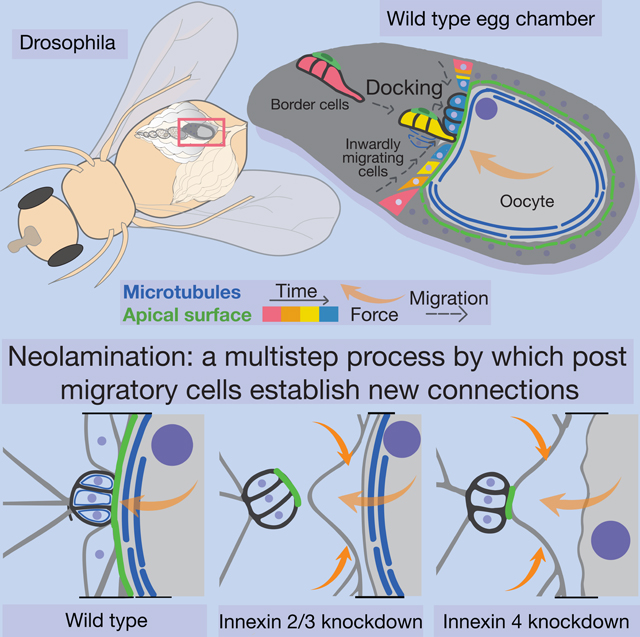
Summary
During embryonic development and cancer metastasis, migratory cells must establish stable connections with new partners at their destinations. Here we establish the Drosophila border cells as a model for this multistep process. During oogenesis, border cells delaminate from the follicular epithelium and migrate. When they reach their target, the oocyte, they undergo a stereotypical series of steps to adhere to it, then connect with another migrating epithelium. We identify gap-junction-forming Innexin proteins as critical. Surprisingly, the channel function is dispensable. Instead, Innexins 2 and 3 function within the border cells, and Innexin 4 functions within the germline, to regulate microtubules. The microtubule-dependent border cell-oocyte interaction is essential to brace the cells against external morphogenetic forces. Thus, we establish an experimental model and use genetic, thermogenetic, and live imaging approaches to uncover the contributions of Innexins and microtubules to a cell biological process important in development and cancer.
Keywords: morphogenesis, neolamination, cell adhesion, cell migration, gap junction, channel-independent, innexin, microtubule, Drosophila, oogenesis, border cells
eTOC Blurb
In normal development and cancer, migratory cells must establish stable connections with new partners at their destinations. Here, Miao et al. establish the Drosophila border cells as a model for this multistep process and uncover essential channel-independent contributions of innexins and microtubules.
Graphical abstract
Introduction
As embryos develop, cells move as individuals, in small clusters, or by the hundreds in fluid like masses (reviewed in Scarpa and Mayor, 2016). Despite this churning, selective associations allow cells to connect dynamically with some cells during their migrations, and stably with specific partners when they reach their destinations (Mishra et al., 2019). The molecular programs that activate motility and govern chemotaxis have been elucidated in detail (Devreotes and Horwitz, 2015). Less clear are the mechanisms by which migratory cells integrate selectively into tissues once they reach their destinations. In most cases, including gastrulation, neural crest development, and primordial germ cell migration, amongst many others, specific cell-cell contacts must be established with select partners. Once established, new contacts must also be maintained despite ongoing pushing, pulling, stretching, and shearing associated with continued morphogenesis. Establishment of new cell-cell contacts is also an essential step when tumor cells spread to new sites (Massagué and Obenauf, 2016; Obenauf and Massagué, 2015). However, this critical step in metastasis is extremely challenging to image live at high resolution as it is not known in advance precisely where cells will settle.
Here we use the border cells in the Drosophila ovary, a pioneer model of collective chemotaxis, and establish it as an experimental model system to study the multistep process by which they establish new and stable connections after they migrate. In addition to describing the cell biological steps, we report the identification of gap junction forming proteins - Innexins 2, 3, and 4 – as critical for the process. Multiple lines of evidence suggest that the channel functions are dispensable and identify microtubule stabilization as a necessary function of these proteins. The results support a model whereby a stable border cell/oocyte interaction braces the cell-cell interface against external morphogenetic forces, which in the absence of innexins dislodge the border cells from their new partners.
Results
Border cell neolamination is a multi-step process
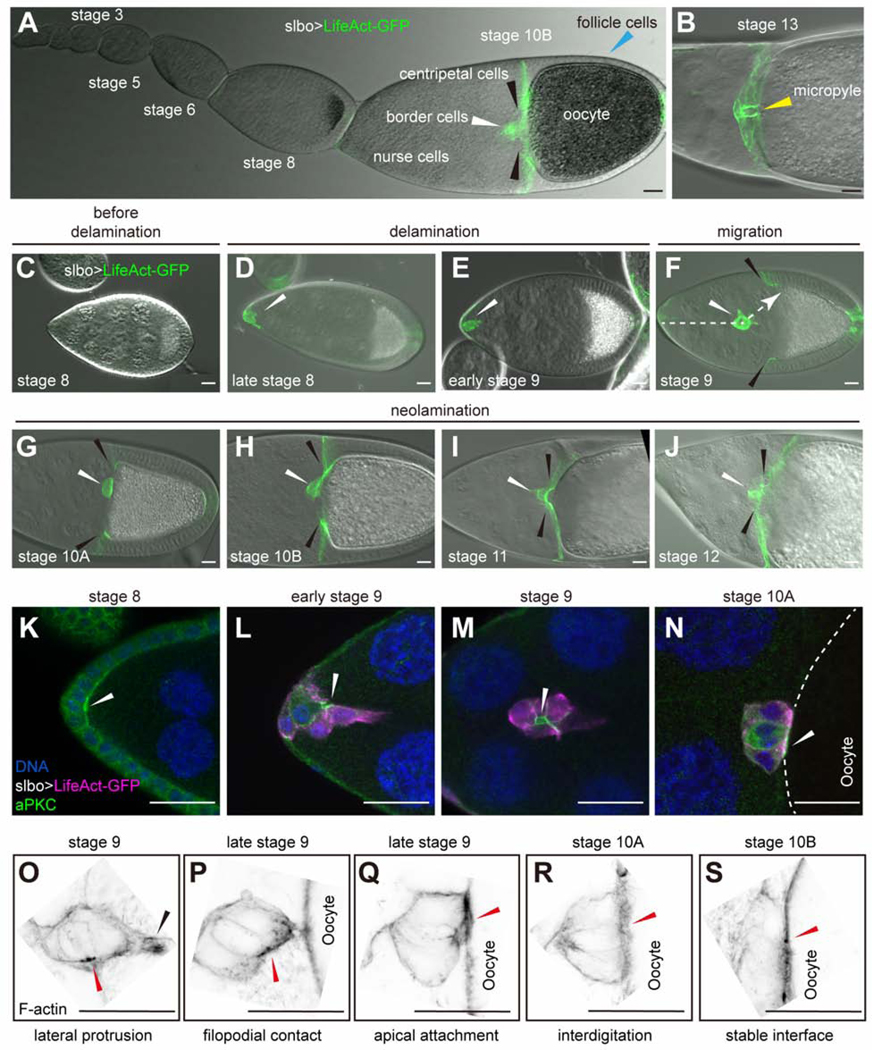
Border cells in the Drosophila ovary have long served as a pioneering model for collective chemotaxis (Montell et al., 1992, 2012), a process now appreciated to drive metastasis of a variety of cancers (Cheung and Ewald, 2016; Friedl et al., 2004). The Drosophila ovary is composed of strings of egg chambers of increasing maturity (King, 1970). Egg chambers are composed of 16 germline cells - 15 nurse cells and one oocyte - surrounded by a monolayer of epithelial follicle cells (Figures 1A, 1B and S1A). During stage 9, a group of 6–8 follicle cells, out of ~850, delaminates from the anterior epithelium, migrates ~150 μm in between the nurse cells (Figures 1C-1F). The cluster arrives at the dorsal/anterior oocyte border by stage 10A (Figure 1G, S1B and S1C). During stage 10B, outer follicle cells at the oocyte/nurse cell boundary begin migrating inward, or centripetally, and are thus called centripetal follicle cells (cfcs) (Figures 1H-1J and S1A-S1E). In cross-section, as they migrate inward, cfcs resemble a closing diaphragm (Figures S1C’-S1E”‘). The oocyte and nurse cells derive from a common precursor cell that undergoes four rounds of cell division with incomplete cytokinesis to produce a 16-cell cyst. Thus, the ring canals - F-actin-rich, stabilized cleavage furrows - connect the nurse cells to each other and to the oocyte (Figure S1). As the cfcs migrate inward, the four ring canals between the oocyte and adjacent nurse cells slide inward (Figures S1C-S1E”‘).
Figure 1.
Normal ovarian development, border cell migration, and neolamination
Images of slbo-Gal4, UAS-LifeAct-GFP egg chambers. (A) An ovariole showing stages of egg chamber development through stage 10B. (B) Stage 13 egg chamber showing the micropyle (yellow arrowhead). (C-J) Stages 8–12 egg chambers including before (C) and after (D) border cell specification, during delamination (E), migration (F) and neolamination (G-J). White arrowheads indicate border cell clusters. Black arrowheads indicate centripetal cells. Dashed arrow indicates the migration path. (K-N) Anti-aPKC staining (green) labels apical surfaces. White arrowheads indicate the apical side of the border cell cluster. (O-S) Super-resolution images of stage 9–10B egg chambers showing the initial steps of neolamination. F-actin staining (black) of border cells attaching to the oocyte. Black arrowhead indicates the border cell protrusion. Red arrowheads indicate the apical side of border cell cluster. Scale bars: 20 μm.
Border cells retain epithelial character as they migrate (Niewiadomska et al., 1999) (Figures 1K-1M). Notably, as they delaminate, they undergo a ~90° turn so that their apical surfaces orient roughly perpendicular to their direction of migration (Figures 1K and 1L). When they reach the oocyte, they turn again (Niewiadomska et al., 1999; Figures 1M and 1N) to attach their apical surfaces to the oocyte (Figure 1N and Figure S1B).
Airyscan confocal microscopy revealed a multistep process by which border cells connect to the oocyte. As the cluster approaches the oocyte, a single border cell protrudes with its lateral surface (Figure 1O) and makes initial contact (Figure 1P), then each border cell docks its apical surface onto the oocyte (Figure 1Q) to establish a stable interface (Figures 1R, 1S and S1B). During cfc migration, one border cell first makes contact with one dorsal cfc (Figures 1H and S1D-S1D”). Upon completion of cfc migration, border cells and cfcs form a continuous epithelial layer and cooperate to build the eggshell structure known as the micropyle (Figure 1B), which is essential for sperm entry and so for fertility (King, 1970; Montell et al., 1992). To be concise, we refer to the process by which the border cells establish and maintain contacts with the oocyte and then cfcs as neolamination, complementary to the established term delamination.
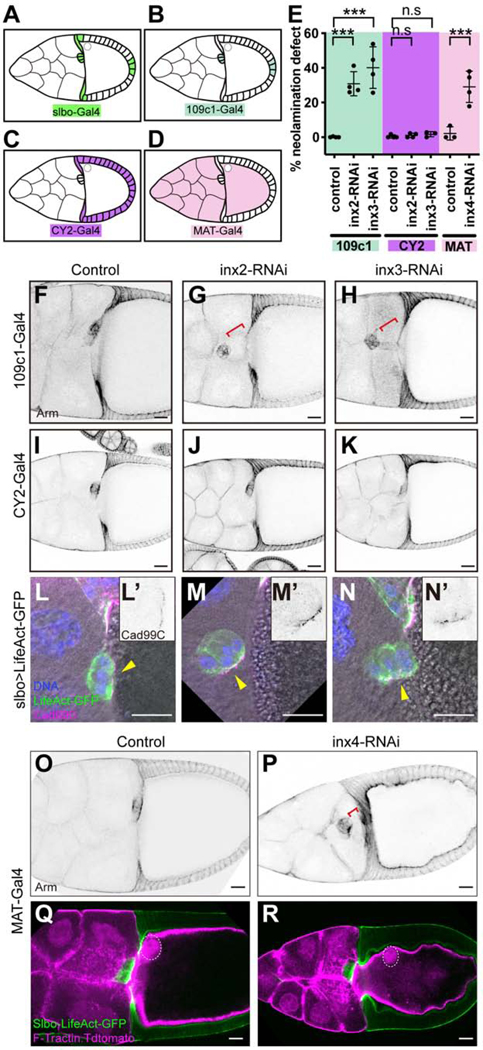
Innexins 2 and 3 are required for neolamination
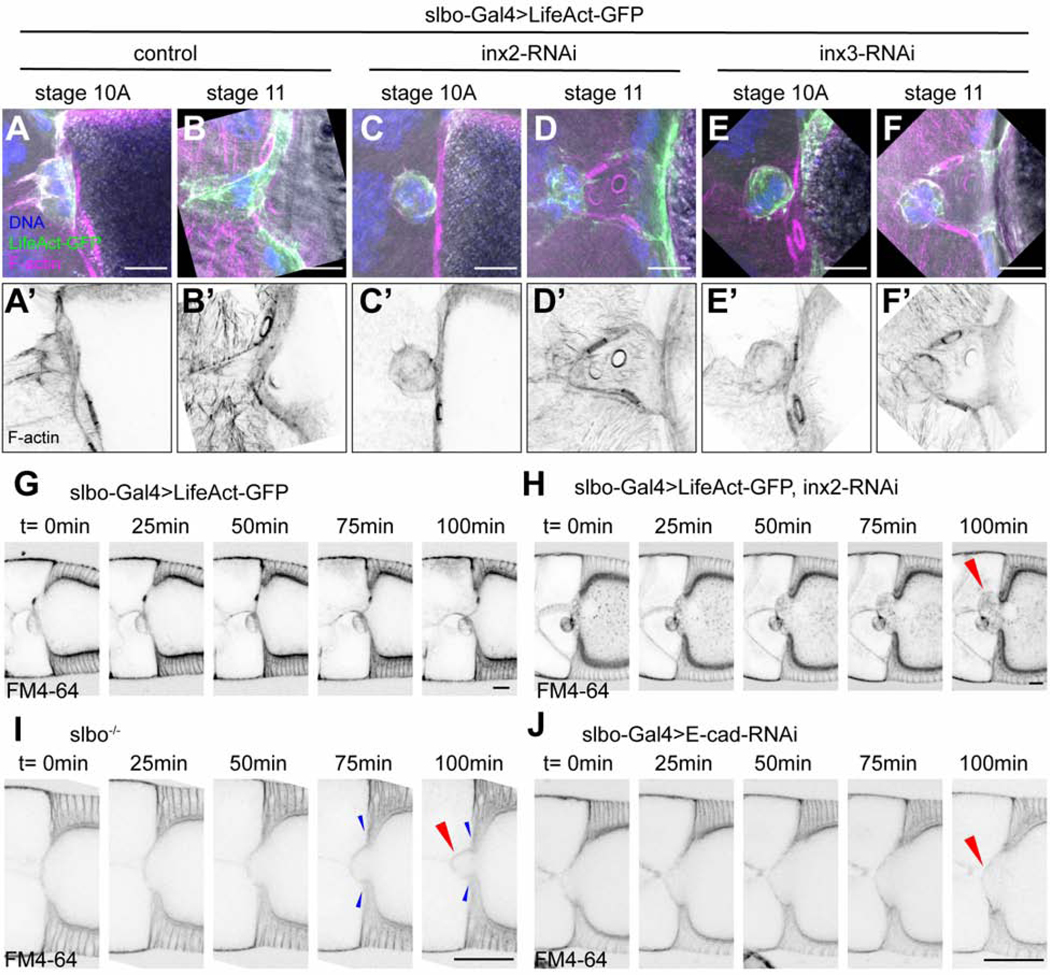
By microarray analysis, we previously identified mRNAs enriched in border cells and/or cfcs (Wang et al., 2006). To elucidate their functions, we screened corresponding UAS-RNAi lines for effects on border cells, using slbo-Gal4, which expresses in border cells and cfcs (Figures 1A-1J and 2A). We noticed a striking neolamination defect upon knockdown of Innexin (Inx)2 or Inx3 (Figures 2A-2I), but not other Innexins (Figure S2A). Innexins are gap junction forming proteins and the functional analogs of vertebrate connexins [reviewed in (Skerrett and Williams, 2017); Figures 2K-2N]. As in controls (Figure 2A), slbo-Gal4 egg chambers expressing either UAS-inx2-RNAi or UAS-inx3-RNAi, border cells migrated normally and reached the oocyte border by stage 10A (Figures 2B and 2C). However, as development proceeded Inx2- or Inx3-depleted border cells failed to connect to cfcs and eventually ended up dislodged from the oocyte (Figures 2D-2J). Live imaging confirmed that border cells reached the oocyte normally but failed to connect with cfcs, and were subsequently displaced (Figure S2B; Video S1).
