Abstract
The development of more affordable high-throughput DNA sequencing technologies and powerful bioinformatics is making of shotgun metagenomics a common tool for effective characterization of microbiomes and robust functional genomics. A shotgun metagenomic approach was applied in the characterization of microbial communities associated with plasticized fabric materials exposed to a harsh tropical environment for 14 months. High-throughput sequencing of TruSeq paired-end libraries was conducted using a whole-genome shotgun (WGS) approach on an Illumina HiSeq2000 platform generating 100 bp reads. A multifaceted bioinformatics pipeline was developed and applied to conduct quality control and trimming of raw reads, microbial classification, assembly of multi-microbial genomes, binning of assembled contigs to individual genomes, and prediction of microbial genes and proteins. The bioinformatic analysis of the large 161 Gb sequence dataset generated 3,314,688 contigs and 120 microbial genomes. The raw metagenomic data and the detailed description of the bioinformatics pipeline applied in data analysis provide an important resource for the genomic characterization of microbial communities associated with biodegraded plastic fabric materials. The raw shotgun metagenomics sequence data of microbial communities on plastic fabric materials have been deposited in MG-RAST (https://www.mg-rast.org/) under accession numbers: mgm4794685.3–mgm4794690.3. The datasets and raw data presented here were associated with the main research work “Metagenomic characterization of microbial communities on plasticized fabric materials exposed to harsh tropical environments” (Radwan et al., 2020).
Keywords: Metagenomics, Microbiome, Bioinformatics, High-throughput sequencing, Biodegradation, Plastics
Specifications Table
| Subject | Environmental Science |
|---|---|
| Specific subject area | Environmental Microbiology and Metagenomics |
| Type of data | Tables, Figures |
| How data were acquired | Illumina HiSeq2000 instrument was used for high-throughput sequencing of six genomic DNA libaries. |
| Data format | Raw and Analyzed |
| Parameters for data collection | Qiagen DNeasy UltraClean Microbial extraction kit (Cat# 12224-250) was used for DNA extraction from the six fabric materials for library preparation and DNA sequencing. |
| Description of data collection | A high-throughput sequencing of TruSeq paired-end libraries was conducted using whole-genome shotgun (WGS) approach on an Illumina HiSeq2000 platform generating 100 bp reads. |
| Data source location | Fabric samples were exposed to the harsh tropical environment of Panama. |
| Data accessibility | Raw data of shotgun metagenomics of microbial communities on plastic fabric materials have been deposited in MG-RAST (https://www.mg-rast.org/mgmain.html?mgpage=project&project=mgp85570) and can be retrieved using accession numbers: mgm4794685.3–mgm4794690.3. |
| Related research article | O. Radwan, J. S. Lee, R. Stote, K. Kuehn, O. N. Ruiz. Metagenomic Characterization of Microbial Communities on Plasticized Fabric Materials Exposed to Harsh Tropical Environments. International Biodeterioration & Biodegradation 154, 2020, 105061. |
Value of the Data
-
•
Raw metagenomic data of microbial communities could be an asset dataset to provide genomic information related to the structure and composition of microbial communities associated with biodegraded plastic fabric materials.
-
•
Draft genomes identified from the dataset can be used to understand the underlying mechanisms by which microorganisms biodegrade plastics, and may help in development of biodegradation resistant materials and new plastic bioremediation approaches.
-
•
These metagenomic data are valuable genomic sources for comparative metagenomics and can be exploited as a reference for other research teams interested in better understanding pathways and mechanisms involved in biodeterioration of plastic materials.
-
•
Functional annotation of sequenced reads from the six different plastic fabric materials will help in elucidating the true composition and behavior of the complex microbiomes associated with environmentally exposed fabrics.
