Abstract
Deglycosylated, live-attenuated SIV vaccines elicited protective immune responses against heterologous SIVsmE543–3, which differs from the vaccine strain SIVmac239 to levels similar to those across HIV-1 clades. 2/3 of the vaccinees contained the chronic SIVsmE543–3 infection (controllers), whereas 1/3 did not (non-controllers). In this study, we investigated immune correlates of heterologous challenge control in rhesus macaques of Burmese origin. Since depletion of CD8+ cells in the controllers by administration of anti-CD8α antibody abrogated the control of viral replication, CD8+ cells were required for the protective immune response. However, classical SIV specific CD8+ T cells did not account for the protective immune response in all controllers. Instead, IL-15 responding CD8α+ cells, including CD8+ T and NK cells, were significantly higher in the controllers than those in the non-controllers, before and after vaccination with deglycosylated SIV. It is well established that IL-15 signal transduction occurs through “trans-presentation” in which IL-15 complexed with IL-15Rα on monocytes, macrophages and dendritic cells binds to IL-15 Rβ/γ expressed on CD8+ T and NK cells. Accordingly, levels of IL-15 stimulation was strongly affected by the depletion of monocytes from PBMCs, implying key roles of innate immune cells. These results suggest that intrinsic IL-15 responsiveness may dictate the outcome of protective responses and may lead to optimized formulations of future broadly protective HIV vaccines.
INTRODUCTION
Remarkable progress has been made in achieving suppression of HIV replication at levels undetectable by conventional methods with the use of anti-retroviral chemotherapy (ART) (1). Such therapy, however, does not eliminate HIV infection and has thus led to studies coined “HIV cure therapy” aimed at the eradication of HIV reservoirs and/or the implementation of immune based therapies capable of containing HIV infection following ART discontinuation (2). It is well established that adaptive immunity plays a key role to protect the host from microbial infections. Thus, the current clinically available vaccines have been shown to elicit highly effective humoral and/or cellular responses against targeted pathogens (3). These facts served to guide the formulation of most HIV vaccine studies (4). However to date, besides the RV144 clinical trial that showed limited efficacy (5), there has been no clinical study that has shown efficacy in inducing protective immune responses against HIV infection (4). On the other hand, there have been several studies that have reported containment of HIV/SIV infections, that include post-exposure prophylaxis in SHIV infections(6, 7), functional cures in clinical studies(2, 8), exposed uninfected cohorts(9), elite controllers of HIV/SIV infections(10), and natural SIV infections of a number of nonhuman primate species in Africa(11, 12). In addition, studies of select live-attenuated SIV infections have also shown a degree of protection in rhesus macaques, with the common theme that once the host abrogates pathogenic infection, they develop protective immune responses(6, 13, 14). Importantly, one of the conclusions from these clinical and nonhuman primate model studies suggest that aside from adaptive immune responses, other host responses, such as innate immune cells may play an important role in the containment of infection(15–18).
We previously reported that a quintuple deglycosylated mutant derived from SIVmac239, named Δ5G, possesses live-attenuated vaccine properties associated with nearly sterilizing protective responses against homologous SIVmac239 challenge infections(19). Based on these studies, we reasoned that similar live-attenuated vaccine approaches may have the potential to elicit protective immune responses against highly diverse HIV-1. This is the rationale for a more detailed examination of the mechanisms involved by which the deglycosylated live-attenuated SIV vaccine mediates its protective effect and is reported herein. It is important to note that the heterologous SIVsmE543–3 utilized in the present studies differs from SIVmac239 to levels equivalent to HIV-1 inter-subtypes. In addition, the use of rhesus macaques with diverse MHC genotypes (see Table S1) makes it more relevant to studies in human (20). The candidate vaccine exhibits protective effects against the heterologous SIV infection in two-thirds of the vaccinees (controllers). However, the remaining one-third had persistent productive infections during the chronic-phase (non-controllers) with an emergence of escape mutants including recombinant viruses between challenge and vaccine virus. In this study, we found no detectable difference in the SIV specific cellular responses between the controller and non-controller groups of monkeys. Instead, there were significantly higher frequencies of IL-15 responding CD8+ T and NK cells in the controllers. Our findings suggest a contribution of innate immunity to enhance and broaden the range of protective immune responses to suppress heterologous SIV infections, knowledge which may contribute to the containment of infection with diverse HIV.
MATERIALS AND METHODS
Viruses
Deglycosylated, live-attenuated SIV vaccines derived by site-directed mutagenesis of an SIVmac239 nef/open(21) DNA clone and challenge viruses, SIVmac239 nef/open and SIVsmE543–3, were prepared and propagated in phytohemagglutinin-stimulated PBMCs from rhesus macaques as previously described(20).
Animals and ethics statement
Juvenile male rhesus macaques 3–5 years old and of Burmese origin were screened and found negative for SIV, simian T-cell lymphotropic virus, herpes B virus, and type D retrovirus. The animals were housed individually and cared for according to the rules and guidelines for experimental animal welfare by National Institute of Infectious Diseases and National Institute of Biomedical Innovation, Health and Nutrition, Japan. The study was reviewed and approved by the Institutional Animal Care and Use Committees at National Institute of Infectious Diseases and National Institute of Biomedical Innovation, Health and Nutrition (Protocol #606009), in accordance with the recommendations of the Weatherall report.
The animals were fed monkey diet supplemented with fresh fruit and water, and animal health was monitored daily and documented by animal care and veterinary staff. All efforts were expended to minimize suffering. These included improvement of housing conditions, whenever possible, adoption of early endpoints, frequent monitoring of viral loads and immunological parameters, and humane euthanasia by barbiturate overdose once clinical AIDS or signs of fatal disease were noted.
Vaccination of rhesus macaques with deglycosylated, live-attenuated SIV vaccines, and challenge infection with SIVmac239 and SIVsmE543–3
12 animals were used for vaccination with four vaccines with different deglycosylated SIV constructs (quintuple deglycosylated mutants of SIVmac239: Δ5G, Δ5GV1, Δ5GV2, and a triple deglycosylated mutant: Δ3G) as reported (20). The timeline of the vaccination and challenge infections with SIVmac239 and SIVsmE543–3 are shown in Fig. S1. The monkeys Mm0301, Mm0409, and Mm0517 were vaccinated with Δ5G, the monkeys Mm0303, Mm0511, and Mm0513 were vaccinated with Δ5GV1, the monkeys Mm0307, Mm0512, and Mm0518 were vaccinated with Δ5GV2. Mm0304, Mm0515, and Mm0516 were vaccinated with Δ3G. One vaccinee in each group, total four vaccinees (Mm0301, Mm0513, Mm0307, and Mm0304) were challenged with SIVmac239 at 40 weeks post-vaccination. All vaccinees (except Mm0307 who died from a cause unrelated to SIV infection), were challenged with a heterologous SIVsmE543–3 as previously described(20).
Depletion of CD8+ cells in the vaccinees by in vivo administration of anti-CD8 monoclonal antibody
Mm0512 and Mm0517 were infused with the in vivo depleting anti-rhesus CD8α monoclonal antibody (M-T807, NIH nonhuman primate reagent resource) (CD8 Ab) at a dose of 50 mg/Kg intravenously (1/4 of total) and subcutaneously (3/4 of total). Mm0301, Mm0303, Mm0511, Mm0515 and Mm0516 were infused with the CD8 Ab in multiple doses: 10 mg/Kg intramuscularly (i.m.) at day 0, followed by 5 mg/Kg i.m. at day 3 and 7. The treatments were done at the following time points: 79 wpc (Mm0301 and Mm0515), 81 wpc (Mm0516), 86 wpc (Mm0512 and Mm0517), 138 wpc (Mm0303) and 149 wpc (Mm0511).
Depletion of monocytes from PBMCs
Monocytes in PBMCs were depleted by using immune-magnetic beads coated with anti-CD11b monoclonal antibody (non-human primate, Miltenyi biotec) utilizing a protocol provided by the vendor. The efficiency of monocyte depletion was monitored on an aliquot of cells prior to and post depletion using standard polychromatic flow cytometric techniques.
Viral loads
The levels of SIV infection were monitored by measuring the plasma viral RNA loads using a highly sensitive quantitative real-time RT-PCR as described(20). Briefly, viral RNA was isolated from plasma using a MagNA PureCompact Nucleic Acid Isolation Kit (Roche Diagnostics). Real-time RT-PCR was performed using a QuantiTec Probe RT-PCR kit (Qiagen) and a Sequence detection system SDS7000 (Applied Biosystems). To detect SIVmac239 gag and SIVsmE543–3 gag separately, primers and probe sets were synthesized as follow; SIVsmE543–3 gag specific primers: 5’- FAM-GCAGAGGAGGAAATTACCCAGTGC-3’, 5’-CAATTTTACCCAAGCATTTAATGTT- TAMRA- 3’ and probe 5’-TGTCCACCTACCCTTAAGTCCAA-3’, SIVmac239 specific gag primers: 5’-GCAGAGGAGGAAATTACCCAGTAC-3’, 5’-CAATTTTACCCAGGCATTTAATGTT-3’ and probe 5’-FAM-TGTCCACCTGCCATTAAGTCCCGA-TAMRA-3’. These primers and probes are unique for reactivity with SIVmac239 RNA and SIVsmE543–3 RNA and do not cross react. The detection sensitivity of plasma viral RNA by this method was calculated to be 100 viral RNA copies per ml of plasma.
Detection of common γ chain cytokine (IL-2, IL-7 and IL-15), or IL-12 responding cells
Cryopreserved PBMC were thawed, resuspended at 2 × 106 cells/ml in R10 (RPMI1640 supplemented with 10 % heat-inactivated FBS, 55 μM 2-mercaptoethanol, 50 U/ml penicillin and 50 μg/ml streptomycin), and rested for 2 h at 37˚C. The cells were washed and stimulated with IL-15 (PeproTech, 10 ng/ml, final concentration) and/or IL-2 (WAKO, 10 ng/ml, final concentration), IL-7 (R&D,10 ng/ml, final concentration) IL-12 (R&D, 10 ng/ml, final concentration) overnight. The stimulated cells were analyzed using a commercial ELISPOT kit (U-CyTech Bioscience) or a commercial intracellular staining kit (BioLegend). Phenotypic properties of the stimulated cells were identified by intracellular staining reagents and analyzed by standard flowcytometry described below.
