Abstract
Despite significant progress in understanding the genetic landscape of T-cell acute lymphoblastic leukemia (T-ALL), the discovery of novel therapeutic targets has been difficult. Our results demonstrate that the levels of PIM1 protein kinase is elevated in early T-cell precursor ALL (ETP-ALL) but not in mature T-ALL primary samples. Small molecule PIM inhibitor (PIMi) treatment decreases leukemia burden in ETP-ALL. However, treatment of animals carrying ETP-ALL with PIMi was not curative. To model other pathways that could be targeted to complement PIMi activity HSB-2 cells, previously characterized as a PIMi sensitive T-ALL cell line, were grown in increasing doses of PIMi. Gene Set Enrichment Analysis of RNA-seq data and functional enrichment of network modules demonstrated that the HOXA9, mTOR, MYC, NF-κB, and PI3K-AKT pathways were activated in HSB-2 cells after long-term PIM inhibition. Reverse phase protein array-based pathway activation mapping demonstrated alterations in the mTOR, PI3K-AKT, and NF-κB pathways, as well. PIMi tolerant HSB-2 cells contained phosphorylated RelA-S536 consistent with activation of the NF-κB pathway. The combination of NF-κB and PIM inhibitors markedly reduced the proliferation in PIMi resistant leukemic cells showing that this pathway plays an important role in driving the growth of T-ALL. Together these results demonstrate key pathways that are activated when HSB-2 cell line develop resistance to PIMi and suggest pathways that can be rationally targeted in combination with PIM kinases to inhibit T-ALL growth.
Introduction
T-ALL is an aggressive hematopoietic malignancy that is treated by intensive chemotherapy (1). To improve risk stratification, T-ALL patients are classified into distinct subtypes via immunophenotyping and gene expression analysis (2,3). ETP-ALL is a high-risk subtype of T-ALL with a unique immunophenotype including myeloid and early progenitor in addition to T-cell lineage markers (4). Whole-genome and transcriptome sequencing of ETP-ALL reveals several activating gene fusions, alterations, and mutations within IL7R and JAK-STAT pathways that could result in constitutively activated tyrosine kinases (5–7), whereas non-ETP or mature ALL often display mutations in PI3K/AKT and NOTCH signaling pathways (8). Despite the advances that have been made in decoding the genetic background of T-ALL, the translation of novel targeted therapies towards clinical practice has remained elusive (8).
Our group and others have shown that the PIM (Provirus Integration sites for Moloney murine leukemia virus) kinase is a potential target in T-ALL (9–13). Recently, two studies identified a similar TCRβ-PIM1 translocation in T-ALL patients that is essential for aberrant activation of PIM1 kinase (12,13). The PIM kinases are a highly conserved family of three serine/threonine kinases involved in cell-cycle, apoptosis, transcription, translation, cell metabolism, and drug resistance through phosphorylation of downstream targets (14–16). PIM kinase levels are increased by cytokines that stimulate JAK-STAT signaling (17). Previously, we reported that PIM1 is overexpressed in ETP-ALL and a small percentage non-ETP ALL patient-derived xenografts (PDXs) (10), suggesting that this kinase may be an ideal therapeutic target. Using ETP-ALL PDXs, we demonstrate the PIMi activity in this disease. To learn more about the signal transduction pathways that are essential for T-ALL cells to survive and potentially complement PIMi, we have modeled the pathways that are activated using both proteomic and transcriptomic approaches when cells develop resistance to these inhibitors. A key finding from these analyses is that PIMi resistant cells contain an activated NF-κB pathway. The combination of NF-κB and PIM inhibitors therapy suppresses T-ALL growth, suggesting an approach that can be investigated to improve therapy for a subset of T-ALL patients with increased PIM kinase activity.
Materials and Methods
T-ALL PDXs engraftment in NCG (NOD CRISPR Prkdc Il2r γ) mice
All in vivo experiments were conducted on protocols approved by the Institutional Animal Care and Use Committee of the University of Arizona. De-identified T-ALL PDXs were supplied by Children’s Cancer Institute, Children’s Oncology Group, and St. Jude Children’s Research Hospital. These PDX samples had previously been characterized by gene expression profiling, whole exome or genome sequencing, or targeted sequencing of a gene panel (7,18). NCG mice were irradiated and inoculated by tail-vein injection with 5 million cells per 100 μL PBS. Mice were monitored for leukemia engraftment by flowcytometric analysis of peripheral blood using antibodies against human and mouse CD45. At necropsy, spleens were minced, and mononuclear cells were cryopreserved for subsequent experiments.
Human T-ALL cell lines and cell culture
T-ALL cell lines including HSB-2, SUP-T1, DU.528, CUTLL1, KOPT-K1 and HPB-ALL were a kind gift of Dr. Jon C. Aster (Brigham and Women’s Hospital, Harvard University) and cultured in RPMI supplemented with 2 mmol/L Glutamax and 10% fetal bovine serum. All cell lines were maintained at 37°C in 5% CO2 and were authenticated by short tandem repeat DNA profiling performed by the University of Arizona Genetics Core Facility. The cell lines were routinely tested for mycoplasma and used for fewer than 50 passages.
Derivation of PIMi Resistant HSB-2 cells
PIMi resistance in HSB-2 cells was induced by growing cells in the presence of increasing concentrations of AZD1208 (AZDR1 and AZDR2) or LGB321 (LGBR2) over a 4-month period until resulting cell populations were able to maintain their growth in 1 μM AZD1208 or LGB321. While the HSB-2 naïve cells had an IC50 < 1μM, all resistant populations had IC50s > 10 μM when incubated with AZD1208 or LGB321 over the 3-day XTT growth study.
Establishment of xenograft models
Xenograft models were established as described previously (7). All experiments were conducted on protocols approved by the Institutional Animal Care and Use Committee of the University of Arizona. For efficacy studies, sub lethally irradiated (2.0 Gy) NCG mice (Charles River) were randomized to treatment or vehicle (5 mice per arm) once xenografts had engrafted with sufficient disease burden to detect 1% peripheral human CD45 positive cells. PIMi (AZD1208; 30 mg/kg) or Vehicle was administered for the entire 3-week treatment period by oral gavage. AZD1208 was formulated using a Cremophore/Ethanol/PBS – 24/6/70 ratio. Studies were terminated, and mice were euthanized after 3 weeks of treatment. Peripheral blood (PB), spleen and bone marrow (BM) were harvested at the time of necropsy. Disease burden was assessed every 4–5 days by flow cytometric measurement of human CD45 positive cells in PB, and at necropsy by measuring leukemia burden by analyzing the percent of human CD45 positive cells in PB, Spleen, and BM by flow cytometry.
Cell viability assay
Cryopreserved PDX cells were thawed at 37°C and washed in RPMI-1640 media supplemented with 10% FBS, 100 mg/mL streptomycin, 100 U/mL penicillin, and 2 mmol/L L-glutamine. After centrifugation (500 × g for 5 min) and a second wash, cell viability was determined using Trypan blue exclusion assay. Cells were resuspended at the appropriate cell density in QBSF-60/F [containing QBSF-60 media (Quality Biological) supplemented with 20 ng/mL Fms-like tyrosine kinase 3 ligand (Flt-3L; ProSpec), 100 U/mL penicillin, 100 mg/mL streptomycin, and 2 mmol/L L-glutamine]. T-ALL PDX cells (5×104 cells/well) were seeded into 96-well plates and treated with PIMi (AZD1208 and LGB321) as indicated. DMSO was added to keep concentrations of DMSO (<0.1%) equal in all wells. After 72 h, cell viability was measured by using an adenosine triphosphate-based luminescence assay (ATPlite 1-step; PerkinElmer) according to the manufacturer’s protocol. Because PDX cells do not proliferate in these conditions, this assay reflects cell survival only. The survival of DMSO control cells was considered as 100% and percent of cell death after individual treatments is reported relative to the DMSO control.
Quantitative Real Time-PCR
To assess the differential expression of genes related to development of PIMi-resistance in HSB-2 cells, total RNA was isolated the RNeasy kit (Qiagen, Cat#74104). cDNA was synthesized from 1 μg RNA using the iScript cDNA synthesis kit (Bio-Rad, Cat#1708890) according to the manufacturer’s instructions. Real-time PCR amplification and analysis were conducted using the SsoAdvanced™ Universal SYBR® Green Supermix (Bio-RAD, Cat#1725271) and CFX96 Real-Time System. Samples were assayed in triplicate and RNA levels were normalized to 18S expression levels. Primer sequences for real-time PCR are provided in the Supplementary Table S1.
