Abstract
Cancers induced by human papillomaviruses should, in principle, be responsive to immunotherapy by virtue of expressing the immunogenic oncoproteins E6/E7 that drive malignancy. Rather, advanced forms of cervical cancer are poorly responsive to immune response-enhancing treatments involving therapeutic vaccination against these viral neo-antigens. Leveraging a transgenic mouse model for HPV-derived cancers, termed K14HPV16/H2b, we demonstrate that a potent nanoparticle-based E7 vaccine but not a conventional ‘liquid’ vaccine can induce E7 tumor-antigen-specific CD8+ T cells in cervical tumor-bearing mice. However, vaccination alone or in combination with anti-PD-1/anti-CTLA4 does not elicit tumor regression nor increase the minimal CD8+ T cell infiltrates in the tumor microenvironment (TME), suggesting the presence of immunosuppressive barriers. It has been reported that cervical cancer patients characteristically have poor dendritic cell functions, weak cytotoxic lymphocyte responses, and evidence an accumulation of myeloid cells in the periphery. Here we illustrate that myeloid cells in K14HPV16/H2b mice possess potent immunosuppressive activity for both antigen presenting cells and CD8+ T cells that dampens the anti-tumor immune responsiveness. These immune-inhibitory effects demonstrably overrule the otherwise synergistic effects of combining the oncoprotein vaccine with immune checkpoint blocking antibodies. Our data highlight a link between HPV-induced cancers, systemic amplification of myeloid cells, and the detrimental effects of these cells on CD8+ T cell activation and recruitment into the TME. The results establish immunosuppressive myeloid cells in lymphoid organs as an HPV+ cancer-induced means of circumventing tumor immunity that will require targeted abrogation to enable the induction of efficacious anti-tumor immune responses.
Keywords: Human papillomavirus, Cervical cancer, Cancer immunotherapy, Myeloid cells, Systemic immunosuppression
Introduction
Oncogenic Human Papillomaviruses (HPVs) are the etiological cause of various malignancies, including cervical cancer and oral/head & neck cancer (1). Cancer progression in infected cells is driven by the viral proteins E6 and E7 that interfere with the activity of the p53 and pRB tumor suppressors (2, 3) eventually leading to malignancy. The vast majority of HPV infections are eradicated by the immune system (4). However, in a small percentage of infected individuals, the virus persists and elicits pre-malignant proliferative lesions - intraepithelial neoplasias - (5) that, if not eliminated by a natural immune response or therapeutic intervention, can eventually evolve to invasive squamous cell carcinomas (6–8).
Immunotherapy has recently taken center stage in cancer treatment (9), and immune checkpoint blocking antibodies are regularly receiving approval for new types of cancer (10). However, the fractions of non-responders to such therapies in most indications remain significant (11), and current efforts are focusing on understanding the underlining innate and acquired resistance mechanisms (10, 12).
Another complementary effort is focused on neo-antigen vaccines designed to stimulate tumor-specific immune responses, including in particular for HPV-induced malignancies, leveraging the E7 oncoprotein (13–17). Surprisingly, approaches based on immunization with E7 peptides have shown only modest success as monotherapy against invasive cancers (14, 16). Combinations with other treatments have shown more encouraging results, but the percentage of responding patients remains low (17).
It has been reported that cervical cancer patients exhibit systemic alterations in their immune system - beyond the tumor microenvironment (TME) - including low numbers of circulating dendritic cells (DC), poor antigen-presenting cell (APC) functions, weak cytotoxic lymphocyte (CTL) responses, and accumulation of myeloid cells in the periphery (18–22). Collectively, these deficiencies may be pertinent to the poor efficacy of immunotherapy-based treatments, including therapeutic vaccination, against cervical carcinomas. Although some reports suggest that myeloid cells might be playing an immunosuppressive role in this disease (20, 22), their activity remains poorly characterized, and a comprehensive analysis of the systemic immune response following immunotherapy in the setting of HPV-related cancers is lacking. Moreover, the mechanisms responsible for the various immune-related defects observed in patients with HPV-induced malignancies remain obscure.
Herein we show that a genetically engineered mouse model (GEMM) of HPV-induced malignancies, K14HPV16/H2b, recapitulates the aforementioned alterations in immune cell functions and defects in anti-tumor immune responses, enabling investigation of its underlying basis, which has implicated systemic immunosuppression manifested by an over-accumulation of myeloid cells in lymphoid organs.
Materials and methods
Mice, Tumor Cells, and Antibody Treatments
For all experiments, K14HPV16/H2b mice or their FVBN/H2b littermates were used. A preliminary characterization of the K14HPV16/H2b model was reported previously (23) now substantiated by considerable further analyses presented in herein. The K14HPV16/H2b line was generated by crossing K14HPV16/FVBN (H2q) mice (24, 25) with C57BL/6 (H2b) mice to introduce the entire H2b locus, followed by backcrossing to FVBN to render the mice congenic for H2b but otherwise genetically FVBN. F1 mice were backcrossed for 11 generations to FVBN, selecting for the H2b locus in every generation by flow cytometry analyses of H2Kb and H2Db. Afterwards, mice were intercrossed to generate homozygous H2b mice, expressing H2Kb and H2Db, but not H2Kq and H2D/Lq. This genetic configuration allows K14HPV16/H2b mice to present E7-derived peptides on MHCI molecules while maintaining the FVBN background that is permissive for squamous carcinogenesis. All the mice were bred in-house and kept under pathogen-free conditions at the animal facility of Ecole Polytechnique Fédérale de Lausanne. Females were subcutaneously implanted with estrogen-releasing pellets at age 1, 3, and 5 months. Five months old females or three months old males were used for the experiments. The TC-1 and SC-1 experiments were performed as previously described (26). All experiments were performed in accordance with Swiss law and with the approval of the Cantonal Veterinary Office of Canton de Vaud, Switzerland (Licenses VD2814 and VD3256). Antibodies and other treatment doses and frequency of treatment are summarized below (treatments were administered in PBS or saline):
Reagents
CpG-B 1826 oligonucleotide (5’-TCCATGAGCTTCCTGACGTT-3’ as phosphorothioated DNA bases) was purchased from Microsynth and used as adjuvant in various vaccine formulations or to activate BMDCs. The full length E7 protein was purchased from the Protein and Peptide Chemistry Facility, UNIL and used for NP conjugation and immunization. HPV16 E7 long peptide (aa 43–77, purity>90%) was purchased from Think Peptides and the Protein and Peptide Chemistry Facility, UNIL and used for NP conjugation, immunization and restimulation of CD4 T cells. The HPV16 E7 CD8 peptide RAHYNIVTF was purchased from Think Peptides and used for restimulation. OVA used for immunization and the OVA CD8 peptide SIINFEKL used for restimulation were provided by Prof. Melody Swartz and Prof. Jeffrey Hubbell labs. The LCMV CD8 peptide KAVYNFATC used for both immunization and restimulation was purchased from the University of Lausanne (UNIL). Ultrapure LPS, used for BMDCs activation was purchased by InvivoGen.
Immunization
Mice were immunized as stated in the text with a total amount 15 μg of E7LP or 40 μg of full length E7 protein, either unconjugated (“free”) or in the NP-bound form, and 40μg of CpG was used as adjuvant. The NP formulations were prepared as indicated below. For the unconjugated formulation, the E7 protein or E7LP were first dissolved in DMSO and then diluted in PBS prior to immunization. The DC-vaccine was prepared as described below. Non-immunized mice were treated with PBS. All the mice from one experiment were immunized together on the same day. Mice received either 1 or 2 shots of vaccine as indicated in the text. Subcutaneous (s.c.) immunizations of mice were performed in the four limbs using the Hock method. Intravenous (I.V.) immunizations were performed in the tail vein.
Nanoparticle (NP) synthesis and Conjugation
NPs were synthesized, functionalized and characterized as previously described (26). For antigen conjugation, the HPV16 E7 long peptide or the full length E7 protein were dissolved in DMSO and incubated for 12h in endotoxin-free water in the presence of NPs and guanidine hydrochloride (AppliChem) at room temperature. NP-E7LP was purified by size-exclusion chromatography using CL-6B matrix (Sigma-Aldrich), eluted and stored in PBS at room temperature. The size of NP before and after conjugation was determined by dynamic light scattering and remained around 30 nm. E7LP loading on the nanoparticles was measured by BCA assay (Thermo Fisher Scientific). NPs alone have been shown to have no adjuvant activity.
Measurements of cervical tumors size
Serial 10 μm sections of the OCT-embedded cervices were taken and stained with H&E. An image of the whole tissue was taken every 100 μm. Tumor area on each of the imaged sections was quantified manually. Tumor volume (V) was calculated using the following formula V = 2/3 × A × Z, where A is the maximal tumor area measured on a section and Z is the depth of tumor determined by the number of sections containing tumors.
Bone marrow dendritic cells (BMDCs) preparation
Femurs and tibiae of 4–6 weeks old mice were collected and cleaned. The bone marrow was extracted by flushing with a syringe, filtered and plated in RPMI medium (Gibco), 10% FBS (Gibco), P/S (100Units/ml penicillin, 100 μg/ml streptomycin; Gibco), 50 μM Beta-mercaptoethanol (Gibco) and 20 ng/ml murine GM-CSF (Peprotech). New media was added at day 3, and half of it was replaced at day 6 (the cells contained in the replaced media were recovered and put back in culture). At day 8 the BMDCs in suspension were collected and used for the different assays.
DC vaccine (DC-VAX) preparation
BMDCs were collected at day 8 of culture as previously illustrated. Cells were incubated with E7LP for 3h at 37°C for antigen loading. E7LP-loaded BMDCs were activated with 20 ng/ml LPS for 12h. After the activation, the cells were extensively washed to remove the excess of LPS, harvested, and mixed with CpG. For certain experiments, when stated in the text, BMDCs were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE, 2μM) prior to mixing with CpG. Each mouse received a total of 2 million BMDCs and 40 μg of CpG either s.c. or I.V. as indicated.
