Abstract
Due to an outbreak of COVID-19, the number of research papers devoted to in-silico drug discovery of potential antiviral drugs is increasing every day exponentially. Still, there is no specific drug to prevent or treat this novel coronavirus (SARS-CoV-2) disease. Thus, the screening for a potential remedy presents a global challenge for scientists. Up to date over a hundred crystallographic structures of SARS-CoV-2 Mpro have been deposited to Protein Data Bank. With many known proteins, the demand for a reliable target has become higher than ever, so as the choice of an efficient computational methods. Therefore, in this study comparative methods have been used for receptor-based virtual screening, targeting 9 selected structures of viral Mpro. Reliability analyses followed by re-docking of the specific co-crystallized ligand provided the best reproductivity for structures with PDB ID 6LU7, 6Y2G and 6Y2F. The influence of crystallographic water on an outcome of a virtual screening against selected targets was also investigated. Once the most reliable targets were selected, the library of easy purchasable natural compounds were retrieved from the MolPort database (10,305 compounds) and docked against the selected Mpro proteins. To ensure the efficiency of the selected compounds, binding energies for top-15 hit ligands were calculated using Molecular Mechanics as well as their absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties were predicted. Based on predicted binding energies and toxicities, top-5 compounds were selected and subjected to Molecular Dynamics simulation and found to be stable in complex to act as possible inhibitors for SARS-CoV-2.
Communicated by Ramaswamy H. Sarma
Keywords: Protein reliability, docking, molecular dynamics, virtual screening, natural compounds, SARS-CoV-2
1. Introduction
Severe Acute Respiratory Syndrome - Coronavirus-2 (SARS-CoV-2) is the causative virus for the pandemic Coronavirus Disease or COVID-19 (Zhou et al., 2020). It has spread more rapidly and with more potency than its two well-known precursors, SARS-CoV and MERS-CoV (Gorbalenya et al., 2020). The infected people produce an array of flu-like symptoms from cough to fever but the novel coronavirus is, also, characterized by extensive dermal and cardiac complications (Joob & Wiwanitkit, 2020; Zheng et al., 2020). Currently, no specific therapies for COVID-19 are available and a treatment of patients is limited to medications that can ease the symptoms. Antiviral and antimalarial drugs are undergoing clinical studies along with the clinical trial of multiple vaccines as potential treatments for COVID-19 (Gautret et al., 2020).
Largest portion of research employing computational drug design approaches targeting SARS-CoV-2 Mpro structure with PDB ID 6LU7, which was a first Mpro structure in complex with covalently linked N3 ligand deposited to PDB (Jin et al., 2020). Zhang, Lin, et al. (2020) deposited crystal to PDB structures of Mpro in its apo (6Y2E), as well as monoclinic (6Y2F), and orthorhombic (6Y2G) forms in complex with α-ketoamide inhibitor. Its Apo form was used along with 6LU7 (Sampangi-Ramaiah et al., 2020) for molecular docking analysis of natural compounds from plants. Must be noted that binding affinities of selected ligands targeting 6LU7 and 6Y2E had a poor correlation (R2 = 0.236), and binding poses were slightly different, which suggest an importance of target selection for identification of potential drugs. Cocrystalized ligand forms of 6Y2G and 6Y2F were used as a target for virtual screening and molecular docking of selected FDA-approved drugs (Kumar & Singh, 2020). SARS-CoV-2 Mpro unliganded structure 6Y84, deposited by Owen et al. (2020) was used for another drug-repurposing blind molecular docking analyses of selected known drugs and bioactive natural compounds (Das et al., 2020). Apo form 6MO3 (Zhang, Zhao, et al., 2020) was used as a target for molecular docking and MD simulation of five natural marine compounds as potential inhibitors (Khan et al., 2020). Up to date over a hundred crystallographic structures of Mpro are available in PDB and this number continues to grow. Due to an increasing number of known protein structures, the demands for a reliable target and efficient computational methods have become more urgent than ever. Interestingly, not many research papers have been devoted to investigation of structure reliability. Palese (2020) in his work analyzed 78 structures of SARS-CoV-2 and 26 structures of SARS-CoV using a principal component analysis (PCA). Interestingly, 6LU7 was detected outside the 99% confidence ellipse and it even showed a bigger similarity to structures of SARS-CoV than to others from its own strain.
Not only a target selection but also identification of an appropriate method for molecular docking is crucial for an achievement of trustable results. Large number of research papers carries out molecular docking using AutoDock Vina software (Kumar & Singh, 2020; Palese, 2020; Sampangi-Ramaiah et al., 2020). Less often Schrodinger Glide (Kandeel & Al-Nazawi, 2020; Ton et al., 2020), Molecular Operating Environment (Farag et al., 2020; Haider et al., 2020; Khan et al., 2020) or other programs are used. Important marker of selected methods’ reproductively is a positive control method, based on re-docking of co-crystallized ligand. Thereby re-docking of N3 co-crystallized ligand of 6LU7 was carried out in order to compare the performance of AutoDock Vina and SMINA (Talluri, 2020). Obtained results indicated a similar performance for both programs. The results obtained by different levels of precision were compared for molecular docking performed using Glide (Ton et al., 2020), and showed Standard Precision (SP) as a fast and at the same time accurate enough method for this task. While some research papers are limited to molecular docking or virtual screening exclusively, the others provide more comprehensive investigation by including prediction of an absorption, distribution, metabolism, and excretion properties (ADME) (Das et al., 2020; Enmozhi et al., 2020; Khan et al., 2020) and toxicity profile. l very much important for timely drug discovery maintaining the safety issue of new drugs. The MD simulations have been mostly performed when investigating a small number of ligands and researchers had used simulation time vary from 1 to 100 ns (Khan et al., 2020; Muralidharan et al., 2020).
Combination of all the techniques described above can provide a state-of-art methodology for a rational in-silico drug design, though currently, the number of such research papers is rather limited. Therefore, herein comparative methods have been used as standard techniques for receptor-based virtual screening. In this study, the reliability of Mpro for total pool of 9 structures retrieved from the PDB was assessed first. It was then followed by evaluation of potential targets achieved by re-docking of co-crystallized ligands. Thereafter, virtual screening of the library of natural compounds (MolPort) against the most reliable Mpro structures of SARS-CoV-2 was performed. At the next step binding energies were calculated using MM-GBSA and ADMET profiling and predicted for top-15 hit ligands. The selected ligands were also docked into an active site of SARS-CoV Mpro to check whether the selected ligands were also active for previous strain. Finally, the MD simulation for 30 ns was carried out for top-5 compounds in order to determine the most suitable drug-like molecule that can become a potential therapeutic against SARS-CoV family of viruses.
2. Materials and methods
2.1. Protein reliability and preparation
Recently deposited 3D structures of the SARS-CoV-2 virus Mpro have been retrieved from the PDB (Table 1). The Schrodinger software package was used for all the simulations study (Sastry et al., 2013). To compare reliability of retrieved structures, a structure analysis panel was used to obtain a Protein Reliability Report. The diffraction data was downloaded from PDB along with the protein structures and was used for reliability analysis.
Table 1.
Selected Mpro protein structures of SARS-COV-2 retrieved from the PDB.
| PDB Code | Form | Resolution | Date released | Reference |
|---|---|---|---|---|
| 6LU7 | in complex with N3 | 2.16 Å | 2/5/2020 | Jin et al., 2020 |
| 6Y2E | unliganded form | 1.75 Å | 3/4/2020 | Zhang, Lin, et al., 2020 |
| 6Y2F | monoclinic form in complex with α-ketoamide | 1.95 Å | 3/4/2020 | Zhang, Lin, et al., 2020 |
| 6Y2G | orthorhombic form in complex with α-ketoamide | 2.20 Å | 3/4/2020 | Zhang, Lin, et al., 2020 |
| 6M03 | unliganded form | 2.00 Å | 3/11/2020 | Zhang, Zhao, et al., 2020 |
| 6Y84 | unliganded form | 1.39 Å | 3/11/2020 | Owen et al., 2020 |
| 5RF8 | in complex with Z271004858 | 1.44 Å | 3/25/2020 | Fearon et al., 2020a |
| 5RG0 | in complex with PCM-0102535 | 1.72 Å | 3/25/2020 | Fearon et al., 2020b |
| 5R8T | unliganded form | 1.27 Å | 4/1/2020 | Fearon et al., 2020c |
Each of the target proteins were prepared in the presence and absence of crystallographic waters using Protein Preparation Wizard (Sastry et al., 2013) implemented in the Schrodinger software package. The bond orders were assigned, and hydrogens were added after removal of the original hydrogens. Missing loops and side chains were filled in using Prime (Jacobson et al., 2004), and the protonation states were generated using Epik (Shelley et al., 2007) with pH value 7.0 ± 2.0. Hydrogen bond networks were optimized, and their restrained minimization was carried out using OPLS3e (Roos et al., 2019) force fields. The SiteMap module (Halgren, 2009) was used to detect possible binding sites. Five top-ranking fine grids have been created for each protein with and without water molecules. The grid was centered on the co-crystallized ligands (centered on x: 14.0, y: 1.0, z: 24.0; length − 20 Å) with a scaling factor value 1.
2.2. Ligand preparation and filtration
The MolPort Database (2020) includes information on 10,305 unique purchasable natural compounds originating in plants, animals, and microbial and marine sources. The structures have been downloaded and subjected to LigPrep (2020). Ionization states for the compounds have been generated at target pH of 7.0 ± 2.0 using Epik.