Antibodies and Reagents
The following antibodies were purchased from Cell Signaling Technology: anti-phospho- IRS1 (S1101 Cat # 2385), anti-phospho-S6 (S240/244, Cat # 5364), anti-phospho-TSC2 (S939 Cat # 3615), anti-TSC2 (Cat # 3990), anti-PTEN (Cat # 138G6), anti-Phospho-NF-kB p65 (S536 Cat # 93H1), anti-NF-kB p65 (Cat # D14E12), anti-phospho-4EBP1 (S65 Cat # 13443), anti-4EBP1(Cat # 9644), and anti-phospho-GSK3β (S9, Cat # 5558). Anti-IRS1 (Cat # 06–248) antibody was purchased from Merck Millipore. Anti-DDIT4 (Cat#10638–1-AP) and anti-SESN2 (Cat # 10795–1-AP) antibodies were purchased from ProteinTech Group. HRP conjugated anti-β-actin (Cat # A3854) was purchased from Sigma. HRP-linked mouse IgG (Cat # NA931V) and rabbit IgG (Cat # NAV934V) were purchased from GE Healthcare Life Sciences. IgG (Isotype control Cat # 400107), FITC conjugated anti-human CD45 (Cat # 304006), and APC conjugated anti-mouse CD45 (Cat # 103112) were purchased from BioLegend. Anti-S6 (Cat # SC74459) antibody was purchased from Santa Cruz laboratories. LGB-321 (Cat # A14420), AZD1208 (Cat # A13203), and IKK16 (# A12836) were purchased from Adooq bioscience. TNF-α (Cat # PHC3015) was purchased from Fisher scientific.
Supplementary Methods include a description of the Western Blot, RNA-Seq data analysis, GSEA analysis, RPPA, Active module analysis, Lentiviral production, the NF-κB reporter assay, and Statistics.
Results
PIM kinase inhibition decreases leukemia burden in ETP-ALL PDXs
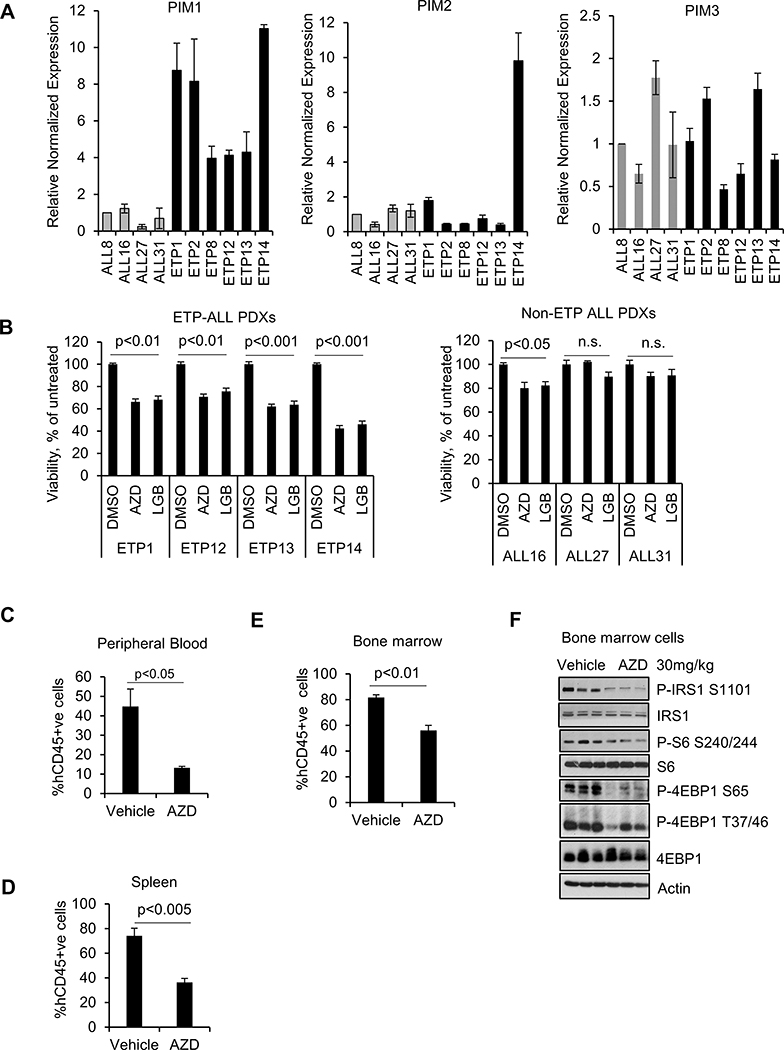
We have examined the level of three PIM isoforms in six ETP-ALL and four mature T-ALL PDXs. Importantly, qRT-PCR of these ten PDXs demonstrated that PIM1 expression, but not PIM2 or PIM3, was elevated specifically in ETP-ALL and not in the mature T-ALL PDXs (Figure 1A). PIM1 protein expression was high in ETP-ALL as compared to mature T-ALL PDXs (Supplementary Figure S1A) and PIM1 expression was correlated with STAT5 activation. This result suggested that the PIM1 could be targeted to inhibit ETP-ALL growth. To examine whether these PDXs are sensitive to PIMi, the cells were incubated with two pan-PIM inhibitors AZD1208 (19) or LGB321 (20). LGB321 is a tool compound related to Pim447, which is being used to treat patients (21). PIMi suppressed ETP-ALL growth by 25 to 50% while the effects on more mature T-ALL PDXs was significantly less (Figure 1B).
Figure 1. Pan-PIM kinase inhibitors reduces leukemia burden in ETP-ALL PDX models.
(A) Relative mRNA expression of PIM1, PIM2, and PIM3 in T-ALL PDX cells. (B) T-ALL PDX cells were incubated with the AZD1208 or LGB321 (AZD 3μM or LGB 1μM) for 72h and then the percentage of viable cells was quantified by the ATPlite assay. The growth of DMSO control cells was considered 100% and percent cell growth after individual treatment is reported relative to the DMSO. The data shown are the average +/− standard deviation (S.D.) of three independent experiments. (C-E) The PB, spleen, and bone marrow (BM) were harvested and analyzed hCD45+ve cells by flow cytometry. % hCD45+ve data shown are the average +/− S.D. from five samples per treatment. (F) Western blot (WB) analysis of lysates collected from BM cells using the specified antibodies. Each lane on the WB represents an individual tumor.
To evaluate the PIMi ability to block leukemic growth in vivo, ETP13 PDXs were injected intravenously into NCG mice. ETP13 PDX was selected for testing because of the ability of this PDX to rapidly engraft in the mice (Supplementary Figure S1B), and its intermediate PIM1 expression. Mice were treated with AZD1208 (10) or vehicle once daily for a duration of 3 weeks Supplementary Figure S1C) without significant change in body weight (Supplementary Figure S1D). Our results demonstrate markedly decreased hCD45 positive leukemic cells in the peripheral blood (PB) (~ 70% decrease; Figure 1C and Supplementary Figure S1E), spleen (~ 50% decrease; Figure 1D), and bone marrow (BM) (30% decrease; Figure 1E) from the AZD1208 treated mice when compared to the vehicle. The modest response of PIMi in BM suggest a possible nutrient rich microenvironment in the BM that might activate compensatory signaling pathways in these leukemic cells (22) and protect from Pimi. In addition, AZD1208 treated mice showed reduced spleen size (Supplementary Figure S1F) and a red colored BM (Supplementary Figure S1G) indicating re-establishment of normal hematopoiesis and decreased leukemic burden. Western blot analysis of total BM cells collected after 3 weeks of drug treatment (Figure 1F) showed a significant decrease in IRS1 phosphorylation, a PIM kinase substrate, demonstrating PIMi activity in the bone marrow cells of these mice (23). As expected from our previous results (10) PIMi treatment also decreased mTORC1 signaling as shown by decreased S6 and 4EBP1 phosphorylation levels. These results supported the potential use of PIMi treatment to decrease tumor load in ETP-ALL patients.
