Abstract
Introduction: The Pals1-associated tight junction (PATJ) is a Crumbs (CRB) complex component that regulates epithelial cell apico-basal polarity and directional migration. This study assessed PATJ expression in clear cell renal cell carcinoma (ccRCC) vs. normal tissues and associated with ccRCC progression and prognosis.
Methods: The effects of PATJ knockdown were investigated on regulation of normal kidney epithelial cell viability and protein expression in vitro. The PATJ mRNA data in ccRCC were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases and analyzed with UALCAN, LinkedOmics, Kaplan-Meier Plotter, GEPIA, and SurvExpress tools. Immunohistochemistry was performed for PATJ in tissue microarray sections (n = 150 ccRCC and 30 normal renal specimens). Normal human kidney tubular epithelial cell (HKC) cells were transfected with PATJ and negative control siRNA for cell viability CCK-8 assay, flow cytometry, and western blots.
Results: The data showed that PATJ mRNA and protein were downregulated in ccRCC tissues and cell lines. Downregulation of PATJ mRNA was associated with male patients, advanced tumor stages, grades, and ccB subtypes as well as poorer overall and disease-free survival of patients. Furthermore, PATJ protein was also significantly downregulated in ccRCC tissues and associated with advanced tumor pathologic, TNM stages and poorer overall. In vitro, knockdown of PATJ expression promoted HKC proliferation and the activation of mitogen-activated protein kinases (MAPK) pathway proteins.
Conclusions: This study revealed that a decrease of PATJ in ccRCC, which was associated with male patients, advanced tumor, and poorer survival, suggesting that PATJ may be a useful prognostic biomarker and therapeutic target for ccRCC.
Keywords: cell polarity protein Pals1-associated tight junction, clear cell renal cell carcinoma, prognosis, tumor cell proliferation, mitogen-activated protein kinases
Introduction
Renal cell carcinoma (RCC) is a frequently diagnosed kidney cancer in men 65 years or older and clear cell RCC (ccRCC) is the most common histologic subtype (Cohen and McGovern, 2005; Hsieh et al., 2017), counting for approximately 90% of kidney cancers and up to 4% of all newly diagnosed cancer cases in 2018 worldwide (Siegel et al., 2018). Clinically, RCC is usually symptomless and at advanced stages, patients have the classic triad of hematuria, flank pain, and flank mass (Cohen and McGovern, 2005; Hsieh et al., 2017). Approximately 25–30% of RCC patients had metastatic renal cell carcinoma (mRCC) at the time of tumor diagnosis (Lam et al., 2005a), and 70–80% patients are treated with nephrectomy for curative intent or tumor removal. Indeed, for patients with a localized disease within the kidney, surgery alone may be sufficient to cure and the recurrence rate is usually less than 25%. However, patients with advanced tumor progression (tumor has spread outside of the kidney or the locoregional disease) have a great risk of recurrence (more than 40%) after initial treatment (Lam et al., 2005b; Gupta et al., 2008), leading to a 12% of the 5-year survival rate (Joosten et al., 2017). RCC is usually resistant to radiation and chemotherapy (Campbell et al., 2016), as well as to recent treatment options include immunomodulatory and targeted therapies. For example, a combination approach to target tumor angiogenesis using sunitinib, bevacizumab, or pazopanib and the mammalian target of rapamycin (mTOR) pathway using temsirolimus has been approved as a front-line treatment option for mRCC, whereas PD-1/PD-L1 is a more recently developed anti-RCC agent (Barata and Rini, 2017; Posadas et al., 2017; Rodriguez-Vida et al., 2017; Zarrabi and Wu, 2018). However, treatment effectiveness, treatment-related toxicities, drug resistance, and financial burden are all major concerns for RCC patients (Hsieh et al., 2017). Thus, the identification and evaluation of tumor markers for preliminary screening, early metastasis detection, and monitoring of treatment responses and prognosis are sorely needed.
Toward this end, a better understanding of RCC pathogenesis and tumor development and progression can help us effectively control RCC initiation and metastasis. ccRCC originates from the proximal renal tubular epithelium. Cell polarity is a key feature of the epithelium (Halaoui and McCaffrey, 2015), which is maintained through the tight junctions, desmosomes, and adherens junctions (Anderson and Van Itallie, 2009). Molecularly, cell polarity is maintained through the expression of specific proteins in specific areas of the cell membrane. Crumbs (CRB), PAR, and SCRIB polarity complexes specifically regulate the apical and basolateral membrane domains via the organization of intracellular signaling pathways for epithelial polarity (Halaoui and McCaffrey, 2015). Dysregulation of these cell polarity proteins could rewire oncogenic and tumor suppressor signaling pathways and promote cell proliferation but inhibit apoptosis and induce tumor cell invasion, metastasis, and differentiation (Halaoui and McCaffrey, 2015; Li et al., 2017). Recent studies have revealed that Pals1-associated tight junctions [PATJ; also called InaD-like protein (INADL)] is a CRB complex component and regulates epithelial cell apico-basal polarity and directional migration by regulating the localization of atypical protein kinase C (aPKC) and PAR3 to the leading edge in Madin-Darby canine kidney II cells (Shin et al., 2007), while the directional migration of blood capillaries during embryonic development was regulated by patj/amot/syx signaling to control local GTPase activity in zebrafish (Ernkvist et al., 2009).
In this study, we analyzed expression of PATJ mRNA using The Cancer Genome Atlas (TCGA) and GEO databases and PATJ protein expression in our retrospective cohort of 150 ccRCC tissues vs. 30 normal renal specimens. We then evaluated the association of PATJ expression with the clinicopathological features and survival of ccRCC patients. After that, we investigated the effect of PATJ expression on the regulation and proliferation of human renal tubular epithelial cells (HKC). We show the utility of PATJ as a diagnostic and prognostic biomarker for ccRCC.
Materials and Methods
Retrieval and Analysis of TCGA Database Data
We utilized a web tool (the UALCAN at http://ualcan.path.uab.edu/index.html) to retrieve and analyze tumor transcriptome data on changes in PATJ mRNA and protein levels in ccRCC tissues in TCGA database and Clinical Proteomic Tumor Analysis Consortium (CPTAC) Confirmatory/Discovery dataset. This web tool provided us (1) publicly accessible cancer transcriptome data (like TCGA) and (2) The graphs and plots depicting gene expression and patients’ survival data (Chandrashekar et al., 2017). We retrieved data on level of PATJ mRNA and protein in ccRCC vs. normal tissues and ccRCC pathological features (gender, stages, grades, and subtypes) from patients from TCGA and CPTAC, and then analyzed using the UALCAN web tool.
