Abstract
Problematic alcohol use (PAU) is a leading cause of death and disability worldwide. Although genome-wide association studies (GWASs) have identified PAU risk genes, the genetic architecture of this trait is not fully understood. We conducted a proxy-phenotype meta-analysis of PAU combining alcohol use disorder and problematic drinking in 435,563 European-ancestry individuals. We identified 29 independent risk variants, 19 of them novel. PAU was genetically correlated with 138 phenotypes, including substance use and psychiatric traits. Phenome-wide polygenic risk score analysis in an independent biobank sample (BioVU, n=67,589) confirmed the genetic correlations between PAU and substance use and psychiatric disorders. Genetic heritability of PAU was enriched in brain and in conserved and regulatory genomic regions. Mendelian randomization suggested causal effects on liability to PAU of substance use, psychiatric status, risk-taking behavior, and cognitive performance. In summary, this large PAU meta-analysis identified novel risk loci and revealed genetic relationships with numerous other traits.
Introduction
Alcohol use and alcohol use disorder (AUD) are leading causes of death and disability worldwide [1]. Genome-wide association studies (GWAS) of AUD and problematic drinking measured by different assessments have identified potential risk genes primarily in European populations [2–5]. Quantity-frequency measures of drinking, for example the Alcohol Use Disorders Identification Test–Consumption (AUDIT-C), which sometimes reflect alcohol consumption in the normal range, differ genetically from AUD and measures of problematic drinking (e.g., the Alcohol Use Disorders Identification Test–Problems [AUDIT-P]), and show a divergent set of genetic correlations [3, 4]. The estimated SNP-based heritability (h2) of AUD ranges from 5.6% to 10.0% [2–5]. To date, more than 10 risk variants have been significantly associated with AUD and AUDIT-P (p < 5 × 10−8). Variants that have been mapped to several risk genes in multiple studies include ADH1B (Alcohol Dehydrogenase 1B (class I), Beta Polypeptide), ADH1C (Alcohol Dehydrogenase 1C (class I), Gamma Polypeptide), ALDH2 (Aldehyde Dehydrogenase 2 Family Member, only in some Asian samples), SLC39A8 (Solute Carrier Family 39 Member 8), GCKR (Glucokinase Regulator), and CRHR1 (Corticotropin Releasing Hormone Receptor 1). In the context of the known extensive polygenicity underlying AUD and AUDIT-P, we anticipate that additional significant risk loci can be identified by increasing sample size; this is the pattern for GWAS of heterogenous complex traits in general also. We characterize both AUD itself and AUDIT-P, as “problematic alcohol use” (PAU). To identify additional risk variants and enhance our understanding of the genetic architecture of PAU, we conducted genome-wide meta-analysis of AUD and AUDIT-P in 435,563 individuals of European ancestry. Our understanding of the genetic architecture of PAU in African populations lags far behind that in Europeans; the largest sample of African ancestry individuals published so far is 56,648 in the Million Veteran Program (MVP) [3] and results have not moved beyond a single genomic region that includes ADH1B. We limited the focus here to European samples because we could not achieve a substantial increment in African-ancestry subjects over previous studies.
Results
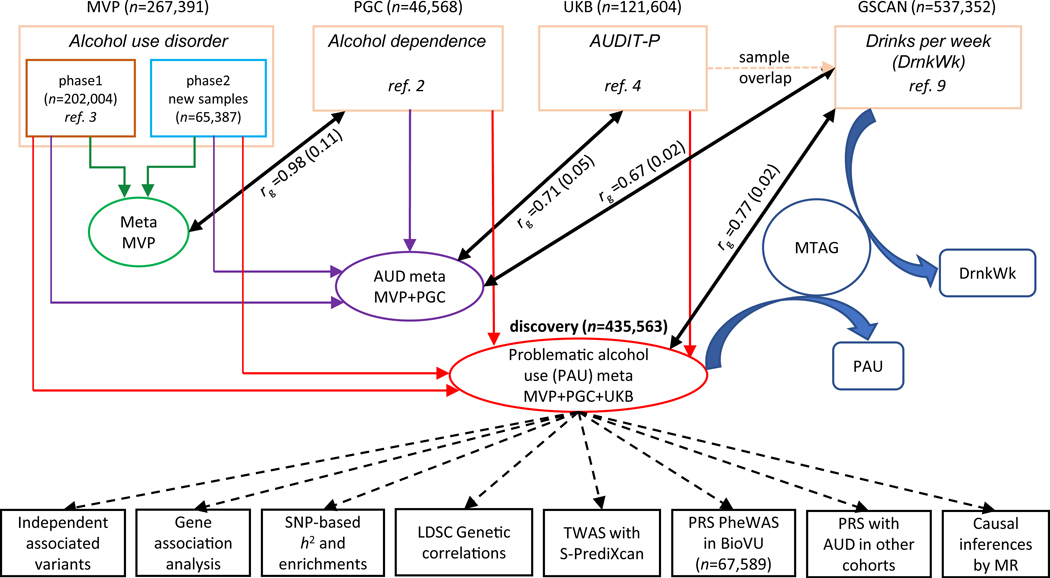
Figure 1 provides an overview of the meta-analysis of the 4 major datasets. The first is the GWAS of AUD in European Americans (EA) from MVP [6] (herein designated “MVP phase1”), comprised of 202,004 individuals phenotyped for AUD (ncase = 34,658, ncontrol = 167,346, neffective = 114,847) using International Classification of Diseases (ICD) codes [3]. The second, MVP Phase2, included an additional 65,387 EA individuals from MVP (ncase = 11,337, ncontrol = 54,050, neffective = 37,485) not previously analyzed. The third dataset is a GWAS of DSM-IV alcohol dependence (AD) from the Psychiatric Genomics Consortium (PGC), which included 46,568 European participants (ncase = 11,569, ncontrol = 34,999, neffective = 26,853) [2]. The fourth dataset is a GWAS of Alcohol Use Disorders Identification Test–Problems (AUDIT-P; a measure of problematic drinking) scores from a UK Biobank sample (UKB) [7] that included 121,604 European participants [4].
Figure 1. Overview of the analysis.
The four datasets that were meta-analyzed as the discovery sample for problematic alcohol use (PAU) included MVP phase1, MVP phase2, PGC, and UK Biobank (UKB). MVP phase1 and phase2 were meta-analyzed, and the result was used to test the genetic correlation with PGC alcohol dependence. An intermediary meta-analysis (AUD meta) combining MVP phase1, phase2, and PGC was then conducted to measure the genetic correlation with UKB AUDIT-P. Due to the sample overlap between UKB and GSCAN, we used the AUD (intermediary) meta-analysis for Mendelian Randomization (MR) analysis rather than the PAU (i.e., from the final) meta-analysis. MTAG, which used the summary data from PAU and DrnkWk (drinks per week) in GSCAN (without 23andMe samples, as those data were not available) as input to increase the power for each trait without introducing bias from sample overlap, returned summary results for PAU and DrnkWk separately.
The genetic correlation (rg) between MVP phase1 AUD and PGC AD was 0.965 (se = 0.15, p = 1.21 × 10−10) [3]. The rg between the entire MVP (meta-analysis of phase1 and phase2) and PGC was 0.98 (se = 0.11, p = 1.99 × 10−19), justifying the meta-analysis of AUD across the three datasets (neffective = 179,185). We detected 24 risk variants in 23 loci in this intermediary meta-analysis (Figure 2a, Supplementary Table 1). The rg between UKB AUDIT-P and AUD (MVP+PGC) was 0.71 (se = 0.05, p = 8.15 × 10−52), and the polygenic risk score (PRS) of AUD was associated with AUDIT-P in UKB (best p-value threshold PTbest = 0.001, R2 = 0.25%, p = 3.28 × 10−41, Supplementary Table 2, Supplementary Figure 1), justifying the proxy-phenotype meta-analysis of problematic alcohol use (PAU) across all four datasets. (AUD and AUDIT-P, though highly correlated genetically, are not identical traits). The total sample size was 435,563 in the discovery analysis (neffective = 300,789).
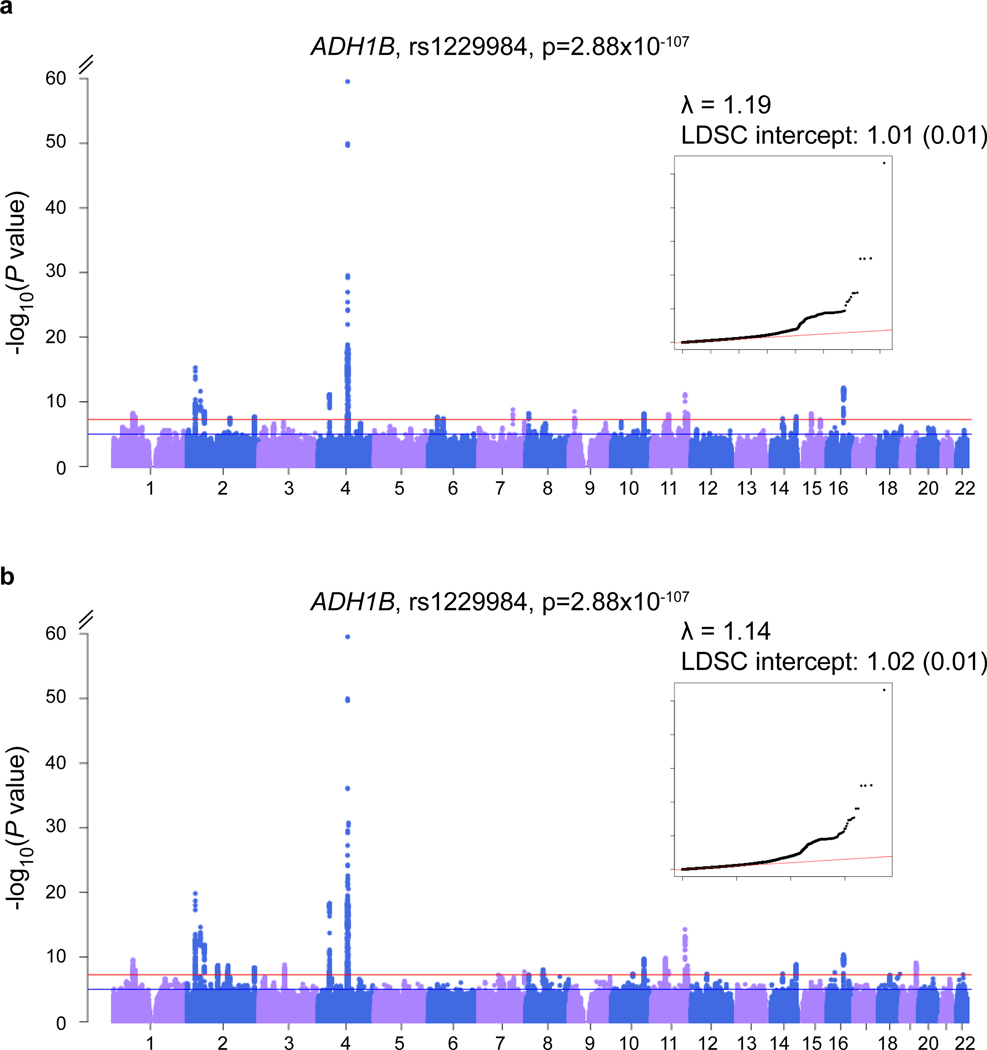
Figure 2. Association results for AUD and PAU meta-analyses.
a. Manhattan and QQ plots for AUD (MVP+PGC), ncase=57,564, ncontrol=256,395, neffective=179,185; b. Manhattan and QQ plots for PAU, n=435,563, neffective=300,789. Effective sample size weighted meta-analyses were performed using METAL. Red lines indicate GWS after correction for multiple testing (p < 5×10–8).