Long-term PIM inhibition in HSB-2 cells exhibit altered gene and protein expression
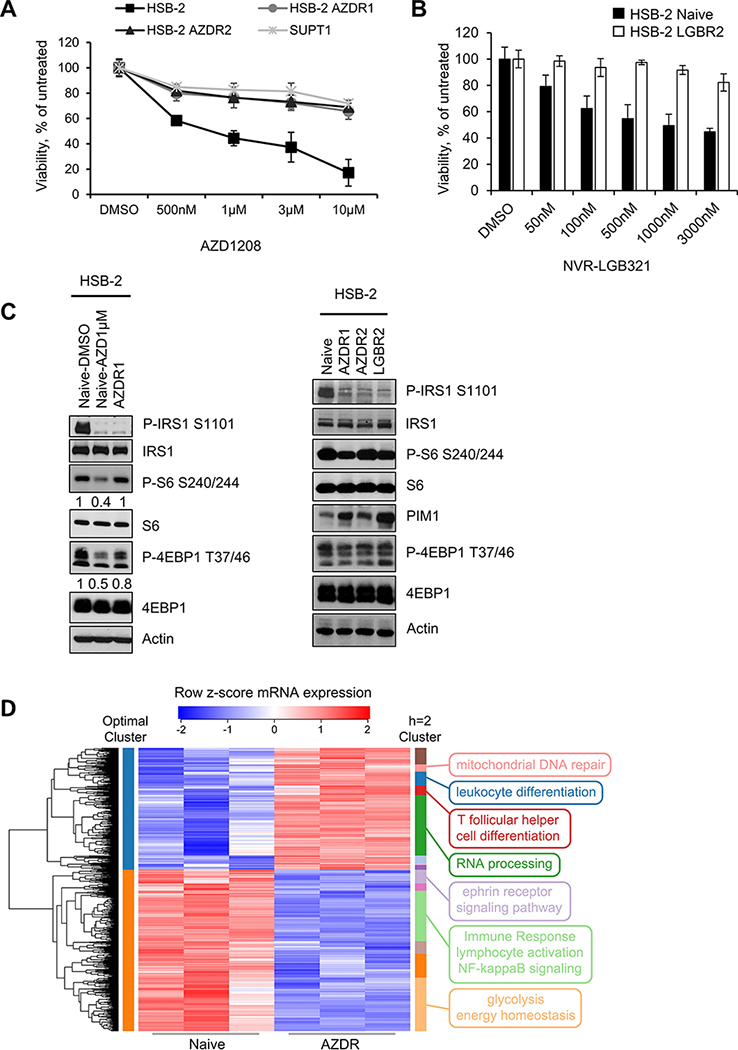
The observation that after three weeks of treatment, leukemic cells remained in the BM and spleen suggested that there were additional pathways being stimulated, which continued to support T-ALL growth even when the PIM kinases were inhibited. To model the biochemical nature of these pathways, previously characterized PIMi sensitive T-ALL cell line HSB-2 (10) was grown in the presence of increasing concentrations of AZD1208 or LGB321 until the cell populations grew continuously in 1μM AZD1208 or LGB321. Previously, we have shown this cell line has gene-expression signature that is consistent with other ETP-ALL cells (10). The approach used to develop PIMi resistance cells was based on published study examining resistance to a NOTCH inhibitor in T-ALL (24). Elevated levels of LCK expression secondary to t(1;7)(p34;q34) chromosomal translocation is found in the HSB-2 cells (25). However, using six T-ALL cell lines, we showed that the PIMi sensitivity was independent of LCK expression (Supplementary Figure S2A-B). Sensitivity to PIMi in HSB-2 cells is comparable to that found in DU.528 and KOPT-K1 cell lines that do not have LCK translocation. Our group and others have shown (10,26,27) in various cancers that PIMi sensitivity is correlated with the level of PIM expression. We did not observe any correlation between PIMi sensitivity (Fig. 1B) and LCK expression in T-ALL PDXs as shown in Supplementary Figure S2C. While the HSB-2 naïve cells had an IC50 of less than 1μM for these drugs, the drug-tolerant cell lines had IC50 values greater than 10μM with either resistance to AZD1208 (AZDR1 and AZDR2) or LGB321 (LGBR2) (Figure 2A and 2B). The drug-tolerant cell lines behaved similarly to the mature T-ALL cell line, SUP-T1 which is in-sensitive to PIMi (10), suggesting that they likely had alternate pathways activated that maintained their growth even when incubated with the inhibitor. Like in HSB-2 naïve and SUPT1 cells incubated short term with PIM kinase inhibitor (24h), PIMi resistant cell lines demonstrated a complete blockade of IRS1 phosphorylation. Importantly, this indicated that incubation with AZD1208 or LGB321 either short or long term blocked Pim kinase activity (28), thus demonstrating that the drug was entering into these resistant cells. However, PIMi treatment reduced mTORC1 activity only in HSB-2 naïve cells where growth is inhibited, but not in PIMi resistant cell lines or the naturally insensitive SUPT1 cells (Figure 2C, Supplementary Figure S2D). These results suggest that mTORC1 plays a role in resistance to the PIMi while IRS1, although a good PIM kinase substrate did not regulate cell growth when the Pim pathway was blocked. This result demonstrates that, when Pim is inhibited the mTORC1 is critical to maintain cell growth and thus could be targeted as part of a long-term combination treatment strategy.
Figure 2. Characterization of HSB-2 naïve and PIMi resistant cells:
(A and B) Using an XTT assay, the percentage of viable cells was measured after each cell line was exposed to increasing doses of AZD or LGB over 72h and compared to vehicle. Data are represented as average +/− S.D of three independent experiments. (C) WB comparing the mTOR signaling activity in HSB-2 naïve (AZD1μM; 24h) and PIMi resistant cell populations. (D) Hierarchical clustering on the differentially expressed genes with high-significance (FDR<0.01) from AZDR RNA-seq data reveals significantly enriched biological pathways. The heat map represents the row z-score of log2 transformed RNA-seq count values. Optimally clustered (by silhouette width) membership is shown with colors on the left side of the heat map while clusters cut at height h=2 are shown with colors on the right side of the heat map. Selected GO term annotations of each cluster are represented on the right side of the heat map. Total of 2225 genes with n=3 in each group were analyzed for the heat map clustering analysis.
To further understand the differences between naïve and resistant cell lines, we performed RNA-sequencing on HSB-2 naïve and AZDR1 cells finding 3,346 genes differentially expressed (FDR<0.05). To evaluate the function of these expression changes, we used the 2225 genes with highest significance (FDR<0.01) as input for hierarchical clustering (Figure 2D). Silhouette width analysis on the resulting dendrogram indicated that the optimal number of clusters is 2 (Supplementary Figure S3A-C). These clusters were enriched for genes in RNA processing, immune response, inflammatory response, and cell migration (Supplementary Table S2). Further cutting the tree down (at height of 2) led to 14 clusters, and Gene Ontology (GO) analysis showed that upregulated genes in AZDR1 cells were enriched for mitochondrial DNA repair, leukocyte differentiation, T follicular helper cell differentiation, and RNA processing genes. In contrast, genes involved in ephrin receptor signaling pathway, immune response, lymphocyte activation, NF-κB signaling, and glycolysis were down-regulated (Figure 2D and Supplementary Table S3). We note that GO gene sets include both activators and inhibitors of the annotated process, so up- or down-regulation of the mRNA expression values does not necessarily correspond to activation or repression of the corresponding signaling pathway but suggests these pathways are modified in cells after long-term PIM inhibition. We validated the RNA-seq results by performing qPCR on genes with the most significant (FDR<1×10−17) change in mRNA expression and demonstrated that BMPR1B, WNK2, SCEL, and DEFA6 genes were upregulated, whereas HKDC1 and DDIT4 were downregulated in AZDR1 compared to naïve HSB-2 cells (Supplementary Figure S3D).