2.3. Virtual screening
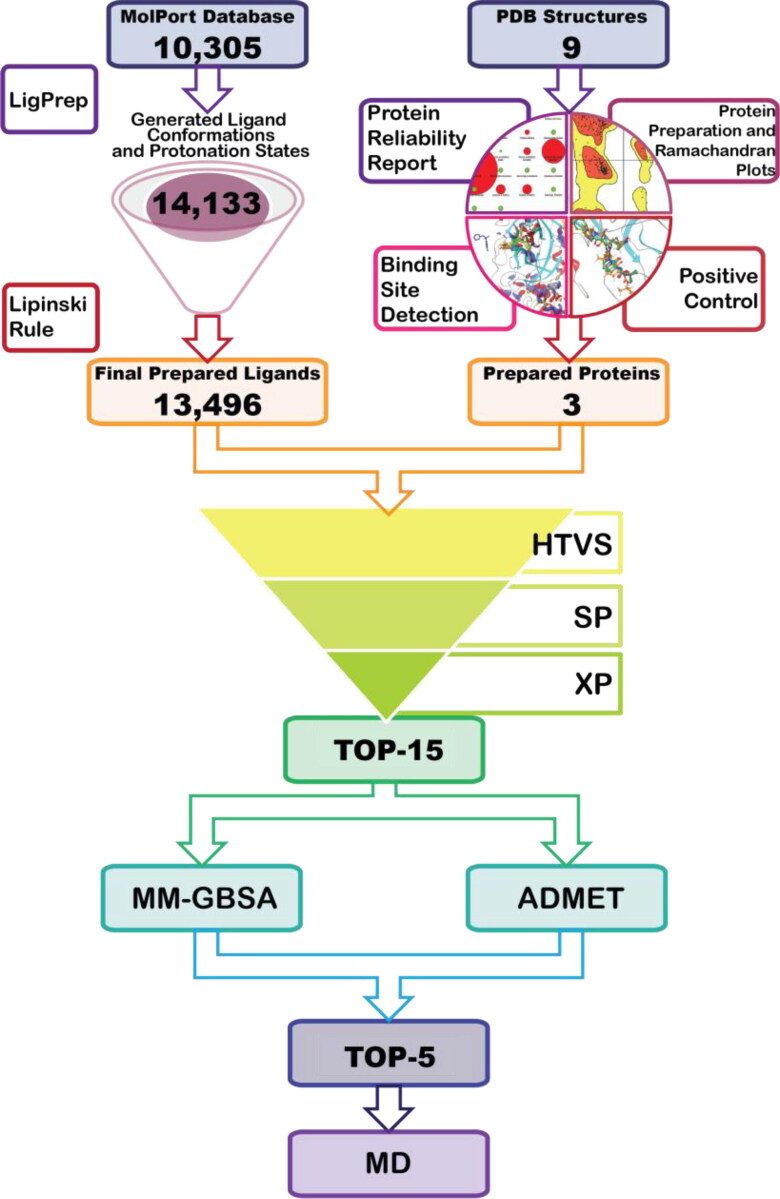
The Virtual Screening Workflow of Glide (Friesner et al., 2006) was used for screening the compounds’ library against the SARS-CoV-2 virus Mpro. During the first step of virtual screening compounds that did not satisfy Lipinski’s rule of 5 were eliminated and filtration was performed using the QikProp (2020). The selected 13,496 compounds were subjected to virtual screening with Glide HTVS (high throughput virtual screening), followed by SP (standard precision) and XP (extra precision) docking calculation (each of methods keeping the top 10% of compounds). Virtual screening was performed for flexible ligands, while a protein structure was kept rigid. Planarity of conjugated π groups was enhanced and Epik state penalties of ligands were added to docking score. The scaling factor was set to 1 and calculations were performed for all the target proteins with water molecules being eliminated and remained using OPLS3e force fields. A description of Ligand Preparation and Virtual screening steps is shown in Figure 1.
Figure 1.
Graphical representation of performed workflow.
The top hits from each XP simulation in total of 15 compounds were used to perform postprocessing with Prime molecular mechanics/generalized born surface area (MM-GBSA) calculation implemented Virtual Screening Workflow of Schrodinger software package. Settings for MM-GBSA are not available to change in Virtual Screening Workflow, thus default settings were used. Estimated free binding energies have been calculated based on the following equation:
| (1) |
Followed by, the protein-ligand complexes were ranked based on their binding free energy calculation.
2.4. Positive control molecular docking
The positive control was performed with the same settings as virtual screening. Co-crystalized ligands of proteases 6LU7, 6Y2F, 6Y2G, 5RF8, and 5RG0 were docked into all Mpro structures in order to verify whether chosen settings of virtual screening workflow can reproduce the experimental data.
2.5. Admet prediction
Once the drug molecule is synthesized and showed its desired activity for anticipated disease, it must go through preclinical and clinical trials (Phase I to Phase IV) for the evaluation of absorption, distribution, metabolism, elimination, and toxicity (ADMET) properties. There should be a fine tuning between drug-likeness and ADMET profiling of a new drug candidate to be a successful one. Thus, early prediction of drug-likeness and ADMET of probable drug molecules can help to avoid costly late-stage drug failure in the drug discovery process (Kar & Leszczynski, 2017). Many of new potential drugs even failed at the last stage ‘Market surveillance’ of the clinical trial. In the initial screening and ligand preparation, we have already filtered molecules with Lipinski’s rule of 5 and Qikprop to pass for further investigations only those ligands which showed the drug-likeness properties like lipophilicity, solubility etc. Therefore, top-15 molecules from docking study were further analyzed to determine their ADMET profile. All ADME properties were determined using pkCSM software (http://biosig.unimelb.edu.au/pkcsm/) (Pires et al., 2015) providing SMILES of the molecules. Meanwhile multiple organ toxicities were predicted employing pkCSM and ProTox-II (http://tox.charite.de/protox_II/) (Banerjee et al., 2018) software. The idea to use both software packages is to predict all possible sources of toxicity along with important toxicological pathways like 7 Nuclear receptor signaling pathways and 5 Stress response pathways with prediction of major toxicity target (checked from 16 major toxicity targets), toxicity value in form of LD50 and toxicity class. In the era of green chemistry and concerning possible ecotoxic effect of pharmaceuticals lead us to check the toxicity towards to major species (T. pyriformis and fathead minnow) for ecotoxicity determination of these drug candidates using pkCSM.
2.6. Molecular dynamics simulation
Selected top-5 compounds became a subject to MD simulation implemented in Desmond module (Bowers et al., 2006). The MM-GBSA output files have been used as an input for MD studies. An orthorhombic box with water molecules has been used to build a model. The system has been neutralized with sodium ions. Temperature and pressure were maintained at 300 K and 1.01325 bar, respectively. Trajectories have been calculated using a simple point charge solvent model. Simulation time was set to 100 ns with a time step of 25 ps. Obtained trajectories were analyzed using Simulation Interaction Diagram.
3. Results
3.1. Proteins’ reliability analysis
Relevant information on protein reliability is illustrated in Figure 1S (Supporting Information file 1). All the Mpro structures have a problem with steric clashes except for 5RF8, 5RGO and 5R8T. However, these three proteases have been detected to possess peptide planarity problems. The 6Y2G displayed the worst reliability report with 552 steric clashes, 12 waters with no hydrogen binding (HB) partners, 12 problems with a peptide planarity, and a poor average B-factor as well as problem with protein packing. Meanwhile, the 6Y2F showed to have similar to previous one’s reliability report. Even though it possesses fewer problems, it contrary to 6Y2G, has buried unsatisfied acceptors. Based on overall analysis, 5R8T, 5RF8, and 6LU7 exhibited the best reliability when compared to the other Mpro structures.
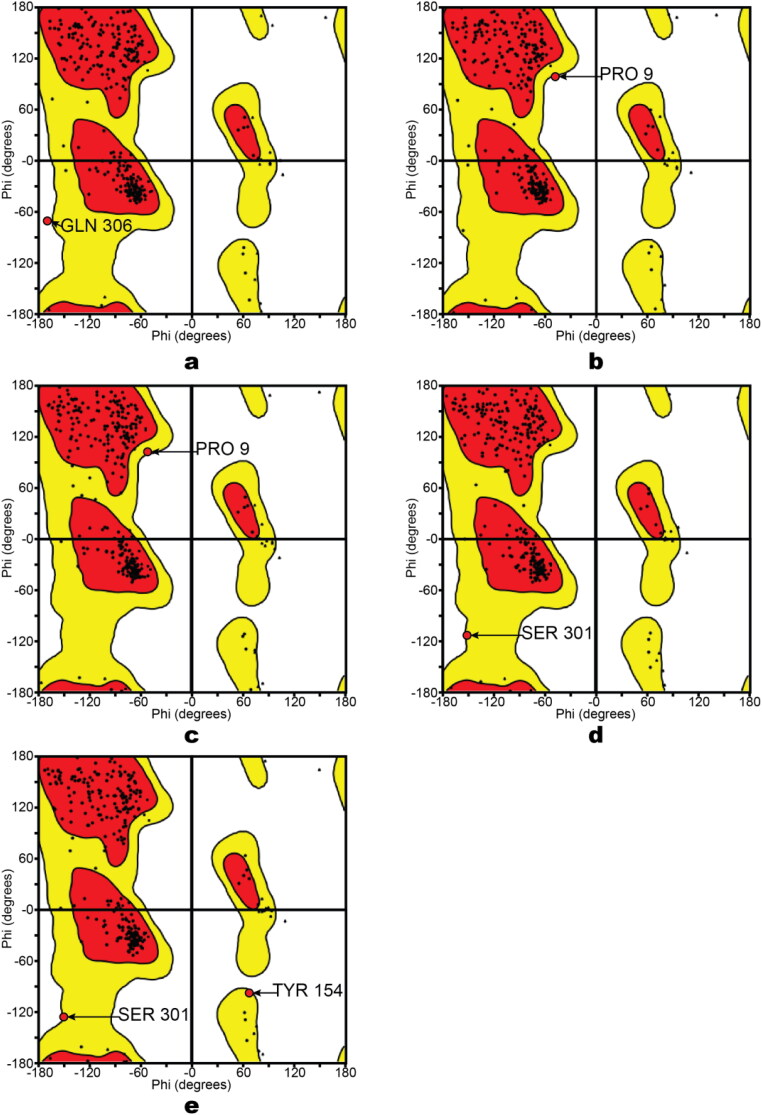
Since we encountered problems with the water molecules with no HB partners, each protein was generated and minimized in two forms (crystallographic water molecules remained and another form with waters being removed). Steric clashes seem to serve as an important contributor to the reliability of some proteases studied here. The verification of steric clashes was carried out by building a Ramachandran plot (Figure 2S, Supporting Information file 1). Each plot illustrates whether any residues of a protein have torsion angle values that are not possible because of steric hindrance in the absence or presence of water molecules. Out of the minimized protease structures, 6Y2E with water molecules has GLN306 inside prohibited region, where only glycine is allowed (Figure 2a). However, in the absence of water molecules 6Y2E has PRO9 in the prohibited region (Figure 2b). Therefore, it explains why the formation of steric clashing occurs. For 6Y2F with no water molecules (Figure 2c) and 6Y2G under both water conditions (Figure 2d-e) showed amino acid residues PRO9, SER301, TYR154 that were very close to prohibited region. Above-mentioned residues were at significant distance from the binding pocket proposed in literature, hence, problems with steric clashes of 6Y2F, 6Y2G, and 6Y2E may have insignificant impact on virtual screening quality.
