Abstract
After T cell receptor (TCR) engagement, the CARD11-Bcl10-Malt1 (CBM) complex oligomerizes to transduce NF-κB activating signals. Bcl10 is then degraded to limit NF-κB activation. The cDNA AK057716 (BinCARD-1) was reported to encode a novel CARD protein that interacts with Bcl10 and modestly inhibits NF-κB activation. In a later study, a second isoform, BinCARD-2, was identified. Here, we report that the cDNA AK057716 (BinCARD-1) is an incompletely spliced derivative of the gene product of C9orf89, whereas CARD19 (BinCARD-2) represents the properly spliced isoform, with conservation across diverse species. Immunoblotting revealed expression of CARD19 in T cells, but no evidence of BinCARD-1 expression, and microscopy demonstrated that endogenous CARD19 localizes to mitochondria. Although we confirmed that both BinCARD-1 and CARD19 can inhibit NF-κB activation and promote Bcl10 degradation when transiently overexpressed in HEK293T cells, loss of endogenous CARD19 expression had little effect on Bcl10-dependent NF-κB activation, activation of Malt1 protease function, or Bcl10 degradation after TCR engagement in primary murine CD8 T cells. Together, these data indicate that the only detectable translated product of C9orf89 is the mitochondrial protein CARD19, which does not play a discernible role in TCR-dependent, Bcl10-mediated signal transduction to Malt1 or NF-κB.
Keywords: BinCARD, CARD19, CARD proteins, Bcl10, T cell signaling, T cell receptor, CBM complex, NF-κB
1.1. Introduction
Caspase activation and recruitment domains (CARDs) are conserved binding motifs that mediate protein-protein interactions; CARD-containing proteins play key roles in both innate and adaptive immunity, including the intermediate steps of cellular signaling cascades that occur downstream of T cell and B cell activation [1]. T cell receptor (TCR) and B cell receptor (BCR) engagement activates a cytosolic signaling cascade that promotes the association between the multidomain scaffold protein, CARD11, and a pre-existing complex of the small adaptor protein Bcl10 and the protease Malt1, thereby forming the “CBM” complex [2, 3]. Assembly of the CBM complex recruits E3 ubiquitin ligases that polyubiquinate IKKγ; when coupled with the phosphorylation of IKKβ, the terminal portion of the NF-κB signaling cascade is activated, resulting in the translocation of NF-κB heterodimers to the nucleus with concomitant transcriptional induction of target genes [4, 5]. NF-κB signaling is crucial for sustained T cell proliferation and effector T cell function. After NF-κB activation, Bcl10 is targeted for degradation to limit T cell responses [6-9]. However, the mechanism by which Bcl10 is degraded after CBM complex formation remains only partially understood, with both macroautophagy and a proteasomal mechanism implicated [10].
CARD19 is a CARD-containing protein which has been minimally studied to date. A CARD19 cDNA was identified via a bioinformatics search of EST databases. The protein product of the originally published cDNA (NCBI accession number AK057716) was named Bcl10-interacting CARD (BinCARD) after mammalian two-hybrid expression assays indicated that this protein interacts only with Bcl10, among the 26 tested CARD proteins [11]. Comparison of the sequence of cDNA AK057716 with the human genome sequence led to the identification of 4 exons. Alignments with other CARD proteins demonstrated that the N terminus of BinCARD contained sequence homology to CARD motifs of ced-3, caspase-1, caspase-2, caspase-4, caspase-5, and RAIDD [11]. Furthermore, the authors reported that BinCARD had a cytosolic and nuclear distribution and appeared to modestly inhibit NF-κB activation after Bcl10-BinCARD co-transfection in HEK293T cells and upon overexpression in PMA and ionomycin-stimulated of Jurkat T cells [11]. They additionally reported a reduction in the amount of phosphorylated Bcl10 HEK293T cells upon Bcl10-BinCARD co-transfection as compared to Bcl10 transfection alone [11].
A 2013 paper identified BinCARD-2, described as a second BinCARD isoform that was transcribed at significantly higher levels than BinCARD-1 (the protein product of AK057716) [12]. Ectopically expressed BinCARD-2 was shown to localize to the endoplasmic reticulum and the mitochondria in HEK293T cells [12]. X-ray crystallography of the CARD-containing N-terminus revealed an asymmetric CARD with some sequence identity to other CARD proteins as well as three fully oxidized cysteines, suggesting this protein may be sensitive to the local redox state [12]. Furthermore, a recent report presented evidence that siRNA silencing of BinCARD-2 reduced IFNβ transcription induced by overexpression of MAVS, TBK1, or RIG-I-like receptor (RLR) ligands in HEK293T cells, and siRNA knock-down of BinCARD-2 also reduced IFNβ and IL-6 transcription induced by vesicular stomatitis virus (VSV) infection in A549 cells [13]. These authors reported that BinCARD-2 interacted with MAVS, a CARD-containing protein localized to the outer mitochondrial membrane [13]. Based on these observations, the authors suggested that BinCARD-2 may be a positive regulator of IFNβ. The results reported in the above three studies represent the only CARD19/BinCARD-focused publications to date; there are no reports thus far assessing CARD19-mediated inhibition of Bcl10 signaling to NF-κB in primary cells. Importantly, there are also no studies definitively establishing the molecular weight(s) of the endogenous protein by immunoblotting or subcellular localization by microscopy or subcellular fractionation methods. Thus, there is almost no published information to date regarding basic characteristics of endogenous CARD19.
Here, we report that the cDNA AK057716 (BinCARD-1) was derived from the incompletely spliced mRNA of human gene C9orf89 , which consists of 6 exons. We also demonstrate that BinCARD-2 (henceforth referred to as CARD19) promotes Bcl10 degradation via proteolysis when overexpressed HEK293T cells. However, in primary CD8 T cells, the absence of CARD19 does not appreciably impact either NF-κB activation or degradation of endogenous Bcl10 after TCR engagement. Furthermore, we show that CARD19 is a mitochondrial protein in mouse primary CD8+ T cells, and that endogenous Bcl10 and CARD19 do not coexist in the same subcellular compartments before or after TCR engagement. Additionally, in both human and mouse T cells, by immunoblotting, we detect only a single protein species with a molecular weight consistent with CARD19/BinCARD-2, and not the larger BinCARD-1. Together, the above data demonstrate that the AK057716 cDNA represents an RNA intermediate of the fully mature CARD19 transcript, which encodes a putative protein product that is not produced in meaningful quantities, if at all, in the Jurkat human T cell line or in primary murine CD8 T cells. Furthermore, although the originally proposed function of BinCARD in limiting Bcl10-mediated NF-κB activation can be demonstrated when CARD19 is overexpressed in HEK293T cells, there is no evidence of an effect on NF-κB activation or Bcl10 degradation upon ex vivo antigen receptor stimulation of CARD19−/− CD8 T cells. Throughout this manuscript, we will refer to the originally reported BinCARD transcript as AK057716 (also called BinCARD-1) and the fully spliced, bona fide gene product as CARD19 (previously called BinCARD-2), in order to dissociate this novel protein from its spurious counterpart.
1.2. Materials and Methods
1.2.1. Identification and cloning of CARD19
The GenBank mouse EST database was searched via nucleotide BLAST, using the BinCARD ORF (accession number AK057716 [11]; nt 130-816) as the query sequence. Many EST clones were identified and found to have high homology to the BinCARD N-terminal CARD, but only sporadic homology downstream of the CARD. A murine EST, CB193298, which included a complete ORF, was used to design nested PCR primers to clone the ORF from primary T cell cDNA. The murine CARD19 ORF was also used to search the non-redundant GenBank nucleotide database, identifying the human homolog of CARD19 as the fully spliced product of gene C9orf89, and identifying highly conserved homologs in seven other vertebrate species. Clustal-W alignment of the AK057716 cDNA with the C9orf89 gene was used to locate exons and intronic regions within this EST clone, using Accelrys Gene 2.5 software (Accelrys, Inc.). Multispecies alignments were generated using Clustal-X2 [14]. Human CARD19 and the AK057716 (BinCARD) ORFs were PCR amplified from I.M.A.G.E. consortium EST clones with the appropriate splicing pattern.
1.2.2. PCR analysis of cDNA-
cDNA was generated by random hexamer priming of Jurkat total RNA using standard methodology. PCR amplification for agarose gel analysis was performed using primers for HPRT, Sense: 5’- CTGAACGTCTTGCTCGAGATGT-3’, Antisense: 5’-CTTGCGACCTTGACCATCTTTG-3’; C9orf89 exons 2-6, Sense: 5’-CCTCCAGCTGAACCGTTACTA-3’, Antisense: 5’- GCTGACATGTCTGTCGATGAT-3’; AK057716 splice pattern (exons 2-3+intron 3), Sense: (identical to C9orf89 sense), Antisense: 5’- GGGAGACAGAGACCAACATCT-3’. To determine transcript levels in Jurkat T cells, 5’ nuclease (TaqMan) real-time PCR methodology was used. 1×107 Jurkat T cells (clone E6-1, ATCC) were stimulated with 5 μg/ml anti CD3 (OKT3, eBioscience), 1 μg/ml anti CD28 (CD28.2, eBioscience), crosslinked with 10 μg/ml anti-mouse IgG (Jackson Immunoresearch) and incubated at 37°C for the times indicated. A small portion of the t=0 and t=30 min samples was analyzed by flow cytometry for IκBα degradation [2], to confirm robust activation of NF-κB. Total RNA was isolated and transcribed into cDNA using Superscript II (Invitrogen). Quantitative PCR was performed using an iQ5 Real Time PCR detection system (Bio-Rad) using 2× Real Time PCR PreMix with ROX (Syd Labs). Primer sets were designed using Primer Express 3.0 software (Applied Biosystems) and selected to cross at least one utilized splice site. Primer sets were C9orf89 sense: 5’-TTCTACCGAGCCCTGTATATCCA-3’ (exon 3), probe: 5’-CCAGCCGCCACGCTCTGCA-3’, antisense: 5’GAGTCTAGCTCTGTGCAATCTGAGTT-3’ (exon 4); AK057716 sense: 5’-TACCAACAAGGAGGCGGAAA-3’ (exon 2), probe: 5’-CGTGTGCGGCTCTGTGACCTCC-3’, antisense: 5’-CATGGTTGGTGATGTGGAACTT-3’ (intron 3); β2-microglobulin sense: 5’- CGTGTGCGGCTCTGTGACCTCC-3’, probe: 5’-TGCTCGCGCTACTCTCTCTTTCTGGC-3’, antisense: 5’-CGTGAGTAAACCTGAATCTTTGGA-3’. Probes were 5’-FAM labeled with ZEN+Iowa Black FQ double-quenching (IDT), to minimize background fluorescence. The threshold cycle number (Ct) corresponding to the point in which DNA amplification was detectable above background was used to determine quantitative values. The mRNA levels were normalized using β2-microglobulin Ct values. A ten-fold dilution series of known quantities of C9orf89 and AK057716 plasmids was analyzed and linear regression analysis of log starting copy number versus Ct value was performed to generate the following line equation: Ct = m(log copy number) + b. Reaction efficiency and absolute copy number was calculated using the following equations: E = 10 – (1/m) % Efficiency = (E-1) x 100% Quantity = 10((Ct – b)/m).
