Abstract
Cancer cells, including melanoma, often metastasize regionally through lymphatics before metastasizing systemically through the blood1–4; however, the reason for this is unclear. Here we show that melanoma cells in lymph experience less oxidative stress and form more metastases than melanoma cells in blood. Immunocompromised mice with patient-derived melanomas and immunocompetent mice with mouse melanomas had more melanoma cells per microliter of tumor-draining lymph than tumor-draining blood. Cells metastasizing through blood, but not lymph, became dependent on the ferroptosis inhibitor GPX4. Cells pre-treated with chemical ferroptosis inhibitors formed more metastases than untreated cells after intravenous, but not intralymphatic, injection. We observed multiple differences between lymph fluid and blood plasma that may contribute to decreased oxidative stress and ferroptosis in lymph, including higher levels of glutathione and oleic acid, and less free iron, in lymph. Oleic acid protected melanoma cells from ferroptosis in an Acsl3-dependent manner and increased their capacity to form metastatic tumors. Melanoma cells from lymph nodes were more resistant to ferroptosis and formed more metastases after intravenous injection than melanoma cells from subcutaneous tumors. Exposure to the lymphatic environment thus protects melanoma cells from ferroptosis and increases their ability to survive during subsequent metastasis through the blood.
One sentence summary
Lymph protects melanoma cells from the oxidative stress and ferroptotic cell death that occurs in the blood during metastasis.
Melanomas and epithelial cancers commonly form metastases in draining lymph nodes before forming distant metastases1–4. Genetic studies in human and mouse cancers, including melanoma, have shown that regional lymph node metastases sometimes give rise to distant metastases5–8. Consistent with this, cancer cells in lymph nodes can migrate into blood vessels and then metastasize through the blood9,10. Nonetheless, some distant metastases arise from clones that are distinct from those in sampled lymph nodes5,7. In these cases, it is unclear whether the distantly metastasizing cells entered the blood directly from primary tumors or whether they first migrated through lymphatics before entering the blood.
In patients with cutaneous melanomas, regional lymph node metastasis is one of the most important predictors of distant metastasis and death11. Complete lymph node dissection does not improve survival12,13; however, this does not necessarily mean that distant metastasis occurs independent of lymphatics as melanoma cells from lymph nodes may commonly enter the blood and disseminate before regional metastases are even detected.
There is increasing evidence that lymphatics promote the migration and survival of cancer cells. Some cancer cells form more tumors after injection into lymph nodes (intranodal) as compared to intravenous injection14. VEGF-C and various chemokines promote the migration of cancer cells into lymphatic vessels15. The capacity to metabolize fatty acids promotes the survival of cancer cells in lymphatics16 and the formation of metastatic tumors17. However, circulating cancer cells have been characterized mainly in the blood, with little information about how these cells compare to cancer cells in lymphatics. While some cancer cells migrate through lymphatics before entering the blood and forming distant metastases5–10,18, it is unknown whether exposure to lymph influences the subsequent survival of these cells in the blood.
Metastasis through the blood is a highly inefficient process in which few cancer cells survive19. One factor that limits survival is oxidative stress20,21; however, it is unknown how oxidative stress kills metastasizing cells. Lipid oxidation can induce ferroptosis—a mode of cell death in which polyunsaturated fatty acids in phospholipids are oxidized by redox-active iron22. Although cancer cells can undergo ferroptosis in vivo23–26, most ferroptosis studies have been performed in culture, limiting our understanding of the circumstances in which ferroptosis affects cancer growth or progression in vivo.
Increased melanoma survival in lymph
We injected efficiently (obtained from patients M405, M481, and UT10) and inefficiently (obtained from patients UM17, UM43, and UM47) metastasizing melanomas subcutaneously, intravenously, and intranodally (into the popliteal lymph node) in NOD/SCID IL2Rγnull (NSG) mice (Table 1). Efficient metastasizers spontaneously give rise to circulating cancer cells in the blood and distant metastases after xenografting in NSG mice while inefficient metastasizers do so to a more limited extent27 (Extended Data Table 1a). All of the melanomas were tagged with constitutive DsRed and luciferase, unambiguously distinguishing these cells from recipient mouse cells (Extended Data Fig. 1a–d for flow cytometry gating strategies) and enabling the quantification of metastasis by bioluminescence imaging (Extended Data Fig. 1e–g).
Table 1.
Human and mouse melanoma cells are more likely to form distant metastases after intralymphatic as compared to intravenous injection.
| Frequency of cells that formed tumors |
|||||
|---|---|---|---|---|---|
| Melanoma | Subcutaneous injection | Intravenous injection | Intranodal injection |
||
| lymph node tumor | distant mets | ||||
| PDXs Efficient | M405 | 1 in 11†§ | 1 in 271*‡§ | 1 in 6†§ | 1 in 63*†‡ |
| M481 | 1 in 6†§ | 1 in 335*‡§ | 1 in 24†§ | 1 in 51*†‡ | |
| UT10 | 1 in 11†§ | 1 in 512*‡ | 1 in 144†§ | 1 in 182*‡ | |
| AVERAGE | 1 in 9†§ | 1 in 373*‡§ | 1 in 58†§ | 1 in 99*†‡ | |
| PDXs Inefficient | UM17 | 1 in 9† | 1 in 2,238*‡ | 1 in 124†§ | 1 in 643‡ |
| UM43 | 1 in 14† | 1 in 2,225*‡ | 1 in 235†§ | 1 in 722‡ | |
| UM47 | 1 in 11†§ | 1 in 512*‡ | 1 in 62†§ | 1 in 127*‡ | |
| AVERAGE | 1 in 11†§ | 1 in 1,658*‡ | 1 in 140†§ | 1 in 497*‡ | |
| Mouse melanomas | YUMM1.7 | 1 in 1,241†‡§ | 1 in 144,270*‡§ | 1 in 622*†§ | 1 in 4,522*†‡ |
| YUMM3.3 | 1 in 3,499†‡§ | 1 in 72,245*‡§ | 1 in 590*†§ | 1 in 1,286*†‡ | |
| AVERAGE | 1 in 2,370†‡§ | 1 in 108,258*‡§ | 1 in 606*†§ | 1 in 2,904*†‡ | |
Limiting dilution analysis of the frequency of human or mouse melanoma cells that formed tumors after subcutaneous, intravenous, or intranodal injection. The data for intranodal injections reflect the frequency of cells that formed lymph node tumors at the injection site as well as the frequency of cells that formed distant metastatic tumors. The minimum frequency of cells that formed tumors was determined using ELDA software. Statistical significance was assessed using generalized linear models for multiple logistic regression followed by Tukey’s multiple comparisons adjustment per melanoma (p<0.05 relative to *subcutaneous, †intravenous, ‡intranodal (lymph node tumor), or §intranodal (distant metastasis) injection). The data reflect a total of 520 mice. Exact numbers of mice for each injection site/melanoma and exact p values for each comparison are in source data files.
Consistent with a prior study20, limiting dilution analysis showed that 1 in 6 to 1 in 14 human melanoma cells formed tumors after subcutaneous injection in NSG mice (Table 1). Also consistent with this study, many fewer cells formed metastatic tumors after intravenous injection: an average of 1 in 373 cells from efficient and 1 in 1,658 cells from inefficient metastasizers. Significantly fewer cells were required to form tumors after injection into popliteal lymph nodes: an average of 1 in 58 cells from efficient metastasizers and 1 in 140 cells from inefficient metastasizers. Moreover, these intranodal injections gave rise to more distant metastases than intravenous injections: an average of 1 in 99 cells from efficient metastasizers and 1 in 497 cells from inefficient metastasizers.
We independently tested this in immunocompetent C57BL mice by subcutaneously transplanting mouse melanomas. We used two YUMM lines28, each of which spontaneously metastasized (Extended Data Table 1b): YUMM1.7 (BrafV600E/+;PTEN–/–;Cdkn2–/–) and YUMM3.3 (BrafV600E/+;Cdkn2–/–). An average of 1 in 2,370 cells formed tumors after subcutaneous injection and 1 in 108,258 cells formed metastatic tumors after intravenous injection (Table 1). After injection into popliteal lymph nodes, an average of 1 in 606 cells formed lymph node tumors, significantly more than after subcutaneous or intravenous injection. Moreover, these intranodal injections gave rise to more distant metastases than intravenous injections (1 in 2,904 cells). Therefore, in both immunocompromised and immunocompetent mice, melanoma cells were significantly better at surviving and forming metastatic tumors after intranodal as compared to intravenous injection.
To compare the numbers of spontaneously metastasizing melanoma cells in lymph versus blood, we injected NSG or C57BL mice subcutaneously with human or mouse melanomas, respectively. Once the subcutaneous tumors reached ~2.0 cm in diameter, we determined the numbers of melanoma cells by flow cytometry in tumor-draining blood and lymph, as well as in distant blood and contralateral lymph from the same mouse (Extended Data Fig. 1h). We consistently observed significantly more melanoma cells per microliter in tumor-draining lymph as compared to tumor-draining blood in human (Fig. 1a) and mouse (Fig. 1b) melanomas. We were generally able to detect circulating human and mouse melanoma cells in blood obtained by cardiac puncture, though they were less frequent than in tumor-draining blood. We were rarely able to detect melanoma cells in contralateral lymph. Melanoma cells are thus significantly more abundant in draining lymph than draining blood.
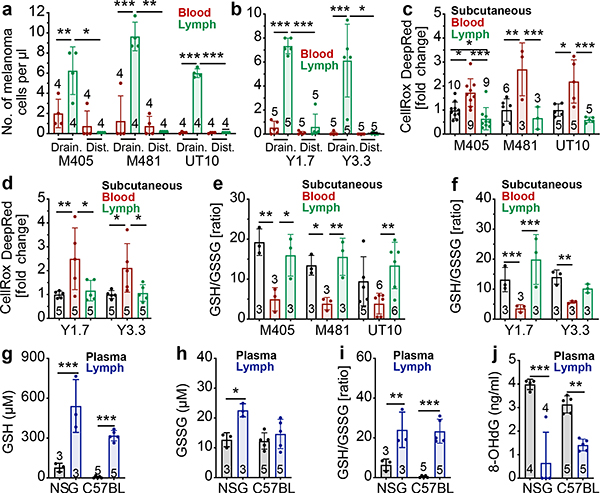
Figure 1. Melanoma cells in lymph experience less oxidative stress as compared to melanoma cells in the blood.
a-b, Efficiently metastasizing human (a) or mouse (b) melanomas were transplanted subcutaneously into NSG (a) or C57BL (b) mice. After the tumors reached ~2 cm in diameter the numbers of melanoma cells per microliter of tumor-draining blood or lymph (Drain.) or blood from cardiac puncture or contralateral lymph (Dist.) were quantified by flow cytometry. c-f, ROS levels (c, d) or GSH/GSSG ratios (e, f) in melanoma cells from subcutaneous tumors, the blood, and the lymph of NSG mice transplanted with patient-derived melanomas (c, e) or C57BL mice with mouse melanomas (d, f). g-j, GSH concentration (g), GSSG concentration (h), GSH/GSSG ratio (i), and 8-OHdG concentration (j) in plasma or lymph fluid from NSG or C57BL mice. The number of replicates (each replicate was pooled from 6–10 mice) is indicated in each panel and the number of independent experiments is shown in Supplementary Data, ‘Statistics and Reproducibility’. All data represent mean ± s.d. Statistical significance was assessed using correlated-samples two-way ANOVAs, paired t-tests or Wilcoxon tests (a, b), correlated-samples two-way ANOVAs (g-j), or two-way ANOVAs (c-f). Multiple comparisons were adjusted using Holm-Sidak’s (a, b) or Tukey methods (c-f). For all panels, statistical tests were two-sided where applicable and *p<0.05, **p<0.01, ***p<0.001. Exact p-values are in source data files.
We also detected inefficiently metastasizing melanoma cells in tumor-draining lymphatics but rarely in the blood (Extended Data Fig. 1i). This raised the possibility that melanoma cells survive better in lymph than in blood.
