Abstract
BACKGROUND AND PURPOSE:
Mutations in isocitrate dehydrogenase (IDH) have a direct effect on gliomagenesis. The purpose of this study is to quantify differences in brain metabolites due to IDH mutations.
METHODS:
Magnetic Resonance Spectroscopic Imaging (MRSI) was performed in 35 patients with gliomas of different grade and varied IDH mutation status. Volumes of interest (VOIs) for active tumor (tVOI), peritumoral area (pVOI), and contralateral normal-appearing white matter (cVOI) were created. Metabolite ratios of Choline (Cho) to both N-acetylaspartate (NAA) and Creatine (Cr) were estimated. Ratios of Glutamate/Glutamine complex (Glx) and myoinositol (mIno) to Cr were also quantified. General linear models (GLMs) were used to estimate the effects of IDH mutation on metabolite measures, with age, gender, and tumor grade used as covariates.
RESULTS:
GLM analysis showed that maximum Cho/NAA and Cho/Cr in the tVOI were significantly (P < .05) higher in IDH mutant lesions as compared to wild-type. In the pVOI, mean Cho/Cr was found to be significantly different among IDH mutant and wild-type gliomas. Mean Cho/NAA (P = .306) and Cho/Cr (P = .292) within the tVOI were not significantly different. Ratios of Glx/Cr and mIno/Cr in any region showed no significant differences between IDH mutant and wild-type gliomas. No significant differences in metabolite ratios were seen in the cVOI between IDH mutants and wild-types.
CONCLUSION:
IDH mutation’s effect in gliomas show an increase in Cho in the tumor and perilesional regions as compared to wild-type lesions but do not show widespread changes across the brain.
Keywords: choline, glioma, isocitrate dehydrogenase (IDH), MRSI, mutation
Introduction
Management of glioma patients is becoming more personalized and based on molecular tumor characteristics in different patients. One such example is that patients with IDH mutant glioma are more sensitive to treatment-induced oxidative cellular damage compared with wild-type gliomas.1
Mutations in isocitrate dehydrogenase (IDH) are prevalent in approximately 80% of low-grade gliomas and secondary high-grade gliomas.2–4 Through its role in the citric acid cycle, IDH functions to maintain the cellular redox state and, hence, plays an important role in cellular defense against oxidative stress5 and is a direct driver of oncogenesis, causing increased proliferation, increased colony formation, and inability to differentiate.2,6 However, IDH mutation is not seen as a driver of tumor initiation in primary glioblastomas.7
IDH mutation in gliomagenesis changes the function of the enzymes, causing the production of 2-hydroxyglutarate (2-HG), a possible oncometabolite,8,9 that can be detected in vivo using magnetic resonance spectroscopy (MRS).10 Measurement of 2-HG using the commonly available 3T MRI systems is challenging, and benefits from optimized acquisition methods and large volume measurements to address the low detection sensitivity.10–13 These limitations have led to an interest in using more conventional MRS measurements to test whether the metabolic alterations may also be associated with changes in metabolites that are relatively easier to detect including choline-containing compounds (Cho).13–15 Previous studies indicated that elevated Cho levels in IDH mutant lesions indicate the increased cell density of these mutant malignancies, due to increased cell proliferation as a direct effect of IDH mutation.16 A few studies have also reported a decrease in intracellular lactate, glutamate, and phosphocholine in IDH mutant cells relative to wild-type.17,18 Glutamate and Glutamine changes, although reported, have not been robustly verified in IDH mutant gliomas due to difficulties in spectral analysis of these compounds as a result of significant overlap or low signal to noise at 3T.
Magnetic Resonance Spectroscopic Imaging (MRSI) is an effective technique for noninvasive quantification of brain metabolite distributions.19–21 MRSI has the distinct advantage of being able to map larger regions of the brain as compared to single-voxel spectroscopy (SVS), which quantifies metabolites in an operator-selected region that can be affected by selection bias.22 A majority of studies that have looked at the association of MRI-observable brain metabolites with IDH status have used SVS based techniques that limit the mapping of brain metabolites to a small region of the tumor and are impaired by the limited localization, thereby, including to some extent normal brain tissue within the SVS volume. To better map changes in brain metabolites in gliomas, this study aims to employ whole-brain volumetric spectroscopic imaging to map the gross tumor areas in IDH mutant and wild-type lesions. An additional goal of this study is to map neurometabolite changes in the peritumoral regions and the contralateral “normal-appearing” white matter in IDH mutant patients to ascertain if any global changes in brain metabolites are evident.
Observations obtained via MRSI may be applied to further our understanding of the underlying mechanisms of the pathology, due to the effect of IDH on cell membrane formation and proliferation. Thus, MRSI may play an important role in precision medicine strategies for glioma patients including early diagnosis, guiding treatment, and evaluating response to therapy.23
Methods
Study Population
Thirty-five subjects with diagnosed glioma (20 males and 15 females), mean age 50 ± 13 years, were recruited for the study. Seventeen subjects (48%) had undergone surgical intervention prior to imaging and testing for IDH status was obtained from tissue removed during surgery or biopsy. T-cell antigen receptor (TCR) sequencing (TCR-seq) was used to identify gene mutations in patients under 55 years of age, whereas immunohistochemistry assay based mutation-specific testing was used in older individuals. Only patients with a definitive IDH mutation status were included in the study, with 20 gliomas showing IDH1 mutations compared to 15 containing wild-type IDH. Glioma grade was histopathologically confirmed using the World Health Organization (WHO) scale and 9 (6 IDH mutant, 3 IDH wild-type), 12 (8 IDH mutant, 4 IDH wild-type) and 14 (6 IDH mutant, 8 IDH wild-type) patients were, respectively, classified as Grade II, III, and IV using histology.