1.2.3. Cell Lines
HEK293T and Jurkat T cells were maintained as previously described [2, 15]. Transient transfection of HEK293T cells and luciferase assays were performed as described [15]. For transient overexpression, cDNAs were cloned into pcDNA3 using standard methods. In the indicated experiments, transfectants were treated with MG132 (Calbiochem) and Bafilomycin A1 (Axxora) for the final 24 hr prior to cell harvest.
1.2.4. Western blotting and immunoprecipitation
Transiently transfected HEK-293T cells were lysed and analyzed by western blotting as previously described [15]. The following antibodies were used: rabbit anti-HA (Y-11); rabbit anti-Bcl10 (H-197); mouse anti-GAPDH (6C5); mouse anti-A20 (A12); rabbit anti-TOMM20; mouse anti-β-actin (C-4); (all from Santa Cruz), rabbit anti-FLAG; mouse anti-FLAG (M2) (Sigma-Aldrich); rabbit anti-phospho-IκBα (Ser32) (14D4); rabbit anti- IκBα (Cell Signaling Technologies); rabbit anti-CARD19 (HPA010990) (Atlas Antibodies); mouse anti-MCPIP1 (anti-Regnase) (MAB7875) (R&D Systems); and mouse anti-α-tubulin (purified from DSHB hybridoma AA4.3 supernatant; AA4.3 Hybridoma deposited by Walsh, C). Immunoprecipitation of Bcl10-GFP protein complexes was performed with 1 μg/mL mouse anti-GFP clone 3E6 (MP Biomedicals), as previously described [15].
1.2.5. CARD19−/− mice and isolation and culture of CD8 T cells
The CARD19−/− strain was purchased from Lexicon Pharmaceuticals and extensively backcrossed to C57BL/6. In this strain, CARD19 was inactivated by replacement of exons 3-6 (encoding amino acids 51-183, encoding the C-terminal half of the CARD and the entire CARD19 C-terminus) with a Bgeo-puro cassette. Characterization of immune cell development and mature immune cell subsets by Lexicon Pharmaceuticals and our group has revealed no significant differences vs WT controls.
To isolate CD8 T cells, Lymph nodes and spleen were harvested from CARD19+/+ and CARD19−/− mice between 6 to 10 weeks of age. Organs were transferred to isolation buffer (2% FBS, 0.6% sodium citrate, 1X PBS), followed by dissociation through 70 μm mesh filters. Spleen samples were centrifuged in RBC Lysis buffer for 5 min and washed twice in isolation buffer. Lymph node and spleen samples were then consolidated and pipetted gently through a 40 μm filter to remove DNA released by lysed red blood cells. Lymphocytes were quantified and CD8 T cells were isolated using the CD8 T cell Dynabeads kit (Thermofisher) according to the manufacturer’s instructions. Isolated CD8 T cells were then seeded on anti-CD28 and anti-TCRβ or anti-CD3ε (1 μg /ml and 100 μg/ml, respectively) wells in a 24 well plate, at a density of 1.3×106 cells per well in Eagle’s Ham’s Amino Acids (EHAA) supplemented with 10% fetal bovine serum. After 48 h of stimulation, T cell blasts were harvested from the wells and cultured as described [16]. In indicated experiments, CARD19−/− CD8 T cells were harvested from wells after 48 hours of stimulation and subjected to retroviral transduction for 2 hours at 1200 × g at 32°C. Polybrene was added to the retroviral supernatant (final concentration 10 μg/ml) to increase the efficiency of the transduction. 24 h after transduction, the transduced cells were passaged in zeocin (50 μg/ml) for 5 days to select cells stably expressing 3×FLAG-CARD19. The Institutional Animal Care and Use Committee at Uniformed Services University (USU) approved all animal procedures.
1.2.6. TCR stimulation of mouse CD8 T cell blasts
CD8 T cell blasts (see above) were seeded in coated with either anti-CD3ε or anti-CD43 (non-stimulatory control) for the times indicated. T cells were cooled on ice, collected and centrifuged at 4°C, and pellets were resuspended in 1× Laemmli buffer. Whole cell lysates were boiled for 5 min and subjected to sonication to shear genomic DNA. Samples were quantified via Nanodrop One/One using the Pierce 660nm Protein Assay Reagent supplemented with IDCR. Equal amounts of total protein were loaded for each sample.
1.2.7. Microscopy
Primary CD8 T cells from WT or KO mice were isolated and cultured as described above. Cells were seeded for 20 min on glass coverslips coated with 100 μg/ml poly-D-lysine and 100 μg/ml anti-CD3ε or anti-CD43 for 20 min. Cells were fixed and prepared for microscopy as described in [16]. Mouse anti-ATP Synthase β (4.3E8.D10) (Thermo Fisher); rabbit anti-CARD19 (HPA010990) (Atlas Antibodies); mouse anti-FLAG (M2) (Sigma Aldrich); mouse-anti Mfn2 (XX-1); mouse anti-Bcl10 (331.3), mouse anti-Calnexin (E-10) and mouse anti-MAVS (E3) (all from Santa Cruz). Goat anti-rabbit (H+L) Alexa Fluor 647, Goat anti-mouse (IgG1) Alexa Fluor 488 and Goat anti-mouse (IgG2a) Alexa Fluor 488 (Thermo Fisher) were used at a 1:200 dilution. For confocal microscopy, coverslips were mounted in Fluoromount G and imaged using a Zeiss 710 2P Confocal Microscope or a Zeiss LSM 980 Confocal Microscope with a 63× oil objective. Images were processed using Zeiss Zen Blue software; representative insets of individual cells were selected and single representative z-planes were exported as TIFFs. For SIM, coverslips were mounted in a glycerol-based mounting medium and imaged using a Zeiss ELYRA PS.1 with a 63× oil objective. Images were processed and aligned with Zeiss Zen Black software. Representative insets of individual cells and single representative z-planes were exported as TIFFs. For insets, maximum intensity projections were generated from select z-plane ranges with ImageJ. Merged images were prepared using Adobe Photoshop.
1.2.8. Subcellular Fractionation
Cell fractions were prepared using a Qiagen Mitochondria isolation kit. CD8 T cell blasts (7.5×106) were untreated or stimulated with 100 μg/ml anti-CD3ε in one well of a 6-well plate per condition. Mitochondria were isolated according to the manufacturer’s protocol, and the cytosolic fraction and microsomal fractions were saved. The subcellular fractions were then diluted 1:1 in 2× Laemmli buffer, boiled, and sonicated. Total protein in all saved fractions was quantified via Pierce 660 nm Protein Assay Reagent supplemented with IDCR. Equal quantities of total protein from each subcellular fraction were analyzed via western blotting as described above.
1.3. Results
1.3.1. The BinCARD/AK057716 cDNA has an intron-encoded (spurious) C-terminus.
Data published in a 2004 paper [11] identified BinCARD, the 228 amino acid (aa) protein product of human EST AK057716. The BinCARD ORF includes an N-terminal CARD (aa 8-97) that binds to Bcl10, and a C-terminal domain with no identified functional motifs. Overexpression of the BinCARD ORF was reported to reduce TCR-induced phosphorylation of Bcl10 and to modestly inhibit Bcl10-mediated activation of NF-κB by an unknown mechanism. A later publication [12] identified a distinct spliced product of C9orf89 that the authors called BinCARD-2, with the initially identified isoform referred to as BinCARD-1. As stated above, we will refer to these variants as AK057716 and CARD19. While AK057716 was reported to have a cytosolic and nuclear distribution, the CARD19 ORF encoded a predicted transmembrane domain in the C-terminal portion of the protein, and ectopically expressed CARD19 was reported to localize to mitochondria [12]. To determine if the findings with human AK057716 could be generalized to murine cells, we performed a bioinformatics search to identify a murine homolog. BLAST screening of the murine EST database, using the AK057716 open reading frame (ORF) as the query sequence, identified many murine cDNAs encoding a 183 aa ORF with high amino acid conservation in the N-terminal (95%), but only sporadic (14%) amino acid conservation in the C-terminus (Fig. 1A). BLAST was then used to query the human EST database using the murine 183 aa ORF. Interestingly, many human EST clones were identified which had high homology to the murine 183 aa ORF through the entire sequence (90% identical). We found that these ESTs were fully homologous to an NCBI gene referred to as C9orf89 (NCBI Gene ID:84270; nucleotide reference sequence NM_032310). Moreover, cDNAs highly homologous to the C9orf89 transcript were identified in eight additional vertebrate species (Fig. 1B).
Figure 1. The C-terminus of BinCARD is not conserved across species.