We tested if melanoma cells experience less oxidative stress in lymph as compared to blood. Human and mouse melanoma cells were subcutaneously transplanted into NSG or C57BL mice, then allowed to spontaneously metastasize. Melanoma cells from subcutaneous tumors, the lymph, and the blood of the same mice were stained with CellRox DeepRed, which detects reactive oxygen species (ROS) including hydroxyl radicals. For both patient-derived xenografts in immunocompromised mice (Fig. 1c) and mouse melanomas in immunocompetent mice (Fig. 1d), melanoma cells consistently exhibited significantly higher ROS levels in the blood than in subcutaneous tumors and lymph. Treatment of cells with the reducing agent 2-mercaptoethanol upon isolation largely rescued this increase in ROS levels (Extended Data Fig. 2a). Dissemination of melanoma cells into the blood was thus associated with a significant increase in ROS while dissemination into lymph was not.
Human (Fig. 1e) and mouse (Fig. 1f) melanoma cells from the blood usually exhibited significantly lower glutathione to oxidized glutathione (GSH/GSSG) ratios than melanoma cells from subcutaneous tumors or lymph. GSH/GSSG ratios did not significantly differ among melanoma cells from lymph versus subcutaneous tumors. Consistent with this, we observed significantly higher GSH concentrations (Fig. 1g–h) and GSH/GSSG ratios (Fig. 1i), in lymph fluid as compared to blood plasma. Lymph fluid also had lower concentrations of the 8-hydroxy-2’-deoxyguanosine oxidative product (Fig. 1j). The blood thus appears to be a more oxidative environment than lymph, potentially explaining the increased ROS levels in melanoma cells from blood. The reason for the more oxidative environment in blood is unclear but could reflect higher oxygen levels than in lymph29 or differences in cellular composition/density.
Melanomas undergo ferroptosis in blood
Human and mouse melanomas were subcutaneously transplanted into NSG and C57BL mice, respectively, allowed to spontaneously metastasize, and then lipid ROS levels in melanoma cells were compared by flow cytometric staining for BODIPY-C11. For most human (Fig. 2a) and mouse (Fig. 2b) melanomas, cells in the blood exhibited significantly higher lipid ROS levels as compared to cells in subcutaneous tumors or lymph.
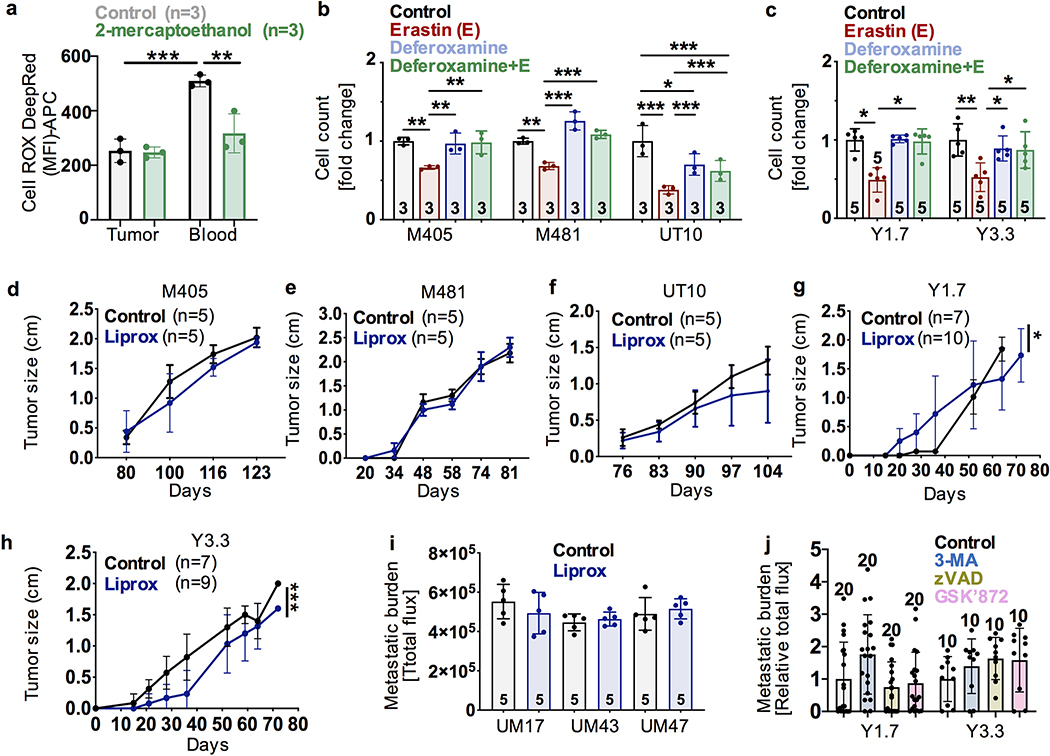
Figure 2. Melanoma cells undergo increased ferroptosis in blood as compared to lymph.
a-b, Lipid ROS (BODIPY-C11Oxidized/C11Oxidized+C11Non-oxidized ratio) levels in melanoma cells from subcutaneous tumors, blood, and lymph of NSG mice with patient-derived melanomas (a) or C57BL mice with mouse melanomas (b). c-d, Human or mouse melanomas were treated in culture with the ferroptosis promoter, Erastin, and/or the ferroptosis inhibitor, Liproxstatin-1 (Liprox). e, Free iron concentrations in plasma or lymph fluid from NSG or C57BL mice. f-i, Human (f, h) or mouse (g, i) melanomas were pre-treated with Liproxstatin-1 then injected intravenously (f, g) or intranodally (h, i) into NSG (f, h) or C57BL (g, i) mice and metastatic disease burden was assessed 1–3 months later by bioluminescence imaging. j-k, Mouse melanomas were pre-treated with N-acetyl-L-cysteine (NAC) or Trolox then injected intravenously (j) or intranodally (k) into C57BL mice and metastatic disease burden was assessed 1–2 months later by bioluminescence imaging. l-n, Percentage of mice transplanted with parental or Gpx4-deficient melanomas that formed metastatic tumors after subcutaneous (l), intravenous (m), or intranodal (n) injection. The number of replicates is indicated in each panel and the number of independent experiments is shown in Supplementary Data, ‘Statistics and Reproducibility’. All data represent mean ± s.d. Statistical significance was assessed using two-way ANOVAs (a, d, f), correlated-samples two-way ANOVAs (e), Kruskal-Wallis tests (b, c, j, k), t-tests (g-i), or multiple logistic regressions (l-n). Multiple comparisons were adjusted using Tukey’s (a, d, l-n) or Dunn’s (b, c, j, k) tests. No statistically significant differences were observed in panels h, i, k, or n. For all panels, statistical tests were two-sided where applicable and *p<0.05, **p<0.01, ***p<0.001. Exact p-values are in the source data files.
To test if the melanomas we studied can undergo ferroptosis, we cultured them with Erastin, which promotes ferroptosis by inhibiting cystine uptake, and/or Liproxstatin-1 (Liprox), which inhibits ferroptosis by reducing lipid peroxide accumulation22. Erastin significantly reduced the numbers of cells relative to control cultures and Liprox blocked this effect (Fig. 2c and 2d).
Iron, which promotes the generation of lipid ROS, is necessary for ferroptosis22. We observed much higher concentrations of free iron in blood plasma than in lymph fluid (Fig. 2e). Treatment with deferoxamine, an iron chelator, blocked the ability of Erastin to deplete melanoma cells in culture (Extended Data Fig. 2b and 2c), consistent with ferroptosis.
To determine if ferroptosis limits the growth of subcutaneous tumors, human and mouse melanoma cells were subcutaneously transplanted into NSG and C57BL mice, respectively. Once the tumors were palpable, mice were treated with daily i.p. injections of Liprox or vehicle. Liprox had little or no effect on the subcutaneous growth of most melanomas (Extended Data Fig. 2d–h), consistent with a prior report23. This suggests ferroptosis does not limit the growth of established subcutaneous tumors, consistent with the lack of oxidative stress in these tumors.
To determine if ferroptosis limits the survival of melanoma cells in the blood, melanoma cells were pre-treated with vehicle or Liprox, then intravenously transplanted into mice. Pre-treatment with Liprox substantially increased metastatic disease burden relative to control mice in most efficiently-metastasizing human (Fig. 2f) and mouse (Fig. 2g) melanomas. Liprox pre-treatment of inefficiently metastasizing melanomas was not sufficient to increase metastatic disease burden (Extended Data Fig. 2i). Ferroptosis thus appears to limit the survival of efficiently metastasizing melanomas in the blood but other mechanisms may contribute to the reduced metastasis of inefficient metastasizers, consistent with a recent study30.
To test if ferroptosis limits the survival of melanoma cells in lymph, melanoma cells were pre-treated with vehicle or Liprox and transplanted into the popliteal lymph nodes of mice. Pre-treatment with Liprox did not significantly affect metastatic disease burden with human (Fig. 2h) or mouse (Fig. 2i) melanomas after intranodal injection. This suggests melanoma cell survival in lymph was not limited by ferroptosis. Pre-treatment of mouse melanoma cells with the antioxidants N-acetyl-L-cysteine or Trolox (a vitamin E analog) also significantly increased metastatic disease burden after intravenous (Fig. 2j), but not intranodal, injection (Fig. 2k).
We also pre-treated mouse melanomas with inhibitors of autophagy (3-Methyladenine, a PI3K inhibitor), apoptosis (VAD-FMK, a pan caspase inhibitor), or necroptosis (GSK’872, a RIP kinase inhibitor) before intravenously injecting the cells. These treatments did not significantly affect metastatic disease burden (Extended Data Fig. 2j).
To genetically test the importance of ferroptosis, we used CRISPR/Cas9 to delete Glutathione peroxidase 4 (Gpx4) from two mouse melanomas (Extended Data Fig. 3a). GPX4 inhibits ferroptosis by reducing the accumulation of lipid ROS31, 32. GPX4 was expressed by all of the human and mouse melanomas we studied (Extended Data Fig. 3a and 3b). Gpx4-deficiency did not significantly affect the proliferation or survival of mouse melanoma cells cultured at physiological oxygen levels but did reduce the survival/proliferation of melanoma cells cultured in atmospheric oxygen (Extended Data Fig. 3c and 3d), raising the possibility that GPX4 becomes more necessary in melanoma cells as oxidative stress increases.
In vivo, Gpx4-deficiency significantly increased lipid ROS levels in subcutaneous tumors relative to parental controls (Extended Data Fig. 3e). Nonetheless, Gpx4-deficiency only slightly reduced the growth of primary subcutaneous tumors (Extended Data Fig. 3f and 3g), while substantially reducing the frequency of melanoma cells in the blood (Extended Data Fig. 3h and 3i), and the percentage of mice that spontaneously formed metastatic tumors in visceral organs (Figure 2l). Total metastatic disease burden could not be quantified by bioluminescence imaging in these experiments because the melanomas spontaneously lost luciferase expression. These data suggest melanoma cells become more dependent upon GPX4 during metastasis.
Gpx4-deficient and parental control melanomas were also transplanted intravenously and intranodally into mice. Gpx4-deficiency reduced the percentage of mice that formed metastatic tumors in visceral organs after intravenous (Fig. 2m), but not intranodal (Fig. 2n), injection. GPX4 is, therefore, required for the survival of metastasizing cells in the blood but not lymph, further suggesting that ferroptosis limits the survival of melanoma cells in the blood. The context-dependence of the effects of GPX4 deficiency on melanoma cells emphasizes the value of studying mechanisms that influence oxidative stress responses in vivo.
Oleic acid protects against ferroptosis
To better understand why melanoma cells undergo ferroptosis in the blood but not lymph, we subcutaneously transplanted melanomas, allowed them to spontaneously metastasize, then flow cytometrically isolated cells from blood and lymph of the same mice for metabolomic analysis. Principal component analysis showed that melanoma cells from lymph clustered together, apart from melanoma cells from blood (Fig. 3a). Melanoma cells from lymph and blood differed more from each other than melanoma cells derived from different patients.
