Abstract
The recent new contagion coronavirus 2019 (COVID-19) disease is a new generation of severe acute respiratory syndrome coronavirus-2 SARS-CoV-2 which infected millions confirmed cases and hundreds of thousands death cases around the world so far. Molecular docking combined with molecular dynamics is one of the most important tools of drug discovery and drug design, which it used to examine the type of binding between the ligand and its protein enzyme. Global reactivity has important properties, which enable chemists to understand the chemical reactivity and kinetic stability of compounds. In this study, molecular docking and reactivity were applied for eighteen drugs, which are similar in structure to chloroquine and hydroxychloroquine, the potential inhibitors to angiotensin-converting enzyme (ACE2). Those drugs were selected from DrugBank. The reactivity, molecular docking and molecular dynamics were performed for two receptors ACE2 and [SARS-CoV-2/ACE2] complex receptor in two active sites to find a ligand, which may inhibit COVID-19. The results obtained from this study showed that Ramipril, Delapril and Lisinopril could bind with ACE2 receptor and [SARS-CoV-2/ACE2] complex better than chloroquine and hydroxychloroquine. This new understanding should help to improve predictions of the impact of such alternatives on COVID-19.
Communicated by Ramaswamy H. Sarma
Keywords: Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2, molecular docking, molecular dynamincs simulation, global reactivity
1. Introduction
In late 2019, a new generation of coronavirus appeared in Wuhan City in the Hubei Province in central China (Wang, Horby, et al., 2020; Zhu et al., 2020). This virus causes severe acute respiratory syndrome. The first case was reported on the 8th of December 2019 for many patients lived around the local Huanan Seafood Wholesale Market (Chan et al., 2020). The novel coronavirus was identified from the throat swab sample of a patient (Wang, Hu, et al., 2020). World Health Organization has abbreviated this novel coronavirus as 2019-nCoV then the pathogen was renamed to SARS-CoV-2(WHO, 2020). After that, World Health Oorganization declared the pandemic when the virus hit many other countries.
Human infections by the SARS coronavirus are known to be closely associated with interactions between the viral spike protein (S-protein) which has favorable binding affinity for the human Angiotensin-Converting Enzyme 2 (ACE2) (Böhm & Schneider, 2005; Li et al., 2005; Prabakaran et al., 2004; Veeramachaneni et al., 2020). Several studies have also provided evidence of the COVID-19 S-protein binding to the ACE2 receptor (Hoffmann et al., 2020; Lu et al., 2020; Wan et al., 2020).
Angiotensin-converting enzyme (ACE)-related carboxypeptidase is a zinc metallopeptidase ectoenzyme, which is predominantly found in the lungs (Skeggs et al., 1956). ACE2, is a type I integral membrane protein, which it consists of 805 amino acid residues with one Zn2+ essential for enzyme activity. ACE2 was implicated in the regulation of heart function and as a functional receptor for the coronavirus, which is linked to the severe acute respiratory syndrome (SARS). ACE2 is the cellular receptor for the new coronavirus (SARS-CoV-2) which is causing the serious pandemic COVID-19 (Hasan et al., 2020; Li et al., 2003; Towler et al., 2004; Yan et al., 2020).
In a recent study, it was suggested that the 2019-nCoV binds to the human ACE2 receptor via densely glycosylated spike (S) protein as the initiation step of the entry mechanism to human cells (Basit et al., 2020; Boopathi et al., 2020; Hoffmann et al., 2020). The entry of the virus depends on its binding with the cell surface units at site 1 and site 2 S1/S2 that contains Zn+2, an important cofactor for numerous viral proteins as well (Te Velthuis et al., 2010). Existence of this metallic ion facilitates the viral attachment to the surface of target cells. It is well known that zinc ions serve as intracellular second messenger and may trigger apoptosis or efficiently impair replication of a number of viruses and this effect may be based on direct inhibition (Alirezaei et al., 1999; Frederickson et al., 2005; Lazarczyk & Favre, 2008; Te Velthuis et al., 2010).
ACE2 exists in every human body but in different quantities (Gurley & Coffman, 2008). Patients, who suffer from hypertension, diabetes or cardiovascular diseases, have high concentration of ACE2 enzyme in their bodies (Fang et al., 2020; Gurley & Coffman, 2008; Zhou et al., 2020). These categories of people can be easily infected by coronavirus compared with children who have low concentration of ACE2 enzyme, their infection percentage is only 2% (Bunyavanich et al., 2020).
Blocking the active site of ACE2 by suitable pharmaceutical compound will prevent the virus entering to the human cells. Therefore, synthesis of such pharmaceutical compound is in great demand. Many scientists worldwide are trying to synthesise new drugs to stop spreading the new infectious disease. We think that this route takes a long time, at least 18 months, until the new vaccine will be available in the markets. Thus, using medicaments already exist is the shortcut to tackle such issue. In 2005, chloroquine was found as a potent inhibitor of SARS coronavirus infection and it was suggested to treat the new novel coronavirus SARS-CoV-2 with hydroxychloroquine (Adeoye et al., 2020; Amin & Abbas, 2020; Böhm & Schneider, 2005; Smith & Smith, 2020; Vincent et al., 2005). However, due to its cardiotoxicity hydroxychloroquine has been red flagged by USFDA for use as a prophylactic measure.
In this study, 18 drugs were selected to evaluate their binding with two receptors ACE2 and SARS-CoV-2 binding with ACE2 ([SARS-CoV-2/ACE2] complex). These drugs were chosen due to their similarities in structure with chloroquine and hydroxychloroquine in order to find an alternative drug for COVID-19.
2. Materials and methods
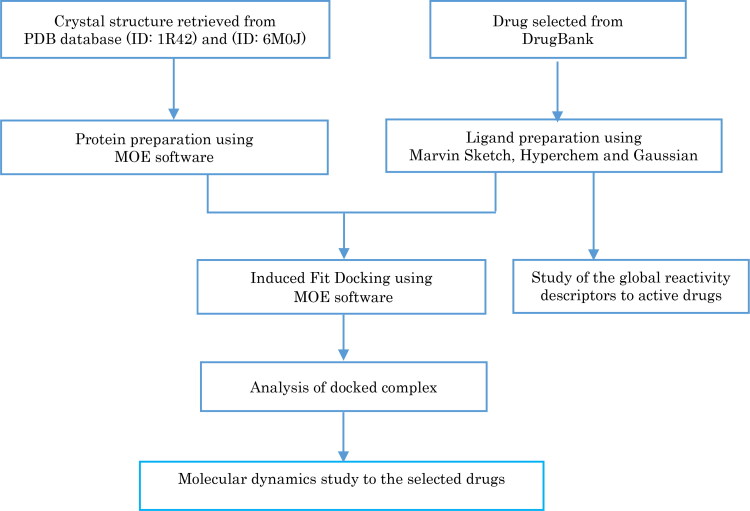
Molecular docking and molecular dynamics simulation was applied to the drugs selected from the DrugBank database (Wishart et al., 2018) to study their affinity with coronavirus antibody ACE2 receptor (PDB ID: 1R42) (Towler et al., 2004) and also study their affinity with the crystal structure of [SARS-CoV-2/ACE2] complex (PDB ID: 6M0J) (Lan et al., 2020) to select the most active drugs that inhibit COVID-19. Global reactivity descriptors of the selected drugs were calculated to understand their structures, stability and reactivity. The methodology of this work is illustrated in Figure 1.
Figure 1.
Schematic representation of the docking procedure, analysis of drugs and reactivity.
2.1. Molecule library preparation
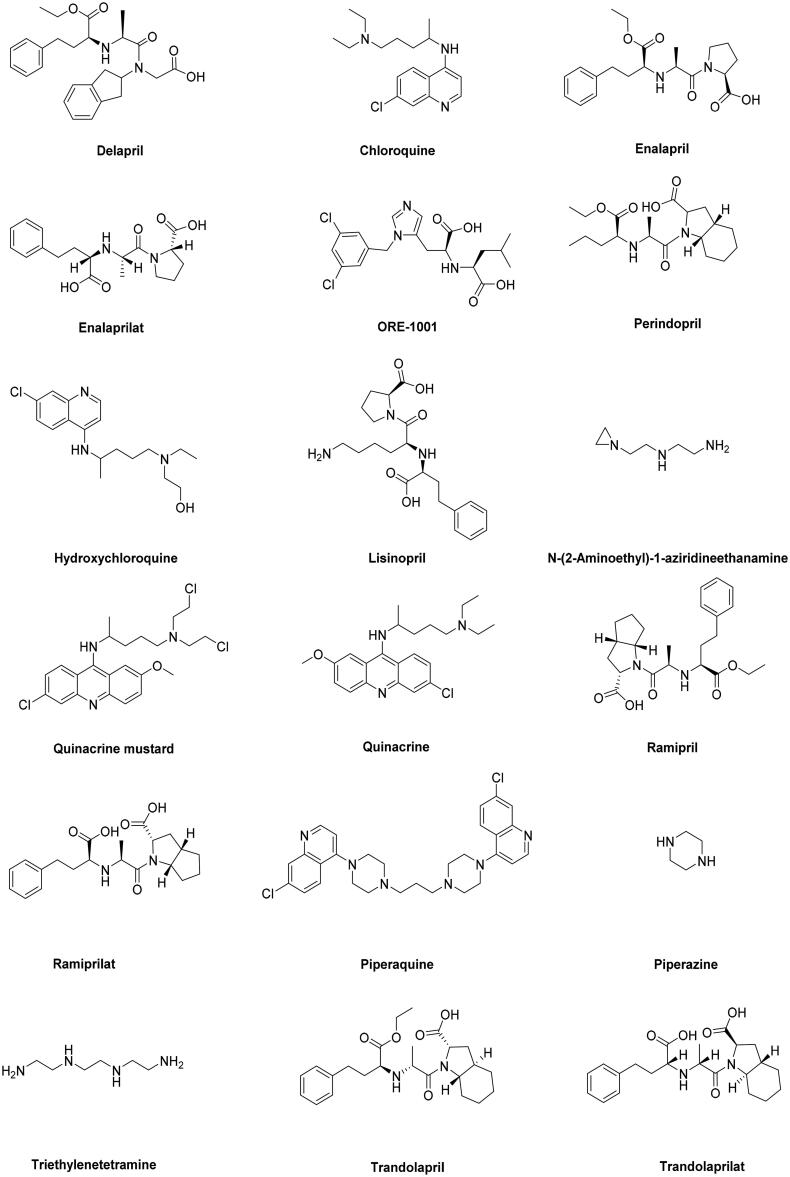
The chemical structure of drugs inhibitors of ACE2 and similar structures were extracted from the DrugBank database (Wishart et al., 2018) in MDL Mol format and converted to 3 D format using Mervin Sketch (MarvinSketch, 2019). The structures were pre-optimized with semi-empirical AM1 method (Stewart, 2013) using Hyperchem 8.08 software (HyperChem, 2009). The structures were optimized using density functional theory DFT method by employing the B3LYP/6-31G basis set (Becke, 1997; Frisch et al., 2009) to obtain the most stable conformation, which was also used to calculate the global reactivity descriptors through Gaussian 09 (Frisch et al., 2009). The convergent value of maximum force, root-mean-square (RMS) force, maximum displacement and RMS displacement are set by default and achieved “YES”. All values are positive after calculation vibrational frequencies to drugs, those results indicate that the drugs are stable (Cavalli et al., 2006). The optimized structures were combined in one database on MOE software (Molecular Operating Environment (MOE), 2015) in order to study the affinity of ligands (Figure 2 and Table 1).
