Abstract
The SRF/MRTF and upstream signaling cascade play key roles in actin cytoskeleton organization and myocyte development. To date, how this signaling axis may function in brown adipocyte lineage commitment and maturation has not been delineated. Here we report that MRTF-SRF signaling exerts inhibitory actions on brown adipogenesis, and suppressing this negative regulation promotes brown adipocyte lineage development. During brown adipogenic differentiation, protein expressions of SRF, MRTFA/B and its transcription targets were down-regulated, and MRTFA/B shuttled from nucleus to cytoplasm. Silencing of SRF or MRTF-A/MRTF-B enhanced two distinct stages of brown adipocyte development, mesenchymal stem cell determination to brown adipocytes and terminal differentiation of brown adipogenic progenitors. We further demonstrate that the MRTF-SRF axis exerts transcriptional regulations of the TGF-β and BMP signaling pathway, critical developmental cues for brown adipocyte development. TGF-β signaling activity was significantly attenuated, whereas that of the BMP pathway augmented by inhibition of SRF or MRTF-A/MRTF-B, leading to enhanced brown adipocyte differentiation. Our study demonstrates the MRTF-SRF transcriptional cascade as a negative regulator of brown adipogenesis, through its transcriptional control of the TGF-β/BMP signaling pathways.
Keywords: Serum Response Factor, Myocardin-related Transcription Factors, Actin Cytoskeleton, Brown Adipogenesis
Graphical Abstract
1. INTRODUCTION
Serum response factor (SRF) is a muscle-enriched transcription factor essential for the embryonic development of myocyte lineages, including cardiac, skeletal and smooth muscle (Pipes, Creemers and Olson, 2006). It belongs to the MADS (MCM1, AGAMOUS, DEFICIENS and SRF)-box family of transcription factors, acting through binding to a consensus DNA sequence known as the CArG box [CC(A/T)6GG](Posern and Treisman, 2006). SRF exerts its transcriptional activity through specific interactions with key cofactors in a cell- and context-specific manner (Wang, Li, Hockemeyer et al., 2002,Miano, 2003). Myocardin-related transcription factors A and B (MRTF-A and MRTF-B, also known as MKL1/MAL and MKL2), are co-activators for SRF that are selectively recruited to activate SRF-mediated in a transcription signal-dependent manner (Posern and Treisman, 2006,Olson and Nordheim, 2010). Through interactions with the monomeric globular actin (G-actin), the MRTFs are sequestered in the cytosol without signal stimulation. Extracellular stimuli, including growth factors and G-protein receptor ligands etc. that converging on intracellular actin dynamics, are ultimately transduced via MRTF nuclear translocation to activate SRF activity, thereby eliciting specific cellular behaviors. In response to extracellular signals, cell-matrix interaction or cell-cell interactions, activated Rho-GTPase activity leads to actin polymerization, MRTFA/B release from G-actin and subsequent nuclear entry. The gene transcription activated by the recruitment of MRTFs to SRF DNA-binding domain drives myriads of fundamental cellular processes involving actin cytoskeleton organization, including cellular morphology, adhesion, migration, proliferation and differentiation (Olson and Nordheim, 2010).
Cell shape and cytoskeletal structure dictated by physical or physiological cues confer instructive signals to cellular proliferation and differentiation behaviors. Cell shape drives mesenchymal stem cell lineage commitment between adipocyte and osteoblast through modulation of cytoskeletal signaling activity (McBeath, Pirone, Nelson et al., 2004). SRF is known to be required for mesoderm lineage development (Arsenian, Weinhold, Oelgeschlager et al., 1998). SRF and MRTF signaling cascade play instructive roles in cardiac, skeletal and smooth muscle myocyte lineage development (Li, Czubryt, McAnally et al., 2005,Parlakian, Tuil, Hamard et al., 2004). Interestingly, only limited studies to date have addressed the involvement of SRF related signaling in adipocytes. Notably, adipogenic initiation in 3T3-L1 cells requires dissolution of actin cytoskeleton architecture accompanying the dramatic cell shape change, a process that precedes cellular lipid synthesis and accumulation in the differentiating adipocytes (Spiegelman and Ginty, 1983). More recently, a epigenomic screen to identify open chromatin-associated transcription regulators reveal SRF as a negative regulator of adipogenesis (Mikkelsen, Xu, Zhang et al., 2010). However, whether the SRF regulatory cascade functions in adipocyte lineage commitment stage remains unclear.
Recent developments revealed surprising functional and developmental heterogeneities within the adipocyte lineage, including the classical white visceral fat adipocytes, beige subcutaneous fat adipocytes or the interscapular fat brown adipocytes (Wu, Jun and McDermott, 2015,Sanchez-Gurmaches, Hung and Guertin, 2016). Although these distinct types of adipocytes arise from a shared mesenchymal progenitor lineage with myocytes, their developmental origins differ (Sanchez-Gurmaches et al., 2016). Brown adipocytes originate from a shared Myf5+ dermomyotome mesenchymal lineage with myogenic precursors (Seale, Bjork, Yang et al., 2008). Given the essential role of SRF signaling in myocyte development and involvement in adipogenesis, it is intriguing to postulate that the MRTF-SRF circuit may influence mesenchymal precursor fate determination between the myogenic vs. thermogenic adipocyte lineage. However, to date, the role of MRTF-SRF in thermogenic adipocyte commitment or terminal differentiation has not been clearly defined. Employing complementary cellular model systems, our current study revealed strong suppressive effects of the MRTF-SRF signaling on thermogenic adipocyte lineage determination and maturation.
2. MATERIALS AND METHODS
2.1. Animal
Wild type male C57BL/6J (Stock No: 000664) were obtained from Jackson laboratories and maintained in City of Hope vivarium. The mice were maintained under a constant 12:12 light dark cycle and fed standard irradiated rodent chow ad libitum. All experiments were approved by the IACUC committee of the Beckman Research Institute of City of Hope. Mouse tissues were harvested between 9–10 AM and flash frozen at −800C till processed.
2.2. Cell culture and differentiation
C3H10T1/2 cell line and HIB1B cells (ATCC) were maintained and differentiated as previously described (Nam, Guo, Chatterjee et al., 2015). Before adipogenic differentiation, cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37°C in 5% CO2. For HIB1B brown preadipocyte differentiation, cells at 100% confluence were treated with induction media containing 1μM dexamethasone, 0.5mM 3-isobutyl-1-methylxanthine, 20nM insulin and 2nM T3 for 2 days followed by maintenance medium containing 20 nM insulin and 2nM T3 for 2–4 days. For C3H10T1/2 differentiation, 1mM rosiglitazone was additionally supplemented in the induction media for 3 days, then replaced with maintenance medium for 6–9 days. 5’-azacytidine treatment of C3H10T1/2 cells was started at low confluency of 20–30% at 20 μM for 5 weeks with media change every 3 days.
2.3. Lentivirus preparation, infection and stable cell line selection
The shRNA constructs for MRTF-A/B (pLKO.1-MRTF-A/B, #27161) and scramble control (pLKO.1-Scr, #8453) were purchased from Addgene. The shRNA constructs for SRF (psiHIV-Srf, MSH032352) and scramble control (psiHIV-Scr) were purchased from Genecopoeia. Three shRNAs were tested for SRF or MRTFA/B for their gene silencing efficiency. The stable clones were generated similarly as previously described(Chatterjee, Nam, Guo et al., 2013). Briefly, lentiviral vectors (pLKO.1-Scr, pLKO.1-MRTF-A/B, psiHIV-Scr or psiHIV-Srf), pSPAX2 and pMD2G were co-transfected into 293T cells using Fugene 6 reagent (Roche). Supernatants were collected 48 hours after transfection, centrifuged at 27,000g for 90 min, and the resultant pellet dissolved in appropriate volume of DMEM. Target cells were infected for 24 hours, and 2μg/ml puromycin (Invitrogen) was applied for stable pooled selection. Knockdown efficiency of stably selected clones was verified by Western blot.