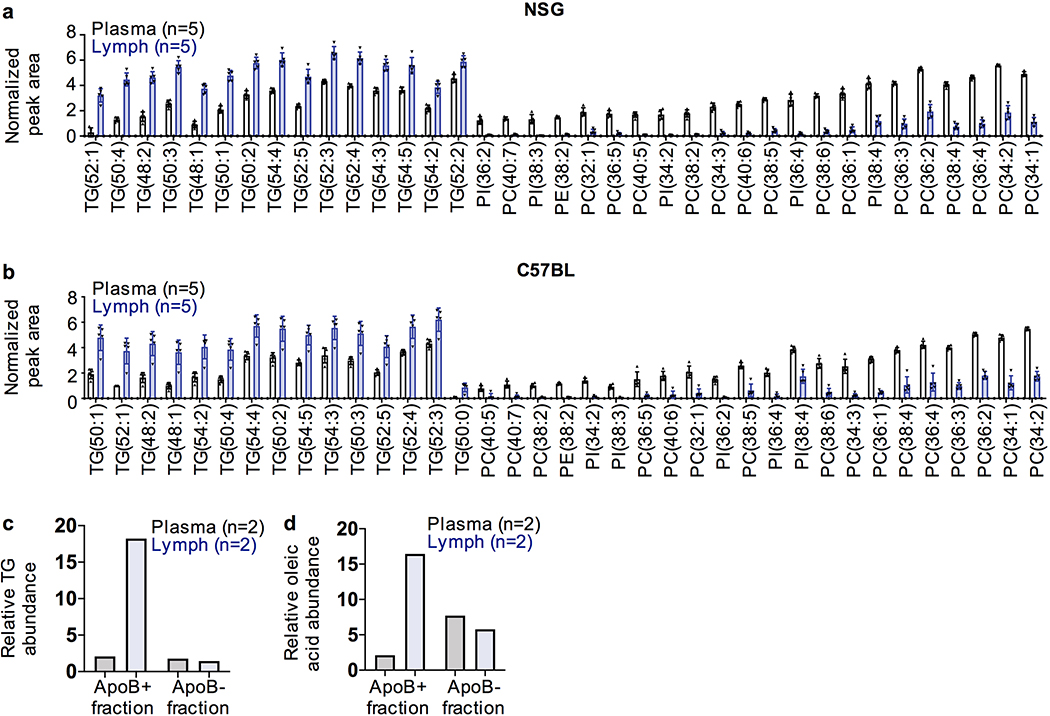
Figure 3. Oleic acid levels are higher in lymph, and in melanoma cells from lymph, and oleic acid protects against ferroptosis.
(a) Principal component analysis of metabolomic profiling of melanoma cells from blood or lymph of the same mice xenografted with M481, M405, or UT10 melanomas. (b) The most enriched pathways among metabolites that significantly differed (p<0.001 and >1.5-fold change) between melanoma cells from blood and lymph. (c, d) Human (c) and mouse (d) melanomas were cultured for 12 hours with or without oleic acid or linoleic acid before adding Erastin for 24 hours and counting cells. (e, f) Human (e) and mouse (f) melanomas were pre-treated for 12 hours with vehicle (control), oleic acid, or linoleic acid then intravenously injected into NSG (e) or C57BL (f) mice and metastatic disease burden was assessed 1–3 months later by bioluminescence imaging. g-h, Relative oleic acid abundance in the plasma and lymph of NSG (g) and C57BL (h) mice (TGs, triacylglycerols, PLs, phospholipids). i-j, Acsl3-deficient mouse melanomas were cultured as in 3d (parental control cell data) for 12 hours with vehicle, oleic acid, and/or linoleic acid before adding Erastin for 24 hours and counting cells. (k) Mouse melanoma cells from primary subcutaneous tumors or lymph node metastases were injected intravenously (IV) or subcutaneously followed by intravenous retransplantation (SQ to IV) in C57BL mice and the frequency of cells that formed metastatic tumors was determined by limiting dilution analysis. Human (l) or mouse (m) melanoma cells were isolated from primary subcutaneous tumors or lymph node metastases of the same mice, then treated with Erastin for 24 hours in culture to assess their sensitivity to ferroptosis. The number of replicates in each treatment is indicated in each panel and the number of independent experiments is shown in Supplementary Data, ‘Statistics and Reproducibility’. All data represent mean ± s.d. Statistical significance was assessed using MetaboAnalyst’s clustering analysis (a) and metabolite set enrichment analysis (MSEA) (b), Kruskal-Wallis tests (c, e(M405), i, j), Welch’s one-way ANOVA (d), one-way ANOVAs (e(M481, UT10), l, m), two-way ANOVAs (f, k), or correlated samples two-way ANOVAs (g, h). Multiple comparisons were adjusted using Dunn’s (c, e(M405), i, j), Dunnett’s T3 (d), Tukey’s (e(M481, UT10), f), or Sidak’s (l, m) tests. For all panels statistical tests were two-sided where applicable and *p<0.05, **p<0.01, ***p<0.001. Exact p-values are in source data files.
We performed pathway enrichment analysis of the metabolites that differed between melanoma cells in lymph versus blood. Three of the top five pathways were related to lipid metabolism (Fig. 3b). The fatty acid composition of phospholipids influences sensitivity to ferroptosis in cultured cancer cells33. Of the 57 lipid species we detected, the lipid that most differed between melanoma cells in lymph versus blood was oleic acid. Oleic acid is abundant in lymph and blood34 and was 8.7±0.9 fold enriched (p<0.05) in melanoma cells from lymph. Oleic acid is a monounsaturated fatty acid that inhibits ferroptosis in culture by reducing the amount/density of polyunsaturated fatty acids available for oxidation in membranes35,36.
We treated mouse and human melanoma cells with oleic acid or linoleic acid (a polyunsaturated fatty acid) for 12 hours, then Erastin for 24 hours, and counted live cells. Oleic acid or linoleic acid, by themselves, had little effect on cell counts while Erastin significantly reduced cell counts in human (Fig. 3c) and mouse (Fig. 3d) melanomas. Oleic acid, but not linoleic acid, protected the cells from Erastin, suggesting oleic acid can protect melanoma cells from ferroptosis.
To test if oleic acid can promote the survival of melanoma cells during metastasis, we pretreated melanoma cells with oleic acid, linoleic acid, or vehicle for 12 hours in culture and then injected equal numbers of live cells intravenously. We observed a significantly higher metastatic burden in mice that received oleic acid treated cells as compared to vehicle or linoleic acid treated cells in 2 of 3 human melanomas (Fig. 3e) and in both mouse melanomas (Fig. 3f). Pre-treatment with oleic acid thus promoted the ability of most melanomas to survive during metastasis through the blood.
We performed a targeted lipidomic analysis of the most abundant lipids in plasma37 (Extended Data Table 2). Of the 121 lipid species we analyzed, 37 significantly differed between lymph fluid and blood plasma (p<0.01; Extended Data Fig. 4a and 4b). These included 15 triacylglycerols that were higher in lymph and 22 phospholipids that were higher in plasma. Free oleic acid was at extremely low levels, consistent with a prior study37, and did not significantly differ between lymph and plasma. However, the total levels of oleic acid in triacylglycerols and phospholipids were 1.9±0.3 fold higher in lymph as compared to blood in NSG mice and 1.6±0.3 fold higher in C57BL mice (Fig. 3g and 3h). Moreover, the oleic acid in lymph was overwhelmingly in triacylglyerols, in contrast to plasma, where it was often in phospholipids. Most circulating triacylglycerols are contained within ApoB+ vesicles, which include chylomicrons38. To test if oleic acid-containing triacylglycerols were contained within ApoB+ vesicles, we obtained plasma or lymph fluid from mice and isolated ApoB+ vesicles by magnetic selection. Almost all of the triacylglycerols and oleic acid were contained within the ApoB+, rather than the ApoB negative, fraction. Oleic acid was more abundant in the ApoB+ fraction of lymph than in the ApoB+ fraction of plasma (Extended Data Fig. 4c and 4d). Therefore, the oleic acid in lymph is mainly within triacylglycerols in ApoB+ vesicles, consistent with the observation that most circulating lipids are transported by these vesicles38.
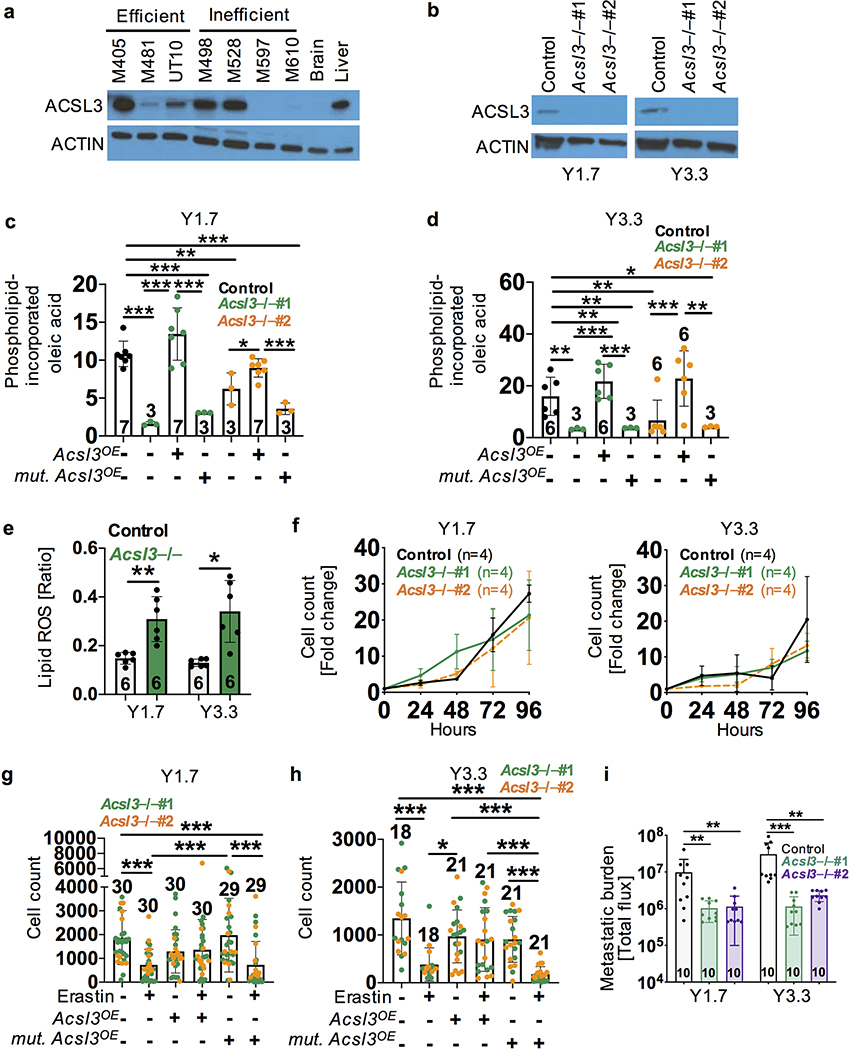
To test if membrane incorporation is required for the protective effect of oleic acid in melanoma cells, we tested if this requires Acyl-CoA synthetase long-chain family member 3 (ACSL3). ACSL3 converts fatty acids into fatty acyl-CoA esters for incorporation into membrane phospholipids39, protecting cells from ferroptosis in culture35. Higher ACSL3, but not ACSL4, expression in melanomas is associated with significantly worse overall survival in patients40. ACSL3 was expressed by most human and mouse melanomas, including all of the efficient metastasizers (Extended Data Fig. 5a and 5b). Inefficient metastasizers sometimes expressed little ACSL3 (Extended Data Fig. 5a), raising the possibility that they might be less sensitive to the effects of oleic acid.
We used CRISPR/Cas9 to delete Acsl3 from two mouse melanomas (two independently targeted clones per line; Extended Data Fig. 5b). Acsl3 deficiency significantly reduced oleic acid incorporation into phospholipids and this was rescued by over-expression of wild-type, but not mutant Acsl3 encoding a catalytically dead enzyme (Extended Data Fig. 5c and 5d). Acsl3-deficiency significantly increased lipid ROS levels in subcutaneous tumors but did not significantly affect proliferation or survival in culture (Extended Data Fig. 5e and 5f).
To test if Acsl3 is required for oleic acid to protect cells against ferroptosis, we treated Acsl3-deficient or parental melanomas with oleic acid for 12 hours, then Erastin for 24 hours, and counted live cells. Oleic acid blocked the effects of Erastin on parental control cells (Fig. 3d) but did not protect Acsl3-deficient cells from Erastin (Fig. 3i and 3j). The protective effect of oleic acid was restored by over-expression of wild-type but not mutant Acsl3 (Extended Data Fig. 5g and 5h). Oleic acid thus blocked the effects of Erastin on melanoma cells in an Acsl3-dependent manner. Acsl3-deficient cells gave rise to significantly reduced metastatic disease upon intranodal injection as compared to parental control cells (Extended Data Fig. 5i).