Figure 2.
The structures of selected drugs.
Table 1.
Names, accessions numbers and clinical indication of drugs.
| Drugs names | Accessions Numbers | Clinical Indication |
|---|---|---|
| Chloroquine | DB00608 (APRD00468) | Anti-malarial Anti-inflammatory Anti-parasitic |
| Hydroxychloroquine | DB01611 | Anti-malarial Anti-parasitic Anti-rheumatic Anti-infective |
| Quinacrine | DB01103 (APRD00317) | Anti-infective Anti-malarial Anti-parasitic |
| Quinacrine mustard | DB02240 (EXPT02733) | Anti-parasitic |
| Piperaquine | DB13941 | Anti-infective Anti-malarial Anti-parasitic |
| Ramipril | DB00178 (APRD00009) | Angiotensin-Converting Enzyme
inhibitors Anti-hypertensive Cardiovascular |
| Trandolapril | DB00519 (APRD01269) | Angiotensin-Converting Enzyme
inhibitors Anti-hypertensive Cardiovascular |
| Ramiprilat | DB14208 | Angiotensin-Converting Enzyme
inhibitors Anti-hypertensive Cardiovascular |
| Enalapril | DB00584 (APRD00510) | Angiotensin-Converting Enzyme
inhibitors Anti-hypertensive Cardiovascular |
| Trandolaprilat | DB14209 | Angiotensin-Converting Enzyme inhibitors |
| Lisinopril | DB00722 (APRD00560) | Angiotensin-Converting Enzyme
inhibitors Anti-hypertensive Cardiovascular |
| Perindopril | DB00790 (APRD01178) | Angiotensin-Converting Enzyme
inhibitors Anti-hypertensive Cardiovascular |
| Enalaprilat | DB09477 | Angiotensin-Converting Enzyme
inhibitors Anti-hypertensive Cardiovascular Decreased blood pressure |
| Delapril | DB13312 | Angiotensin-Converting Enzyme
inhibitors Anti-hypertensive Cardiovascular |
| ORE-1001 | DB12271 (DB06387) | Angiotensin-Converting Enzyme inhibitors |
| N-(2-Aminoethyl)-1-aziridineethanamine | DB15643 | Angiotensin-Converting Enzyme inhibitors |
| Triethylenetetramine | DB06824 | Copper chelator agent |
| Piperazine | DB00592 (APRD00225, DB11514) | Anti-parasitic Anti-infective |
2.2. Receptor preparation
The crystal structure of the angiotensin-converting enzyme related carboxypeptidase ACE2 receptor (PDB ID: 1R42) (Towler et al., 2004) and Crystal structure [SARS-CoV-2/ACE2] complex (PDB ID: 6M0J) (Lan et al., 2020) were found in the Protein Data Bank. The enzymes were prepared by removing the N-acetyl-D-glucosamine in sequence editor. Because the water molecule in the active site of the target enzyme plays an important role, it was inserted in the active sites to ensure making a hydrogen bond between the ligand and the target (Böhm & Schneider, 2005; Klebe, 2006; Marechal, 2007).
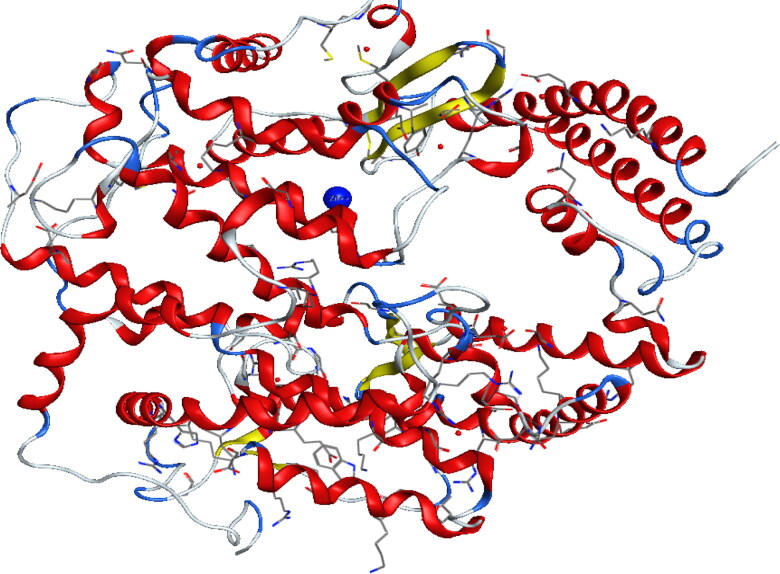
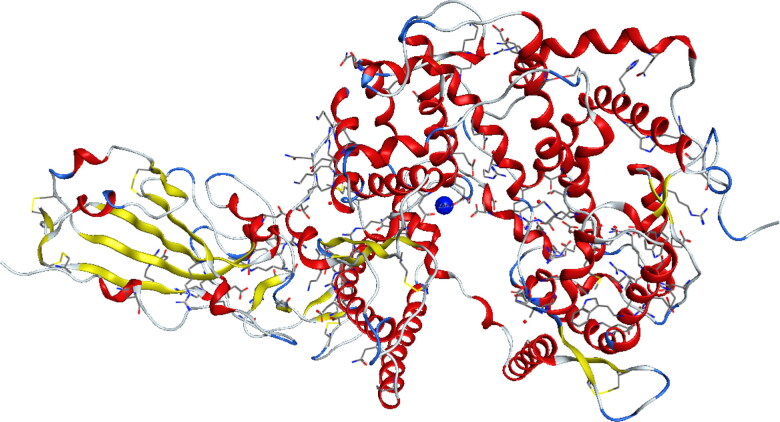
Because Zn2+ is an important cofactor for many viral proteins, Zn2+ can inhibit the replication of ARN polymerase, two active sites containing zinc (Zn2+) in 1R42 and 6M0J enzymes were chosen as shown in Figures 3 and 4 respectively (Te Velthuis et al., 2010). After that, the protein structure was prepared by correcting the missing bonds, which were broken in X-ray diffraction, and then the hydrogen atoms were added (Table 2).
Figure 3.
Crystal structure of native human Angiotensin Converting Enzyme-related carboxypeptidase (ACE2) (PDB ID: 1R42).
Figure 4.
Crystal structure of [SARS-CoV-2/ACE2] complex (PDB ID: 6M0J).
Table 2.
Binding sites residues used as input for receptor grid generation during Induced Fit Docking.
| Receptors | Sites | Residues |
|---|---|---|
| 1R42 | Site 1 | 1: (Arg73, Phe274, Pro346, Asp367, Leu370, Thr371, His374, Glu375, Glu402, Glu406, Ser409, Leu410, Ala413, Phe438, Gln442, Thr445, Ile446, Thr449, Thr453, Phe512, Tyr515, Arg518, Thr519, Gln522) 2 : (Zn804) |
| Site 2 | 1 : (Phe40, Pro346, Thr347, Ala348, Asp350, Gly352, His374, Glu375, His378, Asp382, Tyr385, Phe390, Arg393, Asn394, His401, Glu402) 2: (Zn804) | |
| 6M0J | Site 1 | 1: (Tyr127, Asn149, Asp269, Trp271, Arg273, Phe274, Thr276, Tyr279, Lys288, Pro289, Asn290, Ile291, Asp292, Thr294, His345, Pro346, Thr365, Met366, Asp367, Leu370, Thr371, His374, Glu375, Glu402, Glu406, Ser409, Leu410, Ala413, Thr414, Pro415, Leu418 Phe428, Glu430, Asp431, Thr434, Glu435, Asn437, Phe438, Lys441, Gln442, Thr445, Ile446, Thr449, Leu503, Phe504, His505, Tyr515, Arg518, Thr519, Gln522, Phe523, His540) 3 :(Zn901) |
| Site 2 | 1: (His345, Pro346, Thr347, Ala348, Glu375, His378, Asp382, His401, Glu402) 3 :(Zn901) |
3. Molecular docking
All the docking and scoring calculations were performed using the molecular operation environment software (MOE) (Molecular Operating Environment (MOE), 2015). The crystal structure of human angiotensin converting enzyme (PDB entry: 1R42) (Towler et al., 2004) at a resolution of 2.20 Å and the crystal structure of [SARS-CoV-2/ACE2] complex (PDB entry: 6M0J) (Lan et al., 2020) at a resolution of 2.45 Å were obtained from the Protein Data Bank (Berman et al., 2020) .A resolution between 1.5 and 2.5 Å is considered as a good quality for docking studies (Didierjean & Tête-Favier, 2016; Venugopal et al., 2008). It is known that the best score of RMSD values should be near to 2 Å with an energy score less or equal to −7 Kcal/mol (Kellenberger et al., 2004; Ramalho et al., 2009). These two values are often used as criterion to validate the result of the molecular docking.