Activation of mTOR, MYC, and disruption of NF-κB and HOXA9 signaling in long-term PIMi treated HSB-2 cells
In addition to clustering the differentially expressed genes, we used Gene Set Enrichment Analysis (GSEA) to compare global gene expression in HSB-2 naïve and AZDR1 cells. Genes were ranked by differential expression p-value and by up- or down-regulation and assessed for enrichment in curated canonical pathways and oncogenic signatures (Supplementary Table S4). In agreement with published studies (13,29), in AZDR1 cells glycolysis was significantly changed while cell cycle related processes were up-regulated (Supplementary Figure S4 and S5A, and Supplementary Table S4). Similarly, expression of genes in pathways such as JAK-STAT, mTOR, MAPK, and cytokine signaling were significantly altered in AZDR1 cells suggesting that continued PIMi treatment is associated with major changes in the signal transduction pathways that regulate cell survival and growth. GSEA identifies these pathways as key to the survival of HSB-2 cells.
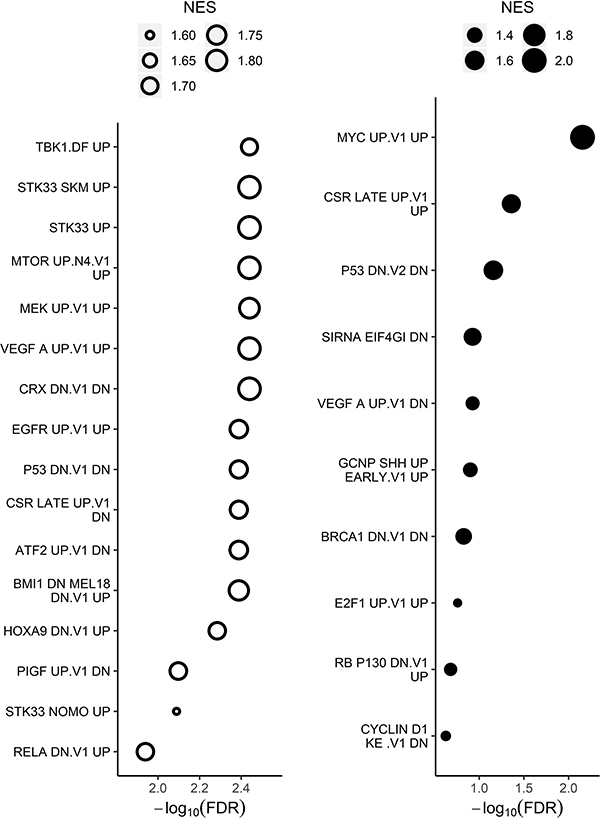
To discover cancer drivers that may not themselves be altered at the mRNA level, but might cause the observed changes in gene expression, we carried out GSEA using the “oncogenic perturbation” gene sets in MSigDB (30). We found evidence that several oncogenic pathways were activated in AZDR1 cells, including mTOR, HOXA9, RelA and MYC. (Figure 3, Supplementary Figure S5B-E, and Supplementary Table S5). A recent study showed increased HOXA9 expression in T-ALL cases with JAK-STAT activation, and in these cases PIM kinase was identified as a possible drug target (11). RelA (NF-κB P65) is a major subunit of NF-κB and its activation has been shown to promote survival in leukemic cells (31). MYC and PIM kinase activities are known to be regulated similarly. MYC overexpression is a common driver of many hematological cancers (32,33). Together these results identify multiple compensatory pathways that are activated in cells surviving after prolonged PIMi treatment.
Figure 3. Oncogenic signatures enriched in transcriptome of long-term PIMi treated cells compared with HSB-2 naïve cells.
Genes were ranked by differential expression and analyzed using GSEA testing against curated oncogenic signatures. The size of the bubbles in the bubble plots represents the normalized enrichment score (NES), which summarizes the number and differential expression intensity of enriched genes. The absolute value of the NES generally varies between 0 (no enrichment) and 5 (extremely high enrichment). The significance of the pathway enrichment is measured by the FDR (x-axis). Open bubbles indicate downregulated pathways and closed indicate upregulated pathways. All oncogenic signatures shown have FDR<0.25 and GSEA significance is defined as FDR<0.25.
Active network module analysis reveals PIMi resistant cells have an active protein complex enriched with cancer-associated signaling pathways
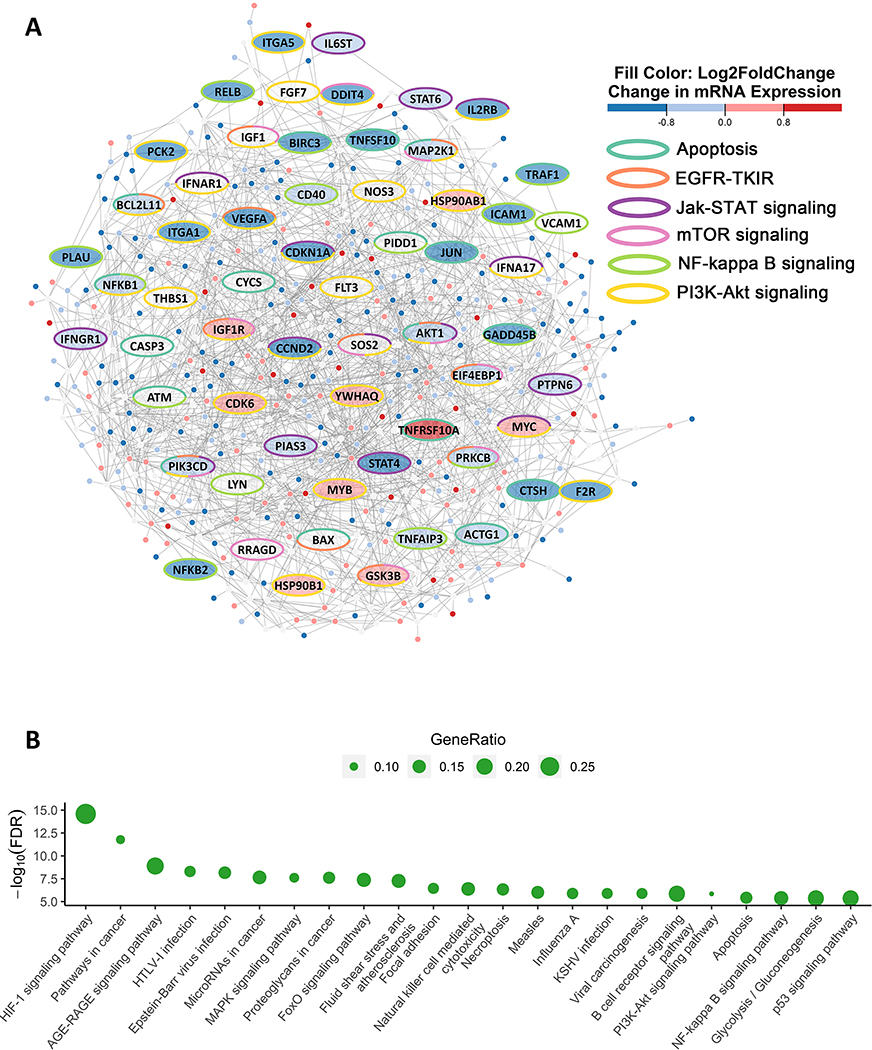
We identified “modules” of interacting proteins that are transcriptionally dysregulated in AZDR cells. Using the active network modules approach (29), we overlaid adjusted p-values from the RNA-seq differential expression results comparing drug resistant and vehicle treated HSB-2 cells over the STRING protein-protein interaction network (34). We then applied the jActiveModules algorithm to find differentially expressed protein complexes (35). Figure 4A shows the statistically significant ANM that was identified.
Figure 4. ANM analysis on long-term PIMi treated cells finds changes in protein complex enriched for signaling and growth pathways.
(A) ANM analysis is shown with nodes representing genes and edge length representing the strength of the protein-protein interaction, with shorter edge lengths indicating higher-weight interactions according to STRING database. The ellipse shaped nodes represent genes in either apoptosis, epidermal growth factor receptor tyrosine kinase inhibitor resistance (EGFR-TKIR), JAK/STAT, mTOR, NF-kB, or PI3K-AKT signaling pathways. The color of the nodes (fill color) indicates the log2(fold change) of the RNA-seq expression in AZDR1 cells compared to HSB-2 naive cells. The red fill indicates significantly increased (FDR<0.05) proteins and blue fill indicates significantly decreased (FDR<0.05) proteins while grey fill indicates not significant (FDR≥0.05). The border color of each node represents pathway membership (e.g. yellow bordered genes are part of the PI3K-AKT signaling pathway). (B) Enrichment of KEGG pathways in active module. The size of the bubble represents gene ratio calculated by taking the number of pathway related genes within the active modules divided by the total number of genes in the pathway. The significance of the pathway enrichment is measured by the FDR (y-axis). Significance is defined as FDR<0.05.