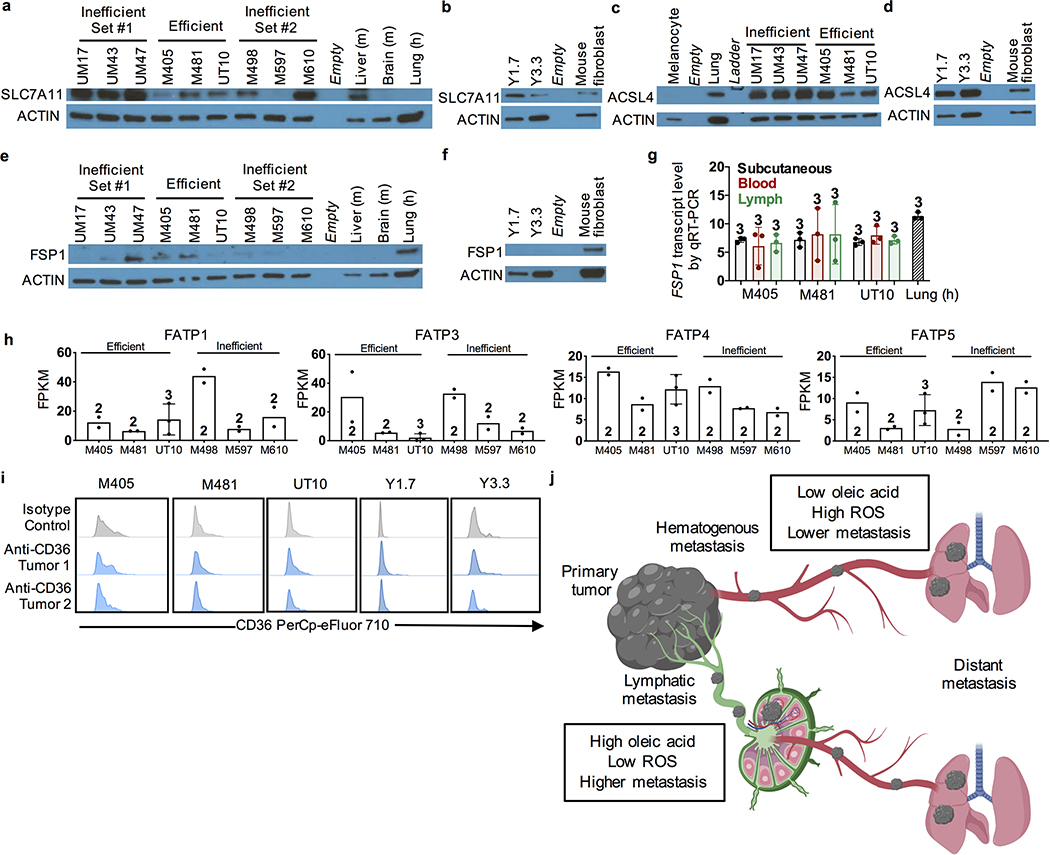
We also tested the expression of SLC7A11, a cystine/glutamate transporter, ACSL4, which esterifies long-chain fatty acids for lipid biosynthesis, and Ferroptosis Suppressor Protein 1 (FSP1), each of which can influence ferroptosis sensitivity33,41–43. SLC7A11 and ACSL4 were expressed at variable levels in all of the melanomas we studied (Extended Data Fig. 6a–d). FSP1 was expressed by a subset of melanomas (e.g. M405 and M481) but other melanomas had little expression (UT10, YUMM1.7, YUMM3.3; Extended Data Fig. 6e and 6f). We observed no difference in FSP1 transcript levels between melanoma cells from lymph versus blood by qRT-PCR (Extended Data Fig. 6g). Differences in SLC7A11, ACSL4, and FSP1 expression levels among melanomas may influence sensitivity to ferroptosis.
Melanomas expressed multiple fatty acid transporters capable of transporting oleic acid including FATP1, FATP3, FATP4, and FATP5 (Extended Data Fig. 6h). FATP2 protects myeloid cells from lipid ROS, altering immune function and melanoma progression44. Another fatty acid transporter, CD36, can promote the metastasis of oral carcinomas by promoting palmitic acid uptake17, though we did not detect CD36 in the melanomas we studied (Extended Data Fig. 6i).
Lymph protects cells from ferroptosis
These results raised the possibility that lymph node metastasis often precedes distant metastasis because exposure to lymph allows cells to incorporate oleic acid and other antioxidants that subsequently protect the cells from ferroptosis during dissemination through the blood. To test this, we isolated mouse melanoma cells from primary subcutaneous tumors and lymph node metastases from the same donor mice and transplanted them intravenously into secondary recipients. Melanoma cells from lymph nodes were more likely to form metastatic tumors as compared to melanoma cells from subcutaneous tumors (Fig. 3k).
To test whether this difference was reversible or irreversible we isolated melanoma cells from primary subcutaneous tumors and lymph node metastases, then serially transplanted the cells first subcutaneously and then intravenously. Transient subcutaneous growth eliminated the metastatic advantage of the lymph node-derived cells over subcutaneous tumor-derived cells (Fig. 3k). Moreover, melanoma cells from lymph node metastases were usually more resistant to Erastin in culture than cells from subcutaneous tumors (Fig. 3l and Fig. 3m).
The intravenous injection experiments suggest it is possible to metastasize through blood, apparently independent of lymph, but that this is a much less efficient route of metastasis than through lymph, particularly in immunocompetent hosts (Table 1). Most, but not all, of the melanomas we studied formed more metastases after oleic acid treatment. This suggests there are mechanisms that protect some melanomas from oxidative stress in the lymph that are independent of exogenous oleic acid, potentially including increased levels of other antioxidants, endogenous fatty acid production, or FSP1-mediated mechanisms. Overall, lymph appears to protect metastasizing cells from ferroptosis by reducing oxidative stress, increasing their survival during subsequent migration through the blood (Extended Data Fig. 6j).
METHODS
Melanoma specimen collection and enzymatic tumor disaggregation
Melanoma specimens were obtained with informed consent from all patients according to protocols approved by the Institutional Review Board (IRB) of the University of Michigan Medical School (IRBMED approvals HUM00050754 and HUM00050085; see ref27) and the University of Texas Southwestern Medical Center (IRB approval 102010–051). Materials used in the manuscript are available, either commercially or from the authors, though there are restrictions imposed by IRB requirements and institutional policy on the sharing of materials from patients. Single cell suspensions were obtained by dissociating tumors mechanically with a scalpel on ice. Cells were filtered through a 40-μm cell strainer to remove clumps.
Cell culture
YUMM1.7 (BrafV600E/+; PTEN−/−; Cdkn2−/−) and YUMM3.3 (BrafV600E/+; Cdkn2−/−) cell lines28 were obtained from and authenticated by ATCC and cell lines were confirmed to be mycoplasma free using the MycoAlert detection kit (product LT07–318, Lonza). YUMM1.7 and YUMM3.3 were transfected with dsRed2 and luciferase (dsRed2-P2A-Luc) for bioluminescence imaging. Cells were cultured in DMEM low-glucose (product 11885076, Thermo Fisher Scientific) supplemented with 10% FBS and 1% pen/strep. Cells were trypsinized using Trypsin-EDTA (0.05%; product 25300054, Thermo Fisher Scientific) for 2 to 3 minutes. For most culture experiments, melanoma cells were grown in regular incubators with 6% carbon dioxide (atmospheric oxygen) with the exception of some studies of Gpx4-deficient melanomas. For these experiments, melanoma cells were cultured in gas-tight chambers flushed with 1% O2/6% CO2/balance N2 to achieve more physiological oxygen levels.
Mouse studies and xenograft assays
All mouse experiments complied with all relevant ethical regulations and were performed according to protocols approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center (protocol 2016–101360). No formal randomization techniques were used; however, animals were allocated randomly to treatment groups and specimens were processed in an arbitrary order. During the linoleic and oleic acid pre-treatment experiments in vivo (Fig. 3e and 3f), the mice were injected and data on tumor formation were collected in a manner blinded to sample identity or treatment. For all experiments, the maximum permitted tumor diameter was 2.5 cm and this limit was not exceeded in any experiment. For all experiments, mice were kept on normal chow and fed ad-libitum. Melanoma cell suspensions were prepared for injection in staining medium (L15 medium containing bovine serum albumin (1 mg/ml), 1% penicillin/streptomycin, and 10 mM HEPES (pH 7.4) with 25% high-protein Matrigel (product 354248; BD Biosciences)).
Patient-derived melanomas were transplanted into 4 to 8-week-old male and female NOD.CB17-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Mouse melanomas were transplanted into 6 to 8-week-old male and female C57BL/Ka mice. Subcutaneous injections were performed in the right flank of mice in a final volume of 50 μl using 100 cells per injection for human melanoma cells and 50,000 cells per injection for mouse melanoma cells unless otherwise specified. Subcutaneous tumor diameters were measured weekly with calipers until any tumor in the mouse cohort reached 2.5 cm in its largest diameter. At that point, all mice in the cohort were killed, per approved protocol, for analysis of subcutaneous tumor diameter, circulating melanoma cell frequency, and metastatic disease burden. Metastatic disease burden was evaluated by bioluminescence imaging of visceral organs (see details below).
Intravenous injections were performed by injecting 1,000 human melanoma cells or 100,000 mouse melanoma cells into the tail vein of mice in 100 μl of PBS. To perform injections into lymph nodes, the lymphatics were first traced by injecting 2% Evans Blue Dye (product E2129, Sigma-Aldrich) into the foot pedal 5 minutes before performing intranodal injections. After injecting Evans Blue Dye, the mice were anesthetized using isoflurane and a small (5–10 mm) incision was made in the region of the popliteal lymph node. The lymph node was located based on Evans Blue staining, immobilized with forceps, and cells suspended in PBS were injected in a volume of 10 μl into the popliteal lymph node using a 27 gauge Hamilton syringe. Injection into the lymph node was confirmed by visible swelling of the lymph node. The incision was closed using surgical glue (product 1469SB, 3M VetBond Tissue Adhesive) and the mice were closely monitored for signs of pain or distress.
Testing the effects of ferroptosis-modulating agents on melanoma cells in vivo
To test the effect of ferroptosis-modulating drugs on the ability of melanoma cells to form subcutaneous tumors, human or mouse melanoma cells were injected subcutaneously into mice and when the tumors became palpable the mice were treated daily with intraperitoneal injections of 100 μl of vehicle (2% DMSO in saline) or Liproxstatin-1 (20mg/kg, product 17730, Cayman Chemical). To test the effect of inhibitors of other pathways on the ability of melanoma cells to survive and form distant metastases, mouse melanoma cells (YUMM1.7 and YUMM3.3) were incubated on ice with vehicle (2% DMSO in saline), Z-VAD-FMK (50 μM, product V116, Sigma Aldrich), 3-Methyladenine (10 μM, 3-MA, product 5142, Sigma-Aldrich), or GSK’872 (10 μM, product 50260836, Sigma-Aldrich) and then injected intravenously in C57BL mice.
To test the effect of ferroptosis-modulating drugs on the ability of melanoma cells to survive and form tumors after intravenous or intranodal injection, human or mouse melanoma cells were incubated on ice with vehicle (2% DMSO in saline), Liproxstatin-1 (1 μM, product 17730, Cayman Chemical), N-acetyl-L-cysteine (5 mM, product A7250, Sigma-Aldrich), or Trolox ((+/−)-6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid, 97%, 0.3 mM, product 238813, Sigma-Aldrich) for 10 minutes prior to injection.
To test the effect of oleic acid on metastasis, 1 × 107 melanoma cells derived from patients or mouse cell lines were incubated in vehicle (2% DMSO in saline), linoleic acid (500 μM, product 90150, Cayman Chemical), or oleic acid (500 μM, product 90260, Cayman Chemical) in DMEM low-glucose (product 11885076, Thermo Fisher Scientific) supplemented with 10% FBS and 1% pen/strep for 12 hours. Then the cells were trypsinized, washed once with PBS, and filtered through a 40-μM mesh. Live cells were counted on a hemocytometer using trypan blue exclusion then injected intravenously or intranodally.