Figure 2.
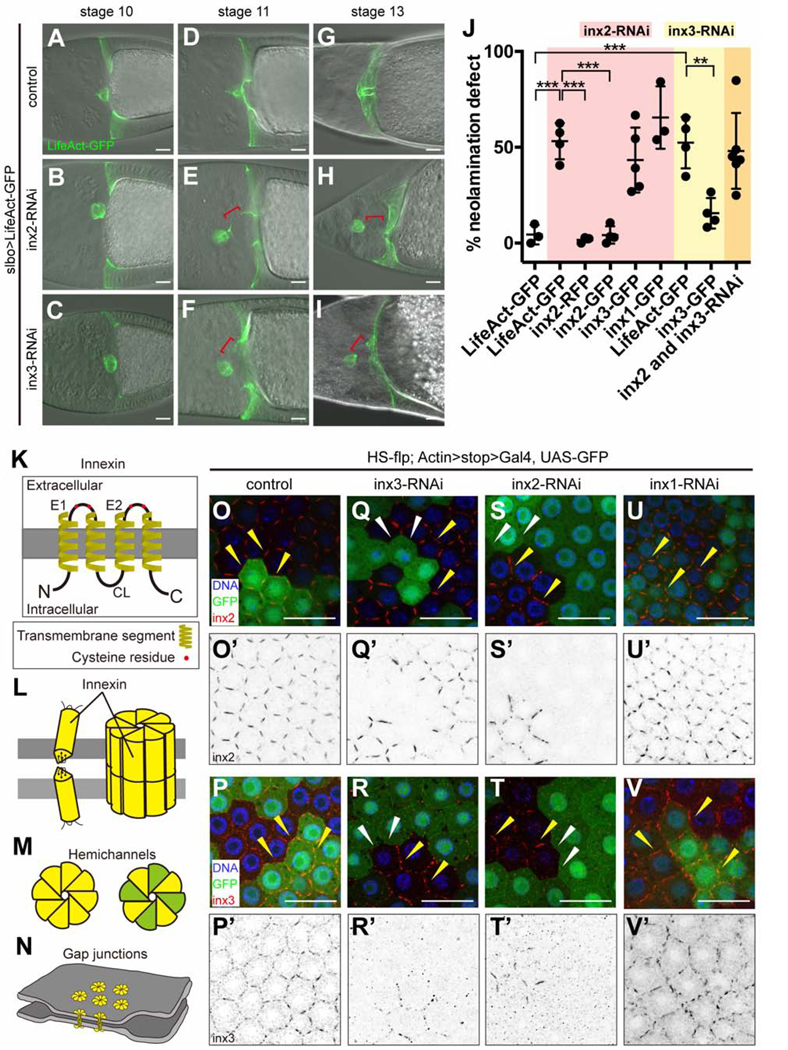
Neolamination defect caused by Inx2/3-RNAi
(A-I) Images of egg chambers in which slbo-Gal4 drives UAS-LifeAct-GFP alone (slbo>LifeAct-GFP crossed to w1118 is the control) (A-C); in combination with inx2-RNAi (D-F); or in combination with inx3-RNAi (G-I). Red brackets indicate abnormal spaces between border cells and cfcs. (J) Quantification of the frequency of neolamination defects in inx2/3-RNAi and the rescue effect of inx2-GFP in stage 10 egg chambers. Each dot represents an independent experiment (see Methods). **p < 0.01; ***p < 0.001. (K-N) Schematic of the structure of innexin monomers, which span the membrane four times (K); monomers assemble into gap junction channels (L); innexin hemichannels can be homomeric (all yellow) or heteromeric (yellow and green) (M); gap junctions between cells pull the cells into close association (N). (O-V) HS-flp-out clones showing inx2/3-RNAi efficiency in follicle cells. GFP labels innexin-RNAi-expressing cells, except in the control. Anti-inx2 staining of control (w1118) (O-O’), inx3-RNAi (Q-Q’), inx2-RNAi (S-S’) and inx1-RNAi (U-U’). Anti-inx3 staining of control (w1118) (P-P’), inx3-RNAi (R-R’), inx2-RNAi (T-T’) and inx1-RNAi (V-V’). Yellow arrowheads indicate the presence of Inx2/3 staining. White arrowheads indicate the loss of Inx2/3 staining. Scale bars: 20 μm.
Co-expression of UAS-inx2-RFP or UAS-inx2-GFP rescued the UAS-inx2-RNAi phenotype (Figures 2J and S2C-C”). Neither UAS-inx3-GFP nor UAS-inx1-GFP rescued UAS-inx2-RNAi (Figure 2J). The rescue specificity demonstrates that Inx2 and Inx3 have separable functions; it is not simply the overall level of Innexin proteins that is important. Innexins, like the vertebrate connexins, are proteins that span the plasma membrane four times, have conserved cysteine residues in the extracellular loops, and cytosolic tails that interact with other proteins (Figure 2K). Innexins, like connexins, assemble into multimeric hemichannels (Skerrett and Williams, 2017), which can be homomeric (composed of one type of Innexin subunit) or heteromeric (Figures 2L and 2M). Hemichannels on adjacent cells can interact to form homotypic or heterotypic gap junction channels (Figures 2L-2N). Knockdown of either Inx2 or Inx3 caused a similar defect in ~ 50% of egg chambers (Figure 2J). Co-expression of both RNAi lines together did not significantly worsen the phenotype (Figure 2J), suggesting that knockdown of either protein eliminates the function of both, which is consistent with the known ability of these two proteins to co-assemble (Lehmann et al., 2006).
To validate the knockdown efficiency and characterize the expression patterns of Innexins 2 and 3, we used antibodies (Lehmann et al., 2006; Smendziuk et al., 2015) to stain mosaic clones in the follicular epithelium, and compared neighboring RNAi-expressing and non-expressing cells. In control clones that expressed GFP but no RNAi, clonal and non-clonal cells showed uniform staining for Inx2 (Figures 2O and 2O’) and Inx3 (Figures 2P and 2P’) at cell-cell boundaries (Figure 2N). In contrast, cells expressing the inx3-RNAi line (BDSC:60112) showed little if any staining for either Inx2 (Figures 2Q and 2Q’) or Inx3 (Figures 2R and 2R’) at cell-cell boundaries. Similarly, inx2-RNAi resulted in loss of both Inx2 (Figures 2S and 2S’) and Inx3 (Figures 2T and 2T’) from cell-cell contacts. In contrast, an inx3 RNAi line that did not cause a phenotype (BDSC:60887) caused less effective knockdown than the phenotypic line (Figure S2D-S2E’). These results show that the Inx2 and Inx3 RNAi lines that caused neolamination defects effectively knocked down expression.
Inx2 co-assembles with Inx3 in some cells and with Inx1 in others (Giuliani et al., 2013; Holcroft et al., 2013; Richard et al., 2017; Spéder and Brand, 2014; Stebbings et al., 2000). However, Inx1 RNAi did not affect Inx2 or Inx3 localization or abundance (Figures 2U-2V’) even though the RNAi was effective (Figure S2F-S2G’). These results indicate that Inx2 and Inx3, but not Inx1, co-assemble in follicle cells, consistent with an earlier report showing that Inx1 localizes basally, whereas Inx2 localizes apico-laterally in follicle cells (Bohrmann and Zimmermann, 2008).
At boundaries between GFP+ (RNAi-expressing) and GFP− (wild-type) cells, staining for Inx2 and Inx3 was also absent, even from wild-type cells (Figures 2Q-2T’, white arrowheads), which had normal staining at contacts with other wild-type cells (Figures 2Q-2T’, yellow arrowheads). Inx1 RNAi did not have the same effect (Figures 2U-2V’). These results show that Inx2 and Inx3 require one another, but not Inx1, both cell-autonomously and on adjacent cells.
Inx2 and Inx3 function in the border cells not cfcs during neolamination
Since Slbo-Gal4 is expressed in both border cells and cfcs (Figures 1, 2 and 3A), a simple hypothesis would be that gap junction formation between these two epithelial cell types is an essential step in neolamination. If so, Innexins 2 and 3 should be required in both border cells and cfcs. To address which cells require the Innexins, we identified a Gal4 line, 109c1-Gal4, which expresses in the border cells but not cfcs, beginning just when border cells are specified and continuing throughout their migration and neolamination (Figures 3B and S3A). When combined with UAS-inx2-RNAi or UAS-inx3-RNAi, 109c1-Gal4 caused two types of border cell defects. First, 50–60% of egg chambers showed incomplete migration to the oocyte at late stage 9/early stage 10, compared to controls (Figure S3E), presumably due to the earlier onset of 109c1-Gal4 expression compared to slbo-Gal4. Second, at stage 10B, few border cell clusters (6% for inx2-RNAi and 19% for inx3-RNAi) associated with the oocyte (Figure S3F), indicating that most clusters that had reached the oocyte by late stage 9/early stage 10 were subsequently displaced (Figures 3E-3H). We confirmed the neolamination defect by live imaging (Figure S4A; Video S2). Border cells that reached the oocyte failed to connect with cfcs. In contrast, when Inx2 or Inx3 was knocked down in cfcs but not border cells using CY2-Gal4 (Figures 3C and S3B), which we confirmed by antibody staining (Figure S3D), no neolamination defect was evident (Figures 3E and 3I-3K). We conclude that Innexins 2 and 3 function in the border cells, but not in cfcs, to promote neolamination.
Figure 3.
Cell type specific knockdown of Innexins 2, 3 and 4
(A-D) Schematic of the expression pattern of slbo-Gal4 (A), 109c1-Gal4 (B), CY2-Gal4 (C) and MAT-α-tub-Gal4 (D). (E) Quantification of the frequency of neolamination defect in CY2-Gal4>inx2/3-RNAi, 109c1-Gal4>inx2/3-RNAi and MAT-α-tub-Gal4>inx4-RNAi stage 10B egg chambers. Each dot is a result of an independent experiment. ***p < 0.001 (F-K) Images of 109c1-Gal4 (F-H) and CY2-Gal4 (I-K) driven LifeAct-GFP (control), inx2-RNAi and inx3-RNAi egg chambers. (L-N) Cad99C is a marker of apical microvilli (Glowinski et al., 2014). Staining shows normal apical/basal polarity in border cells lacking Inx2 or Inx3 but a failure of the cluster to turn the apical surface toward the oocyte (yellow arrowheads). (L’-N’) Cad99C single channel shows the localization of Cad99C in the border cell cluster. (O-P) Images of MAT-α-tub-Gal4 driven UAS-inx4-RNAi egg chambers (w1118 was used as the control). (Q-R) Images of MAT-α-tub-Gal4>F-Tractin.Tdtomato, slbo-LifeAct-GFP driven inx4-RNAi egg chambers showing inx4-RNAi phenotypes (w1118 was used as the control). Oocyte nuclei are marked with white dashed circles. Red brackets indicate neolamination defects. Scale bars: 20 μm.
Consistent with this result, high magnification imaging revealed an abnormality prior to the border cell-cfc interaction. In cells with slbo-Gal4 driving inx2 or 3 RNAi, the cluster lateral surface made an initial contact with the oocyte, but failed subsequently to turn and dock the apical cell surfaces, labeled with Cadherin 99C (Cad99C) (Figures 3L-3N’). These results suggest that Inx2/3 knockdown perturbs the border cell-oocyte interaction first.