Cell Culture and PATJ siRNA Transfection
Human renal tubular epithelial cell line HKC and human ccRCC cell lines Caki-1, Caki-2, and 786-O were obtained from the National Infrastructure of Cell Line Resource (Beijing, China). HKC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 at 1:1 (Coring, Corning, NY, USA) supplemented with NEAA (Gibco, Carlsbad, CA) and 5% fetal bovine serum (FBS; Hyclone, Logan, UT, USA). Caki-1 and Caki-2 cells were grown in the McCoy’5A Medium from Hyclone and supplemented with 10% FBS (Hyclone), while 786-O cells were cultured in Roswell Park Memorial Institute medium-1640 (RPMI1640; Coring) and supplemented with 10% FBS (Hyclone). All cell lines were maintained in a humidified incubator containing 5% CO2 at 37°C.
Two PATJ siRNA constructs were used to knockdown PATJ expression in cells in vitro and their sequences were PATJ-1, 5′-GGA UGU CAA UAC UGA AGA ATT-3′ and 5′-UUC UUC AGU AUU GAC AUC CTT-3′; PATJ-2, 5′-GCA GAU GGU GUA GCA GAA ATT-3′ and 5′-UUU CUG CUA CAC CAU CUG CTT-3′. These PATJ siRNA constructs and a negative control siRNA were purchased from GenePharma Company (Shanghai, China) and used to transiently transfect into HKC cells in 6-cm plates for 48 h [the transfection conditions were 166 pmol siRNA and 17 μl Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) in 250 μl of Opti-MEM medium (Invitrogen)].
Quantitative Reverse Transcriptase-Polymerase Chain Reaction
Total cellular RNA was isolated using the RNA Fast 200 (Cat. #220010; Fastagen Biotech, Shanghai, China) and reversely transcribed into cDNA using the PrimeScript™ RT Master Mix (cat. #RR036A, TaKaRa Biotechnology (Dalian) Co., Ltd., China) according to the manufacturers’ protocols. PCR was performed using the TB Green™ Premix Ex Taq™ II (Cat. #RR820A, TaKaRa) in Bio-Rad CFX96 system (Hercules, CA, USA) according to the manufacturer’s instructions. The primers used were PATJ, 5′-AAG GGT GAC ACG TCG CAG AA-3′ and 5′-GGC TGA ACA ATC TGA GGG TAT ATG G-3′; β-actin, 5′-CAT GTA CGT TGC TAT CCA GGC-3′ and 5′-CTC CTT AAT GTC ACG CAC GAT-3′. The experiment was performed in triplicate and level of PATJ mRNA was normalized to β-actin.
Western Blot
The whole cell lysates were prepared using a modified radioimmunoprecipitation assay (RIPA) buffer and quantified using the Bradford Protein Assay (Bio-Rad). After that, these protein samples were separated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred onto the polyvinylidene fluoride membranes (PVDF; Millipore, Billerica, MA, USA) for a standard western blot protocol according to a previous study (Kim, 2017). The primary antibodies used were an anti-PATJ antibody (A12063; 1:500; ABclonal, Wuhan, China), the MAPK Family Antibody Sampler Kit (#9926; 1:1000; Cell Signaling Technology, Danvers, MA, USA), Phospho-MAPK Family Antibody Sampler Kit (#9910; 1:1000; Cell Signaling Technology), or anti-β-actin antibody (#AC026; 1:10000; ABclonal), while the secondary antibody was the horseradish peroxidase (HRP)-conjugated IgG from Cell Signaling Technology, the positive protein signals were visualized by using the enhanced chemiluminescence (ECL) Plus kit (Millipore).
LinkedOmics
We also utilized a web tool (the LinkedOmics at http://www.linkedomics.org/login.php) to analyze the multi-omics data from TCGA database. We were able to identify and analyze mRNA signatures, biomarkers of clinical attributes, and putative target genes of the transcriptional factors, microRNAs, or protein kinases using this web tool. The analytical results were exported as graphic plots (Vasaikar et al., 2018). Thus, we obtained the levels of PATJ mRNA associated with ccRCC TNM stages.
Kaplan-Meier Plotter
After that, we used another web tool known as the Kaplan-Meier Plotter at http://kmplot.com/analysis/ to assess the association of PATJ mRNA with overall survival (OS) of ccRCC patients. This web tool contains expression data of 54,675 genes in 18,674 cancer samples for examining their potential association with the prognosis of patients with 5,143 breast, 1,816 ovarian, 2,437 lung, 364 liver, and 1,065 gastric cancers. Moreover, the web tool also contains the miRNA subsystems for an additional 11,456 samples of 20 different cancer types. This web tool enables us to perform a meta-analysis of different genes for biomarker assessment (Lánczky et al., 2016).
GEPIA
GEPIA at http://gepia.cancer-pku.cn/ is an interactive web tool for us to analyze data on the RNA sequencing expression of 9,736 tumors vs. 8,587 normal samples from TCGA and the GTEx projects and this web tool uses a standard data processing pipeline to provide customizable function analyses, like differential expression of a given gene in tumor vs. normal tissues or profiling of differentially expressed genes according to cancer types or pathological stages, or survival data, etc., which was developed by Dr. Zefang Tang, Dr. Chenwei Li, and Dr. Boxi Kang in Dr. Zhang’s Lab in Peking University (Tang et al., 2017). We utilized this to associate PATJ mRNA levels with disease free survival (DFS) of ccRCC patients.
SurvExpress
The SurvExpress at http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp is a versatile online biomarker validation tool to assess the significance of gene expression in prognosis of various cancers (Aguirre-Gamboa et al., 2013). The data on renal clear cell carcinoma tissues were from TCGA (n = 468) and in this study, we validated PATJ mRNA as a prognostic marker using this web tool.
Tissue Microarrays and Immunohistochemistry
This study was approved by the Local Ethics Committee of Human Research at the First Affiliated Hospital, Xi'an Jiaotong University (Shaanxi, China). Tissue Microarray (TMA) sections were obtained from Shanghai Outdo Biotech Co., Ltd. (Cat. #HkidE180Su02; Shanghai, China) and contained 150 cases of ccRCC tissues and 30 normal renal specimens. We retrospectively retrieved the medical records and reviewed and analyzed clinicopathological data vs. PATJ expression (Table 1).