Testing the effects of ferroptosis-modulating agents in culture
To test if melanoma cells were able to undergo ferroptosis in culture, 1,000 human or mouse melanoma cells were cultured in each well of 96 well plates containing DMEM low-glucose supplemented with 10% FBS and 1% pen/strep. Liproxstatin-1 (1 μM) or vehicle (0.5 μM DMSO) were added to some wells for 10 minutes, then ferroptosis was induced by treating with Erastin (1 μM) for 24 hours, according to a previously published protocol35, and the numbers of viable cells in the cultures were counted. To test the effect of fatty acids on ferroptosis by melanoma cells in culture, 1,000 human or mouse melanoma cells were cultured in each well of 96 well plates containing DMEM low-glucose supplemented with 10% FBS and 1% pen/strep. First, vehicle (0.5 μM DMSO), linoleic acid (500 μM) or oleic acid (500 μM) were added to the cultures for 12 hours, then Erastin (1 μM) was added for 24 hours, and the numbers of viable cells in the cultures were counted. To test the effect of iron chelation, we cultured 1,000 human or mouse melanoma cells per well in DMEM low-glucose supplemented with 10% FBS and 1% pen/strep, then added deferoxamine (1 μM, product D9533, Sigma-Aldrich) for 10 minutes followed by Erastin (1 μM) for 24 hours prior to cell counting.
Bioluminescence imaging
Metastatic disease burden was assessed by bioluminescence imaging (all melanomas stably expressed luciferase). Five minutes before performing luminescence imaging, mice were injected intraperitoneally with 100 μl of PBS containing d-luciferin monopotassium salt (40 mg/ml; Biosynth, L8220) and mice were anaesthetized with isoflurane 2 minutes prior to imaging. The mice were imaged using an IVIS Imaging System 200 Series (Caliper Life Sciences). After whole-body imaging, the visceral organs were surgically removed and imaged. The exposure time ranged from 10 to 60 s, depending on the maximum signal intensity, to avoid saturation. To measure background, a control mouse not transplanted with melanoma cells was imaged. The bioluminescence signal (total photon flux) was quantified with ‘region of interest’ measurement tools in Living Image software (Perkin Elmer). Metastatic disease burden was calculated as observed total photon flux across all organs minus background total photon flux in negative control mice. Negative values were set to 1 for statistical analysis.
Isolation and analysis of melanoma cells by flow cytometry
To isolate melanoma cells from blood and lymph, mice were transplanted subcutaneously with human or mouse melanoma cells and the cells were allowed to spontaneously metastasize until the subcutaneous tumor reached ~2 cm. Evan’s Blue dye (2%; Sigma-Aldrich, cat. No E2129) was injected into the edge of subcutaneous melanomas to identify tumor-draining blood and lymphatic vessels. Lymphatic fluid was obtained by carefully sliding a 27 gauge Hamilton syringe into a tumor-draining lymphatic vessel. We could collect 8–15 μl of lymph from 4–10 mice from which samples were pooled to gain enough cells for analysis. Tumor-draining blood vessels were identified by injecting Evan’s blue directly into a blood vessel emerging from a tumor so as to observe the directionality of blood flow. Blood was then drawn from the vessel using a 27 gauge Hamilton syringe. To obtain blood that is distant from the tumor, blood was collected by cardiac puncture with a syringe pretreated with citrate-dextrose solution (product sc-2141744, Santa Cruz).
Specimens were first sedimented using Ficoll, according to the manufacturer’s instructions (product 17144002, Ficoll Paque Plus, GE Healthcare), to eliminate red blood cells. The cells that remained in suspension were washed with Hank’s Balanced Buffered Solution (product 14025076, HBSS, Gibco) and then stained with antibodies prior to flow cytometric analysis and sorting. All antibody labeling was performed for 20 minutes on ice, followed by washing with HBBS and centrifugation at 200×g for 5 min. Cells were stained with directly conjugated antibodies against mouse CD45 (violetFluor 450, eBiosciences), mouse CD31 (390-eFluor450, Biolegend), mouse Ter119 (eFluor450, eBiosciences) and human HLA-A, B, C (G46–2.6-FITC, BD Biosciences). Human melanoma cells were isolated as cells that were positive for HLA and negative for mouse endothelial and hematopoietic markers. Mouse melanoma cells were isolated as cells that were positive for dsRed and CD146 (FITC, product 134708, BioLegend) and negative for mouse CD45, Ter119, and CD31. Cells were washed with staining medium and resuspended in 4’,6-diamidino-2-phenylindole (DAPI; 1 μg/ml; Sigma) to eliminate dead cells from sorts and analyses. Cells were analyzed using an LSRFortessa cell analyzer (Becton Dickinson) or sorted on a FACS Fusion Cell Sorter (Becton Dickinson).
To analyze ROS levels, dissociated cells were stained for 30 min at 37°C with 5 mM CellROX Green (product C10444, Life Technologies) or CellROX DeepRed (product C10422, Life Technologies) in HBSS-free (Ca2+ and Mg2+-free) and DAPI (to distinguish live from dead cells). The cells were then washed and analyzed by flow cytometry using either a FACS Fusion or FACS Fortessa (BD Biosciences) to assess ROS levels in live human or mouse melanoma cells. To perform BODIPY-C11 staining for lipid ROS levels, cells were resuspended in 1 ml HBSS containing 5 μM BODIPY 665/676 (product B3832, Thermo Fisher Scientific) and incubated for 15 min at 37°C. Cells were washed and resuspended in fresh HBSS, then analyzed immediately by flow cytometry. For BODIPY staining, the shift in mean fluorescence intensity from the non-oxidized (665 nm) to the oxidized channel (676 nm) was analyzed. To analyze CD36 levels, melanoma cells were stained with anti-CD36 antibody (PerCp-eFluor™ 710, product 46–0369-41, Invitrogen (eBioscience)) then analyzed by flow cytometry.
8-hydroxy-2’-deoxyguanoside (8-OHdG) levels
8-hydroxy-2’-deoxyguanosine levels were quantified by ELISA (8-OHdG ELISA kit; product ab201734, Abcam) using 10 μl of blood plasma or lymph fluid obtained from non-tumor bearing C57BL mice according to the manufacturer’s instructions. This assay detects both free 8-OHdG and cell-free DNA-incorporated 8-OHdG.
Iron levels
Iron levels in blood plasma and lymph fluid from normal NSG or C57BL mice were assayed by spectrophotometry using VITROS Fe Slides (product 1924547, VITROS Chemistry Products), according to the manufacturer’s instructions. A drop of plasma or lymph was added to these slides, which dissociate iron from transferrin at acidic pH, then the iron was reduced to ferrous iron, which bound a dye that was quantitated by spectrophotometry.
Genetic modification of mouse melanomas
Acsl3 and Gpx4 deficient melanoma cell lines were generated using CRISPR. Gpx4 sgRNA #1 (5’– AAA ACT GAG TGT CCT AAC CC –3’) and Gpx4 sgRNA #2 (5’– TTG ACA GTT GAG CAC CAG CG –3’) were used to delete exon 2 of Gpx4 and to introduce a frameshift. Acsl3 sgRNA #1 (5’–AAA ACT GAG TGT CCT AAC CCA GG –3’) and Acsl3 sgRNA #2 (5’–AAA GGG GCG TAC TTA ATG GAA GG–3’) were used to delete exon 2 of Acsl3 and to introduce a frameshift. All sgRNAs were cloned into the U6-driven Cas9 expression vector (pX458-pSpCas9(BB)-2AGFP; product 48318, Addgene). Approximately 1×105 YUMM1.7 or YUMM3.3 mouse melanoma cells were plated adherently in tissue-culture-treated 6-well plates containing DMEM low glucose plus 10% FBS and 1% penicillin/streptomycin. 1 μg of each of the sgRNA constructs was co-transfected into the cells using polyjet (product SL100688, SignaGen) according to the manufacturer’s instructions. After 48 hours, one GFP+ cell was sorted into each well of a 96-well plate containing DMEM low glucose plus 10% FBS and 1% penicillin/streptomycin. The clones were then grown in culture, and genomic DNA was screened to confirm the desired deletions. Two independently targeted clones containing the confirmed deletions were then used in experiments for each melanoma, along with parental control cells.
Acsl3-overexpressing melanomas were generated using lentiviral vectors. Virus was produced using Acsl3 (product 17295D, GenScript) or mutant Acsl3 (GenScript, NM_004457.5, ORF Clone), a catalytically-dead gene product in which amino acids 314–339 were deleted45. Wild-type and mutant ORFs were inserted into the lentiviral vector pLVX-EF1a-IRES-ZsGreen1 (product 631982, Clonetech). For virus production, 293T cells were transfected using polyjet (SignaGen) with 0.9 mg of each plasmid and 1 μg of helper plasmid (0.4 μg pMD2G and 0.6 μg of psPAX2). Viral supernatants were collected 48 hours later and filtered through a 0.45-μm filter. 300,000 melanoma cells were infected with viral supernatants and 10 μg/ml polybrene (Sigma) for 6 hours. Cells were then washed twice with staining medium (L15 medium with 1 mg/ml bovine serum albumin, 1% penicillin/streptomycin, and 10 mM Hepes (pH 7.4)). Transfected cells (GFP+) were isolated by flow cytometry and used in experiments.
qRT-PCR analyses
To compare FSP1 transcript levels, human melanoma cells were isolated by flow cytometry from subcutaneous tumors, the blood, and the lymph of the same NSG mice bearing ~2.0 cm subcutaneous tumors. Samples from 5–8 mice were pooled for each replicate to gain enough cells for quantitative reverse transcription PCR (qRT-PCR) analysis. RNA was extracted from the cells using an RNeasy Mini Kit (product 74106, Qiagen) according to the manufacturer’s instructions. RNA was reverse transcribed using SuperScript III (product 18080044, Invitrogen). qRT-PCR was performed using a Roche LightCycler 480. The primers used for qRT-PCR analysis of RNA were hFSP1-Fwd AGACAGGGTTCGCCAAAAAGA and hFSP1-Rev CAGGTCTATCCCCACTACTAGC.
Western blot analysis
Melanomas were excised and quickly snap frozen in liquid nitrogen. Tumor lysates were prepared in Kontes tubes with disposable pestles using RIPA Buffer (product 9806, Cell Signaling Technology) supplemented with phenylmethylsulphonyl fluoride (product 329–98-6, Sigma), and protease and phosphatase inhibitor cocktail (product 05892970001, Roche). The bicinchoninic acid protein assay (product 23225, Thermo Fisher Scientific) was used to quantify protein concentrations. Equal amounts of protein (10–20 μg) were loaded into each lane and separated on 4–20% polyacrylamide tris glycine SDS gels (product 4568086, BioRad), then transferred to polyvinylidene difluoride membranes (product 1620177, BioRad). The membranes were blocked for 1 hour at room temperature with 5% BSA in TBST (containing 0.1% Tween20) and then incubated with primary antibodies overnight at 4°C.
After washing, then incubating with horseradish peroxidase conjugated secondary antibody (product 7074, Cell Signaling Technology), signals were developed using SuperSignal West (product 34580, Thermo Fisher Scientific). Blots were sometimes stripped using Restore stripping buffer (product 21059, Thermo Fisher Scientific) and restained with other primary antibodies. The following antibodies were used for western blots: anti-ACSL3 (product ab80675, Abcam), anti-GPX4 (product 144–01933-50, RayBiotech), anti-SLC7A11 (product 12691S, Cell Signaling Technology), anti-ACSL4 (product SAB2100035, Sigma-Aldrich), anti-FSP1 (product sc-377120, Santa Cruz) and anti-β-Actin (product 12620S, Cell Signaling Technologies). For all western blots, the beta-actin loading controls were run on the same gel as the protein of interest. For all western blots, the positions of bands in the size ladder were indicated in marker on the film. These markings are visible on some of the western blot scans shown in the figures. Uncropped western blots are in Supplementary Figure 1.
