Abstract
Recent studies have revealed that microRNAs (miRs) are involved in the pathogenesis of colorectal cancer (CRC); however, the roles of miR-590-5p in CRC are not completely understood. Therefore, the present study investigated the expression of miR-590-5p and programmed cell death 4 (PDCD4) in CRC tissues and healthy adjacent tissues via reverse transcription-quantitative PCR. Furthermore, human CRC cells were cultured in vitro and transfected with miR-590-5p inhibitor. CRC cell viability, migration and invasion were evaluated by conducting MTT, wound healing and Transwell assays, respectively. Additionally, the relative expression of PDCD4 and phosphorylated-Smad2/3 was analyzed via western blotting. miR-590-5p was significantly upregulated and PDCD4 was significantly downregulated in CRC tissues compared with healthy adjacent tissues. Moreover, the results indicated that miR-590-5p knockdown inhibited cell viability and migration by altering the expression of PDCD4, transforming growth factor-β and phosphorylated-Smad2/3. PDCD4 was identified as a direct target of miR-590-5p. In conclusion, the results of the present study suggested that miR-590-5p may regulate CRC cell viability and migration, indicating that miR-590-5p may serve as a potential therapeutic target for CRC.
Keywords: microRNA-590-5p, colorectal cancer, programmed cell death 4, Smad2/3, transforming growth factor-β
Introduction
Cancer is one of the most serious threats to human health worldwide and affects a variety of organs and tissues (1). Colorectal cancer (CRC) is a global public health concern (2). The most common treatment strategies for CRC include surgery, chemotherapy and radiotherapy; however, these strategies often do not completely eliminate the disease (3,4). The targeted treatment of tumors has attracted increasing attention and several targeted drugs are extensively used in the clinic (4) with demonstrated effectiveness (5). An increased understanding of the molecular mechanism underlying the pathogenesis of CRC may aid with the identification of effective therapeutic targets and novel biomarkers for the risk assessment and early diagnosis of CRC (6-8).
MicroRNAs (miRNAs/miRs) are small non-coding RNAs that are ~22 nucleotides in length (9,10). Previous studies have revealed that miRNAs may negatively regulate gene expression by binding to the 3'-untranslated region (UTR) of target mRNAs to silence gene expression (9-11). It has been widely established that miRNAs serve critical roles in the regulation of several biological process, including proliferation, differentiation, apoptosis, tumorigenesis and metastasis (6,12,13). Furthermore, miRNAs may predict overall prognosis in certain types of cancer, including lung cancer (14), gastric cancer (15) and CRC (16). Previous studies suggested that miR-590 is abnormally expressed in CRC (5,17); however, the mechanism underlying the action of miR-590-5p in the pathogenesis of CRC, as well as its potential gene target have not been identified.
The present study investigated the role of miR-590-5p, as well as its underlying mechanism, in CRC. The expression of miR-590-5p in CRC tissues were compared with healthy adjacent tissues. Furthermore, in vitro studies were designed to examine the effect of miR-590-5p knockdown on CRC cell viability and invasion.
Materials and methods
Patients and clinical tissue samples
A total of 30 CRC and healthy adjacent tissue samples (distance from tumour margin, 3 cm) were collected from patients with CRC (age, 48-65 years; mean age, 52.32±7.41; 17 male patients and 13 female patients) at Taizhou People's Hospital between May 2017 and February 2019. The tissue samples were obtained from patients who had undergone resection but had not received radiotherapy and/or chemotherapy. All tissues were pathologically confirmed. The present study was approved by the Ethical Review Board of Taizhou People's Hospital and written informed consent was obtained from each patient.