SIV specific T cells
Cryopreserved PBMC were thawed, resuspended at 2 × 106 cells/ml in R10, and rested for 2 h at 37˚C. The cells were washed and stimulated with SIV peptides (2 μg/ml, final concentration) in the presence of anti-CD28 antibodies (BD Biosciences,2 μg/ml, final concentration) and anti-CD49d antibodies (BD Biosciences, 2 μg/ml, final concentration) at 37˚C for 6h, then add BFA (BioLegend, 5 μg/ml, final concentration) and Monensin (BioLegend, 2 μM, final concentration) were then added and the cells incubated overnight. The battery of SIV peptides based on sequences of SIVmac239 viral proteins were synthesized by the Microchemical Facility, Emory University School of Medicine.
Flow cytometry
Cryo-preserved PBMCs from the animals were analyzed and sorted by flow cytometry using a FACS Aria III (BD Biosciences) using FITC/Alexa Fluor 488, PE, ECD, PerCP5.5, PE-Cy7, APC, Alexa Fluor 700, APC-Cy7, Pacific Blue, and BD Horizon V500 as fluorescent probes. Fluorochrome-conjugated mAb against CD4 (L200), CD3 (SP34–2), CD8 (SK1), CD95 (DX2), CD20 (L27), CD122 (mlk-beta2), CCR5 (3A9), CD16 (3G8), IFNγ (Β27) ΤΝFα (ΜΑΒ11), IL-2 (MQ1–17H12) and MHC-II DR (L243) were obtained from BD Biosciences. mAb RMO52 (CD14), B9E9 (CD20), CD28.2 (CD28), TP1.55.3 (CD69) and Z199 (NKG2A) were obtained from Beckmann Coulter, while mAb H4A3 (CD107a) and M1/70 (CD11b) were obtained from BioLegend. A fixable-dead-cells stain kit was obtained from Invitrogen. Data were analyzed using FlowJo ver. 9.9.6 software. Absolute counts of immune cells in blood were determined on an F820 automated hematology analyzer (Sysmex, Japan).
Measurement of ability of PBMCs and CD8+ T cells to inhibit SIV infection of CD4+ T cells
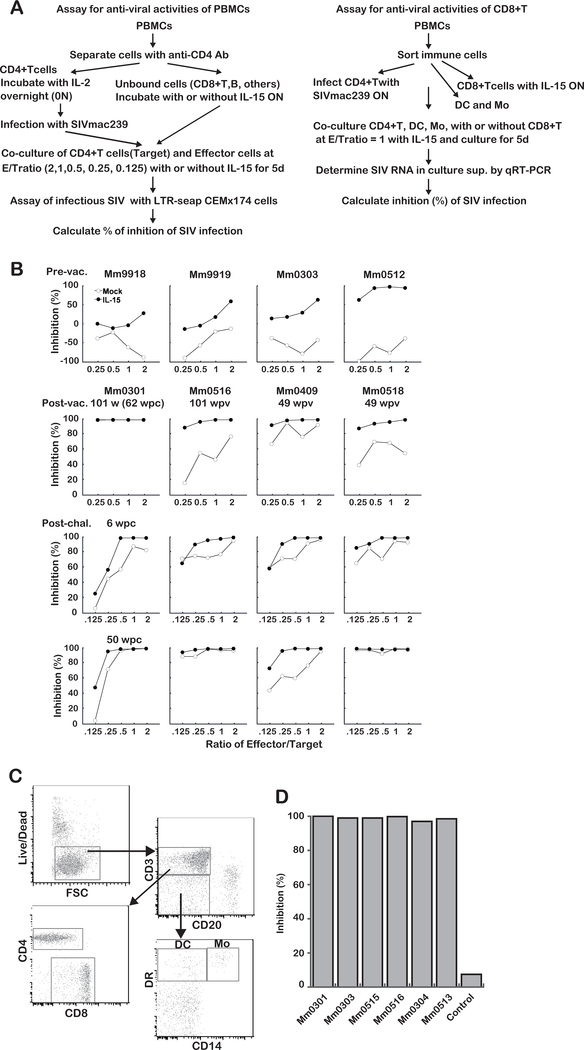
The procedure of the assay for PBMCs and CD8+ T cells are shown in Fig. 7A right and left, respectively. Assay for PBMCs: Cryo-frozen PBMC samples were thawed, resuspended with R10, rested at 37˚ C for 2h, and subjected to a magnetic separation of CD4+ cells from PBMCs with anti-CD4 Abs conjugated beads (BD). CD4+ T cells were incubated with IL-2 (20 U/ml) supplemented R10 (IL2+R10). The remaining fraction (CD8+ T, B, NK, monocytes, DC and other cells) were incubated with R10 with or without IL-15 (10 ng/ml). Next day, CD4+ T cells were infected with SIVmac239 (MOI = 0.1) for 2h. Infected cells (Target) were co-cultured with the remaining fraction (Effector) at ET ratio indicated. After incubation for 5d, levels of infectious SIV were measured using LTR-seap (secreted alkaline phosphatase) expressing CEMx174 cells (22). Assay for CD8+ T cells: Cryo-frozen PBMCs were thawed and sorted into CD4+ T, CD8+ T, DC and monocytes (Mo). CD4+ T cells were re-suspended with IL-2+R10 and infected with or without SIVmac239 at MOI = 0.1 overnight. CD8+ T cells re-suspended with IL-2+ R10 were incubated with IL-15 (50 ng/ml). Monocytes and DC were re-suspended and incubated with R10. Next day, after washing out the virus, CD4+ T cells were co-cultured with Mo and DC with the presence or absence of CD8+ T cells. SIV RNA concentrations in culture supernatant were measured by qRT-PCR described in the method section. Inhibitory activities of CD8+ T cells from the vaccinees were calculated by SIV concentrations of the infected cells (CD4+ T cells with Mo and DC) and those of the infected cells co-cultured with CD8+ T cells at 5d culture. PBMCs from an uninfected animal were used to examine the inhibitory activity with no SIV specific adaptive immune response as a control.
Fig. 7. IL-15 enhances immune cell functions to inhibit SIV infection.
A: Experimental outline of the ex vivo assay to examine the ability of the immune cells to inhibit SIV replication. Anti-viral activity of PBMCs (right) and CD8+T cells (left)
B: Inhibition of SIV replication by immune cells from the vaccinees. First panels: Inhibition by cells from uninfected control animals (Mm9918 and Mm9919) and from two vaccinees (Mm0303 and Mm0512) before vaccination. Inhibition by cells from four of the vaccinees (controllers: Mm0301, Mm0516 and non-controllers: Mm0518 and Mm0409 at three time points are shown as follows. Second panels: before heterologous challenge with SIVsmE543–3 (101 or 49 w post-vaccination); please note that the PBMCs from Mm0301were collected at 62 w post-challenge with SIVmac239 (see Fig. S1). Third panels: 6 w post heterologous challenge with SIVsmE543–3. Bottom panels: 50 w post heterologous challenge with SIVsmE543–3. LTR-seap: SIV long-terminal repeat driving the expression of secreted alkaline phosphatase protein.
C: Cryo-frozen PBMCs were thawed and sorted into CD4+ T, CD8+ T, DC and monocytes (Mo).
D: Inhibitory activities of CD8+ T cells from the vaccinees were calculated by SIV concentrations of the infected cells (CD4+ T cells with Mo and DC) and those of the infected cells co-cultured with CD8+ T cells at 5d culture. PBMCs from an uninfected animal were used to examine the inhibitory activity with no SIV specific adaptive immune response as a control.
Statistical analysis
Statistical analyses were performed using Graphpad Prism software. Two group comparisons were performed using Mann-Whiney U test. Correlations were performed by Spearman test. Significant p values < 0.05 (*), < 0.01 (**), < 0.001(***) are indicated.
RESULTS
CD8+ cell mediated immune responses are associated with containment of heterologous SIV infection
Deglycosylated SIV vaccines induced a near sterile protective immune responses against homologous SIVmac239. We reported that three animals vaccinated with Δ5G were fully protected from homologous SIVmac239 (19). Four animals with one of the other deglycosylated vaccines (Δ5G, Δ5Gv1, Δ5Gv2 or Δ3G) similarly contained SIVmac239 infection for more than 60 weeks post-challenge (wpc) as reported (20). Collectively, deglycosylated SIV vaccine elicited protective immune responses against homologous challenge virus in 7/7 vaccinees. In contrast, the vaccine elicited limiting protective effects on challenge experiments with a heterologous SIVsmE543–3 as previously reported (20) and summarized in Table 1. Whereas seven vaccinees controlled viremia up to 80 weeks post-challenge (wpc) (controllers), four failed to control viral replication (non-controller). These results implied that the immune responses required for the protection from the infection with the heterologous SIV differed from those with the homologous virus. More importantly, these results also indicated differences in the immune responses elicited in the seven controllers as compared with those in four non-controllers against the heterologous SIV, providing us the rationale for defining those differences.
TABLE 1.
Outcomes of heterologous challenge experiments
| Vaccine aΔNG sites in Env |
Δ5G 79, 146, 171, 460, 479 |
Δ5GV1 70, 79, 146, 460, 479 |
Δ5GV2 79, 146, 377, 460, 479 |
Δ3G 146, 171, 460 |
| Animal ID | Mm0301, Mm0409, Mm0517 | Mm0303, Mm0511, Mm0513 | Mm0307d, Mm0512, Mm0518 | Mm0304, Mm0515, Mm0516 |
| bController (N=7) | Mm0301, Mm0517 | Mm0303, Mm0511 | Mm0512 | Mm0515, Mm0516 |
| cNon-controller (N=4) | Mm0409, | Mm0513 | Mm0518 | Mm0304 |
The position of N-glycosylation sites mutated to remove the specific glycans for the vaccines (20).
Vaccinee controlled SIVsmE543-3 replication up to 80 weeks pc (20).
Vaccinee failed to control SIVsmE543-3 replication (20).
Mm0307 died with a SIV-infection-unrelated cause before heterologous challenge infection(20).