Figure 2.
Ramachandran Plots. a – 6Y2E; b – 6Y2E (no water); c – 6Y2F (no water); d – 6Y2G; e – 6Y2G (no water).
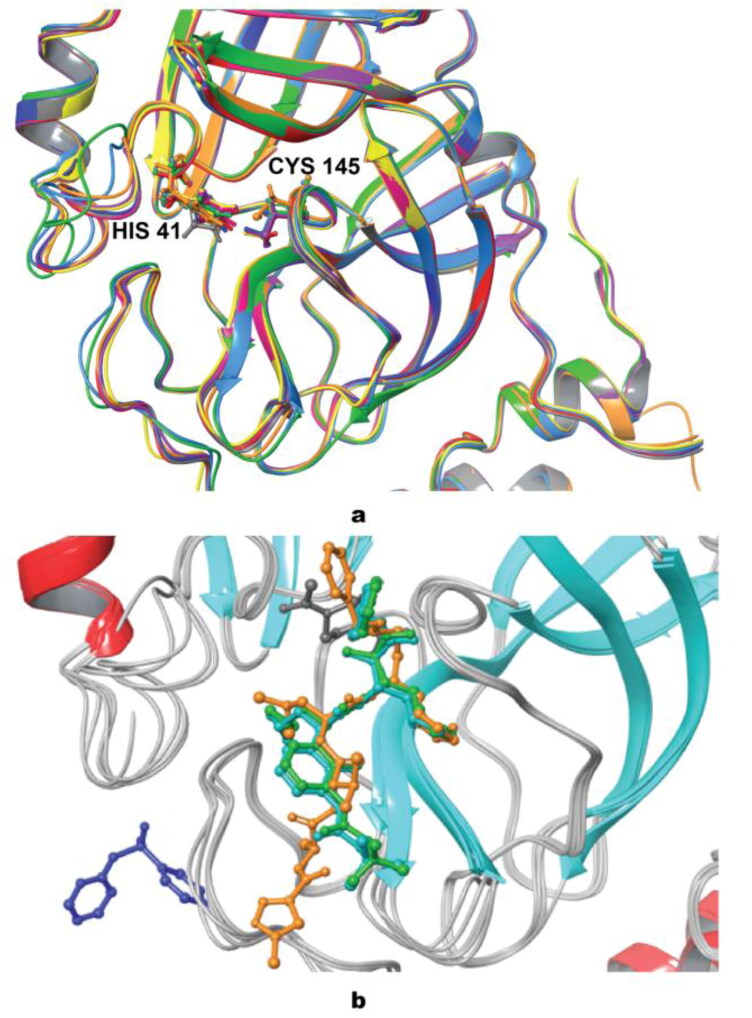
The difference between geometries for all of nine proteins in terms of the root mean square deviation (RMSD) did not exceed 0.792 Å. A significant difference near the binding site was detected with the biggest deviation being found between residues CYS44-TYR54 (Figure 3a). Mpro structure with code 6Y2F (colored in green) has a slight mismatch compared to the other geometries, as well as 6Y2G (colored in light blue) having a slight deviation. In addition to this, segment GLN189-ASP197 was also altered. Since these fragments are close to the binding site, the difference in geometries may play a significant role in the results of virtual screening. Likely, most of the proteins retain similar positioning with HIS41 and CYS145 serving, as the most important residues forming a catalytic dyad. Though, in the case of 5RG0 (colored in gray), the imidazole ring of HIS41 is located perpendicular to the other proteins, while for 6LU7, (colored in orange) CYS145 is slightly moved toward the center of the protease when compared to other structures. The superposition of co-crystallized ligands for 6LU7, 6Y2F, 6Y2G, 5RF8, and 6Y2F is illustrated in Figure 3b. The co-crystallized ligand of 6LU7 has a very similar binding mode as co-crystallized ligand of protein structures 6Y2F and 6Y2G, while the ligand of 5RF8 is bound to different place in protein (with binding pocket including PRO184, VAL186, ARG188, THR190, ALA191, GLN192).
Figure 3.
a - Superposition of all studied proteins, with catalytic dyad ball-and-stick representation; b - superposition of five studied proteins with their co-crystallized ligands. (6LU7 – orange; 6Y2E –yellow; 6Y2F – green; 6Y2G – light blue; 6M03 – purple; 6Y84 – magenta; 5RF8 – dark blue; 5RG0 – grey; 5R8T – red).
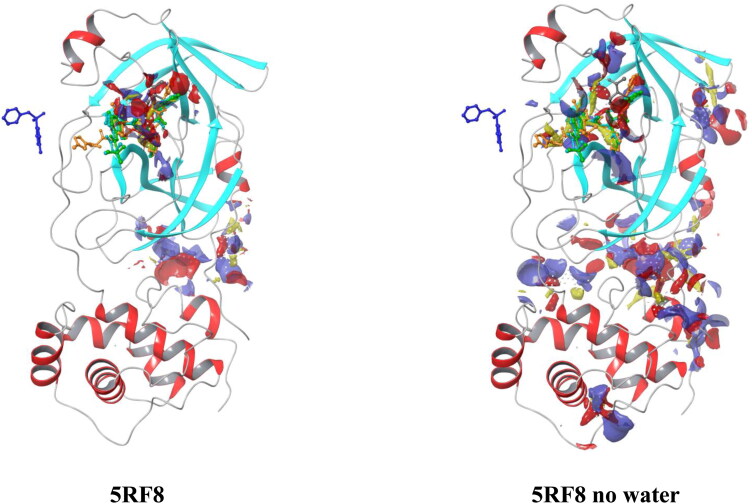
For verification of proper ligand binding, a SiteMap module of Schrodinger was used to calculate the top-ranked binding sites. Unfortunately, for the protein 5RF8, none of five top-ranked binding sites were predicted to be at the same place where co-crystallized ligand is (Figure 4). It should also be noted that for all protein structures in the presence of water molecules the most favorable binding site is the one proposed in literature (Figure 3S, Supporting Information file 1). Meanwhile, when water molecules are removed an additional cavity appears near residues PHE8, GLN110-THR111, GLN127, ASN151-TYR154, GLY302-THR304, thus making this area to be classified as the top-ranked binding site for 6Y2E, 6Y2G, 6Y84, 5RG0, and 5R8T, while binding site proposed in literature was ranked as the second one. To further support these findings, the binding site’s scores were reviewed (Table 1S, Supporting Information file 1). Although both sites indicated not a significant difference in SiteScores and Dscores, it is important to pay close attention to positioning of a grid box for virtual screening.
Figure 4.
Top-ranked binding sites for 5RF8 protein.