Inx4 is required in the oocyte for normal neolamination
Since the interaction between border cells and the oocyte was disrupted in the Inx2 and Inx3 knockdowns, we thought that border cells might form gap junctions with the oocyte. Multiple innexin mRNAs are expressed in the germline of stage 9/10 egg chambers (Stebbings et al., 2002), although Inx4 [aka zero population growth (zpg)], is the only one with an established function (Tazuke et al., 2002). It is required early in oogenesis for germline stem cell differentiation (Gilboa et al., 2003) and egg chamber formation (Mukai et al., 2011). To determine if Inx4 was required for neolamination, it was necessary to knock it down after egg chambers formed. We used Mat-α-tub-Gal4 to drive UAS-inx4-RNAi from stage ~3 (Figures 3D and S3C) and observed a phenotype that included neolamination defects (Figures 3E). Border cells migrated normally to the oocyte by stage 10A (Figure S3E); but at stage 10B, they were mispositioned in ~30% of Mat-α-tub-Gal4; UAS-inx4-RNAi egg chambers, indicative of a neolamination defect (Figures 3E, 3O, 3P, S3F and S4B; Video S3). In addition, Inx4 knockdown caused mispositioning of the oocyte nucleus and irregularities in the oocyte cortex (Figures 3Q and 3R). The oocyte nucleus is normally found in the dorsal/anterior region of the oocyte, close to the plasma membrane and adjacent to the border cells (Figure 3Q). However, in ~40% of Mat-α-tub-Gal4; UAS-inx4-RNAi egg chambers, the nucleus was displaced (Figure 3R). In addition, the normally smooth oocyte cortex (Figures 3O and 3Q), appeared irregular (Figures 3P and 3R). We conclude that Inx4 is required in the germline for border cell neolamination, oocyte nucleus positioning, and maintenance of the oocyte cortex. No other Inx RNAi line caused any detectable defect (Figure S3G) even when expressed with TripleGal4, which expresses in the germline at high levels and early in development.
Structural and functional analysis of gap junctions in egg chambers
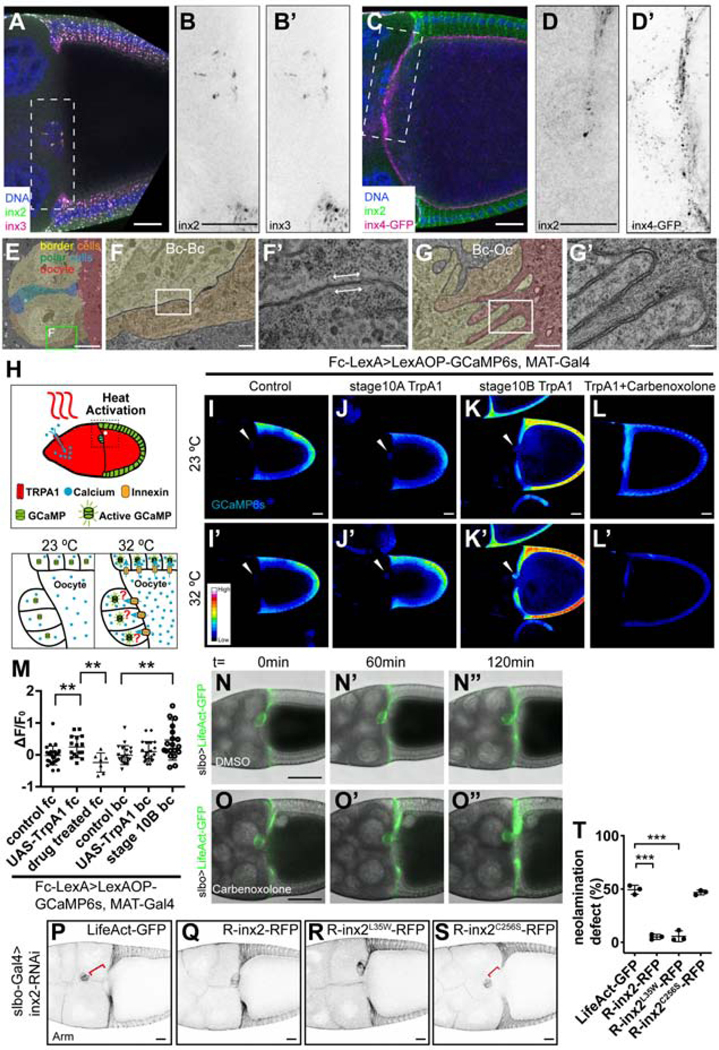
To determine where Innexins 2, 3, and 4 localize in stage 10 egg chambers, we used immunolocalization. Innexins 2 and 3 were enriched at follicle cell/follicle cell contacts (Figures 4A and S5A-S5A”), including within the border cell cluster (Figures 4A, 4B, 4B’ and S5B; Video S4). Double labeling experiments showed significant co-localization between Inx2 and Inx3 in all follicle cells (Pearson’s R value: 0.78±0.09, n=4), including border cells (Pearson’s R value: 0.77±0.08, n=4). Innexin 4 localized on all germ cell membranes (Figures 4C and S5C-S5C”; Video S4), but showed little co-localization with Inx2 either at the oocyte-border cell interface (Pearson’s R value: 0.09±0.06, n=5) (Figures 4D, 4D’ and S5D) or at the contact zone between the follicular epithelium and the oocyte (Pearson’s R value: 0.02±0.13, n=4).
Figure 4.
Distribution of structural and functional gap junctions between egg chamber cells
(A) Image of a stage 10 w1118 egg chamber co-stained with anti-inx2 and inx3. (B-B’) Magnified view of the region outlined in(A). (B) Inx2 single channel. (B’) Inx3 single channel. (C) Inx4-GFP-expressing egg chamber stained with anti-inx2. (D-D’) Magnified view of the region outlined in (C). (D) Inx2 single channel. (D’) Inx4-GFP single channel. Scale bars: 20 μm. (E-G’) TEM images of a stage 10 egg chamber. (E) Border cells are pseudo-colored yellow/orange, polar cells green and blue, and the oocyte red. (F) Magnified views of the box in (M). (F’-G’) Magnified view of (F-G). (F) Gap junction between two border cells. (G) Plasma membrane interdigitations between border cells and oocyte from a parallel section to the one shown in (M). TEM magnifications: (E) x1.2k, (F) x7k, (G) x12k, (F’,G’) x30k. Scale bars: (E) 5 μm (F-G) 500 nm (F’-G’) 200 nm. (H) Schematic of the thermo-genetics experiments. (I-L’) Heat map images showing results of thermo-genetics experiments. Images are pseudo-colored (using 16 color RGB in FIJI) to show the level of GCaMP6s fluorescence. (I-I’) Stage 10A control(w1118), (J-J’) Stage 10A TrpA1, (K-K’) Stage 10B TrpA1, (J-J’) Stage 10B TrpA1 with carbenoxolone treatment. White arrowheads indicate the border cell clusters. (M) Quantification of the level of GCaMP6s in thermo-genetics experiments in follicle cells (fc) and border cells (bc). Each dot is an egg chamber. **p < 0.01. Scale bars: 20 μm. (N-O”) Snapshots of DMSO (N-N”) and carbenoxolone treated (O-O”) slbo-Gal4>LifeAct-GFP egg chambers. Time (t) is relative to the start of live imaging. Scale bars: 50 μm. (P-S) Anti-Arm staining egg chambers show the rescue effects of UAS-R-inx2-RFP constructs. (P) LifeAct-GFP (control); (Q) UAS-R-inx2-RFP; (R) UAS-R-inx2L35W-RFP; (Q) UAS-R-inx2C256S-RFP. Scale bars: 20 μm. (T) Quantification of the neolamination defect in (P-S). ***p < 0.001.
Gap junctions provide close contact between adjacent cells, leaving only a 30–40Å gap. In case the light-microscopy did not provide sufficient resolution, we carried out transmission electron microscopy of stage 10 egg chambers (Figures 4E-4G’ and S5E-S5G’). Gap junctions were identified as zones 100–200nm long, where adjacent plasma membranes were tightly apposed. We readily detected gap junctions at border cell-border cell contacts (Figures 4F and 4F’), between border cells and polar cells, which are a pair of non-migratory cells in the center of the border cell cluster (Figures S5E, S5F and S5F’), and between other cell types (Figures S5E, S5G and S5G’). We detected no gap junctions between the border cells and oocyte, despite the presence of clear interdigitations there (Figures 4G and 4G’).
Gap junction proteins are best-known for their ability to form constitutively open channels between cells allowing passage of molecules <1–3 kilodaltons (Krysko et al., 2005; Skerrett and Williams, 2017). To examine where functional channels between cells could be detected, we took a “thermogenetics” approach. We expressed the thermosensitive, Ca2+-permeable, cation channel TrpA1-A in the germline, under the control of Mat-α-tub-Gal4. We also expressed GCaMP6s, a fluorescent [Ca2+] sensor, in all follicle cells including border cells using the LexA/LexAop system (Figure 4H). The TrpA1-A channel opens at temperatures >27°C (Viswanath et al., 2003). Therefore, we expected a temperature-dependent influx of cations including Ca2+ into the germline from the culture medium in Mat-α-tub-Gal4; UAS-trpA1 expressing egg chambers. Since gap junction channels have been reported to exist between the oocyte and follicle cells based on dye transfer (Bohrmann and Zimmermann, 2008), we expected a temperature-dependent rise in GCaMP fluorescence in Mat-α-tub-Gal4;UAS-trpA1 expressing LexAop-GCaMP under the control of a follicle cell LexA driver (Figure 4H). In control egg chambers lacking TrpA1 expression, no increase in GCaMP fluorescence occurred upon temperature shift from 23°C (Figure 4I) to 32°C (Figure 4I’). As predicted, in the presence of germline TrpA1 expression, GCaMP fluorescence increased in the follicular epithelium upon temperature shift from 23°C (Figure 4J) to 32°C (Figure 4J’; Video S5). However, we did not detect any [Ca2+] rise in border cells at stage 10A, despite their close contact with the oocyte (Figures 4I-4I’ and 4J-4J’, white arrowheads). Interestingly, at stage 10B, when border cells connected to cfcs, we detected a temperature- and TrpA1-dependent [Ca2+] rise in the border cells (Figures 4K-4K’ and 4M, white arrowheads), demonstrating that we were able to detect changes in [Ca2+] in border cells using GCaMP6s. The temperature- and TrpA1-dependent [Ca2+] rise in the follicular epithelium was eliminated upon treatment with the gap junction inhibitor carbenoxolone (Figures 4L and 4L’). Together these results, quantified in Figure 4M, suggest that border cells do not form functional gap junctions with the oocyte at the time of border cell/oocyte neolamination, though the rest of the follicular epithelium does; the border cells eventually do form functional gap junctions either with the oocyte or with cfcs but only later.
Although border cells do not form detectable structural or functional gap junctions with the oocyte as they form a stable connection, we wondered whether gap junction channels between individual border cells might be required for neolamination or whether hemi-channels might play a role. Hemichannels allow passage of ions or small molecules like ATP between cells and the extracellular space. So, we treated egg chambers with carbenoxolone, a known blocker of both Innexin and Connexin gap junction channels and hemichannels (Manjarrez-Marmolejo and Franco-Pérez, 2016; Sahu et al., 2017) and assessed neolamination by live imaging. We confirmed, as previously reported (Sahu et al., 2017), that carbenoxolone eliminates spontaneous [Ca2+] fluxes that travel between follicle cells prior to stage 8 (Video S6). Yet neolamination occurred normally (Figures 4N-4O”; Video S6). This result suggests that although Innexins are required for neolamination, gap junction channels are not required.
Although best known for forming channels, gap junction proteins also have channel-independent functions (Dbouk et al., 2009; Elias and Kriegstein, 2008; Kameritsch et al., 2013; Leo-Macias et al., 2016; Wei et al., 2004; Zhou and Jiang, 2014). For example, migration of neural progenitors during mammalian neocortical development requires a channel-independent function of Connexin (Cx) 26 and Cx43 (Elias et al., 2007). This was demonstrated by making a point mutation that disrupts channel and hemi-channel functions but leaves channel-independent functions intact. Remarkably, the protein lacking channel and hemi-channel functions rescued neuronal migration. Similarly, mutation of the highly conserved lysine 35 in Innexins to tryptophan (L35W) eliminates channel activity but leaves channel-independent functions such as adhesion intact (Baker et al., 2013; Depriest et al., 2011). Therefore, we made the L35W mutation in Inx2 and tested for rescue. First, we introduced silent mutations to render the UAS-R-Inx2-RFP transgene RNAi-resistant. Then we made transgenic flies expressing either wild type UAS-R-Inx2-RFP or UAS-R-Inx2L35W-RFP and confirmed that the transgenes were expressed (Figures S5H-S5I” and S5K-S5L”). R-Inx2-RFP and R-Inx2L35W-RFP rescued the neolamination defect (Figures 4P-4R). In contrast, mutating an extracellular cysteine, which blocks all known Innexin functions, failed to rescue (Figures 4S, S5J-S5J” and S5M-S5M”). Remarkably, R-Inx2L35W-RFP rescued as well as R-Inx2-RFP (Figure 4T).
