Abstract
Early-onset breast cancer is the most common malignancy in women with Li-Fraumeni syndrome, caused by germline TP53 pathogenic variants. It has repeatedly been suggested that breast tumors from TP53 carriers are more likely to be HER2+ than those of non-carriers, but this information has not been incorporated into variant interpretation models for TP53. Breast tumor pathology is already being used quantitatively for assessing pathogenicity of germline variants in other genes, and it has been suggested that this type of evidence can be incorporated into current ACMG/AMP guidelines for germline variant classification. Here, by reviewing published data and using internal datasets separated by different age-groups, we investigated if breast tumor HER2+ status has utility as a predictor of TP53 germline variant pathogenicity, considering age at diagnosis. Overall, our results showed that the identification of HER2+ breast tumors diagnosed before the age of 40 can be conservatively incorporated into the current TP53-specific ACMG/AMP PP4 criterion, following a point system detailed in this manuscript. Further larger studies will be needed to re-assess the value of HER2+ breast tumors diagnosed at a later age.
Keywords: TP53, HER2, variant classification, ACMG
Graphical Abstract
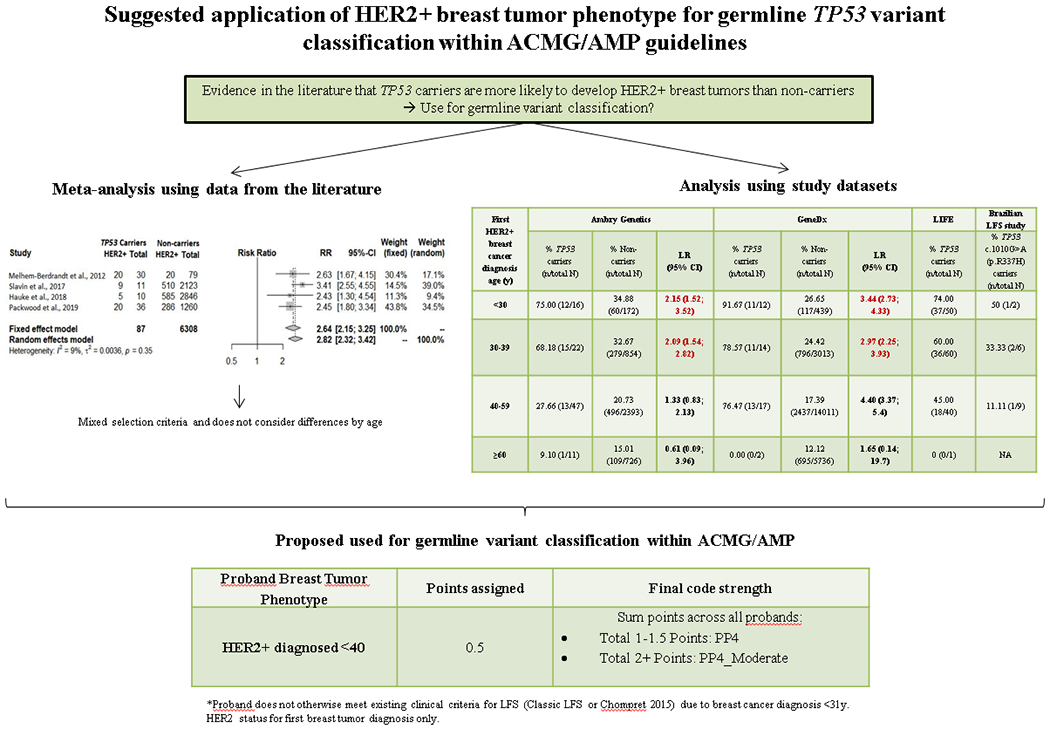
Introduction
Germline pathogenic variants in the TP53 gene are associated with Li-Fraumeni syndrome (LFS), a disorder characterized by a predisposition to a wide range of cancers, especially early onset breast tumors (Bougeard et al., 2015). Multiple studies in the literature have proposed that breast tumors from women carrying a TP53 (likely) pathogenic germline variant (here referred to as “TP53 carriers”) are more likely to be HER2+ than those of women without TP53 pathogenic germline variants (here referred to as “non-carriers”) (Bougeard et al., 2015; Eccles et al., 2016; Hu et al., 2020; Khincha et al., 2019; Masciari et al., 2012; Melhem-Bertrandt et al., 2012; Packwood et al., 2019; Rath et al., 2013; Slavin et al., 2017; Wilson et al., 2010). However, these previous studies were diverse in terms of ascertainment, and the sample sizes for TP53 carriers were typically small. A recent study has reported that TP53 pathogenic variants in cancer cells induce HER2 over-expression through transcriptional activation of the HER2 protein (Roman-Rosales, Garcia-Villa, Herrera, Gariglio, & Diaz-Chavez, 2018), providing a biological explanation for previously mentioned associations. Of note, it has also been suggested that breast tumors from carriers of the NM_000546.5(TP53):c.1010G>A (p.R337H) Brazilian founder variant are less likely to be HER2+ compared to carriers of other TP53 pathogenic variants (Fitarelli-Kiehl et al., 2015). A number of other published studies have assessed the population-based proportion of HER2+ breast tumors, with estimates of 19.9% when diagnosed <40 years and <13% when diagnosed after age 40 (Lund et al., 2010), and 19% when diagnosed before 50 years and 15% after age 50 (Cronin, Harlan, Dodd, Abrams, & Ballard-Barbash, 2010), illustrating how the proportion of HER2+ breast tumors decreases with increasing age at diagnosis.