3.2. Positive control
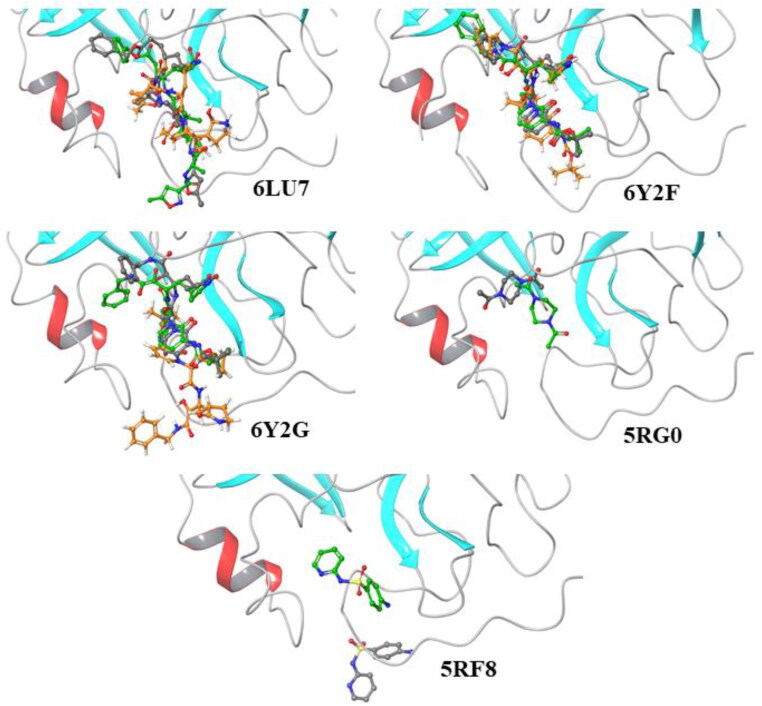
To further validate that our current findings could serve as a reliable method for virtual screening, the co-crystalized ligands of each of the five Mpro were docked into the rigid protein structures under various water conditions (with or without water) and compared in relation to its reference co-crystallized ligand’s binding mode. 6LU7, 6Y2F, and 6Y2G reflected their reference ligand binding mode when docked against corresponding targets without water molecules (green), while docking against targets in the presence of crystallographic waters (orange) did not yield satisfactory results (Figure 5). Insufficient results obtained with water molecules being kept can be explained by the immobility of these molecules during a virtual screening, which causes a steric hindrance. Interestingly, the binding modes of both 5RGO and 5RF8 against their corresponding proteins led to significantly different results from their reference ligands (Figure 5). In the presence of water, these remaining two ligands did not provide any poses. Whereas, upon removal of crystallographic waters the binding modes did not reflect its reference structures. According to Wong and Lightstone (2011), it has been reported that achieving high affinity for a protein target in drug design can become complex with the presence of water, followed by complicating the process of finding poses and calculating the binding affinity, even when water molecules are set to be flexible. Thus, further all the calculations were performed with crystallographic water being removed. Free binding energies from MM-GBSA were calculated and illustrated in Table 2. Results of cross-docking of co-crystallized ligands against all targets indicate that Mpro structures 6LU7, 6Y2F, and 6Y2G can reproduce binding modes of their co-crystallized ligands during virtual screening without the presence of water molecules, whereas, 5RG0 and 5RF8 may not be suitable using proposed methods. Interestingly, re-docking of α-ketoamide inhibitor into 6Y2F performed using AutoDock Vina reproduced binding mode of reference ligand worse then re-docking performed here. Thus, selection of Schrodinger software for calculations carried out here is justified.
Figure 5.
Superposition of ligands docked into protein without water (green), protein with water (orange), and the reference co-crystalized ligand (grey).
Table 2.
Binding energies for cross-docking of co-crystallized ligands against all targets.
| Protein | ΔGBind, kcal/mol |
||||
|---|---|---|---|---|---|
| Ligand 6LU7 | Ligand 6Y2F | Ligand 6Y2G | Ligand 5RF8 | Ligand 5RG0 | |
| 6LU7 no water | −70.60 | −40.19 | −40.19 | −31.73 | −33.63 |
| 6LU7 | −35.45 | −52.22 | −52.22 | −28.96 | −22.09 |
| 6Y2E no water | −31.81 | −35.24 | −35.24 | −31.68 | −27.89 |
| 6Y2E | −30.59 | −12.26 | −12.26 | −12.29 | N/A |
| 6Y2F no water | −86.34 | −55.94 | −55.94 | −24.98 | −30.79 |
| 6Y2F | −36.39 | −42.77 | −42.77 | −31.04 | −34.45 |
| 6Y2G no water | −73.20 | −57.60 | −57.60 | −33.33 | −24.06 |
| 6Y2G | −49.07 | −19.71 | −19.71 | −27.50 | −21.09 |
| 6M03 no water | −38.43 | −49.68 | −49.68 | −29.21 | −27.07 |
| 6M03 | −35.77 | −13.43 | −13.43 | −24.15 | −17.64 |
| 6Y84 no water | −18.38 | −29.56 | −29.56 | −27.46 | −22.04 |
| 6Y84 | −29.77 | −24.31 | −24.31 | −18.07 | N/A |
| 6RF8 no water | −54.89 | −49.13 | −49.13 | −31.42 | −23.44 |
| 5RF8 | −6.50 | −15.84 | −15.84 | N/A | N/A |
| 5RG0 no water | −59.12 | −18.89 | −18.89 | −25.83 | −33.39 |
| 5RG0 | −20.04 | −17.61 | −17.61 | −15.45 | N/A |
| 5R8T no water | −31.95 | −36.13 | −36.13 | −31.37 | −30.63 |
| 5R8T | 6.12 | −4.43 | −4.43 | N/A | N/A |
* N/A – no binding pose were produced.
3.3. Virtual screening
Considering all the information extracted from the protein reliability analysis and the positive control results, 6LU7, 6Y2F, and 6Y2G were selected as final targets for virtual screening. A library of natural compounds were prepared, protonation states and conformations were generated. A set of compounds was filtered, and final prepared set total of 13,496 compounds was screened against the selected protein structures. As a result, fifteen drug-like molecules (Figure 4S, Supporting Information file 1) were selected as hits with consequent calculation of their free binding energies (ΔGbind) (Table 3). The binding modes for the potentially successful candidates were compared to the one of reference ligands.
Table 3.
Virtual screening results.
| Ligand | ΔGbind, kcal/mol |
|||
|---|---|---|---|---|
| 6LU7 | 6Y2F | 6Y2G | Average | |
| MolPort-001-739-296 | −43.52 | −36.89 | −43.68 | −41.36 |
| MolPort-005-944-636 | −47.82 | −59.74 | −56.27 | −53.78 |
| MolPort-005-945-584 | −34.67 | −44.74 | −35.82 | −39.71 |
| MolPort-005-945-924 | −47.70 | −45.04 | −45.54 | −46.09 |
| MolPort-028-754-113 | −37.44 | −46.87 | −61.60 | −48.64 |
| MolPort-035-706-028 | −54.30 | −49.85 | −38.56 | −47.57 |
| MolPort-039-052-338 | −59.51 | −30.96 | −45.17 | −45.21 |
| MolPort-039-052-665 | −47.00 | −56.24 | −42.67 | −48.64 |
| MolPort-039-063-560 | −54.91 | −49.24 | −60.78 | −54.98 |
| MolPort-039-337-168 | −49.94 | −47.27 | −51.55 | −49.59 |
| MolPort-039-338-330 | −60.38 | −53.77 | −58.03 | −57.39 |
| MolPort-039-338-602 | −36.79 | −43.90 | −44.65 | −41.78 |
| MolPort-039-338-717 | −40.37 | −48.27 | −49.44 | −46.03 |
| MolPort-039-338-733 | −41.90 | −44.05 | −34.64 | −40.20 |
| MolPort-044-754-193 | −35.09 | −45.05 | −39.56 | −39.90 |
| Ligand 6LU7 | -70.60 | -86.34 | -73.20 | -76.71 |
| Ligand 6Y2G/6Y2F | -40.19 | -55.94 | -57.60 | -51.24 |
| Ligand 5RF8 | -31.73 | -24.98 | -33.33 | -30.01 |
| Ligand 5RG0 | -33.63 | -30.79 | -24.06 | -29.49 |
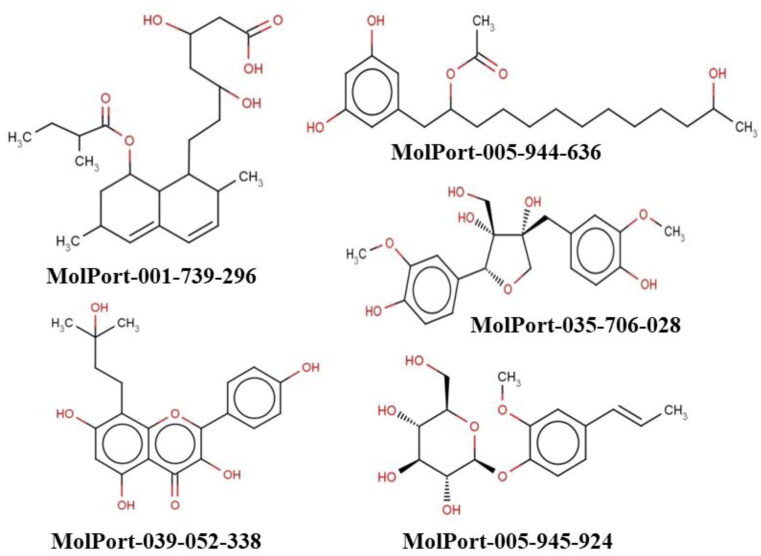
Co-crystallized ligand of 6LU7, N3 exhibited highest average binding affinity (-76.71 kcal/mol) among all the compounds proposed here, followed by MolPort-039-338-330 ((-)-Dehydrodiconiferyl Alcohol, a natural product isolated from several plant species including Aglaia foveolata, Viburnum erosum and Rosa multiflora), MolPort-039-063-560 (8-Lavandulylkaempferol, compound derived from the Sophora flavens), and MolPort-005-944-636 (1-(3,5-Dihydroxyphenyl)-12-hydroxy-2-tridecanyl acetate). Average binding energies of those ligands were varying from −57.39 to −54.61 kcal/mol. The co-crystallized ligand of 6Y2F/6Y2G, α-ketamide, exhibited an average binding energy of −51.24 kcal/mol.