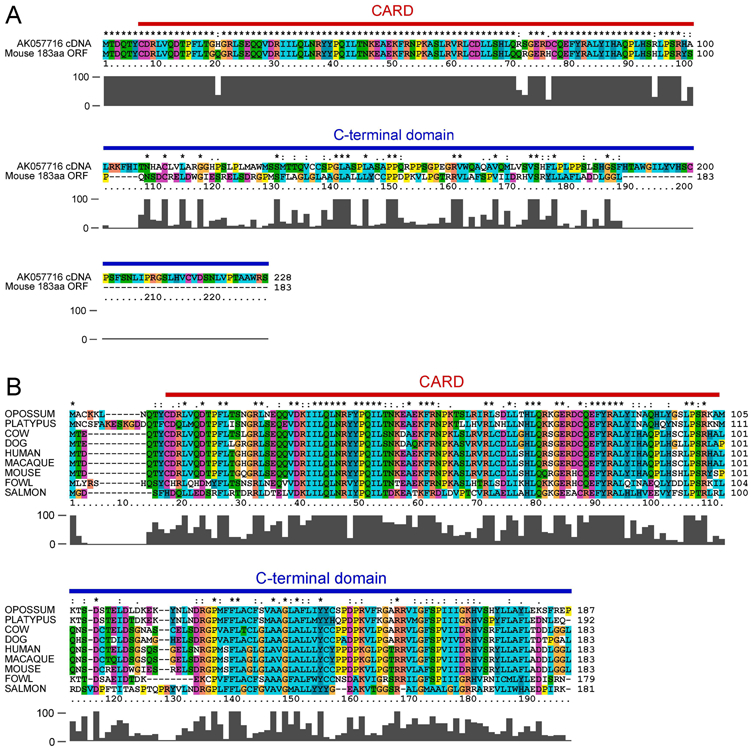
A, Clustal-X2 alignment of the translated product of the AK057716 cDNA (BinCARD-1), and a mouse homolog (CARD19, also known as BinCARD-2) identified via nucleotide BLAST shows a high degree of amino acid identity in the N-terminal 98 amino acids, but only sporadic identity in the C-terminal 130 amino acids. B, Clustal-X2 alignment of the CARD19 gene product with homologous proteins from eight other species.
Alignment of the sequence of the AK057716 cDNA with the human genomic C9orf89 nucleotide sequence demonstrated that AK057716 is a cDNA copy of an incompletely spliced mRNA, in which introns 3 and 4 were not spliced out of the mRNA. In this partially processed transcript, the CARD is encoded by the fully-spliced exons 1-3, but the C-terminus is encoded within intron 3 (Fig. 2A). Alignment of the exon 3-intron 3 genomic region from human C9orf89 and homologs identified in three additional vertebrate species demonstrated that the in-frame translatable sequence in the mouse, opossum, and cow introns is considerably shorter than the human sequence. Furthermore, there is no appreciable homology beyond the two amino acids closest to the splice junction (Fig. 2B). To further investigate conservation of the intron 3 ORF representing the C-terminus of AK057716, we aligned the sequences of 15 distinct primate orthologs of CARD19 (C9orf89), from six different primate orders (Fig S1A). This alignment showed high conservation (>85% amino acids were 100% conserved across these 15 species). However, aside from gorillas, there was no appreciable conservation of the intron 3 ORF (i.e., the AK057716 C-terminus), with analysis demonstrating considerable heterogeneity in both the length of this sequence and the amino acids encoded at each position (Fig S1B). Indeed, even when aligning sequences from only the closest primate relatives to humans (order Hominidae), only the first 60 of 127 (47%) amino acids of the AK057716 C-terminus are well conserved in this order (Fig S1C), in contrast to 181 of 183 (99%) amino acids completely conserved in CARD19, when comparing these same species (Fig S1A). Thus, the intron 3 translation is not conserved between species, including species within the same order of primates as humans. Collectively, the bioinformatics data in Figs. 1, 2B and S1 show that CARD19 is very well conserved across species, but that the intronic ORF representing the reported C-terminus of AK057716 is poorly conserved, even among the closest primate relatives to humans.
Figure 2. The AK057716 cDNA clone is the product of an incompletely spliced CARD19 mRNA.
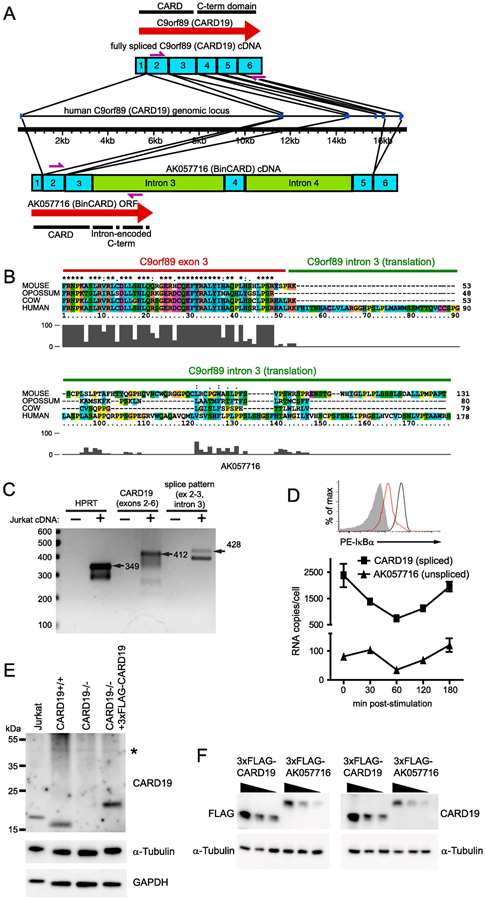
A, Diagram comparing the structures of the fully spliced CARD19 transcript and the partially spliced AK057716 transcript. The human CARD19 (c9orf89) gene structure is indicated in the middle of the panel, with transcript structures shown above and below the gene. Exons are designated by numbered blue boxes and unspliced introns are green. Locations of ORFs are designated by red arrows and primers used for PCR in C are indicated by purple arrows. B, Translated sequences from the intron 3/exon 3 boundary region of the human CARD19 gene and homologues from mouse, opossum, and cow. Clustal-X2 alignment shows an intronic translation of variable length with almost no homology beyond the splice junction. C, cDNA from the Jurkat T cell line was amplified by PCR to detect HPRT, exons 2-6 (fully spliced) of CARD19, and the exon 2-exon 3-intron 3 splicing pattern of the AK057716 clone (see A for locations of primers for CARD19 and AK057716). Expected products of the indicated lengths were detected with each of the primer sets. D, Jurkat T cells were stimulated with anti-CD3 plus anti-CD28 and RNA was harvested at the indicated times post-stimulation. Cells from the 30 min time point were analyzed by intracellular flow cytometry to confirm efficient degradation of IκBα at 30 min post-stimulation, indicative of efficient activation of the TCR (top). Real-time PCR analysis was performed on each sample using primers to specifically detect the CARD19 splicing pattern (exon 3-exon 4) and the AK057716 splicing pattern (exon 2-exon 3-intron 3), and signals were compared to plasmid standard curves to determine per cell copy number at each post-stimulation time point (bottom). E, anti-CARD19 was used in an immunoblotting experiment to detect endogenous CARD19, α-tubulin, and GAPDH in Jurkat T-cells, murine CARD19+/+ and CARD19−/− CD8 T cells, and ectopic 3×FLAG-CARD19 in retrovirally-transduced CARD19−/− CD8 T cells. F, HEK293T cells were transfected with decreasing concentrations of 3xFLAG-CARD19 or 3×FLAG-AK057716 plasmid. Lysates were separated via SDS-PAGE and probed with either anti-FLAG (left) or anti-CARD19 (right). Both sets of lysates were probed with anti-α-tubulin.
To demonstrate that the fully spliced CARD19 mRNA is present in human T cells, total RNA was isolated from Jurkat T cells and reverse transcribed using random hexamer primers. PCR was used to amplify exons 2-6 of the fully spliced CARD19 cDNA and the partially spliced exons 2- 3/intron 3 transcript (characteristic of the AK057716 cDNA). As shown in Figure 2B, both the fully spliced and partially spliced forms of the C9orf89 transcript are present in total RNA of Jurkat T cells (Fig. 2C). This result was not surprising, since various splicing intermediates should be present at detectable levels in total RNA. The 412 bp PCR product (fully-spliced C9orf89 exons 2-6) was also sequenced, confirming the exon 2-3-4-5-6 spliced structure (data not shown). Additionally, we determined the abundance of the exon 3-exon 4 spliced transcripts (representative of the CARD19 mRNA) and the exon 3-intron 3 unspliced transcripts (representative of the AK057716 cDNA) via TaqMan real-time PCR. Total RNA was harvested from an anti-CD3/anti-CD28 stimulation time course in Jurkat T cells, to assess whether either transcript shows induction or repression in response to stimulation. As shown in Figure 2D (top), FACS analysis showed efficient IκBα degradation at 20 min post-stimulation, verifying effective activation [2]. Importantly, the exon 3/exon 4 spliced (CARD19) transcripts were consistently 20-fold more abundant than the exon 3/intron 3 unspliced (AK057716) messages. Thus, the C9orf89 spliced message is much more abundant than the partially spliced transcripts representative of the AK057716 EST clone, and transcription of C9orf89 is transiently reduced in response to anti-CD3/anti-CD28.
We then determined the relative protein expression of the translated products of AK057716 and CARD19 via western blot, using a polyclonal antibody raised against a human CARD19 fragment containing the complete CARD (Atlas Antibodies HPA010990). The predicted molecular weight of the translated product of AK057716 is 25.6 kDa, whereas the predicted molecular weight of the fully spliced CARD19 gene product is 20.6 kDa. Jurkat T cells and murine primary CARD19+/+ CD8 T cells expressed a protein product of approximately 20 kDa when probed with anti-CARD19 antibody. Murine primary CARD19−/− CD8 T cells transduced with 3×FLAG-CARD19 expressed a protein of approximately 24 kDa, consistent with the predicted increase in molecular weight via addition of the 3×FLAG epitope tag (Fig. 2E). In all cases, only a single CARD19 isoform was detected and no protein with a molecular weight of 25.6 kDa was evident (a faint 50 kDa band in the murine T cell lysates is non-specific). To ensure that the rabbit anti-CARD19 antibody is equally capable of recognizing CARD19 and the reported AK057716 ORF, we transfected HEK293T cells with 3×FLAG-CARD19 and 3×FLAG-AK057716, using three dilutions of each plasmid to enable accurate assessment of antibody detection efficacy. Duplicate sets of lysates were separated by SDS-PAGE, with one set probed with anti-FLAG and the other set probed with anti-CARD19. The band intensity pattern upon probing with each antibody was highly similar (Fig. 2F), demonstrating that the rabbit CARD19 antibody detects CARD19 and the AK057716 translated product with equivalent efficiency. Thus, the failure to detect the AK057716 ORF in Fig. 2D clearly correlates with an absence of detectable AK057716 protein in T cells.