There is precedence for use of breast tumor phenotype in quantitative models for assessing pathogenicity of variants in BRCA1 and BRCA2 (Parsons et al., 2019) based on formal analysis of tumor predictors of BRCA1/2 pathogenic variant status (Spurdle et al., 2014). In this study, we used published data and assembled the largest series of TP53-associated breast cancer cases to investigate if breast tumor HER2+ status has utility as a predictor of TP53 variant pathogenicity.
Materials and methods
Literature datasets (summary information)
Data from independent published studies, selected because HER2 status of breast tumors was available for both TP53 carriers and non-carriers, was extracted (see Table 1 for details).
Table 1.
Characteristics of TP53 carriers and non-carriers from published studies reporting HER2 breast tumor status
| Reference | Country of origin | Reported genetic testing information | Reported patient selection criteria | Total N | Purpose for this study |
|---|---|---|---|---|---|
| Melhem-Bertrandt et al., 2012 | USA | Sequencing done at outside CLIA certified laboratories | Women with suspected LFS due to personal and/or family history and diagnosed with breast cancer between 2000 to 2011 | 119 | Meta-analysis of available summary data to compare with results from our individual level study datasets (shown in Table 2) |
| Slavin et al., 2017 | USA | Sequencing done for 26 known or proposed breast cancer susceptibility genes | BRCA1/2-negative women with familial breast cancer from four academic health centers in the USA | 2134 | |
| Hauke et al., 2018 | Germany | Sequencing done at each participating center using Illumina sequencing platforms | Patients meeting the inclusion criteria of the German Consortium for Hereditary Breast and Ovarian Cancer for germ line testing (familial cancer cases, patients with early onset breast cancer, bilateral breast cancer, or both breast and ovarian cancer) | 2856 | |
| Packwood et al., 2019* | UK | Unreported | Women affected by early onset breast cancer | 1296 |
Overlaps with Wilson et al., 2010, which reported on a subset of 9 TP53 carriers and 161 non-carriers from the same cohort.
Study datasets (individual-level information)
We first used external data from non-carrier women from the Variants in Practice (VIP) study (E. R. Thompson et al., 2016), to define age strata for subsequent analysis. We then used data from breast cancer affected women, both TP53 carriers and non-carriers, identified by multigene panel testing carried out by two commercial companies (Ambry Genetics (March 2012-April 2017) and GeneDx (September 2013- December 2018)) to perform age-stratified HER2 analyses. In addition, we compared the proportion of HER2+ breast tumors among germline heterozygote TP53 carriers (excluding known or suspected mosaics) from Ambry Genetics and GeneDx to that observed in TP53 carriers from the Li-Fraumeni Exploration (LiFE) consortium (Ballinger et al., 2017), and from a Brazilian LFS study including only carriers of the NM_000546.5(TP53):c.1010G>A (p.R337H) variant. Details of these datasets can be seen in Table 2.
Table 2.