Innexins regulate microtubule abundance
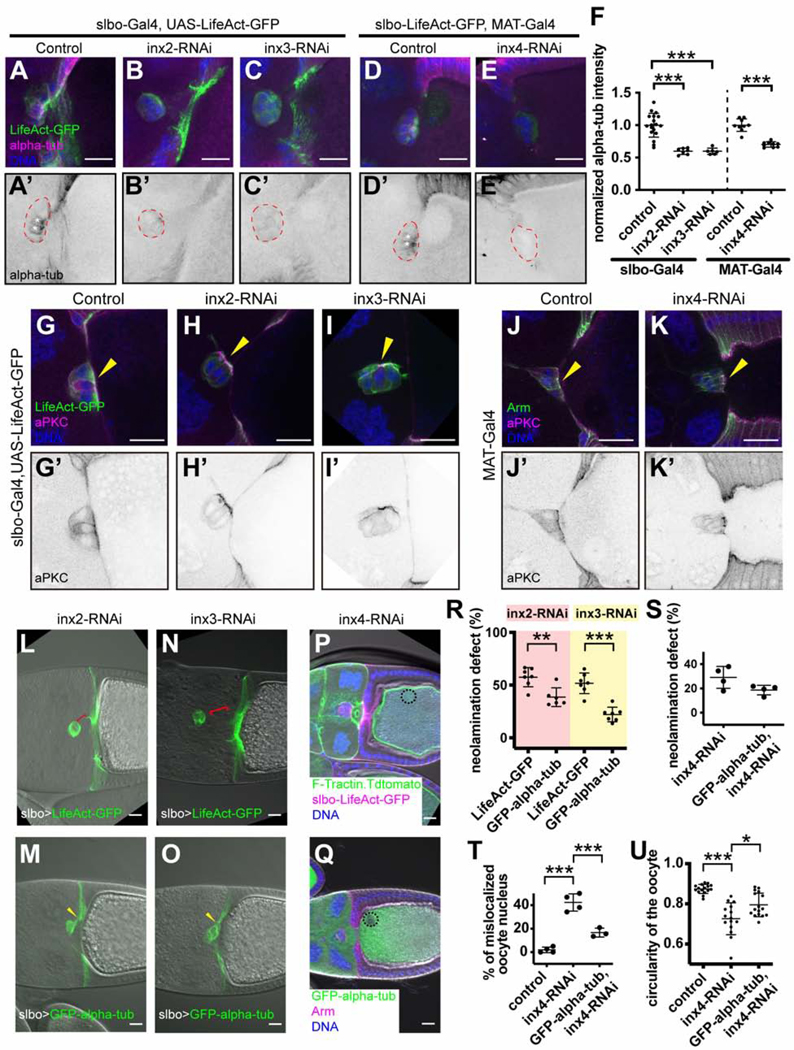
Since Inx4 knockdown affects oocyte nucleus position (Figures 3Q and 3R), which depends on microtubules (Januschke et al., 2006; Zhao et al., 2012), we examined the effects of Innexin knockdowns on microtubules. In contrast to wild type (Figures 5A and 5A’), knockdown of either Inx2 (Figures 5B and 5B’) or Inx3 (Figures 5C and 5C’) with slbo-Gal4 reduced α-tubulin staining in border cells and cfcs by ~2-fold (Figure 5F). This effect was also evident in clones of epithelial follicle cells where wild type and RNAi-expressing cells could be observed side-by side (Figures S6A-S6F’). Expression of inx4-RNAi in the germline also reduced border cell α-tubulin staining (Figures 5D-5E’ and 5F), suggesting that the border cell-oocyte interaction was critical. In contrast to the effects on tubulin, there was no detectable effect on F-actin accumulation or aPKC abundance or localization, upon knockdown of Inx2 or Inx3 in border cells (Figures 5G-5I’) or Inx4 in the germline (Figures 5J-5K’). The aPKC highlighted a difference between the neolamination defects caused by border cell knockdown of Inx-2 or −3 compared to germline knockdown of Inx-4. Border cell knockdown of Inx-2 or −3 caused a defect in docking of the apical surfaces to the oocyte whereas this step occurred normally when Inx-4 was knocked down in the germline. Thus, loss of innexins and microtubules from border cells impeded neolamination at an earlier step than loss of Inx4 from the germline.
Figure 5.
Effect of Innexin knockdowns on the microtubule cytoskeleton
(A-E’) Anti-alpha-tub staining of the indicated genotypes. w1118 was used as the control (A-A’). (F) Quantification of anti-alpha-tub staining in border cells. Each dot represents data from one border cell cluster. ***p < 0.001. (G-I’) aPKC staining of slbo-Gal4, UAS-LifeAct-GFP driven inx2-RNAi and inx3RNAi, showing that both F-actin and the apical marker are normal in border cells expressing inx2 or inx3 RNAi. w1118 serves as the control. (J-K’) aPKC staining of MAT-α-tub-Gal4 driven inx4-RNAi egg chambers. w1118 serves as the control. Yellow arrowheads indicate the apical side of border cell clusters. (L-O) Overexpression of GFP-alpha-tubulin ((M) and (O)) rescued the neolamination defects caused by inx2-RNAi (L) or inx3-RNAi (N). Red brackets indicate the abnormal spaces between border cells and cfcs in inx2-RNAi and inx3-RNAi. Yellow arrowheads indicate the contacts between border cells and cfcs that are restored by GFP-alpha-tub overexpression. (P-Q) Overexpression of GFP-alpha-tub (Q) but not F-Tractin (P) rescued inx4-RNAi phenotypes. Oocyte nuclei are marked with black circles. (K-U) Quantification of GFP-alpha-tub rescue effect. (K) Rescue of inx2-RNAi and inx3-RNAi neolamination defects by GFP-alpha-tubulin compared with the LifeAct-GFP control. (S) Slight but not statistically significant (p=.08) rescue of inx4-RNAi border cell neolamination defect by co-expression of GFP-alpha-tubulin. (T) Significant rescue of mislocalized nucleus phenotype. (U) Significant rescue of the irregularities in the oocyte cortex. In (K) and (S), each dot represents an independent experiment. In (P), each dot represents an egg chamber. *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars: 20 μm.
To test the functional significance of the effects of Innexin knockdown on microtubules, we over-expressed GFP-α-tubulin in egg chambers with Inx knockdowns. About 50% of clusters expressing inx2-RNAi-and co-expressing UAS-LifeAct-GFP as a negative control, showed neolamination defects (Figures 5L and 5R), whereas co-expressing UAS-GFP-α-tubulin significantly reduced the incidence of neolamination defects (Figures 5M and 5R). Co-expression of GFP-α-tubulin similarly reduced the incidence of neolamination defects caused by Inx3 knockdown (Figures 5N, 5O and 5R).
Expression of GFP-α-tubulin together with inx4-RNAi significantly rescued oocyte nucleus position and the irregularity of the oocyte cortex (Figures 5P, 5Q, 5T and 5U). Germline expression of GFP-α-tubulin also reduced the incidence of neolamination defects from ~30 to ~20% of inx4-RNAi-expressing egg chambers, though the P value did not reach the commonly used threshold for significance of P<.05 (Figure 5S). The combined observations that staining of microtubules was reduced upon Innexin knockdown and that over-expression of α-tubulin ameliorated the Innexin knockdown phenotypes indicate that a major function of Innexins in border cells is to regulate the abundance of α-tubulin and/or microtubules (see discussion).
Innexins required to brace the border cell-oocyte interface against external morphogenetic movements
To gain further insight into the cause of the neolamination defect, we carried out fixed and live imaging. Super-resolution, Airyscan confocal imaging of the border cell-oocyte interface during neolamination revealed that as cfcs migrate inward during stage 10B, they appear to squeeze the anterior of the oocyte, causing the cortex and cytoplasm (but not yolk) to bulge (Figures 6A-6A’ and 6B-6B’); this normally resolves quickly, and border cells remain attached to the oocyte and centripetal cells as nurse cells rapidly transfer cytoplasm into the oocyte through the ring canals during stage 11 (Video S7). Knockdown of either Inx2 or Inx3 with slboGal4 caused a more extreme herniation of the oocyte cortex and cytoplasm during stage 10B, which pushed the border cells back into the nurse cells and persisted into stage 11 (Figures 6C-6F’).
Figure 6.
External morphogenetic forces disrupt border cell/oocyte interactions following Inx knockdown
(A-F’) Super-resolution Airyscan images of slbo-Gal4, UAS-LifeAct-GFP crosses to control (w1118) (A-B’), inx2-RNAi (C-D’) and inx3-RNAi (E-F’) egg chambers. (G-J) Snapshots of time lapse movies of slbo-gal4>UAS-LifeAct-GFP (G), slbo-gal4>UAS-LifeAct-GFP, inx2-RNAi (H), slbo−/− (I) and slbo-gal4>Ecad-RNAi (J) egg chambers. FM4–64 dye staining shows the membrane structure. Red arrowheads indicate buckling of the anterior oocyte cortex. Blue arrowheads indicate inward migration of cfcs. Time (t) is relative to the start of live imaging. Scale bars: 20 μm.
Live imaging of control egg chambers stained with the membrane dye FM4–64 revealed the origin of the abnormal herniation. As cfcs migrate inward, they squeeze the growing oocyte causing the slight, temporary bulge (Figure 6G). Normally border cells associate stably with the anterior oocyte boundary during this process (Figure 6G; Video S8). However, upon Inx2 or Inx3 knockdown by slboGal4, cfc migration caused a more severe and lasting herniation of the anterior oocyte cytoplasm (Figure 6H; Video S8). These results suggest that the border cell/oocyte interaction normally braces the anterior of the oocyte, preventing this deformation.
If the border cells serve this function, then a similar effect should occur in homozygous slbo mutants, in which border cells never reach the oocyte (Montell et al., 1992). As predicted, slbo mutant egg chambers exhibit a similar defect in oocyte morphology (Figure 6I; Video S8). To test the hypothesis that cfc migration caused the oocyte phenotype, we knocked E-cadherin down using slbo-Gal4 and Ecad-RNAi, which blocked border cell migration and partially inhibited cfc migration (Oda et al., 1997). The cfcs initiated their inward migration normally causing the oocyte cytoplasm to bulge; however, as the RNAi took effect and their migration slowed, the deformation relaxed rather than worsening (Figure 6J; Video S8). Therefore, we conclude that one normal function of the border cell/oocyte interaction is to brace the boundary against external morphogenetic forces in an Innexin- and microtubule-dependent manner (Figure 7).
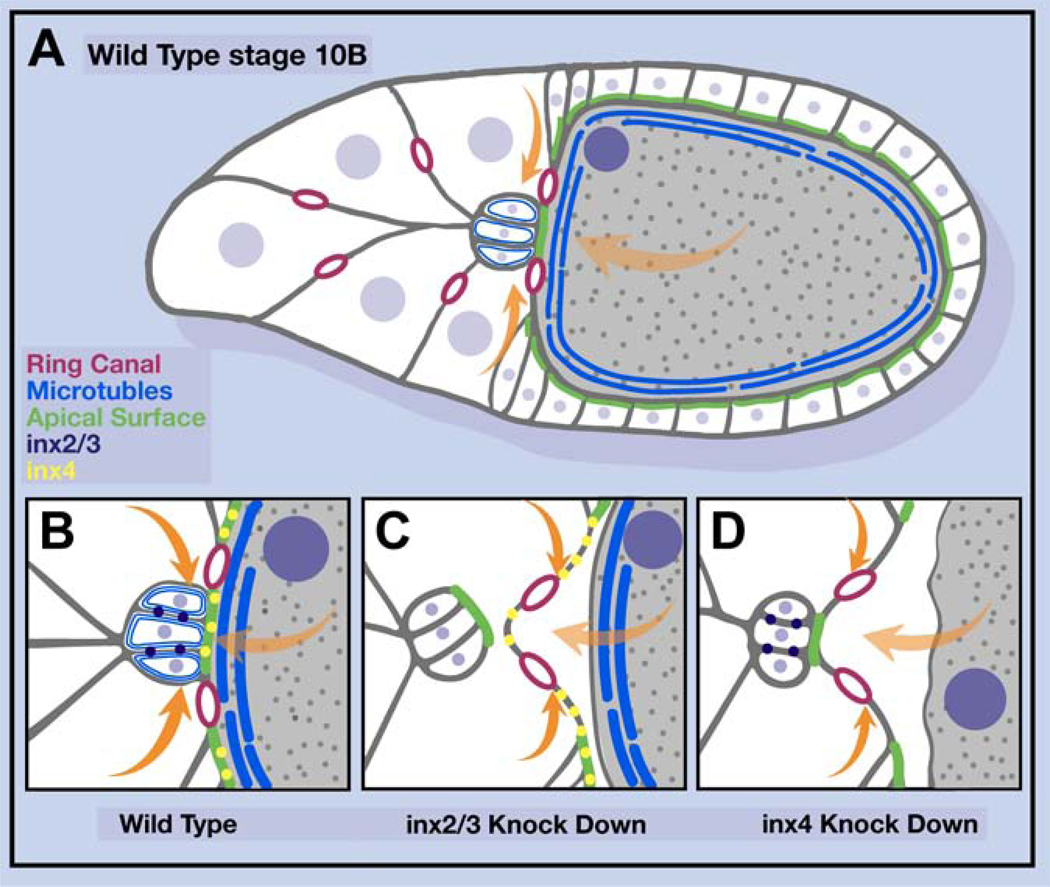
Figure 7.
Working model
(A) Schematic drawing of a wild-type egg chamber. (B-D) Detail of the border cell/oocyte interface in the indicated conditions. Magenta rings represent ring canals, orange arrows indicate the force of cfc migration, light orange arrows indicate the force caused by volume expansion of the oocyte, green lines mark apical domains of cells.
Discussion
Border cells as a model for study of neolamination: a multistep process
Here we establish border cells as a simple model amenable to genetics and live imaging to study the process of neolamination, which we define as the establishment of new, selective, and stable cell-cell contacts after migration. We found that border cell neolamination is a multistep process. We further found that Innexins 2 and 3 are required autonomously for border cells to turn and dock their apical surfaces onto the oocyte. Inx 4 is required subsequently in the germline, to maintain the border cell/oocyte boundary against external morphogenetic forces, consistent with the multistep nature of the process.
At each stage of this project, the results surprised us. When we first noticed the neolamination defect upon Inx2 or Inx3 knockdown, the simple hypothesis was that establishment of gap junctions between border cells and cfcs would be required because epithelial follicle cells are normally connected by gap junctions. Yet we discovered that, it is the border cell/oocyte association that initially requires innexins. Then, it seemed likely to be a channel-dependent function as gap junction channels have been proposed to mediate interactions between germ cells and somatic support cells in prior studies (Gilboa et al., 2003; Mukai et al., 2011; Starich et al., 2014; Tazuke et al., 2002) as well as between glial cells of the fly “blood/brain barrier” (Spéder and Brand, 2014). Yet multiple lines of evidence including electron microscopy, thermogenetics, pharmacological blockade, and selective point mutations suggest that the channel function is dispensable. Transmission electron microscopy provides evidence of structural gap junctions between nearly all cell types in the egg chamber, except between the border cells and the oocyte. The analysis revealed that border cells and the oocyte are intimately connected via interdigitations between the apical surfaces of border cells and the anterior oocyte cortex. Innexin knockdowns did not perturb the apicobasal polarity of the border cell cluster or the abundance or distribution of the cell-cell adhesion protein Armadillo; rather Innexins 2 and 3 are essential for the border cells to dock their apical surfaces to the oocyte and establish a stable connection.