Cell culture and transfection
Human CRC cell lines (HCT116, SW480, SW620 and LoVo) and human normal colon epithelial cells (FHC) were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C in a humidified incubator with 5% CO2. Cells were plated in 24-well plates (1x105 cells/well) and transfected with miR-590-5p mimic, miR-590-5p inhibitor, miR-590-5p mimic negative control (NC), miR-590-5p inhibitor NC, PDCD4 small interfering (si)RNA NC or PDCD4 siRNA (all, 100 pmol) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The sequences of the oligonucleotides were: miR-590-5p mimic, 5'-GAGCUUAUUCAUAAAAUGCAG-3'; miR-590-5p mimic NC, 5'-CGCCAAUAUCAUUAUACCUC-3'; miR-590-5p inhibitor, 5'-AAAUAUGCUGUAUGUCAUGUGUU-3'; miR-590-5p inhibitor NC, 5'-GUCCAGUGAAUUCCCAG-3'; PDCD4 siRNA, 5'-GGAGGTGGATGTGAAAGAT-3'; and PDCD4 siRNA NC, 5'-GGCCTTGCGATAGCGTAGC-3'. At 48 h post-transfections, cells were used for subsequent experiments.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells or clinical tissue samples using TRIzol® reagent (cat. no. 15596026; Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using PrimeScript RT Master mix (Takara Biotechnology, Co., Ltd.) at 37˚C for 15 min followed by 85˚C for 5 sec. Subsequently, qPCR was performed using SYBR premix Ex Taq (Takara Biotechnology, Co., Ltd.) and a thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions used for qPCR were as follows: Initial denaturation 95˚C for 30 sec; followed by 40 cycles at 95˚C for 5 sec and 60˚C for 30 sec. The sequences of the primers used for qPCR were as follows: miR-590-5p forward, 5'-GAGCTTATTCATAAAAGT-3' and reverse, 5'-TCCACGACACGCACTGGATACGAC-3'; PDCD4 forward, 5'-GCAAAAAGGCGACTAAGGAAAAA-3' and reverse, 5'-TAAGGGCGTCACTCCCACT-3'; U6 forward, 5'-GTGCTCGCTTCGGCAGCACAT-3' and reverse, 5'-TACCTTGCGAAGTGCTTAAAC-3'; and GAPDH forward, 5'-TGTGGGCATCAATGGATTTGG-3' and reverse, 5'-ACACCATGTATTCCGGGTCAAT-3'. miRNA and mRNA levels were quantified using the 2-ΔΔCq method (18) and normalized to the internal reference genes U6 and GAPDH, respectively.
Cell viability analysis
At 48 h post-transfection, the effect of miR-590-5p on cell viability was measured using an MTT assay. Briefly, cells were washed with PBS (pH 7.4), harvested by trypsinization and seeded (5x105 cells/well) into a 96-well plate. The plate was incubated in humidified incubator at 37˚C with 5% CO2. Subsequently, 10 µl MTT solution was added to each well and incubated for 1 to 4 h at 37˚C. DMSO was added to each well to dissolve the formazan crystals. The absorbance of each well was measured at a wavelength of 490 nm using a microplate reader.
Wound healing assay
HCT116 and SW480 cells were seeded (8x105 cells/well) into 6-well plates. At 100% confluence, 8 µg/ml mitomycin C (Beyotime Institute of Biotechnology) was added to each well for 3 h at 37˚C to inhibit cell proliferation. Subsequently, the cell monolayer was scratched with a 10 µl pipette tip and incubated in RPMI-1640 medium supplemented with 1% FBS at 37˚C for 24 h. Images of the wounds were captured using an inverted light microscope at 0 and 24 h (magnification, x200).
Transwell assay
The transwell plates were pre-coated with Matrigel® at room temperature for 30 min. Cells were seeded (5x104 cells/well) into the upper chamber of the Transwell plate (24-well plates; Corning, Inc.) with serum-free RPMI-1640 medium. The lower chamber was treated with RPMI-1640 medium containing 10% FBS Cells were incubated at 37˚C for 24 h. Invading cells were fixed with 100% methanol for 20 min at room temperature, stained with crystal violet for 10 min at room temperature and visualized under a light microscope (magnification, x200).
Western blotting
Total protein was isolated from cells using a protease inhibitor cocktail (RIPA; Beyotime Institute of Biotechnology). Total protein was quantified using a bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Proteins (30 µg) were separated via 10% SDS-PAGE, transferred onto PVDF membranes and blocked with 5% non-fat dry milk in TBST (pH 7.4; 0.05% Tween 20) at room temperature for 2 h. The membranes were incubated at 4˚C overnight with the following primary antibodies: Anti-PCDC4 (cat. no. ab80590; 1:1,000), anti-p-Smad2/3 (cat. no. ab63399; 1:1,000), anti-TGF-β (cat. no. ab92486; 1:1,000) and anti-GAPDH (cat. no. ab9485; 1:2,000). Following primary antibody incubation, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (cat. no. ab6721; 1:5,000) at room temperature for 45 min. All antibodies were purchased from Abcam. Proteins were visualized using the Pierce™ ECL Western Blotting Substrate (Pierce; Thermo Fisher Scientific, Inc.) and chemiluminescence signals were detected using a Tanon-5200 Imaging system (Tanon Science and Technology, Co., Ltd.). Protein expression levels were quantified using ImageJ software (version 1.47; National Institutes of Health) with GAPDH as the loading control.
