Abstract
Ferroptosis is a newly defined form of cell death induced by iron-dependent accumulation of lethal lipid peroxidation. Ferroptosis represent a therapeutic strategy to suppress therapy-resistant cancer cells with more property of epithelial-mesenchymal transition (EMT). However, epigenetic reprogramming of EMT has been rarely studied in the context of ferroptosis susceptibility. Therefore, we examined the therapeutic potentiality of EMT epigenetic reprogramming in promoting ferroptosis in head and neck cancer (HNC) cells. The effects of ferroptosis inducers and EMT inhibition or induction were tested in HNC cell lines and mouse tumor xenograft models. These effects were analyzed concerning cell viability and death, lipid reactive oxygen species and iron production, labile iron pool, glutathione contents, NAD/NADH levels, and mRNA/protein expression. Cell density and the expression levels of E-cadherin, vimentin, and ZEB1 were associated with the different susceptibility to ferroptosis inducers. CDH1 silencing or ZEB1 overexpression increased the susceptibility to ferroptosis, whereas CDH overexpression or ZEB1 silencing decreased the susceptibility, in vitro and in vivo. Histone deacetylase SIRT1 gene silencing or pharmacological inhibition by EX-527 suppressed EMT and consequently decreased ferroptosis, whereas SIRT inducers, resveratrol and SRT1720, increased ferroptosis. MiR-200 family inhibitors induced EMT and increased ferroptosis susceptibility. In HNC cells with low expression of E-cadherin, the treatment of 5-azacitidine diminished the hypermethylation of CDH1, resulting in increased E-cadherin expression and decreased ferroptosis susceptibility. Our data suggest that epigenetic reprogramming of EMT contributes to promoting ferroptosis in HNC cells.
Keywords: Ferroptosis, Epithelial-mesenchymal transition, E-cadherin, ZEB1, Epigenetic reprogramming
Graphical abstract
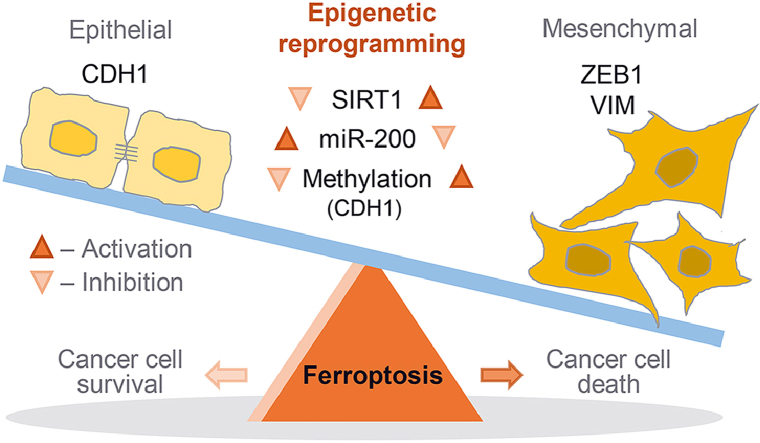
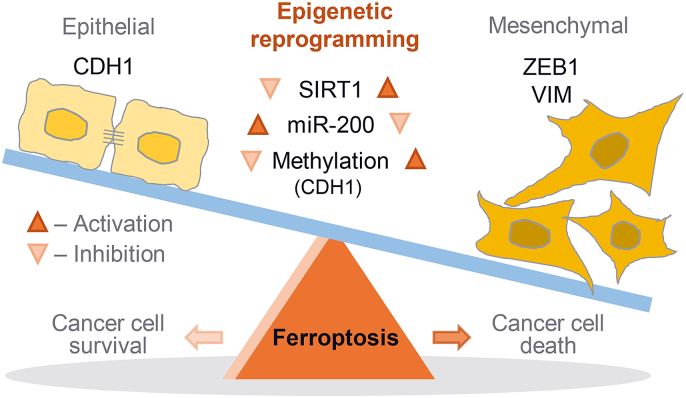
Epigenetic reprogramming of epithelial-mesenchymal transition (EMT) promotes ferroptosis of cancer cells. The EMT markers of E-cadherin (CDH1) and ZEB1 are closely related to ferroptosis sensitivity in cancer cells. Epigenetic reprogramming of EMT to gain a mesenchymal phenotype, such as SIRT1 activation or miR-200 family inhibition, promotes ferroptosis in HNC cells retaining relatively high epithelial traits and low ferroptosis sensitivity. However, 5-azacitidine induces CDH1 demethylation that contributed to reducing EMT and decreasing ferroptosis.
Highlights
-
•
The EMT markers of E-cadherin and ZEB1 were closely related to ferroptosis susceptibility.
-
•
Transition to epithelial traits by CDH overexpression or ZEB1 silencing decreased ferroptosis.
-
•
Gaining of mesenchymal traits by CDH1 silencing or ZEB overexpression increased ferroptosis.
-
•
SIRT1 activation or miR-200 family inhibition shifted HNC to EMT and increased ferroptosis.
-
•
5-Azacitidine induced CDH1 demethylation that contributed to reducing EMT and decreasing ferroptosis.
Condensed Abstract: Expression of epithelial-mesenchymal transition (EMT) markers was closely related to susceptibility to ferroptosis inducers. Epigenetic reprogramming of EMT to gain a mesenchymal phenotype, such as SIRT1 activation or miR-200 family inhibition, promoted ferroptosis in head and neck cancer cells retaining relatively high epithelial traits and low sensitivity to ferroptosis inducers.
Abbreviations
- CC
cystine;
- CCK-8
counting kit-8
- CDH1
cadherin-1
- EMT
epithelial-mesenchymal transition
- GPX4
glutathione peroxidase 4
- GSH
glutathione
- HNC
head and neck cancer
- LIP
labile iron pool
- miR
microRNA
- MSP
methylation-specific PCR
- NAD
nicotinamide adenine dinucleotide;
- PCR
polymerases chain reaction
- PI
propidium iodide;
- ROS
reactive oxygen species
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- siRNA
short-interfering RNA
- shRNA
short hairpin RNA
- SAS
sulfasalazine;
- SIRT1
sirtuin 1
- TGF-β
transforming growth factor-beta
- VIM
vimentin
- ZEB1
zinc finger E-box-binding homeobox 1
1. Introduction
Epithelial-mesenchymal transition (EMT) is a dynamic process that epithelial cells lose their junctions and polarity as changing their form easy to move. It is an essential process of epithelial cells to break free from their state of cell-to-cell and cell-to-extracellular matrix adhesion. Loss of epithelial traits allows cancer cells to easily invade adjacent tissues, destroy their functions, and spread to remote sites. Further, EMT endows cancer cells with chemoresistance reducing the efficiency of anti-cancer drugs [1]. Zinc finger E-box-binding homeobox 1 (ZEB1) is a key molecule of EMT playing a significant role in invasion and metastasis. ZEB1 downregulates E-cadherin known as an epithelial marker through a close relationship with SIRT1 [2]. ZEB1 also enhances the mesenchymal characteristics of cancer cells related to chemoresistance and leads to poor clinical outcomes in cancer patients [3].
Ferroptosis is a newly defined form of cell death that is induced by iron-dependent accumulation of lethal lipid peroxidation [4]. Ferroptosis shows distinct differences from other forms of cell death in terms of its molecular process and morphology [5]. Recent studies have shown that therapy-resistant or -persistent cancer cells are associated with a mesenchymal or metastatic property that is apt to be more susceptible to ferroptosis inducers [6,7]. Ferroptosis is also regulated by cadherin-mediated intercellular interaction: E-cadherin activates intracellular merlin and Hippo signaling pathway, resulting in suppressing ferroptosis [8]. This suggests that E-cadherin inhibition or EMT induction in cancer cells might contribute to enhance ferroptosis.
Ferroptosis can be controlled by epigenetic regulation. Histone demethylase KDM3B is a potential epigenetic regulator of ferroptosis by upregulating the expression of SLC7A11, a cystine-glutamate antiporter [9]. EMT can be also epigenetically modulated, which drives cellular plasticity to increase or decrease the sensitivity of chemotherapeutic agents with genetic or pharmacological control [10]. Previous studies have focused on the inhibition of EMT to enhance the effects of anti-cancer therapeutics [3,11]. Epigenetic reprogramming of EMT in cancer cells to gain more mesenchymal property might increase their susceptibility to ferroptosis inducers, which has been rarely studied. The present study has newly found the therapeutic possibility of EMT epigenetic reprogramming sensitizing head and neck cancer (HNC) cells to ferroptosis. Here, we examined the therapeutic potentiality of EMT reprogramming in promoting ferroptosis in HNC cells.
2. Materials and methods
2.1. Cell culture and reagents
Head and neck cancer (HNC) cell lines, namely AMC HN3, HN4, HN5, HN6, HN9, and HN10 [12], were used for our experiments. These cell lines were authenticated by short tandem repeat-based DNA fingerprinting and multiplex polymerase chain reaction (PCR). The cells were cultured in Eagle's minimum essential medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum at 37 °C in a humidified atmosphere containing 5% CO2. The cells were also cultured in the conditioned media with no cysteine and cystine (cyst(e)ine), with exposure to (1S, 3R)-RSL3 (Cayman Chemical Co., Ann Arbor, MI, USA), or sulfasalazine (Sigma-Aldrich).
2.2. Cell viability and death assays
The cells were subjected to cystine deprivation, RSL3, or sulfasalazine treatment for indicated dose and time. Control cells were cultured in cyst(e)ine-free media or exposed to RSL3, sulfasalazine, or an equivalent amount of dimethyl sulfoxide (DMSO). After exposure, cell viability was assessed using counting kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc., Tokyo, Japan) according to the manufacturer's protocol. The cells were incubated with the CCK-8 solution for 1 h, and then the cell viability was measured at the absorbance of 450 nm using a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Cell death after cyst(e)ine deprivation, RSL3, or sulfasalazine treatment was assessed via propidium iodide (PI) staining. Control cells were exposed to an equivalent amount of DMSO. The samples were washed three times with phosphate-buffered saline (PBS), followed by staining of cells in each plate with 2.5 μg/ml PI (Sigma-Aldrich, St. Louis, MO, USA) in PBS for 20 min. The stained cells were analyzed using a CytoFLEX flow cytometer equipped with CytExpert software (Beck Coulter, Brea, CA, USA) and observed using a ZEISS fluorescent microscope (Oberkochen, Germany). The mean PI-positive fractions were compared with those of the control group.