Although Innexins and Connexins are best known for their gap junction channel forming ability, they also exhibit channel-independent functions in specific contexts. For example, Cx43 - the most widely expressed connexin – and Cx26 mediate channel-independent interactions between migrating neuronal cells and radial glia during establishment of the laminar structure of the neocortex during mammalian development (Elias et al., 2007). In addition, in zebrafish, mutations in two different connexins were identified in a screen for mutations affecting development of pigmentation patterns (Watanabe, et al., 2006; Irion, et al., 2014). Pigmentation patterns are established by neural crest-derived, pigment-forming cells. Zebrafish are so-named because they have different types of pigment cells that tile the skin in a striped pattern. In mutants disrupting Cx41.8 and Cx39.4, pigment cells form and migrate normally but do not adopt the typical pattern of connections. So, instead of continuous stripes of dark pigment, intermittent patches appear. The Cx41.8 mutant is called leopard as the pattern resembles leopard spots rather than zebra stripes. It is interesting to speculate that this phenotype could be due to a failure of pigment cell neolamination. Experiments to distinguish between channel-dependent or channel-independent functions have not been reported in this context.
Effects of Innexins 2, 3, and 4 on the microtubule cytoskeleton
Knockdown of Inx2 or 3 in the border cells reduced the overall staining for α-tubulin, which could be an effect on α-tubulin protein abundance or – more likely based on the staining pattern - on microtubule abundance. A handful of studies have demonstrated interactions between both vertebrate and invertebrate gap junction proteins and microtubules. One functional interaction is that microtubules facilitate transport of gap junction hemichannels to adherens junctions (Shaw et al., 2007). Another study showed that the C-terminus of Cx43 binds to tubulin directly (Giepmans et al., 2001). In addition, Cx43 and Cx26 are required for stability of the microtubule-rich leading process and microtubule-dependent nuclear translocation in neurons migrating on radial glia, through a channel-independent function (Elias et al., 2007). Although the authors provided data supporting a role in adhesion between the neuronal process and radial glia, a role in microtubule stabilization was not investigated. The results presented here suggest that microtubule stabilization might contribute to cell-cell contact stabilization. Further work will be required to elucidate the mechanism by which Innexins and possibly Connexins affect tubulin abundance and/or microtubule stability.
It is also striking that knockdown of Inx4 in the oocyte caused a non-cell-autonomous effect on tubulin staining in the border cells. We propose that stable cortical microtubules are required in both the border cells and the oocyte to establish and maintain the boundary between the two cell types. Although not extensively studied, coordination of microtubule cytoskeletons across newly formed cell-cell contacts is not unprecedented (Stramer et al., 2010; Davis et al., 2015).
Knockdown of either Inx2 or Inx3 at the time of initiation of border cell migration also caused a defect in their ability to complete migration while later knockdown caused only the neolamination defect. A previous study also reported, and we have confirmed, that even earlier knockdown of Inx2 resulted in a failure of border cell specification (Sahu et al., 2017). This function is likely distinct from the requirement for Inx2 in border cell migration and neolamination as it is blocked by carbenoxolone. It is striking that Innexins serve so many distinct and essential functions, at every stage of development. The availability of Gal4 lines with spatially - and importantly - temporally defined expression patterns was essential for disentangling each of these functions. Analysis of loss-of-function mutants, even in mosaic clones, does not afford the necessary degree of temporal control.
Mis-localization of the oocyte nucleus was our first clue that the microtubule cytoskeleton might be affected by the Innexin knockdowns, a phenotype that was noticeable because of the large size and normally asymmetric location of the nucleus within the oocyte. Fortunately, prior studies had established the importance of the microtubule cytoskeleton to oocyte nucleus localization. Microtubule polymerization appears to push the oocyte nucleus from a posterior to a dorsal/anterior position in the oocyte (Zhao et al., 2012) and maintain it there (Guichet et al., 2001). Thus, microtubules within the oocyte are able to provide resistance to cytoplasmic movements that otherwise displace the nucleus. Similarly, we propose that the microtubule cytoskeleton is critical for the border cell/oocyte interface to resist cfc-generated forces and cytoplasmic flows that cause the oocyte to deform and push the border cells out of position in the absence of Innexins. Striking confirmation of the importance of the microtubule cytoskeleton to the neolamination phenotype emerged when we found that over-expression of α-tubulin was sufficient to rescue border cell neolamination defects and germline oocyte nucleus positioning.
The most parsimonious model consistent with all the results is that Innexins 2 and 3 function autonomously within the border cell cluster to regulate microtubule abundance or dynamics, which is essential for the border cells to turn and dock their apical surfaces toward the oocyte and form a connection to the oocyte cortex that is stable to outside pushing forces exerted by cfcs, cytoplasmic flows through ring canals that connect the germ cells, and rapid growth of the oocyte (Figures 7A-7C). Inx4 in the oocyte similarly regulates the microtubule cytoskeleton, holding the oocyte nucleus in place and stabilizing the oocyte cortex. In the absence of Inx4 the cortex becomes wavy, the border cell microtubule cytoskeleton is destabilized, and the border cell/oocyte contact does not resist cfc pushing and cytoplasmic flows (Figure 7D). These phenotypes highlight an under-appreciated feature of embryo and organ development: as everything moves around, selective cell-cell interactions must resist external forces. Since Innexins and connexins are large protein families, it is interesting to speculate that combinations of interactions could play an important role in stabilizing specific cell-cell contacts.
Other neolamination-like events in development
Here we coin the term neolamination as a concise way of describing the process by which migratory cells establish connections with new partners when they reach their destinations. Neolamination parallels the term delamination, which describes the process by which cells exit an epithelium. For epithelial cells that acquire motility and delaminate by undergoing epithelial to mesenchymal transitions (EMTs), it has been proposed that at least some cells reverse that process at the end of migration by undergoing mesenchymal to epithelial transitions (METs) (Pei et al., 2018). For border cells the neolamination process is distinct from MET because border cells do not undergo an EMT when they delaminate. They never lose apical/basal polarity or adherens junction connections during migration (Niewiadomska et al., 1999). Additionally, they do not, as far as we know, require EMT transcription factors like Twist and Snail. They arrive at the oocyte with their epithelial character intact and so do not need to undergo MET. Even in the absence of innexins, apical/basal polarity is unaffected as evidenced by normal polarity and abundance of apical markers like aPKC and Cadherin99. While MET may contribute to neolamination of some cell types, neolamination is a more general phenomenon that refers to establishment of new, selective, and stable cell-cell contacts, independent of the epithelial, mesenchymal, or any other morphological state of the partnering cells.
Other developmental events that resemble neolamination include formation of mesoderm and definitive endoderm during gastrulation in vertebrate embryos. These cells delaminate from the epiblast layer, move through the primitive streak, and then make specific homotypic associations, while sorting out from each other. Both mesoderm and endoderm undergo EMT and MET, and advances in imaging of mouse embryos has allowed some live imaging of these stages of development (Kang et al., 2017); yet the mechanisms by which selective cell-cell interactions are achieved and maintained despite widespread ongoing morphogenetic movements remain to be clarified. Similarly, neural crest cells delaminate from the border between epidermal ectoderm and neural ectoderm, migrate in diverse streams, and eventually assimilate into many different organs. While the mechanisms of neural crest cell delamination and migration have been the subject of intense study, post-migratory development has received less attention. Some neural-crest-derived cells undergo MET, but others, like neurons, cardiac crest cells, and melanocytes do not. Yet each of these cell types must make stable contacts with specific partners at the destination despite ongoing morphogenetic movements.
Neolamination-like events in cancer
Selective cell-cell associations are important at many stages in tumor metastasis. Tumor cells bind to endothelial cells during the processes of intra- and extra-vasation, and tumor cell clusters lodge in small vessels to form micrometastases. Connexins have been implicated in tumor cell/endothelial cell interactions (Elzarrad et al., 2008), though again whether channel-dependent or channel-independent functions are required is not known. To form a distant colony, tumor cells establish connections with resident cells. Intriguingly, connexins mediate interaction of breast and lung tumor cells with astrocytes in brain metastases in mice (Chen et al., 2016). In the absence of Connexins, metastasis is much reduced. In this experimental model, the effect is channel-dependent. On the other hand Cx43 appears to promote the spread of glioma cells in a gap junction independent manner (Lin et al., 2002). Therefore, there likely exist multiple mechanisms by which proteins with gap junction forming potential promote cell-cell interactions and enhance metastasis.
To what extent diverse neolamination events that occur during development and metastasis share common molecular mechanisms is unknown, but gap junction forming proteins have been implicated in a number of intriguing processes including breast cancer metastasis to lung and brain, tumor-endothelial cell interactions, and pigment cell patterning in zebrafish. Further elucidation of the molecular mechanisms responsible for neolamination-like events is an exciting frontier.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Denise Montell (dmontell@ucsb.edu).
Materials Availability
Drosophila lines and other reagents generated in this study will be available upon request.
Data and Code Availability
Data including all imaging datasets produced in this study will be made available upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Fly husbandry
Fly strains were raised in vials containing a standard cornmeal-yeast food (https://bdsc.indiana.edu/information/recipes/molassesfood.html) which contains 163g yellow cornmeal, 33g dried yeast, 200mL molasses and 16g agar with 2.66L water. All flies were raised in vials containing 5mL fly food.
Flies were kept in incubators at 25°C, 80% humidity and on a 12 hr light/dark cycle unless otherwise noted. For thermo-genetics experiments, flies were kept at 18°C, 80% humidity and on a 12 hr light/dark cycle.
For the RNAi knockdown experiments (except for MAT>inx4-RNAi), 2–4 day-old females were kept in 29°C for 2 days, then transferred to a vial with dry yeast and at 29°C overnight before dissection. Because the phenotype of MAT>inx4-RNAi was very severe, those flies and control were kept at 25°C and transferred to a vial with dry yeast at 29°C overnight before dissection.
For clonal analyses (flip-out clones), 2–4 days females were heat-shocked for one hour at 37°C to induce clones, and were then kept at 25°C for 3 days with dry yeast until dissection.
For the thermo-genetics experiments, 2–4 days females were transferred to a vial with dry yeast and kept at 18°C for 2 days before dissection.
Detailed fly genotypes in each figure are listed below.
| Figure | genotypes |
|---|---|
| Fig 1 | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig S1 | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig 2A-2C | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig 2D-2F | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx2-RNAi/+ |
| Fig 2G-2I | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx3-RNAi/+ |
| Fig 2O-2P | HSflp/+; AY-Gal4, UAS-GFP/+ |
| Fig 2S-2T | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-inx2-RNAi/+ |
| Fig 2Q-2R | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-inx3-RNAi/+ |
| Fig 2U-2V | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-inx1-RNAi/+ |
| Fig S2B | control: slbo-Gal4, UAS-LifeAct-GFP/+ |
| inx2-RNAi: slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx2-RNAi/+ | |
| Fig S2C | HS-flp; AY-Gal4, UAS-GFP/UAS-inx2-RNAi; UAS-inx2-RFP/+ |
| Fig S2D | HSflp/+; AY-Gal4, UAS-GFP/+ |
| Fig S2E | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-inx1-RNAi/+ |
| Fig S2F | HSflp/+; AY-Gal4, UAS-GFP/+ |
| Fig S2G | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-inx3-RNAi/+ (BDSC: 60887) |
| Fig 3F | 109c1-Gal4/+;; UAS-LifeAct-GFP/+ |
| Fig 3G | 109c1-Gal4/+; UAS-inx2-RNAi/+ |
| Fig 3H | 109c1-Gal4/+;; UAS-inx3-RNAi/+ |
| Fig 3I | CY2-Gal4/UAS-LifeAct-GFP |
| Fig 3J | CY2-Gal4/+; UAS-inx2-RNAi/+ |
| Fig 3K | CY2-Gal4/+; UAS-inx3-RNAi/+ |
| Fig 3L | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig 3M | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx2-RNAi/+ |
| Fig 3N | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx3-RNAi/+ |
| Fig 3O | UAS-F-Tractin.Tdtomato/+; MAT-α-tub-Gal4/+ |
| Fig 3P | MAT-α-tub-Gal4/inx4-RNAi |
| Fig 3Q | slbo-LifeAct-GFP/UAS-F-Tractin.Tdtomato; MAT-α-tub-Gal4/+ |
| Fig 3R | slbo-LifeAct-GFP/UAS-F-Tractin.Tdtomato; MAT-α-tub-Gal4/inx4-RNAi |
| Fig S3A | 109c1-Gal4/+; UAS-LifeAct-GFP/+ |
| Fig S3B | CY2-Gal4/UAS-LifeAct-GFP |
| Fig S3C | Ftractin.tdTomato/+; MAT-α-tub-Gal4/+ |
| Fig S3D | CY2-Gal4/+ |
| CY2-Gal4/+; UAS-inx2-RNAi/+ | |
| CY2-Gal4/+; UAS-inx3-RNAi/+ | |
| Fig S4A | control: 109c1-Gal4/+; UAS-LifeAct-GFP/+ |
| inx2-RNAi: 109c1-Gal4/+; UAS-LifeAct-GFP/+; UAS-inx2-RNAi/+ | |
| Fig S4B | control: MAT-α-tub-Gal4/+; slbo-LifeAct-GFP/+ |
| inx4-RNAi: MAT-α-tub-Gal4/+; slbo-LifeAct-GFP/UAS-inx4-RNAi | |
| Fig 4A-4B | w1118 |
| Fig 4C-4D | zpg-zpg::GFP (inx4-GFP) |
| Fig 4E-4G | Oregon-R-C |
| Fig 4I | LexAOP-GCaMP6s/+; FC-LexA/MAT-α-tub-Gal4 |
| Fig 4J-4L | LexAOP-GCaMP6s/UAS-TrpA1; FC-LexA/MAT-α-tub-Gal4 |
| Fig 4N-4O | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig 4P | slbo-Gal4, UAS-LifeAct-GFP/UAS-inx2-RNAi |
| Fig 4Q | slbo-Gal4/UAS-inx2-RNAi; UAS-R-inx2-RFP/+ |
| Fig 4R | slbo-Gal4/UAS-inx2-RNAi; UAS-R-inx2L35W-RFP/+ |
| Fig 4S | slbo-Gal4/UAS-inx2-RNAi; UAS-R-inx2C256S-RFP/+ |
| Fig S5A-S5B | w1118 |
| Fig S5C-S5D | zpg-zpg::GFP (inx4-GFP) |
| Fig S5E-S5G | Oregon-R-C |
| Fig S5H-S5H” | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-R-inx2-RFP/+ |
| Fig S5I-S5I” | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-R-inx2L35W-RFP/+ |
| Fig S5J-S5J” | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-R-inx2C256S-RFP/+ |
| Fig S5K-S5K” | HSflp/+; AY-Gal4, UAS-GFP/UAS-inx2-RNAi; UAS-R-inx2-RFP/+ |
| Fig S5L-S5L” | HSflp/+; AY-Gal4, UAS-GFP/UAS-inx2-RNAi; UAS-R-inx2L35W-RFP/+ |
| Fig S5M-S5M” | HSflp/+; AY-Gal4, UAS-GFP/UAS-inx2-RNAi; UAS-R-inx2C256S-RFP/+ |
| Fig 5A, 5G | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig 5B, 5H, 5L | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx2-RNAi/+ |
| Fig 5C, 5I, 5N | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx3-RNAi/+ |
| Fig 5D | slbo-LifeAct-GFP/+; MAT-α-tub-Gal4/+ |
| Fig 5E | slbo-LifeAct-GFP/+; MAT-α-tub-Gal4/inx4-RNAi |
| Fig 5J | MAT-α-tub-Gal4/+ |
| Fig 5K | MAT-α-tub-Gal4/UAS-inx4-RNAi |
| Fig 5M | slbo-Gal4/+; UAS-GFP-alpha-tub/UAS-inx2-RNAi |
| Fig 5O | slbo-Gal4/+; UAS-GFP-alpha-tub/UAS-inx3-RNAi |
| Fig 5P | UAS-Ftractin.Tdtomato/+; MAT-α-tub-Gal4/UAS-inx4-RNAi |
| Fig 5Q | UAS-GFP-alpha-tub/+; MAT-α-tub-Gal4/UAS-inx4-RNAi |
| Fig S6A, S6D | HSflp/+; AY-Gal4, UAS-GFP/+ |
| Fig S6B, S6E | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-inx2-RNAi/+ |
| Fig S6C, S6F | HSflp/+; AY-Gal4, UAS-GFP/+; UAS-inx3-RNAi/+ |
| Fig S6G | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig S6H | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx2-RNAi/+ |
| Fig S6I | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx3-RNAi/+ |
| Fig 6A-6B | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig 6C-6D | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx2-RNAi/+ |
| Fig 6E-6F | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx3-RNAi/+ |
| Fig 6G | slbo-Gal4, UAS-LifeAct-GFP/+ |
| Fig 6H | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-inx2-RNAi |
| Fig 6I | slboe7b/slboPZ1310 |
| Fig 6J | slbo-Gal4, UAS-LifeAct-GFP/+; UAS-Ecad-RNAi |
METHOD DETAILS
Generation of RNAi-resistant UAS-inx2-RFP point mutation transgenic lines
Inx2-RFP fragment was PCR out from UAS-inx2-RFP flies (inx2-N-F:5’-ATGTTTGATGTCTTTGGGTCCGTC-3’ RFP-C-R: 5’-TCACGTGGACCGGTGGGCGC-3’) and subcloned to pUAST-attB vector with EcoRI and XbaI sites. UAS-inx2-RFP vector was digested with EcoRI and BsrGI to remove original inx2 cDNA fragment. RNAi-resistant inx2, inx2L35W,inx2C256S cDNA fragments were synthesized (Integrated DNA Technologies, Inc) and subcloned to UAS-RFP vector with EcoRI and BsrGI sites. The clones were sequence-verified and transgenic lines were established through ΦC-31 integrase mediated transformation (Bestgene). attP sites used was attP2 (BDSC: 8622). RNAi-resistant inx2 cDNA sequence is showed below. For inx2L35W point mutation, codon CTG is changed to TGG. For inx2C256S point mutation, codon TGC is changed to TCC.