1. Data Description
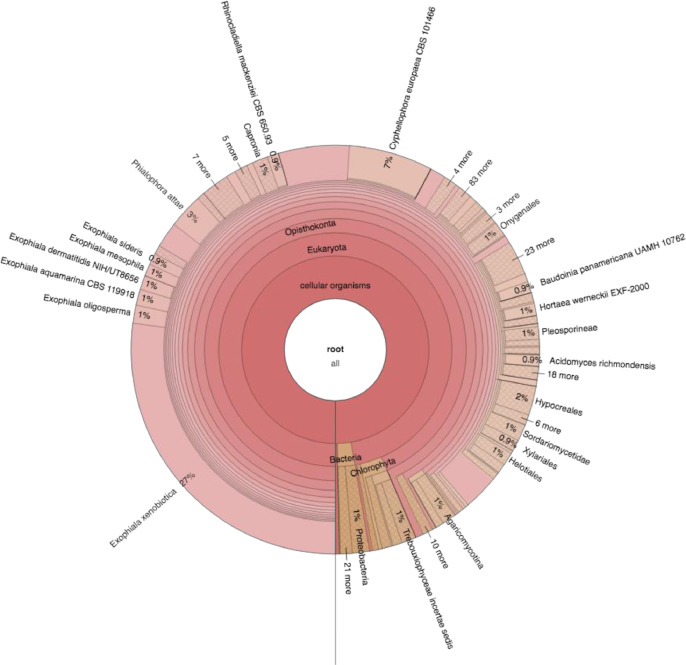
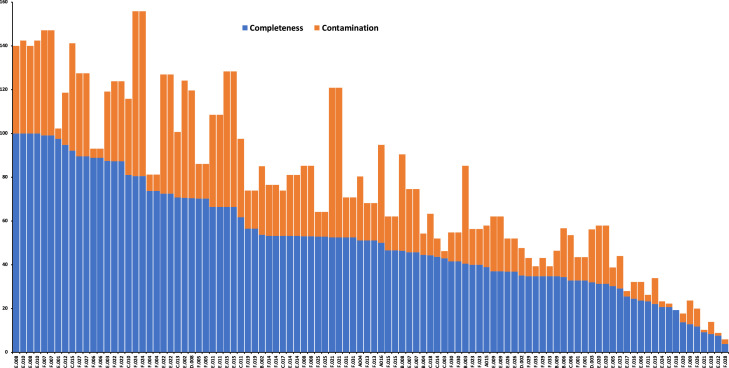
The datasets presented in this article are the raw sequences of pair-end reads with 100 bp length generated by Illumina HiSeq2000 platform. Shotgun metagenomics of six plastic fabric materials exposed to a harsh tropical environment produced 1.61 Gb of raw reads with a total of 161 Gb of 100 bp sequences [1]. The data files in FASTQ format were deposited in MG-RAST (https://www.mg-rast.org/) and can be retrieved using accession numbers: mgm4794685.3–mgm4794690.3. In this article, Fig. 1 provides a summary of the in-house pipeline that was established for bioinformatics analysis of metagenomic data. Table 1, contains a summary of raw reads, trimmed reads and total sequences (bp) from each sample. Table 1 also presents the number of sequences after trimming and the percent of surviving reads compare with the raw reads. Surviving reads from paired-end are reads after applying the trimming procedure. Table 2 summarizes the results of genomic assembled contigs generated by the MEGAHIT assembler program using trimmed sequences from the six fabrics. The sum (Mb), number of contigs > 500 bp, L50, N50 and the longest contig from each sample are presented in Table 2. N50 is the number of contigs whose length when summed up covers 50% or more of the genome assembly while L50 is the length of the smallest contig in the N50 set. Fig. 2 shows an overall summary of microbial distribution and taxon paths in one of the six plastic fabric materials. The data shown in Fig. 2 have been generated by KAIJU [2], a bioinformatic pipeline that is rapid and sensitive for taxonomic classification of short predicted proteins from metagenomic reads. Tables 3–8 summarize the results from MaxBin analysis that provided the different microbial genomes in each of the six plastic fabrics that were exposed to a harsh tropical environment for 14 months. Those microbial genomes are initially classified to different species of algae, black yeast, fungi and bacteria using KAIJU [2]. Tables 3–8 also show the genome size (Mb), GC content, classification and genome identification for each identified microbial genome. Fig. 3 shows the percent of completeness and contamination of each microbial genome presented in Tables 3–8 calculated using CheckM bioinformatic program [3]. A functional annotation summary of proteins predicted with MG-RAST from the sequenced reads is presented in Table 9. Table 9 shows twenty-eight functional categories with Carbohydrates; Amino Acids and Derivatives; Protein Metabolism; Cofactors, Vitamins, Prosthetic Groups, Pigments; and Respiration being the foremost categories in the sequences from the six platic fabric samples.
