Abstract
The goal of this study was to compare the microbiota in different pig-present settings in China. Bioaerosol samples from pig farms and slaughterhouses and nasal samples from pig farmers and slaughterhouse workers were collected in Guangdong, southern China. The bacterial genomic DNA was isolated and subjected to 16S sequencing. The data were analyzed using QIIME2 with the DADA2 pipeline. A total of 14,923,551 clean reads and 2785 operational taxonomic units (OTUs) were obtained, which were mostly grouped into 4 phyla (Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria) and 220 families. The microbiota richness of nasal samples in pig-present workers was higher than that of bioaerosols collected in the vicinity of the pig enclosures. There were 31.7% (620/1954) shared OTUs between pig farm bioaerosols and pig farmers which was higher than that between pig slaughterhouses and slaughterhouse workers (23.4%, 364/1553) (p < 0.001). Acinetobacter and Pseudomonas were the most abundant in pig-present bioaerosols, and Staphylococcus, Pseudomonas, and Corynebacterium were dominant bacterial genus in pig farmers. The bacterial patterns are also specific to the location of sample collected. The results suggest that bioaerosol microbiota interact with human nasal microbes in the vicinity of the pig farm enclosures, providing the basis for further analysis of microbial transmission across hosts in pig-present settings.
Electronic Supplementary Material
The online version of this article (10.1007/s00284-020-02187-w) contains supplementary material, which is available to authorized users.
Introduction
The interaction between human and animals is critically important for transmission of zoonotic pathogens from vertebrate animals to humans. Zoonotic pathogens account for an estimated 70% of all human-related infectious diseases [1, 2], which are still a major threat to public health as illustrated by recent COVID-19 outbreak, along with the epidemic of swine influenza virus, Streptococcus suis, and so on [3–5]. Pig industry comprises a big portion of animal-related work. Pig workers tend to face a greater risk being infected by zoonotic disease than others, and would be susceptible to active infection from pig herds, which helps to facilitate transmission from pigs to pig workers [6, 7]. Further bilateral transmission may cause an amplification effect, leading to high levels of infection in both pigs and pig industry workers [8]. Therefore, revealing the bacterial diversity of nasal and bioaerosol samples in pig-present settings is important for better understanding the transmission patterns of pig-borne pathogens.
Bioaerosol is a collection of airborne particles of fungi, bacteria, viruses, and their metabolic products. Bioaerosols can connect human and animal hosts, and understanding the roles of bioaerosols can help to understand the transmission patterns of airborne pathogens [9, 10]. Corzo et al. have investigated the transmission of influenza A virus through air and detected viral genomes 2.1 km away from pig farms [11]. Another study has detected Reproductive and Respiratory Syndrome virus in 26% of pigs exposed to bioaerosol samples [12]. Clearly, bioaerosols can facilitate pathogen survival and spread.
The recent development of Next-Generation Sequencing (NGS) methods has provided a culture-independent approach to characterize microbial communities, resulting in significant enhancement of our understanding for the diverse microbial communities in specific habitats of interest. For example, Lee, et al. have analyzed airborne microbiota of 108 samples collected from Beijing, Seoul, and Nagasaki, and determined that Beijing had the most diverse bacterial community and was influenced by meteorological factors [13]. Other studies found that bacterial structure was associated with airborne particulate matter as well as spatial and seasonal variation [14, 15]. However, the microbiota in occupational settings with animal exposure has never been investigated in China. In this study, we aimed to compare the difference of the bacterial diversity, abundance, and characteristics in pig industry samples, which would be a helpful reference for potential public health problem and pathogen surveillance in the pig industry.
Material and Methods
Ethical Statement
Ethical approval was obtained from the ethics committee of the School of Public Health, Sun Yat-Sen University (2014-18).
Study Population
The surveys were conducted between May and Oct 2017. The pig-present farmers (pig farmer and slaughterhouse workers) and bioaerosols were sampled at pig farms and slaughterhouses in seven cities of Guangdong, China. Briefly, human subjects were recruited through face-to-face interactions with study personnel during visits. The inclusion criteria were (1) had worked or lived in pig farm or the slaughterhouse for more than one year with more than 28 h a week of exposure time to pigs; and (2) at least 16 years old. We excluded subjects who were (1) suffering from immune damage or acute respiratory tract infection; (2) pregnant female subjects; or (3) suffering immunosuppressive or immunodeficiency disease (including HIV infection) or receiving immunosuppressive therapy.
Sample Collection
With consent of written informed, a nasal swab sample was drawn from each participant. A cotton swab with sterile physiological saline was used to wipe the left and right nasal cavities for 30 s and then placed in a collection tube containing 3 ml of 0.5% bovine serum albumin (BSA)–phosphate-buffered saline (PBS) solution. At each site, samples were collected from 5–10 subjects and pooled. Samples from each pig farm were combined into a single pool, and samples from each slaughterhouse were combined into two pools.
