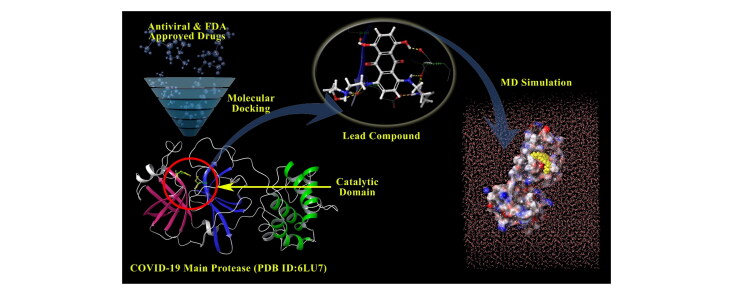
Abstract
The outbreak of novel coronavirus (COVID-19), which began from Wuhan City, Hubei, China, and declared as a Public Health Emergency of International Concern by World Health Organization (WHO) on 30th January 2020. The present study describes how the available drug candidates can be used as a potential SARS-CoV-2 Mpro inhibitor by molecular docking and molecular dynamic simulation studies. Drug repurposing strategy is applied by using the library of antiviral and FDA approved drugs retrieved from the Selleckchem Inc. (Houston, TX, http://www.selleckchem.com) and DrugBank database respectively. Computational methods like molecular docking and molecular dynamics simulation were used. The molecular docking calculations were performed using LeadIT FlexX software. The molecular dynamics simulations of 100 ns were performed to study conformational stability for all complex systems. Mitoxantrone and Leucovorin from FDA approved drug library and Birinapant and Dynasore from anti-viral drug libraries interact with SARS-CoV-2 Mpro at higher efficiency as a result of the improved steric and hydrophobic environment in the binding cavity to make stable complex. Also, the molecular dynamics simulations of 100 ns revealed the mean RMSD value of 2.25 Å for all the complex systems. This shows that lead compounds bound tightly within the Mpro cavity and thus having conformational stability. Glutamic acid (Glu166) of Mpro is a key residue to hold and form a stable complex of reported lead compounds by forming hydrogen bonds and salt bridge. Our findings suggest that Mitoxantrone, Leucovorin, Birinapant, and Dynasore represents potential inhibitors of SARS-CoV-2 Mpro.
Keywords: SARS-CoV-2 main protease (Mpro), drug repurposing, molecular docking, molecular dynamic simulation
GRAPHICAL ABSTRACT
1. Introduction
The world witnessed the deadliest outbreak of coronavirus disease (COVID-19) which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (World Health Organization, 2020b). It shows various symptoms such as difficulty in breathing, fever, pneumonia, sore throat, and lung infection (Adhikari et al., 2020). The origin of Coronavirus disease is from Wuhan City, Hubei, China has rapidly spread to many other countries (Chen, Liu, et al., 2020). The outbreak of COVID-19 is declared as a Public Health Emergency of International Concern by the World Health Organization (WHO) on 30th January 2020 (Sohrabi et al., 2020). According to WHO, till 22nd June 2020, there were 8,860,331 confirmed cases and 465,740 deaths across the globe (World Health Organization, 2020a). Initially, coronavirus was named as the 2019 novel coronavirus(2019-nCoV) by WHO on 12th January 2020 but later on 11th February 2020, WHO officially declared its name as coronavirus disease 2019 (COVID19). Also on 11 February 2020, SARS-CoV-2 was proposed as the name for coronavirus by Coronavirus Study Group (CSG) of the International Committee of Taxonomy of Viruses (Chen, Liu, et al., 2020; Guo et al., 2020).
COVID-19 is enveloped, positive-sense, a single-stranded large RNA virus. There are 4 different genera of coronaviruses-Alpha, beta, gamma, and delta. Alpha- and beta-coronaviruses found to be originated from bats, while gamma- and delta-viruses originated from pigs and birds (He et al., 2020; Velavan & Meyer, 2020). Different coronaviruses have been identified- Human coronavirus OC43 (HCoV-OC43), Human coronavirus HKU1, Human coronavirus NL63 (HCoV-NL63), Human coronavirus 229E (HCoV-229E), Middle East respiratory syndrome-related coronavirus (MERS-CoV), Severe acute respiratory syndrome coronavirus (SARS-CoV) and now Severe acute respiratory syndrome coronavirus 2. Severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome-related coronavirus (MERS-CoV) emerged as an epidemic in 2003 and 2012 respectively (Corman et al., 2018). According to taxonomic classification, COVID-19 belongs to the family of Coronaviridae, genus- betacoronavirus, subgenus-Sarbacovirus. This virus is identified as the strain which belongs to the species- Severe acute respiratory syndrome-related coronavirus (NCBI Taxonomy Browser, 2020). SARS-CoV and MERS CoV6 also belong to betacoronavirus genus and upon a comparison of the genome sequence, COVID-19 showed a good sequence identity of 82% with SARS-CoV as compared to MERS CoV6 (Kannan et al., 2020; Zhang & Liu, 2020). The genome size of coronaviruses ranges between approximately 26 to 32 kb which has open reading frames (ORFs) ranging from 6 to 11. The first ORF is 70% of the entire genome and has16 non-structural proteins (nsps). The main protease (Mpro), also known as chymotrypsin-like cysteine protease (3CLpro), is encoded by nsp5 which plays an important role in replication and gene expression of the virus. Thus, Mpro is found to be an attractive target so as to design effective drugs for COVID-19. The first released crystal structure of SARS-C0V-2 Mpro with PDB ID: 6LU7 was used for designing drugs (Bzówka et al., 2020).
COVID-19 is spreading easily and rapidly through human to human transmission via respiratory droplets produced by an infected person while coughing or sneezing thus compulsory precautions should be taken by social distancing, frequently washing hands and by avoiding large public gatherings. People with medical conditions like lung or heart disease, diabetes, and also old age have more risk of developing COVID-19 (Centers for Disease Control & Prevention, 2020; Cascella et al., 2020). Currently, there are no specific antiviral vaccines or therapies to treat COVID-19 (Centers for Disease Control & Prevention, 2020). Drug repurposing is the technique of searching for new benefits or purposes of the existing drugs which are approved drugs and investigational drugs, thus reduces the cost and time of drug development and also lowers the risk of unexpected side effects of the drug (Pushpakom et al., 2019; Simsek et al., 2018). In this research work, drug repurposing strategy is used by approaching computational methods like molecular docking and molecular dynamics simulation is performed to find the probable drug for COVID-19 by using the library of antiviral and FDA approved drugs. Molecular docking helps to predict the binding affinity between protein and ligand by using scoring functions (Wang & Zhu, 2016). Molecular dynamics simulation is a computational method that gives insights into the interaction and motion of molecules and atoms according to Newton’s physics. In molecular dynamics simulations, there is the integration of Newton’s laws of motions which helps to generate successive configurations of the evolving system that provides trajectories that specify the velocities and positions of the particles over time (De Vivo et al., 2016).
2. Materials and methods
2.1. The sequence and x-ray crystal structure of the main protease from SARS-CoV-2
The X-ray crystal structure of the main protease (Mpro) (PDB ID: 6LU7) from SARS-CoV-2 complexed with inhibitor N3 was retrieved from the Protein Data Bank (Jin et al., 2020). The homodimeric Mpro contains Chain A and B. The chain A was prepared using the Protein Preparation Wizard of Schrodinger tool and used for further study. The preparation of the receptor input data requires the definition of the receptor atoms, the determination of the positions of the essential hydrogen atoms, and the definition of the active site atoms.