We next tested the genes in the ANM for functional enrichment in KEGG pathways (34). Using this approach many pathways that are found activated in tumor cells, including MAPK, PI3K-AKT, NF-κB, p53, glycolysis, and apoptosis were among the most enriched pathways (Figure 4B; Supplementary Table S6). This set of pathway enrichments in the ANM is consistent with results in different tumor types (13,29,36,37). It is possible that the protein-protein network from any literature-based database, including STRING, could be biased towards known cancer pathways. To check for such a bias, we simulated data by randomly assigning p-values to sets of genes in the RNA-seq data and performed jActiveModules search to show that the results above could not have arisen through intrinsic bias in the PPI network (Supplementary Figures S6A-D and Supplementary Tables S7-10). The ANM approach allows the discovery of not only enriched pathways, but also reveals new pathway members that are not altered at the mRNA level but interact strongly with genes that are differentially expressed. These proteins identified in HSB-2 cells could be important candidates for further study of essential signaling in T-ALL with high PIM1 expression (Figure 4A).
Long-term PIM inhibition in cells exhibit protein level alterations in mTOR, PI3K-AKT, and NF-κB signaling
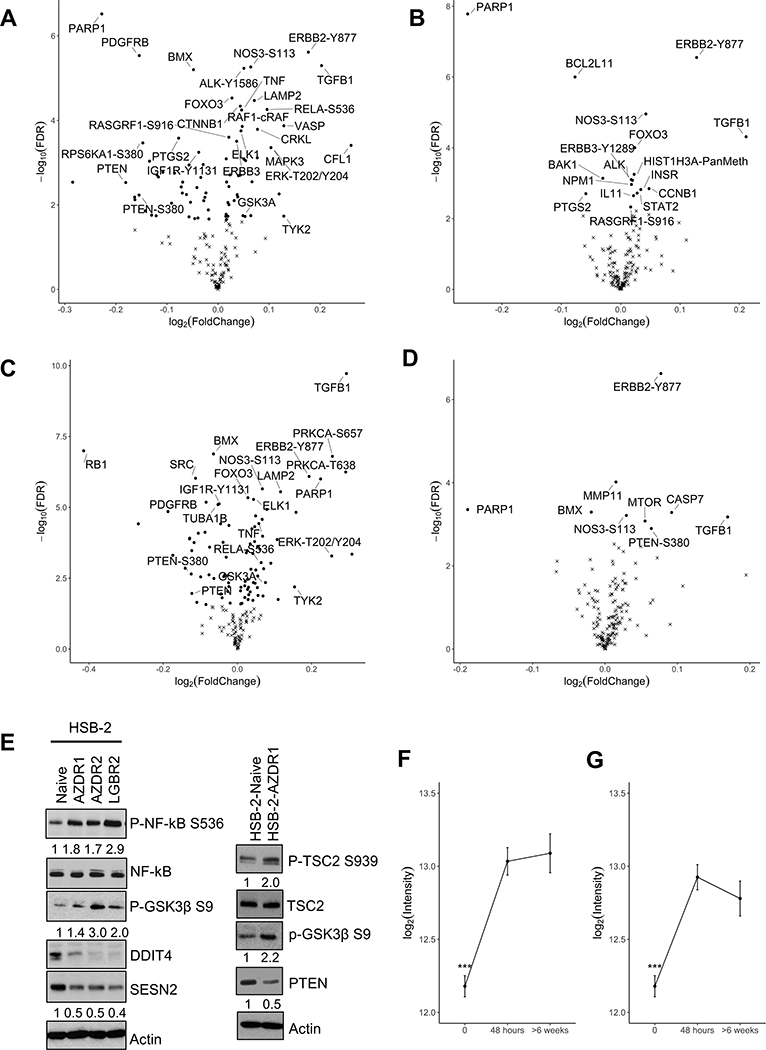
To gain a deeper insight into the functional relationship between PIMi-dependent tumor suppression and the development of drug-tolerance, we measured the relative expression/activation levels of 200 proteins and phosphoproteins using Reverse Phase Protein Arrays (RPPA). A pair-wise differential abundance analysis between five treatment groups compared HSB-2 naive or short-term (AZD1μM / LGB1μM) to long-term (AZDR1 / LGBR2) PIMi treated cells (Figures 5A-D). When compared to naïve cells, the long-term PIMi treated cells exhibited differential abundance of 79 proteins in AZDR1 and 95 proteins in LGBR2 (Figure 5A, C). When compared to the short-term PIMi treated cells, only 17 proteins in AZDR1 and 9 proteins in LGBR2 were differentially abundant (FDR<0.05; Figure 5B, D). This pattern was visualized by volcano plots (Figures 5A-D and Supplementary Tables S11-14).
Figure 5. AZD1208 and LGB321 resistant HSB-2 cells exhibit significant changes in protein expression.
(A-D) Volcano plot (log2(fold change) as x-axis and −log10(p-value) as y-axis) of RPPA differential expression comparing HSB-2 (A) Naïve vs AZDR1, (B) AZD 1μM (short-term) vs AZDR1 (long-term), (C) Naïve vs LGBR2, and (D) LGB 1μM (short-term) vs LGBR2 (long-term) indicate that many proteins have significant changes in expression. Dots indicate Benjamin and Hochberg FDR < 0.05 and x-mark indicates FDR ≥ 0.05. (E) Validation of RPPA data generated from comparing HSB-2 naïve and AZDR1 or LGBR2 cells by WB. (F & G) RelA-S536 abundance is shown on the y-axis and time of treatment with PIMi on the x-axis. Total of 200 proteins were analyzed with n=6 for each sample. Results are expressed as mean+/−S.E.M; ***p-value<0.001; ****p-value < 0.0001.
In comparison to naive cells, long-term PIMi treated cells demonstrate significant changes in the abundance/activation state of various phospho-proteins, for example GSK3, NF-κBp65 (RelA), Tyk2, Rb, and ErbB2, and changes in total proteins including LAMP2, TGFβ, PTEN, and TNFα. As predicted from our previous studies and those of others, ERK phosphorylation at the T202/Y204 sites (9,10) was significantly upregulated in HSB-2 cells after long-term PIM inhibition. Both TNF-α and TGF-β protein levels were increased in AZDR1 and LGBR2 cells (Figure 5A, C and Supplementary Tables S11, S13), and TGFβ levels were also induced in HSB-2 cells after short-term PIMi treatment (Figure 5B, D and Supplementary Tables S12, S14), suggesting a possible compensatory mechanism that might maintain PIM kinase levels.
We then validated RPPA data comparing HSB-2 naïve and PIMi short and long term treated cells by Western blot (Figure 5E). We found an increased expression of NF-κB and activation of the AKT pathway possibly induced by decreased PTEN levels. Activated AKT after long-term PIM inhibition caused an increase in GSK3β phosphorylation in these resistant HSB-2 cells. Activation of the AKT pathway after long-term PIM inhibition has been noted previously in other cell types (36). The protein levels of DDIT4 and SESN2 (both negative regulators of mTORC1) were downregulated and TSC2 phosphorylation was increased in long-term PIMi treated cells. The observation that the mTORC1 pathway was activated as part of PIMi resistance was consistent with our finding in cell culture and on RNA-seq. In comparison, short-term PIMi treatment of naïve HSB-2 cells showed significant suppression of mTORC1 signaling (Figure 2C) while in AZDR1, AZDR2, and LGBR2 cells PIMi treatment showed no effect on this pathway.
We observed that the differentially abundant proteins in long-term PIMi treated cells found by RPPA were significantly enriched in the ANM constructed from transcriptomic data (p-value = 0.0211 by hypergeometric test), with the overlapping proteins including NOS3, IGF1R, PDK1, PRKCD, STAT4, and CASP3. Therefore, both the gene and protein expression changes in long-term PIMi treated cells converge on common biological functions.
