Abstract
Purpose of Review:
The purpose of the review is to discuss recent advances in microRNA (miRNA) regulation of lipid metabolism and highlight the importance of miRNA-mediated gene regulation in dyslipidemia and fatty liver disease. This article reviews examples of miRNAs that bridge disparate metabolic pathways in the liver. For example, we highlight miRNAs that are regulated by the sterol-sensing pathway in the liver that in turn regulate cellular or systemic cholesterol, fatty acid, and glucose levels.
Recent Findings:
The most widely-studied of these miRNAs are miR-33a/b; however, we recently reported that miRNAs in the miR-183/96/182 cluster are also likely regulated by hepatic cholesterol content and mediate the observed glucose-lowering effects of the bile acid sequestrant colesevelam through the sterol-sensing pathway. In addition, several other hepatic and adipose miRNAs have been recently demonstrated to be key regulators of cellular lipid synthesis, storage, and catabolism, as well as systemic lipid metabolism. Moreover, many of these miRNAs are altered in fatty liver disease and dyslipidemia.
Summary:
miRNAs are not just fine-tuners of lipid metabolism, but critical regulatory factors in lipid homeostasis and health. Loss of these miRNA regulatory modules very likely contributes to the underlying metabolic defects observed in lipid disorders.
Keywords: microRNA, lipids, cholesterol, liver, adipose
Introduction:
The study of microRNAs (miRNA) is as vibrant as ever and many new functions for miRNAs in lipid-related mechanisms and disorders have been recently reported. Currently, miRbase.org (v22.1), the universal database of miRNAs, contains 1,917 human miRNAs; however, only 26% of these (505 miRNAs) are considered to be high confidence miRNAs1. Nevertheless, key biological functions have been reported for most of the broadly conserved mammalian miRNAs in humans, and many of these miRNAs regulate genes previously linked to lipid metabolism.
miRNAs are a class of small non-coding RNAs approximately 22 nucleotides (nt) in length which serve to post-transcriptionally regulate mRNA expression and stability through the RNA-Induced Silencing Complex (RISC)2. miRNAs are present in all tissues, and most, if not all, mammalian biological processes are regulated by miRNAs through either direct or indirect mechanisms3. miRNAs are transcribed by polymerase II as long pri-miRNAs that are processed in the nucleus by the RNAse III enzyme Drosha and its double-stranded RNA binding cofactor DiGeorge syndrome critical region 8 (DGCR8)4. This microprocessor complex cleaves the pri-miR structure into an approximately 60nt pre-miR, which is exported from the nucleus to the cytoplasm by exportin-5 5. In the cytoplasm, the pre-miRNA is further processed by the RNAse III endonuclease Dicer to generate a small double-stranded RNA duplex of approximately 22 nts in length containing a 2 nt 3’ overhang5. The miRNA duplex is then loaded into the RISC through an ATP-dependent process facilitated by chaperones6. RISC structure and function are conferred by members of the Argonaute (AGO) family of proteins 1–4. Either strand of the miRNA duplex can be loaded into AGO-RISC pocket and recognize mRNAs and other transcripts through a critical seed region (bases 2–7) on the 5’ end of the mature miRNA. Most miRNA target pairing occurs through imperfect matching conferred by Watson-Crick pairing between the miRNA seed region and sites within the 3’ untranslated region (3’ UTR) of the mRNA7. Most miRNA target sites are found in the 3’ UTRs; however, miRNAs can also bind and repress mRNAs through sites in the open reading frame and 5’ UTR of the mRNA targets, although these sites occur less frequently. Due to the imperfect base pairing between miRNAs and target mRNAs, one miRNA can potentially silence hundreds of genes, and multiple miRNAs can target the same mRNA. miRNAs repress target mRNA expression by interfering with translational initiation, preventing elongation, and/or destabilizing the transcript through mRNA decay4. mRNA decay after miRNA recognition involves the recruitment of the glycine-tryptophan protein of 182kDa (GW182), which interacts with the polyadenylate-binding protein and promotes deadenylation of the mRNA transcript8. The current models and state-of-the-art of miRNA processing, mRNA target recognition, and miRNA targeting mechanisms were recently reviewed here9.