Association results for PAU
Of 42 lead variants (mapping to 27 loci, Figure 2b, and Supplementary Table 3) that were genome-wide significant (GWS) for PAU, 29 were independently associated after conditioning on lead SNPs in the regions (see below and Table 1). Ten variants were previously identified through the same index SNPs or tagged SNPs, located in or near the following genes: GCKR, SIX3, KLB, ADH1B, ADH1C, SLC39A8, DRD2, and FTO [2–5]. Thus, 19 variants reported here are novel, of which 11 were located in gene regions, including PDE4B Phosphodiesterase 4B), THSD7B (Thrombospondin Type 1 Domain Containing 7B), CADM2 (Cell Adhesion Molecule 2), ADH1B (different from the locus identified previously), DPP6 (Dipeptidyl Peptidase Like 6), SLC39A13 (Solute Carrier Family 39 Member 13), TMX2 (Thioredoxin Related Transmembrane Protein 2), ARID4A (AT-Rich Interaction Domain 4A), C14orf2 (Chromosome 14 Open Reading Frame 2), TNRC6A (Trinucleotide Repeat Containing Adaptor 6A), and FUT2 (Fucosyltransferase 2). A novel rare ADH1B variant, rs75967634 (p = 1.07 × 10−9, with a minor allele frequency of 0.003), which causes a substitution of histidine for arginine, is in the same codon as rs2066702 (a well-known variant associated with AUD in African populations [3, 8], but not polymorphic in European populations).This association is independent of rs1229984 in ADH1B and rs13125415 (a tag SNP of rs1612735 in MVP phase1 [3]) in ADH1C. The identification of rs75967634 demonstrates the present study’s greater power to detect risk variants in this region, beyond the frequently reported ADH1B*rs1229984.
Table 1. Genome-wide significant associations for PAU.
The total sample size is 435,563, effective sample size from each cohort was used for sample size weighted meta-analyses (neffective=300,789) using METAL.
| Chr | Pos (hg19) | rsID | Gene | A1 | A2 | EAF | Z | P | Direction |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 66419905 | rs6421482 | PDE4Ba | A | G | 0.4363 | −6.315 | 2.7×10−10 | ---- |
| 1 | 73848610 | rs61767420 | [] | A | G | 0.3999 | 5.714 | 1.11×10−8 | ++++ |
| 2 | 27730940 | rs1260326 | GCKRa | T | C | 0.4033 | −9.296 | 1.45×10−20 | --+- |
| 2 | 45141180 | rs494904 | SIX3b | T | C | 0.5961 | −7.926 | 2.26×10−15 | ---- |
| 2 | 58042241 | rs1402398 | VRK2b | A | G | 0.6274 | 7.098 | 1.27×10−12 | ++++ |
| 2 | 104134432 | rs9679319 | [] | T | G | 0.4797 | −6.01 | 1.86×10−9 | ---- |
| 2 | 138264231 | rs13382553 | THSD7Ba | A | G | 0.766 | −6.001 | 1.97×10−9 | ---- |
| 2 | 227164653 | rs2673136 | IRS1b | A | G | 0.6387 | −5.872 | 4.31×10−9 | ---- |
| 3 | 85513793 | rs62250713 | CADM2a | A | G | 0.368 | 6.049 | 1.46×10−9 | ++++ |
| 4 | 39404872 | rs13129401 | KLBb | A | G | 0.4532 | −8.906 | 5.29×10−19 | ---- |
| 4 | 100229016 | rs75967634 | ADH1Ba | T | C | 0.003 | −6.098 | 1.07×10−9 | --?- |
| 4 | 100239319 | rs1229984 | ADH1Ba | T | C | 0.0302 | −22 | 2.9×10−107 | ---? |
| 4 | 100270452 | rs13125415 | ADH1Ca | A | G | 0.5849 | −9.073 | 1.16×10−19 | ---- |
| 4 | 103198082 | rs13135092 | SLC39A8a | A | G | 0.9192 | 11.673 | 1.75×10−31 | ++++ |
| 7 | 153489074 | rs2533200 | DPP6a | C | G | 0.5163 | −5.631 | 1.79×10−8 | ---- |
| 8 | 57424874 | rs2582405 | PENKb | T | C | 0.237 | 5.751 | 8.86×10−9 | ++++ |
| 10 | 72907951 | rs7900002 | UNC5Bb | T | G | 0.6012 | −5.503 | 3.74×10−8 | --+- |
| 10 | 110537834 | rs56722963 | [] | T | C | 0.2551 | −6.374 | 1.85×10−10 | ---- |
| 11 | 47423920 | rs10717830 | SLC39A13a | G | GT | 0.674 | 6.422 | 1.34×10−10 | ++++ |
| 11 | 57480623 | rs576859 | TMX2a | A | C | 0.3272 | 5.67 | 1.43×10−8 | +++? |
| 11 | 113357710 | rs138084129 | DRD2b | A | AATAT | 0.6274 | 7.824 | 5.13×10−15 | ++++ |
| 11 | 113443753 | rs6589386 | DRD2b | T | C | 0.4323 | −7.511 | 5.88×10−14 | ---- |
| 11 | 121542923 | rs1783835 | SORL1b | A | G | 0.4569 | −5.979 | 2.24×10−9 | ---- |
| 12 | 51903860 | rs12296477 | SLC4A8b | C | G | 0.5469 | 5.484 | 4.15×10−8 | ++++ |
| 14 | 58765903 | rs61974485 | ARID4Aa | T | C | 0.2646 | 5.506 | 3.67×10−8 | ++++ |
| 14 | 104355883 | rs8008020 | C14orf2a | T | C | 0.4175 | 6.062 | 1.35×10−9 | ++++ |
| 16 | 24693048 | rs72768626 | TNRC6Aa | A | G | 0.9448 | 5.591 | 2.26×10−8 | ++++ |
| 16 | 53820813 | rs9937709 | FTOa | A | G | 0.585 | 6.602 | 4.06×10−11 | ++++ |
| 19 | 49206417 | rs492602 | FUT2a | A | G | 0.5076 | −6.143 | 8.08×10−10 | ---- |
Listed are the 29 independent variants that were genome-wide significant. Variants labeled in bold are novel associations with PAU. A1, effect allele; A2, other allele; EAF, effective allele frequency. Directions are for the A1 allele in MVP phase1, MVP phase2, PGC, and UKB datasets.
Protein-coding gene contains the lead SNP,
Protein-coding gene nearest to the lead SNP.
Moderate genetic correlation between AUD and alcohol consumption and pervasive pleiotropic effects of SNPs were demonstrated previously [2–4]. Some of the novel variants (10 of 19) identified in this study were also associated with other alcohol-related traits, including AUDIT-C score [3], total AUDIT score [4], and drinks per week (DrnkWk) from the GSCAN (GWAS & Sequencing Consortium of Alcohol and Nicotine use) study [9] (described below and in Supplementary Table 3). Rs1402398, close to VRK2, was associated with AUDIT-C score (tagged by rs2683616) [3]; rs492602 in FUT2 was associated with DrnkWk [9] and total AUDIT score [4]; and rs6421482, rs62250713, rs2533200, rs10717830, rs1783835, rs12296477, rs61974485, and rs72768626 were associated with DrnkWk directly or through tag SNPs in high linkage disequilibrium (LD) [9]. Analysis conditioned on DrnkWk shows that 11 of the 29 independent variants were independently associated with PAU (i.e., not mediated by DrnkWk) (Supplementary Table 3).
Gene-based association analysis identified 66 genes that were associated with PAU at GWS (p < 2.64 × 10−6, Supplementary Table 4). DRD2, which has been extensively studied in many fields of neuroscience, was among these genes and was previously reported in both UKB [4] and MVP phase1 [3]. Among the 66 genes, 46 are novel, including ADH4 (Alcohol Dehydrogenase 4 (class II), Pi polypeptide), ADH5 (Alcohol Dehydrogenase 5 (class III), Chi Polypeptide), and ADH7 (Alcohol Dehydrogenase 7 (class IV), Mu or Sigma Polypeptide), extending alcohol metabolizing gene associations beyond the well-known ADH1B and ADH1C; SYNGAP1 (Synaptic Ras GTPase Activating Protein 1), BDNF (Brain-Derived Neurotrophic Factor), and others. Certain genes show associations with multiple traits including previous associations with AUDIT-C (4 genes in MVP phase1, 12 genes in UKB), total AUDIT score (19 genes in UKB), and DrnkWk (46 genes in GSCAN, which includes results for DrnkWk after MTAG (multi-trait analysis of GWAS) [10] analysis).
Examination of the 66 associated genes for known drug-gene interactions through the Drug Gene Interaction Database v3.0.2 [11] showed 327 interactions between 16 genes and 325 drugs (Supplementary Table 5). Of these 16 genes with interactions, DRD2 had the most drug interactions (n = 177), followed by BDNF (n = 68) and PDE4B (n = 36).
SNP-based h2 and partitioning heritability enrichment
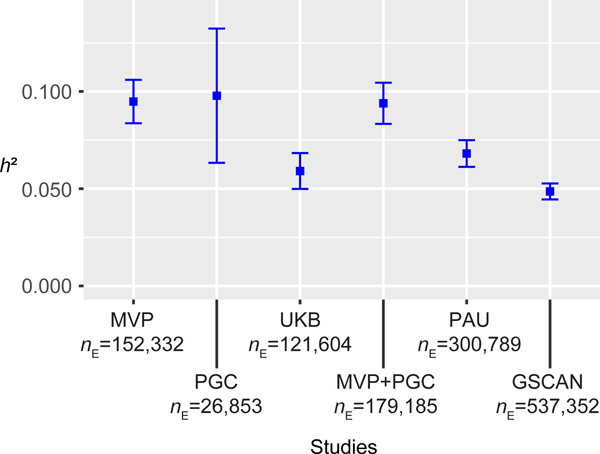
We used LD Score Regression (LDSC) [12] to estimate SNP-based h2 in the different datasests and the meta-analyses (Figure 3). Because of the unbalanced case/control ratio, we used effective sample size instead of actual sample size in MVP (following the PGC AD GWAS [2]). The h2 of PAU (the meta result) was 0.068 (se = 0.004). The h2 of AUD in the MVP metaanalysis (phases 1 and 2) was 0.095 (se = 0.006) and 0.094 (se = 0.005) in the meta-analysis that combined MVP and PGC.