Interestingly, from a pool of natural compounds found in roots of Sophora flavescens Ait not only MolPort-039-063-560, but also MolPort-028-754-113 (Leachianone A) and MolPort-039-338-717 (Kushenol X) exhibited high binding affinity to the Mpro of SARS-CoV-2. Other compounds, that showed to be potentially active included MolPort-039-337-168 (Magnolignan C, compound found in barks of Magnolia officinalis), MolPort-039-052-665 (Dihydroxybergamottin, found and different citruses), MolPort-035-706-028 (Massoniresinol, found in Pinus massoniana), MolPort-005-945-924 (2-Methoxy-4-[(1E)-1-propen-1-yl]phenyl hexopyranoside), and MolPort-039-052-338 (Noricaritin, derived from Epimedium brevicornu Maxim). Even though all the compounds indicated a similar binding pose to the one of reference ligands, not all of them satisfied another criterion (interaction with HIS41 and CYS145). Most of them indicated a contact only with one out of two important residues, while MolPort-005-945-924, MolPort-039-052-338, MolPort-039-063-560, MolPort-039-337-168, and MolPort-044-754-193 showed no such interactions at all, making instead contacts with other residues inside a binding pocket. The best interaction pattern was shown by MolPort-039-052-665 forming both H-bond with CYS145 and π-π stacking with HIS41 of 6Y2F.
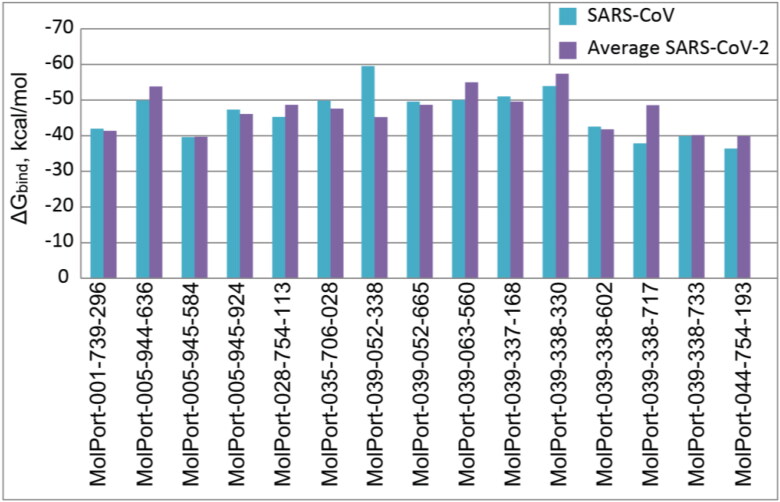
In order to verify whether selected compounds can also be efficient against SARS-CoV strain, XP docking with consequent MM-GBSA post-screening was performed against Mpro of SARS-CoV (PDB Code: 1W0F). The comparison of top-15 ligands’ binding affinities towards SARS-CoV Mpro 1W0F and average binding affinities towards SARS-CoV-2 Mpro proteins 6LU7, 6Y2F, and 6Y2G is illustrated in Figure 6. One can see that most of the selected ligands have similar potential for inhibition of both viral Mpro. Meanwhile, Noricaritin (MolPort-039-052-338) showed significantly stronger affinity towards SARS-CoV Mpro. The best binding affinity correlation was shown between structures 1W0F and 6LU7 with correlation coefficient of R = 0.89, while the worst correlation (R = 0.05) is between 1W0F and 6Y2F. This finding indicates that due to the similarity of a binding pocket, potential drugs targeting Mpro of SARS-CoV-2 can possibly be efficient against all family of SARS-CoV viruses.
Figure 6.
Binding energies of top-15 ligands with SARS-CoV Mpro protein 1W0F and average binding energies with SARS-CoV-2 Mpro proteins 6LU7, 6Y2F, and 6Y2G.
3.4. Admet properties
To compare the ADMET profile of the selected 15 ligands from the docking results, we have considered Co-crystalized ligand N3 of 6LU7 protein and α-ketamide of 6Y2F/6Y2G. The complete ADMET results for 15 ligands can be found in supplementary information. The top-5 ligands were identified based on the desired ADME profile and toxicity to multiple organs as well as effect on different major toxicity targets of human body. The top-5 screened ligands are MolPort-001-739-296, MolPort-005-944-636, MolPort-005-945-924, MolPort-035-706-028, and MolPort-039-052-338. The details can be found in Table 4. Regarding organs toxicity, except MolPort-001-739-296 (weak carcinogenic), all remaining four ligands reported as much safer drug compare to N3 and α-ketoamide co-crystalized ligands of all three selected proteins (both N3 and α-ketoamide ligands are weak inactive for hepatotoxicity, carcinogenicity and mutagenicity, while N3 showed weak inactive nature to cytotoxicity and α-ketoamide reported weak active profile). In case of Nuclear receptor signaling pathways, except MolPort-005-944-636, all four ligands showed strong inactive affinity to major target domain. The MolPort-005-944-636 showed weak active binding to Estrogen Receptor Alpha (ER-Alpha). Meanwhile, in case of stress response pathways prediction, MolPort-039-052-338 reported weak active effect on mitochondrial membrane potential (MMP). Remaining four ligands had strong inactive effect in majority of the cases except, MolPort-001-739-296 and MolPort-005-944-636 had weak inactive effect on MMP, and MolPort-039-052-338 had weak inactive effect on Phosphoprotein tumor suppressor (p53). The MolPort-005-945-924 and MP-035-706-028 showed excellent toxicity profile among all the studied ones. The ADMET profile based on predicted toxicity class, all ligands are residing under toxicity classes of 4 or 5 which supports their safe nature as drug candidate if considered for human usage. The detailed parameters predicted under ADMET profiling for all top-15 drug candidates can be found in Table 2S (Supporting information file 2).
Table 4.
ADMET profiling of top-5 drug candidates.
| ADMET | Property/ Parameter |
MP-001-739-296 | MP-005-944-636 | MP-005-945-924 | MP-035-706-028 | MP-039-052-338 | N3 | α-ketoamide |
|---|---|---|---|---|---|---|---|---|
| Absorption | Water solubility (log mol/L) |
−3.52 | −4.74 | −2.00 | −3.39 | −3.29 | −3.85 | −3.84 |
| Caco2 permeability (log Papp, 10-6 cm/s) |
0.66 | 0.54 | 0.50 | −0.19 | −0.35 | 0.56 | 0.38 | |
| Intestinal absorption (% Absorbed) |
49.15 | 85.64 | 46.12 | 58.25 | 68.87 | 53.99 | 65.54 | |
| Skin Permeability (log Kp) |
−2.73 | −2.87 | −2.89 | −2.74 | −2.74 | −2.74 | −2.75 | |
| P-glycoprotein sub | No | Yes | No | Yes | Yes | Yes | Yes | |
| P-glycoprotein I inh | No | Yes | No | No | No | Yes | Yes | |
| P-glycoprotein II inh | No | Yes | No | No | Yes | No | Yes | |
| Distribution | VD (human) (log L/kg) |
−0.84 | 0.19 | −0.21 | −0.22 | 0.56 | −0.49 | 0.09 |
| Fraction unbound (human) (Fu) |
0.20 | 0.22 | 0.50 | 0.17 | 0.09 | 0.19 | 0.05 | |
| BBB permeability (log BB) |
−0.54 | −1.07 | −0.99 | −1.29 | −1.18 | −1.41 | −1.07 | |
| CNS permeability (log PS) |
−2.98 | −2.84 | −3.57 | −3.72 | −3.19 | −3.97 | −3.52 | |
| Metabolism | CYP2D6 sub | No | No | No | No | No | No | No |
| CYP3A4 sub | Yes | Yes | No | Yes | Yes | Yes | Yes | |
| CYP1A2 inh | No | No | No | No | Yes | No | No | |
| CYP2C19 inh | No | Yes | No | No | No | No | No | |
| CYP2C9 inh | No | No | No | No | Yes | No | No | |
| CYP2D6 inh | No | No | No | No | No | No | No | |
| CYP3A4 inh | No | Yes | No | No | Yes | Yes | Yes | |
| Excretion | Total Clearance (log ml/min/kg) |
1.24 | 1.32 | 0.20 | 0.12 | 0.47 | 0.44 | 0.16 |
| Renal OCT2 sub | No | No | No | No | No | No | No | |
| Organ Toxicity to human | Hepatotoxicity# | 0.81 | 0.8 | 0.84 | 0.88 | 0.73 | 0.64 | 0.66 |
