Abstract
Background:
Malignant melanoma has a propensity for development of hepatic and pulmonary metastases. MicroRNAs (miRs) are small, non-coding RNA molecules containing about 22 nucleotides that mediate protein expression and can contribute to cancer progression. We aim to identify clinically useful differences in miR expression in metastatic melanoma tissue.
Methods:
RNA was extracted from formalin fixed, paraffin embedded samples of hepatic and pulmonary metastatic melanoma, benign nevi, and primary cutaneous melanoma. Assessment of miR expression was performed on purified RNA using the NanoString nCounter miRNA assay. miRs with >2 fold change in expression when compared to other tumor sites (p-value ≤0.05, modified t-test) were identified as dysregulated. Common gene targets were then identified among dysregulated miRs unique to each metastatic site.
Results:
Melanoma metastatic to the liver had differential expression of 26 miRs compared to benign nevi and 16 miRs compared to primary melanoma (P<0.048). Melanoma metastatic to lung had differential expression of 19 miRs compared to benign nevi and 10 miRs compared to primary melanoma (P<0.024). Compared to lung metastases, liver metastases had greater than 2-fold up-regulation of 4 miRs, and 4.2-fold down-regulation of miR-200c-3p (P<0.0081).
Conclusions:
These findings indicate that sites of metastatic melanoma have unique miR profiles that may contribute to their development and localization. Further investigation of the utility of these miRs as diagnostic and prognostic biomarkers and their impact on development of metastatic melanoma is warranted.
Keywords: malignant melanoma, miRNA, miR, metastasis, biomarker
Background
Melanoma remains the deadliest form of skin cancer and was responsible for an estimated 9,320 deaths in the United States in 2018.[1] Approximately 5% of patients will present with distant metastasis, for which 5-year survival is below 15%. Of those melanoma patients diagnosed with a high-risk primary tumor at the time of surgery, 13% will ultimately develop recurrent disease.[2] Furthermore, over 10% of patients with a negative sentinel lymph node biopsy at the time of surgery will also experience recurrence, with nearly 40% of recurrent tumors arising at distant sites.[3] Distant metastasis is reported most commonly to the lungs (64%), extra-regional lymph nodes (46%), brain (45%), and liver (41%).[4] Survival varies depending on the site of metastasis. This variability has led to further classification of M1 disease in AJCC melanoma TNM staging into four distinct categories: M1a (skin, subcutaneous tissue, or distant lymph nodes), M1b (lung), M1c (other visceral organs), and M1d (CNS)).[5] Resection of recurrent distant metastasis (with or without systematic therapy) results in a median survival >60 months for M1a, while M1b and M1c have poorer outcomes with survival at 17.9 and 15.0 months following resection, respectively.[6]
microRNAs (miRs) are small 19–22 nucleotide long, non-coding RNA molecules that inhibit protein translation through binding to mRNA targets.[7] miR dysregulation has been discovered in several neoplastic settings and can promote tumorigenesis through its inhibitory effect on target genes and their downstream pathways leading to: unlimited cell replication, avoidance of anti-proliferation signals, evasion of apoptosis, genomic instability, induction of angiogenesis, tissue invasion, metastasis and evasion of the immune system.[8] Thus, dysregulated miR expression provides a potential target for diagnostic and therapeutic exploitation.[7,9,10] Over 2654 different mature miRs that have been identified in humans, with patterns of expression that may differ across different cancer types and according to the stage of disease.[8,11]
The potential for alteration of the miR profile in tissue from distant metastatic melanoma sites remains incompletely understood. To our knowledge, no other group has investigated how the miR transcriptome may vary at different sites of melanoma metastasis, or how this compares to primary cutaneous melanoma. Herein, we hypothesize that pulmonary and hepatic distant metastatic melanoma lesions have distinct miR profiles, and differ from primary tumor controls. The results of this work may further the understanding of miR dysregulation at different sites of metastasis and potentially identify novel diagnostic and therapeutic targets. Furthermore, site-specific alterations in miR expression may be informative of tumor behavior and predilection to specific metastatic sites.
Methods
Patient Samples
Patients with melanoma metastases to either the lung or liver that had undergone metastasectomy at the Ohio State University Wexner Medical Center were selected at random. Formalin fixed, paraffin embedded biopsy tissue from sites of metastatic melanoma in the liver or lung were collected under the auspices of an Institutional Review Board (IRB) approved protocol (No. 2007C0015) from cases encountered between 2009 and 2014. Samples consisted of 6 benign nevi, 6 primary cutaneous non-metastatic melanoma, 6 pulmonary metastatic melanoma sites, and 6 hepatic metastatic melanoma sites, all originating from unique patients.
RNA Isolation
Samples from each biopsy were shaved at 10 μm from each paraffin block for use in RNA isolation. Total RNA was harvested from sections of paraffin-embedded tumors using the RecoverAll Total Nucleic Acid Isolation Kit® (Ambion, Inc., Foster City, CA, USA) according to the manufacturer’s directions. Resultant RNA was assessed for quality and concentration using a Nanodrop spectrophotometer (ThermoFisher Scientific, Waltham, MA, results shown in Supplemental Table 1).
NanoString
Isolated and purified RNA (100 ng) was loaded onto a NanoString nCounter (NanoString Technologies, Seattle, WA) platform and miR expression quantification was carried out as previously described [12]. Normalization of melting temperatures and miR identification was facilitated by ligation of individual miRs within the sample to DNA tags. Excess tags were washed away. The tagged microRNA products were hybridized to capture reporter probes at 64°C for 18 hours. Notably, the reporter probes contain unique fluorescent signals, thereby permitting downstream identification of individual miRs. Hybridized probes were immobilized onto a streptavidin coated cartridge via nCounter Prep Station (NanoString Technologies, Seattle, WA) and the florescence of each hybridized miR was analyzed by an nCounter Digital analyzer (NanoString Technologies, Seattle, WA). A high-density scan containing 600 fields of view was performed. The investigation included 5 positive, 5 negative, and 5 housekeeping genes.