Cell sorting for metabolite extraction and metabolomics
Cells were isolated for metabolomics using previously published methods46. These methods were optimized for speed and to keep cells cold at all times, to minimize metabolic changes during isolation. Blood or lymph cells were filtered through a 40-μm cell strainer or a 45-μm nylon mesh into 5 ml tubes on ice. Paramagnetic MicroBeads against CD45 (product 130–052-301, Miltenyi) were used to pre-enrich the CD45− melanoma cells using a QuadroMACS manual separator (product 130–090-976, Miltenyi) in the cold room. Upon eluting the CD45− cells from the column, they were stained with the other antibodies required to isolate melanoma cells by flow cytometry as HLA-A,B,C+CD31-Ter119-CD45- cells (Extended Data Figure 1). Antibody staining was for 20 minutes on ice in a 4°C cold room and the cells were resuspended in HBSS + 0.5% BSA before sorting.
Cells were sorted using a FACSAria flow cytometer. The sheath fluid was 0.5x phosphate buffered saline (PBS) prepared using Milli-q water (Millipore). A 70 μm nozzle was used in 4-way purity sort mode to minimize the volume of sorted drops. This was done to reduce ion suppression of mass spectrometry signals from salt in the sheath fluid. The FACSAria sheath tank was washed with Milli-q deionized water before the experiment to reduce contamination and dedicated glassware was used to make all buffers. Cell suspensions were kept at 4°C during the sorts, which took about 20 minutes per sample. Equal numbers of melanoma cells from blood and lymph (usually 5,000 cells) were sorted directly into 40 μl of cold 100% methanol pre-chilled on dry ice and maintained at 4°C during the sort. After sorting, the samples were cooled on dry ice, then vortexed to ensure cell lysis, and then stored at −80°C until being analyzed by mass spectrometry.
Prior to analysis, the samples were vortexed again for 1 minute, then centrifuged at 17,000×g for 15 minutes at 4°C. For metabolomic analysis, the supernatant was transferred to autosampler vials and analyzed by LC-MS/MS (see below for details). For GSH/GSSG analysis, the supernatant was transferred to a new tube and lyophilized using a Speedvac (Thermo). The dried metabolites were reconstituted in 100 μl of 0.1% formic acid to inhibit spontaneous oxidation, vortexed, and centrifuged at 17,000×g for 15 minutes at 4°C. The sample was transferred into autosampler vials and analyzed by LC-MS/MS (see below for details).
Mass spectrometry for metabolomic analysis
Metabolomic data were acquired with a Thermo Scientific (Bremen, Germany) QExactive HF-X mass spectrometer (LC-MS/MS). A MS1 polarity-switching acquisition method was set up for individual metabolites. Spectra were acquired with a resolving power of 120,000 full width at half maximum (FWHM) with a scan range set to 80–1,200 daltons; the automatic gain control (AGC) target was set to 1,000,000 with a maximum inject time of 50 ms. MS2 spectra were acquired with a data dependent method on pooled samples in individual polarities and precursor MS data were acquired with identical settings. Product ion MS data were acquired with a resolving power of 15,000 FWHM; the AGC target was set to 200,000, with a maximum inject time of 150 ms. A top-10 data dependent MS schema was used with an isolation window of 2 Da and an isolation offset of 1.5 daltons. Analytes were fragmented with a stepped collision energy of 30, 50 and 70 normalized collision energy (NCE). The minimum AGC target was set to 8,000 with a dynamic exclusion of 30 seconds. Naturally-abundant isotopes were excluded. Metabolites were chromatographically-resolved using published methods46.
GSH/GSSG analysis by LC-MS/MS
GSH/GSSG analysis was performed using a SCIEX 6500+ Q-Trap mass spectrometer coupled to a Shimadzu LC-20A UHPLC system. Chromatographic separation was carried out with a Waters HSS T3 column and a binary solvent gradient of water with 0.1% formic acid (solvent A) and acetonitrile with 0.1% formic acid (solvent B). The following gradient was used for separation: 0–3 minutes, isocratic flow of 0% B; 3–8 minutes, 0–100% B; 8–13 minutes, isocratic flow of 100% B; 13–13.1 minutes, 100–0% B; 13.1–18 minutes, isocratic flow of 0% B. The flow rate was held constant at 0.2 ml/minute. The MS was operated in MRM mode monitoring the following transitions for GSH, GSSH and their respective internal standards in positive mode: GSH 308/162; GSSG 613/355; GSH internal standard 311/165; GSSG internal standard 619/165. Transitions and source parameters were optimized by infusion of 13C-labelled standards prior to analysis. GSH and GSSG molar concentrations were individually determined in each sample using a standard curve and the 13C-labelled internal standards.
Isolation of the ApoB+ and ApoB- fractions from blood plasma and lymph fluid
Blood or lymph were pooled from 5 mice and the cells were eliminated by centrifuging for 5 minutes at 6,000 × g. Dynabeads® Protein G for Immunoprecipitation (product 10003D, Thermo Fisher Scientific) were coated for two hours with anti-ApoB48/100 antibody (product K2330R, Meridian Life Sciences), then the coated beads were incubated with the blood plasma or lymph fluid for 30 minutes. ApoB+ vesicles were isolated by magnetic positive selection and the unbound supernatant was taken as the ApoB negative fraction. The samples were used for lipidomic analysis by mass spectrometry as described in the Lipidomics section below.
Lipidomics
To prepare blood plasma and lymph fluid for lipidomic analysis samples were transferred to glass vials for Bligh-Dyer lipid extraction using methanol:PBS:chloroform (1:1:2) containing 0.1% formic acid. Ten μl of a mix of internal standards were added to each extraction mixture. The internal standards included known quantities of 13:0 lyso-phosphatidyl choline, 12:0–13:0 phosphatidyl choline, and 12:0–13:0 phosphatidyl inositol, purchased as solutions from Avanti Polar Lipids (Alabaster, AL). After vortexing for 1 minute, the samples were centrifuged at 1500xg for 15 minutes. Using a glass Hamilton syringe, the bottom (chloroform) layer of solvent was removed and transferred to a clean screw-cap glass vial. Lipid extracts were concentrated by drying under a compressed air stream (to reduce lipid oxidation) and then reconstituting in 100 μl of chloroform prior to liquid chromatography and mass spectrophotometry.
Mass spectrometry for lipidomics
Lipid extracts were separated using a Phenomenex (Torrance, CA) Luna C5 column (50 × 4.6 mm, 5 μm bead size) with a binary solvent gradient. Solvent A is composed of a mixture of water and methanol (95:5) containing 5mM ammonium acetate, 0.1% (v/v) ammonium hydroxide and 0.1% (v/v) formic acid. Solvent B is composed of a mixture of isopropanol, methanol and water (65:35:5) containing 0.1% ammonium hydroxide (v/v) and 0.1% formic acid (v/v). The solvent gradient used for separation was: 0–5 minutes, 0% solvent B; 5–45 minutes solvent B, increased linearly from 0% to 100%; from 45–55 minutes, 100% solvent B; 55–60 minutes, solvent B was decreased linearly from 100% to 0%; from 60–65 minutes, the column was re-equilibrated with 0% solvent B. Flow rate was held at a constant 400 μl per minute throughout the gradient and increased from 400 μl per minute to 500 μl per minute from 45–55 minutes, then held at 500 μl per minute for 55–60 minutes, and decreased to 400 μl per minute from 60–65 minutes.
Lipidomics data were acquired using two parallel reaction monitoring (PRM) methods on a QExactive HF-X. The first method acquired data in negative ionization mode to measure fatty acid incorporation into phospholipids and the second acquired data in positive mode to measure fatty acid incorporation into triacylglycerols. Precursor ion data were acquired with a resolving power of 120,000 full width at half maximum (FWHM); the automatic gain control (AGC) target was set to 1,000,000 with a maximum inject time of 50 ms and the scan range was set to 150–2000. PRM data was acquired with a resolving power of 7,500 FWHM. The AGC target was set to 200,000 with a maximum inject time of 30 ms. The loop count was set to 5 (5 product ion scans for every one precursor ion scan), with an isolation window of 1.2 daltons and an isolation offset of 0.5 Da. Analytes were fragmented with a stepped collision energy of 30, 50 and 70 normalized collision energy (NCE).
The lipid identities of mass spectrometry peaks were verified using an exploratory data-dependent acquisition method as described above. Isobaric masses are a potential problem when analyzing lipid species by mass spectrometry. To ensure the accurate detection of lipid species, we used a combination of positive- and negative-ionization product ion spectra generated with the exploratory data-dependent method. These spectra ensured the correct identification of both fatty acyl chains and head groups in phospholipids. These analyses were conducted manually in FreeStyle 1.6 (Thermo Scientific). A total of 121 lipid species were validated in this way and associated with appropriate precursor ions. The targeted lipid species were based on an inclusion list of the most abundant lipid species in human plasma identified by the LIPID MAPS consortium37 (Extended Data Table 2).
The oleic acid content of each lipid species was determined by analyzing oleic acid ions from parallel reaction monitoring spectra searched at a 20 ppm tolerance using TraceFinder 1.4 (Fisher Scientific). These sources of oleic acid were then summed to determine the total amount of oleic acid in the triacylglycerol and phospholipid pools.
Statistical methods
In each type of experiment, multiple melanomas from different patients were tested in multiple independent experiments performed on different days. Mice were allocated to experiments randomly and samples processed in an arbitrary order, but formal randomization techniques were not used. Prior to analyzing the statistical significance of differences among treatments, we tested whether the data were normally distributed and whether variance was similar among treatments. To test for normal distribution, we performed the Shapiro–Wilk test when 3≤n<20 or the D’Agostino Omnibus test when n≥20. To test if variability significantly differed among treatments we performed F-tests (for experiments with two treatments) or Levene’s median tests (for more than two treatments). When the data significantly deviated from normality or variability significantly differed among treatments, we log2-transformed the data and tested again for normality and variability. Fold change data were always log2-transformed. If the transformed data no longer significantly deviated from normality and equal variability, we performed parametric tests on the transformed data. If log2-transformation was not possible or the transformed data still significantly deviated from normality or equal variability, we performed non-parametric tests on the non-transformed data.
All of the statistical tests we used were two-sided, where applicable. To assess the statistical significance of a difference between two treatments, we used Student’s t-tests/paired t-tests (when a parametric test was appropriate) or Wilcoxon tests (when a non-parametric paired test was appropriate). Multiple t-tests (parametric or non-parametric) were followed by Holm-Sidak’s multiple comparisons adjustment. To assess the statistical significance of differences between more than two treatments, we used one-sided or two-sided ANOVAs (when a parametric test was appropriate) followed by Tukey’s, Dunnet’s, Dunnett’s T3, or Sidak’s multiple comparisons adjustment or Kruskal-Wallis tests (when a non-parametric test was appropriate) followed by Dunn’s multiple comparisons adjustment.
To assess the statistical significance of differences between time-course data, we used repeated-measures two-sided ANOVAs (when a parametric test was appropriate and there were no missing data) or mixed-effects analyses (when a parametric test was appropriate and there were missing data points) followed by Dunnett’s multiple comparisons adjustment, or nparLD, a statistical tool for the analysis of non-parametric longitudinal data, followed by the Benjamini-Hochberg method for multiple comparisons adjustment. Multiple comparisons were adjusted per melanoma as a family. Therefore, when multiple comparisons were due to only two treatments applied to multiple melanomas, ANOVAs were followed by uncorrected Fisher’s LSD test and Kruskal-Wallis tests were followed by uncorrected Dunn’s test.
To assess the statistical significance of overall differences between the percentages of mice that developed tumors in different treatments/cell doses in all melanomas, we used multiple logistic regressions. The fraction of cells that formed tumors was estimated using the Extreme Limiting Dilution Analysis (ELDA) tool47. To assess the statistical significance of metabolic differences between treatments, we imputed zero values with half of the minimum non-zero intensity of each metabolite, and used general linear modeling with log-transformed data followed by Benjamini-Hochberg’s multiple comparison adjustment. All statistical analyses were performed with Graphpad Prism 8.1 or R 3.5.1 with the stats, fBasics, car, emmeans, and nparLD packages. Principle component and pathway enrichment analysis of metabolomics data was performed using the MetaboAnalyst 4.0 tool48. RNA-seq data were processed using the next generation sequencing analysis pipeline from the high-performance computing core (BioHPC) at the University of Texas Southwestern Medical Center. All data represent mean ± standard deviation.
