Abstract
Background
Myositis, or idiopathic inflammatory myopathy (IIM), is a group disorders of unknown etiology characterized by the inflammation of skeletal muscle. The role of T cells and their antigenic targets in IIM initiation and progression is poorly understood. T cell receptor (TCR) repertoire sequencing is a powerful approach for characterizing complex T cell responses. However, current TCR sequencing methodologies are complex, expensive, or both, greatly limiting the scale of feasible studies.
Methods
Here we present Framework Region 3 AmplifiKation sequencing (“FR3AK-seq”), a simplified multiplex PCR-based approach for the ultra-efficient and quantitative analysis of TCR complementarity determining region 3 (CDR3) repertoires. By using minimal primer sets targeting a conserved region immediately upstream of CDR3, undistorted amplicons are analyzed via short read, single-end sequencing. We also introduce the novel algorithm Inferring Sequences via Efficiency Projection and Primer Incorporation (“ISEPPI”) for linking CDR3s to their associated variable genes.
Findings
We find that FR3AK-seq is sensitive and quantitative, performing comparably to two different industry standards. FR3AK-seq and ISEPPI were used to efficiently and inexpensively characterize the T cell infiltrates of surgical muscle biopsies obtained from 145 patients with IIM and controls. A cluster of closely related TCRs was identified in samples from patients with sporadic inclusion body myositis (IBM).
Interpretation
The ease and minimal cost of FR3AK-seq removes critical barriers to routine, large-scale TCR CDR3 repertoire analyses, thereby democratizing the quantitative assessment of human TCR repertoires in disease-relevant target tissues. Importantly, discovery of closely related TCRs in muscle from patients with IBM provides evidence for a shared antigen-driven T cell response in this disease of unknown pathogenesis.
Funding
This work was supported by NIH grant U24AI118633 and a Prostate Cancer Foundation Young Investigator Award.
Keywords: TCR repertoire, Myositis, Idiopathic Inflammatory myopathy, Autoimmunity
1. Introduction
Research in Context.
Evidence before this study
T cells are an essential component of the adaptive immune system and are characterized by antigen specific receptors formed by recombination of diverse genetic elements. Sequencing-based quantification of this receptor population provides unprecedented insight into human immune responses. Interrogation of T cell receptors (TCRs) in the muscle of patients with IIMs can provide essential insights into disease pathogenesis. However, the cost and complexity of existing assays have impeded their use in studies involving large numbers of samples.
Added value of this study
Added value of this study: Here we present an ultra-efficient alternative to TCR repertoire sequencing and demonstrate its utility by characterizing muscle-infiltrating T cells in a large cohort of patients with IIMs. We found a cluster of closely related TCR sequences in muscle from patients with sporadic inclusion body myositis, suggesting the presence of a previously unknown antigen-driven T cell response at the site of pathology.
Implications of all the available evidence
Implications of all the available evidence: Our novel approach democratizes T cell receptor repertoire sequencing-based studies, as demonstrated by the IIM cohort presented here. This will enable large-scale analyses of T cell responses in human health and disease, thereby accelerating the identification and interrogation of disease-relevant TCRs in autoimmunity, infectious disease, transplantation medicine, and cancer.
Alt-text: Unlabelled box
Myositis collectively refers to a heterogeneous group of disorders that includes autoimmune and inflammatory muscle disease, commonly known as idiopathic inflammatory myopathies (IIMs). While muscle inflammation and weakness are classic features of these disorders, IIMs are often systemic, commonly involving other organ systems including the skin, joints, and lungs. The IIMs include dermatomyositis (DM), polymyositis (PM), anti-synthetase syndrome (ASyS), immune-mediated necrotizing myopathy (IMNM), and inclusion body myositis (IBM) [1]. The diagnosis of IIM is often complicated by the diversity within and between its various subgroups, some of which have overlapping clinical and histopathological features [2]. Myositis-specific autoantibodies (MSAs) are detected in 60-70% of patients with IIMs and have proven useful for delineating distinct IIM subtypes [3,4].
Discrete pathological features are apparent in muscle biopsies from patients with IIMs. Perifascicular atrophy of myofibers and perivascular cellular infiltrates including CD4 T cells are characteristic of DM. ASyS muscle also shows perifascicular atrophy, but may have more necrotic fibers than DM. In contrast, IMNM muscle shows scattered necrotic myofibers with minimal lymphocytic infiltration, while IBM muscle usually demonstrates endomysial infiltration of CD8 T cells and rimmed vacuoles within myocytes [1]. Importantly, the role of muscle-infiltrating CD4 and CD8 T cells in IIM disease initiation and progression is largely unknown [5]. Examination of T cell receptor (TCR) repertoires within the muscle of patients with IIM presents a unique opportunity to elucidate the T cell-mediated immunopathogenesis of these diseases.
TCR repertoire analysis has emerged as a powerful tool for examining adaptive immune responses. TCR repertoires, generated by the process of V(D)J recombination, encompass the T cell clones within a given individual or sample. The unique TCR of each clone defines its antigen specificity, and TCRs can be associated with distinct cellular phenotypes and tissue occupancy. Notably, TCR repertoires encompass dynamic populations of cells that represent past and current immune exposures and reactivities [6]. Development of next generation sequencing (NGS)-based technologies over the last decade has enabled the unprecedented analysis of TCR repertoires [7], [8], [9], [10].
Examination of TCR repertoires both within and between individuals can be indicative of disease status, prior infections or immunizations, and individual-specific attributes of epitope selection [11], [12], [13], [14], [15], [16], [17]. Clonally expanded or tissue-infiltrating T cell clones can also reveal the nature of local immune responses, while integrating TCR repertoire analyses with phenotypic measurements (e.g. single cell transcriptional profiling [18] or flow cytometric sorting of T cell subpopulations [19,20]) can further illuminate the nature of T cell responses (Fig 1a). A variety of methods have been used to generate TCR libraries for NGS, including multiplex PCR, 5’-RACE, and target enrichment [[7], [8], [9], [10],21,22]. Generally, these technologies prioritize sequencing of the hypervariable complementarity determining region 3 (CDR3) of the TCR beta (TCRB) chain. The TCRB CDR3 harbors the greatest amount of sequence diversity, typically confers most of the antigen specificity to a TCR, and is often used as a surrogate for T cell clonal identity [23], [24], [25]. Despite their utility, however, current repertoire sequencing approaches remain complex, expensive, or both. These limitations have greatly hindered the utilization of TCR repertoire-based analyses, particularly for studies involving large numbers of samples. To overcome these current barriers to discovery, we developed a simple and quantitative multiplex PCR-based approach that prioritizes the ultra-efficient analysis of TCRB CDR3 sequences.
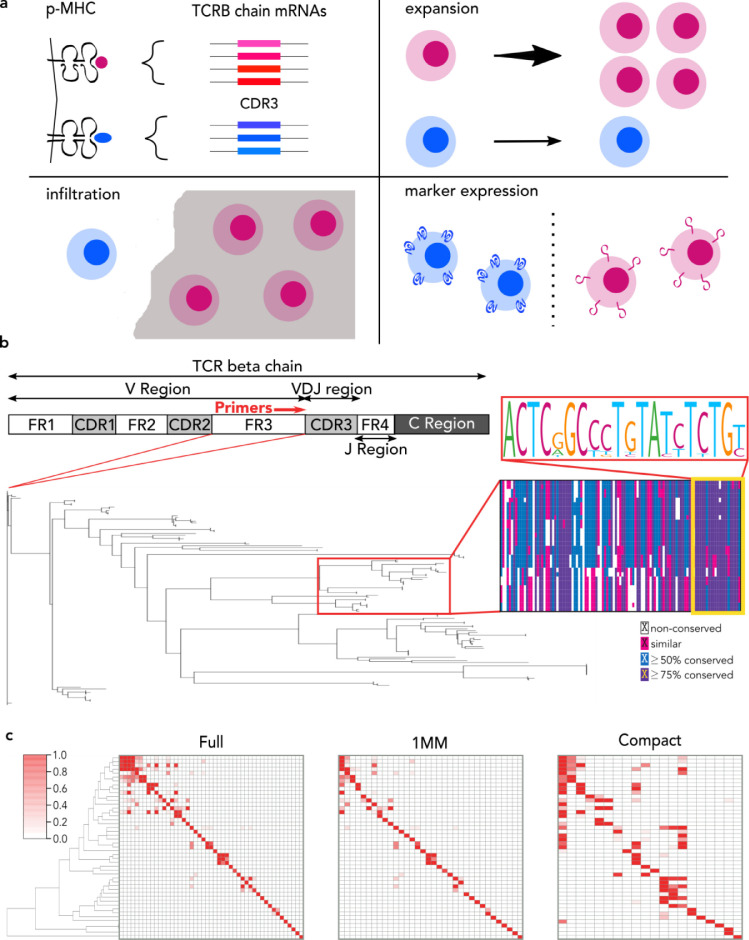
Fig. 1.
