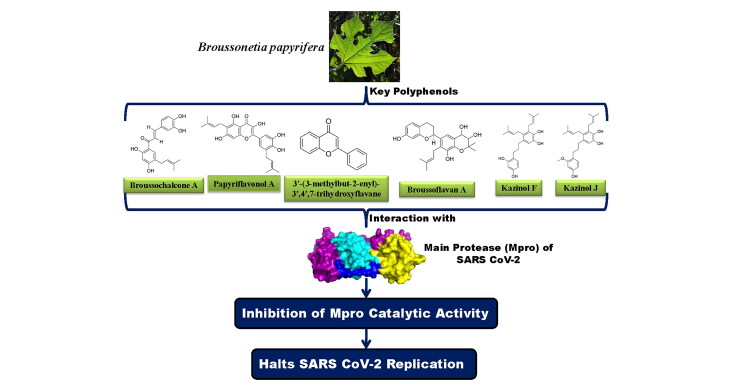
Abstract
The current COVID-19 pandemic is caused by SARS CoV-2. To date, ∼463,000 people died worldwide due to this disease. Several attempts have been taken in search of effective drugs to control the spread of SARS CoV-2 infection. The main protease (Mpro) from SARS CoV-2 plays a vital role in viral replication and thus serves as an important drug target. This Mpro shares a high degree of sequence similarity (>96%) with the same protease from SARS CoV-1 and MERS. It was already reported that Broussonetia papyrifera polyphenols efficiently inhibit the catalytic activity of SARS CoV-1 and MERS Mpro. But whether these polyphenols exhibit any inhibitory effect on SARS CoV-2 Mpro is far from clear. To understand this fact, here we have adopted computational approaches. Polyphenols having proper drug-likeness properties and two repurposed drugs (lopinavir and darunavir; having binding affinity −7.3 to −7.4 kcal/mol) were docked against SARS CoV-2 Mpro to study their binding properties. Only six polyphenols (broussochalcone A, papyriflavonol A, 3'-(3-methylbut-2-enyl)-3',4',7-trihydroxyflavane, broussoflavan A, kazinol F and kazinol J) had interaction with both the catalytic residues (His41 and Cys145) of Mpro and exhibited good binding affinity (−7.6 to −8.2 kcal/mol). Molecular dynamic simulations (100 ns) revealed that all Mpro-polyphenol complexes are more stable, conformationally less fluctuated; slightly less compact and marginally expanded than Mpro-darunavir/lopinavir complex. Even the number of intermolecular H-bond and MM-GBSA analysis suggested that these six polyphenols are more potent Mpro inhibitors than the two repurposed drugs (lopinavir and darunavir) and may serve as promising anti-COVID-19 drugs.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, SARS CoV-2 main protease, docking, molecular dynamics simulation, Broussonetia papyrifera polyphenols
Graphical Abstract
1. Introduction
COVID-19 accounted for ∼8,760,000 infected cases worldwide while ∼463,000 people died between January to mid-June 2020. This highly contagious febrile respiratory illness was declared as a pandemic disease on January 30 2020, by the World Health Organization (WHO) (Cucinotta & Vanelli, 2020). China was the epicenter of this disease, but it rapidly spread throughout the globe (Zhu et al., 2020). The United States remains the most affected country with ∼2,300,000 infected cases and out of which ∼122,000 people died due to COVID-19. Fever, cough, sore throat, runny nose and difficulty in breathing remain the main symptoms but it has been reported to be asymptotic for some individuals which in turn, accelerates the spread of this disease (N. Chen et al., 2020; Ren et al., 2020; Yu & Yang, 2020; Zhu et al., 2020). The unavailability of suitable drugs or therapies for effective treatment until now has transformed this disease into a dangerous and life-threatening.
A novel coronavirus, severe acute respiratory syndrome corona virus-2 (SARS CoV-2) has been identified as the etiological agent of the disease which belongs to the genus β-coronavirus (Zheng, 2020). The whole-genome sequence of this RNA virus revealed that it is highly similar to that of SARS CoV-1 with a 79.6% sequence identity (Zhou et al., 2020). However, the sequence similarities vary significantly for different viral proteins (Lu et al., 2020). For example, the sequence of spike proteins (S-protein) is quite divergent throughout different coronavirus species (Li, 2016). This may be a consequence of rapid mutations and recombination across the species. Besides this, the binding propensities of these spike proteins towards the host receptors vary across the species (Lan et al., 2020). For instance, both SARS CoV-1 and SARS CoV-2 use the same host receptor (ACE2) and show affinity to the same binding site but their binding affinities to ACE2 vary due to slight interface sequence variations (Lan et al., 2020). On the other hand, the sequence of some proteins such as the main protease (Mpro) is highly conserved throughout coronavirus species (Mirza & Froeyen, 2020). The Mpro from SARS CoV-2 shares more than 96% sequence similarity with the same protease from SARS CoV-1 and MERS (Supplementary Figure 1). This makes Mpro an ideal target for broad-spectrum anti-CoV therapy. Mpro [also known as 3CLpro (chymotrypsin-like protease)] is a cysteine protease, which is an analog to the main picornavirus 3C protease (Rota et al., 2003). Mpro plays an important role in the replication process of single-stranded RNA from SARS CoV-2. It helps in the proteolytic cleavage at 11 sites involving the Leu-Gln↓(Ser, Ala, Gly) sequence of the viral polyprotein and resulting in the release of a total number of 16 nonstructural proteins (nsps) (Fan et al., 2004; Rota et al., 2003). Each of the protomers of the homodimeric SARS CoV-2 Mpro protein consists of three domains (Supplementary Figure 1). Domain I (amino acid residues 8-101) and domain II (amino acid residues 102-184) form a chymotrypsin-like architecture and these two domains are connected to the domain III (amino acid residues 201-303) via a long loop (Jin et al., 2020). Among them, domain I and II are essentially β-barrels while, domain III mainly consists of α-helices (Jin et al., 2020). The catalytic site/active site/substrate binding site comprising of cysteine (Cys145) and histidine (His41) amino acid moiety is located at the cleft of domain I and domain II (Jin et al., 2020). Cysteine145 serves as a common nucleophile and plays a vital role in the proteolytic functioning of Mpro (Anand et al., 2003; Chou et al., 2003; Hsu et al., 2005). Deprotonation of Cys-thiol followed by nucleophilic attack of resulting anionic sulfur on the substrate carbonyl carbon is the first step in the proteolytic process of Mpro (Hsu et al., 2005). As a result, a peptide product having an amine terminus is released whereas the deprotonated form of histidine is restored. Subsequently, the thioester product is hydrolyzed to produce a carboxylic acid and the free enzyme is generated in the final step (Hsu et al., 2005). Therefore, proteolytic processing as the functional significance of Mpro in viral propagation makes this protease an important drug target. Besides this, lack of any human homolog of Mpro makes it an ideal target for the development of drugs against COVID-19 infection (Kim et al., 2016). In recent times many computational studies have been carried out to repurpose various FDA approved antiviral drugs for COVID-19 treatment by targeting SARS CoV-2 Mpro (Arun et al., 2020; Elmezayen et al., 2020; Kandeel & Al-Nazawi, 2020; Mahanta et al., 2020; Muralidharan et al., 2020). Apart from these drugs, various medicinal phytochemicals have been proposed as potent SARS CoV-2 Mpro inhibitors (Aanouz et al., 2020; Bhardwaj, Singh, Sharma, Rajendran, et al., 2020; Das et al., 2020; Enmozhi et al., 2020; Gyebi et al., 2020; Islam et al., 2020; Joshi et al., 2020; Umesh et al., 2020).
Figure 1.
