Abstract
The complement system may be crucial during dengue virus infection and progression to severe dengue. This study investigates the role of MBL2 genetic variants and levels of MBL in serum and complement proteins in Vietnamese dengue patients. MBL2 genotypes (− 550L/H, MBL2 codon 54), MBL2 diplotypes (XA/XO, YA/XO) and MBL2 haplotypes (LXPB, HXPA, XO) were associated with dengue in the study population. The levels of complement factors C2, C5, and C5a were higher in dengue and dengue with warning signs (DWS) patients compared to those in healthy controls, while factor D levels were decreased in dengue and DWS patients compared to the levels determined in healthy controls. C2 and C5a levels were associated with the levels of AST and ALT and with WBC counts. C9 levels were negatively correlated with ALT levels and WBC counts, and factor D levels were associated with AST and ALT levels and with platelet counts. In conclusions, MBL2 polymorphisms are associated with dengue in the Vietnamese study population. The levels of the complement proteins C2, C4b, C5, C5a, C9, factor D and factor I are modulated in dengue patients during the clinical course of dengue.
Subject terms: Genetics, Viral infection, Immunology, Immunogenetics, Immunopathogenesis, Infection
Introduction
Dengue is a mosquito-borne viral infection caused by dengue virus (DENV) with an estimated 390 million dengue infections worldwide every year and over 2.5 billion people at risk of the infection1. Dengue cases in South-East Asian countries are exponentially increasing with an annual average of 386,000 cases between 2001 and 20102. Vietnam is a country with high dengue incidence and hyperendemic transmission of DENV. The outbreaks are generally larger and more common in southern provinces of Vietnam, with incidence peaking between June and October3,4.
Four distinct serotypes of the virus cause dengue (DENV-1, DENV-2, DENV-3 and DENV-4)5. DENVs circulate in Vietnam with a predominance of the DENV-1 and DENV-2 serotypes and DENV-2 appear to cause more severe conditions than the other serotypes6. The clinical manifestation of an acute DENV infection varies from mild febrile illness lasting approximately 4–7 days to life-threatening severe dengue haemorrhagic fever and dengue shock syndrome (DHF/DSS), characterized by plasma leakage, fluid accumulation, respiratory distress, severe bleeding, or organ impairment. The complex interplay of human innate and adaptive cellular immune responses with the virus is believed to influence the clinical course of DENV infection7. Recovery from infection provides lifelong immunity against the serotype which has caused the disease episode. However, cross-immunity to the other serotypes after recovery is only partial and short-term. Subsequent infections by other serotypes increase the risk of developing dengue haemorrhagic fever and such secondary infections are mediated by antibody-dependent immune enhancement (ADE)7,8.
Complement proteins, innate immune recognition molecules are activated either by classical, alternative or the lectin complement pathways during the early phase of infection9. The lectin complement pathway is triggered when the lectin proteins, namely the mannose-binding lectin (MBL) and/or the ficolins (FCN) bind to the pathogen recognition receptors on the surface of pathogens. Subsequently, a sequential enzyme cascade is triggered where MBL-associated protein 2 (MASP2) and other complement proteins (C5b-9) are activated leading to the formation of the membrane attack complex which eventually ends with opsonization, lysis, activation of inflammatory responses and/or clearance of immune complexes9. Human MBL encoded by MBL2 gene is located on chromosome 10 and three single nucleotide polymorphisms (SNPs) in exon1 at codons 52 (MBL2*D), 54 (MBL2*B), and 57 (MBL2*C) respectively, interfere with the formation of higher MBL oligomers, causing alterations in functional activity of the protein and their circulating levels10–12. In addition, three promoter polymorphisms (positions, − 550, − 221 and + 4) were shown to regulate plasma MBL levels12. The MBL2*B, MBL2*C or MBL2*D mutant alleles are referred to as MBL2*O, whereas the wildtype is addressed as MBL2*A. MBL deficiency and MBL2 polymorphisms have been shown to be associated with several infectious and autoimmune diseases13,14.
MBL was shown to neutralize DENV via complement-dependent and independent mechanisms, suggesting a significant role of MBL in the pathogenesis of dengue15. A previous study has indicated direct complement restriction of DENV infection via MBL recognition of the high-mannose glycans of DENV envelope proteins16. In addition, MBL2 polymorphisms have been shown to be associated with dengue severity in Brazilian and Thai populations17–19. The above studies have investigated only the contribution of MBL2 polymorphisms and MBL serum levels. The present study utilizing a Vietnamese study group has investigated not only MBL serum levels and MBL2 variants but has also looked at associations of several downstream complement proteins of the complement pathway, namely C2, C4b, C5, C5a, C9, factor D and factor I with DENV infection.
Materials and methods
Study participants
A total of 279 unrelated Vietnamese patients (aged 34 ± 13; male/female, 130/149) with fever and symptoms of DENV infection admitted to the 103 Military Hospital of the Vietnam Military Medical University, Hanoi, collected between 2016 and 2018 were included. Patients were diagnosed based on the diagnostic criteria for dengue according to the World Health Organization (WHO)20. Patients were confirmed positive for NS1 antigen or/and positive for anti-DENV IgG and IgM as well as the clinical presentation of dengue. The patients were classified into two different groups according to the 2009 WHO dengue classification20. The first group included patients with dengue without warning signs (n = 172) and the second group comprised of dengue patients with warning signs (DWS, n = 107). DWS is characterized with fever, positive tourniquet test, petechiae or ecchymosis, maculopapular rash, myalgia/arthralgia, lymphadenopathy, leukopenia, thrombocytopenia and gastrointestinal bleeding with supportive serology of IgG and IgM titers and positive for NS1 antigen. Patients with dengue are usually characterized by a rapid and weak pulse (narrow pulse pressure < 20 mm Hg) with fever or a history of acute fever lasting 2–7 days (occasionally biphasic), haemorrhagic tendencies, evidenced by at least one of the following: a positive tourniquet test, petechiae, ecchymosis or purpura, bleeding from the mucosa, gastrointestinal tract, haematemesis or melaena20. Exclusion criteria were children below 12 and adults over 82 years, and patients who had any underlying chronic diseases and blood disorders. The demographic and clinical data of the study individuals are summarized in Table 1. Blood samples were collected on the day of admission and were used for measuring the levels of complement proteins. Serum samples were separated and stored at − 70 °C until further use. The control group included 200 healthy blood donors (aged 19.6 ± 1.2; male/female, 122/78) recruited in the same hospital and belonging to the Kinh ethnicity.
Table 1.
Characteristics of study subjects.
| Characteristics | Dengue (n = 172) | DWS (n = 107) | P value |
|---|---|---|---|
| Age (years) | 31 (14–81) | 31 (13–68) | NS |
| Gender | NS | ||
| Male n (%) | 82 (47.7) | 48 (44.9) | |
| Female n (%) | 90 (52.3) | 59 (55.1) | |
| DENV NS1 | NS | ||
| Positive n (%) | 153 (89.0) | 93 (86.9) | |
| Negative n (%) | 16 (9.3) | 10 (9.3) | |
| Undetermined n (%) | 3 (1.7) | 4 (3.7) | |
| DENV specific IgM | NS | ||
| Positive n (%) | 29 (16.9) | 27 (25.2) | |
| Negative n (%) | 138 (80.2) | 75 (70.1) | |
| Undetermined n (%) | 5 (2.9) | 5 (4.7) | |
| DENV specific IgG | 0.024 | ||
| Positive n (%) | 30 (17.4) | 31 (29) | |
| Negative n (%) | 137 (79.7) | 71 (66.4) | |
| Undetermined n (%) | 5 (2.9) | 5 (4.7) | |
| DENV serotype | |||
| DENV-1 n (%) | 45 (26.2) | 20 (18.7) | Reference |
| DENV-2 n (%) | 8 (4.6) | 13 (12.1) | 0.019 |
| DENV-1 and -2 n (%) | 1 (0.6) | 1 (0.9) | NS |
| Undetermined n (%) | 118 (68.6) | 69 (64.5) | |
| DENV Viremia (1/Ct × 100) | 3 (2–6)a | 3.1 (2–5)b | NS |
| Day of illness at hospital admission | 5 (1–13) | 5 (1–12) | NS |
| Day of illness | 9 (4–20) | 10 (6–16) | 0.001 |
| Bleeding | < 0.001 | ||
| Yes n (%) | 138 (80.2) | 46 (43) | |
| No n (%) | 34 (19.8) | 61 (57) | |
| Liver function test | |||
| AST (IU/ml) | 61.3 (18–369.2) | 108.3 (20.5–2,322) | < 0.001 |
| ALT (IU/ml) | 39 (8–301) | 73.8 (8.6–992.3) | < 0.001 |
| Blood count test | |||
| RBC (× 103/ml) | 4.6 (2.8–6.81) | 4.75 (2.88–7.17) | 0.048 |
| WBC (× 106/ml) | 3.39 (1–14.6) | 3.38 (1.19–17.29) | NS |
| PLT (× 103/ml) | 86.5 (3.2–316) | 60 (8–1,100) | < 0.001 |
| HCT (%) | 40 (27–53) | 40.7 (24–52) | NS |
Data presented as median (min–max).