To identify the conserved domain and hierarchical classifications of the SARS-CoV-2 Mpro structure among the various distributed protein domain families, we uploaded the fasta sequence at the NCBI CD database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Lu et al., 2020), which reveals the conserved site residues which are associated with the SARS-CoV-2 Mpro molecular function. To define the binding site of the Mpro, all amino acids are selected that are located within 6.5 Å from N3 atom at its crystalline position. The binding site of Mpro is occupied with Thr24, Thr26, Leu27, His41, Cys44, Met49, Pro52, Ser139, Phe140, Leu141, Asn142, Gly143, His164, Glu166, His172, Phe181, Gln189, Thr190 and Gln192 amino acids, and also this binding site was confirmed with the literature study (Chen, Yiu, et al., 2020).
2.2. Retrieval of antiviral and FDA approved drug library
The 2-dimensional structures of Antiviral Drugs and FDA approved drugs were obtained from Selleckchem Inc. (Houston, TX, https://www.selleckchem.com/h) and Drug Bank database (Wishart et al., 2018) respectively. The antiviral compound library contains medicinally active, structurally diverse, and cell-permeable compounds some of them have been FDA approved. These antiviral compounds target HIV Protease, HCV Protease, Reverse Transcriptase, and Integrase, etc. FDA-approved drug repurposing is an urgent need for a patient's treatment if there is no medicine of the particular disease. The collection of unique 348 antiviral compounds and 2454 FDA approved drugs were energy minimized using OPLS-2005 force field (Kollar & Frecer, 2017) using Maestro program of Schrodinger until an energetically stable conformation is obtained. These energetically stable conformations were used for molecular docking study with SARS-CoV-2 Mpro.
2.3. Molecular docking studies
Molecular docking techniques dock small molecules into the protein binding site. To understand how these drugs bind to the SARS-CoV-2 Mpro, docking studies were performed on Antiviral Drugs and FDA approved drugs using FlexX (Rarey et al., 1996) software. Receptor atoms that are taken from the PDB database i.e. PDB files of SARS-CoV-2 Mpro (PDB ID: 6LU7) are treated as rigid during the docking calculations. The binding cavity in the receptor is defined using the receptor preparation wizard in FlexX. To define the binding cavity of the receptor all amino acids are selected that are located within 6.5 Å from N3 at its crystalline position in 6LU7. All compounds are then docked into the binding site of the SARS-CoV-2 Mpro to predict binding poses for the retrieved drugs. The docking procedure begins with the systematic conformational expansion of the ligand followed by placement in the receptor site. A collection of poses was generated from the pool of ligand conformations using an alpha triangle that yields several possible conformations depends on the quantity of the rotatable bonds present in the structure, in FlexX maximum number of solutions i.e. per iteration and fragmentation is 2000. The used software i.e. FlexX in this study gives binding energy not only depending on H-bond interaction but also includes terms accounting for short-ranged van der Waals and electrostatic interaction, loss of entropy upon ligand binding, hydrogen bond and solvation energy (Bursulaya et al., 2003). The best poses for a given compound are determined by the docking energy and binding interaction.
2.4. Molecular dynamic (MD) studies
To evaluate the binding strength of crystal ligand N3 and screened lead drugs after docking study in the binding pocket of Mpro as revealed under conformational dynamics. We have performed 100 ns full-scale molecular dynamic simulation for lead antiviral compounds viz. Birinapant, Dynasore, and lead FDA approved drugs viz. Mitoxantrone and Leucovorin complexed with the crystal structure of SARS-CoV-2 Mpro using Desmond (Bowers et al., 2006). The process of MD simulations helps to calculate forces and compute the motion of amino acid’s atoms. However, Desmond incorporates a more detailed temperature, pressure, volume system (Bowers et al., 2006) and has more functionality built-in for executing protein-ligand interactions. Using the system builder of Desmond in the Maestro program, the system for complexes are immersed in a water-filled cubic box containing 1 Å spacing water molecules using an extended simple point charge (SPC), a three-point water model with periodic boundary conditions. The total charge of the solvent system is neutralized by adding appropriate counter ion randomly in the solvated complex system. Energy minimization is a very important step in MD and it is done using the steepest descent method. Cubic box type (with box size 0.9) is considered for minimizing edge effects in a finite system to apply periodic boundary conditions. The atoms of the system to be simulated are put into the space-filling box, which is surrounded by translated copies of itself. The OPLS_2005 force field (parameters used to describe the potential energy of a system) (Kollar & Frecer, 2017) is chosen in this work which is an improved force field suited for molecular dynamics simulation of proteins. The molecular dynamics studies are done by taking into consideration certain parameters as input such as constraints set as all-bonds, integrator as MD, Nose Hoover chain thermostat method and it uses Martyna-Tobias-Klein barostat method, temperature 300k.
After the system gets equilibrated, the stable conformation trajectories are captured and analyzed to explore interaction stability. The conformational change of the C-alpha backbone of the SARS-CoV-2 Mpro crystal structure has been compared with initial conformations. Besides, we have performed MD trajectory clustering analysis to determine the binding mode of the selected ligand within the binding cavity of Mpro using trajectories generated during 100 ns MD simulation.
2.5. MD trajectory analysis and prime MM/GBSA calculations
To understand H-bond contribution in the protein-ligand stability, the trajectories generated from the 100 ns MD simulation subjected to the H-bond monitoring using a simulation event analysis panel of Maestro software. Also, the Prime [Jacobson et al., 2004] module was used to calculate the binding free energies of the complex system. The MM/GBSA (Molecular Mechanics, The Generalized Born Model, and Solvent Accessibility) was performed to calculate the ligand binding free energies and ligand strain energies for docked lead compounds with SARS-CoV-2 Mpro. The binding free energy comprises the non-polar solvation energies, polar solvation energies and potential energy (Bhardwaj et al., 2020). Prime MM-GBSA works with the combination of advanced OPLS-2005 force field, SGB solvation model for polar solvation (GSGB), non-polar solvation (GNP), and Molecular Mechanics Energies (EMM) that compiled different nonpolar solvent accessible surface area and van der Waals interactions. The free energy changes upon ligand binding were calculated using the following equations.
The Gcomplex represents complex energy, Gprotein is the receptor energy and Gligand is the unbound ligand energy. EMM represents molecular mechanics energies, GSGB is an SGB solvation model for polar solvation and GNP is a nonpolar solvation term.