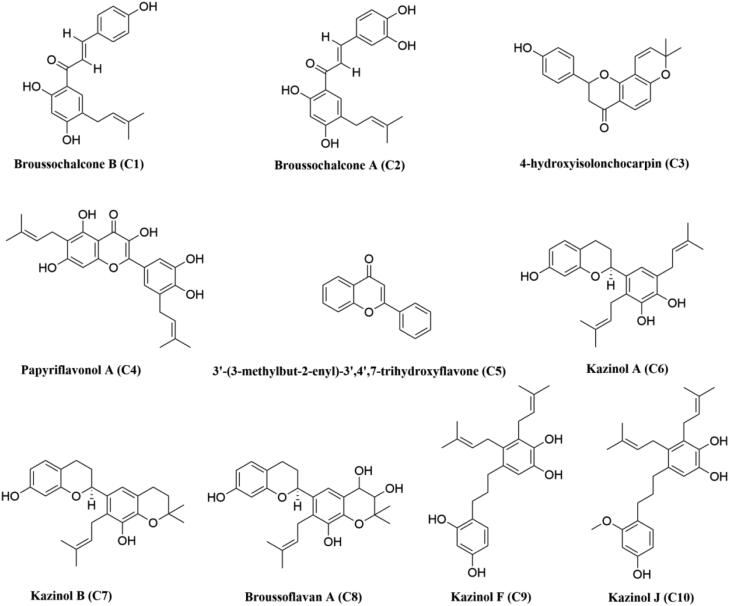
Chemical structures of B. papyrifera polyphenols. The two-dimensional structures of ten polyphenols from B. papyrifera with their respective names are shown. The identification number of each polyphenol is mentioned inside the bracket.
Even many plant-derived natural polyphenols (Oolonghomobisflavan-A from the tea plant Camellia sinensis L., epigallocatechin gallate, epicatechingallate and gallocatechin-3-gallate from green tea Camellia sinensis) having antiviral properties are effective SARS CoV-2 Mpro inhibitors (Bhardwaj, Singh, Sharma, Rajendran, et al., 2020; Ghosh et al., 2020). Polyphenols having glucosidase inhibitory properties are reported to show anti-viral activity against various coronaviruses such as SARS CoV-1 and MERS (Dan et al., 2019; Williams & Goddard-Borger, 2020; Zhao et al., 2015). A group of polyphenols from Broussonetia papyrifera (B. papyrifera) are known to possess α-glucosidase inhibition activity (H. W. Ryu et al., 2010; Hyung Won Ryu et al., 2012). In 2017, Lee and coworkers have found that ten of them [broussochalcone B or bavachalcone (C1), broussochalcone A (C2), 4-hydroxyisolonchocarpin (C3), papyriflavonol A (C4), 3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane (C5), kazinol A (C6), kazinol B (C7), broussoflavan A (C8), kazinol F (C9) and kazinol J (C10)] (structures mentioned in Figure 1) show inhibitory effect against Mpro from SARS CoV-1 and MERS (Park et al., 2017). In the same study, investigators have also experimentally revealed the mode of enzymatic inhibitionand calculated the IC50 values of these polyphenols against Mpro (Park et al., 2017). But whether these polyphenols exhibit any antiviral activity against SARS CoV-2 by inhibiting the enzymatic/proteolytic activity of Mpro is far from clear. Therefore, in this study, we have examined the inhibitory potency of these ten polyphenols from B. papyrifera against SARS CoV-2 Mpro with the aid of in-silico docking studies, molecular dynamics simulations and MM-GBSA analysis. This study has revealed that six polyphenols [broussochalcone A, papyriflavonol A, 3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane, broussoflavan A, kazinol F and kazinol J] exhibit stronger binding affinity with Mpro and are possibly effective Mpro inhibitors.
2. Materials and methods
2.1. Preparation of the ligands
The structures of ten B. papyrifera polyphenols [broussochalcone B or bavachalcone (C1), broussochalcone A (C2), 4-hydroxyisolonchocarpin (C3), papyriflavonol A (C4), 3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane (C5), kazinol A (C6), kazinol B (C7), broussoflavan A (C8), kazinol F (C9) and kazinol J (C10)] were downloaded from PubChem database server in SDF format (https://pubchem.ncbi.nlm.nih.gov). Using the PyMol (DeLano, 2002), all the SDF files were converted to PDB format and each of the polyphenols structures was optimized with B3LYP/6-31G* basis set by using Gaussian09 software (Frisch & Clemente, 2009). Each of the optimized polyphenols structure was then inserted to AutoDock Tools and standard processes were used to obtain the pdbqt files.
2.2. Preparation of Mpro
The crystal structure of the SARS CoV-2 Mpro was taken from the RCSB Protein Data Bank (http://www.rcsb.org) (PDB ID: 6LU7) (Jin et al., 2020). After correcting the improper bonds, missing hydrogens, side-chain anomalies etc (if any), pdbqt file for Mpro was made through AutoDock Tools (Morris et al., 2009; Morris et al., 2008).
2.3. Molecular docking
The docking of Mpro with two anti-HIV drugs and ten polyphenols from B. papyrifera was performed with the aid of AutoDock Vina (Morris et al., 2009; Morris et al., 2008). The binding affinities of polyphenols-Mpro were determined and analyzed using it. As per the position of active site region, the grid box was assigned with a 10.0 Å radius throughout the initial inhibitor, where the ligands can easily be fitted and which covers the entire active site pocket. The same grid box size and other parameters were used for docking studies of two anti-HIV drugs along with all the ten polyphenols to obtain different docked conformations. The best-suited conformations with the lowest root mean square deviation (RMSD) values along with the highest Vina score were selected for Mpro and B. papyrifera polyphenols complexes. The output from AutoDock Vina was rendered with PyMOL and DS visualizer softwares (Biovia, 2017; DeLano, 2002).
2.4. Molecular dynamics simulation
The molecular dynamics (MD) simulations were performed using the GROningen MAchine for Chemical Simulations GROMACS 2019 (Abraham et al., 2015). The GROMOS9653a6 force field and SPC water model were used for all the MD simulations (Oostenbrink et al., 2004). The ligand topologies were obtained from the PRODRG server (Schuttelkopf & van Aalten, 2004). All bond lengths of protein and anti-HIV drugs/polyphenols were constrained using the LINCS algorithm, while water molecules were restrained by SETTLE algorithm (Hess et al., 1997; Miyamoto & Kollman, 1992). The system was accommodated in a cubic box with a total number of 30226, 30199, 30198, 30204, 30203, 30202, 30200, 30198, 30196 water molecules containing the unligated Mpro, Mpro-C2, Mpro-C4, Mpro-C5, Mpro-C8, Mpro-C9, Mpro-C10, Mpro-darunavir and Mpro-lopinavir complexes, respectively. Each system was energy-minimized using the steepest descent algorithm and equilibrated to achieve the appropriate volume. The leapfrog algorithm with time step 2 fs was used and at every 5 steps, the neighbor list was updated. The Particle Mesh Ewald method is used to treat the Long-range electrostatics with cut off 1.2 nm and with a Fourier grid spacing of 1.2 nm (Essmann et al., 1995). Periodic boundary conditions were applied in all three directions. Equilibration of the systems was carried out in two main stages. First, the system was allowed to heat gradually to 300 K in NVT ensemble using the v-rescale algorithm for 10 ns. Then NPT ensemble was employed for 10 ns by positional restraining of the complexes (unligated Mpro, Mpro-darunavir, Mpro-lopinavir and Mpro-polyphenol complexes) slowly allowing the solvent molecules to relax around it. We have used the Parrinello-Rahman method and Berendsen barostat to maintain the pressure and temperature, respectively (Berendsen et al., 1984; Parrinello & Rahman, 1981). For each system, the average temperature and pressure values remained close to the desired values. The equilibrated systems were then subjected to unrestrained production MD simulations of 100 ns each, maintaining target pressure (1 bar) and temperature (300 K). The root mean square deviation (RMSD), the total number of hydrogen bonds, root mean square fluctuation (RMSF), the radius of gyration (Rg), solvent accessible surface area (SASA) for each system were calculated from the MD trajectories (Bhardwaj & Purohit, 2020; Bhardwaj, Singh, Sharma, Das, et al., 2020; Bhardwaj, Singh, Sharma, Rajendran, et al., 2020; Ghosh et al., 2020; Kamaraj & Purohit, 2016; Rajendran, 2016; Rajendran et al., 2018).
2.5. MM-GBSA
To evaluate the theoretical free energies of binding of ligands to the receptor, generally, two methods are commonly used (a) the molecular mechanics generalized Born surface area (MM-GBSA) and (b) molecular mechanics Poisson-Boltzmann surface area (MM-PBSA). These two methods are equally efficient in predicting the correct binding affinities (Jianzhong Chen, 2016; J. Chen et al., 2015; Hou et al., 2011; Venugopal et al., 2020). Here we have used the MM-GBSA method to calculate the relative binding free energies of anti-HIV drugs and B. papyrifera polyphenols to Mpro. The free energy of binding can be calculated as ΔGbind=ΔH − TΔS.