Samples sizes were not pre-determined based on statistical power calculations but were based on our experience with these assays. For assays in which variability is commonly high, we typically used n>10. For assays in which variability is commonly low, we typically used n<10. No data were excluded; however, mice sometimes died during experiments, presumably due to the growth of metastatic tumors. In those instances, data that had already been collected on the mice in interim analyses were included (such as subcutaneous tumor growth measurements over time) even if it was not possible to perform the end-point analysis of metastatic disease burden due to the premature death of the mice.
Extended Data
Extended Data Figure 1. Representative flow cytometry gates for the isolation of melanoma cells from the blood and lymph and representative bioluminescence images of visceral organs to quantitate metastatic disease burden. Related to Table 1, Figures 1, 2 and 3.
a-d, Flow cytometry plots showing the gating strategies used to identify human melanoma cells in the blood (a) or lymph (b) of NSG mice or mouse melanoma cells in the blood (c) or lymph (d) of C57BL mice. In all cases, cells were gated on forward scatter area versus side height (FSC-H vs. FSC-A) to exclude red blood cells and cell clumps. Mouse hematopoietic and endothelial cells were excluded by gating out cells that stained positively for anti-mouse CD45, CD31, or Ter119. Human melanoma cells were selected by including cells that stained positively for HLA-ABC and mouse melanoma cells were selected by including cells that stained positively for CD146. Melanoma cells were also identified in these studies based on DsRed, which was stably expressed in all melanomas along with luciferase. e-g, Representative bioluminescence imaging of visceral organs dissected from a negative control mouse (e), and mice transplanted with luciferase-expressing human (f) or mouse (g) melanomas. (h) Evan’s blue dye was injected into a subcutaneous melanoma to expose the tumor-draining blood (white arrow) and lymphatic (black arrow) vessels. (i) Inefficiently metastasizing human melanomas were transplanted subcutaneously into NSG mice and the number of melanoma cells per microliter of blood and tumor-draining lymph were determined (n=4 or 5 mice per melanoma from two independent experiments). Statistical significance was assessed using a Kruskal-Wallis test (*p<0.05, ***p<0.001). Exact p-values are in source data files.
Extended Data Figure 2. The effect of Liproxstatin-1 on the growth of subcutaneous tumors and the effect of other inhibitors on metastasis. Related to Figures 1 and 2.
(a) Addition of 2-mercaptoethanol (2ME) to M405 melanoma cells isolated from subcutaneous tumors or the blood blunted the increase in ROS levels in melanoma cells from the blood. b-c, Human (b) or mouse (c) melanomas were treated in culture with Erastin and/or Deferoxamine. d-h, Treatment of mice with Liproxstatin-1 had little or no effect on the growth of subcutaneous tumors formed by human (d-f) or mouse (g, h) melanomas. (i) Pre-treatment of inefficiently metastasizing human melanoma cells with Liproxstatin-1 did not significantly affect metastatic disease burden after intravenous injection into NSG mice. (j) Mouse melanomas were pre-treated with autophagy (3-MA), apoptosis (ZVAD), or necroptosis (GSK’872) inhibitors then injected intravenously into C57BL mice and metastatic disease was assessed 1 month later by bioluminescence imaging. The number of replicates is indicated in each panel and the number of independent experiments is shown in Supplementary Data, ‘Statistics and Reproducibility”. All data represent mean ± s.d. Statistical significance was assessed using a correlated-samples two-way ANOVA followed by Sidak’s multiple comparisons adjustment (a), two-way ANOVA (i) followed by Tukey’s multiple comparison adjustment (b), Kruskal-Wallis tests followed by Dunn’s multiple comparisons adjustment (c (Y1.7), j), one-way ANOVA followed by Tukey’s multiple comparisons adjustment (c (Y3.3)), or nparLD tests (d-h). No statistically significant differences were observed in panels d-f, i, or j. For all panels, statistical tests were two-sided where applicable and *p<0.05, **p<0.01, ***p<0.001. Exact p-values are in source data files.
Extended Data Figure 3. The effect of Gpx4 deficiency on the survival and proliferation of mouse melanomas in culture. Related to Figure 2.
(a) Western blot analysis of GPX4 in parental or Gpx4-deficient mouse melanomas (representative of 2 independent experiments). (b) Western blot analysis of GPX4 in efficiently and inefficiently metastasizing melanomas from patients as well as normal mouse brain and liver tissue. Actin was used as a loading control (representative of 2 independent experiments). Uncropped western blots are in Supplementary Figure 1. c, d, Gpx4-deficiency did not significantly affect the growth of melanoma cells cultured in low oxygen (c) but did significantly reduce the growth of some melanomas cultured at atmospheric oxygen levels (d) (n=6 replicate cultures per melanoma; data reflect one representative experiment of two conducted). (e) Lipid ROS levels in melanoma cells from subcutaneous tumors formed by Gpx4-deficient or parental control melanomas cells (n=6 mice per melanoma in two independent experiments). f-i, Growth of primary subcutaneous tumors (f, g) and frequency of circulating melanoma cells in the blood (h, i) of NSG mice transplanted with parental or Gpx4-deficient melanomas (n=4–5 mice per melanoma per experiment from two independent experiments). All data represent mean ± s.d. Statistical significance was assessed using repeated measures two-way ANOVAs (c (Y1.7), d) or mixed-effects analysis (c (Y3.3)) followed by Dunnett’s multiple comparisons adjustment (c, d), correlated-samples two-way ANOVA (e), nparLD tests (f, g), Fisher’s LSD test (e), Welch’s one-way ANOVA followed by Dunett’s T3 multiple comparisons adjustment (h), or Kruskal-Wallis tests followed by Dunn’s multiple comparisons adjustment (i). For all panels, statistical tests were two-sided where applicable and **p<0.01, ***p<0.001. Exact p-values are in the source data files.
Extended Data Figure 4. Lipid species in plasma and lymph. Related to Figure 3.
Lipid species that significantly differed in abundance between the plasma and lymph of NSG (a) or C57BL (b) mice (p<0.01). (c) Relative triacylglycerol (TG) content in the ApoB+ and ApoB- fractions of blood plasma or lymph fluid (after cells were removed) from C57BL mice (two independent samples per treatment). (d) Relative oleic acid abundance in the ApoB+ and ApoB- fractions of blood plasma or lymph fluid from C57BL mice (two independent samples per treatment). Statistical significance was assessed using generalized linear modeling with log-transformed, half-min imputed data replacing zeros followed by the Benjamini-Hochberg multiple comparisons adjustment using two-sided t-statistics (a and b). The number of replicates is indicated in each panel. Each panel reflects two independent experiments. All data represent means and, when present, error bars reflect s.d. Exact p-values are in the source data files.
Extended Data Figure 5. Acsl3 is required for oleic acid incorporation into phospholipids and the protective effect of oleic acid against ferroptosis. Related to Figure 3.
(a) Western blot analysis of ACSL3 in efficiently and inefficiently metastasizing melanomas from patients as well as normal mouse brain and liver tissue. Actin was used as a loading control (representative of 2 independent experiments). (b) Western blot analysis of ACSL3 in parental control and Acsl3 deficient mouse melanomas (representative of 2 independent experiments). Uncropped western blots are in Supplementary Figure 1. c-d, Relative levels of oleic acid in phospholipids from Acsl3-deficient and parental control melanomas. In some cases, wild-type (Acsl3OE) or catalytically dead mutant Acsl3 (mut. Acsl3OE) were over-expressed in YUMM1.7 (c) or YUMM3.3 (d) mouse melanomas. The number of replicates per treatment is indicated in each panel (two independent experiments). (e) Lipid ROS (BODIPY-C11Oxidized/C11Oxidized+C11Non-oxidized ratio) levels in mouse melanoma cells from subcutaneous tumors in C57BL mice. The number of replicates per melanoma is indicated in each panel (two independent experiments). The data from parental controls cells in this experiment are also shown in Extended Data Fig. 3e. (f) Growth of Acsl3-deficient and parental control melanomas in culture (4 replicate cultures per melanoma per experiment, representative of two independent experiments; no differences were statistically significant). g-h, Growth of Acsl3 deficient melanomas in culture with oleic acid and with or without Erastin. In some cases, wild-type (Acsl3OE) or catalytically dead mutant Acsl3 (mutant Acsl3OE) were over-expressed in YUMM1.7 (g) or YUMM3.3 (h) melanomas. (i) Metastatic disease burden in mice intranodally injected with Acsl3-deficient or control melanomas. All data represent mean ± s.d. Statistical significance was assessed using one-sided ANOVAs followed by Sidak’s and Dunnett’s (c, d) multiple comparisons adjustments, paired t-tests (e), repeated measures two-sided ANOVAs followed by Dunnett’s multiple comparisons adjustment (f), Kruskal-Wallis tests followed by Dunn’s multiple comparisons adjustment (g, h, I (Y3.3)), or Welch’s one-way ANOVA followed by Dunnett’s T3 multiple comparisons adjustment (I (Y1.7). For all panels, statistical tests were two-sided where applicable and *p<0.05, **p<0.01, ***p<0.001. Exact p-values are in source data files.
Extended Data Figure 6. Expression of potential ferroptosis regulators by melanoma cells. Related to Figure 3.
(a-f) Western blot analysis of SLC7A11 (a, b), ACSL4 (c, d), and FSP1 (e, f) in efficiently and inefficiently metastasizing human melanomas (a, c, e) as well as mouse melanomas (b, d, f) (representative of two experiments). Normal mouse liver and brain, human lung, and mouse fibroblasts were sometimes included as positive or negative controls. Uncropped western blots can be found in Supplementary Figure 1. (g) FSP1 transcript levels by qRT-PCR in human melanoma cells isolated from subcutaneous tumors, blood, or lymph of xenografted mice (3 replicates (each replicate was pooled from 4–5 mice) per melanoma). (h) RNAseq analysis of the fatty acid transporters FATP1, FATP3, FATP4, and FATP5 in efficiently and inefficiently metastasizing human melanomas (2 to 3 replicates per melanoma). (i) Staining with anti-CD36 or isotype control antibody in human and mouse melanomas. Data are representative of 2 tumors per melanoma. (j) Elevated levels of oleic acid in lymph suppress lipid ROS accumulation and ferroptosis in metastasizing melanoma cells, increasing their ability to survive during subsequent dissemination through the blood. Image created with BioRender. No statistically significant differences were observed in panels g and h. All data represent mean ± s.d. Statistics were determined repeated measures two-sided ANOVA followed by Tukey’s multiple comparisons adjustment (g). Exact p-values are in source data files.
Extended Data Table 1. Characterization of human and mouse melanomas. Related to Table 1.
(a) Summary of the human melanomas used in this study and their spontaneous metastatic behavior after subcutaneous transplantation into NSG mice. Melanomas were characterized as inefficient or efficient metastasizers. Before subcutaneous tumors grew to 2.5 cm in diameter, inefficient metastasizers rarely formed macrometastases or micrometastases beyond the lung whereas efficient metastasizers commonly formed macrometastases as well as micrometastases in multiple organs. Some of these data have been published previously27. (b) Summary of the mouse melanomas used in this study and their spontaneous metastatic behavior after subcutaneous transplantation into C57BL mice. For each melanoma, the number of mice analyzed to assess metastatic behavior is indicated in parentheses within each column. The data reflect 1 to 5 independent experiments per melanoma.
| a | Metastatic potential in mice | Melanoma ID | Age/sex | AJCC clinical stage | Tumor site | Known driver mutations | % of mice with macromets | % of mice with micrometastasis only in the lung (BLI) | % of mice with micrometastasis in organs other than the lung (BLI) |
| Inefficient | M597 | 61/M | IIIA | Primary cutaneous | p53 | 0% (0/22) | 10% (1/10) | 30% (3/10) | |
| Inefficient | UM17 | 72/F | IIA | Lymph node | N/A | 0% (0/10) | 50% (5/10) | 0% (0/10) | |
| Inefficient | UM43 | 64/M | IIIB | Lymph node | BRAF V600E BRAF V600M |
0% (0/17) | 0% (0/17) | 0% (0/17) | |
| Inefficient | UM47 | Unk./M | IV | Lymph node | N/A | 0% (0/19) | 0% (0/19) | 0% (0/19) | |
| Inefficient | M498 | 83/M | IIIB | Primary cutaneous | p53 | 0% (0/17) | 0% (0/8) | 0% (0/8) | |
| Inefficient | M610 | 76/F | IIA | Primary cutaneous | BRAF V600E | 0% (0/27) | 43% (3/7) | 0% (0/7) | |
| Efficient | M405 | 29/M | IIIB | Lymph node | NRAS Q61R | 70% (52/74) | 0% (0/23) | 96% (22/23) | |
| Efficient | M481 | 67/M | IIIB | Primary cutaneous | BRAF V600E | 56% (75/135) | 22% (4/18) | 78% (14/18) | |
| Efficient | UT10 | 65/M | IIIC | Lymph node | NRAS Q61K | 28% (15/54) | 76% (41/54) | 61% (33/54) | |
| b | Metastatic potential in mice | Melanoma cell line | Mutations | % of mice with mets | % of mice with micrometastasis only in the lung (BLI) | % of mice with micrometastasis in organs other than the lung (BLI) | |||
| Efficient | YUMM1.7 | BrafV600E/+, PTEN−/−, Cdkn2−/− | 65% (13/20) | 45% (9/20) | 20% (4/20) | ||||
| Efficient | YUMM3.3 | BrafV600E/+, Cdkn2−/− | 53% (10/19) | 0% (0/19) | 53% (10/19) |
Extended Data Table 2. List of lipid species analyzed by mass spectrometry. Related to Extended Data Figure 4.