3. Result and discussion
3.1. Catalytic/active site of the SARS-CoV-2 mpro
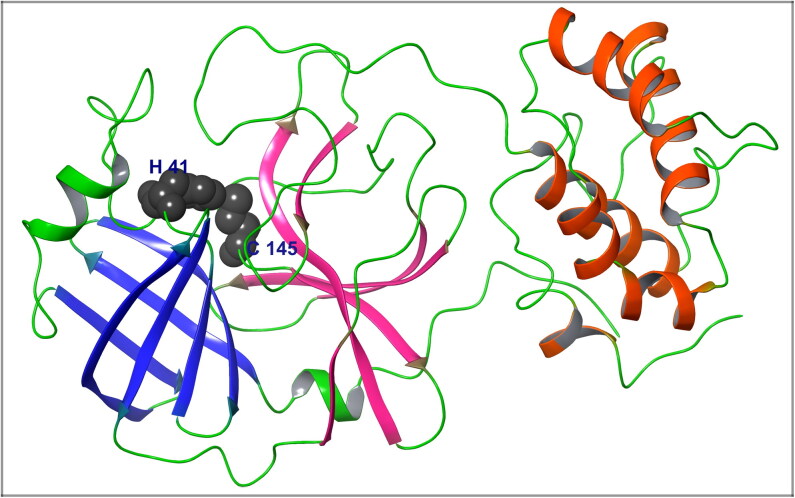
The conserved domain search result shows that SARS-CoV-2 Mpro belongs to the MEROPS peptidase C30 family, where the catalytic site residues i.e. His41 and Cys145 form a catalytic dyad. The structures of Mpro consist of three domains with the first two containing anti-parallel beta barrels and the third consisting of an arrangement of alpha-helices. The catalytic residues are found in a cleft between the first two domains (Figure 1).
Figure 1.
The catalytic residues i.e.His41 and Cys145of COVID-19 Mpro, form a catalytic dyad. The crystal structure of COVID-19 Mpro represented in ribbon form and the residue His41 and Cys145 shown in the gray-coloured molecular surface.
3.2. Interaction patterns of lead compounds with SARS-CoV-2 mpro
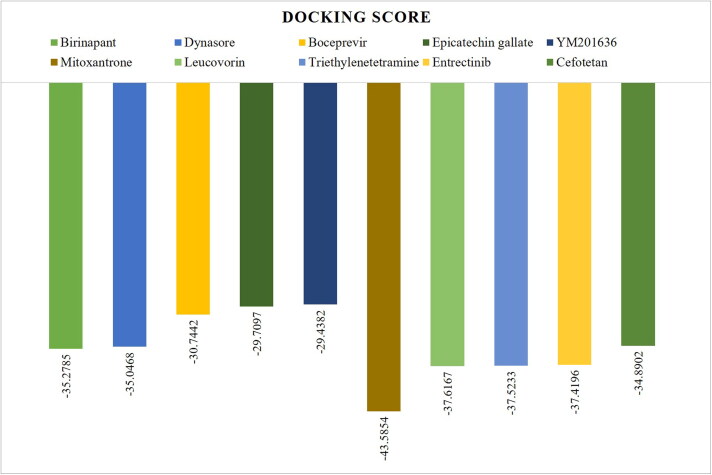
The molecular docking of the anti-viral drugs and FDA approved drugs with SARS-CoV-2 Mpro crystal structure were performed. The docking score of the top five lead compounds from each category (anti-viral and FDA approved drugs) were presented in Figure 2. The lead compounds Mitoxantrone and Leucovorin from FDA approved drug library and Birinapant and Dynasore from anti-viral drug libraries were selected for further study. The interaction pattern and binding mode of these lead compounds and crystallized ligand N3 with the crystal structure of Mpro were analyzed in Maestro. The detailed docking analysis with their interaction pattern shown in Table 1. Crystalized ligand with SARS-CoV-2 Mpro shows −18.5693 docking score which suggest, N3 forms 2-3 fold weaker complex with SARS-CoV-2 Mpro as compared to selected lead compounds.
Figure 2.
The FlexX docking score of the top five lead compounds from anti-viral and FDA approved drugs which are showing the highest binding affinity towards COVID-19 Mpro.
Table 1.
Docking analysis of lead compounds with SARS-CoV-2 Mpro.
| Drug Library | Lead Compound | Docking Score | Interacting Amino Acid | Types of Bond | Bond Distance (Å) |
|---|---|---|---|---|---|
| Crystalized Ligand with Mpro (PDB ID: 6LU7) | N3 | −18.5693 | Phe140 | Aromatics HB | 1.70 |
| Gly143 | H-Bond | 2.01 | |||
| His164 | H-Bond | 2.29 | |||
| Glu166 | 2 H-Bond | 1.82, 1.89 | |||
| Gln189 | 2 H-Bond | 2.04, 2.04 | |||
| FDA Approved | Mitoxantrone | −43.5854 | Phe140 | 3 H-Bond | 1.56, 1.64, 2.42 |
| Leu141 | H-Bond | 1.47 | |||
| His164 | H-Bond | 2.25 | |||
| Glu166 | 4 H-Bond | 1.90, 1.94, 2.05, 2.24 | |||
| Glu166 | Salt Bridge | 2.84 | |||
| Leucovorin | −37.6167 | Thr24 | H-Bond | 1.95 | |
| Thr26 | H-Bond | 1.85 | |||
| His41 | H-Bond | 2.23 | |||
| Gln189 | H-Bond | 2.21 | |||
| Antiviral | Birinapant | −35.2785 | Phe140 | H-Bond | 1.60 |
| Asn142 | 2 H-Bond | 2.16, 2.57 | |||
| His164 | H-Bond | 2.12 | |||
| Glu166 | 3 H-Bond | 1.62, 1.77, 1.83 | |||
| Glu166 | Salt Bridge | 2.82 | |||
| Dynasore | −35.0468 | Leu141 | H-Bond | 2.06 | |
| Glu166 | 2 H-Bond | 1.88. 2.13 | |||
| Thr190 | Aromatics HB | 2.03 |
The hydrochloride salt of Mitoxantrone used as an anti-cancer agent (Malakar et al., 2018) also, this potential compound has shown great antiviral agents in many studies (Huang et al., 2019). In this study, Mitoxantrone has shown potent lead compounds to inhibit SARS-CoV-2 Mpro as it shows greater binding affinity than other compounds.
The docking score of Mitoxantrone is −43.5854 which suggests strong binding of these compounds within the binding cavity of the SARS-CoV-2 Mpro structure. The binding mode of Mitoxantrone within the SARS-CoV-2 Mpro binding cavity is shown in Figure 3(a). Mitoxantrone forms nine hydrogen bonds and one salt bridge with Mpro. The hydrogen bond is an intermolecular force that kept two or more molecules together. The amino acids Phe140, Leu141, His164, and Glu166 involved in hydrogen bonding with Mitoxantrone, and it also formed a salt bridge with Mitoxantrone. The Phe140 forms 3-hydrogen bonds with Mitoxantrone with bond distances of 1.56 Å, 1.64 Å, and 2.42 Å. Another hydrogen bond forms with Mitoxantrone by amino acid Leu141, His164 at a 1.47 Å, and 2.25 Å bond distance respectively. Glu166 is an important residue from SARS-CoV-2 Mpro which forms four hydrogen bonds with Mitoxantrone at 1.90 Å, 1.94 Å, 2.05 Å, 2.24 Å bond distances. Also, these residues were involved in non-covalent interaction by forming a salt bridge with Mitoxantrone with 2.84 Å bond distance. This non-covalent interaction gives more stability of Mitoxantrone conformation with SARS-CoV-2 Mpro.
Figure 3.
(a) Binding pose of Mitoxantrone within the COVID-19 Mpro binding pocket (b) The Binding pose of Leucovorin within the COVID-19 Mro binding pocket. Lead compounds are represented in the ball and stick model, COVID-19 Mprobackbone shown in ribbon, and interacting amino acids represented in the wire-frame. Black color dashed lines indicated hydrogen bonds and the salt bridge is shown in pink color dashed lines.