Co-culture experiments with BMDCs
BMDCs were collected at day 8 of culture as previously illustrated. Cells were activated for 12h with either LPS (20 ng/ml) or CpG (0,1 μM) as indicated in the text. After the activation, the cells were extensively washed to remove the excess of LPS and CpG and plated together with Tregs, CD4+ non Treg cells or CD11b+ cells isolated from K14HPV16/H2b or FVBN/H2b mice as indicated. The ratio of the co-culture was 1 BMDC to 10 other cells. BMDCs were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE, 2μM) prior to mixing with CD11b+ cells. After 24h of co-culture, cells were harvested and processed for Flow cytometry analysis.
Cell preparation for flow cytometry, antigen-specific in-vitro restimulation and magnetic isolation
Spleens or lymph nodes were harvested and gently disrupted through a 40-μm filter (Fisher Scientific). Red blood cells were lysed using ACK lysis buffer (not for magnetic isolation), and cells were filtered again through a 40-μm filter before use. For CD8+ T cell antigen-specific restimulation, cell suspensions from either spleen or lymph nodes were cultured at 37°C for 6h in a 96 well plate in the presence of 1μg/ml of the HPV16 CD8 peptide RAHYNIVTF. After the first 3h of culture, brefeldin A (Sigma-Aldrich) was added to a final concentration of 5 μg/ml. All the cells were cultured in IMDM medium (Gibco) supplemented with 10% FBS (Gibco) and 1x penicillin/streptomycin (100 Units/ml penicillin, 100 μg/ml streptomycin; Gibco).
Co-culture experiments with CD8+ T cells
CD8+ T cells were isolated from naïve mice or from mice that were previously immunized against E7 as stated in the text. CD8+ T cells isolation was performed on the spleen using the Easysep Mouse CD8+ T cell Isolation Kit (Stemcell Technologies) for magnetic isolation according to the manufacturer’s instructions. CD8+ T cells purity was above 90% in all the experiments. For proliferation assays, CD8+ T cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE, 2μM) and added to a plate that has previously been incubated for 12h with 2 μg/ml anti-CD3/anti-CD28 in PBS at 4°C. CD8+ T cells were harvested 48h later, stained with a live-dead dye and analyzed by flow cytometry to assess their proliferation. For restimulation experiments, isolated CD8+ T cells were added to a plate, and the assay was performed as previously illustrated. When indicated, CD8+ T cells were co-cultured with other cell types in a 1 CD8 to 10 other cell ratio.
Cell isolations for co-culture experiments and neutrophils sorting
In order to obtain enough cells to perform the various assays, cell isolations were performed on the spleen. Tregs were isolated using a double step magnetic isolation CD4+CD25+ Regulatory T Cell Isolation Kit (Miltenyi). Cells purity was around 70% in all the experiments. CD11b+ cells were isolated using the magnetic isolation positive selection Easysep Mouse CD11b positive selection kit II (Stemcell Technologies). Collectively, monocytes, neutrophils, macrophages and DCs accounted for more than 80% of the isolated cells. Monocytes were isolated using the magnetic isolation negative selection Easysep Mouse Monocyte Isolation Kit (Stemcell Technologies), purity was higher than 80%. Macrophages were isolated using the magnetic isolation Easysep PE positive selection kit with an F4/80 PE antibody (clone BM8, eBioscience), purity was higher than 90%. All the procedures were carried out according to the manufacturer’s instructions. CD11b+Ly6G+Ly6C− Neutrophils used for RNAseq were sorted using a MoFlo Astrios.
RNAseq on neutrophils
Live CD11b+Ly6G+Ly6C− neutrophils were FACS sorted, and total RNA was isolated with the miRNeasy micro kit (Qiagen). The quality and quantity of the RNA was determined by a Fragment Analyzer (Advanced Analytical). The RQN for the RNA ranged from 9.6 to 10. Library preparation and sequencing was performed at the Lausanne Genomic Technologies Facility (University of Lausanne, Switzerland). Double stranded cDNA for RNA-seq library preparation was generated using SMART-Seq v4 Ultra Low Input RNA reagents (Catalog Number 634888, Clontech, Mountain View, California, USA) according to the protocol provided with the reagents beginning with 4ng of total RNA and using 9 cycles of PCR. 300 pg of the resulting cDNA were used for library preparation with the Illumina Nextera XT DNA Library reagents (Catalog Number 15032354, Illumina, San Diego, California, USA) using the single cell RNA-seq library preparation protocol developed for the Fluidigm C1 (Fluidigm, South San Francisco, California, USA). Cluster generation was performed with the libraries using the Illumina SR Cluster Kit v4 reagents and sequenced on the Illumina HiSeq 2500 using SBS Kit v4 reagents. Sequencing data were demultiplexed using the bcl2fastq Conversion Software (v. 2.20, Illumina; San Diego, California, USA). Single 100 nucleotide long reads were mapped to mouse genome build mm10 using HISAT2 (v 2.0.3beta) aligner. Counts obtained with FeatureCounts (v 1.4.4) were normalized for library size using TMM method from EdgeR and voom from limma. Differential expression was computed with limma, after filtering out genes with average TPM (transcripts per kilobase million) < 1. Genes with significant FDR adjusted P-value < 0.05 were considered differentially expressed. Gene Set Enrichment Analysis (27) in K14HPV16/H2b-derived neutrophils versus FVBN/H2b-derived neutrophils was performed using gene sets in the Hallmark and KEGG collections of MSigDB v5.0 with 100’000 permutations and P-values were FDR adjusted. Gene sets with FDR < 0.1 were considered significant. All statistical analyses were performed using the free high-level interpreted statistical language R (version 3.5.1) and various Bioconductor packages (http://bioconductor.org). Data is publically available in GEO database (GSE126913)
Protein extractions and western blot
Popliteal and brachial lymph nodes were harvested from three months old naïve untreated mice. Lymph nodes were snap frozen upon collection. Lymph nodes were put in RIPA buffer with protease (cOmplete Mini, Roche) and phosphatase (PhosSTOP, Roche) inhibitors and mechanically lysed. CD11b+ cells were isolated from the spleen as previously described and the CD11b− splenocytes were also recovered from the kit. Cells were re-suspended in RIPA buffer. Samples were centrifuged at high speed, and the supernatant was collected and used for western blot. Protein concentration was determined by Bradford assay (BioRad) according to the manufacturer’s instructions and 10–20 μg of total protein were used for the western blots. The same amount of total protein was loaded in each well. The following antibodies were used: Beta-actin (cat. 4867S, Cell Signaling), G-CSF (clone EPR3203(N)(B), Abcam), GM-CSF (clone MP122E9, R&D), M-CSF (cat. AB99178, Abcam), IL10 (clone JES052A5, R&D), IDO (clone mIDO-48, Biolegend), IL1beta (cat. AB9722, Abcam), HSP90 (clone F-8, Santa Cruz), and C-reactive protein (CRP, cat. AF1829, R&D). Western blot bands were visualized on a Fusion Fx7 using WesternBright Sirius (advansta) and quantified using Fiji (ImageJ).
Reactive oxygen species (ROS) production assay
CD11b+ cells were isolated as previously described and cultured in a 96 wells plate for 1h at 37°C in complete RPMI. Media was then removed, and the ROS-reactive dye 2′,7′-Dichlorofluorescin diacetate (DCFDA, Sigma) was added at a final concentration of 10 μg/ml in PBS, and the cells were left for 30 min at 37°C. Phorbol 12-myristate 13-acetate (PMA) at a final concentration of 1 μg/ml was used as positive control for ROS production. At the end of the experiment, cells were harvested, stained with a live/dead dye and analyzed by flow cytometry.
Flow cytometry
For blocking and surface staining, cells were incubated for 15min on ice with the antibodies diluted in PBS 2% FBS. Before tetramer and antibody staining, all cell suspensions from tumor or spleen were blocked with anti CD16/32 (BioLegend). Cells were then labeled with fixable live/dead cell viability reagent (Invitrogen) diluted in PBS for 15 min on ice. Staining with a tetramer recognizing HPV16 E7 peptide 49–57 presented by H2Db (University of Lausanne, UNIL) was performed before antibody staining for 30min at room temperature. Tetramer staining for OVA-specific and LCMV-specific CD8+ T cells was performed similarly using tetramers recognizing the SIINFEKL (Proimmune) and KAVYNFATC (University of Lausanne, UNIL) peptide-MHC complexes. If no intracellular staining was performed after surface staining, cells were fixed with 2% PFA in PBS for 15min on ice. For intracellular staining, cells were permeabilized and fixed with the Foxp3/Transcription Factor Staining Buffer Set Kit (eBioscience) following the manufacturer instructions and then incubated overnight with the antibodies diluted in 1x Permeabilization buffer provided with the aforementioned kit. After staining, cells were washed and re-suspended in PBS 2% FBS for analyses. Samples were acquired on a Cyan, or Gallios analyzer (Beckman Coulter) and data were analyzed using FlowJo software (Tree Star Inc.). Antibodies used for flow cytometry: CD3 (clone 145–2C11, ThermoFisher), CD4 (clone RM4–5, BioLegend), CD8a (clone 5H10, ThermoFisher), B220 (clone RA3–6B2, ThermoFisher), IFNγ (clone XMG1.2, BioLegend), TNFα (clone MP6-XT22, ThermoFisher), Foxp3 (clone FJK-16s, ThermoFisher), CD25 (clone PC61.5, ThermoFisher), CD11b (clone M1/70, ThermoFisher), CD11c (clone N418, BioLegend), Ly6G (clone 1A8, BioLegend), Ly6C (clone HK1.4, ThermoFisher), MHCII (clone M5/114.15.2, BioLegend), F4/80 (clone BM8, eBioscience), CD80 (clone 16–10A1, Biolegend), CD86 (clone GL-1, Biolegend), MHCII (clone M5/114.15.2, Biolegend), CD40 (clone 3/23, BD), CD127 (clone A7R34, Biolegend), KLRG1 (clone 2F1/KLRG1, Biolegend), CD44 (clone IM7, eBioscience), CD62L (clone MEL-14, Biolegend), H-2Db (clone KH95, BD Biosciences), H-2Kb (clone AF6–88.5, BD Biosciences), H-2Kq (clone KH114, Biolegend), H-2Dq/H2Lq (clone KH117, BD Biosciences).