Characteristics of TP53 carriers and non-carriers from our own datasets with HER2 breast tumor status available*
| Dataset | Country of origin | Details for TP53 carriers | Details for non-carriers | Total N | Purpose for this study |
|---|---|---|---|---|---|
| VIP (E. R. Thompson et al., 2016) | Australia | NA | Probands with features of hereditary cancer (family history, early onset, triple-negative) who tested negative for all genes included in the panel used | 2719 | External dataset for determination of age groupings |
| Ambry Genetics | USA | Probands carrying a high-risk (likely) pathogenic TP53 variant identified through multigene panel testing, excluding those who were mosaic (defined as allele fraction of <30% confirmed by Sanger) and carriers of (likely) pathogenic variants in other genes | Probands who tested negative for all genes included in the panel used** | 4240*** | Derivation of tumor pathology likelihood ratios towards pathogenicity |
| GeneDx | USA | Probands carrying a high-risk (likely) pathogenic TP53 variant identified through multigene panel testing, excluding those who were mosaic (defined as allele fraction of <35% or lack of heterozygous appearance (uneven sequencing peaks) on Sanger confirmation) | Probands who tested negative for TP53 | 23244*** | |
| LiFE (Ballinger et al., 2017) | International, incorporating patients from City of Hope, Dana Farber Cancer Institute, National Cancer Institute, Manchester Universities Foundation Trust, Hospital Sirio-Libanes, St Jude Medical Center, University of Pennsylvania, and Huntsman Cancer Institute | Probands with a pathogenic TP53 variant, ascertained in a clinic-based setting for testing due to personal and/or family history, and diagnosed with breast cancer | NA | 138 | Carrier-only studies for comparison of HER2+ breast tumor proportions |
| LiFE/Brazilian LFS study | International, incorporating patients from LiFE centers and Hospital A.C. Camargo Cancer Center | Carriers of the Brazilian TP53 c.1010G>A (p.R337H) founder variant | NA | 40 |
TP53 pathogenic variant carrier status was used as reported by the data contributors at the time of initiation of this study (May 2019). It is possible that there is certain overlap between the literature and study datasets, or even within our own study datasets. However, the Ambry Genetics and GeneDx datasets used for the main analyses (derivation of LR of pathogenicity according to age) are expected to be entirely independent, since to the best of our knowledge they capture probands presenting for testing by only one of these laboratories. Only 7 TP53 carriers were identified in VIP, thus only non-carriers were used for analysis. Note, the VIP dataset has expanded in size since 2006 due to ongoing recruitment. HER2 status was not available for probands with TP53 c.1010G>A (p.R337H) identified in the Ambry dataset. Variant-level nomenclature was not provided for the GeneDx, precluding separate assessment of HER2 status for carriers of this variant. Variant-level information was available for a subset of LiFE participants; 13 probands from the LiFE dataset known to carry c.1010G>A (p.R337H) were excluded from the main LiFE dataset.
A subset 346 non-carriers were matched on the basis of gene panel 4:1 to TP53 carriers. Since the combined dataset did not include all individuals found to be negative for TP53, it does not represent actual carrier rates for Ambry diagnostic testing.
Breast cancer accounted for approximately 90% of all first cancer diagnoses for both TP53 carriers and non-carriers.
Statistics
A likelihood ratio (LR) towards pathogenicity for a TP53 variant found in a patient with a HER2+ breast tumor was calculated by comparing the proportion of HER2+ breast tumors between TP53 carriers and non-carriers, when both were available within the same dataset or study, following approaches used previously (Spurdle et al., 2014). Calculated LRs towards pathogenicity from the literature were meta-analyzed using the metabin function in RStudio (Version 0.99.903). Calculated LRs towards pathogenicity from Ambry Genetics and GeneDx were analyzed independently. LRs were then translated to the corresponding ACMG/AMP strength level following a published Bayesian reanalysis of these guidelines (Tavtigian et al., 2018).
Differences in the proportion of HER2+ breast tumors across datasets were assessed using the Fisher’s exact probability test.
Results and Discussion
Evidence from literature datasets
Using the literature datasets, it was observed that the proportion of HER2+ breast tumors was higher in TP53 carriers than non-carriers in all studies, resulting in a combined LR towards pathogenicity of 2.82 (2.32; 3.42) (Figure 1), considered equivalent to ACMG/AMP Supporting strength level.
Figure 1.
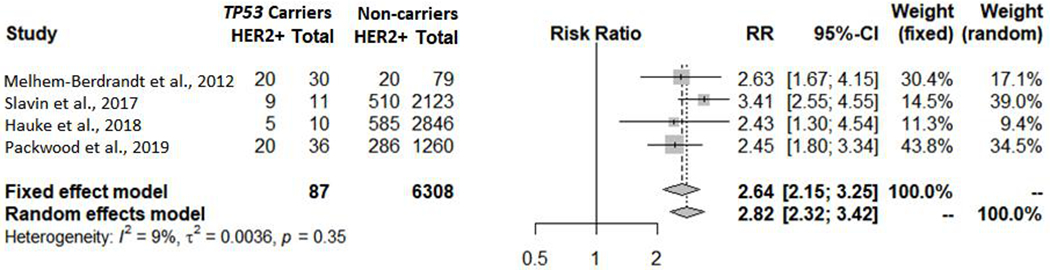
HER2+ breast tumor meta-analysis using data from the literature
Differences in HER2+ proportion by age
The proportion of HER2+ breast tumors by 5 year age-group were assessed in the VIP dataset as an external reference group (Table 3). Based on these results, the following age cut-offs were selected for subsequent analysis: <30, 30-39, 40-59, and ≥60.