RNAi-resistant inx2 cDNA sequence:
GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATTCGAGCTCGGTACCCGGGGATCCGA TTATGTTCGACGTGTTCGGTTCCGTGAAAGGTTTGCTAAAAATTGATCAAGTCTGTATTGATAATAAC GTGTTCAGGATGCATTATAAAGCAACTGTTATTATCCTGATAGCGTTTTCCTTATTGGTCACGTCGAG GCAGTATATTGGAGATCCAATTGACTGCATCGTCGATGAAATACCTCTCGGAGTAATGGATACATATT GTTGGATATATTCTACATTCACAGTCCCTGAACGACTCACAGGTATTACGGGCCGAGACGTAGTACA ACCAGGAGTTGGGTCCCACGTTGAAGGGGAAGATGAAGTAAAATATCATAAATATTATCAATGGGTC TGTTTTGTTTTGTTTTTTCAAGCGATATTGTTTTATGTGCCACGTTACTTATGGAAATCCTGGGAGGGA GGCCGTCTTAAAATGCTAGTAATGGACTTGAATAGTCCGATCGTCAATGACGAATGTAAAAATGACC GAAAGAAAATTCTAGTTGATTATTTTATAGGAAATCTCAATCGTCATAACTTTTATGCGTTTCGTTTTTT TGTATGTGAGGCACTCAATTTCGTCAACGTAATAGGTCAAATATATTTCGTCGATTTTTTTCTGGATGG TGAATTTAGTACGTATGGAAGCGACGTACTCAAATTTACCGAACTAGAACCAGACGAACGAATAGAC CCGATGGCTAGGGTATTCCCTAAAGTGACAAAGTGCACTTTTCATAAGTATGGACCGTCCGGAAGCG TTCAAACGCATGATGGGCTATGCGTCCTCCCGCTCAATATCGTGAATGAGAAAATATATGTCTTTCTT TGGTTTTGGTTTATAATTTTGAGTATAATGTCGGGTATCTCGTTGATATATCGCATTGCAGTCGTCGC CGGCCCAAAACTTCGACACTTGCTTCTTCGGGCGCGCTCCCGACTCGCGGAGAGTGAAGAAGTAGA GCTCGTCGCTAATAAATGTAATATTGGAGACTGGTTTCTACTTTACCAACTAGGTAAAAATATAGACC CCCTTATATATAAAGAAGTAATATCGGATCTATCCCGTGAGATGAGTGGTGACGAACACAGTGCGCA TAAACGTCCATTTGATGCGCACCCAGCTTTCTTGTACAAAGTGGTGAGCTCCGCCACCATGGCCTCC TCCGAGGACGTCATCAAGGAGTTCATGCGCTT
Immunohistochemistry
Adult female ovaries were dissected in Schneider’s Drosophila medium (Thermo Fisher Scientific, Waltham, MA) with 20% fetal bovine serum. Ovarioles were immediately fixed for 20 min in 4% paraformaldehyde at room temperature. After fixation, ovarioles were washed with PBS/0.1% Triton X-100 (PBST) for 4 times (15min each time), and then incubated with primary antibodies overnight at 4°C. The following day, ovarioles were washed with PBST 4 times (10min each) before incubation in secondary antibody for 2 hours at room temperature. After removal of secondary antibodies, samples were washed with PBST 4 times (10min each) and then stored in Vectashield (Vector Laboratories, Burlingame, CA) at 4°C before mounting.
For alpha-tub staining, ovaries were dissected in BRB80 buffer (80mmol/l PIPES pH6.8, 1mmol/l MgCl2, 1mmol/l EGTA), containing 1% Triton X100. Ovaries were then incubated in BRB80 buffer for one hour without agitation and fixed in MeOH at −20 °C for 15 minutes. Then ovaries were rehydrated overnight at 4 degree in PBS with 0.1% Tween, then blocked for one hour in PBS with 0.1% Triton X100 containing 3%(w/v) bovine serum albumin (BSA) before incubation with anti-alpha-tub overnight at 4 °C.
The following antibodies were used in this study: rat anti-E-cadherin (1:50, DCAD2, DSHB), mouse anti-Armadillo (1:75, N2.7A1, DSHB), guinea pig anti-inx2 (1:1000), rabbit anti-inx3(1:75), rabbit anti-inx4(1:20000), rabbit anti-Cad99C (1:1000) rabbit anti-aPKC (1:200, Santa Cruz), rabbit anti-GFP (1:300, lifetech), mouse anti-alpha-tub (1:100, sigma). Hoechst (1:1000), Alexa 488, 568, 633 (1:300, lifetech), phalloidin 488, 568 (1:300, lifetech).
Fixed sample imaging and image processing
Samples were mounted on a glass slide in Vectashield. Images were taken on a Zeiss LSM 780 or 800 confocal microscope, using a 20×1.2 N.A. objective or 40×1.4 N.A. water objective. Z-stacks covering the egg chambers were taken with a 1 μm step size for border cell clusters. For the cross view of egg chambers, laser power corrections were applied by increasing the laser power as the objective scans from the top of the sample to the bottom of the sample, so that the signal on the bottom did not appear weaker than the top. Cross views of the egg chambers were visualized in Imaris (Bitplane, South Windsor, CT). Representative images were exported from Imaris using Easy 3D view. Exported images were rotated and cropped in Adobe Illustrator (Adobe, San Jose, CA). To obtain super-resolution images, Airyscan images were taken on a Zeiss LSM 800 confocal microscope, using 63x, 0.8 NA oil objective. Three stacks of horizontal plane images (1120 pixels X 1120 pixels corresponding to 33.8 μm X 33.8 μm) with a z-step of 0.16 μm.
Live imaging
For live imaging of neolamination, ovaries were dissected in Schneider’s Drosophila medium (Thermo Fisher Scientific, Waltham, MA) with 20% fetal bovine serum. Individual ovarioles were carefully pulled out and stage 14 egg chambers were removed. The egg chambers were collected in a 1.7 mL tube and washed with dissecting medium twice, then added 200 μL dissecting medium with insulin (200 μg/mL) and 1% low melt agarose. 90 μL medium with the egg chambers then were mounted on a 50 mm Lumox dish. Time-lapse imaging was performed using a 20×1.2N.A. objective or 40×/1.1 NA water immersion objective lens. The 1-μm-thick z-sections ranging the entire border cell cluster were collected at 2-min or 4-min intervals.
For thermo-genetics, ovaries were dissected in 18°C dissecting medium. The egg chambers were collected in a 1.7 mL tube and washed with dissecting medium twice, then added 200 μL dissecting medium with insulin (200 μg/mL). 90 μL medium with the egg chambers then were mounted on a 50 mm Lumox dish. The dish was placed on ice before imaging. Time-lapse imaging was performed on Zeiss LSM780 using a 20x objective lens. 1-μm-thick z-sections covering the entire border cell cluster were collected at 1-min intervals. The LSM 780 temperature module was applied to control the temperature during time-lapse imaging. The initial imaging temperature setting was 23°C, then the temperature shifted to 32°C after 5 minutes of time-lapse imaging. The temperature increased from 23°C to 32°C within one minute for the sample stage and within five minutes for the whole imaging chamber.
For carbenoxolone treatment, DMSO and Carbenoxolone (final concentration:0.25 μM) were added to the medium before mounting.
Transmission electron microscopy (TEM)
Fixation and embedding of egg chambers and the preparation of sections followed the protocol described in detail in Glowinski et al. (2014). In short, ovaries were dissected in Schneider’s medium, fixed (1) in 2.5% glutaraldehyde, 2% para-formaldehyde, 2% acrolein, (2) 2.5% glutaraldehyde, (3) 1% OsO4 and 0.1% sorbitol, using 75 mM cacodylate pH 7.4 as a buffer. Ovaries were dehydrated in an ethanol series and then propylene oxide and embedded in araldite-502/Embed-812. 100-nm-thick sagittal sections of egg chambers were contrasted with uranyl acetate and lead citrate. Images were taken at x1K-30K with a Hitachi HT-7700 TEM (Hitachi High-Technologies, Etobicoke, ON, Canada). Plasma membrane interfaces between (1) border cells, (2) border and nurse cells, (3) border cells and oocyte (4) border and polar cells, (5) polar cells (6) nurse cells, and (7) nurse cells and oocyte were analyzed for gap junctions in 5 sections. 1-μm-thick toluidine/methylene blue-stained sections were used to determine the stage of egg chambers, following criteria of King (1970).
QUANTIFICATIONS AND STATISTICAL ANALYSES
Neolamination defect quantification
For quantification of neolamination defects, stage 10B egg chambers were imaged at 20x magnification. Z-stacks Projection images of LifeAct-GFP or anti-Arm channel were used to analyze the border cell-centripetal cell interaction. If the border cells maintained their interaction with centripetal cells, that egg chamber would be scored as a normal for neolamination. If the border cells failed to interact with the centripetal cells, the egg chamber would be counted as a neolamination defective egg chamber. In 109c1-Gal4 experiments, neolamination defect= percentage of egg chambers complete migration in stage 10A - percentage of egg chambers complete migration in stage 10B.
Inx2, inx3 and inx4 colocalization quantification
For quantification of inx2/3/4 colocalization, z-stack projection images of the egg chamber were imaged at 40x magnification. Border cells and follicle cells were selected to analyze the colocalization using FIJI coloc2 function (FIJI function: Analyze > Colocalization Analysis > Coloc 2). Pearson’s correlation, r was used as the readout of colozalization.
GCaMP6s quantifications
For quantification of GCaMP6s level, a single section of the egg chamber was imaged at 20x magnification. GCaMP6s level was quantified using FIJI in areas of identical size in follicle cells or border cells across all genotypes. Threshold was adjusted for the GFP channel (FIJI function: Image > Adjust > Threshold) to subtract background. Changes in GCaMP6s intensity were calculated and expressed as ΔF/F0. ΔF= F1-F0, F1 was the GCaMP6s intensity after temperature shift to 32°C for 10 minutes, F 0 was the GCaMP6s intensity before temperature shift.
Alpha-tub quantifications
For quantification of alpha-tub level, 40x magnification sum intensity projections images of anti-alpha-tub channel were measured in FIJI. Threshold was adjusted for the anti-alpha-tub channel (ImageJ function: Image > Adjust > Threshold) to subtract background. The fluorescence intensity was normalized by the means of control staining.