Informed consent was acquired from each patient, and the protocol was approved by the Review Board for Protection of Human Subjects in Research.
Imaging Protocol
MRI and MRSI data were acquired at 3T (Siemens Medical Solutions, Erlangen, Germany). T1-weighted imaging (T1w) was carried out using a 3D Magnetization Prepared Rapid Acquisition Gradient Echo (MPRAGE) sequence with 1.0 mm isotropic or .9 × .9 × .7 mm3 resolution; TR/TE/TI = 2,300/2.41/930 milliseconds; flip angle, 9°; NEX, 1; image matrix, 256 × 256 or 320 × 216. Imaging also included Fluid-attenuated inversion recovery (FLAIR) (TR/TE = 9,000/106 milliseconds, resolution = .36 × .36 × 3 mm3, flip angle = 120°), and T2-weighted (TR/TE = 4,810/76 milliseconds, resolution = .45 × .45 × 3 mm3, flip angle = 160°) images. Whole-brain MRSI was acquired using echo-planar acquisition with spin-echo excitation;TR/TE = 1,551/17.6 milliseconds; nonselective lipid inversion-nulling with TI = 198 milliseconds; a field-of-view of 280 × 280 × 180 mm3; matrix size of 50 × 50 with 18 slices; echo train length of 1,000 points; bandwidth of 2,500 Hz; and a nominal voxel volume of .313 cc and acquisition time of 15 minutes. A water reference MRSI dataset was obtained using an acquisition interleaved with the metabolite signal acquisition.
MRSI Processing
MRSI data were processed using the MIDAS package (http://mrir.med.miami.edu).20,24 This included B0 and phase correction using the water reference data prior to any further processing in the frequency domain. The relative gray- and white-matter tissue and cerebrospinal fluid (CSF) contents in each SI voxel were estimated by downsampling the tissue segmentation maps, which were obtained using FSL/FAST algorithm,22 using the spatial response function of the MRSI acquisition. Additional processing included generating masks for brain and lipid regions, k-space extrapolation to reduce the contribution of extracranial lipid into the brain,25 linear registration between the T1-weighted MR and MRSI, and signal intensity normalization following the creation of individual metabolite maps. The spectral datasets were interpolated to 64 × 64 × 32 points and spatial smoothing was applied after B0 correction, resulting in an effective voxel volume of 1.55 mL.
Automated spectral analysis was carried out for N-acetylaspartate (NAA), creatine and phosphocreatine (Cr), choline, glycerophosphocholine, and phosphocholine (Cho), myoinositol (mIno), lactate and Glutamate and Glutamine complex (Glx).26 Additional maps were generated for the fitted spectral linewidth (LW) and the Cramer-Rao lower bounds (CRLB) of fitting for each metabolite.
Normalization of metabolite signal across subjects remains a challenging task due to variations in tissue T1, water content, and bias fields especially in the presence of lesions. In order to account for these variations between subjects, metabolite ratios are reported in the analysis.
Image Processing
Three volumes of interest (VOIs) were created for each patient, over which mean values of the detected metabolites were evaluated. These included the active tumor (tVOI), a surrounding perilesional region (pVOI), and a region of normal-appearing white matter on the contralateral side of the tumor (cVOI). These regions were initially created based on FLAIR images using a Gaussian mixture model to automatically threshold regions of hyperintensity. FLAIR images were segmented using a mixture of three Gaussian distributions and the component with the largest mean value selected as the initial tumor region. Non-tissue regions and CSF were excluded using CSF labeled segmentation maps derived from T1w using FSL FAST.27 Regions of interest were finally edited manually to limit the region of interest to the gross tumor region, which is inclusive of the solid tumor and any surrounding edema. The tVOI was identified as regions within the gross tumor region that had a Choline/N-Acetylaspartate ratio greater than 1.5 relative to the mean of the Cho/NAA value in contralateral normal-appearing white matter (Cho/NAANAWM > 1.5).28 A VOI for the peritumoral region (pVOI) was defined as a 1 cm wide band around the tVOI.29 Additionally, VOIs were filtered using spectral quality criteria to remove any voxels that have a LW> 13 Hz and a creatine CRLB for the spectral fitting of greater than 20%.
cVOI regions were delineated using a semiautomatic segmentation approach using T1w and FLAIR images.29 Steps included estimation of the midsagittal plane of the brain on T1w,30 detection of the contralateral brain hemisphere, and delineation of a large contiguous region of white matter (WM) in the contralateral hemisphere. The contralateral brain hemisphere was identified as the hemisphere of the brain with a smaller gray matter volume. Tissue classification on T1w images using FAST generally classifies tumor regions as gray matter (GM) rendering a higher WM volume on the contralateral side of the lesion. Finally, cVOI volume was selected as a contiguous WM region in the supraventricular region of the brain (top 6 cm of the brain). All cVOI regions were visually verified. cVOI regions were also filtered to remove voxels with LW> 13 Hz and a creatine CRLB of greater than 20%.