Table 3.
Proportion of HER2+ breast tumors in TP53 non-carriers by 5 year age-group in VIP
| Age group | HER2+ breast tumor proportion (%) |
|---|---|
| <30 | 36.45 |
| 30-34 | 30.60 |
| 35-39 | 27.76 |
| 40-44 | 17.09 |
| 45-49 | 16.18 |
| 50-54 | 15.92 |
| 55-59 | 17.67 |
| ≥60 | 13.47 |
HER2+ proportion by age-group across multiple TP53 carrier and non-carrier datasets
The frequency of HER2+ breast tumors in TP53 carriers and non-carriers in the remaining groups, using the selected age cut-offs, is shown in Table 4, as well as the proposed use for variant classification.
Table 4.
Proportion of HER2+ breast tumors by age-group across different datasets
| First HER2+ breast cancer diagnosis age (y) | Ambry Genetics | GeneDx | LiFE | LiFE/Brazilian LFS study | Proposed use for TP53 variant classification | ||||
|---|---|---|---|---|---|---|---|---|---|
| % TP53 carriers (n/total N) | % Non-carriers (n/total N) | LR (95% CI) | % TP53 carriers (n/total N) | % Non-carriers (n/total N) | LR (95% CI) | % TP53 carriers (n/total N) | % TP53 c.1010G>A (p.R337H) carriers (n/total N) | ||
| <30 | 75.00 (12/16) | 34.88 (60/172) | 2.15 (1.52; 3.52) | 91.67 (11/12) | 26.65 (117/439) | 3.44 (2.73; 4.33) | 74.00 (37/50) | 50 (1/2) | Apply PP4 or PP4_Moderate (if proband does not otherwise already meet Classic LFS or Chompret 2015 criteria). See point system in Table 5 |
| 30-39 | 68.18 (15/22) | 32.67 (279/854) | 2.09 (1.54; 2.82) | 78.57 (11/14) | 24.42 (796/3013) | 2.97 (2.25; 3.93) | 56.60 (30/53) | 47.06 (8/17) | |
| 40-59 | 28.26 (13/46) | 20.73 (496/2393) | 1.36 (0.81; 2.30) | 76.47 (13/17) | 17.39 (2437/14011) | 4.40 (3.37; 5.4) | 47.06 (16/34) | 15.00 (3/20) | N/A. Unexplained heterogeneity across carrier datasets. Further analyses needed |
| ≥60 | 9.10 (1/11) | 15.01 (109/726) | 0.61 (0.09; 3.96) | 0.00 (0/2) | 12.12 (695/5736) | 1.65 (0.14; 19.7) | 0 (0/1) | 0 (0/1) | N/A. Carrier count too low for meaningful comparison. |
We highlight several observations. For both TP53 carriers and non-carriers, the proportion of HER2+ tumors decreased with increasing age at breast cancer diagnosis. However, very few carriers were diagnosed ≥60y, precluding meaningful comparisons for this age-group, as indicated by the extreme 95% confidence intervals around LR estimates. For all other age-groups, the proportion of HER2+ breast tumors was higher in TP53 carriers than non-carriers, in agreement with findings from the literature. The proportion of HER2+ breast tumors among NM_000546.5(TP53):c.1010G>A (p.R337H) carriers was noticeably lower than that observed for TP53 carriers from all other datasets, but this difference was not significant except for the GeneDx age group 40-49y (p<0.01) and LiFE age group 40-49y (p=0.02).
The proportion of HER2+ breast tumors from carriers from LiFE was generally comparable to that observed for carriers from Ambry Genetics and GeneDx, with no significant differences. However, TP53 carriers diagnosed at age 40-59y had an elevated frequency of HER2+ in the GeneDx dataset (76%) compared to those from Ambry Genetics (28%, p<0.01).