ΔH = ΔEelec+ΔEvdW+ΔGpolar+ΔGnon-polar, where Eelec and EvdW are the electrostatic and van der Waal’s contributions, and Gploar and Gnon-polar are the polar and non-polar solvation terms, respectively. The polar contribution of the free energy is estimated by a generalized Born model with an external dielectric constant of 80 and an internal dielectric constant of 1, while the non-polar energy contribution is calculated from the solvent accessible surface area (SASA). As similar types of ligands bind to the receptor, the entropic contribution is neglected here. Therefore, our calculated values referred to as relative binding free energies (ΔGbind).
MM-GBSA is a popular method to calculate binding energy, which uses energy properties of free ligand, free receptor and receptor-ligand complex for binding affinity calculation. The prime module of the Schrodinger suite (Schrödinger Release 2020-1: Prime, Schrödinger, LLC, New York, NY, 2020) was used for all MM-GBSA calculations.
2.6. Pharmacokinetic properties analysis
Swiss ADME and pkCSM-pharmacokinetics online softwares were used for the prediction of different pharmacokinetic properties of ten polyphenols from B. papyrifera (Daina et al., 2017; Pires et al., 2015). Levels of toxicity along with the drug-likeness properties of these ten polyphenols such as absorption, distribution, metabolism and excretion parameters were mainly scrutinized.
3. Result and discussion
The resolved crystal structure of Mpro in complex with i) a Michael inhibitor N3 and ii) inhibitor alpha-ketoamide provide useful information about the structural integrity of Mpro which we have elaborately discussed in the introduction section (Jin et al., 2020; Zhang et al., 2020). These two crystal structures (PDB ID 6LU7 and 6Y2E) also laid foundation towards the structure-based drug design against Mpro. Several small molecules including different plant polyphenols are being proposed as effective SARS CoV-2 Mpro inhibitor (Aanouz et al., 2020; Bhardwaj, Singh, Sharma, Rajendran, et al., 2020; Das et al., 2020; Enmozhi et al., 2020; Gyebi et al., 2020; Islam et al., 2020; Joshi et al., 2020; Umesh et al., 2020). Many anti-HIV drugs (darunavir, lopinavir, atazanavir, etc) also have a good binding affinity towards the active site of Mpro (Beck et al., 2020). In recent past, many investigators have chosen darunavir and lopinavir (structures mentioned in Supplemental Figure 2) as standard substrates and have compared the binding affinity and/or binding modes between various small molecules with that of “Mpro-darunavir/lopinavir complex” (Bhardwaj, Singh, Sharma, Rajendran, et al., 2020; Gyebi et al., 2020; Mahanta et al., 2020). Thus, we have also decided to take these two anti-HIV drugs as standard Mpro inhibitors for this study.
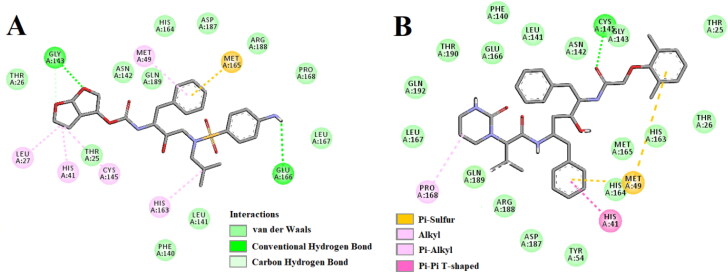
Figure 2.
Molecular docking of anti-HIV drugs with Mpro. Interactions of various amino acids of Mpro with darunavir (panel A) and lopinavir (panel B) are presented with the best docking pose.
3.1. Pharmacokinetics property analysis
Prior to conduct molecular docking studies, the polyphenols of B. papyrifera (structures shown in Figure 1) were screened based on their drug-likeness characteristics. Pharmacokinetics analysis revealed that the molecular weight (MW) of these seven polyphenols were less than 500 (ranging from 322 to 438) which suggested that all the polyphenols may easily transported, diffused and absorbed inside the body. The number of hydrogen bond donors (H-Do) was less than 5 and the number of hydrogen bond acceptors (H-Ac) was in range from 4 to 7 for these polyphenols, which are in accordance with Lipinski’s rules. Furthermore, the topological polar surface area (TPSA) of all the polyphenols was found in the range of 55.76 to 131.36 Å2 indicating good bioavailability of these polyphenols. The calculated intestinal absorption (IA) varies between 84 to 94% which signified good cell membrane permeability and oral bioavailability. All the polyphenols were negative towards AMES test and none of these polyphenols had shown hepatoxicity. However, pharmacokinetics analysis indicated a negative tolerance dose of three polyphenols (C1, C3 and C7) for humans (Table 1). Such negative dose tolerance makes these three polyphenols susceptible to human use. As the rest of the polyphenols (C2, C4, C5, C6, C8, C9 and C10) harbor good and favorable pharmacokinetic properties (Table 1), we selected them for testing their inhibition potency against Mpro.
Table 1.
Pharmacokinetic properties of Broussonetia papyrifera polyphenols.
| Compound | MW | H-Ac | H-Do | Nrot | TPSA | LogP | IA | TC | LD50 | HT | AT | MTD | NLV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | 324.37 | 4 | 3 | 5 | 77.76 | 4.2082 | 90.04 | 0.12 | 2.014 | No | No | −0.048 | 0 |
| C2 | 340.37 | 5 | 4 | 5 | 97.99 | 3.9138 | 74.186 | 0.049 | 2.181 | No | No | 0.168 | 0 |
| C3 | 322.35 | 4 | 1 | 1 | 55.76 | 4.2829 | 93.773 | −0.095 | 2.523 | No | No | −0.149 | 0 |
| C4 | 438.47 | 7 | 5 | 5 | 131.36 | 5.0054 | 88.145 | 0.265 | 2.655 | No | No | 0.656 | 0 |
| C5 | 398.41 | 7 | 3 | 5 | 109.36 | 4.1027 | 84.297 | 0.486 | 2.334 | No | No | 0.297 | 0 |
| C6 | 394.50 | 5 | 3 | 5 | 69.92 | 5.8871 | 89.903 | 0.578 | 2.108 | No | No | 0.029 | 0 |
| C7 | 392.49 | 4 | 2 | 3 | 58.92 | 5.857 | 91.551 | 0.382 | 2.643 | No | No | −0.545 | 0 |
| C8 | 426.50 | 6 | 4 | 3 | 99.38 | 4.2381 | 71.921 | 0.253 | 2.518 | No | No | 0.373 | 0 |
| C9 | 396.52 | 4 | 4 | 8 | 80.92 | 5.7017 | 89.18 | 0.478 | 2.215 | No | No | 0.501 | 0 |
| C10 | 410.55 | 4 | 3 | 9 | 69.92 | 6.0047 | 89.174 | 0.71 | 1.994 | No | No | 0.415 | 1 |
MW = Molecular weight (g/mol); H-Ac = No. of hydrogen bond acceptor; H-Do = No. of hydrogen bond donors; Nrot = No. of rotatable bonds; TPSA = Topological polar surface area (Å2); LogP = Predicted octanol/water partition coefficient; IA = Intestinal absorption (% Absorbed); TC = Total clearance (log ml/min/kg); LD50 = Oral rat acute toxicity; HT = Hepatoxicity; AT = AMES toxicity; MTD = Maximum tolerated dose for human (log mg/kg/day); NLV = No. of Lipinski rule violation.
3.2. Molecular docking studies
All these seven polyphenols and two anti-HIV drugs (darunavir and lopinavir) were subjected to molecular docking studies to assess the polyphenol(s) exhibiting the higher or comparable binding energy to that of “Mpro-darunavir/lopinavir interaction”. Darunavir interacted with Mpro via two hydrogen bonds [Gly143 (2.3 Å) and Glu166 (2.4 Å)], one Pi-sulfur bond (Met165) and multiple alkyl/Pi-alkyl bonds (Leu27, His41, Met49, Cys145 and His163) (Figure 2A and Table 2). It also formed many van der Waals interactions with different amino acid residues of Mpro (Figure 2A and Table 2). Lopinavir formed only one hydrogen bond with Cys145 and several other non-covalent bonds with various important amino acid residues (such as Thr26, His41, Met49, Phe140, Glu166, Leu167, etc) within the active site of Mpro (Figure 2B and Table 2). The binding affinity of darunavir and lopinavir towards Mpro was −7.4 and −7.3 kcal/mol, respectively (Table 2). Interestingly, all the polyphenols (C2, C4, C5, C6, C8, C9 and C10) exhibited higher binding affinity (−7.6 to −8.2 kcal/mol) towards Mpro than that of darunavir and lopinavir (Table 2). The highest binding affinity was observed for C5 (−8.2 kcal/mol) and the lowest one is for C6 (−7.6 kcal/mol).