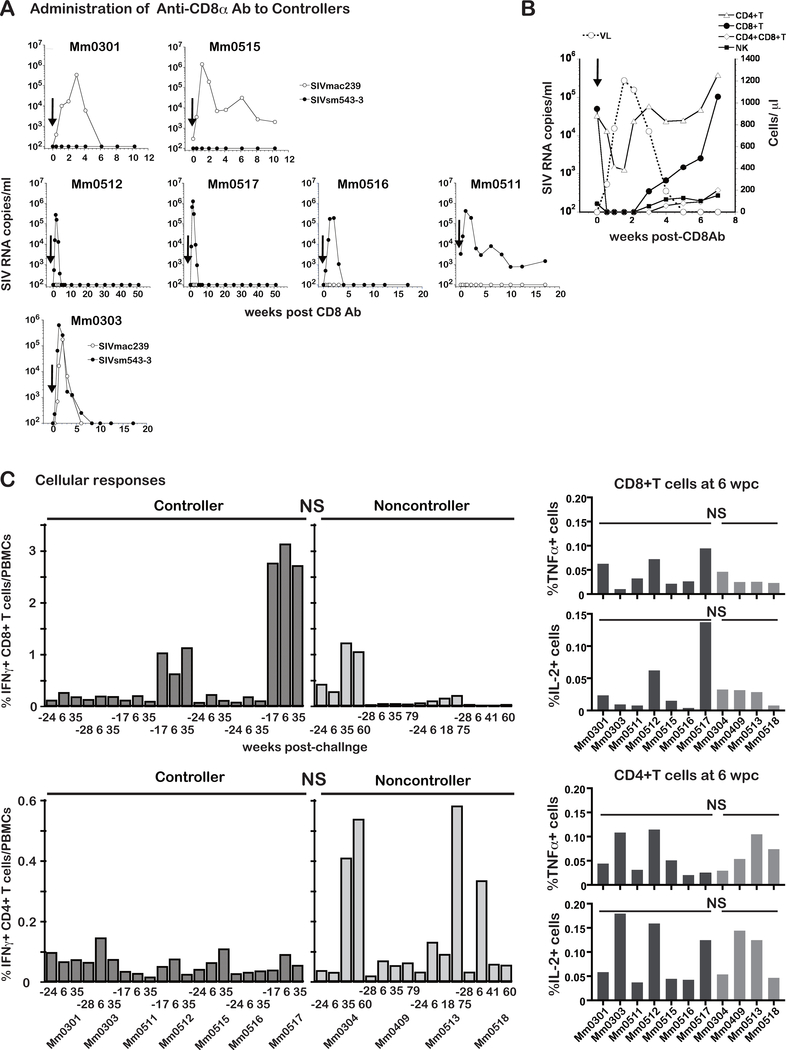
In macaque AIDS models, the administration of an in vivo depleting anti-CD8α monoclonal antibody (CD8 Ab) was shown to reverse the containment of SIV infection, and viral replication resumed concurrent with depletions of CD8+ T cells in blood and lymphoid tissues emphasizing the role of CD8+ T cells (23). Based on these studies, we investigated whether CD8 Ab treatment would also reverse the control of SIV infection in our model. We therefore administrated CD8 Ab to the seven controllers Mm0301, Mm0303, Mm0511, Mm0512, Mm0515, Mm0516 and Mm0517 (Fig. 1A). The CD8 Ab treatment led to SIV replication in each of the controller animals but showed three different patterns of viral rebounds (Fig. 1A). VLs of the vaccine but not the challenge virus was detected in Mm0301 and Mm0515 (Fig. 1A upper). VLs of the challenge but not vaccine virus was detected in Mm0512, Mm0517, Mm0516, and Mm0511 (Fig. 1A middle) and VLs of both viruses were detected in Mm0303 (Fig. 1A bottom), confirming the lasting presence of the latent vaccine virus. Since no SIVsmE543–3 RNA was detected in Mm0301 and Mm0515 after the challenge infection as previously reported (20), they seemed protected from the challenge infection. Whereas the remaining five vaccinees from this group had detectable acute viremia, they also elicited immune responses able to contain the virus replication. Since we detected the vaccine virus with SIVmac239 gag sequences in Mm0301, Mm0515 and Mm0303, we examined the Env sequences to determine if any mutations occurred resulting in changes of N-glycosylation of Env in these animals (Table 2). The results indeed showed mutations associated with new N-glycosylation sites (NGS) at 148, 248 or 417, together with those associated with loss of NGS, resulting in the mutated vaccine viruses with increased total NGS in Mm0301 and Mm0515. These results demonstrated the emergence of vaccine viruses after the CD8 mAb treatment potentially due to increases of N-glycans as an adaptive mechanism in these vaccinees. The vaccine viruses in Mm0303 had experienced both loss and gain of N-glycosylation sites that seemed associated with gain of fitness compared to the original vaccine despite the equal total NGS.
Fig. 1. CD8+ cells mediated immune responses are required for containment of SIV infection.
A: Plasma viral loads of seven controller animals following the in vivo administration of an anti-CD8α antibody (CD8 Ab) administration (shown by arrows). Two of the animals (Mm0301 and Mm0515) had viremia due to the vaccine but not challenge virus. Four animals (Mm0512, Mm0517, Mm0516 and Mm0511) had viremia of the heterologous challenge but not vaccine virus. Mm0303, however, had viremia due to both challenge and vaccine virus origin.
B: Correlation of the absolute number of CD4+ T, CD8+ T, CD4+CD8+ T and NK cells with plasma viral load kinetics in a representative animal (Mm0512) following the in vivo administration of CD8 Ab.
C: SIV specific cellular responses. PBMCs from the controller (n = 7) and non-controller animals (n = 4) were incubated with a pool of overlapping SIV peptides encompassing the viral proteins (Gag, Pol, Env, Vif, Vpr, Vpx, Tat, Rev, and Nef) of SIVmac239 and the frequencies of IFNγ expressing cells are shown. The PBMC samples analyzed were collected pre-challenge (−28, −24 or −17 weeks prior to the challenge), and at 6, 35, 41, 60, and 75 weeks post-challenge with SIVsmE543–3. Responses of SIV specific CD8+ T cells (left upper panel) and SIV specific CD4+ T cells (left lower panel) are shown for the frequencies of IFNγ expressing cells. Responses of SIV specific CD8+ T cells (right upper panel) and SIV specific CD4+ T cells (right lower panel) at 6 wpc are shown by the frequencies of TNFα and IL-2 expressing cells. Differences in the cellular responses of the controller and non-controller were not significant (NS) (p >0.05) by Mann-Whitney t test.
TABLE 2.
Mutations associated with changes in N-glycosylation in SIV vaccine
| VL (SIV RNA copies/ml) | original | vaccine Env | SIV543 Env | ||||||
|---|---|---|---|---|---|---|---|---|---|
| vaccinee | days post treatment | SIV239 gag | SIV543 gag | vaccine | new NGS (a.a)a | lost NGS (a.a) | total #NGS | deglycosylationb | |
| Mm0301 | 8 | 10125 | NDc | Δ5G | 417 | 19 | −4 | ND | |
| Δ5G | 417 | 19 | −4 | ND | |||||
| 21 | 340000 | ND | Δ5G | 148 | 19 | −4 | ND | ||
| Δ5G | 417 | 19 | −4 | ND | |||||
| Δ5G | 148 | 19 | −4 | ND | |||||
| Δ5G | 148, 417 | 20 | −3 | ND | |||||
| Mm0303 | 15 | 173750 | 258125 | Δ5GV1 | 417 | 247 | 18 | −5 | Dd |
| Δ5GV1 | 417 | 247 | 18 | −5 | D | ||||
| Δ5GV1 | 417 | 247 | 18 | −5 | D | ||||
| Δ5GV1 | 417 | 247 | 18 | −5 | D | ||||
| Δ5GV1 | 417 | 247 | 18 | −5 | D | ||||
| Δ5GV1 | 417 | 247 | 18 | −5 | D | ||||
| Δ5GV1 | 417 | 247 | 18 | −5 | D | ||||
| Δ5GV1 | 417 | 247 | 18 | −5 | D | ||||
| Mm515 | 8 | 1406250 | ND | Δ3G | 148, 248 | 202 | 21 | −2 | ND |
| ND | Δ3G | 148, 248 | 202 | 21 | −2 | ND | |||
| ND | Δ3G | 148, 248 | 202 | 21 | −2 | ND | |||
The position of new N-glycosylation site (NGS: NXS/T) in amino acid residue in Env (a.a) of the vaccine virus.
Number of N-glycosylation sites mutated to reduce N-glycosylation sites relative to SIVmac239.
Not detected.
Detected.
Despite differences in SIV replication during CD8 depletion, VLs returned to the levels measured before the depletion by 10 weeks after CD8 Ab administration in all controllers (Fig. 1A), indicating that the containment of SIV infection was only transiently reversed by depletion of CD8+ cells. Since CD8α is expressed by variety of CD8+ cells such as classical CTLs that are CD3+ CD8+ T cells, they are also expressed by NK cells and CD4+/CD8+ (DP) T cells. We thus monitored these different lineages following the administration of the CD8 Ab. None of the CD8+ cell lineages were detectable from day 3 to 2 weeks post-treatment (pt), associated with VLs rebounds with a peak VL (4 × 105 copies/ml) at 11 days pt (Mm0512, Fig. 1B). CD8+ T cells were detectable at 3 weeks pt, and the relative number of all except CD3+/CD8+ T cells returned to levels prior to the treatment by 4 weeks pt, with the levels of CD8+ T cells returning to pre-treatment levels much later. Similar results were obtained in other animals as well (data not shown). These results suggest a role for not only CD8+ T cells but also the other CD8 expressing lineages in the containment of SIV infection.
Next, we examined SIV specific CD8+ and CD4+ T cell responses in PBMCs by intracellular staining of IFNγ in cells restimulated in vitro from the weeks −24, 6 and later post challenge. Levels of SIV specific CD8+T cells varied markedly from animal to animal, in both controllers and non-controllers, indicating no statistical difference between controllers and non-controllers (Fig. 1C left upper). Likewise, levels of SIV specific CD4+ T cells were variable among the vaccinees with notably high levels in two non-controllers (Mm0304 and Mm0513), but once again, without statistical difference between controllers and non-controllers (Fig. 1C left lower). We examined SIV specific CD8+ and CD4+ T cell responses by the expression of TNFα and IL-2, confirming the absence of statistical difference (Fig. 1C right panels).