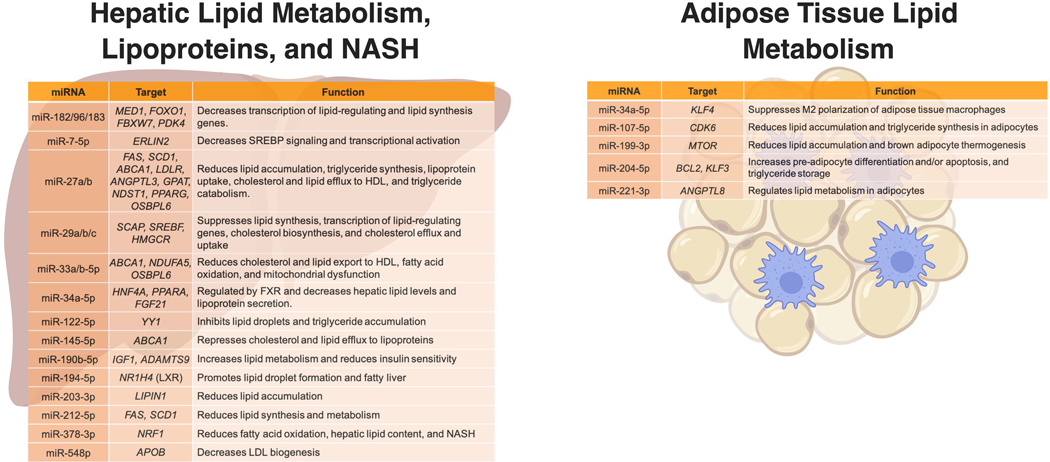
The miRNA interactome network for lipid disorders was recently reported based on text mining and many exciting results emerged from this study10. Approximately 150 distinct miRNAs were found to represent the 227 miRNA-lipid disease networks and the top 20 miRNA networks were further resolved10. These include experimentally validated regulatory modules of lipid genes for miR-33a/b, miR-223–3p, miR-375–3p, miR-144, miR-122, miR-103/107, miR-30c, miR-145, miR-146a, miR-29, and miRNAs in the miR-17–92 and miR-183/96/182 clusters. This study is a great resource for future studies investigating the mechanisms and consequences of miRNA-mediated lipid regulation. Here, we will discuss these key lipid miRNAs and their role in lipid diseases (Figure 1).
Figure 1. miRNA regulation of lipid metabolism in the liver and adipose.
Created using BioRender.
Sterol-sensing miRNAs bridge metabolic pathways
Like many genes that regulate cholesterol and lipid homeostasis, the expression of specific miRNAs is sensitive to cellular cholesterol levels. Although a comprehensive study of sterol-regulated miRNAs in metabolic cell-types remains to be completed, specific examples of miRNAs that harbor sterol-response elements in their promoters or host gene promoters have been identified. miR-33, the most widely-studied miRNA in lipid metabolism, was discovered through a screen of cholesterol-induced miRNAs in macrophages11. miR-33a and miR-33b are co-transcribed with host genes SREBF2 and SREBF1, respectively12. While humans have two copies of miR-33 (miR-33a and miR-33b), rodents only have one copy, miR-33a, as miR-33b cannot be expressed due to a deletion in its encoding sequence in Srebf1. Cells respond to elevated cellular cholesterol (and other sterol) levels through a series of biological processes within the sterol-sensing pathway. Briefly, excess cellular cholesterol content causes a decrease in the transcription of genes that promote cellular cholesterol content, including critical cholesterol biosynthesis enzymes, cholesterol transporters, and lipoprotein uptake receptors13. When cellular cholesterol levels are low, cells respond by increasing cholesterol biosynthesis and lipoprotein uptake through increased transcriptional activity of sterol regulatory element-binding proteins (SREBPs) and activation of critical cholesterol-linked genes14. Part of this mechanism is the control of SREBP expression, which are the key transcription factors that control cellular cholesterol homeostasis. For example, when cellular cholesterol levels are low, SREBF1 and SREBF2 genes are turned on with miR-33b and miR-33a, which are harbored within the respective introns and co-transcribed with the host genes11. miR-33a/b directly targets and inhibits ATP-binding cassette transporter A1 (ABCA1), a key transporter of cellular cholesterol to lipid-poor apolipoprotein A-I and nascent high-density lipoproteins (HDL)11. miR-33 has been extensively investigated in most lipid pathways and miR-33a/b-5p has been reviewed exhaustively prior15. Nonetheless, recent studies have demonstrated new functions for miR-33 in lipid metabolism16, 17. One of the most exciting studies found that miR-33a/b-5p regulates key features of the NLR Family Pyrin Domain Containing 3 (NLRP3) inflammasome complex in macrophages, and thus, provides a further link between cellular cholesterol metabolism, immune cell activation, and vascular inflammation18.