HN3 or HN4 cells were cultured with the indicated concentrations of SRT1720 for 24 h and then, was added with the indicated concentrations of RSL3, sulfasalazine, or DMSO. The combination matrix and deviation from the additive effect were calculated assuming a Loewe additivity model for compound interactions [13]: colors indicates synergistic (blue), additive (yellow), and less than additive (orange-red) effects.
2.3. Measurement of GSH synthesis and ROS production
Intracellular glutathione (GSH) levels in HNC cell lysates and cancer tissue lysates after harvesting tumors transplanted and grown in nude mice were measured using a GSH/GSSG assay kit (BioAssay Systems, Hayward, CA, USA). After RSL3 or sulfasalazine treatment, cellular reactive oxygen species (ROS) generation was measured by adding 10 μM 2ʹ,7ʹ-dichlorofluorescein diacetate (DCF-DA) (cytosolic ROS; Enzo Life Sciences, Farmingdale, NY, USA) or 5 μM C11-BODIPY C11 (lipid peroxidation; Thermo Fisher Scientific) for 30 min at 37 °C. The ROS levels were analyzed using a CytoFLEX flow cytometer (Beckman Coulter).
2.4. Labile iron pool assay
Labile iron pool (LIP) assay was measured by using calcein acetoxymethyl ester (Corning Inc., Corning, NY, USA) and iron chelator, deferoxamine (Abcam, Cambridge, UK). The cells were loaded with calcein (8 μg/ml) for 30 min at 37 °C and then washed with Hanks’ balanced salt solution without calcium and magnesium (HBSS) (Thermo Fisher Scientific). Deferoxamine was added at a final concentration of 100 μM to remove iron from calcein, causing dequenching. The change in fluorescence following the addition of deferoxamine was used as an indirect measure of the LIP. Fluorescence was measured at 485 nm excitation and 535 nm emission with a VICTOR X3 microplate reader (PerkinElmer, Waltham, MA, USA).
2.5. RNA interference and gene transfection
HN3, HN4, HN6, and HN9 cells were seeded for gene silencing or overexpression. Cells were transfected 24 h later with 10 nmol/L small-interfering RNA (siRNA) targeting human CDH1, ZEB1, SIRT1, or scrambled control siRNA (Integrated DNA Technologies, Coralville, IA, USA) using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific). To generate cells that overexpress CDH1 or ZEB1, HN3, HN4, HN6, and HN9 cells were seeded and stably transfected with a control plasmid (pBABE-puro, Addgene, Watertown, MA, USA), ZEB1, or CDH1 (Applied Biological Materials Inc., Richmond, BC, Canada) plasmid by using Lipofectamine 3000 reagent (Thermo Fisher Scientific). The levels of CDH1, ZEB1, and SIRT1 expression were confirmed via western blotting.
2.6. Reverse transcription-quantitative PCR, methylation-specific PCR, and immunoblotting
Cells were plated and grown with 70% confluence, and then treated with indicated drugs or not. Total RNA from HNC cells was extracted using an RNA extraction kit (Thermo Fisher Scientific) according to the manufacturer's instructions. A reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was conducted using SensiFAST™ SYBR® No-ROX Kit (Bioline International, Toronto, Canada) after performing cDNA synthesis using SensiFAST™ cDNA Synthesis Kit (Bioline International). CDH1, ZEB1, SIRT1, VIM, SLC7A11, and ACTB were amplified, and the relative target mRNA levels were determined using the 2−(ΔΔCt) method and normalized against ACTB mRNA levels.
Methylation-specific PCR (MSP) was conducted to measure the methylation level of genes. Cells were seeded and grown with 70% confluence. Genomic DNA from HNC cells was obtained using a genomic DNA extraction kit (Real Biotech Corporation), and then genomic DNA was converted into bisulfite form using a BisulFlash DNA Modification Kit (EpiGentek, Farmingdale, NY, USA). The level of methylation was confirmed by RT-qPCR using a Methylamp MS-qPCR Fast Kit (EpiGentek) and a ViiA™ 7 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
For immunoblotting, cells were lysed at 4 °C in a cell lysis buffer (Cell Signaling Technology, Danvers, MA, USA) with protease/phosphatase inhibitor cocktail (Cell Signaling Technology). A total of 20–25 μg protein was resolved by SDS-PAGE on 10%–15% gels; the resolved proteins were then transferred to nitrocellulose or polyvinylidene difluoride membranes and probed with primary and secondary antibodies. The following primary antibodies were used: E-cadherin (13-1700; Thermo Fisher Scientific), vimentin (SC-6260; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), ZEB1 (ab203829; Abcam, Cambridge, UK), SIRT1 (sc74465; Santa Cruz Biotechnology), xCT (ab37185; Abcam), Gpx4 (ab125066; Abcam), GSH (ab19534; Abcam), 4-HNE (MA5-27570; Thermo Fisher Scientific), PTGS2 (35-8200; Thermo Fisher Scientific) and Nrf2 (ab62352; Abcam). β-actin (BS6007 M; BioWorld, Atlanta, GA, USA) served as the total loading control. All antibodies were diluted to concentrations between 1:500 and 1:10000.
2.7. NAD/NADH ratio measurement
HN3, HN4, HN6, and HN9 cells were plated and grown with 70% confluence. Cancer tissues transplanted in nude mice were harvested, homogenized, and sonicated. Intracellular nicotinamide adenine dinucleotide (NAD)/NADH ratio of the cells and cancer tissues were measured using a NAD/NADH assay kit (Abcam) according to the manufacturer's instructions. NAD/NADH ratio was determined by subtracting NADH from total NAD followed by dividing the product by NADH. The relative quantities of the NAD/NADH ratio among groups were normalized against the control.
2.8. Tumor xenograft
All animal study procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC). Six-week-old athymic BALB/c male nude mice (nu/nu) were purchased from OrientBio (Seoul, Republic of Korea). HN9 cells with transfection of CDH1 or control vector or HN4 cells with ZEB1 or control vector were subcutaneously injected into the bilateral flank of nude mice. From the day when gross nodules were detected in tumor implants, mice were subjected to different treatments: vehicle or sulfasalazine (250 mg/kg daily per intraperitoneal route) [14]. Each group included six mice. Tumor size and weight of each mouse were measured twice a week, and tumor volume was calculated as (length × width2)/2. After the scarification of mice, tumors were isolated and analyzed by measuring GSH contents, NAD/NADH ratio, and molecular levels. The values were compared among differently treated tumors.
2.9. Statistical analysis
Data were presented as mean ± standard error of the mean. The statistically significant differences between the treatment groups were assessed using Mann–Whitney U test or analysis of variance (ANOVA) with Bonferroni post-hoc test. All statistical tests were two-sided and a P value of <0.05 was considered to be statistically significant. The statistical tests were performed using IBM SPSS Statistics version 22.0 (IBM, Armonk, NY, USA).
3. Results
3.1. Cell density and EMT markers are related to ferroptosis sensitivity
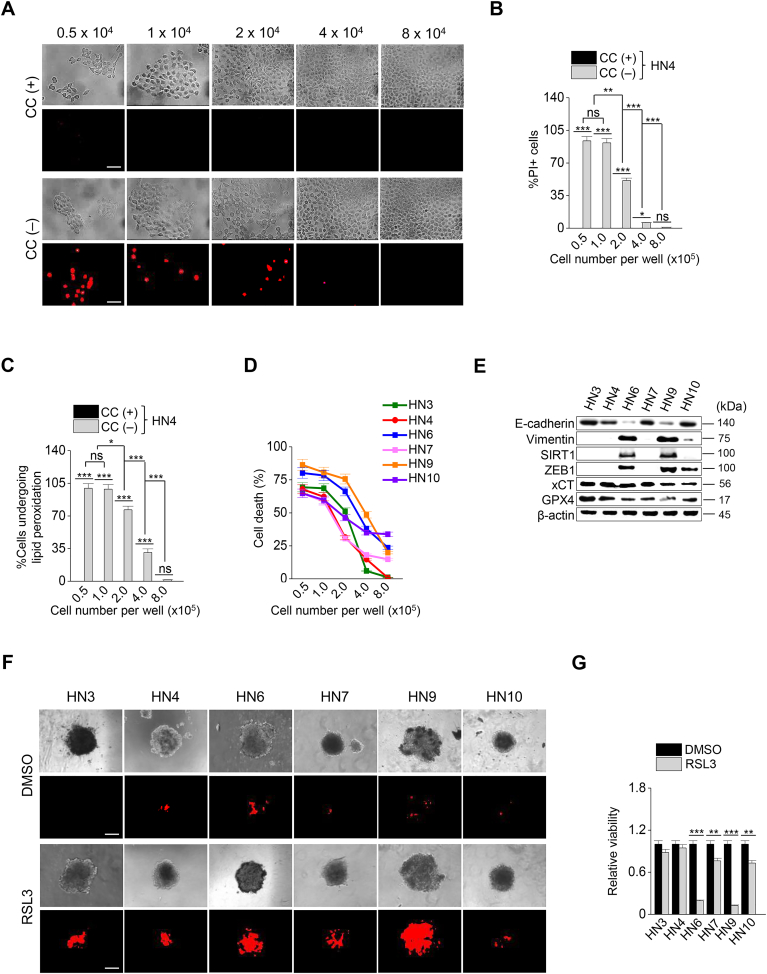
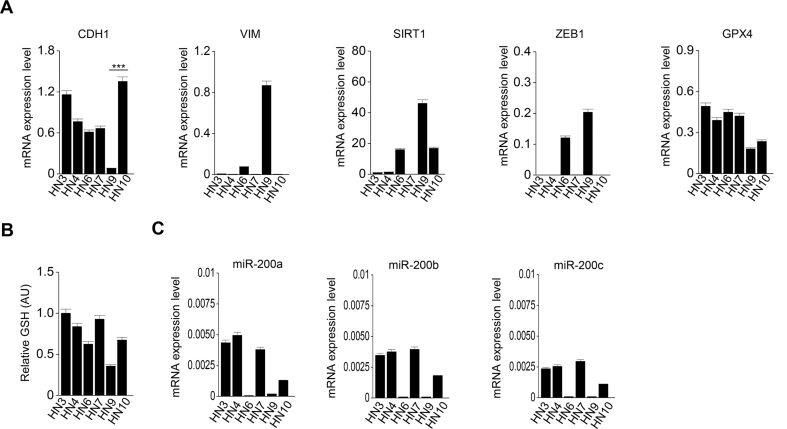
To determine whether cell density affects the sensitivity to ferroptosis in HNC cells, we first performed cell death assay in a culture condition of cyst(e)ine deprivation. Interestingly, PI-positive cell fractions significantly decreased in the HN4 cancer cells in a manner of the number of seeding cells per well (P < 0.01) (Fig. 1A and B). The levels of cellular lipid peroxidation also decreased along with the increasing population of HN4 (P < 0.01) (Fig. 1C). The results suggested that cell density was related to ferroptosis sensitivity. To further determine whether the same event occurred in other HNC cells, cell death was examined in six HNC cell lines. It was also confirmed that the denser cancer cells were, the more they survived in a condition of cyst(e)ine deprivation, as shown in HN4 cancer cells (Fig. 1D). Notably, three HNC cell lines, HN6, HN9, and HN10, showed higher percentages of cell death than others. Thus, we screened the expression of several proteins related to cell adhesion and EMT; E-cadherin, vimentin, ZEB1, and SIRT1. Immunoblotting and RT-qPCR results showed that the epithelial marker of E-cadherin was highly expressed in HN3, HN4, HN7, and HN10, whereas the mesenchymal markers of vimentin and ZEB1 were highly expressed in HN6 and HN9 (Fig. 1E and Supplementary Fig. S1A). Relative GSH contents and miR-200 family levels were higher in HN3, HN4, and HN7 cells than HN6, HN9, and HN10 cells (Supplementary Figs. S1B–C). This suggested that the expression level of EMT markers was correlated with the sensitivity to ferroptosis in cyst(e)ine-free media (Fig. 1D and E). Further, the association between cell-to-cell adhesion and ferroptosis was also examined in the spheroid culture of HNC cell lines. Cell death to exposure of RSL3, a ferroptosis inducer, was significantly higher in the HN6 and HN9 with mesenchymal traits than other HNC cell lines with epithelial traits (P < 0.001) (Fig. 1F and G). Taken together, these data suggested that the epithelial characteristics of cancer cells might be involved in the resistance to ferroptosis. In other words, the transition to gain a mesenchymal property might make cancer cells more vulnerable to ferroptosis.