Immunofluorescence staining
Cervices were harvested, embedded in OCT (Sakura) and snap frozen on dry ice. 10 μm thick sections were cut from OCT-embedded samples using a cryostat and collected on Superfrost Plus glass slides (Thermo Scientific). Tissue sections and OCT embedded samples were stored at −80°C. For immunofluorescence staining, sections were fixed in ice-cold methanol (Fisher Scientific) for 10min before proceeding. Slides were washed with PBS to remove the remaining OCT and then blocked for 45min at room temperature with PBS + 5% BSA + 2.5% FBS and then stained with primary antibodies diluted in PBS + 1% BSA overnight at 4° in a humidified chamber. On the following day, slides were washed with PBS and stained with secondary antibodies diluted in PBS + 1% BSA for 1h at room temperature. Before mounting, slides were washed again in PBS and then covered with mounting media (Dako) containing DAPI (Roche, 5 μg/ml). Coverslips (Menzel Glaser) were applied to the slides and sealed using nail polish. Images were acquired using a Leica DM5500B or an Olympus Slide Scanner VSL120-L100 and processed using Fiji (ImageJ). Antibodies used for immunofluorescence staining: CD8 (clone 53–6.7, eBioscience), keratin 14 (clone poly19053, BioLegend), CD3 (clone 500A2, BD), PD-L1 (clone MIH5, eBioscience).
TCRβ deep sequencing of E7-specific CD8 T cells
Lymph nodes were processed as described above to obtain a single cell suspension. Cells were stained with APC tetramers recognizing HPV16 E7 peptide 49–57 presented by H2Db (University of Lausanne, UNIL) and E7 specific CD8 T cells were isolated using the EasySep APC positive selection kit II (Stemcell) following the manufacturer’s instructions. Total genomic DNA was extracted using the QIAamp DNA Micro Kit (QIAGEN). TCRβ sequencing was performed using the 2-stage PCR method as previously described (28). Briefly, the 1st stage involved amplifying the gDNA and the synthetic TCR template (internal controls) using a multiplex PCR with 20 Vβ forward and 13 Jβ reverse primers using Qiagen multiplex PCR kit. Using 2.0 % of purified PCR product from stage 1 as template, we performed a 2nd stage PCR with universal and indexed Illumina adaptors. TCRβ library was prepared by pooling equal volumes of the 2nd stage PCR products. The library was profiled on a 2200 TapeStation (Agilent) and the concentration was determined by real time PCR using a StepOne Real Time Workstation (ABI/Thermo) with a commercial library quantification kit (Kapa Biosystems). Using a Midoutput 300 sequencing kit on an Illumina NextSeq 500 platform, paired-end (2 × 150 bp) sequencing was performed with a target clustering of about 160 million clusters per run. Raw data were processed and clonotypes were identified from purified sequences using the MiXCR tool suite. Normalized clonotype counts were exported in tabular format for use in downstream analysis. TCR repertoire metrics, including clonality, maximum clonal frequency, and the Shannon diversity index were calculated using the methods described in Medler et al., (28).
Statistical analyses
Statistical analyses were performed in GraphPad Prism 7. Flow cytometry, tumor size and western blot quantification data were compared using Student t test. TCRβ deep sequencing results were compared using Mann-Whitney test. Statistical significance is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant.
Results
The K14HPV16/H2b mouse model
Notably, the previously described, prototypical K14HPV16 transgenic mouse model of cervical carcinogenesis (24, 25) had an immunological bias that qualified its utility as a relevant model to assess immunotherapeutic strategies: the model was generated in the FVBN genetic background that, while susceptible to the development of invasive squamous cell carcinomas in skin and cervix (24, 25), contained Class I MHC alleles that were incapable of presenting peptides from the HPV16 E7 and E6 oncoproteins to CD8 T cells (29). To rectify this bias, the line was rendered congenic for the entire H2b MHC, while retaining the FVBN background that conveys genetic susceptibility to squamous carcinogenesis. Similarly to the original K14HPV16 mouse (24, 25, 30, 31), this refined model expresses the early region of HPV16 under the human keratin 14 promoter (Fig. 1A) and, upon continuous estrogen treatment (Fig 1B), female mice develop de novo cervical carcinomas (Fig 1C) via progressive cervical intra-epithelial neoplasias that phenocopy the multi-stage progression implicated in human cervical carcinogenesis (31). Because of the widespread expression of HPV16 genes in basal keratinocytes of the skin, the prototypical K14HPV16 mice also develop skin lesions characterized by hyperkeratosis and dysplasia, and squamous cell carcinomas later in life (24, 25, 32), phenotypes that are also observed in K14HPV16/H2b mice. As shown in Fig 1D, the new line expresses the H2Db and H2Kb class I MHC molecules, as a result of introducing the H2b locus from the C57Bl/6 background.
Figure 1.
The K14HPV16/H2b mouse model. A) Schematic of the HPV16 transgene in K14HPV16/H2b mice, showing the human keratin 14 gene transcriptional regulatory region and the early region genes of HPV16. B) Timeline (in months) of cervical cancer development in K14HPV16/H2b mice. C) Representative histologic section of the cervix of a K14HPV16/H2b mouse (left) bearing squamous cell carcinoma (right). D) Representative flow cytometry analysis of H2Db, H2Kb, H2Kq and H2Dq/Lq expression on CD45+ cells in the blood of C57BL/6, FVBN and K14HPV16/H2b mice.
To assess the capability of this congenic line to mount a CD8+ T cell response toward HPV16 neoantigens, we immunized the mice against E7. As previously reported, antigen conjugation to 30-nm in diameter ultra-small nanoparticles (NP) can boost the efficacy of an anti-E7 therapeutic vaccine – compared to a classical unconjugated formulation - against transplantable tumors derived from two cell lines expressing the HPV16 E6-E7 oncoproteins, TC-1 and SC1, leading to long-term survival of a significant percentage of tumor-bearing mice (26). To assess the ability of K14HPV16/H2b mice to generate E7-specific immune responses, we immunized mice with the full length E7 protein. Five-month old K14HPV16/H2b female mice received the vaccine at day 0 and 14 and were sacrificed at day 19 to assess the immune response in lymph nodes. The unconjugated (‘liquid’) formulation was unable to elicit a measurable response in transgenic mice, whereas the NP-E7 vaccine produced detectable albeit modest numbers of E7-specific CD8 T cells (Fig S1). No spontaneous response was evident in unvaccinated mice. Thus E7-specific CD8+ T cells can be generated in K14HPV16/H2b mice.
Immunotherapy is ineffective in K14HPV16/H2b mice
We next assessed the therapeutic efficacy of the NP vaccine alone or in combination with checkpoint-blocking antibodies against cervical tumors in K14HPV16/H2b mice. In these and subsequent experiments, we switched to a vaccine formulation containing an E7-synthetic long peptide (E7LP) conjugated to nanoparticles (NP-E7LP) (26), instead of the full length E7 protein, so as to model published and ongoing studies and clinical trials using similar formulations involving this particular peptide(13–17).
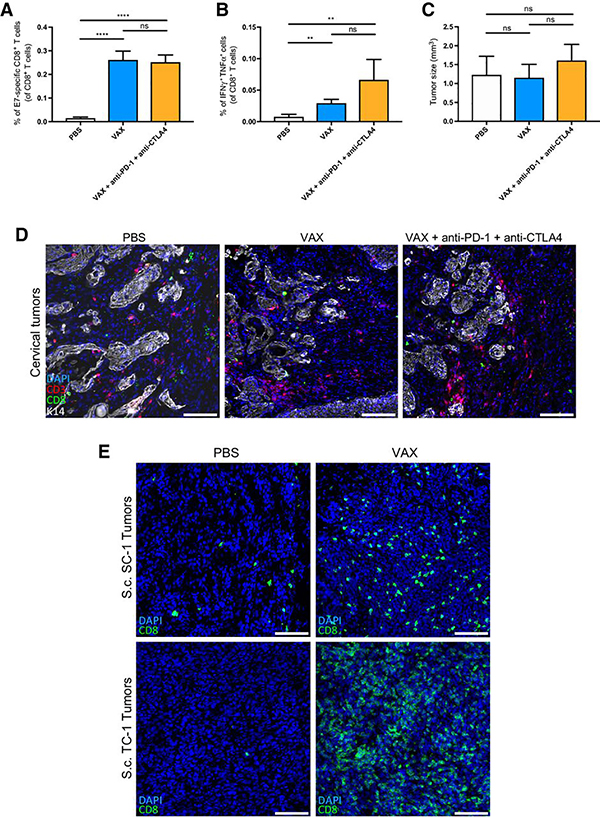
Five-month old female mice with cervical carcinomas were immunized with the therapeutic vaccine alone or in combination with anti-PD-1 (given that PD-L1 expression was detected on cervical tumors, Fig S2) and anti-CTLA4. Antibody treatment started on day 0 concomitant with the vaccination. Flow cytometry analysis of the immune response was performed 24 days later, which revealed that the checkpoint-blocking antibodies were unable to increase the abundance of E7-specific CD8+ T cells (Fig 2A). Upon ex-vivo restimulation, cytokine production was also similar between the two treatment groups (Fig 2B). In addition, neither treatment had an impact on the tumor size (Fig 2C) and we observed no obvious changes in CD3+ or CD8+ T cell infiltrates in the TME of the cervix (Fig 2D (representative field) and Fig S3 (entire cervical tumor). These results stand in marked contrast to those seen previously in mice bearing transplanted SC-1 and TC-1 tumors, in which CD8+ T cell infiltrates were robustly increased upon vaccination with NP-E7LP (Fig 2E, and (26)). This dichotomy implicates immunosuppressive barriers in K14HPV16/H2b mice capable of blocking the effects of the oncoprotein vaccine, alone and in combination with immune checkpoint inhibitors. Similarly to what has been observed in patients (14, 17, 33), these data reveal that immunotherapy is poorly effective against de novo formed tumors present in K14HPV16/H2b mice.