Dual-luciferase reporter assay
Wild-type PDCD4 3'-UTR (PDCD4-3'UTR), containing the miR-590-5p binding site, and mutant PDCD4 3'UTR (PDCD4-MUT) were cloned into the p-MIR-reporter plasmid (Thermo Fisher Scientific, Inc.). 293T cells (Cell Bank of Type Culture Collection of the Chinese Academy of Sciences) were seeded onto 6 well plates (8x105 cells/well), co-transfected with PDCD4-3'UTR or PDCD4-MUT (100 pmol) and miR-590-5p mimic or miR-590-5p mimic NC (each, 50 pmol) using Lipofectamine® RNAi Max (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, luciferase activities were detected using the Dual Luciferase Reporter Assay kit (Beyotime Institute of Biotechnology). Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
All experiments were repeated at least three times. Statistical analyses were performed using SPSS software (version 17.0; SPSS, Inc.). Data are presented as the mean ± standard deviation. Comparisons between two groups were analyzed using the paired Student's t-test. Comparisons among >2 groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-590-5p expression is increased and is negatively associated with PDCD4 in human CRC tissues
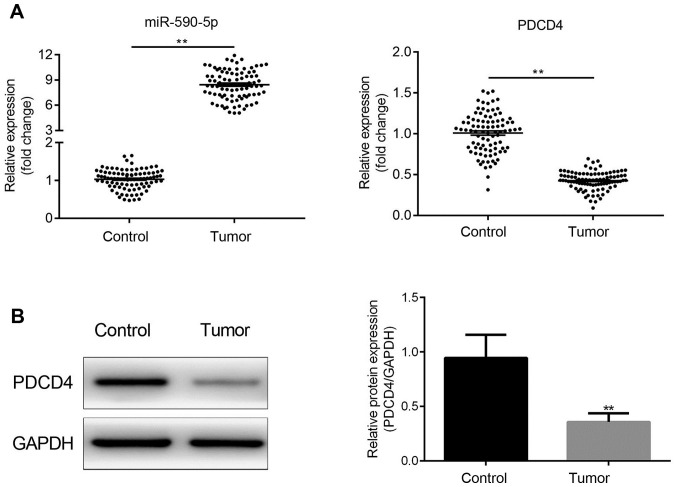
miR-590-5p and PDCD4 expression levels were detected in all 30 paired samples of human CRC and adjacent healthy tissues. The results indicated that miR-590-5p expression levels were significantly increased in CRC tissues compared with adjacent healthy tissues (P<0.01; Fig. 1A). By contrast, PDCD4 expression levels were significantly decreased in CRC tissues compared with adjacent healthy tissues (P<0.01; Fig. 1A and B).
Figure 1.
miR-590-5p is upregulated and negatively associated with PDCD4 in CRC tissues. (A) miR-590-5p and PDCD4 mRNA expression levels in human CRC and healthy adjacent tissue samples. **P<0.01. (B) PDCD4 protein expression levels were determined by western blotting. **P<0.01. miR, microRNA; PDCD4, programmed cell death 4 programmed cell death 4; CRC, colorectal cancer.
miR-590-5p inhibitor decreases human CRC HCT116 and SW480 cell viability
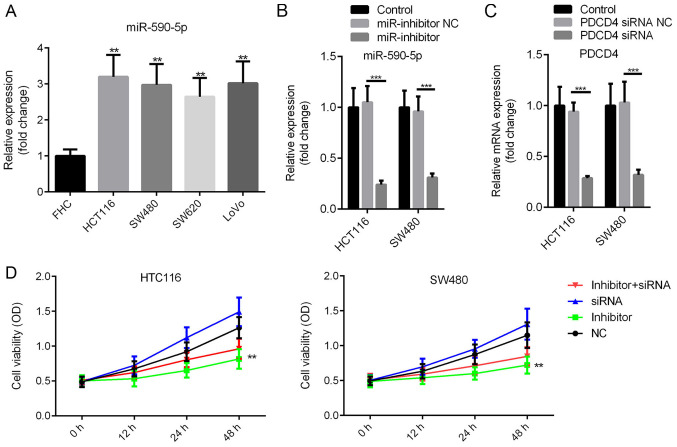
miR-590-5p expression levels in CRC cells and human normal colon epithelial cells (FHC) were compared. miR-590-5p expression levels were significantly increased in CRC cells compared with FHC cells (P<0.01; Fig. 2A). miR-590-5p inhibitor or miR-590-5p inhibitor NC were transfected into HCT116 and SW480 cells. The MTT assay was conducted to assess the effect of miR-590-5p on cell viability. miR-590-5p inhibitor and PDCD4 siRNA significantly decreased the expression levels of miR-590-5p and PDCD4, respectively, in HCT116 and SW480 cells (P<0.001; Fig. 2B and C). Moreover, compared with the NC group, miR-590-5p inhibitor decreased cell viability at 12 and 24 h post-transfection, and significantly decreased cell viability at 48 h post-transfection (Fig. 2D; P<0.01). Additionally, co-transfection of miR-590-5p inhibitor and PDCD4 siRNA partially reversed the effects of miR-590-5p inhibitor on cell viability (Fig. 2D).