Another critical miRNA, or cluster of miRNAs, to lipid metabolism is the miR-183/96/182 poly-cistronic cluster19. This miRNA cluster has previously been shown to be a transcriptional target of SREBPs and harbor sterol response elements in its promoter, thus this miRNA cluster is intricately linked to cellular cholesterol levels20. Therefore, it is not surprising that miR-182–5p expression was reported to be down-regulated in white adipose tissue, skeletal muscle, liver, pancreas, and blood from rats on a high-fat diet (HFD) which may have included increased cholesterol content in the diet21. For example, miR-182–5p expression would be predicted to be suppressed in cells with excess cholesterol gained through the HFD. In agreement, miR-182–5p levels were also found to be decreased in blood from non-human primates (NHP) that were fed a HFD21. Like miR-33a/b-5p, miRNAs in this cluster likely serves as a bridge between cellular cholesterol content and systemic energy control, e.g. glucose and triglyceride homeostasis. For example, miR-96–5p was recently found to target FOXO1, a critical regulator of insulin signaling, gluconeogenesis, and adipogenesis; critical processes in systemic energy and lipid metabolism22–24. Recently, we reported that bile acid sequestrants, specifically the drug colesevelam (brand names Welchol, Cholestagel, or Lodalis), increased hepatic expression of the miR-183/96/182 cluster in rats and mice. Since this miRNA cluster is under regulation of SREBPs20, and thus cellular cholesterol levels, the observed increase of this miRNA cluster in the liver is likely due to the transcriptional response of SREBPs to reduced hepatic cholesterol content due to the conversion and replenishment of bile acids lost to intestinal sequestration and reduced bile acid reuptake25. Colesevelam has previously been shown to promote SREBP2 transcriptional activity to stimulate hepatic cholesterol biosynthesis in response to reduced cellular cholesterol levels due to the increased bile acid conversion from cholesterol26, 27. Collectively, colesevelam creates a cholesterol sink in the liver which increases hepatic uptake of circulating LDL particles to replenish hepatic cholesterol lost to bile secretion. As a consequence, colesevelam lowers circulating LDL-cholesterol levels, the primary risk factor for cardiovascular disease (CVD). Colesevelam is the most-effective drug in the bile acid sequestrant class for binding to bile acids and is indicated for primary hyperlipidemia; however, one remarkable pleiotropic effect of colesevelam is its glucose-lowering capacity. Therefore, it is prescribed to patients with type 2 diabetes mellitus to improve glycemic control28–32. We posit that miRNAs in the miR-183/96/182 cluster mediate, in part, colesevelam’s glucose lowering effects, and thus, further links hepatic cholesterol metabolism and systemic lipid and glucose metabolism. Recently, we found that inhibition of miR-182–5p partially reversed the observed improvements in glucose tolerance observed with colesevelam in db/db mice, a model of diabetic dyslipidemia25. Paradoxically, the miR-183/96/182 cluster is also induced with statins20, which contrary to colesevelam, has been reported to worsen glucose tolerance33. Therefore, it is possible that the hepatic gene regulatory networks that are altered with bile acid sequestrants are distinct from those altered with statins despite the likely shared utilization of the SREBP2 sterol-sensing pathway and transcriptional response. We recently demonstrated that miRNAs in the miR-183/96/182 directly target and suppress Med1, a gene that links nuclear receptor activity to Pol II transcription25. In addition to Med1 and Foxo1, miRNAs in the miR-183/96/182 cluster also likely regulate Pyruvate Dehydrogenase Kinase 4 (Pdk4) another key gene in glucose and lipid metabolism34. It should be noted that colesevelam very likely alters the expression of other miRNAs in the liver, particularly miRNAs that are regulated by farnesoid X receptor (FXR). For example, normally 95% of bile acids are reabsorbed by the intestine and transported back to the liver through enterohepatic circulation and FXR is the primary sensor for endogenous bile acid feedback35. Colesevelam inhibits bile acid reabsorption and promotes their excretion, resulting in decreased FXR signaling36. One of these FXR-regulated miRNAs is likely miR-34a-5p. Fxr knockout mice were shown to have increased levels of miR-34a-5p37, and