Figure 2.
Immunotherapy has no anti-tumor efficacy against cervical cancer in K14HPV16/H2b mice. A) Flow cytometry analysis of E7-specific CD8+ T cells in the spleen (Groups: PBS, n=6, VAX, n=4; VAX aPD-1 aCTLA4, n=5). B) Flow cytometry analysis of IFNγ and TNFα production from CD8+ T cells after in vitro restimulation with the HPV16 E7 CD8 peptide RAHYNIVTF (Groups: PBS, n=6, VAX, n=4; VAX aPD-1 aCTLA4, n=5). C) Cervical tumor size measured in K14HPV16/H2b mice (Groups: PBS, n=5; VAX, n=9; VAX aPD-1 aCTLA4, n=5). D) Representative immunofluorescent staining for CD8 (green), CD3 (red), keratin 14 (K14, white) and nuclei (blue) on frozen OCT–embedded cervical tumors. Samples from 3 mice per group were stained, and a representative field of the tissue is shown. E) Representative immunofluorescent staining for CD8 (green), keratin 14 (K14, white, SC-1 only) and nuclei (blue) on frozen OCT–embedded subcutaneous SC-1 and TC-1 tumors. Scale bars, 100 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P<0.0001; n.s., not significant.
The immune response to vaccination is weaker in K14HPV16/H2b mice compared to FVBN/H2b littermates
Given the relatively widespread expression of the HPV oncoproteins mediated by the keratin 14 gene regulatory region in K14HPV16/H2b transgenic mice, it could be envisaged that these mice might exhibit partial tolerance towards E7, leading to the generation of T cells with reduced effector functions and impaired ability to migrate into and accumulate within tumors. To investigate this possibility, we immunized five-month old females and three-month old males, comparing in each case K14HPV16/H2b mice with their WT FVBN/H2b littermates. Mice were vaccinated once, and the immune response was measured 9 days later in the lymph nodes draining the vaccination site. Flow cytometry analysis showed similar immune responses regardless of the sex and age of the mice, with K14HPV16/H2b mice exhibiting fewer E7 specific CD8+ T cells compared to FVBN/H2b controls (Fig 3A–D). These results indicate that an impaired immune response is characteristic of the HPV transgenic mice and is not solely caused by the cervical tumors present only in female mice. This impairment might be resultant to the widespread expression of E7 in K14+ keratinocytes of the skin, where hyperplasias and dysplasias but not overt squamous carcinomas arise in mice at these ages.
Figure 3.
Transgenic K14HPV16/H2b mice have a weaker systemic immune response to therapeutic vaccination compared to their FVBN/H2b wild type littermates. Flow cytometry analysis of E7-specific CD8+ T cells in the vaccination-site draining lymph nodes after immunization of five month old females (A) or three month old males (B). Representative flow cytometry plots of tetramer staining for E7-specific CD8+ T cells gated on single cell live B220−CD3+CD8+ lymphocytes in female (C) or male (D) mice. E) Flow cytometry analysis of CD62L+CD44+ memory, CD62L−CD44+ effector and CD44+KLRG1+ terminal effector E7-specific CD8+ T cells in female mice. F) Flow cytometry analysis of IFNγ and TNFα production from CD8+ T cells from female mice after in vitro restimulation with different concentrations of the HPV16 E7 CD8 peptide RAHYNIVTF. G) Flow cytometry analysis of IFNγ and TNFα production from CD4+ T cells from female mice after in vitro restimulation with the HPV16 E7 long peptide used in the vaccine. H) Clonal index measured from TCRβ deep sequencing of E7-specific CD8 T cells. Cumulative frequencies of hyperexpanded (> 0.01) (I) and large (0.001 – 0.01) (J) clones indicated as percentage of total TCRβ repertoire. K) Flow cytometry analysis of CFSE low cells after 48h in-vitro proliferation assay of CD8+ T cells isolated from the spleen in the presence of anti-CD3/anti-CD28. L) Flow cytometry analysis of IFNγ and TNFα production after 48h anti-CD3/anti-CD28 -mediated activation of CD8+ T cells isolated from the spleen. M) Flow cytometry analysis of OVA-specific CD8+ T cells in the vaccination-site draining lymph nodes. N) Flow cytometry analysis of IFNγ and TNFα production from CD8+ T cells after in vitro restimulation with the OVA CD8 peptide SIINFEKL. O) Flow cytometry analysis of LCMV-specific CD8+ T cells in the vaccination-site draining lymph nodes. P) Flow cytometry analysis of IFNγ and TNFα production from CD8+ T cells after in vitro restimulation with the LCMV CD8 peptide KAVYNFATC. Groups: females, n=6 (A); males, n=4 (B, E-G); E7-specific CD8 TCR sequencing, n=8 (H-J); CD8+ T cell proliferation and cytokine production, n=5 (K,L); OVA and LCMV immunization, n=5 (M-P). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P<0.0001; n.s., not significant.
We went on to further characterize the impaired immune response, finding that the effector/memory phenotype of E7 specific CD8+ T cells was altered, with fewer CD62L−CD44+ effector and CD44+KLRG1+ terminal effector cells, and a relatively higher percentage of CD62L+CD44+ memory cells in K14HPV16/H2b mice compared non-transgenic FVBN/H2b controls (Fig 3E). These differences suggest that antigen-specific CD8+ T cells undergo limited expansion and differentiation into effector cells upon encounter with their cognate antigen in the transgenic mice. After ex-vivo restimulation, cytokine production from E7 specific T cells was lower in K14HPV16/H2b mice for both CD8+ (Fig 3F and S4A) and CD4+ (Fig 3G and S4B) T cells as compared to T cells from FVBN/H2b mice. Of note, cytokine production from CD8+ T cells in K14HPV16/H2b mice did not increase, even upon raising the concentration of the cognate peptide (Fig 3F).
Deep sequencing of beta chain of the T cell receptor (TCRβ) from E7-specific CD8+ T cells revealed a lower clonality (Fig 3H) in K14HPV16/H2b mice as compared to FVBN/H2b mice, which was associated with fewer hyperexpanded and large frequency clones (Fig 3I,J and S5A). The clonal frequency of the most highly expanded clonotype was also significantly lower in the transgenic mice (Fig S5B), while no differences were apparent in Shannon diversity index (Fig S5C) or in number of unique clones (Fig S5D). Although some hyperexpanded clones present in FVBN/H2b mice were also identified in the K14HPV16 mice, their frequencies were generally lower in transgenic mice (Fig S5E), indicating that, in addition to E7-specific clones that underwent limited expansion, other clones were evidently unable to expand to sufficient numbers in transgenic mice to be detected.
To exclude the possibility that this impaired T cell phenotype is a cell-intrinsic defect, we isolated CD8+ T cells from K14HPV16/H2b and FVBN/H2b mice and performed in-vitro proliferation and cytokine production assays. CD8+ T cell proliferation, measured by CFSE dilution upon anti-CD3/anti-CD28 activation, was similar between the two strains (Fig 3K), whereas IFNγ/TNFα production from anti-CD3/anti-CD28 activated CD8+ T cells was notably higher (3x) in the cells isolated from K14HPV16/H2b mice (Fig 3L). These data indicate that CD8+ T cells from transgenic K14HPV16/H2b mice have no cell-intrinsic defects in their proliferation and function, suggesting that their impaired expansion and activity is the result of external factors.
Systemic immunosuppression in K14HPV16/H2b mice also affects immune responses against foreign antigens
A priori, the above analyses are consistent with a condition of partial tolerance. If so, then the impairment would be specific to the E7 neo-self-antigen, whereas the immune response to bona fide non-self-antigens should be similar in transgenic and non-transgenic mice. To address this possibility, we immunized three-month old K14HPV16/H2b or FVBN/H2b male mice with two non-self-antigens: chicken ovalbumin (OVA) and an LCMV-derived peptide recognized by CD8+ T cells. K14HPV16/H2b mice exhibited fewer OVA-specific CD8+ T cells (Fig 3M) and lower cytokine production upon ex-vivo restimulation with the OVA-derived CD8-specific peptide SIINFEKL (Fig 3N) compared to their WT FVBN/H2b littermates. As for the immunization with the LCMV-derived peptide, although the abundance of LCMV-specific CD8+ T cells was similar between the two strains (Fig 3O), cytokine production after ex-vivo restimulation was severely impaired in K14HPV16/H2b mice (Fig 3P).
Thus, although we cannot completely exclude the existence of a certain degree of partial self-tolerance to the E7 protein, it is evident that the impaired immune response seen in K14HPV16/H2b mice is not restricted to the E7 neo-antigen. The results rather suggest that a systemic immunosuppression mechanism is operative, affecting immune responses against different antigens in these mice, and likely impairing the anti-tumor response as well.
Activation of antigen presenting cells is suppressed in K14HPV16/H2b mice
Since antigen presentation by dendritic cells and other antigen-presenting cells (APC) is the first step in generating an immune response, we sought to determine whether APCs were directly affected by the immunosuppressive mechanism evident in K14HPV16/H2b mice. To do so, we subcutaneously immunized K14HPV16/H2b mice and their FVBN/H2b littermates with the NP-E7LP vaccine, and after 24 hours harvested DCs from the lymph nodes draining the vaccination site, and assessed their activation by flow cytometry. CD11b−CD11c+ DCs from FVBN/H2b mice showed a significant upregulation of all the analyzed markers, whereas DCs from K14HPV16/H2b mice only upregulated CD86 (Fig 4A), albeit to a significantly lower extent compared to the FVBN/H2b counterpart. No upregulation of CD80, CD40, and MHCII was detected in K14HPV16/H2b mice (Fig 4A), indicating that DC activation is markedly impaired in HPV transgenic mice. Interestingly, and similarly to cervical cancer patients (21), K14HPV16/H2b mice had decreased numbers of DCs in the spleen, as measured by flow cytometric analysis (Fig S6A); in contrast, there was no difference in the lymph nodes (Fig S6B), suggesting that the weaker immune response measured in the GEMM cannot be explained by insufficient DC abundance there.