Figure 3. Estimated SNP-based h2.
h2 results for single datasets or meta-analysis between datasets, from published studies or analyzed here. MVP is the phase1-phase2 MVP metaanalysis, PAU is the discovery meta-analysis. Effective sample sizes (nE) were used in LDSC. Center values are the estimated h2 and error bars indicate 95% confidence intervals.
Partitioning heritability enrichment analyses using LDSC [13, 14] showed the most significantly enriched cell type group to be central nervous system (CNS, p = 3.53 × 10−9), followed by adrenal and pancreas (p = 1.89 × 10−3), and immune and hematopoietic (p = 3.82 × 10−3, Supplementary Figure 2). Significant enrichments were also observed in six baseline annotations, including conserved regions, conserved regions with 500bp extended (ext), fetal DHS (DNase I hypersensitive sites) ext, weak enhancers ext, histone mark H3K4me1 ext, and TSS (transcription start site) ext (Supplementary Figure 3). We also investigated heritability enrichments using Roadmap data, which contains six annotations (DHS, H3K27ac, H3K4me3, H3K4me1, H3K9ac, and H3K36me3) in a subset of 88 primary cell types and tissues [14, 15]. Significant enrichments were observed for H3K4me1 and DHS in fetal brain, and H3K4me3 in fetal brain and in brain germinal matrix (Supplementary Table 6). Although no heritability enrichment was observed in tissues using gene expression data from GTEx [16], the top nominally enriched tissues were all in brain (Supplementary Figure 4).
Functional enrichments
MAGMA tissue expression analysis [17, 18] using GTEx showed significant enrichments in several brain tissues including cerebellum and cortex (Supplementary Figure 5). Although no enrichment was observed via MAGMA gene-set analysis using gene-based p-values of all protein-coding genes, the 152 genes prioritized by positional, expression quantitative trait loci (eQTL), and chromatin interaction mapping were enriched in several gene sets, including ethanol metabolic processes (Supplementary Table 7).
Genetic correlations with other traits
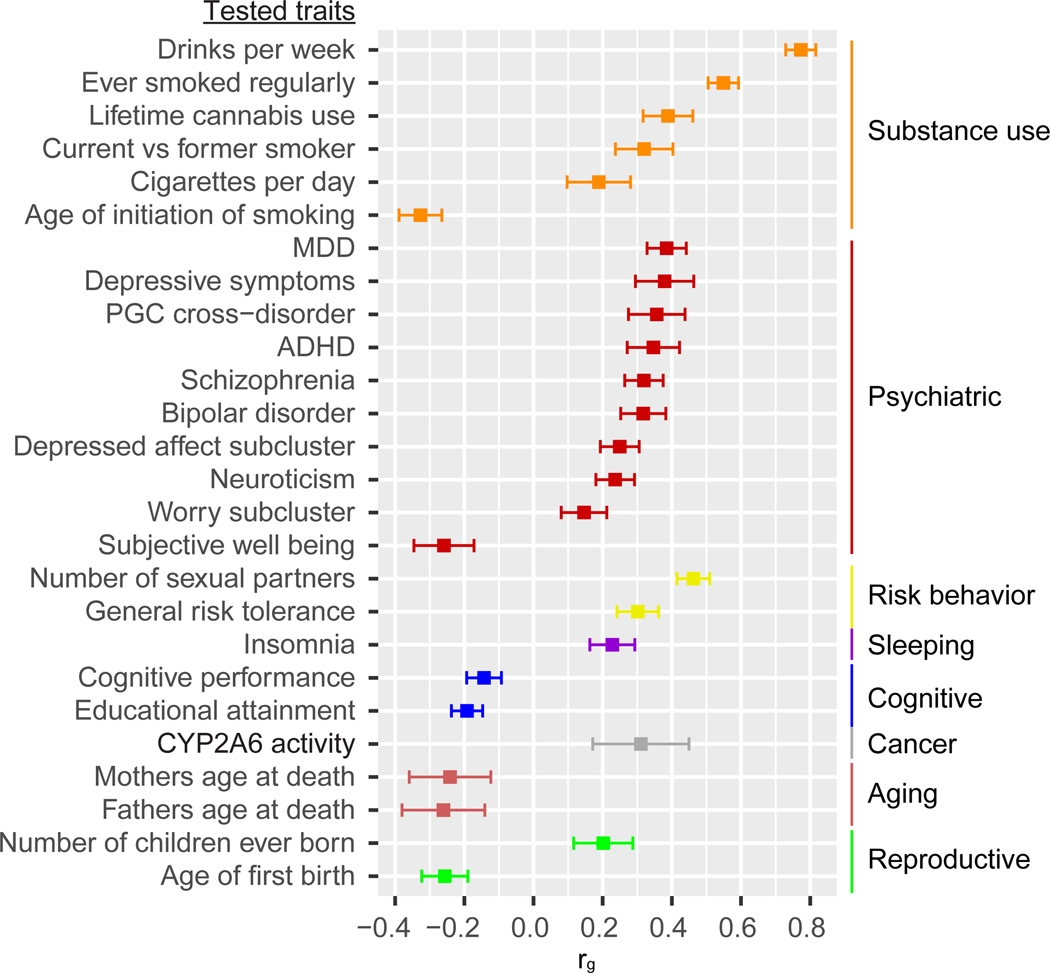
We estimated the genetic correlations between PAU and 715 publicly available sets of GWAS summary statistics, which included 228 published sets and 487 unpublished sets from the UK Biobank. After Bonferroni correction (p < 6.99 × 10−5), 138 traits were significantly correlated with PAU (Supplementary Table 8). Among the 26 published correlated traits, drinks per week showed the highest correlation with PAU (rg = 0.77, se = 0.02, p = 3.25 × 10−265), consistent with the overall quantity of alcohol consumed being a key domain of PAU [5, 19]. Several smoking traits and lifetime cannabis use were positively genetically correlated with PAU, consistent with the high comorbidity between alcohol and other substance use disorders in the general population [20]. Among psychiatric disorders, major depressive disorder (MDD, rg = 0.39, se = 0.03, p = 1.43 × 10−40) showed the highest genetic correlation with PAU, extending the evidence for a shared genetic contribution to MDD and alcohol-related traits [21, 22]. PAU was positively correlated with risk-taking behavior, insomnia, CYP2A6 activity, and other traits, and negatively correlated with cognitive traits and parents’ age at death. These findings are in line with the known adverse medical, psychiatric, and social consequences of problem drinking (Figure 4).
Figure 4. Genetic correlations with published traits.
LDSC was applied to test genetic correlation between PAU and 715 traits. Of 228 published traits, 26 were genetically correlated with PAU after Bonferroni correction (p < 6.99×10−5). MDD, major depressive disorder; ADHD, attention deficit hyperactivity disorder. Center values are the estimated genetic correlation and error bars indicate 95% confidence intervals.
Transcriptomic analyses
We used S-PrediXcan [23] to predict gene expression and the mediating effects of variation on gene expression on PAU. Forty-eight tissues from GTEx [16] release v7 and whole blood samples from the Depression Genes and Networks study (DGN) [24] were analyzed as reference transcriptomes (Supplementary Table 9). After Bonferroni correction, 103 gene-tissue associations were significant, representing 39 different genes, some of which were identified in multiple tissues (Supplementary Table 10). For example, C1QTNF4 (C1q and TNF Related 4) was detected in 18 tissues, including brain, gastrointestinal, adipose, and liver. None of the four significant alcohol dehydrogenase genes (ADH1A, ADH1B, ADH4, and ADH5) was associated with expression in brain tissue, but they were associated with expression in other tissues -- adipose, thyroid, gastrointestinal and heart. These cross-tissue associations indicate that there are widespread functional consequences of PAU-risk-associated genetic variation at the expression level.
Although the sample size for tissues used for eQTL analysis limits our ability to detect associations, there are substantial common eQTLs across tissues [16]. Integrating evidence from multiple tissues can increase power to detect genes relative to the tissues tested individually, at least for shared eQTLs. We applied S-MultiXcan [25] to the summary data for PAU using all 48 GTEx tissues as reference transcriptomic data. The expression of 34 genes was significantly associated with PAU, including ADH1B, ADH4, ADH5, C1QTNF4, GCKR, and DRD2 (Supplementary Table 11). Among the 34 genes, 27 overlapped with genes detected by S-PrediXcan.
PAU PRS for phenome-wide associations
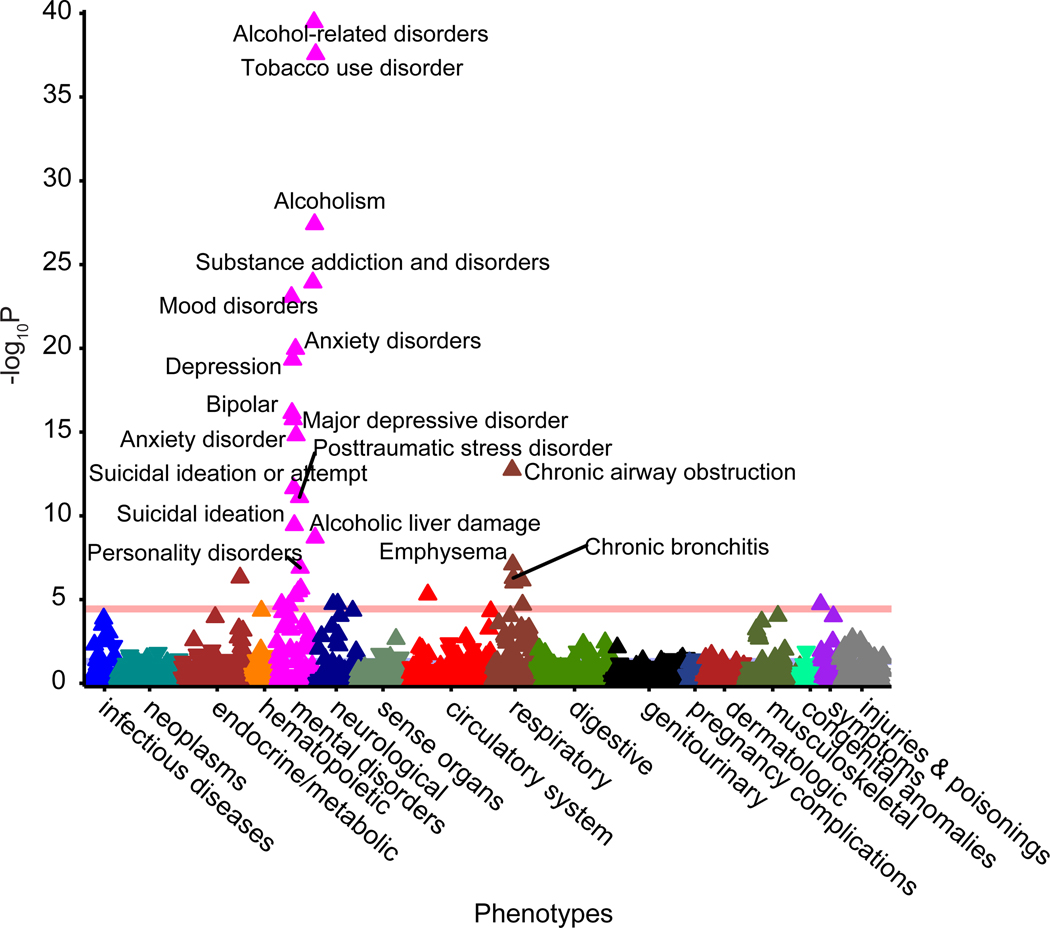
We calculated PRS for PAU in 67,589 individuals of European descent from the Vanderbilt University Medical Center’s biobank, BioVU. We conducted a phenome-wide association study (PheWAS) of PRS for PAU adjusting for sex, age (calculated as the median age across an individual’s medical record), and the top 10 principal components of ancestry. We standardized the PRS so that the odds ratios correspond to a standard deviation increase in the PRS. After Bonferroni correction, 31 of the 1,372 phenotypes tested were significantly associated with PAU PRS, including alcohol-related disorders (OR = 1.46, se = 0.03, p = 3.34 × 10−40), alcoholism (OR = 1.33, se = 0.03, p = 3.85 × 10−28), tobacco use disorder (OR = 1.21, se = 0.01, p = 2.71 × 10−38), 6 respiratory conditions, and 17 additional psychiatric conditions (Figure 5, Supplementary Table 12).