Cyp7a1 overexpressing mice were also reported to have increased levels of miR-33a-5p38. Therefore, it is possible that loss of Fxr or induction of Cyp7a1 by colesevelam may also contribute to some of the observed effects of colesevelam in hepatic and systemic lipid homeostasis, as well as explain some of the discrepancies between statins and BAS drug classes on hepatic miRNA gene regulation and lipid metabolic outcomes. miRNA regulation of hepatic cholesterol metabolism is an important aspect of hepatic lipid homeostasis as many metabolic pathways are inter-connected through the sterol-sensing pathway. In addition to miR-33a/b and miRNAs in the miR-183/96/182 cluster, miR-29a/b/c was demonstrated to regulate SREBF1/2 and SREBF Chaperone (SCAP), and thus lipid metabolism, through a SREBP feedback loop39. Like miR-33a-5p, miR-29c was also identified in a screen for miRNAs that are regulated (significantly decreased) by cellular cholesterol11. miR-29a-5p, the miRNA processing enzyme Dicer, and HMG Co-reductase (HMGCR) were also reported to form a novel axis that governs cholesterol accumulation in hepatocytes which contributes to fatty liver disease40. LDL-cholesterol is a causal factor in atherosclerosis and CVD and is secreted from the liver in the form of very low-density lipoproteins (VLDL). Apolipoprotein B (APOB), the key structural and functional protein of VLDL and LDL, is generally considered to be regulated at the protein level during VLDL assembly and biogenesis. Strikingly, miR-548p was recently reported to target and suppress APOB in hepatocytes which is a remarkable advancement41. Both miR-33a/b-5p and the miRNAs in the miR-183/96/182 cluster represent sterol-sensing responsive miRNAs that bridge cellular cholesterol metabolism and systemic lipid homeostasis, including glucose pathways. These miRNAs are not likely the only miRNAs that harbor SREs in their promoters and are transcriptional targets of SREBPs. Moreover, these miRNAs are not the only miRNAs that link cellular cholesterol metabolism to other metabolic networks that have been recently investigated. For example, miR-7–5p was found to be a hepatic peroxisome proliferator activated receptor-α (PPAR-α)-dependent miRNA that regulates SREBP signaling, as opposed to being regulated by SREBPs, through targeting and repression of endoplasmic reticulum lipid raft associated 2 (ERLIN2), which also provides a link between metabolic pathways in the liver42.
miRNA regulation of hepatic lipid metabolism
The liver is one of, if not, the most important regulatory tissue in systemic lipid homeostasis and considerable advances in miRNA regulation of lipid-regulating genes in the liver have been made in the last few years. One of the most well-studied lipid-associated miRNAs in the liver is miR-34a-5p. Recently, miR-34a-5p has been reported to mediate the hepatic response to metabolic stress through regulation of HFN4A (hepatocyte nuclear factor 4 alpha), which is a critical hepatic transcription factor for lipid metabolism and lipoprotein secretion43. miR-34a antagonizes VLDL secretion and likely regulates plasma lipoprotein levels. miR-34a also suppresses NAFLD through targeting and suppression of PPARA44. Moreover, miR-34a, via PPAR-α, was recently found to contribute to systemic energy homeostasis through indirect regulation of FGF21, a key hormone in lipid metabolism45. The roles of miRNAs in post-transcriptional gene regulation underlying non-alcoholic fatty liver disease (NAFLD) and/or nonalcoholic steatophepatitis (NASH) was one of the more active areas of research for hepatic miRNAs in recent years. For example, miR-378–3p was reported to promote NASH in response high-fat diet (HFD) feeding through direct targeting of Nrf146. miR-212–5p was also found to inhibit lipid accumulation in hepatocytes through suppression of fatty acid synthase (FAS) and stearoyl-CoA desaturase (SCD1), two critical fatty acid regulators in the liver47. Most interestingly, miR-194 likely is a key lipid-regulating miRNA in liver, as inhibition of miR-194 was reported to improve fatty liver disease through regulation of FXR48. Hepatic miRNAs were also found to contribute to alcohol-induced steatohepatitis, as miR-203–3p was found to target and repress Lipin149. We have previously reported that miR-27b-3p is key regulatory hub for lipid metabolism and directly regulates critical lipid genes, including angiopoietin-like 3 (ANGPTL3), N-Deacetylase And N-Sulfotransferase 1 (NDST1), and Glycerol-3-phosphate acyltransferase 1 (GPAM)50. miR-27b-3p has also been reported to regulate ABCA1 and low-density lipoprotein receptor (LDLR), which are essential for cholesterol and lipid efflux and lipoprotein uptake in the liver51. Recently, a group reported that miR-27a-3p also regulates hepatic lipids and fatty liver disease through regulation of FAS and SCD152. Moreover, Ouimet et al. reported that oxysterol-binding protein-like 6 (OSBPL6) was found to be a target of both miR-27b-3p and miR-33a/b53. miR-122–5p is the most abundant miRNA in the liver and has been extensively studied in hepatic lipid metabolism. Recently, miR-122–5p was reported to antagonize lipid droplet formation through regulation of the transcription factor Yin Yang 1 (YY1) which may confer some of the previously identified links between miR-122–5p and hepatic lipid control54. Another key lipid miRNA in liver is miR-145–5p, and recently, statin treatments, e.g. Atorvastatin, were found to promote miR-145–5p expression in hepatocytes through activation of the PI3K/AKT pathway55. miR-145–5p has previously been found to regulate ABCA1 in the liver and pancreatic islets and like many of the miRNAs listed in this review bridge multiple metabolic pathways in the liver that contribute to hepatic and systemic lipid homeostasis56. miR-145 is also one of multiple miRNAs (listed here in this review) that regulate hepatic lipid metabolism but have been found to regulate lipid metabolism in other tissues and organs. For example, miR-33a/b-5p was also found to control lipid raft cholesterol content in the heart57.
miRNA regulation of adipose lipid metabolism
In addition to the liver, adipose is another major tissue of systemic lipid regulation. miR-107,−5p which is was previously found to be a critical miRNA in glucose metabolism and insulin sensitivity58, was found to inhibit cyclin dependent kinase 6 (CDK6) expression in adipocytes, which regulates adipogenesis and lipid storage59. In the last few years, multiple miRNAs have been found to be critical regulators of adipogenesis and lipid metabolism. Among the many studies, miR-204–5p was found to regulate adipocyte differentiation and adiposity60, miR-199a-3p was demonstrated to regulate brown adipogenesis via mTOR61, and miR-221–3p was shown to regulate angiopoietin-like 8 (ANGPTL8) in adipocytes62. Moreover, miR-34a-5p was found to be packaged and secreted from adipocytes in extracellular vesicles and was found to suppress anti-inflammatory phenotypes (M2-like) in adipose tissue macrophages through regulation of Kruppel-like factor 4 (KLF4), and this communication network likely promotes obesity-related adipose inflammation63.
Conclusions:
In summary, numerous miRNAs have and continue to emerge as critical regulators in all facets of lipid biology. Despite the unfortunate designation as fine-tuners of gene expression, loss-of-function studies in both cells and animal models clearly reveal critical roles for miRNAs in cellular and animal phenotypes, metabolism, and disease. For example, one can quickly examine the list of specific miRNA-deficient mice (and other animal models) to easily grasp that miRNAs indeed play key roles in regulating gene expression in mammals9. In this review, we highlight key sterol-sensing miRNAs, such as miR-33 and the miR-183/96/182 cluster, that bridge the gap of metabolic pathways such as cellular cholesterol metabolism, immune cell activation and glucose and triglyceride homeostasis. Furthermore, we discuss miRNAs that play a key role in hepatic and adipose lipid metabolism. Therefore, it is likely unwise to discount miRNAs due to any perceived lack of regulator strength. Here we show there is clearly nothing micro about the level of regulation miRNAs have on the expression of lipid metabolism genes.
Key Points:
miRNAs are important regulators of all facets of lipid metabolism in multiple cell-types and tissues, including lipid synthesis, storage, circulation, and catabolism.
Sterol-sensing miRNAs in the miR-183/96/182 cluster bridge metabolic pathways, including hepatic cholesterol, fatty acid, and glucose metabolism.
miRNAs in the liver and adipose serve to control systemic energy homeostasis
Acknowledgements:
We would like to acknowledge Meaghan E. Kuzmich for editorial assistance with this article.
Financial Support: This work was funded and supported though awards from the National Institutes of Health (USA); HL128996, HL127173, and HL116263.