Table 4.
Summary of MaxBin results showing the different genomes belonging to algae, black yeast, fungi, and bacteria from fabric sample B.
| Genome code | Genome size (Mb) | GC content | Classification | Genome identification |
|---|---|---|---|---|
| A18 | 29.31 | 51.1 | Algae | Coccomyxa sp. |
| A20 | 17.33 | 50.4 | Algae | Coccomyxa sp. |
| A14 | 66.73 | 46.5 | Algae | Coccomyxa sp. |
| A15 | 28.91 | 55.1 | Algae | Coccomyxa sp. |
| A16 | 26.21 | 56.1 | Algae | Coccomyxa sp. |
| A05 | 0.13 | 41.1 | Algae | Trebouxia sp. |
| A02 | 0.17 | 38.4 | Algae | Watanabea sp. |
| A06 | 0.16 | 33.7 | Algae | Watanabea sp. |
| A07 | 0.16 | 40.4 | Algae | Watanabea sp. |
| A11 | 19.52 | 49.9 | Black yeast | Baudoinia sp. |
| A19 | 13.29 | 46.2 | Black yeast | Cladophialophora sp. |
| A09 | 10.95 | 48.1 | Black yeast | Cyphellophora sp. |
| A10 | 10.03 | 54 | Black yeast | Cyphellophora sp. |
| A08 | 0.16 | 42.4 | Black yeast | Exophiala sp. |
| A03 | 0.24 | 43.1 | Black yeast | Exophiala sp. |
| A01 | 0.13 | 32.5 | Black yeast | Exophiala sp. |
| A04 | 33.34 | 51.6 | Black yeast | Exophiala sp. |
| A17 | 23.88 | 52.6 | Black yeast | Hortaea sp. |
| A12 | 17.27 | 55.1 | Black yeast | Phialophora sp. |
| A13 | 17.95 | 65.1 | Fungi | Melampsora sp. |
Table 5.
Summary of MaxBin results showing the different genomes belonging to algae, black yeast, fungi, and bacteria from fabric sample C.
| Genome code | Genome size (Mb) | GC content | Classification | Genome identification |
|---|---|---|---|---|
| C03 | 2583.65 | 65.8 | Acidobacteria | Granulicella sp. |
| C04 | 2445.965 | 60.9 | Acidobacteria | Bryocella sp. |
| C13 | 4205.182 | 64.9 | Acidobacteria | Terriglobus sp. |
| C14 | 2733.709 | 60.7 | Acidobacteria | Terriglobus sp. |
| C18 | 3185.587 | 64.8 | Acidobacteria | Terriglobus sp. |
| C12 | 4845.024 | 71.5 | Actinobacteria | Jatrophihabitans sp. |
| C16 | 11235.87 | 46.8 | Algae | Coccomyxa sp. |
| C07 | 5678.483 | 38.3 | Algae | Watanabea sp. |
| C01 | 3120.5 | 28.9 | Black yeast | Zasmidium sp. |
| C02 | 29277.454 | 56.9 | Black yeast | Cyphellophora sp. |
| C05 | 386.73 | 52.2 | Black yeast | Verruconis sp. |
| C06 | 14141.935 | 49.7 | Black yeast | Cyphellophora sp. |
| C08 | 3408.081 | 60.2 | Black yeast | Verruconis sp. |
| C09 | 9494.505 | 45.7 | Black yeast | Neonectria sp. |
| C10 | 5638.762 | 72 | Proteobacteria | Gluconacetobacter sp. |
| C11 | 4430.702 | 68.5 | Proteobacteria | Methylobacterium sp. |
| C15 | 4963.268 | 72.6 | Proteobacteria | Caulobacteraceae sp. |
| C17 | 3677.465 | 68.7 | Proteobacteria | Methyloferula sp. |
| C19 | 1212.841 | 65.6 | Proteobacteria | Methylobacterium sp. |
| C20 | 934.412 | 64.4 | Proteobacteria | Methyloferula sp. |
Table 6.
