Abstract
In an effort to provide new treatments for the global crisis of bacterial resistance to current antibiotics, we have used a rational approach to design several new antimicrobial peptides (AMPs). The present study focuses on 24-mer WLBU2 and its derivative, D8, with the amino acid sequence, RRWVRRVRRWVRRVVRVVRRWVRR. In D8, all of the valines are the D-enantiomer. We use X-ray low- and wide-angle diffuse scattering data to measure elasticity and lipid chain order. We show a good correlation between in vitro bacterial killing efficiency and both bending and chain order behavior in bacterial lipid membrane mimics; our results suggest that AMP-triggered domain formation could be the mechanism of bacterial killing in both Gram-positive and Gram-negative bacteria. In red blood cell lipid mimics, D8 stiffens and orders the membrane, while WLBU2 softens and disorders it, which correlate with D8’s harmless vs. WLBU2’s toxic behavior in hemolysis tests. These results suggest that elasticity and chain order behavior can be used to predict mechanisms of bactericidal action and toxicity of new AMPs.
1. Introduction
The world is now facing a growing threat from bacteria that are resistant to available antibiotics.1,2 Several strategies are being utilized to combat this threat, including eliminating antibiotics from animal feed, increasing antibiotic stewardship, reducing transmission in hospital settings and designing new types of antibiotics. Most current classes of antibiotics inhibit a specific metabolic pathway and typically require multiple doses to eradicate bacterial infections, during which time a resistance phenotype can occur.3 As a result, a class of pathogens termed ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) has been identified as the most common multidrug-resistant (MDR) bacteria.4 As a component of the host innate immune system, natural antimicrobial peptides5 (AMPs) such as human cathelicidin LL-376 have inspired the rational design of novel synthetic AMPs. Some of us have developed new AMPs that were inspired by both natural AMPs and the lytic peptide, LLP1, part of the C-terminus of the HIV-1 gp41 fusion protein.7 These peptides contain a cationic amphipathic motif with only 2 or 3 types of amino acids compared to natural peptides. WLBU2 is a 24-mer containing 3 types of amino acids: arginine, valine and tryptophan, with the primary sequence shown in Fig. 1. It was rationally designed, taking into account hydrophobicity, hydrophobic moment, helicity and overall net charge.8 Some of us previously demonstrated that WLBU2 was more effective than LL37 (made of 14 different types of amino acids) in animal models.9–11 In a recent study, it was demonstrated that WLBU2 is able to overcome resistance from 92% of 142 clinical isolates representing the ESKAPE pathogens, including those resistant to colistin and LL-37.12 In addition, WLBU2 effectively treated a variety of clinical methicillin-resistant S. aureus surgical implant biofilms.13
Fig. 1.
Primary structures of WLBU2 and D8. The underlined valines in D8 indicate D-Val substitutions. The net charge of both peptides is +13.
An important consideration is that the activity of AMPs measured as MIC/MBC (minimum inhibitory/bactericidal concentration) must be weighed against their toxicity. The latter can be assessed through hemolytic activity14 or a combination of hemolytic activity and lymphocyte viability.15 The ratio between a maximum nontoxic dose/minimum effective therapeutic dose in animals is called the Therapeutic Index (TI). The activity of WLBU2 under physiological conditions, including salt, improved when tryptophan was included in the sequence along with arginine and valine.11 The typical MBC (which reduces bacterial viability by 3 logs) of WLBU2 is lower than that of LL-37.11 Similarly, the TI of WLBU2, 3–5,11 based on efficacy in a murine model of P. aeroginosa bacteremia, is far superior to that of LL-37,11 although it is not optimal. Another consideration is protease digestion of the AMPs, which reduces their efficacy. In an older work, Wade et al. found that the D-enantiomer of the frog-skin derived AMP, temporin A, was as effective as the L-form, indicating that AMPs kill bacteria at the level of the lipid membrane, rather than in protein-protein interactions.16 In this work, in addition to WLBU2, we utilize a second form, D8, in which all of the L-valines in WLBU2 are replaced with their D-enantiomer. This substitution acts to prolong the peptide’s lifetime in the bloodstream17 and also reduces its in vitro toxicity.
AMPs share an amphipathic character and are thought to act directly and rapidly on bacterial membranes.18 In many cases antimicrobial activities are attained by the nonspecific interactions of AMPs with membranes, which lead to membrane disruption. The mechanism of action of AMPs is traditionally attributed to the formation of pores in the lipid cell membranes of pathogens.19 More recently AMPs have been shown to produce perturbations without membrane lysis, e.g., by changing lipid ordering, membrane curvature and thickness, forming of specific lipid-peptide domains, lateral lipid phase segregation and even forming non-lamellar lipid phases.19–22 Such perturbations may be manifested in the elastic properties of lipid membranes; these can be extracted using X-ray scattering-based approaches.
This work aims to delineate the role of lipids and D-enantiomeric amino acids in the interactions between WLBU2 and D8 with bacterial and eukaryotic membrane mimics, and compare to their efficacy and toxicity. It is of interest to determine if the reason WLBU2 has higher toxicity than D8 is because it has a different effect on membrane elastic properties. By means of X-ray diffuse scattering we measure the elastic parameter bending modulus (KC), which yields information about membrane stiffening/softening. In addition, an order parameter (SX-ray) that assesses lipid acyl chain ordering is obtained. Our results will be discussed in terms of mechanisms of action of antimicrobial peptides and toxicity through nonspecific as well as specific interactions with lipid components of the bacterial/eukaryotic membrane mimics.