Table 1.
Association of PATJ expression with clinicopathological parameters from ccRCC patients.
| PATJ level | ||||
|---|---|---|---|---|
| Variables | No. | − (n = 80) | + (n = 70) | p |
| Histologic grade | ||||
| 1–2 | 103 | 50 (48.5%) | 53 (51.5%) | <0.0001 |
| 3–4 | 47 | 30 (63.8%) | 17 (36.2%) | |
| TNM stage | ||||
| I | 122 | 60 (49.2%) | 62 (50.8%) | 0.037 |
| II–IV | 28 | 20 (71.4%) | 8 (28.6%) | |
For immunohistochemistry, we stained PATJ protein using the TMA sections using an immunohistochemical staining kit (PV-6001; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China) according to the manufacturer’s recommended protocol. After deparaffinization and rehydration, TMA sections in the HIER solution (pH 6) were placed into a microwave oven and heated at 100 W for 6 min and 50 W for 13 min for antigen recapture, and then incubated in 3% H2O2 at room temperature for 10 min for inactivation of potential endogenous peroxidase activity. The sections were then incubated in a blocking solution containing a normal goat serum (1:4) for 15 min and in a primary anti-PATJ antibody (Cat. #HPA066352; Sigma-Aldrich, St Louis, MO, USA; 1:100) at 4°C overnight. On the next day, the sections were washed with PBS thrice, and then further incubated with a secondary antibody at the room temperature for 30 min and subsequently with the ABC reagent in the dark for 30 min. After that, the TMA sections were subjected to a brief color reaction using the 3,3′-diami-nobenzidine (DAB; Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China), and then counterstaining with hematoxylin. The immunostained TMA sections were finally reviewed and scored under a Leica microscope (Model #SCN 400; Mannheim, Germany) by an experienced pathologist in a blind fashion to the patients’ clinical data.
The immunostaining score system was developed according to the staining intensity (0, negative; 1, weakly positive; 2, moderately positive; and 3, strongly positive) and the percentage of stained cells per field (0, no staining; 1, 1–25% of tumor cell stained; 2, 26–50%; 3, 51–75%; and 4, 76–100%). The sum score of the expression level of 0–2 was considered a negative case, whereas the score of 3–12 was considered as a positive case.
Cell Viability Assay
Cells were seeded into 96-well plates at a density of 104 cells/well and transiently transfected with PATJ siRNA constructs and a negative control siRNA for up to 48 h. At different periods of time, 10 μl of the CCK-8 reagent was added (Saint-Bio, Shanghai, China) and cells were further incubated at 37°C for 3 h and the optical density (OD) values were then measured using a microplate reader (PerkinElmer, Waltham, MA, USA) at the wavelength of 490 nm. Each experiment was in triplicate and repeated at least three times.
Fluorescence-Activated Cell Sorting
To assess the effects of PATJ siRNA, we sorted cells after gene transfection using Fluorescent-activated cell sorting (FACS) and the PI and carboxyfluorescein diacetate succinimidyl ester (CFSE) staining. Briefly, cells were grown and transiently transfected with PATJ and negative control siRNA (see above for the details) for 48 h, and then stained with 0.5 mL PI/RNase Staining Buffer (#550825, BD, San Jose, CA) for 15 min at room temperature or 5 μM CFSE for 15 min at 37°C. The cells were then sorted by using FACS (BD).
Statistical Analysis
All statistical analyses were performed using GraphPad Prism Version 7.0 (GraphPad Software, La Jolla, CA, USA), i.e., to compare the differences between two groups, we carried out the paired t test or Mann-Whitney test, while to compare the differences among multiple groups, we performed the one-way analysis of variance (ANOVA) followed by using Dunnett’s multiple comparisons test. The association between PATJ expression and clinicopathological characteristics was assessed using the chi-square test or Fisher’s exact test. All statistical tests were assessed for the two-sided and a value of p < 0.05 was considered as statistically significant.
Results
Downregulation of PATJ mRNA and Protein in ccRCC Tissues and Cell Lines
In this study, we first retrieved PATJ data on ccRCC vs. normal tissues from the TCGA database and analyzed their expression using the UALCAN web tool. We revealed 533 cases of ccRCC and 72 normal kidney tissues and found that PATJ mRNA levels were significantly (p = 1.625e-12) reduced in ccRCC tissues compared to normal tissues (Figure 1A). After that, we performed a bioinformatic analysis of PATJ expression in 72 ccRCC patients using the GSE53757 data set and revealed a significant decrease in PATJ mRNA levels in ccRCC tissues compared with matched normal tissues (p < 0.0001; Figure 1B).
Figure 1.
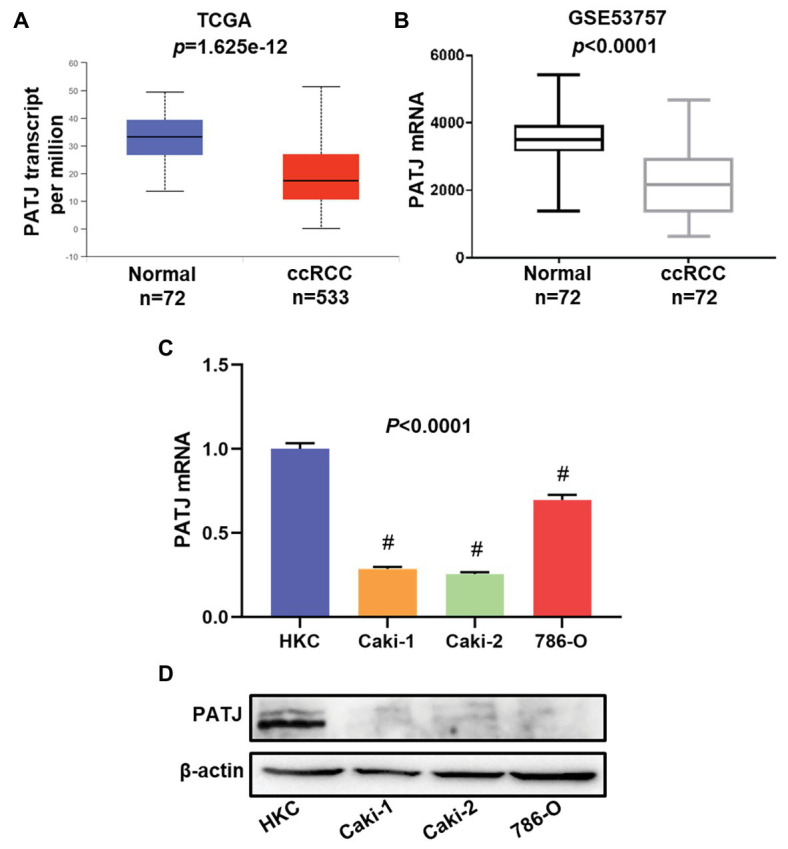
Downregulation of PATJ mRNA in clear cell renal cell carcinoma (ccRCC) tissues and cell lines. (A) The Cancer Genome Atlas (TCGA) database data. The box plots and p were generated using The UALCAN (http://ualcan.path.uab.edu/index.html). (B) GSE53757 data on PATJ mRNA in 72 ccRCC patients. (C) Expression analysis of PATJ mRNA in a human renal tubular epithelial cell HKC line and three ccRCC cell lines detected by using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). # p < 0.01 and compare to (HKC) cell. (D) Western blot. A HKC cell line and three ccRCC cell lines were grown and subjected to Western blot analysis of PATJ protein.