The bioaerosol samples were collected from pig farms in Heyuan, Yangjiang, Shaoguan, Jiangmen, and Maoming, and from slaughterhouses in Guangzhou, China (Fig. 1). The bioaerosol sampling method was carried out as previously described [16]. Briefly, 9600-L volumes of bioaerosols were collected for each sample during 48 h of sampling, and six samples were collected from each site. All the collected samples were then transported and shipped to the laboratory in sterile collection tubes packed in dry ice. All samples were stored at − 80 °C until further testing.
Fig. 1.
Locations of pig farms and slaughterhouses in this study. The regions where samples were collected are marked as chocolate; black triangles indicate where pig farms are located; black dots indicate where slaughterhouses are located. GZ Guangzhou, HY Heyuan, JM Jiangmen, MM Maoming, SG Shaoguan, YJ Yangjiang
DNA Extraction and Amplicon Sequencing
Total genomic DNA was extracted from bioaerosols and nasal swabs using the PowerSoil DNA Isolation Kit (MoBio, USA) following the protocol’s instructions. These DNA extracts were then sent to BGI Co., Ltd (Wuhan, China) for library construction and amplicon sequencing. Sequencing analysis was performed by amplification of the V4 region of the 16S rRNA gene using the previously described specific primers: forward (5′-GTGCCAGCMGCCGCGGTAA-3′) and reverse (5′-GGACTACHVGGGTWTCTAAT-3′) [17]. After amplification, sequencing libraries were prepared and submitted for sequencing on the Illumina HiSeq platform using the Next-Generation Sequencing Platform for indexing and paired-end 2 × 250 bp sequencing (San Diego, USA). The obtained raw Illumina amplicon sequencing files for the amplified 16S rRNA V4 regions were deposited with GenBank accession number PRJNA605910 in the Sequence Read Archive (SRA) database (https://submit.ncbi.nlm.nih.gov/subs/bioproject/SUB6954900/overview).
Data Analysis
Sequencing was performed at BGI Co., Ltd (Wuhan, China). The obtained reads were cleaned and then analyzed using the Quantitative Insights into Microbial Ecology bioinformatic pipeline, QIIME2, which was designed for analysis of microbial communities. The DADA2 package from Bioconductor was used with the default parameter setting to correct and analyze the amplicon data. We then constructed a feature table, which describes the abundances of the individual OTU or sequence variants for each sample. The OTU identified by DADA2 exceeded the resolution of the QIIME 1 default of 97% OTU. QIIME2 includes improved quality control steps, so the OTUs identified in this analysis were of higher quality than what would be obtained using QIIME 1. Due to the improved quality of reads, this analysis should allow more accurate diversity and taxonomic composition determinations than previous analysis efforts based on QIIME 1.
To quantitatively measure sample diversity, Alpha diversities, Chao1 Diversity index, and Faith’s Phylogenetic Diversity index were calculated for each sample. The significance of differences among different groups was evaluated by Kruskal–Wallis test, a non-parametric method to assess whether samples originate from the same distribution.
To quantitatively measure Beta diversities, the Jaccard distance and weighted Unifrac values were calculated. The pairwise distances between samples were computed using the vegdist function in R. The metaMDS function was used to take the resulting matrices of data and generate non-metric multidimensional scaling (NMDS) plots. Dissimilarity boxplots were also constructed. Permutational multivariate analysis of variance (PERMANOVA) was used to assess the significance of sample groupings using 1000 Monte Carlo simulations. Annotation was performed using the q2-feature-classifier plugin starting with a pre-trained classifier (targeting the V4 region, with endpoints defined by the 515F and 806R primers, with PE250 reads). Bar plots were constructed to visualize the community composition of each sample. The differentially abundant OTUs (i.e., OTU with different abundances in different sample groups) were detected across all groups at the level of family, genus, and species using the q2-composition-ancom plugin.
Results
Characterization of Sampling Population
39 samples (including 15 pig farm bioaerosols, 12 slaughterhouse bioaerosols, 6 slaughterhouse workers, and 6 pig farmers) were collected from nine pig-present settings (including 6 pig farms and 3 pig slaughterhouses) located in Jiangmen, Heyuan, Maoming, Shaoguan, Yangjiang, and Guangzhou cities of Guangdong, Southern China, between May 2017 and Oct 2017 (Table 1).
Table 1.