2. Experimental section
2.1. Materials
The synthetic lyophilized lipids 1-palmitoy-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′rac-glycerol) sodium salt (POPG), 1′,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol sodium salt (TOCL, i.e., cardiolipin), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1,2-dioleoyl-3-trimeathylammonium-propane chloride salt (DOTAP) were purchased from Avanti Polar Lipids (Alabaster, AL) and used as received. Cholesterol was from Nu-Chek-Prep (Waterville, MN). HPLC grade organic solvents were purchased from Sigma-Aldrich (St. Louis, MO). The 24-mer peptides WLBU2 and D8 (chemical structures shown in Fig. 1) were synthesized either by the Peptide Synthesis Facility (University of Pittsburgh, Pittsburgh, PA) or Genscript (Piscataway, NJ). Purity was ~98% as shown by mass spectroscopy analysis. While their amino acid sequence is identical, all of the valines in D8 are the D-enantiomer.
2.1.a. LPS purification and lipid A isolation.
Large-scale P. aeruginosa (PA01) LPS preparations were isolated using a hot phenol/water extraction method after growth in lysogenic broth supplemented with 1 mM MgCl2 at 37 °C.23 Subsequently, LPS was treated with RNase A, DNase I and proteinase K to ensure purity from contaminating nucleic acids and proteins.24 Individual LPS samples were additionally extracted to remove contaminating phospholipids25 and TLR2 contaminating proteins.26 Finally, individual LPS preparations were resuspended in 500 μl of water, frozen on dry ice and lyophilized.
To confirm the structure of the lipid A component of the purified LPS, lipid A was isolated after hydrolysis in 1% SDS at pH 4.5 as described.27 Briefly, 500 μl of 1% SDS in 10 mM Na-acetate, pH 4.5 was added to a lyophilized sample. Samples were incubated at 100 °C for 1 h, frozen, and lyophilized. The dried pellets were resuspended in 100 μl of water and 1 ml of acidified ethanol (100 μl 4N HCl in 20 ml 95% EtOH). Samples were centrifuged at 5000 rpm for five minutes. The lipid A pellet was further washed (3×) in 1 ml of 95% EtOH. The entire series of washes was repeated twice. Samples were resuspended in 500 μl of water, frozen on dry ice and lyophilized.
2.1.b. Mass spectrometry procedures.
Negative ion matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) experiments were performed as described for the analysis of LPS or lipid A preparations with the following modifications.28 Lyophilized lipid A was dissolved with 10 μl 5-chloro-2-mercaptobenzothiazole (CMBT) (Sigma-Aldrich, St. Louis, MO) MALDI matrix in chloroform/methanol, 1 : 1 (v/v), and then applied (1 μl) onto the sample plate. All MALDI-TOF experiments were performed using a Bruker Autoflex II MALDI- TOF mass spectrometer (Bruker Daltonics Inc., Billerica, MA). Each spectrum was an average of 200 shots. ES Tuning Mix (Agilent, Palo Alto, CA) was used to calibrate the MALDI-TOF MS.
2.2. Sample preparation
Membrane mimics were prepared by first dissolving lyophilized lipids in chloroform or LPS in trifluoroethanol (TFE)/H2O (4:1, v/v). These lipid stock solutions were combined to create lipid mixtures in molar ratios mimicking bacterial membranes: Gram-negative inner membrane G(−) (IM), POPE/POPG/TOCL (7 : 2 : 1 molar ratio), Gram-positive membrane, G(+) POPG/DOTAP/POPE/TOCL (6 : 1.5 : 1.5 : 1); eukaryotic membrane POPC/POPE/cholesterol (5 : 1 : 1.8) (23 mole% cholesterol, Euk23), and red blood cell (RBC) eukaryotic membrane (5 : 1 : 6) (50 mole% cholesterol, Euk50). For the outer membrane (OM) of G(−) bacteria, a mixture of LPS/DLPG (1 : 9) (LPS model) was used instead of pure LPS because LPS alone did not undergo thermal fluctuations when fully hydrated. Composition of bacterial membrane mimics was based on ref. 29 and eukaryotic mimics was based on ref. 30. Our data analysis relies on membrane fluctuations that produce diffuse X-ray scattering as described below. WLBU2 and D8 stock solutions were prepared by mixing lyophilized powder in hexafluoroisopropanol (HIP).
Multilamellar stacked samples for X-ray scattering were prepared by mixing 4 mg of the lipid mixtures plus WLBU2 or D8 into glass test tubes in molar ratios from 1000 : 1 to 75 : 1, lipid/peptide. When calculating the molecular weight of the peptides, 13 trifluoroacetate counter-ions were included for each peptide for a total gram-molecular weight of 4865. Solvents were removed by evaporation under vacuum and samples were redissolved in appropriate HPLC-grade solvents for spreading (v/v ratios): G(−) IM, HIP/chloroform (2 : 1); G(+), HIP/chloroform (1 : 1); LPS/DLPG, TFE/H2O (10 : 1); eukaryotic with 23 mole% cholesterol, TFE/chloroform (1 : 1); and eukaryotic with 50 mole% cholesterol, chloroform. These mixtures were plated onto silicon wafers (1 × 15 × 30 mm) via the rock-and-roll method31 to produce stacks of ~1800 well-aligned bilayers, where the silicon wafer is rocked continuously during solvent evaporation. Plating occurred in the fume hood instead of the usual glove box due to the toxicity of HIP. Once immobile, the thin film was additionally evacuated for at least 2 h. The sample was trimmed to a central 5 mm wide strip parallel to the long-edge of the wafer.31 Hydration occurred through the vapor in a thick-walled X-ray hydration chamber.32
2.3. Low-angle X-ray scattering