Fig. 1.
Cell density and the expression of epithelial-mesenchymal transition (EMT) markers control ferroptosis. (A–C) Cyst(e)ine (CC) deprivation induced ferroptosis in head and neck cancer (HNC) cells. Cells were seeded in 5.0–8.0 × 105 cells per well. Cell viability and death were measured by counting kit-8 (CCK-8) and propidium iodide (PI) staining, respectively, and lipid ROS was measured using flow cytometry after cyst(e)ine deprivation for 30 h. Original magnification, × 200. Scale bar, 50 μm. The error bars represent standard errors from three technical replicates. NS indicates statistically non-significant, *P < 0.05, **P < 0.01, ***P < 0.001 between different cell density. (D) Cell death was measured in the six different HNC cell lines after cyst(e)ine deprivation for 30 h. (E) Western blots of E-cadherin, vimentin, SIRT1, ZEB1, xCT, and GPX4 expression in the six HNC cell lines. (F, G) Spheroids generated from the six HNC cell lines were cultured with 2 μM RSL3 for 72 h. Cell death was measured by PI staining. Original magnification, × 40. Scale bar, 50 μm.
3.2. Expression of E-cadherin regulates the induction of ferroptosis
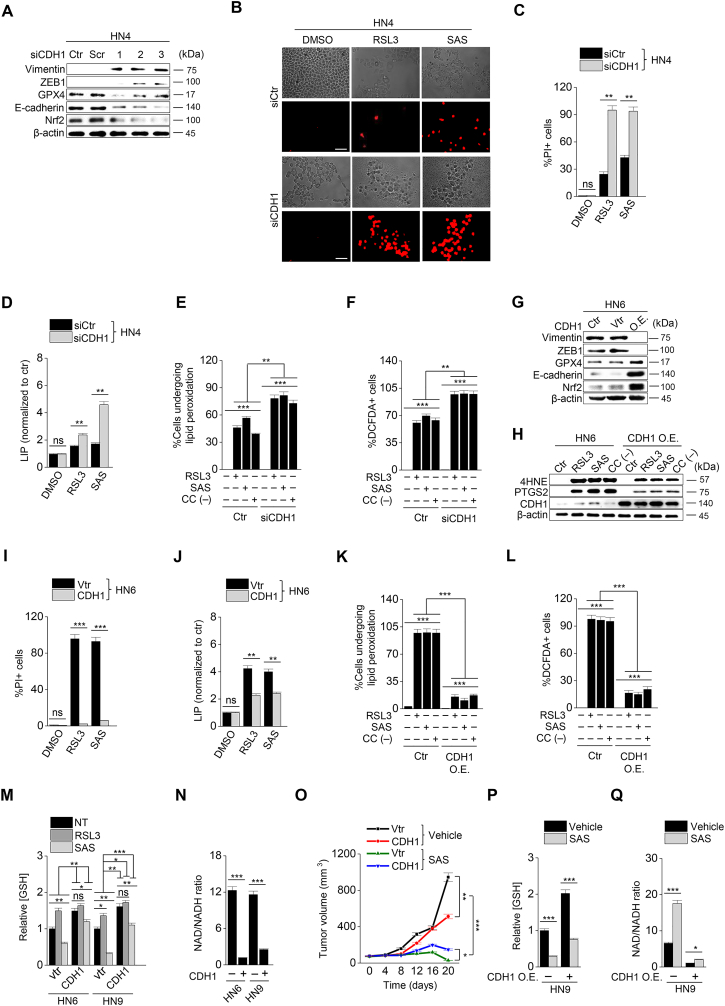
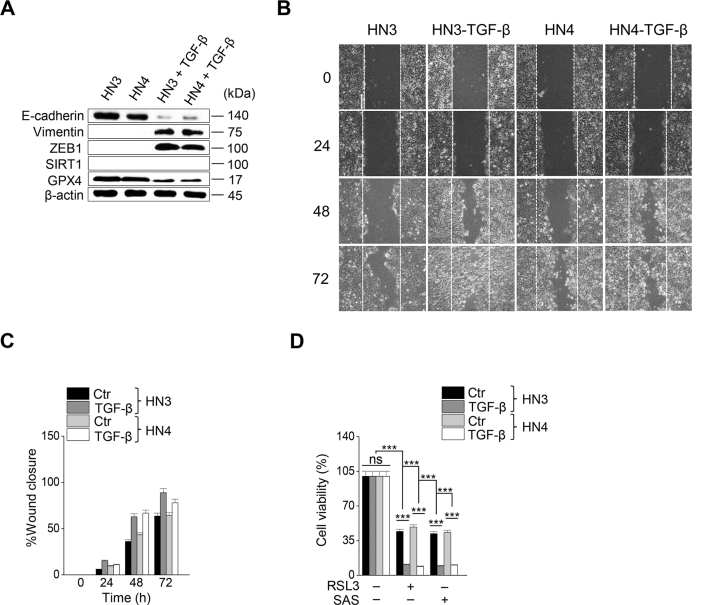
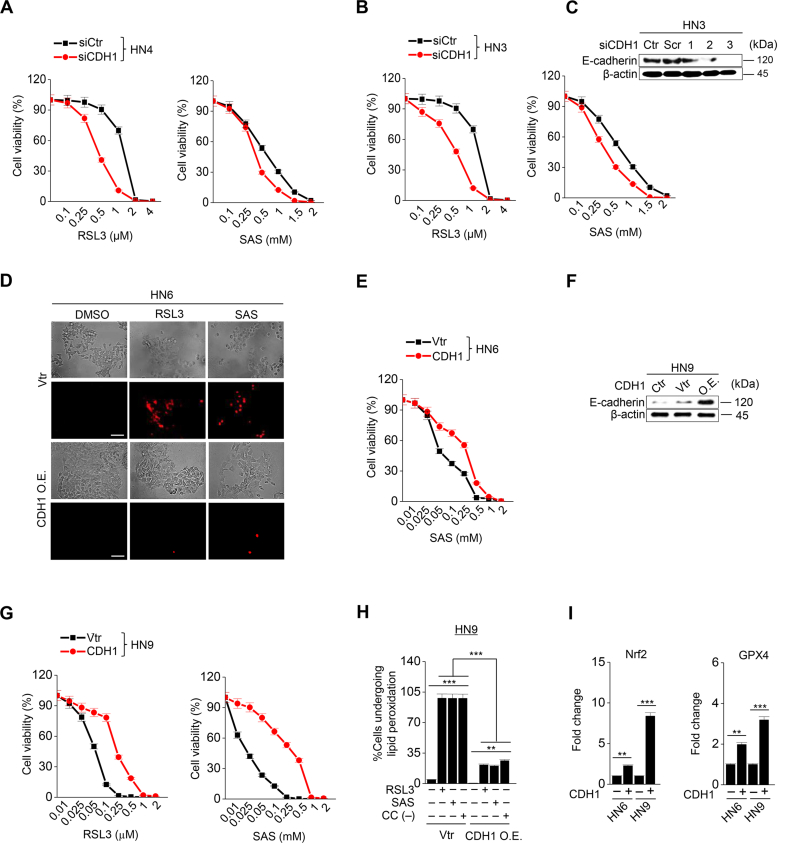
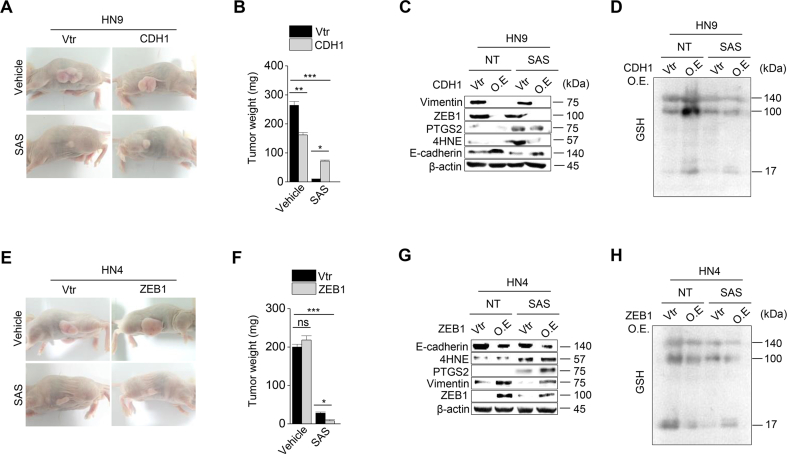
Next, we tested whether the regulation of mesenchymal expression made HNC cells susceptible to ferroptosis. First, transforming growth factor-beta (TGF-β) was treated to HN3 and HN4 cancer cells with epithelial traits to induce EMT (Supplementary Figs. S2A–C). Cell viability was significantly lower in the TGF-β plus RSL3 treatment group than in the only RSL3 treatment group (P < 0.001) (Supplementary Fig. S2D). Second, we inhibited CDH1 in HN3 and HN4 using an RNA silencing system. CDH1 silencing induced ZEB1 and vimentin expression (Fig. 2A). When HN3 or HN4 cells were treated with RSL3 or sulfasalazine, the silencing of CDH1 made the cells more vulnerable to the ferroptosis inducers. The inhibition of CDH1 led to a significant decrease in cell viability compared with the control (P < 0.001) (Supplementary Figs. S3A–C). Moreover, PI-positive cell fraction considerably increased in the CDH1-silenced group (P < 0.001) (Fig. 2B and C). The increased cell death enhanced by CDH1 silencing occurred in accompany with the increased level of labile iron in cancer cells exposed to RSL3 or sulfasalazine (Fig. 2D). Further, the fractions of cells undergoing lipid peroxidation and total ROS generation more increased in the CDH1-silenced cells than the control when treated with RSL3, sulfasalazine, or cyst(e)ine deprivation (P < 0.001) (Fig. 2E and F). Lastly, we examined the effect of CDH1 overexpression on the induction of ferroptosis in HNC cells with mesenchymal traits. Both HN6 and HN9 were transfected with CDH1 overexpression vector, and then cell death and ROS generation after exposure to RSL3, sulfasalazine, or cyst(e)ine deprivation were examined. CDH1 overexpression reduced ZEB1 and vimentin expression in HN6 cells (Fig. 2G). 4-HNE and PTGS2 expression increased when HN6 cells were treated with RSL3, sulfasalazine, or cyst(e)ine deprivation, but the increased level was reduced in the cells with CDH1 overexpression (Fig. 2H). The increase of the PI-positive fraction in the CDH1-overexpressing group was considerably less than in the vector control when treated with RSL3 or sulfasalazine (P < 0.001) (Fig. 2I and Supplementary Fig. S3D). The cell viability after exposure to RSL3 or sulfasalazine also significantly increased in HN6 and HN9 cells with CDH1 overexpression (Supplementary S3E–G). The increase in intracellular labile iron in the CHD1-overexpressing cancer cells was also significantly less than the vector control (P < 0.01) (Fig. 2J). The increase in cellular lipid peroxidation or cytosolic ROS generation was also less than the vector control (P < 0.001) (Fig. 2K–L and Supplementary Fig. S3H). According to a previous report [6], we performed GSH and NAD/NADH assays to examine the efficiency of antioxidant systems. The level of intracellular GSH in the CDH1 overexpression group increased by that cellular antioxidant systems were enhanced (Fig. 2M). On the contrary, the NAD/NADH ratio of the CDH1 overexpression group decreased more than that of the vector control (Fig. 2N). These suggest that CDH1 overexpression in HN6 and HN9 might enhance the antioxidant systems of cancer cells with mesenchymal traits (Supplementary Fig. 3I). Based on the in vitro findings, we expanded to the in vivo experiments by using a model of mice with transplantation of CDH1 or vector-transfected HN9 and then exposure to sulfasalazine or vehicle. Tumor volume was less significantly reduced in the CDH1 overexpression than the vector control by the treatment of sulfasalazine (Fig. 2O and Supplementary Figs. S4A–B). The GSH content decreased but NAD/NADH ratio increased by sulfasalazine treatment (Fig. 2P–Q and Supplementary Figs. S4C–D). Taken together, the results suggested that the control of E-cadherin expression in cancer cells was able to modulate the susceptibility of ferroptosis.