Statistical Analysis
The raw NanoString data was first technically normalized using nSolver Analysis software (NanoString Technologies, Seattle, WA). In order to limit the potential batch effect associated with inter-chip variability, the expression of positive controls was used to adjust the dataset generated from the second cartridge to the first. Normalized data was then filtered using the negative controls. The cutoff value was defined as the maximum normalized expression count among the negative controls (72 for this data set). Individual miRs were removed from further analysis if 90% of the samples had expression levels lower than this cutoff value, resulting in reduction of the number of miRs for assessment from 800 to 249. Fold change was then calculated between all experimental groups to determine which miRs were relatively increased or decreased in each comparison of tumor source (benign nevi vs. primary melanoma, benign nevi vs. hepatic melanoma, benign nevi vs. pulmonary melanoma, primary melanoma vs. hepatic melanoma, primary melanoma vs. pulmonary melanoma, and hepatic vs. pulmonary melanoma). A t-test was then performed with an alpha value of 0.05 to determine statistical significance of each miR in the comparisons. In order to account for multiple comparisons and control for false positives, a Benjamini-Hochberg correction procedure was performed.[13] miRs of potential significance were selected for downstream analysis using the following criteria: 1) a 2-fold increase or decrease in the comparison and 2) a p-value less than or equal to 0.05.
Gene Target Analysis
For each experimental comparison, the miRs that fit this criteria were run through a network enrichment analysis using the miRNet software tool to identify miR gene targets.[14] Gene targets of greatest potential biologic significance were identified as those with a minimum degree of three (i.e., interact with a minimum of three dysregulated miRs identified in the comparison of two tissue groups). Thus, there may be altered protein expression of these gene targets based on the composition of associated miRs with significant dysregulated expression in the examined tissue site of metastatic melanoma.
Results
In our analysis of miR expression in primary and metastatic melanocytic lesions isolated from unique patients, a comparison of benign nevi to primary cutaneous non-metastatic melanoma revealed no miRs with up or down-regulation of significance. Of note, no miRs were identified as having at least a 2-fold increased expression in primary cutaneous melanoma relative to benign nevi. This was not an unexpected finding given the approach being taken was an exploratory one. These benign nevi and primary cutaneous non-metastatic melanoma samples used as comparators were from unrelated patients. The miR profile of pulmonary metastatic melanomas was then assessed relative to benign nevi and primary cutaneous melanoma. These comparisons revealed differential expression of 19 miRs in pulmonary sites of melanoma metastasis compared to benign nevi (P<0.016, Fig. 1A), and 10 miRs when compared to primary cutaneous melanoma based on evaluation by t-test with a Benjamini-Hochberg correction (P<0.025, Fig. 1B). Upregulation of four miRs was associated with pulmonary metastatic melanoma when compared to both benign nevi and primary cutaneous melanoma, including miR-4286, miR-30b-5p, miR-146a and miR-324–5p (in order of decreasing fold change in expression). Additionally, three miRs (in order of decreasing fold change in expression), namely miR-205–5p, miR-203a-3p and miR-1246, were downregulated in pulmonary sites of metastatic melanoma relative to both benign nevi and primary cutaneous melanoma.
Figure 1.
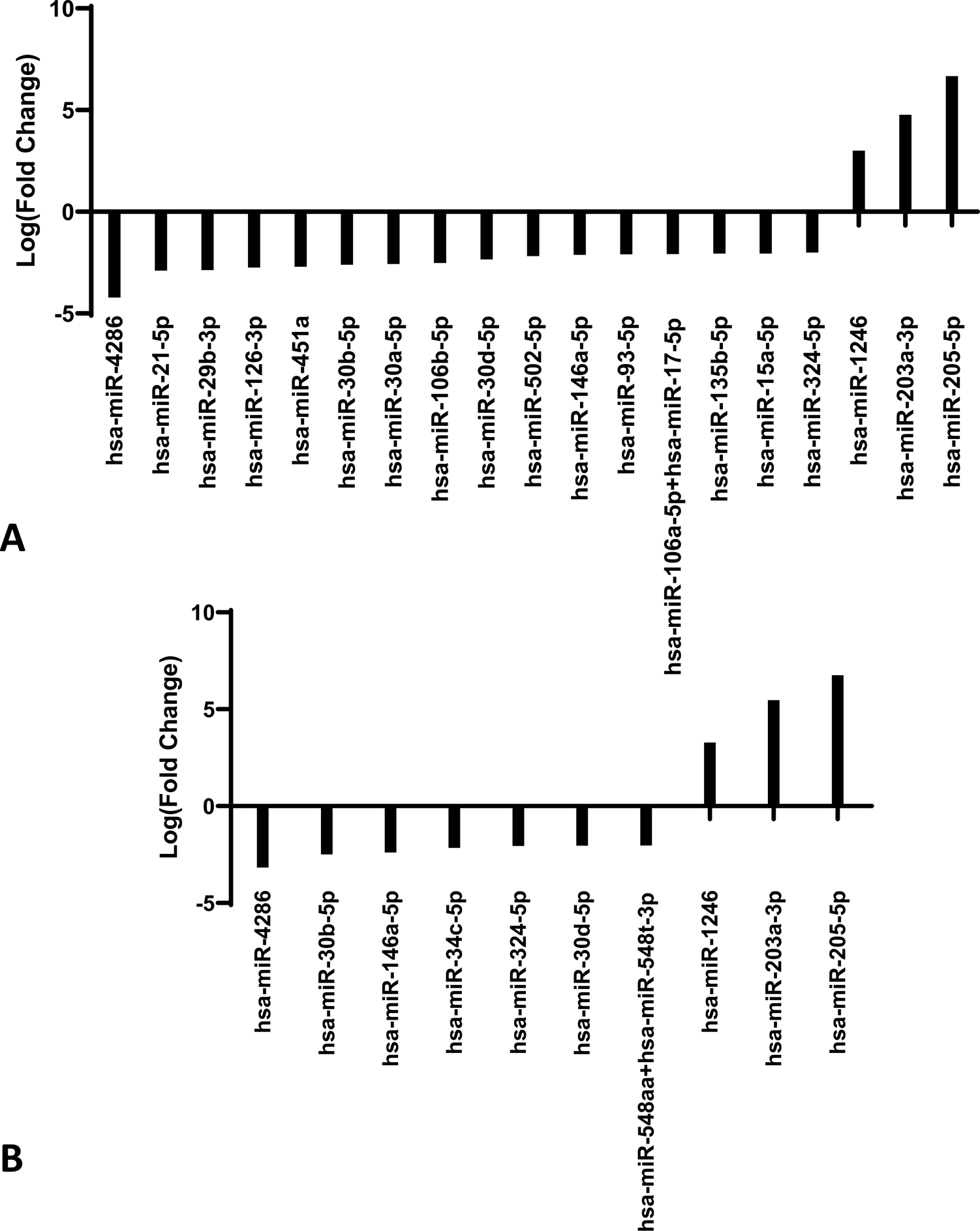
(A) MicroRNA profile of benign nevi compared to pulmonary metastatic melanoma. NanoString expression of pulmonary metastatic melanoma (n=6) compared to benign nevi (n=6). Expression of statistically significant differentially expressed microRNAs are shown as log(fold change) in expression (p< 0.016). (B) MicroRNA profile of cutaneous primary melanoma compared to pulmonary metastatic melanoma. NanoString expression of pulmonary metastatic melanoma (n=6) compared to non-metastatic cutaneous primary melanoma (n=6). Expression of statistically significant differentially expressed microRNAs are shown as log(fold change) in expression (p< 0.025).