Figure 4.
Dendritic cell activation is actively suppressed and immunization with a DC vaccine fails to rescue the immune response in K14HPV16/H2b mice. A) Flow cytometry analysis of CD80, CD86, CD40 and MHCII on CD11b−CD11c+ DCs in the vaccination site draining lymph nodes 24h after immunization with the NP-E7 vaccine. B) Flow cytometry analysis of E7-specific CD8+ T cells in the vaccination-site draining lymph nodes after immunization with a DC vaccine. C) Flow cytometry analysis of CD62L+CD44+ memory, CD62L−CD44+ effector and CD44+KLRG1+ terminal effector E7-specific CD8+ T cells. D) Flow cytometry analysis of IFNγ and TNFα production from CD8+ T cells after in vitro restimulation with the HPV16 E7 CD8 peptide RAHYNIVTF. Groups: DC activation, n=4; E7 specific CD8 T cells, phenotype, and cytokine production, n=5. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P<0.0001; n.s., not significant.
We next generated DCs from the bone marrow of K14HPV16/H2B mice or from their FVBN/H2b littermates, aiming to assess their functionality. In-vitro activation experiments with either LPS or CpG revealed that upregulation of activation markers was similar between BMDCs from either strain (Fig S6C), indicating that the impaired DC activation is not cell intrinsic and is not related to the use of CpG, whose receptor, TLR9, can be downregulated by HPVs (34).
Given that the endogenous DCs were functionally impaired, we reasoned that it might be possible to rescue the immune response by administering antigen-loaded, activated dendritic cells as an anti-E7 DC-vaccine (Fig S6D). To avoid any possible interference from the HPV16 transgenes, we generated BMDCs from the bone marrow of a syngeneic FVBN/H2b mouse. BMDCs were loaded with E7LP, activated with LPS, and injected into K14HPV16/H2b or FVBN/H2b mice. The administration was performed either subcutaneously or intravenously to trigger the generation of an anti-E7 response in lymph nodes or spleen, respectively. Additional groups of mice received CFSE-labeled BMDCs and were sacrificed 24h after the administration to assess their activation (Fig S6D). Regardless of the administration route, the expression of CD86, CD40 and MHCII showed a trend pointing to an impaired activation status of the transferred BMDCs in K14HPV16/H2b mice (Fig S6E and F), suggesting that they were being actively suppressed. When mice that received the unlabeled BMDC vaccine were sacrificed 9 days after immunization, we found a situation similar to what was described above for NP-E7LP vaccination. Namely, there were fewer E7 specific CD8+ T cells (Fig 4B and S6G), with an altered phenotype in both lymph nodes and spleen (Fig 4C and S6H respectively) and lower cytokine production by CD8+ T cells after restimulation (Fig 4D). These data indicate that adoptively transferred dendritic cells are being actively suppressed in both lymph nodes and spleen of K14HPV16/H2b mice, much as endogenous DCs.
Myeloid cells are orchestrating systemic immunosuppression in K14HPV16/H2b mice
The data collectively indicate that the T cell responsiveness in K14-HPV16/H2b mice is being suppressed (in paracrine fashion) in lymphoid organs. We reasoned that an immunosuppressive immune cell population could be mediating the observed effects and sought to investigate the possibility.
Previous reports have revealed that T regulatory cells (Tregs) might play a role in HPV E7-mediated immunosuppression (23, 35). Flow cytometry analysis of K14HPV16/H2b mice and their FVBN/H2b littermates showed that the number of Tregs was similar in spleen and slightly higher in lymph nodes of K14HPV16/H2b mice (Fig S7A and B). However, their phenotype was substantially different: CD4+CD25+Foxp3+ Treg cells expressing the IL-7 receptor CD127 were increased in K14HPV16/H2b mice (Fig S7C and D). Notably, CD127+ Tregs are reportedly more efficient at sequestering IL-2, a cytokine required for optimal T cell expansion during immune responses (36).
To test the suppressive activity of Tregs on CD8+ T cells and APCs, we set up a series of in-vitro experiments to assess their impact on CD8 T cell proliferation and cytokine production, and APC activation. Naïve CD8+ T cells were isolated from the spleen of an FVBN/H2b mouse, labeled with CFSE and co-cultured in the presence of anti-CD3/anti-CD28 with either K14HPV16/H2b or FVBN/H2b mouse spleen-derived Treg cells (Fig S7E). As expected, CD8+ T cell proliferation was suppressed by Tregs compared to conventional CD4+ T cells; notably, however, we detected no difference in suppression between Tregs derived from K14HPV16/H2b vs. FVBN/H2b mice (Fig S7F). To ascertain the effects of Tregs on cytokine production from E7-specific CD8+ T cells, we immunized an FVBN/H2b mouse with the NP-E7LP vaccine and, 9 days later, isolated CD8+ T cells (Fig S7G). Similar to the previous experiment, isolated CD8+ T cells were then re-stimulated in the presence of Treg cells or conventional CD4+ T cells isolated from either K14HPV16/H2b or FVBN/H2b mice (Fig. S6G). Once again, Tregs from both strains suppressed cytokine production in a similar manner compared to conventional CD4+ T cells (Fig S7H).
To assess the effects of Tregs on APCs, we generated BMDCs from an FVBN/H2b mouse, loaded them with E7LP, and activated them with LPS. On the following day, BMDCs were mixed with Treg cells or with conventional CD4+ T cells, and co-cultured for 24h (Fig S7I). The expression of activation markers was similar between BMDCs cultured with K14HPV16/H2b or FVBN/H2b-derived cells, both for Treg or conventional CD4+ T cells (Fig S7J), with no sign of increased suppression from the K14HPV16/H2b-derived populations. Collectively, these results indicate that Tregs have similar activity in both strains; as such, the evidence suggests Tregs are not responsible for the selectively impaired immune response seen in K14HPV16/H2b transgenic mice.
We next focused on myeloid cells, since so called ‘myeloid-derived suppressor’ cells (MDSCs) within the broad umbrella of myeloid cell subtypes have been recognized to have immunosuppressive and tumor-promoting capabilities, although most studies have focused on their presence and functions within the TME. Consistent with the enlarged spleen and lymph nodes (Fig S8), all major myeloid cell populations were significantly more abundant in lymphoid organs of K14HPV16/H2b mice, exhibiting increases of several fold (Fig 5A–D). In order to determine if myeloid cells from K14HPV16/H2b mice possess immunosuppressive capabilities, we performed, much as previously described, an in-vitro co-culture experiments with CD11b+ myeloid cells in place of Tregs. Both CD8+ T cell proliferation (Fig 5E and S9A and B) and cytokine production (Fig 5F and S9C) were significantly lower when FVBN/H2b-derived CD8+ T cells (from untreated mice and after NP-E7LP immunization, respectively) were co-cultured with CD11b+ myeloid cells isolated from the spleens of K14HPV16/H2b mice. Of note, when CFSE labeled CD8+ T cells were co-cultured in the presence of myeloid cell-depleted CD11b− splenocytes (the cell fraction recovered after CD11b+ cell isolation), the inhibition of their proliferation was almost entirely absent (Fig 5E and S9B). These results indicate that active immunosuppression is conveyed by CD11b+ myeloid cells present in the splenocyte mix. BMDC activation (Fig S9D) was also lower in cells that were cultured in the presence of CD11b+ myeloid cells derived from K14HPV16/H2b mice compared to the co-culture with FVBN/H2b-derived CD11b+ cells (Fig 5G). Collectively, these data demonstrate that myeloid cells derived from lymphoid organs of K14HPV16/H2b mice have the capability to impair the functions of both CD8+ T cells and DC/APCs, and are therefore likely responsible for the observed systemic suppression of immune responses seen in the HPV transgenic mice.
Figure 5.
Myeloid cells are expanded in lymphoid organs and possess suppressive activity on CD8+ T cells and DCs in K14HPV16/H2b mice. Flow cytometry analysis on lymph nodes (A) and spleen (B) of total CD11b+ myeloid cells, CD11b+Ly6G+Ly6C− G-MDSCs/neutrophils, CD11b+Ly6G−Ly6C+ Mo-MDSCs/monocytes and CD11b+Ly6G−Ly6C−F4/80+ macrophages (three month old males, n=6). Representative flow cytometry plots of CD11b+ cells gated on single cell live lymphocytes in lymph nodes (C) and spleen (D). E) Flow cytometry analysis of CFSE low cells after 48h in-vitro proliferation assay of CD8+ T cells isolated from the spleen of an FVBN/H2b mouse in the presence of anti-CD3/anti-CD28 and CD11b+ cells or myeloid cell-depleted CD11b− cells isolated from the spleen of K14HPV16/H2b or FVBN/H2b mice as indicated (n=4). F) Flow cytometry analysis of IFNγ and TNFα production from CD8+ T cells isolated from the spleen of an NP-E7LP immunized FVBN/H2b mouse after in vitro restimulation with the HPV16 E7 CD8 peptide RAHYNIVTF in the presence of CD11b+ myeloid cells isolated from the spleen of K14HPV16/H2b or FVBN/H2b mice as indicated (n=5). G) Flow cytometry analysis of CD80, CD86, CD40 and MHCII on FVBN/H2b mouse-derived LPS-activated BMDCs after a 24h co-culture with CD11b+ cells isolated from the spleen of K14HPV16/H2b or FVBN/H2b mice as indicated (Groups: non-activated BMDCs, BMDCs only, FVBN/H2b CD11b+ cells, n=4; K14HPV16/H2b CD11b+ cells, n=6). H) Flow cytometry analysis of CFSE low cells after 48h in-vitro proliferation assay of isolated CD8+ T cells in the presence of anti-CD3/anti-CD28 and monocytes (CD11b+Ly6C+Ly6G− and CD11b+Ly6C−Ly6G−F4/80+ cells) isolated from the spleen of K14HPV16/H2b or FVBN/H2b mice using magnetic beads as described in the methods (Groups: FVBN/H2b monocytes, n=5; K14HPV16/H2b monocytes, n=6). I) Flow cytometry analysis of CFSE low cells after 48h in-vitro proliferation assay of isolated CD8+ T cells in the presence of anti-CD3/anti-CD28 and macrophages (F4/80+ cells) isolated from the spleen of K14HPV16/H2b or FVBN/H2b mice using magnetic beads as described in the methods (n=6). The ratio of CD8/BMDCs to CD11b+ cells/monocytes/macrophages was 1:10 for all the experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P<0.0001; n.s., not significant.