IL-15 responding CD8+ T and NK cells in PBMCs from vaccinees
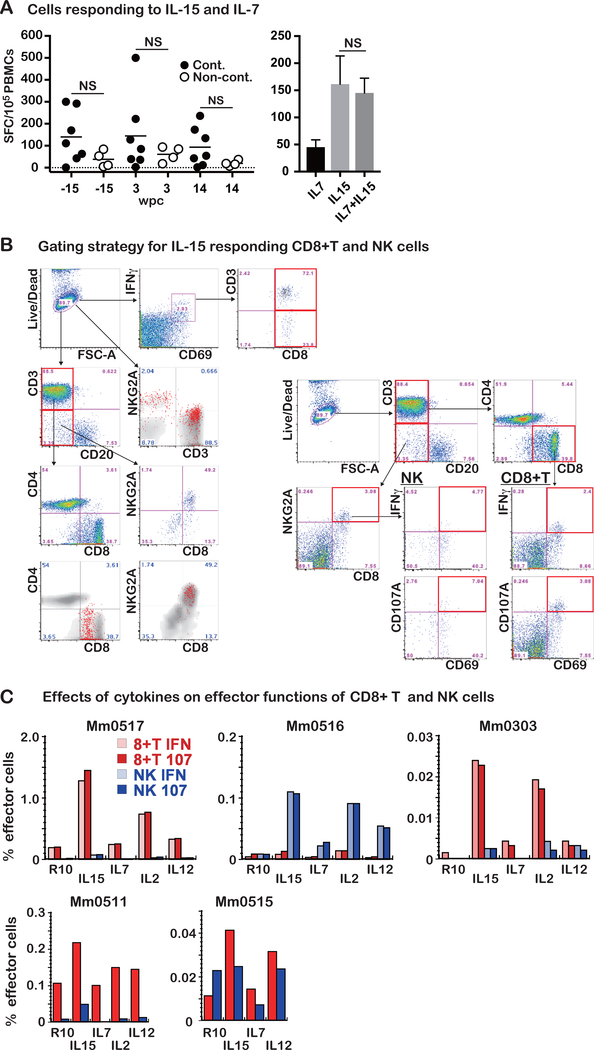
IL-7 and IL-15 are key cytokines for homeostatic proliferation and maintenance of naïve and memory T cells (24). They are also useful adjuncts for the detection of low levels of antigen specific T cells by ex vivo assays (25). Thus, we examined the effects of these cytokines on the quantitation of SIV specific T cells in PBMCs by the ELISPOT assay. A mixture of these cytokines stimulated T cells to express IFNγ even in the absence of SIV peptides (Fig. 2A, left). Strikingly, the frequencies of IFNγ expressing cells from the controllers were higher than those in the non-controllers. Next, we examined which cytokine was required for the IFNγ expression by using PBMCs from one of the controllers. Whereas IL-7 exhibited marginal effects, IL-15 alone induced stimulatory activity equivalent to that noted with the combined use of IL-7 and IL-15 (Fig. 2A right). These results indicated that PBMCs from controllers consist of the cells that synthesized significantly higher levels of IFNγ upon IL-15 stimulation than non-controllers.
Fig. 2. CD8+ T and NK cells are main cells stimulated with IL-15.
A. Left panel: Stimulation of PBMCs from the seven controller and four non-controller animals were incubated in vitro with a mixture of rIL-7 and rIL-15 overnight and the frequencies of IFNγ expressing cells using a standard ELISPOT assay. Data shown are results obtained from PBMC samples collected at −15, 3, and 14 wpc. As noted, the frequencies of IFNγ synthesizing cells were higher in the controllers (n =7) compared with those in the non-controllers (n = 4), although the difference was not statistically significant (NS, p > 0.05). Right panel: Comparison of the effects of rIL-7 and rIL-15 alone or in combination. Frequencies of IFNγ expressing cells in PBMCs from the animals (n = 4) following in vitro incubation overnight with either rIL-7, rIL-15, or a cocktail of rIL-7 and rIL-15. The stimulation with IL-15 resulted in similar SFC to that with the cocktail: correlation (r = 0.905) and no statistical difference (p > 0.05). The results are shown as mean and SD.
B. Left panel: Results from the flow cytometric analyses of PBMCs from the controller (Mm0517, 45 wpc) show that the IFNγ−synthesizing cells following incubation overnight with rIL-15 in vitro comprised both CD8+ T cells and CD3-CD8+NKG2A+ (NK) cells. To identify phenotypic properties of IFNγ−synthesizing cells (shown in red), IFNγ+CD69+ cells, IFNγ+CD69+CD3+CD8+ cells, and IFNγ+CD69+CD3−CD8+ cells were overlapped to the relevant gating panels. Right panel: The frequencies of IFNγ− and CD107− expressing CD8+ T and CD3-CD8+NKG2A+ NK cells were analyzed by the gating strategy as shown.
C: The in vitro stimulatory effects of the common γ chain cytokines (IL-2, IL-7 and IL-15) and IL-12 were compared. The degree of stimulation was rank ordered as: IL-15 >IL-2 >IL-12≧IL-7. The frequencies of CD8+ T and CD3-CD8+NKG2A+ NK cells stimulated with these cytokines varied among animals. However, the predominant phenotypes stimulated were CD8+ T and CD3-CD8+NKG2A+ NK cells in PBMC samples from Mm0517, Mm0516, Mm0303, Mm0511 and Mm0515. The frequencies of IFNγ- and/or CD107a-expressing cells were similar as shown, since most stimulated cells expressed both IFNγ and CD107a (data not shown). R10: RPMI1640 media supplemented with 10% FCS
In efforts to further investigate the effects of IL-15, we studied the cell lineages responding to IL-15 by flow cytometric assisted intracellular staining assays with a focus on CD8α expressing cells since their depletion induced viral rebounds. As seen in Fig. 2B left, both CD3+/CD8+ and CD3-/CD8+ cells expressed IFNγ and CD69 (upper panels). Further analysis showed that the IFNγ+ cells were both CD3-/NKG2A+ (NK cells) and CD3+/ NKG2A dim (CD8+ T cells) (The second panel). The cell lineage assignment of IFNγ+ synthesizing cells was confirmed by noting that these were indeed CD3+/CD8+ (bottom left) and CD3-/CD20-/NKG2A+/CD8+ (bottom right). Based on these results, we measured levels of IL-15 responding CD8+ T cells and NK cells by expression of IFNγ and CD107a (lysosome-associated membrane protein 1, a marker of degranulation) using the gating strategy shown in Fig. 2B right.
Besides IL-7 and IL-15, effector functions of CD8+ T and NK cells are regulated by IL-2 and IL-12. Thus, we stimulated PBMCs from five controllers (Mm0303, Mm0511, Mm0515, Mm0516 and Mm0517) with IL-15, IL-7, IL-2, or IL-12, and measured the frequencies of IFNγ+ and CD107a+ responding cells. The frequencies of IFNγ- and CD107a-expressing cells were similar as shown (Fig. 2C upper panels), since most stimulated cells expressed both IFNγ and CD107a (data not shown). These cytokines stimulated different levels of CD8+ T or NK cells with the following hierarchy: IL-15>IL-2>IL-12≧IL-7 (Fig. 2C). Of note, while the predominant IL-15 stimulated cells were CD8+ T cells in Mm0517, Mm0303 and Mm0511, NK cells were the predominant cells stimulated in Mm0516.
Frequencies of IL-15 responding cells in controllers are significantly higher than those in non-controllers
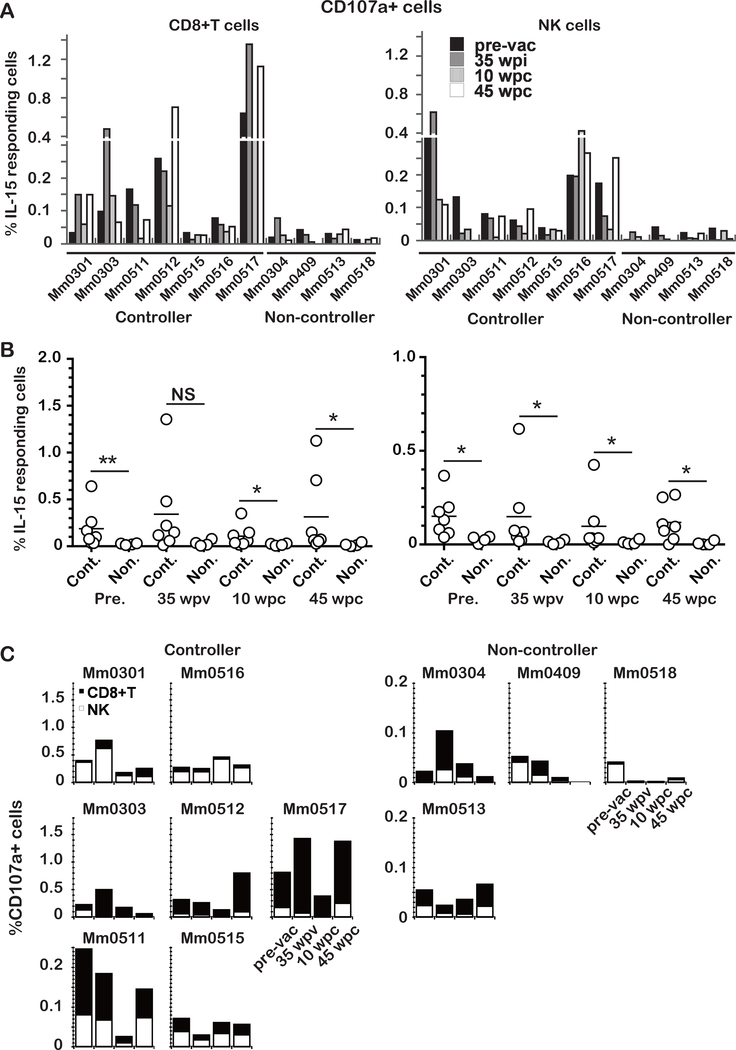
Different frequencies of IL-15 responding cells were observed between controllers and non-controllers before and after the challenge infection and they were composed of CD8+ T and NK cells (Fig. 2). Thus, we examined the frequencies of CD107a+ or IFNγ+ CD8+ T and NK cells by the intracellular staining assay (Fig. 2B right) at 0 or −13 weeks prior to the vaccination (pre-vac), 35 weeks post-vaccination (35 wpv), 10 and 45 weeks post-challenge infection with SIVsmE543–3 (10, 45 wpc). Since the IL-15 responding cells co-expressed CD107a and IFNγ, similar results were obtained for both markers (data not shown), those of CD107a are shown in Fig. 3. Frequencies of IL-15 responding cells were consistently lower in non-controllers for each of the time points compared to those measured in the controllers (Fig. 3A). Statistical differences in the frequencies of IL-15 responding cells between the controllers and non-controllers were examined (Fig. 3B). Frequencies of IL-15 responding CD8+ T cells were significantly higher in controllers than those in non-controllers at each time point examined except 35 wpv (left). Similarly, frequencies of IL-15 responding NK cells in controllers were significantly higher at all time points (right). These results suggest that both IL-15 responding CD8+ T and NK cells played important roles in the containment of the heterologous SIV infection. Of note, the difference was found even before the vaccination, implying an intrinsic individually set of innate immune responses regulating the levels of IL-15 responses.