Figure 5. Phenome-Wide associations with PAU PRS in BioVU.
Polygenic score for PAU was calculated in 67,588 participants in BioVU (Vanderbilt University Medical Center’s biobank) using PRS-CS. 1,372 phenotypes were tested and Bonferroni correction (p < 3.64×10−5) was applied.
PAU PRS with AD in independent samples
We tested the association between PAU PRS and alcohol dependence in 3 independent samples: the iPSYCH group (ncase = 944, ncontrol = 11,408, neffective = 3,487); University College London (UCL) Psych Array (ncase = 1,698, ncontrol = 1,228, neffective = 2,851); and UCL Core Exome Array (ncase = 637, ncontrol = 9,189, neffective = 2,383). The PAU PRSs were significantly associated with AD in all three samples, with the most variance explained in the UCL Psych Array sample, which includes the most alcohol dependence cases (PTbest = 0.001, R2 = 2.12%, p = 8.64 × 10−14). In the iPSYCH group and UCL Core Exome Array samples, the maximal variance explained was 1.61% (PTbest = 0.3, p = 1.87 × 10−22), and 0.77% (PTbest = 5 × 10−8, p = 1.65 × 10−7), respectively (Supplementary Table 13).
Mendelian Randomization
We tested the bi-directional causal effects between other traits and AUD (MVP+PGC), rather than PAU; the UKB AUDIT-P GWAS sample was excluded to minimize overlap with other GWAS for putative exposures. (When we refer to exposure having causal effect on outcome, this should be understood to mean susceptibility or liability to exposure having causal effect on susceptibility or liability to outcome.) We limited the exposures to those genetically correlated with PAU, and which yielded >10 available instruments to have a robust causal estimate. Among the 15 tested exposures on AUD, seven showed evidence of a causal effect on liability to AUD (Table 2). DrnkWk and ever smoked regularly have a positive causal effect on AUD risk by all four methods, without violating MR assumptions through horizontal pleiotropy (MR-Egger intercept p > 0.05). General risk tolerance was causally related to AUD risk, and the estimate was robust after correction for horizontal pleiotropy. The “worry” sub-cluster of neuroticism and number of sexual partners show evidence of positive causal effects on liability to AUD with at least one method, while cognitive performance and educational attainment show evidence of negative causal effects. As an exposure, AUD has a positive causal effect on DrnkWk, and a negative causal effect on educational attainment, indicating bi-directional causality. There is no evidence of a causal effect of AUD on other traits (Table 3).
Table 2.
Causal effects on AUD (MVP+PGC) by MR.
| Exposure (#instruments) | Ref | IVW [27] | Weighted median [28] | MR-Egger [29] | MR-Egger intercept p | MR-PRESSO [30] | GSMR [31] | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (se) | p | β (se) | p | β (se) | p | #outlier | β (se) | p | #HEIDI-outlier | β (se) | p | |||
| DrnkWk (58) | [9] | 0.89 (0.06) | 1.80×10−46 | 0.89 (0.08) | 2.89×10−26 | 0.91 (0.20) | 3.80×10−6 | 0.898 | 0 | 0.89 (0.06) | 1.58×10−20 | 2 | 0.92 (0.05) | 6.37×10−79 |
| Ever smoked regularly (199) | [9] | 0.32 (0.02) | 8.72×10−51 | 0.33 (0.02) | 4.20×10−43 | 0.26 (0.08) | 1.21×10−3 | 0.471 | 3 | 0.33 (0.02) | 1.34×10−37 | 6 | 0.34 (0.01) | 1.84×10–115 |
| Current vs former smoker (12) | [9] | 0.04 (0.09) | 0.678 | 0.00 (0.06) | 0.978 | −0.33 (0.22) | 0.140 | 0.078 | 5 | 0.02 (0.04) | 0.692 | 0 | 0.04 (0.03) | 0.292 |
| Cigarettes per day (33) | [9] | 0.04 (0.06) | 0.475 | −0.10 (0.04) | 0.010 | −0.18 (0.09) | 0.034 | 1.27×10−3 | 5 | 0.09 (0.06) | 0.151 | 4 | 0.01 (0.03) | 0.643 |
| MDD (78) | [32] | 0.14 (0.03) | 8.42×10−6 | 0.14 (0.03) | 2.79×10−6 | −0.17 (0.20) | 0.390 | 0.113 | 5 | 0.14 (0.03) | 3.73×10−6 | 1 | 0.15 (0.02) | 1.65×10−18 |
| Schizophrenia (110) | [33] | 0.04 (0.01) | 2.47×10−6 | 0.04 (0.01) | 4.96×10−6 | −0.05 (0.04) | 0.202 | 0.016 | 4 | 0.04 (0.01) | 6.03×10−8 | 5 | 0.06 (0.01) | 4.65×10−26 |
| Bipolar disorder (23) | [34] | 0.03 (0.01) | 0.012 | 0.03 (0.02) | 0.049 | −0.05 (0.07) | 0.423 | 0.120 | 0 | 0.03 (0.01) | 0.020 | 0 | 0.03 (0.01) | 6.56×10−3 |
| Depressed affect sub-cluster (56) | [35] | 0.19 (0.06) | 1.75×10−3 | 0.24 (0.05) | 5.44×10−6 | −0.20 (0.28) | 0.462 | 0.147 | 7 | 0.23 (0.04) | 1.12×10−6 | 5 | 0.26 (0.04) | 6.80×10−13 |
| Neuroticism (131) | [35] | 0.20 (0.04) | 1.10×10−7 | 0.20 (0.04) | 1.10×10−7 | −0.26 (0.16) | 0.097 | 2.64×10−3 | 6 | 0.19 (0.03) | 5.83×10−8 | 4 | 0.17 (0.02) | 3.44×10−12 |
| Worry sub-cluster (61) | [35] | 0.13 (0.06) | 0.020 | 0.17 (0.05) | 8.06×10−4 | 0.04 (0.26) | 0.890 | 0.702 | 7 | 0.19 (0.04) | 8.64×10−5 | 5 | 0.21 (0.03) | 7.40×10−11 |
| Number of sexual partners (64) | [36] | 0.31 (0.04) | 3.27×10−12 | 0.36 (0.05) | 9.00×10−16 | 0.51 (0.20) | 0.011 | 0.309 | 4 | 0.33 (0.04) | 1.14×10−12 | 3 | 0.34 (0.03) | 6.13×10−28 |
| General risk tolerance (64) | [36] | 0.26 (0.06) | 7.37×10−6 | 0.28 (0.07) | 5.93×10−5 | 0.88 (0.25) | 3.69×10−4 | 9.62×10−3 | 0 | 0.26 (0.06) | 3.18×10−5 | 0 | 0.28 (0.05) | 1.91×10−9 |
| Insomnia (159) | [37] | 0.05 (0.01) | 1.90×10−5 | 0.03 (0.01) | 5.31×10−3 | −0.00 (0.05) | 0.993 | 0.288 | 7 | 0.04 (0.01) | 3.89×10−4 | 8 | 0.04 (0.01) | 3.51×10−6 |
| Cognitive performance (134) | [38] | −0.08 (0.02) | 1.03×10−3 | −0.05 (0.03) | 0.044 | −0.21 (0.12) | 0.086 | 0.282 | 4 | −0.08 (0.02) | 4.21×10−3 | 3 | −0.09 (0.02) | 6.20×10−8 |
| Educational attainment (570) | [38] | −0.22 (0.02) | 1.32×10−25 | −0.21 (0.02) | 1.45×10−17 | −0.24 (0.08) | 2.21×10−3 | 0.781 | 4 | −0.21 (0.02) | 1.37×10−23 | 16 | −0.23 (0.02) | 1.69×10−51 |
P-values labeled in bold are significant after multiple testing correction (p < 1.32×10−3). Traits labeled in bold are those having a causal effect on AUD by at least one method and consistent for the direction of effect by all 5 methods. IVW: inverse-variance weighted (IVW) linear regression. #outlier: number of pleiotropic variants which are removed from the MR estimate. #HEIDI-outlier: number of pleiotropic variants which are removed from the MR estimate. DrnkWk: drinks per week. MDD: major depressive disorder.
Depressed affect sub-cluster: depressed affect sub-cluster of neuroticism. Worry sub-cluster: worry sub-cluster of neuroticism.
Outliers are variants showing evidence of horizontal pleiotropy, which were removed before the causal estimate was made.
Table 3.