We next sought to determine which of the major cell sub-populations contained within the CD11b+ umbrella possess suppressive activity. We focused on monocytes, macrophages, and neutrophils. As above, we isolated the different cell populations and performed the co-culture assay scoring for effects on CD8+ T cell proliferation. Monocytes (Fig 5H) but not macrophages (Fig 5I) showed suppressive activity. Since we could not perform in-vitro assays with neutrophils due to their very brief survival after isolation, we performed RNAseq to compare K14HPV16/H2b-derived with FVBN/H2b-derived neutrophils. Sorted CD11b+Ly6G+Ly6C− neutrophils from K14HPV16/H2b or FVBN/H2b mice clustered separately, with 761 genes significantly differentially expressed, indicating that neutrophil transcriptome profiles are substantially different between the two strains (Fig S10). Pathway analyses using the KEGG and Hallmark genesets (Fig S11) revealed an upregulation of several inflammatory and metabolic pathways in neutrophils from K14HPV16/H2b transgenic mice including the IL6-JAK-STAT3 signaling pathway (Fig S12). Notably, immune cell signaling via STAT3 has previously been proposed to have immunosuppressive activity in cervical cancer patients (22), and both IL-6 and STAT3 have been linked with the suppressive functions of MDSCs (37, 38), implicating neutrophils, together with monocytes, in the systemic immunosuppression evident in K14HPV16/H2b mice.
Increased immunosuppressive and myeloid cell regulatory factors in K14HPV16 mice
We then analyzed the draining lymph nodes to vaccination site, looking for additional factors that could be involved in the underlaying suppressive mechanism. We reasoned that analyzing lymph nodes might implicate local factors involved in impairing the generation of anti-E7 immune responses, in addition to the systemic factors suspected to mediate the expansion/alteration of the myeloid cell compartment.
To this end, protein expression of candidate immunosuppressive factors in the vaccination site draining lymph nodes of untreated K14HPV16/H2b and their FVBN/H2b littermates was assessed. IL-10 and IDO were both found increased in the LNs of K14HPV16/H2b mice (Fig 6A and S13). Analyses of protein extracts from CD11b+ cells isolated from K14HPV16/H2b and FVBN/H2b mice, as well as from total CD11b− cells, confirmed that these factors were produced by K14HPV16/H2b myeloid cells (Fig 6B and S14). Additionally, we observed a significant increase in ROS production by CD11b+ cells in K14HPV16/H2b mice compared to FVBN/H2b (Fig 6C). Thus, myeloid cells from K14HPV16/H2b mice express a number of immunosuppressive factors, and are more abundant, implicating them in the immune suppression.
Figure 6.
Immunosuppressive factors produced by K14HPV16/H2b myeloid cells are increased in lymph nodes together with myeloid cell growth factors and proteins associated with inflammation. A) Western blot analysis of whole lymph node protein extracts from K14HPV16/H2b or FVBN/H2b mice. B) Western blots of protein extracts from total CD11b+ myeloid cells or myeloid cell-depleted CD11b− cells isolated from the spleen of K14HPV16/H2b or FVBN/H2b mice. C) Flow cytometry analysis of ROS production by CD11b+ cells measured in-vitro with DCFDA (Groups: FVBN/H2b, K14HPV16/H2b, n=5; positive controls with PMA, n=4). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P<0.0001; n.s., not significant.
Concurrently, we analyzed the lymph nodes for factors known to be involved in the regulation of myeloid cell expansion. Notably, we observed a significant increase in GM-CSF and G-CSF but not M-CSF in K14HPV16/H2b mice (Fig 6A and S13). Interestingly, we also observed a general increase in the inflammation-related factors C-reactive protein, IL1-beta and HSP90 (Fig 6A and S13), suggestive of a chronic inflammatory state, which is also implicated by the IFNγ transcriptome signature in neutrophils (Fig S11). The upregulation of these immuno-regulatory factors seems likely to be involved in the myeloid cell expansion, and may link our findings with previous reports of local inflammation in the skin of a different immunological sub-strain of K14HPV16 transgenic mice as well as SCC patients (39–41). These results further suggest that the effects of local inflammation of the skin could be transmitted systemically to drive the expansion of myeloid cells, mediated by G-CSF and GM-CSF along with other factors, known and unknown.
Myeloid cell depletion in K14HPV16 mice by available agents is too short-lived to rescue the immune response
We next conducted a series of drug trials wherein we sought to eliminate the immunosuppressive myeloid cells in combination with therapeutic vaccination, aiming to disrupt the systemic immunosuppression and enhance the anti-tumor immune response in K14HPV16/H2b mice. To do so, we applied previously documented approaches aimed at depleting or reprogramming myeloid cells. Namely, we tested the depleting antibodies anti-Ly6G, anti-Ly6C, and combination of anti-GR1, anti-GM-CSF and anti-G-CSF, as well as chemotherapy treatments with 5-fluorouracil (42), gemcitabine (43) and a combination of carboplatin and paclitaxel (CarboTaxol) (20). In supplementary figure S15, to investigated the effects of anti-Ly6G treatment we performed two inoculums of vaccine (day zero and day 14) to assess both lymph node and tumor site for E7 specific CD8+ T cell production, cytokine production and tumor infiltration, respectively, in the context of myeloid depletion. At day 19, 5 days after the second vaccination, neither the generation of E7-specific CD8 T cells in the lymph nodes or the number of CD8 T cells in the tumor microenvironment were increased following anti-Ly6G treatment. Subsequently, we chose (Figures S16–19) a shorter schedule (day 9), following a single vaccination at day zero, and focused on the impact of myeloid cell depletion on the generation of E7-specific CD8 T cells in the lymph nodes.
Although some of the treatments caused a transient decrease in certain myeloid cell populations (Fig S15–19), their effects were usually short lasting and typically induced a compensatory increase in other myeloid populations. Indeed, the transitory depletion elicited by these agents was unable to provide a sufficient temporal window of intervention to relieve the predominant systemic immunosuppression of APCs and CD8+ T cells. Ultimately, none of the treatments was capable of appreciably boosting the immune response to the E7 vaccine (Fig S15–19).
Beneficial effects of checkpoint blockade immunotherapy are masked by immunosuppressive cells in K14HPV16
Having identified myeloid cells as the dominant immunosuppressive population in K14HPV16/H2b mice, we revisited the failure of the immune checkpoint inhibitors to boost the production of antigen-specific CD8+ T cells following vaccination with the NP-E7LP formulation. Notably, in an effort to improve upon the efficacy, we employed ‘next generation’ isotype switched versions, using both an anti-PD-1 that does not activate FC receptor functions (44), and an anti-CTLA4 that possesses Treg depleting activity (45). Antibody treatment was initiated at the same time as vaccination. Analysis of the immune response 9 days later showed no increase in E7-specific CD8+ T cells compared to the vaccine alone (Fig S20A and B). As expected, cytokine production measured after restimulation of the whole splenocyte or lymph node cells also showed no improvement upon antibody treatment (Fig 7 and S20C).
Figure 7.
Myeloid cells from K14HPV16/H2b mice mask the synergistic effects of immune checkpoint blockade and therapeutic vaccination. Flow cytometry analysis of IFNγ and TNFα production from CD8+ T cells after in vitro restimulation with the HPV16 E7 CD8 peptide RAHYNIVTF. The assay was performed in parallel on total splenocytes and spleen-derived isolated CD8+ T cells. Groups: total splenocytes, n=6; CD8 T cell only, n=5. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P<0.0001; n.s., not significant.
To determine if cytokine production by CD8+ T cells elicited in the vaccine- plus checkpoint inhibitor-treated K14-HPV16/H2b mice was intrinsically defective, or rather repressed in paracrine fashion by the immunosuppressive cell milieu of the spleen, we isolated CD8+ T cells from treated mouse spleens, and performed ex-vivo restimulation assays. Interestingly we found that cytokine production was significantly higher in CD8+ T cells isolated from the spleens of HPV transgenic mice that received anti-PD-1 and anti-CTLA4 in combination with the NP-E7LP vaccine (Fig 7). This result indicates that the addition of checkpoint inhibitors to the therapeutic vaccination was indeed enhancing the CD8 response, resulting in superior cytokine production. However, such benefits were nullified and undetectable when CD8+ T cells were immersed in the total splenocyte cell milieu, as revealed when the CD8+ T cells were separated from these cells, presumably due to the presence of immunosuppressive factors produced by CD11b+ myeloid cells that overruled upregulation of the cytokines.
Interestingly, similar results were obtained with CD8+ T cells isolated from mice treated with a combination of anti-GM-CSF, anti-G-CSF and anti-GR1. Namely, E7-specific CD8+ T cells isolated from mice that received the myeloid cell-targeting combination showed a significant increase in cytokine production compared to mice that only received the vaccine (Fig S21), despite the lack of measurable depletion at the observed timepoint (Fig S19); thus, we infer transitory inhibition of their immune-inhibitory activity without myeloid cell depletion. Although we did not investigate the possibility of brief depletion of myeloid cell numbers at earlier timepoints, this result further supports both the conclusion that myeloid cells are suppressing immune responses in K14HPV16/H2b mice and the proposition that their sustainable abrogation would improve anti-tumor immune responses.