Fig. 2.
Inhibition of CDH1 increases the susceptibility of HNC cells to ferroptosis inducers. (A) Immunoblotting of vimentin, ZEB1, GPX4, Nrf2, and E-cadherin with or without CDH1 silencing in HN4 cancer cells. Ctr, control; scr, scrambled. (B–C) Cell death was measured using PI staining, in HN4 with or without CDH1 gene silencing and exposure to 1 μM RSL3 or 0.5 mM sulfasalazine (SAS) treatment for 72 h. Original magnification, × 200. Scale bar, 50 μm. (D) Labile iron pool (LIP) was measured by calcein-AM (8 μg/ml) after 1 μM RSL3 or 0.5 mM SAS; all data were quantified by the Image J software. (E,F) Lipid and cytosolic ROS were measured using flow cytometry with staining BODIPY C11 and DCFDA, respectively, after 1 μM RSL3 or 0.5 mM SAS treatment, or cyst(e)ine deprivation for 24 h. (G) Immunoblotting of vimentin, ZEB1, GPX4, Nrf2, and E-cadherin with or without transfection of CDH1 overexpression or control vector. (H) Immunoblotting of 4-HNE, PTGS2, and CDH1 in HN6 cells with or without CDH1 overexpression. (I–L) PI staining, LIP assay, lipid peroxidation (BODIPY C11), and total ROS (DCFDA) measurements in HN6 cells transfected with or without CDH-1 overexpression and then exposure to 1 μM RSL3 or 0.5 mM SAS, or cyst(e)ine deprivation. Scale bar 50 μm. The error bars represent standard errors from three replicates. **P < 0.01, ***P < 0.001 between the control and CDH1 overexpression. (M–N) Tumor volume, glutathione (GSH) content, and NAD/NADH ratio were measured in HN6 and HN9 cells with CDH1 overexpression or control vector. (O–Q) Tumor volume, GSH, and NAD/NADH ratio were assessed in HN9 tumors with or without CDH1 overexpression that were transplanted and grown in nude mice. *P < 0.05, **P < 0.01, ***P < 0.001 between the vector control and CDH overexpression or between the vehicle control and SAS treatment groups.
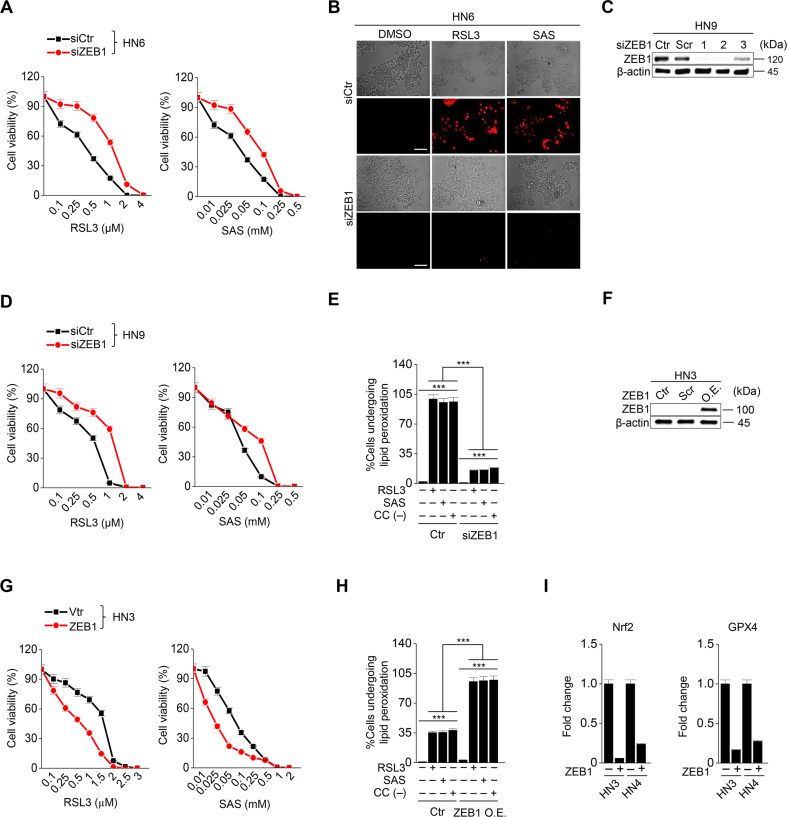
3.3. Expression of ZEB1 alters cancer cell susceptibility to ferroptosis
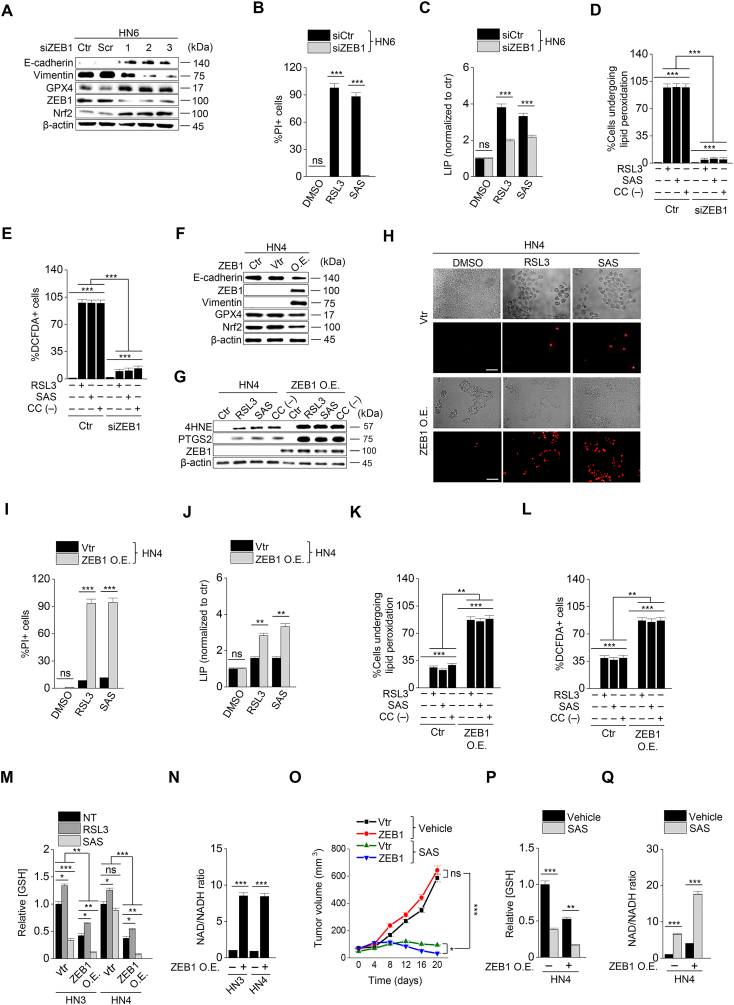
Considering that ZEB1 is a key molecule to induce EMT [15], we confirmed whether the regulation of ZEB1 expression resulted in the same as that of CDH1. First, ZEB1 was silenced in HN6 and HN9 cells with the transfection of ZEB1 siRNA (Fig. 3A). ZEB1 silencing induced E-cadherin expression and inhibited vimentin (Fig. 3A). After exposure to RSL3 or sulfasalazine, the PI-positive cell fraction of cancer cells considerably less increased but cell viability less decreased in the ZEB1-silenced cells compared with those of the control (P < 0.001) (Fig. 3B and Supplementary Fig. 5A–D). The increases in LIP, lipid peroxidation, and cytosolic ROS generation in the ZEB1-silenced cancer cells were also less than the control when treated with RSL3, sulfasalazine, or cyst(e)ine deprivation (P < 0.001) (Fig. 3C–E and Supplementary Fig. S5E). Second, ZEB1 was overexpressed in HN3 and HN4 cells with the transfection of ZEB1 cDNA. As previously reported [16], the overexpression of ZEB1 in HN4 cells induced vimentin expression and inhibited E-cadherin (Fig. 3F).). 4-HNE and PTGS2 expression increased when HN4 cells were treated with RSL3, sulfasalazine, or cyst(e)ine deprivation, but the increased level was enhanced in the cells with ZEB1 overexpression (Fig. 3G). ZEB1-overexpressing cells showed significantly more increase of PI-positive cells and more decrease of cell viability compared with the control when treated with RSL3 or sulfasalazine (P < 0.01) (Fig. 3H and I and Supplementary Figs. S5F–G). Also, the amounts of LIP, lipid ROS, and cytosolic ROS after RSL3, sulfasalazine, or cyst(e)ine deprivation increased significantly more than the control (P < 0.01) (Fig. 3J–L and Supplementary Fig. S5H). In HN3 and HN4, the level of cellular GSH was lower in the ZEB1 overexpression group than the vector control when exposed to RSL3 or sulfasalazine (Fig. 3M). Moreover, the cellular NAD/NADH ratio was higher in the ZEB1 overexpression group than in the vector control because the antioxidant function was weakened (Fig. 3N and Supplementary Fig. 5I). Furthermore, the results of in vivo experiment showed that tumor volume was reduced in the ZEB1 overexpression group more than the vector control when treated with sulfasalazine (Fig. 3O and Supplementary Figs. S4E–F). The GSH contents decreased and NAD/NADH ratio increased by sulfasalazine treatment in both ZEB1 overexpression and control groups (Fig. 3P and Q and Supplementary Figs. S4G–H). Taken together, the results suggested that the regulation of ZEB1 in cancer cells was able to modulate the susceptibility of ferroptosis.