Low-angle X-ray scattering (LAXS) data from oriented, fully hydrated samples were obtained at the G1 line at the Cornell High Energy Synchrotron Source (CHESS, Ithaca, NY) on two separate trips using X-ray wavelengths of 1.096 and 1.098 Å and sample-to-detector (S)-distances of 397 and 417 mm. In addition, a laboratory X-ray source RUH3R rotating anode X-ray generator (Rigaku, Tokyo, Japan) with a FOX 2D focusing collimator (Xenocs, Sassenage, France) and a Mercury CCD detector (Rigaku) was used with an X-ray wavelength of 1.5418 Å and S-distance of 283 mm. Full hydration was judged by no further increase in lamellar D-spacing after equilibration. Measurements were carried out in the fluid phase at 37 °C. The flat silicon wafer is rotated from −1.6 to 7 degrees during the data collection to equally sample all scattered X-rays (30 s dezingered scans at CHESS or 10 minute dezingered scans at CMU). The background was collected by setting the X-ray angle of incidence = −2.4°, where sample scattering does not contribute to the image. For data analysis, backgrounds were first subtracted to remove extraneous air and mylar scattering and the images were laterally symmetrized to increase the signal- to-noise ratio. As the sample nears full hydration, membrane fluctuations occur which produce “lobes” of diffuse X-ray scattering data.33,34 The fluctuations are quantitated by measuring the fall-off in lobe intensity in the lateral qr direction. The fitting procedure is a non-linear least squares fit that uses the free energy functional from liquid crystal theory
| (1) |
where N is the number of bilayers in the Z (vertical) direction, Lr is the domain size in the r (horizontal) direction, KC is the bending modulus, un is the vertical membrane displacement, and B is the compressibility modulus. We are able to obtain KC and B independently, where KC describes the bending of a single bilayer.35 A higher KC indicates a stiffer membrane, while a lower KC indicates a softer membrane, since KC is the bending modulus. This method has been used to obtain bending moduli of pure lipid membranes,36–43 membranes with peptides,44–52 cholesterol,53–55 sugar,56 drugs57 and bioflavinoids.58
2.4. Wide-angle X-ray scattering
Wide-angle X-ray scattering (WAXS) was obtained at CHESS (S-distance = 180 and 173 mm) and at CMU (S-distance = 123 mm). In order to obtain WAXS data, the same sample that was hydrated for LAXS is X-rayed with the CCD detector closer to the sample. Two exposures are taken at angle of X-ray incidence α = +0.5° and α = −0.5°, which are then subtracted from each other. Both are dezingered, 30 second scans (CHESS), or dezingered 10 minute scans (CMU). In some cases at CMU, multiple scans at α = +0.5° were averaged before subtracting the background scan to enhance the sample signal-to-noise ratio. The subtraction procedure removes extraneous scatter due to the mylar chamber windows and shadows. The chain-chain correlation appears as strong diffuse scatter projecting upwards circularly from the equator; the fall-off in intensity yields information about chain order. In order to obtain an SX-ray order parameter the intensity is first integrated along its trajectory, then fit with a wide-angle liquid crystal theory.59 The chain scattering model assumes long thin rods that are locally well aligned along the local director nL, with orientation described by the angle β. While acyl chains from lipids in the fluid phase are not long cylinders, the model allows the cylinders to tilt (β) to approximate chain disorder. From the fit of the intensity data, we obtain SX-ray using eqn (2) and a Matlab computer program.59
| (2) |
We also obtain the RMSE (root mean square error), which indicates the goodness of the fit. This method has been used to identify liquid-liquid domains in samples containing cholesterol,60 and chain order in many of the same references as for LAXS scattering (see above).
2.5. Bacterial growth inhibition assays
Antibacterial activity was examined by a standard growth inhibition assay endorsed by the Clinical and Laboratory Standards Institute (CLSI) with minor modifications as follows.61 Bacteria: P. aeruginosa (PA01); Methicillin-resistant Staphylococcus aureus (MRSA), USA300, were incubated at 37 °C with each of the indicated peptides in cation-adjusted Muller-Hinton broth (MHB; Sigma-Aldrich, St. Louis, MO) for 18 h, at which time A600 (absorbance at 600 nm) values were measured to examine growth inhibition using a BioTek microplate reader (BioTek Instruments).8 MICs were defined as the peptide concentration completely preventing detectable growth. Peptide concentrations up to 32 μM were evaluated for antibacterial activity.
3. Results & discussion
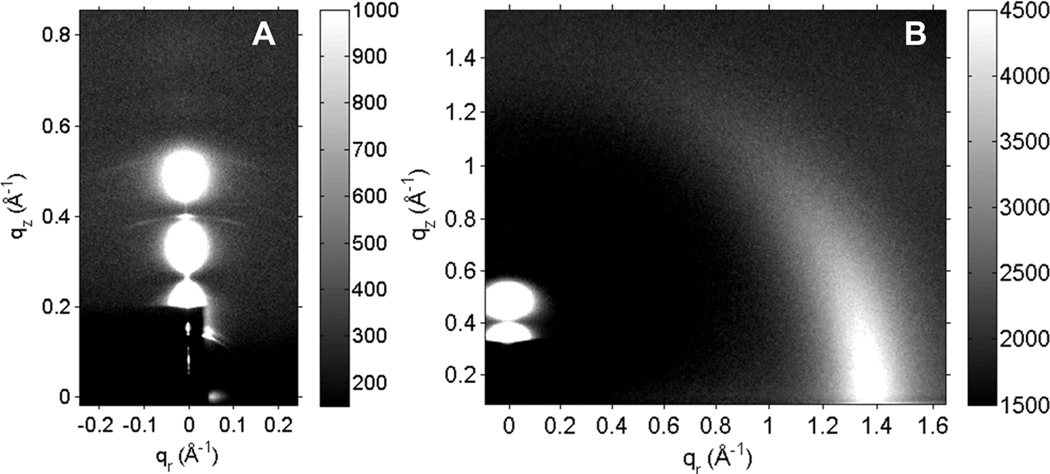
Fig. 2A shows typical background-subtracted LAXS data from oriented, fully-hydrated stacks of G(−) IM mimics containing WLBU2 in a 250 : 1 molar ratio. In addition to the usual white round “lobes” of diffuse data shown in Fig. 2A that are due to fluctuating bilayers, there are faint arcs with a much lower, desiccated D-spacing. These arcs were present in all G(−) IM/peptide samples with a concentration of either WLBU2 or D8 ≥ 500 : 1, and at ≥ 100 : 1 in G(+)/peptide samples, but not in either eukaryotic mimic (23 or 50 mole% cholesterol) or in the LPS model mimic. Since the G(−) IM mimic displays the desiccated D-spacing at a lower concentration of peptide, it suggests that the higher content of PE in this mimic could be interacting with some of the peptide creating a phase-separated domain with a small D-spacing. As the amount of PE is comparable in G(+) and Euk23 but the arcs are only present in the former, it is perhaps that even a small cholesterol concentration inhibits this phase separation. PE bilayers are known to take up very little water and we have shown previously that fluctuations of PE are inhibited when a positively charged peptide encounters it.44 Fig. 2B shows typical raw WAXS data of the same sample as in Fig. 2A, collected at a 0.5° glancing angle of X-ray incidence. Background collected at −0.5° was subtracted. The wide-angle correlation between chains is strongest near the equator at qr ~ 1.4 Å−1, and then tapers off azimuthally due to chain disorder, which is quantitated using liquid crystal theory as described in Materials and Methods. LAXS and WAXS images for the rest of the samples in this work were similar.