One possible explanation for this observation is the difference in allele read fraction used to define potential mosaicism for GeneDx compared to Ambry Genetics (<35% vs <30%). TP53 variants detected in blood from women diagnosed at later age are more likely to arise from somatic causes than when diagnosed at an earlier age (Coffee et al., 2017; Weitzel et al., 2017), and intuitively it is expected that tumors from such individuals would be less likely to be HER2+. This may explain in part the higher HER2+ proportion in individuals considered to be non-mosaic from GeneDx vs Ambry (information on read count and potential mosaicism was not available for LiFE “carriers”). However, the relatively small cell sizes for TP53 carrier groups in particular would suggest that the finding most likely reflects random error.
Use of breast tumor HER2+ status as a predictor of TP53 variant pathogenicity
It has already been proposed that tumor pathology could potentially be incorporated under the original ACMG/AMP PP4 criterion (“Patient’s phenotype and/or family history is highly specific for a disorder with a single genetic etiology”), from a study aimed to integrate somatic variant data and other biomarkers in germline variant classification (Walsh et al., 2018). An initial version of TP53-specific ACMG/AMP guidelines has been developed (https://clinicalgenome.org/site/assets/files/3876/clingen_tp53_acmg_specifications_v1.pdf, manuscript in preparation), but does not consider tumor pathology.
Considering LRs by age-group, the LR towards pathogenicity was significant for women diagnosed <40 years of age with a HER2+ breast tumor, using data from both Ambry Genetics (LR 2.09) and GeneDx (LR 3.44). These LRs are considered equivalent to the ACMG/AMP Supporting strength level for pathogenicity (Tavtigian, 2018), and we propose that HER2+ status may be considered as additional evidence for variant interpretation under this ACMG/AMP PP4 criterion for future versions of the TP53-specifiic guidelines. To avoid putting too much emphasis on a single data point, we suggest the conservative requirement of having 2–3 HER2+ carriers to apply PP4, and four or more to upgrade to Moderate (PP4_Moderate), following the point system shown in Table 5. As noted above and in Table 4, the utility of HER2+ breast tumor phenotype for classification of TP53 variants identified in women diagnosed ≥40 years is currently unclear, and the higher rate of likely somatic variants in blood in older age groups (Coffee et al., 2017; Weitzel et al., 2017) make analyses more challenging. We excluded individuals with known or suspected somatic TP53 variants from our analysis datasets, but acknowledge that unrecognized somatic variants may be present (Coffee et al., 2020; Mester et al., 2020). However, based on the report of median age at diagnosis of 44 years for individuals with confirmed somatic TP53 variants (Coffee et al., 2020), we believe that this phenomenon is unlikely to have confounded our LR estimates for women aged under 40 years.
Table 5.
Proposed point system for the incorporation of breast tumor pathology into the existing ACMG/AMP PP4 criterion for TP53*
| Proband breast tumor phenotype | Points assigned | Final code strength |
|---|---|---|
| HER2+ diagnosed <40 | 0.5 | Sum points across all probands: • Total 1-1.5 Points: PP4 • Total 2+ Points: PP4_Moderate |
Proband does not otherwise meet existing clinical criteria for LFS (Classic LFS or Chompret 2015) due to breast cancer diagnosis <31y. HER2 status for first breast tumor diagnosis only.
We also note need for caution in considering overlap with application of other clinical criteria used for TP53 variant interpretation. In particular, the PS4 criterion is already used for patients meeting Classic Li-Fraumeni syndrome or Chompret 2015 clinical criteria, one component of which is breast cancer diagnosis <31y (https://clinicalgenome.org/site/assets/files/3876/clingen_tp53_acmg_specifications_v1.pdf, manuscript in preparation). Given the correlation between age at breast cancer onset and HER2 status seen in this study and reported previously, these factors cannot be considered as independent. We therefore propose that PP4 for breast tumor HER2+ status can only be applied for a TP53 germline variant carrier in addition to PS4 for clinical criteria if the clinical criteria are met for reasons other than breast cancer diagnosis <31y. The proposed use of PP4 for breast tumor HER2+ status depending on these clinical criteria is further clarified in Figure 2. Further, we note, as has been highlighted previously for use of tumor pathology information in interpretation of variants in other cancer predisposition genes (Spurdle et al., 2014; B. A. Thompson et al., 2014), care should be taken to exclude the possibility that breast tumor pathology features predictive of variant pathogenicity were not used as an indicator for TP53 testing for a given patient. A remaining question is if HER2- status could be used as evidence against TP53 variant pathogenicity, however no current ACMG/AMP rule allows for the use of benign “personal” cancer history features.
Figure 2.