Based on the aggregate data from Figures 1-2, we conclude that the AK057716 cDNA is the product of an incompletely spliced mRNA intermediate from the C9orf89 gene. Because the translated (intronic) C-terminus of the AK057716 cDNA is not conserved between species, the AK057716 transcript pattern is present within cells at very low levels, and a protein corresponding to the predicted molecular weight of only the fully-spliced transcript is apparent, we furthermore conclude that the previously identified AK057716 ORF is almost certainly spurious and unlikely to be translated at biologically meaningful levels.
1.3.2. Ectopic 3×FLAG-CARD19 enhances Bcl10 degradation and NF-κB activation in HEK293T cells
To assess the effect of the C9orf89 (CARD19) protein on Bcl10 activity in vitro, we co-transfected HEK293T cells with an NF-κB reporter vector, 3×HA-Bcl10, and either 3×FLAG-CARD19, or 3×FLAG-AK057716. We observed dose-dependent repression of Bcl10-stimulated NF-κB activation by both 3×FLAG-CARD19 and 3×FLAG-AK057716, but the potency of 3×FLAG-CARD19 was greater at the two highest plasmid doses (Fig. 3A-B). Additionally, we probed the lysates from these same cells with anti-FLAG to detect the CARD19 and AK057716 translated products, and with anti-HA to detect Bcl10; we then quantified the relative expression of the translated products and normalized these values to GAPDH. These data showed that as CARD19 protein levels increase, there is a corresponding decrease in levels of Bcl10 protein (Fig 3C-D); the reduction in Bcl10 levels is less robust upon expression of the AK057716 ORF (Fig. 3C-D). Notably, whereas the predicted protein species of approximately 24 kDa was detected for 3×FLAG-CARD19, the anti-FLAG antibody detected 50 kDa and 20 kDa forms of 3×FLAG-AK057716 in addition to the predicted 29 kDa product (Fig 3C). Furthermore, expression of CARD19 and AK057716 increased with increasing plasmid concentration, with peak expression observed at the 200 ng dose of plasmid. Increases of plasmid amount above 200 ng did not correspond to an increase in either AK057716 or CARD19 expression (Fig. 3C-D). Interestingly, despite equal amounts of plasmid used for transfection, protein expression of 3×FLAG-CARD19 was always less than that of 3×FLAG-AK057716 (Fig. 3C-D). Finally, we transfected HEK293T cells with a constant amount of the irrelevant protein, GFP, and increasing amounts of 3×FLAG-CARD19. We then probed whole cell lysates for GFP and FLAG. These data show that CARD19 expression only reduces production of GFP at the highest transfected dose of CARD19 (Fig. 3E). Together, these data show that the CARD19 protein product is a more potent inhibitor of Bcl10-dependent NF-κB activation than the translated AK057716 product. Moreover, NF-κB inhibition by expression of the 3×FLAG-CARD19 product correlates with a dose-dependent reduction in Bcl10 protein levels, and this reduction in protein levels is specific, except at the highest transfected dose, where a significant non-specific effect may be a component of reduced Bcl10 expression.
Figure 3. CARD19 inhibits NF-κB activation and reduces Bcl10 protein levels when overexpressed in HEK293T cells.
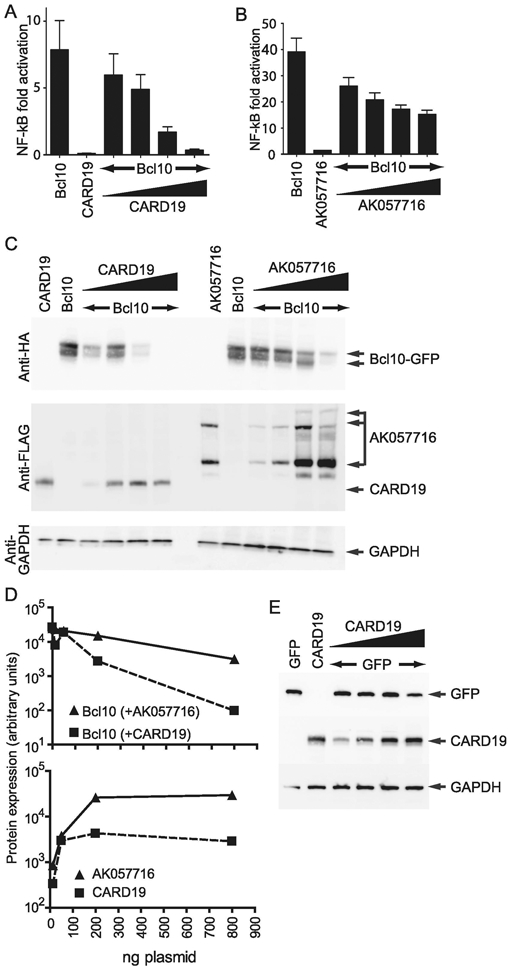
A, An NF-κB luciferase assay was performed in HEK293T cells by transfecting expression vectors for 3×HA-Bcl10-GFP (alone), 3×FLAG-CARD19 (alone), or constant amounts of 3×HA-Bcl10-GFP plus increasing amounts of 3×FLAG-CARD19. B, The experiment is the same as that in A, except cells were transfected with 3×FLAG-AK057716 rather than 3×FLAG-CARD19. C, Whole cell lysates from transfections in A and B were analyzed by immunoblotting, using the indicated antibodies. D, Protein levels of 3×HA-Bcl10-GFP and 3×FLAG-CARD19 or 3×FLAG-AK057716 from C were quantified and normalized to GAPDH expression. E, HEK293T were transfected with expression vectors for GFP (alone), 3×FLAG-CARD19 (alone), or constant amounts of GFP plus increasing amounts of 3×FLAG-CARD19. Whole cell lysates were analyzed by immunoblotting, using antibodies against GFP, FLAG (for detection of CARD19) and GAPDH.
1.3.3. Reduction of Bcl10 protein levels and NF-κB inhibition by ectopically expressed CARD19 requires the C-terminal domain.
AK057716 and CARD19 are identical through the CARD but have divergent C-termini (Figs. 1-2). Based on the more potent NF-κB-inhibitory activity of CARD19 vs. AK057716 (Fig. 3), we hypothesized that the repression of NF-κB activity and the reduction of Bcl10 protein levels are properties of the unique C-terminus of CARD19. We therefore made four progressive deletions of the CARD19 C-terminus: Δ155, Δ146, Δ130, and Δ105. Δ155 removes most of the C-terminal domain downstream of the predicted transmembrane region, and Δ146 deletes the entire C-terminal domain; Δ130 removes the entire distal C-terminus and half of the transmembrane domain, and Δ105 removes the entire transmembrane domain and distal C-terminus. We also made a point mutant, G73R, in a glycine in the helix 4-5 turn [12] which is conserved in CARD19 across diverse species (Fig. 1B). Moreover, this residue is conserved in many CARD proteins because it is an important determinant of the helix-turn-helix structure of the CARD. Mutation of this conserved CARD glycine to arginine in both CRADD and CASP2 (G65R and G78R, respectively) has been shown to disrupt CARD-CARD interactions, most likely by interfering with the normal helix-turn-helix CARD confirmation [17]. Thus, CARD19-G73R is predicted to be deficient in CARD-CARD interactions. A diagram indicating the position of the above mutants in the CARD19 protein is shown in Fig. 4A.
Figure 4. The CARD19 C-terminus and a properly folded CARD are essential for Bcl10 degradation in HEK293T cells.
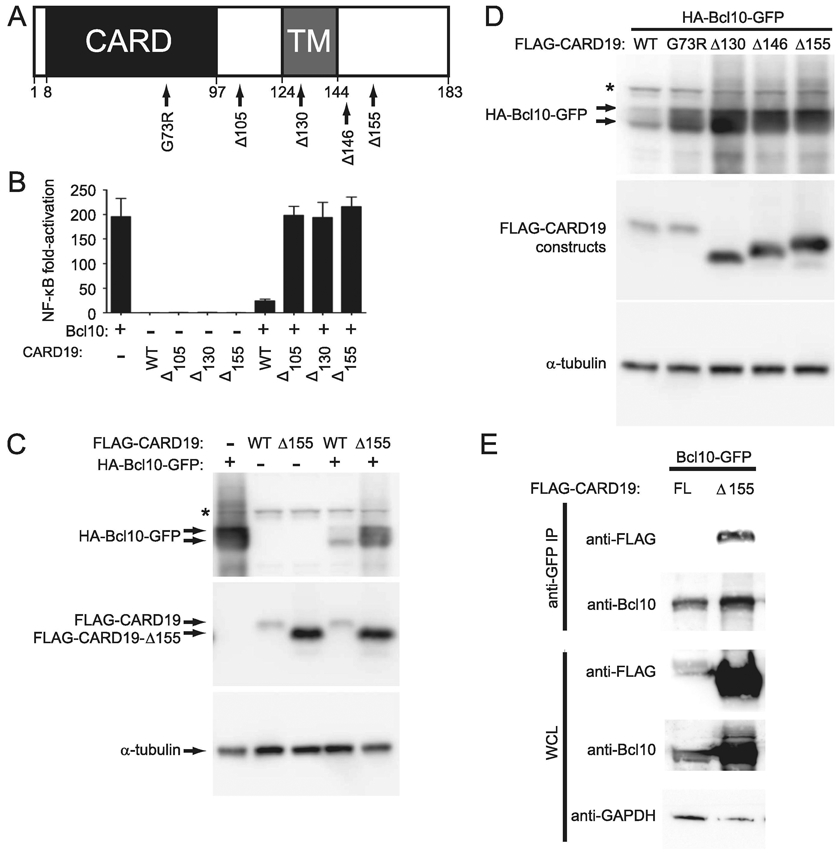
A, Graphical representation of the G73R point mutation and progressive CARD19 deletions Δ105, Δ130, Δ146 and Δ155. The locations of the CARD and the predicted transmembrane domain (TM) are indicated. B, HEK293T cells were transfected with the indicated combinations of 3×FLAG-CARD19 and 3×HA-Bcl10-GFP expression vectors and NF-κB activity was assessed via luciferase assay. C and D, HEK293T cells were transfected with 3×HA-Bcl10-GFP and the indicated 3×FLAG-CARD19 constructs. 3×FLAG-CARD19 and 3×HA-Bcl10-GFP proteins were detected by anti-FLAG and anti-HA western blotting, respectively. E, Requirements for physical interaction between Bcl10 and CARD19 were assessed by co-immunoprecipitation of proteins from transfected HEK-293T cells. Bcl10-GFP was immunoprecipitated with an anti-GFP monoclonal antibody and detected with a polyclonal anti-Bcl10 antibody. Co-precipitated 3×FLAG-CARD19 or 3×FLAG-CARD19-Δ155 was detected via anti-FLAG. Whole cell lysates were probed with anti-FLAG (to detect CARD19 constructs), anti-Bcl10, and anti-GAPDH.