Figure 3(b) shows the intermolecular binding interactions of another FDA approved drug i.e. Leucovorin with the crystal structure of SARS-CoV-2 Mpro. Leucovorin, also known as folinic acid has shown a promising antagonistic effect of Methotrexate in many cancer cells (Beck et al., 2019). We observed that Leucovorin forms only hydrogen bonds with SARS-CoV-2 Mpro, interestingly amino acids which are involved in bonding with Mitoxantrone are not interacted with Leucovorin, because of their different molecular structures. Amino acids viz. Thr24, Th426, His41, and Gln189 were found to make hydrogen bonds with Leucovorin at 1.95 Å, 1.85 Å, 2.23 Å, and 2.21 Å bond distances respectively. The docking score of Leucovorin is −37.6167 which suggests that this compound is also having the capability to inhibit SARS-CoV-2 Mpro.
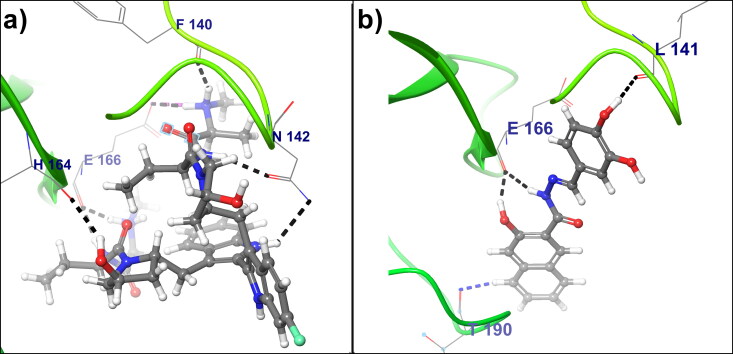
On the other hand, Birinapant and Dynasore from the category of the antiviral drug show a good binding affinity towards SARS-CoV-2 Mpro. Antiviral activity of Birinapant against chronic hepatitis B virus (HBV) infection by induction of apoptosis by inhibiting cellular inhibitor of apoptosis proteins (cIAPs) has been reported in the literature (Alonso et al., 2017; Ebert et al., 2015). The second antiviral lead compound i.e. Dynasore is known as GTPase inhibitor, which prevents endocytosis by rapidly inhibiting Dynamin activity (Preta et al., 2015). Also, the antiviral activity of Dynasore against the dengue virus (DENV) (Carro et al., 2018) and it is showing a vaccine adjuvant or antiviral therapeutic strategy by activating mitochondrial antiviral signaling protein (Ailenberg et al., 2015).
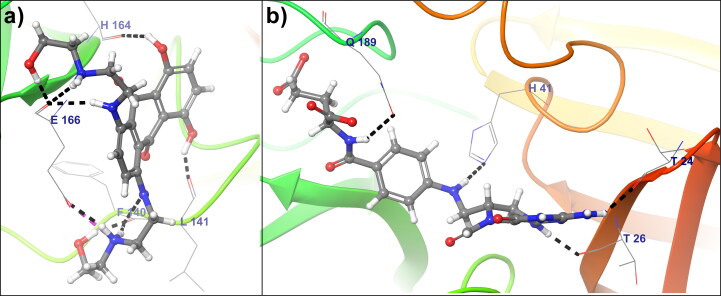
Figure 4(a) shows, the binding mode of Birinapant within the SARS-CoV-2 Mpro cavity. Birinapant forms seven hydrogen bonds and one salt bridge with −35.2785 docking score. The amino acid Phe140 from SARS-CoV-2 Mpro forms a hydrogen bond with Birinapant at 1.60 Å bond distance, also Asn142 forms two hydrogen bonds with Birinapant. Another residues i.e. His164 forms one and Glu166 forms three hydrogen bond at 2.12 Å, 1.62 Å, 1.77 Å, 1.83 Å bond distances respectively. Glu166 is also involved in the formation of a salt bridge with Birinapant at 2.82 Å bond distance. The lead compound Dynasore shows a docking score of −35.0468 with three hydrogen bonds and one aromatic hydrogen bond to the crystal structure of SARS-CoV-2 Mpro. Figure 4(b) shows intermolecular hydrogen bonding between residue Leu142 and Glu166 with Dynasore at 2.06 Å, 1.88 Å, and 2.13 Å bond distance respectively. Similarly, Figure 4(b) shows that, aromatic hydrogen bond formation between Thr190 and Dynasore, which forms a stable complex between lead compound Dynasore and Mpro.
Figure 4.
(a) Binding pose of Birinapant within the COVID-19 Mpro binding pocket (b) The Binding pose of lead compound Dynasore within the COVID-19 Mpro binding pocket. Lead Compounds are represented in the ball and stick model, COVID-19 Mprobackbone shown in ribbon, and interacting amino acids represented in the wire-frame. Black color dashed lines indicated hydrogen bonds, aromatic hydrogen bond shown in blue dashed line and the salt bridge is shown in pink color dashed line.
From this docking study, we observed that acidic amino acid i.e. Glutamic acid (Glu166) is a key residue to hold and form a stable complex of reported lead compounds by forming hydrogen bonds and salt bridge. Also, an aliphatic amino acid (Leu141), the aromatic amino acid (Phe140), and basic amino acid (His164) play a critical role to form a stable complex. To know the structural similarity between selected lead compounds, we have used the Calculate Similarity Panel of Maestro software for atom-pair (AP) similarity between selected leads. The maximum AP similarity and mean AP Similarity (is the mean of the similarities to each of the probes) was reported in Table 2. By defaults, the AP similarities are calculated on a scale of 0.0 (no structural similarity) to 1.0 (Maximum structural similarity). The similarity results suggest that the selected lead compounds having maximum structural similarity and averaged mean AP similarity.
Table 2.
The atom-pair similarity between the lead compounds (Max AP), and the atom-pair similarities to each of the probes (Mean AP).
| Lead Compound | Max AP Similarity | Mean AP similarity |
|---|---|---|
| Mitoxantrone | 1 | 0.434 |
| Leucovorin | 1 | 0.442 |
| Birinapant | 1 | 0.409 |
| Dynasore | 1 | 0.411 |
3.3. RMSD and RMSF calculation
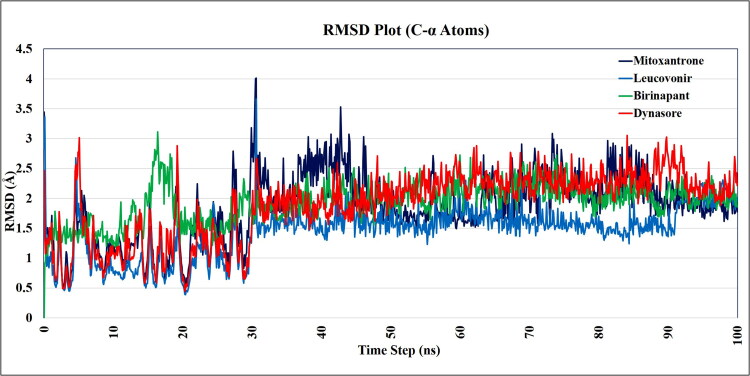
Since molecular docking calculations were done using the rigid crystal structure of SARS-CoV-2 Mpro, we have analyzed target receptor and lead compounds interactions in the dynamic behavior of both receptor and ligand using molecular dynamic simulation to probe the stability of bound conformation after binding of lead compounds within the cavity of Mpro. To study the conformational stability of protein-ligand complexes and their changes, we have simulated the systems up to 100 ns each. In this study, we used the period of 100 ns which is adequate time for the rearrangements of Cα atoms of SARS-CoV-2 Mpro in complexes with lead compounds.
The Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) values of the C-α atoms of Mpro assessment were done to reveal thermodynamic conformational stability during 100 ns period. Further, 1000 trajectories generated during the MD simulation were captured and superimposed on the initial crystal structure of SARS-CoV-2 Mpro with the help of Simulation Event Analysis (SEA) panel of Schrodinger which gives the output in “.dat” format. This collected data of RMSF and RMSD values were used to plot RMSD and RMSF graph. The RMSD plot (Figure 5) suggests that all the lead compounds i.e. Mitoxantrone, Leucovorin, Birinapant, and Dynasore are within the acceptable range of RMSD with a mean RMSD value of 2.25 Å, indicates that all the reported lead compounds bound tightly within the cavity of SARS-CoV-2 Mpro. If we look into the RMSD graph (Figure 5) all the complex systems have starting equilibrating from 33 ns to 100 ns within the RMSD range of 1.5 Å to 3 Å which gives averaged RMSD value of 2.25 Å.
Figure 5.
Time dependence of root mean square deviations (RMSDs) of the C-α backbone of COVID-19 Mprorepresented after binding with lead compounds viz. Mitoxantrone, Leucovorin, Birinapant, and Dynasore.
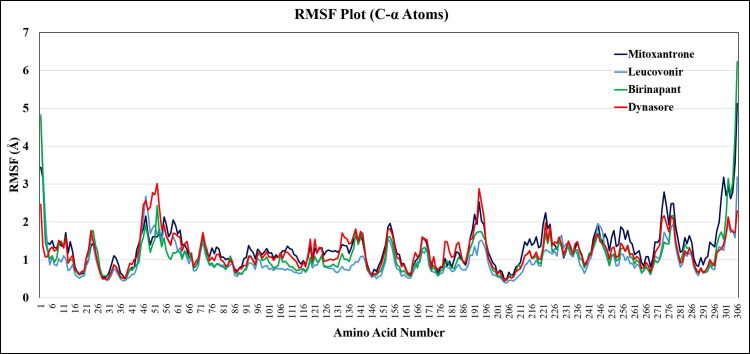
.On the other hand, to examine the binding efficiency of lead compounds with SARS-CoV-2 Mpro, the root mean square fluctuation (RMSF) values for C-α atoms of all the residues were measured based on 100 ns trajectory data. The average RMSFs measured for Mpro upon binding of Mitoxantrone, Leucovorin, Birinapant, and Dynasore is 1.5 Å as all the residues fluctuated within the RMSF range of 0.5 Å to 3 Å (Figure 6), which reveals the minimum fluctuation and relative secondary conformational stability of SARS-CoV-2 Mpro upon binding of reported lead compounds. Therefore, the MD studies showed that the lead Mitoxantrone, Leucovorin, Birinapant, and Dynasore were more potential towards SARS-CoV-2 Mpro.
Figure 6.
Time dependence of root mean square fluctuation (RMSFs) of the C-α backbone of COVID-19 Mprorepresented after binding with lead compounds viz. Mitoxantrone, Leucovorin, Birinapant, and Dynasore.
Besides, during the 100 ns period of MD simulation, we have explored the binding pocket stability after ligand binding to the SARS-CoV-2 Mpro by calculating RMSF values of all the residues which come under the binding pocket of SARS-CoV-2 Mpro. The 1000 MD trajectories were captured during the 100 ns MD simulation and superimposed on initial crystal structure to calculate the RMSF of C-α atoms of the binding pocket residues, all the RMSF values in Å of binding pocket residues are listed in Table 3. The RMSF of SARS-CoV-2 Mpro binding pocket residues, after binding to lead compounds i.e. Mitoxantrone, Leucovorin, Birinapant, and Dynasore is lower than 2.0 Å, which suggests that the binding pocket of SARS-CoV-2 Mpro is quite stable during the given period of MD simulation.
Table 3.
RMSF values of SARS-CoV-2 Mpro binding pocket residues (C-α atoms) after binding of lead compounds.
| Residues | Mitoxantrone | Leucovorin | Birinapant | Dynasore |
|---|---|---|---|---|
| Thr24* | 1.428 | 1.509 | 1.774 | 1.505 |
| Thr26* | 0.859 | 0.857 | 1.222 | 0.919 |
| Leu27 | 0.666 | 0.671 | 0.768 | 0.656 |
| His41* | 0.794 | 0.615 | 0.717 | 0.876 |
| Cys44 | 1.425 | 1.223 | 0.927 | 1.275 |
| Met49 | 1.398 | 1.694 | 1.204 | 2.395 |
| Pro52 | 1.604 | 1.656 | 2.434 | 3.013 |
| Ser139 | 1.622 | 0.956 | 1.745 | 1.816 |
| Phe140* | 1.477 | 0.941 | 1.410 | 1.479 |
| Leu141* | 1.651 | 1.079 | 1.573 | 1.751 |
| Asn142 | 1.498 | 1.094 | 1.709 | 1.651 |
| Gly143 | 1.147 | 0.982 | 1.240 | 1.141 |
| His164* | 0.806 | 0.664 | 0.767 | 0.964 |
| Glu166* | 1.024 | 0.944 | 0.787 | 1.065 |
| His172 | 0.923 | 0.700 | 0.712 | 0.780 |
| Phe181 | 0.795 | 0.694 | 0.780 | 1.064 |
| Gln189* | 1.557 | 1.128 | 1.296 | 1.535 |
| Thr190* | 1.801 | 1.113 | 1.451 | 1.557 |
| Gln192 | 1.940 | 1.125 | 1.721 | 2.062 |
indicates interacting residues with SARS-CoV-2 Mpro.
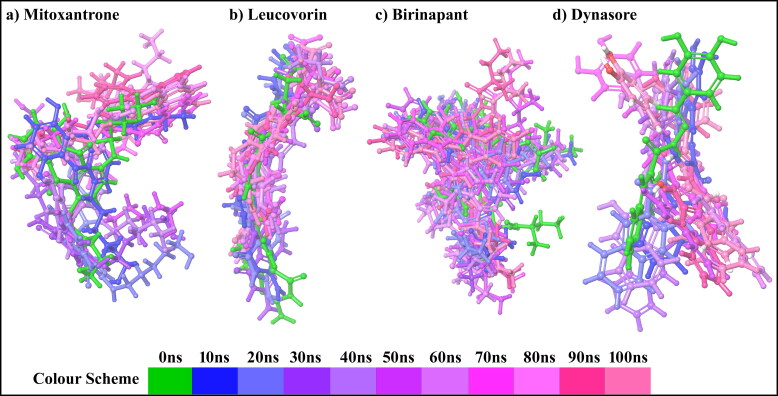
Also, we have done the superimposition of docked ligand binding pose with poses obtained during the 100 ns MD simulation for each complex. The Figure 7 illustrates the superimposition of lead compound poses at a 10 ns intervals with their docked binding pose within the SARS-CoV-2 Mpro cavity. This study also supports the stability of lead compounds during the 100 ns MD simulation with SARS-CoV-2 Mpro.