Discussion
Despite the entrée of immunotherapies into clinical practice and their unprecedented successes (9), a significant percentage of cancer patients receive limited or no benefit from such treatments (11). Elucidating the operative mechanisms leading to the failure of current immunotherapies is a crucial step towards the development of more broadly efficacious therapeutic strategies. Potentially immunosuppressive myeloid cells have been observed in patients with cervical cancer (20, 22) but their importance has remained unclear.
In this study we leveraged a genetically engineered, fully immunocompetent mouse model that phenocopies characteristics observed in cervical cancer patients that may contribute to their poor responsiveness to tumor vaccines and immune checkpoint inhibitors. Namely, these mice evidence impaired activation of CTLs, fewer and poorly functional dendritic cells, and a systemic amplification of myeloid cells with immuno-suppressive functions (18–22).
By comparing K14HPV16/H2b mice with their non-transgenic FVBN/H2b littermates and focusing our attention on the lymphoid organs where immune responses are orchestrated rather than on the tumor microenvironment, we have uncovered an expansion in myeloid cells that are capable of suppressing both APCs and CD8+ T cells, abrogating both the effects of therapeutic vaccination as well as the synergistic effects of checkpoint blockade, thereby rendering these therapeutic approaches ineffective.
Although others have reported a role for Tregs in suppressing anti-tumor responses in other mouse models (23, 35), our analyses do not implicate these cells in the systemic immunosuppressive mechanism described herein, leading us to conclude that myeloid cells are the culprits.
Currently, clinical trials involving therapeutic vaccination with E7 long peptides are showing little or no benefit (14, 16), much as we have observed in our mouse model. While a ‘solid-phase’ nanoparticle-based formulation induced the development of detectable numbers of E7-specific CD8+ T cells in spleen and LN, their abundance was low, their ability to secrete inflammatory cytokines was severely impaired, and there was no appreciable accumulation proximal to or inside carcinomas in the cervix.
In addition to direct CD8+ T cell suppression, an ancillary factor is likely defective T cell priming consequent to myeloid cell-mediated suppression of dendritic cell/APC activation. In K14HPV16/H2b mice, both endogenous DCs as well as antigen-loaded and pre-activated BMDCs were unable to robustly upregulate a variety of co-stimulatory molecules required for optimal T cell priming, concordant with a defective activation state that was unable to produce fully functional and abundant effector CD8+ T cells capable of migrating from the spleen and lymph nodes to infiltrate the tumors and attack cancer cells.
Our results support the hypothesis that neoplasia-induced systemic expansion of immunosuppressive myeloid cells can establish a first line of defense against efficacious tumor immunity and, as such, these cells or their suppressive functions will need to be disrupted to allow for the generation of a strong and fully functional anti-tumor immune response. Accumulation of myeloid cells has been reported for cervical and a wide variety of other cancers where they might be suspected to cause potent systemic immunosuppression (20, 22, 38, 46). Such a bottleneck, operative upstream in the cancer-immunity cycle (47), poses an important limitation to all immunotherapy approaches both in humans and in mice, potentially explaining the failure of immunotherapies in a certain patients.
In addition to limiting the abundance and functions of tumor-antigen-specific CD8 T cells induced by the tumor vaccine, CD11b+ myeloid cells dominantly suppress the otherwise demonstrably beneficial effects that anti-PD-1 and anti-CTLA4 have on CTL activity. Notably, significantly increased cytokine production by cells in the dual checkpoint treatment group was only measurable when CD8+ T cells were separated from the immunosuppressive myeloid cells in the splenocyte mix.
Despite considerable efforts aimed at targeting myeloid cells in K14HPV16/H2b mice, all the approaches that we tested proved to be inadequate. Since both male and female mice show a systemically immunosuppressed phenotype, it is reasonable to assume that the increase in myeloid cells is caused by the expression of HPV oncoproteins in basal keratinocytes of skin (and cervix). Given the broad extent of the epidermis and the consequent widespread presence of the viral oncogenes, it is possible that the regulation of myeloid cell expansion by HPV-expressing keratinocytes might be exacerbated in our mouse model compared to the case of more restricted expression in infected cervical keratinocytes in human females. Although this hyper-expansion of the systemic myeloid compartment could render the condition particularly hard to counteract, it presents this model as an attractive platform to elucidate the details of the mechanisms responsible for the expansion and immunosuppression, thereby guiding the design of next-generation targeting approaches.
Clinically feasible, specific and effective treatments aimed at targeting myeloid cells are still lacking. Although certain chemotherapeutic agents have shown the ability to decrease myeloid cell numbers and produced promising results in combination with therapeutic vaccination (20), new myeloid cell-targeting strategies are clearly needed. The development of approaches guided by a deeper understanding of myeloid cell regulation and suppressive functions hold promise to identify treatments that can break down the barrier they manifest, facilitating efficacious immunotherapy.
This study has clarified the significance of systemic myeloid-cell mediated immunosuppression and its link to the failure of immunotherapy, heretofore underappreciated compared to the well-established roles of myeloid and other immunoregulatory cells in the TME (48). The abundance of immunosuppressive myeloid cells in both spleen and lymph nodes, and their ability to repress CTLs and DCs that are critical for the generation of a strong systemic antitumor response, explains the anemic immune response to the HPV16 E7 neoantigen in this mouse model. Importantly, similar suppressive features have been reported for myeloid cells isolated from patients with HPV-related cancers (20, 22), encouraging the relevance of these findings and presenting clues into why immunotherapy has been disappointingly ineffective to date in in cervical cancer patients, and potentially, in patients with other types of cancer associated with increased MDSCs (46, 49, 50) in spleen and LN. As such, it seems increasingly likely that immunosuppressive myeloid cells will need to be neutralized to achieve successful immunotherapy in cancer patients with systemic myeloid expansion.
Supplementary Material
Table 1.
Treatements
| Treatment | Dosage | Frequency | Route | Volume | Source |
|---|---|---|---|---|---|
| Anti-PD-1 (Fig. 2) | 200 µg/mouse | 2x week | IP | 200 µL | Bioxcell, clone RMP1-14 |
| Anti-CTLA4 (Fig. 2) | 200 µg/mouse | 2x week | IP | 200 µL | Bioxcell, clone 9D9 |
| Anti-PD-1 (mPD1-4H2-mIgG1-D265A, clone 6A1_RAS_Ab; Fig. 7 and Supplementary Fig. S28) | 200 µg/mouse | 2x week | IP | 200 µL | Bristol-Myers Squibb |
| Anti-CTLA4 (mCTLA4-9D9-mIgG2c, clone 2B7_RAS_Ab_02; Fig. 7 and Supplementary Fig. S28) | 200 µg/mouse | 2x week | IP | 200 µL | Bristol-Myers Squibb |
| Anti-Ly6G | 400 µg/mouse | 3x week | IP | 200 µL | Bioxcell, clone RB6-8C5 |
| Anti-Ly6C | 400 µg/mouse | 3x week | IP | 200 µL | Bioxcell, clone Monts 1 |
| 5-Fluorouracil | 50 mg/kg | 1 shot | IP | 200 µL | Sigma |
| Carboplatin | 40 mg/kg | 1x week for 2 weeks | IP | 200 µL | Bristol-Myers Squibb |
| Paclitaxel | 20 mg/kg | 2x week for 2 weeks | IP | 200 µL | Labatec |
| Anti-GM-CSF | 100 µg/mouse | 3x week | IP | 200 µL | Bioxcell, clone MP1-22E9 |
| Anti-G-CSF | 100 µg/mouse | 3x week | IP | 200 µL | Bio-Techne, cat. MAB414 |
| Anti-GR1 | 200 µg/mouse | 3x week | IP | 200 µL | Bioxcell, clone RB6-8C5 |
Abbrevation: IP, intraperitoneal.
Acknowledgments
We thank B. Torchia, M. Wen-Peng, and M.A. Gaveta for technical support; Chris Chiu for the backcrossing and initial analysis of K14HPV16/H2b mice, and for scientific advice; Mark Selby from Bristol-Myers Squibb for helpful discussion and support, the Histology and Flow cytometry facilities at EPFL; the protein and peptide chemistry and genomic technologies facilities at the University of Lausanne (UNIL), in particular C. Servis and K. Harshman, respectively. This work was variously supported by a SNSF Sinergia grant (#160742) and by a sponsored research agreement with Bristol-Myers Squibb. We further acknowledge the Swiss Bridge Foundation (Zurich, Switzerland), and in particular thank Philipp Lücke and Thomas Hoepli for their support and encouragement.
Financial support: This work was supported by SNF Sinergia, Bristol-Myers Squibb and the Swiss Bridge Foundation. LMC acknowledges financial support from the Brenden-Colson Center for Pancreatic Health, and a Stand Up To Cancer – Lustgarten Foundation Pancreatic Cancer Convergence Dream Team Translational Research Grant (SU2C-AACR-DT14-14).
Conflict of interest statement: The Swiss Federal Institute of Technology Lausanne (EPFL), has filed for patent protection on the nanoparticle platform described herein and M.A.S. is named as an inventor. The other authors declare no potential conflicts of interest. This work was partially supported by a sponsored research agreement form Bristol Myers Squibb; A.K was employee of BMS during this research project. L.M.C. is a paid consultant for Cell Signaling Technologies, received reagent and/or research support from Plexxikon Inc., Pharmacyclics, Inc., Acerta Pharma, LLC, Deciphera Pharmaceuticals, LLC, Genentech, Inc., Roche Glycart AG, Syndax Pharmaceuticals Inc., and NanoString Technologies, and is a member of the Scientific Advisory Boards of Syndax Pharmaceuticals, Carisma Therapeutics, Zymeworks, Inc and Verseau Therapeutics.