Characteristics of the study population
| Region | N of bioaerosols sampled (pool) | N of workers sampled (pool) | Farm size | N of pigsty | |
|---|---|---|---|---|---|
| Pig farm | |||||
| 1 | Heyuan | 3 | 1 | Mediuma | 38 |
| 2 | Maoming | 3 | 1 | Medium | 27 |
| 3 | Jiangmen | 3 | 1 | Largea | 58 |
| 4 | Jiangmen | 3 | 1 | Large | 47 |
| 5 | Shaoguan | 3 | 1 | Smalla | 17 |
| 6 | Yangjiang | 3 | 1 | Small | 4 |
| Slaughterhouse | |||||
| 1 | Guangzhou | 3 | 2 | NA | 38 |
| 2 | Guangzhou | 3 | 2 | NA | 24 |
| 3 | Guangzhou | 3 | 2 | NA | 27 |
aSmall, N < 750; medium, 750 < N < 2499; large, N > 2500
NA data not available
Sequence Analysis
A total of 14,923,551 clean reads were obtained, with an average of clean reads of 382,655 ± 84,547, which accounted for 97.7% of the raw data. Of these reads, 7,329,892 clean reads were from pig farm bioaerosols, with an average of 407,216 ± 77,324 clean reads, accounting for 98.5% of the raw data. For slaughterhouse bioaerosols, 3,432,275 clean reads were obtained, with an average of 381,364 ± 89,214 clean reads, accounting for 98.5% of the raw data. From nasal samples of pig farmers, 1,834,688 clean reads were obtained, with an average of 307,615 ± 99,936 clean reads, accounting for 95.7% of the raw data. From slaughterhouse workers, 2,315,696 clean reads were obtained, with an average of 385,949 ± 48,293 clean reads, accounting for 95.6% of the raw data (Table 2).
Table 2.
Cleaned data and OTUs of pig-present bioaerosol and workers
| Sample type | Sample size (N) | Clean reads (N) | Clean reads (N ± SD) | OTUs (N) |
|---|---|---|---|---|
| Pig farm bioaerosols | 18 | 7,329,892 | 407,216 ± 77,324 | 1222 |
| Slaughterhouse bioaerosols | 9 | 3,432,275 | 381,364 ± 89,214 | 1043 |
| Pig farmer nasal samples | 6 | 1,834,688 | 307,615 ± 99,936 | 1352 |
| Slaughterhouse worker nasal samples | 6 | 2,315,696 | 385,949 ± 48,293 | 874 |
| Total | 39 | 14,923,551 | 382,655 ± 84,547 | 2785 |
SD standard deviation, OTUs operational taxonomic units
Taxonomy Assignment at Phylum Level
The reads were grouped into a total of 2,785 OTUs (with ≥ 97% mean relative abundance for all sample groups), including 1222, 1043, 1352, and 874 OTUs from pig farm bioaerosols, slaughterhouse bioaerosols, pig farmer nasal samples, and slaughterhouse worker nasal samples, respectively. All 2,785 OTUs were grouped into 4 phyla and 220 families. The composition of bacterial community was highly diverse, and was dominated at the phylum level by Proteobacteria (45.8 ± 32.1%), Bacteroidetes (23.1 ± 29.3%), Firmicutes (22.2 ± 28.6%), and Actinobacteria (9.6 ± 14.4%). Other phyla were rarely represented, including Tenericutes, Cyanobacteria, Fusobacteria, Chloroflexi, and Euryarchaeota (Table S1 and S2).
Differences and Similarities of the Microbiota
Phyla with sequences of more than 10,000 reads, including Proteobacteria, Bacteroidetes, and Firmicutes, and Actinobacteria, were used to construct a phylogenetic tree (Fig. S1). Proteobacteria was the most diversified including Acinetobacter, Moraxella, Psychrobacter, Escherichia, Pseudomonas, and Stenotrophomonas. This was followed by Firmicutes, including Staphylococcus, Exiguobacterium, Alloiococcus, Lactobacillus, Streptococcus, Anaerococcus, Finegoldia, and SMB53. Next was Bacteroides, including Wautersiella, Chryseobacterium, and Myroides. Finally, Actinobacteria was the least diverse, including Micrococcus, Arthrobacter, and Corynebacterium. Furthermore, the most abundant genera in the pig-present bioaerosols were Acinetobacter and Pseudomonas, and the pig worker nasal samples were dominated by Staphylococcus, Pseudomonas, and Corynebacterium (Fig. 2).
Fig. 2.