4. Global reactivity descriptors
Global reactivity indices are the most relevant traits, which can be derived from the conceptual density functional theory (DFT). They have important properties which enable us to understand the chemical reactivity and kinetic stability of compounds (Shahab et al., 2016). The global reactivity descriptors can be described by energy of the highest occupied molecular orbital (EHOMO), energy of the lowest unoccupied molecular orbital (ELUMO), energy gap (ΔE), global electrophilicity (ω), chemical potential (µ), chemical hardness (η), chemical softness (S) and nucleophilicity (N) (Defranceschi & C. Le Bris, 2000; Domingo et al., 2016; Harkati et al., 2017; Zekri et al., 2020). Those descriptors were calculated at B3LYP/6-31G using the following formulas:
(ΔE = ELUMO-EHOMO), (ω = µ2/2η), (µ = (ELUMO+EHOMO)/2), (η = (ELUMO- EHOMO)/2), (S = 1/(2 η)), (N = EHOMO (Nucleophile) – EHOMO (TCE)).
In this study, the global reactivity descriptors were calculated to compounds that have best result in docking with ACE2 and [SARS-CoV-2/ACE2] complex.
5. Molecular dynamics simulation
The molecular dynamics (MD) simulation study was carried out for the most promising drugs Delapril, Lisinopril and Ramipril to target [SARS-CoV-2/ACE2] complex (6M0J) using standard default parameter setting in the MOE software (Molecular Operating Environment (MOE), 2015).
There are four algorithms implemented in MOE software for MD simulations; the Nosé-Poincaré-Andesen (NPA), the Nosé-Hoover-Andersen (NHA), Berendsen velocity/position (BER) and Nanoscale Molecular Dynamics (NAMD). In this study, the NPA is: the most precise and the most sensitive, was used to study the molecular dynamics of ligands (Sturgeon & Laird, 2000). In MD calculations, MMFF94x force field, sphere shape, water as a solvent, six margins and delete far existing solvent with distance greater than four Å were selected to optimize the system.
6. Results and discussion
6.1. Molecular docking
Molecular docking was run for 18 ligands against the [SARS-CoV-2/ACE2] complex and the ACE2 receptor.
6.1.1. The binding affinities of the drugs into ACE2 active sites
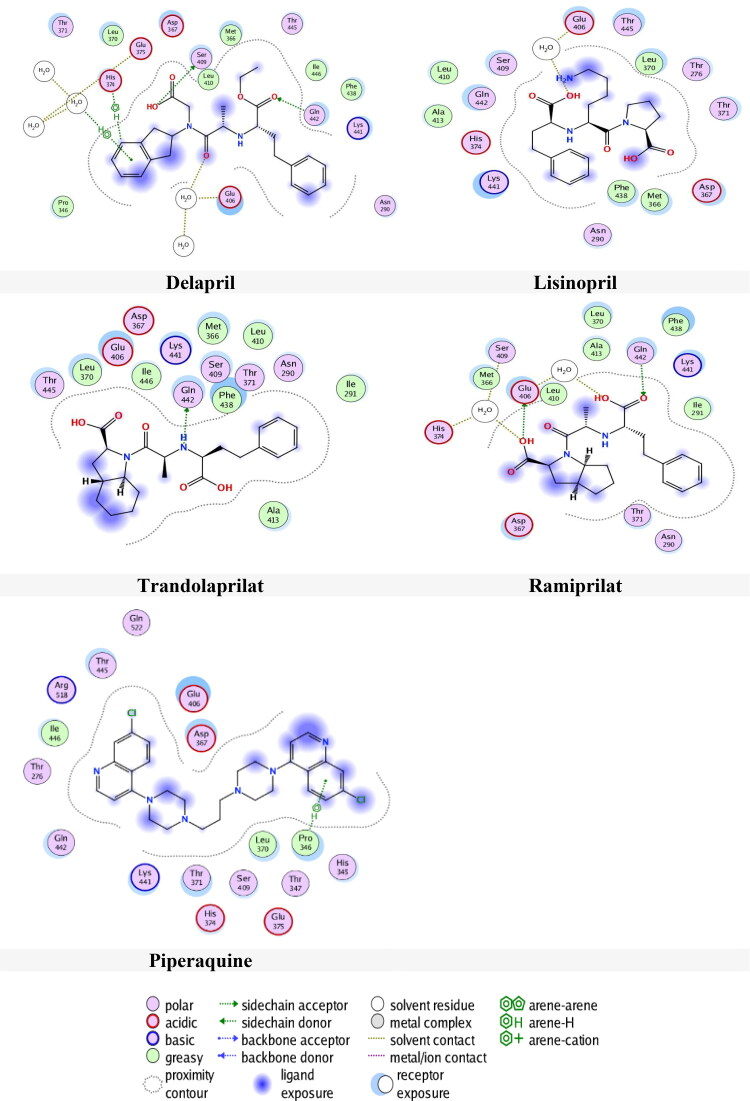
Tables 3 and 4 present the results of docking the drugs in 1R42 at two selected pockets S1 and S2 respectively. The results, as shown in Table 3, indicate that only seven ligands have an interaction with the receptor in pocket S1. Delapril has the best docking score (-6.9809 kcal/mol) followed by Lisinopril (-6.6886 kcal/mol)) with RMSDs 2.2570 Å and 1.5417 Å respectively. On the other hand, Ramiprilat and Piperaquine had RMSDs more than 3 Å and Trandolaprilat, Chloroquine and Perindopril had RMSDs less than 1.5 Å, which this is inadequate .
Table 3.
The results obtained from docking of Drugs with 1R42 in site 1.
| Drugs | S score (kcal/mol) | RMSD (Å) | Bonds
between atoms of compounds and residues of active site 1 of 1R42 |
|||||
|---|---|---|---|---|---|---|---|---|
| Atom of compound | Atom of receptor | Involved receptor residues | Type of interaction bond | Distance (Å) | E (kcal/mol) | |||
| Chloroquine | −6.1074 | 1.1063 | N-1 | O | H2O 932 | H-acceptor | 2.79 | −1 |
| Delapril | −6.9809 | 2.2570 | O-31 | OG | Ser 409 | H-donor | 3.08 | −0.7 |
| O-24 | O | H2O 932 | H-acceptor | 2.84 | −1.3 | |||
| O-25 | NE2 | Gln 442 | 3.16 | −1.7 | ||||
| C-43 | 5-ring | His 374 | H-pi | 3.71 | −1 | |||
| 6-ring | O | H2O 932 | pi-H | 4.08 | −1.2 | |||
| Lisinopril | −6.6886 | 1.5417 | O-5 | O | H2O 932 | H-donor | 3.24 | −0.6 |
| Perindopril | −6.5856 | 1.1260 | O-42 | NE2 | Gln 442 | H-acceptor | 3.3 | −0.8 |
| Piperaquine | −6.6531 | 3.2826 | 6-ring | CD | Pro 346 | pi-H | 4.35 | −0.8 |
| Ramiprilat | −6.6703 | 4.3112 | O-46 | O | H2O 1075 | H-donor | 2.98 | −1.6 |
| O-51 | OE1 | Glu 406 | 2.9 | −2.3 | ||||
| O | H2O 1099 | 2.89 | −1.1 | |||||
| O-45 | NE2 | Gln 442 | H-acceptor | 3 | −1 | |||
| Trandolaprilat | −6.7507 | 1.4433 | N-45 | OE1 | Gln 442 | H-donor | 3.09 | −1.6 |
Table 4.