Table 2.
Binding energy and hydrogen bond interactions of Darunavir and different polyphenols of Broussonetia papyrifera with the active site of SARS CoV-2 Mpro.
| Compounds interact with Mpro | Binding energy (kcal/mol) | Amino acids of Mpro involved in H-bonding |
|---|---|---|
| Darunavir | −7.4 | Gly143 (2.3 Å), Glu166 (2.4 Å) |
| Lopinavir | −7.3 | Cys145 (2.3Å) |
| C2 | −8.1 | Thr26 (2.8 Å), Gly143 (2.5 Å), Ser144 (3.0 Å), Cys145 (3 Å), Glu166 (2.3 Å) |
| C4 | −7.9 | Leu141 (2.3 Å), Cys145 (2.3 Å), Arg188 (2.1 Å) |
| C5 | −8.2 | Leu141 (2.7 Å), Asn142 (2.2 Å), Gly143 (2.3 Å), Cys145 (2.3 Å), Glu166 (3.2 Å) |
| C6 | −7.2 | Ser144 (2.4 Å, 2.5 Å) |
| C8 | −7.8 | Gly143 (2.3 Å, 2.4 Å), Glu166 (2.4 Å) |
| C9 | −8.1 | Leu141 (2.4 Å), Gly143 (2.5 Å), Met165 (2.4 Å) |
| C10 | −8.0 | Ser144 (2.3 Å), His163(2.4 Å), Thr190(2.1 Å) |
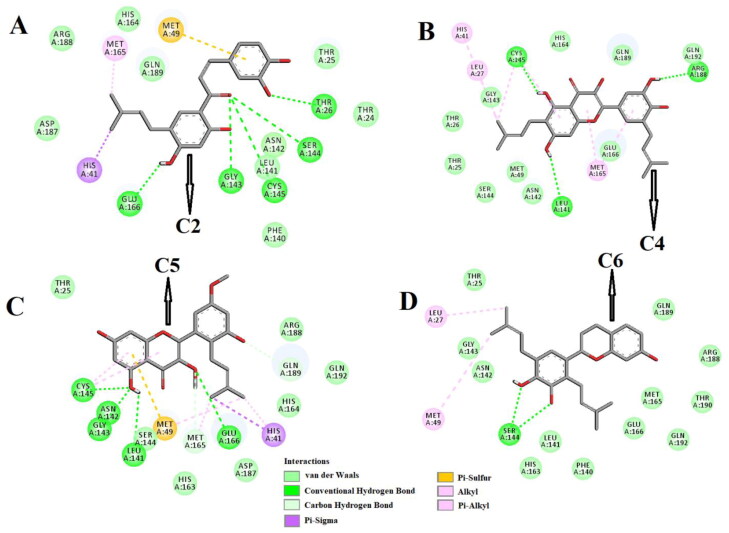
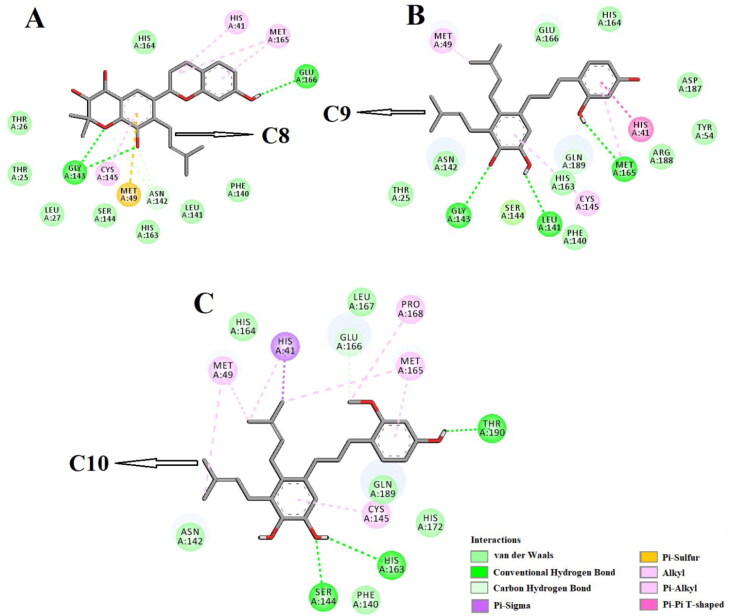
Molecular docking studies further depicted that all these polyphenols efficiently interacted with different amino acid residues of domain I and II of Mpro (Figure 3-4 and Table 2). C2 formed hydrogen bonds with five amino acid residues (Thr26, Gly143, Ser144, Cys145 and Glu166) of Mpro (Figure 3A, Table 2). Many other amino acid residues including His41 were also involved in different non-covalent interactions (van der Waals, Pi-sulfur, Pi-sigma and alkyl/Pi-alkyl) with C2 (Figure 3A). When C4 was docked into the active site of Mpro, three hydrogen bondinteractions [Leu141 (2.3 Å), Cys145 (2.3 Å) and Arg188 (2.1 Å)] and thirteen other non-covalent interactions (van der Waals and alkyl/Pi-alkyl) were evidenced (Figure 3B and Table 2) The Mpro-C5 complex was stabilized by one Pi-sigma interaction (His41), seven van der Waals interactions (Thr25, Ser144, His163, His164, Asp187, Arg188 and Gln192), two C-H bond interactions (Met165 and Gln189), one Pi-sulfur interaction (Met49) and five hydrogen bonds [Leu141 (2.7 Å), Asn142 (2.2 Å), Gly143 (2.3 Å), Cys145 (2.3 Å) and Glu166 (3.2 Å)] (Figure 3C and Table 2). In case of Mpro-C6 complex, we observed two hydrogen bond interactions [Ser144 (2.4 Å, 2.5 Å)], twelve van der Waals interactions (Thr25, Phe140, Leu141, Asn142, Gly143, His163, Met165, Glu166, Arg188, Gln189, Thr190 and Gln192) and two other types of non-covalent interactions (alkyl and Pi-alkyl) (Figure 3D and Table 2). When the other three polyphenols (C8, C9 and C10) were docked individually to Mpro, these complexes were stabilized by three number of hydrogen bond interactions and many non-covalent (C-H bond, van der Waals, Pi-alkyl etc) interactions (Figure 4A-C and Table 2). Gly143 of Mpro formed two and Glu166 formed one hydrogen bond with C8 (Figure 4A, Table 2). However, C9 formedhydrogen bonds with Leu141, Gly143 and Met165 of Mpro and the amino acid residues of Mpro engaged in the hydrogen bonding with C10 were Ser144, His163 and Thr190, respectively (Figure 4B-C, Table 2). C10 interacted with His41 and Cys145 of Mpro via Pi-sigma and Pi-alkyl interactions, respectively (Figure 4B). These two amino acid residues of Mpro formed Pi-Pi (His41) and Pi-alkyl (Cys145) bonds with C9 (Figure 4A). It was also evidenced that the other four polyphenols (C2, C4, C5, and C8) were involved in interactions with His41 and Cys145 via hydrogen bonding or other non-covalent forces (Figure 3 and 4). Only polyphenol C6 had no interaction with these two key catalytic residue(s) of Mpro protease (Figure 4C). Thus, it can be concluded that all these polyphenols (except C6) may possibly inhibit the proteolytic activity of Mpro and may be effective candidates for the treatment of COVID-19 disease.
Figure 3.
Molecular docking of B. papyrifera polyphenols C2, C4, C5 and C6 with Mpro. The docked conformation of the Mpro-C2 complex (panel A), Mpro-C4 complex (panel B), Mpro-C5 complex (panel C) and Mpro-C6 complex (panel D) depicting the possible interactions with various amino acids of Mpro. All these polyphenolic compounds (except C6) interact with various amino acid residues including His41 and Cys141 of Mpro.