Figure 7.
The superimposition of lead compound poses at a 10 ns intervals with their docked binding pose within the SARS-CoV-2 Mpro cavity.
3.4. Prime MM/GBSA energies for lead compounds complexed with SARS-CoV-2 mpro
MM/GBSA has been considered the most suitable procedure to calculate free binding energies (ΔG Bind) which states the results in terms of hydrophobic, VDW, or solvation components. The screened compounds (Mitoxantrone, Leucovorin, Birinapant, and Dynasore), serving as ligands to SARS-CoV-2 Mpro were submitted to ensemble-averaged Prime MM/GBSA method for a long time MD simulation. The calculated binding free energies of four complexes using ensemble-averaged MM/GBSA are reported in Table 4. The protein-ligand complex is stronger when the binding energy is less (more negative value) (Bhardwaj et al., 2020). The data denotes Mitoxantrone to have the most negative binding free energy (-171.316 kcal/mol) which confers the binding stability of the Mitoxantrone to be more potent. Whereas, other lead compounds i.e. Leucovorin, Dynasore, and Birinapant also show favorable binding free energy with SARS-CoV-2 Mpro, suggesting that all the lead molecules make a strong complex with the SARS-CoV-2 Mpro. These MM/GBSA results suggest that the lead compounds satisfy the prime MM/GBSA approach to achieve a stable complex with SARS-CoV-2 Mpro and all the energies predicted from the prime MM/GBSA are thermodynamically favorable.
Table 4.
The ensemble-averaged prime binding free energies (kcal/mol) of docked complexes during 100 ns MD simulation.
| Lead Compounds complexed with Mpro | ΔG Binda (kcal/mol) | ΔG Coulombb (kcal/mol) | ΔG Bind vdWc (kcal/mol) | ΔG Solv GBd (kcal/mol) | Complex Energye (kcal/mol) |
|---|---|---|---|---|---|
| Mitoxantrone | −171.316 ± 7.035 | 81.802 ± 10.088 | 76.694 ± 2.814 | 94.726 ± 12.234 | −15350.07 ± 67.527 |
| Leucovorin | −168.920 ± 10.166 | 89.722 ± 5.802 | 74.912 ± 4.443 | 99.93 ± 7.911 | −15330.41 ± 68.109 |
| Birinapant | −161.870 ± 3.643 | 64.414 ± 7.948 | 76.162 ± 1.685 | 75.768 ± 4.678 | −15351.44 ± 45.620 |
| Dynasore | −159.376 ± 3.348 | 51.428 ± 7.928 | 77.474 ± 2.219 | 55.082 ± 4.948 | −15407.90 ± 63.901 |
MM/GBSA binding free energy.
Coulomb Coulomb energy.
Van der Waals energy.
GB Generalized Born electrostatic solvation energy.
Energy of protein-ligand complex.
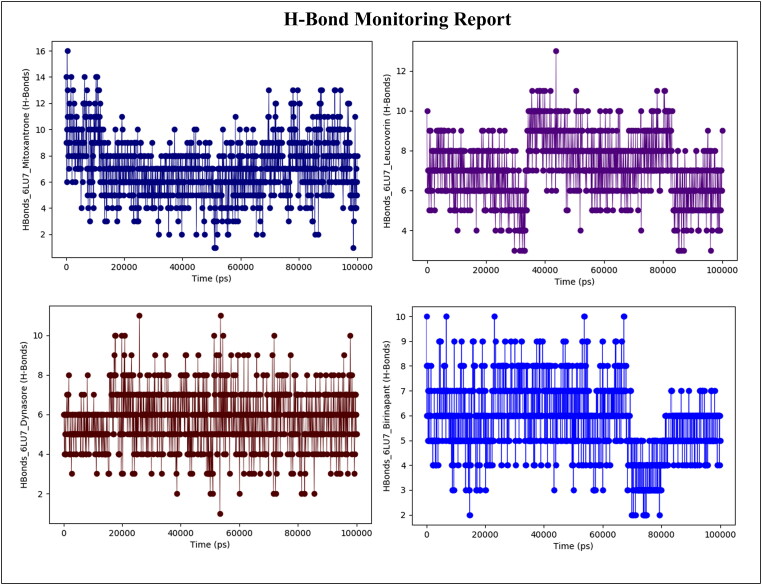
3.5. H-bond monitoring during 100 ns MD simulation
To explore the binding stability between lead compounds and SARS-CoV-2 Mpro, the intermolecular hydrogen bond (H-bond) interactions of lead compounds with SARS-CoV-2 Mpro were evaluated using 1000 frames generated from 100 ns MD simulation trajectory data and was used to generate intermolecular H-bonding pattern through simulation event analysis application in Maestro. Figure 8 shows the existence of intermolecular H-bonding between lead compounds and SARS-CoV-2 Mpro. In the initial time (10 ns) of MD, the complex of SARS-CoV-2 Mpro-Mitoxantrone formed up to 14 hydrogen bonds, after 10 ns the average number hydrogen bonds formed by these complex were 10. It was noted that at last phase of dynamics has reached 12 hydrogen bonds. Figure 8(a) were reached to 12 in the last phase of dynamics Figure 8(a).
Figure 8.
H-bond monitoring report of (a) Mitoxantrone-SARS-CoV-2 Mpro, (b) Leucovorin-SARSCoV-2 Mpro, (c) Dynasore- SARS-CoV-2 Mpro, and (d) Birinapant-SARS-CoV-2 Mpro complexes during 100 ns MD simulation.
Figure 8(b) illustrates that other lead compounds i.e. Leucovorin forms 8-10 hydrogen bonds during the 100 ns simulation run. However, in the complex of SARS-CoV-2 Mpro-Dynasore, it seems that only 8 hydrogen bonds are formed in the intimal 10 ns time of MD, afterward, the few trajectories are found to be forms up to 11 hydrogen bonds during 100 ns (Figure 8(c)). Figure 8(d) shows the hydrogen bonds between Birinapant and SARS-CoV-2 Mpro, suggest that stability of the complex is maintained up to 70 ns by forming 8-10 hydrogen bonds but after 70 ns the hydrogen bonding by Birinapant decreases and forms up to 5 hydrogen bonds during 70 ns-80ns, and then 5-7 hydrogen bonds formed during 80 ns-100ns of MD simulation. From these results, we can conclude that the screened lead compounds form a stable complex with SARS-CoV-2 Mpro and thus obtain the complex stability during the 100 ns MD simulation.