Relative abundance of different phyla. CBY-1, CBY-2, CBY-3, CHD-1, CHD-2, CHD-3, CXH-1, CXH-2, CXH-3, CHY-1, CHY-2, CHY-3, CJM-1, CJM-2, CJM-3, CKP-1, CKP-2, CKP-3, CHZ-1, CHZ-2, CHZ-3, CSG-1, CSG-2, CSG-3, CYC-1, CYC-2, CYC-3, RBY-1, RBY-2, RHD-1, RHD-2, RHY, RHZ, RJM, RKP, RSG, RXH-1, RXH-2, RYC indicated different sample numbers
Differentially Abundant Taxa at the Genus Level
At the genus level, the bacterial abundances in farm samples differed, with farm and slaughterhouse specificity. For different locations and regions, Actinobacteria, Pseudomonas, and Wautersiella were significantly abundant in pig-present bioaerosols. Staphylococcus, Corynebacterium, Pseudomonas, and Lactobacillus were most abundant in pig workers. The genus Psychrobacter was more abundant in slaughterhouse workers than that in pig farmers, and was also detected in pig-present bioaerosols. In addition, the genera Alloiococcus, Peptoniphilus, and Anaerococcus, were significantly more abundant in pig workers than in pig-present bioaerosols. To better understand the different abundances of bacterial genera in different setting and regions, a hierarchical clustering heat map of bacterial genera was constructed, as shown in Fig. 3.
Fig. 3.
Hierarchical clustering heat map of bacterial genera in the pig-present bioaerosols and farmers. Not_Assigned indicates the sequence reads that were not assigned at the genus level but were assigned at the higher taxonomic level. CBY-1, CBY-2, CBY-3, CHD-1, CHD-2, CHD-3, CXH-1, CXH-2, CXH-3, CHY-1, CHY-2, CHY-3, CJM-1, CJM-2, CJM-3, CKP-1, CKP-2, CKP-3, CHZ-1, CHZ-2, CHZ-3, CSG-1, CSG-2, CSG-3, CYC-1, CYC-2, CYC-3, RBY-1, RBY-2, RHD-1, RHD-2, RHY, RHZ, RJM, RKP, RSG, RXH-1, RXH-2, RYC indicated different sample numbers
Alpha and Beta Diversity Analysis
The alpha diversity analysis was performed using Chao1 and Faith's phylogenetic diversity index (Faith's PDI). Nasal samples from pig farmers showed the highest Chao1, and Faith's PDI values, whereas slaughterhouse bioaerosols displayed the lowest values. Because multiple samples were collected at sampling sites, we also performed a linear mixed regression with the location as a random effect to compare any differences between groups. The bacterial richness in nasal samples from pig farm and slaughterhouse workers was significantly higher than that of pig-present bioaerosols (Chao1, p < 0.05; Faith's PDI, p < 0.05). The abundance of pig farmer’s nasal samples was higher than that in slaughter nasal samples, and pig farm bioaerosol samples exhibited higher abundances than slaughter bioaerosol samples, as shown by Chao1 and Faith's phylogenetic diversity (Faith's PD) indices (p < 0.05 or p < 0.01) (Fig. 4a, b; Table S3).
Fig. 4.
Alpha and beta diversity of samples from pig-present bioaerosols and farmers. For the alpha diversity, the differences in Chao1 diversity (observed OTU) (a) and phylogenetic diversity indices based on sample types (b) are shown. For beta diversity, the confidence ellipse is also shown for the group centroid. c and d Beta dispersion based on weighted UniFrac (c) and Jaccard (d) dissimilarity indices in each sample type. The boxplots represent median (midline), interquartile ranges (shaded boxes), and ranges (whiskers). a slaughterhouse bioaerosol samples; b pig farm bioaerosol samples; c nasal samples from slaughterhouse workers; d pig farmer nasal samples. Significant differences within panels a, b, c, and d are indicated by asterisks (*p < 0.05; **p < 0.01; ***p < 0.001)
Ordination method-based non-metric multidimensional scaling (NMDS) plots with weighted Unifrac and Jaccard values (Fig. S2) showed a distinct clustering of farm bioaerosols, pig farmers, slaughterhouse workers, and slaughterhouse bioaerosol samples, which was confirmed by permutational multivariate analysis of variance (PERMANOVA). Profiles of nasal samples from pig farmers, nasal samples from slaughterhouse workers, pig farm bioaerosol samples, and slaughterhouse bioaerosol samples were similar, indicating an effect of pig-present setting factors on the human nasal microbiota. Interestingly, microbial communities isolated from pig industry workers seemed to exhibit significantly lower beta diversity values than microbial communities isolated from pig-present bioaerosols (weighted distances from the centroid; p < 0.01 or 0.001; Fig. 4c, d), indicating a more homogeneous microbial community structure in pig industry workers (Table 3).
Table 3.
