid cells is required for the glioblastoma growth. Sci Rep, 2017. 7(1): p. 13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui Y, et al. , Upregulation of glucose metabolism by NF-kappaB2/p52 mediates enzalutamide resistance in castration-resistant prostate cancer cells. Endocr Relat Cancer, 2014. 21(3): p. 435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salminen A, Kauppinen A, and Kaarniranta K, AMPK activation inhibits the functions of myeloid-derived suppressor cells (MDSC): impact on cancer and aging. J Mol Med (Berl), 2019. 97(8): p. 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu YP, et al. , Adenosine 5’-monophosphate-activated protein kinase regulates IL-10-mediated anti-inflammatory signaling pathways in macrophages. J Immunol, 2015. 194(2): p. 584–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huttemann M, et al. , Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochim Biophys Acta, 2007. 1773(12): p. 1701–20. [DOI] [PubMed] [Google Scholar]
- 32.Bader DA, et al. , Mitochondrial pyruvate import is a metabolic vulnerability in androgen receptor-driven prostate cancer. Nature Metabolism, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bricker DK, et al. , A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science, 2012. 337(6090): p. 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grivennikov SI, Greten FR, and Karin M, Immunity, inflammation, and cancer. Cell, 2010. 140(6): p. 883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becerra-Diaz M, et al. , Androgen and Androgen Receptor as Enhancers of M2 Macrophage Polarization in Allergic Lung Inflammation. J Immunol, 2018. 201(10): p. 2923–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin TH, et al. , Anti-androgen receptor ASC-J9 versus anti-androgens MDV3100 (Enzalutamide) or Casodex (Bicalutamide) leads to opposite effects on prostate cancer metastasis via differential modulation of macrophage infiltration and STAT3-CCL2 signaling. Cell Death Dis, 2013. 4: p. e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster WR, et al. , Drug safety is a barrier to the discovery and development of new androgen receptor antagonists. Prostate, 2011. 71(5): p. 480–8. [DOI] [PubMed] [Google Scholar]
- 38.Jurmeister S, et al. , Transcriptomic analysis reveals inhibition of androgen receptor activity by AMPK in prostate cancer cells. Oncotarget, 2014. 5(11): p. 3785–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majumder A, Singh M, and Tyagi SC, Post-menopausal breast cancer: from estrogen to androgen receptor. Oncotarget, 2017. 8(60): p. 102739–102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svoronos N, et al. , Tumor Cell-Independent Estrogen Signaling Drives Disease Progression through Mobilization of Myeloid-Derived Suppressor Cells. Cancer Discov, 2017. 7(1): p. 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M, et al. , Metabolic rewiring of macrophages by CpG potentiates clearance of cancer cells and overcomes tumor-expressed CD47-mediated ‘don’t-eat-me’ signal. Nat Immunol, 2019. 20(3): p. 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.