2.4. Immunofluorescence staining
Cells were grown in four-well cell culture chamber slides were fixed with 4% paraformaldehyde, followed by permeabilization in 0.5% Triton X-100 and blockade in 2% BSA prior to staining. Cells were incubated with primary antibodies: SRF (Abcam, ab155013, 1:200), MRTF-A (Sigma, HPA030782, 1:100) or MRTF-B (Bethyl Lab., A302–768A, 1:200) overnight at 4°C, followed by Alexa Fluor 594 conjugated anti-rabbit IgG antibody (Invitrogen, A11012, 1:200) for 1 hour at room temperature. Phalloidin (Invitrogen; A12379 or A12380, 1:1,000) or BODIPY® lipid probe (4,4-difluoro-3a,4adiaza-s-indacene, Invitrogen; D-3922, 1 mg/ml) were applied directly after Triton X-100 incubation. Slides were mounted with anti-fade mounting medium with DAPI. MitoTracker Deep Red (Invitrogen; M22426, 100 nM) incubation in live cells was used for mitochondria staining, as described (Nam et al., 2015,Nam, Chatterjee, Yin et al., 2015). Immunofluorescent images were captured using a Nikon 80i microscope with a color camera and processed using Nikon NIS Elements acquisition software.
2.5. RNA extraction and real-time PCR
Total RNAs were extracted using TRIzol reagent (Invitrogen, 15596–018) according to manufacturer’s protocol. Complementary DNA were synthesized using q-Script cDNA Supermix kit (Quanta Biosci. 101414–106). Realtime PCR was performed using Perfecta SYBR Green Supermix (Quanta Biosci. 101414–152) in a Roche 480 Light Cycler machine. Relative mRNA expression levels were determined by normalization of target genes to 36B4 as internal control. Primer were designed using PrimerBank experimentally validated sequences, as previously described (Nam et al., 2015,Nam et al., 2015).
2.6. Western blot analysis
Protein samples were extracted using ice-cold RIPA buffer (Thermo Scientific), supplemented with protease inhibitor and phosphatase inhibitor (Roche) when necessary. 20–40 μg of protein were fractionated by SDS-PAGE gel, transferred to PVDF membranes (Millipore) and blocked in 5% non-fat milk-PBS. Primary and HRP-conjugated secondary antibodies were applied followed by enhanced chemiluminescence substrate (Supersignal, Thermo Sci.) for detection. The antibody dilutions used were: SRF 1:2,000 (Santa Cruz, sc-13029), HSP90 1:2,000 (Cell signal, #4874), MRTF-A 1:500 (Sigma, HPA030782), MRTF-B 1:500 (Bethyl Lab., A302–768A), Vinculin 1:10,000 (Millipore, FAK100), C/EBPα 1:1,000 (Santa Cruz, sc-61), C/EBPβ 1:1,000 (Santa Cruz, sc-150), PRDM16 1:500 (Santa Cruz, sc-130243), GAPDH 1:50,000 (Millipore, MAB374), aP2 (R&D, AF1443), UCP1 1:500 (Millipore, AB3038), Smads 1:1,000 (Cell Signaling, #9963), α-SMA 1:1,000 (A5228, Sigma), α-tubulin 1:1,000 (Santa Cruz, sc-8035), TBP (Santa Cruz, sc-204).
2.7. TGFβ and BMP signaling luciferase reporter assay
TGF-β and BMP pathway signaling activities were assessed as previously described (Nam et al., 2015,Nam et al., 2015). Briefly, C3H10T1/2 cells were seeded in 24-well plates at 30–40% confluence. Luciferase construct plasmids, pGL3-Smad-binding element (SBE4-Luc, Addgene, #16495) or pGL3-BMP-response elements (BRE2Luc, kindly provided by P. Dijke) 200ng and pRL-null (Promega) 5ng were co-transfected using Lipofectamin 2000 (Invitrogen). Cells were starved overnight 24 hours transfection, prior to treatment with TGF-β1 or BMP-4 (ProSpec) for 24 hours at indicated concentration. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer’s protocol. Relative luciferase unit was determined by normalization to Renilla activity as internal control.
2.8. Statistical analysis
Data presented are representative of at least three independent experiments. Values are expressed as mean ± SEM with minimum of three biological replicates. The differences between groups were determined by two-sided, unpaired Student’s t test. P values less than 0.05 were considered statistically significant.
3. RESULTS
3.1. Suppression of SRF-MRTF signaling cascade expression during brown adipocyte differentiation
To determine the role of SRF signaling pathway in thermogenic adipocytes, we first screened protein expressions of the key transcription regulators in adult mesoderm-derived tissues including heart, skeletal muscle, white and brown adipose depots (Fig. 1A). Surprisingly, moderate SRF level was detected in the interscapular brown adipose tissue (BAT) as compared to classic epidydimal white adipose tissue (WAT), albeit much lower than that of skeletal muscle or heart (Fig. 1A & 1B). SRF co-activators, MRTF-A and -B display distinct expression patterns in adipose depots. While MRTF-A was barely expressed in most adult mice tissues examined, this protein exhibit slightly higher but variable levels in BAT similar to that of skeletal muscle. Most notably, MRTF-B was significantly enriched in both WAT and BAT, a potential dominant SRF co-activator present in adipose depots that was largely not detectable in other tissues. Furthermore, examination of SRF level among mesodermal cell types revealed its highest expression in C3H10T1/2 (10T1/2) mesenchymal stem cells (Fig. 1B), which was comparable to that of lineage-committed myogenic precursor C2C12 myoblasts. In contrast, SRF is more enriched in 3T3-L1 white preadipocytes than HIB1B brown preadipocytes, distinct from its distribution in brown vs. white fat depots in vivo. Brown adipogenic induction of the 10T1/2 mesenchymal progenitors is a well-established multipotent mesenchymal stem cell model capable of lineage commitment and differentiation into brown adipocytes (Tseng, Kokkotou, Schulz et al., 2008). Using this brown adipocyte differentiation model, we next examined the expression dynamics of key mediators of the MRTF-SRF signaling cascade along the time course of brown adipogenesis. As shown in Fig. 1C, within the first 6 days of brown adipogenic differentiation, brown lineage marker PRDM16 and adipogenic transcription factors C/EBPα and C/EBPβ were robustly induced as expected, indicative of brown adipocyte induction. Accompanying the brown lineage development in 10T1/2 cells, there was a rapid reduction of SRF protein, along with its up-stream transcription coactivator proteins, MRTF-A and MRTF-B. The drop in MRTF-A level was the most pronounced in the early stage at one day after adipogenic induction and nearly absent after day 2. In addition, clear gradual declining of protein expressions of MRTF-SRF downstream signaling target cytoskeleton components were evident during the induction time course, including α-smooth muscle actin (SMA), vinculin and α-tubulin (Fig. 1D). As monitored by lipid accumulation using BODIPY staining together with co-staining of F-actin by Phalloidin to identify actin cytoskeleton, brown adipogenic conversion in these cells led to marked reductions of intracellular actin cytoskeleton network with accumulating lipid droplets formation (Fig. 1E). These results demonstrate that SRF signaling was suppressed during brown adipocyte lineage development from its mesenchymal precursor, implicating its potential function in this process.
Figure 1. Suppression of SRF pathway in brown adipocyte differentiation.