Fig. 3.
ZEB-1 regulates ferroptosis sensitivity. (A) Immunoblotting of E-cadherin, vimentin, GPX4, Nrf2, and ZEB1 in HN6 cells with or without ZEB1 silencing. (B–E) Cell death, labile iron pool (LIP), lipid peroxidation, and total ROS assays were measured in HN6 cancer cells with or without ZEB1 gene silencing after 1 μM RSL3 or 0.5 mM SAS or cyst(e)ine deprivation. (F) Immunoblotting of E-cadherin, ZEB1, vimentin, Nrf2 and GPX4 in HN4 cells with or without ZEB1 overexpression. (G) Immunoblotting of 4-HNE, PTGS2, and ZEB1 in HN4 cells with or without ZEB1 overexpression. (H–L) Cell death, LIP, lipid peroxidation, and total ROS assays were also measured in HN4 cancer cells with or without ZEB1 overexpression after exposure to 1 μM RSL3 and 0.5 mM SAS. Original magnification, × 200. Scale bar, 50 μm. The error bars represent standard errors from three technical replicates. **P < 0.01, ***P < 0.001 between the control and ZEB1 overexpression. (M–N) GSH content and NAD/NADH ratio were measured in HN3 and HN4 cells with ZEB1 overexpression or control vector. (O–Q) Tumor volume, GSH content, and NAD/NADH ratio were assessed in HN4 tumors with or without ZEB overexpression that were transplanted and grown in nude mice. *P < 0.05, **P < 0.01, ***P < 0.001 between the vector control and ZEB1 overexpression or between the vehicle control and SAS treatment groups.
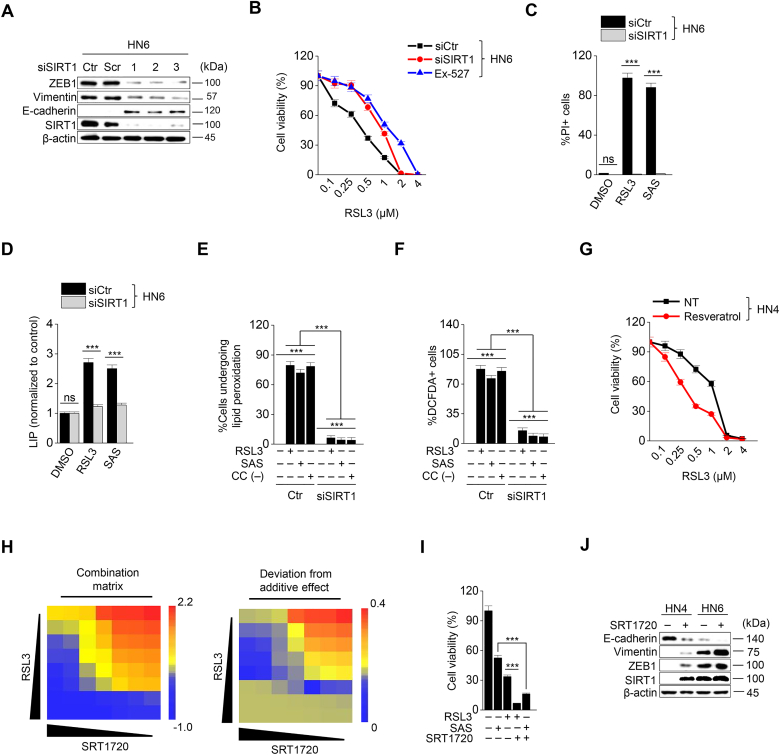
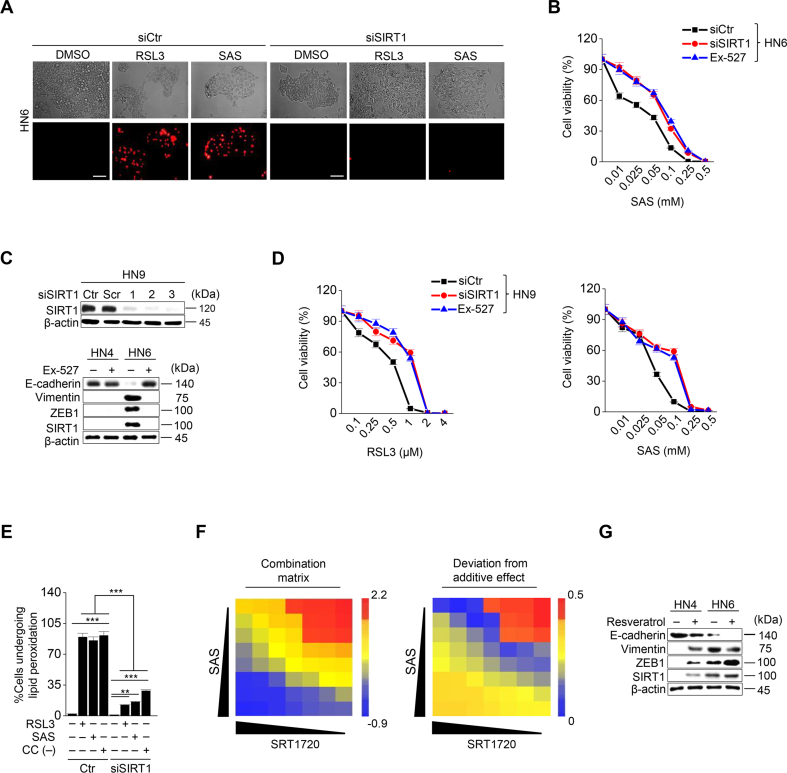
3.4. SIRT1 activation promotes ferroptosis in HNC cells
Next, we examined whether epigenetic reprogramming of EMT in cancer cells, particularly targeting ZEB1, regulated ferroptosis. Since histone deacetylase SIRT1 is known to modulate EMT by activating the function of ZEB1 [16,17], we tested the effect of SIRT1 inhibition and activation on ferroptosis. SIRT1 was genetically inhibited by an RNA silencing tool or pharmacologically by Ex-527, a SIRT1 specific inhibitor, in HN6 and HN9 cancer cells with mesenchymal traits. SIRT1 silencing or Ex-527 treatment decreased ZEB1 and vimentin expression but increased E-cadherin expression (Fig. 4A and Supplementary Fig. S6C). Cell viability significantly increased and cell death decreased in cancer cells with SIRT1 silencing or Ex-527 treatment compared with the control after exposure to RSL3 or sulfasalazine (P < 0.01) (Fig. 4B and C and Supplementary Figs. S5A–D). Moreover, labile iron as well as the amount of lipid ROS and cytosolic ROS increased less than the control cells (Fig. 4D–F and Supplementary Fig. S6E). However, resveratrol, a SIRT1 inducer, significantly decreased the survival of HN3 and HN4 when the cells treated with RSL3 or sulfasalazine (P < 0.01) (Fig. 4G). In HN3 and HN4 with epithelial traits, the combination of RSL3 or sulfasalazine with SRT1720, a SIRT1 specific inducer, significantly decreased cell viability more than the control with the increased combination matrix (P < 0.01), particularly in the combination of high concentrations (Fig. 4H and I and Supplementary Fig. S6F). SIRT1720 or resveratrol treatment increased ZEB1 and vimentin expression but decreased E-cadherin expression (Fig. 4J and Supplementary Fig. S6G). Taken together, the results suggested that the control of SIRT1 in HNC cancer cells may have an influence on the induction of ferroptosis by a weakened anti-oxidant ability.
Fig. 4.
SIRT1 activation promotes ferroptosis. (A) Immunoblotting of ZEB1, vimentin, E-cadherin, and SIRT1 in HN6 cancer cells with or without SIRT1 silencing. (B) Cell viability in HN6 with or without SIRT1 silencing or exposure to 10 μM Ex-527 was measured by CCK-8 assay after RSL3 treatment for 72 h. (C–F) Cell death, LIP, lipid peroxidation, and total ROS generation were examined in HN6 cancer cells with or without SIRT1 silencing after 1 μM RSL3, 0.5 mM SAS, or cyst(e)ine deprivation. The error bars represent standard errors from three technical replicates. ***P < 0.001 between the control and SIRT1 silencing. (G) The combination effect of resveratrol and RSL3 was measured in HN4 cancer cells by cell viability using CCK-8 assay after RSL3 treatment plus or minus 20 μM resveratrol for 72 h. (H) The combination matrix was created through the cell viability of HN4 cells using SRT1720 and RSL3 for 72 h. (I) Cell viability was measured using CCK-8 assay in HN4 cancer cells with exposure to 1 μM RSL3 and 0.5 mM SAS in combination with or without 5 μM SRT1720 for 72 h. (J) Immunoblotting of E-cadherin, vimentin, ZEB1, and SIRT1 in HN4 and HN6 cells treated with or without SRT1720.