The miR profile of metastatic melanoma of the liver was then assessed relative to benign nevi and primary cutaneous melanoma. Hepatic metastases had differential expression of 26 miRs relative to benign nevi (P<0.048, Fig. 2A) and 16 miRs relative to primary cutaneous melanoma based on evaluation by t-test with a Benjamini-Hochberg correction (P<0.035, Fig. 2B). Comparison of hepatic metastatic melanoma to both benign nevi and primary melanoma revealed upregulation of miR-122–5p, miR-194–5p, miR-4286, miR-885–5p, miR-30b-5p, miR-363–3p, and miR-192–5p (in order of decreasing fold change in expression) in hepatic sites of metastatic melanoma relative to both benign nevi and primary cutaneous melanoma. Of the miRs identified to have upregulated expression in hepatic sites of melanoma metastases, miR-4286 and miR-30b-5p were also upregulated in pulmonary sites of metastasis relative to benign nevi and primary cutaneous melanoma. In order of decreasing fold change in expression, downregulation of miR-200c-3p and miR-199b-5p was observed specifically in hepatic sites of metastasis relative to benign nevi and primary cutaneous melanoma. Additionally, the three miRs found to have significantly downregulated expression in pulmonary metastatic melanoma relative to benign nevi and primary cutaneous melanoma (miR-205–5p, miR-203a-3p and miR-1246) were also significantly downregulated in hepatic melanoma metastases relative to benign nevi and primary cutaneous melanoma. Thus, the consistent pattern of dysregulated expression of decreased miR-205–5p, miR-203a-3p, and miR-1246, and increased miR-4286 and miR-30b-5p suggests that alteration of expression of these miRs is common and specific to metastatic sites of melanoma.
Figure 2.
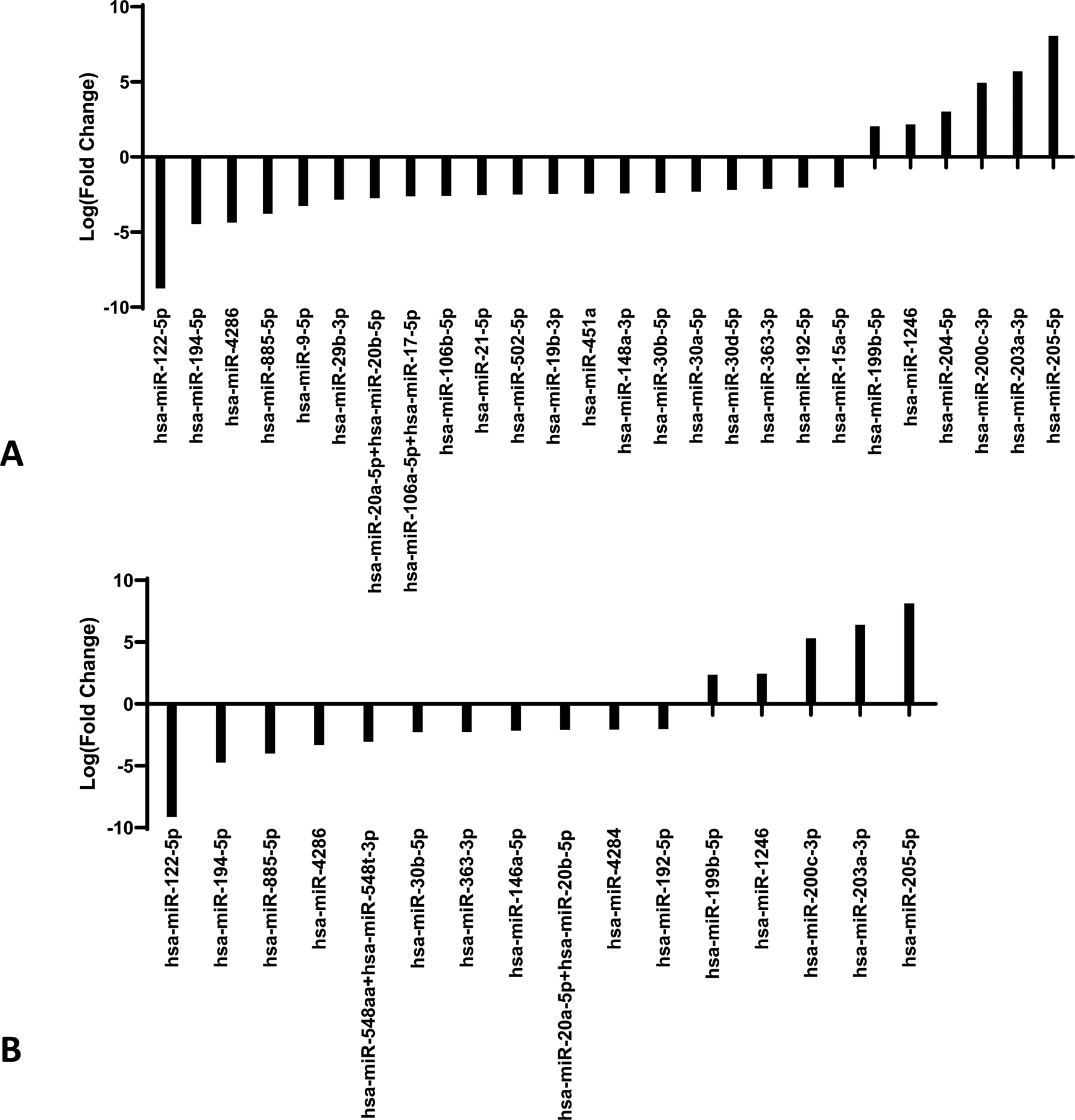
(A) MicroRNA profile of benign nevi compared to hepatic metastatic melanoma. NanoString expression of hepatic metastatic melanoma (n=6) compared to benign nevi (n=6). Expression of statistically significant differentially expressed microRNAs are shown as log(fold change) in expression (p< 0.048). (B) MicroRNA profile of cutaneous primary melanoma compared to hepatic metastatic melanoma. NanoString expression of hepatic metastatic melanoma (n=6) compared to non-metastatic cutaneous primary melanoma (n=6). Expression of statistically significant differentially expressed microRNAs are shown as log(fold change) in expression (p<0.035).