(A, B) Immunoblot analysis of SRF protein tissue distribution (A), and quantification with normalization to heat shock protein 90 (HSP90) as internal control (B) in 3-weeks old mice (n=3/lane). (C) SRF protein levels in mesodermal lineage cell lines, C2C12 murine myoblasts, HIB1B brown preadipocyte, 3T3-L1 white preadipocytes and C3H10T1/2 mesenchymal stem cells. (D, E) Dynamic protein expression of SRF signaling components and brown adipogenic differentiation markers (C/EBPα, C/EBPβ, Prdm16) (D), and SRF transcriptional targets involved in cytoskeleton organization (smooth muscle α-actin SMA, vinculin and α-tubulin) along the indicated time course of C3H10T1/2 differentiation induced by brown adipogenic cocktail (E), with Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal control. (F) Fluorescence staining of F-actin stress fibers by Alexa 546-conjugated phalloidin and BODIPY 493/503 labeling of lipid droplets before and at day 8 of brown adipogenic differentiation. Arrowheads: Bodipy-stained lipid droplets. Scale bar: 50μm.
3.2. Dynamic regulation of MRTFA/B nuclear translocation during brown differentiation
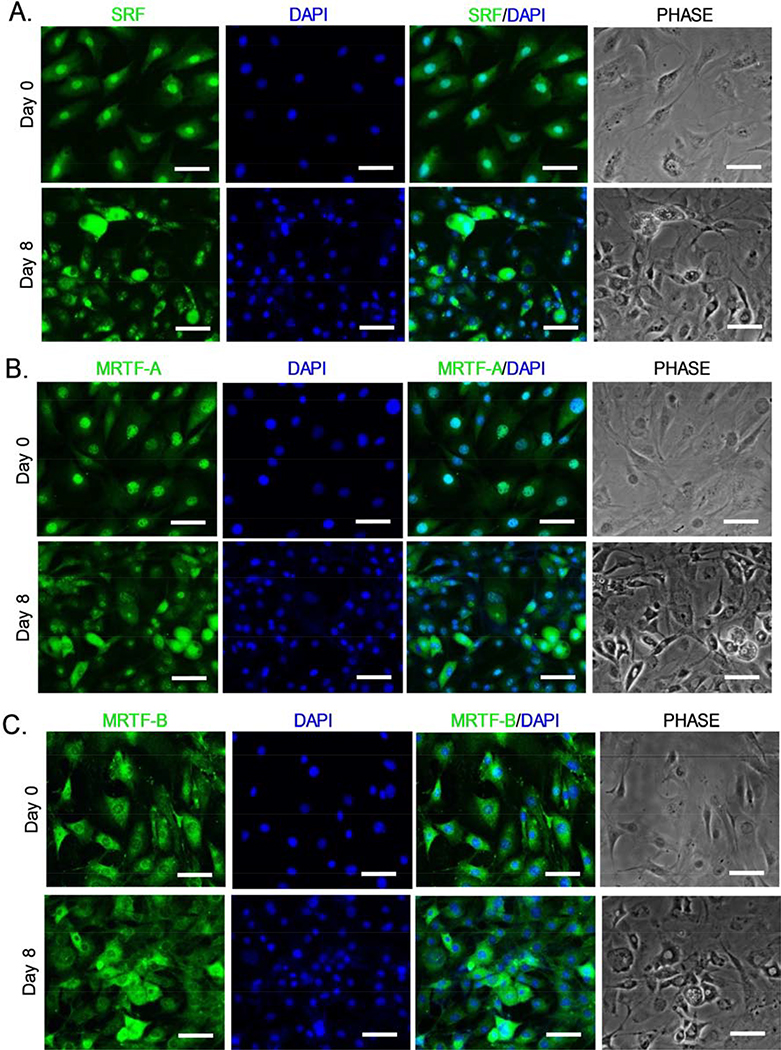
As transcription co-activators of SRF, MRTFA/B translocation from cytosol to the nucleus activates SRF-mediated transcription involved in actin cytoskeleton remodeling (Olson and Nordheim, 2010). We thus examined SRF and MRTF nuclear localization to monitor MRTF-SRF signaling activity during brown adipogenic day 0 and day 8 by immunofluorescence staining. Cells were seeded at 20–30% confluency at time of adipogenic induction to avoid overlap immunofluorescence signals. In undifferentiated state, SRF was largely nuclear in 10T1/2 cells as expected with the presence of 10% serum in the culture media (Fig. 2A). Upon differentiation, SRF shuttles from the predominantly nuclear localization to largely cytoplasmic in the day 8-differnetiated cells. Similarly, MRTF-A also displayed nuclear exclusion upon differentiation, as shown in Fig. 2B. In contrast, MRTF-B localization at day 0 was distinct from that of SRF or MRTF-A, which was largely cytoplasmic (Fig. 2C), and this cytoplasmic accumulation was further enhanced in readily differentiating cells following 8 days of adipogenic induction. These findings further validated the inhibition of MRTF-SRF signaling activity during brown adipocyte maturation, in line with the observation of the dissolution of actin filaments in this process.
Figure 2.
Dynamic regulation of SRF and MRTF cytoplasmic to nuclear shuttling during C3H10T1/2 brown adipogenesis. Representative confocal images of immunofluorescence staining of MRTF-A (A), MRTF-B (B) and SRF (C) identify subcellular localization between cytoplasm and nucleus upon brown adipogenic induction. Immunofluorescence staining for MRTF-A, MRTFB and SRF before and at day 8 of differentiation were shown. Scale bar: 50μm.
3.3. Loss of SRF or MRTFs promotes beige adipogenesis
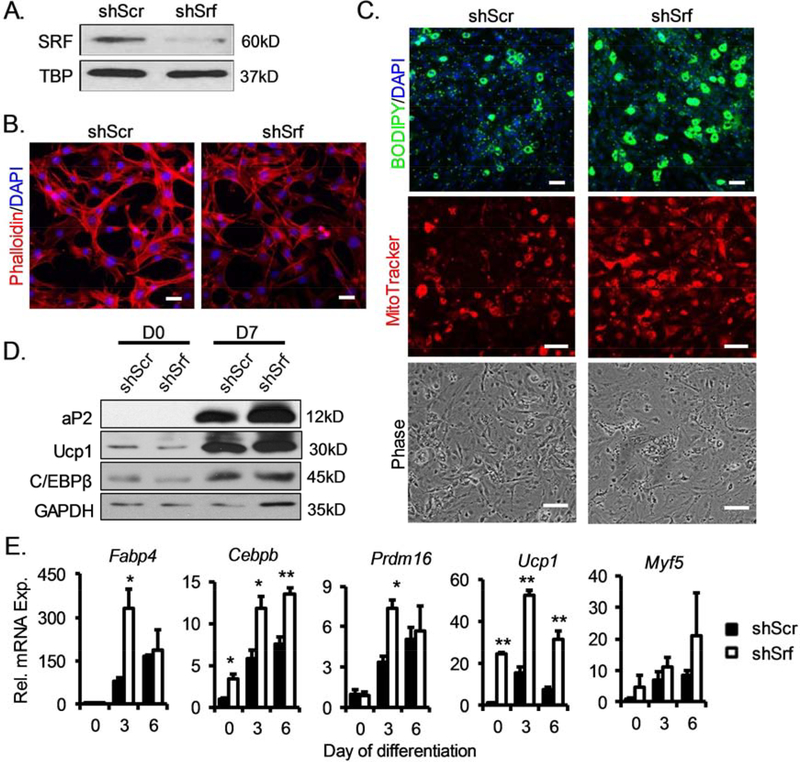
To investigate the specific functions of MRTF-SRF signaling in brown adipocyte lineage determination and differentiation, we generated stable C3H10T1/2 lines with shRNA-mediated silencing of SRF or MRTFA/B. Srf shRNA knockdown largely abolished SRF protein expression, as shown by immunoblot analysis (Fig. 3A). As a result, actin cytoskeleton staining was markedly diminished in the knockdown cells as compared to that of the controls with stable expression of scrambled shRNA (shScr), indicating inhibition of SRF activity (Fig. 3B). Upon brown adipogenic induction, differentiation of shSrf 10T1/2 cells were markedly enhanced, as shown by increased lipid accumulation and mitochondrial formation than the controls (Fig. 3C). Consistent with the enhanced morphological differentiation, protein expression of adipogenic factor C/EBPβ, mature adipocyte marker (aP2, or FABP4) and themogenic marker (Ucp1) were markedly upregulated at 7 days upon differentiation, indicating an accelerated brown adipogenic program (Fig. 3D). RT-qPCR analysis of the differentiation program further confirmed augmented adipogenic maturation (Cebpb, Fabp4) along with marked inductions of brown lineage commitment and thermogenic program (Myf5, Prdm16, and Ucp1) as a result of SRF inhibition (Fig. 3D). We next investigated whether inhibition of the upstream co-activators of SRF, MRTF-A and MRTF-B, exert similar effects on promoting brown differentiation as seen with inhibition of SRF function. Knock-down of MRTFA and MRTFB using a common shRNA consteuct targeting both regulators effectively abolished these proteins in 10T1/2 cells (Fig. 4A). As indicated by phalloidin staining, the loss of MRTFs resulted in a near complete dissolution of actin cytoskeleton at Day 7 upon differentiation, an effect stronger than that of SRF inhibition (Fig. 4B). In line with the observed SRF effect on suppressing brown adipogenesis, loss of these SRF coactivators resulted in robustly augmented adipogenic and thermogenic induction, as shown by lipid and mitochondrial staining (Fig. 4C). Analyses of the thermogenic and adipogenic program along the differentiation time course further corroborated these findings (Fig. 4D).