Utility of T cell receptor beta (TCRB) chain sequencing and FR3AK-seq multiplex PCR primer design. a. TCRB chain sequencing can be used to (i) identify related CDR3s that may share antigen specificity, (ii) detect expanded clones, (iii) identify tissue infiltrating clones, and (iv) link CDR3 sequences with T cell phenotypes. b. A schematic of the TCRB chain. A neighbor joining phylogenetic tree of FR3s extracted from the 128 functional human TCRBV alleles in the IMGT/GENE-DB reference directory and multiple sequence alignment from a dominant FR3 sequence cluster (red box on tree) identifies homology within the 3’ ~20 nucleotides (yellow box). c. Clustered heatmaps for the Full, 1MM, and Compact primer sets showing primer amplification efficiencies calculated using the R package DECIPHER [38] for each primer across each of the 47 unique 3’ 20 nucleotides of the human TCRBV FR3 region. Inosines were considered exact matches for the Compact set. Y axis: 47 unique TCRBV FR3s. X axis: primers. The order of the primers in each heatmap can be found in Table S1. The order of unique TCRBV FR3s is identical to the order of the Full primers for all heatmaps.
The sequence diversity of the TCRB chain variable domain, which in humans is encoded by >120 unique TCRB variable (TCRBV) alleles, necessitates the use of complex primer pools for multiplex PCR amplification. Primer competition and differential amplification efficiencies distort clonal amplicon abundance, confounding quantification using high-throughput DNA sequencing [26]. Sophisticated techniques have been developed to computationally correct amplification bias, such as the use of spike-in library standards or the incorporation of unique molecular identifiers (UMIs) [27], [28], [29]. Adaptive Biotechnologies has become an industry leader by offering a spike-in standard corrected multiplex PCR-based assay. At a current cost of $500-$1100 per sample, however, the scale of feasible studies using this assay has been greatly constrained. UMI-based approaches, including the ArcherDx Immunoverse assay, provide quantitation while resolving amplification and sequencing errors [28], [29], [30], [31], [32], [33]. However, UMI approaches typically require multi-day protocols, complicated analytical pipelines, and deeper sequencing to obtain sufficient sampling of distorted libraries [22,34].
By designing maximally compact primer sets and a streamlined workflow, we have essentially eliminated PCR amplification bias while maintaining high sequence level accuracy. Lower sequencing depth requirements, coupled with CDR3 analysis via single-end, short-read (100 nucleotide) sequencing, dramatically reduces the cost associated with TCR repertoire analysis. We benchmark this simplified multiplex PCR-based assay, Framework Region 3 AmplifiKation Sequencing (“FR3AK-seq”), against two different industry standards for quantitative TCR repertoire sequencing: Adaptive Biotechnologies’ multiplex PCR-based hsTCRB immunoSEQ assay and ArcherDX's UMI-based Immunoverse HS TCR assay. We found that FR3AK-seq data, which was analyzed using open-source software, was in excellent agreement with both. Finally, we illustrate the ability of FR3AK-seq to democratize TCR repertoire sequencing by efficiently characterizing T cell responses within muscle tissue from 145 patients with IIMs and controls. We identified a cluster of related TCR sequences from patients with sporadic IBM, suggesting the presence of an antigen-driven T cell response within the muscle of these donors.
2. Materials and Methods
2.1. FR3 primer design
FR3 sequences from all functional human TCRBV alleles were downloaded from the IMGT/LIGM-DB reference directory (non-functional and pseudogenes were excluded) [35]. Multiple sequence alignment (MSA) was performed using MUSCLE. UPGMA (Unweighted Pair Group Method with Arithmetic Mean) and neighbor-joining phylogenetic trees were generated via the R package msa [36]. The 3’ 20 nucleotides of the TCRB chain FR3s were subsequently used to design three sets of primers as described in the text. We automated design of the 1MM and Compact primer sets with an algorithm as outlined in Fig S1 and available on Github (https://github.com/jmmontagne/FR3AK-seq). Human TCRB primer sequences can be found in Table S1.
2.2. PBMC preparation, T cell purification, RNA extraction, and cDNA synthesis
PBMCs from Donors A and B were isolated by Ficoll-Paque (GE Healthcare) gradient centrifugation and cryopreserved. T cells were purified from thawed PBMCs using the EasySep Human T cell enrichment kit (STEMCELL Technologies). RNA was extracted from purified human T cells using the RNeasy Plus Minikit (Qiagen). 0.5 μg Donor B RNA was mixed with 4.5 μg Donor A RNA to create sample C for quantitation experiments. 4 μg each of Donor A, Donor B, and sample C RNA was reverse transcribed using a TCRB chain constant region reverse primer with Superscript III First-Strand Synthesis System (Invitrogen). cDNA was column purified with the Oligo Clean and Concentrator Kit (Zymo Research). 100 ng of purified cDNA from each sample was sent to Adaptive Biotechnologies for the immunoSEQ hsTCRb Deep sequencing service or used for FR3AK-seq PCR. For the ArcherDx Immunoverse analysis, RNA was extracted from matched Donor A and B PBMCs and mixed at the same 9:1 ratio to create sample C. 800 ng of RNA from each sample was input into the Immunoverse assay. The concordance between FR3AK-seq and Immunoverse remained high despite the use of PBMC RNA rather than T cell RNA in the Immunoverse assay. Primer sequences are provided in Table S1.
2.3. Polymerase Chain Reaction (PCR) and sequencing library preparation
100 ng of TCRB chain cDNA was used as template for PCR with KAPA2G Fast Multiplex Mix (Roche). Forward TCRB FR3 primers from the Full, 1 MM, or Compact primer sets were used with a single nested TCRB constant region reverse primer at 0.2 μM per primer. Samples underwent 30 cycles of PCR for visualization by agarose gel electrophoresis, or 20 cycles for NGS library preparation (“PCR1”) using the following cycling conditions: 1) 3 min at 95˚C, 2) 15 s at 95˚C, 3) 30 s at 52˚C (Full) or 47˚C (1 MM) or 42˚C (Compact), 4) 30 s at 72˚C, 5) Back to step 2 × 20 or 30 total cycles (as noted), 6) 1 min at 72˚C, 7) Hold at 4˚C.
A range of acceptable annealing temperatures that maximizes the number of clones detected was found for each primer set, and we selected an annealing temperature from these ranges. For the Full set, we detected 129251, 130486, and 129026 clones at annealing temperatures of 53.9˚C, 47.3˚C, and 45.2˚C, respectively. Using the 1MM set we detected 126164, 127399, and 125948 clones at 50.3˚C, 47.3˚C, and 45.2˚C, respectively. For the Compact set, we detected 112609, 112048, and 111546 clones at 45.8˚C, 42.3˚C, and 37.2˚C, respectively. Other annealing temperatures within these ranges are therefore also acceptable. We also compared the performance of Herculase II Fusion DNA polymerase (Agilent) and KAPA2G Fast Multiplex Mix for PCR1 and found that both enzymes perform comparably (ρ = 0.966, Fig S2). Herculase II is therefore an acceptable enzyme alternative for PCR1 if desired.
20 cycles of PCR2 were performed on PCR1 product (2 μL of PCR1 added to 18 μL of PCR2 master mix, which contained 0.25 μM each i5 and i7 sample barcoding primers) to incorporate sample barcodes and Illumina sequencing adaptors using Herculase II Fusion DNA Polymerase (Agilent) and the following cycling conditions: 1) 2 min at 95°C, 2) 20 s at 95°C, 3) 20 s at 58°C, 4) 30 s at 72°C, 5) Back to 2 × 20 total cycles, 6) 3 min at 72°C, 7) Hold at 4°C. Equal volumes of barcoded PCR2 products (5 μL each) were pooled and PCR column purified using QIAquick PCR Purification Kit (Qiagen). Libraries were quantified using KAPA Library Quantification Kit for Illumina Platforms (Roche). Barcoding primer sequences can be found in Supplementary File 1. See Fig S3 for a schematic of our sequencing strategy.
2.4. Sequencing and CDR3 analysis
Sequencing was performed on an Illumina NextSeq 500 (immunoSEQ benchmarking comparisons), MiSeq (Immunoverse samples), or HiSeq 2500 (IBM muscle biopsy analysis). CDR3s were identified and quantified using MiXCR v2.1.11 software [37] with default parameters except as noted in Table S2. Data obtained from Adaptive Biotechnologies’ immunoSEQ assay were re-analyzed using MiXCR v2.1.11 and identical parameters for comparison to our own data. For reanalysis, full nucleotide sequences for each CDR3 from the Adaptive Biotechnologies ImmunoSEQ dataset were expanded to repeat as many times as indicated by the corresponding “count” column. This file, with each clonal nucleotide sequence represented as many times as its “count” column, was converted to FASTA format for compatibility with MiXCR. The sequences obtained from ArcherDX's Immunoverse assay were expanded to repeat as many times as their pre-deduplicated clone counts and then matched back to their deduplicated clone counts after reanalysis with MiXCR v2.1.11. Clone count cutoffs for each analysis are as described in the figure legends and refer to the pre-deduplicated counts for the Immunoverse assay (Note: deduplicated clone counts were used for all comparisons, while pre-deduplicated counts were only used to filter the dataset to ≥ 10 counts). Information regarding the total number of clones, the number of singletons, and the number of clones occurring at ≥10 counts for FR3AK-seq, immunoSEQ, and Immunoverse can be found in Table S3.