Lastly, melanoma metastasis of the lung were compared to metastasis of the liver (Fig. 3). Melanoma tissue samples from the lung and from the liver were isolated from unique patients and thus no single patient is represented in both groups of metastatic lesions. Five miRs were differentially expressed in hepatic metastasis relative to pulmonary metastasis based on evaluation by t-test with a Benjamini-Hochberg correction (P<0.0081). There was 4.2-fold downregulation of miR-200c-3p, and at least 2-fold upregulation of (in descending order of fold change in expression) miR-122–5p, miR-885–5p, miR-194–5p and miR-192–5p in hepatic metastatic melanoma as compared to pulmonary metastatic melanoma tissue. All four of these miRs that were upregulated in hepatic sites of melanoma metastasis had increased expression specifically in this tissue site when compared to benign nevi, primary cutaneous melanoma and pulmonary sites of melanoma metastasis. These findings suggest that unique miR profiles are associated with melanoma metastases in specific tissue sites.
Figure 3. MicroRNA profile of hepatic metastatic melanoma compared to pulmonary metastatic melanoma.
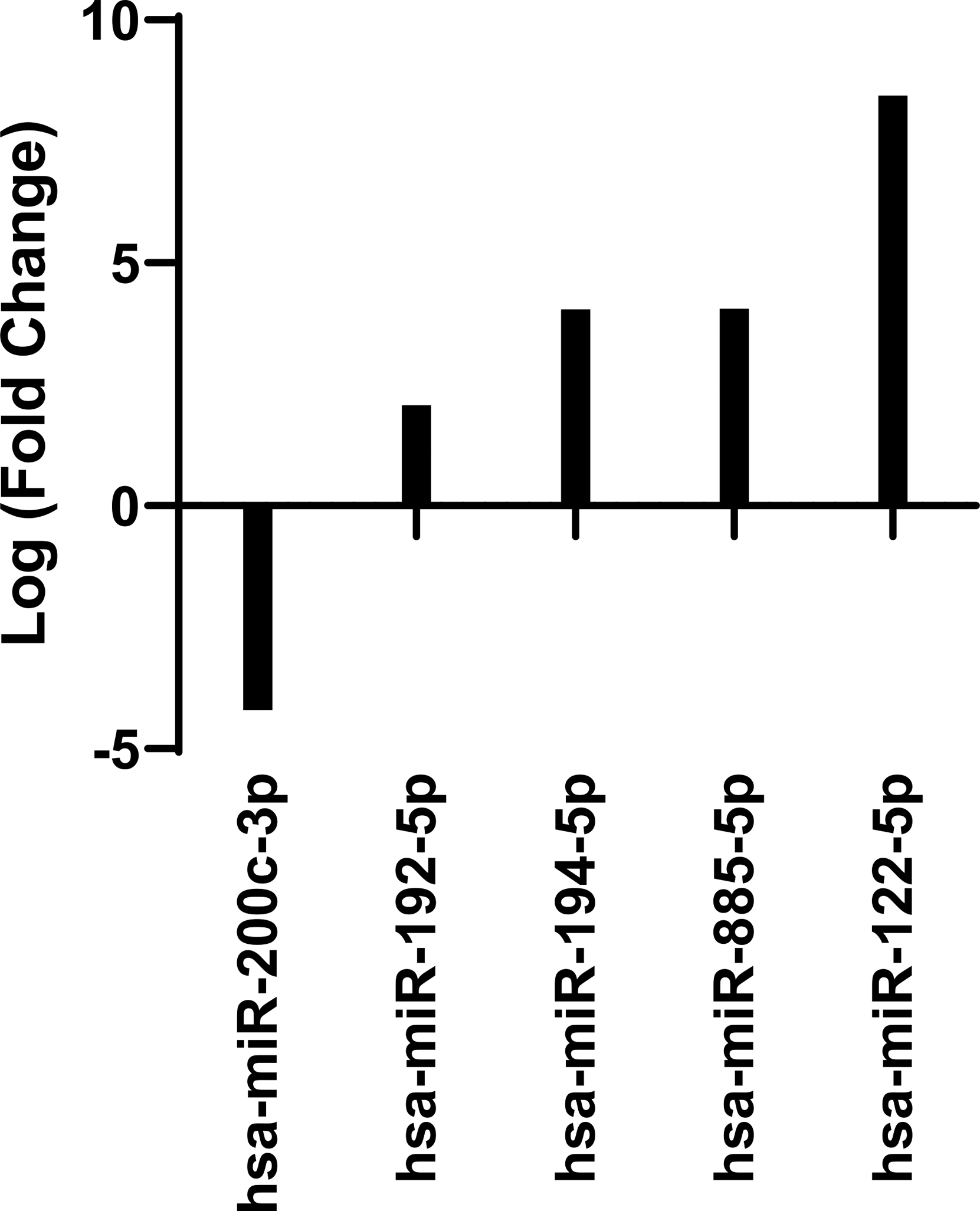
NanoString expression of hepatic metastatic melanoma (n=6) compared to pulmonary metastatic melanoma (n=6). Expression of statistically significant differentially expressed microRNAs are shown as log (fold change) in expression (p< 0.0081).
The biological impact of miR expression is a function of the gene products that the miRs are able to inhibit. Two multiple interaction models were generated in order to predict the potential effects on target genes associated with the miR profiles of each metastatic site. These models were used to prioritize which miRs and associated gene targets would be of most interest in hepatic or pulmonary sites of melanoma metastasis (Fig. 4). First, miRs shown to have consistently decreased expression in both hepatic and pulmonary sites of metastasis relative to primary cutaneous melanoma and benign nevi were examined. These criteria restricted the miRs for analysis to three, namely miR-205–5p, miR-203a-3p, and miR-1246. Generation of a multiple interaction model to highlight common gene targets among these miRs (shown in Fig. 4) revealed one common gene target among these three miRs, PLAGL2. A multiple interaction model was not generated for common miRs with increased expression specific to both hepatic and pulmonary metastatic melanoma, as the number of miRs available for construction of the model was below the degree filter threshold applied of three miR interactions with each potential gene of interest.
Figure 4. Multiple-Interaction gene target model for microRNAs with decreased expression in both hepatic and pulmonary metastases relative to benign nevi and primary cutaneous melanoma.