Figure 3.
Inhibition of SRF promotes brown adipogenic differentiation of mesenchymal stem cells. (A) Loss of SRF protein expression by stable expression of Srf shRNA (psiHIV-shSrf) in C3H10T1/2 as compared to scrambled control shRNA (psiHIV-shScr) by immunoblot analysis. (B) Phalloidin staining of F-actin stress fiber in stable knockdown of Srf. Scale bar: 50μm. (C) Brown adipogenic differentiation of shSrf and shScr C3H10T1/2 cells after 6 days of induction as assessed by lipid accumulation by BODIPY and mitochondrial staining by Mitotracker Red (Scale bar: 50μm). Phase-contrast images are shown for cell morphology. (D, E) Immunoblot (D), and RT-qPCR (E) analysis of adipogenic factors and mature brown adipocyte marker gene expression in Srf knockdown cells compared with scramble controls. n=3/group. *: p≤ 0.05 or **: p≤0.01 by Student’s T test shSrf vs. shScr.
Figure 4. Loss of MRTFs promotes mesenchymal stem cell brown adipogenesis.
(A) Inhibition of MRTF-A and MRTF-B protein level by stable knockdown of MRTF1/2-trageting shRNA (pLKO.1-shMrtf) in C3H10T1/2 cells. (B) Reduction of intracellular cytoskeleton as indicated by phalloidin staining in shMrtf and control (shScr) C3H10T1/2 cells (Scale bar: 50μm). (C) Enhanced brown adipogenic differentiation as assessed by lipid accumulation by BODIPY staining and mitochondrial staining by Mitotracker Red in shMrtf cells as compared shScr after 6 days of brown induction (Scale bar=50μm). Phase-contrast images are shown for cell morphology. Scale bar: 50μm. (D) RT-qPCR analysis of adipogenic and thermogenic gene program in shMrtf C3H10T1/2 as compared shScr cells (n=3/group). *P < 0.05, ** P < 0.01, by un-paired Student’s T test shMrtf vs. shScr.
3.4. SRF-MRTF inhibits lineage commitment of mesenchymal progenitors to brown adipocyte lineage
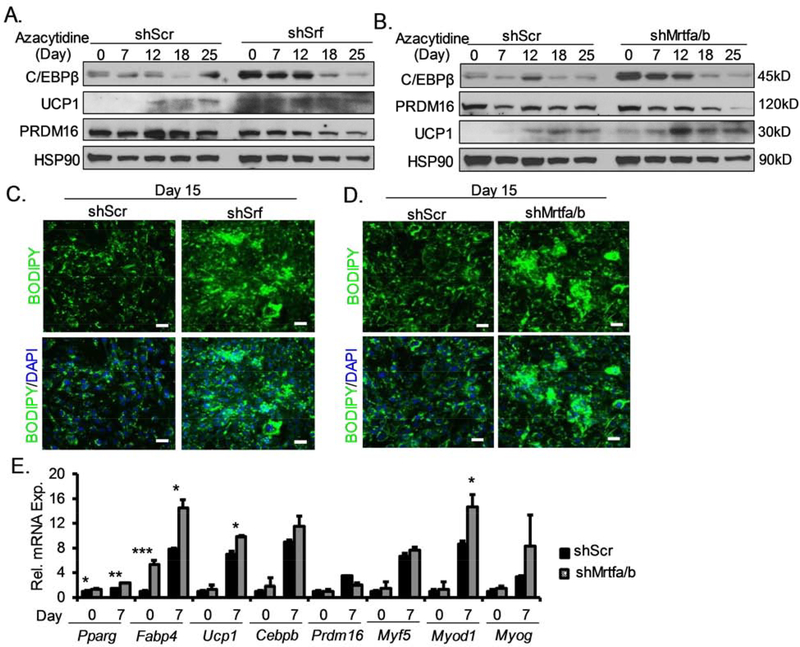
Based on MRTF-SRF inhibition of brown adipocyte development, we further tested whether this pathway functions to suppress the initial commitment of mesenchymal progenitor to the brown adipocyte lineage. We tested this by subjecting the 10T1/2 to 5-Azacytidine (AZA) treatment, a DNA methyltransferase inhibitor known to induce distinct mesodermal lineage determination in this multipotent mesenchymal precursor cell type. The 10T1/2 cells can be induced to commit and differentiate into mesodermal cell types including adipocytes, myoblasts, chondrocytes and osteoblasts under AZA without lineage-specific differentiation cocktail (Xie, Qin, Lin et al., 2011). In control shScr-containing cells, examination of lineage-specific gene induction time-course revealed robust upregulation of adipogenic marker C/EBPβ only at day 25, while thermogenic gene Ucp1 was not induced until after 12 days (Fig. 5A, B). The key brown adipocyte determination factor from myogenic precursors Prdm16 remained at constant level in the controls. Interestingly, the loss of either SRF (Fig. 5A), or MRTFs (Fig. 5B), markedly increased C/EBP-β protein even prior to AZA induction and remained highly elevated till day 18 of treatment with tapering off till day 25. Significant elevations of Ucp1 protein were observed in both SRF and MRTF knockdown cells, indicating enhanced lineage determination and differentiation into thermogenic adipocytes. Interestingly, Prdm16 level in either SRF- or MRTF-silenced 10T1/2 cells did not differ significantly from that of the controls. Following 15 days of AZA treatment, enhanced adipogenic commitment in these cells were confirmed by lipid staining (Fig. 5C and Fig. 5D). In cells with MRTF silencing, analysis of brown adipocyte lineage during the initial 7 days revealed significantly elevated transcript levels of brown adipogenic markers, Pparg, Cebpb, and Ucp1, consistent with the protein analyses (Fig. 5E). In addition, a tendency toward augmented myogenic lineage gene expression in the early time course was also observed, as suggested by increased levels of Myod1 and a tendency for Myogenin. Thus, the inhibition of MRTF-SRF signaling potentially shifted mesenchymal lineage allocation toward a common early brown/muscle progenitor with consequent differentiation into thermogenic adipocytes.
Figure 5. Loss of SRF or MRTFs induces commitment of mesenchymal stem cells toward brown adipocyte lineage under 5-Azacytidine treatment.