Targeting NF-κB signaling overcomes resistance to PIM inhibitor treatment
Computational analysis of both RNA-seq and RPPA data indicated activation of signaling pathways including EGFR, HOXA9, mTOR, MYC, NF-κB, and PI3K-AKT after long-term PIM inhibition in HSB-2 cells. In other cell types, the EGFR, mTOR, and PI3K-AKT pathway activity have been closely associated with PIMi resistance (36–38). However, the NF-κB activation has not previously been identified. After 48h PIMi treatment, HSB-2 cells exhibited a significant increase in the phosphorylated form of RelA (NF-κB P65) on S536. This modification denotes transcription factor activation (Figure 5F, G). As seen on the RPPA, the heightened level of NF-κB-S536 phosphorylation was maintained during long-term PIM inhibition with either AZD1208 (F2,15=24.57, p-value=1.85×10−5) or LGB321 (F2,15=17.43, p-value=0.00012) (Figure 5F, G). This result is consistent with the RNA-seq analysis which also identified increases in RelA in HSB-2-AZDR cells.
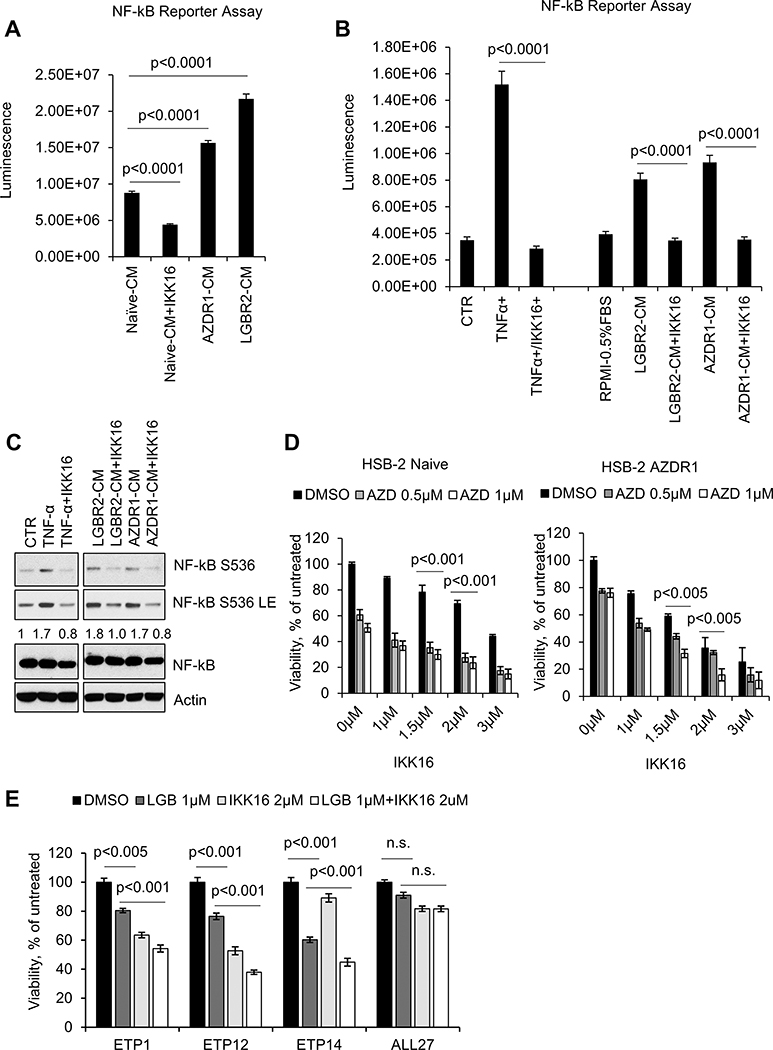
A possible mechanism that could maintain NF-κB activity in PIMi treated HSB-2 cells is the autocrine secretion of growth factors that activate signaling pathways that stimulate NF-κB. To test this possibility, conditioned media (CM) from HSB-2 naïve or AZDR1 or LGBR2 cells was incubated with HEK-293T cells stably expressing lenti-NF-κB-luc-GFP (39). The CM from long-term PIMi treated cells showed a two to three-fold increase in NF-κB activation compared to CM from naïve cells (Figure 6A and Supplementary Figure S7A). Addition of a selective IκB kinase (IKK) inhibitor, IKK16 (40,41) to the CM, inhibited activation of this reporter. TNF- α addition to these 293T cells increased NF-κB driven luciferase activity while the addition of IKK16 inhibits TNFα, AZDR1, and LGBR2 cells CM induced NF-κB reporter activity (Figure 6B). Importantly, NF-κB reporter activity was highly correlated with NF-κΒ-S536 phosphorylation levels in these experiments (Figure 6C). This result was consistent with our observation that a 60-minute treatment with a PIMi did not induce RelA-S536 phosphorylation, while overnight treatment with inhibitors did, suggesting that sufficient secretion of growth factors occurred after 18 h to activate this pathway (Supplementary Figure S7B, C). In a PIMi insensitive mature T-ALL cell line, SUP-T1 (10), AZD1208 treatment had no effect on S536 phosphorylation and did not activate the NF-κB pathway. Due to the LCK translocation in HSB-2 cells, we also validated the activation of NF-κB using PIM kinase dependent DU.528 and KOPT-K1 cell lines with no LCK translocation (Supplementary Figure S7D), further establishing that the NF-κB activation after PIM kinase inhibition is independent of LCK translocation or expression. These results suggest that, PIMi treatment in HSB-2 cells appears to be associated with the increased secretion of growth factors that in an autocrine fashion activate NF-κB signaling.
Figure 6. NF-κB activation in PIMi treated HSB-2 cells.
(A) HEK-293T-NF-κB-luc cells were incubated with conditioned media from HSB-2 naïve or AZDR1 and LGBR2 cells with or without IKK16 (2 μM) for 6h. (B & C) HEK-293T-NF-κB-luc cells were incubated with media containing TNF-α (20 ng/mL) alone or in combination with IKK16 (2 μM) or CM from HSB-2 LGBR2 or AZDR1 with or without IKK16 (2 μM) for 6h. NF-κB mediated luciferase activity was measured using One-Glo Luciferase Assay System. Cell lysates were Western blotted with the specified antibodies. LE: long exposure. (D) Cell viability of HSB-2 naïve and AZDR1 cells treated with indicated concentrations of AZD alone and/or in combination with IKK16 for 72h. (E) T-ALL PDX cells were incubated with the indicated concentrations of LGB321 (LGB) alone or in combination with IKK16 for 72h and then the percentage of viable cells was quantified by the ATPlite assay. For (D and E), the growth of DMSO control cells was considered 100% and percent cell growth after individual treatment is reported relative to the DMSO. The data shown are the average +/− S.D. of three independent experiments.
To evaluate whether the NF-κB pathway inhibition could enhance T-ALL growth suppression by PIMi, increasing doses of IKK16 were added to HSB-2 naïve or AZDR cells with and without AZD1208. IKK16 alone suppressed the growth of both naive and AZDR cells in a dose-dependent fashion (Figure 6D) but was significantly more effective in the AZDR cells. This result is consistent with the NF-κB pathway activation in long-term PIMi treated cells. In addition, when combined with AZD1208, IKK16 was able to inhibit the growth of AZDR cells to a greater extent than the naïve cells, suggesting that the role of NF-κB in driving cell growth had been increased by PIMi treatment. Notably, Combosyn analysis demonstrated that the AZD1208 and IKK16 combination was highly synergistic in suppressing the growth of AZDR as compared to naïve HSB-2 cells (Supplementary Figure S7E). Finally, the combination of PIM and NF-κB inhibitors treatment lead to a moderate but significant decrease in the viability of ETP-ALL PDXs when compared to mature T-ALL PDXs (Figure 6E). Thus, inhibition of NF-κB by small molecules can be used to enhance the growth inhibitory effect of PIMi in ETP-ALL cells.