Summary of MaxBin results showing the different genomes belonging to algae, black yeast, fungi, and bacteria from fabric sample D.
| Genome code | Genome size (Mb) | GC content | Classification | Genome identification |
|---|---|---|---|---|
| D02 | 5.96 | 61.3 | Acidobacteria | Terriglobus sp. |
| D10 | 1.43 | 65 | Acidobacteria | Terriglobus sp. |
| D06 | 0.69 | 68.9 | Actinobacteria | Jatrophihabitans sp. |
| D03 | 31.56 | 46.5 | Black yeast | Cyphellophora sp. |
| D04 | 5.73 | 56.1 | Black yeast | Verruconis sp. |
| D05 | 7.14 | 56.4 | Black yeast | Cyphellophora sp. |
| D01 | 2.56 | 60.7 | Fungi | Melampsora sp. |
| D08 | 19.60 | 40.1 | Fungi | Ceraceosorus sp. |
| D07 | 0.75 | 68.7 | Proteobacteria | Methylobacterium sp. |
| D09 | 0.97 | 67.5 | Proteobacteria | Methylobacterium sp. |
Table 7.
Summary of MaxBin results showing the different genomes belonging to algae, black yeast, fungi, and bacteria from fabric sample E.
| Genome code | Genome size (Mb) | GC content | Classification | Genome identification |
|---|---|---|---|---|
| E02 | 6.31 | 71 | Actinobacteria | Williamsia sp. |
| E03 | 5.69 | 74.7 | Actinobacteria | Jatrophihabitans sp. |
| E04 | 1.36 | 74.6 | Actinobacteria | Actinomycetospora sp. |
| E05 | 1.40 | 74.8 | Actinobacteria | Actinomycetospora sp. |
| E06 | 4.39 | 77.6 | Actinobacteria | Actinomycetospora sp. |
| E07 | 7.24 | 71.9 | Actinobacteria | Actinomycetospora sp. |
| E13 | 12.37 | 74.2 | Actinobacteria | Geodermatophilus sp. |
| E14 | 5.27 | 71.6 | Actinobacteria | Jatrophihabitans sp. |
| E15 | 5.96 | 71.9 | Actinobacteria | Actinomycetospora sp. |
| E19 | 3.62 | 67.3 | Actinobacteria | Jatrophihabitans sp. |
| E20 | 2.72 | 69.8 | Actinobacteria | Actinomycetospora sp. |
| E21 | 2.67 | 69 | Actinobacteria | Geodermatophilus sp. |
| E17 | 8.85 | 65.3 | Alphaproteobacteria | Methylobacterium sp. |
| E01 | 7.99 | 48.9 | Bacteroidetes | Spirosoma sp. |
| E11 | 11.70 | 69.1 | Bacteroidetes | Parafilimonas sp. |
| E12 | 2.50 | 29.1 | Black yeast | Zasmidium sp. |
| E16 | 0.95 | 44.9 | Black yeast | Pyrenochaeta sp. |
| E18 | 1.53 | 47.4 | Black yeast | Cyphellophora sp. |