Fig. 2.
(A) LAXS and (B) WAXS data collected at CHESS at 37 °C of the G(−) IM mimic with 250 : 1 molar ratio lipid/WLBU2. In 2A, the beam stop (dark rectangle) covers the beam and the first and second lamellar orders. The thin, white vertical line in 2A is the X-ray reflectivity from the underlying silicon wafer. This sample is fully hydrated with a D-spacing of 85 Å. The faint arcs seen at ~0.4 Å−1 and ~0.5 Å−1 in qz are consistent with a second desiccated D-spacing of ~48 Å. In 2B, the chain correlation is the strong intensity centered qr ~1.4 Å−1. In 2B, the LAXS pattern is partially visible above the dark beam stop in the lower left hand corner.
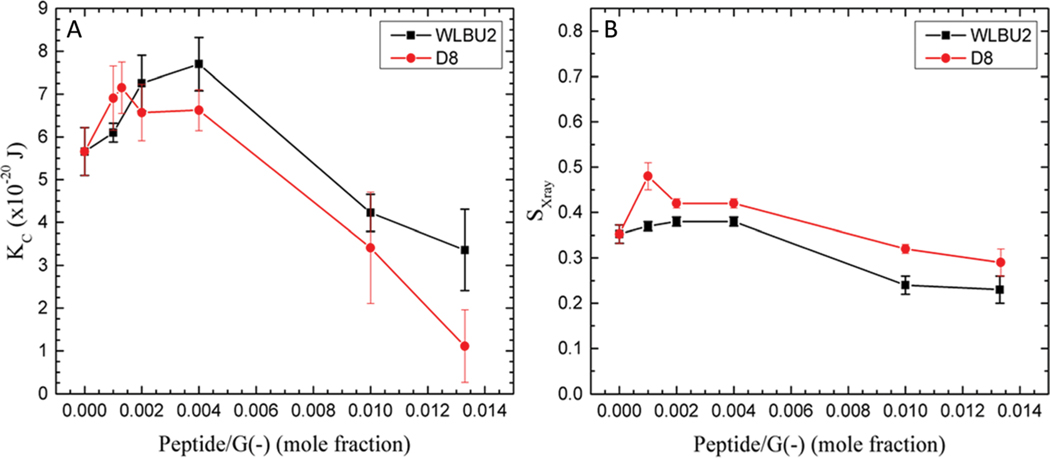
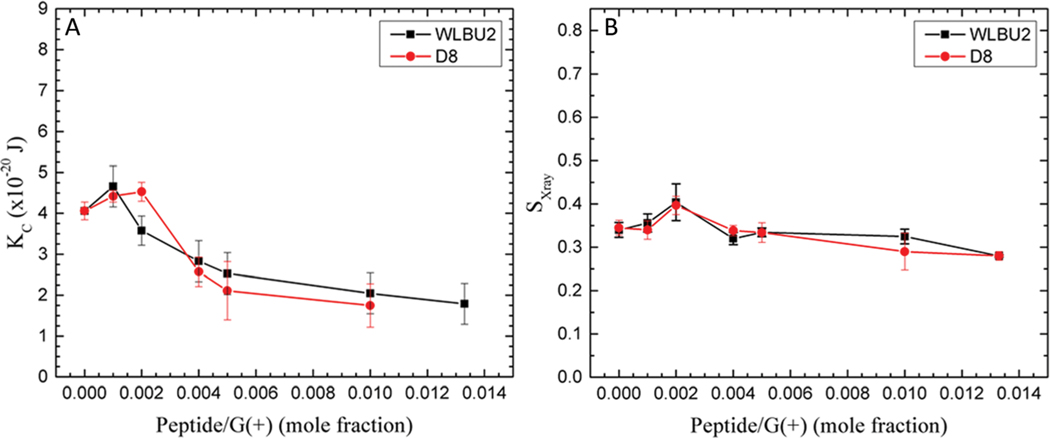
There is an increase in KC as low concentrations of the peptides are added to the G(−) IM (Fig. 3) and to the G(+) mimics (Fig. 4). This indicates that small amounts of both peptides cause stiffening of both mimics. The stiffening is more pronounced for G(−) IM mimics, which continue to be stiffened at 250 : 1 lipid : peptide molar ratio. Notice that the G(−) control is stiffer than the G(+) control, as we have seen previously,44 which is due to the higher PE content. The chain order results paralleled the KC behavior, indicating that the stiffening also involves ordering of lipid chains. We have previously observed that another cationic antimicrobial peptide, colistin, caused loss of fluctuations when added to POPE, due to extreme stiffening of the pure POPE membrane.44 At higher concentrations of both WLBU2 and D8, there is a softening below the control value. The parallel behavior of both peptides with G(−) IM and G(+) membrane mimics is an important result since both peptides efficiently kill both kinds of bacteria (see Table 1), indicating that modulation of elastic properties of membranes should be involved in the antimicrobial mechanism of action.