The results obtained from docking of Drugs with 1R42 in site 2.
| Drug | S score (kcal/mol) | RMSD (Å) | Bonds
between atoms of compounds and residues of active site 2 of 1R42 |
|||||
|---|---|---|---|---|---|---|---|---|
| Atom of compound | Atom of receptor | Involved receptor residues | Type of interaction bond | Distance (Å) | E (kcal/mol) | |||
| Chloroquine | −5.5271 | 1.3462 | N-17 | O | Ala 348 | H-donor | 3.05 | −2 |
| 6-ring | 6-ring | Trp 349 | pi-pi | 3.96 | 0 | |||
| Delapril | −6.5831 | 2.0115 | O-25 | O | H2O 894 | H-acceptor | 2.9 | −0.8 |
| Enalapril | −6.1282 | 2.6836 | C-28 | 5-ring | Trp 349 | H-pi | 3.86 | −0.7 |
| Enalaprilat | −5.9910 | 1.2547 | O-40 | N | Asp 350 | H-acceptor | 3.34 | −1.3 |
| C-45 | 5-ring | Trp 349 | H-pi | 3.46 | −2.6 | |||
| Hydroxychloroquine | −5.6369 | 1.8041 | O-2 | O | Arg 393 | H-donor | 2.99 | −0.8 |
| N-7 | N | Asp 350 | H-acceptor | 3.13 | −1.3 | |||
| Lisinopril | −5.6358 | 1.7176 | O-5 | O | Arg 393 | H-donor | 3.19 | −2.4 |
| Perindopril | −6.2821 | 1.1895 | O-23 | 5-ring | His 401 | H-pi | 3.51 | −0.7 |
| Piperazine | −3.4925 | 2.5032 | C-5 | 5-ring | Trp 349 | H-pi | 3.86 | −0.9 |
| Quinacrine | −5.9184 | 1.1669 | C-37 | 6-ring | Trp 349 | H-pi | 4.42 | −0.6 |
| C-37 | 5-ring | Trp 349 | 3.8 | −1.4 | ||||
| Ramipril | −6.1181 | 1.5054 | O-46 | N | Asp 350 | H-acceptor | 3 | −3.2 |
| O-58 | O | H2O 892 | 3.07 | −1 | ||||
| Ramiprilat | −5.8613 | 1.8268 | O-51 | O | Leu 391 | H-donor | 2.92 | −1.4 |
| O-46 | ND2 | Asn 394 | H-acceptor | 3.02 | −0.8 | |||
| O-49 | NZ | Lys 562 | 3.01 | −5.7 | ||||
| O-54 | ND2 | Asn 394 | 2.85 | −0.9 | ||||
| Trandolaprilat | −5.7171 | 2.8424 | O-53 | O | H2O 952 | H-donor | 2.97 | −2.2 |
Interactions were further examined for bond lengths and hydrogen bonds in site 1 and were illustrated in Figure 5. The results from this Figure 5 showed that Delapril interacts with three amino acids residues in three different interactions; H-donor with amino acid Ser409, H-acceptor with Gln442, H-pi with His374 as well as two H-acceptor and pi-H interactions with the water. The distance and energy binding of interaction are listed in Table 3.
Figure 5.
Compounds binding with 1R42 in site 1.
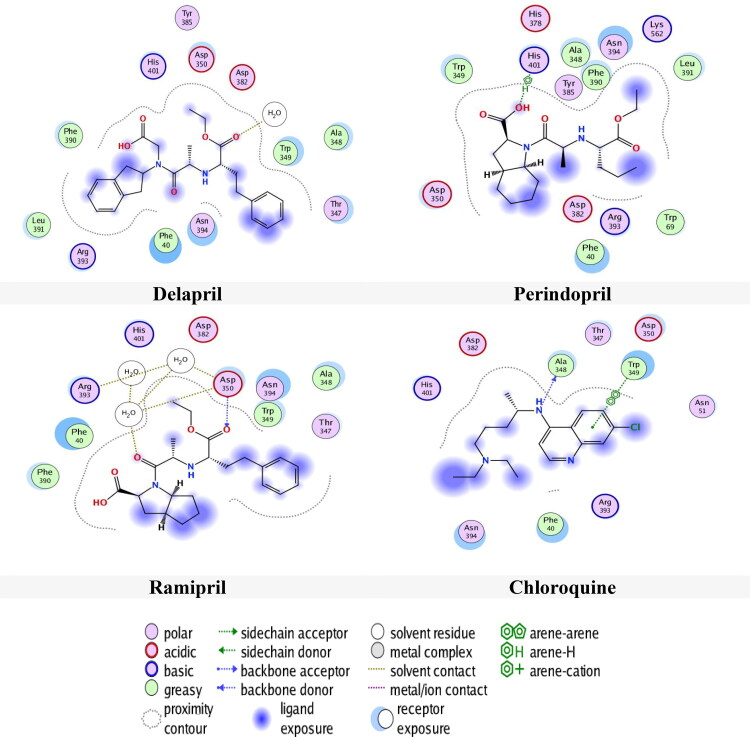
From Table 4, the docking results in pocket S2, it can be noticed that Delapril had the lowest docking score (-6.5831 kcal/mol) with RMSD (2.0115 Å) followed by Perindopril, Ramipril and Chloroquine with docking score and RMSD values of (-6.2821 Kcal/mol, 1.1895 Å), (-6.1181 Kcal/mol, 1.5054 Å) and (-5.5271 Kcal/mol, 1.3462 Å) respectively. Even in this site, Chloroquine had a good score but actually it had an inadequate RMSD value (1.3462 Å), which is less than the accepted limit 1.5 Å. The same things can be said for Enalaprilat, Perindopril and Quinacrine.
The interactions of drugs with site 2 were also examined and depicted in Figure 6. Figure 6 shows that Delapril had H-acceptor interaction with water, while Perindopril had H-pi interaction with amino acid His401. Meanwhile, Ramipril had H-acceptor interaction with amino acid Asp350 and H-acceptor with water and Chloroquine had H-donor interaction with amino acid Ala348 and pi-pi interaction with Trp349. The distance and the energy binding are presented in Table 4.
Figure 6.
Compounds binding with 1R42 in site 2.
6.1.2. The binding affinities of the drugs into [SARS-CoV-2/ACE2] complex active sites
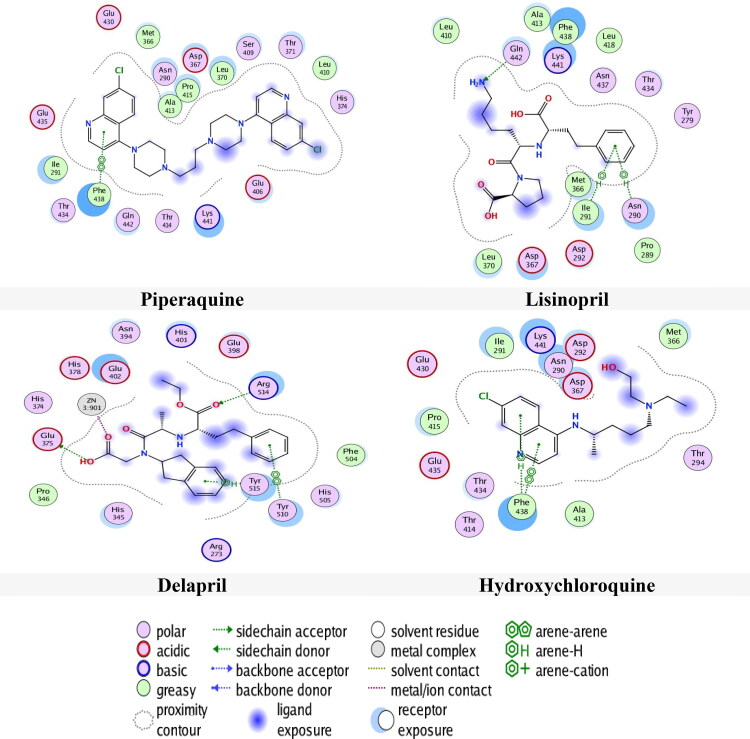
Tables 5 and 6 show the results of docking of the drugs in 6M0J at two selected pockets S1 and S2 respectively. The results in pocket S1 revealed that Piperaquine had the lowest docking score (-8.6132 Kcal/mol) and RMSD (2.3325 Å) compared with Delapril and Hydroxychloroquine, which they had energy scores and RMSD values of (-7.5271 Kcal/mol, 2.1735 Å) and (-7.2272 Kcal/mol, 2.1035 Å) respectively. In spite of Delapril and Hydroxychloroquine did not have the lowest score, they have the best RMSD values. Lisinopril and Quinacrine Mustard had RMSD value less than 1.5 Å.
Table 5.
The results obtained from docking of Drugs with 6M0J in site 1.
| Drugs | S score (kcal/mol) | RMSD (Å) | Bonds
between atoms of compounds and residues of active site 1 of 6M0J |
|||||
|---|---|---|---|---|---|---|---|---|
| Atom of compound | Atom of receptor | Involved receptor residues | Type of interaction bond | Distance (Å) | E (kcal/mol) | |||
| Chloroquine | −6.8442 | 1.9853 | 6-ring | 6-ring | Phe 438 | pi-pi | 3.37 | 0 |
| Delapril | −7.5271 | 2.1735 | O-31 | OE2 | Glu 375 | H-donor | 3.01 | −4.5 |
| O-25 | NH2 | Arg 514 | H-acceptor | 3.04 | −1.4 | |||
| O-26 | ZN | Zn 901 | metallic | 1.96 | −2.1 | |||
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| 6-ring | OH | Tyr 515 | Pi-H | 3.38 | −0.9 | |||
| 6-ring | Tyr 510 | pi-pi | 3.93 | 0 | ||||
| Enalapril | −7.8671 | 1.9897 | O-22 | O | Pro 289 | H-donor | 3.39 | −0.8 |
| Enalaprilat | −6.9279 | 1.8459 | O-44 6-ring |
NZ | Lys 441 | H-acceptor | 3.16 | −8.4 |
| 6-ring | Phe 438 | pi-pi | 3.73 | 0 | ||||
| Hydroxychloroquine | −7.2272 | 2.1035 | 6-ring | CB | Phe 438 | pi-H | 3.82 | −0.8 |
| 6-ring | 6-ring | Phe 438 | pi-pi | 3.81 | 0 | |||
| Lisinopril | −7.5918 | 1.3368 | N-11 | NE2 | Gln 442 | H-acceptor | 3.18 | −2.8 |
| 6-ring 6-ring |
CA | Asn 290 | Pi-H | 4.07 | −0.8 | |||
| N | Ile 291 | 4.22 | −0.9 | |||||
| ORE-1001 | −7.3872 | 1.5557 | Cl | O | Leu 410 | H-donor | 3.49 | −0.8 |
| 5-ring | CB | Phe 438 | pi-H | 4.43 | −0.7 | |||
| 6-ring | 6-ring | Phe 438 | pi-pi | 3.37 | 0 | |||
| Perindopril | −6.4327 | 2.4655 | N-26 | O | Ile 291 | H-donor | 3.21 | −0.8 |
| Piperaquine | −8.6132 | 2.3325 | 6-ring | 6-ring | Phe 438 | pi-pi | 3.35 | 0 |
| Quinacrine | −8.2350 | 1.6346 | 6-ring 6-ring | N | Ile 291 | pi-H | 4.81 | −0.6 |
| N | Ile 291 | 3.98 | −1.1 | |||||
| Quinacrine Mustard | −7.8570 | 1.4398 | Cl-58 | SD | Met 366 | H-donor | 3.74 | −0.4 |
| 6-ring 6-ring |
N | Ile 291 | pi-H | 3.98 | −1.4 | |||
| 6-ring | Phe 438 | pi-pi | 3.58 | 0 | ||||
| Ramipril | −7.7464 | 1.6166 | O-58 | N | Ile 291 | H-acceptor | 3.47 | −0.8 |
| Ramiprilat | −6.9943 | 2.4607 | O-49 | Zn | Zn 901 | metallic | 2.01 | −3.9 |
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
Table 6.