Pairwise PERMANOVA results based on Jaccard and weighted Unifrac index
| Compared to sample types | Weighted UniFrac | Jaccard dissimilarity | ||
|---|---|---|---|---|
| F value | P value | F value | P value | |
| Slaughterhouse bioaerosols vs. pig farm bioaerosols | 1.23 | 0.275 | 1.60 | 0.042 |
| Slaughterhouse bioaerosols vs. slaughterhouse workers | 10.47 | < 0.001 | 4.35 | < 0.001 |
| Slaughterhouse bioaerosols vs. pig farmers | 13.37 | < 0.001 | 3.66 | < 0.001 |
| Pig farm bioaerosols vs. pig farmers | 10.82 | < 0.001 | 3.83 | < 0.001 |
| Pig farm bioaerosols vs. slaughterhouse workers | 8.39 | < 0.001 | 4.74 | < 0.001 |
| Pig farmers vs. slaughterhouse workers | 6.65 | 0.007 | 1.94 | 0.004 |
PERMANOVA permutational multivariate analysis of variance, vs. versus
Analysis of Shared OTUs
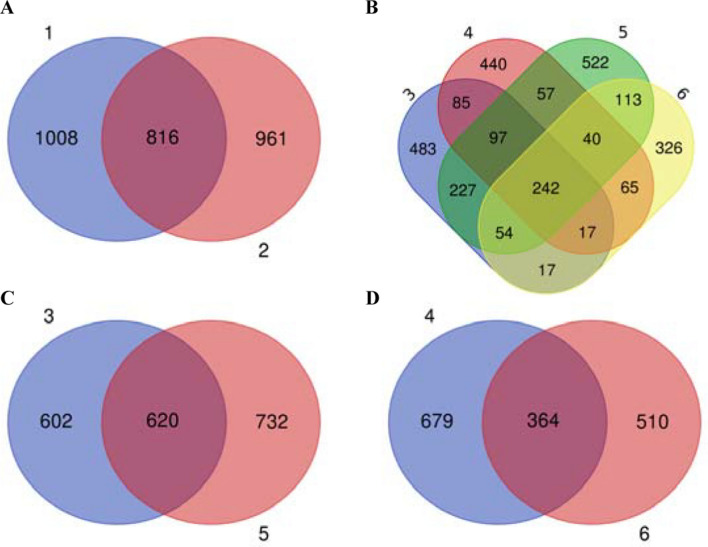
The ratio of shared OTUs between microbial communities isolated from pig-present bioaerosols and microbial communities isolated from farmers in the pig-present settings was 29.3% (816/2785). The sequencing of microbial communities from bioaerosols at pig farm and ones isolated from pig farmers shared 31.7% (620/1954) of OTUs. The ratio of shared OTUs between the microbial communities of the slaughterhouse and the slaughterhouse workers was 23.4% (364/1553), with statistically significant difference (p < 0.001). Only 8.7% (242/2785) OTUs were shared between pig farmers, slaughterhouse workers, pig farm bioaerosols, and slaughterhouse bioaerosols. The number of shared OTUs in pig farms and pig-present farmers was higher than that between slaughterhouses and slaughterhouse workers, suggesting that pig farm bioaerosols have a greater impact on the microbial colonization in noses of farmers (Fig. 5; Fig. S3; Table S4).
Fig. 5.
Venn diagram showing unique and shared OTUs. 1, pig-present bioaerosol samples; 2, pig-present farmer nasal samples; 3, pig farm bioaerosol samples; 4, slaughterhouse bioaerosol samples; 5, pig farmer nasal samples; 6, slaughterhouse worker nasal samples. a Venn diagram showing the numbers of shared OTU between pig-present farmers and Bioaerosols; b Venn diagram showing the numbers of shared OTU between pig farmers, slaughterhouse workers, pig farms, and slaughterhouse. c Venn diagram showing the numbers of shared OTU between pig farmers and pig farms; d Venn diagram showing the numbers of shared OTU between the slaughterhouse workers and the slaughterhouse samples
Discussion
The animal-exposed occupational population potentially has a high risk of susceptivity in zoonotic infection, and assessing the transmission risk and achieving early prevention is critical [18–20]. The high density of animals feeding in large-scale farms could create the conditions required for pathogen survival and spread [21, 22], those who work for significant amounts of time in such environments might have latent infections, and could act as a carrier to introduce zoonotic pathogens into the general population and lead to a serious public health crisis [23–25]. Here, this study was conducted to reveal the microbiota associated with difference pig-present settings and it is also beneficial for future prevention and control of zoonotic pathogens.
Our study collected about 9600 L of bioaerosol for each animal-exposed bioaerosol samples, which was similar to the volume of air that humans inhale each day about 10,000 L [26]. The 16S rRNA analyses results revealed Proteobacteria as the dominant phylum, followed by Bacteroidetes and Firmicutes in pig farm and slaughterhouse bioaerosols. These phyla were consistent with those identified in inhalable microbial communities in Beijing, China [27]. However, Dueker et al. [28] analyzed waterfront sites air samples collected from New York, USA, and identified Verrucomicrobia, Proteobacteria, and Firmicutes as the dominant bacteria. The proportions of Proteobacteria in this study varied between individuals, and this variation may be associated with multiple factors, such as temperature, humidity, surroundings, or biases caused by 16S rRNA primer selection [29, 30].