2.5. Inferring Sequences via Efficiency Projection and Primer Incorporation (ISEPPI)
Primer amplification efficiencies for ISEPPI analysis were calculated for the 47 unique FR3 20-mers for the 1MM and Full primer sets using the R package DECIPHER and its CalculateEfficiencyPCR function [38]. For primer usage vectors, we searched for each relevant read associated with a given CDR3 nucleotide sequence obtained from MiXCR within the FASTQ sequencing file and extracted the first 20 nucleotides from each matching read. These sequences were then compared to the corresponding primer sequences and use of each primer was tabulated to generate a vector of length N, where N equals the number of primers used for amplification (1 MM: N = 34, Full: N = 47). Sequences that were not an exact match to a primer sequence were presumed sequencing errors and excluded from analysis. All vectors were converted to unit vectors prior to distance measurements. Each CDR3 was assigned to the FR3 that had a unit efficiency vector nearest (minimum Euclidean distance) to the primer usage unit vector. For assessment of FR3-assignment accuracy using the immunoSEQ data as ground truth, we extracted all sequences upstream of CDR3, including the first 3 nucleotides of CDR3 (which is included as FR3 in our primer design), from the immunoSEQ dataset. We then trimmed this FR3 sequence to the 3’ 20 nucleotides and assigned its corresponding IMGT reported FR3 20-mer as ground truth. For this analysis, we only considered CDR3s with clone counts of ≥10 in both data sets. The code for ISEPPI is available on Github (https://github.com/jmmontagne/ISEPPI-1.0).
2.6. IIM muscle biopsy cohort
Muscle biopsies were obtained from 145 patients with IIMs (124) as well as healthy controls [9] and non-IIM controls [12]. One non-IIM control muscle biopsy was sequenced in duplicate, making the total number of samples 146. IIM patients included those with dermatomyositis (DM, 40), immune-mediated necrotizing myopathy (IMNM, 49), sporadic inclusion body myositis (IBM, 14), and anti-synthetase syndrome (ASyS, 21) (Table S4). All patients provided informed consent and were not selected to be naïve to treatment [39]. All subjects were enrolled in institutional review board (IRB)-approved longitudinal cohorts from the National Institutes of Health (IRB number 91-AR-0196), the Johns Hopkins Myositis Center (IRB number NA_00007454), the Clinic Hospital (Barcelona; IRB number HCB/2015/0479), and the Vall d'Hebron Hospital (Barcelona; IRB number PR (AG) 68/2008).
2.7. Repertoire analysis using muscle biopsies from patients with idiopathic inflammatory myopathies (IIMs)
RNA isolated from muscle biopsies taken from 145 IIM patients or controls was reverse transcribed, PCR amplified, sequenced, and analyzed as described above. Healthy control muscle biopsies were obtained from healthy volunteers at the Skeletal Muscle Biobank of the University of Kentucky, while non-IIM controls were obtained for clinical purposes from the Johns Hopkins Neuromuscular Pathology Laboratory and were normal after pathologic evaluation. We added 1,000 cell equivalents of RNA from a clonal tumor infiltrating T cell line as a spike-in to each sample [40]. Sequencing of 6 spike-in only samples provided the CDR3 sequences to remove from downstream analyses of samples containing muscle biopsy RNA. The spike-in counts were used for cell number quantification within patient samples, which distinguished ‘bystander’ versus ‘non-bystander’ T cell clones as described in the text. Using the spike-in also provides template to ensure generation of PCR products in samples with few infiltrating T cells and thus few TCR RNA templates. This approach enabled the detection of a single T cell clone in one of these samples. Supplementary File 2 summarizes the number of reads, T cell clones, estimated number of T cells, and clonality for each sample. Clonality was calculated as (1-Shannon's equitability), computed as:
where pi is the proportion of the ith clone from a repertoire of n clones [41]. Clonality values range from 0-1, with 0 indicating equal representation of clones within a sample (lower clonality).
CDR3 sequences obtained from these samples were analyzed for disease-specific clusters using the GLIPH 1.0 group discovery algorithm (see Table S2 for parameters) [15]. To determine the statistical significance of each cluster, we performed a chi-square tests on all clusters with at least three contributing individuals, followed by Benjamini-Hochberg multiple comparison correction. This analysis was performed for both the number of sequences contributed by each disease subgroup to each cluster, as well as the number of patients in each disease group represented within each cluster. p-values ≤ 0.05 after multiple test correction were considered statistically significant.
2.8. Statistical analysis
Statistical analyses were performed using R software (www.r-project.org). For comparisons of TCR repertoires, Lin's concordance correlation coefficients[42] were computed as:
where μX and μY are the means, and are the variances, and ρ(X, Y) is the Pearson correlation coefficient for the variables X and Y. Coefficients of variation were calculated across triplicates for the Jurkat spike-in experiments. Kruskal-Wallis p-values were calculated to compare absolute T cell number and clonal diversity between IIM subgroups. To test the statistical significance of GLIPH clusters, chi-square analysis with Benjamini-Hochberg multiple comparison correction was performed on clusters with sequences contributed by at least three individuals. A Mann-Whitney p-value was calculated to compare GLIPH clustered to unclustered CDR3s.
2.9. Role of funding sources
This work was supported by NIH grant U24AI118633 and a Prostate Cancer Foundation Young Investigator Award. The funders had no role in the design of this study, data collection, data analyses, data interpretation, or writing of this manuscript.
3. Results
3.1. Minimal primer sets targeting the TCRB framework region 3 (FR3) efficiently amplify CDR3 sequences
We hypothesized that minimizing the number of potentially competing variable (V) gene primers, while also minimizing amplicon length, would maximize the efficiency of TCRB CDR3 region amplification. To identify candidate primer binding sequences upstream and proximal to CDR3, we constructed a phylogenetic tree of all functional TCRB FR3 regions available from the Immunogenetics (IMGT/GENE-DB) reference directory [35]. This revealed a high degree of sequence conservation within the 3’ terminus of FR3 (Fig 1b). This homology may reflect a critical role for these sequences in the optimal presentation of the CDR3 loop. The 3’ 20 nucleotides of these IMGT FR3s were therefore extracted and used as target primer binding sequences. Remarkably, the 128 functional TCRBV alleles reported by IMGT are represented by only 47 distinct 3′-terminal 20-mers. Beyond their sequence homology, additional advantages of targeting these sequences include minimal CDR3 amplicon length (and in turn optimal amplification efficiency) and the ability to sequence using short (100 nucleotide) single end reads initiated proximal and upstream of CDR3. We refer to this approach to TCRB CDR3 analysis as FR3 AmplifiKation sequencing, or “FR3AK-seq".
We developed a greedy algorithm to automate the design of three candidate sets of primers, each targeting the 47 distinct TCRB FR3 3′ 20-mers (Fig S1): one primer set lacks any universal bases or mismatches ("Full" set), one primer set lacks universal bases but allows a single mismatch to occur ("1MM" set), and one primer set contains parsimonious incorporation of up to 3 universal (inosine) bases and allows one mismatch ("Compact" set). No mismatches were permitted within the five nucleotides of the 3′ end of the primers, as a precaution to minimize interference with polymerase extension [38,43,44]. The Full set consists of 47 primers (equal to the number of unique FR3 3’ 20-mers), the 1MM set consists of 34 primers, and the Compact set consists of 20 primers (Table S1). We calculated the expected efficiency for each primer to amplify each FR3 sequence; these efficiencies are visualized in the form of a clustered heatmap (Fig 1c) and are used later for inference of FR3 utilization.
Using DNA as a template for repertoire sequencing is appealing because of its stability, and also because it allows for precise quantification of T cell clones as exactly one functionally rearranged template should be present per cell. However, we found using RNA as starting material was more advantageous than DNA for several reasons. First, RNA is more abundant than genomic DNA and thus requires less input material. Additionally, one copy of the TCRB chain will be nonfunctional at the DNA level, while the RNA products from this template are less likely to be sampled because of nonsense-mediated decay or allelic exclusion. Furthermore, a single TCRB chain reverse primer can be used to amplify all CDR3 regions from cDNA, reducing the number of primers required versus DNA (which requires multiplexed joining (J) region priming). Although RNA expression will vary between clones based on their phenotype and activation state, we can infer that the more abundant and activated T cells will contribute more RNA transcripts than rare, less activated clones [22]. Additionally, it has been shown that the number of TCRB RNA molecules per cell is equivalent between naïve, activated, and memory CD8 T cells [45], and therefore RNA may potentially be used as a surrogate for cellular abundance.