Figure 1 shows the T1 postcontrast image (A), FLAIR image (B), illustrative VOI of the tVOI (C), peritumoral region (D), contralateral VOI (E), and MRSI maps for Cho/NAA (F), NAA (G), creatine (H), and choline (I) in a 70-year-old female IDH wild-type subject. Figure 2 shows example of integrated spectra from the VOI of tVOI, pVOI, and cVOI for a 70-year-old female IDH wild-type subject with imaging data is shown in Figure 1 (left panel) and in a 53-year-old female IDH mutant subject (right panel).
Fig 1.
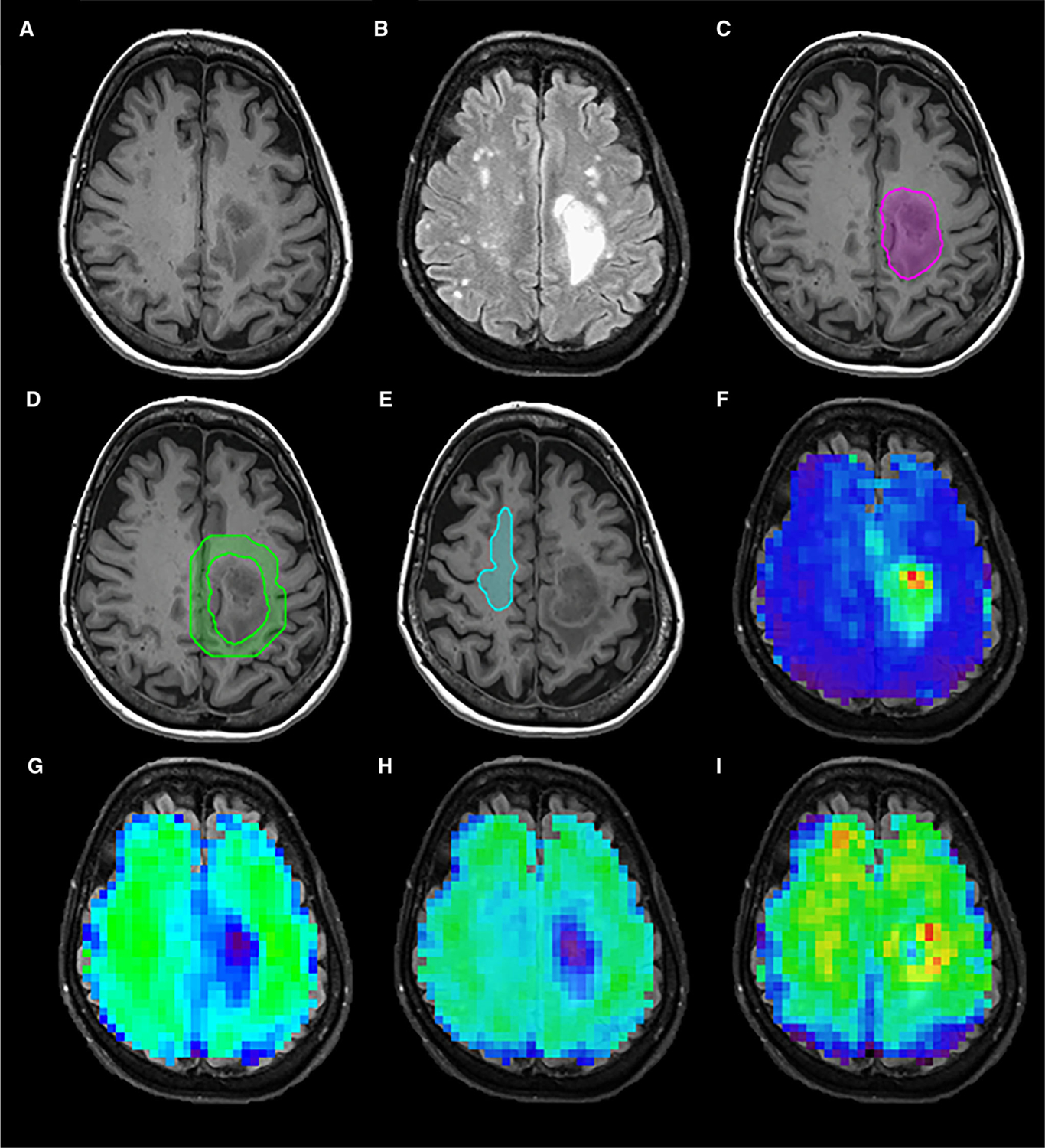
T1 postcontrast image (A), Fluid-attenuated inversion recovery image (B), volume of interests for active tumor (C), peritumoral region (D), contralateral normal-appearing white matter (E), and MRSI metabolite maps for Cho/NAA (F), NAA (G), Creatine (H), and Choline (I) in a 70-year-old female IDH wild-type subject.
Fig 2.