| E23 | 8.95 | 57.2 | Black yeast | Hortaea sp. |
| E26 | 12.28 | 44.8 | Black yeast | Exophiala sp. |
| E27 | 1.67 | 52.8 | Black yeast | Baudoinia sp. |
| E28 | 1.34 | 51.9 | Black yeast | Dothistroma sp. |
| E08 | 4.95 | 65.7 | Proteobacteria | Methylobacterium sp. |
| E09 | 4.84 | 73.8 | Proteobacteria | Methylobacterium sp. |
| E10 | 8.26 | 42.3 | Proteobacteria | Methylobacterium sp. |
| E22 | 5.79 | 68.8 | Proteobacteria | Aureimonas sp. |
| E24 | 9.86 | 60.5 | Proteobacteria | Methylobacterium sp. |
| E25 | 1.08 | 44.9 | Acidobacteria | Acidobacterium sp. |
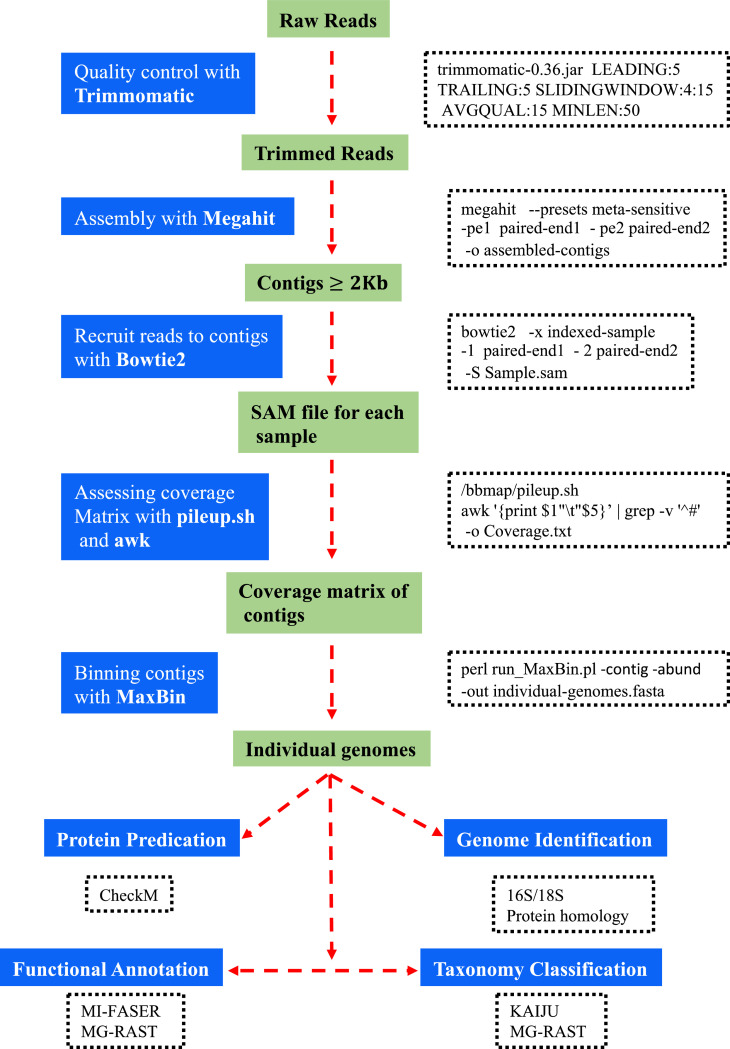
Fig. 1.
Schematic summary of the bioinformatics pipeline used to analyze shotgun metagenomic datasets.
Table 1.
Summary of raw reads, trimmed reads, and total sequence reads (bp) from each sample. Also, the percentage of surviving pair end reads after applying the trimming procedure is provided.
| Raw reads | Sequence (bp) | Trimmed reads | % Surviving reads | |
|---|---|---|---|---|
| Sample A | 290,535,652 | 29,191,462,169 | 273,136,734 | 94.01 |
| Sample B | 242,732,424 | 24,394,813,486 | 228,956,282 | 94.32 |
| Sample C | 280,501,300 | 28,198,140,693 | 257,866,956 | 91.93 |
| Sample D | 283,123,548 | 28,452,082,418 | 261,796,312 | 92.47 |
| Sample E | 245,775,804 | 24,706,114,436 | 223,055,718 | 90.76 |
| Sample F | 264,529,024 | 26,594,662,007 | 247,595,802 | 93.60 |
| Total | 1,607,197,752 | 161,537,275,209 | 1,492,407,804 |
Table 2.
Summary of sequence assembly from the different samples using Megahit assembly program.
| Sample | Sum (Mb) | # Contigs > 500 bp | L50 | N50 | Max (bp)* |
|---|---|---|---|---|---|
| Sample F⁎⁎ | 1128 | 787,368 | 106,600 | 1902 | 184,053 |
| Sample D | 1120 | 611,503 | 57,474 | 3290 | 584,409 |
| Sample A | 1052 | 589,392 | 59,676 | 3150 | 977,595 |
| Sample C | 1006 | 718,706 | 105,794 | 1774 | 126,842 |
| Sample B | 925.5 | 492,053 | 52,173 | 3251 | 1,435,925 |
| Sample E | 802 | 615,666 | 11,915 | 1544 | 740,880 |
| Total | 6033.5 | 3,814,688 |
N50 is the number of contigs whose length when summed up covers 50% or more of the genome assembly.