3.2. TCRB FR3 primers amplify CDR3 sequences with minimal bias
Each primer was tested separately and within its set for the ability to produce amplicon from peripheral blood mononuclear cell (PBMC) cDNA. Agarose gel electrophoresis revealed a PCR product at the expected size of ~220 base pairs (Fig S4a). Importantly, incorporation of the inosine base did not prevent amplification by the KAPA Fast HotStart Taq DNA Polymerase. While lower PCR primer annealing temperature might increase off-target priming, it may also reduce unwanted bias in amplification from primers containing a single nucleotide mismatch[46]. We therefore utilized gradient PCR to assess the effect of annealing temperature on the number of unique CDR3 sequences detected using the Compact primer set. As expected, more unique CDR3 clones were recovered at lower annealing temperatures (Fig S4b), and a similar trend was observed for the 1MM primer set. A range of amenable annealing temperatures was detected for each primer set, and we selected the following for each from these ranges: Compact: 42°C, 1MM: 47°C, Full: 52°C. Use of a hemi-nested RT-PCR strategy (Fig S3) resulted in minimal amplification of non-TCRB sequences.
We next quantified PCR amplification bias inherent to the three primer sets. cDNA first underwent 20 cycles of PCR using each primer set separately. An aliquot of this product was diluted 210-fold and then subjected to 10 more cycles of PCR; sequencing was then performed on both amplicons and the resulting data sets compared to each other. In this experiment, significant per-cycle amplification bias would manifest as discordance. However, the concordance correlation coefficient remains high for all three primer sets, suggesting that FR3AK-seq primers negligibly bias the CDR3 repertoire during cycles of PCR amplification (ρ = 0.997 for the Full primer set and ρ = 0.998 for the 1 MM primer set, Fig 2a; ρ = 0.997 for the Compact primer set, Fig S4c).
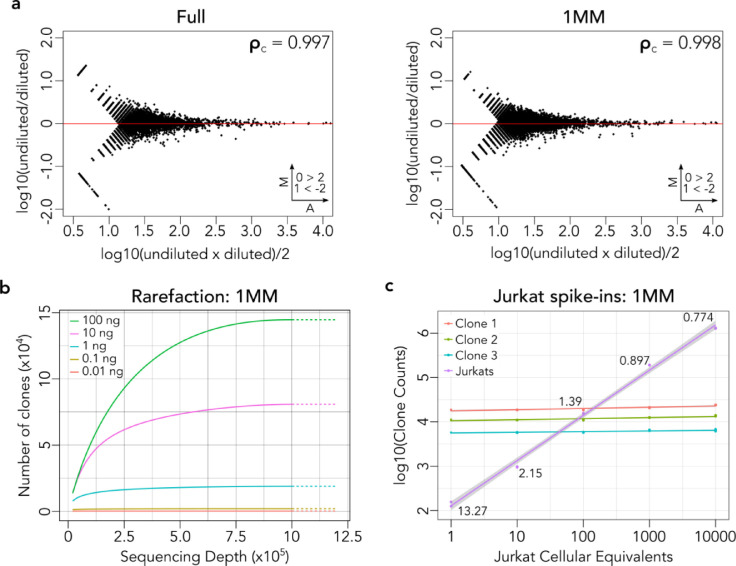
Fig. 2.
FR3AK-seq multiplex PCR performance. a. MA plots of 210 dilution experiments for the Full and 1 MM primer sets demonstrated high linear concordance (Full: 0.997, 1 MM: 0.998, Compact in Fig S3c), indicating negligible PCR bias. Clones with ≥10 counts in either dataset were compared. Insets indicate number of clones above or below Y axis limits (set to 2 and –2 for visualization). b. Rarefaction analysis using T cell RNA purified from PBMCs showed the relationship between RNA input, number of clones detected, and sequencing depth using FR3AK-seq. c. The spiked-in Jurkat CDR3 sequence was detected at the expected abundances using FR3AK-seq (performed in triplicate). Estimated abundances of the top three T cell clones were consistent among triplicates and across spiked-in samples. 95% confidence intervals for regression lines and coefficients of variation for the cellular equivalents calculated for the T cell clones are shown.
We then performed rarefaction analysis to determine the relationship between RNA input amount, sequencing depth, and the number of unique clones detected using FR3AK-seq. We found that diversity discovery is saturated at ~1 × 106 reads for 100 ng of purified peripheral blood T cell RNA (Fig 2b), providing the rationale for RNA input amounts and sequencing depths used in subsequent analyses. We also assessed the sensitivity of FR3AK-seq to detect rare clones by spiking varying amounts of monoclonal Jurkat RNA (from 1 up to 10,000 cellular equivalents) into a background of 400 ng PBMC RNA (~20% T cells). We found that FR3AK-seq detected the Jurkat CDR3 sequence at all input amounts at the expected abundances, while the abundance of the top three PBMC T cell clones remained constant across the dilution series (Fig 2c). These Jurkat sequences were used to calculate cellular equivalents for the top three PBMC clones in each sample. Notably, the coefficient of variation for these cellular equivalents was below 1% in the 1,000 Jurkat cell equivalent spike-in samples (CV = 0.897%). This provided the basis for the subsequent use of 1,000 cellular equivalents of spike-in cells.
3.3. FR3AK-seq performs comparably to a multiplex PCR-based industry standard
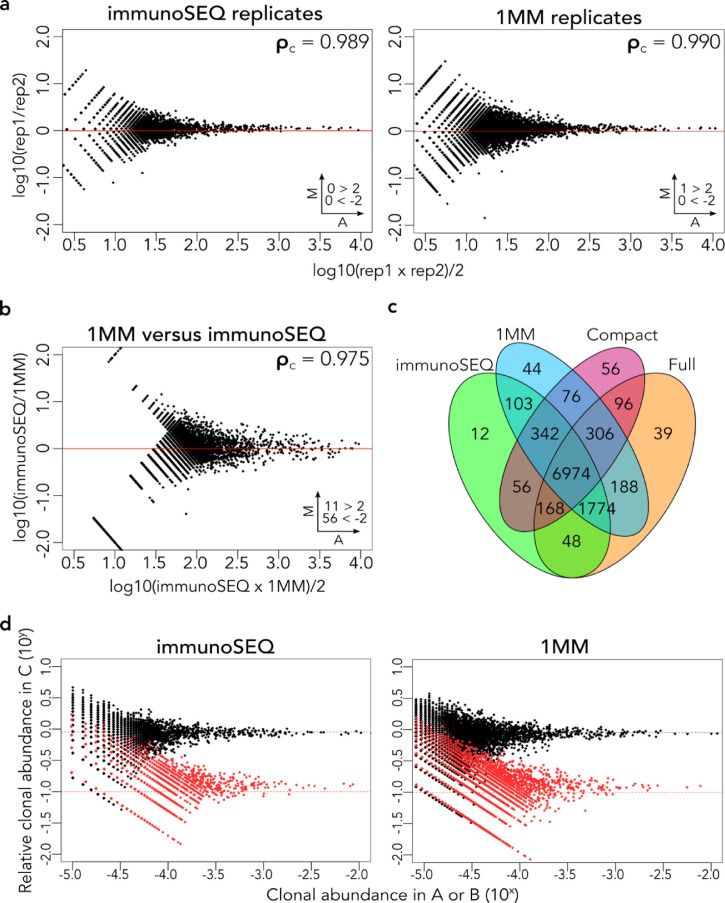
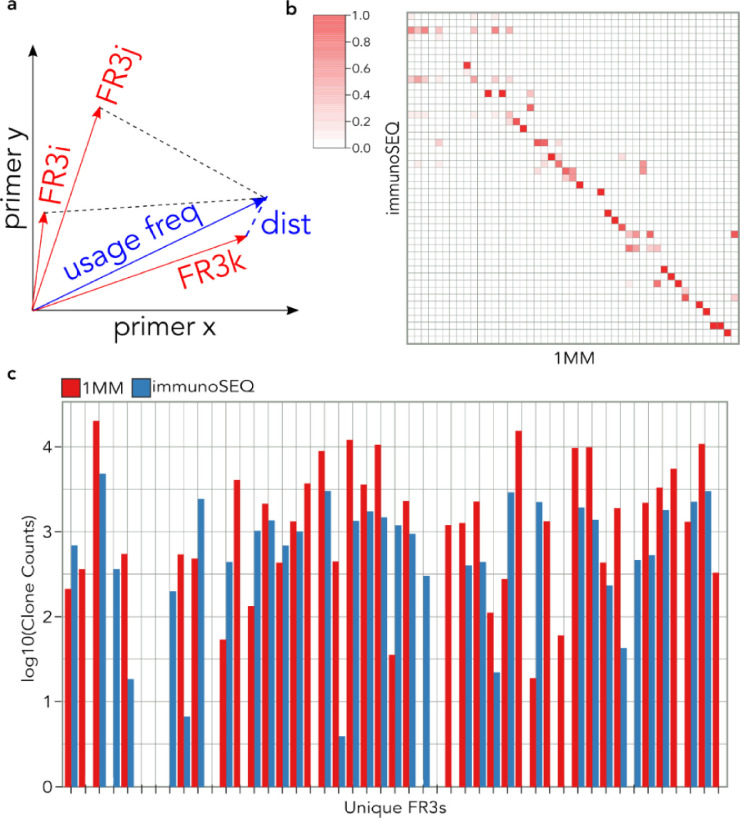
T cell RNA from two donors, A and B, as well as a third sample comprised of 90% Donor A RNA and 10% Donor B RNA (sample “C”), were used to generate cDNA for benchmarking studies against the current multiplex PCR-based industry standard, the immunoSEQ hsTCRB “Deep Resolution” sequencing service offered by Adaptive Biotechnologies. Separate cDNA aliquots were also subjected to FR3AK-seq analysis using each of the three primer sets. The open source software MiXCR v2.1.11 was used to define and quantify CDR3 sequences from both assays for direct comparison [37]. The technical reproducibility of the immunoSEQ assay was quantified by comparing the abundances of Donor A's unique CDR3 sequences against their corresponding abundances in sample C (concordance correlation coefficient, ρ = 0.989, left panel Fig 3a). The same analysis was applied to the FR3AK-seq data sets, which were generated using each of the three FR3 primer sets. Equally high measures of internal concordance were observed (ρ = 0.990 for the 1 MM primer set, right panel Fig 3a; ρ = 0.990 for the Full primer set and ρ = 0.987 for the Compact primer set, Fig S5a). Importantly, the concordance between immunoSEQ and FR3AK-seq measurements of CDR3 abundance was also very high (ρ = 0.975, Fig 3b, Fig S5b), providing further evidence that FR3AK-seq amplifies CDR3s with negligible bias. The majority of even moderately abundant immunoSEQ-defined CDR3 sequences were also detected using any of the three FR3 primer sets, although the Compact set detected ~15% fewer CDR3s as compared to the rest (Fig 3c). Additionally, the 9:1 mixture composed of cells from Donor A and Donor B (sample C) was used to determine quantification accuracy of the FR3 primer sets relative to the immunoSEQ assay (Fig 3d, Fig S5c). These data indicate that the full and 1MM FR3 primer sets provide CDR3 quantification comparable to the industry standard immunoSEQ assay, with the 1MM primer set performing most accurately.