Fig. 3. Different frequencies of CD107a expressing CD8+ T and CD3-CD8+NKG2A+ NK cells by ex-vivo IL-15 stimulation between controllers (n =7) and non-controllers (n = 4).
A: CD107a expressing CD8+ T and CD3-CD8+NKG2A+ NK cells in PBMCs following incubation overnight with rIL-15 from the vaccinees collected at pre-vaccination (pre-vac), 35 weeks post-infection with vaccine (wpv), 10 and 45 weeks post-challenge infection (wpc).
B: Statistical analysis of the data presented in A. NS (not significant) p >0.05, *p < 0.05, ** p <0.01.
C: Comparison of the frequencies of CD107a-expressing CD8+ T and CD3-CD8+NKG2A+ NK cells in response to rIL-15 in vitro among the different group of animals. While the frequencies of the two cell lineages varied, the predominant cells were NK cells in Mm0301 and Mm0516, the predominant responding cells were CD8+ T cells in Mm0303, Mm0512 and Mm0517. In addition, while both CD8+ T and NK cells were stimulated, the frequencies were lower among controllers (Mm0511 and Mm0515). The response to rIL-15 by PBMCs from the non-controllers (Mm0304, Mm0409 and Mm0518) decreased to or remained at very low levels following challenge infection. In contrast, PBMCs from animal Mm0513 showed an increase in the frequencies of rIL-15 responding cells.
Since frequencies of IL-15 responding CD8+ T versus those of IL-15 responding NK cells were variable among individual animals (Fig. 3A), the cumulative frequencies of IL-15 responding CD8+ T and NK cells were examined (Fig. 3C). The controllers were sorted into three groups from the results. 1) Animals that showed NK cells as the major IL-15 responding cells (Mm0301 and Mm0516; upper left). 2) Animals that showed CD8+ T cells as the major IL-15 responding cells (Mm0303, Mm0512, and Mm0517; second left). 3) Animals that showed the major IL-15 responding cells to be CD8+ T and NK cells, but the frequencies of IL-15 responding cells in these animals (Mm0511 and Mm0515; bottom left, please note the Y axis) were significant lower. The non-controller animals showed two different patterns. 1) The animals in whom the frequencies of IL-15 responding cells declined to low levels after 10 wpc and developed AIDS(20) (Mm0304, Mm0409 and Mm0518; upper right). 2) One animal in which the frequencies of IL-15 responding cells increased after 10 wpc, consistent with the control of viral replication previously reported(20) (Mm0513; bottom right).
Frequencies of NK cells in controllers and non-controllers
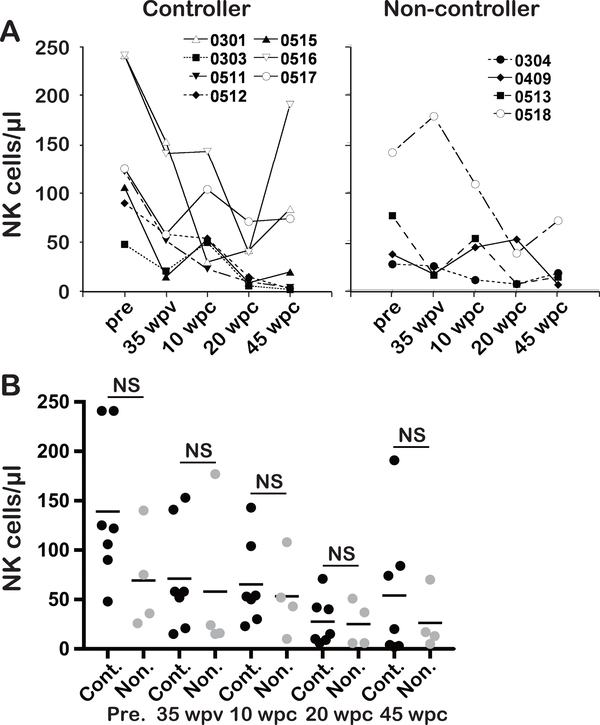
To investigate whether the higher frequencies of IL-15 responding NK cells were due to higher overall frequencies of NK cells in the two controllers (Mm0301and Mm0516) among the vaccinees, we examined the frequencies of NK cells. As shown in Fig. 4A, Mm0301 and Mm0516 indeed had higher frequencies of NK cells at most time points studied. However, similar high frequencies of NK cells were observed in the non-controllers (e.g. Mm0409). Furthermore, there was no statistical difference in frequencies of NK cells between controllers and non-controller at any time point (Fig. 4B). Together, these findings suggest a qualitative rather than a quantitative NK function in the control of heterologous SIV infection.
Fig. 4. Frequencies of NK cells in PBMCs in controllers and non-controllers.
A: The frequencies of NK (CD3- CD8+ NKG2A+) cells in PBMCs from the controllers (Mm0301, Mm0303, Mm511, Mm0512, Mm0515, Mm0516 and Mm0517) and the non-controllers (Mm0304, Mm0419, Mm0513 and Mm0518) at pre-vac., 35 wpv, 10, 45 wpc as determined by flow cytometric analyses are shown.
B: Statistical analysis of the data presented in A. NS (not significant) p > 0.05.
Monocytes are required for IL-15 stimulation of CD8+ T and NK cells
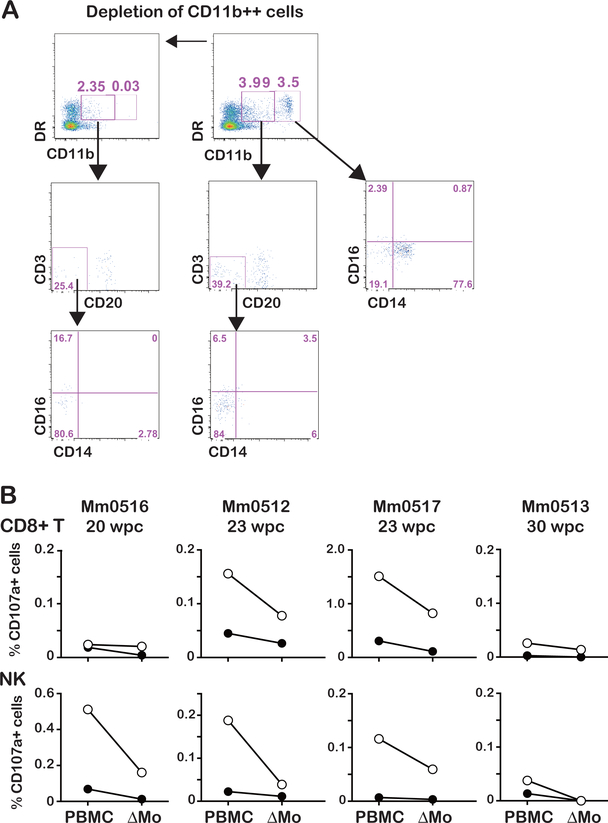
IL-15 stimulates cells by a unique process, termed trans-presentation(26–28), in which IL-15 complexed to its cognate IL-15α-receptor expressed on monocytes (Mo)/macrophages (Mf)/dendritic cells (DC) binds to the IL-2/IL-15Rβ (CD122)/common γ chain (CD132) receptor dimers expressed on CD8+ T and NK cells. Thus, we reasoned that the IL-15 stimulation of the cells in PBMCs as reported above should require Mo. We thus examined if the depletion of Mo from PBMCs could affect IL-15-induced stimulation of CD8+ T and NK cells. As seen in Fig. 5A, incubation of PBMC with anti-CD11b antibody coated beads led to >95% depletion of Mo (CD11b++) and partial depletion of DC (CD11b dim). In the intracellular staining assay described above, Mo depleted PBMCs showed a marked reduction in the frequencies of CD107a expressing CD8+ T and NK cells in response to IL-15 in the animals (Mm0512, and Mm0517) or NK cells in animal Mm0516 (Fig. 5B). Since the depletion of Mo reduced CD107a expressing cells in PBMCs even in the absence of IL-15-stimulation, these findings indicate that the endogenous IL-15/IL-15Rα-stimulation was affected. Of note, frequencies of CD107a expressing cells in IL-15-stimulated Mo-depleted PBMCs were higher than those in the un-stimulated PBMCs from Mm0512, Mm0516 and Mm0517, suggesting IL-15-trans-presentation may occur also by cells other than Mo, such as DC as reported(29). In contrast, the effects of Mo depletion in the non-controller Mm0513 were much lower than those in the controller animals.
Fig. 5. Monocytes are involved in IL-15-stimulation of CD8+ cells.
A: Efficiency of monocyte depletion: monocytes were depleted from PBMCs with anti-CD11b Ab coated beads (Miltenyi Biotec). Note, the flow cytometric profile prior to (top panel) and post-depletion (bottom panel) showing > 95 % of depletion of CD11b++ expressing monocytes and remaining >50% of CD11b+ DC (DR+CD20-CD3-CD14- cells).