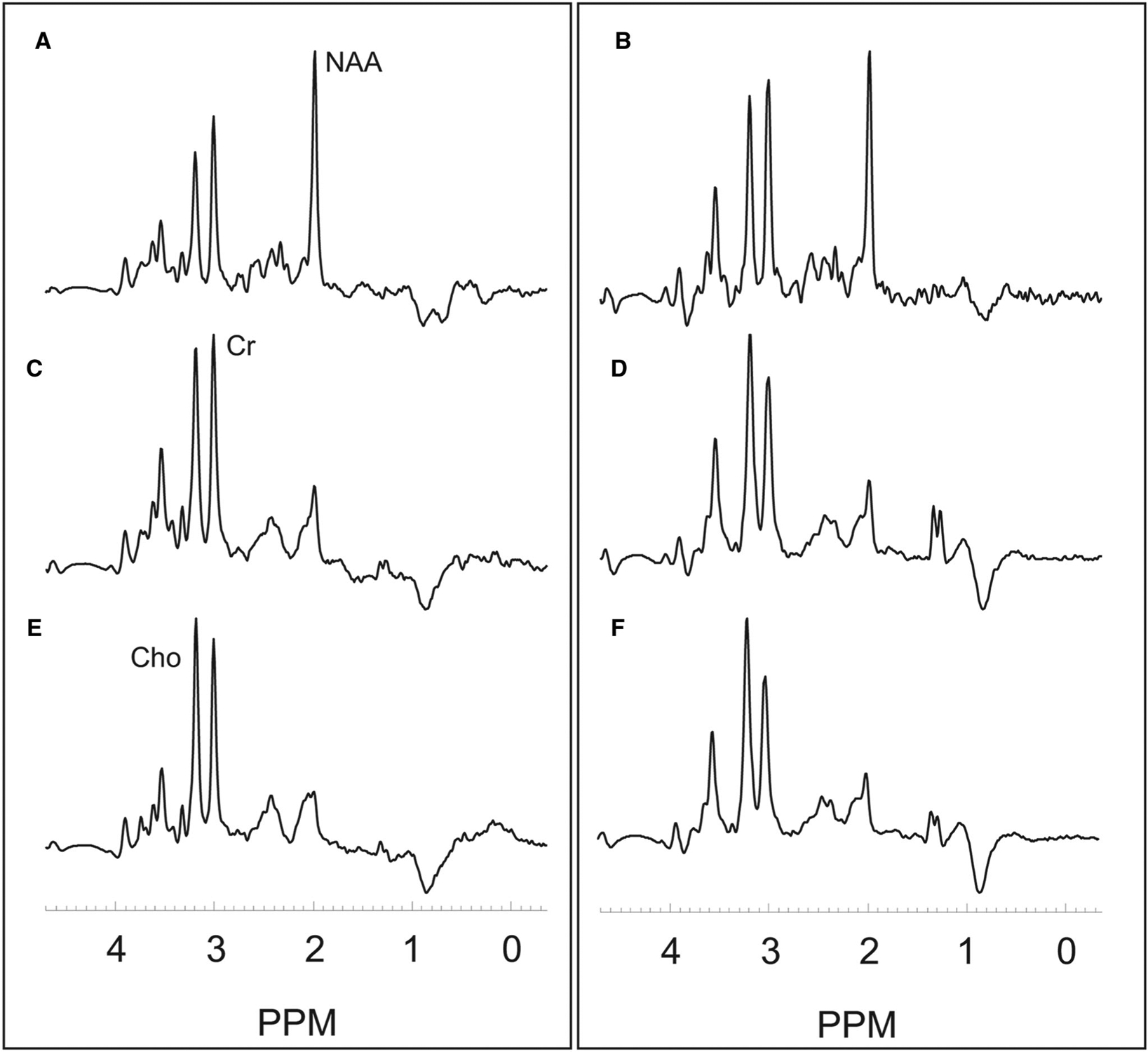
Example integrated spectra for the volume of interests for contralateral normal-appearing white matter (A,B), peritumoral region (C,D), and active tumor (E,F) in a 70-year-old female with an IDH wild-type Grade 4 glioma with imaging data shown in Figure 1 (left panel) and in a 53-year-old female with an IDH mutant Grade 3 oligodendroglioma (right panel). ppm denotes parts per millions with NAA at 2.0 ppm, Cho at 3.2 ppm, and Cr at 3.0 ppm.
Statistical Analysis
Cho is widely considered to be a marker of tumor proliferation, therefore, for each VOI the mean and maximum Cho/NAA and Cho/Cr ratios are calculated. Additionally, ratios of mean values for Glx/Cr and myo-inositol/Cr were also estimated for all the VOIs of interest.
General linear models (GLMs) were used to estimate the effects of IDH mutation on neurometabolite measures in the tumor (tVOI), peritumoral (pVOI), and contralateral (cVOI) WM of the patients, with age, gender, and tumor grade used as covariates in the GLM analyses. For each GLM model, the eta-squared (η2) was calculated to provide an estimate of the effect size for each metabolite ratio in differentiating between IDH positive and wild-type tumors.
Results
Figure 3 shows the ratios for maximum (Fig 3A) and mean (Fig 3B) Cho/NAA in the active tumor VOI (tVOI) separated by tumor grade. GLM analysis showed that maximum Cho/NAA in the tVOI was significantly higher in IDH positive lesions as compared to wild-type lesions (P = .03). Moreover, the grade of the tumors also significantly affected the maximal Cho/NAA in the tVOI. Mean Cho/NAA in the tVOI was found to be not significantly (P = .069) different between the different IDH groups. Maximum and mean Cho/NAA ratios show an effect size of 18.03 and 12.96%, respectively, in differentiating IDH positive and wild-type lesions.
Fig 3.