Fig. 3.
FR3AK-seq performs comparably to the multiplex PCR-based immunoSEQ assay from Adaptive Biotechnologies. a. MA plots comparing technical replicates of the immunoSEQ and 1MM FR3AK-seq assays. b. MA plot comparing the 1MM FR3AK-seq assay against the immunoSEQ assay. Clones with ≥10 counts in either dataset were included. c. A Venn diagram shows overlapping and non-overlapping CDR3 sequences detected using immunoSEQ and all three FR3AK-seq primer sets. CDR3 sequences were included in this analysis if they had a clone count ≥10 in any of the four data sets. d. Relative frequencies of Donor A (black points) and Donor B (red points) in sample C determined using the immunoSEQ assay or the 1MM FR3AK-seq assay. Insets in a and b indicate number of clones above or below Y axis limits (set to 2 and -2 for visualization).
3.4. FR3AK-seq performs comparably to a unique molecular identifier (UMI)-based industry standard
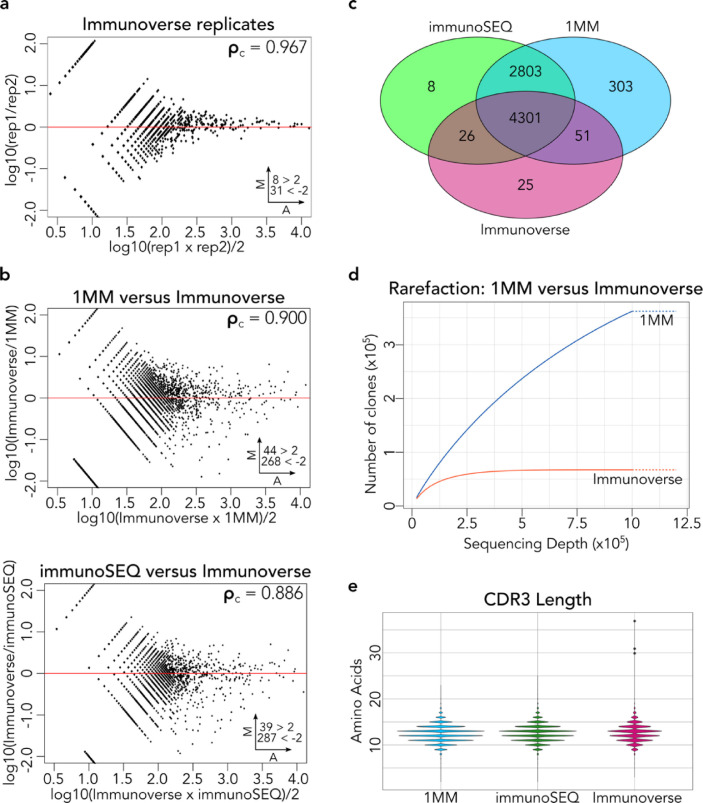
To assess whether FR3AK-seq achieves results comparable to UMI-based methodologies, we compared the FR3AK-seq 1MM and immunoSEQ datasets to one obtained using ArcherDX's UMI-based Immunoverse HS TCR assay. PBMC RNA from the same samples A, B, and C described above were subjected to Immunoverse analysis for comparison with the FR3AK-seq and immunoSEQ datasets. The technical reproducibility of the Immunoverse assay was similar to that of both immunoSEQ and FR3AK-seq (ρ = 0.967, Fig 4a). Additionally, concordance between FR3AK-seq and Immunoverse was high (ρ = 0.900, upper panel Fig 4b). Relatively high concordance was also observed when comparing immunoSEQ to Immunoverse (ρ = 0.886, lower panel Fig 4b). Crucially, the majority of clones detected by the Immunoverse assay were also detected by both immunoSEQ and FR3AK-seq (Fig 4c). These data demonstrate that FR3AK-seq has a high sensitivity for detecting clones of the correct sequence (4352/4403, 98.8% of total clones detected by Immunoverse).
Fig. 4.
FR3AK-seq performs comparably to the UMI-based ArcherDx Immunoverse assay. a. MA plot comparing Immunoverse technical replicates. b. MA plots comparing the 1MM FR3AK-seq assay against the Immunoverse assay (top panel) and Adaptive Biotechnologies’ immunoSEQ assay against the Immunoverse assay (bottom panel). Clones with ≥10 counts in either dataset were included (using pre-deduplicated counts for the Immunoverse dataset). c. A Venn diagram shows overlapping and non-overlapping CDR3 sequences detected using Immunoverse, immunoSEQ, and the 1MM FR3AK-seq primer set. CDR3 sequences were included in this analysis if they had a clone count of ≥10 in any of the three data sets. d. Rarefaction analysis comparing the 1MM FR3AK-seq assay to the Immunoverse assay using 100 ng of T cell RNA (~250,000 cells) for 1MM FR3AK-seq analysis and 200ng of T cell RNA (~500,000 cells) for Immunoverse analysis. MM: ~1.2 × 106 reads, Immunoverse: ~1.4 × 106 reads. e. Violin plots comparing CDR3 lengths captured by the short-read 1MM FR3AK-seq versus longer-read immunoSEQ and Immunoverse assays. Insets in a and b indicate number of clones above or below Y axis limits (set to 2 and -2 for visualization).
Strikingly, a large number (2803) of clones were detected by both immunoSEQ and FR3AK-seq but not Immunoverse. Rarefaction analysis of the FR3AK-seq and Immunoverse datasets explained this difference in clonal detection sensitivity, in that FR3AK-seq sampled more clones at all sequencing depths (Fig 4d). Although FR3AK-seq achieves better coverage of the repertoire than Immunoverse across sequencing depths, we wanted to quantify how many of the FR3AK-seq detected clones may be the result of false discovery. To address this question, we performed FR3AK-seq 1MM analysis of 1,000 cellular equivalents of Jurkat cell RNA in triplicate. Since the Jurkat cell line is monoclonal, the frequencies of subdominant clones, presumably generated by PCR or sequencing errors, can be used to assess the rate of false positive discovery. 99.98% of MiXCR-defined CDR3s mapped to the correct reported Jurkat TCRB sequence in each sample. We found an average of 50 non-Jurkat clones across triplicates, of which only one was moderately abundant (≥10 clone counts) in each. This secondary clone was the same between replicates and represented on average 0.0067% of total MiXCR-defined CDR3s. Using default MiXCR settings, FR3AK-seq analysis is therefore associated with a very low rate of false discovery due to PCR or sequencing errors. Modification of MiXCR thresholds can be used to adjust the stringency with which these subdominant clones are grouped. We also compared the lengths of CDR3 clones detected using each sequencing platform and found that FR3AK-seq's short (100 nucleotide) reads did not bias the distribution of CDR3 lengths recovered (Fig 4e).