Footnotes
Conflicts of Interest: The authors have no real or perceived conflicts of interests to disclose.
References:
- 1.Kozomara A, Birgaoanu M and Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47: D155–D62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 3.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushati N and Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007; 23: 175–205. [DOI] [PubMed] [Google Scholar]
- 5.Ha M and Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014; 15: 509–24. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki S, Kobayashi M, Yoda M, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010; 39: 292–9. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136: 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebert LFR and MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. Metazoan MicroRNAs. Cell. 2018; 173: 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandhro AH, Shoombuatong W, Nantasenamat C, et al. The MicroRNA Interaction Network of Lipid Diseases. Front Genet. 2017; 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rayner KJ, Suarez Y, Davalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010; 328: 1570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore KJ, Rayner KJ, Suarez Y, et al. The role of microRNAs in cholesterol efflux and hepatic lipid metabolism. Annu Rev Nutr. 2011; 31: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown MS and Goldstein JL. Sterol regulatory element binding proteins (SREBPs): controllers of lipid synthesis and cellular uptake. Nutr Rev. 1998; 56: S1–3; discussion S54–75. [DOI] [PubMed] [Google Scholar]
- 14.Edwards PA, Tabor D, Kast HR, et al. Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta. 2000; 1529: 103–13. [DOI] [PubMed] [Google Scholar]
- 15.Aryal B, Singh AK, Rotllan N, et al. MicroRNAs and lipid metabolism. Curr Opin Lipidol. 2017; 28: 273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishino T, Horie T, Baba O, et al. SREBF1/MicroRNA-33b Axis Exhibits Potent Effect on Unstable Atherosclerotic Plaque Formation In Vivo. Arterioscler Thromb Vasc Biol. 2018; 38: 2460–73. [DOI] [PubMed] [Google Scholar]
- 17.Nie H, Yu X, He H, et al. Hepatocyte miR-33a mediates mitochondrial dysfunction and hepatosteatosis by suppressing NDUFA5. J Cell Mol Med. 2018; 22: 6285–93.*This study further links the sterol-sensing pathway and miR-33 to hepatic lipid storage and ultilization.
- 18.Xie Q, Wei M, Zhang B, et al. MicroRNA33 regulates the NLRP3 inflammasome signaling pathway in macrophages. Mol Med Rep. 2018; 17: 3318–27.*This study describes the role of the sterol-sensing pathway and autophagy in macropahges as well as other macrophage processes related to lipid metabolism.
- 19.Xu S, Witmer PD, Lumayag S, et al. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. The Journal of biological chemistry. 2007; 282: 25053–66. [DOI] [PubMed] [Google Scholar]
- 20.Jeon TI, Esquejo RM, Roqueta-Rivera M, et al. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013; 18: 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Meng Y, Tian S, et al. Comparative MicroRNA Expression Profiles of Cynomolgus Monkeys, Rat, and Human Reveal that miR-182 Is Involved in T2D Pathogenic Processes. J Diabetes Res. 2014; 2014: 760397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y, Liu H, Zhang H, et al. miR-96 regulates FOXO1-mediated cell apoptosis in bladder cancer. Oncol Lett. 2012; 4: 561–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guttilla IK and White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009; 284: 23204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song HM, Luo Y, Li DF, et al. MicroRNA-96 plays an oncogenic role by targeting FOXO1 and regulating AKT/FOXO1/Bim pathway in papillary thyroid carcinoma cells. Int J Clin Exp Pathol. 2015; 8: 9889–900. [PMC free article] [PubMed] [Google Scholar]
- 25.Sedgeman LR, Beysen C, Allen RM, et al. Intestinal bile acid sequestration improves glucose control by stimulating hepatic miR-182–5p in type 2 diabetes. Am J Physiol Gastrointest Liver Physiol. 2018.**This study demonstrates that miRNAs in the miR-183/96/182 cluster mediate the glucose lowering effects of colesevelam, a bile acid sequestrant that lowers LDL-cholesterol through creating a cholesterol sink in the liver due to loss of BA reabsorption.