The alpha diversity values of pig-present bioaerosols were lower than those of pig-present workers, and the abundance of bacteria in bioaerosols was relatively low, indicating that not all microorganisms were continuously discharged and suspended in the bioaerosols. These alpha diversity differences between pig farmers and slaughterhouse workers suggest that pig farming leads to an increased bacterial diversity in the noses of workers compared to those of individuals working in slaughterhouses. The high concentration of diverse air-borne bacteria present in pig farms may lead to a modification and an enrichment of the farmer’s nasal microbiota [31]. We also found significant variation in samples from farmers working on the same farm, with Chao1 diversity index values ranging from 64 to 1024 in Heyuan. Additionally, there was significant variation in samples from slaughterhouses in Baiyun district, with Faith’s phylogenetic diversity index values ranging from 3.22 to 55.44, likely due to the influence on bioaerosols on pig-present settings by dust, bacteria, and fungi [32]. Factors such as temperature, humidity, and surroundings can also obviously impact the alpha diversity.
Our results revealed a high number of shared OTUs between communities isolated from pig-present bioaerosols and pig workers, indicating the frequent exchange of microorganisms. The result suggests an important role of bioaerosols in the transmission of animal-associated bacteria to pig workers [31]. However, there were more shared OTUs between pig farm bioaerosol samples and pig farmer nasal samples than between slaughterhouse bioaerosol samples and nasal samples from slaughterhouse workers, and the ratio of shared OTUs between slaughterhouse bioaerosol samples and nasal samples from slaughterhouse workers was similar to that in cow-exposed or non-exposed control as described by Kraemer et al. [17], and indicated that exposed factor of bioaerosol in slaughterhouse had poor impact on microbial diversity of slaughterhouse worker nasal cavity. This might be explained by a more extensive exposure time to pigs of the pig farmers compared to that of slaughterhouse workers, with long-lasting exposure to relatively stable pig populations as pig farmers in China mostly live and work at the pig farms, with almost full-time exposure to pigs. In contrast, the slaughterhouse workers generally have a relatively fixed working time with more time without pig exposure compared to pig farmers. To counter this great exposure of pig farmers, it is necessary to improve daily protective measures for the farmers, including to put on mask while working, to avoid unnecessary direct exposure to pigs, and to keep pigsty with safe distance from human living areas.
Although we only examined microbiota from pig-present bioaerosols and pig worker nasal samples, however, common microorganisms, such as Staphylococcus and Escherichia, can be carriers of drug resistance genes. These bacteria easily colonize the nasal cavity of animal-exposed workers, and further work should classify resistance genes. Increasing awareness of the potential interactions between humans, animals, and the environment underscores the need for better surveillance of zoonotic pathogens at human–animal–environment interfaces [18, 19].
This study also had some limitations worth noting as follows: (1) this study only used amplicon sequencing, and did not isolate or verify microorganisms, preventing collection of more detailed data from bacterial classification or identification of antimicrobial resistance genes; and (2) sampling was performed during the summer of Guangdong, China, so there was no information collected about samples in winter, when there might be a higher incidence of infectious diseases. Thus, the influence of seasonal factors on microbial diversity was not evaluated.
In conclusion, respiratory bacterial colonization in the pig farmers was strongly associated with airborne bacteria. There was a significantly higher proportion of shared OTUs and bacterial diversity between pig farms and pig farmers than that between slaughterhouses and slaughterhouse workers. Strengthening the prevention activity in pig farms is important to reduce pathogen dissemination and accumulation and long-term surveillance should be performed to allow monitoring of zoonotic pathogen circulation.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by The National Key Research and Development Program of China (Grant No. 2018YFD0500500), The Guangdong Provincial Science and Technology Project (Grant No. 2018B020241002), The National Science and Technology Major Project (Grant No. 2018ZX10101002), The National Natural Science Fund of China (Grant No. 31872499), and The Guangdong Provincial Science and Technology Project (Grant No. 2018B020207013). We would like to thank profs. Changxu Song and Leyi Zhang from South China Agricultural University for their kind help of contacting with the pig farms and collecting samples.
Author Contributions
JHL designed the experiments; JYW and YSZ conducted most of the experiments; YX and JYW analyzed data; JYW, CG, ZMG, and QLL drafted the paper. All authors read, commented, and approved the final version of the manuscript.
Data Availability
The obtained raw amplicon sequencing files for the amplified 16S rRNA V4 regions were deposited with GenBank accession number PRJNA605910 in the Sequence Read Archive (SRA) database.
Compliance with Ethical Standards
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical Approval
Ethical statement was obtained from the ethics committee of the School of Public Health, Sun Yat-Sen University (2014-18).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jian-Yong Wu, Yan-Shan Zhu, Cheng Guo and Yao Xia have contributed equally as co-first authors.