DENV dengue virus; DWS dengue with warning signs; NS1 non-structural antigen 1; AST, ALT aspartate and alanine amino transferase; WBC white blood cells; RBC red blood cells; PLT platelets; HCT haematocrit; IU international unit; NS not significant; NA not applicable.
aDone in 65 samples.
bDone in 35 samples.
MBL2 genotyping
Genomic DNA was extracted from blood samples using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instruction. MBL2 genotyping was performed as described previously21. Briefly, the MBL2 promoter polymorphisms at − 550G>C, − 221G>C and 5′UTR + 4C>T and exon1 polymorphisms at codons 52C>T, 54G>A and 57G>A were amplified by PCR using primer pairs: PromF: 5′-GCC AGA AAG TAG AGA GGT ATT TAG CAC-3′, and Exon1R: 5′-CCA ACA CGT ACC TGG TTC CC-3′. The internal primer Exon1F: 5′-CAG GTG TCT AGG CAC AGA TGA ACC-3′ was used for Sanger sequencing of PCR products. The PCR assays were run in a 20 µl volume of reaction mixture containing 1× PCR buffer, 0.125 mM of dNTPs, 0.25 mM of each primer, 1U Taq DNA polymerase and 10 ng of genomic DNA. Thermal cycling conditions were: initial denaturation at 94 °C for 5 min, followed by 35 cycles of 30 s at 94 °C for denaturation, 30 s at 68 °C for annealing, 1 min 30 s at 72 °C for extension, followed by a final extension step of 2 min at 72 °C. PCR products were cleaned with Exo-SAP-IT and 1 µl of the purified product was used as templates for sequencing of the PCR products using the BigDye terminator v.1.1 (Applied Biosystems, Waltham, MA, USA) on an ABI 3130XL DNA sequencer. Polymorphisms were identified by assembling the sequences with reference sequences of the MBL2 gene (NG_008196.1) using the Bioedit 7.2.6 software (https://www.mbio.ncsu.edu/BioEdit/bioedit.html) and were reconfirmed visually from their respective electropherograms.
Quantification of plasma MBL and complement proteins
Levels of MBL and of the complement proteins C2, C4b, C5, C5a, C9, adipsin (factor D) and factor I were measured in serum samples from patients and healthy individuals using the MILLIPLEX MAP human Complement Magnetic Bead Panel 1 (Millipore Corporation, Billerica, MA, USA; Cat. Number: HCMP1MAG-19K), according to the manufacturer’s instructions. Briefly, after washing the plate with washing buffer, 25 μl of each diluted standard, control and diluted plasma samples (1:100) were added to the wells and 25 μl of assay buffer were added to the sample wells. 25 μl of the mixed beads were added to each well; the plate was covered with a sealer and wrapped with foil and then incubated with agitation on a plate shaker overnight at 2–8 °C. The well contents were removed and the plates were washed three times. Subsequently, 50 μl of detection antibodies were added to each well. Plates were sealed and incubated under agitation for 1 h at ambient temperature. 50 μl of streptavidin–phycoerythrin were added to each well and the sealed plates were incubated again on a shaker for 30 min at room temperature. Next, the well contents were removed and plates were washed three times. 150 μl of sheath fluid were added to all wells and the beads were resuspended on a plate shaker for 5 min. Plates were subjected to a Luminex 200 system and median fluorescent intensity (MFI) data were analyzed using a 5-parameter logistic method for calculating protein concentrations in the samples.
Statistical analysis
Quantitative variables were compared using Kruskal–Wallis or Mann–Whitney U tests and were given as medians with range. Categorical variables were compared using Chi-square or Fisher’s exact tests and given as percentages. Genotype and allele frequencies were analyzed by simple gene counting. MBL2 haplotypes were analysed using the expectation-maximum (EM) algorithm implemented in the Arlequin v. 3.5.1.2 software (https://cmpg.unibe.ch/software/arlequin3512/) and deviations from Hardy–Weinberg (HW) equilibrium were tested. Linkage disequilibrium (LD) analysis was performed using the Haploview v. 3.2 program (https://www.broadinstitute.org/haploview/haploview). Binary logistic regression model adjusted for confounding factors (age and gender) was used to analyze the association of MBL2 polymorphisms (genotypes, alleles, haplotypes and diplotypes) with dengue fever and disease severity. Odds ratios (ORs) and their 95% confidence intervals (CIs) were calculated. Kruskal–Wallis test or Mann–Whitney U test was used to analyze the association of serum MBL levels with MBL2 gene variants. All statistical analyses were performed with Stata v.14 (StataCorp, College Station, Texas, USA) and SPSS software version 22.0 (SPSS Statistics, IBM, Armonk, NY, USA) and the level of significance was set to a P value of < 0.05.
Ethics statement
Patients and healthy individuals were informed in detail about the study and written consent was given by all participants or by a parent of patients if the patients were under 18 years old. All clinical procedures and experiments were performed in accordance with international and national guidelines. The study was approved by the Institutional Review Board of the Vietnam Military Medical University, Hanoi, Vietnam (103MCH/RES/DENV-GER_V-D1-2016).
Results
Baseline characteristics of study groups
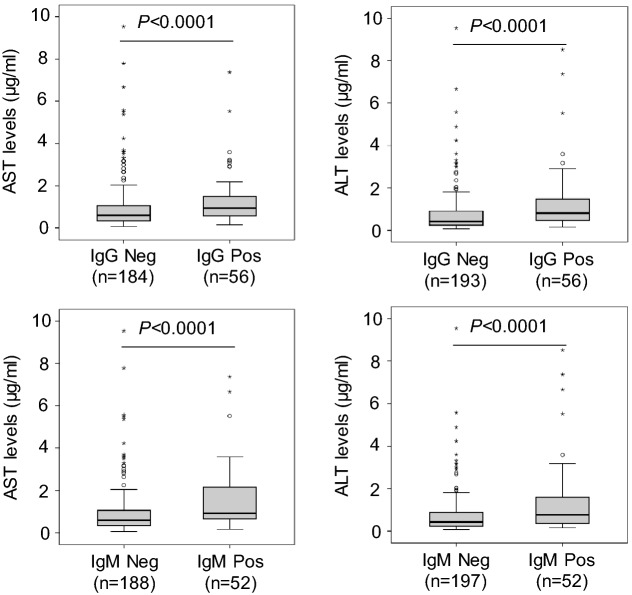
Demographic and clinical characteristics of dengue patients are summarized in Table 1. There was no difference of mean age, gender distribution, positivity rate of DENV NS1 and DENV-specific IgM positivity between the dengue and DWS groups. The DENV-specific IgG positivity rate was significantly higher in the DWS compared to the dengue group (P = 0.024). The DENV-1 serotype was predominant in the study cohort and DENV-2 was observed more frequently in DWS patients compared to dengue patients (P = 0.019) indicating the association of DENV-2 with more severity. No significant difference in DENV viremia was observed. Days of illness at hospital admission were not different between dengue and DWS patients (P > 0.05). Nevertheless, days of illness (from onset of symptoms to hospital discharge) in DWS patients were longer compared to dengue patients (P = 0.001). The levels of AST and ALT were significantly increased in patients with DWS compared to dengue patients (P < 0.001), indicating that DWS is associated with increased liver injuries. Although the HCT and WBC and WBC counts did not differ between the dengue and DWS groups, the platelet counts were significantly lower in patients with DWS compared to those with dengue (P < 0.001) (Table 1). We compared AST and ALT levels between patient subgroups positive and negative for DENV-specific IgG and IgM antibodies. AST and ALT levels were significantly increased in patients positive for DENV-specific IgG and IgM compared to those negative for DENV-specific IgG and IgM (P < 0.0001) (Fig. 1). These results indicate that DENV infection is associated with liver injury.