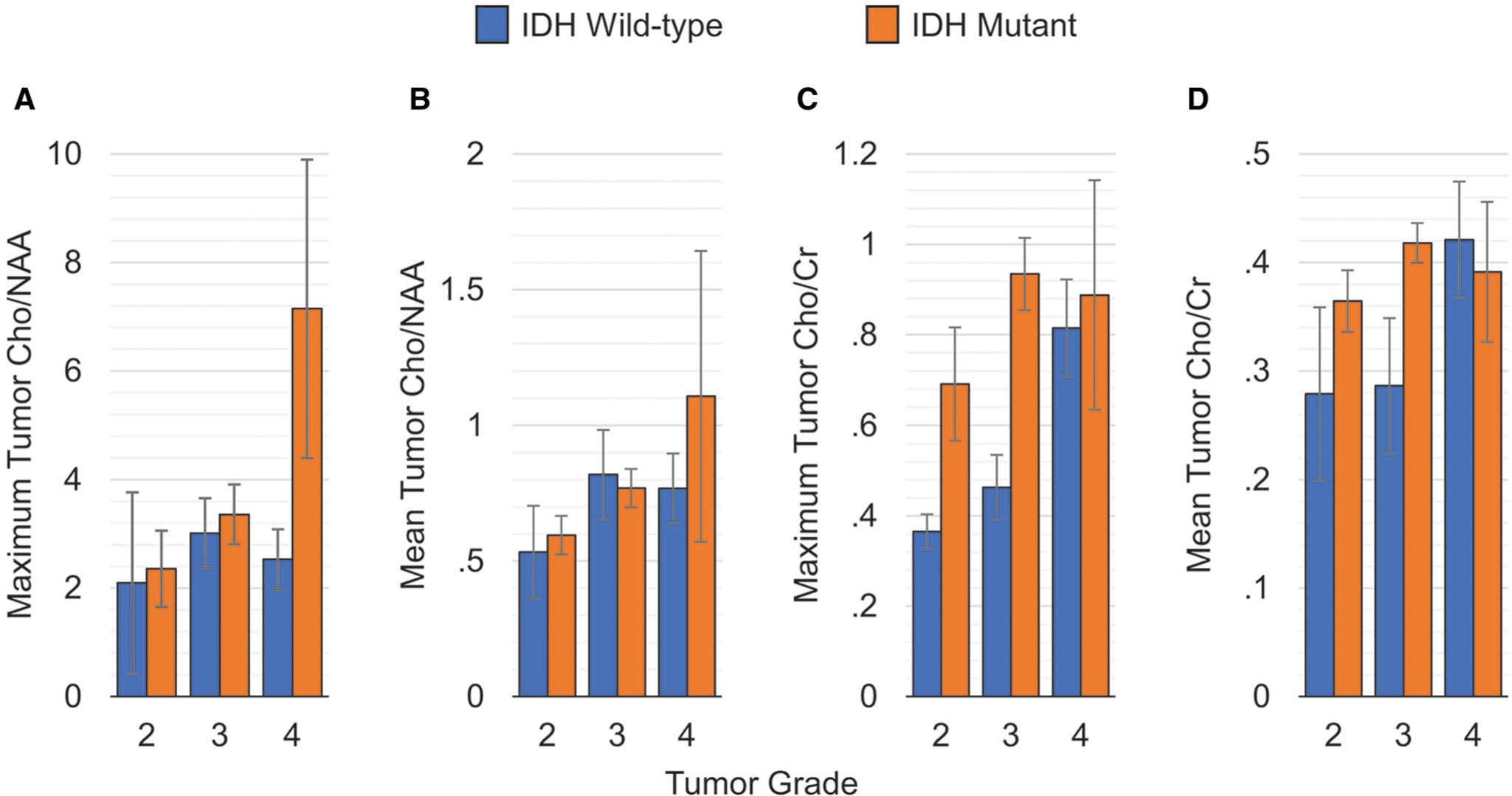
Cho/NAA and Cho/Cr measurements in the active tumor volume-of-interest differentiated by tumor grade.
In Figure 3 (C,D), the results for Cho/Cr are shown. GLM analysis showed that maximum Cho/Cr (Fig 3C) was significantly (P < .05) different between IDH positive and wild-type mutant gliomas, with tumor grade also significantly (P < .05) impacting the maximum Cho/Cr ratio in the tVOI. Similar to that seen for the mean Cho/NAA ratio, mean tumor Cho/Cr (Fig 3D) was not different between different IDH status (P = .292). Maximum and mean Cho/Cr ratios show an effect size of 16.31 and 3.58%, respectively, in differentiating IDH positive and wild-type lesions.
Glx/Cr (P = .943) and mIno/Cr (P = .239) were not found to be significantly different between IDH mutant and wild-type lesions in the tVOI. Further, Glx/Cr (P = .155) and mIno/Cr (P=.650) were also not found to be significantly different across tumor grades.
Figure 4 shows the maximum and mean Cho/NAA and Cho/Cr measures in the peritumoral areas of the tumor. Maximum Cho/NAA (Fig 4A) and Cho/Cr (Fig 4C) in the peritumoral areas were not significantly different in IDH mutants as compared to IDH wild-type lesions (P = .089 and P = .084, respectively). In contrast, mean Cho/Cr (Fig 4D) was found to be significantly (P = .021) different with an effect size (η2) of 15.4%. Similar to the active tumor region, no significant differences were found for Glx/Cr (P = .154) and mIno/Cr (P = .959), nor were these significantly different across tumor grades (P = .067 and .103, respectively).