L50 is the length of the smallest contig in the N50 set.
The longest contig (bp) in each sample.
Samples are ordered descending based on to their sum (Mb).
Fig. 2.
Diagram of the microbial composition and classification of the microbiome associated with fabric sample A developed by the KIAJU database.
Table 3.
Summary of MaxBin results showing the different genomes belonging to algae, black yeast, fungi, and bacteria from fabric sample A.
| Genome code | Genome size (Mb) | GC content | Classification | Genome identification |
|---|---|---|---|---|
| B05 | 4.865 | 35.7 | Algae | Watanabea sp. |
| B09 | 3.511 | 51.2 | Algae | Coccomyxa sp. |
| B01 | 0.63 | 29 | Black yeast | Exophiala sp. |
| B02 | 2.69 | 43.9 | Black yeast | Rhinocladiella sp. |
| B03 | 3.212 | 49.7 | Black yeast | Baudoinia sp. |
| B04 | 2.212 | 54.4 | Black yeast | Cyphellophora sp. |
| B06 | 2.83 | 45.8 | Black yeast | Exophiala sp. |
| B07 | 1.89 | 57.4 | Black yeast | Cyphellophora sp. |
| B08 | 4.20 | 53.8 | Black yeast | Phialophora sp. |
Table 8.
Summary of MaxBin results showing the different genomes belonging to algae, black yeast, fungi, and bacteria from fabric sample F.
| Genome code | Genome size (Mb) | GC content | Classification | Genome identification |
|---|---|---|---|---|
| F04 | 6.98 | 66.9 | Actinobacteria | Gordonia sp. |
| F05 | 4.59 | 71.3 | Actinobacteria | Williamsia sp. |
| F08 | 3.44 | 74.1 | Actinobacteria | Actinomycetospora sp. |
| F15 | 4.86 | 74.9 | Actinobacteria | Actinomycetospora sp. |
| F18 | 2.31 | 71.1 | Actinobacteria | Actinomycetospora sp. |
| F23 | 4.65 | 69.5 | Actinobacteria | Nakamurella sp. |
| F25 | 2.81 | 71.1 | Actinobacteria | Micrococcales sp. |
| F31 | 1.91 | 70.6 | Actinobacteria | Jatrophihabitans sp. |
| F01 | 39.98 | 51.3 | Alage | Coccomyxa sp. |
| F02 | 5.33 | 37.1 | Alage | Watanabea sp. |
| F06 | 3.89 | 35.6 | Bacteroidetes/ | Parafilimonas sp. |
| F07 | 6.04 | 51.5 | Bacteroidetes | Mucilaginibacter sp. |
| F27 | 6.53 | 44.7 | Bacteroidetes | Parafilimonas sp. |
| F03 | 2.20 | 27.3 | Black yeast | Cladophialophora sp. |
| F10 | 1.07 | 33 | Black yeast | Zasmidium sp. |
| F11 | 7.73 | 42.6 | Black yeast | Cyphellophora sp. |
| F17 | 0.31 | 39 | Black yeast | Coniochaeta sp. |
| F21 | 26.81 | 57.8 | Black yeast | Hortaea sp. |
| F22 | 22.34 | 55.3 | Black yeast | Cyphellophora sp. |
| F26 | 5.31 | 52.3 | Black yeast | Hortaea sp. |
| F32 | 0.65 | 44.9 | Black yeast | Hortaea sp. |
| F09 | 0.74 | 30.5 | Chlorophyta | Cephaleuros sp. |
| F28 | 0.58 | 26.6 | Chlorophyta | Cephaleuros sp. |
| F12 | 16.85 | 72.9 | Proteobacteria | Methylobacterium sp. |
| F13 | 4.23 | 69.7 | Proteobacteria | Methylobacterium sp. |
| F14 | 3.23 | 70.7 | Proteobacteria | Methylobacterium sp. |
| F16 | 1.21 | 66.5 | Proteobacteria | Methylobacterium sp. |
| F19 | 2.91 | 69.2 | Proteobacteria | Sphingomonas sp. |
| F20 | 0.81 | 67.1 | Proteobacteria | Methylobacterium sp. |
| F24 | 10.18 | 66.7 | Proteobacteria | Xylophilus sp. |
| F29 | 1.73 | 65 | Proteobacteria | Sphingomonas sp. |
| F30 | 5.67 | 62 | Proteobacteria | Methylobacterium sp. |
| F33 | 4.16 | 61.4 | Proteobacteria | Rhodospirillales sp. |
Fig. 3.