Moreover, we determined the level of PATJ mRNA and protein in normal human renal tubular epithelial HKC cell vs. ccRCC cell lines using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) and western blots. Our data showed that HKC cells expressed the highest level of PATJ mRNA and protein among these cell lines, whereas the level of PATJ was much lower in all ccRCC cell lines (Figures 1C,D). These data indicate that the expression of PATJ mRNA and protein was downregulated in different sets of ccRCC samples (both TCGA and GEO databases) and cell lines.
Association of PATJ mRNA and Pathological Features in ccRCC
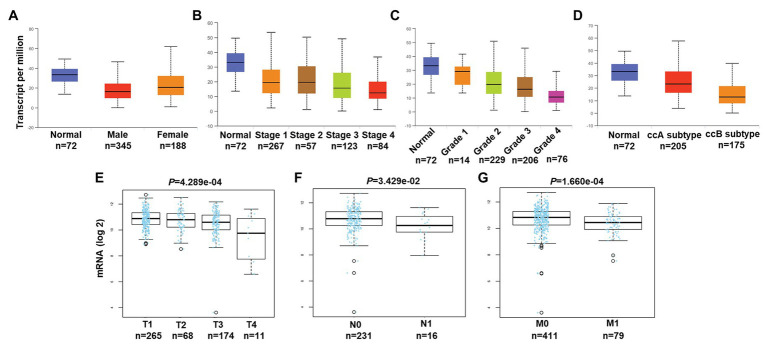
We then associated levels of PATJ mRNA with pathological features from ccRCC patients using the UALCAN web tool. The results showed that the decrease in PATJ mRNA is associated with males, advanced tumor stages, grades, and ccB subtypes (Figures 2A–D) and the values of p are shown in Table 2.
Figure 2.
Association of PATJ mRNA level with pathological features from ccRCC patients. (A) Association of PATJ mRNA with patients’ gender. (B) PATJ mRNA with ccRCC stages. (C) PATJ mRNA with ccRCC grades. (D) PATJ mRNA with ccRCC subtypes. The box plots and p (Table 2) in A–D were generated by using the UALCAN (http://ualcan.path.uab.edu/index.html). (E) PATJ mRNA with ccRCC T stage. (F) PATJ mRNA with ccRCC N stage. (G) PATJ mRNA with ccRCC M stage. The box plots and values p (Table 2) in E–G were obtained from the LinkedOmics (http://www.linkedomics.org/login.php).
Table 2.
Association of PATJ mRNA and ccRCC pathological features.
| Comparison groups | p |
|---|---|
| Normal-vs.-Male | <1E-12 |
| Normal-vs.-Female | 2.169E-07 |
| Male-vs.-Female | 2.101E-07 |
| Normal-vs.-Stage 1 | 4.908E-12 |
| Normal-vs.-Stage 2 | 1.921E-05 |
| Normal-vs.-Stage 3 | 1.776E-15 |
| Normal-vs.-Stage 4 | 1.625E-12 |
| Stage 1-vs.-Stage 3 | 5.997E-03 |
| Stage 1-vs.-Stage 4 | 1.336E-06 |
| Stage 2-vs.-Stage 4 | 2.839E-03 |
| Normal-vs.-Grade 2 | 8.160E-11 |
| Normal-vs.-Grade 3 | 1.625E-12 |
| Normal-vs.-Grade 4 | 1.624E-12 |
| Grade 1-vs.-Grade 2 | 3.920E-02 |
| Grade 1-vs.-Grade 3 | 3.003E-03 |
| Grade 1-vs.-Grade 4 | 1.799E-04 |
| Grade 2-vs.-Grade 3 | 1.664E-02 |
| Grade 2-vs.-Grade 4 | 1.657E-12 |
| Grade 3-vs.-Grade 4 | 2.960E-08 |
| Normal-vs.-ccA subtype | 5.605E-05 |
| Normal-vs.-ccB subtype | 1.624E-12 |
| ccA subtype-vs.-ccB subtype | 1.624E-12 |
The value of p was generated by using the UALCAN (http://ualcan.path.uab.edu/index.html).
Moreover, we also analyzed the association between level of PATJ mRNA and pathological features from ccRCC patients using the LinkedOmics web tool. Our results further indicated that PATJ mRNA was correlated with advanced T stage (p = 4.289e-04; n = 533; Figure 2E), N stage (p = 3.429e-04; n = 256; Figure 2F), and M stage (p = 1.660e-04; n = 501; Figure 2G). These results indicate that decreases in PATJ mRNA were related to ccRCC progression.