3.5. MiR-200 family inhibitors increase ferroptosis susceptibility
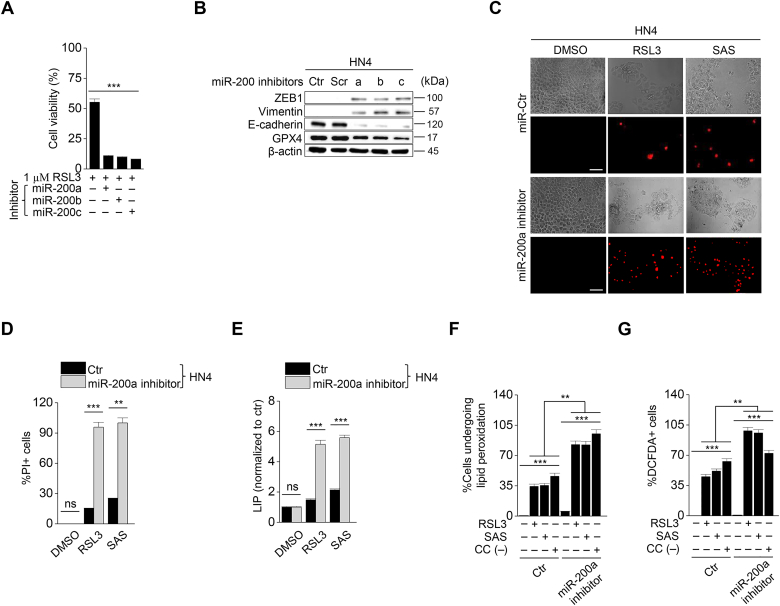
Considering various mechanisms to induce EMT, we additionally used miR-200 family inhibitors to induce EMT. The miR-200 family is known to inhibit EMT by repressing ZEB1 [18,19]. Therefore, we examined whether inhibition of miR-200 family caused high sensitivity to ferroptosis by diverting cancer cells to EMT tendency. To confirm the effect of miR 200 inhibition on ferroptosis, we first transfected miR-200 family inhibitors (Fig. 5A) and then examined cell viability, LIP, and ROS generation after exposure to RSL3, sulfasalazine, or cyst(e)ine deprivation in HNC cells. MiR-200 family inhibitors increased ZEB1 and vimentin expression but decreased E-cadherin expression (Fig. 5B). The cell viability of HN3 and HN4 significantly decreased, whereas the PI-positive cell fraction increased more in the miR-200a-inhibiting cells than in the control (P < 0.01) (Fig. 5C and D). Levels of intracellular labile iron, lipid peroxidation, and cytosolic ROS significantly increased in the cells with a miR-200a inhibitor than the control (Fig. 5E–G). Taken together, the results suggested that EMT induced by miR-200 family inhibitors in cancer cells might enforce the increased susceptibility to ferroptosis.
Fig. 5.
Suppression of miR-200 family increases the sensitivity to ferroptosis. (A) Cell viability in HN4 with or without miR-200a, b, c transfection was measured using CCK-8 assay after 1 μM RSL3 treatment for 72 h. (B) Immunoblotting of ZEB1, vimentin, E-cadherin, and GPX4 in HN4 cancer cells with or without inhibition of miR-200 family. (C–G) Cell death, LIP, lipid peroxidation, and total ROS generation were measured in HN4 cancer cells after 1 μM RSL3, 0.5 mM SAS, or cyst(e)ine deprivation. Original magnification, × 200. Scale bar, 50 μm. The error bars represent standard errors from three technical replicates. **P < 0.01, ***P < 0.001 between the control and miR-200a silencing.
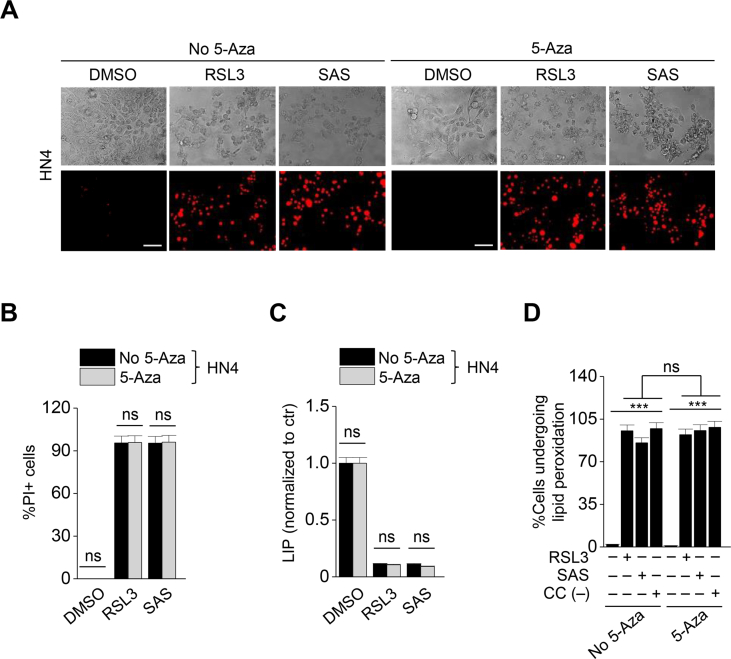
3.6. 5-Azacitidine diverts to an epithelial property and weakens ferroptosis susceptibility
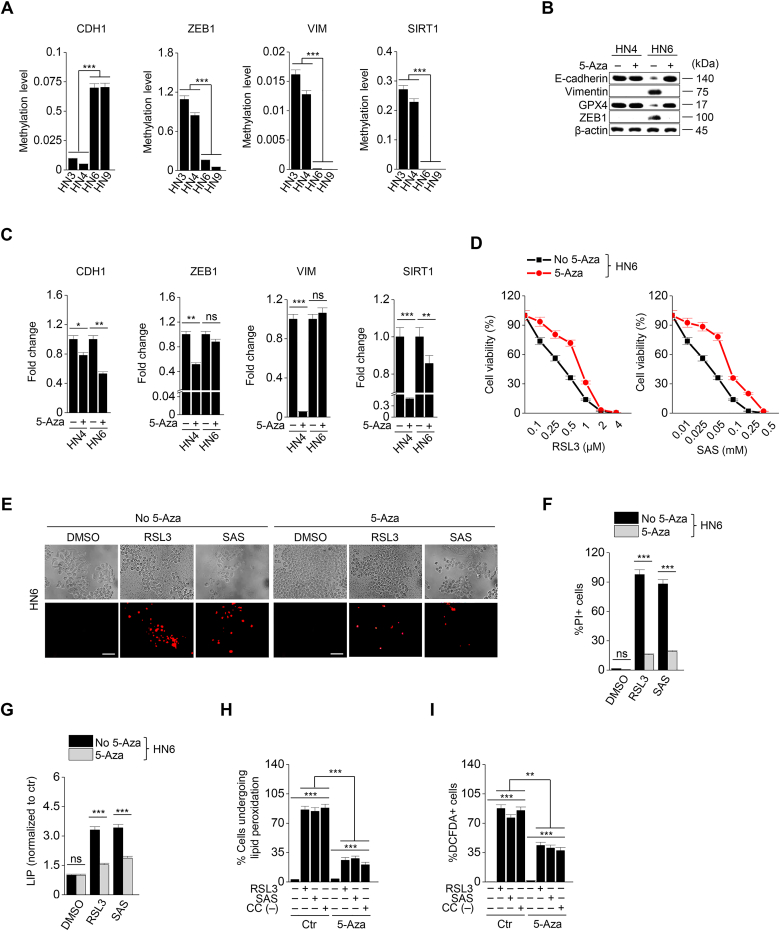
Taking into account that EMT is epigenetically regulated, we performed methylation-specific PCR to confirm the methylation levels of EMT-related genes. The methylation levels of genes were the opposite of the expression of proteins that were shown in Fig. 1E. The methylation level of CDH1 was relatively high in HN6 and HN9 with mesenchymal traits, while those of ZEB1, VIM, and SIRT1 were relatively high in HN3 and HN4 with epithelial traits (Fig. 6A). Next, we examined the effect of demethylation on inducing ferroptosis. A demethylating agent, 5-azacitidine is known to prevent EMT [18]. 5-Azacitidine was treated to HN4 and HN6 with RSL3 or sulfasalazine. 5-azacitidine treatment induced the decreased methylation level of CDH1 in HN6 cells and increased ZEB1 and vimentin expression (Fig. 6B and C). This also resulted in a decrease in ferroptosis to RSL3 or sulfasalazine treatment (Fig. 6D–F). Moreover, the increases of labile iron, lipid peroxidation, and cytosolic ROS generation after exposure to RSL3, sulfasalazine, or cyst(e)ine deprivation were lessened when co-treated with 5-azacitidine in HN6 cells (Fig. 6G–I). However, the levels of cell death and viability, labile iron, and lipid peroxidation did not significantly differ between HN4 cells (predominantly epithelial maker expression) with and without 5-azacitidine treatment when exposed to RSL3 or sulfasalazine (Supplementary Figs. S7A–D) (P > 0.05). Taken together, the data suggested that 5-azacitidine might decrease the sensitivity of ferroptosis inducers by demethylating an epithelial marker, CDH1.
Fig. 6.
5-Azacitidine decreases the sensitivity of ferroptosis inducers. (A) Methylation-specific PCR (MSP) of ZEB1, CDH1, VIM, and SIRT1 was performed in HN3, HN4, HN6, and HN9 cancer cells, and the relative target methylation levels were quantified by the 2−(ΔΔCt) method. (B) Immunoblotting of E-cadherin, vimentin, ZEB1, and GPX4 in HN6 cells treated with or without 5-azacitidine (5-aza). (C) The methylation level was measured by using MSP after 5 μM 5-aza for 48 h, and the methylation levels were quantified relative to the control. (D) Cell viability in HN6 with or without 5 μM 5-aza treatment was measured by CCK-8 assay after RSL3 or SAS for 72 h. (E–I) Cell death, LIP, lipid peroxidation, and total ROS generation were measured in HN6 cancer cells with or without 5 μM 5-aza after 1 μM RSL3, 0.5 mM SAS, or cyst(e)ine deprivation. Original magnification, × 200. Scale bar, 50 μm. The error bars represent standard errors from three technical replicates. **P < 0.01, ***P < 0.001 between the control and 5-aza treatment group.