Figure 1.
Liver enzyme levels in patients positive and negative with DENV-specific antibodies. Pos positive, Neg negative. P values were calculated by Mann–Whitney U test.
Association of MBL2 polymorphisms with dengue
The genotype frequencies of MBL2 SNPs in the control group were in Hardy–Weinberg equilibrium (P > 0.05). The distribution of MBL2 genotype and allele frequencies is shown in Table 2. SNPs in the promoter region were in strong linkage disequilibrium both among patients and controls. The frequency of the promoter allele − 550H was significantly higher in controls compared to all dengue patients (dengue and DWS), suggesting a relative protective effect (OR = 0.6, 95%CI = 0.37–0.95, P = 0.03). The frequency of the minor allele MBL2*A at codon 54 was significantly higher in all dengue patients (dengue and DWS) compared to controls (dengue plus DWS: OR = 2.1, 95% CI 1.2–3.8, P = 0.012; dengue: OR = 1.99, 95% CI 1.0–4.1, P = 0.05; DWS: OR = 2.2, 95% CI 1.05–4.8, P = 0.037), indicating that MBL2*A at codon 54 may confer an increased risk of dengue infection. Similarly, the frequencies of allele MBL2*O at exon1 (codons 52 + 54 + 57) were also higher among all patients (dengue plus DWS) compared to healthy controls (dengue plus DWS: OR = 1.97, 95% CI 1.1–3.5, P = 0.021; DWS: OR = 2.1, 95% CI 1.0–4.5, P = 0.05), indicating an increased risk of dengue (Table 2). No significant differences were observed for other MBL2 variants (− 221, + 4, codons 52 and 57) in comparisons of dengue patients with controls. We observed the variant at codon 52 with a low allele frequency (1.4%) in the control group but not in the patient group. The variant at codon 57 was not observed in our study population. Furthermore, there was no significant difference in genotype and allele frequencies of MBL2 variants in the comparison of dengue with DWS patients.
Table 2.
Genotype and allele distribution of MBL2 polymorphisms in dengue patients and healthy controls.
| MBL2 polymorphisms | All patients n (%) | Dengue n (%) | DWS n (%) | Healthy controls n (%) | All patients vs. healthy controls | Dengue vs. healthy controls | DWS vs. healthy controls | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |||||
| rs11003125 (− 550C/G) | ||||||||||
| CC (LL) | 77 (33.0) | 53 (37.3) | 24 (26.4) | 40 (23.5) | Reference | Reference | Reference | |||
| GC (HL) | 95 (40.8) | 52 (36.6) | 43 (47.3) | 76 (44.7) | NS | NS | NS | |||
| GG (HH) | 61 (26.2) | 37 (26.1) | 24 (26.4) | 54 (31.8) | NS | NS | NS | |||
| C (L) | 249 (53.4) | 158 (55.6) | 91 (50) | 156 (45.9) | Reference | Reference | Reference | |||
| G (H) | 217 (46.6) | 126 (45.4) | 91 (50) | 184 (54.1) | 0.6 (0.37–0.95) | 0.03 | NS | NS | ||
| rs7096206 (− 221 G/C,A) | ||||||||||
| CC (XX) | 152 (56.5) | 97 (59.1) | 55 (52.4) | 99 (55.6) | Reference | Reference | Reference | |||
| CG(A) (XY) | 94 (34.9) | 53 (32.3) | 41 (39.0) | 66 (37.1) | NS | NS | NS | |||
| GG (YY) | 23 (8.6) | 14 (8.5) | 9 (8.6) | 13 (7.3) | NS | NS | NS | |||
| C (X) | 398 (74) | 247 (75.3) | 151 (71.9) | 264 (74.9) | Reference | Reference | Reference | |||
| G (Y) | 140 (26) | 81 (24.7) | 59 (28.1) | 92 (25.1) | NS | NS | NS | |||
| rs7095891 (+ 4G/A) | ||||||||||
| GG (PP) | 233 (84.1) | 143 (83.6) | 90 (84.9) | 169 (86.2) | Reference | Reference | Reference | |||
| GA (PQ) | 42 (15.1) | 28 (16.4) | 14 (13.1) | 25 (12.8) | NS | NS | NS | |||
| AA (QQ) | 2 (0.7) | 0 (0) | 2 (1.9) | 2 (1.0) | NS | NS | NS | |||
| G (P) | 508 (91.7) | 314 (91.8) | 194 (91.5) | 363 (92.6) | Reference | Reference | Reference | |||
| A (Q) | 46 (8.3) | 28 (8.2) | 18 (8.5) | 29 (7.4) | NS | NS | NS | |||
| MBL2*Exon1 (codon 54) G>A | ||||||||||
| GG | 161 (59.7) | 96 (58.2) | 64 (62.1) | 140 (78.2) | Reference | Reference | Reference | |||
| GA | 103 (38.8) | 67 (40.6) | 37 (35.9) | 38 (21.2) | 2.6 (1.4–5.1) | 0.004 | 2.4 (1.1–5.5) | 0.034 | 2.9 (1.2–6.9) | 0.018 |
| AA | 4 (1.5) | 2 (1.2) | 2 (1.9) | 1 (0.6) | NS | NS | NS | |||
| G | 425 (79.3) | 260 (78.8) | 165 (80.1) | 318 (88.8) | Reference | Reference | Reference | |||
| A | 111 (20.7) | 70 (21.2) | 41 (19.9) | 40 (11.2) | 2.1 (1.2–3.8) | 0.012 | 1.99 (1–4.1) | 0.05 | 2.2 (1.05–4.8) | 0.037 |
| MBL2*Exon1 codons 52 + 54 + 57 | ||||||||||
| AA | 161 (59.7) | 96 (58.2) | 64 (62.1) | 137 (76.5) | Reference | Reference | Reference | |||
| AO | 103 (38.8) | 67 (40.6) | 37 (35.9) | 41 (22.9) | 2.4 (1.3–4.7) | 0.008 | 2.2 (1.0 − 5.1) | 0.05 | 2.7 (1.1–6.4) | 0.027 |
| OO | 4 (1.5) | 2 (1.2) | 2 (1.9) | 1 (0.6) | NS | NS | NS | |||
| A | 425 (79.3) | 260 (78.8) | 165 (80.1) | 315 (88) | Reference | Reference | Reference | |||
| O | 111 (20.7) | 70 (21.2) | 41 (19.9) | 43 (12) | 1.97 (1.1–3.5) | 0.021 | NS | 2.1 (1–4.5) | 0.05 | |
Bold values indicate statistical significance.
DWS dengue with warning signs, NS not significant.
We reconstructed MBL2 haplotypes based on the − 221Y/X promoter variant and three structural exon1 SNPs (MBL2*A/O), which mainly regulate MBL serum levels. The frequency of the diplotype XA/XO was significantly higher in patients compared to healthy controls (dengue plus DWS: OR = 3.3, 95% CI 1.4–8.1, P = 0.007; dengue: OR = 3.1, 95% CI 1.0–9.3, P = 0.044; DWS: OR = 3.7, 95% CI 1.1–12.1, P = 0.029), suggesting an increased risk of dengue fever. In addition, the frequency of haplotype XO was significantly higher in patients compared to healthy controls (dengue plus DWS: OR = 2.2, 95% CI 1.2–4.2, P = 0.015; DWS: OR = 2.7, 95% CI 1.2–5.9, P = 0.015). There was no significant difference in the comparison of dengue with DWS patients (Table 3).