| Carcinogenicity# | 0.5 | 0.72 | 0.85 | 0.8 | 0.66 | 0.52 | 0.6 | |
| Mutagenicity# | 0.85 | 0.9 | 0.77 | 0.72 | 0.51 | 0.59 | 0.63 | |
| Cytotoxicity# | 0.61 | 0.78 | 0.78 | 0.79 | 0.86 | 0.69 | 0.51 | |
| AMES toxicity | No | No | Yes | No | No | No | No | |
| hERG I inhibitor | No | No | No | No | No | No | No | |
| hERG II inhibitor | No | No | No | No | Yes | Yes | Yes | |
| Oral Rat AT (LD50) (mol/kg) |
2.08 | 2.23 | 1.97 | 2.54 | 2.53 | 3.538 | 2.155 | |
| Oral Rat CT (LOAEL) (log mg/kg_bw/day) |
0.68 | 1.994 | 3.489 | 3.154 | 2.418 | 3.417 | 2.142 | |
| Skin Sensitization | No | Yes | No | No | No | No | No | |
|
Tox21-Nuclear receptor signaling pathways |
AhR# | 0.98 | 0.89 | 0.85 | 0.8 | 0.83 | 0.91 | 0.91 |
| AR# | 0.93 | 0.98 | 0.96 | 0.95 | 0.92 | 0.95 | 0.97 | |
| AR-LBD# | 0.94 | 1 | 0.97 | 0.97 | 0.94 | 0.97 | 0.98 | |
| Aromatase# | 0.85 | 0.97 | 0.96 | 0.81 | 0.74 | 0.95 | 0.91 | |
| ER-Alpha# | 0.85 | 0.58 | 0.82 | 0.71 | 0.6 | 0.88 | 0.87 | |
| ER- LBD# | 0.92 | 0.59 | 0.96 | 0.92 | 0.84 | 0.92 | 0.96 | |
| PPAR-Gamma# | 0.98 | 0.91 | 0.98 | 0.9 | 0.81 | 0.92 | 0.96 | |
| Tox21-Stress response pathways | nrf2/ARE | 0.83 | 0.78 | 0.96 | 0.86 | 0.8 | 0.93 | 0.93 |
| HSE | 0.83 | 0.78 | 0.96 | 0.86 | 0.8 | 0.93 | 0.93 | |
| MMP | 0.67 | 0.51 | 0.92 | 0.7 | 0.64 | 0.88 | 0.69 | |
| p53 | 0.91 | 0.82 | 0.9 | 0.72 | 0.69 | 0.88 | 0.83 | |
| ATAD5 | 0.99 | 0.97 | 0.96 | 0.93 | 0.88 | 0.95 | 0.91 | |
|
Toxicity target* |
Receptor# | AR (75.1%) |
NF | PG/H S1 (94.3%) | GR (81.9%) |
AR (72.3%) | N/A | N/A |
| Predicted Toxicity | LD50# mg/kg |
3000 | 2000 | 4000 | 1500 | 3919 | 4000 | 675 |
| Toxicity Class | Class# | 4 | 4 | 5 | 4 | 5 | 5 | 4 |
| Toxicity to environment | T. Pyriformis (log ug/L) |
0.39 | 1.10 | 0.29 | 0.29 | 0.36 | 0.285 | 0.291 |
| Minnow toxicity (log mM) |
1.04 | −0.49 | 3.80 | 3.20 | 3.57 | 5.647 | 2.122 | |
| Strong Active | Strong Inactive | Weak Active | Weak Inactive | |||||
MolPort is designated as MP; The number inside the colored boxes are probability of correctness of the prediction; #These parameters are computed using ProTox-II and remaining ones are employing pkCSM;* Possible toxicity targets out of 16 major toxicity targets in human body where the value represents % Avg Similarity Known Ligands; VD: Volume distribution; BBB: Blood brain barrier; Substrate: Sub; Inhibitor: Inh; Acute toxicity: AT; Chronic toxicity: CT; LOAEL: Lowest-observed-adverse-effect level; AhR: Aryl hydrocarbon Receptor; AR: Androgen Receptor; Androgen Receptor Ligand Binding Domain (AR-LBD); ER-Alpha: Estrogen Receptor Alpha; ER-LBD: Estrogen Receptor Ligand Binding Domain; PPAR-Gamma: Peroxisome Proliferator Activated Receptor Gamma; nrf2/ARE: Nuclear factor (erythroid-derived 2)-like 2/antioxidant responsive element; HSE: Heat shock factor response element; MMP: Mitochondrial Membrane Potential; p53: Phosphoprotein (Tumor Suppressor); ATAD5: ATPase family AAA domain-containing protein 5; PG/H S1: Prostaglandin G/H Synthase 1; GR: Glucocorticoid Receptor; NF: Not found under tested 16 toxicity target
3.5. Molecular dynamics
Considering binding energies, interactions with amino acid residues HIS41 and CYS145, and ADMET properties of 15 selected ligands, top-5 were selected for further elucidation through MD simulation (Figure 7). Selected ligands included MolPort-001-739-296, MolPort-005-944-636, MolPort-005-945-924, MolPort-035-706-028, and MolPort-039-052-338.
Figure 7.
2D Structures and unique smiles of five top hit ligands.
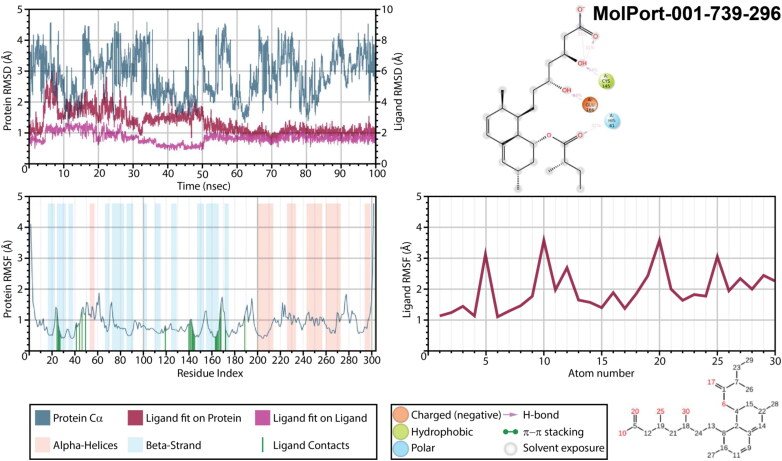
Simulations were performed for all these complexes for 100 ns, and obtained trajectories were analyzed using Simulation Interaction Diagram. Mpro complexes with MolPort-001-739-296 (Figure 8) showed very high values of protein’s root mean square deviation (RMSD) after equilibration (3.5-4.0 Å) and did not stabilized during all 100 ns, indicating an unstable protein conformation after binding. The RMSD of ligands fit on a protein and fit on itself were also high (around 4.0 and 2.0 Å, respectively), though, ligand seamed to stabilized after 50 ns of simulation. During 35% and 31% of simulation time both oxygen atoms of a carboxylic group served as acceptors of intramolecular hydrogen bond from the nearest hydroxyl group, while an oxygen of this hydroxyl group accepted hydrogen from CYS145 (34% of a simulation time). The second hydroxyl group interacted with Mpro residue GLU166 through hydrogen bonding during 44% of trajectory simulation time. Interaction with HIS41 was also detected (32%) through the alkoxyl group of a MolPort-001-739-296. After stabilization (after 50 ns) all those interactions were enhanced and additional interactions between carboxylic group and ASN142 and GLY143 were formed. While root mean square fluctuations (RMSF) of a protein did not exceed 2.0 Å, on the ligands RMSF one can find fluctuations over 3.0 Å (carboxylic group).
Figure 8.
Protein and ligand RMSD, 2D interaction diagram, protein and ligand RMSF of Mpro structures in complex with MolPort-001-739-296 as a result of 100 ns molecular dynamic simulation.
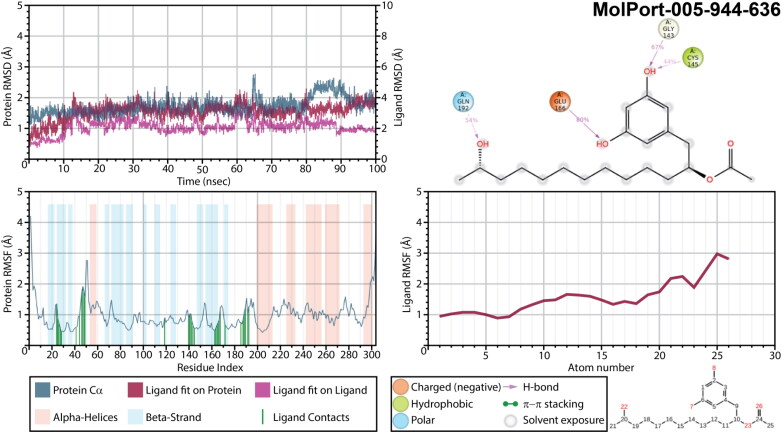
The least conformational changes were detected for a complex of Mpro with MolPort-005-944-636 (Figure 9), while changes of protein’s RMSD varied within 1 Å. Larger deviations were observed only for a trajectory segment near 65 ns and the one between 80 and 90 ns. The ligand’s deviations were also not significant. Oxygen atoms of both hydroxyl groups attached to MolPort-005-944-636 phenyl ring served as acceptor of hydrogen bond from GLU166, GLY143 and CYS145 during 80%, 67%, and 44% of a simulation time, respectively. Meanwhile, a long hydrocarbon chain is fixed with its hydroxyl group interacting with GLN192 (54%). This fixation resulted in relatively low RMSF of a ligand’s hydrocarbon chain not exceeding 2.0 Å, except for the last methyl in a chain and hydroxyl group, which fluctuations were just slightly above. The highest fluctuations in MolPort-005-944-636 were detected for acetoxyl group that did not exhibit any interactions with a main protease, thus remained flexible inside a binding pocket.
Figure 9.
Protein and ligand RMSD, 2D interaction diagram, protein and ligand RMSF of Mpro structures in complex with MolPort-005-944-636 as a result of 100 ns molecular dynamic simulation.