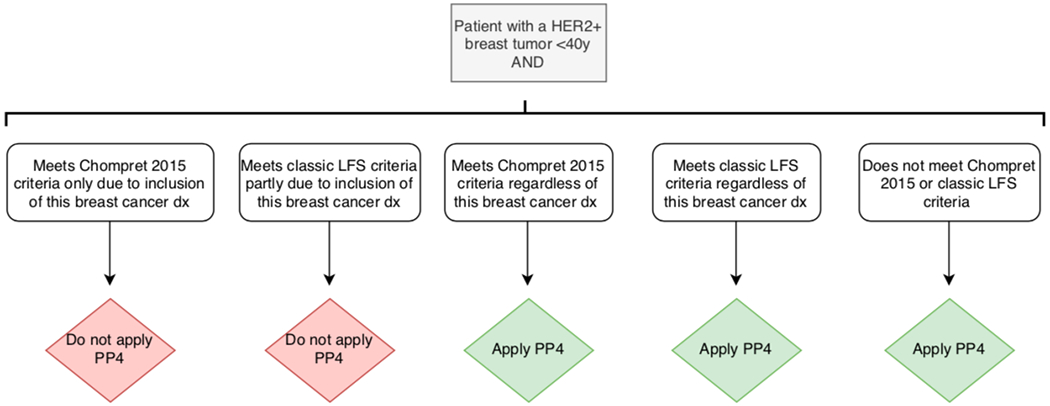
Instances in which PP4 is applied depending on patient’s clinical criteria. See point system in Table 5 for further details. For use of PS4, please refer to the TP53-specific ACMG/AMP guidelines ((https://clinicalgenome.org/site/assets/files/3876/clingen_tp53_acmg_specifications_v1.pdf, manuscript in preparation)
Dx = diagnosis
Conclusion
Our work confirms the previously observed association between germline TP53 pathogenic variants and the development of HER2 amplified breast cancer. Further, to our knowledge, this is the first study that has assessed the proportion of HER2+ breast tumors in TP53 carriers in comparison to non-carriers as a quantified measure for use in germline variant classification.
Overall, according to our results, a high risk pathogenic TP53 variant observed in a woman with a HER2+ breast tumor would have an LR towards pathogenicity of 2.82-fold when age at diagnosis is not considered, as determined from the literature. However, we expect these previously published studies to be enriched for women with early onset breast cancer and/or classic LFS presentation. Further age-stratified analysis indicates that LR towards pathogenicity is at least 2.09 for women with a HER2+ breast cancer diagnosis under age 40y. That is, while breast tumors from TP53 carriers will not all exhibit HER2+ pathology, these observations justify consideration of HER2+ breast tumor status in women with diagnosis under 40y as an additional source of clinical evidence towards pathogenicity within comprehensive TP53 variant classification strategies.
Further larger studies will be useful to re-assess findings for women diagnosed with HER2+ breast cancer at or after 40 years), before completely discarding this criterion as evidence of pathogenicity. Further, the lower proportions of HER2+ tumors observed for carriers of the well-characterized reduced penetrance pathogenic variant NM_000546.5(TP53):c.1010G>A (p.R337H), for all age-groups, would seem to indicate that HER2+ status may be less predictive of reduced penetrance TP53 variants. However, the relationship between HER2+ status and reduced penetrance TP53 variants as a group remains unclear, and therefore additional studies with carriers of these variants would be necessary to identify if HER2+ status can be used as predictor (and at what level of strength) to aid classification of reduced penetrance variants.
The findings from this study, based on formal analysis of multiple datasets, provide a strategy for use of breast tumor HER2+ status for future TP53 variant interpretation within ACMG/AMP guidelines, for cases diagnosed <40 years that are not selected for testing on the basis of HER2+ tumor phenotype. Finally, another application of this study is to include the LRs towards pathogenicity calculated as an additional component in quantitative statistical modelling to predict pathogenicity of p53 missense variants (Fortuno et al., 2019).
Acknowledgements
C.F is supported by a University of Queensland (UQ) International Scholarship from the UQ School of Medicine. J.N.W is supported by a grant from the National Cancer Institute (1R01CA242218-01). A.B.S received Fellowship funding from NHMRC (ID1061779, ID1177524). D.G.E is supported by the NIHR Manchester Biomedical Research Centre (IS-BRC-1215-20007). The work of S.A.S and P.P.K is supported by the Intramural Research Program of the National Institutes of Health. This research was supported by NHMRC Project Grant ID 1161589. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA242218. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Footnotes
Conflict of Interests
Jessica Mester is an employee of GeneDx, Inc., a wholly-owned subsidiary of OPKO Health, Inc. Tina Pesaran, Jill Dolinsky, Amal Yussuf, and Kelly McGoldrick are paid employees of Ambry Genetics. All other authors have declared no conflicts of interest.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The proband data is not publicly available due to privacy or ethical restrictions. Variant classifications from Ambry Genetics and GeneDx are available in the ClinVar database.