Fig. 3.
G(−) IM mimic. (A) Elasticity (KC) results for WLBU2 (black squares) and D8 (red circles). (B) SX-ray order parameter results for WLBU2 and D8, symbols as in (A). Standard deviations represent averages of three or more samples.
Fig. 4.
G(+) mimic. (A) Elasticity (KC) results for WLBU2 and D8 in G(+) mimic. (B) SX-ray order parameter results for WLBU2 and D8, same symbols as in Fig. 3. The data point at 75 : 1 G(+)/D8 in (A) was not successful after several attempts. Standard deviations represent averages of three or more samples.
Table 1.
Comparison of bacterial killing efficacy with toxicity
| Peptide | PA01 MIC (μM) | MRSA MIC (μM) | %RBC lysisa | %PBMC toxicityb |
|---|---|---|---|---|
| WLBU2 | 3.5 ± 1 (N = 4) | 3.2 ± 1.1 (N = 5) | l4 ± 1 | 23 ± 1 |
| D8 | 3 ± 1.2 (N = 4) | 3.2 ± 1.1 (N = 5) | 0 ± 1 | 8 ± 1 |
Red blood cell lysis measured at 50 μM peptide.
Peripheral blood mononuclear cell (leukocyte) toxicity measured by propidium iodide incorporation using flow cytometry, measured at 50 μM peptide.
Different elastic behavior at lower and higher concentration of peptides implies a possibility of peptides interacting with particular lipid components to a different degree, also evident from the appearance of desiccated lamellar orders mentioned above. Consequently, addition of peptide could initiate or enhance lipid demixing. Therefore, in the bacterial cell membrane there could be two or more different local concentrations of WLBU2 or D8, which could juxtapose domains with different material moduli, as we have suggested previously for the interaction of colistin with G(−) IM mimics.44 At the interface of these domains, defects could form allowing release of ions, water and perhaps proteins, thus killing the bacteria. The idea for juxtaposed domains with different material moduli causing defects along the domain walls was proposed by López Cascales, inspired by the results of their MD simulation.71 Existence of domains and lipid demixing as well as domain boundary effects triggered by specific lipid-peptide interactions have been shown experimentally.63,64 For example, Epand and Epand have called attention to indirect experiments using DSC, FTIR and NMR that have demonstrated a phase coexistence in model membrane mimics induced by cationic AMPs forming domains with negatively charged lipids.65 Our present results support a mode of action for both WLUB2 and D8 with both G(−) and G(+) bacterial cell membranes of domain formation with different elasticity that could lead to leakage along domain walls.
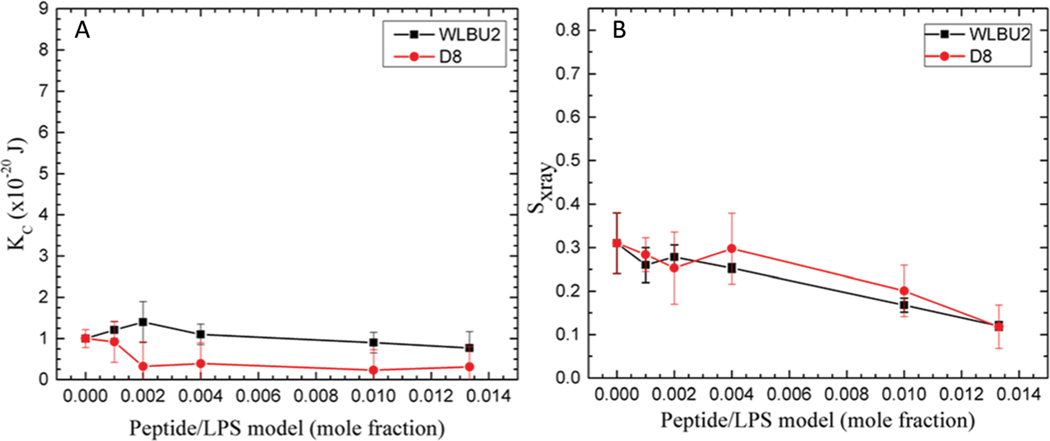
Before AMPs can reach the cell IM in G(−) bacteria, they must first bind to the outer LPS leaflet of the outer membrane, and then traverse the periplasmic space. Fig. 5 shows elasticity and chain order results for the LPS membrane mimic (DLPG/LPS 9 : 1) with increasing amounts of WLBU2 or D8. As shown in Fig. 5A, KC is quite low compared to G(−) IM or G(+) membranes, indicating a much softer membrane, which could allow the peptides to pass through. As we published previously, it is the LPS component, and not the DLPG that lowers the bending modulus, presumably due to the high carbohydrate content of LPS.44 The marked stiffening and softening that occurred with G(−) IM and G(+) membranes are not observed with the LPS membrane mimic. Indeed, the small apparent decrease in chain order caused by both peptides could be within the error of these measurements.
Fig. 5.
LPS mimic. (A) Elasticity (KC) results for WLBU2 and D8. (B) SX-ray order parameter results for WLBU2 and D8, same symbols as in Fig. 3. Standard deviations represent averages of three or more samples.