Figure 2.
miR-590-5p inhibitor suppresses HCT116 and SW480 cell viability. (A) miR-590-5p expression levels in CRC cell lines and normal human normal colon epithelial cells (FHC). Transfection efficiency of (B) miR-590-5p inhibitor and (C) PDCD4 siRNA. (D) The MTT assay was performed to examine cell viability. **P<0.01 vs. control. ***P<0.001 as indicated. miR, microRNA; CRC, colorectal cancer; PDCD4, programmed cell death 4 programmed cell death 4; siRNA, small interfering RNA; NC, negative control; OD, optical density.
miR-590-5p inhibitor suppresses human CRC HCT116 and SW480 cell migration and invasion
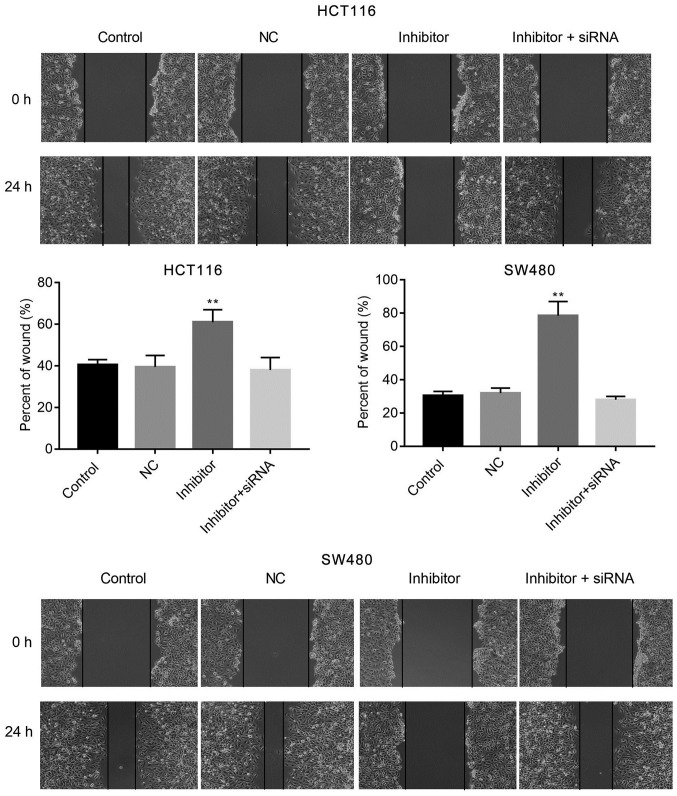
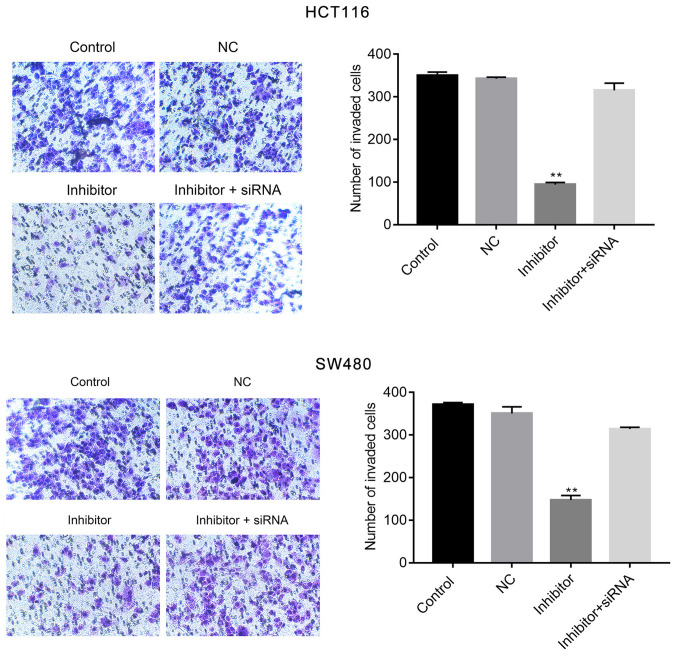
The effects of miR-590-5p inhibitor on cell migration and invasion were examined by conducting wound healing and Transwell assays, respectively. miR-590-5p inhibitor significantly increased cell migration at 24 h post-transfection (P<0.01; Fig. 3) when compared with the control. Moreover, the Transwell assay indicated that miR-590-5p inhibitor significantly reduced HCT116 and SW480 cell invasion compared with the control (P<0.01; Fig. 4). Additionally, co-transfection of miR-590-5p inhibitor and PDCD4 siRNA reversed the effects of miR-590-5p inhibitor on HCT116 and SW480 cell migration and invasion (Figs. 3 and 4).