We first compared WT CARD19 to three deletion mutants, Δ105, Δ130, and Δ155, for their ability to suppress Bcl10-mediated activation of NF-κB (Fig. 4B). While WT 3xFLAG-CARD19 robustly inhibited NF-κB activation by Bcl10, the three 3×FLAG-CARD19 deletion mutants showed no ability to repress Bcl10 dependent activation of NF-κB (Fig. 4B). We then compared the effect of WT or Δ155 3×FLAG-CARD19 on levels of co-expressed Bcl10-GFP protein. Whereas levels of Bcl10-GFP protein were highly reduced by co-expression of 3×FLAG-CARD19-WT, levels of Bcl10-GFP were reduced to a lesser extent by co-transfection of the 3×FLAG-CARD19-Δ155 (Fig. 4C). We next compared co-expression of 3xHA-Bcl10-GFP with 3×FLAG-CARD19-WT, the Δ130, Δ146, and Δ155 C-terminal mutants and the G73R point mutant. Notably, 3×HA-Bcl10-GFP was more highly expressed in combination with each C-terminal deletion and the G78R point mutation, as compared to co-expression with 3×FLAG-CARD19-WT (Fig. 4D). Thus, both C-terminal sequences downstream of residue 155 and a properly folded CARD19 CARD are required for efficient CARD19-mediated reduction in levels of overexpressed 3×HA-Bcl10-GFP. Notably, deletion of CARD19 C-terminal sequences also resulted in higher levels of CARD19 expression.
To rule out the possibility that a failure to bind to Bcl10-GFP may underlie the inability of CARD19 C-terminal mutants to efficiently reduce Bcl10-GFP protein levels, we performed an immunoprecipitation experiment. We transfected HEK293T cells with 3×HA-Bcl10-GFP plus either WT 3×FLAG-CARD19 or Δ155 3×FLAG-CARD19. Although the association between full-length 3×FLAG-CARD19 and 3×HA-Bcl10 could not be detected in this assay (presumably due to low steady-state levels of the 3×FLAG-CARD19 protein), we detected robust association between 3×FLAG-CARD19-Δ155 and 3×HA-Bcl10-GFP (Fig. 4E). Thus, deletion of the distal C-terminus (aa 156-183) does not prevent physical association between CARD19 and Bcl10. Collectively, the data in Figure 4 show that 3×FLAG-CARD19-WT reduces both Bcl10-GFP-mediated NF-κB activation and levels of overexpressed Bcl10-GFP protein. A point mutant predicted to interfere with proper folding of the CARD19 CARD or deletions that remove CARD19 C-terminal regions (minimally including residues 156-183) greatly diminish the ability of CARD19 to reduce levels of overexpressed Bcl10-GFP. However, our data show that deletion of aa 156-183 does not prevent physical association between 3×FLAG-CARD19 and Bcl10-GFP, suggesting that this region of CARD19 plays a mechanistic role in Bcl10-GFP codegradation.
1.3.4. Ectopically expressed CARD19 reduces Bcl10 protein levels primarily via proteasome-dependent degradation of CARD19-Bcl10 complexes.
We hypothesized that association between overexpressed CARD19 and Bcl10 results in targeting of Bcl10 for degradation via a proteasomal or lysosomal mechanism in HEK293T cells. We therefore performed Bcl10 and CARD19 co-transfection experiments in which cells were incubated with either MG132 to inhibit proteasomal degradation or bafilomycin A1(BafA1) to inhibit lysosomal degradation. Constant levels of Bcl10-GFP were co-transfected with increasing amounts of 3×FLAG-CARD19, and each transfection condition received either no treatment or treatment with either MG132 or BafA1. Treatment with MG132 effected increased levels of 3×FLAG-CARD19 protein at all doses of transfected 3×FLAG-CARD19 expression vector relative to untreated controls, whereas MG132-dependent increases in Bcl10-GFP protein levels were detected at all but the highest dose of transfected CARD19 expression vector (Fig. 5A). Very little 3×FLAG-CARD19 protein expression was detected in samples not treated with MG132, even at very high doses of transfected 3×FLAG-CARD19, suggesting that transfected, WT 3×FLAG-CARD19 is rapidly turned over, consistent with data shown in Figure 4. BafA1 treatment had a minimal effect on 3×FLAG-CARD19 and Bcl10-GFP protein levels (Fig. 5B). Together, these data suggest that CARD19 reduces Bcl10 levels via co-degradation of CARD19-Bcl10 complexes. Furthermore, inhibition of proteasomal degradation is more effective at blocking 3×FLAG-CARD19 and Bcl10-GFP degradation than inhibition of lysosomal degradation, suggesting that CARD19-Bcl10 codegradation occurs primarily via a proteasomal mechanism in heterologous transfection-overexpression experiments.
Figure 5. CARD19 promotes degradation of Bcl10 in heterologous overexpression experiments primarily via a proteasomal mechanism.
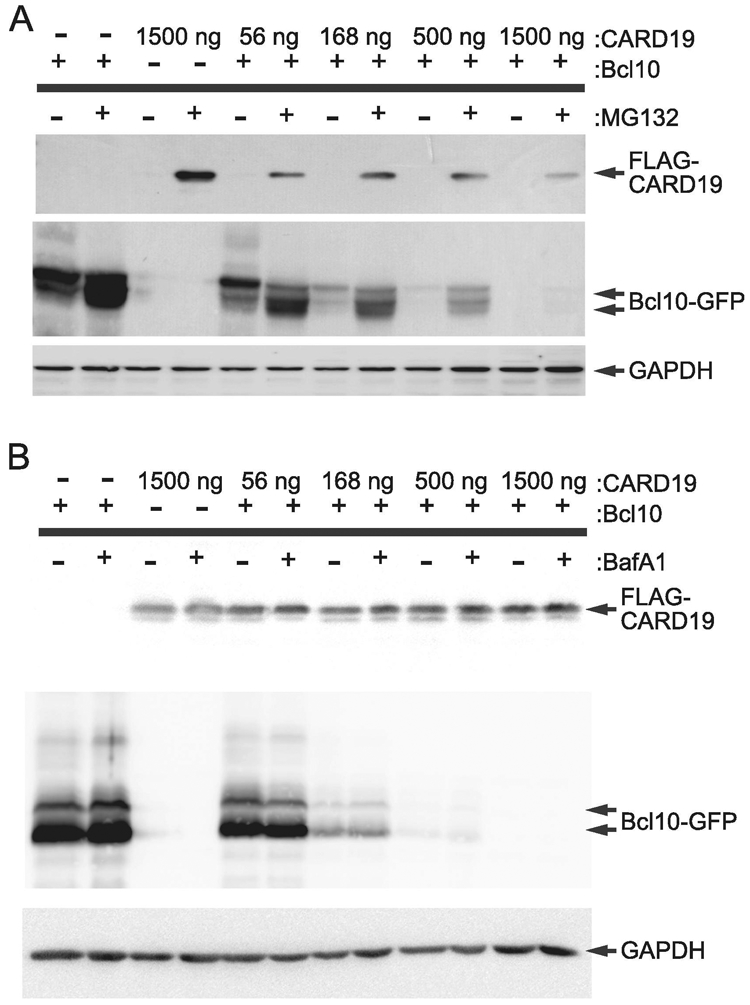
A, HEK293T cells were transfected with the indicated amounts of 3×FLAG-CARD19 expression vector, and with 1500 ng (+) or 0 ng (−) of 3×HA-Bcl10-GFP expression vector. Cells were treated with 25 μM MG132 (+) or vehicle only (−) for 24 hr prior to harvest. Whole cell lysates were probed with anti-FLAG or anti-HA. B, Experiment is as in A, except cells were treated with 25 nM bafilomycin A1 (+) or vehicle only (−).
1.3.5. Absence of CARD19 does not affect NF-κB activation in murine primary CD8 T cells
Collectively, the data in Figures 4-5 show that ectopically expressed FLAG-tagged CARD19 inhibits NF-κB by associating with Bcl10 and promoting proteasomal degradation of Bcl10. To determine whether or not endogenously expressed murine CARD19 promotes Bcl10 degradation and TCR-dependent NF-κB activation, we obtained a mouse model with germline inactivation of the CARD19 gene (see Materials and Methods). We then produced CARD19−/− and CARD19+/+ CD8 T-cell blasts, which were stimulated with anti-CD3ε over a 120 minute time course. Immunoblotting of whole cell lysates from these cells demonstrated that Bcl10 was readily degraded in both cell types after TCR stimulation (Fig 6A). Although there were minor differences in Bcl10 degradation over time, by 120 min post-stimulation, both populations of CD8 T cell blasts exhibited similar reductions in Bcl10 (Fig. 6B). To assess NF-κB activation, we measured IκBα degradation, resynthesis, and phosphorylation via western blot. Surprisingly, each of these CD8 T cell populations exhibited very similar NF-κB activation kinetics (Fig 6A-B). Furthermore, A20, another protein induced as a consequence of NF-κB activation, exhibited similar increases in expression at 120 min post-activation in CARD19+/+ and CARD19−/− CD8 T cells. Additionally, Regnase, a substrate of Malt1 paracaspase cleavage dependent on Bcl10, was cleaved to similar degrees in CARD19+/+ and CARD19−/− CD8 T cells. Combined with the similar degree of IκBα resynthesis and phosphorylation across genotypes, these data demonstrate that there was similar Bcl10-dependent activation of NF-κB and the protease function of Malt1 regardless of whether or not CARD19 was expressed (Fig. 6A-B).
Figure 6. Loss of endogenous CARD19 does not affect NF-κB activation or decrease Bcl10 degradation in primary murine CD8 T cells.