While the results so far have referred to bactericidal activity, experiments with neutral phospholipids, more typical of eukaryotic cells (POPC: POPE 5 : 1), were also carried out to compare to AMP toxicity. Toxicity results of WLBU2 and D8 with two types of eukaryotic cells, leukocytes and red blood cells (RBCs), are shown in Table 1. In hemolysis tests in RBCs, WLBU2 caused significant hemolysis (14%) at 14X MIC, while D8 displayed essentially no hemolysis at 16X MIC. In leukocytes, WLBU2 was also more toxic (23%) than D8 (8%). One of the main differences between these two cell types is the percentage of cholesterol. Leukocytes have ~ 33 mole% cholesterol,66 while RBCs have ~48 mole% cholesterol.67 Therefore we tested two eukaryotic membrane mimics with 23 and 50 mole% cholesterol to bracket the in vivo cholesterol content to investigate how cholesterol affects the elastic moduli and lipid chain ordering.
As shown in Fig. 6, both peptides perturb lipid membranes as seen from a marked softening (A) and disordering of chains (B) when added to the 23% cholesterol eukaryotic mimic. Within the standard deviations there was no difference between WLBU2 and D8. The mechanism could be a generalized perturbation, similar to the carpet model68 or detergent model69 where either a reorientation of lipid headgroups (carpet model) or micelle formation (detergent model) causes perturbation leading to leakage of contents through the cell membrane. Both of these mechanisms would be promoted by a softer and more disordered membrane. Therefore, we predict that eukaryotic cells containing 23 mole% cholesterol would be equally perturbed by both peptides. On the other hand, leukocytes containing ~ 33 mole% cholesterol,66 do show some differences in toxicity (see Table 1). This suggests that the higher amount of cholesterol in leukocytes could offer a protection from D8, but not from WLBU2. As a comparison to literature studies, Golbek et al. added WLBU2 to the subphase of a condensed phase of DPPC monolayers at room temperature and used sum frequency generation spectroscopy to find an increase in chain order due to WLBU2.70 However, given the lack of cholesterol and PE and different experimental conditions, this comparison may not be meaningful.
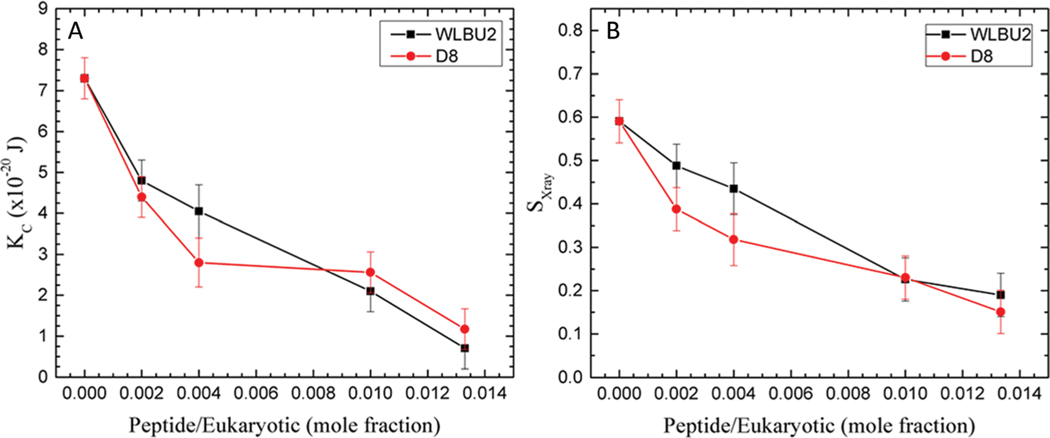
Fig. 6.
Euk 23 mimic. (A) Elasticity (KC) results for WLBU2 and D8. (B) SX-ray order parameter results for WLBU2 and D8, same symbols as in Fig. 3. Standard deviations represent duplicate samples.
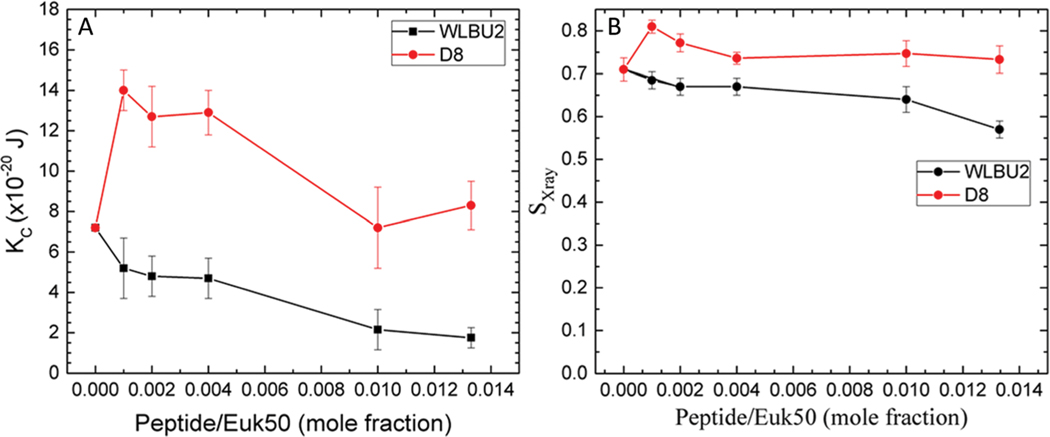
When we tested the RBC mimic with 50% cholesterol, the differences in bending modulus with peptide were very dramatic. Notice that the scale of the Y-axis in Fig. 7 is double that in Fig. 3–6. Fig. 7A shows that D8 increased KC (stiffened the membrane) by a factor 2, even at very low concentration (1000 : 1). At higher concentrations, the KC value declined back towards the control value. By contrast, WLBU2 softened the membrane even at low concentration, and increased softening as its concentration increased. Differences in chain order were also observed (7B). Chain order increased above control when D8 was added to the 50% cholesterol mimic, while it decreased with WLBU2. While these are real differences, they are not as dramatic as the KC behavior. Chain order does not always parallel membrane stiffness48 since it involves only the lipid acyl chains, while KC results from changes in material moduli of both headgroup and lipid chains.38 When we compare these results to the dramatic differences in RBC hemolysis (see Table 1), this suggests that D8 stabilizes membranes containing ~50% cholesterol which protects them from hemolysis. With stiffer membranes and more ordered chains than even the control, D8 does not cause lysis of RBCs, even at high concentration. By contrast, lysis in RBCs by WLBU2 is correlated with an increased softening of RBCs by WLBU2, and increased chain disorder compared to controls, similar to toxicity in leukocytes. In a literature study that used MD simulations, López Cascales et al. explored membrane perturbations caused by two 9-mer peptides with a charge of +4 or +7 in a fluid DPPC bilayer.62 They found that the +4 peptide caused larger membrane perturbations, especially an enhanced entropic component in the center of the bilayer that correlated with its greater microbicidal activity. While we do not measure entropy, we similarly find that the softening perturbation seen in Fig. 7 corresponds to WLBU2’s toxic effect in RBC membrane mimics, while D8’s stiffening effect may protect the membrane from degradation.