Discussion
In this study, we demonstrate that PIM1 expression is markedly increased in ETP-ALL but not in mature T-ALL PDXs, suggesting that this kinase could be a therapeutic target to inhibit the growth of ETP-ALL. The crucial role of PIM kinases in hematopoietic tumors has fueled the development of a number of pan-PIM inhibitors which are in phase IB/II trials for multiple myeloma, myelodysplastic syndrome, myelofibrosis and lymphoma (21). Independent studies (11–13) demonstrate that PIM1 function is necessary to support T-ALL growth. In both pediatric and adult T-ALL, activation of PIM1 is driven by a rare but recurrent TCRβ-PIM1 translocation (12,13), and plays a clear role in tumor growth. In vitro and in vivo results demonstrate that PIMi reduces T-ALL growth. These results suggest that PIMi could be used to treat chemotherapy resistant ETP-ALL patients with high PIM1 expression. However, PIMi alone is not completely effective in ETP-ALL PDX models, suggesting that other agents are needed to hit complementary targets. To help define those signaling pathways, we derived PIMi resistant cells using HSB-2 cell line and analyzed the pathways that were enhanced. In addition, our previous work demonstrated that the genes expressed in the PIMi sensitive cells, including HSB-2 were highly correlated with the gene expression profile of ETP-ALL PDXs rather than the mature non-ETP ALL PDXs (10), suggesting that this cell line could serve as a model for PIMi resistant ETP-ALL.
To investigate these pathways after long-term PIMi treatment, we first analyzed the transcriptomes of PIMi resistant HSB-2 cells. RNA-seq oncogenic enrichment analysis demonstrated activation of mTOR, MYC, NF-κB, and HOXA9 pathways in long-term PIMi treated cells. PIM kinases have been implicated in the control of mTOR pathway (9,10). mTOR plays a key role in early T-lymphopoiesis and eradicating mTORC1 blocks the development of T-ALL (42). Short-term treatment of HSB-2 cells with PIMi blocks mTORC1 activation as demonstrated by S6 and 4EBP1 phosphorylation, while mTOR is continuously activated in long-term PIMi treated cells. In AML, AZD1208 resistance occurs through feedback activation of mTOR signaling, which is mediated by reactive oxygen species, p38, and AKT (37). One possible mechanism accounting for mTOR activation in PIMi resistant HSB-2 cells is the compensatory decrease in DDIT4 (REDD1) and Sestrin2 (SESN2) expression. Decreases in these proteins lead to activation of the two GTP binding proteins, Rheb and Rag, which regulate mTOR and control the activation of protein synthesis. MYC plays an important role in driving T-ALL growth and is increased by NOTCH activation. PIM kinase regulates MYC levels through phosphorylation dependent activation and PIM inhibitors are known to decrease MYC levels (43). MYC levels are also regulated by mTOR, and the constitutive activation of this enzyme in long-term PIMi treated cells could maintain the activation of this signaling pathway. NF-κB and HOXA9, positive regulators of PIM kinase expression, are often abnormally activated in AML (44,45). The activation of NF-κB and HOXA9 signaling pathways appear to be a compensatory response to the prolonged inhibition of PIM kinases.
Although many pathways are involved in the development of PIMi resistance in HSB-2 cells, this is the first demonstration of the contribution of the NF-κB pathway after long term PIMi treatment. Through RNA-seq and RPPA profiling, we found compelling evidence that PIM inhibition activates RelA (NF-kB P65) in AZDR cells compared to naïve HSB-2. PIM kinase facilitates the activation of the NF-κB pathway through phosphorylation dependent stabilization of RelA (46). The sustained elevation of phosphorylated NF-κB S536 in HSB-2 cells during PIM kinase inhibition indicates a dysfunctional regulation of the NF-κB pathway where PIM kinase inhibition fails to deactivate NF-κB. Because mTOR is an important regulator of NF-κB (47), it is possible that this kinase plays a role in the aberrant regulation. Alternatively, we demonstrate using conditioned medium that long-term PIMi treated cells produce growth factors that activate NF-κB. It is possible that one or more interleukins known to activate NF-κB, e.g. IL-6, could be secreted in increased quantities (48). The observation that concurrent inhibition of PIM kinase and NF-κB leads to reduced viability in ETP-ALL PDXs suggests that activation of this pathway is important to the survival of ETP-ALL cells. Considering that the effect of PIM and NF-κB inhibitors when administered together is modest, targeting additional pathways like PI3K/AKT and mTOR activated along with long-term PIM inhibition will be needed to completely kill T-ALL. Future in vivo PDX studies using a triple kinase inhibitor IBL-302, a compound that hits PIM/PI3K/mTOR (49) in combination with NF-kB inhibitor could potentially show a broader response.
Furthermore, our network-based analysis identified NF-κB pathway members such as ATM (ataxia telangiectasia mutated), a tyrosine kinase whose protein-level alteration has been linked extensively to leukemia (50). Likewise, the ANM suggests that LYN, PIDD1, and VCAM1 are additional NF-κB associated protein markers that differentiate between long-term and short-term PIM kinase inhibition provides further evidence that NF-κB signaling plays a significant role in PIMi resistance. Future studies focused on proteins discovered in the ANM could provide additional insight into the role of the NF-κB signaling in modulating ETP-ALL growth.
Our unbiased RNA-seq analysis and targeted RPPA assay of long-term PIMi treated HSB-2 cells demonstrated that key survival pathways are turned on with prolonged incubation with this inhibitor. These results obtained in HSB-2 cells suggest the potential of combining NF-κB and PIM inhibitors to treat a subset of T-ALL with high PIM1 expression.
Supplementary Material
Acknowledgements
The authors acknowledge the Experimental Mouse Shared Resource and Cytometry Core facility at the University of Arizona Cancer Center (UACC) for helping with in vivo experiments and flow cytometry respectively. The authors acknowledge Sammed Mandape in the UACC Bioinformatics Shared Resource for providing preliminary data analysis. The authors wish to thank the University of Arizona Genetics Core (UAGC) for providing RNA sequencing services. This research was supported by University of Arizona Cancer Center support grant P30CA023074, NIH award R01CA173200, and DOD award W81XWH-12–1-0560 (to A.S.K.).
Footnotes
Competing Interests
The authors declare no potential conflicts of interest
Data Availability
RNA-seq data is available in the Gene Expression Omnibus under accession number GSE135545.
References
- 1.Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children’s oncology group. J Clin Oncol 2012;30(14):1663–9 doi 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 2002;1(2):133–43. [DOI] [PubMed] [Google Scholar]
- 3.Cleaver AL, Beesley AH, Firth MJ, Sturges NC, O’Leary RA, Hunger SP, et al. Gene-based outcome prediction in multiple cohorts of pediatric T-cell acute lymphoblastic leukemia: a Children’s Oncology Group study. Mol Cancer 2010;9:105 doi 10.1186/1476-4598-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol 2009;10(2):147–56 doi 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481(7380):157–63 doi 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neumann M, Heesch S, Schlee C, Schwartz S, Gokbuget N, Hoelzer D, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood 2013;121(23):4749–52 doi 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 7.Maude SL, Dolai S, Delgado-Martin C, Vincent T, Robbins A, Selvanathan A, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood 2015;125(11):1759–67 doi 10.1182/blood-2014-06-580480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017;49(8):1211–8 doi 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin YW, Beharry ZM, Hill EG, Song JH, Wang W, Xia Z, et al. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood 2010;115(4):824–33 doi 10.1182/blood-2009-07-233445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padi SKR, Luevano LA, An N, Pandey R, Singh N, Song JH, et al. Targeting the PIM protein kinases for the treatment of a T-cell acute lymphoblastic leukemia subset. Oncotarget 2017;8(18):30199–216 doi 10.18632/oncotarget.16320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bock CE, Demeyer S, Degryse S, Verbeke D, Sweron B, Gielen O, et al. HOXA9 Cooperates with Activated JAK/STAT Signaling to Drive Leukemia Development. Cancer Discov 2018;8(5):616–31 doi 10.1158/2159-8290.CD-17-0583. [DOI] [PubMed] [Google Scholar]