Figure 3.
miR-590-5p inhibitor suppresses HCT116 and SW480 cell migration. The wound healing assay was conducted to examine cell migration. Magnification, x200. **P<0.01 vs. control. miR-590-5p, microRNA-590-5p; NC, negative control; siRNA, small interfering RNA.
Figure 4.
miR-590-5p inhibitor suppresses HCT116 and SW480 cell invasion. The Transwell assay was performed to examine cell migration in each group. Magnification, x200. **P<0.01 vs. control. miR-590-5p, microRNA-590-5p; NC, negative control; siRNA, small interfering RNA.
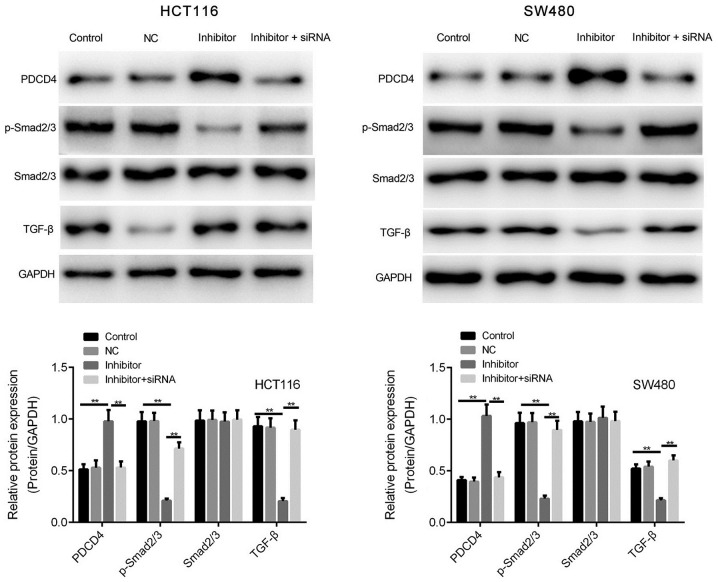
miR-590-5p inhibitor enhances the expression of PDCD4 and TGF-β/Smad2/3 signaling
Based on the association between miR-590-5p and PDCD4 expression observed in human CRC tissues, HCT116 and SW480 cells were transfected with miR-590-5p inhibitor to investigate whether PDCD4 expression was enhanced by miR-590-5p inhibitor. miR-590-5p inhibitor significantly increased PDCD4 protein expression levels compared with the control group in HCT116 and SW480 cells (P<0.01; Fig. 5).
Figure 5.
miR-590-5p inhibitor increases the expression of PDCD4, TGF-β and p-Smad2/3 in HCT116 and SW480 cells. PDCD4, p-Smad2/3, Smad2/3 and TGF-β protein expression levels were determined by western blotting. **P<0.01. miR-590-5p, microRNA-590-5p; PDCD4, programmed cell death 4 programmed cell death 4; TGF-β, transforming growth factor-β; p-Smad2/3, phosphorylated-Smad2/3; NC, negative control; siRNA, small interfering RNA.
The effect of miR-590-5p on the phosphorylation of Smad2/3 was investigated. Compared with the control group, miR-590-5p inhibitor significantly decreased the expression levels of TGF-β and p-Smad2/3 (P<0.01; Fig. 5). However, co-transfection of miR-590-5p inhibitor and PDCD4 siRNA significantly reversed the effects of miR-590-5p inhibitor on p-Smad2/3 expression levels (P<0.01; Fig. 5).