Fig. 7.
Euk50 mimic. (A) Elasticity (KC) results for WLBU2 and D8. (B) SX-ray order parameters results for WLBU2 and D8, same symbols as in Fig. 3. Standard deviations represent duplicate samples.
In our previous study of the AMP colistin interacting with membrane mimics,44 we found a correlation between irregular elasticity and chain order in G(−) IM mimics and killing of G(−) bacteria, which we interpreted as domain formation. We did not observe irregular changes in elasticity with G(+) mimics and colistin. Since colistin kills G(−) but not G(+) bacteria, we suggested that domain boundaries between the domains formed by colistin in G(−) bacteria allow leakage of bacterial contents. Since WLBU2 and D8 both kill G(−) and G(+) bacteria, we expected that their elastic and chain order behavior would be similar, which they were. We suggest that studies of membrane elastic behavior and chain order can be predictive of the efficiency of bacterial killings as well as of the toxicity towards eukaryotic cells when new AMPs are designed. Studies are now underway to decipher the structure and membrane location of WLBU2 and D8 when interacting with several membrane mimics.
4. Conclusions
In this work we show similar elastic behavior of WLBU2 and D8 with the G(−) IM and G(+) bacterial membrane mimics that increase stiffness and chain order at low concentrations, and soften and disorder these membranes at higher concentrations. This could lead to a juxtaposition of membrane domains with different stiffness and order, which could lead to leakage between the domains along their domain boundaries, similar to our proposed interaction of colistin with G(−) IM mimics. Both peptides slightly softened an already soft LPS mimic of the G(−) OM, which could facilitate transport of these AMPs across the OM to the IM of G(−) bacteria. While no differences in elastic and chain order behavior were observed between WLBU2 and D8 in eukaryotic membrane mimics containing 23% cholesterol, dramatic differences occurred with 50% cholesterol, a similar amount as present in an RBC membrane, correlating with their hemolytic activity. These results suggest that elasticity and chain order behavior can be used to predict mechanisms of bactericidal action and toxicity of new AMPs.
Acknowledgements
The authors would like to acknowledge Belita Opene (U Maryland-Baltimore) for purifying the PA01 LPS, and John F. Nagle for help with data collection at CHESS. Support for this work was from the Carnegie Mellon University SURF (AK, DM), HL125128 (YPD), AI133351 (STN, YPD, BD), Programa de Becas Externas of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina) (FGD), the Slovenian Research Agency Research Program (No. P1-0060) and the Fulbright Visiting Scholar Program (ZA), NIH R01-GM111066 (RKE), Center for Vaccine Research (RCM). This work is based upon research conducted at Carnegie Mellon University and at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation (NSF) under award no. DMR-1332208.
Footnotes
Conflicts of interest
R. C. M. holds stock in and serves on an advisory board for Peptilogics. Although a financial conflict of interest was identified based on the author’s relationship with Peptilogics, the research findings included in this publication may not necessarily be related to the interests of Peptilogics. All other authors: none to declare.
References
- 1.Talbot GH, Bradley J, Edwards JE, Gilbert D, Scheld M. and Bartlett JG, Clin. Infect. Dis, 2006, 42, 657–668. [DOI] [PubMed] [Google Scholar]
- 2.Rolain JM, Canton R. and Cornaglia G, Clin. Microbiol. Infect, 2012, 18, 615–616. [DOI] [PubMed] [Google Scholar]
- 3.Opatowski L, Mandel J, Varon E, Boelle PY, Temime L. and Guillemot D, Antimicrob. Agents Chemother, 2010, 54, 2330–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT and Monnet DL, Clin. Microbiol. Infect, 2012, 18, 268–281. [DOI] [PubMed] [Google Scholar]
- 5.Hancock RE, Falla T. and Brown M, Adv. Microb. Physiol, 1995, 37, 135–175. [DOI] [PubMed] [Google Scholar]
- 6.Gennaro R. and Zanetti M, Biopolymers, 2000, 55, 31–49. [DOI] [PubMed] [Google Scholar]
- 7.Tencza SB, Douglass JP, Creighton DJ, Montelaro RC and Mietzner TA, Antimicrob. Agents Chemother, 1997, 41, 2394–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Mietzner TA and Montelaro RC, Antimicrob. Agents Chemother, 2013, 57, 2511–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deslouches B, Islam K, Craigo JK, Paranjape SM, Montelaro RC and Mietzner TA, Antimicrob. Agents Chemother, 2005, 49, 3208–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deslouches B, Gonzalez IA, DeAlmeida D, Islam K, Steele C, Montelaro RC and Mietzner TA, J. Antimicrob. Chemother, 2007, 60, 669–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deslouches B, Phadke SM, Lazarevic V, Cascio M, Islam K, Montelaro RC and Mietzner TA, Antimicrob. Agents Chemother, 2005, 49, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Burns JL and Montelaro RC, Antimicrob. Agents Chemother, 2015, 59, 1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandell JB, Deslouches B, Montelaro RC, Shanks RMQ, Doi Y. and Urish KL, Sci. Rep, 2017, 7, 18098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YX, Mant CT, Farmer SW, Hancock REW, Vasil ML and Hodges RS, J. Biol. Chem, 2005, 280, 12316–12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bacalum M. and Radu M, Int. J. Pept. Res. Ther, 2015, 21, 47–55. [Google Scholar]