- 12.La Starza R, Messina M, Gianfelici V, Pierini V, Matteucci C, Pierini T, et al. High PIM1 expression is a biomarker of T-cell acute lymphoblastic leukemia with JAK/STAT activation or t(6;7)(p21;q34)/TRB@-PIM1 rearrangement. Leukemia 2018;32(8):1807–10 doi 10.1038/s41375-018-0031-2. [DOI] [PubMed] [Google Scholar]
- 13.De Smedt R, Peirs S, Morscio J, Matthijssens F, Roels J, Reunes L, et al. Pre-clinical evaluation of second generation PIM inhibitors for the treatment of T-cell acute lymphoblastic leukemia and lymphoma. Haematologica 2019;104(1):e17–e20 doi 10.3324/haematol.2018.199257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan B, Zemskova M, Holder S, Chin V, Kraft A, Koskinen PJ, et al. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem 2003;278(46):45358–67 doi 10.1074/jbc.M307933200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang F, Beharry ZM, Harris TE, Lilly MB, Smith CD, Mahajan S, et al. PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells. Cancer Biol Ther 2009;8(9):846–53 doi 10.4161/cbt.8.9.8210. [DOI] [PubMed] [Google Scholar]
- 16.Warfel NA, Kraft AS. PIM kinase (and Akt) biology and signaling in tumors. Pharmacol Ther 2015;151:41–9 doi 10.1016/j.pharmthera.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen XP, Losman JA, Cowan S, Donahue E, Fay S, Vuong BQ, et al. Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc Natl Acad Sci U S A 2002;99(4):2175–80 doi 10.1073/pnas.042035699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolai S, Sia KC, Robbins AK, Zhong L, Heatley SL, Vincent TL, et al. Quantitative phosphotyrosine profiling of patient-derived xenografts identifies therapeutic targets in pediatric leukemia. Cancer Res 2016;76(9):2766–77 doi 10.1158/0008-5472.CAN-15-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keeton EK MK, Dillman KS, Palakurthi S, Cao Y, Grondine MR, Kaur S, Wang S, Chen Y, Wu A, Shen M, Gibbons FD, Lamb ML, Zheng X, Stone RM, Deangelo DJ, Platanias LC, Dakin LA, Chen H, Lyne PD, Huszar D. AZD1208, a potent and selective pan-Pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia PD, Langowski JL, Wang Y, Chen M, Castillo J, Fanton C, et al. Pan-PIM kinase inhibition provides a novel therapy for treating hematologic cancers. Clin Cancer Res 2014;20(7):1834–45 doi 10.1158/1078-0432.CCR-13-2062. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Song M, Kundu JK, Lee MH, Liu ZZ. PIM Kinase as an Executional Target in Cancer. J Cancer Prev 2018;23(3):109–16 doi 10.15430/JCP.2018.23.3.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiarini F, Lonetti A, Evangelisti C, Buontempo F, Orsini E, Evangelisti C, et al. Advances in understanding the acute lymphoblastic leukemia bone marrow microenvironment: From biology to therapeutic targeting. Biochim Biophys Acta 2016;1863(3):449–63 doi 10.1016/j.bbamcr.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Song JHP SKR; Luevano LA; Minden MD; DeAngelo DJ; Hardiman G; Ball LE; Warfel NA; Kraft AS Insulin Receptor Substrate 1 Is a Substrate of the Pim Protein Kinases. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 2014;46(4):364–70 doi 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright DD, Sefton BM, Kamps MP. Oncogenic activation of the Lck protein accompanies translocation of the LCK gene in the human HSB2 T-cell leukemia. Mol Cell Biol 1994;14(4):2429–37 doi 10.1128/mcb.14.4.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horiuchi D, Camarda R, Zhou AY, Yau C, Momcilovic O, Balakrishnan S, et al. PIM1 kinase inhibition as a targeted therapy against triple-negative breast tumors with elevated MYC expression. Nat Med 2016;22(11):1321–9 doi 10.1038/nm.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braso-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, et al. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med 2016;22(11):1303–13 doi 10.1038/nm.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song JH, Padi SK, Luevano LA, Minden MD, DeAngelo DJ, Hardiman G, et al. Insulin receptor substrate 1 is a substrate of the Pim protein kinases. Oncotarget 2016;7(15):20152–65 doi 10.18632/oncotarget.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee S, Chakraborty P, Daenthanasanmak A, Iamsawat S, Andrejeva G, Luevano LA, et al. Targeting PIM Kinase with PD1 Inhibition Improves Immunotherapeutic Antitumor T-cell Response. Clin Cancer Res 2019;25(3):1036–49 doi 10.1158/1078-0432.CCR-18-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bueso-Ramos CE, Rocha FC, Shishodia S, Medeiros LJ, Kantarjian HM, Vadhan-Raj S, et al. Expression of constitutively active nuclear-kappa B RelA transcription factor in blasts of acute myeloid leukemia. Hum Pathol 2004;35(2):246–53. [DOI] [PubMed] [Google Scholar]
- 32.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol 2007;9(8):932–44 doi 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 33.Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst 2018;6(3):282–300 e2 doi 10.1016/j.cels.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607–D13 doi 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics 2002;18 Suppl 1:S233–40 doi 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 36.Silverbush D, Grosskurth S, Wang D, Powell F, Gottgens B, Dry J, et al. Cell-Specific Computational Modeling of the PIM Pathway in Acute Myeloid Leukemia. Cancer Res 2017;77(4):827–38 doi 10.1158/0008-5472.CAN-16-1578. [DOI] [PubMed] [Google Scholar]
- 37.Brunen D, Garcia-Barchino MJ, Malani D, Jagalur Basheer N, Lieftink C, Beijersbergen RL, et al. Intrinsic resistance to PIM kinase inhibition in AML through p38alpha-mediated feedback activation of mTOR signaling. Oncotarget 2016;7(25):37407–19 doi 10.18632/oncotarget.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aziz AUR, Farid S, Qin K, Wang H, Liu B. PIM Kinases and Their Relevance to the PI3K/AKT/mTOR Pathway in the Regulation of Ovarian Cancer. Biomolecules 2018;8(1) doi 10.3390/biom8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson AA, Kwok LW, Porter EL, Payne JG, McElroy GS, Ohle SJ, et al. Lentiviral delivery of RNAi for in vivo lineage-specific modulation of gene expression in mouse lung macrophages. Mol Ther 2013;21(4):825–33 doi 10.1038/mt.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duckworth C, Zhang L, Carroll SL, Ethier SP, Cheung HW. Overexpression of GAB2 in ovarian cancer cells promotes tumor growth and angiogenesis by upregulating chemokine expression. Oncogene 2016;35(31):4036–47 doi 10.1038/onc.2015.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waelchli R, Bollbuck B, Bruns C, Buhl T, Eder J, Feifel R, et al. Design and preparation of 2-benzamido-pyrimidines as inhibitors of IKK. Bioorg Med Chem Lett 2006;16(1):108–12 doi 10.1016/j.bmcl.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 42.Hoshii T, Kasada A, Hatakeyama T, Ohtani M, Tadokoro Y, Naka K, et al. Loss of mTOR complex 1 induces developmental blockage in early T-lymphopoiesis and eradicates T-cell acute lymphoblastic leukemia cells. Proc Natl Acad Sci U S A 2014;111(10):3805–10 doi 10.1073/pnas.1320265111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beharry Z, Mahajan S, Zemskova M, Lin YW, Tholanikunnel BG, Xia Z, et al. The Pim protein kinases regulate energy metabolism and cell growth. Proc Natl Acad Sci U S A 2011;108(2):528–33 doi 10.1073/pnas.1013214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu N, Ramirez LM, Lee RL, Magnuson NS, Bishop GA, Gold MR. CD40 signaling in B cells regulates the expression of the Pim-1 kinase via the NF-kappa B pathway. J Immunol 2002;168(2):744–54 doi 10.4049/jimmunol.168.2.744. [DOI] [PubMed] [Google Scholar]
- 45.Hu YL, Passegue E, Fong S, Largman C, Lawrence HJ. Evidence that the Pim1 kinase gene is a direct target of HOXA9. Blood 2007;109(11):4732–8 doi 10.1182/blood-2006-08-043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nihira K, Ando Y, Yamaguchi T, Kagami Y, Miki Y, Yoshida K. Pim-1 controls NF-kappaB signalling by stabilizing RelA/p65. Cell Death Differ 2010;17(4):689–98 doi 10.1038/cdd.2009.174. [DOI] [PubMed] [Google Scholar]
- 47.Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes Dev 2008;22(11):1490–500 doi 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 2001;98(8):2301–7 doi 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- 49.Mohlin S, Hansson K, Radke K, Martinez S, Blanco-Aparicio C, Garcia-Ruiz C, et al. Anti-tumor effects of PIM/PI3K/mTOR triple kinase inhibitor IBL-302 in neuroblastoma. EMBO Mol Med 2020;12(1):e11749 doi 10.15252/emmm.201911749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 2006;311(5764):1141–6 doi 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.