Causal effects of AUD (MVP+PGC) on other traits by MR.
| Outcome (#instruments) | Ref | IVW [27] | Weighted median [28] | MR-Egger [29] | MR-Egger intercept p | MR-PRESSO [30] | GSMR [31] | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (se) | p | β (se) | p | β (se) | p | #outlier | β (se) | p | #HEIDI-outlier | β (se) | p | |||
| DrnkWk (17) | [9] | 0.34 (0.05) | 3.16×10−10 | 0.31 (0.04) | 1.62×10−12 | 0.61 (0.39) | 0.117 | 0.479 | 2 | 0.30 (0.04) | 1.31×10−6 | 1 | 0.28 (0.03) | 1.72×10−25 |
| Ever smoked regularly (20) | [9] | 0.08 (0.04) | 0.021 | 0.04 (0.03) | 0.186 | −0.04 (0.06) | 0.544 | 0.032 | 4 | 0.07 (0.03) | 0.028 | 2 | 0.08 (0.02) | 6.94×10−6 |
| Lifetime cannabis use (21) | [39] | 0.05 (0.17) | 0.763 | −0.32 (0.13) | 0.013 | −0.44 (0.27) | 0.100 | 0.027 | 3 | 0.17 (0.17) | 0.320 | 2 | −0.07 (0.08) | 0.345 |
| Current vs former smoker (24) | [9] | 0.05 (0.03) | 0.113 | 0.03 (0.03) | 0.374 | 0.01 (0.07) | 0.917 | 0.482 | 1 | 0.04 (0.03) | 0.197 | 1 | 0.04 (0.02) | 0.061 |
| Cigarettes per day (23) | [9] | 0.06 (0.04) | 0.125 | 0.05 (0.04) | 0.185 | −0.06 (0.08) | 0.431 | 0.073 | 0 | 0.06 (0.04) | 0.139 | 0 | 0.06 (0.02) | 0.011 |
| Age of initiation of smoking (24) | [9] | −0.05 (0.03) | 0.065 | −0.06 (0.04) | 0.109 | 0.07 (0.05) | 0.147 | 0.004 | 1 | −0.11 (0.03) | 0.001 | 0 | −0.05 (0.02) | 0.027 |
| MDD (23) | [32] | 0.11 (0.11) | 0.320 | 0.04 (0.09) | 0.646 | −0.81 (0.51) | 0.112 | 0.064 | 10 | 0.14 (0.08) | 0.118 | 5 | 0.00 (0.05) | 0.914 |
| Depressive symptom (23) | [40] | 0.01 (0.05) | 0.794 | −0.04 (0.05) | 0.402 | −0.26 (0.21) | 0.207 | 0.177 | 1 | −0.02 (0.04) | 0.673 | 0 | 0.01 (0.04) | 0.736 |
| PGC Cross-disorder (22) | [41] | 0.31 (0.18) | 0.086 | 0.16 (0.19) | 0.382 | −2.28 (1.10) | 0.038 | 0.017 | 0 | 0.31 0.18 | 0.100 | 0 | 0.31 (0.12) | 0.010 |
| ADHD (24) | [42] | 0.25 (0.17) | 0.132 | −0.14 (0.16) | 0.405 | −0.44 (0.29) | 0.122 | 0.005 | 1 | 0.18 (0.14) | 0.220 | 1 | 0.18 (0.11) | 0.101 |
| Schizophrenia (21) | [33] | 0.45 (0.20) | 0.026 | 0.21 (0.10) | 0.045 | 0.00 (0.29) | 0.999 | 0.047 | 6 | 0.24 (0.08) | 0.009 | 6 | 0.24 (0.08) | 0.004 |
| Bipolar disorder (22) | [34] | −0.06 (0.18) | 0.732 | −0.03 (0.14) | 0.812 | −0.20 (0.31) | 0.511 | 0.569 | 2 | −0.02 (0.14) | 0.893 | 2 | −0.01 (0.11) | 0.931 |
| Depressed affect sub-cluster (22) | [35] | 0.02 (0.04) | 0.650 | −0.02 (0.03) | 0.594 | −0.08 (0.08) | 0.313 | 0.131 | 4 | 0.02 (0.03) | 0.508 | 1 | 0.00 (0.02) | 0.845 |
| Neuroticism (22) | [35] | 0.01 (0.04) | 0.840 | −0.01 (0.03) | 0.641 | −0.06 (0.07) | 0.388 | 0.234 | 4 | −0.02 (0.03) | 0.591 | 3 | −0.03 (0.02) | 0.112 |
| Worry sub-cluster (24) | [35] | 0.03 (0.04) | 0.393 | 0.01 (0.03) | 0.754 | −0.04 (0.07) | 0.591 | 0.239 | 4 | 0.01 (0.03) | 0.820 | 3 | −0.01 (0.02) | 0.777 |
| Subjective well-being (22) | [40] | −0.02 (0.05) | 0.70 | −0.05 (0.05) | 0.264 | 0.03 (0.27) | 0.921 | 0.860 | 3 | −0.06 (0.04) | 0.132 | 1 | −0.05 (0.03) | 0.092 |
| Number of sexual partners (23) | [36] | 0.09 (0.05) | 0.058 | −0.00 (0.03) | 0.941 | −0.00 (0.09) | 0.966 | 0.219 | 7 | 0.05 (0.04) | 0.225 | 4 | 0.02 (0.02) | 0.266 |
| General risk tolerance (24) | [36] | 0.05 (0.03) | 0.096 | −0.03 (0.03) | 0.323 | −0.06 (0.06) | 0.251 | 0.015 | 3 | 0.07 (0.03) | 0.053 | 0 | 0.05 (0.02) | 0.002 |
| Insomnia (24) | [37] | 0.08 (0.06) | 0.157 | 0.06 (0.06) | 0.367 | −0.04 (0.11) | 0.744 | 0.196 | 1 | 0.12 (0.06) | 0.050 | 2 | 0.10 (0.04) | 0.020 |
| Cognitive performance (22) | [38] | −0.03 (0.0) | 0.460 | −0.08 (0.03) | 0.021 | −0.09 (0.09) | 0.295 | 0.440 | 3 | −0.08 (0.04) | 0.054 | 1 | −0.05 (0.02) | 0.030 |
| Educational attainment (20) | [38] | −0.06 (0.03) | 0.055 | −0.10 (0.02) | 7.38×10−6 | −0.12 (0.06) | 0.024 | 0.152 | 3 | −0.07 (0.02) | 6.04×10−3 | 5 | −0.08 (0.02) | 3.12×10−7 |
| Mothers age at death (24) | [43] | −0.03 (0.04) | 0.424 | −0.02 (0.06) | 0.692 | −0.01 (0.08) | 0.886 | 0.764 | 0 | −0.03 0.03 | 0.342 | 0 | −0.03 (0.04) | 0.424 |
| Fathers age at death (24) | [43] | −0.05 (0.05) | 0.352 | −0.09 (0.06) | 0.113 | −0.08 (0.10) | 0.408 | 0.671 | 1 | −0.03 (0.05) | 0.523 | 0 | −0.05 (0.04) | 0.206 |
P-values labeled in bold are significant after multiple testing correction (p < 1.32×10−3). Traits labeled in bold are those having a causal effect from AUD by at least one method and consistent for the directions of effect by all 5 methods.
Joint Analysis of PAU and DrnkWk Using MTAG
We conducted a joint analysis of PAU and DrnkWk using MTAG, which can increase the power for each trait without introducing bias from sample overlap [10]. MTAG analysis increased the GWAS-equivalent sample size (nEq) for PAU to 514,790, i.e., a 71.1% increase from the original effective sample size (nE = 300,789, n = 435,563). In this analysis, we observed an increase in the number of independent variants for PAU to 119, 76 of which were conditionally independent (Supplementary Figure 6a, Supplementary Table 14). For DrnkWk, the MTAG analysis increased the nEq to 612,968 from 537,352, which yielded 141 independent variants, 86 of which were conditionally independent (Supplementary Figure 6b, Supplementary Table 15).
The MTAG analysis also increased the power for the functional enrichment analysis. MAGMA gene set analysis for PAU after MTAG analysis detected 10 enriched Gene Ontology terms, including ‘regulation of nervous system development’ (pBonferroni = 8.80 × 10−4), ‘neurogenesis’ (pBonferroni = 0.010), and ‘synapse’ (pBonferroni = 0.046) (Supplementary Table 16).
Discussion
We report here a genome-wide meta-analysis of PAU in 435,563 individuals of European ancestry from the MVP, PGC, and UKB datasets. MVP is a mega-biobank that has enrolled >750,000 subjects (for whom genotype data on 313,977 subjects were used in this study), with rich phenotype data assessed by questionnaires and from the EHR. Currently, MVP is the largest single cohort available with diagnostic information on AUD [3, 6]. PGC is a collaborative consortium that has led the effort to collect smaller cohorts with DSM-IV AD [2]. UKB is a population-level cohort with the largest available sample with AUDIT-P data [4].
Our discovery meta-analysis of PAU yielded 29 independent variants, of which 19 were novel, with 0.059 to 0.113 of the phenotypic variance explained in different cohorts or meta-analyses. The h2 in the Phase1-Phase2 MVP meta-analysis was 0.095 (se = 0.006), which was higher than MVP phase1: 0.056 (se = 0.004, in MVP phase1 where only the actual (as opposed to effective) sample size was used) [3]. The h2 of AD in PGC was 0.098 (se = 0.018), comparable to the reported liability-scale h2 (0.090, se = 0.019) [2]. Functional and heritability analyses consistently showed enrichments in brain regions and gene expression regulatory regions, providing biological insights into the etiology of PAU. Variation associated with gene expression in the brain is central to PAU risk, a conclusion that is also consistent with our previous GWASs in MVP of both alcohol consumption and AUD diagnosis [3]. The enrichments in regulatory regions point to specific brain tissues relevant to the causative genes; the specific interactions between 16 genes and 325 drugs may provide targets for the development of medications to manage PAU. Potential targets identified include the D2 dopamine receptor (encoded by DRD2) and phosphodiesterase 4B (encoded by PDE4B). The presence of risk variation at these loci also suggests that they may be “precision medicine” targets as well.
We also found that PAU was significantly genetically correlated with 138 other traits. The top correlations were with substance use and substance-related disorders, MDD, schizophrenia, and several other neuropsychiatric traits. In a conceptually similar analysis, we performed a PheWAS of PAU PRS in BioVU, which confirmed in an independent sample the genetic correlations between PAU and multiple substance use disorders, mood disorders, and other psychiatric traits. We also used MR to infer causal effects of the above traits on liability to AUD (we tested AUD excluding UKB samples to avoid sample overlap) using selected genetic instruments. We found evidence of positive causal relationships from DrnkWk (bi-directional), ever smoked regularly, worry sub-cluster, and number of sexual partners, while cognitive performance and educational attainment (bi-directional) showed protective effects on liability to AUD. In comparison, we detected few causal effects from AUD to other traits, possibly because of lack of power since there are fewer instrumental variants for AUD available in our study than for many comparison GWAS.
The study has other limitations. First, only European populations were included; therefore, the genetic architecture of PAU in other populations remains largely unknown. To date, the largest non-European sample to undergo GWAS for alcohol-related traits is African American (AA), which was reported in the MVP phase1 sample (17,267 cases; 39,381 controls, an effective sample size of 48,015), with the only associations detected on chromosome 4 in the ADH gene locus (where several ADH genes map) [3]. The collection of substantial numbers of non-European subjects will require a concerted effort by investigators in our field. Second, despite the high genetic correlation between AUD and AUDIT-P, they are not identical traits. We conducted a meta-analysis of the two traits to increase the power for the association study of PAU, consequently, associations specific to AUD or AUDIT-P could have been attenuated. Third, there was no opportunity for replication of the individual novel variants. Because the variants were detected in more than 430,000 subjects and have small effect sizes, a replication sample with adequate power would also have to be very large, and no such sample is currently available. To validate the findings, we conducted PRS analyses in three independent cohorts, which showed strong association with AUD. Although this indicates that our study had adequate power for variant detection, it does not address the validity of the individual variants discovered.