- 26.Watanabe M, Morimoto K, Houten SM, et al. Bile acid binding resin improves metabolic control through the induction of energy expenditure. PLoS One. 2012; 7: e38286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meissner M, Wolters H, de Boer RA, et al. Bile acid sequestration normalizes plasma cholesterol and reduces atherosclerosis in hypercholesterolemic mice. No additional effect of physical activity. Atherosclerosis. 2013; 228: 117–23. [DOI] [PubMed] [Google Scholar]
- 28.Beysen C, Murphy EJ, Deines K, et al. Effect of bile acid sequestrants on glucose metabolism, hepatic de novo lipogenesis, and cholesterol and bile acid kinetics in type 2 diabetes: a randomised controlled study. Diabetologia. 2012; 55: 432–42. [DOI] [PubMed] [Google Scholar]
- 29.Davidson MH, Dicklin MR, Maki KC, et al. Colesevelam hydrochloride: a non-absorbed, polymeric cholesterol-lowering agent. Expert Opin Investig Drugs. 2000; 9: 2663–71. [DOI] [PubMed] [Google Scholar]
- 30.Fonseca VA, Handelsman Y and Staels B. Colesevelam lowers glucose and lipid levels in type 2 diabetes: the clinical evidence. Diabetes Obes Metab. 2010; 12: 384–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nwose OM and Jones MR. Atypical Mechanism of Glucose Modulation by Colesevelam in Patients with Type 2 Diabetes. Clin Med Insights Endocrinol Diabetes. 2013; 6: 75–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smushkin G, Sathananthan M, Piccinini F, et al. The effect of a bile acid sequestrant on glucose metabolism in subjects with type 2 diabetes. Diabetes. 2013; 62: 1094–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aiman U, Najmi A and Khan RA. Statin induced diabetes and its clinical implications. J Pharmacol Pharmacother. 2014; 5: 181–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D, Li Y, Yao X, et al. miR-182 Regulates Metabolic Homeostasis by Modulating Glucose Utilization in Muscle. Cell Rep. 2016; 16: 757–68. [DOI] [PubMed] [Google Scholar]
- 35.Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013; 3: 1191–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potthoff MJ, Potts A, He T, et al. Colesevelam suppresses hepatic glycogenolysis by TGR5-mediated induction of GLP-1 action in DIO mice. American journal of physiology Gastrointestinal and liver physiology. 2013; 304: G371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, Padhye A, Sharma A, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. The Journal of biological chemistry. 2010; 285: 12604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T, Francl JM, Boehme S, et al. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology. 2013; 58: 1111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ru P and Guo D. microRNA-29 mediates a novel negative feedback loop to regulate SCAP/SREBP-1 and lipid metabolism. RNA Dis. 2017; 4.*This study demonstrate a critical role for miR-29 in the sterol-sensing pathway and lipid metabolism.
- 40.Liu MX, Gao M, Li CZ, et al. Dicer1/miR-29/HMGCR axis contributes to hepatic free cholesterol accumulation in mouse non-alcoholic steatohepatitis. Acta Pharmacol Sin. 2017; 38: 660–71.*This article describes a novel miRNA feedback network that controls sterol-sensing and cholesterol biosynthesis.
- 41.Zhou L and Hussain MM. Human MicroRNA-548p Decreases Hepatic Apolipoprotein B Secretion and Lipid Synthesis. Arterioscler Thromb Vasc Biol. 2017; 37: 786–93.*This article describes one of the few examples of a miRNA regulating APOB.
- 42.Singaravelu R, Quan C, Powdrill MH, et al. MicroRNA-7 mediates cross-talk between metabolic signaling pathways in the liver. Sci Rep. 2018; 8: 361.**This study presents a compelling argument that miRNAs bridge metabolic networks in the liver.
- 43.Xu Y, Zalzala M, Xu J, et al. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat Commun. 2015; 6: 7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding J, Li M, Wan X, et al. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Sci Rep. 2015; 5: 13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han HS, Choi BH, Kim JS, et al. Hepatic Crtc2 controls whole body energy metabolism via a miR-34a-Fgf21 axis. Nat Commun. 2017; 8: 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T, Zhao X, Steer CJ, et al. A negative feedback loop between microRNA-378 and Nrf1 promotes the development of hepatosteatosis in mice treated with a high fat diet. Metabolism. 2018; 85: 183–91.*This study describes a miR-378 regulatory module that mediates, in part, the hepatic repsonse to dietary lipid and lipid storage in th eliver.