Lipid species analyzed in the lipidomic analysis in Extended Data Figure 4a–b. All of these lipid species are capable of containing oleic acid except for those highlighted in grey.
| Phospholipid Species Monitored | Monoacyl-glycerols (MGs) | Diacyl-glycerols (DGs) | Triacyl-glycerols (TGs) | |||
|---|---|---|---|---|---|---|
| Phosphatidyl-inositol (PI) | Phosphatidyl-choline (PC) | Phosphatidyl-ethanolamine (PE) | Phosphatidyl-serine (PS) | |||
| PI(32:1) | PC(30:1) | PE(32:1) | PS(36:1) | MG(16:0) | 1,2-DG(30:0) | TG(48:1) |
| PI(34:0) | PC(32:0) | PE(34:0) | PS(36:2) | MG(18:0) | 1,2-DG(30:1) | TG(48:2) |
| PI(34:1) | PC(32:1) | PE(34:1) | PS(38:4) | MG(18:1) | 1,2-DG(30:2) | TG(50:0) |
| PI(34:2) | PC(32:2) | PE(34:2) | PS(38:5) | MG(18:2) | 1,2-DG(32:0) | TG(50:1) |
| PI(36:0) | PC(34:0) | PE(36:0) | MG(20:0) | 1,2-DG(32:1) | TG(50:2) | |
| PI(36:1) | PC(34:1) | PE(36:1) | MG(20:4) | 1,2-DG(32:2) | TG(50:3) | |
| PI(36:2) | PC(34:2) | PE(36:2) | 1,2-DG(32:3) | TG(50:4) | ||
| PI(36:3) | PC(34:3) | PE(36:3) | 1,2-DG(34:0) | TG(52:1) | ||
| PI(36:4) | PC(36:0) | PE(36:4) | 1,2-DG(34:1) | TG(52:2) | ||
| PI(36:5) | PC(36:1) | PE(36:5) | 1,2-DG(34:2) | TG(52:3) | ||
| PI(38:2) | PC(36:2) | PE(38:1) | 1,2-DG(34:3) | TG(52:4) | ||
| PI(38:3) | PC(36:3) | PE(38:2) | 1,2-DG(34:4) | TG(52:5) | ||
| PI(38:4) | PC(36:4) | PE(38:3) | 1,2-DG(36:0) | TG(54:2) | ||
| PI(38:5) | PC(36:5) | PE(38:4) | 1,2-DG(36:1) | TG(54:3) | ||
| PI(38:6) | PC(38:2) | PE(38:5) | 1,2-DG(36:2) | TG(54:4) | ||
| PI(40:3) | PC(38:4) | PE(38:6) | 1,2-DG(36:3) | TG(54:5) | ||
| PI(40:4) | PC(38:5) | PE(40:1) | 1,2-DG(36:4) | TG(54:6) | ||
| PI(40:5) | PC(38:6) | PE(40:4) | 1,2-DG(36:5) | TG(56:6) | ||
| PI(40:6) | PC(40:2) | PE(40:5) | 1,2-DG(38:0) | |||
| PC(40:4) | PE(40:6) | 1,2-DG(38:1) | ||||
| PC(40:5) | PE(40:7) | 1,2-DG(38:2) | ||||
| PC(40:6) | PE(42:1) | 1,2-DG(38:3) | ||||
| PC(40:7) | PE(42:5) | 1,2-DG(38:4) | ||||
| 1,2-DG(38:5) | ||||||
| 1,2-DG(38:6) | ||||||
| 1,2-DG(40:4) | ||||||
| 1,2-DG(40:6) | ||||||
| 1,2-DG(40:7) | ||||||
Supplementary Material
ACKNOWLEDGEMENTS
S.J.M. is a Howard Hughes Medical Institute (HHMI) Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, the Kathryn and Gene Bishop Distinguished Chair in Pediatric Research, the director of the Hamon Laboratory for Stem Cells and Cancer, and a Cancer Prevention and Research Institute of Texas Scholar. The research was supported by the Cancer Prevention and Research Institute of Texas (RP170114 and RP180778) and by the National Institutes of Health (U01 CA228608). A.T. was supported by the Leopoldina Fellowship (LPDS 2016-16) from the German National Academy of Sciences and the Fritz Thyssen Foundation. B.S. was supported by a Ruth L. Kirschstein National Research Service Award Postdoctoral Fellowship from the National Heart, Lung, and Blood Institute (F32 HL139016). M.L.M. and D.R.S. were supported by NIH grants P01 (CA217797) and P30 (CA086862). We would like to thank M. Dellinger for comments on the manuscript. We thank N. Meireles (University of Michigan) for collecting human melanomas, M. Nitcher for mouse colony management, the BioHPC (High Performance Computing) core for data storage, as well as N. Loof and the Moody Foundation Flow Cytometry Facility.
Footnotes
COMPETING INTEREST DECLARATION
S.J.M. is an advisor for Frequency Therapeutics and Protein Fluidics as well as a stockholder in G1 Therapeutics and Mereo Biopharma. Correspondence and requests for materials should be addressed to S.J.M. (Sean.Morrison@UTSouthwestern.edu).
DATA AVAILABILITY
Source Data for Table 1, Figures 1–3 and Extended Data Figures 1–6 are provided with the paper. Uncropped western blots are in Supplementary Figure 1. All other data are available from the corresponding author upon request.
ADDITIONAL INFORMATION
Supplementary Information is available for this paper.
Reprints and permissions information is available at www.nature.com/reprints.
REFERENCES
- 1.Sleeman J, Schmid A & Thiele W Tumor lymphatics. Semin Cancer Biol 19, 285–297, (2009). [DOI] [PubMed] [Google Scholar]
- 2.Alitalo A & Detmar M Interaction of tumor cells and lymphatic vessels in cancer progression. Oncogene 31, 4499–4508, (2012). [DOI] [PubMed] [Google Scholar]
- 3.Leong SP et al. Cutaneous melanoma: a model to study cancer metastasis. J Surg Oncol 103, 538–549, (2011). [DOI] [PubMed] [Google Scholar]
- 4.Willis RA The spread of tumours in the human body. 3d edn, (Butterworths, 1973). [Google Scholar]
- 5.Naxerova K et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gundem G et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanborn JZ et al. Phylogenetic analyses of melanoma reveal complex patterns of metastatic dissemination. Proc Natl Acad Sci U S A 112, 10995–11000, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McFadden DG et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 156, 1298–1311, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira ER et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science 359, 1403–1407, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown M et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 359, 1408–1411, (2018). [DOI] [PubMed] [Google Scholar]
- 11.Morton DL et al. Final trial report of sentinel-node biopsy versus nodal observation in melanoma. N Engl J Med 370, 599–609, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiter U et al. Complete lymph node dissection versus no dissection in patients with sentinel lymph node biopsy positive melanoma (DeCOG-SLT): a multicentre, randomised, phase 3 trial. Lancet Oncol 17, 757–767, (2016). [DOI] [PubMed] [Google Scholar]
- 13.Faries MB et al. Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. N Engl J Med 376, 2211–2222, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace AC & Hollenberg NK The Transplantability of Tumours by Intravenous and Intralymphatic Routes. Br J Cancer 19, 338–342, (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshida T et al. Imaging steps of lymphatic metastasis reveals that vascular endothelial growth factor-C increases metastasis by increasing delivery of cancer cells to lymph nodes: therapeutic implications. Cancer Res 66, 8065–8075, (2006). [DOI] [PubMed] [Google Scholar]
- 16.Lee CK et al. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science 363, 644–649, (2019). [DOI] [PubMed] [Google Scholar]
- 17.Pascual G et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45, (2017). [DOI] [PubMed] [Google Scholar]
- 18.Ma Q et al. Unexpected contribution of lymphatic vessels to promotion of distant metastatic tumor spread. Sci Adv 4, eaat4758, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanharanta S & Massague J Origins of metastatic traits. Cancer Cell 24, 410–421, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piskounova E et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 527, 186–191, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Gal K et al. Antioxidants can increase melanoma metastasis in mice. Sci Transl Med 7, 308re308, (2015). [DOI] [PubMed] [Google Scholar]
- 22.Dixon SJ and Stockwell BR The hallmarks of ferroptosis. Annu. Rev. Cancer Biol. 3, 35–54, (2019). [Google Scholar]
- 23.Wang W et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y et al. Imidazole Ketone Erastin Induces Ferroptosis and Slows Tumor Growth in a Mouse Lymphoma Model. Cell Chem Biol 26, 623–633 e629, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J et al. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature 572, 402–406, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viswanathan VS et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintana E et al. Human melanoma metastasis in NSG mice correlates with clinical outcome in patients. Sci Transl Med 4, 159ra149, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meeth K, Wang JX, Micevic G, Damsky W & Bosenberg MW The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res 29, 590–597, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hangai-Hoger N, Cabrales P, Briceno JC, Tsai AG & Intaglietta M Microlymphatic and tissue oxygen tension in the rat mesentery. Am J Physiol Heart Circ Physiol 286, H878–883, (2004). [DOI] [PubMed] [Google Scholar]
- 30.Tasdogan A et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedmann Angeli JP et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 16, 1180–1191, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seiler A et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab 8, 237–248, (2008). [DOI] [PubMed] [Google Scholar]
- 33.Doll S et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol 13, 91–98, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kayden HJ, Karmen A & Dumont A Alterations in the Fatty Acid Composition of Human Lymph and Serum Lipoproteins by Single Feedings. J Clin Invest 42, 1373–1381, (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magtanong L et al. Exogenous Monounsaturated Fatty Acids Promote a Ferroptosis-Resistant Cell State. Cell Chem Biol 26, 420–432 e429, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spitz DR, Kinter MT, Kehrer JP & Roberts RJ The effect of monosaturated and polyunsaturated fatty acids on oxygen toxicity in cultured cells. Pediatr Res 32, 366–372, (1992). [DOI] [PubMed] [Google Scholar]
- 37.Bowden JA et al. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in Frozen Human Plasma. J Lipid Res 58, 2275–2288, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher EA & Ginsberg HN Complexity in the secretory pathway: the assembly and secretion of apolipoprotein B-containing lipoproteins. J Biol Chem 277, 17377–17380, (2002). [DOI] [PubMed] [Google Scholar]
- 39.Tang Y, Zhou J, Hooi SC, Jiang YM & Lu GD Fatty acid activation in carcinogenesis and cancer development: Essential roles of long-chain acyl-CoA synthetases. Oncol Lett 16, 1390–1396, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen WC et al. Systematic Analysis of Gene Expression Alterations and Clinical Outcomes for Long-Chain Acyl-Coenzyme A Synthetase Family in Cancer. PLoS One 11, e0155660, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon SJ et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doll S et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698, (2019). [DOI] [PubMed] [Google Scholar]
- 43.Bersuker K et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veglia F et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature 569, 73–78, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
SUPPLEMENTAL REFERENCES
- 45.Kassan A et al. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol 203, 985–1001, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agathocleous M et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Y & Smyth GK ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347, 70–78, (2009). [DOI] [PubMed] [Google Scholar]
- 48.Chong JW, D.S. and Xia J Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Current Protocols in Bioinformatics 68 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.