PATJ mRNA as an Independent Favorable Prognostic Factor for ccRCC
We next associated PATJ mRNA levels with survival of ccRCC patients using Kaplan-Meier curves and the log-rank test in the Kaplan-Meier Plotter 1 . We found that ccRCC patients with low PATJ mRNA expression had a significantly shorter OS than patients with high PATJ mRNA expression [p = 2.2e-09; HR = 0.4 (0.29–0.55); Figure 3A]. We analyzed PATJ mRNA expression vs. disease-free survival (DFS) of ccRCC patients using the GEPIA 2 . Our data showed that patients with low PATJ mRNA expression had a significantly poorer DFS than patients with high expression (p = 3.1e-06; HR = 0.26; Figure 3B). In addition, the SurExpress database data showed that levels of PATJ was low in high-risk ccRCC patients using COX regression analysis (n = 468; p = 1.1e-05; CI = 64.4; HR = 2.1; Figure 3C), while a bioinformatic analysis of PATJ expression in 46 patients with kidney transplants using the GSE22229 data set showed a significant decrease in PATJ mRNA in standard immunotherapeutic tissues compared with tolerant tissues (p = 0.020; Figure 3D), indicating that patients with high PATJ expression were more tolerant to kidney transplantation.
Figure 3.
Association of PATJ mRNA levels with ccRCC prognosis. (A) Kaplan-Meier curves of 530 ccRCC patients stratified by PATJ mRNA level analyzed by using Kaplan-Meier Plotter (http://kmplot.com/analysis/). (B) Kaplan-Meier curves of 258 ccRCC patients stratified by PATJ mRNA level analyzed by using the GEPIA (http://gepia.cancer-pku.cn/). (C) Kaplan-Meier curves of 468 ccRCC patients stratified by PATJ mRNA level analyzed by using the SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). (D) PATJ expression data from GSE22229 were stratified by patients with kidney transplantation.
Expression and Association of PATJ Protein in ccRCC vs. Adjacent Normal Renal Tissues With Patients’ Pathological Data
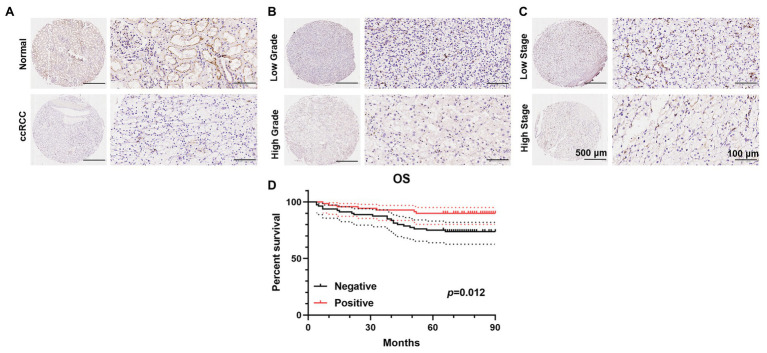
To further validate the decrease in PATJ expression in ccRCC and its association with patients’ pathological data, we performed immunohistochemistry of tissue microarrays containing 150 cases of ccRCC and 30 adjacent normal renal tissues. We found that PATJ protein was predominantly expressed in the membrane of renal tubular epithelial cells, but was reduced in ccRCC tissues (Figure 4A), i.e., 53.3% of ccRCC tissues lost PATJ expression (p = 0.027, Fisher’s exact test; Table 3). The loss of PATJ protein was significantly associated with advanced histologic grades and TNM stages (Figures 4B,C and Table 1).
Figure 4.
Immunohistochemical detection of PATJ protein in 150 ccRCC vs. normal tissues. The TMAs containing 150 ccRCC and 30 normal renal tissue specimens were immunostained with an anti-PATJ antibody and quantified. Representative fields of view (FOV) show the TMA cores of PATJ expression. (A) PATJ expression in normal renal specimens vs. ccRCC. (B) PATJ expressions in low grade vs. high grade ccRCC. (C) PATJ expressions in low stage vs. high stage ccRCC. (D) Patients with negative PATJ expression showed a significantly shorter OS than patients with positive PATJ expression (Log-rank test).
Table 3.
Association of PATJ protein and ccRCC pathological features.
| Comparison groups | p |
|---|---|
| Normal-vs.-Primary | 1.765E-53 |
| Normal-vs.-Male | 1.253E-48 |
| Normal-vs.-Female | 3.659E-17 |
| Normal-vs.-Stage 1 | 2.116E-34 |
| Normal-vs.-Stage 2 | 1.003E-06 |
| Normal-vs.-Stage 3 | 4.255E-23 |
| Normal-vs.-Stage 4 | 1.258E-09 |
| Stage 1-vs.-Stage 4 | 2.071E-02 |
| Stage 2-vs.-Stage 4 | 3.490E-02 |
| Normal-vs.-Grade 1 | 7.409E-05 |
| Normal-vs.-Grade 2 | 2.121E-34 |
| Normal-vs.-Grade 3 | 5.408E-26 |
| Normal-vs.-Grade 4 | 4.228E-07 |
The value of p were generated by using the UALCAN (http://ualcan.path.uab.edu/index.html).
Of these 150 patients, 70 patients showed positive PATJ expressed tumor, whereas 80 patients showed a negative PATJ expressed tumor. Patients with a negative PATJ expressed tumor had a significantly shorter OS than patients with a positive expressed tumor (p = 0.012 analyzed by using the log-rank test; Figure 4D). The result indicated that PATJ may be a prognostic indicator for ccRCC patients.
Expression and Association of PATJ Protein in ccRCC vs. Adjacent Normal Renal Tissues From CPTAC Dataset
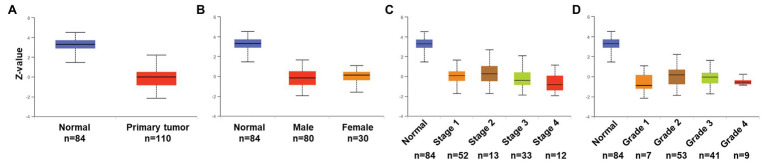
To validate expression and association of PATJ protein in ccRCC with patients’ pathological data, we utilized the UALCAN web tool and found that PATJ protein was reduced in ccRCC tissues vs. normal renal tissues (Figure 5A) and decrease in PATJ protein was associated with advanced tumor stages (Figure 5C; Table 4).
Figure 5.
Association of PATJ protein level with pathological features from ccRCC patients. (A) Downregulation of PATJ in ccRCC tissues. (B) Association of PATJ with patients’ gender. (C) PATJ with ccRCC stages. (D) PATJ with ccRCC grades. The box plots and values of p (Table 3) in A–D were generated by using the UALCAN (http://ualcan.path.uab.edu/index.html).
Table 4.