Figure 4.
Molecular docking of B. papyrifera polyphenols C8, C9 and C10 with Mpro. The docked conformation of the Mpro-C8 complex (panel A), Mpro-C9 complex (panel B) and Mpro-C10 complex (panel C) depicting the possible interactions with various amino acids of Mpro. All these polyphenolic compounds interact with various amino acid residues including His41 and Cys141 of Mpro.
We selected six Mpro-polyphenol complexes (Mpro-C2, Mpro-C4, Mpro-C5, Mpro-C8, Mpro-C9 and Mpro-C10) for performing the subsequent studies.
3.3. Molecular dynamics simulation studies
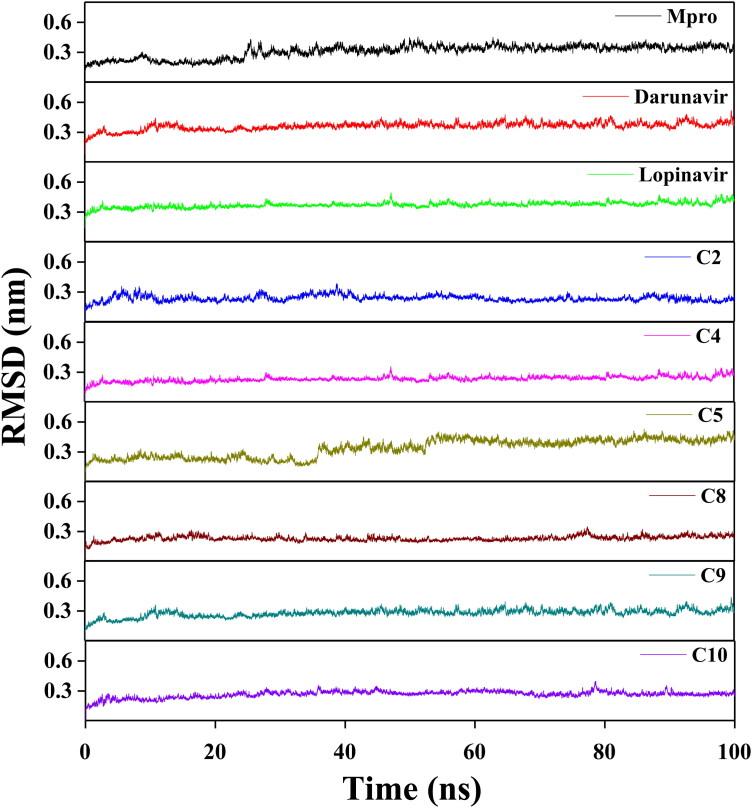
In order to get an idea about the structural stability, conformational fluctuations, compactness and folding behavior of Mpro alone/Mpro (unligated) and Mpro complexed with two anti-HIV drugs as well as six polyphenols, we performed MD simulations for 100 ns using GROMOS9653a6 force field. The analysis of RMSD usually provides important information about the stability of the protein-ligand complex. Thus, we first estimated the RMSD of backbone alpha carbon atoms of all these systems (Figure 5). The RMSD of Mpro (unligated) maintained a constant value (∼0.21–0.22 nm) from 2 ns to 17 ns. Thereafter the RMSD value gradually increased till 25 ns and reached ∼0.35 nm. Then, the value .was slightly decreased and persisted at ∼0.31 nm from 65 ns till the end of the MD run. The RMSD values for both Mpro-darunavir and Mpro-lopinavir complexes were found to remain almost constant (∼0.36–0.37 nm) from 10 ns to 100 ns with some marginal fluctuations (Figure 5). The RMSD profiles of Mpro-polyphenol complexes revealed that most of the complexes were stabilized quite quickly (Figure 5) The magnitude of RMSD corresponding to three Mpro-polyphenol complexes (Mpro-C2, Mpro-C9 and Mpro-C10) attained an equilibrium value after 40 ns (∼0.24 nm for Mpro-C2 and ∼0.26-0.27 nm for other two systems) and remained almost the same throughout the 100 ns simulation time span. On the contrary, the saturation of the RMSD curve for two other Mpro-polyphenol complexes (Mpro-C4 and Mpro-C8) was observed after 30 and 20 ns, respectively (Figure 5). For Mpro-C5 complex, the RMSD value from 2 ns to 34 ns oscillated between ∼0.19 and 0.24 nm. Then, within next 8 ns, the value progressively increased to ∼0.35 nm and remained almost the same till 50 ns, Again the value started increasing and reached to ∼0.46 nm after 58 ns and remained almost the same till the end of MD run with some fluctuations (Figure 5). The average RMSD values for Mpro (unligated), Mpro-darunavir complex and Mpro-lopinavir complex were found to be ∼0.31 nm, ∼0.36 nm and ∼0.37 nm, respectively, which are in agreement with previously reported values (Table 3) (Bhardwaj, Singh, Sharma, Rajendran, et al., 2020). While the same for all the Mpro-polyphenol complexes were ranging from ∼0.23 nm to ∼0.34 nm with the lowest RMSD for the Mpro-C8 system and the highest one for Mpro-C5 system (Table 3). Thus, it can be concluded that all Mpro-polyphenol complexes are stable. However, the stability of Mpro-C5 is least among all of them. These findings additionally indicated that the stability of these six Mpro-polyphenol complexes is comparable or relatively more than that of the Mpro-darunavir/Mpro-lopinavir complex.
Figure 5.
RMSD plots of Mpro (unligated), Mpro-darunavir, Mpro-lopinavir and six Mpro-polyphenol complexes. The MD simulations for Mpro (unligated), Mpro-C2, Mpro-C4, Mpro-C5, Mpro-C8, Mpro-C9 and Mpro-C10 complexes were performed for 100 ns. All these MD trajectories were analyzed with the aid of RMSD.
Table 3.
Average values of the RMSD, RMSF, Rg, SASA and the total number of intermolecular hydrogen bond formed for the simulated systems.
| System | RMSD (nm) | RMSF (nm) | Rg (nm) | SASA (nm2) | Total Number of H-bonds formed |
|---|---|---|---|---|---|
| Mpro (unligated) | 0.309 | 0.1937 | 2.195 | 151.4483 | 547 |
| Mpro-darunavir | 0.361 | 0.1952 | 2.197 | 151.1540 | 550 |
| Mpro-lopinavir | 0.371 | 0.1948 | 2.196 | 151.2825 | 551 |
| Mpro-C2 | 0.237 | 0.1568 | 2.229 | 154.0753 | 554 |
| Mpro-C4 | 0.232 | 0.1405 | 2.215 | 156.3992 | 558 |
| Mpro-C5 | 0.335 | 0.2069 | 2.214 | 154.1175 | 552 |
| Mpro-C8 | 0.229 | 0.1454 | 2.225 | 152.7752 | 564 |
| Mpro-C9 | 0.278 | 0.1587 | 2.208 | 154.3884 | 555 |
| Mpro-C10 | 0.266 | 0.1541 | 2.210 | 155.7089 | 552 |
The conformational stability of these six Mpro-polyphenol complexes was further analyzed by estimating the total number of intermolecular hydrogen bonds formed during the entire 100 ns simulation time span (Table 3). The average number of intermolecular hydrogen bonds in the Mpro (unligated) system was 547. In Mpro-darunavir and Mpro-lopinavir complex, the existence of more number of intermolecular hydrogen bonds was evidenced (550 and 551, respectively). We also noticed a higher number of intermolecular hydrogen bonds in all selected Mpro-polyphenol complexes (552-564) (Table 3). Among these six complexes, the highest number of intermolecular hydrogen bonds (564) was observed when C8 was complexed with Mpro. These results support the RMSD data obtained from MD simulations. Based on these findings we can convincingly say that the complex originated from the binding of each selected polyphenol to Mpro is quite stable.