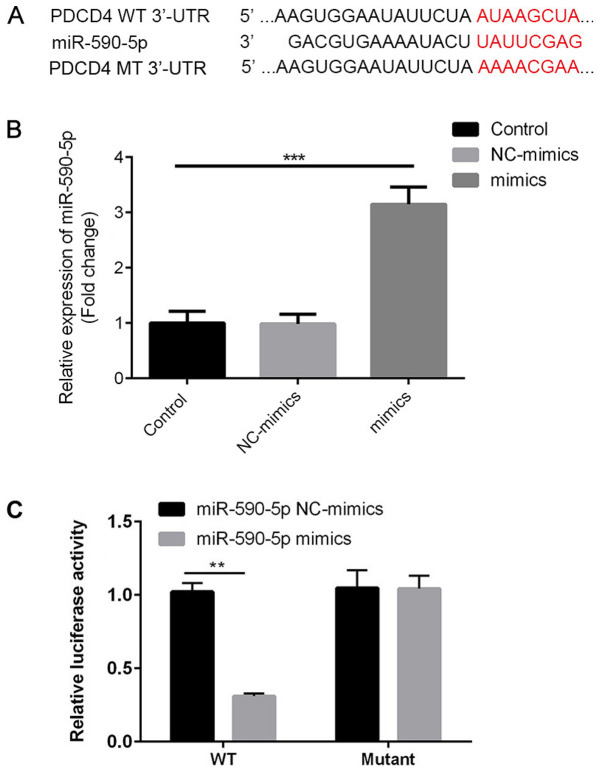
PDCD4 is a direct target of miR-590-5p
The present study investigated whether the observed reduction in PDCD4 was caused by direct binding of miR-590-5p to the 3'UTR of PDCD4. Therefore, PDCD4-3'UTR containing the putative binding site for miR-590-5p and the mutated segment were cloned in a firefly luciferase reporter vector (Fig. 6A). miR-590-5p mimic significantly increased the expression levels of miR-590-5p in 293T cells compared with the control group (Fig. 6B). miR-590-5p mimic significantly decreased the luciferase activity of PDCD4-3'UTR WT compared with miR-590-5p mimic NC (P<0.01), but did not significantly alter the luciferase activity of PDCD4-3'UTR MUT (Fig. 6C). The results suggested that miR-590-5p directly bound to PDCD4-3'UTR, resulting in reduced PDCD4 protein expression levels.
Figure 6.
PDCD4 is a direct target of miR-590-5p. (A) The binding site between miR-590-5p, PDCD4-3'UTR and PDCD4-MUT. (B) Transfection efficiency of miR-590-5p mimic. ***P<0.001. (C) Luciferase activities of PDCD4-3'UTR and PDCD4-MUT. **P<0.01. PDCD4, programmed cell death 4; miR-590-5p, microRNA-590-5p; 3'UTR, 3'-untranslated region; MUT, mutant; NC, negative control; WT, wild-type.
Discussion
miRNAs display great potential for the diagnosis and treatment of CRC (19-21). miR-590-5p has been reported to be strongly associated with CRC progression (17,22). The present study indicated that miR-590-5p expression levels were significantly increased in human CRC tissues and cells compared with healthy adjacent tissues and FHC cells, suggesting that miR-590-5p could function as an oncogene.
The present study investigated the roles of miR-590-5p in CRC pathogenesis by performing in vitro studies in human CRC HCT116 and SW480 cells. Cell proliferation and metastasis are important events associated with tumor progression (23,24). The results of the present study suggested that miR-590-5p inhibitor reduced HCT116 and SW480 cell viability compared with the control. Furthermore, when compared with the control group, miR-590-5p inhibitor decreased cell migration and invasion in vitro, which was consistent with the results obtained in a previous study (17). Of note, miR-590-5p inhibitor suppressed the TGF-β/Smad2/3 signaling pathway, which regulates CRC cell viability and migration (23,25,26). Collectively, the results indicated that miR-590-5p knockdown inhibited the TGF-β/Smad2/3 signaling pathway in HCT116 and SW480 cells.
Bioinformatics analysis identified PDCD4 as one of the direct targets of miR-590-5p. PDCD4 expression levels were significantly lower in human CRC tissues compared with healthy adjacent tissues, which was consistent with previous studies (27-30). Therefore, it was hypothesized that miRNA-590-5p may directly negatively regulate the expression of PDCD4. To investigate the hypothesis, the present study investigated the effects of miR-590-5p inhibitor in HCT116 and SW480 cells. The results suggested that miR-590-5p inversely regulated the expression of PDCD4. Subsequently, the direct binding of miR-590-5p to the PDCD4-3'UTR was confirmed by conducting a dual-luciferase reporter assay. The results indicated that co-transfection of miR-590-5p inhibitor and PDCD4 siRNA partially reversed the antitumor effects of miR-590-5p inhibitor. The results suggested that miR-590-5p may exert its carcinogenic behaviors by downregulating the expression of PDCD4.