B: (Top panel) frequencies of CD107a expressing CD8+ T cells in PBMCs from three controller monkeys (Mm0516, Mm0512, and Mm0517) and one non-controller monkey (Mm0513) in the absence (closed circles) or presence (open circles) of IL-15. (Bottom panel), frequencies of CD107a+ NK cells (CD3-CD8+NKG2A+) in PBMC or monocyte depleted PBMCs (ΔMo) in the absence (closed circles) and presence (open circles) of IL-15.
Increases of IL-15 responding cells were associated with renewed control of viremia post administration of anti-CD8α Ab
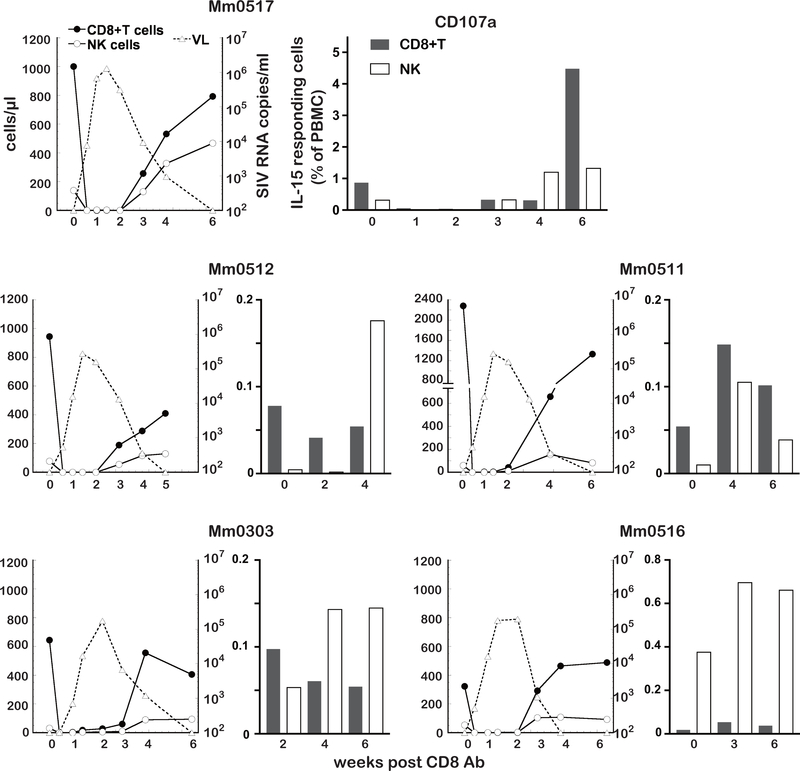
As shown in Fig.1A, the in vivo administration of the CD8 Ab led to rapid SIV rebound but the rebound was transient. Viral replication was gradually controlled which appeared to correlate with the restoration of CD8+ cells. Based on the data above, we carried out studies to determine if there was a role for IL-15 responding CD8 expressing cells in the re-control of viremia in such animals. Mm0517 was administered with CD8 Ab at 86 wpc that, as expected, led to the depletions of CD8+ cells followed by rapid viral rebound that continued up to 6 weeks pt (Fig. 6, upper left). Thus, the CD8 expressing cells became undetectable from day 3 pt and up to 2 weeks pt. VL were detected at 3 days pt, peaked at 10 days pt, then decreased to undetectable levels at 6 weeks pt. As noted, IL-15 responding CD8+ T and NK cells were detectable as early as 3 weeks pt, and increased with distinct kinetics (Fig. 6, upper right). The level of IL-15 responding NK cells in Mm0517 at 4 weeks pt was four-fold higher than that at pre-treatment. A similar increase in the levels of IL-15 responding CD8+ T cells was observed but only at 6 weeks pt. These results indicate earlier increase of IL-15 responding NK cells compared to IL-15 responding CD8+T cells, suggesting a more important role of the former than the latter cells. Similar earlier increases of IL-15 responding NK cells were observed in three controllers (Mm0512, Mm0511 and Mm0303). And these four controllers maintained higher levels of IL-15 responding CD8+T cells than IL-15 responding NK cells (Fig. 3C). Mm0516 was administered CD8 Ab at 81 wpc that resulted in the depletion of CD8+ cells followed by viral rebound starting day 3 that was controlled again at 4 weeks pt (Fig. 6, bottom left). The rebound of NK and CD8+ T cells appeared at 3 weeks pt with IL-15 responding NK cells that were higher than those at pre-treatment (the results at 1, 2, 4 weeks pt were not available due to low viability of the PBMCs). The frequencies of IL-15 responding CD8+ T cells were much lower at pre-treatment and 3 and 6 weeks pt, compared to those of IL-15 responding NK cells, consistent with the results that the predominant IL-15 responding cells were NK cells in Mm0516 (Fig. 3). Taken together, these results suggest IL-15 responding NK cells are key effector cells to suppress viral rebound caused by CD8+ cell depletion.
Fig. 6. IL-15 responding cells increase consistent with control of viral rebound by CD8 Ab administration.
To examine levels of IL-15 responding cells, PBMC samples collected from the CD8 Ab treated animals were subjected to the ex-vivo assay described.
Left panels: Relation between the kinetics of depletion of CD8+ T cells (closed circle) and NK cells (open circle) and viral rebound (open triangle) in 5 controller animals (Mm0517, Mm0512, Mm0511, Mm0303 and Mm0516).
Right panels: Frequencies of CD107a+CD8+ T cells (gray) and CD107a+NK (white) cells in PBMCs from the same 5 controller monkeys obtained at various times post depletion of CD8- expressing cells using the CD8 Ab.
IL-15 enhances immune cells’ functions to inhibit SIV infection
Since depletion of CD8+ cells by the administration of the CD8 Ab disrupted the containment of SIV infection, we examined the ability of immune cells from PBMCs, which contain CD8+ T and NK cells, to inhibit SIV infection in an ex vivo assay as shown in Fig. 7A. The cells from uninfected (Mm9918, Mm9919) and before vaccination (Mm0303 and Mm0512) did not inhibit SIV infection except for cells stimulated with IL-15 at high E/T ratios (Fig. 7B first panel). In contrast, the cells from the vaccinated animals (Mm0301, Mm0516, Mm0409 and Mm0518) inhibited SIV infection (Fig. 7B second to fourth), that indicated a critical role of SIV specific adaptive immune responses elicited by the vaccination. Of note, the infections of CD4+ T cells with SIVmac239 were almost completely suppressed in the presence of IL-15 stimulated cells not only from the controllers (Mm0301 and Mm0516) but also those from the non-controllers (Mm0409 and Mm0518). These results confirmed the protective immune responses in the controllers and non-controllers were enough to contain the homologous SIV infection (19, 20).
Since the immune cells consist of cells other than CD8+ T, such as NK, B, Mo, DC and innate lymphoid cells, we evaluated the inhibitory activity of CD8+ T cells by the ex vivo assay using sorted cells (Fig. 7D). Again, CD8+ T cells from the controllers or non-controllers strongly inhibited SIVmac239 infection in the presence of IL-15 stimulation. Collectively, these results demonstrate a key role of adaptive immunity and the importance of IL-15 responses for the containment of SIV infection via this arm of immunity.
DISCUSSION
In this study, we investigated immune correlates in the protection from the challenge infection with heterologous SIVsmE543–3 in rhesus macaques vaccinated with the deglycosylated, live-attenuated SIV. The vaccine induced near sterile protective immunity against homologous SIVmac239 in all vaccinees as reported (19, 20), but the protective immune responses against a heterologous challenge were varied among the vaccinees resulting the controllers and non-controllers, that enabled us to investigate immune components associated with the protective immunity against diverse SIV. It is widely accepted that CD8 expressing cell lineages play critical roles in the protective immune response against viral pathogens including HIV/SIV. Indeed, depletion of CD8+ cells in the controllers by the administration of anti-CD8α Ab resulted in rebound of SIV replication, clearly demonstrating that CD8+ cells were required for the containment of heterologous SIV infection. However, unexpectedly, levels of SIV specific CD8+ T cells could not account for the differences in the containment among the vaccinees. Instead, we identified frequencies of IL-15 responding CD8+ T and NK cells to be correlated, suggesting critical roles of this cytokine pathway and effector cells in the containment of the infection. In addition, the rapid increases of the IL-15 responding cells concomitant with the decreases of VL following viral rebound caused by the administration of the CD8 Ab, further supports important roles of these IL-15 responding cells in the suppression of viremia. However, we also demonstrated SIV specific CD8+ T cells were required in the protective immunity in the vaccinees in ex vivo studies (Fig. S2). Collectively, we submit that the live-attenuated, deglycosylated SIV vaccine induces potent protective immune responses consist of adaptive and innate immune responses, though host innate responses contribute to the breadth or magnitude of this protection, whereby IL-15 mediated effector cells play critical roles in the containment of infection with diverse SIV.