Inx4 KD phenotype quantification
For quantification of the oocyte nuclei mislocalization defect, stage 10A/10B/11 egg chambers were imaged at 20x magnification. The bright field, Hoechst staining and Ftractin.tdtomato channel were used to identify the localization of oocyte nuclei. For quantification of the irregularities in the oocyte plasma membrane, stage 10B egg chambers were imaged at 20x magnification. The oocyte membrane of control and inx4-RNAi egg chambers were selected to analyze the circularity of oocyte in FIJI. In a normal egg chamber, the oocyte membrane is smooth and without wrinkles, so its circularity is close to a circle. In the inx4-RNAi egg chamber, the oocyte shrinks and oocyte membrane has a lot of wrinkles. Its circularity hence reduces.
Statistics and data presentation
Standard statistical tests were performed using Prism 6. Unpaired t-test (two-tailed) was used for comparing two groups with similar variance as determined by F-test. Mann–Whitney nonparametric test (two-tailed) was used for comparing two groups with different variance. Ordinary one-way ANOVA, followed by Tukey’s multiple comparisons test was used for comparing multiple groups with similar variance as determined by Brown–Forsythe test.
All graphs were generated using Prism 6. All confocal images belonging to the same experiment were acquired using the exact same settings. For visualization purposes, brightness adjustments were applied using FIJI to the confocal images shown in the figure panels. All quantitative analyses were carried out on unadjusted raw images or sum intensity projections. All fly crosses were repeated at least twice and ovary dissections and staining were repeated at least three times. The exact sample size (n) is listed below, representing biological replicates. Sample size was not predetermined by statistical methods but we used prior knowledge to estimate minimum sample size. The experiments were not randomized. Investigators were not blinded.
The sample numbers (N) for each figure are listed below.
| Figure | egg chamber numbers(N) (From top to bottom in the graphics) |
|---|---|
| Fig 2J | LifeAct-GFP: 30,29,23 |
| LifeAct-GFP, inx2-RNAi: 40, 142, 56, 64 | |
| inx2-RNAi, inx2-RFP: 34, 41, 38 | |
| inx2-GFP, inx2-RNAi: 28, 29, 33, 32 | |
| inx3-GFP, inx2-RNAi: 24, 22, 23, 27, 26 | |
| inx1-GFP, inx2-RNAi: 38, 53, 50 | |
| LifeAct-GFP, inx3-RNAi: 52, 52, 56, 23 | |
| inx2-RNAi, inx3-RNAi: 33, 45, 84, 46, 29, 36 | |
| Fig 3E | 109c1-Gal4, UAS-LifeAct-GFP: stage 10A: 82, 38, 61, 34; stage 10B: 56, 36, 36, 11 |
| 109c1-Gal4, UAS-inx2-RNAi: stage 10A: 46, 41, 66, 54; stage 10B: 43, 19, 42, 47 | |
| 109c1-Gal4, UAS-inx3-RNAi: stage 10A: 42, 45, 43, 74; stage 10B: 47, 37, 21, 56 | |
| CY2-Gal4, UAS-LifeAct-GFP: 56, 21, 44, 32 | |
| CY2-Gal4, UAS-inx2-RNAi: 44, 56, 70, 59 | |
| CY2-Gal4, UAS-inx3-RNAi: 36, 43, 44 | |
| MAT-Gal4, UAS-F-Tractin.tdTomato: 30, 47, 39 | |
| MAT-Gal4, UAS-inx4-RNAi: 50, 29, 50, 28 | |
| Fig 4T | LifeAct-GFP: 47, 34, 29 |
| R-inx2-RFP: 27, 39, 39 | |
| R-inx2L35W-RFP: 35, 33, 42 | |
| R-inx2C256S-RFP: 39, 55, 36 | |
| Fig 5R | LifeAct-GFP, inx2-RNAi: 40, 142, 56, 64(previous experiments used in Fig 2J), 45, 32, 31(new experiments) |
| GFP-alpha-tub, inx2-RNAi: 27, 45, 74, 24, 48, 60 | |
| LifeAct-GFP, inx3-RNAi: 52, 52, 56, 23(previous experiments used in Fig 2J), 24, 50, 54(new experiments) | |
| GFP-alpha-tub, inx3-RNAi: 23, 49, 46, 52, 44, 35, 44 | |
| Fig 5S | MAT-Gal4, UAS-inx4-RNAi: 50, 29, 50, 28(previous experiments used in Fig 3E) |
| MAT-Gal4, UAS-inx4-RNAi, UAS-GFP-alpha-tub: 69, 58, 31, 31 | |
| Fig 5T | MAT-Gal4, UAS-F-Tractin.tdTomato: 20, 37, 25, 23 |
| MAT-Gal4, UAS-inx4-RNAi: 20, 32, 21, 38 | |
| MAT-Gal4, UAS-inx4-RNAi, UAS-GFP-alpha-tub: 43, 45, 37 |
Supplementary Material
Video S1 Related to Figure 2 and Figure S2 Time lapse movies of slbo-Gal4, UAS-LifeAct-GFP/+ (control, left) and slbo-Gal4, UAS-LifeAct-GFP/inx2-RNAi (right). Scale bars: 20 μm.
Video S2 Related to Figure 3 and Figure S4 Time lapse movies of 109c1-Gal4, UAS-LifeAct-GFP/+ (control, left) and slbo-Gal4, UAS-LifeAct-GFP/inx2-RNAi (right). Scale bars: 20 μm.
Video S3 Related to Figure 3 and Figure S4 Time lapse movies of MAT-α-tub-Gal4, slbo-LifeAct-GFP/+ (control, left) and MAT-α-tub-Gal4, slbo-LifeAct-GFP/inx4-RNAi (right). Scale bars: 20 μm.
Video S4 Related to Figure 4 Z-projection movies of w1118 co-stained with anti-Inx2 (green) and anti-Inx3 (magenta) (left) and inx4-GFP (magenta) stained with anti-Inx2 (green) (right). Scale bars: 20 μm.
Video S5 Related to Figure 4 Time lapse movies of MAT-α-tub-Gal4, FC-LexA>LexAOP-GCaMP6s (control, left) and MAT-α-tub-Gal4>UAS-TrpA1, FC-LexA>LexAOP-GCaMP6s (right) egg chambers. 0 min indicates when the temperature shifted from 23°C to 32°C. There is a brief, transient increase in fluorescence in the control but a sustained increase follicle cells of the TrpA1-expressing egg chamber. Scale bars: 20 μm.
Video S6 Related to Figure 4 Time lapse movies of DMSO (upper left) and carbenoxolone treatment (upper right) of ubi-Gal4>GCaMP6s and DMSO (bottom left) and carbenoxolone treatment (bottom right) of slbo-Gal4>LifeAct-GFP egg chambers. Scale bars: 20 μm.
Video S7 Related to Figure 6 Time lapse movies of late stages slbo-Gal4, UAS-LifeAct-GFP/+ (left side) and slbo-Gal4, UAS-LifeAct-GFP/inx2-RNAi (right side). Scale bars: 20 μm.
Video S8 Related to Figure 6 Time lapse movies of FM4–64 staining egg chambers. The genotypes from left to right are slbo-Gal4, UAS-LifeAct-GFP/+ (control); slbo-Gal4, UAS-LifeAct-GFP/inx2-RNAi; slbo−/− and slbo-Gal4>E-cad-RNAi. Scale bars: 20 μm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 568 Phalloidin | ThermoFisher | Cat#A12380 |
| Mouse monoclonal anti-Arm (1:75 dilution) | Developmental Studies Hybridoma Bank |
DSHB Cat#N2.7A1;RRID: AB_528089 |
| Rat monoclonal anti-Ecad (1:50 dilution) | Developmental Studies Hybridoma Bank |
DSHB Cat#DCAD2;RRID: AB_528120 |
| Guinea Pig polyclonal anti-Inx2 (1:1000 dilution) | Smendziuk et al., 2015 | N/A |
| Rabbit anti-Inx3 (1:75 dilution) | Lehmann et al., 2006 | N/A |
| Rabbit polyclonal anti-Inx4 (1:20000 dilution) | Smendziuk et al., 2015 | N/A |
| Rabbit polyclonal anti-Cad99C (1:1000 dilution) | Glowinski et al. (2014) | N/A |
| Mouse monoclonal anti-Tubulin (clone DM1A) (1:100 dilution) | Sigma-Aldrich | Cat#T9026; RRiD: AB_477593 |
| Rabbit anti-aPKC (1:200 dilution) | Santa Cruz Biotechnology | Cat#sc-216 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hoechst 33342 | Sigma-Aldrich | Cat#14533 |
| FM4–64 | ThermoFisher | Cat#T3166 |
| Carbenoxolone | Sigma-Aldrich | Cat#C4790 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: slbo-Gal4 | Rorth. P., et al.,1998 | FBtp0002716 |
| D. melanogaster: y[1] w[*]; P{y[+t*] w[+mC]=UAS-Lifeact-GFP}ViE-260B | Hatan, M., etal., 2011 | FBtp0064437 |
| D. melanogaster: w[111S] | Bloomington Drosophila Stock Center |
RRID:BDSC_3605 FBst0003605 |
| D. melanogaster: RNAi of inx2 y[1] v[1]; P{y[+t7.7] v[+t1.S]=TRiP.JF02446}attP2 | Bloomington Drosophila Stock Center |
BDSC_29306 |
| D. melanogaster: RNAi of inx2 P{KK111067}VIE-260B | Vienna Drosophila Resource Center |
FBst0474063 |
| D. melanogaster: RNAi of inx3 y1 sc* v1 sev21; P{TRiP.HMC05106}attP2 | Bloomington Drosophila Stock Center |
BDSC_60112 FBst0060112 |
|
D. melanogaster: RNAi of inx1 y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.HMS02764}attP2 |
Bloomington Drosophila Stock Center |
BDSC_44048 FBst0044048 |
|
D. melanogaster: RNAi of inx4 y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.GL00447}attP2 |
Bloomington Drosophila Stock Center |
BDSC_35607 FBst0035607 |
| D. melanogaster: w[1118]; P{y[+t7.7] w[+mC]=20XUAS-IVS-GCaMP6s}attP40 | Bloomington Drosophila Stock Center |
BDSC_42746 FBst0042746 |
| D. melanogaster: w[*]; P{w[+mC]=matalpha4-GAL-VP16}V37 | Bloomington Drosophila Stock Center |
BDSC_7063 FBst0007063 |
| D. melanogaster: MTD-Gal4 (Triple-Gal4) P{w[+mC]=otu-GAL4::VP16.R}1, w[*]; P{w[+mC]=GAL4-nos.NGT}40; P{w[+mC]=GAL4::VP16- nos.UTR}CG6325[MVD1] | Bloomington Drosophila Stock Center |
RRID:BDSC 31777 FBst0031777 |
| D. melanogaster: slbo[e7b]/CyO; ry[506] | Bloomington Drosophila Stock Center |
BDSC_58686 FBst0058686 |
|
D. melanogaster: P{ry[+t7.2]=PZ}slbo[01310] cn[1]/CyO; ry[506] |
Bloomington Drosophila Stock Center |
BDSC_12227 FBst0012227 |
|
D. melanogaster: RNAi of E-cad y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00693}attP2 |
Bloomington Drosophila Stock Center |
BDSC_32904 FBst0032904 |
| D. melanogaster: CY2-gal4 | D. Montell lab stock, UCSB | FBti0007266 |
| D. melanogaster: 109c1-Gal4 | Bloomington Drosophila Stock Center |
BDSC_7020 FBst0007020 |
| D. melanogaster: w[*]; P{w[+mC]=UASp-F- Tractin.tdTomato}15A/SM6b; MKRS/TM2 | Bloomington Drosophila Stock Center |
BDSC_58989 FBst0058989 |
| D. melanogaster: w[*]; P{w[+mC]=slbo- Lifeact-GFP}2M/CyO; MKRS/TM6B, Tb[1] | Bloomington Drosophila Stock Center |
BDSC_58364 FBst0058364 |
| D. melanogaster: FC-LexA | Laboratory of Xiaobo Wang, University of Toulouse | N/A |
|
D. melanogaster: [1118]; P{y[+t7.7] w[+mC]=13XLexAop2-IVS-GCaMP6s- SV40}su(Hw)attP5 |
Bloomington Drosophila Stock Center |
BDSC_44589 FBst0044589 |
| D. melanogaster: w[*]; P{y[+t7.7] w[+mC]=UAS-TrpA1 (B).K}attP16 | Bloomington Drosophila Stock Center |
BDSC_26263 FBst0026263 |
| D. melanogaster: UAS-inx2-RFP | Spéder and Brand, 2014 | FBal0338200 |
| D. melanogaster: UAS-inx2-GFP | Bauer et al.,2004 | FBal0179392 |
| D. melanogaster: UAS-inx3-GFP | Lehmann et al., 2006 | FBal0288436 |
| D. melanogaster: w[*]; P{w[+m*]=Ubi- GAL4.U}2/CyO | Bloomington Drosophila Stock Center |
BDSC_32551 FBst0032551 |
| D. melanogaster: y[1] w[*]; P{w[+mC]=UASp-GFPS65C- alphaTub84B}14–6-II/CyO, P{ry[+t7.2]=sevRas1.V12}FK1 | Bloomington Drosophila Stock Center |
BDSC_7374 FBst0007374 |
| D. melanogaster: w[*]; P{w[+mC]=UASp- GFPS65C-alphaTub84B}3/TM3, Sb[1] | Bloomington Drosophila Stock Center |
BDSC_7373 FBst0007373 |
| D. melanogaster: Zpg::zpg-GFP | Smendziuk et al., 2015 | FBal0318355 |
| D. melanogaster: Oregon-R-C | Bloomington Drosophila Stock Center |
BDSC_5 FBsn0000277 |
| D. melanogaster: UAS-R-inx2-RFP (RNAi- resistant inx2) | This paper | N/A |
| D. melanogaster: UAS-R-inx2L35W-RFP (RNAi-resistant inx2 point mutation) | This paper | N/A |
| D. melanogaster: UAS-R-inx2C256S-RFP (RNAi-resistant inx2 point mutation) | This paper | N/A |
| Oligonucleotides | ||
| Primers for PCR out inx2-RFP: inx2-N-F: 5’- ATGTTTGATGTCTTTGGGTCCGTC-3’ |
This paper | N/A |
| Primers for PCR out inx2-RFP: RFP-C-R: 5’- TCACGTGGACCGGTGGGCGC-3’ |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pUAST-attB | www.flyc31.org | GenBank EF362409.1 |
| Software and Algorithms | ||
| Fiji | PMID: 22743772 | https://fiji.sc/ |
| Adobe Illustrator CS6 | Adobe.com | N/A |
| Prism 6 GraphPad | GraphPad Software |
https://www.graphpad.com/scientific-software/prism/ |
| Imaris | Bitplane | https://imaris.oxinst.com/ |
Highlights.