Table 3.
MBL2 diplotypes in dengue patients and healthy controls.
| MBL2 (− 221) + Exon1 | All patients | Dengue | DWS | Healthy controls | All patients vs. healthy controls | Dengue vs. healthy controls | DWS vs. healthy controls | |||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Diplotypes | n = 260 | n = 158 | n = 102 | n = 161 | ||||||
| XA/XA | 82 (31.5) | 52 (32.9) | 30 (29.4) | 68 (42.2) | Reference | Reference | Reference | |||
| XA/XO | 59 (22.7) | 38 (24.1) | 21 (20.6) | 21 (13.0) | 3.3 (1.4–8.1) | 0.007 | 3.1 (1.0–9.3) | 0.044 | 3.7 (1.1–12.1) | 0.029 |
| XA/YA | 58 (22.4) | 33 (20.9) | 25 (24.5) | 46 (28.6) | NS | NS | NS | |||
| YA/XO | 35 (13.5) | 20 (12.7) | 15 (14.7) | 13 (8.1) | NS | NS | 3.8 (1–14.9) | 0.05 | ||
| YA/YA | 19 (7.3) | 11 (7.0) | 8 (7.8) | 10 (6.2) | NS | NS | NS | |||
| YA/YO | 3 (1.2) | 2 (1.3) | 1 (1.0) | 2 (1.2) | NS | NS | NA | |||
| XO/XO | 4 (1.5) | 2 (1.3) | 2 (2.0) | 1 (0.6) | NS | NS | NS | |||
| Haplotype | n = 520 | n = 316 | n = 204 | n = 322 | ||||||
| XA | 281 (54.0) | 175 (55.4) | 106 (52.0) | 203 (63.0) | Reference | Reference | Reference | |||
| YA | 134 (25.8) | 77 (24.4) | 57 (27.9) | 81 (25.2) | NS | NS | NS | |||
| XO | 102 (19.6) | 62 (19.6) | 40 (19.6) | 36 (11.2) | 2.2 (1.2–4.2) | 0.015 | NS | 2.7 (1.2–5.9) | 0.015 | |
| YO | 3 (0.6) | 2 (0.6) | 1 (0.5) | 2 (0.6) | NS | NS | NS | |||
Bold values indicate statistical significance.
DWS dengue with warning signs, NS not significant, NA not applicable.
We reconstructed MBL2 haplotypes based on three promoter SNPs and three exon 1 SNPs. The frequency of the haplotype LXPB was significantly higher in patients compared to controls (dengue plus DWS: OR = 4.1, 95% CI 1.2–14.3, P = 0.027; DWS: OR = 5.6, 95% CI 1.2–26, P = 0.028), suggesting an increased risk of dengue fever. The HXPA haplotype occurred less frequently among patients compared with controls (dengue plus DWS: OR = 0.3, 95% CI 0.1–0.9, P = 0.025; DWS: OR = 0.2, 95% CI 0.03–0.98, P = 0.047), suggesting a protective role in DENV infection. However, no significant difference in haplotype frequencies was observed between dengue and DWS patients (Table 4).
Table 4.
MBL2 haplotypes in dengue patients and healthy controls.
| MBL2 haplotypes | All patients | Dengue | DWS | Healthy controls | All patients vs. healthy controls | Dengue vs. healthy controls | DWS vs. healthy controls | |||
|---|---|---|---|---|---|---|---|---|---|---|
| n = 452 (%) | n = 274 (%) | n = 176 (%) | n = 276 (%) | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| LXPA | 201 (44.7) | 127 (46.4) | 74 (42.0) | 120 (43.5) | Reference | Reference | Reference | |||
| HYPA | 116 (25.8) | 67 (24.5) | 49 (27.8) | 70 (25.4) | NS | NS | NS | |||
| HXPB | 62 (13.8) | 37 (13.5) | 25 (14.2) | 23 (8.3) | NS | NS | NS | |||
| LXPB | 29 (6.4) | 19 (6.9) | 10 (5.7) | 6 (2.2) | 4.1 (1.2–14.3) | 0.027 | NS | 5.6 (1.2–26) | 0.028 | |
| HXPA | 24 (5.3) | 16 (5.8) | 8 (4.5) | 39 (14.1) | 0.3 (0.1–0.9) | 0.025 | NS | 0.2 (0.03–0.98) | 0.047 | |
| HXQA | 5 (1.1) | 1 (0.4) | 4 (2.3) | 6 (2.2) | NS | NS | NS | |||
| LXQA | 6 (1.3) | 3 (1.1) | 3 (1.7) | 4 (1.4) | NS | NS | NS | |||
| LYPA | 4 (0.9) | 2 (0.7) | 2 (1.1) | 3 (1.1) | NS | NS | NS | |||
| HYPB | 3 (0.7) | 2 (0.7) | 1 (0.6) | 0 (0.0) | NA | NA | NA | |||
| LYPB | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.7) | NA | NA | NA | |||
| HXPD | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.7) | NA | NA | NA | |||
| LXPD | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.4) | NA | NA | NA | |||
Bold values indicate statistical significance.
DWS dengue with warning signs, NS not significant, NA not applicable.
Serum levels of MBL and complement proteins in patients with Dengue
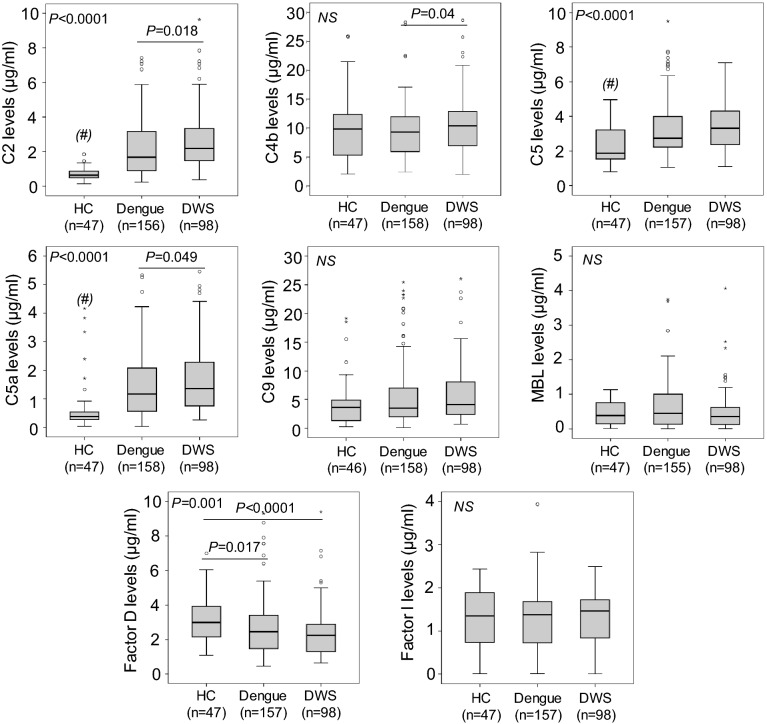
We measured the levels of MBL and the complement proteins C2, C4b, C5, C5a, C9, adipsin (factor D) and factor I in serum samples of dengue and DWS patients and healthy controls. The levels of C2, C5, and C5a were significantly higher in dengue and DWS patients compared to those in healthy controls (P < 0.0001) and also higher in DWS patients compared to those in dengue patients (P = 0.018, < 0.0001 and 0.049, respectively). The levels of factor D were decreased in dengue and DWS patients compared to those in controls (P = 0.017 and < 0.0001, respectively). No difference of the levels of C4b, C9, MBL and factor I was observed among dengue and DWS patients and healthy controls (P > 0.05) (Fig. 2). These results indicate that the levels of C2, C5, C5a and factor D are modulated during DENV infection and progression of dengue.