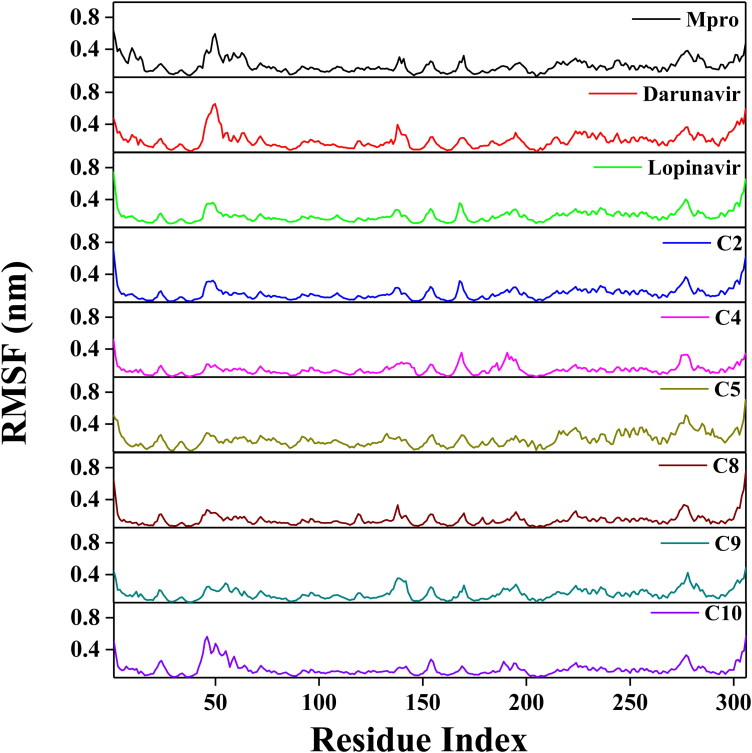
Next, we looked into the flexibility of different regions of Mpro by calculating the RMSF of alpha carbon atoms for all systems (Figure 6). It was quite evident from the RMSF profiles that all systems experience higher conformational fluctuations in domain III. Moreover, the fluctuations for the amino acid residues pertaining to domain III of the Mpro-C5 complex were highest among all the studied systems. In the case of Mpro (unligated) system, we additionally observed higher fluctuations (up to ∼0.6 nm) in a certain portion of domain I (residues 45-60). In fact, most of the amino acid residues within the domain I and II of this system had RMSF fluctuation below 0.3 nm. The average RMSF value for Mpro (unligated) system was 0.194 nm (Table 3). The Mpro-darunavir and Mpro-lopinavir system experienced more or less similar conformational fluctuations to that of Mpro (unligated) system (Figure 6). In fact, the fluctuations for the residues 45-60, were reduced upon the binding of lopinavir to Mpro (up to 0.35 nm). For both Mpro-darunavir and Mpro-lopinavir complexes, the average RMSF value was 0.195 nm (Table 3). Furthermore, it was evidenced after analyzing the RMSF profiles of four Mpro-polyphenol systems (Mpro-C2, Mpro-C4, Mpro-C8 and Mpro-C10) that these complexes exhibited lower fluctuations (especially in domain I and II) as compared to the Mpro (unligated) system. The average RMSF values of these Mpro-polyphenol complexes were ranging between ∼0.141 nm to ∼0.157 nm (Table 2). Even in Mpro-C9 complexes, the fluctuations of most of the amino acid residues (residues 134-144 in Mpro-C9 complex) residing at the domain I and II were reduced (Figure 6). The average RMSF value of this complex is ∼0.159 nm (Table 3). The RMSF plot of the Mpro-C5 complex reflected that very few amino acid residues within domain I and II have an RMSF value of more than 0.25 nm (Figure 6). Interestingly, the RMSF values of several stretches within these two domains of this Mpro-polyphenol system (residues 21-24, 85-109 and 130-136) were more compared to that of Mpro (unligated) system, The average RMSF value of Mpro-C5 complex was ∼0.207 nm (Table 3). Most importantly, the fluctuations of many key residues of the binding region of Mpro were ceased down after binding to these six polyphenols. These findings suggested that the conformational fluctuations of these six Mpro-polyphenol complexes are comparable or relatively less than that of the Mpro-darunavir/Mpro-lopinavir complex.
Figure 6.
RMSF profiles of Mpro (unligated), Mpro-darunavir, Mpro-lopinavir and six Mpro-polyphenol complexes. The RMSF values for Mpro (unligated) and Mpro-anti-HIV drug complexes as well as six Mpro-polyphenol complexes were estimated from the respective 100 ns MD trajectories. The values were then plotted against the amino acid residues of Mpro.
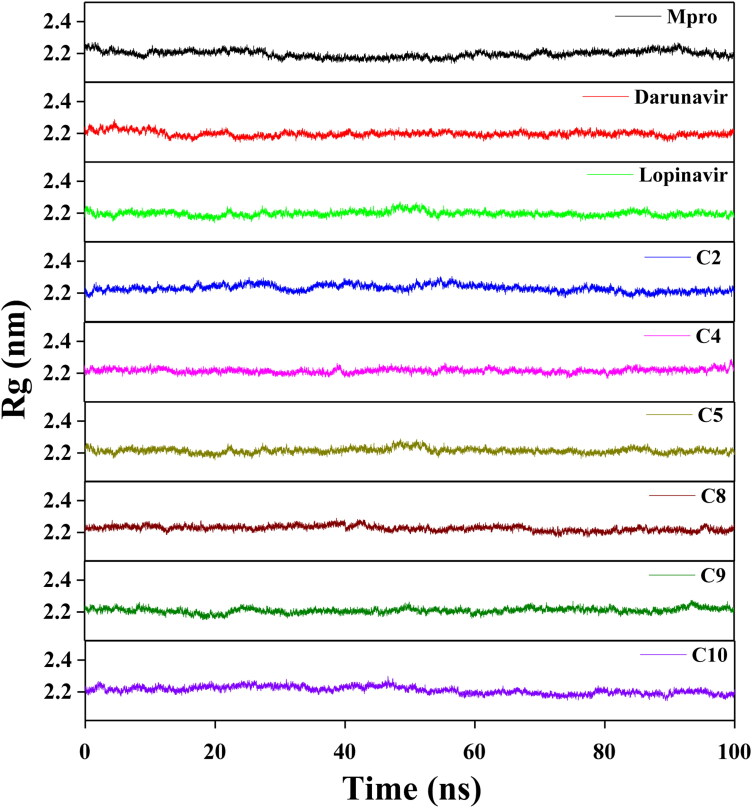
Afterward, we estimated the Rg to assess the compactness of all the complexes (Figure 7 and Table 3). The average Rg value for Mpro (unligated) and the other two complexes (Mpro-darunavir and Mpro-lopinavir) was almost identical (∼2.20 nm). We observed a slightly higher average Rg value for six Mpro-polyphenol systems (∼2.21–2.23 nm) (Table 3). Thus, it can be suggested that all these Mpro-polyphenol complexes were slightly less compact in comparison to free Mpro/Mpro (unligated) and other two Mpro-HIV drug complexes. Thereafter, SASA was employed to assess the extent of expansion of protein volume in each system (Figure 8 and Table 3). The average SASA values of Mpro-darunavir complex (151.154 nm2) and Mpro-lopinavir complex (∼151.283 nm2) were found to be in the similar range with Mpro (unligated) (∼151.448 nm2) (Table 3). Higher SASA values were observed in all the Mpro-polyphenol complexes (∼152.775–∼156.399 nm2). This increased SASA values indicated marginal expansion of the Mpro upon interaction with these six polyphenols.
Figure 7.
Determination of Rg values of Mpro (unligated), two Mpro-anti-HIV drugs and six Mpro-polyphenol complexes. The MD simulations for Mpro (unligated), Mpro-darunavir, Mpro-lopinavir, Mpro-C2, Mpro-C4, Mpro-C5, Mpro-C8, Mpro-C9 and Mpro-C10 complexes were performed for 100 ns. All these MD trajectories were analyzed with the aid of Rg.
Figure 8.
Estimation of SASA values of Mpro (unligated), two Mpro-anti-HIV drugs and six Mpro-polyphenol complexes. The MD simulations for Mpro (unligated), Mpro-darunavir, Mpro-lopinavir, Mpro-C2, Mpro-C4, Mpro-C5, Mpro-C8, Mpro-C9 and Mpro-C10 were performed for 100 ns. All these MD trajectories were analyzed with the aid of SASA.