Percent of completeness and contamination of each microbial genome generated by CheckM bioinformatic program.
Table 9.
Functional annotation showing the percentage of sequence reads containing predicated proteins of known functions.
| Functional categories | A | B | C | D | E | F | Average |
|---|---|---|---|---|---|---|---|
| Carbohydrates | 15.18 | 16.22 | 15.87 | 16.03 | 15.55 | 15.78 | 15.77 |
| Amino acids and derivatives | 12.92 | 14.33 | 11.48 | 12.61 | 11.24 | 11.28 | 12.31 |
| Protein metabolism | 10.32 | 9.20 | 8.02 | 8.12 | 6.44 | 6.72 | 8.14 |
| Cofactors, vitamins, prosthetic groups, pigments | 7.52 | 7.29 | 6.78 | 6.88 | 6.83 | 6.72 | 7.00 |
| Respiration | 8.11 | 9.02 | 4.64 | 5.95 | 3.23 | 3.45 | 5.73 |
| Fatty acids, lipids, and isoprenoids | 4.40 | 4.80 | 4.16 | 4.56 | 4.83 | 4.56 | 4.55 |
| RNA metabolism | 4.98 | 4.59 | 4.74 | 4.65 | 3.90 | 4.17 | 4.50 |
| Nucleosides and nucleotides | 3.21 | 3.37 | 2.78 | 3.12 | 2.45 | 2.52 | 2.91 |
| Stress response | 2.85 | 2.99 | 2.85 | 2.85 | 2.69 | 2.61 | 2.81 |
| Metabolism of aromatic compounds | 2.76 | 3.29 | 1.98 | 2.29 | 2.20 | 2.14 | 2.44 |
| Cell wall and capsule | 1.37 | 1.12 | 2.82 | 2.31 | 3.11 | 3.25 | 2.33 |
| DNA metabolism | 1.06 | 0.75 | 2.69 | 2.13 | 3.35 | 3.28 | 2.21 |
| Virulence, disease and defense | 0.96 | 0.75 | 2.14 | 1.75 | 2.49 | 2.39 | 1.75 |
| Membrane transport | 0.71 | 0.42 | 1.61 | 1.27 | 2.13 | 2.17 | 1.39 |
| Sulfur metabolism | 1.14 | 1.10 | 1.09 | 1.08 | 1.22 | 1.17 | 1.13 |
| Regulation and cell signaling | 0.69 | 0.58 | 1.03 | 0.93 | 1.15 | 1.14 | 0.92 |
| Cell division and cell cycle | 0.74 | 0.65 | 0.91 | 0.84 | 0.97 | 0.93 | 0.84 |
| Phosphorus metabolism | 0.39 | 0.35 | 0.81 | 0.68 | 1.03 | 0.95 | 0.70 |
| Nitrogen metabolism | 0.58 | 0.56 | 0.65 | 0.65 | 0.91 | 0.77 | 0.69 |
| Photosynthesis | 1.87 | 1.10 | 0.46 | 0.15 | 0.17 | 0.24 | 0.67 |
| Phages, prophages, transposable elements, plasmids | 0.33 | 0.18 | 0.79 | 0.63 | 1.10 | 0.95 | 0.66 |
| Secondary metabolism | 0.81 | 1.08 | 0.51 | 0.68 | 0.37 | 0.37 | 0.64 |
| Motility and chemotaxis | 0.15 | 0.03 | 0.80 | 0.60 | 0.91 | 0.86 | 0.56 |
| Iron acquisition and metabolism | 0.16 | 0.14 | 0.42 | 0.38 | 0.73 | 0.72 | 0.43 |
| Potassium metabolism | 0.12 | 0.09 | 0.30 | 0.24 | 0.37 | 0.38 | 0.25 |
| Dormancy and sporulation | 0.02 | 0.01 | 0.10 | 0.07 | 0.13 | 0.13 | 0.08 |
| Clustering-based subsystems | 9.39 | 8.65 | 12.08 | 11.12 | 13.40 | 13.13 | 11.30 |
| Miscellaneous | 7.26 | 7.34 | 7.48 | 7.41 | 7.08 | 7.22 | 7.30 |
2. Experimental design, materials and methods
2.1. Samples and exposure environments
The U.S. Army Research, Development and Engineering Center (Natick, MA) provided six plastic fabric materials after 14 months of exposure to harsh tropical environment in the Republic of Panama [1]. The plastic fabric samples were used for DNA extraction, library preparation of genomic DNA, high-throughput sequencing, and bioinformatic analysis.