3.5. Inference of FR3 from primer usage patterns
V gene usage is often of interest, for example to detect usage skewing and for functional studies involving TCR cloning. FR3AK-seq prioritizes small amplicons to reduce both PCR bias and cost. Therefore, by design, amplicons do not contain easily recoverable V gene sequence information. Each of the 47 unique 3’ FR3 20-mers, however, is associated with a specific subset of V alleles. Therefore, we wondered how accurately we could assign a unique FR3 sequence (linked to a subset of V alleles) to each FR3AK-seq generated CDR3 sequence.
As visualized in Fig 1c, some primers can efficiently amplify multiple unique FR3 sequences. Based on this, we hypothesized that specific patterns of FR3AK-seq primer usage would indicate which of the underlying FR3 20-mers was associated with each CDR3 sequence. We first tabulated the proportional usage of each primer (determined using the first 20 nucleotides of each sequencing read) by each CDR3. Primer usage can be represented as a “usage vector” in N-dimensional space (“primer space”), where N is equal to the total number of primers contained in the primer set. Each primer usage vector can then be compared against 47 “efficiency vectors”, which are defined by calculating the efficiency with which each primer will amplify each FR3 20-mer. Under the assumption that for a given CDR3, its primer usage vector should be most closely aligned with its corresponding efficiency vector, we developed the Inferring Sequences via Efficiency Projection and Primer Incorporation (“ISEPPI”) procedure to link CDR3s with their most likely associated FR3 20-mer (Fig 5a). ISEPPI was performed for Donor A's CDR3-associated 1MM primer usage vectors and compared to the immunoSEQ-defined FR3 20-mer, which we considered ground truth (Fig 5b, Full primer set Fig S6a). For moderately abundant CDR3 sequences (≥10 clone counts), ISEPPI correctly assigned 62.9% of the CDR3 clone counts to the correct FR3 20-mer (Fig 5c). For clones with a higher abundance of ≥50 counts or ≥100 counts, 66.1% and 67.9%, respectively were assigned to the same FR3 20-mer as in the immunoSEQ data set. The accuracy of ISEPPI was slightly improved with use of the Full primer set: ≥10: 67.7%, ≥50: 71.4%, ≥100: 72.4% (Fig S6b). Interestingly, ~25% of abundant (≥10 clone counts) unique CDR3s were associated with more than one FR3 20-mer in the immunoSEQ dataset. We found that this reduced the accuracy of ISEPPI in our dataset, as this obscures primer usage patterns and FR3 assignment for these CDR3s.
Fig. 5.
Inferring Sequences via Efficiency Projection and Primer Incorporation (ISEPPI). a. Each primer's amplification efficiency is calculated for each of the 47 unique TCRB FR3 20-mer sequences, thereby forming each FR3 amplification efficiency vector (red vectors, FR3i-k). A primer usage vector for each CDR3 is also determined (blue vector). Each CDR3 is assigned to a FR3 sequence by finding the nearest FR3 amplification efficiency vector (Euclidean ‘dist’, blue dashed line). b. Heatmap comparing FR3 assignments to CDR3s using ISEPPI analysis of 1MM FR3AK-seq data (X axis) versus immunoSEQ data (Y axis, ground truth). CDR3s were included in the analysis if they had a clone count of at least 10 in both datasets. The order of FR3s is the same as in Fig 1c; heatmap columns are normalized to 1. c. ISEPPI correctly assigned 62.9% of clone counts (for CDR3s with a clone count of at least 10). The order of unique TCRBV FR3s is identical to the order of the Full primer set for all heatmaps as in Table S1.
3.6. FR3AK-seq enables inexpensive cohort-scale repertoire studies and identifies shared T cell responses in muscle from patients with IIM
Infiltrating and perivascular T cells are common features in muscle from patients with IIMs. However, the precise role of T cells in the initiation and progression of IIMs is unclear[5]. Sequencing the TCR repertoires within muscle enables the discovery of IIM-relevant T cell responses but requires the analysis of many samples with associated clinical and histopathological data. Therefore, muscle biopsies from 145 patients with IIMs (124) as well as healthy controls[9] and non-IIM controls [12] were analyzed using the FR3AK-seq 1MM primer set at a total cost of ~$20 per sample (Tables S4, S5).
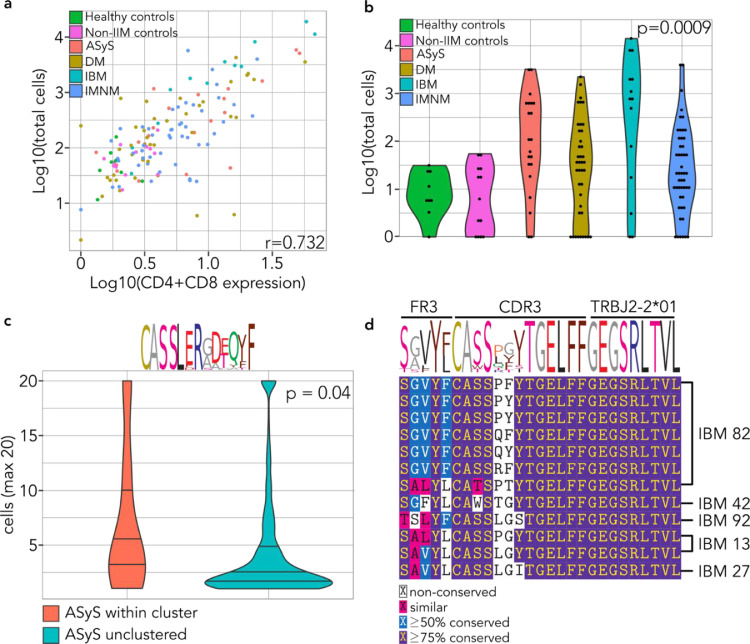
We anticipated that these biopsies would have varying numbers of infiltrating T cells, and potentially very few. To address this, 1000 cell equivalents of RNA derived from a clonal T cell line[40] were spiked into each sample after RNA purification. This enabled the absolute estimation of T cells in each sample, while also providing RNA templates to ensure PCR product generation in samples with few or no infiltrating T cells. Aggregated CD4 and CD8 sequencing reads from RNA-seq analysis [39] of the same samples tightly correlated with a FR3AK-seq based estimate of cellular equivalents (Fig 6a). In subsequent analyses, CDR3 sequences present at levels at or below one cell equivalent per biopsy were considered ‘bystander’ T cell clones, unlikely to be involved in the disease process. By this metric, the number of non-bystander T cells were elevated in each disease subgroup versus the controls (Fig 6b). In agreement with previous immunohistochemical analyses, most IBM biopsies contained particularly high levels of T cell infiltrates [47], [48], [49]. We determined this to be the case both in terms of absolute T cell number and clonal diversity (Fig S7a). No distinct patterns of FR3 or J gene usage were found among patient subgroups (Fig S8a-b), and CDR3 hydrophobicity and lengths were similar between patient subgroups (Fig S8c-d).
Fig. 6.
FR3AK-seq detects related T cell responses in subsets of patients with idiopathic inflammatory myopathies. a. Scatterplot of estimated T cell counts per patient versus CD4 and CD8 expression levels obtained from bulk mRNA-seq. Each patient data point is colored according to their IIM association. Pearson correlation coefficient (r) is shown. b. Distributions of non-bystander cell counts per patient for each IIM subgroup and controls. Bystander cell counts were set to 0 (p = 0.0009 [Kruskal-Wallis]). c. A CDR3 cluster enriched for sequences from a single patient with ASyS was identified using GLIPH software (p = 1.4 × 10−7 [chi-square test with Benjamini-Hochberg multiple comparison correction]). The amino acid sequence logo of the CDR3 region is shown above the violin plot. The clones within this cluster are more expanded than corresponding unclustered clones from the same patient (p = 0.04 [Mann-Whitney]). Cell counts were set to a maximum of 20 for visualization. d. An IBM-exclusive cluster was also identified using GLIPH, encompassing 5 of 12 IBM patients who had non-bystander T cell infiltrates (41.7%, p = 1.4 × 10−6 [chi-square test with Benjamini-Hochberg multiple comparison correction]). Multiple sequence alignment (using MUSCLE) is shown for the corresponding ISEPPI-defined FR3s, MiXCR-defined CDR3s, and the sequenced J chain allele (TRBJ2-2*01).
Assessing muscle-resident T cell responses across patients with IIM is crucial for understanding disease pathogenesis and progression. Public T cell clones, defined as clones with identical amino acid CDR3 sequences present in two or more individuals, indicate responses to shared antigenic targets [25]. We therefore summarized the presence of public T cell clones within the aggregated set of non-bystander CDR3s from the IIM cohort. We found 80 public clones, of which 76 were shared by only two individuals (Supplementary File 3). The remaining four public clones were shared by either 3 or 4 individuals, suggesting that most muscle-resident T cell clones in this cohort are private.