3.4. MM-GBSA
Finally, we estimated the binding energy of “Mpro-polyphenols interaction” as well as “Mpro-HIV drugs interaction” using the MM-GBSA method. We utilized the docking conformation having the highest AutoDock Vina energy values. Table 4, illustrated very high MM-GBSA binding free energies of our Mpro-polyphenol complexes, which are comparable with the docking results. These higher MM-GBSA free energy results signify greater stability of these Mpro-polyphenol complexes. The MM-GBSA free energy values of Mpro-darunavir and Mpro-lopinavir complexes were found to be −35.65 kcal/mol and −40.39 kcal/mol, respectively (Table 4). On the contrary, all the six Mpro-polyphenol complexes exhibited higher MM-GBSA binding energy than that of Mpro-darunavir complex and Mpro-lopinavir complex. The MM-GBSA free energies of six Mpro-polyphenol complexes were ranging from −41.32 kcal/mol to −56.23 kcal/mol (Table 4).
Table 4.
MM-GBSA values of different Mpro-anti-HIV drugs and Mpro-polyphenol complexes.
| System | Binding Free Energy (kcal/mol ) |
|---|---|
| Mpro-darunavir | −35.65 |
| Mpro-lopinavir | −40.39 |
| Mpro-C2 | −50.91 |
| Mpro-C4 | −47.28 |
| Mpro-C5 | −51.59 |
| Mpro-C8 | −41.32 |
| Mpro-C9 | −56.23 |
| Mpro-C10 | −41.98 |
Among all of them, the Mpro-C9 complex exhibited the highest binding free energies, while the Mpro-C8 system showed the lowest binding free energies. It is quite evident from these MM-GBSA values that all six polyphenols efficiently interacted with Mpro with higher binding free energy than that of “Mpro-darunavir/lopinavir interaction.” The higher MM-GBSA values (ΔGbind) in the case of these six Mpro-polyphenol complexes were mostly contributed by the coulombic interactions, SASA and hydrophobic interactions. Among these six Mpro-polyphenol complexes, Mpro-C2 showed the highest, whereas Mpro-C10 showed the lowest coulombic interactions. The maximum free energy contribution from SASA was experienced by Mpro-C9 and Mpro-C10 complexes, whereas the rest of the Mpro-polyphenol complexes showed similar SASA values. The Mpro-C9 showed the highest hydrophobic interactions, whereas the lowest hydrophobic interactions was observed in the case of Mpro-C2 complex. Other Mpro-polyphenol complexes showed similar hydrophobic interactions.
4. Conclusion
This study is aimed to test the inhibition potency of B. papyrifera polyphenols against SARS CoV-2 Mpro using a computational approach. The polyphenols which possess favorable drug-likeness characteristics [broussochalcone A (C2), papyriflavonol A (C4), 3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane (C5), kazinol A (C6), broussoflavan A (C8), kazinol F (C9) and kazinol J (C10)] including two anti-HIV drugs (darunavir and lopinavir) were subjected to molecular docking studies. All these polyphenols had higher AutoDock Vina energy values than the darunavir and lopinavir. Among them, six polyphenols (C2, C4, C5, C8, C9 and C10) had interaction with both the key catalytic residues (His41 and Cys145) of Mpro. The RMSD and RMSF profiles corresponding to these six Mpro-polyphenol complexes clearly suggested that they (complexes) are highly stable and experience less conformational fluctuations. The Rg and SASA analysis revealed that all Mpro-polyphenol complexes are slightly less compact and expand marginally. The existence of a higher number of intermolecular hydrogen bonds in the complexes with B. papyrifera polyphenols (C2, C4, C5, C8, C9 and C10) than in Mpro-darunavir/lopinavir complex suggesting greater stability of these polyphenols in the binding pockets of Mpro. These findings were further corroborated by MM-GBSA analysis. This analysis revealed that all Mpro-polyphenol complexes were more stable than Mpro-darunavir and Mpro-lopinavir complex. Therefore, it can be concluded that broussochalcone A, papyriflavonol A, 3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane, broussoflavan A, kazinol F and kazinol J were more effective Mpro inhibitors than earlier recommended anti-HIV drugs (darunavir and lopinavir). However, their inhibitory effectiveness and usage as anti-COVID-19 drugs should be thoroughly examined using various experimental studies.
Supplementary Material
Acknowledgements
RG acknowledges IIT Bhubaneswar for providing fellowship. The authors thank IIT Delhi HPC facility for computational resources.
Glossary
Abbreviations
- COVID-19
corona virus disease 2019
- SARS CoV-2
severe acute respiratory syndrome corona virus-2
- Mpro
Main protease
- MD
Molecular dynamics
- RMSD
Root mean square deviation
- RMSF
Root mean square fluctuation
- Rg
Radius of gyration
- SASA
Solvent accessible surface area
Disclosure statement
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., & Bouachrine, M. (2020). Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.).), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Arun, K. G., Sharanya, C. S., Abhithaj, J., Francis, D., & Sadasivan, C. (2020). Drug repurposing against SARS-CoV-2 using E-pharmacophore based virtual screening, molecular docking and molecular dynamics with main protease as the target. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1779819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, B. R., Shin, B., Choi, Y., Park, S., & Kang, K. (2020). Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Computational and Structural Biotechnology Journal, 18, 784–790. 10.1016/j.csbj.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., Gunsteren, W. F. v., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Bhardwaj, V. K., & Purohit, R. (2020). A new insight into protein-protein interactions and the effect of conformational alterations in PCNA. International Journal of Biological Macromolecules, 148, 999–1009. 10.1016/j.ijbiomac.2020.01.212 [DOI] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Das, P., & Purohit, R. (2020). Structural based study to identify new potential inhibitors for dual specificity tyrosine-phosphorylation- regulated kinase. Computer Methods and Programs in Biomedicine, 194, 105494. 10.1016/j.cmpb.2020.105494 [DOI] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia, D. S. (2017). Discovery studio modeling environment. Release.
- Chakraborty, A., Nandi, S. K., Panda, A. K., Mahapatra, P. P., Giri, S., & Biswas, A. (2018). Probing the structure-function relationship of Mycobacterium leprae HSP18 under different UV radiations . International Journal of Biological Macromolecules, 119, 604–616. 10.1016/j.ijbiomac.2018.07.151 [DOI] [PubMed] [Google Scholar]
- Chen, J. (2016). Drug resistance mechanisms of three mutations V32I, I47V and V82I in HIV-1 protease toward inhibitors probed by molecular dynamics simulations and binding free energy predictions. RSC Advances, 6(63), 58573–58585. 10.1039/C6RA09201B [DOI] [Google Scholar]
- Chen, J., Wang, X., Zhu, T., Zhang, Q., & Zhang, J. Z. (2015). A comparative insight into amprenavir resistance of mutations V32I, G48V, I50V, I54V, and I84V in HIV-1 protease based on thermodynamic integration and MM-PBSA methods. Journal of Chemical Information and Modeling, 55(9), 1903–1913. 10.1021/acs.jcim.5b00173 [DOI] [PubMed] [Google Scholar]
- Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., Qiu, Y., Wang, J., Liu, Y., Wei, Y., Xia, J., Yu, T., Zhang, X., & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet (London, England), 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, K. C., Wei, D. Q., & Zhong, W. Z. (2003). Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochemical and Biophysical Research Communications, 308(1), 148–151. 10.1016/S0006-291X(03)01342-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta, D., & Vanelli, M. (2020). WHO declares COVID-19 a pandemic. Acta Bio-Medica: Atenei Parmensis, 91(1), 157–160. 10.23750/abm.v91i1.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717. 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan, W. J., Zhang, Q., Zhang, F., Wang, W. W., & Gao, J. M. (2019). Benzonate derivatives of acetophenone as potent α-glucosidase inhibitors: synthesis, structure-activity relationship and mechanism. Journal of Enzyme Inhibition and Medicinal Chemistry, 34(1), 937–945. 10.1080/14756366.2019.1604519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics., 1–11. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W. (2002). The PyMOL Molecular Graphics System (2002) DeLano Scientific, Palo Alto, CA, USA. http://www.pymol.org.