(A and B) Induction of adipogenic and thermogenic genes in C3H10T1/2 cells with inhibition of SRF (A) or MRTF (B), as assessed by immunoblot analysis during lineage commitment and differentiation induced by 5-Azacytidine (20μM) for 25 days. (C, D) Enhanced lipid accumulation as assessed by Bodipy staining in Srf (C) or Mrtf1/2 knockdown cells (D) at 15 days of azacytidine induction (Scale bar=50μm). (E) RT-qPCR analysis of brown adipocyte lineage markers expression at day 0 and 7 of azacytidine treatment (n=3/group). Data are displayed as mean ± SEM. *P < 0.05, ** P < 0.01, *** P < 0.001.
3.5. Silencing of SRF or MRTFA/B promotes brown preadipocyte terminal differentiation
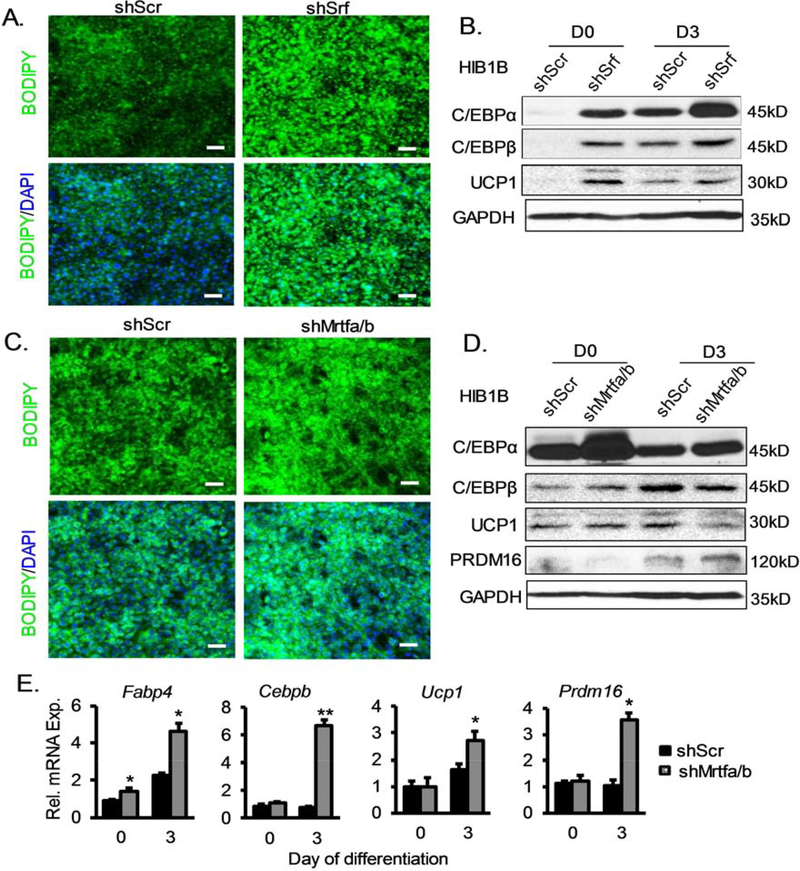
Based on earlier findings, we further tested whether MRTF-SRF cascade modulates the terminal differentiation of committed preadipocytes into mature brown adipocyte using the HIB1B brown preadipocytes. We generated stable knockdowns of SRF or MRTFs in HIB1B cells similarly as in 10T1/2 cells, and determined their differentiation into brown adipocytes after 4 days of brown adipogenic induction. Compared to the scrambled controls, silencing of SRF in HIB1B cells led to markedly enhanced differentiation, as indicated by Bodipy staining (Fig. 6A) together with significant elevations of brown adipocyte markers, C/EBPα, C/EBPβ and UCP1 (Fig. 6B). Surprisingly, significant inductions of these brown adipogenic genes occurred even prior to induction in shSrf knockdown cells, suggesting significant spontaneous differentiation with inhibition of SRF function (Fig. 6B). In comparison, loss of MRTFA/B in brown preadipocytes resulted in a similarly enhanced terminal differentiation phenotype, as indicated by lipid accumulation (Fig. 6C), and markedly higher levels of C/EBPβ and PRDM16 proteins (Fig. 6D). Detailed analysis of the brown adipocyte genes program by RT-qPCR further validated the augmented terminal differentiation toward mature brown adipocytes in Mrtfa/b-deficient HIB1B cells (Fig. 6E).
Figure 6. Inhibition of SRF or MRTFs promotes terminal differentiation of brown preadipocyte.
(A) Marked induction of adipogenic differentiation as indicated by BODIPY lipid staining in HIB1B committed brown preadipocytes with of SRF silencing by stable expression of Srf shRNA (psiHIV-shSrf), as compared to scramble shRNA control (psiHIV-shScr). (B) Immunoblot analysis of adipogenic and thermogenic gene expression in shSrf vs. shScr HIB1B cells. (C, D) Enhanced terminal differentiation of HIB1B cells with stable inhibition of MRTFs in shMrtf-expressing vs. shScr controls, as assessed by lipid accumulation by BODIPY (C, day 4), immunoblot blot (D) or RT-qPCR analysis (E, n=3) of adipogenic and thermogenic marker gene expression at day 0 and day 3 of differentiation. *P < 0.05, ** P < 0.01, by un-paired Student’s T test shMrtf vs. shScr.
3.6. SRF-MRTF regulation of TGFβ and BMP signaling pathway activity
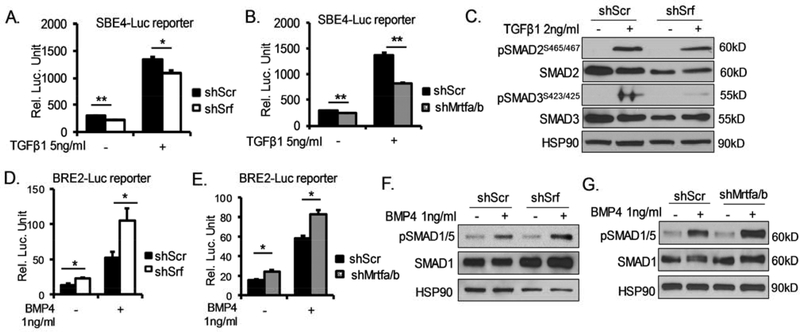
Tgf-β and BMPs are critical developmental signals that exert antagonistic regulations in brown fat development. While Tgf-β signaling inhibits brown adipogenesis, BMPs promotes brown lineage development. To test whether these signals are involved in mediating MRTF-SRF effects on suppressing brown adipogenic commitment and differentiation, we determined Tgf-β and BMP signaling activities in 10T1/2 cells with SRF or MRTF silencing using established luciferase reporters, the Smad-binding elements (SBE) and BMP response element (BRE) luciferase reporters, respectively. As shown in Fig. 7A, SRF inhibition resulted in a significant reduction of SBE-driven luciferase activity under either basal or Tgf-β-stimulated conditions. Loss of MRTFs led to even greater attenuation of Tgf-β signaling activity as compared to SRF inhibition (Fig. 7B), in agreement with their relative degree of regulations of actin cytoskeleton and brown adipogenesis. Furthermore, upon Tgf-β stimulation, phosphorylation of Smad3, the key Tgf-β signaling transducer, was markedly reduced in cells with SRF knockdown (Fig. 7C) as compared to controls. Total Smad2 protein was also significantly lower in shSrf cells, although its phosphorylation was not altered. In contrast, BMP signaling as shown by BRE-driven luciferase activity was substantially increased by either SRF (Fig. 7D) or MRTF silencing (Fig. 7E), as compared to respective scramble controls. In agreement with elevated luciferase activity, BMP-induced phosphorylation of its key signaling mediator, Smad1/5, were significantly higher in SRF (~105%, Fig. 7F) or MRTF knockdown cells (~54%, Fig. 7G) than that of the controls. Taken together, these data indicate that SRF/MRTF deficiency resulted in impaired Tgf-β but augmented BMP activity. The combined effects may collectively contribute to the enhanced thermogenic adipocyte development.