This is the largest GWAS study of PAU so far. Previous work has shown that the genetic architecture of AUD (and PAU) differs substantially from that of alcohol consumption [2–4]. There have been larger studies of alcohol quantity-frequency measures [9, 26]; alcohol consumption data are available in many EHRs, thus they were included in many studies of other primary traits, like cardiac disease. AUD diagnoses are collected much less commonly. The 3item AUDIT-C is a widely used measure of alcohol consumption that is often available in EHRs, but the full 10-item AUDIT, which allows the assessment of AUDIT-P, is not as widely available. Despite the high genetic correlation between, for example, PAU and DrnkWk (rg=0.77), very different patterns of genetic correlation and pleiotropy have been observed via LDSC and other methods for these different kinds of indices of alcohol use [2–5]. PAU captures pathological alcohol use: physiological dependence and/or significant psychological, social or medical consequences. Quantity/frequency measures may capture alcohol use that is in the normal, or anyway nonpathological, range. As such, we argue that although quantity/frequency measures are important for understanding the biology of habitual alcohol use, PAU is the more clinically important trait. Thus, we did not meta-analyze PAU with DrnkWk directly, but used MTAG analysis instead, recognizing that they are different traits. These circumstances underscore the need to assemble a large GWAS sample of PAU to inform its biology, and our study moves towards this goal via the identification of numerous previously-unidentified risk loci – we increased known PAU loci from 10 to 29, nearly tripling our knowledge of specific risk regions. Similarly, we identified 66 gene-based associations, of which 46 were novel – again roughly tripling current knowledge. MTAG analysis increased locus discovery to 119, representing 76 independent loci, by leveraging information from DrnkWk [9]. By the same token, we provide a major increment in information about the biology of PAU, providing considerable fodder for future studies that will be required to delineate the biology and function associated with each risk variant. We anticipate that knowledge of the functional effects of the variants will contribute eventually to personalized treatment of PAU, facilitating identification of individuals with PAU who may be most treatment responsive or for whom a specific medication may be most efficacious.
Methods
MVP datasets.
The MVP is a mega-biobank supported by the U.S. Department of Veterans Affairs (VA), enrollment for which began in 2011 and is ongoing. Phenotypic data were collected using questionnaires and the VA electronic health records (EHR), and a blood sample was obtained from each participant for genetic studies. Two phases of genotypic data have been released and were included in this study. MVP phase1 contains 353,948 subjects, of whom 202,004 European Americans (EA) with AUD diagnoses were included in a previous GWAS and the summary statistics were used in this study [3]. MVP phase2 released data on another 108,416 subjects, of whom 65,387 EAs with AUD diagnosis information were included in this study. Following the same procedures as for MVP phase1, participants with at least one inpatient or two outpatient alcohol-related ICD-9/10 codes from 2000 to 2018 were assigned a diagnosis of AUD.
Ethics statement:
The Central VA Institutional Review Board (IRB) and site-specific IRBs approved the MVP study. All relevant ethical regulations for work with human subjects were followed in the conduct of the study and informed consent was obtained from all participants.
Genotyping for both phases of MVP was performed using a customized Affymetrix Biobank Array. Imputation and quality control methods for MVP phase1 were described in detail in Kranzler et al. [3]. Similar methods were used for MVP phase2. Before imputation, phase2 subjects or SNPs with genotype call rate < 0.9 or high heterozygosity were removed, leaving 108,416 subjects and 668,324 SNPs. Imputation for MVP phase2 was done separately from phase1; both were performed with EAGLE2 [44] and Minimac3 [45] using 1000 Genomes Project phase 3 data [46] as the reference panel. Imputed genotypes with posterior probability ≥ 0.9 were transferred to best-guess genotypes (the rest were treated as missing genotype calls). A total of 6,635,093 SNPs with INFO scores > 0.7, genotype call rates or best guess rates > 0.95, Hardy-Weinberg Equilibrium (HWE) p value > 1 × 10−6, minor allele frequency (MAF) > 0.001 were remained for GWAS.
We removed subjects with mismatched genotypic and phenotypic sex and one subject randomly from each pair of related individuals (kinship coefficient [47] threshold = 0.0884), leaving 107,438 phase2 subjects for subsequent analyses. We used the same processes as MVP phase1 to define EAs. First, we ran principal components analysis (PCA) on 74,827 common SNPs (MAF > 0.05) shared by MVP and the 1000 Genomes phase 3 reference panels using FastPCA [48]. Then we clustered each participant into the nearest reference population according to the Euclidean distances between the participant and the centers of the 5 reference populations using the first 10 PCs. A second PCA was performed for participants who were clustered to the reference European population (EUR), and outliers were removed if any of the first 10 PCs were > 3 standard deviations from the mean, leaving 67,268 EA subjects.
Individuals < 22 or > 90 years of age and those with a missing AUD diagnosis were removed from the analyses, leaving 65,387 phase2 EAs (11,337 cases; 54,050 controls). GWAS was then performed on the MVP phase2 dataset. We used logistic regression implemented in PLINK v1.90b4.4 [49] for the AUD GWAS correcting for age, sex, and the first 10 PCs. The mean age is 63.2 (SD=13.4) in the entire MVP sample and 92.5% are males. Data collection and analysis were not performed blind to the conditions of the experiments.
PGC summary statistics.
We used the 46,568 European ancestry subjects (11,569 cases and 34,999 controls) from 27 cohorts that were analyzed by the Psychiatric Genomics Consortium (PGC). The phenotype was lifetime DSM-IV diagnosis of alcohol dependence (AD). The summary data were downloaded from the PGC website (https://www.med.unc.edu/pgc/) with full agreement to the PGC conditions. Allele frequencies were not reported in the summary data. We used allele frequencies from the 1000 Genome European sample as proxy measures in PGC for some downstream analyses.
UK Biobank summary statistics.
The UK Biobank (UKB) included 121,604 White-British unrelated subjects with available AUDIT-P scores. Past-year AUDIT-P was assessed by 7 questions: 1). Frequency of inability to cease drinking; 2). Frequency of failure to fulfil normal expectations due to drinking alcohol; 3). Frequency of needing a morning drink of alcohol after a heavy drinking session; 4). Frequency of feeling guilt or remorse after drinking alcohol; 5). Frequency of memory loss due to drinking alcohol; 6). Been injured or injured someone else through drinking alcohol; 7). Had a relative, friend, or health worker who was concerned about or suggested a reduction in alcohol consumption. The AUDIT-P was log10-transformed for GWAS (see ref [4] for details). We removed SNPs with INFO < 0.7 or call rate < 0.95.
Meta-analyses.
Meta-analyses were performed using METAL [50]. The meta-analysis within MVP (for the purpose of genetic correlation analysis with PGC AD) was conducted using an inverse variance weighted method because the two subsets were from the same cohort. The meta-analyses for AUD (MVP+PGC) and PAU (MVP+PGC+UKB) were performed using the sample size weighted method. Given the unbalanced ratios of cases to controls in MVP samples, we calculated effective sample sizes for meta-analysis following the approach used by the PGC:
The calculated effective sample sizes in MVP and reported effective sample sizes in PGC were used in meta-analyses and all downstream analyses. AUDIT-P in UKB is a continuous trait, so we used actual sample sizes for that trait. For the AUD meta-analysis, variants present in only one sample (except MVP phase1 which is much larger than the others) or with heterogeneity test p-value < 5 × 10−8 were removed, leaving 7,003,540 variants. For the PAU meta-analysis, variants present in only one sample (except MVP phase1 or UKB) or with heterogeneity test pvalue < 5 × 10−8 and variants with effective sample size < 45,118 (15% of the total effective sample size) were removed, leaving 14,069,427 variants.
AUD polygenic risk score in UKB.
We calculated AUD polygenic risk scores (PRS) for each of the 82,930 unrelated subjects in UKB (application number 41910) who had non-missing AUDITP information [7]. A PRS was calculated as the sum of the number of effective alleles with pvalues less than a given threshold, weighted by the effect sizes from AUD meta-analysis (MVP+PGC). We analyzed 10 p-value thresholds: 5 × 10−8, 1 × 10−7, 1 × 10−6, 1 × 10−5, 1 × 10−4, 0.001, 0.05, 0.3, 0.5, and 1, and clumped the AUD summary data by LD with r2 < 0.3 in a 500-kb window. Then we tested the association between AUD PRS and AUDIT-P, corrected for age, sex, and 10 PCs. The analysis was performed using PRSice-2 [51].
Independent variants and conditional analyses.
We identified the independent variant (p < 5 × 10−8) in each locus (1 Mb genomic window) based on the smallest p value and r2 < 0.1 with other independent variants and assigned these variants to the independent variant’s clump. Any two independent variants less than 1 Mb apart whose clumped regions overlapped were merged into one locus. Given the known long-range LD for the ADH gene cluster on chromosome 4, we defined chr4q23–q24 (~97.2 Mb – 102.6 Mb) as one locus. When multiple independent variants were present in a locus, we ran conditional analyses using GCTA-COJO [52] to define conditionally independent variants. For each variant other than the most significant one (index), we tested the marginal associations conditioning on the index variant using Europeans (n = 503) from the 1000 Genomes as the LD reference sample. Variants with significant marginal associations (p < 5 × 10−8) were defined as conditionally independent variants (i.e., independent when conditioned on other variants in the region) and subject to another round of conditional analyses for each significant association.
For the conditionally independent variants for AUD or PAU, we also conducted a multitrait analysis conditioning on GSCAN drinks per week [9] using GCTA-mtCOJO [31] to identify variants associated with AUD or PAU, but not drinks per week, i.e., not alcohol consumption alone. Europeans from the 1000 Genomes were used as the LD reference. For variants missing in GSCAN, we used proxy variants (p < 5 × 10−8) in high LD with the locus for analyses. Whereas conditional analyses require the beta (effect size) and standard error, we calculated these using Z-scores (z), allele frequency (p) and sample size (n) from the meta-analyses [53]:
Gene-based association analysis.
Gene-based association analysis for PAU was performed using MAGMA implemented in FUMA [17, 18], which uses a multiple regression approach to detect multi-marker effects that account for SNP p-values and LD between markers. We used default settings to analyze 18,952 autosomal genes, with p < 2.64 × 10−6 (0.05/18,952) considered GWS.
Drug-gene interaction.
For the genes identified as significant by MAGMA, we examined druggene interaction through Drug Gene Interaction Database (DGIdb) v3.0.2 [11] (http://www.dgidb.org/), a database of integrated drug–gene interaction information based on 30 sources.
SNP-based h2 and partitioning heritability enrichment.