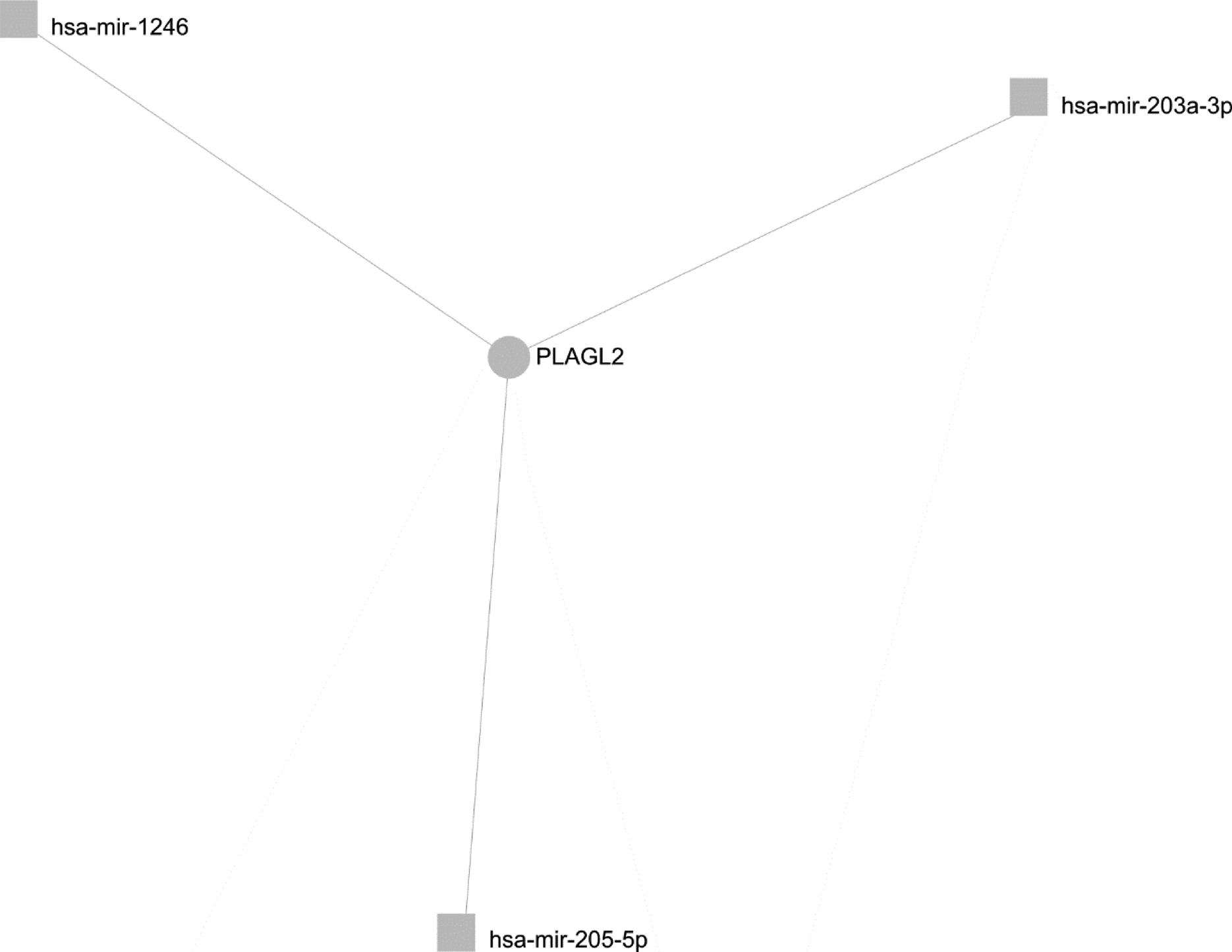
Gene targets (circles) of miRs (squares) with significantly decreased expression in pulmonary and hepatic melanoma relative to benign nevi and primary cutaneous melanoma, identified by miRNet with a minimum degree (number of associations with dysregulated microRNAs) of 3. This multiple interaction model is used to demonstrate the potential biological impact of loss of expression of these miRs in metastatic melanoma.
In Figure 5, assessment of common gene targets was performing using only miRs with relatively increased expression in each metastatic site relative to the other in order to make comparisons between pulmonary and hepatic metastases. In this comparison, miRs with decreased relative expression in one site have relatively increased expression in the other. This model revealed three genes of interest, each targeted by at least three miRs with significantly increased expression in hepatic metastatic sites of melanoma relative to pulmonary sites, namely RAC1, DUSP18, and IGF1R. The targeting of these gene transcripts by multiple miRs could theoretically have a combinatorial effect and lead to a significant decrease in the expression of these genes. Since only one miR (miR-200c-3p) was found to be significantly upregulated in pulmonary sites of melanoma metastasis relative to hepatic sites, a network could not be generated for this metastatic site. Therefore, the gene targets of miR-200c-3p, identified using miRNet, were considered for their potential effect on the pulmonary metastatic tumor microenvironment. Recognition of a common gene target using a multiple interaction model provides compelling insight to how the combination of miRs with increased and decreased expression in each site of melanoma metastasis can contribute to disease progression.
Figure 5. Multiple-Interaction gene target model for microRNAs with elevated expression in hepatic metastases relative to pulmonary metastases.