Figure 7. SRF/MRTF regulation of Tgf-β and BMP pathway signaling activity.
(A, B) Reduced Tgf-β signaling by Srf (A), or Mrtf1a/b silencing (B), as assessed by Tgf-β-responsive Smad-binding-element (SBE)-luciferase reporter assay (n=4/group). *P < 0.05, ** P < 0.01, *** P < 0.001. (C) Immunoblot analysis of Tgf-β-induced Smad2 and Smad3 phosphorylation (2ng/ml, 1hr) in shSrf-expressing 10T1/2 cells as compared to shScr control. *P < 0.05, ** P < 0.01. (D, E) Induction of BMP signaling in 10T1/2 cells with inhibition of Srf (D), or Mrtfa/b (E), as assessed by BMP-responsive luciferase reporter assay (n=4/group). (F, G) Immunoblot analysis of Smad1/5 phosphorylation upon BMP4 treatment (1ng/ml, 40min) in shSrf-expressing (F), or shMrtf-expressing (G), as compared to shScr 10T1/2 cells.
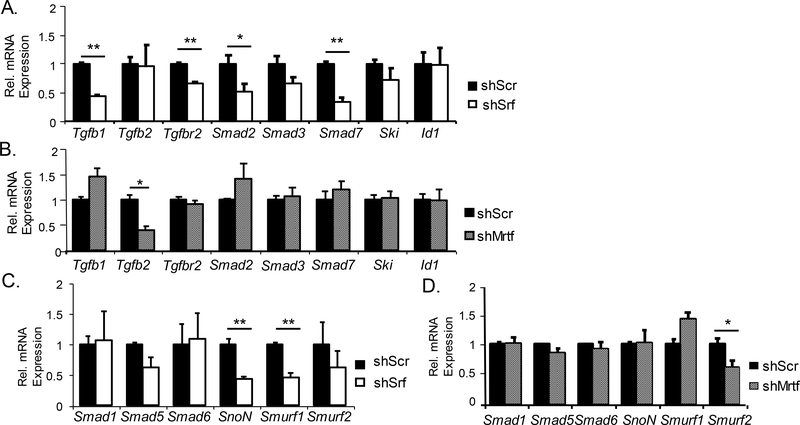
To determine whether MRTF-SRF modulates Tgf-β or BMP pathway through transcriptional regulation, we analyzed gene expression levels of key steps of the TGF-β and BMP signaling cascade. As shown in Fig. 8A in 10T1/2 cells with shSrf silencing, Tgfb1 ligand, Tgf-β receptor Tgfbr2, transcription mediator Smad2, and signaling modulator Smad7 were markedly down-regulated. In comparison, MRTF transcriptional regulation of the Tgf-β pathway was largely limited to Tgfb2 gene expression (Fig. 8B). On the other hand, major positive signal transducers of the BMP signaling pathway, R-Smads (Smad1 and 5) or Co-Smad (Smad6), were not altered by Srf knockdown, whereas the negative regulators, Smurf1 and SnoN, were significantly suppressed (Fig. 8C). Similar to MRTF limited effects on Tgf-β pathway, only Smurf2, another negative modulator of BMP signaling was down-regulated in shMrtf 10T1/2 cells (Fig. 8D). Thus, although precise transcriptional targets may differ, SRF and MRTF exert positive regulations of Tgf-β signaling genes but inhibits BMP gene pathway. Further analysis of SRF ChIP-Seq datasets obtained from 10T1/2 cells (GSE40369, GSM992343) identified SRF binding peaks within proximal promoter regions of Tgfb1 and Smurf2 (Suppl Fig. 1), indicating potential direct SRF transcriptional control of these signaling components in TGF-β and BMP cascades. These findings of SRF/MRTF positive regulation of the Tgf-β signaling components and negative control of the BMP pathway were in line with functional assays (Fig. 7).
Figure 8. SRF/MRTF regulation of Tgf-β and BMP signaling pathway components.
(A, B) RT-qPCR analysis of expression of genes involved in Tgf-β signaling pathway in shSrf-expressing (A) or shMrtf-expressing (B) 10T1/2 cells (n=3). (C, D) RT-qPCR analysis of expression of genes of the BMP signal transduction pathway in shSrf-expressing (C) or shMrtf-expressing (D) 10T1/2 cells (n=3). *P < 0.05, ** P < 0.01.
4. DISCUSSION
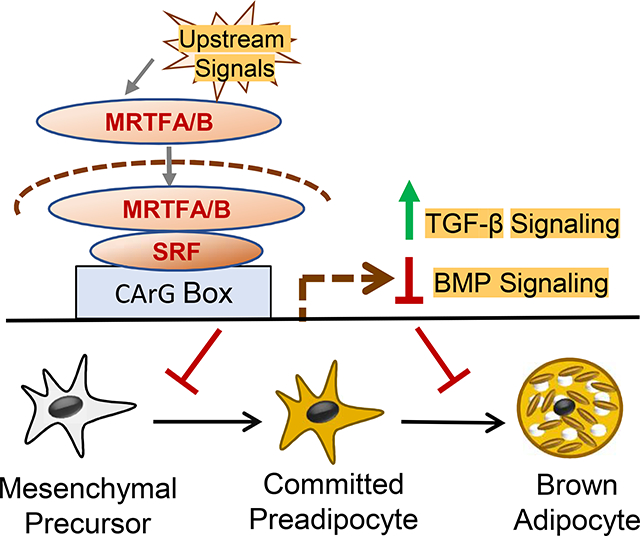
Utilizing multiple cellular models, our study established that the MRTF-SRF signaling axis suppresses distinct steps of brown fat cell lineage development, lineage determination of mesenchymal precursor and terminal differentiation. In addition, we provide new evidence that the MRTF/SRF pathway exerts transcriptional control of TGFβ-BMP signaling components, critical developmental signals driving brown/beige adipogenesis.
Cell shape and its upstream signaling events involving cytoskeleton remodeling play critical roles in cell fate decisions among mesenchymal stem cell lineages (McBeath, Pirone, Nelson et al., 2004). An intriguing finding from our study is that the activity of the MRTF-SRF signaling cascade decreases with brown adipocyte maturation, as revealed by down-regulation of protein expression, nuclear exclusion, and marked reduction of actin cytoskeletal indictive of MRTF transcriptional activity. SRF-MRTFs shuttled from predominantly nuclear localizations to cytoplasm upon differentiation. The changes in MRTF-SRF signaling is concomitant with the dissolution of actin cytoskeleton architecture in brown adipocyte progenitors, a major MRTF-SRF-controlled downstream gene transcription program. These observations are also in line with observations of the loss of actin architecture in 3T3-L1 adipogenic differentiation, a model of white adipocyte development, which precedes lipid accumulation and morphological transformation (Spiegelman and Ginty, 1983). Actin cytoskeleton and upstream RhoA-ROCK signaling has long been known to drive mesenchymal cell fate decisions between adipocyte and osteoblast (McBeath et al., 2004,Huang, Hu, Song et al., 2011). Interestingly, the suppression of MRTF-SRF during brown adipogenic differentiation was in contrast to the moderate high expression in adult brown fat, albeit at much lower than that of skeletal muscle and heart. As brown adipocytes arise from Myf5+ progenitors (Seale et al., 2008), the moderately high SRF expression in BAT may reflect its shared developmental origin with the myogenic lineage. In addition, distinct cell populations present in adult BAT may contribute to higher SRF expression observed. Most interestingly, MRTF-B was found to be highly enriched in both adipose depots examined, suggesting a potential dichotomy of SRF co-activators in adipose tissues. Notably, a recent study found that SRF binding sites were associated with active chromatin marks during early stages of adipogenesis, suggesting modulation of SRF signaling as a potential chromatin remodeling event that triggers adipogenic precursor lineage determination (Mikkelsen et al., 2010). As suggested by our current findings, MRTF-SRF may function as transcriptional mediators in thermogenic adipocyte lineage commitment involving epigenomic chromatin state regulation. Exploration of the developmental cues and signaling events driving MRTF-SRF transcriptional response in these processes may yield better understandings of the in vivo inductive events in thermogenic fat development. In vascular smooth muscle cells, cAMP-PKA signaling has been reported to inhibit SRF and MRTF (Blaker, Taylor and Mack, 2009,Davis, Hogarth, Fernandes et al., 2003,Smith, Hudson, Kimura et al., 2017). Within the context of brown adipogenic induction, as IBMX in the media raises cAMP, it may trigger the down-regulation of SRF/MRTF activity or expression through a related mechanism. Of note, microRNAs are important posttranscriptional mechanisms modulating SRF expression (Park, Hennig, Sanders et al., 2011,Miano and Long, 2015). Whether microRNA-mediated SRF inhibition may function in the brown adipogenic program warrants future investigations.