4. Discussion
The present study showed that the shifting of HNC cells to gain a motile mesenchymal phenotype promoted ferroptosis susceptibility (Fig. 7). Cancer cells with an epithelial property or a dense cellular population had relatively less sensitivity to ferroptosis than those with a mesenchymal property or a low cellular density. Decreased E-cadherin expression or increased ZEB1 expression in cancer cells led to increase the sensitivity to ferroptosis. Epigenetic reprogramming of EMT, such as SIRT1 induction or miR-200 family inhibition, resulted in shifting cancer cells to have a mesenchymal property and increase the susceptibility to ferroptosis inducers. On the contrary, 5-azacitidine induced the demethylation of CDH1, resulting in holding epithelial traits and decreased the sensitivity to ferroptosis. Therefore, our study showed a new therapeutic potentiality of EMT reprogramming to promote ferroptosis in cancer cells.
Fig. 7.
An illustration showing the epigenetic reprogramming of EMT for promoting ferroptosis. The EMT markers of E-cadherin (CDH1) and ZEB1 were closely related to ferroptosis sensitivity in cancer cells. Epigenetic reprogramming of EMT to gain a mesenchymal phenotype, such as SIRT1 activation or miR-200 family inhibition, promoted ferroptosis in HNC cells retaining relatively high epithelial traits and low ferroptosis sensitivity. However, 5-azacitidine induced CDH1 demethylation that contributed to reducing EMT and decreasing ferroptosis.
E-cadherin expression in cancer cells is closely related to the decreased sensitivity to ferroptosis in a manner dependent on cell density. In epithelial cells, E-cadherin suppresses ferroptosis by mediating intracellular interaction with merlin and Hippo signaling, whereas antagonizing the E-cadherin-mediated signaling axis promotes ferroptosis by allowing yes-associated protein (YAP) [8]. In non-epithelial cells, cell density also affects the susceptibility to ferroptosis by regulating the Hippo-YAP/TAZ pathway and epithelial membrane protein 1 [20]. This suggests that regulation of the epithelial markers and relevant pathways might modulate the sensitivity of ferroptosis inducers in cancer cells of both epithelial and non-epithelial origins. Our data also supported the previous findings showing the relationship between ferroptosis and cell density or E-cadherin expression in HNC cells. HNC commonly arise in the epithelium of the upper aerodigestive tract. Epithelial cancers are relatively less dependent on lipid peroxidase pathway (GPX4) and less sensitive to ferroptosis inducers than other types of human cancers in a mesenchymal state [7]. Since HNC frequently expresses epithelial markers, the downregulation of CDH1 increases ferroptosis susceptibility, while the overexpression of CDH1 in HNC cells decreases ferroptosis susceptibility. Our results underline the role of CDH1 as a defender in ferroptosis and the potential application of a therapeutic strategy using CDH1 suppression to promote ferroptosis in cancer cells.
ZEB1 is a crucial element controlling the transition from an epithelial to a mesenchymal state [21]. The EMT process activated by ZEB1 mitigates the sensitivity to conventional and targeted therapies in cancer cells [3]. High mesenchymal cell state in human cancers is commonly associated with therapy resistance but prompts a vulnerability to ferroptosis [6,7]. Drug-tolerant persister cancer cells are characterized by the increased expression of mesenchymal and stem cell markers and a disabled antioxidant program that can be vulnerable to GPX4 inhibition [6]. High expression of ZEB1 correlates with the sensitivity to GPX4 inhibitors, such as RSL3, ML210, and ML162 [7]. TGF-β-induced ZEB1 activation drives a mesenchymal state in cancer cells with epithelial traits while increasing the susceptibility to GPX4 inhibitors and statins [7]. The present study showed that ZEB1 inhibition had the same effects as CDH1 overexpression, lessening ferroptosis susceptibility. In contrast, the mesenchymal transition by ZEB1 overexpression in HNC cells with an epithelial property increased the ferroptosis susceptibility. Our results imply that EMT regulation in cancer cells is a promising therapeutic strategy to promote the anticancer effects of ferroptosis inducers.
Cellular plasticity involving EMT can be driven by the interplay between epigenetic regulators and EMT transcriptional factors [18]. Three main families of EMT-transcriptional factors, SNAIL, TWIST, and ZEB interact with proteins involving in several layers of epigenetic modification: histone modifications, RNA interference, and DNA methylation [18]. NAD-dependent deacetylase sirtuin 1 promotes EMT process and metastasis via a SIRT1/ZEB1-positive feedback loop [22]. The present study showed that genetic or pharmacological inhibition of SIRT1 decreased ZEB1 expression and EMT phenotype, mitigating the sensitivity to ferroptosis inducers. Resveratrol, a type of natural phenol, can induce the SIRT1 signaling [23]. The present study also confirmed the pharmacological activation of SIRT1 and its relevant mesenchymal phenotype that causes the increased sensitivity to ferroptosis inducers. Of multiple non-coding RNAs controlling EMT, members of the miR-200 family induce epithelial differentiation and suppress invasion and metastasis via a miR-200/ZEB-negative feedback loop [24,25]. In the context of ferroptosis, the present study showed that the inhibition of miR-200 family members induced transition to a motile mesenchymal phenotype and increased the sensitivity to ferroptosis inducers. The core EMT decision-making circuit with SIRT1 or miR-200/ZEB1 might be modulated through GRHL2, a phenotypic stability factor of hybrid epithelial-mesenchymal phenotype [26,27]. Focal hypermethylation of the CpG islands in the CDH1 promoter is often observed in cancer cells with a mesenchymal phenotype [28]. 5-Azacytidine blocks the activity of DNA methyltransferase, leading to hypomethylation and gene de-repression and preventing EMT [29,30]. The present study also indicates the role of 5-azacytidine inhibiting EMT by the de-repression of CDH1, resulting in the decreased ferroptosis susceptibility in HNC cells with a mesenchymal phenotype.
Modifications of a variety of other epigenetic regulators involving EMT might contribute to the changes of ferroptosis susceptibility, which might be elucidated by further studies. Nonetheless, our study has appealed the necessity of epigenetic reprogramming of EMT to promote the anticancer effects of ferroptosis inducers. The induction of EMT is still a major problem to cause the increased probability of cancer invasion and metastasis. This might be solved by a therapeutic strategy of epigenetic reprogramming transiently shifting to reversible EMT to boost up ferroptosis in epithelial cancer cells with relatively high E-cadherin expression and low sensitivity to ferroptosis inducers. This needs further investigations for improving the therapeutic success of ferroptosis induction in resistant epithelial cancers.
5. Conclusion
This study suggests that the cell density and expression of epithelial-mesenchymal markers are closely related to ferroptosis susceptibility. These might be useful biomarkers to predict the sensitivity of ferroptosis inducers in cancer cells. Induction to gain a mesenchymal phenotype leads to promote ferroptosis in HNC cells retaining relativity high epithelial traits and low sensitivity to ferroptosis inducers. Epigenetic reprogramming of EMT contributes to promoting the ferroptosis susceptibility in HNC cells, which might be recommended as a promising combination therapy in combating cancers resistant to ferroptosis.
Author contributions
J.L. and J.-L.R. conceived and designed the experiments. J.L., J.H.Y., M.-S.K., and J.-L.R. performed the experiments. J.L. and J.-L.R. analyzed the data. J.L., J.H.Y., and M.-S.K. contributed reagents/materials/analysis tools; J.L. and J.-L.R. wrote the draft, and checked and revised. All authors approved to submit this version to this publication.
Declaration of competing interest
All authors declare no conflict of interests.
Acknowledgements
This study was supported by the National Research Foundation of Korea (NRF) grant, funded by the Ministry of Science and ICT (MSIT), The Government of Korea (No. 2019R1A2C2002259).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101697.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
The mRNA expression levels of epithelial-mesenchymal transition (EMT) or ferroptosis-related genes (A), glutathione (GSH) content (B), and mRNA levels of miR-200 family (C) in head and neck cancer (HNC) cells. CDH1, VIM, SIRT1, ZEB1, GPX4, and miR-200a, b, c were amplified from six HNC cell lines, and the relative target mRNA levels were determined using the 2−(ΔΔCt) method and normalized against ACTB mRNA levels.
Fig. S2.
TGF-β promotes the susceptibility to ferroptosis inducers by inducing EMT in cancer cells. (A) Immunoblotting of E-cadherin, vimentin, ZEB1, SIRT1 and GPX4 in HN3 and HN4 cells with or without TGF-β treatment. (B–D) Wound closure and cell viability in HN3 and HN4 cells with or without treatment of TGF-β (10 ng/ml) and then exposed to ferroptosis inducers, 1 μM RSL3 and 0.5 mM sulfasalazine (SAS). Original magnification, × 100. Scale bar 100 μm. The error bars represent standard errors from three technical replicates. ***P < 0.001 between the control (ctr) and TGF-β treatment.
Fig. S3.
CDH1 silencing enhances ferroptosis in HNC cells. (A) Cell viability of HN4 cells with or without CDH1 silencing. The cells were transfected with CDH siRNA or scrambled control and then exposed to after RSL3 or SAS for 72h. (B–C) Cell viability and immunoblotting of E-cadherin in HN3 with or without CDH1 silencing and exposure to RSL3. (D–E) Cell death and viability of HN6 cells with or without CDH1 overexpression. The cells were transfected with CDH1 cDNA or control vector (vtr) and then exposure to RSL3 or SAS for 72 h. Original magnification, × 200. Scale bar, 50 μm. (F–H) Immunoblotting of E-cadherin, cell viability, and lipid peroxidation in HN9 cells with or without CDH overexpression and exposure to RSL3, SAS, or cyst(e)ine deprivation. (I) The mRNA expression levels of Nrf2 and GPX4 genes in HN6 and HN9 cells with or without CDH1 overexpression vector. The error bars represent standard errors from three technical replicates. **P < 0.01, ***P < 0.001 between the vector control and CDH1 overexpression.
Fig. S4.
CDH1 or ZEB1 regulation modulates the susceptibility to sulfasalazine in vivo. (A–D) Representative images, weights, and immunoblotting of tumors grown in mice with HN9 cells transfected with CDH1 overexpression or control vector. The mice were subjected to vehicle or SAS (250 mg/kg daily per intraperitoneal route) for 21 days. Immunoblotting was performed from tumors extracted at 21 days with the primary antibodies of vimentin, ZEB1, PTGS2, 4HNE, E-cadherin, GSH, and β-actin. (E–H) Representative images, weights, and immunoblotting of tumors harvested in mice HN4 cells with or without ZEB1 overexpression or control vector. The mice were also subjected to SAS or vehicle for 21 days. *P < 0.05, *P < 0.01, ***P < 0.001 between the vector control and CDH1 or ZEB1 overexpression or between vehicle or SAS treatment.