References
- Ballinger ML, Best A, Mai PL, Khincha PP, Loud JT, Peters JA, … Savage SA. (2017). Baseline Surveillance in Li-Fraumeni Syndrome Using Whole-Body Magnetic Resonance Imaging: A Meta-analysis. JAMA Oncol. doi: 10.1001/jamaoncol.2017.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougeard G, Renaux-Petel M, Flaman JM, Charbonnier C, Fermey P, Belotti M, … Frebourg T. (2015). Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. J Clin Oncol, 33(21), 2345–2352. doi: 10.1200/JCO.2014.59.5728 [DOI] [PubMed] [Google Scholar]
- Coffee B, Cox HC, Bernhisel R, et al. A substantial proportion of apparently heterozygous TP53 pathogenic variants detected with a next-generation sequencing hereditary pan-cancer panel are acquired somatically. Hum Mutat. 2020;41(1):203–211. doi: 10.1002/humu.23910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Cox HC, Kidd J, Sizemore S, Brown K, Manley S, & Mancini-DiNardo D (2017). Detection of somatic variants in peripheral blood lymphocytes using a next generation sequencing multigene pan cancer panel. Cancer Genet, 211, 5–8. doi: 10.1016/j.cancergen.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Cronin KA, Harlan LC, Dodd KW, Abrams JS, & Ballard-Barbash R (2010). Population-based estimate of the prevalence of HER-2 positive breast cancer tumors for early stage patients in the US. Cancer Invest, 28(9), 963–968. doi: 10.3109/07357907.2010.496759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles DM, Li N, Handwerker R, Maishman T, Copson ER, Durcan LT, … Campbell I. (2016). Genetic testing in a cohort of young patients with HER2-amplified breast cancer. Ann Oncol, 27(3), 467–473. doi: 10.1093/annonc/mdv592 [DOI] [PubMed] [Google Scholar]
- Fitarelli-Kiehl M, Giacomazzi J, Santos-Silva P, Graudenz MS, Palmero EI, Michelli RA, … Ashton-Prolla P. (2015). The breast cancer immunophenotype of TP53-p.R337H carriers is different from that observed among other pathogenic TP53 mutation carriers. Fam Cancer, 14(2), 333–336. doi: 10.1007/s10689-015-9779-y [DOI] [PubMed] [Google Scholar]
- Fortuno C, Cipponi A, Ballinger ML, Tavtigian SV, Olivier M, Ruparel V, … James PA. (2019). A quantitative model to predict pathogenicity of missense variants in the TP53 gene. Hum Mutat, 40(6), 788–800. doi: 10.1002/humu.23739 [DOI] [PubMed] [Google Scholar]
- Hauke J, Horvath J, Gross E, Gehrig A, Honisch E, Hackmann K, … Hahnen E. (2018). Gene panel testing of 5589 BRCA1/2-negative index patients with breast cancer in a routine diagnostic setting: results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med, 7(4), 1349–1358. doi: 10.1002/cam4.1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Polley EC, Yadav S, et al. The contribution of germline predisposition gene mutations to clinical subtypes of invasive breast cancer from a clinical genetic testing cohort [published online ahead of print, 2020 Feb 24]. J Natl Cancer Inst. 2020;djaa023. doi: 10.1093/jnci/djaa023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khincha PP, Best AF, Fraumeni JF Jr., Loud JT, Savage SA, & Achatz MI (2019). Reproductive factors associated with breast cancer risk in Li-Fraumeni syndrome. Eur J Cancer, 116, 199–206. doi: 10.1016/j.ejca.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund MJ, Butler EN, Hair BY, Ward KC, Andrews JH, Oprea-Ilies G, … Eley JW (2010). Age/race differences in HER2 testing and in incidence rates for breast cancer triple subtypes: a population-based study and first report. Cancer, 116(11), 2549–2559. doi: 10.1002/cncr.25016 [DOI] [PubMed] [Google Scholar]