The TGF-β and BMP signaling pathways are key developmental signals influencing brown and beige adipocyte development (Nam, Yechoor and Ma, 2016,Zamani and Brown, 2011). TGF-β transduction cascade inhibits whereas the BMP signaling promotes brown or beige adipogenesis. Various BMP ligands, including BMP4, 6 and 7, induce brown fat development by functioning as a major cell fate commitment switch (Tseng et al., 2008,Townsend, Suzuki, Huang et al., 2012,Xue, Wan, Zhang et al., 2014,Sharma, Huard, Vernochet et al., 2014,Qian, Tang, Li et al., 2013). In contrast, Tgf-β-induced signal transduction strongly suppresses thermogenic adipocyte formation, with its inhibition promoting white fat browning (Tseng et al., 2008,Sassmann-Schweda, Singh, Tang et al., 2016,Yadav, Quijano, Kamaraju et al., 2011,Wankhade, Lee, Dagur et al., 2018). As the MRTF-SRF cascade selectively inhibits TGF-β signaling while augmenting BMP activity in mesenchymal precursors, these effects may synergistically drive mesenchymal lineage specification toward the thermogenic adipocyte fate. Screening of SRF ChIP-Seq dataset from 10T1/2 cells indeed identified binding peaks that align with active chromatin remodeling marks in proximal promoters of specific components of these pathways, suggesting the involvement of SRF in chromatin remodeling events in transcriptional control (Suppl. Fig. 1). Consistent with our current finding, MRTF-B was found to control TGF-β signaling and Tgfb2 gene transcription though a conserved SRF binding site (Li, Bowens, Cheng et al., 2012). Additional experimental screening of SRF/MRTF occupancy of transcriptional targets in these pathways in the future may uncover additional direct transcriptional targets involved. Intriguingly, TGF-β ligand is known to induce actin filament assembly and MRTF-A/B nuclear transport that augments SRF signaling (Crider, Risinger, Haaksma et al., 2011). In 10T/2 cells, BMP2 and 4, on the other hand, are capable of suppressing F-actin cytoskeleton formation to induce adipocyte lineage commitment (Huang et al., 2011). Collectively, these studies together with our findings suggest a potential regulatory feedforward loop between MRTF-SRF and TGF-β/BMP that may help to maintain a developmental drive for lineage specification and differentiation in thermogenic adipocytes.
A few studies to date examining MRTF or SRF modulation of thermogenic adipocytes are in agreement with our findings. However, the lineage stage-specific effects our study revealed have yet to be investigated. Using the mesenchymal stem cell model under azacytidine-induced lineage specification into muscle, adipogenic, osteogenic or chondrogenic cell types, we specifically tested whether the MRTF-SRF cascade influence thermogenic adipocyte lineage induction. Under this condition without specific adipogenic induction, loss of SRF or MRTF led to remarkably elevated C/EBP-β along with UCP1 indicative of thermogenic adipocyte lineage specification. Mikkelsen et al. identified SRF to be associated with early chromatin remodeling events in adipogenesis, suggesting its potential involvement in chromatin dynamics accompanying adipogenic commitment (Mikkelsen et al., 2010). A recent study by Rosenwald et al. revealed similar findings using isolated adipocyte progenitors (Rosenwald, Efthymiou, Opitz et al., 2017), although the underlying mechanisms investigated were distinct and lineage determination effects were not examined. Our study provides a systematic analysis of the MRTF-SRF signaling dynamic and function in brown adipocyte development. The current findings could be corroborated by in vivo validations through targeted genetic ablation or gain-of-function studies in brown or beige fat. Consistent with this notion, McDonald et al. found that the loss of MRTF-A induces browning of white adipocyte in mice, supporting the physiological relevance of MRTF-SRF cascade in modulating thermogenic fat development (McDonald, Li, Bian et al., 2015).
Since the discovery of functional thermogenic brown fat capable of dissipating energy in humans (Virtanen et al., 2009), its thermogenic capacity has become a desirable therapeutic target to counter over-nutrition-induced metabolic abnormalities including obesity and Type II diabetes (Stanford et al., 2013). It is possible that MRTF-SRF effects on brown adipogenesis could be targeted to combat metabolic disorders. Evidence has emerged that inhibiting the MRTF-SRF pathway may improve glucose homeostasis. Increased SRF transcriptional activity in skeletal muscle was found to be associated with insulin resistance, while pharmacological inhibition of SRF improved insulin sensitivity (Jin, Goldfine, Boes et al., 2011). As the findings from our study suggest, insulin-sensitizing effects of SRF pharmacological inhibition in vivo may involve recruitment of thermogenic adipocytes to promote energy expenditure. Targeting the suppressive effects of the MRTF-SRF pathway in brown lineage commitment and maturation may have potential applications in metabolic disease treatment or prevention.
Supplementary Material
Highlights.
SRF-MRTF pathway expression and activity are down-regulated during brown adipocyte maturation.
Silencing of SRF or MRTF-A/MRTF-B promotes mesenchymal stem cell commitment to the brown adipocyte lineage.
Inhibition of SRF or MRTF-A/MRTF-B enhances brown preadipocyte terminal differentiation.
Suppressing SRF/MRTF attenuates TGF-β signaling while augmenting BMP pathway activity.
ACKNOWLEDGEMENTS
We thank the Shared Resources Core Laboratory at the Beckman Research Institute of City of Hope for their expert technical support. This project was supported by grants from National Institute of Health 1R01DK112794 and American Heart Association 17GRNT33370012 to K.M; American Heart Association 19CDA34770034 to R.L, and National Institute of Health grant DK097160-01 to VY.
Non-standard Abbreviations
- SRF
serum response factor
- MRTF
Myocardin-related transcription factor
- TGF-β
transforming growth factor-β
- BMP
bone morphogenic protein
Footnotes
DECLARATION OF INTEREST
I certify that neither I nor my co-authors have a conflict of interest as described above that is relevant to the subject matter or materials included in this work.