- 47.Guo Y, Yu J, Wang C, et al. miR-212–5p suppresses lipid accumulation by targeting FAS and SCD1. J Mol Endocrinol. 2017; 59: 205–17. [DOI] [PubMed] [Google Scholar]
- 48.Nie H, Song C, Wang D, et al. MicroRNA-194 inhibition improves dietary-induced non-alcoholic fatty liver disease in mice through targeting on FXR. Biochim Biophys Acta Mol Basis Dis. 2017; 1863: 3087–94. [DOI] [PubMed] [Google Scholar]
- 49.Cheng XY, Liu JD, Lu XY, et al. miR-203 Inhibits Alcohol-Induced Hepatic Steatosis by Targeting Lipin1. Front Pharmacol. 2018; 9: 275.*This study highlights the role of miRNA-mediated gene regulation and lipid storage in alcohol-induced steatohepatosis.
- 50.Vickers KC, Shoucri BM, Levin MG, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013; 57: 533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goedeke L, Rotllan N, Ramirez CM, et al. miR-27b inhibits LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels in mice. Atherosclerosis. 2015; 243: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang M, Sun W, Zhou M, et al. MicroRNA-27a regulates hepatic lipid metabolism and alleviates NAFLD via repressing FAS and SCD1. Sci Rep. 2017; 7: 14493.*This study further establishes miR-27 as an essential regulator of hepatic lipid metabolism and fatty liver disease.
- 53.Ouimet M, Hennessy EJ, van Solingen C, et al. miRNA Targeting of Oxysterol-Binding Protein-Like 6 Regulates Cholesterol Trafficking and Efflux. Arterioscler Thromb Vasc Biol. 2016; 36: 942–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu GY, Rui C, Chen JQ, et al. MicroRNA-122 Inhibits Lipid Droplet Formation and Hepatic Triglyceride Accumulation via Yin Yang 1. Cell Physiol Biochem. 2017; 44: 1651–64.*This article links the most abundant miRNA in the liver to lipid droplet formation and hepatic lipid storage.
- 55.Docrat TF, Nagiah S, Krishnan A, et al. Atorvastatin induces MicroRNA-145 expression in HEPG2 cells via regulation of the PI3K/AKT signalling pathway. Chem Biol Interact. 2018; 287: 32–40.*This study reports that miR-145, a miRNA that regulates cholesterol efflux from hepatocytes to HDL, is itself controlled by the AKT signaling pathway.
- 56.Kang MH, Zhang LH, Wijesekara N, et al. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arterioscler Thromb Vasc Biol. 2013; 33: 2724–32. [DOI] [PubMed] [Google Scholar]
- 57.Nishiga M, Horie T, Kuwabara Y, et al. MicroRNA-33 Controls Adaptive Fibrotic Response in the Remodeling Heart by Preserving Lipid Raft Cholesterol. Circ Res. 2017; 120: 835–47. [DOI] [PubMed] [Google Scholar]
- 58.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011; 474: 649–53. [DOI] [PubMed] [Google Scholar]
- 59.Ahonen MA, Haridas PAN, Mysore R, et al. miR-107 inhibits CDK6 expression, differentiation, and lipid storage in human adipocytes. Mol Cell Endocrinol. 2019; 479: 110–6.*This study links miR-107, a critial miRNA for glycemic control, to adipose lipid storage.
- 60.Du J, Zhang P, Gan M, et al. MicroRNA-204–5p regulates 3T3-L1 preadipocyte proliferation, apoptosis and differentiation. Gene. 2018; 668: 1–7.*This article highights the role of miR-204–5p in adiposity.
- 61.Gao Y, Cao Y, Cui X, et al. miR-199a-3p regulates brown adipocyte differentiation through mTOR signaling pathway. Mol Cell Endocrinol. 2018; 476: 155–64.*This study demonstrates the role of miR-199a-3p in cell signaling and browning of adipose.
- 62.Mysore R, Ortega FJ, Latorre J, et al. MicroRNA-221–3p Regulates Angiopoietin-Like 8 (ANGPTL8) Expression in Adipocytes. J Clin Endocrinol Metab. 2017; 102: 4001–12.*This study establishes the role of miR-221–3p in adipose lipid metabolism.
- 63.Pan Y, Hui X, Hoo RLC, et al. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J Clin Invest. 2019; 129: 834–49.*This study demonstrates the role of extracellular miR-34a in promoting specific macrophage phenotypes in adipose which contribute to obesity-associated adipose inflammation.