- Masciari S, Dillon DA, Rath M, Robson M, Weitzel JN, Balmana J, … Garber JE. (2012). Breast cancer phenotype in women with TP53 germline mutations: a Li-Fraumeni syndrome consortium effort. Breast Cancer Res Treat, 133(3), 1125–1130. doi: 10.1007/s10549-012-1993-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack VA, Joffe M, van den Berg E, Broeze N, Silva Idos S, Romieu I, … Cubasch H (2013). Breast cancer receptor status and stage at diagnosis in over 1,200 consecutive public hospital patients in Soweto, South Africa: a case series. Breast Cancer Res, 15(5), R84. doi: 10.1186/bcr3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhem-Bertrandt A, Bojadzieva J, Ready KJ, Obeid E, Liu DD, Gutierrez-Barrera AM, … Arun BK. (2012). Early onset HER2-positive breast cancer is associated with germline TP53 mutations. Cancer, 118(4), 908–913. doi: 10.1002/cncr.26377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester JL, Jackson SA, Postula K, et al. Apparently Heterozygous TP53 Pathogenic Variants May Be Blood Limited in Patients Undergoing Hereditary Cancer Panel Testing. J Mol Diagn. 2020;22(3):396–404. doi: 10.1016/j.jmoldx.2019.12.003 [DOI] [PubMed] [Google Scholar]
- Packwood K, Martland G, Sommerlad M, Shaw E, Moutasim K, Thomas G, … Eccles DM. (2019). Breast cancer in patients with germline TP53 pathogenic variants have typical tumour characteristics: the Cohort study of TP53 carrier early onset breast cancer (COPE study). J Pathol Clin Res, 5(3), 189–198. doi: 10.1002/cjp2.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MT, Tudini E, Li H, Hahnen E, Wappenschmidt B, Feliubadalo L, … Spurdle AB. (2019). Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum Mutat. doi: 10.1002/humu.23818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath MG, Masciari S, Gelman R, Miron A, Miron P, Foley K, … Garber JE. (2013). Prevalence of germline TP53 mutations in HER2+ breast cancer patients. Breast Cancer Res Treat, 139(1), 193–198. doi: 10.1007/s10549-012-2375-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Rosales AA, Garcia-Villa E, Herrera LA, Gariglio P, & Diaz-Chavez J (2018). Mutant p53 gain of function induces HER2 over-expression in cancer cells. BMC Cancer, 18(1), 709. doi: 10.1186/s12885-018-4613-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavin TP, Maxwell KN, Lilyquist J, Vijai J, Neuhausen SL, Hart SN, … Couch FJ. (2017). The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer, 3, 22. doi: 10.1038/s41523-017-0024-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Couch FJ, Parsons MT, McGuffog L, Barrowdale D, Bolla MK, … kConFab I. (2014). Refined histopathological predictors of BRCA1 and BRCA2 mutation status: a large-scale analysis of breast cancer characteristics from the BCAC, CIMBA, and ENIGMA consortia. Breast Cancer Res, 16(6), 3419. doi: 10.1186/s13058-014-0474-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Greenblatt MS, Harrison SM, Nussbaum RL, Prabhu SA, Boucher KM, & Biesecker LG (2018). Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet Med. doi: 10.1038/gim.2017.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Spurdle AB, Plazzer JP, Greenblatt MS, Akagi K, Al-Mulla F, … InSiGht. (2014). Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet, 46(2), 107–115. doi: 10.1038/ng.2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ER, Rowley SM, Li N, McInerny S, Devereux L, Wong-Brown MW, … Campbell IG (2016). Panel Testing for Familial Breast Cancer: Calibrating the Tension Between Research and Clinical Care. J Clin Oncol, 34(13), 1455–1459. doi: 10.1200/jco.2015.63.7454 [DOI] [PubMed] [Google Scholar]
- Walsh MF, Ritter DI, Kesserwan C, Sonkin D, Chakravarty D, Chao E, … Plon SE. (2018). Integrating somatic variant data and biomarkers for germline variant classification in cancer predisposition genes. Hum Mutat, 39(11), 1542–1552. doi: 10.1002/humu.23640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel JN, Chao EC, Nehoray B, Van Tongeren LR, LaDuca H, Blazer KR, … Jasperson K. (2017). Somatic TP53 variants frequently confound germ-line testing results. Genet Med. doi: 10.1038/gim.2017.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, Bateman AC, Hanson H, An Q, Evans G, Rahman N, … Eccles DM (2010). A novel HER2-positive breast cancer phenotype arising from germline TP53 mutations. J Med Genet, 47(11), 771–774. doi: 10.1136/jmg.2010.078113 [DOI] [PubMed] [Google Scholar]