It has recently been shown that TCR sequence features can be used to define clusters of related TCRs predicted to bind the same antigen [15,16,50]. The open source software GLIPH (grouping of lymphocyte interactions by paratope hotspots) [15] was therefore used to extend our analysis beyond public clones to search for clusters of related CDR3 sequences within the aggregated set of non-bystander CDR3s (Table S6). 143 clusters composed of sequences contributed by at least 3 individuals were identified. We first examined clusters that had a disproportionate number of sequences contributed by a single individual. Fig 6c provides an example of a cluster dominated by related CDR3 sequences contributed by a single ASyS patient. Compared to CDR3 sequences from the same patient which were not in any cluster, the clonal abundances of these CDR3s were significantly increased, suggestive of a polyclonal, shared antigen-driven expansion. These results demonstrate that GLIPH is likely able to cluster functionally related CDR3 sequences that have been detected and quantified using FR3AK-seq.
We next determined whether any of the GLIPH clusters were composed of sequences contributed disproportionately by individuals from one of the IIM subgroups. Indeed, we found a GLIPH cluster that was composed exclusively of sequences contributed by IBM patients. Specifically, 5 of the 12 IBM patients with non-bystander T cells contributed at least one sequence to this cluster (p = 1.4 × 10−6 [chi-square test with Benjamini-Hochberg multiple comparison correction], Fig 6d). ISEPPI analysis revealed related FR3 usage within this cluster, as visualized in the sequence logo. Remarkably, all CDR3 sequences in this IBM cluster are linked to a single J-chain (TRBJ2-2*01), providing additional support for shared antigen recognition. Taken together, this study demonstrates the biomedical utility of FR3AK-seq and ISEPPI in probing patients’ T cell responses within large cohorts.
4. Discussion
Although the utility of TCR repertoire sequencing to characterize T cell responses has been well established, the lack of a streamlined, quantitative, inexpensive, and non-proprietary assay has limited the scope and scale of feasible studies. By rationally designing minimal sets of primers that target the 3′ terminus of FR3, we have developed a multiplex PCR-based approach for ultra-efficient library preparation and sequencing of TCRB CDR3 repertoires. By minimizing amplification bias (via reduction of primer number and amplicon length), the resulting sequencing libraries quantitatively capture clonal abundance distributions with similar accuracy as multiplex PCR-based and UMI-based industry standards. While the library preparation and sequencing strategy presented here already bring the per-sample cost to ~$20 (Table S5), we expect that alternative approaches to library preparation, reduced sequencing depth, and declining sequencing costs will likely enable another ~10-fold reduction in per-sample cost. In addition to reducing the overall cost of obtaining CDR3 sequences, we have developed the FR3AK-seq workflow to require minimal expertise, effort, and time. Quantitative sequencing libraries can be readily prepared within a single day. Comparisons of protocols and cost between FR3AK-seq, immunoSEQ, and Immunoverse are summarized in Table S7.
Our approach to TCRB repertoire analysis should be generalizable to additional immune receptor types, as well as to those of non-human species (see Table S8 for Mus musculus TCRB primers). Although these mouse primers have not been as extensively validated as the human set, they have been confirmed to efficiently amplify CDR3 sequences from mouse tissues. We have additionally used the same FR3 primer design and analysis principles to target human TCR alpha, gamma, and delta repertoires, although these primers have not yet been validated (Table S9). We have also designed a set of TCRB J region reverse primers (Table S10) that perform comparably to our constant region reverse primer using cDNA templates (ρ = 0.949, Fig S9). These will be potentially useful for analysis of DNA from FFPE samples which suffer from extensive RNA degradation. Interestingly, during the preparation of this manuscript, the Euroclonality Group (creators of the BIOMED2 primer set[51]) published a new set of TCR primers for next-generation sequencing, including some that are designed in the 5’ portion of FR3[52], providing additional validation of an FR3-targeted approach to TCR repertoire sequencing.
It is possible to make inferences on each CDR3’s associated V genes based on the pattern of FR3 primer usage, as demonstrated with the ISEPPI procedure presented here. ISEPPI is based on simple Euclidean distances measured in primer space, and successfully assigns up to 72.4% of T cell clone counts to their correct FR3 20-mer sequence. Future iterations of ISEPPI will be enhanced by incorporating more sophisticated classification strategies. In addition, it is likely that the ISEPPI principle may find applications beyond TCR repertoire studies, in settings where deconvolution of multiplex PCR primer usage is important. When comprehensive analysis of V gene usage is desired, complementary techniques (e.g. 5’RACE) can be parsimoniously utilized to capture this information. One may use FR3AK-seq to track unique CDR3 sequences of interest over time, across tissues, and/or after FACS analysis – these FR3AK-seq detected CDR3 sequences can then be readily associated with a V gene by merging data sets.
Large-scale TCR repertoire studies enable the interrogation of the complex role of T cells in human disease. To demonstrate the utility of FR3AK-seq for efficient large-scale analysis of TCRB CDR3 repertoires, we characterized the muscle-infiltrating T cells present within biopsies obtained from 145 inflammatory muscle disease patients and controls. TCRB CDR3 sequence clustering indicated both donor and disease specific antigen-driven T cell responses. Importantly, the majority of T cell clones detected in IBM muscle biopsies were not hyperexpanded, in contrast to the hypothesis that infiltrating IBM T cells originate from a clonal T cell large granular lymphocytic leukemia (T-LGL) (Fig S7b) [53,54]. Future studies will utilize FR3AK-seq in combination with in vitro stimulation to characterize the antigenic determinants of these clonal clusters. Further comparisons will be made to TCRs with known antigen specificities within public databases including VDJdb [55,56] and McPAS-TCR [57]. Presence of disease relevant antigen-specific clones in the periphery will also be explored, as this could be valuable for diagnostics and disease stratification of IIM.
Data sharing statement
Data are available upon request.
Author Contributions
Conceptualization and experimental design: J.M.M., H.B.L.; performing experiments: J.M.M.; data analysis and software: J.M.M., X.A.Z.; human specimen procurement and analysis: I.P-F., J.C.M., L.C-S., T.E.L., A.L.M., H.B.L.; data interpretations: J.M.M., X.A.Z., T.E.L., A.L.M., H.B.L.; paper writing: J.M.M., H.B.L. All authors reviewed the paper.
Declaration of Competing Interest
H. Benjamin Larman is a consultant for Tscan Therapeutics, which seeks to develop T cell receptor based cellular therapies. Andrew Mammen and Lisa Christopher-Stine are listed as inventors on a patent (JHU C-11077) for a myositis antibody biomarker licensed to Inova Diagnostics. The remaining authors declare no competing interests.
Acknowledgements
We would like to thank the Johns Hopkins Medical Institute Deep Sequencing and Microarray Core Facility and the Johns Hopkins Institute of Genetic Medicine Genetics Resources Core Facility.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102972.