Differential expression of PATJ protein between ccRCC and adjacent normal kidney tissues.
| PATJ level | ||||
|---|---|---|---|---|
| Variables | n | − (n = 89) | + (n = 91) | p |
| Normal | 30 | 9 (30%) | 21 (70%) | 0.027 |
| ccRCC | 150 | 80 (53.3%) | 70 (46.7%) | |
Promotion of HKC Cell Proliferation After Knockdown of PATJ Expression in vitro
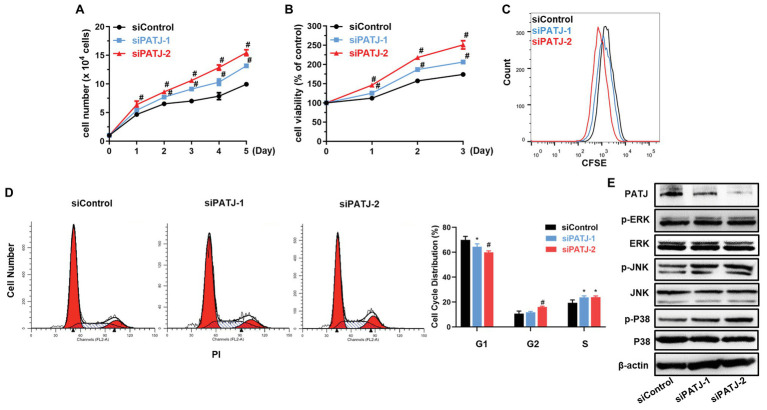
Next, we further investigated the effect of PATJ knockdown on the regulation of human renal tubular epithelial cell viability. As shown in Figure 6A, growth of PATJ-downregulated KHC cells was induced compared with that of control cells, which were consistent to cell viability (Figure 6B) and the CFSE and flow cytometry (Figure 6C). PATJ-knockdown HKC cells appeared to mainly accumulate in the S phase of the cell cycle, whereas the number of cells in the G1 phase was decreased significantly (Figure 6D).
Figure 6.
Promotion of human renal tubular epithelial cell HKC proliferation after knockdown of PATJ protein. (A) Growth curves of siPATJ transfected HKCs. (B) Cell viability using the CCK-8 assay. The percentage of cell viability was calculated from the OD values of the test groups normalized to the control group. (C) The CFSE and FACS assay. (D) Flow cytometric analysis of cell cycle. (E) Western blot. * p < 0.05; and # p < 0.01.
After that, we assessed levels of proteins that may contribute to the increase in HKC viability, such as the mitogen-activated protein kinases (MAPK) pathway [the extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and P38]. We found that levels of phospho-ERK, phospho-JNK, and phospho-P38 were all increased after knockdown of PATJ expression, whereas levels of their total proteins remained unchanged (Figure 6E), indicating that PATJ downregulation potentially promotes HKC cell viability by activation of the MAPK pathway.
Discussion
Dysregulation of cell polarity and cell polarity protein expression has been well-documented in the literature as a key step in cancer development and progression (Apodaca, 2018). The mammalian CRB polarity complex containing the integral membrane protein CRB3, PATJ, and PALS1 plays a key role in the establishment and maintenance of epithelial apicobasal polarity and tissue morphogenesis (Michgehl et al., 2017). Our previous study revealed that CRB3 was a favorable independent prognostic factor in ccRCC (Mao et al., 2015). In this study, we assessed the expression of PATJ, a CRB component, for association with ccRCC clinicopathological features and prognosis. We found that levels of both PATJ mRNA and protein were downregulated in ccRCC tissues and cell lines compared with healthy controls and cell lines. Reduced PATJ mRNA was associated with male patients, advanced tumor stages, grades, and ccB subtypes as well as poorer overall and DFS of patients. Our own experimental data showed that PATJ protein was also significantly downregulated in ccRCC vs. normal tissues, which was associated with advanced tumor pathologic stage and TNM stage, and patients with a negative PATJ expressed tumor had a significantly shorter OS. Moreover, knockdown of PATJ expression induced HKC cell proliferation and activation of MAPK pathway proteins. In conclusion, this study demonstrated that PATJ expression was downregulated in ccRCC and reduced PATJ level was associated with male patients and advanced tumor and poorer survival. Future studies will assess PATJ levels as a ccRCC prognostic biomarker and therapeutic target, such as, increase in PATJ expression in ccRCC cell lines by PATJ cDNA.
Indeed, the advantage of the UALCAN web, as an integrated data-mining platform, contains data of the potential cancer transcriptome using the TCGA level 3 RNA-seq and patients’ clinicopathological features on 31 cancer types. This website has been now visited over 300,000 times by cancer researchers from over 100 countries and cited over 500 times (Chandrashekar et al., 2017). However, the UALCAN web tool does not provide the original data for users to further analyze; thus, it is not possible for us to correct any of the p-values for the false-discovery rate (FDR).
PATJ protein was originally identified localized in the tight junctions of epithelial cells (Bhat et al., 1999; Céline et al., 2002; Yoko et al., 2002) and was a novel regulatory element of the polycystin-2 (PC2) channel and involved in autosomal dominant polycystic kidney disease (ADPKD) (Duning et al., 2010). A previous study reported a PATJ somatic mutation in two of the three PD-L1-positive ccRCC cases (Wang et al., 2018); thus, suggesting that PATJ might be a promising predictive factor for PD-L1 expression in ccRCC cells. However, to date, there is no study reporting PATJ expression in ccRCC, to the best of our knowledge. In the current study, we observed high PATJ expression in kidney epithelial cells but PATJ expression was reduced in ccRCC tissues and cell lines. Previous studies showed that epithelial-mesenchymal transition (EMT) markers SANIL and ZEB1 could transcriptionally inhibit PATJ expression in MDCK and MDA-MB-231 cells (Aigner et al., 2007; Whiteman et al., 2008). PATJ SNP variants were associated with worse outcomes in ischemic stroke patients (Mola-Caminal et al., 2019). However, the cause of PATJ downregulation in ccRCC requires future investigation. Indeed, ccRCC frequently occurs in aged male patients (Cohen and McGovern, 2005), indicating that patients may have an aggressive ccRCC, further confirmed in our current study of PATJ expression. It could be obvious that PATJ, a protein at tight junction, does function to maintain and regulates the epithelial cell polarity and decrease in PATJ expression promotes ccRCC progression, which is supported by our current data. However, to date, it is still unknown how PATJ expression is dysregulated in human cancers, including ccRCC and whether restoration of PATJ expression could suppress tumor progression as a novel therapeutic target.