2.2. Library preparation and DNA sequencing for metagenomic study
DNA from the six plastic fabric materials was extracted with the Qiagen DNeasy UltraClean Microbial extraction kit (Cat# 12224-250), and then used for library preparation and DNA sequencing. A 300 ng of DNA from each fabric sample was used for the preparation of the genomic library using the PrepX DNA Library kit and Apollo 324 NGS automatic library prep system (WaferGen, Fremont, CA). A high-throughput sequencing of TruSeq paired-end libraries was conducted using a whole-genome shotgun (WGS) approach on an Illumina HiSeq2000 platform generating 100 bp reads. A TruSeq SBS kit v3 for 2 × 101 cycles of Incorporation Reagent (ICR) was used for read sequencing (Illumina, Inc. San Diego, CA).
2.3. Bioinformatics analysis for metagenomics study
An in-house multifaceted bioinformatics pipeline (Fig. 1) was established for the stepwise processing of sequence data required for completion of the metagenomic study. Quality control of raw reads was performed by Trimmomatic version 0.36 [4], which allowed trimming low quality reads and short reads from raw reads. Trimmed reads were sorted by BBtools (https://jgi.doe.gov/data-and-tools/bbtools/) “bbnorm.sh” to ensure the compatibility and normalization of paired-end before mapping to different contigs using MEGAHIT assembler program [5]. Bowtie2 [6] was employed for mapping raw reads to contigs produced by MEGAHIT, and the BAM file from each fabric sample was used for generating the coverage matrix and abundance files. Binning of individual genomes in each fabric sample was performed by MaxBin bioinformatic program [7] using the abundance file and fasta contigs generated by MEGAHIT.
2.4. Functional annotation of metagenomic reads and genome identification
The functional annotation of metagenomic reads of each fabric sample exposed to the tropical environment was extracted from the MG-RAST analysis (https://www.mgrast.org/mgmain.html?mgpage=project&project=mgp85570). Both RNAmmer [8] and CheckM [3] programs were used for ribosomal RNA identification of each binned genome generated by the MaxBin bioinformatic program. KAIJU [2], a fast and sensitive bioinformatic pipeline, was used for taxonomic classification of predicted proteins from metagenomic reads. Additionally, CheckM was used for assessing the completeness and presence contamination of microbial genomes generated by MaxBin.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This material is based on research sponsored by AFRL/RQTF under agreement number FA8650-16-2-2605. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of AFRL/RQTF or the U.S. Government.
References
- 1.Radwan O., Lee J.S., Stote R., Kuehn K., Ruiz O.N. Metagenomic characterization of microbial communities on plasticized fabric materials exposed to harsh tropical environments. Int. Biodeter. Biodegr. 2020;154 doi: 10.1016/j.ibiod.2020.105061. [DOI] [Google Scholar]
- 2.Menzel P., Ng K.L., Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016;7:11257. doi: 10.1038/ncomms11257. volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. Assessing the quality microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2014;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;15:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 6.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2013;4:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y-W., Tang Y-H., Tringe S.G., Simmons B.A., Singer S.W. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome. 2014;2:26. doi: 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H-H, Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]