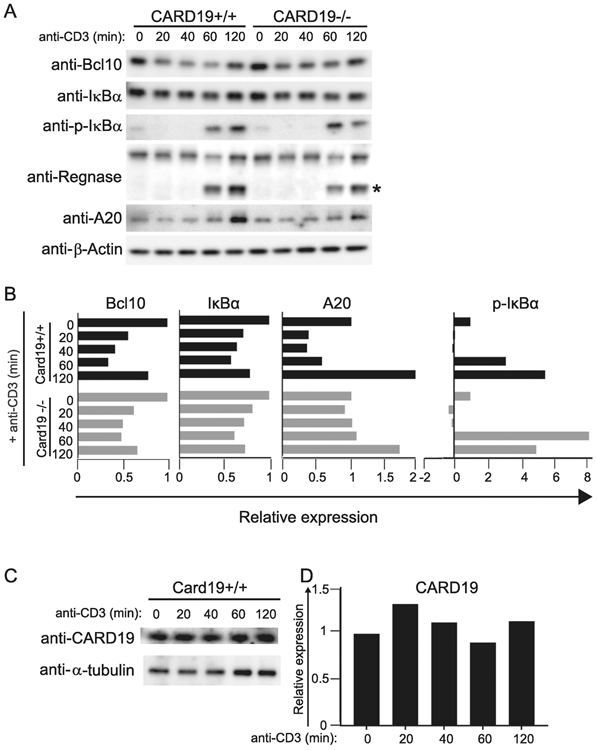
A, CARD19+/+ and CARD19−/− CD8 T cells were stimulated with anti-CD43 (t=0) or anti-CD3ε for the indicated times (t=20-120) and whole cell lysates were analyzed via western blot for expression of Bcl10, IκBα, p-IκBα, Regnase, A20, and β-actin. * indicates the Malt1-cleaved form of Regnase. B, Band intensities of Bcl10, IκBα, A20 and p-IκBα were quantified; samples were expressed as intensities relative to the untreated sample in each sample group, following normalization to β-actin. C, Whole cell lysates from CARD19+/+ CD8 T cells were analyzed via western blot for expression of CARD19. D, Band intensities of α-tubulin and CARD19 were quantified; intensities were normalized to α-tubulin expressed on a relative scale, with the untreated sample defined as 1.0.
Additionally, we examined CARD19 expression in CARD19+/+ CD8 T cell lysates and observed no indication of degradation in response to anti-CD3ε stimulation (Fig. 6C-D). Together, the data shown in Figure 6 indicate that endogenous levels of CARD19 neither regulate Bcl10 signaling nor contribute appreciably to Bcl10 degradation in response to TCR stimulation of primary CD8 T cell blasts. These data thus contrast markedly with the results of the ectopic expression studies in Figures 3-5.
1.3.6. CARD19 is a mitochondrial protein that does not interact with Bcl10 in T cells
Because a previous study presented evidence that overexpression of V5/His-tagged CARD19 overexpressed in HeLa cells results primarily in mitochondrial localization [12], we performed microscopy analyses to determine the subcellular localization of endogenous CARD19 in murine CD8 T-cells. We labeled CARD19+/+ CD8 T cells with anti-CARD19 and either the outer mitochondrial membrane (OMM) markers anti-Mitofusin 2 (Mfn2) and anti-MAVS, or the inner mitochondrial membrane (IMM) marker anti-ATP Synthase β (Fig. 7A-C). Endogenous CARD19 was detected as cytoplasmic punctae and these punctae colocalized with all mitochondrial markers; these CARD19 punctae were not detected in CARD19−/− CD8 T cells labeled with anti-CARD19 and anti-Mfn2 (Fig S2A) or anti-MAVS (Fig S2B). Notably, unlike the reported localization for ectopically expressed AK057716 [11], we failed to detect cytosolic or nuclear CARD19 via confocal microscopy (Fig. 7A-C). To better define the localization of CARD19, we also performed Structured Illumination Microscopy (SIM) (which has at least twice the maximal hypothetical resolution of confocal microscopy) on CARD19−/− CD8 T-cells stably expressing 3×FLAG-CARD19 and stained with anti-FLAG and anti-ATP Synthase β. We found again that CARD19 was distributed to cytosolic punctae generally overlapping with and/or surrounding the IMM marker, ATP Synthase β (Fig S3B). Because it was previously reported that CARD19 was also detected in the ER [12], we labeled CARD19+/+ CD8 T cells with ER-marker Calnexin and CARD19 (Fig S4). While CARD19 and Calnexin were in close proximity, and perhaps make contact in some areas of the cell, overall, CARD19 and Calnexin exhibit distinct and mostly non-overlapping distribution patterns.
Figure 7. CARD19 colocalizes with mitochondrial markers MAVS and ATP Synthase β in murine CD8 T cells.
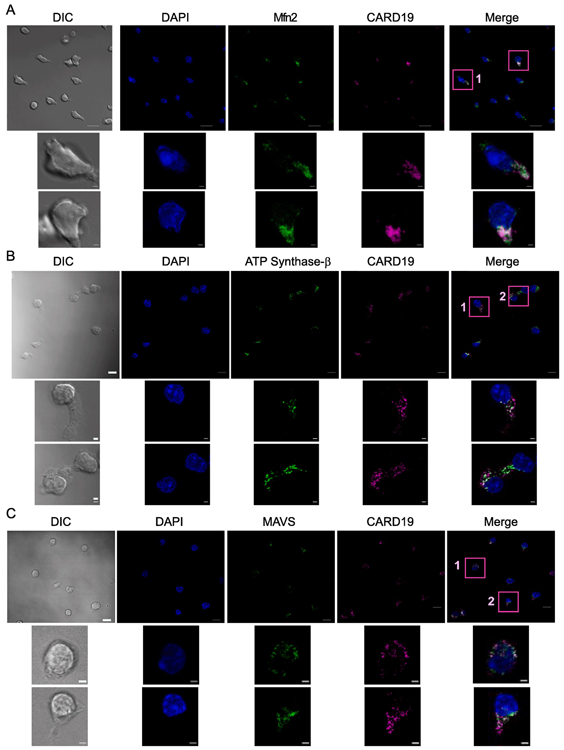
CD8 T cells isolated from CARD19+/+ mice were stimulated with anti-TCRβ/anti-CD28 and allowed to expand and differentiate for one week. Cells were seeded for 20 min on anti-CD43-coated coverslips and fixed. Samples were incubated with DAPI to stain nuclear DNA, anti-CARD19, and: A, anti-Mfn2, a marker of the outer mitochondrial membrane (OMM); B, anti-ATP Synthase β, an inner mitochondrial membrane (IMM) marker; and C, anti-MAVS, an OMM marker. Scale bars in fully-sized images are 10 μm. Scale bars in inset images are 2 μm.
Given the transmembrane domain and mitochondrial localization of CARD19, Bcl10 would most likely have to be recruited from the cytosol to mitochondria for a Bcl10-CARD19 interaction to occur. To determine if there is evidence of subcellular colocalization of Bcl10 and CARD19 after T cell activation, we visualized Bcl10 and CARD19 subcellular distribution via confocal microscopy in CARD19+/+ primary murine T-cells. Following activation of CD8 T-cells with anti-CD3ε, Bcl10 punctae were visible; however, we did not observe frequent Bcl10 colocalization with endogenous CARD19. Although the microscopy data show occasional juxtaposition of Bcl10 and CARD19 staining (Fig. 8A), these observations may simply represent a consequence of the relatively low cytosol volume in T cells and the limitations of the resolution of confocal microscopy.
Figure 8. CARD19 and Bcl10 do not co-fractionate in murine CD8 T cells after anti-CD3ε stimulation.
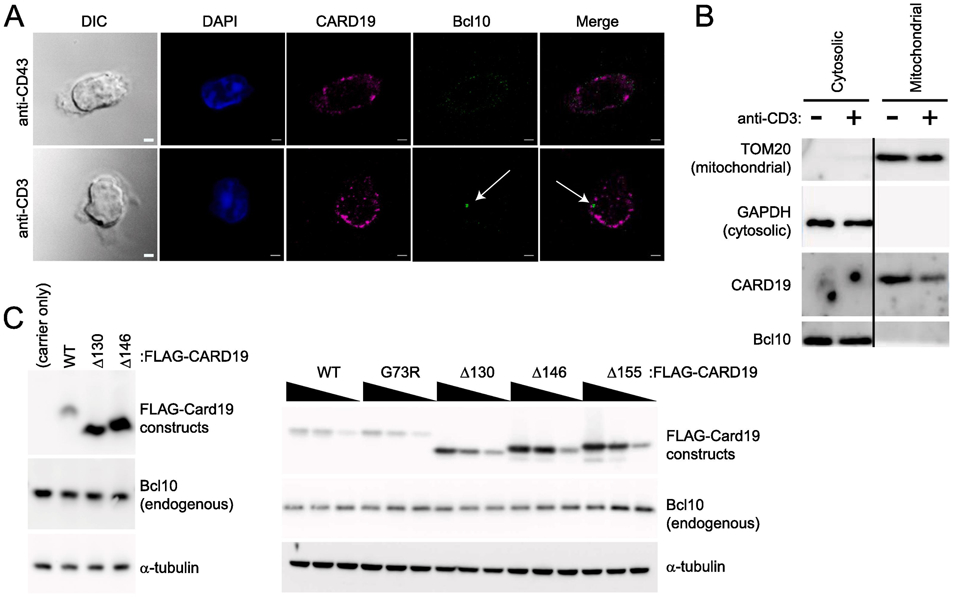
A, CD8 T cells isolated from CARD19+/+ mice were stimulated with anti-TCRβ/anti-CD28 and allowed to expand and differentiate for one week. Cells were seeded for 20 min on anti-CD43 or anti-CD3ε coated coverslips and fixed. Samples were stained with anti-CARD19 and anti-Bcl10. Bcl10 punctae were clearly present in activated T cells, but did not colocalize with CARD19, although there was some juxtaposition of Bcl10 and CARD19 punctae. Scale bars are 2 μm. B, CD8 T cells from the same isolation as A were stimulated with anti-CD43 or anti-CD3ε. Mitochondria and the cytosolic fraction were isolated via the Qproteome Mitochondrial Isolation Kit (Qiagen) and analyzed via immunoblotting for detection of TOMM20 (a mitochondrial marker) and GAPDH (a cytosolic marker), as well as CARD19 and Bcl10. C, 3×FLAG-CARD19-WT and the indicated CARD19 mutants were overexpressed at a single concentration (left) or decreasing concentrations (right) in HEK293T cells, and lysates were separated via SDS-PAGE. Immunoblotting was performed to detect ectopic CARD19 (anti-FLAG), endogenous Bcl10, and α-tubulin.