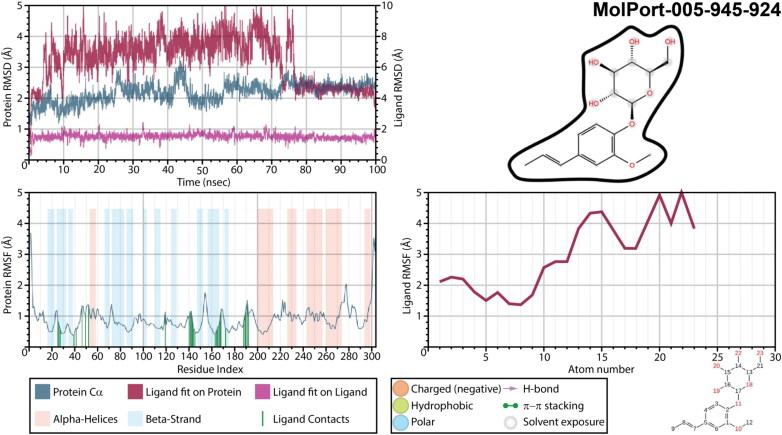
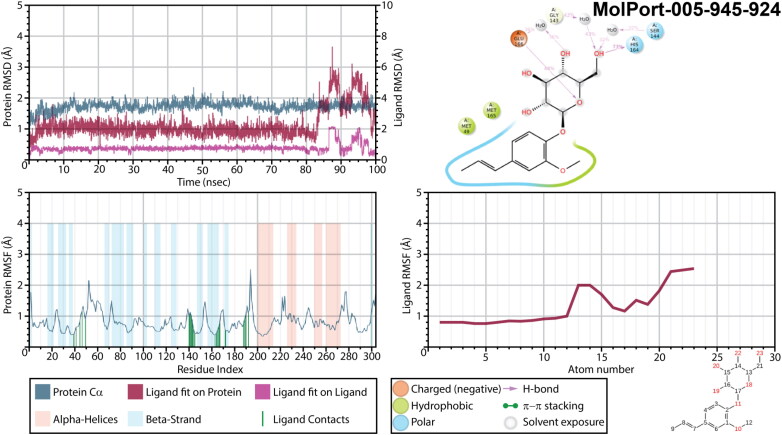
Insufficient results were obtained for a Mpro complex with MolPort-005-945-924 (Figure 10). It exhibited an extremely high ligand’s fluctuations and RMSD of a ligand fit on protein. Moreover, no interactions, which would remain for more than 30% of a simulation time, were detected for this complex. It is possible that for this complex 100 ns simulation is not enough, since after 75 ns of simulation ligand’s RMSD was stabilized, and two H-bonds were formed between GLU166 residue of a main protease and two oxygen atoms of hexopyranosyl group, complimented by numerous water bridges. Must be noted, that unsuccessful results could be obtained due to either a ligand’s failure or due to an unsuitable initial binding mode selected for molecular dynamic calculation. Therefore, in order to clarify it, trajectory after 75 ns was clustered, and the binding mode of the most populated cluster with the largest number of interactions was subjected to an additional 100 ns of molecular dynamic simulations. Interestingly with a new initial geometry the results of a simulation improved drastically (Figure 11). Changes of protein’s RMSD did not exceed 1.0 Å, while ligand’s RMSD remained extremely stable and ligand’s RMSF lower than 2.5 Å until after 83 ns of a simulation time. Phenyl ring was strongly kept inside a binding pocket within hydrophobic/polar cavity. Oxygen atom of a hexopyranosyl ring was an acceptor of an H-bond from GLU166 (48% of a simulation time), and hydroxymethyl group interacted with HIS164 (73% of a time). Even though these two interactions were broken after 83 ns, phenyl ring remained inside a cavity. An interesting observation is that when compare protein RMSF of this MD simulation with all the other, one can notice the difference in a secondary structure of main proteases. While residues ASN53-LEU57 and ASP295-GLN295 represented as α-Helices in all the other structures, a complex of a Mpro with MolPort-005-945-924 during a second MD simulation revealed this region being neither α-Helices nor a β-strand. Presumably, it may indicate that binding of this ligand to SARS-CoV-2 main protease structure significantly changes its conformation.
Figure 10.
Protein and ligand RMSD, 2D interaction diagram, protein and ligand RMSF of Mpro structures in complex with MolPort-005-945-924 as a result of a first 100 ns molecular dynamic simulation.
Figure 11.
Protein and ligand RMSD, 2D interaction diagram, protein and ligand RMSF of Mpro structures in complex with MolPort-005-945-924 as a result of an additional 100 ns molecular dynamic simulation.
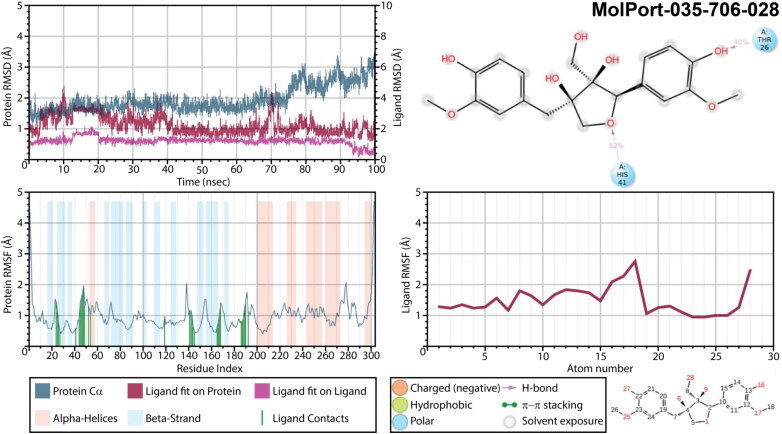
As for the complex with Massoniresinol (MolPort-035-706-028), the protein seemed to be very stable until after 75 ns of a simulation time, while a ligand’s RMSD varied up to 3.0 Å (Figure 12). During first 12 ns both phenyl rings hydroxyl groups formed interactions THR26 and ARG188, hydroxymethyl group of a furan ring interacted with GLU47, while the oxygen atom of a furan ring accepted a H-bond from HIS41. After 12.5 ns and until 20 ns, ligand underwent a conformational change by breaking most of interactions except for the one with ARG188. During a simulation time between 21 and 91 ns ligand regained interactions with HIS41 and THR26, and only after 91 ns strong interactions fixing both phenyl rings and the central furan ring occurred. For this complex, similarly to previous one, trajectory clustering was performed after 91 ns of a simulation time. The binding mode of the most populated cluster that exhibited interactions of both phenyl rings and the central furan ring with a protein was used as an initial frame for 100 ns of molecular dynamic simulations. Though, even during additional simulation ligand remained extremely flexible and no improvement of interactions was detected, making Massoniresinol being the least enough among 5 selected ligands.
Figure 12.
Protein and ligand RMSD, 2D interaction diagram, protein and ligand RMSF of Mpro structures in complex with MolPort-035-706-028 as a result of 100 ns molecular dynamic simulation.
According to the results of MD simulations the most stable interactions occurred between Noricaritin (MolPort-039-052-338) and 6LU7 (Figure 13). This ligand showed to be very stable inside a binding pocket during all 100 ns simulation with very the lowest values of ligand’s RMSD fit on itself among all complexes studied here. One can see a significant change in proteins’ RMSD. Interestingly, a change of ligand’s RMSD fit on protein in time repeated the one of proteins’ RMSD. Moreover, it did not reflect anyhow in ligand’s binding pose. Hydroxyl group of phenyl ring formed an H-bond with TYR54 as a donor (97% of a simulation time) and accepted hydrogen from CYS44. Formation of intramolecular H-bond between the hydrogen of a flavonoid A ring’s hydroxyl and double-bonded oxygen of a C ring enhanced the capability of A ring’s hydroxyl to accept the H-bond from GLU166. Meanwhile, hydroxyl of a C ring served as a donor of H-bond interacting with GLN189. While results of MM-GBSA indicate no interactions with HIS41 and CYS145, molecular dynamic simulation shows ring B forming π − π stacking with HIS41 during 57% of a simulation time.
Figure 13.
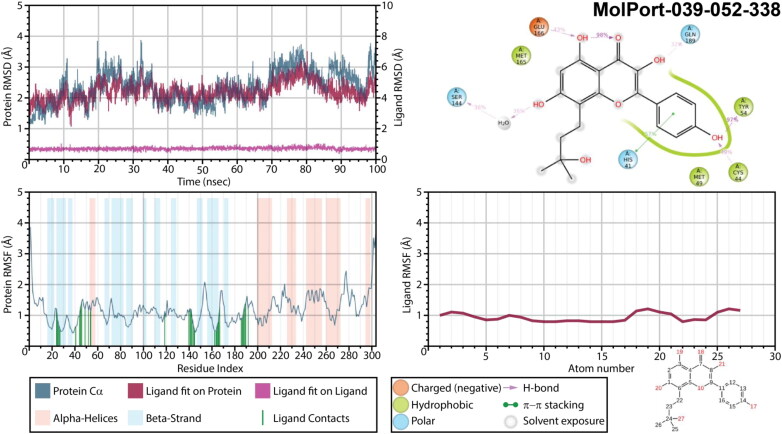
Protein and ligand RMSD, 2D interaction diagram, protein and ligand RMSF of Mpro structures in complex with MolPort-039-052-338 as a result of 100 ns molecular dynamic simulation.
An additional information as for analyses of MD trajectories, including ligand RMSD, Radius of Gyration, Intramolecular Hydrogen Bonds, Molecular Surface Area, Solvent Accessible Surface Area, Polar Surface Area can be found in Figure 5S (Supporting Information file 1).
4. Conclusions
A comprehensive reliability analysis was performed for nine selected SARS-CoV-2 Mpro protein structures. While most of the reliability analyses approaches have shown structures deposited recently by Fearon et al. (2020a; 2020b; 2020c) being the most reliable, we concluded that a positive control docking provided significantly better reproductivity for structures with PDB ID 6LU7, 6Y2G and 6Y2F. Thus, pursuant to results of our investigation, these structures were selected as potent targets for in-silico drug discovery of SARS-CoV-2 inhibitors. Benchmarking performed here also showed that crystallographic water has a negative impact on an outcome of a virtual screening against selected targets, therefor it is suggested, that water molecules must be deleted before the virtual screening.