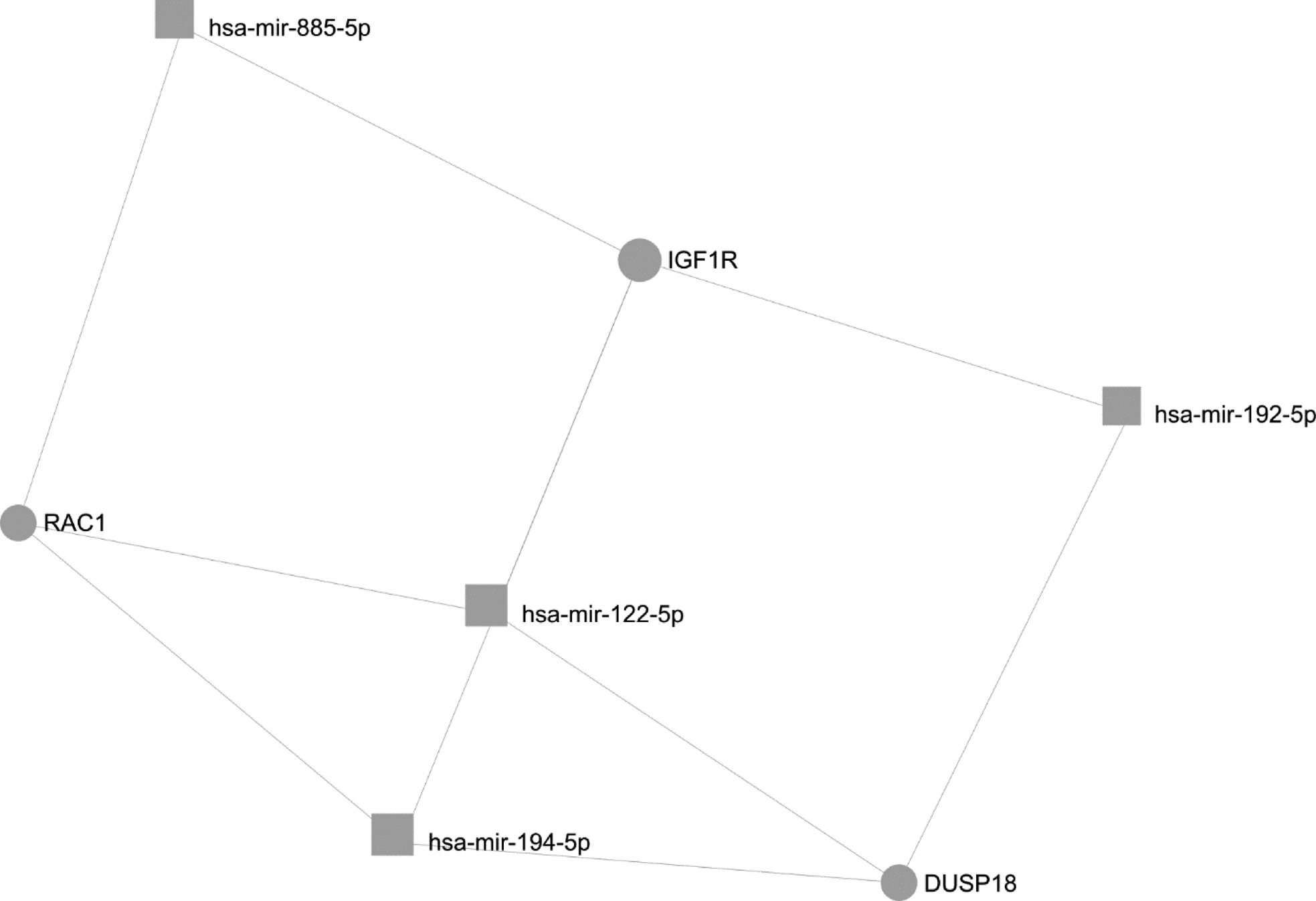
Gene targets (circles) of significantly dysregulated microRNAs (squares) in pulmonary vs hepatic melanoma identified by miRNet with a minimum degree (number of associations with dysregulated microRNAs) of 3. miRs with significantly increased expression in hepatic metastatic melanoma relative to pulmonary metastatic melanoma are included. This multiple interaction model is used to demonstrate the biological significance of these miRs in melanoma, and helps prioritize which miRs and gene targets are of the greatest potential relevance in the development of metastatic melanoma at hepatic vs. pulmonary sites.
Discussion
Using a NanoString approach to evaluate the miR expression in different sites of metastatic melanoma, we have discovered a unique miR profile for melanoma metastatic to different anatomic sites. In particular, metastatic melanoma to the liver has a unique miR profile when compared to metastatic melanoma to the lung, with 4.2-fold downregulation of miR-200c-3p, and upregulation of miR-192–5p (2.07-fold), miR-194–5p (4.04-fold), miR-885–5p (4.05-fold) and miR-122–5p (8.44 fold) associated with hepatic metastatic melanoma relative to pulmonary metastatic melanoma tissue (p<0.0081 for all down- and upregulated miRs).
The potential biologic effects associated with downregulation of miR-203a-3p, miR-205–5p, and miR-1246, as was detected in both hepatic and pulmonary sites of melanoma metastasis relative to primary cutaneous melanoma and benign nevi, are indicated in a gene target interaction network shown in Figure 4. Notably, all three miRs with significantly downregulated expression in sites of metastasis share pleiomorphic adenoma gene-like 2 (PLAGL2) as a common gene target. Thus, PLAGL2 is expected to have increased expression in the setting of metastatic melanoma based on the decreased expression of multiple regulatory miRs that target this gene. PLAGL2 is a zinc finger protein known to contribute to epithelial-mesenchymal transition (EMT) through promotion of the stability of β-catenin.[15] PLAGL2 is also involved in regulation of actin cytoskeleton rearrangement and cellular migration.[16] Elevated expression of PLAGL2 has also been shown to correlate with high stage colorectal adenocarcinoma relative to benign and low stage or borderline malignancies.[17] Thus, loss of inhibition of PLAGL2 expression may promote development of metastatic melanoma given the reduced expression of miR-203a-3p, miR-205–5p, and miR-1246 in these sites relative to primary tumor tissue. Furthermore, miR-203 has recently been identified as a miR with decreased expression in sites of metastatic melanoma that is correlated with decreased survival. In vitro and in vivo studies investigating the role of miR-203 have demonstrated restoration of miR-203 expression significantly reduces tumor growth, metastasis, tumor cell motility, and angiogenesis, among other functions. Thus, while downregulation of miR-203 may not be specific to a certain metastatic site, it remains of significant interest in melanoma for its clear role in mediating melanoma metastasis.[18]
The effect of dysregulated expression of the miRs detected specifically in hepatic metastatic melanoma relative to pulmonary sites is made apparent in the gene target interaction network shown in Figure 5. This network highlights the frequency of common specific miR gene targets among those miRs with significantly increased expression in hepatic metastases relative to pulmonary metastases. Based on this network analysis, the genes RAC1, DUSP18, and IGF1R are predicted to be downregulated in hepatic sites of metastasis, as these genes are each targeted by three or more upregulated miRs.
RAC1, a member of the RAS superfamily, is targeted by miR-122–5p, miR-885–5p, and miR-194–5p, all of which have significantly increased expression in hepatic melanoma metastases relative to pulmonary sites of metastasis. Therefore, decreased RAC1 expression would be predicted for hepatic melanoma metastases as compared to pulmonary sites. Downregulation of RAC1 expression by miR-122 has been associated with mesenchymal-epithelial transition (MET) in the setting of hepatocellular carcinoma.[19] MET is hypothesized to be a late step in the establishment of metastatic colonies of tumor cells.[20] Therefore, inhibition of RAC1 protein expression in melanoma by miR-122–5p, miR-885–5p, and miR-194–5p may contribute to metastatic colonization of melanoma tumor cells in the liver.