In our current study, we found that PATJ was predominantly expressed in the nuclei of ccRCC cells. A previous study reported by Cho et al. showed PATJ localization in the nuclei of the apically located subset of early embryonic retinal progenitor cells (Kim et al., 2015). Nuclear PATJ localization in subsets of mitotic cells, if proven, might indicate a different function of PATJ protein. As progenitor cells possess stem cell characteristics, it may be that nuclear PATJ localization in progenitor cells may underlie its function in maintaining stemness. Furthermore, our current study revealed that downregulated PATJ expression was associated with poorer overall and DFS of ccRCC patients. These data indicate that PATJ expression could block ccRCC progression and be associated with treatment responses, although future study is needed to confirm this speculation. Indeed, our current data revealed that knockdown of PATJ expression enhanced HKC proliferation and activity of the MAPK pathway proteins in vitro, which indirectly supports the role of PATJ in suppression of ccRCC progression. Furthermore, the MAPK pathway contains a cascade of protein kinases including ERK, c-Jun N-terminal kinase (JNK), and p38. The latter is a highly conserved signal transduction in eukaryotic cells and one of the best-characterized signaling cascades to regulate cell proliferation (Achkar et al., 2018; Liu et al., 2018; Liang and Yang, 2019). A number of studies have demonstrated a role for the increased activity of MAPK signaling cascades in RCC cells and activation of the MAPK pathway was generally recognized for RCC growth (Wu et al., 2018; Zhao et al., 2018). Our current data further support this notion.
In conclusion, this study demonstrated a reduction of PATJ mRNA and protein in ccRCC tissues and cell lines, downregulation of which was associated with male patients and advanced tumors as well as poorer overall and DFS of patients. Our in vitro data further support the role of PATJ in ccRCC progression. Our current finding suggests that detection of PATJ expression may be used as an independent favorable prognostic factor for ccRCC patients and that PATJ should be further evaluated as a novel target for future control of ccRCC.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of human subject research of the First Affiliated Hospital, Xi’an Jiaotong University (Xi’an, China). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
PLi analyzed and interpreted the patient data regarding the database. PLa, SL, and YW analyzed the data and given some scientific advices. PLi and PLiu wrote the manuscript, and PLiu supervised all experiments. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was financially supported by grants from the National Natural Science Foundation of China (#81702631 and #81672810), Natural Science Basic Research Project of Shaanxi Province (2020JM-362) and the Institutional foundation of The First Affiliated Hospital of Xi’an Jiaotong University (2019ZYTS-19).
References
- Achkar I. W., Abdulrahman N., Al-Sulaiti H., Joseph J. M., Uddin S., Mraiche F. (2018). Cisplatin based therapy: the role of the mitogen activated protein kinase signaling pathway. J. Transl. Med. 16:96. 10.1186/s12967-018-1471-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Gamboa R., Gomez-Rueda H., Martinez-Ledesma E., Martinez-Torteya A., Chacolla-Huaringa R., Rodriguez-Barrientos A., et al. (2013). SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One 8:e74250. 10.1371/journal.pone.0074250, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner K., Dampier B., Descovich L., Mikula M., Sultan A., Schreiber M., et al. (2007). The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 26, 6979–6988. 10.1038/sj.onc.1210508, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. M., Van Itallie C. M. (2009). Physiology and function of the tight junction. Cold Spring Harb. Perspect. Biol. 1:a002584. 10.1101/cshperspect.a002584, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodaca G. (2018). Role of polarity proteins in the generation and organization of apical surface protrusions. Cold Spring Harb. Perspect. Biol. 10:a027813. 10.1101/cshperspect.a027813, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barata P. C., Rini B. I. (2017). Treatment of renal cell carcinoma: current status and future directions. CA Cancer J. Clin. 67, 507–524. 10.3322/caac.21411, PMID: [DOI] [PubMed] [Google Scholar]
- Bhat M. A., Izaddoost S., Lu Y., Cho K. O., Choi K. W., Bellen H. J. J. C. (1999). Discs lost, a novel multi-PDZ domain protein, establishes and maintains epithelial polarity. Cell 96, 833–845. 10.1016/S0092-8674(00)80593-0, PMID: [DOI] [PubMed] [Google Scholar]
- Campbell M. T., Jonasch E., Wood C. G., Tannir N. M. (2016). Renal Cell Carinoma. Vol. 733-752 New York: McGraw Hill Education. [Google Scholar]
- Céline L., Emmanuelle M., Marie-Hélène D., Didier M., Jean-Pierre A., André L. (2002). hINADl/PATJ, a homolog of discs lost, interacts with crumbs and localizes to tight junctions in human epithelial cells. J. Biol. Chem. 277, 25408–25415. 10.1074/jbc.M202196200 [DOI] [PubMed] [Google Scholar]
- Chandrashekar D. S., Bashel B., Sah B., Creighton C. J., Poncerodriguez I., Bvsk C., et al. (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658. 10.1016/j.neo.2017.05.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H. T., McGovern F. J. (2005). Renal-cell carcinoma. N. Engl. J. Med. 353, 2477–2490. 10.1056/NEJMra043172, PMID: [DOI] [PubMed] [Google Scholar]
- Duning K., Rosenbusch D., Schluter M. A., Tian Y., Kunzelmann K., Meyer N., et al. (2010). Polycystin-2 activity is controlled by transcriptional coactivator with PDZ binding motif and PALS1-associated tight junction protein. J. Biol. Chem. 285, 33584–33588. 10.1074/jbc.C110.146381, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernkvist M., Luna Persson N., Audebert S., Lecine P., Sinha I., Liu M., et al. (2009). The Amot/Patj/Syx signaling complex spatially controls RhoA GTPase activity in migrating endothelial cells. Blood 113, 244–253. 10.1182/blood-2008-04-153874, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K., Miller J. D., Li J. Z., Russell M. W., Charbonneau C. (2008). Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat. Rev. 34, 193–205. 10.1016/j.ctrv.2007.12.001, PMID: [DOI] [PubMed] [Google Scholar]