The results obtained from docking of Drugs with 6M0J in site 2.
| Drugs | S score (kcal/mol) | RMSD (Å) | Bonds
between atoms of compounds and residues of active site 2 of 6M0J |
|||||
|---|---|---|---|---|---|---|---|---|
| Atom of compound | Atom of receptor | Involved receptor residues | Type of interaction bond | Distance (Å) | E (kcal/mol) | |||
| Chloroquine | −5.4920 | 2.3627 | C-45 | 5-ring | His 401 | H-pi | 4.25 | −0.9 |
| Delapril | -8.1604 | 1.5603 | O-26 | ZN | Zn 901 | metallic | 2.13 | −3.6 |
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| His 378 | 2.27 | −5.7 | ||||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| Enalapril | −6.7570 | 2.6763 | O-14 | ZN | Zn 901 | metallic | 2 | −2.5 |
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| C-52 | 5-ring | His 378 | H-pi | 3.88 | −1 | |||
| Hydroxychloroquine | −6.3125 | 1.8513 | O-2 | OE2 | Glu 375 | H-donor | 2.86 | −1.9 |
| O-2 | ZN | Zn 901 | metallic | 2 | −2.6 | |||
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| C-47 | 5-ring | His 378 | H-pi | 4.12 | −0.6 | |||
| Lisinopril | −6.6966 | 1.9981 | O-5 | O | H2O 1004 | H-donor | 2.97 | −2 |
| O-1 | ZN | Zn 901 | metallic | 2.06 | −2.3 | |||
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| 6-ring | N | Ile 291 | pi-H | 3.98 | −1.1 | |||
| ORE-1001 | −6.2755 | 2.5319 | N-6 | OH | Tyr 515 | H-acceptor | 3.09 | −2.1 |
| O-25 | ZN | Zn 901 | metallic | 2.09 | −2.3 | |||
| O-31 | ZN | Zn 901 | 2.31 | −0.9 | ||||
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| Perindopril | −6.7968 | 2.2965 | O-23 | O | Glu 398 | H-donor | 2.84 | −3.1 |
| N-26 | OE1 | Glu 402 | 3.11 | −1.4 | ||||
| C-46 | OE2 | Glu 375 | 3.49 | −0.6 | ||||
| O-16 | O | H2O 1033 | H-acceptor | 2.86 | −1.9 | |||
| O-25 | NH2 | Arg 514 | 2.91 | −1.9 | ||||
| O-42 | ZN | Zn 901 | metallic | 1.97 | −2.9 | |||
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| Ramipril | −7.6305 | 2.4853 | O-53 | ZN | Zn 901 | metallic | 2.13 | −1.7 |
| O-58 | ZN | Zn 901 | 2.44 | −1.4 | ||||
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| Ramiprilat | −7.1864 | 1.7252 | O-45 | Zn | Zn 901 | metallic | 1.94 | −2.9 |
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| Trandolapril | −7.1160 | 1.9818 | O-1 | O | H2O 1030 | H-acceptor | 3.04 | −1 |
| O-4 | ZN | Zn 901 | metallic | 2.07 | −3.8 | |||
| Zn-901 | NE2 | His 374 | 2.4 | −3.2 | ||||
| NE2 | His 378 | 2.27 | −5.7 | |||||
| OE1 | Glu 402 | 2.1 | −5.6 | |||||
| NE2 | His 378 | ionic | 2.27 | −11.7 | ||||
| OE1 | Glu 402 | 2.1 | −14.4 | |||||
| OE2 | Glu 402 | 3.13 | −3.7 | |||||
| 6-ring | CA | Glu 398 | pi-H | 3.63 | −0.6 | |||
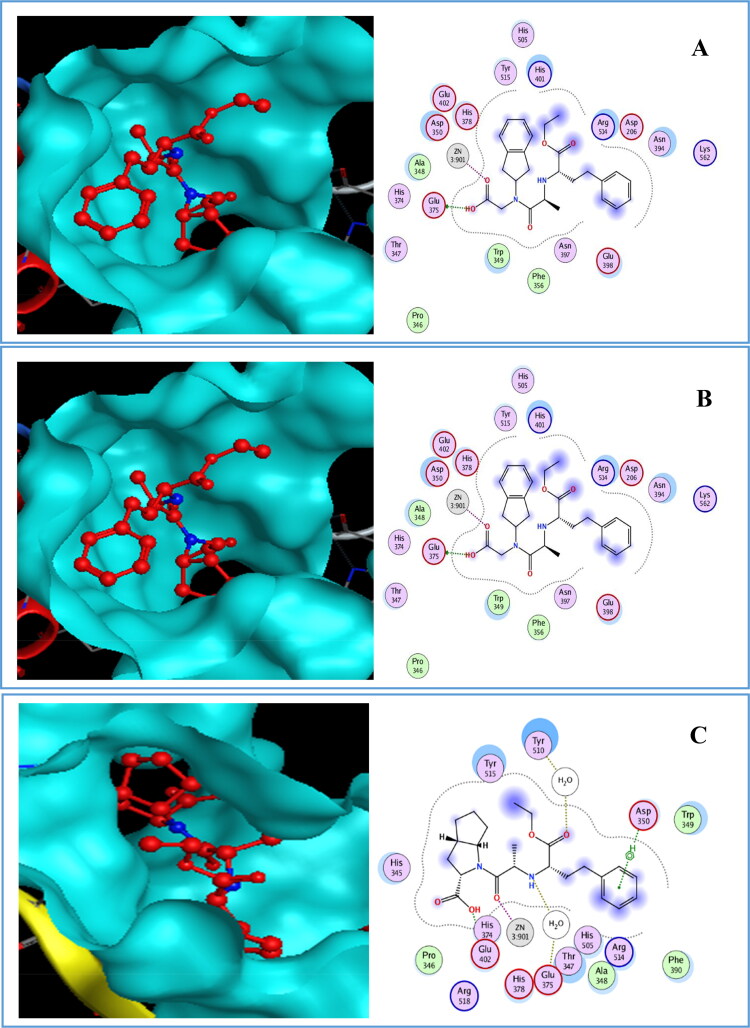
The results of the binding of drugs with 6M0J in site 1 are shown in Figure 7. From the Figure 7, It is apparent that Piperaquine had pi-pi interaction with amino acid Phe438, whereas Hydroxychloroquine had pi-H and pi-pi interactions with amino acid Phe438 and Delapril had numerous interactions; H-donor interaction with amino acid Glu375, H-acceptor with Arg514 and metallic interaction with zinc.
Figure 7.
Compounds binding with 6M0J in site 1.
The interaction of carboxylic functional group in Delapril with zinc motivates the zinc to interact with His374 by metallic interaction and with His378 and Glu402 by ionic and metallic interactions respectively. As mentioned above, zinc had an antiviral activity and this type of interaction may inhibit the COVID-19.
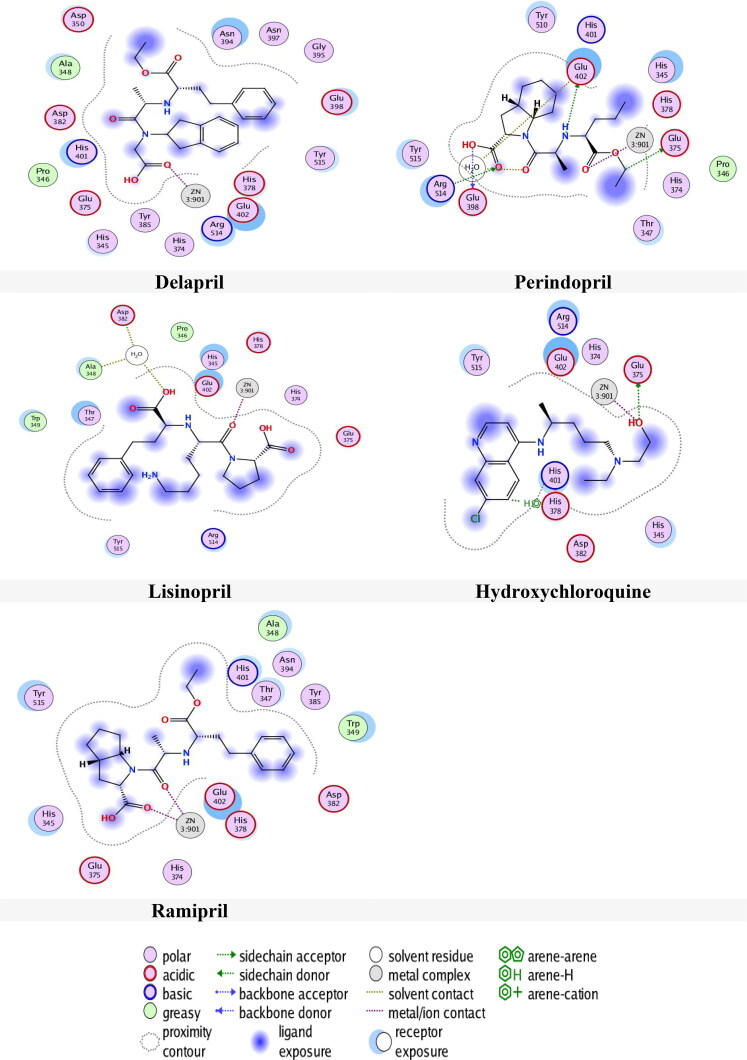
The results of docking of drugs with 6M0J in site 2 are shown in Table 6. According to the results in this site 2, almost all drugs make interacted in pocket S2 via zinc. Delapril showed excellent docking score −8.1604 Kcal/mol and RMSD 1.5603 Å compared with Perindopril, Lisinopril, Hydroxychloroquine and Ramipril with energy scores and RMSD values of (-6.7968 kcal/mol, 2.2965 Å), (-6.6966 Kcal/mol, 1.9981 Å), (-6.3125 Kcal/mol, 1.8513 Å) and (-7.6305 kcal/mol, 2.4853 Å) respectively.