We used LDSC [12] to estimate the SNP-based h2 for common SNPs mapped to HapMap3 [54], with Europeans from the 1000 Genomes Project [46] as the LD reference panel. We excluded the major histocompatibility complex (MHC) region (chr6: 26–34Mb).
We conducted portioning h2 enrichment analyses for PAU using LDSC in different models [13, 14]. First, we analyzed a baseline model consisting of 52 functional categories that included genomic features (coding, intron, UTR etc), regulatory annotations (promoter, enhancer etc), epigenomic annotations (H3K27ac, H3K4me1, H3K3me3 etc) and others (see ref [13] for details, Supplementary Figure 3). We then analyzed cell type group h2 enrichments with 10 cell types: central nervous system (CNS), adrenal and pancreas, immune and hematopoietic, skeletal muscle, gastrointestinal, liver, cardiovascular, connective tissue and bone, kidney, and other (see ref [13] for details, Supplementary Figure 2). Third, we used LDSC to test for enriched heritability in regions surrounding genes with the highest tissue-specific expression using 53 human tissue or cell type RNA-seq data from the Genotype-Tissue Expression Project (GTEx) [16], or enriched heritability in epigenetic markers from 396 human epigenetic annotations (six features in a subset of 88 primary cell types or tissues) from the Roadmap Epigenomics Consortium [15] (see ref [14] for details, Supplementary Figure 4, Supplementary Table 6). For each model, the number of tested annotations was used to calculate a Bonferroni corrected p-value < 0.05 as a significance threshold.
Gene-set and functional enrichment.
We performed gene-set analysis for PAU for curated gene sets and Gene Ontology (GO) terms using MAGMA [17, 18]. We then used MAGMA for gene-property analyses to test the relationships between tissue-specific gene expression profiles and PAU-gene associations. We analyzed gene expression data from 53 GTEx (v7) tissues. We also performed gene-set analysis on the 152 prioritized genes using MAGMA. Gene sets with adjusted p-value < 0.05 were considered as significant.
Genetic correlation.
We estimated the genetic correlation (rg) between traits using LDSC [55]. For PAU, we estimated the rg with 218 published traits in LD Hub [56], 487 unpublished traits from the UK Biobank (integrated in LD Hub), and recently published psychiatric and behavioral traits [9, 32, 34–39, 42, 57, 58], bringing the total number of tested traits to 715 (Supplementary Table 8). For traits reported in multiple studies or in UKB, we selected the published version of the phenotype or used the largest sample size. Bonferroni correction was applied and correlation was considered significant at a p-value threshold of 6.99 × 10-5.
S-PrediXcan and S-MultiXcan.
To perform transcriptome-wide association analysis, we used S-PrediXcan [23] (a version of PrediXcan that uses GWAS summary statistics [59]) to integrate transcriptomic data from GTEx [16] and the Depression Genes and Networks study (DGN) [24] to analyze the summary data from the PAU meta-analysis. Forty-eight tissues with sample size > 70 from GTEx release v7 were analyzed, totaling 10,294 samples. DGN contains RNA sequencing data from whole blood of 992 genotyped individuals. The transcriptome prediction model database and the covariance matrices of the SNPs within each gene model were downloaded from the PredictDB repository (http://predictdb.org/, 2018–01-08 release). Only individuals of European ancestry in GTEx were analyzed. S-PrediXcan was performed for each of the 49 tissues (48 from GTEx and 1 from DGN), for a total of 254,345 gene-tissue pairs. Significant association was determined by Bonferroni correction (p < 1.97 × 10−7).
Considering the limited eQTL sample size for any single tissue and the substantial sharing of eQTLs across tissues, we applied S-MultiXcan [25], which integrates evidence across multiple tissues using multivariate regression to improve association detection. Forty-eight tissues from GTEx were analyzed jointly. The threshold for condition number of eigenvalues was set to 30 when truncating singular value decomposition (SVD) components. In total, 25,626 genes were tested in S-MultiXcan, leading to a significant p-value threshold of 1.95 × 10−6 (0.05/25,626).
PAU PRS for phenome-wide associations.
Polygenic scores were generated using PRS-CS [60] on all genotyped individuals of European descent (n = 67,588) in Vanderbilt University Medical Center’s EHR-linked biobank, BioVU. PRS-CS uses a Bayesian framework to model linkage disequilibrium from an external reference set and a continuous shrinkage prior on SNP effect sizes. We used 1000 Genomes Project Phase 3 European sample [46] as the LD reference. Additionally, we used the PRS-CS-auto option, which allows the software to learn the continuous shrinkage prior from the data. Polygenic scores were constructed from PRS-CS-auto adjusted summary statistics containing 811,292 SNPs. All individuals used for polygenic scoring were genotyped on the Illumina Multi-Ethnic Global Array (MEGA). Genotypes were filtered for SNP (95%) and individual (98%) call rates, sex discrepancies, and excessive heterozygosity. For related individuals, one of each pair was randomly removed (pi_hat > 0.2). SNPs showing significant associations with genotyping batch were removed. Genetic ancestry was determined by principal component analysis performed using EIGENSTRAT [61]. Imputation was completed using the Michigan Imputation Server [45] and the Haplotype Reference Consortium [62] as the reference panel. Genotypes were then converted to hard calls, and filtered for SNP imputation quality (R2 < 0.3), individual missingness (>2%), SNP missingness (>2%), MAF (<1%) and HWE (p < 1 × 10−10). The resulting dataset contained 9,330,483 SNPs on 67,588 individuals of European ancestry.
We conducted a phenome-wide association study (PheWAS) [63] of the PAU PRS by fitting a logistic regression model to 1,372 case/control phenotypes to estimate the odds of each diagnosis given the PAU polygenic score, controlling for sex, median age across the medical record, top 10 principal components of ancestry, and genotyping batch. We required the presence of at least two International Classification of Disease (ICD) codes that mapped to a PheWAS disease category (Phecode Map 1.2) to assign “case” status. A phenotype was required to have at least 100 cases to be included in the analysis. PheWAS analyses were run using the PheWAS R package [64]. Bonferroni correction was applied to test for significance (p < 0.05/1,372).
PAU PRS in independent samples.
We calculated PAU PRS in three independent samples, where we tested the association between PAU PRS and AD, corrected for age, sex, and 10 PCs. Ten p-value thresholds were applied in all samples.
iPSYCH Group.
DNA samples for cases and controls were obtained from newborn bloodspots linked to population registry data [65]. Cases were identified with the ICD-10 code F10.2 (AD; n = 944); controls were from the iPSYCH group (n = 11,408; neffective = 3,487)). The iPSYCH sample was genotyped on the Psych Array (Illumina, San Diego, CA, US). GWAS QC, imputation against the 1,000 Genomes Project panel [46] and association analysis using the Ricopili pipeline [66] were performed. The current study is part of a general study in iPSYCH investigating the comorbidity of alcohol misuse and psychiatric disorders.
UCL Psych Array.
Cases were identified with ICD-10 code F10.2 (n = 1,698) and comprised 492 individuals with a diagnosis of alcoholic hepatitis who had participated in the STOPAH (Steroids or Pentoxifylline for Alcoholic Hepatitis) trial (ISRCTN88782125; EudraCT Number: 2009013897–42) and 1,206 subjects recruited from the AD arm of the DNA Polymorphisms in Mental Health (DPIM) study; controls were UK subjects who had either been screened for an absence of mental illness and harmful substance use (n = 776), or were random blood donors (n-452; total n = 1,228; neffective = 2,851). The sample was genotyped on the Psych Array (Illumina, San Diego, CA, US). GWAS QC was performed using standard methods and imputation was done using the haplotype reference consortium (HRC) panel [67] on the Sanger Imputation server (https://imputation.sanger.ac.uk/). Association testing was performed using Plink1.9 [49].
UCL Core Exome Array.
Cases had an ICD-10 diagnosis of F10.2 (n = 637), including 324 individuals with a diagnosis of alcoholic hepatitis who had participated in the STOPAH trial and 313 subjects recruited from the AD arm of the DPIM study; controls were unrelated UK subjects from the UK Household Longitudinal Study (UKHLS; n = 9,189; neffective = 2,383). The sample was genotyped on the Illumina Human Core Exome Array (Illumina, San Diego, CA, US). GWAS QC was performed using standard methods and imputation was done using the HRC panel [67] on the Sanger Imputation server (https://imputation.sanger.ac.uk/). Association testing was performed with Plink1.9 [49].
Mendelian Randomization.
We used Mendelian Randomization (MR) to investigate the bidirectional causal relationships between PAU liability and traits that were significantly genetically correlated (p < 6.99 × 10−5). However, all or most of the published traits in recent large GWAS include UKB data. To avoid biases caused by overlapping samples in MR analysis, we only tested the relationship between published traits and AUD (MVP+PGC). For robust causal effect inference, we limited the traits studied to those with more than 10 available instruments (association p < 5 × 10−8). For causality on AUD, 15 exposures were analyzed (Table 2), and for causality from AUD on others, 23 traits were tested. We applied Bonferroni correction for the 38 hypotheses, interpreting p-values < 1.32×10−3 (0.05/38) as significant.
Four methods, weighted median [28], inverse-variance weighted (IVW, random-effects model) [27], and MR-Egger [29], implemented in the R package “MendelianRandomization v0.3.0” [68], MR-PRESSO [30], and GSMR [31] were used for MR inference. Evidence of average pleiotropic effects was examined by the MR-Egger intercept test, where a non-zero intercept indicates horizontal pleiotropy [29]. Individual variants with horizontal pleiotropy were detected by MR-PRESSO, and an outlier test was applied to correct horizontal pleiotropy via outlier removal. Pleiotropic variants were also detected by the HEIDI test in GSMR, and removed from causal inference. Instrumental variants that are associated with outcome (p < 5 × 10−8) were removed. For instrumental variants missing in the outcome summary data, we used the results of the best-proxy variant with the highest LD (r2 > 0.8) with the missing variant. If the MAF of the missing variant was < 0.01, or none of the variants within 200 kb had LD r2 > 0.8, we removed the instrumental variant from the analysis. Palindromic SNPs (A/T or G/C alleles) with MAF [0.4, 0.5], which can introduce ambiguity into the identity of the effect allele, were also removed.
MTAG between PAU and drinks per week.
Multiple trait analysis between PAU and drinks per week (DrnkWk) from GSCAN was performed on summary statistics with multi-trait analysis of GWAS (MTAG) v1.0.7 [10]. The summary data of DrnkWk were generated from 537,352 subjects, excluding the 23andMe samples that were not available to us for inclusion. We analyzed variants with a minimum effective sample size of 80,603 (15%) in DrnkWk and a minimum effective sample size of 45,118 (15%) in PAU, which left 10,613,246 overlapping variants.