Collectively, the results of the present study suggested that miR-590-5p increased CRC cell viability and migration, and negatively regulated the expression of PDCD4 by directly binding to it. The results suggested that miR-590-5p may serve an important role in CRC pathogenesis and serve as a novel therapeutic strategy for CRC.
Acknowledgements
Not applicable.
Funding
The present work was funded by Taizhou High Level Talents Research Project in 2015 (grant no. 201529).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TG and JW performed the majority of experiments and wrote the manuscript. GC performed the remaining experiments and statistically analyzed the data. HH designed the present study, provided funding and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethical Review Board of Taizhou People's Hospital and written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jacobsen F, Kraft J, Schroeder C, Hube-Magg C, Kluth M, Lang DS, Simon R, Sauter G, Izbicki JR, Clauditz TS, et al. Up-regulation of biglycan is associated with poor prognosis and PTEN deletion in patients with prostate cancer. Neoplasia. 2017;19:707–715. doi: 10.1016/j.neo.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson JC, Levine JB. Age and crc risk in the serrated pathway. J Clin Gastroenterol. 2018;52:465–467. doi: 10.1097/MCG.0000000000001051. [DOI] [PubMed] [Google Scholar]
- 3.Wang G, Fu Y, Hu F, Lan J, Xu F, Yang X, Luo X, Wang J, Hu J. Loss of BRG1 induces CRC cell senescence by regulating p53/p21 pathway. Cell Death Dis. 2017;8(e2607) doi: 10.1038/cddis.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orang AV, Barzegari A. MicroRNAs in colorectal cancer: From diagnosis to targeted therapy. Asian Pac J Cancer Prev. 2014;15:6989–6999. doi: 10.7314/apjcp.2014.15.17.6989. [DOI] [PubMed] [Google Scholar]
- 5.Sun ZQ, Shi K, Zhou QB, Zeng XY, Liu J, Yang SX, Wang QS, Li Z, Wang GX, Song JM, et al. MiR-590-3p promotes proliferation and metastasis of colorectal cancer via Hippo pathway. Oncotarget. 2017;8:58061–58071. doi: 10.18632/oncotarget.19487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen M, Li D, Gong N, Wu H, Su C, Xie C, Xiang H, Lin C, Li X. miR-133b down-regulates ABCC1 and enhances the sensitivity of CRC to anti-tumor drugs. Oncotarget. 2017;8:52983–52994. doi: 10.18632/oncotarget.17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guglielmo A, Staropoli N, Giancotti M, Mauro M. Personalized medicine in colorectal cancer diagnosis and treatment: A systematic review of health economic evaluations. Cost Eff Resour Alloc. 2018;16(2) doi: 10.1186/s12962-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker E, IJspeert JEG. Serrated pathway: A paradigm shift in CRC prevention. Gut. 2018;67:1751–1752. doi: 10.1136/gutjnl-2017-314290. [DOI] [PubMed] [Google Scholar]
- 9.Park YR, Lee ST, Kim SL, Zhu SM, Lee MR, Kim SH, Kim IH, Lee SO, Seo SY, Kim SW. Down-regulation of miR-9 promotes epithelial mesenchymal transition via regulating anoctamin-1 (ANO1) in CRC cells. Cancer Genet. 2019;231-232:22–31. doi: 10.1016/j.cancergen.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Li J, Huang Y, Peng W, Qian W, Gu J, Wang Q, Hu T, Ji D, Ji B, et al. Upregulated miR-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2F8 in CRC. Cell Prolif. 2018;51(e12505) doi: 10.1111/cpr.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong F, Ying Y, Pan H, Zhao W, Li H, Zhan X. MicroRNA-466 (miR-466) functions as a tumor suppressor and prognostic factor in colorectal cancer (CRC) Bosn J Basic Med Sci. 2018;18:252–259. doi: 10.17305/bjbms.2018.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Cui M. MiRNA-98-5p inhibits the progression of osteosarcoma by regulating cell cycle via targeting CDC25A expression. Eur Rev Med Pharmacol Sci. 2019;23:9793–9802. doi: 10.26355/eurrev_201911_19542. [DOI] [PubMed] [Google Scholar]