References
- 1.Sikkema RS, Freidl GS, de Bruin E, Koopmans M. Weighing serological evidence of human exposure to animal influenza viruses—a literature review. Euro Surveill. 2016;21(44):30388. doi: 10.2807/1560-7917.ES.2016.21.44.30388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klous G, Huss A, Heederik DJJ, Coutinho RA. Human–livestock contacts and their relationship to transmission of zoonotic pathogens, a systematic review of literature. One Health. 2016;2:65–76. doi: 10.1016/j.onehlt.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Samkar A, Brouwer MC, Schultsz C, van der Ende A, van de Beek D. Streptococcus suis meningitis: a systematic review and meta-analysis. Plos Neglect Trop D. 2015;9(10):e0004191. doi: 10.1371/journal.pntd.0004191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;10222(8–14):391–393. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radon K, Weber C, Iversen M, Danuser B, Pedersen S, Nowak D. Exposure assessment and lung function in pig and poultry farmers. Occup Environ Med. 2001;58(6):405–410. doi: 10.1136/oem.58.6.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walser SM, Gerstner DG, Brenner B, Bunger J, Eikmann T, Janssen B, Kolb S, Kolk A, Nowak D, Raulf M, Sagunski H, Sedlmaier N, Suchenwirth R, Wiesmuller G, Wollin KM, Tesseraux I, Herr CE. Evaluation of exposure-response relationships for health effects of microbial bioaerosols—a systematic review. Int J Hyg Environ Health. 2015;218(7):577–589. doi: 10.1016/j.ijheh.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Fragaszy E, Ishola DA, Brown IH, Enstone J, Nguyen-Van-Tam JS, Simons R, Tucker AW, Wieland B, Williamson SM, Hayward AC, Flu Watch G, Wood JL, Combating Swine Influenza C. Increased risk of A(H1N1)pdm09 influenza infection in UK pig industry workers compared to a general population cohort. Influenza Other Respir Viruses. 2016;10(4):291–300. doi: 10.1111/irv.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mainelis G, Masquelier D, Makarewicz A, Dzenitis J. Performance characteristics of the aerosol collectors of the autonomous pathogen detection system (APDS) Aerosol Sci Technol. 2005;39(5):461–471. doi: 10.1080/027868290960382. [DOI] [Google Scholar]
- 10.Hagiwara D, Sakai K, Suzuki S, Umemura M, Nogawa T, Kato N, Osada H, Watanabe A, Kawamoto S, Gonoi T, Kamei K. Temperature during conidiation affects stress tolerance, pigmentation, and trypacidin accumulation in the conidia of the airborne pathogen Aspergillus fumigatus. PLoS ONE. 2017;12(5):e0177050. doi: 10.1371/journal.pone.0177050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corzo CA, Romagosa A, Dee SA, Gramer MR, Morrison RB, Torremorell M. Relationship between airborne detection of influenza A virus and the number of infected pigs. Vet J. 2013;196(2):171–175. doi: 10.1016/j.tvjl.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitkin A, Deen J, Dee S. Use of a production region model to assess the airborne spread of porcine reproductive and respiratory syndrome virus. Vet Microbiol. 2009;136(1–2):1–7. doi: 10.1016/j.vetmic.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Lee JY, Park EH, Lee S, Ko G, Honda Y, Hashizume M, Deng F, Yi SM, Kim H (2017) Airborne bacterial communities in three East Asian Cities of China, South Korea, and Japan. Sci Rep 7(1):5545. 1038/s41598-017-05862-4. [DOI] [PMC free article] [PubMed]
- 14.Li H, Zhou XY, Yang XR, Zhu YG, Hong YW, Su JQ. Spatial and seasonal variation of the airborne microbiome in a rapidly developing city of China. Sci Total Environ. 2019;665:61–68. doi: 10.1016/j.scitotenv.2019.01.367. [DOI] [PubMed] [Google Scholar]
- 15.Yan D, Zhang T, Su J, Zhao LL, Wang H, Fang XM, Zhang YQ, Liu HY, Yu LY. Structural variation in the bacterial community associated with airborne particulate matter in Beijing, China, during Hazy and Nonhazy Days. Appl Environ Microbiol. 2018;84(9):e00004. doi: 10.1128/AEM.00004-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson BD, Ma M, Xia Y, Wang T, Shu B, Lednicky JA, Ma MJ, Lu J, Gray GC. Bioaerosol sampling in modern agriculture: a novel approach for emerging pathogen surveillance? J Infect Dis. 2016;214(4):537–545. doi: 10.1093/infdis/jiw180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraemer JG, Ramette A, Aebi S, Oppliger A, Hilty M. Influence of pig farming on the human nasal microbiota: key role of airborne microbial communities. Appl Environ Microbiol. 