Figure 2.
Complement protein levels in dengue patients and healthy individuals. HC healthy control, DWS dengue patients with warning signs. P values were calculated by Kruskal–Wallis test or Mann–Whitney U test, where appropriate.
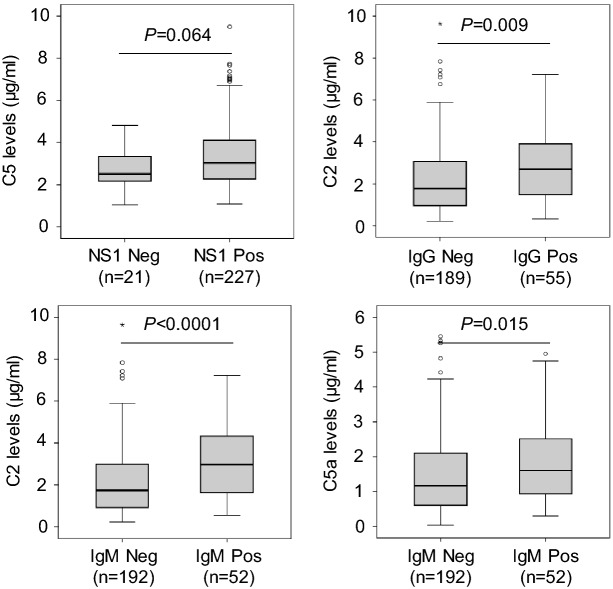
We then segregated the patients into subgroups based on positivity for NS1 and DENV-specific IgG and IgM antibodies and compared the levels of C2, C4b, C5, C5a, C9, factor D and factor I between these subgroups. C5 levels were marginally increased in patients positive compared to those negative for NS1 (P = 0.064). C2 levels were increased in IgG positive patients compared to those negative for IgG antibodies (P = 0.0009). The levels of C2 and C5a were significantly increased in IgM positive compared to patients negative for IgM (P < 0.0001 and P = 0.015, respectively) (Fig. 3). These results indicate that the levels of C2, C5 and C5a might be associated with DENV infection. In addition, we classified the patients into subgroups based on the infection with DENV-1 or DENV-2 and compared the levels of C2, C4b, C5, C5a, C9, factor D and factor I between these classified groups. However, no statistical significance was observed.
Figure 3.
Complement protein levels in patients positive and negative with DENV-specific antibodies. NS1 non-structural antigen 1, Pos positive, Neg negative. P values were calculated by Mann–Whitney U test.
Association of MBL2 polymorphisms with MBL serum levels
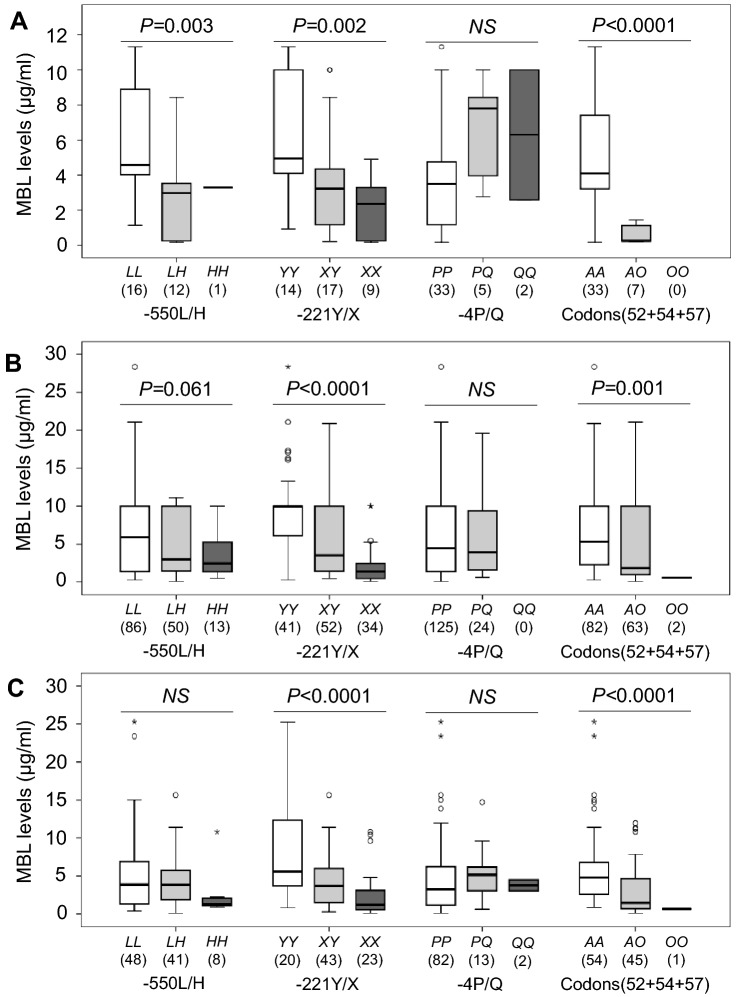
The association of MBL2 polymorphisms with MBL levels has been established in different world population. Therefore, we analyzed the distribution of MBL serum levels across different MBL2 genotypes and haplotypes. Alleles MBL2*L and MBL2*Y at the promoter region and allele MBL2*A at codons 52 + 54 + 57 significantly contributed to higher MBL levels, while alleles MBL2*H MBL2*X and MBL2*O contributed to lower MBL levels in healthy individuals (P < 0.05). MBL2 − 4P/Q polymorphism was not significantly associated with MBL levels in healthy individuals (Fig. 4A). In dengue and DWS patients, allele MBL2*Y at the promoter region and allele MBL2*A at codons 52 + 54 + 57 significantly contributed to higher MBL levels, whereas alleles MBL2*X and MBL2*O contributed to lower MBL levels (P < 0.05). However, significant associations of MBL2 − 550L/H and MBL2 − 4P/Q polymorphisms with MBL levels in dengue and DWS patients were not observed (P < 0.05) (Fig. 4B,C).
Figure 4.
Distribution of MBL levels in individuals with different MBL2 genotypes. (A), healthy individuals; (B), dengue patients; (C), dengue patients with warning signs (DWS). P values were calculated by Kruskal–Wallis test or Mann–Whitney U test, where appropriate.
Correlation of serum levels of complement proteins with clinical parameters
The correlation between a pair of complement protein levels as well as between complement protein levels and clinical parameters in dengue patients was analyzed. In healthy individuals, C2 levels were significantly associated with C4b, C5, C5a and C9 levels (Spearman’s rho = 0.49, 0.7, 0.39, and 0.3; P = 0.0005, < 0.0001, 0.006, and 0.046, respectively). C4b levels were correlated with the levels of C5, C5a, C9 and factor I (Spearman’s rho = 0.64, 0.37, 0.42 and 0.29; P < 0.0001, 0.011, 0.004, and 0.049, respectively). C5 levels were associated with C5a and factor I levels (Spearman’s rho = 0.52 and 0.35; P = 0.0002 and 0.016, respectively) (Table 5).
Table 5.