Dual-specificity phosphatase 18 (DUSP18), a mitochondrial-specific phosphatase that is widely expressed but relatively uncharacterized, is targeted by miR-194–5p, miR-122–5p, and miR-192–5p, all of which are elevated in hepatic sites of melanoma metastasis. Therefore, it is expected that expression of DUSP18 would be downregulated in hepatic sites of metastases relative to pulmonary metastases. DUSP18 is a member of the MAPK phosphatase (MKP) family of proteins, which function to regulate the activity of MAPK and JNK-ERK signaling.[21] Of the MKPs, DUSP18 has not yet been investigated for its role in cancer development, progression, or response to therapy, but is thought to play a role in the regulation of apoptosis upon release from the intermembrane space of mitochondria with cellular stress.[22] Therefore, further investigation of down-regulated DUSP18 in the setting of melanoma is required to determine the role of this MKP in melanoma metastasis.
Insulin-like growth factor 1 receptor (IGF1R) expression is targeted by miR-122–5p, miR-192–5p, miR-194–5p, and miR-885–5p, all of which are upregulated in hepatic sites of melanoma metastasis relative to benign nevi, primary cutaneous melanoma, and pulmonary melanoma metastases. Therefore, one might expect to see decreased expression of IGF1R in melanoma metastatic to the liver. IGF1R is a receptor tyrosine kinase involved in activation of MAPK and PI3K/Akt signaling that is associated with increased tumor growth and metastases when overexpressed, and is upregulated in BRAF inhibitor resistant melanoma.[23,24] Contrary to what may be expected in a metastatic site, the miRs overexpressed in hepatic metastases suggest a protective, anti-tumor effect via the ability of multiple upregulated miRs to inhibit IGF1R expression. Further analysis of IGF1R expression is needed to fully interpret the biologic effect of elevated miR-122–5p, miR-192–5p, miR-194–5p, and miR-885–5p expression in hepatic metastatic melanoma, and how these microRNAs may affect response of hepatic melanoma metastases to current therapies.
While only miR-200c-3p was found to be significantly downregulated in hepatic sites of metastatic melanoma relative to pulmonary sites, this miR can affect many significant gene targets of potential interest, including BMI1, TUBB3 and ZEB2, among others. The following discussion focuses on loss of miR-200c-3p in hepatic metastatic sites (rather than relative upregulated miR-200c-3p expression in pulmonary sites) as miR-200c-3p was also found to be consistently downregulated in hepatic sites of metastasis relative to primary cutaneous melanoma and benign nevi, suggesting a consistent pattern of miR-200c expression in hepatic sites of metastasis relative to all other examined groups. Loss of miR-200c-3p expression in hepatic melanoma metastases may therefore play a significant role in metastasis to the liver based on the function of the predicted target genes.
BMI1 is a core catalytic component of the epigenetic regulator polycomb repressive complex 1 (PRC1) that is regulated by miR-200c.[25] BMI1 overexpression might be expected in the hepatic metastatic samples based on decreased expression of miR-200c. Increased activity of BMI has been associated with enhanced invasion and metastasis via induction of non-canonical Wnt signaling, resulting in polarized actin cytoskeleton rearrangement for directed migration.[25] Therefore, loss of miR-200c expression in melanoma cells may directly contribute to increased tumor invasion and metastasis to hepatic sites. BMI1, as well as Tubulin Beta 3 Class III (TUBB3) and Zinc Finger E-Box Binding Homeobox 2 (ZEB2) all have increased expression in BRAF inhibitor resistant melanoma in association with low miR-200c expression. The expression of these genes has also been significantly correlated with increased N-cadherin and SNAIL expression in melanoma tumor tissue, two markers of epithelial-mesenchymal transition (EMT) that were induced by loss of miR-200c expression in these tissues after BRAF inhibitor treatment.[26] The relative persistence of miR-200c expression in pulmonary sites of metastasis and the important role of the target genes regulated by miR-200c expression in epithelial-mesenchymal transition suggests a differential role for this pathway in development of metastasis to the liver compared to the lung.
There are several limitations to this work. Notably, the small sample size is attributed to the distinct features of the NanoString platform which allow for a maximum of twelve samples to be analyzed simultaneously. The use of multiple cartridges sequentially can overcome this shortcoming, but this comes at the cost of introducing inter-chip variability. These findings need to be validated with greater numbers using accepted practices such as qPCR before in vivo and clinical endeavors can be justified. While the current study provides a unique approach to examination of miR expression patterns in melanoma, additional findings from other groups regarding miR expression in primary and/or metastatic sites provide detailed insight regarding miRs in melanoma that have been summarized in multiple reviews.[27–29]
Due to limitation of tissue and RNA availability for the current samples, qPCR of dysregulated miRs and gene targets was not performed in the present study. Furthermore, documentation of the therapies received by these patients was not available at the time of sample collection for correlation with tissue miR expression. Therefore, the potential effect of specific therapies on tissue miR expression in these patients cannot be determined.
Additionally, the miR expression profile in adjacent normal tissue at the sites of metastasis in the lung and liver was not compared to the miR expression of the metastatic lesion due to a lack of tissue availability. Specifically, hepatic tissue is known to express miR-122 and may provide a significant source of background in our hepatic metastatic melanoma samples, leading to potential elevations in miR-122–5p expression of non-tumor origin in hepatic metastases relative to other groups.[30,31] However, recent literature indicates melanoma can express miR-122 and secrete miR-122–5p in exosomes upon activation of lysophosphatidic acid receptor 3 (LPAR3), a receptor critical to mediation of Wnt signaling and associated with increased melanoma cell survival upon activation.[32] Therefore, consideration of metastatic melanoma tissue as a potential source of increased miR-122–5p within the tissue is warranted. It must also be noted that the primary melanoma and metastatic melanoma samples available for assessment were isolated from unique patients. Thus, differences between these groups must be interpreted with the caveat that these groups do not represent progression of melanoma and associated miR expression within individual patients. Rather, the groups provide a representative sample of metastatic melanoma lesions in specific tissue microenvironments unique from primary cutaneous melanoma. Nonetheless, the present approach is useful as a means of focusing attention on a small group of miRs in each metastatic site for further assessment of their role the setting of melanoma metastasis. This process allows for direction of future efforts to a limited number of attractive miR candidates.
Conclusions
Collectively, this work suggests distinct miR profiles for melanoma metastatic to the lung as compared to liver and indicates that these miR patterns that may play a role in tumor localization and biologic behavior. Further validation studies are necessary to confirm these findings and explore the utility of these miR profiles as site-specific biomarkers of metastatic disease.
Supplementary Material
Funding:
NIH Grants P01 CA095426 (to W.E. Carson), P30 CA16058 (to W.E. Carson), T32 CA090223 (to N. Latchana), T32 CA009338 (to M.J. DiVincenzo), K24 CA093670 (to W.E. Carson), NLM grant T15LM011270 to ZA.