Postmigratory border cells form new contacts in a multistep neolamination process
Innexins 2/3 are required in border cells and Inx4 in the germline for neolamination
Innexins 2/3 and 4 regulate microtubule abundance during neolamination
Innexins and microtubules brace new contacts against ongoing morphogenetic forces
Acknowledgements
We thank James Mondo, Joseph Campanale, Xiaoran Guo and all Montell lab members for discussions. We thank James Mondo for the image in Figure S1B and Wei Dai for the images in Figures S1C-S1E”‘. We thank Guy Tannentzapf for reagents, discussions, and critical reading of the manuscript. We thank Michael Hoch and Andrea Brand for reagents. We thank the Craig Montell and Julie Simpson labs for providing stocks and advice on the thermogenetics experiment. We thank BDSC and VDRC for fly stocks.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baker MW, Yazdani N, and Macagno ER (2013). Gap junction-dependent homolog avoidance in the developing CNS. J Neurosci 33, 16673–16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R, Lehmann C, Martini J, Eckardt F, and Hoch M. (2004). Gap Junction Channel Protein Innexin 2 Is Essential for Epithelial Morphogenesis in the Drosophila Embryo. Molecular Biology of the Cell, 15(8), 2992–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrmann J, and Zimmermann J. (2008). Gap junctions in the ovary of Drosophila melanogaster: localization of innexins 1, 2, 3 and 4 and evidence for intercellular communication via innexin-2 containing channels. BMC Dev Biol 8, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, Jacob L, Patwa R, Shah H, Xu K, et al. (2016). Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, and Ewald AJ (2016). A collective route to metastasis: Seeding by tumor cell clusters. Science 352, 167–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JR, Luchici A, Mosis F, Thackery J, Salazar JA, Mao Y, … Stramer BM (2015). Inter-cellular forces orchestrate contact inhibition of locomotion. Cell, 161(2), 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbouk HA, Mroue RM, El-Sabban ME, and Talhouk RS (2009). Connexins: a myriad of functions extending beyond assembly of gap junction channels. Cell Commun Signal 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depriest A, Phelan P, and Martha Skerrett I. (2011). Tryptophan scanning mutagenesis of the first transmembrane domain of the innexin Shaking-B(Lethal). Biophys J 101, 2408–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P, and Horwitz AR (2015). Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol 7, a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias LAB, and Kriegstein AR (2008). Gap junctions: multifaceted regulators of embryonic cortical development. Trends Neurosci 31, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias LAB, Wang DD, and Kriegstein AR (2007). Gap junction adhesion is necessary for radial migration in the neocortex. Nature 448, 901–907. [DOI] [PubMed] [Google Scholar]
- Elzarrad MK, Haroon A, Willecke K, Dobrowolski R, Gillespie MN, and Al-Mehdi A-B (2008). Connexin-43 upregulation in micrometastases and tumor vasculature and its role in tumor cell attachment to pulmonary endothelium. BMC Med 6, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Hegerfeldt Y, and Tusch M. (2004). Collective cell migration in morphogenesis and cancer. Int J Dev Biol 48, 441–449. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Verlaan I, Hengeveld T, Janssen H, Calafat J, Falk MM, and Moolenaar WH (2001). Gap junction protein connexin-43 interacts directly with microtubules. Curr Biol 11, 1364–1368. [DOI] [PubMed] [Google Scholar]
- Gilboa L, Forbes A, Tazuke SI, Fuller MT, and Lehmann R. (2003). Germ line stem cell differentiation in Drosophila requires gap junctions and proceeds via an intermediate state. Development 130, 6625–6634. [DOI] [PubMed] [Google Scholar]
- Giuliani F, Giuliani G, Bauer R, and Rabouille C. (2013). Innexin 3, a new gene required for dorsal closure in Drosophila embryo. PLoS ONE 8, e69212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowinski C, Liu R-HS, Chen X, Darabie A, and Godt D. (2014). Myosin VIIA regulates microvillus morphogenesis and interacts with cadherin Cad99C in Drosophila oogenesis. J Cell Sci 127, 4821–4832. [DOI] [PubMed] [Google Scholar]
- Guichet A, Peri F, and Roth S. (2001). Stable anterior anchoring of the oocyte nucleus is required to establish dorsoventral polarity of the Drosophila egg. Dev Biol 237, 93–106. [DOI] [PubMed] [Google Scholar]
- Hatan M, Shinder V, Israeli D, Schnorrer F, and Volk T. (2011). The Drosophila blood brain barrier is maintained by GPCR-dependent dynamic actin structures. Journal of Cell Biology, 192(2), 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcroft CE, Jackson WD, Lin W-H, Bassiri K, Baines RA, and Phelan P. (2013). Innexins Ogre and Inx2 are required in glial cells for normal postembryonic development of the Drosophila central nervous system. J Cell Sci 126, 3823–3834. [DOI] [PubMed] [Google Scholar]
- Januschke J, Gervais L, Gillet L, Keryer G, Bornens M, and Guichet A. (2006). The centrosome-nucleus complex and microtubule organization in the Drosophila oocyte. Development 133, 129–139. [DOI] [PubMed] [Google Scholar]
- Kameritsch P, Khandoga N, Pohl U, and Pogoda K. (2013). Gap junctional communication promotes apoptosis in a connexin-type-dependent manner. Cell Death Dis 4, e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Garg V, and Hadjantonakis A-K (2017). Lineage Establishment and Progression within the Inner Cell Mass of the Mouse Blastocyst Requires FGFR1 and FGFR2. Dev Cell 41, 496–510.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC (1970). Ovarian Development in Drosophila melanogaster (New York: Academic Press; ). [Google Scholar]
- Krysko DV, Leybaert L, Vandenabeele P, and D’Herde K. (2005). Gap junctions and the propagation of cell survival and cell death signals. Apoptosis 10, 459–469. [DOI] [PubMed] [Google Scholar]
- Lehmann C, Lechner H, Löer B, Knieps M, Herrmann S, Famulok M, Bauer R, and Hoch M. (2006). Heteromerization of innexin gap junction proteins regulates epithelial tissue organization in Drosophila. Mol Biol Cell 17, 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JHC, Takano T, Cotrina ML, Arcuino G, Kang J, Liu S, Gao Q, Jiang L, Li F, Lichtenberg-Frate H, et al. (2002). Connexin 43 enhances the adhesivity and mediates the invasion of malignant glioma cells. J Neurosci 22, 4302–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo-Macias A, Agullo-Pascual E, and Delmar M. (2016). The cardiac connexome: Non-canonical functions of connexin43 and their role in cardiac arrhythmias. Semin Cell Dev Biol 50, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjarrez-Marmolejo J, and Franco-Pérez J. (2016). Gap Junction Blockers: An Overview of their Effects on Induced Seizures in Animal Models. Curr Neuropharmacol 14, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, and Obenauf AC (2016). Metastatic colonization by circulating tumour cells. Nature 529, 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Campanale JP, Mondo JA, and Montell DJ (2019). Cell interactions in collective cell migration. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell DJ, Rorth P, and Spradling AC (1992). slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell 71, 51–62. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Yoon WH, and Starz-Gaiano M. (2012). Group choreography: mechanisms orchestrating the collective movement of border cells. Nat Rev Mol Cell Biol 13, 631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M, Kato H, Hira S, Nakamura K, Kita H, and Kobayashi S. (2011). Innexin2 gap junctions in somatic support cells are required for cyst formation and for egg chamber formation in Drosophila. Mech Dev 128, 510–523. [DOI] [PubMed] [Google Scholar]
- Niewiadomska P, Godt D, and Tepass U. (1999). DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J Cell Biol 144, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauf AC, and Massagué J. (2015). Surviving at a distance: organ specific metastasis. Trends in Cancer 1, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Uemura T, and Takeichi M. (1997). Phenotypic analysis of null mutants for DE-cadherin and Armadillo in Drosophila ovaries reveals distinct aspects of their functions in cell adhesion and cytoskeletal organization. Genes Cells 2, 29–40. [DOI] [PubMed] [Google Scholar]
- Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, … Cohen SM (1998). Systematic gain-of-function genetics in Drosophila. Development, 125(6), 1049–1057. [DOI] [PubMed] [Google Scholar]
- Richard M, Bauer R, Tavosanis G, and Hoch M. (2017). The gap junction protein Innexin3 is required for eye disc growth in Drosophila. Dev Biol 425, 191–207. [DOI] [PubMed] [Google Scholar]
- Sahu A, Ghosh R, Deshpande G, and Prasad M. (2017). A Gap Junction Protein, Inx2, Modulates Calcium Flux to Specify Border Cell Fate during Drosophila oogenesis. PLoS Genet 13, e1006542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa E, and Mayor R. (2016). Collective cell migration in development. J Cell Biol 212, 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RM, Fay AJ, Puthenveedu MA, von Zastrow M, Jan Y-N, and Jan LY (2007). Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell 128, 547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerrett IM, and Williams JB (2017). A structural and functional comparison of gap junction channels composed of connexins and innexins. Dev Neurobiol 77, 522–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smendziuk CM, Messenberg A, Vogl AW, and Tanentzapf G. (2015). Bi-directional gap junction-mediated soma-germline communication is essential for spermatogenesis. Development 142, 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spéder P, and Brand AH (2014). Gap junction proteins in the blood-brain barrier control nutrient-dependent reactivation of Drosophila neural stem cells. Dev Cell 30, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich TA, Hall DH, and Greenstein D. (2014). Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics 198, 1127–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbings LA, Todman MG, Phelan P, Bacon JP, and Davies JA (2000). Two Drosophila innexins are expressed in overlapping domains and cooperate to form gap-junction channels. Mol Biol Cell 11, 2459–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbings LA, Todman MG, Phillips R, Greer CE, Tam J, Phelan P, Jacobs K, Bacon JP, and Davies JA (2002). Gap junctions in Drosophila: developmental expression of the entire innexin gene family. Mech Dev 113, 197–205. [DOI] [PubMed] [Google Scholar]
- Stramer B, Moreira S, Millard T, Evans I, Huang CY, Sabet O, … Wood W. (2010). Clasp-mediated microtubule bundling regulates persistent motility and contact repulsion in Drosophila macrophages in vivo. Journal of Cell Biology, 189(4), 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazuke SI, Schulz C, Gilboa L, Fogarty M, Mahowald AP, Guichet A, Ephrussi A, Wood CG, Lehmann R, and Fuller MT (2002). A germline-specific gap junction protein required for survival of differentiating early germ cells. Development 129, 2529–2539. [DOI] [PubMed] [Google Scholar]
- Viswanath V, Story GM, Peier AM, Petrus MJ, Lee VM, Hwang SW, Patapoutian A, and Jegla T. (2003). Opposite thermosensor in fruitfly and mouse. Nature 423, 822–823. [DOI] [PubMed] [Google Scholar]
- Wang X, Bo J, Bridges T, Dugan KD, Pan T, Chodosh LA, and Montell DJ (2006). Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev Cell 10, 483–495. [DOI] [PubMed] [Google Scholar]
- Wei C-J, Xu X, and Lo CW (2004). Connexins and cell signaling in development and disease. Annu Rev Cell Dev Biol 20, 811–838. [DOI] [PubMed] [Google Scholar]
- Zhao T, Graham OS, Raposo A, and St Johnston D. (2012). Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science 336, 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JZ, and Jiang JX (2014). Gap junction and hemichannel-independent actions of connexins on cell and tissue functions--an update. FEBS Lett 588, 1186–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1 Related to Figure 2 and Figure S2 Time lapse movies of slbo-Gal4, UAS-LifeAct-GFP/+ (control, left) and slbo-Gal4, UAS-LifeAct-GFP/inx2-RNAi (right). Scale bars: 20 μm.
Video S2 Related to Figure 3 and Figure S4 Time lapse movies of 109c1-Gal4, UAS-LifeAct-GFP/+ (control, left) and slbo-Gal4, UAS-LifeAct-GFP/inx2-RNAi (right). Scale bars: 20 μm.
Video S3 Related to Figure 3 and Figure S4 Time lapse movies of MAT-α-tub-Gal4, slbo-LifeAct-GFP/+ (control, left) and MAT-α-tub-Gal4, slbo-LifeAct-GFP/inx4-RNAi (right). Scale bars: 20 μm.
Video S4 Related to Figure 4 Z-projection movies of w1118 co-stained with anti-Inx2 (green) and anti-Inx3 (magenta) (left) and inx4-GFP (magenta) stained with anti-Inx2 (green) (right). Scale bars: 20 μm.
Video S5 Related to Figure 4 Time lapse movies of MAT-α-tub-Gal4, FC-LexA>LexAOP-GCaMP6s (control, left) and MAT-α-tub-Gal4>UAS-TrpA1, FC-LexA>LexAOP-GCaMP6s (right) egg chambers. 0 min indicates when the temperature shifted from 23°C to 32°C. There is a brief, transient increase in fluorescence in the control but a sustained increase follicle cells of the TrpA1-expressing egg chamber. Scale bars: 20 μm.
Video S6 Related to Figure 4 Time lapse movies of DMSO (upper left) and carbenoxolone treatment (upper right) of ubi-Gal4>GCaMP6s and DMSO (bottom left) and carbenoxolone treatment (bottom right) of slbo-Gal4>LifeAct-GFP egg chambers. Scale bars: 20 μm.
Video S7 Related to Figure 6 Time lapse movies of late stages slbo-Gal4, UAS-LifeAct-GFP/+ (left side) and slbo-Gal4, UAS-LifeAct-GFP/inx2-RNAi (right side). Scale bars: 20 μm.
Video S8 Related to Figure 6 Time lapse movies of FM4–64 staining egg chambers. The genotypes from left to right are slbo-Gal4, UAS-LifeAct-GFP/+ (control); slbo-Gal4, UAS-LifeAct-GFP/inx2-RNAi; slbo−/− and slbo-Gal4>E-cad-RNAi. Scale bars: 20 μm.
Data Availability Statement
Data including all imaging datasets produced in this study will be made available upon request.