Fig 4.
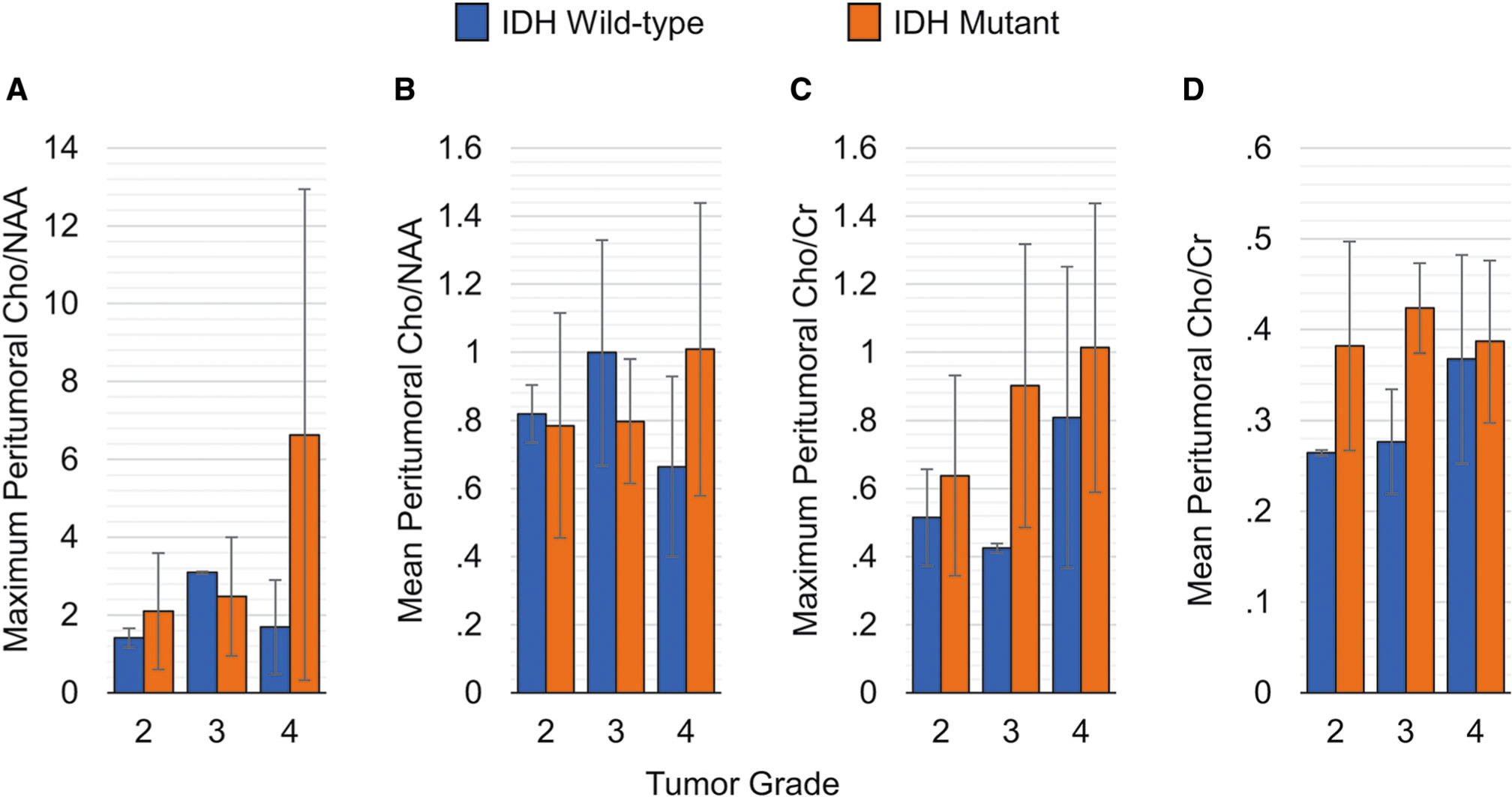
Cho/NAA and Cho/Cr measurements in the peritumoral volume-of-interest differentiated by tumor grade.
In the cVOI, neither metric (maximum or mean) of either Cho/NAA or Cho/Cr showed significant difference with IDH status or tumor grade. Moreover, Glx/Cr (P = .414) and mIno/Cr (P=.681) were also not significantly different between IDH mutants and wild-type individuals.
Discussion
This study evaluated the effects of mutation of the IDH gene on MRSI derived metabolite measures in patients with gliomas in VOI in the metabolically active tumor, surrounding white matter, and contralateral normal-appearing white matter. The aim of this study was to examine localized and widespread effects due to the mutation on metabolites that can be detected using widely available MRS measurement methods. Results found significant changes in choline measurements in the tumor and peritumoral regions indicating localized metabolic changes due to IDH mutation but did not show any differences in the contralateral normal-appearing white matter, indicating an absence of any global brain changes associated with the IDH status.