Correlation of complement proteins in healthy individuals.
| C2 (µg/ml) | C4b (µg/ml) | C5 (µg/ml) | C5a (µg/ml) | C9 (µg/ml) | Factor D (µg/ml) | MBL (µg/ml) | Factor I (µg/ml) | |
|---|---|---|---|---|---|---|---|---|
| C2 (µg/ml) | ||||||||
| rs | 0.49 | 0.7 | 0.39 | 0.3 | 0.14 | − 0.17 | 0.13 | |
| P | 0.0005 | < 0.0001 | 0.006 | 0.046 | 0.35 | 0.26 | 0.37 | |
| C4b (µg/ml) | ||||||||
| rs | 0.49 | 0.64 | 0.37 | 0.42 | 0.013 | − 0.002 | 0.29 | |
| P | 0.0005 | < 0.0001 | 0.011 | 0.004 | 0.93 | 0.99 | 0.049 | |
| C5 (µg/ml) | ||||||||
| rs | 0.7 | 0.64 | 0.52 | 0.27 | 0.09 | − 0.12 | 0.35 | |
| P | < 0.0001 | < 0.0001 | 0.0002 | 0.07 | 0.56 | 0.41 | 0.016 | |
| C5a (µg/ml) | ||||||||
| rs | 0.39 | 0.37 | 0.52 | 0.15 | 0.079 | 0.09 | 0.2 | |
| P | 0.006 | 0.011 | 0.0002 | 0.31 | 0.6 | 0.53 | 0.18 | |
| C9 (µg/ml) | ||||||||
| rs | 0.3 | 0.42 | 0.27 | 0.15 | − 0.18 | 0.004 | 0.15 | |
| P | 0.046 | 0.004 | 0.07 | 0.31 | 0.24 | 0.98 | 0.31 | |
| Factor D (µg/ml) | ||||||||
| rs | 0.14 | 0.013 | 0.09 | 0.079 | − 0.18 | 0.15 | 0.011 | |
| P | 0.35 | 0.93 | 0.56 | 0.6 | 0.24 | 0.32 | 0.94 | |
| MBL (µg/ml) | ||||||||
| rs | − 0.17 | − 0.002 | − 0.12 | 0.09 | 0.004 | 0.15 | − 0.15 | |
| P | 0.26 | 0.99 | 0.41 | 0.53 | 0.98 | 0.32 | 0.32 | |
| Factor I (µg/ml) | ||||||||
| rs | 0.13 | 0.29 | 0.35 | 0.2 | 0.15 | 0.011 | − 0.15 | |
| P | 0.37 | 0.049 | 0.016 | 0.18 | 0.31 | 0.94 | 0.32 | |
Spearman’s rho (rs) calculated by Spearman's rank correlation coefficient. Bold rs and P values indicate a significant and strong correlation between variables.
In dengue patients, C2 levels correlated with the levels of C4b, C5, C5a and factor D (Spearman’s rho = 0.5, 0.39, 0.61, − 0.25; P < 0.0001, < 0.0001, < 0.0001 and = 0.0002, respectively). C4b levels correlated with C5 and C5a levels (Spearman’s rho = 0.52, 0.38; P < 0.0001, respectively). Factor D levels were negatively associated with C5a levels (Spearman’s rho = − 0.34; P < 0.0001), while they positively correlated with C9 levels (Spearman’s rho = 0.21; P = 0.01). Regarding the correlation between complement protein levels and clinical parameters, C4b levels were significantly associated with platelet counts (Spearman’s rho = 0.25; P = 0.002), and factor D levels were positively correlated with RBC counts and HCT values (Spearman’s rho = 0.31, 0.32; P < 0.0001, respectively), while negatively correlated with WBC and platelet counts (Spearman’s rho = − 0.24, − 0.31; P = 0.002 and < 0.0001, respectively) (Table 6, upper-right area).
Table 6.
Correlation of complement proteins with clinical parameters in dengue patients.
| C2 (µg/ml) | C4b (µg/ml) | C5 (µg/ml) | C5a (µg/ml) | C9 (µg/ml) | Factor D (µg/ml) | MBL (µg/ml) | Factor I (µg/ml) | AST (IU/ml) | ALT (IU/ml) | RBC (× 103/ml) | WBC (× 106/ml) | PLT (× 103/ml) | HCT (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C2 (µg/ml) | ||||||||||||||
| rs | 0.5 | 0.39 | 0.61 | 0.052 | − 0.25 | 0.004 | 0.02 | 0.14 | 0.13 | − 0.09 | 0.16 | 0.07 | − 0.12 | |
| P | < 0.0001 | < 0.0001 | < 0.0001 | 0.51 | 0.002 | 0.96 | 0.79 | 0.09 | 0.11 | 0.27 | 0.04 | 0.38 | 0.14 | |
| C4b (µg/ml) | ||||||||||||||
| rs | 0.35 | 0.52 | 0.38 | 0.08 | − 0.15 | − 0.09 | 0.19 | − 0.1 | − 0.04 | − 0.15 | 0.06 | 0.25 | − 0.12 | |
| P | < 0.0001 | < 0.0001 | < 0.0001 | 0.29 | 0.06 | 0.23 | 0.014 | 0.22 | 0.66 | 0.053 | 0.45 | 0.002 | 0.15 | |
| C5 (µg/ml) | ||||||||||||||
| rs | 0.17 | 0.37 | 0.37 | 0.32 | 0.04 | − 0.11 | 0.12 | 0.03 | 0.003 | 0.03 | 0.09 | 0.03 | 0.03 | |
| P | 0.08 | < 0.0001 | < 0.0001 | < 0.0001 | 0.58 | 0.16 | 0.45 | 0.69 | 0.97 | 0.72 | 0.23 | 0.76 | 0.76 | |
| C5a (µg/ml) | ||||||||||||||
| rs | 0.62 | 0.32 | 0.34 | − 0.17 | − 0.34 | 0.07 | − 0.06 | 0.16 | 0.1 | 0.02 | 0.06 | − 0.03 | 0.01 | |
| P | < 0.0001 | 0.001 | 0.001 | 0.03 | < 0.0001 | 0.39 | 0.46 | 0.64 | 0.23 | 0.81 | 0.43 | 0.67 | 0.9 | |
| C9 (µg/ml) | ||||||||||||||
| rs | − 0.24 | − 0.14 | 0.18 | − 0.31 | 0.21 | − 0.05 | 0.12 | − 0.044 | − 0.001 | 0.015 | − 0.02 | 0.09 | 0.005 | |
| P | 0.02 | 0.18 | 0.08 | 0.002 | 0.01 | 0.51 | 0.12 | 0.6 | 0.99 | 0.86 | 0.79 | 0.26 | 0.95 | |
| Factor D (µg/ml) | ||||||||||||||
| rs | − 0.5 | − 0.23 | 0.09 | − 0.53 | 0.49 | − 0.17 | 0.145 | 0.09 | 0.06 | 0.31 | − 0.24 | − 0.31 | 0.32 | |
| P | < 0.0001 | 0.03 | 0.39 | < 0.0001 | < 0.0001 | 0.034 | 0.69 | 0.34 | 0.61 | < 0.0001 | 0.002 | < 0.0001 | < 0.0001 | |
| MBL (µg/ml) | ||||||||||||||
| rs | 0.18 | 0.06 | 0.11 | 0.16 | − 0.17 | − 0.12 | − 0.04 | − 0.01 | 0.01 | − 0.08 | − 0.05 | 0.11 | − 0.02 | |
| P | 0.07 | 0.54 | 0.29 | 0.11 | 0.09 | 0.23 | 0.59 | 0.9 | 0.94 | 0.31 | 0.58 | 0.17 | 0.71 | |
| Factor I (µg/ml) | ||||||||||||||
| rs | 0.12 | 0.29 | 0.24 | − 0.05 | 0.09 | 0.25 | 0.06 | − 0.13 | − 0.15 | − 0.08 | − 0.005 | 0.15 | − 0.1 | |
| P | 0.23 | 0.004 | 0.02 | 0.62 | 0.38 | 0.01 | 0.53 | 0.14 | 0.06 | 0.34 | 0.95 | 0.065 | 0.24 | |
| AST (IU/ml) | ||||||||||||||
| rs | 0.27 | 0.07 | − 0.02 | 0.22 | − 0.18 | − 0.25 | − 0.12 | 0.02 | 0.87 | 0.28 | − 0.03 | − 0.46 | 0.23 | |
| P | 0.012 | 0.54 | 0.88 | 0.04 | 0.09 | 0.02 | 0.29 | 0.8 | < 0.0001 | < 0.0001 | 0.75 | < 0.0001 | 0.005 | |
| ALT (IU/ml) | ||||||||||||||
| rs | 0.32 | 0.14 | − 0.001 | 0.3 | − 0.27 | − 0.25 | − 0.03 | 0.04 | 0.91 | 0.27 | 0.09 | − 0.34 | 0.21 | |
| P | 0.002 | 0.2 | 0.99 | 0.005 | 0.01 | 0.015 | 0.8 | 0.7 | < 0.0001 | 0.001 | 0.27 | < 0.0001 | 0.008 | |
| RBC (× 103/ml) | ||||||||||||||
| rs | 0.11 | − 0.22 | 0.03 | 0.03 | 0.04 | 0.18 | 0.02 | 0.05 | 0.001 | 0.07 | 0.006 | − 0.3 | 0.87 | |
| P | 0.28 | 0.03 | 0.79 | 0.8 | 0.7 | 0.08 | 0.85 | 0.66 | 0.99 | 0.5 | 0.94 | < 0.0001 | < 0.0001 | |
| WBC (× 106/ml) | ||||||||||||||
| rs | 0.24 | 0.04 | 0.02 | 0.25 | − 0.23 | − 0.16 | 0.18 | 0.25 | 0.26 | 0.36 | 0.25 | 0.26 | 0.04 | |
| P | 0.02 | 0.7 | 0.86 | 0.012 | 0.02 | 0.11 | 0.07 | 0.014 | 0.013 | < 0.0001 | 0.009 | < 0.0001 | 0.61 | |
| PLT (× 103/ml) | ||||||||||||||
| rs | − 0.08 | 0.14 | 0.11 | 0.05 | − 0.05 | − 0.22 | − 0.07 | − 0.05 | − 0.1 | − 0.007 | − 0.26 | − 0.09 | − 0.28 | |
| P | 0.43 | 0.14 | 0.29 | 0.6 | 0.6 | 0.026 | 0.5 | 0.63 | 0.32 | 0.94 | 0.006 | 0.36 | < 0.0001 | |
| HCT (%) | ||||||||||||||
| rs | 0.004 | − 0.24 | 0.03 | − 0.07 | − 0.04 | 0.18 | − 0.02 | 0.05 | 0.04 | 0.08 | 0.86 | 0.26 | − 0.32 | |
| P | 0.97 | 0.17 | 0.78 | 0.68 | 0.68 | 0.067 | 0.87 | 0.6 | 0.7 | 0.4 | < 0.0001 | 0.006 | 0.001 | |
Spearman’s rho (rs) calculated by Spearman's rank correlation coefficient. rs and P values in the upper-right area are the correlation between variables in dengue patients. rs and P values in the lower-left area are the correlation between variables in dengue patients with warning signs (DWS).