- Halaoui R., McCaffrey L. (2015). Rewiring cell polarity signaling in cancer. Oncogene 34, 939–950. 10.1038/onc.2014.59, PMID: [DOI] [PubMed] [Google Scholar]
- Hsieh J. J., Purdue M. P., Signoretti S., Swanton C., Albiges L., Schmidinger M., et al. (2017). Renal cell carcinoma. Nat. Rev. Dis. Primers 3:17009. 10.1038/nrdp.2017.9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten S. C., Deckers I. A., Aarts M. J., Hoeben A., van Roermund J. G., Smits K. M., et al. (2017). Prognostic DNA methylation markers for renal cell carcinoma: a systematic review. Epigenomics 9, 1243–1257. 10.2217/epi-2017-0040, PMID: [DOI] [PubMed] [Google Scholar]
- Kim B. (2017). “Western Blot Techniques” in Molecular profiling: Methods and protocols. ed. Espina V. (New York, NY: Springer; ), 133–139. [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Song J. Y., Karnam S., Park J. Y., Lee J. J., Kim S., et al. (2015). Common and distinctive localization patterns of crumbs polarity complex proteins in the mammalian eye. Gene Expr. Patterns 17, 31–37. 10.1016/j.gep.2015.01.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J. S., Leppert J. T., Belldegrun A. S., Figlin R. A. (2005a). Novel approaches in the therapy of metastatic renal cell carcinoma. World J. Urol. 23, 202–212. 10.1007/s00345-004-0466-0, PMID: [DOI] [PubMed] [Google Scholar]
- Lam J. S., Shvarts O., Leppert J. T., Pantuck A. J., Figlin R. A., Belldegrun A. S. (2005b). Postoperative surveillance protocol for patients with localized and locally advanced renal cell carcinoma based on a validated prognostic nomogram and risk group stratification system. J. Urol. 174, 466–472. 10.1097/01.ju.0000165572.38887.da, PMID: [DOI] [PubMed] [Google Scholar]
- Lánczky A., Nagy Á., Bottai G., Munkácsy G., Szabó A., Santarpia L., et al. (2016). miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 160, 439–446. 10.1007/s10549-016-4013-7, PMID: [DOI] [PubMed] [Google Scholar]
- Li P., Wang Y., Mao X., Jiang Y., Liu J., Li J., et al. (2017). CRB3 downregulation confers breast cancer stem cell traits through TAZ/β-catenin. Oncogene 6:e322. 10.1038/oncsis.2017.24, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y. J., Yang W. X. (2019). Kinesins in MAPK cascade: how kinesin motors are involved in the MAPK pathway? Gene 684, 1–9. 10.1016/j.gene.2018.10.042, PMID: [DOI] [PubMed] [Google Scholar]
- Liu F., Yang X., Geng M., Huang M. (2018). Targeting ERK, an Achilles’ Heel of the MAPK pathway, in cancer therapy. Acta. Pharm. Sin. B 8, 552–562. 10.1016/j.apsb.2018.01.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Li P., Ren Y., Li J., Zhou C., Yang J., et al. (2015). Cell polarity protein CRB3 is an independent favorable prognostic factor for clear cell renal cell carcinoma. Int. J. Oncol. 46, 657–666. 10.3892/ijo.2014.2763, PMID: [DOI] [PubMed] [Google Scholar]
- Michgehl U., Pavenstadt H., Vollenbroker B. (2017). Cross talk between the crumbs complex and hippo signaling in renal epithelial cells. Pflugers Arch. 469, 917–926. 10.1007/s00424-017-2004-0, PMID: [DOI] [PubMed] [Google Scholar]
- Mola-Caminal M., Carrera C., Soriano-Tarraga C., Giralt-Steinhauer E., Diaz-Navarro R. M., Tur S., et al. (2019). PATJ low frequency variants are associated with worse ischemic stroke functional outcome. Circ. Res. 124, 114–120. 10.1161/CIRCRESAHA.118.313533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posadas E. M., Limvorasak S., Figlin R. A. (2017). Targeted therapies for renal cell carcinoma. Nat. Rev. Nephrol. 13, 496–511. 10.1038/nrneph.2017.82, PMID: [DOI] [PubMed] [Google Scholar]
- Rodriguez-Vida A., Hutson T. E., Bellmunt J., Strijbos M. H. (2017). New treatment options for metastatic renal cell carcinoma. ESMO Open 2:e000185. 10.1136/esmoopen-2017-000185, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K., Wang Q., Margolis B. (2007). PATJ regulates directional migration of mammalian epithelial cells. EMBO Rep. 8, 158–164. 10.1038/sj.embor.7400890, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2018). Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30. 10.3322/caac.21442, PMID: [DOI] [PubMed] [Google Scholar]
- Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102. 10.1093/nar/gkx247, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasaikar S. V., Straub P., Wang J., Zhang B. (2018). LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963. 10.1093/nar/gkx1090, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xi Z., Xi J., Zhang H., Li J., Xia Y., et al. (2018). Somatic mutations in renal cell carcinomas from Chinese patients revealed by whole exome sequencing. Cancer Cell Int. 18:159. 10.1186/s12935-018-0661-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman E. L., Liu C. J., Fearon E. R., Margolis B. (2008). The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene 27, 3875–3879. 10.1038/onc.2008.9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. W., Wu Y. G., Cheng C., Hong Z. D., Shi Z. M., Lin S. Q., et al. (2018). Interleukin-33 Predicts Poor Prognosis and Promotes Renal Cell Carcinoma Cell Growth Through its Receptor ST2 and the JNK Signaling Pathway. Cell. Physiol. Biochem. 47, 191–200. 10.1159/000489766 [DOI] [PubMed] [Google Scholar]
- Yoko H., Masahiko I., Hiroyuki S., Mikio F., Shoichiro T. (2002). Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J. Biol. Chem. 277, 455–461. 10.1074/jbc.M109005200, PMID: [DOI] [PubMed] [Google Scholar]
- Zarrabi K., Wu S. (2018). Current and emerging therapeutic targets for metastatic renal cell carcinoma. Curr. Oncol. Rep. 20:41. 10.1007/s11912-018-0684-z, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao Y., Tang H., Zeng X., Ye D., Liu J. (2018). Resveratrol inhibits proliferation, migration and invasion via Akt and ERK1/2 signaling pathways in renal cell carcinoma cells. Biomed. Pharmacother. 98, 36–44. 10.1016/j.biopha.2017.12.029, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.