2018 doi: 10.1128/AEM.02470-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SH. Challenge for one health: co-circulation of zoonotic H5N1 and H9N2 avian influenza viruses in Egypt. Viruses. 2018;10(3):121. doi: 10.3390/v10030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma MJ, Wang GL, Anderson BD, Bi ZQ, Lu B, Wang XJ, Wang CX, Chen SH, Qian YH, Song SX, Li M, Lednicky JA, Zhao T, Wu MN, Cao WC, Gray GC. Evidence for cross-species influenza A virus transmission within swine farms, China: a one health, Prospective Cohort Study. Clin Infect Dis. 2018;66(4):533–540. doi: 10.1093/cid/cix823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bird BH, Mazet JAK. Detection of emerging zoonotic pathogens: an integrated one health approach. Annu Rev Anim Biosci. 2018;6:121–139. doi: 10.1146/annurev-animal-030117-014628. [DOI] [PubMed] [Google Scholar]
- 21.Mennerat A, Nilsen F, Ebert D, Skorping A. Intensive farming: evolutionary implications for parasites and pathogens. Evol Biol. 2010;37(2):59–67. doi: 10.1007/s11692-010-9089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones BA, Grace D, Kock R, Alonso S, Rushton J, Said MY, McKeever D, Mutua F, Young J, McDermott J, Pfeiffer DU. Zoonosis emergence linked to agricultural intensification and environmental change. Proc Natl Acad Sci USA. 2013;110(21):8399. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Cleef BAGL, Van Benthem BHB, Verkade EJM, Van Rijen MML, Kluytmans-Van Den Bergh MFQ, Graveland H, Bosch T, Verstappen KMHW, Wagenaar JA, Heederik D, Kluytmans JAJW. Health and health-related quality of life in pig farmers carrying livestock-associated methicillin-resistant Staphylococcus aureus. Epidemiol Infect. 2016;144(8):1774–1783. doi: 10.1017/s0950268815003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dohmen W, Bonten MJM, Bos MEH, van Marm S, Scharringa J, Wagenaar JA, Heederik DJJ. Carriage of extended-spectrum β-lactamases in pig farmers is associated with occurrence in pigs. Clin Microbiol Infec. 2015;21(10):917–923. doi: 10.1016/j.cmi.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 25.Fischer J, Hille K, Ruddat I, Mellmann A, Köck R, Kreienbrock L. Simultaneous occurrence of MRSA and ESBL-producing Enterobacteriaceae on pig farms and in nasal and stool samples from farmers. Vet Microbiol. 2017;200:107–113. doi: 10.1016/j.vetmic.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Camarinha-Silva A, Jáuregui R, Chaves-Moreno D, Oxley APA, Schaumburg F, Becker K, Wos-Oxley ML, Pieper DH. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol. 2014;16(9):2939–2952. doi: 10.1111/1462-2920.12362. [DOI] [PubMed] [Google Scholar]
- 27.Du P, Du R, Ren W, Lu Z, Fu P. Seasonal variation characteristic of inhalable microbial communities in PM2.5 in Beijing city China. Sci Total Environ. 2018;610–611:308–315. doi: 10.1016/j.scitotenv.2017.07.097. [DOI] [PubMed] [Google Scholar]
- 28.Dueker ME, French S, O'Mullan GD. Comparison of bacterial diversity in air and water of a Major Urban Center. Front Microbiol. 2018;9:2868–2868. doi: 10.3389/fmicb.2018.02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai L, Ye L, Tong AHY, Lok S, Zhang T. Biased diversity metrics revealed by bacterial 16s pyrotags derived from different primer sets. PLoS ONE. 2013;8(1):e53649. doi: 10.1371/journal.pone.0053649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunwar A, Tamrakar S, Poudel S, Sharma S, Parajuli P. Bacteriological assessment of the indoor air of different hospitals of Kathmandu district. Int J Microbiol. 2019;2019:9. doi: 10.1155/2019/5320807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraemer JG, Ramette A, Aebi S, Oppliger A, Hilty M. Influence of pig farming on the human's nasal microbiota: The key role of the airborne microbial communities. Appl Environ Microbiol. 2018;84(6):e02470–e2517. doi: 10.1128/AEM.02470-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sowiak M, Bródka K, Buczyńska A, Cyprowski M, Kozajda A, Sobala W, Szadkowska-Stańczyk I. An assessment of potential exposure to bioaerosols among swine farm workers with particular reference to airborne microorganisms in the respirable fraction under various breeding conditions. Aerobiologia. 2012;28(2):121–133. doi: 10.1007/s10453-011-9216-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The obtained raw amplicon sequencing files for the amplified 16S rRNA V4 regions were deposited with GenBank accession number PRJNA605910 in the Sequence Read Archive (SRA) database.