Data Availability:
The full summary-level association data from the meta-analysis are available through dbGaP: [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v3.p1] (accession number phs001672.v3.p1).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability:
Kinship analysis was performed using KING (http://people.virginia.edu/~wc9c/KING/); principal component analyses were performed using EIGENSOFT (https://data.broadinstitute.org/alkesgroup/EIGENSOFT/); imputation was performed using EAGLE2 (https://data.broadinstitute.org/alkesgroup/Eagle/), Minimac3 (https://genome.sph.umich.edu/wiki/Minimac3), Sanger imputation server (https://imputation.sanger.ac.uk/), or RICOPILI (https://data.broadinstitute.org/mpg/ricopili/), depends on the sample; GWAS was performed using PLINK (https://www.coggenomics.org/plink2); meta-analyses was performed using METAL (https://genome.sph.umich.edu/wiki/METAL_Documentation); polygenic risk score analyses were performed using PRSice-2 (https://www.prsice.info/) or PRS-CS (https://github.com/getian107/PRScs); GCTA (https://cnsgenomics.com/software/gcta/#Overview) was used for identifying independent loci (GCTA-COJO), multi-trait conditional analysis (GCTA-mtCOJO), and Mendelian Randomization (GCTA-GSMR); LDSC (https://github.com/bulik/ldsc) was used for heritability estimate, genetic correlation analysis (also used LD-Hub, http://ldsc.broadinstitute.org/), and heritability enrichment analyses; FUMA (https://fuma.ctglab.nl/) was used for gene association, functional enrichment, and gene-set enrichment analyses; transcriptomic analyses were performed using S-PrediXcan and S-MultiXcan (https://github.com/hakyimlab/MetaXcan); PheWAS analyses were run using the PheWAS R package (https://github.com/PheWAS/PheWAS); Mendelian Randomization R Package (https://cran.rproject.org/web/packages/MendelianRandomization/index.html) and MR-PRESSO (https://github.com/rondolab/MR-PRESSO) were used for MR analyses; MTAG (https://github.com/omeed-maghzian/mtag) was used for Multiple trait analysis.
Supplementary Material
Acknowledgements
This research used data from the Million Veteran Program (MVP), and was supported by funding from the Department of Veterans Affairs Office of Research and Development, Million Veteran Program Grant #I01BX003341 and the VA Cooperative Studies Program (CSP) study #575B. This publication does not represent the views of the Department of Veterans Affairs or the United States Government. A list of members and affiliations of MVP appears in the Supplementary Information.Supported also by NIH (NIAAA) P50 AA12870 (JG), a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation (HZ), and NIH grants 5T32GM080178 (JMS) and K02DA32573 (AA); and the NIHR Imperial Biomedical Research Centre (SRA and MRT). This research also used summary data from the Psychiatric Genomics Consortium (PGC) Substance Use Disorders (SUD) working group. The PGC-SUD is supported by funds from NIDA and NIMH to MH109532 and, previously, had analyst support from NIAAA to U01AA008401 (COGA). PGC-SUD gratefully acknowledges its contributing studies and the participants in those studies, without whom this effort would not be possible. This research also used individual-level/summary data from UK Biobank, a population-based sample of participants whose contributions we gratefully acknowledge. We thank the iPSYCH-Broad Consortium for access to data on the iPSYCH cohort. The iPSYCH project is funded by the Lundbeck Foundation (R102-A9118 and R155-2014-1724) and the universities and university hospitals of Aarhus and Copenhagen. Genotyping of iPSYCH samples was supported by grants from the Lundbeck Foundation and the Stanley Foundation, The Danish National Biobank resource was supported by the Novo Nordisk Foundation. Data handling and analysis on the GenomeDK HPC facility was supported by NIMH (1U01MH109514-01 to ADB). High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to ADB). The UCL and STOPAH case samples were genotyped with funding from the NIHR (National Institute for Healthcare Research) Imperial Biomedical Research Centre (BRC). UCL cases and controls were collected with UK Medical Research Council project grants G9623693N, G0500791, G0701007 and G1000708 and with support from the NIHR.
Genotyping of the UCL control sample was supported by grants from the Stanley Foundation. The UK Household Longitudinal Study (Understanding Society) is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. The survey was conducted by NatCen and the genome-wide scan data were analysed and deposited by the Wellcome Trust Sanger Institute. Information on how to access the data can be found on the Understanding Society website https://www.understandingsociety.ac.uk/. Dr McQuillin is supported by the University College London Hospitals NHS Foundation Trust NIHR BRC.
Footnotes
Competing Interests: Dr. Kranzler is a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor, and Amygdala Neurosciences. Drs. Kranzler and Gelernter are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018.
co-senior authors
References
- 1.GBD 2016 Alcohol Collaborators., Alcohol use and burden for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet, 2018. 392(10152): p. 1015–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walters RK, et al. , Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci, 2018. 21(12): p. 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kranzler HR, et al. , Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun, 2019. 10(1): p. 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Roige S, et al. , Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. Am J Psychiatry, 2019. 176(2): p. 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gelernter J, et al. , Genome-wide Association Study of Maximum Habitual Alcohol Intake in >140,000 U.S. European and African American Veterans Yields Novel Risk Loci. Biol Psychiatry, 2019. 86(5): p. 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaziano JM, et al. , Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol, 2016. 70: p. 214–23. [DOI] [PubMed] [Google Scholar]
- 7.Bycroft C, et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature, 2018. 562(7726): p. 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelernter J, et al. , Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry, 2014. 19(1): p. 41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu M, et al. , Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet, 2019. 51(2): p. 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turley P, et al. , Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet, 2018. 50(2): p. 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotto KC, et al. , DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res, 2018. 46(D1): p. D1068–D1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulik-Sullivan BK, et al. , LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet, 2015. 47(3): p. 291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finucane HK, et al. , Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet, 2015. 47(11): p. 1228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finucane HK, et al. , Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet, 2018. 50(4): p. 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roadmap Epigenomics Consortium, et al. , Integrative analysis of 111 reference human epigenomes. Nature, 2015. 518(7539): p. 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GTEx Consortium, Genetic effects on gene expression across human tissues. Nature, 2017. 550(7675): p. 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe K, et al. , Functional mapping and annotation of genetic associations with FUMA. Nat Commun, 2017. 8(1): p. 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Leeuw CA, et al. , MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol, 2015. 11(4): p. e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marees AT, et al. , Potential influence of socioeconomic status on genetic correlations between alcohol consumption measures and mental health. Psychol Med, 2019: p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant BF, et al. , Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA Psychiatry, 2015. 72(8): p. 757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen AM, et al. , Polygenic Scores for Major Depressive Disorder and Risk of Alcohol Dependence. JAMA Psychiatry, 2017. 74(11): p. 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H, et al. , Genetic Risk Variants Associated With Comorbid Alcohol Dependence and Major Depression. JAMA Psychiatry, 2017. 74(12): p. 1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barbeira AN, et al. , Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun, 2018. 9(1): p. 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battle A, et al. , Characterizing the genetic basis of transcriptome diversity through RNAsequencing of 922 individuals. Genome Res, 2014. 24(1): p. 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbeira AN, et al. , Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet, 2019. 15(1): p. e1007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evangelou E, et al. , New alcohol-related genes suggest shared genetic mechanisms with neuropsychiatric disorders. Nat Hum Behav, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowden J, et al. , A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med, 2017. 36(11): p. 1783–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowden J, et al. , Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol, 2016. 40(4): p. 304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowden J, Davey Smith G, and Burgess S, Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol, 2015. 44(2): p. 512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verbanck M, et al. , Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet, 2018. 50(5): p. 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, et al. , Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun, 2018. 9(1): p. 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard DM, et al. , Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci, 2019. 22(3): p. 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schizophrenia Working Group of the Psychiatric Genomics, C., Biological insights from 108 schizophrenia-associated genetic loci. Nature, 2014. 511(7510): p. 421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl EA, et al. , Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet, 2019. 51(5): p. 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagel M, et al. , Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet, 2018. 50(7): p. 920–927. [DOI] [PubMed] [Google Scholar]
- 36.Karlsson Linner R, et al. , Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet, 2019. 51(2): p. 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen PR, et al. , Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet, 2019. 51(3): p. 394–403. [DOI] [PubMed] [Google Scholar]
- 38.Lee JJ, et al. , Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet, 2018. 50(8): p. 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasman JA, et al. , GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci, 2018. 21(9): p. 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okbay A, et al. , Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet, 2016. 48(6): p. 624–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cross-Disorder Group of the Psychiatric Genomics, C., Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet, 2013. 381(9875): p. 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demontis D, et al. , Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet, 2019. 51(1): p. 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilling LC, et al. , Human longevity is influenced by many genetic variants: evidence from 75,000 UK Biobank participants. Aging (Albany NY), 2016. 8(3): p. 547–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loh PR, et al. , Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet, 2016. 48(11): p. 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das S, et al. , Next-generation genotype imputation service and methods. Nat Genet, 2016. 48(10): p. 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.1000 Genomes Project Consortium, A global reference for human genetic variation. Nature, 2015. 526(7571): p. 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manichaikul A, et al. , Robust relationship inference in genome-wide association studies. Bioinformatics, 2010. 26(22): p. 2867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galinsky KJ, et al. , Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am J Hum Genet, 2016. 98(3): p. 456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang CC, et al. , Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 2015. 4: p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willer CJ, Li Y, and Abecasis GR, METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 2010. 26(17): p. 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Euesden J, Lewis CM, and O’Reilly PF, PRSice: Polygenic Risk Score software. Bioinformatics, 2015. 31(9): p. 1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, et al. , Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet, 2012. 44(4): p. 369–75, S1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Z, et al. , Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet, 2016. 48(5): p. 481–7. [DOI] [PubMed] [Google Scholar]
- 54.International HapMap Consortium, et al. , Integrating common and rare genetic variation in diverse human populations. Nature, 2010. 467(7311): p. 52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bulik-Sullivan B, et al. , An atlas of genetic correlations across human diseases and traits. Nat Genet, 2015. 47(11): p. 1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng J, et al. , LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics, 2017. 33(2): p. 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Savage JE, et al. , Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet, 2018. 50(7): p. 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jansen IE, et al. , Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet, 2019. 51(3): p. 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gamazon ER, et al. , A gene-based association method for mapping traits using reference transcriptome data. Nat Genet, 2015. 47(9): p. 1091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge T, et al. , Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun, 2019. 10(1): p. 1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price AL, et al. , Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet, 2006. 38(8): p. 904–9. [DOI] [PubMed] [Google Scholar]
- 62.McCarthy S, et al. , A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet, 2016. 48(10): p. 1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denny JC, et al. , Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol, 2013. 31(12): p. 1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carroll RJ, Bastarache L, and Denny JC, R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics, 2014. 30(16): p. 2375–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pedersen CB, et al. , The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry, 2018. 23(1): p. 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lam M, et al. , RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics, 2020. 36(3): p. 930–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCarthy S, et al. , A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet, 2016. 48(10): p. 1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yavorska OO and Burgess S, MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol, 2017. 46(6): p. 1734–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full summary-level association data from the meta-analysis are available through dbGaP: [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v3.p1] (accession number phs001672.v3.p1).