The result of the virtual screening of the MolPort database of natural compounds against the selected Mpro structures indicated top-15 hit ligands, for which MM-GBSA calculations and ADMET properties prediction were carried out. Interestingly, it was found that these compounds can also inhibit SARS-CoV Mpro. This suggests that these molecules can act as inhibitors for both SARS-CoV strains. Since a potent drug supposed to not only have a high affinity towards a target, but also must be relatively harmless for a human body, the choice of prospective drug-like molecules cannot be limited to the compounds performing well in molecular docking simulation. Taking into account the binding affinities predicted by MM-GBSA, binding modes, interactions with a CYS145 and HIS41, and ligands’ ADMET profiling, five hit molecules were identified: MolPort-001-739-296, MolPort-005-944-636, MolPort-005-945-924, MolPort-035-706-028, and MolPort-039-052-338. Further, 100 ns MD simulation indicated ligands MolPort-001-739-296 and MolPort-035-706-028 being unstable inside a binding pocket of SARS-CoV-2 Mpro, while two natural compounds MolPort-005-944-636 and MolPort-005-945-924, as well as a Noricaritin (MolPort-039-052-338) derived from Epimedium brevicornu Maxim appeared to be the most favorable ones. The analysis offers further investigation of these three potential molecules as probable drug candidates as viral Mpro inhibitors. Considering safety issue of these drugs, all three molecules showed better ADMET profile compare to co-crystalized ligands of all three proteins.
Supplementary Material
Glossary
Abbreviations
- Mpro
Main Protease
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- MM-GBSA
Molecular Mechanics - Generalized Born Surface Area
- ADMET
Absorption, Distribution, Metabolism, Excretion, and Toxicity
- MD
Molecular Dynamics
- PCA
Principal Component Analysis
- HTVS
High Throughput Virtual Screening
- SP
Standard Precision
- XP
Extra Precision
- RMSD
Root Mean Square Deviation
Funding Statement
The authors want to thank the National Science Foundation [Grant: NSF/CREST HRD-1547754] for financial support.
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Banerjee, P., Eckert, A. O., Schrey, A. K., & Preissner, R. (2018). ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Research, 46(W1), W257–W263. 10.1093/nar/gky318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. J., Chow, D. E., Xu, H., Dror, R. O., Eastwood, M. P., Gregersen, B. A., Klepeis, J. L., Kolossvary, I., Moraes, M. A., Sacerdoti, F. D., Salmon, J. K., Shan, Y., & Shaw, D. E. (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters [Paper presentation]. SC'06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL. pp. 43. 10.1109/SC.2006.54 [DOI] [Google Scholar]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag, A., Wang, P., Ahmed, M., & Sadek, H. (2020). Identification of FDA approved drugs targeting COVID-19 virus by structure-based drug repositioning (Version 4). chemrxiv, 10.26434/chemrxiv.12003930.v4 [DOI]
- Fearon, D., Owen, C. D., Douangamath, A., Lukacik, P., Powell, A. J., Strain-Damerell, C. M., Resnick, E., Krojer, T., Gehrtz, P., Wild, C., Aimon, A., Brandao-Neto, J., Carbery, A., Dunnett, L., Skyner, R., Snee, M., London, N., Walsh, M. A., & von Delft, F. (2020. a). PanDDA analysis group deposition – Crystal Structure of SARS-CoV-2 main protease in complex with Z271004858, 10.2210/pdb5RF8/pdb [DOI]
- Fearon, D., Owen, C. D., Douangamath, A., Lukacik, P., Powell, A. J., Strain-Damerell, C. M., Resnick, E., Krojer, T., Gehrtz, P., Wild, C., Aimon, A., Brandao-Neto, J., Carbery, A., Dunnett, L., Skyner, R., Snee, M., London, N., Walsh, M. A., & von Delft, F. (2020. b). PanDDA analysis group deposition – Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102535, 10.2210/pdb5RG0/pdb [DOI]
- Fearon, D., Owen, C. D., Douangamath, A., Lukacik, P., Powell, A. J., Strain-Damerell, C. M., Resnick, E., Krojer, T., Gehrtz, P., Wild, C., Aimon, A., Brandao-Neto, J., Carbery, A., Dunnett, L., Skyner, R., Snee, M., London, N., Walsh, M. A., & von Delft, F. (2020. c). PanDDA analysis group deposition of ground-state model of SARS-CoV-2 main protease screened against DSI poised (Enamine), Fraglites and Peplites (Newcastle university), Mini Frags (Astex). York 3D (York university), electrophile cysteine covalent (Weizman institute) fragment libraries. 10.2210/pdb5R8T/pdb [DOI]
- Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., Sanschagrin, P. C., & Mainz, D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- Gautret, P., Lagier, J.-C., Parola, P., Hoang, V. T., Meddeb, L., Mailhe, M., Doudier, B., Courjon, J., Giordanengo, V., Vieira, V. E., Tissot Dupont, H., Honoré, S., Colson, P., Chabrière, E., La Scola, B., Rolain, J.-M., Brouqui, P., & Raoult, D. (2020). Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. International Journal of Antimicrobial Agents, 56(1), 105949. 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., Gulyaeva, A. A., Haagmans, B. L., Lauber, C., Leontovich, A. M., Neuman, B. W., Penzar, D., Perlman, S., Poon, L. L. M., Samborskiy, D. V., Sidorov, I. A., Sola, I., & Ziebuhr, J. (2020). The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiol, 5(4), 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider, Z., Subhani, M. M., Farooq, M. A., Ishaq, M., Khalid, M., Khan, R. S. A., & Niazi, A. K. (2020). In Silico discovery of novel inhibitors against main protease (Mpro) of SARS-CoV-2 using pharmacophore and molecular docking based virtual screening from ZINC database. Preprints, https://www.preprints.org/manuscript/202002.0431/v1
- Halgren, T. A. (2009). Identifying and characterizing binding sites and assessing druggability. Journal of Chemical Information and Modeling, 49(2), 377–389. 10.1021/ci800324m [DOI] [PubMed] [Google Scholar]
- Jacobson, M. P., Pincus, D. L., Rapp, C. S., Day, T. J. F., Honig, B., Shaw, D. E., & Friesner, R. A. (2004). A hierarchical approach to all-atom protein loop prediction . Proteins , 55(2), 351–367. 10.1002/prot.10613 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Joob, B., & Wiwanitkit, V. (2020). COVID-19 can present with a rash and be mistaken for Dengue. Journal of the American Academy of Dermatology, 82(5), E177. 10.1016/j.jaad.2020.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel, M., & Al-Nazawi, M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, 251, 117627. 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, S., & Leszczynski, J. (2017). Recent advances of computational modeling for predicting drug metabolism: A perspective . Current Drug Metabolism, 18(12), 1106–1122. 10.2174/1389200218666170607102104 [DOI] [PubMed] [Google Scholar]
- Khan, M. T., Ali, A., Wang, Q., Irfan, M., Khan, A., Zeb, M. T., Zhang, Y.-J., Chinnasamy, S., & Wei, D.-Q. (2020). Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1769733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, Y., & Singh, H. (2020). In Silico Identification and Docking-Based Drug Repurposing Against the Main Protease of SARS-CoV-2., Causative Agent of COVID-19. Preprint at 10.26434/chemrxiv.12049590.v1 [DOI]
- LigPrep . (2020). Schrödinger., LLC.
- MolPort Database . (2020). MolPort Database. https://www.molport.com/shop/index
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Owen, C. D., Lukacik, P., Strain-Damerell, C. M., Douangamath, A., Powell, A. J., Fearon, D., Brandao-Neto, J., Crawshaw, A. D., Aragao, D., Williams, M., Flaig, R., Hall, D. R., McAuley, K. E., Mazzorana, M., Stuart, D. I., von Delft, F., & Walsh, M. A. (2020). COVID-19 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, SARSCoV-2). 10.2210/pdb6Y84/pdb. [DOI]
- Palese, L. L. (2020). The Structural Landscape of SARS-CoV-2 Main Protease: Hints for Inhibitor Search. Preprint, at 10.26434/chemrxiv.12209744.v1 [DOI]
- Pires, D. E. V., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- QikProp (2020). Schrödinger., LLC.
- Roos, K., Wu, C., Damm, W., Reboul, M., Stevenson, J. M., Lu, C., Dahlgren, M. K., Mondal, S., Chen, W., Wang, L., Abel, R., Friesner, R. A., & Harder, E. D. (2019). OPLS3e: Extending force field coverage for drug-like small molecules. Journal of Chemical Theory and Computation, 15(3), 1863–1874. 10.1021/acs.jctc.8b01026 [DOI] [PubMed] [Google Scholar]
- Sampangi-Ramaiah, M. H., Vishwakarma, R., & Shaanker, R. U. (2020). Molecular docking analysis of selected natural products from plants for inhibition of SARS-CoV-2 main protease. Current Science, 118(7), 1087–1092. [Google Scholar]
- Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Shelley, J. C., Cholleti, A., Frye, L. L., Greenwood, J. R., Timlin, M. R., & Uchimaya, M. (2007). Epik: A software program for pK(a) prediction and protonation state generation for drug-like molecules. Journal of Computer-Aided Molecular Design, 21(12), 681–691. 10.1007/s10822-007-9133-z [DOI] [PubMed] [Google Scholar]
- Talluri, S. (2020). Virtual Screening Based Prediction of Potential Drugs for COVID-19. Preprint, at 10.20944/preprints202002.0418.v2 [DOI] [PubMed]
- Ton, A., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. (2020). Rapid identification of potential inhibitors of SARS‐CoV‐2 main protease by deep docking of 1.3 billion compounds. Molecular Informatics, 39(8), 2000028–2000026. 10.1002/minf.202000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, S. E., & Lightstone, F. C. (2011). Accounting for water molecules in drug design. Expert Opinion on Drug Discovery, 6(1), 65–74. 10.1517/17460441.2011.534452 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Zhao, Y., Jin, Z., Liu, X., Yang, H., & Rao, Z. (2020). The crystal structure of COVID-19 main protease in apo form, 10.2210/pdb6M03/pdb [DOI]
- Zheng, Y.-Y., Ma, Y.-T., Zhang, J.-Y., & Xie, X. (2020). COVID-19 and the cardiovascular system. Nature Reviews. Cardiology, 17(5), 259–260. 10.1038/s41569-020-0360-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.