To more rigorously investigate possible interactions between CARD19 and Bcl10, we used biochemical fractionation to compare the subcellular distribution of Bcl10 and CARD19 in CD8 T cells in the presence and absence of 20 min stimulation with anti-CD3ε. Clean separation of cytoplasmic and mitochondrial fractions was demonstrated by the markers GAPDH and TOMM20, respectively. In both the presence and absence of anti-CD3ε stimulation, endogenous CARD19 was present in the mitochondrial fraction and absent from the cytoplasmic fraction. By contrast, Bcl10 was present in the cytosolic fraction in both unstimulated and anti-CD3ε-stimulated conditions, and not detected in the mitochondrial fraction. Thus, if Bcl10 was recruited to mitochondria by CARD19 or if CARD19 was recruited to the cytosol after TCR stimulation, we would expect to see enrichment of Bcl10 in the mitochondrial fraction or CARD19 in the cytosolic fraction. However, no such co-fractionation of CARD19 with Bcl10 was observed. (Fig. 8B). Together, these data indicate that CARD19 and Bcl10 do not measurably interact before TCR engagement or after 20 min of TCR engagement in murine CD8 T cell blasts.
Additionally, we overexpressed 3×FLAG-CARD19-WT or various 3×FLAG-CARD19 mutants in HEK293T cells. In contrast to results obtained with overexpression of both 3×FLAG-CARD19 and Bcl10-GFP, in which 3×FLAG-CARD19-WT caused reduced expression of Bcl10-GFP (Figs. 3-5), the expression of 3×FLAG-CARD19-WT showed no effect on expression of endogenous Bcl10 in comparison to cells transfected with carrier DNA only or with several different CARD19 mutants (Fig. 8C). This experiment demonstrates that endogenous Bcl10 protein is not subject to CARD19-dependent degradation. Taken together, the data in Figures 3-8 demonstrate that functional interactions between CARD19 and Bcl10 can only be demonstrated when both proteins are over-expressed. Furthermore, our data fail to provide evidence of physical or functional interaction between endogenous CARD19 and endogenous Bcl10 in murine CD8 T cells.
1.4. Discussion
In this study, we have clarified the splicing pattern and sequence of the mRNA product of the human gene, C9orf89; identified CARD19 (formerly known as BinCARD-2) as the single protein product of this gene; determined that endogenous CARD19 localizes to mitochondria in T cells; and presented strong evidence that CARD19 does not regulate TCR-mediated activation of NF-κB in T cells, despite compelling evidence of such an activity based on heterologous overexpression experiments. Through multiple approaches, we have shown that the originally identified AK057716 cDNA (BinCARD-1) reflects an incompletely spliced transcript. Although this human cDNA encodes a putative protein product, this ORF is not translated at measurable levels in murine primary T cells or a human T cell line. Moreover, in silico analyses of CARD19 cDNAs and gene structures from diverse vertebrate species demonstrated that the C-terminus of the BinCARD-1 ORF is encoded within intron 3 of human gene C9orf89. Clustal W alignment demonstrated that this intronic ORF is not conserved across species, in contrast to the high conservation of the CARD19/BinCARD-2 ORF. Indeed, even among primate species, conservation of the intronic ORF is poor, demonstrating that this putative coding region has not been under selective pressure, even during recent evolutionary time. Although it is not possible to entirely rule out that AK057716/BinCARD-1 is a very recently evolved variant of CARD19 with a distinct function, our bioinformatics data collectively provide very strong evidence that the intron 3 portion of the ORF is spurious, rather than being representative of an alternatively spliced isoform of CARD19.
RT-PCR analysis of Jurkat total RNA demonstrated that the AK057716 splicing pattern (exon 2-exon 3-intron 3) is present only at low levels, whereas the fully spliced CARD19 mRNA splicing pattern (exon 3-exon 4) was at least 20-fold more abundant, consistent with the AK057716 splicing pattern representing a splicing intermediate rather than a mature mRNA. Furthermore, using an antibody equally capable of detecting the properly spliced CARD19 gene product and the putative ORF AK057716/BinCARD-1, we were only able to detect the properly spliced CARD19 isoform via western blot from lysates of T cells, based on the fact that there was a single band, which matched the molecular weight of the translated product of the fully spliced CARD19 mRNA, and not the larger, putative AK057716 protein product. Additionally, three bands were detected in lysates of cells transfected with epitope-tagged AK057716: a band which matched the predicted molecular weight of the translated product of the putative ORF, as well as a smaller band which presumably is a degradation product, and a large 50 kDa band, which may be post-translationally modified. The presence of three discrete bands is also distinct from observations of expression of endogenous CARD19, providing further evidence that the AK057716-encoded protein is not produced at detectable levels. Collectively, the above data provide strong evidence that there is only one isoform of CARD19, equivalent to the protein previously named BinCARD-2.
Exploration of influence of CARD19 on NF-κB signaling revealed a major incongruity between the activity of overexpressed CARD19 in HEK293T cells and endogenous CARD19 in primary murine CD8 T cells. We initially compared NF-κB inhibitory activity of CARD19 vs. the AK057716 ORF (BinCARD-1), by co-transfecting HEK293T cells with 3×FLAG-CARD19 and 3×HA-Bcl10-GFP. We found that 3×FLAG-CARD19 inhibited NF-κB activation and decreased 3×HA-BcL10-GFP protein levels much more potently than cells transfected with 3×FLAG-AK057716, although this differential was only evident at the two highest doses of transfected plasmid. Additional data showed that this degradative activity requires both an intact CARD19 C-terminus and a properly-folded CARD19 CARD, and that the degradative mechanism is primarily proteasomal. Together, these data suggest that overexpressed 3×FLAG-CARD19 inhibits NF-κB activation by interacting with 3×HA-Bcl10-GFP and driving its proteasomal degradation, thus limiting CBM complex formation.
To further investigate the potential functional interaction between endogenous CARD19 and endogenous Bcl10, we performed ex vivo experiments with primary murine CD8 T cells. Our goal was to determine if CARD19 is required for Bcl10 degradation to limit NF-κB activation and/or Malt1 protease function. Based on our heterologous overexpression data, we expected to observe an extended kinetic of TCR-induced degradation of IκBα in CARD19−/− CD8 T cells and/or increased A20 and IκBα expression, Regnase cleavage, and IκBα phosphorylation at 120 min post-anti-CD3ε stimulation in CARD19−/− CD8 T cells relative to CARD19+/+ cells. However, we observed no marked differences in these parameters of Bcl10 signaling between CARD19+/+ and CARD19−/− CD8 T cell blasts. We did observe very slight differences in Bcl10 degradation between CARD19+/+ and CARD19−/− CD8 T cells, with CARD19−/− cells exhibiting proportionally less Bcl10 expression by 120 min of anti-CD3 stimulation, as compared to CARD19+/+ cells. If TCR-mediated Bcl10 degradation was a CARD19-dependent phenomenon, we would expect the opposite result, with higher degradation of CARD19 in CARD19+/+ T cells. We also observed that there was slightly less IκBα phosphorylation and Regnase cleavage in the CARD19−/− CD8 T cells, although this is again the opposite outcome of what we would expect if CARD19 drove TCR-mediated Bcl10 degradation.
Consistent with previous studies that employed heterologous expression of epitope-tagged BinCARD-2 [12, 13], we demonstrated that endogenous CARD19 is a mitochondrial protein in murine T cells. Mitochondrial localization is also consistent with the presence of a consensus transmembrane domain in the CARD19 C-terminal domain (which is notably not present in the AK057716 ORF). Since there is some CARD19 that does not appear to colocalize with Mfn2, MAVS, or ATP Synthase β, it is possible that some CARD19 is present in, or makes contact with, the endoplasmic reticulum. However, the distribution of CARD19 did not generally overlap with Calnexin when co-labeled in CARD19+/+ CD8 T cells. Our interpretation of these data is that CARD19 is not uniformly distributed around the entire mitochondria, and therefore does not completely colocalize with Mfn2, MAVS, or ATP Synthase β. This interpretation is supported by our SIM data, which indicated a strongly punctate CARD19 distribution that appears to encircle ATP Synthase β (an IMM protein).
Furthermore, via microscopy and subcellular fractionation, we demonstrated that mitochondrial CARD19 does not colocalize or cofractionate with cytosolic Bcl10 in murine T lymphocytes, either prior to or after TCR stimulation. These data thus strongly argue that CARD19 and Bcl10 do not interact at physiological levels in unstimulated or TCR-stimulated CD8 T cell blasts. Furthermore, we know of no current evidence that Bcl10 is recruited to mitochondria in T cells after TCR engagement. Finally, the observation that overexpressed 3×FLAG-CARD19 was unable to reduce levels of endogenous Bcl10 strongly suggests that Bcl10 is only able to interact with CARD19 when expressed at supraphysiological levels, presumably at concentrations that drive both proteins out of their normal subcellular niches and into a common compartment.
Clearly, the ectopic expression studies in HEK293T cells and the gene inactivation studies in primary murine CD8 T cells are in conflict. Although we did not investigate these disparities enough to provide a definitive explanation, certain observations are suggestive of a plausible mechanism. Specifically, we found that transient overexpression of wild-type CARD19 in HEK293T cells resulted in very low protein expression relative to C-terminally truncated mutants. Expression of the WT protein was consistent across most of the transfected doses of plasmid, whereas protein expression increased proportional to dose for the mutants. However, treatment with the proteasome inhibitor MG132 resulting in striking accumulation of CARD19 and Bcl10. Although we demonstrated that CARD19-Δ155 interacts with Bcl10, this mutant had little ability to reduce Bcl10 protein levels and had no impact on Bcl10-dependent NF-κB activation. Thus, the distal C-terminus of CARD19 (beyond residue 155) encodes a domain that apparently determines a short protein half-life or limits accumulation of CARD19 protein by some other mechanism, resulting in co-degradation of CARD19 and Bcl10 when these proteins physically interact. However, such interaction apparently only occurs when both proteins are overexpressed, and primarily at two highest amounts of CARD19 plasmid we tested. Together, our observations may suggest that above a certain level of expression, additional CARD19 protein is rapidly degraded, perhaps due to limiting levels of an endogenous chaperone or mitochondrial binding partner that interacts with the CARD19 distal C-terminus. This excess CARD19 presumably cannot access the mitochondria and would thus likely accumulate in the golgi, ER or cytoplasm where it could potentially interact with the overexpressed Bcl10 in these transfection experiments. Overall, based on the presented data, there are a number of plausible mechanisms to explain why the apparent phenotype observed in HEK293T cells was not recapitulated in primary CD8 T cells.