Declaration of Interest: None
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- [1].Pipes GC, Creemers EE and Olson EN, 2006. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis, Genes Dev. 20, 1545–56. [DOI] [PubMed] [Google Scholar]
- [2].Posern G and Treisman R, 2006. Actin’ together: serum response factor, its cofactors and the link to signal transduction, Trends Cell Biol. 16, 588–96. [DOI] [PubMed] [Google Scholar]
- [3].Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A and Olson EN, 2002. Potentiation of serum response factor activity by a family of myocardin-related transcription factors, Proc Natl Acad Sci U S A. 99, 14855–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Miano JM, 2003. Serum response factor: toggling between disparate programs of gene expression, J Mol Cell Cardiol. 35, 577–93. [DOI] [PubMed] [Google Scholar]
- [5].Olson EN and Nordheim A, 2010. Linking actin dynamics and gene transcription to drive cellular motile functions, Nat Rev Mol Cell Biol. 11, 353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McBeath R, Pirone DM, Nelson CM, Bhadriraju K and Chen CS, 2004. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment, Dev Cell. 6, 483–95. [DOI] [PubMed] [Google Scholar]
- [7].Arsenian S, Weinhold B, Oelgeschlager M, Ruther U and Nordheim A, 1998. Serum response factor is essential for mesoderm formation during mouse embryogenesis, EMBO J. 17, 6289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li S, Czubryt MP, McAnally J, Bassel-Duby R, Richardson JA, Wiebel FF, Nordheim A and Olson EN, 2005. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice, Proc Natl Acad Sci U S A. 102, 1082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Parlakian A, Tuil D, Hamard G, Tavernier G, Hentzen D, Concordet JP, Paulin D, Li Z and Daegelen D, 2004. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality, Mol Cell Biol. 24, 5281–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Spiegelman BM and Ginty CA, 1983. Fibronectin modulation of cell shape and lipogenic gene expression in 3T3-adipocytes, Cell. 35, 657–66. [DOI] [PubMed] [Google Scholar]
- [11].Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES and Rosen ED, 2010. Comparative epigenomic analysis of murine and human adipogenesis, Cell. 143, 156–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu J, Jun H and McDermott JR, 2015. Formation and activation of thermogenic fat, Trends Genet. 31, 232–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sanchez-Gurmaches J, Hung CM and Guertin DA, 2016. Emerging Complexities in Adipocyte Origins and Identity, Trends Cell Biol. 26, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR and Spiegelman BM, 2008. PRDM16 controls a brown fat/skeletal muscle switch, Nature. 454, 961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nam D, Guo B, Chatterjee S, Chen MH, Nelson D, Yechoor VK and Ma K, 2015. The adipocyte clock controls brown adipogenesis through the TGF-beta and BMP signaling pathways, J Cell Sci. 128, 1835–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chatterjee S, Nam D, Guo B, Kim JM, Winnier GE, Lee J, Berdeaux R, Yechoor VK and Ma K, 2013. Brain and muscle Arnt-like 1 is a key regulator of myogenesis, J Cell Sci. 126, 2213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nam D, Chatterjee S, Yin H, Liu R, Lee J, Yechoor VK and Ma K, 2015. Novel Function of Rev-erbalpha in Promoting Brown Adipogenesis, Sci Rep. 5, 11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN and Kahn CR, 2008. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure, Nature. 454, 1000–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xie X, Qin J, Lin SH, Tsai SY and Tsai MJ, 2011. Nuclear receptor chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) modulates mesenchymal cell commitment and differentiation, Proc Natl Acad Sci U S A. 108, 14843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huang HY, Hu LL, Song TJ, Li X, He Q, Sun X, Li YM, Lu HJ, Yang PY and Tang QQ, 2011. Involvement of cytoskeleton-associated proteins in the commitment of C3H10T1/2 pluripotent stem cells to adipocyte lineage induced by BMP2/4, Mol Cell Proteomics. 10, M110 002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Blaker AL, Taylor JM and Mack CP, 2009. PKA-dependent phosphorylation of serum response factor inhibits smooth muscle-specific gene expression, Arterioscler Thromb Vasc Biol. 29, 2153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Davis A, Hogarth K, Fernandes D, Solway J, Niu J, Kolenko V, Browning D, Miano JM, Orlov SN and Dulin NO, 2003. Functional significance of protein kinase A activation by endothelin-1 and ATP: negative regulation of SRF-dependent gene expression by PKA, Cell Signal. 15, 597–604. [DOI] [PubMed] [Google Scholar]
- [23].Smith MC, Hudson CA, Kimura TE, White SJ, Sala-Newby GB, Newby AC and Bond M, 2017. Divergent Regulation of Actin Dynamics and Megakaryoblastic Leukemia-1 and −2 (Mkl1/2) by cAMP in Endothelial and Smooth Muscle Cells, Sci Rep. 7, 3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Park C, Hennig GW, Sanders KM, Cho JH, Hatton WJ, Redelman D, Park JK, Ward SM, Miano JM, Yan W and Ro S, 2011. Serum response factor-dependent MicroRNAs regulate gastrointestinal smooth muscle cell phenotypes, Gastroenterology. 141, 164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Miano JM and Long X, 2015. The short and long of noncoding sequences in the control of vascular cell phenotypes, Cell Mol Life Sci. 72, 3457–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nam D, Yechoor VK and Ma K, 2016. Molecular clock integration of brown adipose tissue formation and function, Adipocyte. 5, 243–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zamani N and Brown CW, 2011. Emerging roles for the transforming growth factor-{beta} superfamily in regulating adiposity and energy expenditure, Endocr Rev. 32, 387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Townsend KL, Suzuki R, Huang TL, Jing E, Schulz TJ, Lee K, Taniguchi CM, Espinoza DO, McDougall LE, Zhang H, He TC, Kokkotou E and Tseng YH, 2012. Bone morphogenetic protein 7 (BMP7) reverses obesity and regulates appetite through a central mTOR pathway, FASEB J. 26, 2187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xue R, Wan Y, Zhang S, Zhang Q, Ye H and Li Y, 2014. Role of bone morphogenetic protein 4 in the differentiation of brown fat-like adipocytes, Am J Physiol Endocrinol Metab. 306, E363–72. [DOI] [PubMed] [Google Scholar]
- [30].Sharma A, Huard C, Vernochet C, Ziemek D, Knowlton KM, Tyminski E, Paradis T, Zhang Y, Jones JE, von Schack D, Brown CT, Milos PM, Coyle AJ, Tremblay F and Martinez RV, 2014. Brown fat determination and development from muscle precursor cells by novel action of bone morphogenetic protein 6, PLoS One. 9, e92608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Qian SW, Tang Y, Li X, Liu Y, Zhang YY, Huang HY, Xue RD, Yu HY, Guo L, Gao HD, Liu Y, Sun X, Li YM, Jia WP and Tang QQ, 2013. BMP4-mediated brown fat-like changes in white adipose tissue alter glucose and energy homeostasis, Proc Natl Acad Sci U S A. 110, E798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sassmann-Schweda A, Singh P, Tang C, Wietelmann A, Wettschureck N and Offermanns S, 2016. Increased apoptosis and browning of TAK1-deficient adipocytes protects against obesity, JCI Insight. 1, e81175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yadav H, Quijano C, Kamaraju AK, Gavrilova O, Malek R, Chen W, Zerfas P, Zhigang D, Wright EC, Stuelten C, Sun P, Lonning S, Skarulis M, Sumner AE, Finkel T and Rane SG, 2011. Protection from obesity and diabetes by blockade of TGF-beta/Smad3 signaling, Cell Metab. 14, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wankhade UD, Lee JH, Dagur PK, Yadav H, Shen M, Chen W, Kulkarni AB, McCoy JP, Finkel T, Cypess AM and Rane SG, 2018. TGF-beta receptor 1 regulates progenitors that promote browning of white fat, Mol Metab. 16, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rosenwald M, Efthymiou V, Opitz L and Wolfrum C, 2017. SRF and MKL1 Independently Inhibit Brown Adipogenesis, PLoS One. 12, e0170643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].McDonald ME, Li C, Bian H, Smith BD, Layne MD and Farmer SR, 2015. Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes, Cell. 160, 105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jin W, Goldfine AB, Boes T, Henry RR, Ciaraldi TP, Kim EY, Emecan M, Fitzpatrick C, Sen A, Shah A, Mun E, Vokes V, Schroeder J, Tatro E, JimenezChillaron J and Patti ME, 2011. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance, J Clin Invest. 121, 918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.