Abbreviations
- miR
microRNA
- IGF1R
Insulin-like growth factor 1 receptor
- PRC1
polycomb repressive complex 1
- TUBB3
Tubulin Beta 3 Class III
- EMT
epithelial-mesenchymal transition
- LPAR3
lysophosphatidic acid receptor 3
Footnotes
Ethics approval and consent to participate: Use of human tissue in this study was approved by the Ohio State University Office of Responsible Research Practices Cancer Institutional Review Board under protocol #2007C0015 (OSU-07030). A waiver of patient consent was approved by the IRB for this study as it is exploratory in nature.
Consent to Publish: Not applicable.
Competing Interests: No competing interested are declared.
Availability of data and materials: The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68 (1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.von Schuckmann LA, Hughes MCB, Ghiasvand R, Malt M, van der Pols JC, Beesley VL, et al. Risk of Melanoma Recurrence After Diagnosis of a High-Risk Primary Tumor. JAMA Dermatol 2019; 155 (6):688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas DC, Han G, Leong SP, Kashani-Sabet M, Vetto J, Pockaj B, et al. Recurrence of Melanoma After a Negative Sentinel Node Biopsy: Predictors and Impact of Recurrence Site on Survival. Ann Surg Oncol 2019; 26 (7):2254–2262. [DOI] [PubMed] [Google Scholar]
- 4.Gassenmaier M, Keim U, Leiter U, Eigentler TK, Rocken M, Gesierich A, et al. Age as key factor for pattern, timing, and extent of distant metastasis in patients with cutaneous melanoma: A study of the German Central Malignant Melanoma Registry. J Am Acad Dermatol 2019; 80 (5):1299–1307 e1297. [DOI] [PubMed] [Google Scholar]
- 5.Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017; 67 (6):472–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard JH, Thompson JF, Mozzillo N, Nieweg OE, Hoekstra HJ, Roses DF, et al. Metastasectomy for distant metastatic melanoma: analysis of data from the first Multicenter Selective Lymphadenectomy Trial (MSLT-I). Ann Surg Oncol 2012; 19 (8):2547–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev 2012; 31 (3–4):653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett 2009; 285 (2):116–126. [DOI] [PubMed] [Google Scholar]
- 9.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017; 16 (3):203–222. [DOI] [PubMed] [Google Scholar]
- 10.Thyagarajan A, Shaban A, Sahu RP. MicroRNA-Directed Cancer Therapies: Implications in Melanoma Intervention. J Pharmacol Exp Ther 2018; 364 (1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019; 47 (D1):D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alder H, Taccioli C, Chen H, Jiang Y, Smalley KJ, Fadda P, et al. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis 2012; 33 (9):1736–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaelsen TY, Richter J, Brondum RF, Klapper W, Johnsen HE, Albertsen M, et al. A B-cell-associated gene signature classification of diffuse large B-cell lymphoma by NanoString technology. Blood Adv 2018; 2 (13):1542–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res 2016; 44 (W1):W135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell 2010; 17 (5):497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekiya R, Maeda M, Yuan H, Asano E, Hyodo T, Hasegawa H, et al. PLAGL2 regulates actin cytoskeletal architecture and cell migration. Carcinogenesis 2014; 35 (9):1993–2001. [DOI] [PubMed] [Google Scholar]
- 17.Wang YP, Guo PT, Zhu Z, Zhang H, Xu Y, Chen YZ, et al. Pleomorphic adenoma gene like-2 induces epithelial-mesenchymal transition via Wnt/beta-catenin signaling pathway in human colorectal adenocarcinoma. Oncol Rep 2017; 37 (4):1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohcharoenkal W, Das Mahapatra K, Pasquali L, Crudden C, Kular L, Akkaya Ulum YZ, et al. Genome-Wide Screen for MicroRNAs Reveals a Role for miR-203 in Melanoma Metastasis. J Invest Dermatol 2018; 138 (4):882–892. [DOI] [PubMed] [Google Scholar]
- 19.Wang SC, Lin XL, Li J, Zhang TT, Wang HY, Shi JW, et al. MicroRNA-122 triggers mesenchymal-epithelial transition and suppresses hepatocellular carcinoma cell motility and invasion by targeting RhoA. PLoS One 2014; 9 (7):e101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunasinghe NP, Wells A, Thompson EW, Hugo HJ. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev 2012; 31 (3–4):469–478. [DOI] [PubMed] [Google Scholar]
- 21.Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev 2008; 27 (2):253–261. [DOI] [PubMed] [Google Scholar]
- 22.Rardin MJ, Wiley SE, Murphy AN, Pagliarini DJ, Dixon JE. Dual specificity phosphatases 18 and 21 target to opposing sides of the mitochondrial inner membrane. J Biol Chem 2008; 283 (22):15440–15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Sinnberg T, Niessner H, Dolker R, Sauer B, Kempf WE, et al. PTEN regulates IGF-1R-mediated therapy resistance in melanoma. Pigment Cell Melanoma Res 2015; 28 (5):572–589. [DOI] [PubMed] [Google Scholar]
- 24.Werner H Tumor suppressors govern insulin-like growth factor signaling pathways: implications in metabolism and cancer. Oncogene 2012; 31 (22):2703–2714. [DOI] [PubMed] [Google Scholar]
- 25.Ferretti R, Bhutkar A, McNamara MC, Lees JA. BMI1 induces an invasive signature in melanoma that promotes metastasis and chemoresistance. Genes Dev 2016; 30 (1):18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Tetzlaff MT, Wang T, Yang R, Xie L, Zhang G, et al. miR-200c/Bmi1 axis and epithelial-mesenchymal transition contribute to acquired resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res 2015; 28 (4):431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gajos-Michniewicz A, Czyz M. Role of miRNAs in Melanoma Metastasis. Cancers (Basel) 2019; 11 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thyagarajan A, Tsai KY, Sahu RP. MicroRNA heterogeneity in melanoma progression. Semin Cancer Biol 2019; 59:208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mannavola F, Tucci M, Felici C, Stucci S, Silvestris F. miRNAs in melanoma: a defined role in tumor progression and metastasis. Expert Rev Clin Immunol 2016; 12 (1):79–89. [DOI] [PubMed] [Google Scholar]
- 30.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest 2012; 122 (8):2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J, Xu Y, Hao J, Wang S, Li C, Meng S. MiR-122 in hepatic function and liver diseases. Protein Cell 2012; 3 (5):364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byrnes CC, Jia W, Alshamrani AA, Kuppa SS, Murph MM. miR-122–5p Expression and Secretion in Melanoma Cells Is Amplified by the LPAR3 SH3-Binding Domain to Regulate Wnt1. Mol Cancer Res 2019; 17 (1):299–309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