Although it has been reported recently that Bcl10 can inducibly localize to the mitochondria, these observations were not reflective of events that occur during TCR signaling. In this report [18], Bcl10 was shown to interact with MAVS in HEK239T cells (upon overexpression) and in murine embryonic fibroblastsin response to infection with VSV. In this proposed pathway, viral infection causes Bcl10 to interact with CARMA3, followed by binding to MAVS, thus preventing MAVS aggregation. However, despite intriguing circumstantial evidence that CARD19 and Bcl10 may interact in CARMA3-expressing cells in response to viral infection, we do not find evidence of such an interaction or even of Bcl10 recruitment to mitochondria in murine CD8 T cells with or without TCR stimulation.
Given the conservation of CARD19 across species, we expect CARD19 to function similarly in other vertebrates, including humans. However, the function of CARD19 in T cells remains undetermined. In this study, we have used both microscopy and subcellular fractionation to demonstrate the mitochondrial localization of endogenous CARD19 in T cells. These data will likely serve as a useful lead to elucidate the function of this protein in T cell biology. Mitochondria are well-established as organelles that are essential for multiple cell metabolism and cell death pathways. There is increasing evidence that regulation of metabolism drastically affects the adaptive immune system. It has long been reported that quiescent T cells rely on oxidative phosphorylation, whereas activated T cells are reprogrammed to rely on aerobic glycolysis after stimulation [19-21]. This metabolic reprogramming affects T cell function and sensitivity to cell death [19, 22-24]. Notably, glycolysis occurs in the cytosol, whereas oxidative phosphorylation occurs within the mitochondria. It is possible that CARD19 plays a regulatory role in T cell metabolism that affects proliferation, differentiation, and effector function; these potential functions should be specifically investigated. CARD19 may also have a more significant role outside of T cells: gene expression databases and previous publications suggest that CARD19 is ubiquitously expressed, but highly transcribed in human myeloid cells. Thus, CARD19 may play a particularly important role in macrophages, perhaps impacting innate immunity and inflammation. The investigation of possible immunoregulatory functions of CARD19 in myeloid cells is another intriguing area for future studies.
Supplementary Material
Highlights.
The human gene C9orf89 produces a single protein, CARD19, which is conserved across multiple vertebrate species.
The previously reported product of C9orf89, the ORF encoded by cDNA AK057716 (BinCARD-1), represents an incompletely spliced mRNA with no detectable protein product in human or murine T cells.
When transiently overexpressed with Bcl10 in HEK293T cells, CARD19 can inhibit NF-κB activation by directing proteolysis of BCL10. However, these activities are not observed at endogenous levels of CARD19 expression.
CARD19 is a mitochondrial protein in murine primary CD8 T cells.
Endogenous CARD19 does not affect activation of NF-κB or the protease function of Malt1; endogenous CARD19 does not co-fractionate with endogenous BCL10 in murine primary CD8 T cells.
Acknowledgements:
We thank Dr. Dennis McDaniel for assistance with collection of the microscopy data, and J. Latoche, C. Huaman and B. Akanji for their management and care of the mice used in these experiments. The Hybridoma Product AA4.3 (anti-α-tubulin) developed by C. Walsh was obtained from the Developmental Studies Hybridoma Bank (DSHB), created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Supported by grants from the National Institutes of Health (U01GM109887, R01AI125552) to B.C.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University or any other agency of the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References.
- [1].Bouchier-Hayes L, Martin SJ, CARD games in apoptosis and immunity, EMBO reports, 3 (2002) 616–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC, Cutting Edge: TCR Ligation Triggers Digital Activation of NF-κB, The Journal of Immunology, 185 (2010) 4520–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Thome M, Charton JE, Pelzer C, Hailfinger S, Antigen Receptor Signaling to NF-κB via CARMA1, BCL10, and MALT1, Cold Spring Harbor Perspectives in Biology, 2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sun L, Deng L, Ea C-K, Xia Z-P, Chen ZJ, The TRAF6 Ubiquitin Ligase and TAK1 Kinase Mediate IKK Activation by BCL10 and MALT1 in T Lymphocytes, Molecular Cell, 14 (2004) 289–301. [DOI] [PubMed] [Google Scholar]
- [5].Shambharkar PB, Blonska M, Pappu BP, Li H, You Y, Sakurai H, Darnay BG, Hara H, Penninger J, Lin X, Phosphorylation and ubiquitination of the IκB kinase complex by two distinct signaling pathways, The EMBO Journal, 26 (2007) 1794–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lobry C, Lopez T, Israël A, Weil R, Negative feedback loop in T cell activation through IκB kinase-induced phosphorylation and degradation of Bcl10, Proceedings of the National Academy of Sciences, 104 (2007) 908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D, Degradation of Bcl10 Induced by T-Cell Activation Negatively Regulates NF-κB Signaling, Molecular and Cellular Biology, 24 (2004) 3860–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wegener E, Oeckinghaus A, Papadopoulou N, Lavitas L, Schmidt-Supprian M, Ferch U, Mak TW, Ruland J, Heissmeyer V, Krappmann D, Essential Role for IκBα Kinase β in Remodeling Carma1-Bcl10-Malt1 Complexes upon T Cell Activation, Molecular Cell, 23 (2006) 13–23. [DOI] [PubMed] [Google Scholar]
- [9].Hu S, Du M-Q, Park S-M, Alcivar A, Qu L, Gupta S, Tang J, Baens M, Ye H, Lee TH, Marynen P, Riley JL, Yang X, cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas, The Journal of Clinical Investigation, 116 (2006) 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Paul S, Kashyap Anuj K., Jia W, He Y-W, Schaefer Brian C., Selective Autophagy of the Adaptor Protein Bcl10 Modulates T Cell Receptor Activation of NF-κB, Immunity, 36 (2012) 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Woo H-N, Hong G-S, Jun J.-l., Cho D-H, Choi Hyun-Woo, Lee H-J, Chung C-W, Kim I-K, Jo D-G, Pyo J-O, Bertin J, Jung Y-K, Inhibition of BcllO-mediated activation of NF-κB by BinCARD, a Bcl10-interacting CARD protein, FEBS Letters, 578 (2004) 239–244. [DOI] [PubMed] [Google Scholar]
- [12].Chen K-E, Richards AA, Caradoc-Davies TT, Vajjhala PR, Robin G, Lua LHL, Hill JM, Schroder K, Sweet MJ, Kellie S, Kobe B, Martin J, The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized, Acta Crystallographica Section D, 69 (2013) 774–784. [DOI] [PubMed] [Google Scholar]
- [13].Suzuki H, Kameyama T, Takaoka A, BinCARD2 as a positive regulator of interferon response in innate immunity, Biochemical and Biophysical Research Communications, 511 (2019)287–293. [DOI] [PubMed] [Google Scholar]
- [14].Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG, Clustal W and Clustal X version 2.0, Bioinformatics, 23 (2007) 2947–2948. [DOI] [PubMed] [Google Scholar]
- [15].Langel FD, Jain NA, Rossman JS, Kingeter LM, Kashyap AK, Schaefer BC, Multiple Protein Domains Mediate Interaction between Bcl10 and MALT1, Journal of Biological Chemistry, 283 (2008) 32419–32431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Paul S, Schaefer BC, Visualizing TCR-Induced POLKADOTS Formation and NF-κB Activation in the D10 T-Cell Clone and Mouse Primary Effector T Cells, in: May MJ (Ed.) NF-kappa B: Methods and Protocols, Springer New York, New York, NY, 2015, pp. 219–238. [DOI] [PubMed] [Google Scholar]
- [17].Chou JJ, Matsuo H, Duan H, Wagner G, Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment, Cell, 94 (1998) 171–180. [DOI] [PubMed] [Google Scholar]
- [18].Jiang C, Zhou Z, Quan Y, Zhang S, Wang T, Zhao X, Morrison C, Heise Mark T., He W, Miller Matthew S., Lin X, CARMA3 Is a Host Factor Regulating the Balance of Inflammatory and Antiviral Responses against Viral Infection, Cell Reports, 14 (2016) 2389–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cao Y, Rathmell JC, Macintyre AN, Metabolic Reprogramming towards Aerobic Glycolysis Correlates with Greater Proliferative Ability and Resistance to Metabolic Inhibition in CD8 versus CD4 T Cells, PLOS ONE, 9 (2014) e104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jacobs SR, Herman CE, Maclver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC, Glucose Uptake Is Limiting in T Cell Activation and Requires CD28-Mediated Akt-Dependent and Independent Pathways, The Journal of Immunology, 180 (2008) 4476–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pearce EL, Poffenberger MC, Chang C-H, Jones RG, Fueling Immunity: Insights into Metabolism and Lymphocyte Function, Science, 342 (2013) 1242454.24115444 [Google Scholar]
- [22].Cham CM, Gajewski TF, Glucose Availability Regulates IFN-γ Production and p70S6 Kinase Activation in CD8+ Effector T Cells, The Journal of Immunology, 174 (2005) 4670–4677. [DOI] [PubMed] [Google Scholar]
- [23].Larsen SE, Bilenkin A, Tarasenko TN, Arjunaraja S, Stinson JR, McGuire PJ, Snow AL, Sensitivity to Restimulation-Induced Cell Death Is Linked to Glycolytic Metabolism in Human T Cells, The Journal of Immunology, 198 (2017) 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang R, Green DR, Metabolic reprogramming and metabolic dependency in T cells, Immunological Reviews, 249 (2012) 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