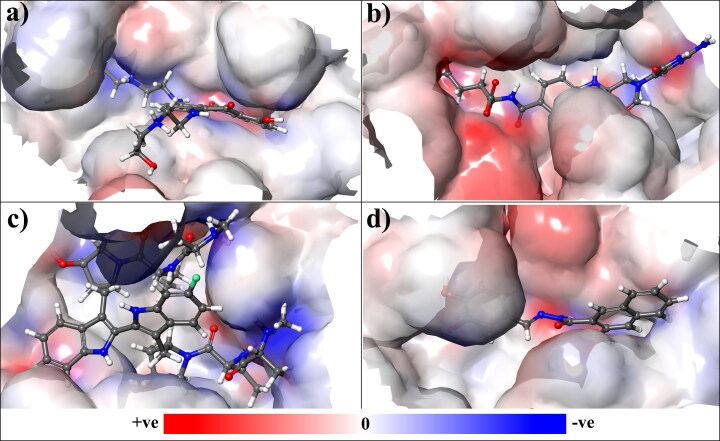
3.6. Electrostatic potential surfaces (EPS) at SARS-CoV-2 mpro binding site
To optimize the electrostatic interactions between the protein and the ligand, the Electrostatic Potential Surface (EPS) are beneficial in computational drug discovery (Agic et al., 2019). These EPS can be very useful technique to compare inhibitors when it binds with its receptor. The relative polarity of the protein-ligand complex system can be illustrated by the molecular electrostatic potential surface. In this study, we have calculated the EPS by using four complex system (i.e. Mpro-Mitoxantrone, Mpro-Leucovorin, Mpro-Birinapant and Mpro-Dynasore) with the help of Maestro software. Figure 9 represents the size, shape, charge density, and the chemical reactivity area of the SARS-CoV-2 Mpro binding site when complexed with the selected lead compounds. The positive, negative, and zero electrostatic potential area of the complex system were represented by red, blue, and white color respectively. The electrostatic potential surface map reveals the selected lead compounds show favorable negative and positive electrostatic potential within the binding cavity to make a strong complex. These electrostatic interactions between the complexes is crucial for molecular identification and are also essential contributors to the protein-ligand complex binding free energy. This study shows EPS can be assessed for the analysis and further lead optimization of electrostatic receptor-ligand interaction, constructing a more potent lead candidate and predict electrostatic target selectivity. Calculating EPS of protein-ligand complexes delivers important findings that, why lead compounds gave binding affinity toward receptor and what functionality can be modified to get better binding.
Figure 9.
The electrostatic potential area of SARS-CoV-2 Mpro binding site when complexed with the selected lead compounds. The positive, negative, and zero electrostatic potential area of the complex system were represented by red, blue, and white color respectively.
4. Conclusion
The World Health Organization (WHO) declared COVID-19 pandemic as a Public Health Emergency of International Concern on 30th January 2020. As there are no available specific therapies or antiviral vaccines until now to treat COVID-19, which leads to urgency and high demand for immediate drug discovery for the current situation. Thus, in this research work, we used the strategy of drug repurposing which can be less time consuming, cost-effective with fewer chances of side effects by using the library of antiviral and FDA approved drugs. The library of 348 antiviral compounds and 2454 FDA approved drugs were retrieved from Selleckchem Inc. (Houston, TX, http://www.selleckchem.com) and DrugBank database respectively and was used to perform molecular docking with SARS-CoV-2 Mpro by using FlexX software. The result which we obtained showed that Mitoxantrone and Leucovorin from FDA approved drug library and Birinapant and Dynasore from anti-viral drug library can be potent inhibitors as they showed a high binding affinity with the crystal structure of SARS-CoV-2 Mpro. Mitoxantrone, Leucovorin, Birinapant, and Dynasore showed a docking score of −43.5854, −37.6167, −35.2785, and −35.0468 respectively.
Further molecular dynamics simulations of 100 ns were carried out for each of these four protein-ligand complexes which revealed mean RMSD value of 2.25 Å. This shows lead compounds bound tightly within the cavity of SARS-CoV-2 Mpro and thus having conformational stability. The RMSF of SARS-CoV-2 Mpro binding pocket residues, after binding to lead compounds was lower than 2.0 Å, thus it reveals that the binding pocket of SARS-CoV-2 Mpro was quite stable during molecular dynamics simulations. Also, it came into light that, Glutamic acid (Glu166) of SARS-CoV-2 Mpro is found to be a key residue to hold and form a stable complex of reported lead compounds by forming hydrogen bonds and salt bridge. Also, the hydrogen bond monitoring and MM/GBSA free energy calculations reveal the lead compounds forms a stable complex with SARS-CoV-2 Mpro. In addition to this, an electrostatic potential surface map can be assessed for the analysis and further lead optimization of electrostatic receptor-ligand interaction, constructing a more potent lead candidate and predict electrostatic target selectivity. Thus, the obtained lead compounds can be further analyzed by using in vitro, in vivo and clinical trial studies to treat COVID-19.
Supplementary Material
Acknowledgements
The authors are thankful to Dr. D. Y. Patil Biotechnology and Bioinformatics Institute, Dr. D. Y. Patil Vidyapeeth, Pune for the physical infrastructure and Department of Science and Technology Science and Engineering Research Board (DST-SERB), Govt. of India, New Delhi, (File Number: YSS/2015/002035) for utilizing an Optimized Supercomputer for docking and dynamics calculations. Senior Research Fellowship awarded to Kiran Bharat Lokhande (Project ID: 2019-3458; File No.: ISRM/11(54)/2019) by the Indian Council of Medical Research, New Delhi is also acknowledged. Authors also acknowledge Ms. Shweta Ashok More, Ph.D. Scholar, Y.B. Chavan College of Pharmacy, Aurangabad, India for technical help during MM/GBSA calculation for this study.
Disclosure statement
The authors declare no conflict of interest.
Data availability statement
The docking structures are available upon request from the corresponding author.
References
- Adhikari, S. P., Meng, S., Wu, Y. J., Mao, Y. P., Ye, R. X., Wang, Q. Z., Sun, C., Sylvia, S., Rozelle, S., Raat, H., & Zhou, H. (2020). Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: A scoping review. Infectious Diseases of Poverty, 9(1), 29. 10.1186/s40249-020-00646-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agic, D., Brkic, H., Kazazic, S., Tomic, A., & Abramic, M. (2019). Aprotinin interacts with substrate-binding site of human dipeptidyl peptidase III. Journal of Biomolecular Structure and Dynamics, 37(14), 3596–3606. 10.1080/07391102.2018.1521343. [DOI] [PubMed] [Google Scholar]
- Ailenberg, M., Di Ciano-Oliveira, C., Szaszi, K., Dan, Q., Rozycki, M., Kapus, A., & Rotstein, O. D. (2015). Dynasore enhances the formation of mitochondrial antiviral signalling aggregates and endocytosis-independent NF-κB activation. British Journal of Pharmacology, 172(15), 3748–3763. 10.1111/bph.13162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, S., Guerra, A. R., Carreira, L., Ferrer, J. Á., Gutiérrez, M. L., & Fernandez-Rodriguez, C. M. (2017). Upcoming pharmacological developments in chronic hepatitis B: Can we glimpse a cure on the horizon? BMC Gastroenterology, 17(1), 168. 10.1186/s12876-017-0726-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, S., Zhu, Z., Oliveira, M. F., Smith, D. M., Rich, J. N., Bernatchez, J. A., & Siqueira-Neto, J. L. (2019). Mechanism of action of methotrexate against Zika virus. Viruses, 11(4), 338. 10.3390/v11040338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure & Dynamics, 1–10. 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. J., Chow, D. E., Xu, H., Dror, R. O., Eastwood, M. P., Gregersen, B. A., Klepeis, J. L., Kolossvary, I., Moraes, M. A., Sacerdoti, F. D., Salmon, J. K., Shan, Y., & Shaw, D. E. (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters, SC '06 [Paper presentation]. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL (pp. 43). doi/proceedings/10.1145/1188455. 10.1145/1188455.1188544 [DOI] [Google Scholar]
- Bursulaya, B. D., Totrov, M., Abagyan, R., & Brooks, C. L. (2003). Comparative study of several algorithms for flexible ligand docking. Journal of Computer-Aided Molecular Design, 17(11), 755–763. 10.1023/B:JCAM.0000017496.76572.6f [DOI] [PubMed] [Google Scholar]
- Bzówka, M., Mitusińska, K., Raczyńska, A., Samol, A., Tuszyński, J. A., & Góra, A. (2020). Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. International Journal of Molecular Sciences, 21(9), 3099. 10.3390/ijms21093099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro, A. C., Piccini, L. E., & Damonte, E. B. (2018). Blockade of dengue virus entry into myeloid cells by endocytic inhibitors in the presence or absence of antibodies. PLoS Neglected Tropical Diseases, 12(8), e0006685. 10.1371/journal.pntd.0006685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S., & Di Napoli, R. (2020). Features, evaluation and treatment coronavirus (COVID-19). StatPearls Publishing. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . (2020). Coronavirus disease 2019 (COVID-19): How to protect yourself & others. Retrieved April 28, 2020, from https://www.cdc.gov/coronavirus/2019-ncov/prepare/prevention.html.