Bold rs and P values indicate a significant and strong correlation between variables.
AST and ALT aspartate and alanine amino transferase, WBC white blood cells, RBC red blood cells, PLT platelets, HCT haematocrit, IU international unit.
In DWS patients, the correlation patterns differed from those observed in dengue patients. In particular, C2 levels were significantly associated with C4b, C5a, C9 and factor D levels. C4b levels were significantly correlated with the levels of C5, C5a, factor D, and factor I. C5 levels significantly correlated with C5a and factor I levels and factor D levels were associated with the C9 and factor I levels. With regard to correlations of complement proteins with laboratory parameters, C2 and C5a levels significantly correlated with AST and ALT levels, and with WBC counts. C9 levels were negatively correlated with ALT levels and WBC counts. Factor D levels were negatively correlated with AST and ALT levels and platelet counts. A negative correlation between C4b levels and RBC counts and a positive correlation between factor I levels and WBC counts was also observed (Table 6, lower-left area).
Discussion
Dengue is a serious health problem in tropical and subtropical areas including Vietnam. Studies have shown that the pathogenesis and severity of the disease are closely associated with distinct host immune responses against DENV, including components of the complement system22,23. Here we show that MBL2 polymorphisms and the levels of the complement proteins C2, C4b, C5, C5a, C9, factor D and factor I are associated with DENV infection and severity of the disease. The results indicate that complement proteins are modulated during DENV infection and play an important role in the innate immune response against DENV infection and in the progression to severe dengue.
The association of MBL2 polymorphisms with infectious diseases has previously been documented13,14. In particular, the MBL2*OO genotype, the MBL2*O allele and MBL2 haplotypes, which correlate with low MBL levels have been shown to be associated with DHF in a Brazilian study group18. A recent study has also shown an association of MBL2 haplotypes with an increased risk of dengue severity in Brazilians19. However, no association between MBL2 polymorphisms and DENV infection was observed in a Thai study group24. A genome-wide association study has identified SNP rs3132468 in the MHC class I polypeptide-related sequence B (MICB) and SNP rs3765524 in the phospholipase C epsilon 1 gene (PLCE1) to be associated with DENV infection and severe dengue in a Vietnamese population25. To the best of our knowledge, our study is the first report showing an association of MBL2 polymorphisms in the promoter and exon 1 regions including MBL2 genotypes (− 550L/H, MBL2 codon 54), MBL2 diplotypes (XA/XO, YA/XO) and MBL2 haplotypes (LXPB, HXPA, XO) with DENV infection and dengue in Vietnam.
In DENV infection, the complement system is consistently activated26. However, the frequencies of MBL2 variants, which are associated with MBL levels, are not significantly different when comparing dengue patients and controls. In addition, the MBL2 OO genotype and the MBL2 O allele, which are involved both in regulation of MBL levels and the recognition of pathogens, differ between dengue patients and controls. The DENV envelope consists of E and M proteins with high-mannose glycans that are recognized and opsonized by MBL16. Although sufficient MBL levels and functions are required to neutralize DENV via complement-mediated mechanisms15, our results show that serum MBL levels do not differ significantly between healthy individuals, dengue and DWS patients. These results suggest that MBL2 variants predominantly contribute to complement activation by recognizing DENV antigens, rather than regulate MBL serum levels.
The complement proteins C2, C4b, C5, C5a, and factor D are significantly modulated during clinical course of DENV infection, suggesting that activation of the complement system through alternative pathways might be more dominant in response to DENV infection and, thus, might contribute to dengue severity. This is supported by a previous study indicating the effect of alternative complement pathway deregulation on dengue severity via imbalance of factor D and factor H levels in DHF patients27. In addition, overactivity of the alternative complement pathway in the cellular microenvironment plays a crucial role in the pathogenesis of dengue, involving increased vascular permeability and plasma leakage28. Notably, the expression of the key regulators of the alternative complement pathway including factor H and factor B is induced by DENV infection in DENV-targeted cells28. Factor B is known as an activator of the alternative pathway and factor D is a serine protease that cleaves factor B and, thus, contributes to activation of the alternative pathway29. Deficiency of factor D has been shown to be associated with increased susceptibility to dengue infection30; factor D levels were decreased in dengue compared to healthy controls27. Similarly, the decrease of factor D levels, but not factor I, in dengue and DWS patients compared to healthy controls as well as their correlation with clinical parameters in both dengue and DWS suggests that lower levels of factor D are associated with an increased risk of DENV infection and disease severity.
Although in the present study C4b levels did not differ significantly between dengue patients and healthy individuals, higher C4b levels in DWS patients compared to dengue patients suggest a functional role of C4b in disease progression. The NS1 protein of flaviviruses has been shown to both interact with C4 to promote degradation of C4 to C4b and to bind to the C4b binding protein (C4BP) to inactivate C4b, and these interactions lead to reduced activation of the classical and the lectin pathway31,32. C2, C5, and C5a levels were significantly increased in dengue patients compared to controls and are highest in DWS patients, indicating that the complement system is strongly activated in order to respond to DENV infection. A previous study has shown that C5a does not only interact with DENV antigens in human hosts but also reduces DENV infection in mosquito cells33. Several host biomarkers including C5a and factor D levels have been suggested to support the progression of dengue34. Our data also indicate that the levels of C2, C4b, C5, C5a, C9, factor D and factor I are significantly associated with clinical parameters, especially with platelet counts, of dengue patients. Therefore, C2, C4b, C5, C5a, and factor D levels may be considered markers for progression of DENV infection and severity of the disease.
Although this study provides interesting results of the functional role of complement proteins, there are several limitations. The complement proteins were not determined at different time points of the clinical course and the activity levels of the complement system during progression of dengue were not assessed continuously. Other complement proteins such as C1, C3 and ficolins were not determined.