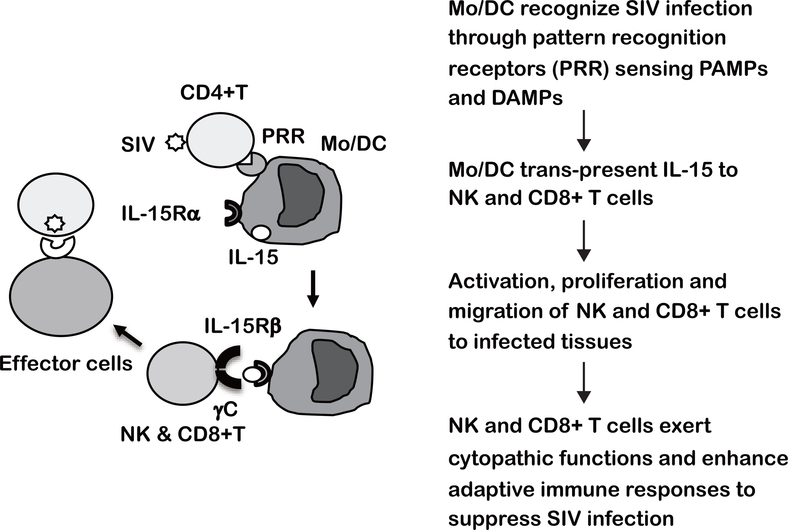
IL-15 primarily acts as a cytokine that stimulates NK and CD8+ T cells through cell to cell interaction, termed “trans-presentation”, in which IL-15/IL-15Rα complexes expressed on myeloid cells (Mo, Mf and DC) bind to IL-2/15Rβ and the common γ receptor (γC) hetero-dimers expressed on NK and CD8+ T cells(26–28). Thus, we speculate such cell to cell interactions are one of the mechanisms by which IL-15 mediated responses contribute to the protective immune response against diverse HIV/SIV as shown in Fig. 8. Innate immune cells such as Mo, Mf and DC sense PAMPs and DAMPs(30, 31) derived from SIV infection and the associated tissue damages, respectively. The activated innate immune cells trans-present IL-15 to NK and CD8+ T cells, that leads to the activation of their effector functions including select chemokine(s) that promote their migration to the infected tissues, such as secondary lymphoid organs (SLO), intestine and other tissues/organs. Collectively, NK and CD8+ T cells stimulated with IL-15 not only exert cytopathic functions against diverse SIV but also contribute to the strengthening of adaptive immune responses that in concert lead to the containment of heterologous SIV infection.
Fig. 8. Working model to illustrate IL-15 mediated immune responses for containment of heterologous SIV infection.
Innate immune cells such as Mo Mf, and DC sense pattern-associated molecular patterns (PAMPs) and/or damage-associated molecular patterns (DAMPs) derived from SIV infection. The activated innate immune cells synthesize IL-15 and present IL-15/IL-15 receptor α complex on the cell surface, and trans-present IL-15 to NK and CD8+ T cells expressing IL-2/15β receptor/IL-15γ (common γ) receptor heterodimer. IL-15 stimulated NK and CD8+ T cells proliferate, are armed with effector functions and migrate to the inflammatory sites caused by SIV infection. IL-15 responding CD8+ T and NK cells not only exert cytopathic function and but also enhance adaptive SIV specific immune responses against diverse SIV.
The anti-microbial functions of IL-15 has been explored in efforts to improve the efficacy of vaccines and immune therapies of HIV infection by several groups but the results obtained have so far been unfortunately inconsistent (32–34). Nevertheless, since the discovery that the predominant in vivo function of IL-15 occurs through trans-presentation, clinical applications with the modified forms of IL-15, such as highly glycosylated heterodimeric IL-15/IL15αR (hetIL-15)(35) and IL-15N72D mutein bound to IL-15RαSU/FC (ALT-803)(36), were investigated. These compounds exhibited superior effects on suppression of SHIV and SIV infections (37, 38). Of note, both studies demonstrated an increase in the frequencies of NK and CD8+ T cells in B-cell follicles of LNs, one of the most studied sites that are thought to serve as potential reservoirs of SIV/HIV infections(39–42). Intriguingly, NK cells are required in B cell follicles for the control of non-pathogenic SIV infection in African green monkeys(43). Thus, accumulating evidence indicates that IL-15 activated CD8+ cells exert pivotal roles in anti-HIV/SIV immune responses, particularly in SLO where latent and persistent productive infections occur during the chronic-phase.
It is tempting to speculate about the parameters that dictate the levels of IL-15 responding cells in the vaccinees. Thus, is it the cells expressing heterodimers of IL-2Rβ/γC or IL-15Rα expressing myeloid cell lineage? The heterodimer-expressing CD8+ T cells are abundant as reported(24). In contrast, levels of NK cells were variable and seemed to influence the levels of IL-15 responding NK cells as shown in Fig. 4. Alternatively, NK functions are regulated by a variety of NK receptors such as killer cell lectin-like receptors and killer cell immunoglobin-like receptors (KIRs)(44). Since KIRs are highly diverse in macaques and their functions are influenced by cognate MHC-1 epitopes (45), the levels of IL-15 responding NK and CD8+ T cells might be influenced by MHC-1 genotypes (Table S1). On the other hand, levels of IL-15Rα expressing Mo and DC are also important factors determining the IL-15 responding cells in the vaccinees. Thus, levels of IL-15Rα expressing myeloid cells in vaccinees might be critical factors that determine the frequencies of IL-15 responding CD8+ cells. Collectively, these points serve as a foundation to investigate and define the protective immune responses against diverse SIV/HIV further.
Mf are the major target cells of HIV/SIV in tissues (brain, lung, intestine and others) and the infections of Mf define distinct diseases manifestations(46–48). Recently, we reported critical roles played by Mo and Mf to define the pathogenic SIVmac239 infection of CXCR3+CD4+ T cells(49). We provided data to support the fact that Mo/Mf expressed high levels of CXCL10 that facilitates the migration of CXCR3+ cells to the inflammatory sites and SLO, where aggregates of activated immune cells localize to the paracortex, consisting of cells enriched for infected CD4+ T cells. Herein, we demonstrate a critical role for Mo and DC by their ability to induce high levels of IL-15 responding CD8+ cells that lead to the containment of heterologous SIV infection. Collectively, it is critical to define what subsets of the myeloid lineage play key roles in the pathogenic infection and protective immunity.
Live-attenuated vaccines have been successfully used to protect from several infectious diseases. Such vaccines have been shown to cause pathogenic side-effects at a very low frequencies, which prompts their use to develop much safer vaccines(50). In case of HIV vaccine studies, several kinds of live-attenuated SIV vaccines showed potent protective effects in macaque AIDS models(13, 14, 19). However the residual pathogenic properties present in such candidate vaccines has prevented advancing the development of live-attenuated HIV vaccines(4). Nevertheless it is clear that unravelling the mechanisms by which such live-attenuated vaccines elicit potent protective immune responses provide an invaluable tool to delineate immune correlates of protection, a subject that remains a main challenge in HIV vaccine research(51). We previously reported that deglycosylated, live-attenuated mutant (Δ5G) targets distinct CD4+ T cells than SIVmac239 which preferentially infects CD4+ T cells in SLO such as Th1 and Tfh (52). In this study, we demonstrate that while levels of SIV specific CD8+ T cells could not account for the differences in the containment of heterologous SIV between controllers and non-controllers, the levels of IL-15 mediated effector cells were closely correlated with this protective immunity. Since the emergence of recombinant viruses between the vaccine and challenge virus was associated with the pathogenic outcomes in the non-controllers (20), IL-15 mediated immune responses may play a key role in the suppression of the highly virulent viruses created by recombination among diverse replicating SIVs. However, the vaccination did not change the intrinsic differences in levels of IL-15 responding cells between controllers and non-controllers (Supplemental Fig. 2), since they seem primarily regulated by innate immune cells such as monocytes and DCs. Thus, understanding the mechanism by which IL-15 mediated immune responses protect host from diverse SIV may contribute to the development of HIV vaccines and immune therapies for “HIV cure”.
Supplementary Material
KEY POINTS.
Deglycosylated SIV vaccine elicited protective response against heterologous SIV.
SIV specific CD8+T cells did not account for the protective response.
IL-15 responding NK and CD8+T cells were associated with the protective response.
ACKNOWLEDGEMENTS
We thank Kayoko Ueda for excellent technical assistance. This study was conducted through Cooperative Research Program in Tsukuba Primate Research Center, National Institutes of Biomedical Innovation, Health and Nutrition. We would like to acknowledge all the animal care and veterinary staff at the Tsukuba Primate Research Center.
Financial support
This work was supported by the Japanese Research Grants from the Ministry of Health, Labor, and Welfare in Japan (11947035 to K.M.), from the Ministry of Education, Culture, Sports, Science and Technology in Japan (15K10043 & 18K08558 to M.F. and 18K05986 to K.H.), and by US NIH grant (RO1AI111907 to F.V.).