Although in site 2, Enalaprilat, N-(2-aminoethyl)-1-aziridineethamine, Piperaquine, Piperazine, Quinacrine Mustard, Trandolaprilat and Quinacrine have interactions with the active site but they have unacceptable RMSD values.
In all pockets, N-(2-aminoethyl)-1-aziridineethamine, Triethylenetetramine and Piperazine had energy docking scores higher than −4 Kcal/mol, they had energy scores out of the accepted limit, therefore these compounds could not be considered. Also, in all results, Chloroquine had energy scores higher than Hydroxychloroquine and Delapril.
Figure 8 presents the interactions of drugs with 6M0J in site 2. From Figure 8, it can be seen that Delapril had a metallic interaction with Zn, meanwhile Zn interacts with three amino acids by two types of interactions. These are: two ionic and one metallic interactions with Glu402, one ionic and one metallic interactions with His378 and ionic interaction with His374.
Figure 8.
Compounds binding with 6M0J in site 2.
Perindopril had many interactions, three H-donor interactions with amino acids Glu398, Glu402 and Glu375, two H-acceptor with water and with amino acid Arg514 as well as metallic interaction with Zn. Meanwhile Zn had two ionic and metallic interactions, with amino acid Glu402, metallic and ionic interactions with amino acid His378 and metallic interaction with amino acid His374.
Hydroxychloroquine had H-donor interaction with amino acid Glu375, metallic interaction with Zn, H-pi interaction with amino acid His378, while Zn had the same interactions with these amino acids. Lisinopril had H-donor interaction with water and metallic interaction with Zn, whereas Zn interacts with the same amino acids. Ramipril had two metallic interaction with Zn. whereas Zn interacts with the same amino acids.
6.2. Global reactivity descriptors
The chemical reactivity descriptors were calculated and presented in Table 7. The EHOMO and ELUMO were obtained from GaussView (Dennington et al., 2016). The results of the global hardness and softness, which they are related to the stability of chemical system, as shown in Table 7, indicate that Ramipril, Chloroquine, ORE-1001 and Delapril are harder than the Hydroxychloroquine and other compounds.
Table 7.
HOMO and LUMO energy, energy gap ΔE and global reactivity indices µ, ω, η and N for drugs.
| Drugs | HOMO (eV) | LUMO (eV) | ΔE (eV) | η (eV) | S (eV) | µ (eV) | ω (eV) | N (eV) |
|---|---|---|---|---|---|---|---|---|
| Chloroquine | −5.4861 | −1.2232 | 4.2629 | 2.1315 | 173.6972 | −3.3546 | 2.6398 | 3.1698 |
| Delapril | −5.9438 | −0.5853 | 5.3585 | 2.6792 | 138.1850 | −3.2646 | 1.9888 | 2.7121 |
| Enalapril | −5.7435 | −0.7380 | 5.0055 | 2.5028 | 147.9282 | −3.2407 | 2.0981 | 2.9124 |
| Hydroxychloroquine | −6.5095 | 0.2797 | 6.7892 | 3.3946 | 109.0637 | −3.1149 | 1.4291 | 2.1464 |
| Lisinopril | −6.6328 | −1.0583 | 5.5745 | 2.7873 | 132.8292 | −3.8455 | 2.6527 | 2.0231 |
| ORE-1001 | −6.9346 | −1.9323 | 5.0023 | 2.5011 | 148.0248 | −4.4334 | 3.9292 | 1.7213 |
| Perindopril | −5.6564 | 0.3793 | 6.0358 | 3.0179 | 122.6789 | −2.6386 | 1.1534 | 2.9995 |
| Piperaquine | −6.9269 | 0.0678 | 6.9947 | 3.4973 | 105.8603 | −3.4296 | 1.6815 | 1.7290 |
| Ramipril | −6.0807 | −3.1299 | 2.9508 | 1.4754 | 250.9350 | −4.6053 | 7.1873 | 2.5752 |
| Ramiprilat | −6.4178 | −0.3420 | 6.0758 | 3.0379 | 121.8712 | −3.3799 | 1.8802 | 2.2381 |
| Trandolapril | −6.1084 | −0.7565 | 5.3519 | 2.6760 | 138.3537 | −3.4324 | 2.2013 | 2.5475 |
Notes: the HOMO energy -8.6559 eV. of the reference system (TCE) had been calculated at DFT/B3LYP 6-31 G.
In addition, Ramipril have the smaller energy gap (ΔE = 2.9508 eV), Delapril and Lisinopril have smaller energy gaps than Hydroxychloroquine. Moreover, Ramipril, Chloroquine, ORE-1001 and Delapril have softness values higher than that of Hydroxychloroquine. These results indicate that Ramipril, Chloroquine, ORE-1001 and Delapril are more stable and more reactive than Hydroxychloroquine.
The electronic chemical potential (µ) for Perindopril (µ= −2.6386 eV) is higher than other compounds followed by Hydroxychloroquine, Enalapril and Delapril. According to these results, these compounds can exchange electron density with the environment efficiently (Azarhazin et al., 2019).
A further classification of organic molecules as strong (N > 3 eV), moderate (2.0 eV ≤ N ≤ 3.0 eV) and marginal nucleophilic (N < 2.0 eV) were obtained by analysis of a series of common nucleophilic species participating in polar organic reaction. Note that nucleophilicity value is referred to tetracyanoethylen (TCE) taken as a reference, because it presents the lowest EHOMO in a large series of molecule already investigated (Jaramillo et al., 2008). According to the results in Table 7, Chloroquine can be classified as strong nucleophile and the others as moderate nucleophile except ORE-1001, which is considered as marginal nucleophile.
The electrophilicity ω had become a potent tool for the study of the reactivity of organic compounds that can participate in polar reaction (Domingo et al., 2016; Srivastava, 2020). Ramipril had the highest electrophilicity value (ω = 7.1873 eV), whereas as Delapril had an electrophilicity value (ω = 1.9888 eV) higher more than that of Hydroxychloroquine (ω = 1.4291 eV).
6.3. Molecular dynamics simulation
In order to examine the conformational flexibilities of docked drug-receptor complexes and to attain dependable drug-receptor–binding affinities, the MD process combined with binding energy (MM-GBSA) (De Vivo et al., 2016; Kerrigan, 2013)calculations was run for 600 ps on the most promising drugs Delapril, Lisinopril and Ramipril to target [SARS-CoV-2/ACE2] complex (6M0J). The evaluated average MM-GBSA binding energies are given in Table 8.
Table 8.
Calculated MM-GBSA binding energies (in kcal/mol) for the Delapril, Lisinopril and Ramipril drugs against 6M0J over MD simulations.
| Drugs | Site 1 | Site 2 |
|---|---|---|
| Delapril | −54 | −45 |
| Lisinopril | −33 | −38 |
| Ramipril | −46 | −42 |
In general, it is apparent from this table that the selected three drugs exhibited considerable binding energies). In site 2, Delapril and Ramipril showed promising binding energies −54 and −46 kcal/mol respectively. On the other hand, Lisinopril showed relatively weak binding energy −33 kcal/mol. Whereas, in site 2, all three drugs Delapril, Lisinopril and Ramipril showed promising binding affinities with binding energies.