The study showed increased choline in the active tumor regions of IDH mutant gliomas as compared to IDH wild-type gliomas. Both Cho/Cr and Cho/NAA ratios were found to be elevated in the active tumor regions. Increased Cho has been reported in other studies using single-voxel PRESS MRS13 or metabolic profiling on sister clones of the human oligodendroglioma cell line in in-vitro experiments.14 This increase in choline can be attributed to reflect the increased cell density of mutant malignancies due to an IDH-mediated increase in cell proliferation.13,16 Moreover, studies have shown higher proliferative rates in IDH wild-type gliomas using Ki-67 expression and the significant increase in Cho in wild-type phenotypes seen in this study may be attributed to this reflect a higher density of viable cells that in wild-type tumors.31
An additional finding reported in this study is the increased Cho ratios in the peritumoral regions IDH mutant tumors as compared to wild-type phenotypes. This is a unique feature afforded due to the use of whole-brain spectroscopy which, unlike single-voxel spectroscopy, permits sampling of the surrounding tumor tissue with increased detail. The increased Cho levels seen in the peritumoral regions around the active tumor are illustrative of the diffuse nature of changes in brain metabolism due to IDH mutation.
In this study, the Cho/NAA ratio in the tumor was found to have a higher effect size in differentiating IDH mutant and wild-type lesions. The increased effect sizes of Cho/NAA measures can be attributed to increased Cho with a corresponding decrease of NAA that has been identified at a greater extent in IDH mutant gliomas.14 Comparing metabolite ratios of Cho in the active tumor regions, it was seen that the maximum of each ratio offered greater effect sizes than the mean of the ratio across the region. This can be attributed to an averaging effect that can occur across the large regions of interest. On the contrary, the maximum Cho ratios target the most active parts of the lesions and offer a better estimate of tumor proliferation. The larger effect sizes show that the maximum Cho/NAA may be more indicative of IDH status when accounting for tumor grade. It is to be noted, however, that the maximum value can be affected due to variations in spectral quality and errors in spectral fitting algorithms. To avoid these errors, spectra used in the quantification of metabolite ratios were filtered using strict quality criteria using linewidth for spectral quality and CRLB for spectral fitting estimates.
In the peritumoral regions, no significant differences of the Cho/NAA ratio were found between IDH mutant and wild-type gliomas; however, the Cho/Cr ratio was significantly different between the different IDH phenotypes. The discrepancy in the findings between Cho/NAA and Cho/Cr may indicate that the large NAA reduction, which is usually seen in active tumor regions but not in peritumoral regions with gliomas, may be the differentiating factor in the active tumor regions. Overall, the study shows that Cho changes due to IDH mutations are more widespread and not limited to the active tumor only.
MRSI and diffusion-weighted MR imaging have shown changes in metabolite concentrations and water diffusivity in normal-appearing brain tissue with an association with tumor grade.29,32 In this study, accounting for tumor grade, no significant differences were seen in the contralateral white matter regions between IDH mutant and wild-type gliomas. This finding suggests that although IDH driven changes are more widespread than only the active tumor, genetic mutation of the IDH gene has little systemic effect on observed metabolite concentrations throughout the brain.
Previous studies that examined associations of glutamate and glutamine with IDH mutation have shown mixed results. A significant decrease of glutamate in IDH mutant cells compared to wild-type has been reported using metabolic profiling14,18 and high-resolution MRS.15 Pope et al reported no changes in glutamate or glutamine in IDH mutant gliomas using a standard single-voxel double echo point-resolved spectroscopy (PRESS) sequence.13 In this study, the glutamine/glutamate complex (Glx) showed no significant differences between IDH mutant and wild-type gliomas. Although quality criteria were applied to filter spectra before any analysis, at 3T strongly overlapping multiplet resonances of glutamate and glutamine with signals from N-acetylaspartate (NAA) and γ-aminobutyric acid (GABA) may play a role in limiting the accuracy for quantification19 of 2-HG, which is elevated with IDH mutation, is also present as a multiplet that overlaps the glutamate, glutamine, and NAA region, such that improved detection requires the use of spectral editing11 or optimized pulse sequences.10 This study used a short TE acquisition at 17.6 milliseconds so as to optimize detection of glutamate and glutamine, limiting its utility in measuring 2-HG.19
A few studies have demonstrated that in a small percentage of subjects molecular biomarkers are changed over time, mainly due to the use of adjuvant treatments with radiotherapy, and IDH status can be altered.33,34 In this study, the mean time between the estimation of IDH status and MRSI imaging was 495 ± 1008 days. The large standard deviation in the duration is because 3 subjects out of the 35, all with low-grade tumors, had undergone genetic testing prior to being seen as a patient at our institution, and the genetic testing was not repeated for logistic reasons. Removing those 3 subjects from the estimation of the duration between genetic testing and MRSI imaging, it was found to be only 45.3 ± 10.5 days. Moreover, since genetic alterations are mainly seen in grade 3 IDH mutant tumors we do not expect the large variation in IDH status between the genetic testing and the imaging study.
Limitations of this study include that the measurements were based on a VOI definition for the active tumor region using a threshold of Cho/NAA ratio of 1.5 times that of the contralateral side. This may not include the whole tumor environment, thereby, possibly introducing a selection bias, especially in low-grade lesions. Furthermore, the imaging measures that report average values for a region of interest may not account for possible heterogeneity within the tumor. An additional limitation is the relatively small number of subjects, which meant that potential age-related influences on parameter estimation could not be specifically investigated; although to ameliorate age effects this study did use age as a covariate in GLM analysis. Moreover, due to the nature of the analysis it can be arqued that increased Cho seen in wild-type lesions is grade dependent. However, the small sample size in the study does not allow us to estimate these differences within each grade separately. Finally, other mutations that affect tumor proliferation, such as epidermal growth factor receptors (EGFR) and phosphatase and tensin homolog (PTEN), were not available for the study population and could not be accounted for.