- Chen, Y. W., Yiu, C. B., & Wong, K. Y. (2020). Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research, 9, 129. 10.12688/f1000research.22457.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Liu, Q., & Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92(4), 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M., Muth, D., Niemeyer, D., & Drosten, C. (2018). Hosts and sources of endemic human Coronaviruses. Advances in Virus Research, 100, 163–188. 10.1016/bs.aivir.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vivo, M., Masetti, M., Bottegoni, G., & Cavalli, A. (2016). Role of molecular dynamics and related methods in drug discovery. Journal of Medicinal Chemistry, 59(9), 4035–4061. 10.1021/acs.jmedchem.5b01684 [DOI] [PubMed] [Google Scholar]
- Ebert, G., Allison, C., Preston, S., Cooney, J., Toe, J. G., Stutz, M. D., Ojaimi, S., Baschuk, N., Nachbur, U., Torresi, J., Silke, J., Begley, C. G., & Pellegrini, M. (2015). Eliminating hepatitis B by antagonizing cellular inhibitors of apoptosis. Proceedings of the National Academy of Sciences of the United States of America, 112(18), 5803–5808. 10.1073/pnas.1502400112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. R., Cao, Q. D., Hong, Z. S., Tan, Y. Y., Chen, S. D., Jin, H. J., Tan, K. S., Wang, D. Y., & Yan, Y. (2020). The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Military Medical Research, 7(1), 11. 10.1186/s40779-020-00240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F., Deng, Y., & Li, W. (2020). Coronavirus disease 2019: What we know? Journal of Medical Virology, 92(7), 719–725. 10.1002/jmv.25766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Q., Hou, J., Yang, P., Yan, J., Yu, X., Zhuo, Y., He, S., & Xu, F. (2019). Antiviral activity of mitoxantrone dihydrochloride against human herpes simplex virus mediated by suppression of the viral immediate early genes. BMC Microbiology, 19(1), 274. 10.1186/s12866-019-1639-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, M. P., Pincus, D. L., Rapp, C. S., Day, T. J. F., Honig, B., Shaw, D. E., & Friesner, R. A. (2004). A hierarchical approach to all-atom protein loop prediction. Proteins, 55(2), 351–367. 10.1002/prot.10613 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Kannan, S., Shaik Syed Ali, P., Sheeza, A., & Hemalatha, K. (2020). COVID-19 (Novel Coronavirus 2019) - recent trends. European Review for Medical and Pharmacological Sciences, 24(4), 2006–2011. 10.26355/eurrev_202002_20378 [DOI] [PubMed] [Google Scholar]
- Kollar, J., & Frecer, V. (2017). How accurate is the description of ligand-protein interactions by a hybrid QM/MM approach? Journal of Molecular Modeling, 24(1), 11. 10.1007/s00894-017-3537-z [DOI] [PubMed] [Google Scholar]
- Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., Gwadz, M., Hurwitz, D. I., Marchler, G. H., Song, J. S., Thanki, N., Yamashita, R. A., Yang, M., Zhang, D., Zheng, C., Lanczycki, C. J., & Marchler-Bauer, A. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Reserach, 48(D1), D265–D268. 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakar, S., Sreelatha, L., Dechtawewat, T., Noisakran, S., Yenchitsomanus, P. T., Chu, J., & Limjindaporn, T. (2018). Drug repurposing of quinine as antiviral against dengue virus infection. Virus Research, 255, 171–178. 10.1016/j.virusres.2018.07.018 [DOI] [PubMed] [Google Scholar]
- NCBI Taxonomy Browser. (2020). Severe acute respiratory syndrome coronavirus 2. Retrieved April 28, from https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=2697049.
- Preta, G., Cronin, J. G., & Sheldon, I. M. (2015). Dynasore - not just a dynamin inhibitor. Cell Communication and Signaling: CCS, 13, 24. 10.1186/s12964-015-0102-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., Doig, A., Guilliams, T., Latimer, J., McNamee, C., Norris, A., Sanseau, P., Cavalla, D., & Pirmohamed, M. (2019). Drug repurposing: Progress, challenges and recommendations. Nature Reviews Drug Discovery, 18(1), 41–58. 10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- Rarey, M., Kramer, B., Lengauer, T., & Klebe, G. (1996). A fast flexible docking method using an incremental construction algorithm. Journal of Molecular Biology, 261(3), 470–489. 10.1006/jmbi.1996.0477 [DOI] [PubMed] [Google Scholar]
- Simsek, M., Meijer, B., van Bodegraven, A. A., de Boer, N., & Mulder, C. (2018). Finding hidden treasures in old drugs: The challenges and importance of licensing generics. Drug Discovery Today, 23(1), 17–21. 10.1016/j.drudis.2017.08.008 [DOI] [PubMed] [Google Scholar]
- Sohrabi, C., Alsafi, Z., O'Neill, N., Khan, M., Kerwan, A., Al-Jabir, A., Iosifidis, C., & Agha, R. (2020). World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). International Journal of Surgery (London, England), 76, 71–76. 10.1016/j.ijsu.2020.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan, T. P., & Meyer, C. G. (2020). The COVID-19 epidemic. Tropical Medicine & International Health: TM & IH, 25(3), 278–280. 10.1111/tmi.13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., & Zhu, W. (2016). Molecular docking for drug discovery and development: A widely used approach but far from perfect. Future Medicinal Chemistry, 8(14), 1707–1710. 10.4155/fmc-2016-0143 [DOI] [PubMed] [Google Scholar]
- Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li, C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu, Y., Maciejewski, A., Gale, N., Wilson, A., Chin, L., Cummings, R., Le, D., … Wilson, M. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research, 46(D1), D1074–D1082. 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2020. a). Naming the coronavirus disease (COVID-19) and the virus that causes it. Retrieved April 28, 2020, from https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it.
- World Health Organization . (2020. b). WHO Coronavirus disease (COVID-19) dashboard. Retrieved June 22, 2020, from https://covid19.who.int/.
- Zhang, L., & Liu, Y. (2020). Potential interventions for novel coronavirus in China: A systematic review. Journal of Medical Virology, 92(5), 479–490. 10.1002/jmv.25707 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The docking structures are available upon request from the corresponding author.