- 13.Ling Z, Guan H, You Z, Wang C, Hu L, Zhang L, Wang Y, Chen S, Xu B, Chen M. Aloperine executes antitumor effects through the induction of apoptosis and cell cycle arrest in prostate cancer in vitro and in vivo. Onco Targets Ther. 2018;11:2735–2743. doi: 10.2147/OTT.S165262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. doi: 10.1080/09168451.2020.1783197. Wang X, Shi J, Niu Z, Wang J and Zhang W: MiR-216a-3p regulates the proliferation, apoptosis, migration, and invasion of lung cancer cells via targeting COPB2. Biosci Biotechnol Biochem: Jul 3, 2020 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 15. doi: 10.1007/s12094-020-02436-1. Gao ZY, Liu H and Zhang Z: miR-144-3p increases radiosensibility of gastric cancer cells by targeting inhibition of ZEB1. Clin Transl Oncol: Jul 1, 2020 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Wang Y, Xu M, Zhang L, Su Y, Wang B, Zhang X. miR-1184 regulates the proliferation and apoptosis of colon cancer cells via targeting CSNK2A1. Mol Cell Probes. 2020;101625 doi: 10.1016/j.mcp.2020.101625. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 17.Zhou Q, Zhu Y, Wei X, Zhou J, Chang L, Sui H, Han Y, Piao D, Sha R, Bai Y. MiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor A axis. Cell Death Dis. 2016;7(e2413) doi: 10.1038/cddis.2016.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Yang W, Luo Y, Hu S, Zhu L. Correlation between miR-21 and miR-145 and the incidence and prognosis of colorectal cancer. J BUON. 2018;23:29–35. [PubMed] [Google Scholar]
- 20.Hu JL, He GY, Lan XL, Zeng ZC, Guan J, Ding Y, Qian XL, Liao WT, Ding YQ, Liang L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7(16) doi: 10.1038/s41389-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao D, Li T, Ye C, Zeng L, Li H, Pu X, Ding C, He Z, Huang GL. miR-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Exp Ther Med. 2018;15:1712–1717. doi: 10.3892/etm.2017.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou C, Sun Z, Li X, Li X, Ren W, Qin Z, Zhang X, Yuan W, Wang J, Yu W, et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53–63. doi: 10.1016/j.canlet.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Lai Q, He J, Li Q, Ding J, Lan Z, Gu C, Yan Q, Fang Y, Zhao X, Liu S. LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-β/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1. Int J Med Sci. 2019;16:51–59. doi: 10.7150/ijms.27359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng H, Wang L, Su Q, Yi K, Du J, Wang Z. MiR-31-5p promotes the cell growth, migration and invasion of colorectal cancer cells by targeting NUMB. Biomed Pharmacother. 2018;109:208–216. doi: 10.1016/j.biopha.2018.10.048. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Z, Cao Q, Dai G, Wang J, Liu C, Lv L, Pan J. Celastrol inhibits colorectal cancer through TGF-beta1/Smad signaling. Onco Targets Ther. 2019;12:509–518. doi: 10.2147/OTT.S187817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y, Chen W, Yang H, Yan Z, Lai Z, Feng J, Peng J, Lin J. Scutellaria barbata D. Don inhibits migration and invasion of colorectal cancer cells via suppression of PI3K/AKT and TGF-β/Smad signaling pathways. Exp Ther Med. 2017;14:5527–5534. doi: 10.3892/etm.2017.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Uzair-Ur-Rehman Guo Y, Liang H, Cheng R, Yang F, Hong Y, Zhao C, Liu M, Yu M, et al. miR-181b functions as an oncomiR in colorectal cancer by targeting PDCD4. Protein Cell. 2016;7:722–734. doi: 10.1007/s13238-016-0313-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YR, Chen SH, Lin CY, Chao WY, Lim YP, Yu HI, Lu CH. In vitro antitumor activity of aloperine on human thyroid cancer cells through caspase-dependent apoptosis. Int J Mol Sci. 2018;19(312) doi: 10.3390/ijms19010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Zhao M, Guo C, Wang G, Zhu F, Wang J, Wang X, Wang Q, Zhao W, Shi Y, et al. PDCD4 deficiency aggravated colitis and Colitis-associated colorectal cancer via promoting IL-6/STAT3 pathway in mice. Inflamm Bowel Dis. 2016;22:1107–1118. doi: 10.1097/MIB.0000000000000729. [DOI] [PubMed] [Google Scholar]
- 30.Lim SC, Hong R. Programmed cell death 4 (Pdcd4) expression in colorectal adenocarcinoma: Association with clinical stage. Oncol Lett. 2011;2:1053–1057. doi: 10.3892/ol.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.