Acknowledgments and Disclosure:
This work was supported by the National Institute of Health (NIH) grants R01CA172210, R01EB016064. The authors have no financial conflicts of interest to report.
Contributor Information
Mohammed Goryawala, Department of Radiology, University of Miami, Miami, FL.
Efrat Saraf-Lavi, Department of Radiology, University of Miami, Miami, FL.
Natalia Nagornaya, Department of Radiology, University of Miami, Miami, FL.
Deborah Heros, Department of Neurology, University of Miami, Miami, FL.
Ricardo Komotar, Department of Neurological Surgery, University of Miami, Miami, FL.
Andrew A. Maudsley, Department of Radiology, University of Miami, Miami, FL
References
- 1.Zhang C, Moore LM, Li X, et al. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro Oncol 2013;15:1114–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep 2013;13:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 2009;118:469–74. [DOI] [PubMed] [Google Scholar]
- 4.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 2009;27:4150–4. [DOI] [PubMed] [Google Scholar]
- 5.Lee SM, Koh HJ, Park DC, et al. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med 2002;32:1185–96. [DOI] [PubMed] [Google Scholar]
- 6.Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 2012;483:484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res 2004;64:6892–9. [DOI] [PubMed] [Google Scholar]
- 8.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2010;462:739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber K Oncometabolite? IDH1 discoveries raise possibility of new metabolism targets in brain cancers and leukemia. J Natl Cancer Inst 2010;102:926–8. [DOI] [PubMed] [Google Scholar]
- 10.Choi C, Ganji SK, DeBerardinis RJ, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 2012;18:624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andronesi OC, Kim GS, Gerstner E, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med 2012;4:116ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emir UE, Larkin SJ, de Pennington N, et al. Noninvasive quantification of 2-hydroxyglutarate in human gliomas with IDH1 and IDH2 mutations. Cancer Res 2016;76:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pope WB, Prins RM, Albert Thomas M, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol 2012;107:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reitman ZJ, Jin G, Karoly ED, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA. 2011;108:3270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viswanath P, Chaumeil MM, Ronen SM. Molecular imaging of metabolic reprograming in mutant IDH cells. Front Oncol 2016;6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leather T, Jenkinson MD, Das K, Poptani H. Magnetic resonance spectroscopy for detection of 2-hydroxyglutarate as a biomarker for IDH mutation in gliomas. Metabolites 2017;7:E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izquierdo-Garcia JL, Viswanath P, Eriksson P, et al. Metabolic reprogramming in mutant IDH1 glioma cells. PLOS One 2015;10:e0118781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohka F, Ito M, Ranjit M, et al. Quantitative metabolome analysis profiles activation of glutaminolysis in glioma with IDH1 mutation. Tumour Biol 2014;35:5911–20. [DOI] [PubMed] [Google Scholar]
- 19.Goryawala MZ, Sheriff S, Maudsley AA. Regional distributions of brain glutamate and glutamine in normal subjects. NMR Biomed 2016;29:1108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maudsley AA, Domenig C, Govind V, et al. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI). Magn Reson Med 2009;61:548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabati M, Sheriff S, Gu M, et al. Multivendor implementation and comparison of volumetric whole-brain echo-planar MR spectroscopic imaging. Magn Reson Med 2015;74:1209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Taub E, Salibi N, et al. Comparison of reproducibility of single voxel spectroscopy and whole-brain magnetic resonance spectroscopy imaging at 3T. NMR Biomed 2018;31:e3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MM, Parolia A, Dunphy MP, Venneti S. Non-invasive metabolic imaging of brain tumours in the era of precision medicine. Nat Rev Clin Oncol 2016;13:725–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maudsley AA, Darkazanli A, Alger JR, et al. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed 2006;19:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haupt CI, Schuff N, Weiner MW, Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med 1996;35:678–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis-III: application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med 1998;40:822–31. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE T Med Imaging 2001;20:45–57. [DOI] [PubMed] [Google Scholar]
- 28.Cordova JS, Kandula S, Gurbani S, et al. Simulating the effect of spectroscopic MRI as a metric for radiation therapy planning in patients with glioblastoma. Tomography 2016;2:366–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goryawala MZ, Heros DO, Komotar RJ, et al. Value of diffusion kurtosis imaging in assessing low-grade gliomas. J Magn Reson Imaging 2018;48:1551–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayasuriya SA, Liew AWC. Symmetry plane detection in neuroimages based on intensity profile analysis. International Symposium on Information Technologies in Medicine and Education (ITME 2012); 2012:599–603. [Google Scholar]
- 31.Zeng A, Hu Q, Liu Y, et al. IDH1/2 mutation status combined with Ki-67 labeling index defines distinct prognostic groups in glioma. Oncotarget 2015;6:30232–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maudsley AA, Roy B, Gupta RK, et al. Association of metabolite concentrations and water diffusivity in normal appearing brain tissue with glioma grade. J Neuroimaging 2014;24:585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harat M, Blok M, Harat A, Soszyñska K. The impact of adjuvant radiotherapy on molecular prognostic markers in gliomas. Onco Targets Ther 2019;12:2215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazor T, Chesnelong C, Pankov A, et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc Natl Acad Sci USA 2017;114:10743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