Appendix. Supplementary materials
References
- 1.Selva-O'Callaghan A, Pinal-Fernandez I, Trallero-Araguás E, Milisenda JC, Grau-Junyent JM, Mammen AL. Classification and management of adult inflammatory myopathies. Lancet Neurol. 2018;17(9):816–828. doi: 10.1016/S1474-4422(18)30254-0. [DOI] [PubMed] [Google Scholar]
- 2.Lundberg IE, de Visser M, Werth VP. Classification of myositis. Nat Rev Rheumatol. 2018 May;14(5):269–278. doi: 10.1038/nrrheum.2018.41. [DOI] [PubMed] [Google Scholar]
- 3.Betteridge Z, McHugh N. Myositis-specific autoantibodies: an important tool to support diagnosis of myositis. J Intern Med. 2016 Jul;280(1):8–23. doi: 10.1111/joim.12451. [DOI] [PubMed] [Google Scholar]
- 4.McHugh NJ, Tansley SL. Autoantibodies in myositis. Nat Rev Rheumatol. 2018 20;14(5):290–302. doi: 10.1038/nrrheum.2018.56. [DOI] [PubMed] [Google Scholar]
- 5.Miller FW, Lamb JA, Schmidt J, Nagaraju K. Risk factors and disease mechanisms in myositis. Nat Rev Rheumatol. 2018 20;14(5):255–268. doi: 10.1038/nrrheum.2018.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attaf M, Huseby E, Sewell AK. αβ T cell receptors as predictors of health and disease. Cell Mol Immunol. 2015 Jul;12(4):391–399. doi: 10.1038/cmi.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009 Oct;19(10):1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009 Nov 5;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009 May 8;324(5928):807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009 Dec 23;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muraro PA, Robins H, Malhotra S, Howell M, Phippard D, Desmarais C. T cell repertoire following autologous stem cell transplantation for multiple sclerosis. J Clin Invest. 2014 Mar;124(3):1168–1172. doi: 10.1172/JCI71691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014 May;124(5):2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beausang JF, Wheeler AJ, Chan NH, Hanft VR, Dirbas FM, Jeffrey SS. T cell receptor sequencing of early-stage breast cancer tumors identifies altered clonal structure of the T cell repertoire. Proc Natl Acad Sci U S A. 2017 28;114(48):E10409–E10417. doi: 10.1073/pnas.1713863114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet. 2017 May;49(5):659–665. doi: 10.1038/ng.3822. [DOI] [PubMed] [Google Scholar]
- 15.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94–98. doi: 10.1038/nature22976. 06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547(7661):89–93. doi: 10.1038/nature22383. 06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeWitt WS, Smith A, Schoch G, Hansen JA, Matsen FA, Bradley P. Human T cell receptor occurrence patterns encode immune history, genetic background, and receptor specificity. eLife. 2018;28:7. doi: 10.7554/eLife.38358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014 Jul;32(7):684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris H, DeWolf S, Robins H, Sprangers B, LoCascio SA, Shonts BA. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med. 2015 Jan 28;7(272):272ra10. doi: 10.1126/scitranslmed.3010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theil A, Wilhelm C, Kuhn M, Petzold A, Tuve S, Oelschlägel U. T cell receptor repertoires after adoptive transfer of expanded allogeneic regulatory T cells. Clin Exp Immunol. 2017 Feb;187(2):316–324. doi: 10.1111/cei.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamedov IZ, Britanova OV, Bolotin DA, Chkalina AV, Staroverov DB, Zvyagin IV. Quantitative tracking of T cell clones after haematopoietic stem cell transplantation. EMBO Mol Med. 2011 Apr;3(4):201–207. doi: 10.1002/emmm.201100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol. 2017 10;17(1):61. doi: 10.1186/s12896-017-0379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 24.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006 Dec;6(12):883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 25.Miles JJ, Douek DC, Price DA. Bias in the αβ T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011 Mar;89(3):375–387. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 26.Kebschull JM, Zador AM. Sources of PCR-induced distortions in high-throughput sequencing data sets. Nucleic Acids Res. 2015 Dec 2;43(21):e143. doi: 10.1093/nar/gkv717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung M-W, Parsons JM. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 28.Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR. Towards error-free profiling of immune repertoires. Nat Methods. 2014 Jun;11(6):653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 29.Peng Q, Vijaya Satya R, Lewis M, Randad P, Wang Y. Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics. 2015 Aug 7;16:589. doi: 10.1186/s12864-015-1806-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vander Heiden JA, Yaari G, Uduman M, Stern JNH, O'Connor KC, Hafler DA. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014 Jul 1;30(13):1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol. 2015 Nov;11(11) doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oakes T, Heather JM, Best K, Byng-Maddick R, Husovsky C, Ismail M. Quantitative Characterization of the T Cell Receptor Repertoire of Naïve and Memory Subsets Using an Integrated Experimental and Computational Pipeline Which Is Robust, Economical, and Versatile. Front Immunol. 2017;8:1267. doi: 10.3389/fimmu.2017.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma K-Y, He C, Wendel BS, Williams CM, Xiao J, Yang H. Immune Repertoire Sequencing Using Molecular Identifiers Enables Accurate Clonality Discovery and Clone Size Quantification. Front Immunol. 2018;9:33. doi: 10.3389/fimmu.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egorov ES, Merzlyak EM, Shelenkov AA, Britanova OV, Sharonov GV, Staroverov DB. Quantitative profiling of immune repertoires for minor lymphocyte counts using unique molecular identifiers. J Immunol Baltim Md. 1950;194(12):6155–6163. doi: 10.4049/jimmunol.1500215. 2015 Jun 15. [DOI] [PubMed] [Google Scholar]
- 35.Giudicelli V, Chaume D, Lefranc M-P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodenhofer U, Bonatesta E, Horejš-Kainrath C, Hochreiter S. msa: an R package for multiple sequence alignment. Bioinforma Oxf Engl. 2015 Dec 15;31(24):3997–3999. doi: 10.1093/bioinformatics/btv494. [DOI] [PubMed] [Google Scholar]
- 37.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015 May;12(5):380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 38.Wright ES, Yilmaz LS, Ram S, Gasser JM, Harrington GW, Noguera DR. Exploiting extension bias in polymerase chain reaction to improve primer specificity in ensembles of nearly identical DNA templates. Environ Microbiol. 2014 May;16(5):1354–1365. doi: 10.1111/1462-2920.12259. [DOI] [PubMed] [Google Scholar]
- 39.Pinal-Fernandez I, Casal-Dominguez M, Derfoul A, Pak K, Miller FW, Milisenda JC. Machine learning algorithms reveal unique gene expression profiles in muscle biopsies from patients with different types of myositis. Ann Rheum Dis. 2020 Jun 16 doi: 10.1136/annrheumdis-2019-216599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng L, Langley RJ, Brown PH, Xu G, Teng L, Wang Q. Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor. Nat Immunol. 2007 Apr;8(4):398–408. doi: 10.1038/ni1447. [DOI] [PubMed] [Google Scholar]
- 41.Shannon CE. A mathematical theory of communication. ACM SIGMOBILE Mob Comput Commun Rev. 2001 Jan 1;5(1):3–55. [Google Scholar]
- 42.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989 Mar;45(1):255–268. [PubMed] [Google Scholar]
- 43.Simsek M, Adnan H. Effect of single mismatches at 3’-end of primers on polymerase chain reaction. J Sci Res Med Sci. 2000 Jan;2(1):11–14. [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J-H, Hong P-Y, Liu W-T. Quantitative effects of position and type of single mismatch on single base primer extension. J Microbiol Methods. 2009 Jun;77(3):267–275. doi: 10.1016/j.mimet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Klinger M, Kong K, Moorhead M, Weng L, Zheng J, Faham M. Combining next-generation sequencing and immune assays: a novel method for identification of antigen-specific T cells. PloS One. 2013;8(9):e74231. doi: 10.1371/journal.pone.0074231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishii K, Fukui M. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl Environ Microbiol. 2001 Aug;67(8):3753–3755. doi: 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arahata K, Engel AG. Monoclonal antibody analysis of mononuclear cells in myopathies. I: Quantitation of subsets according to diagnosis and sites of accumulation and demonstration and counts of muscle fibers invaded by T cells. Ann Neurol. 1984 Aug;16(2):193–208. doi: 10.1002/ana.410160206. [DOI] [PubMed] [Google Scholar]
- 48.Fyhr IM, Moslemi AR, Lindberg C, Oldfors A. T cell receptor beta-chain repertoire in inclusion body myositis. J Neuroimmunol. 1998 Nov 2;91(1–2):129–134. doi: 10.1016/s0165-5728(98)00163-5. [DOI] [PubMed] [Google Scholar]
- 49.Cavazzana I, Fredi M, Selmi C, Tincani A, Franceschini F. The Clinical and Histological Spectrum of Idiopathic Inflammatory Myopathies. Clin Rev Allergy Immunol. 2017 Feb;52(1):88–98. doi: 10.1007/s12016-015-8517-4. [DOI] [PubMed] [Google Scholar]
- 50.Huang H, Wang C, Rubelt F, Scriba TJ, Davis MM. Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat Biotechnol. 2020 Apr 27:1–9. doi: 10.1038/s41587-020-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Dongen JJM, Langerak AW, Brüggemann M, Evans PAS, Hummel M, Lavender FL. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003 Dec;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 52.Brüggemann M, Kotrová M, Knecht H, Bartram J, Boudjogrha M, Bystry V. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia. 2019 Sep;33(9):2241–2253. doi: 10.1038/s41375-019-0496-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greenberg SA, Pinkus JL, Amato AA, Kristensen T, Dorfman DM. Association of inclusion body myositis with T cell large granular lymphocytic leukaemia. Brain. 2016 May 1;139(5):1348–1360. doi: 10.1093/brain/aww024. [DOI] [PubMed] [Google Scholar]
- 54.Greenberg SA, Pinkus JL, Kong SW, Baecher-Allan C, Amato AA, Dorfman DM. Highly differentiated cytotoxic T cells in inclusion body myositis. Brain J Neurol. 2019;142(9):2590–2604. doi: 10.1093/brain/awz207. 01. [DOI] [PubMed] [Google Scholar]
- 55.Shugay M, Bagaev DV, Zvyagin IV, Vroomans RM, Crawford JC, Dolton G. VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res. 2018 Jan 4;46(D1):D419–D427. doi: 10.1093/nar/gkx760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 2020 Jan 8;48(D1):D1057–D1062. doi: 10.1093/nar/gkz874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tickotsky N, Sagiv T, Prilusky J, Shifrut E, Friedman N. McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinforma Oxf Engl. 2017 Sep 15;33(18):2924–2929. doi: 10.1093/bioinformatics/btx286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.