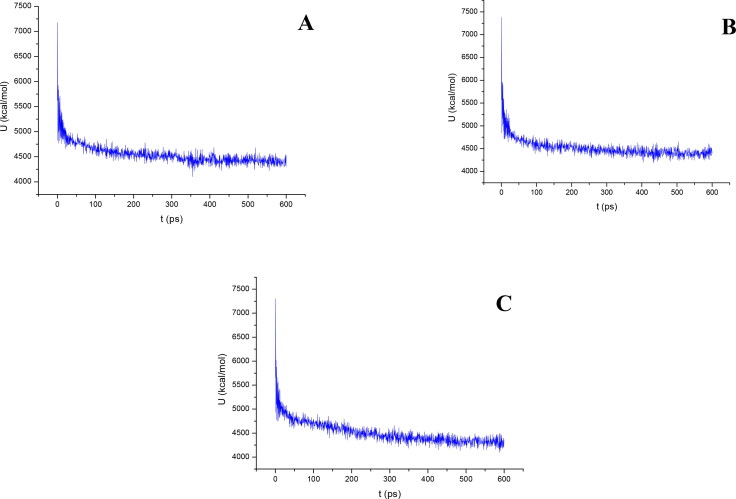
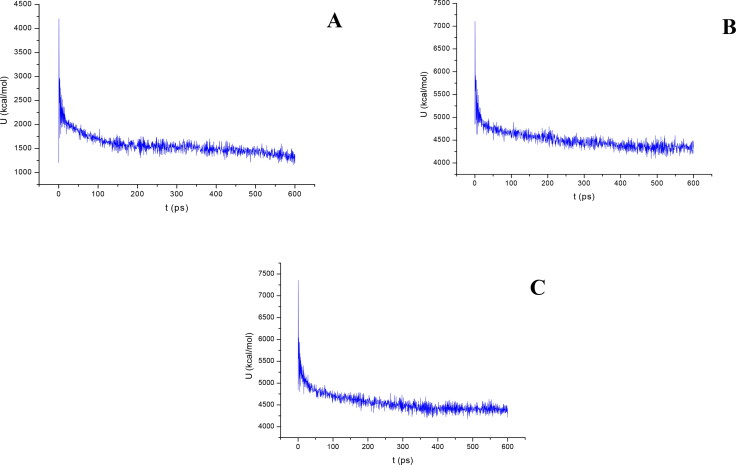
Figures 9 and 10 show the results of the atomic potential energy function during dynamic study calculation for Delapril, Lisinopril and Ramipril in the 6M0J at site 1 and 2 respectively. To explore the dynamic stability of the 6M0J/inhibitor drugs complexes, the time-dependent potential energy of the complex were calculated during MD trajectories. It is apparent in Figure 9, site 1, that complex A (6M0J/Delapril) achieved equilibrium around 300 ps. Meanwhile complex B (6M0J/Lisinopril) achieved the equilibrium around 350 ps. Whereas, complex C (6M0J/Ramipril) achieved the equilibrium stability around 400 ps. It can be seen from Figure 10, site 2, that the complex A achieved the equilibrium stability around 400 ps, complex B achieve the equilibrium stability around 400 ps, meanwhile complex C achieve the equilibrium stability around 350 ps.
Figure 9.
The evaluation of potential energy of complex of (A) Delapril, (B) Lisinopril and (C) Ramipril with 6M0J receptor site 1 as function of time.
Figure 10.
The evaluation of potential energy of complex of (A) Delapril, (B) Lisinopril and (C) Ramipril with 6M0J receptor site 2 as function of time.
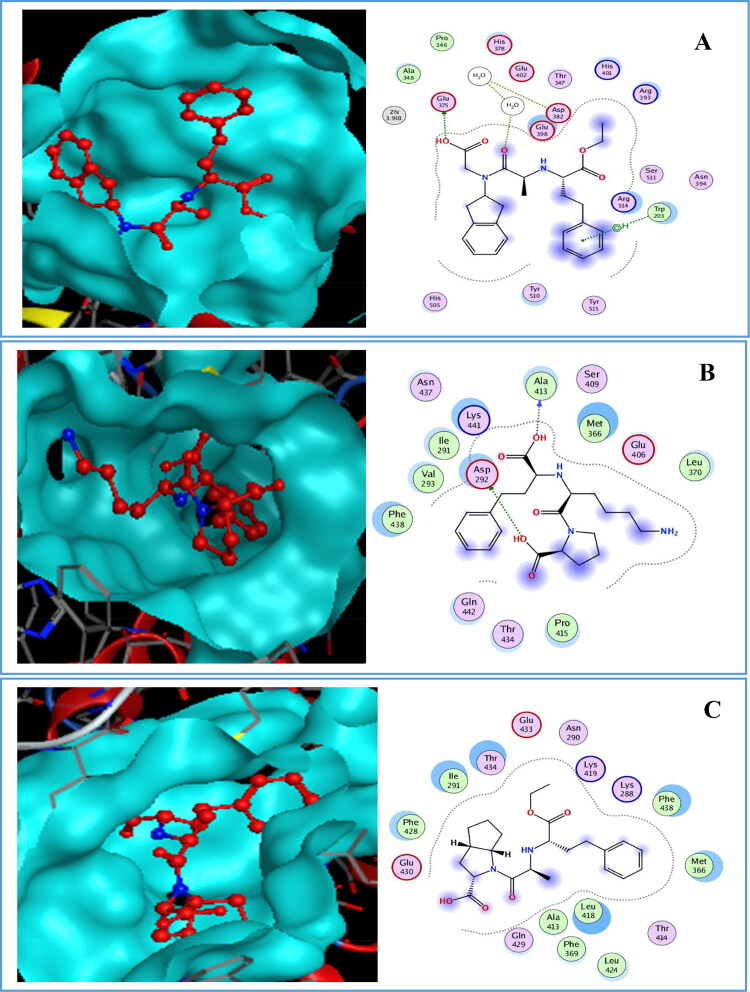
In general, if the interaction energy between a residue and a ligand is lower than −0.8 Kcal/mol, the residue is regarded as an important residue in the molecular recognition of the ligand. For the 6M0J/Delapril complex A (Figure 11), the major favourable energy contributions (-2.2 to −1.4 kcal/mol) originate predominately from Glu375 (-1.4), H2O1030 (-1.5) and Trp203 (-2.2), As shown in Figure 11 the complex B had energy binding with Asp292 (-7.8) and Ala413 (-4.7). However, complex C did not interact in this site.
Figure 11.
Docked pose and binding interaction of (A) Delapril, (B) Lisinopril, (C) Ramipril with 6M0J in site 1.
It is clear from Figure. 12 that complex A had interactions in site 2 of 6M0J with Glu375 (-8.7) and Zn901 (-3.4), while complex B had the major favourable energy contributions (-0.6 to −6.2 kcal/mol) which originate predominately from Glu402 (-2.3), Asp382 (-6.2), H2O1033 (-1.3), Tyr510 (-2.8), H2O1004 (-1.5), His401 (-0.6) and Trp349 (-1). Nevertheless, His401 cannot be considered as an important residue.
Figure 12.
Docked pose and binding interaction of (A) Delapril, (B) Lisinopril, (C) Ramipril with 6M0J in site 2.
Complex C showed more favourable interactions with residues Glu402 (-3.8), H2O1030 (-1.3), H2O1002 (-0.9), Zn901 (-4.1) and Asp350 (-2).
7. Conclusion
The aim of the present research was to examine the binding of eighteen candidate drugs with ACE2 enzyme and [SARS-CoV-2/ACE2] complex using docking analysis. The docking ranking results in this study showed that some of these ligands might have the ability to inhibit SARS-CoV-2. The results of docking these ligands with ACE2 enzyme (1R42) in two pockets indicated that Delapril gave the lowest energy score and good RMSD value followed by Lisinopril (site1) and Ramipril (site 2). In addition, the docking results with 6M0J showed that only Delapril and Ramiprilat interacted with Zn in site 1, while in site 2 Delapril gave the best energy score followed by Ramipril. The drugs mentioned above presented good results with the two chosen enzymes compared with Chloroquine and Hydroxychloroquine. Moreover, the results obtained from global reactivity indices indicated that Ramipril is the most reactive drug, it had the highest electrophilicity value followed by ORE-1001, Chloroquine and Lisinopril. The most obvious finding to emerge from this study is that Ramipril, Delapril and Lisinopril gave good docking results compared with Chloroquine and Hydroxychloroquine. Also, Delapril, Lisinopril and Ramipril showed encouraging binding affinity, MM/GBSA energies, to [SARS-CoV-2/ACE2] complex. Further investigation and experimentation into Delapril, Lisinopril and Ramipril, which they are promising candidate drugs for COVID-19 patients, is strongly recommended.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Adeoye, A. O., Oso, B. J., Olaoye, I. F., Tijjani, H., & Adebayo, A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics–. 10.1080/07391102.2020.1765876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alirezaei, M., Nairn, A. C., Glowinski, J., Prémont, J., & Marin, P. (1999). Zinc inhibits protein synthesis in neurons. Potential role of phosphorylation of translation initiation factor-2alpha. The Journal of Biological Chemistry, 274(45), 32433–32438. 10.1074/jbc.274.45.32433 [DOI] [PubMed] [Google Scholar]
- Amin, M., & Abbas, G. (2020). Docking study of Chloroquine and Hydroxychloroquine interaction with SARS-CoV-2 spike glycoprotein-An in silico insight into the comparative efficacy of repurposing antiviral drugs. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1775703 [DOI] [PubMed] [Google Scholar]
- Azarhazin, E., Izadyar, M., & Housaindokht, M. R. (2019). Drug-DNA interaction, a joint DFT-D3/MD study on safranal as an anticancer and DNA nanostructure model. Canadian Journal of Chemistry, 97(2), 120–130. 10.1139/cjc-2018-0126 [DOI] [Google Scholar]
- Basit, A., Ali, T., & Rehman, S. U. (2020). Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1768150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becke, A. D. (1997). Density-functional thermochemistry. V. Systematic optimization of exchange-correlation functionals. The Journal of Chemical Physics, 107(20), 8554–8560. 10.1063/1.475007 [DOI] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., Bourne, P. E. (2020). The Protein Data Bank. https://www.rcsb.org/pdb. [DOI] [PMC free article] [PubMed]
- Böhm, H. J., & Schneider, G. (2005). Protein-ligand interactions: From molecular recognition to drug design. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA. 10.1002/3527601813 [DOI] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 Coronavirus Structure, Mechanism of Action, Antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunyavanich, S., Do, A., & Vicencio, A. (2020). Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA, 323(23), 2427. 10.1001/jama.2020.8707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli, A., Carloni, P., & Recanatini, M. (2006). Target-related applications of first principles quantum chemical methods in drug design. Chemical Reviews, 106(9), 3497–3519. 10.1021/cr050579p [DOI] [PubMed] [Google Scholar]
- Chan, J. F.-W., Yuan, S., Kok, K.-H., To, K. K.-W., Chu, H., Yang, J., Xing, F., Liu, J., Yip, C. C.-Y., Poon, R. W.-S., Tsoi, H.-W., Lo, S. K.-F., Chan, K.-H., Poon, V. K.-M., Chan, W.-M., Ip, J. D., Cai, J.-P., Cheng, V. C.-C., Chen, H., Hui, C. K.-M., & Yuen, K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vivo, M., Masetti, M., Bottegoni, G., & Cavalli, A. (2016). Role of molecular dynamics and related methods in drug discovery. Journal of Medicinal Chemistry, 59(9), 4035–4061. 10.1021/acs.jmedchem.5b01684 [DOI] [PubMed] [Google Scholar]
- Defranceschi, M., & Le Bris, C. (2000). Mathematical models and methods for ab initio quantum chemistry (Vol. 136). Springer Science & Business Media. https://doi.org/ 10.1016/0166-1280(86)87076-2 [DOI] [Google Scholar]
- Dennington, R., Keith, T. A., & Millam, J. M. (2016). GaussView, version 6.0. 16. Semichem Inc. [Google Scholar]
- Didierjean, C., & Tête-Favier, F. (2016). Introduction to Protein Science. Architecture, Function and Genomics. Third Edition. By Arthur M. Lesk. Oxford University Press, 2016. Pp. 466. Paperback. Price GBP 39.99. ISBN 9780198716846. Acta Crystallographica Section D Structural Biology, 72(12), 1308–1309. 10.1107/S2059798316018283 [DOI] [Google Scholar]
- Domingo, L. R., Ríos-Gutiérrez, M., & Pérez, P. (2016). Applications of the conceptual density functional theory indices to organic chemistry reactivity. Molecules, 21(6), 748. 10.3390/molecules21060748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D. S. , Feunang, Y. D., Guo, A. C, Lo, E. J., Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li, C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu, Y., Maciejewski, A., Gale, N., Wilson, A., Chin, L., Cummings, R., Le, D., … & Wilson M. 2018. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2017 Nov 8. 10.1074/jbc.274.45.32433. [DOI] [PMC free article] [PubMed]
- Fang, L., Karakiulakis, G., & Roth, M. (2020). Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? The Lancet. Respiratory Medicine, 8(4), e21. 10.1016/S2213-2600(20)30116-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson, C. J., Koh, J. Y., & Bush, A. I. (2005). The neurobiology of zinc in health and disease. Nature Reviews. Neuroscience, 6(6), 449–462. 10.1038/nrn1671 [DOI] [PubMed] [Google Scholar]
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloin, O. J., Zheng, G., Sonnenberg, J. L., Hada, M., … Tomasi, J. (2009). Gaussian, Inc., 2009.