- Elmezayen, A. D., Al-Obaidi, A., Sahin, A. T., & Yelekci, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Fan, K., Wei, P., Feng, Q., Chen, S., Huang, C., Ma, L., Lai, B., Pei, J., Liu, Y., Chen, J., & Lai, L. (2004). Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. The Journal of Biological Chemistry, 279(3), 1637–1642. 10.1074/jbc.M310875200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, M., & Clemente, F. Gaussian 09, Revision A. 01, MJ Frisch, GW Trucks, HB Schlegel, GE Scuseria, MA Robb, JR Cheeseman, G. Scalmani, V. Barone, B. Mennucci, GA Petersson, H. Nakatsuji, M. Caricato, X. Li, HP Hratchian, AF Izmaylov, J. Bloino, G. Zhe.
- Ghosh, R., Chakraborty, A., Biswas, A., & Chowdhuri, S. (2020). Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn, 1–13. 10.1074/jbc.M310875200 [DOI] [PMC free article] [PubMed]
- Gyebi, G. A., Ogunro, O. B., Adegunloye, A. P., Ogunyemi, O. M., & Afolabi, S. O. (2020). Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CL(pro)): An in silico screening of alkaloids and terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. G. E. M. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [DOI] [Google Scholar]
- Hou, T., Wang, J., Li, Y., & Wang, W. (2011). Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. Journal of Chemical Information and Modeling, 51(1), 69–82. 10.1021/ci100275a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, M. F., Kuo, C. J., Chang, K. T., Chang, H. C., Chou, C. C., Ko, T. P., Shr, H. L., Chang, G. G., Wang, A. H., & Liang, P. H. (2005). Mechanism of the maturation process of SARS-CoV 3CL protease. The Journal of Biological Chemistry, 280(35), 31257–31266. 10.1074/jbc.M502577200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, M. R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. K., Chugh, A., Giri, A. P., & Kulkarni, M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaraj, B., & Purohit, R. (2016). Mutational analysis on membrane associated transporter protein (MATP) and their structural consequences in Oculocutaeous albinism Type 4 (OCA4)-A molecular dynamics approach. Journal of Cellular Biochemistry, 117(11), 2608–2619. 10.1002/jcb.25555 [DOI] [PubMed] [Google Scholar]
- Kandeel, M., & Al-Nazawi, M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, 251, 117627. 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Liu, H., Galasiti Kankanamalage, A. C., Weerasekara, S., Hua, D. H., Groutas, W. C., Chang, K. O., & Pedersen, N. C. (2016). Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathogens, 12(3), e1005531. 10.1371/journal.ppat.1005531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., & Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581(7807), 215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 3(1), 237–261. 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England), 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanta, S., Chowdhury, P., Gogoi, N., Goswami, N., Borah, D., Kumar, R., Chetia, D., Borah, P., Buragohain, A. K., & Gogoi, B. (2020). Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: An in silico based approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1768902 [DOI] [PubMed] [Google Scholar]
- Mirza, M. U., & Froeyen, M. (2020). Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. Journal of Pharmaceutical Analysis. 10.1016/j.jpha.2020.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S., & Kollman, P. A. (1992). Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. Journal of Computational Chemistry, 13(8), 952–962. 10.1002/jcc.540130805 [DOI] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Olson, A. J. (2008). Using AutoDock for ligand-receptor docking. Curr Protoc Bioinformatics, Chapter 8, Unit 8 14. 10.1002/0471250953.bi0814s24 [DOI] [PubMed]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Nandi, S. K., Chakraborty, A., Panda, A. K., & Biswas, A. (2016). Conformational perturbation, hydrophobic interactions and oligomeric association are responsible for the enhanced chaperone function of Mycobacterium leprae HSP18 under pre-thermal condition [10.1039/C6RA00167J. RSC Advances, 6(67), 62146–62156. 10.1039/C6RA00167J [DOI] [Google Scholar]
- Oostenbrink, C., Villa, A., Mark, A. E., & van Gunsteren, W. F. (2004). A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. Journal of Computational Chemistry, 25(13), 1656–1676. 10.1002/jcc.20090 [DOI] [PubMed] [Google Scholar]
- Park, J. Y., Yuk, H. J., Ryu, H. W., Lim, S. H., Kim, K. S., Park, K. H., Ryu, Y. B., & Lee, W. S. (2017). Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. Journal of Enzyme Inhibition and Medicinal Chemistry, 32(1), 504–515. 10.1080/14756366.2016.1265519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics, 52(12), 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Pires, D. E., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran, V. (2016). Structural analysis of oncogenic mutation of isocitrate dehydrogenase 1. Molecular Biosystems, 12(7), 2276–2287. 10.1039/c6mb00182c [DOI] [PubMed] [Google Scholar]
- Rajendran, V., Gopalakrishnan, C., & Sethumadhavan, R. (2018). Pathological role of a point mutation (T315I) in BCR-ABL1 protein-A computational insight. Journal of Cellular Biochemistry, 119(1), 918–925. 10.1002/jcb.26257 [DOI] [PubMed] [Google Scholar]
- Ren, L. L., Wang, Y. M., Wu, Z. Q., Xiang, Z. C., Guo, L., Xu, T., Jiang, Y. Z., Xiong, Y., Li, Y. J., Li, X. W., Li, H., Fan, G. H., Gu, X. Y., Xiao, Y., Gao, H., Xu, J. Y., Yang, F., Wang, X. M., Wu, C., … Wang, J. W. (2020). Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chinese Medical Journal, 133(9), 1015–1024. 10.1097/CM9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M. H., Tong, S., Tamin, A., Lowe, L., Frace, M., DeRisi, J. L., Chen, Q., Wang, D., Erdman, D. D., Peret, T. C., … Bellini, W. J. (2003). Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science (New York, N.Y.).), 300(5624), 1394–1399. 10.1126/science.1085952 [DOI] [PubMed] [Google Scholar]
- Ryu, H. W., Lee, B. W., Curtis-Long, M. J., Jung, S., Ryu, Y. B., Lee, W. S., & Park, K. H. (2010). Polyphenols from Broussonetia papyrifera displaying potent alpha-glucosidase inhibition. Journal of Agricultural and Food Chemistry, 58(1), 202–208. 10.1021/jf903068k [DOI] [PubMed] [Google Scholar]
- Ryu, H. W., Lee, J. H., Kang, J. E., Jin, Y. M., & Park, K. H. (2012). Inhibition of xanthine oxidase by phenolic phytochemicals from Broussonetia papyrifera. Journal of the Korean Society for Applied Biological Chemistry, 55(5), 587–594. 10.1007/s13765-012-2143-0 [DOI] [Google Scholar]
- Schuttelkopf, A. W., & van Aalten, D. M. (2004). PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography, 60(Pt 8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Umesh, K. D., Selvaraj, C., Singh, S. K., & Dubey, V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal, P. P., Das, B. K., Soorya, E., & Chakraborty, D. (2020). Effect of hydrophobic and hydrogen bonding interactions on the potency of ss-alanine analogs of G-protein coupled glucagon receptor inhibitors. Proteins: Structure, Function, and Bioinformatics, 88(2), 327–344. 10.1002/prot.25807 [DOI] [PubMed] [Google Scholar]
- Williams, S. J., & Goddard-Borger, E. D. (2020). alpha-glucosidase inhibitors as host-directed antiviral agents with potential for the treatment of COVID-19. Biochemical Society Transactions, 10.1042/BST20200505 [DOI] [PubMed] [Google Scholar]
- Yu, X., & Yang, R. (2020). COVID-19 transmission through asymptomatic carriers is a challenge to containment. Influenza and Other Respiratory Viruses, 14(4), 474–475. 10.1111/irv.12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.).), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Guo, F., Comunale, M. A., Mehta, A., Sehgal, M., Jain, P., Cuconati, A., Lin, H., Block, T. M., Chang, J., & Guo, J. T. (2015). Inhibition of endoplasmic reticulum-resident glucosidases impairs severe acute respiratory syndrome coronavirus and human coronavirus NL63 spike protein-mediated entry by altering the glycan processing of angiotensin I-converting enzyme 2. Antimicrobial Agents and Chemotherapy, 59(1), 206–216. 10.1128/AAC.03999-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. (2020). SARS-CoV-2: An emerging coronavirus that causes a global threat. International Journal of Biological Sciences, 16(10), 1678–1685. 10.7150/ijbs.45053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., Si, H. R., Zhu, Y., Li, B., Huang, C. L., Chen, H. D., Chen, J., Luo, Y., Guo, H., Jiang, R. D., Liu, M. Q., Chen, Y., Shen, X. R., Wang, X., … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., Tan, W., China Novel Coronavirus, I., & Research, T. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.