REFERENCES
- 1.Eisele E, and Siliciano RF. 2012. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 37: 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard JP, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, and Rouzioux C. 2013. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 9: e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plotkin SA 2008. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis 47: 401–409. [DOI] [PubMed] [Google Scholar]
- 4.Esparza J 2013. A brief history of the global effort to develop a preventive HIV vaccine. Vaccine 31: 3502–3518. [DOI] [PubMed] [Google Scholar]
- 5.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, and M.-T. Investigators. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361: 2209–2220. [DOI] [PubMed] [Google Scholar]
- 6.Mori K, Yasutomi Y, Sawada S, Villinger F, Sugama K, Rosenwith B, Heeney JL, Uberla K, Yamazaki S, Ansari AA, and Rubsamen-Waigmann H. 2000. Suppression of acute viremia by short-term postexposure prophylaxis of simian/human immunodeficiency virus SHIV-RT-infected monkeys with a novel reverse transcriptase inhibitor (GW420867) allows for development of potent antiviral immune responses resulting in efficient containment of infection. J Virol 74: 5747–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai CC, Follis KE, Sabo A, Beck TW, Grant RF, Bischofberger N, Benveniste RE, and Black R. 1995. Prevention of SIV infection in macaques by (R)-9-(2-phosphonylmethoxypropyl)adenine. Science 270: 1197–1199. [DOI] [PubMed] [Google Scholar]
- 8.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, Deeks SG, Luciw PA, Chipman JG, Beilman GJ, Hoskuldsson T, Khoruts A, Anderson J, Deleage C, Jasurda J, Schmidt TE, Hafertepe M, Callisto SP, Pearson H, Reimann T, Schuster J, Schoephoerster J, Southern P, Perkey K, Shang L, Wietgrefe SW, Fletcher CV, Lifson JD, Douek DC, McCune JM, Haase AT, and Schacker TW. 2017. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med 23: 1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton RE, McLaren PJ, Fowke K, Kimani J, and Ball TB. 2010. Cohorts for the study of HIV-1-exposed but uninfected individuals: benefits and limitations. J Infect Dis 202 Suppl 3: S377–381. [DOI] [PubMed] [Google Scholar]
- 10.Goulder P, and Deeks SG. 2018. HIV control: Is getting there the same as staying there? PLoS Pathog 14: e1007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, and Silvestri G. 2012. Natural SIV hosts: showing AIDS the door. Science 335: 1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenchley JM, Vinton C, Tabb B, Hao XP, Connick E, Paiardini M, Lifson JD, Silvestri G, and Estes JD. 2012. Differential infection patterns of CD4+ T cells and lymphoid tissue viral burden distinguish progressive and nonprogressive lentiviral infections. Blood 120: 4172–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, and Desrosiers RC. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258: 1938–1941. [DOI] [PubMed] [Google Scholar]
- 14.Miller CJ, McChesney MB, Lu X, Dailey PJ, Chutkowski C, Lu D, Brosio P, Roberts B, and Lu Y. 1997. Rhesus macaques previously infected with simian/human immunodeficiency virus are protected from vaginal challenge with pathogenic SIVmac239. J Virol 71: 1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altfeld M, and Gale M Jr. 2015. Innate immunity against HIV-1 infection. Nat Immunol 16: 554–562. [DOI] [PubMed] [Google Scholar]
- 16.Carrington M, and Alter G. 2012. Innate immune control of HIV. Cold Spring Harb Perspect Med 2: a007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ploquin MJ, Jacquelin B, Jochems SP, Barre-Sinoussi F, and Muller-Trutwin MC. 2012. Innate immunity in the control of HIV/AIDS: recent advances and open questions. AIDS 26: 1269–1279. [DOI] [PubMed] [Google Scholar]
- 18.Hertoghs N, Geijtenbeek TBH, and Ribeiro CMS. 2017. Interplay between HIV-1 innate sensing and restriction in mucosal dendritic cells: balancing defense and viral transmission. Curr Opin Virol 22: 112–119. [DOI] [PubMed] [Google Scholar]
- 19.Mori K, Yasutomi Y, Ohgimoto S, Nakasone T, Takamura S, Shioda T, and Nagai Y. 2001. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J Virol 75: 4023–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto C, Watanabe S, Naruse T, Kajiwara E, Shiino T, Umano N, Ueda K, Sato H, Ohgimoto S, Hirsch V, Villinger F, Ansari AA, Kimura A, Miyazawa M, Suzuki Y, Yamamoto N, Nagai Y, and Mori K. 2010. Protection of macaques with diverse MHC genotypes against a heterologous SIV by vaccination with a deglycosylated live-attenuated SIV. PLoS One 5: e11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kestler HW 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, and Desrosiers RC. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65: 651–662. [DOI] [PubMed] [Google Scholar]
- 22.Means RE, Greenough T, and Desrosiers RC. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J Virol 71: 7895–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matano T, Shibata R, Siemon C, Connors M, Lane HC, and Martin MA. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol 72: 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surh CD, and Sprent J. 2008. Homeostasis of naive and memory T cells. Immunity 29: 848–862. [DOI] [PubMed] [Google Scholar]
- 25.Jennes W, Kestens L, Nixon DF, and Shacklett BL. 2002. Enhanced ELISPOT detection of antigen-specific T cell responses from cryopreserved specimens with addition of both IL-7 and IL-15--the Amplispot assay. J Immunol Methods 270: 99–108. [DOI] [PubMed] [Google Scholar]
- 26.Dubois S, Mariner J, Waldmann TA, and Tagaya Y. 2002. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity 17: 537–547. [DOI] [PubMed] [Google Scholar]
- 27.Lodolce JP, Burkett PR, Boone DL, Chien M, and Ma A. 2001. T cell-independent interleukin 15Ralpha signals are required for bystander proliferation. J Exp Med 194: 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stonier SW, and Schluns KS. 2010. Trans-presentation: a novel mechanism regulating IL-15 delivery and responses. Immunol Lett 127: 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koka R, Burkett P, Chien M, Chai S, Boone DL, and Ma A. 2004. Cutting edge: murine dendritic cells require IL-15R alpha to prime NK cells. J Immunol 173: 3594–3598. [DOI] [PubMed] [Google Scholar]
- 30.Iwasaki A, and Medzhitov R. 2015. Control of adaptive immunity by the innate immune system. Nat Immunol 16: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, and Hauser CJ. 2010. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464: 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halwani R, Boyer JD, Yassine-Diab B, Haddad EK, Robinson TM, Kumar S, Parkinson R, Wu L, Sidhu MK, Phillipson-Weiner R, Pavlakis GN, Felber BK, Lewis MG, Shen A, Siliciano RF, Weiner DB, and Sekaly RP. 2008. Therapeutic vaccination with simian immunodeficiency virus (SIV)-DNA + IL-12 or IL-15 induces distinct CD8 memory subsets in SIV-infected macaques. J Immunol 180: 7969–7979. [DOI] [PubMed] [Google Scholar]
- 33.Hryniewicz A, Price DA, Moniuszko M, Boasso A, Edghill-Spano Y, West SM, Venzon D, Vaccari M, Tsai WP, Tryniszewska E, Nacsa J, Villinger F, Ansari AA, Trindade CJ, Morre M, Brooks D, Arlen P, Brown HJ, Kitchen CM, Zack JA, Douek DC, Shearer GM, Lewis MG, Koup RA, and Franchini G. 2007. Interleukin-15 but not interleukin-7 abrogates vaccine-induced decrease in virus level in simian immunodeficiency virus mac251-infected macaques. J Immunol 178: 3492–3504. [DOI] [PubMed] [Google Scholar]
- 34.Mueller YM, Petrovas C, Bojczuk PM, Dimitriou ID, Beer B, Silvera P, Villinger F, Cairns JS, Gracely EJ, Lewis MG, and Katsikis PD. 2005. Interleukin-15 increases effector memory CD8+ t cells and NK Cells in simian immunodeficiency virus-infected macaques. J Virol 79: 4877–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong E, Usiskin IM, Bergamaschi C, Hanlon DJ, Edelson RL, Justesen S, Pavlakis GN, Flavell RA, and Fahmy TM. 2016. Configuration-dependent Presentation of Multivalent IL-15:IL-15Ralpha Enhances the Antigen-specific T Cell Response and Anti-tumor Immunity. J Biol Chem 291: 8931–8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Jones M, Liu B, Zhu X, Johnson CB, Edwards AC, Kong L, Jeng EK, Han K, Marcus WD, Rubinstein MP, Rhode PR, and Wong HC. 2013. Efficacy and mechanism-of-action of a novel superagonist interleukin-15: interleukin-15 receptor alphaSu/Fc fusion complex in syngeneic murine models of multiple myeloma. Cancer Res 73: 3075–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seay K, Church C, Zheng JH, Deneroff K, Ochsenbauer C, Kappes JC, Liu B, Jeng EK, Wong HC, and Goldstein H. 2015. In Vivo Activation of Human NK Cells by Treatment with an Interleukin-15 Superagonist Potently Inhibits Acute In Vivo HIV-1 Infection in Humanized Mice. J Virol 89: 6264–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson DC, Moysi E, Valentin A, Bergamaschi C, Devasundaram S, Fortis SP, Bear J, Chertova E, Bess J Jr., Sowder R, Venzon DJ, Deleage C, Estes JD, Lifson JD, Petrovas C, Felber BK, and Pavlakis GN. 2018. Treatment with native heterodimeric IL-15 increases cytotoxic lymphocytes and reduces SHIV RNA in lymph nodes. PLoS Pathog 14: e1006902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, and Perreau M. 2016. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 22: 754–761. [DOI] [PubMed] [Google Scholar]
- 40.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M Jr., Estes JD, Lifson JD, and Picker LJ. 2015. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 21: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong JJ, Amancha PK, Rogers K, Ansari AA, and Villinger F. 2012. Spatial alterations between CD4(+) T follicular helper, B, and CD8(+) T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J Immunol 188: 3247–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, and Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210: 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huot N, Jacquelin B, Garcia-Tellez T, Rascle P, Ploquin MJ, Madec Y, Reeves RK, Derreudre-Bosquet N, and Muller-Trutwin M. 2017. Natural killer cells migrate into and control simian immunodeficiency virus replication in lymph node follicles in African green monkeys. Nat Med 23: 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanier LL 2008. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 9: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter L, and Ansari AA. 2015. MHC and KIR Polymorphisms in Rhesus Macaque SIV Infection. Front Immunol 6: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desrosiers RC, Hansen-Moosa A, Mori K, Bouvier DP, King NW, Daniel MD, and Ringler DJ. 1991. Macrophage-tropic variants of SIV are associated with specific AIDS-related lesions but are not essential for the development of AIDS. Am J Pathol 139: 29–35. [PMC free article] [PubMed] [Google Scholar]
- 47.DiNapoli SR, Hirsch VM, and Brenchley JM. 2016. Macrophages in Progressive Human Immunodeficiency Virus/Simian Immunodeficiency Virus Infections. J Virol 90: 7596–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sattentau QJ, and Stevenson M. 2016. Macrophages and HIV-1: An Unhealthy Constellation. Cell Host Microbe 19: 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujino M, Sato H, Okamura T, Uda A, Takeda S, Ahmed N, Shichino S, Shiino T, Saito Y, Watanabe S, Sugimoto C, Kuroda MJ, Ato M, Nagai Y, Izumo S, Matsushima K, Miyazawa M, Ansari AA, Villinger F, and Mori K. 2017. Simian Immunodeficiency Virus Targeting of CXCR3(+) CD4(+) T Cells in Secondary Lymphoid Organs Is Associated with Robust CXCL10 Expression in Monocyte/Macrophage Subsets. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minor PD 2015. Live attenuated vaccines: Historical successes and current challenges. Virology 479–480: 379–392. [DOI] [PubMed] [Google Scholar]
- 51.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, McDermott AB, Schultz A, Zamb TJ, Boyle R, and Desrosiers RC. 2006. HIV vaccine design: insights from live attenuated SIV vaccines. Nat Immunol 7: 19–23. [DOI] [PubMed] [Google Scholar]
- 52.Sugimoto C, Nakamura S, Hagen SI, Tsunetsugu-Yokota Y, Villinger F, Ansari AA, Suzuki Y, Yamamoto N, Nagai Y, Picker LJ, and Mori K. 2012. Glycosylation of simian immunodeficiency virus influences immune-tissue targeting during primary infection, leading to immunodeficiency or viral control. J Virol 86: 9323–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.