- Gurley, S. B., & Coffman, T. M. (2008). Angiotensin-converting enzyme 2 gene targeting studies in mice: Mixed messages. Experimental Physiology, 93(5), 538–542. 10.1113/expphysiol.2007.040014 [DOI] [PubMed] [Google Scholar]
- Harkati, D., Belaidi, S., & Saleh, B. A. (2017). A theoretical investigation on the structures, global and local reactivity descriptors of oxazolidine-2,4-dione, imidazolidine-2,4-dione and thiazolidine-2,4-dione. Quantum Matter, 6, 1–5. 10.1166/qm.2017.1441 [DOI] [Google Scholar]
- Hasan, A., Paray, B. A., Hussain, A., Qadir, F. A., Attar, F., Aziz, F. M., Sharifi, M., Derakhshankhah, H., Rasti, B., Mehrabi, M., Shahpasand, K., Saboury, A. A., & Falahati, M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, . 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N. H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HyperChem (8.08). (2009). Molecular modelling system. Hypercube Inc. [Google Scholar]
- Jaramillo, P., Domingo, L. R., Chamorro, E., & Pérez, P. (2008). A further exploration of a nucleophilicity index based on the gas-phase ionization potentials. Journal of Molecular Structure: Theochem, 865(1–3), 68–72. 10.1016/j.theochem.2008.06.022 [DOI] [Google Scholar]
- Kellenberger, E., Rodrigo, J., Muller, P., & Rognan, D. (2004). Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins, 57(2), 225–242. 10.1002/prot.20149 [DOI] [PubMed] [Google Scholar]
- Kerrigan, J. E. (2013). Molecular dynamics simulations in drug design. Methods in Molecular Biology (Clifton, N.J.).), 993, 95–113. 10.1007/978-1-62703-342-8_7 [DOI] [PubMed] [Google Scholar]
- Klebe, G. (2006). Virtual ligand screening: Strategies, perspectives and limitations. Drug Discovery Today, 11(13–14), 580–594. 10.1016/j.drudis.2006.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., & Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581(7807), 215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Lazarczyk, M., & Favre, M. (2008). Role of Zn2+ ions in host-virus interactions. Journal of Virology, 82(23), 11486–11494. 10.1128/JVI.01314-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., Choe, H., & Farzan, M. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Zhang, C., Sui, J., Kuhn, J. H., Moore, M. J., Luo, S., Wong, S. K., Huang, I. C., Xu, K., Vasilieva, N., Murakami, A., He, Y., Marasco, W. A., Guan, Y., Choe, H., & Farzan, M. (2005). Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. The EMBO Journal, 24(8), 1634–1643. 10.1038/sj.emboj.7600640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal, Y. (2007). The hydrogen bond and the water molecule. Elsevier. 10.1016/B978-0-444-51957-3.X5000-8 [DOI] [Google Scholar]
- MarvinSketch (19.25.0). (2019). Calculation Module Developed by ChemAxon.
- Molecular Operating Environment (MOE) (2015.10). (2015). Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910.
- Prabakaran, P., Xiao, X., & Dimitrov, D. S. (2004). A model of the ACE2 structure and function as a SARS-CoV receptor. Biochemical and Biophysical Research Communications, 314(1), 235–241. 10.1016/j.bbrc.2003.12.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho, T. C., Caetano, M. S., da Cunha, E. F. F., Souza, T. C. S., & Rocha, M. V. J. (2009). Construction and assessment of reaction models of class i epsp synthase: Molecular docking and density functional theoretical calculations. Journal of Biomolecular Structure & Dynamics, 27(2), 195–207. 10.1080/07391102.2009.10507309 [DOI] [PubMed] [Google Scholar]
- Shahab, S., Hajikolaee, F. H., Filippovich, L., Darroudi, M., Loiko, V. A., Kumar, R., & Borzehandani, M. Y. (2016, February). Molecular structure and UV–Vis spectral analysis of new synthesized azo dyes for application in polarizing films. Dyes and Pigments, 129, 9–17. 10.1016/j.dyepig.2016.02.003 [DOI] [Google Scholar]
- Skeggs, L. T., Kahn, J. R., & Shumway, N. P. (1956). The preparation and function of the hypertensin-converting enzyme. The Journal of Experimental Medicine, 103(3), 295–299. 10.1084/jem.103.3.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M., & Smith, J. C. (2020). Repurposing therapeutics for COVID-19: Supercomputer-based docking to the SARS-CoV-2 viral spike protein and viral spike protein-human ACE2 interface. ChemRxiv. 10.26434/chemrxiv.11871402.v3 [DOI] [Google Scholar]
- Srivastava, R. (2020). Chemical reactivity theory (CRT) study of small drug-like biologically active molecules. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1725642 [DOI] [PubMed] [Google Scholar]
- Stewart, J. J. P. (2013). Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. Journal of Molecular Modeling, 19(1), 1–32. 10.1007/s00894-012-1667-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgeon, J. B., & Laird, B. B. (2000). Symplectic algorithm for constant-pressure molecular dynamics using a Nosé-Poincaré thermostat. The Journal of Chemical Physics, 112(8), 3474–3482. 10.1063/1.480502 [DOI] [Google Scholar]
- Te Velthuis, A. J. W., van den Worm, S. H. E., Sims, A. C., Baric, R. S., Snijder, E. J., & van Hemert, M. J. (2010). Zn(2+) inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathogens, 6(11), e1001176. 10.1371/journal.ppat.1001176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler, P., Staker, B., Prasad, S. G., Menon, S., Tang, J., Parsons, T., Ryan, D., Fisher, M., Williams, D., Dales, N. A., Patane, M. A., & Pantoliano, M. W. (2004). ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis . Journal of Biological Chemistry, 279(17), 17996–18007. 10.1074/jbc.M311191200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramachaneni, G. K., Thunuguntla, V. B. S. C., Bobbillapati, J., & Bondili, J. S. (2020). Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1773318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal, C., Demos, C. M., Jagannatha Rao, K. S., Pappolla, M. A., & Sambamurti, K. (2008). Beta-secretase: Structure, function, and evolution. CNS & Neurological Disorders Drug Targets, 7(3), 278–294. 10.2174/187152708784936626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., Seidah, N. G., & Nichol, S. T. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology Journal, 2(1), 10–69. 10.1186/1743-422X-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. Journal of Virology, 94(7), e00127-20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Horby, P. W., Hayden, F. G., & Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., Wang, B., Xiang, H., Cheng, Z., Xiong, Y., Zhao, Y., Li, Y., Wang, X., & Peng, Z. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA, 323(11), 1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2020). World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.).), 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri, A., Harkati, D., Kenouche, S., & Saleh, B. A. (2020). QSAR modeling, docking, ADME and reactivity of indazole derivatives as antagonizes of estrogen receptor alpha (ER-α) positive in breast cancer. Journal of Molecular Structure, 1217, 128442. 10.1016/j.molstruc.2020.128442 [DOI] [Google Scholar]
- Zhou, F., Yu, T., Du, R., Fan, G., Liu, Y., Liu, Z., Xiang, J., Wang, Y., Song, B., Gu, X., Guan, L., Wei, Y., Li, H., Wu, X., Xu, J., Tu, S., Zhang, Y., Chen, H., & Cao, B. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. The Lancet, 395(10229), 1054–1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]