References
- 1.Giuliano AR, et al. (2015) EUROGIN 2014 roadmap: Differences in human papillomavirus infection natural history, transmission and human papillomavirus-related cancer incidence by gender and anatomic site of infection: EUROGIN 2014 roadmap. International Journal of Cancer 136(12):2752–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Havre PA, Yuan J, Hedrick L, Cho KR, Glazer PM (1995) p53 inactivation by HPV16 E6 results in increased mutagenesis in human cells. Cancer Res 55(19):4420–4424. [PubMed] [Google Scholar]
- 3.Stirdivant SM, et al. (1992) Human papillomavirus type 16 E7 protein inhibits DNA binding by the retinoblastoma gene product. Mol Cell Biol 12(5):1905–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serrano B, Brotons M, Bosch FX, Bruni L (2018) Epidemiology and burden of HPV-related disease. Best Practice & Research Clinical Obstetrics & Gynaecology 47:14–26. [DOI] [PubMed] [Google Scholar]
- 5.Smola S (2017) Immunopathogenesis of HPV-Associated Cancers and Prospects for Immunotherapy. Viruses 9(9). doi: 10.3390/v9090254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winer RL, et al. (2011) Early Natural History of Incident, Type-Specific Human Papillomavirus Infections in Newly Sexually Active Young Women. Cancer Epidemiology Biomarkers & Prevention 20(4):699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez AC, et al. (2010) Longitudinal Study of Human Papillomavirus Persistence and Cervical Intraepithelial Neoplasia Grade 2/3: Critical Role of Duration of Infection. JNCI Journal of the National Cancer Institute 102(5):315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castle PE, et al. (2017) Treatment of cervical intraepithelial lesions. International Journal of Gynecology & Obstetrics 138:20–25. [DOI] [PubMed] [Google Scholar]
- 9.Mellman I, Coukos G, Dranoff G (2011) Cancer immunotherapy comes of age. Nature 480(7378):480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribas A, Wolchok JD (2018) Cancer immunotherapy using checkpoint blockade. Science 359(6382):1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventriglia J, et al. (2017) Immunotherapy in ovarian, endometrial and cervical cancer: State of the art and future perspectives. Cancer Treatment Reviews 59:109–116. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins RW, Barbie DA, Flaherty KT (2018) Mechanisms of resistance to immune checkpoint inhibitors. British Journal of Cancer 118(1):9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Burg SH, Melief CJ (2011) Therapeutic vaccination against human papilloma virus induced malignancies. Current Opinion in Immunology 23(2):252–257. [DOI] [PubMed] [Google Scholar]
- 14.Kenter GG, et al. (2009) Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. New England Journal of Medicine 361(19):1838–1847. [DOI] [PubMed] [Google Scholar]
- 15.Trimble CL, Frazer IH (2009) Development of therapeutic HPV vaccines. Lancet Oncol 10(10):975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Poelgeest MI, et al. (2013) HPV16 synthetic long peptide (HPV16-SLP) vaccination therapy of patients with advanced or recurrent HPV16-induced gynecological carcinoma, a phase II trial. Journal of translational medicine 11(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massarelli E, et al. (2018) Combining Immune Checkpoint Blockade and Tumor-Specific Vaccine for Patients With Incurable Human Papillomavirus 16–Related Cancer: A Phase 2 Clinical Trial. JAMA Oncology. doi: 10.1001/jamaoncol.2018.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song D, Li H, Li H, Dai J (2015) Effect of human papillomavirus infection on the immune system and its role in the course of cervical cancer. Oncology Letters 10(2):600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, Song Y, Lu Y-L, Sun J-Z, Wang H-W (2013) Increased expression of programmed death (PD)-1 and its ligand PD-L1 correlates with impaired cell-mediated immunity in high-risk human papillomavirus-related cervical intraepithelial neoplasia. Immunology 139(4):513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welters MJ, et al. (2016) Vaccination during myeloid cell depletion by cancer chemotherapy fosters robust T cell responses. Science Translational Medicine 8(334):334ra52. [DOI] [PubMed] [Google Scholar]
- 21.Strickler HD, et al. (2014) The Relation of Plasmacytoid Dendritic Cells (pDCs) and Regulatory T-Cells (Tregs) with HPV Persistence in HIV-Infected and HIV-Uninfected Women. Viral Immunology 27(1):20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez KLF, et al. (2017) Local and systemic immunomodulatory mechanisms triggered by Human Papillomavirus transformed cells: a potential role for G-CSF and neutrophils. Sci Rep 7(1):9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pere H, et al. (2011) A CCR4 antagonist combined with vaccines induces antigen-specific CD8+ T cells and tumor immunity against self antigens. Blood 118(18):4853–4862. [DOI] [PubMed] [Google Scholar]
- 24.Arbeit JM, Howley PM, Hanahan D (1996) Chronic estrogen-induced cervical and vaginal squamous carcinogenesis in human papillomavirus type 16 transgenic mice. Proc Natl Acad Sci USA 93(7):2930–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coussens LM, Hanahan D, Arbeit JM (1996) Genetic predisposition and parameters of malignant progression in K14-HPV16 transgenic mice. Am J Pathol 149(6):1899–1917. [PMC free article] [PubMed] [Google Scholar]
- 26.Galliverti G, et al. (2018) Nanoparticle Conjugation of Human Papillomavirus 16 E7-long Peptides Enhances Therapeutic Vaccine Efficacy against Solid Tumors in Mice. Cancer Immunology Research 6(11):1301–1313. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medler TR, et al. (2018) Complement C5a Fosters Squamous Carcinogenesis and Limits T Cell Response to Chemotherapy. Cancer Cell 34(4):561–578.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herd K, et al. (1997) E7 Oncoprotein of Human Papillomavirus Type 16 Expressed Constitutively in the Epidermis Has No Effect on E7-Specific B- or Th-Repertoires or on the Immune Response Induced or Sustained after Immunization with E7 Protein. Virology 231(1):155–165. [DOI] [PubMed] [Google Scholar]
- 30.Elson DA, et al. (2000) Sensitivity of the cervical transformation zone to estrogen-induced squamous carcinogenesis. Cancer Res 60(5):1267–1275. [PubMed] [Google Scholar]
- 31.Giraudo E, Inoue M, Hanahan D (2004) An amino-bisphosphonate targets MMP-9-expressing macrophages and angiogenesis to impair cervical carcinogenesis. J Clin Invest 114(5):623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erez N, Truitt M, Olson P, Arron ST, Hanahan D (2010) Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell 17(2):135–147. [DOI] [PubMed] [Google Scholar]
- 33.van Poelgeest MIE, et al. (2016) Vaccination against Oncoproteins of HPV16 for Noninvasive Vulvar/Vaginal Lesions: Lesion Clearance Is Related to the Strength of the T-Cell Response. Clinical Cancer Research 22(10):2342–2350. [DOI] [PubMed] [Google Scholar]
- 34.Hasan U (2014) Human papillomavirus (HPV) deregulation of Toll-like receptor 9. Oncoimmunology 3(1):e27257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narayan S, et al. (2009) Epithelial expression of human papillomavirus type 16 E7 protein results in peripheral CD8 T-cell suppression mediated by CD4+CD25+ T cells. European Journal of Immunology 39(2):481–490. [DOI] [PubMed] [Google Scholar]
- 36.Schmaler M, et al. (2015) IL-7R signaling in regulatory T cells maintains peripheral and allograft tolerance in mice. Proc Natl Acad Sci USA 112(43):13330–13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabrilovich DI, Nagaraj S (2009) Myeloid-derived suppressor cells as regulators of the immune system. Nature Reviews Immunology 9(3):162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umansky V, Blattner C, Gebhardt C, Utikal J (2016) The Role of Myeloid-Derived Suppressor Cells (MDSC) in Cancer Progression. Vaccines (Basel) 4(4). doi: 10.3390/vaccines4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Visser KE, Korets LV, Coussens LM (2005) De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell 7(5):411–423. [DOI] [PubMed] [Google Scholar]
- 40.Andreu P, et al. (2010) FcRgamma activation regulates inflammation-associated squamous carcinogenesis. Cancer Cell 17(2):121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thun MJ, Henley SJ, Gansler T (2004) Inflammation and cancer: an epidemiological perspective. Novartis Found Symp 256:6–21; discussion 22–28, 49–52, 266–269. [PubMed] [Google Scholar]
- 42.Vincent J, et al. (2010) 5-Fluorouracil Selectively Kills Tumor-Associated Myeloid-Derived Suppressor Cells Resulting in Enhanced T Cell-Dependent Antitumor Immunity. Cancer Research 70(8):3052–3061. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki E, Kapoor V, Jassar AS, Kaiser LR, Albelda SM (2005) Gemcitabine selectively eliminates splenic Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clin Cancer Res 11(18):6713–6721. [DOI] [PubMed] [Google Scholar]
- 44.Dahan R, et al. (2015) FcγRs Modulate the Anti-tumor Activity of Antibodies Targeting the PD-1/PD-L1 Axis. Cancer Cell 28(3):285–295. [DOI] [PubMed] [Google Scholar]
- 45.Selby MJ, et al. (2013) Anti-CTLA-4 Antibodies of IgG2a Isotype Enhance Antitumor Activity through Reduction of Intratumoral Regulatory T Cells. Cancer Immunology Research 1(1):32–42. [DOI] [PubMed] [Google Scholar]
- 46.Diaz-Montero CM, et al. (2009) Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother 58(1):49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen DS, Mellman I (2013) Oncology meets immunology: the cancer-immunity cycle. Immunity 39(1):1–10. [DOI] [PubMed] [Google Scholar]
- 48.Kumar V, Patel S, Tcyganov E, Gabrilovich DI (2016) The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol 37(3):208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engblom C, Pfirschke C, Pittet MJ (2016) The role of myeloid cells in cancer therapies. Nature Reviews Cancer 16(7):447–462. [DOI] [PubMed] [Google Scholar]
- 50.Toor SM, et al. (2017) Myeloid cells in circulation and tumor microenvironment of breast cancer patients. Cancer Immunol Immunother 66(6):753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.