Fig. S5.
ZEB1 regulation affects the susceptibility to ferroptosis inducers. (A–B) Cell death and viability assays in HN6 cells with or without ZEB1 siRNA or scrambled control and exposure to RSL3 or SAS for 72 h. Original magnification, × 200. Scale bar 50 μm. (C–E) Immunoblotting, cell viability, and lipid peroxidation of HN9 cells with or without ZEB1 silencing and exposure to RSL3 or SAS. (F–H) Immunoblotting, cell viability, and lipid peroxidation of HN3 cells with or without ZEB1 overexpression. The cells were transfected with ZEB1 cDNA or control vector and then exposed to RSL3 or SAS. (I) The changes of Nrf2 and GPX4 mRNA levels in HN3 and HN4 cells according to ZEB1 overexpression.
Fig. S6.
SIRT1 activation promotes ferroptosis susceptibility in HNC cells. (A–C) Cell death, cell viability, and immunoblotting of HN9 cells with or without SIRT1 silencing. The cells were transfected with SIRT1 siRNA or scrambled control or a SIRT1 inhibitor (10 μM Ex-527) treatment and then exposed to RSL3 or SAS for 72 h. Original magnification, × 200. Scale bar 50 μm. (D–E) Cell viability and lipid peroxidation of HN9 cells with or without SIRT1 silencing or EX-527 and exposure to RSL3 or SAS for 72 h. (F) The combination matrix was created through the cell viability of HN4 cells using SRT1720 and SAS. (G) Immunoblotting of E-cadherin, vimentin, ZEB1, and SIRT1 in HN4 and HN6 cells treated with or without 20 μM resveratrol.
Fig. S7.
Effects of 5-azacitidine on ferroptosis susceptibility in HN4 cells. (A) Representative images of PI staining in HN4 cells treated with or without 5-azacitidine (5-aza) plus RSL3 or SAS. Original magnification, × 200. Scale bar 50 μm. (B–D) Cell death and viability, LIP, and lipid peroxidation in HN4 cells with or without 5-azacitidine (5-aza) plus RSL3 or SAS. LIP was measured by calcein-AM (8 μg/ml) after 5 μM 5-azacitidine plus or minus 2 μM RSL3 or 1 mM SAS; all data were quantified by the Image J software. The error bars represent standard errors from three technical replicates. ***P < 0.001.
References
- 1.Ashrafizadeh M., Zarrabi A., Hushmandi K., Kalantari M., Mohammadinejad R., Javaheri T., Sethi G. Association of the epithelial-mesenchymal transition (EMT) with cisplatin resistance. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun L., Kokura K., Izumi V., Koomen J.M., Seto E., Chen J., Fang J. MPP8 and SIRT1 crosstalk in E-cadherin gene silencing and epithelial-mesenchymal transition. EMBO Rep. 2015;16:689–699. doi: 10.15252/embr.201439792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drápela S., Bouchal J., Jolly M.K., Culig Z., Souček K. ZEB1: a critical regulator of cell plasticity, DNA damage response, and therapy resistance. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.00036. 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascon S., Hatzios S.K., Kagan V.E., Noel K., Jiang X., Linkermann A., Murphy M.E., Overholtzer M., Oyagi A., Pagnussat G.C., Park J., Ran Q., Rosenfeld C.S., Salnikow K., Tang D., Torti F.M., Torti S.V., Toyokuni S., Woerpel K.A., Zhang D.D. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou Y., Palte M.J., Deik A.A., Li H., Eaton J.K., Wang W., Tseng Y.Y., Deasy R., Kost-Alimova M., Dancik V., Leshchiner E.S., Viswanathan V.S., Signoretti S., Choueiri T.K., Boehm J.S., Wagner B.K., Doench J.G., Clish C.B., Clemons P.A., Schreiber S.L. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-09277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hangauer M.J., Viswanathan V.S., Ryan M.J., Bole D., Eaton J.K., Matov A., Galeas J., Dhruv H.D., Berens M.E., Schreiber S.L., McCormick F., McManus M.T. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viswanathan V.S., Ryan M.J., Dhruv H.D., Gill S., Eichhoff O.M., Seashore-Ludlow B., Kaffenberger S.D., Eaton J.K., Shimada K., Aguirre A.J., Viswanathan S.R., Chattopadhyay S., Tamayo P., Yang W.S., Rees M.G., Chen S., Boskovic Z.V., Javaid S., Huang C., Wu X., Tseng Y.Y., Roider E.M., Gao D., Cleary J.M., Wolpin B.M., Mesirov J.P., Haber D.A., Engelman J.A., Boehm J.S., Kotz J.D., Hon C.S., Chen Y., Hahn W.C., Levesque M.P., Doench J.G., Berens M.E., Shamji A.F., Clemons P.A., Stockwell B.R., Schreiber S.L. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J., Minikes A.M., Gao M., Bian H., Li Y., Stockwell B.R., Chen Z.N., Jiang X. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature. 2019;572:402–406. doi: 10.1038/s41586-019-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Zhao Y., Wang H., Zhang C., Wang M., Yang Y., Xu X., Hu Z. Histone demethylase KDM3B protects against ferroptosis by upregulating SLC7A11. FEBS Open Bio. 2020;10:637–643. doi: 10.1002/2211-5463.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galle E., Thienpont B., Cappuyns S., Venken T., Busschaert P., Van Haele M., Van Cutsem E., Roskams T., van Pelt J., Verslype C., Dekervel J., Lambrechts D. DNA methylation-driven EMT is a common mechanism of resistance to various therapeutic agents in cancer. Clin. Epigenet. 2020;12 doi: 10.1186/s13148-020-0821-z. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song N., Jing W., Li C., Bai M., Cheng Y., Li H., Hou K., Li Y., Wang K., Li Z., Liu Y., Qu X., Che X. ZEB1 inhibition sensitizes cells to the ATR inhibitor VE-821 by abrogating epithelial-mesenchymal transition and enhancing DNA damage. Cell Cycle. 2018;17:595–604. doi: 10.1080/15384101.2017.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S.Y., Chu K.-C., Lee H.R., Lee K.-S., Carey T.E. Establishment and characterization of nine new head and neck cancer cell lines. Acta Otolaryngol. 1997;117:775–784. doi: 10.3109/00016489709113477. [DOI] [PubMed] [Google Scholar]
- 13.Yadav B., Wennerberg K., Aittokallio T., Tang J. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput. Struct. Biotechnol. J. 2015;13:504–513. doi: 10.1016/j.csbj.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishimoto T., Nagano O., Yae T., Tamada M., Motohara T., Oshima H., Oshima M., Ikeda T., Asaba R., Yagi H., Masuko T., Shimizu T., Ishikawa T., Kai K., Takahashi E., Imamura Y., Baba Y., Ohmura M., Suematsu M., Baba H., Saya H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Canc. Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 15.De Craene B., Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Canc. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 16.Byles V., Zhu L., Lovaas J.D., Chmilewski L.K., Wang J., Faller D.V., Dai Y. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012;31:4619–4629. doi: 10.1038/onc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao C., Zhu P.X., Yang X., Han Z.P., Jiang J.H., Zong C., Zhang X.G., Liu W.T., Zhao Q.D., Fan T.T., Zhang L., Wei L.X. Overexpression of SIRT1 promotes metastasis through epithelial-mesenchymal transition in hepatocellular carcinoma. BMC Canc. 2014;14:978. doi: 10.1186/1471-2407-14-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skrypek N., Goossens S., De Smedt E., Vandamme N., Berx G. Epithelial-to-Mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet. 2017;33:943–959. doi: 10.1016/j.tig.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Title A.C., Hong S.J., Pires N.D., Hasenohrl L., Godbersen S., Stokar-Regenscheit N., Bartel D.P., Stoffel M. Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat. Commun. 2018;9:4671. doi: 10.1038/s41467-018-07130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W.H., Ding C.C., Sun T., Rupprecht G., Lin C.C., Hsu D., Chi J.T. The Hippo pathway effector TAZ regulates ferroptosis in renal cell carcinoma. Cell Rep. 2019;28:2501–2508. doi: 10.1016/j.celrep.2019.07.107. e2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caramel J., Ligier M., Puisieux A. Pleiotropic roles for ZEB1 in cancer. Canc. Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 22.Yu X.J., Guo X.Z., Li C., Chong Y., Song T.N., Pang J.F., Shao M. SIRT1-ZEB1-positive feedback promotes epithelial-mesenchymal transition process and metastasis of osteosarcoma. J. Cell. Biochem. 2019;120:3727–3735. doi: 10.1002/jcb.27653. [DOI] [PubMed] [Google Scholar]
- 23.Chao S.C., Chen Y.J., Huang K.H., Kuo K.L., Yang T.H., Huang K.Y., Wang C.C., Tang C.H., Yang R.S., Liu S.H. Induction of sirtuin-1 signaling by resveratrol induces human chondrosarcoma cell apoptosis and exhibits antitumor activity. Sci. Rep. 2017;7:3180. doi: 10.1038/s41598-017-03635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brabletz S., Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill L., Browne G., Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int. J. Canc. 2013;132:745–754. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 26.Jolly M.K., Tripathi S.C., Jia D., Mooney S.M., Celiktas M., Hanash S.M., Mani S.A., Pienta K.J., Ben-Jacob E., Levine H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7:27067–27084. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Zeng Z., Guan L., Ao R. GRHL2 induces liver fibrosis and intestinal mucosal barrier dysfunction in non-alcoholic fatty liver disease via microRNA-200 and the MAPK pathway. J. Cell Mol. Med. 2020;24:6107–6119. doi: 10.1111/jcmm.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim S.O., Gu J.M., Kim M.S., Kim H.S., Park Y.N., Park C.K., Cho J.W., Park Y.M., Jung G. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology. 2008;135:2128–2140. doi: 10.1053/j.gastro.2008.07.027. 2140.e2121-2128. [DOI] [PubMed] [Google Scholar]
- 29.Bender C.M., Pao M.M., Jones P.A. Inhibition of DNA methylation by 5-aza-2'-deoxycytidine suppresses the growth of human tumor cell lines. Canc. Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 30.Kurkjian C., Kummar S., Murgo A.J. DNA methylation: its role in cancer development and therapy. Curr. Probl. Canc. 2008;32:187–235. doi: 10.1016/j.currproblcancer.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.