In conclusion, our study shows that MBL2 polymorphisms are associated with dengue in a Vietnamese study group and that C2, C4b, C5, C5a and factor D levels are significantly modulated in dengue patients and during the progression of dengue. Our data point to the crucial role of the alternative complement pathway in response to DENV infection and disease severity.
Acknowledgements
We thank all patients and healthy individuals for their participation. This study was funded by the Vietnam National Foundation for Science and Technology Development (NAFOSTED), Grant number 01/2018/ĐX. The funder has no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions
N.T.G. performed the experiments, carried out the statistical analyses and contributed to writing of the manuscript. H.V.T. designed, supervised the study, contributed materials and reagents, carried out the statistical analyses, interpreted data and wrote the manuscript. D.Q. supervised the study, recruited patients, contributed materials and reagents. N.X.H. contributed to performing the experiments. T.H.N., N.M.N., H.V.H., D.T.A., C.V.M., H.A.S. recruited patients, collected samples and clinical data. C.G.M. provided critical comments and revised the manuscript. T.P.V. contributed to study design and revised the manuscript. N.L.T. designed, supervised the study, contributed materials and reagents, revised the manuscript. All authors agreed with the results and conclusions.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ngo Truong Giang, Hoang van Tong and Do Quyet.
Contributor Information
Hoang van Tong, Email: hoangvantong@vmmu.edu.vn.
Nguyen Linh Toan, Email: toannl@vmmu.edu.vn.
References
- 1.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shepard DS, Undurraga EA, Halasa YA. Economic and disease burden of dengue in Southeast Asia. PLoS Negl. Trop. Dis. 2013;7:e2055. doi: 10.1371/journal.pntd.0002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung TM, et al. The estimates of the health and economic burden of dengue in Vietnam. Trends Parasitol. 2018;34:904–918. doi: 10.1016/j.pt.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wartel TA, et al. Three decades of dengue surveillance in five highly endemic South East Asian Countries. Asia Pac. J. Public Health. 2017;29:7–16. doi: 10.1177/1010539516675701. [DOI] [PubMed] [Google Scholar]
- 5.Hasan S, Jamdar SF, Alalowi M, Beaiji SMA. Dengue virus: A global human threat: Review of literature. J. Int. Soc. Prev. Community Dent. 2016;6:1–6. doi: 10.4103/2231-0762.175416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leitmeyer KC, et al. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 1999;73:4738–4747. doi: 10.1128/JVI.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St John AL, Rathore APS. Adaptive immune responses to primary and secondary dengue virus infections. Nat. Rev. Immunol. 2019;19:218–230. doi: 10.1038/s41577-019-0123-x. [DOI] [PubMed] [Google Scholar]
- 8.Martina BE, Koraka P, Osterhaus AD. Dengue virus pathogenesis: An integrated view. Clin. Microbiol. Rev. 2009;22:564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarma JV, Ward PA. The complement system. Cell Tissue Res. 2011;343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swierzko AS, et al. The role of complement activating collectins and associated serine proteases in patients with hematological malignancies, receiving high-dose chemotherapy, and autologous hematopoietic stem cell transplantations (auto-HSCT) Front. Immunol. 2018;9:2153. doi: 10.3389/fimmu.2018.02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garred P, Larsen F, Seyfarth J, Fujita R, Madsen HO. Mannose-binding lectin and its genetic variants. Genes Immun. 2006;7:85–94. doi: 10.1038/sj.gene.6364283. [DOI] [PubMed] [Google Scholar]
- 12.Worthley DL, Bardy PG, Mullighan CG. Mannose-binding lectin: Biology and clinical implications. Intern Med. J. 2005;35:548–555. doi: 10.1111/j.1445-5994.2005.00908.x. [DOI] [PubMed] [Google Scholar]
- 13.Eisen DP, Minchinton RM. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin. Infect Dis. 2003;37:1496–1505. doi: 10.1086/379324. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Ruiz M, et al. Impact of MBL2 gene polymorphisms on the risk of infection in solid organ transplant recipients: A systematic review and meta-analysis. Am. J. Transpl. 2019;19:1072–1085. doi: 10.1111/ajt.15160. [DOI] [PubMed] [Google Scholar]
- 15.Avirutnan P, et al. Complement-mediated neutralization of dengue virus requires mannose-binding lectin. MBio. 2011 doi: 10.1128/mBio.00276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs A, et al. Direct complement restriction of flavivirus infection requires glycan recognition by mannose-binding lectin. Cell Host Microbe. 2010;8:186–195. doi: 10.1016/j.chom.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acioli-Santos B, et al. MBL2 gene polymorphisms protect against development of thrombocytopenia associated with severe dengue phenotype. Hum. Immunol. 2008;69:122–128. doi: 10.1016/j.humimm.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Figueiredo GG, et al. Mannose-binding lectin gene (MBL2) polymorphisms related to the mannose-binding lectin low levels are associated to dengue disease severity. Hum. Immunol. 2016;77:571–575. doi: 10.1016/j.humimm.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Ornelas AMM, et al. Association between MBL2 haplotypes and dengue severity in children from Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. 2019;114:e190004. doi: 10.1590/0074-02760190004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. Dengue guidelines for diagnosis, treatment, prevention and control: new edition. (World Health Organization, 2009). https://apps.who.int/iris/handle/10665/44188. [PubMed]
- 21.Antony JS, et al. Mannose-binding lectin and susceptibility to schistosomiasis. J. Infect. Dis. 2013;207:1675–1683. doi: 10.1093/infdis/jit081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uno N, Ross TM. Dengue virus and the host innate immune response. Emerg. Microbes Infect. 2018;7:167. doi: 10.1038/s41426-018-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giang NT, et al. Association of FCN2 polymorphisms and Ficolin-2 levels with dengue fever in Vietnamese patients. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.02.029. [DOI] [PubMed] [Google Scholar]
- 24.Prommalikit O, Thisyakorn U. Association between Mannose-Binding lectin gene polymorphisms and susceptibility to dengue virus infection: A preliminary report. Southeast Asian J. Trop. Med. Public Health. 2015;46(Suppl 1):29–34. [PubMed] [Google Scholar]
- 25.Khor CC, et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat. Genet. 2011;43:1139–1141. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conde JN, Silva EM, Barbosa AS, Mohana-Borges R. The complement system in flavivirus infections. Front. Microbiol. 2017;8:213. doi: 10.3389/fmicb.2017.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nascimento EJ, et al. Alternative complement pathway deregulation is correlated with dengue severity. PLoS ONE. 2009;4:e6782. doi: 10.1371/journal.pone.0006782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabezas S, et al. Dengue virus induces increased activity of the complement alternative pathway in infected cells. J. Virol. 2018 doi: 10.1128/JVI.00633-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le GT, Abbenante G, Fairlie DP. Profiling the enzymatic properties and inhibition of human complement factor B. J. Biol. Chem. 2007;282:34809–34816. doi: 10.1074/jbc.M705646200. [DOI] [PubMed] [Google Scholar]
- 30.Sprong T, et al. Deficient alternative complement pathway activation due to factor D deficiency by 2 novel mutations in the complement factor D gene in a family with meningococcal infections. Blood. 2006;107:4865–4870. doi: 10.1182/blood-2005-07-2820. [DOI] [PubMed] [Google Scholar]
- 31.Avirutnan P, et al. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J. Exp. Med. 2010;207:793–806. doi: 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avirutnan P, et al. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J. Immunol. 2011;187:424–433. doi: 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Londono-Renteria B, Grippin C, Cardenas JC, Troupin A, Colpitts TM. Human C5a protein participates in the mosquito immune response against dengue virus. J. Med. Entomol. 2016;53:505–512. doi: 10.1093/jme/tjw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conroy AL, et al. Host biomarkers are associated with progression to dengue haemorrhagic fever: A nested case-control study. Int. J. Infect. Dis. 2015;40:45–53. doi: 10.1016/j.ijid.2015.07.027. [DOI] [PubMed] [Google Scholar]