- 16.Wade D, Silberring J, Soliymani R, Heikkinen S, Kilpelainen I, Lankinen H. and Kuusela P, FEBS Lett.,2000, 479, 6–9. [DOI] [PubMed] [Google Scholar]
- 17.Hamamoto K, Kida Y, Zhang Y, Shimizu T. and Kuwano K, Microbiol. Immunol, 2002, 46, 741–749. [DOI] [PubMed] [Google Scholar]
- 18.Malanovic N. and Lohner K, Biochim. Biophys. Acta, Biomembr, 2016, 1858, 936–946. [DOI] [PubMed] [Google Scholar]
- 19.Sharma S, Sahoo N. and Bhunia A, Curr. Top. Med. Chem,2016, 16, 46–53. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen LT, Haney EF and Vogel HJ, Trends Biotechnol., 2011, 29, 464–472. [DOI] [PubMed] [Google Scholar]
- 21.Teixeira V, Feio MJ and Bastos M, Prog. Lipid Res, 2012, 51, 149–177. [DOI] [PubMed] [Google Scholar]
- 22.Kaur R. and Liu S, Prog. Surf. Sci, 2016, 91, 136–153. [Google Scholar]
- 23.Westphal OJK, Methods Carbohydr. Chem, 1965, 43, 83–91. [Google Scholar]
- 24.Fischer W, Koch HU and Haas R, Eur. J. Biochem, 1983, 133, 523–530. [DOI] [PubMed] [Google Scholar]
- 25.Folch J, Lees M. and Stanley GHS, J. Biol. Chem, 1957, 226, 497–509. [PubMed] [Google Scholar]
- 26.Hirschfeld M, Ma Y, Weis JH, Vogel SN and Weis JJ, J. Immunol, 2000, 165, 618–622. [DOI] [PubMed] [Google Scholar]
- 27.Caroff M, Tacken A. and Szabo L, Carbohydr. Res, 1988, 175, 273–282. [DOI] [PubMed] [Google Scholar]
- 28.Hajjar AM, Harvey MD, Shaffer SA, Goodlett DR, Sjostedt A, Edebro H, Forsman M, Bystrom M, Pelletier M, Wilson CB, Miller SI, Skerrett SJ and Ernst RK, Infect. Immun, 2006, 74, 6730–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson SG, Microbial Lipids, Academic Press, San Diego, CA, 1988. [Google Scholar]
- 30.De Gier J. and Van Deenen LLM, Biochim. Biophys. Acta, 1961, 49, 286–296. [DOI] [PubMed] [Google Scholar]
- 31.Tristram-Nagle SA, Methods Mol. Biol, 2007, 400, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kučerka N, Liu YF, Chu NJ, Petrache HI, Tristram-Nagle ST and Nagle JF, Biophys. J, 2005, 88, 2626–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y. and Nagle JF, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys, 2004, 69, 040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyatskaya Y, Liu Y, Tristram-Nagle S, Katsaras J. and Nagle JF, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys,2001, 63, 011907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, PhD thesis, Carnegie Mellon University, 2003. [Google Scholar]
- 36.Tristram-Nagle S. and Nagle JF, Chem. Phys. Lipids, 2004, 127, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan J, Tristram-Nagle S, Kucerka N. and Nagle JF, Biophys. J, 2008, 94, 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagle JF, Jablin MS, Tristram-Nagle S. and Akabori K, Chem. Phys. Lipids, 2015, 185, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu N, Kucerka N, Liu YF, Tristram-Nagle S. and Nagle JF, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys, 2005, 71, 041904. [DOI] [PubMed] [Google Scholar]
- 40.Boscia AL, Treece BW, Mohammadyani D, Klein-Seetharaman J, Braun AR, Wassenaar TA, Klosgen B. and Tristram-Nagle S, Chem. Phys. Lipids, 2014, 178, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guler SD, Ghosh DD, Pan J, Mathai JC, Zeidel ML, Nagle JF and Tristram-Nagle S, Chem. Phys. Lipids, 2009, 160, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kucerka N, Tristram-Nagle S. and Nagle JF, Biophys. J,2009, 90, L83–L85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrache HI, Tristram-Nagle S, Gawrisch K, Harries D, Parsegian VA and Nagle JF, Biophys. J, 2004, 86, 1574–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupuy FG, Pagano I, Andenoro K, Peralta MF, Elhady Y, Heinrich F. and Tristram-Nagle S, Biophys. J, 2018, 114, 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tristram-Nagle S, J. Phys. D: Appl. Phys, 2018, 51, 183001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neale C, Huang K, Garcia AE and Tristram-Nagle S, Membranes, 2015, 5, 473–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akabori K, Huang K, Treece BW, Jablin MS, Maranville A. Woll J. F. Nagle, Garcia AE and Tristram-Nagle S, Biochim. Biophys. Acta, Bioenerg, 2014, 1838, 3078–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boscia AL, Akabori K, Benamram Z, Michel JA, Jablin MS, Steckbeck JD, Montelaro RC, Nagle JF and Tristram-Nagle S, Biophys. J, 2013, 105, 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shchelokovskyy P, Tristram-Nagle S. and Dimova R, New J. Phys, 2011, 13, 25004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tristram-Nagle S. and Nagle JF, Biophys. J, 2007, 93, 2048–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greenwood AI, Pan JJ, Mills TT, Nagle JF, Epand RM and Tristram-Nagle S, Biochim. Biophys. Acta, Bioenerg, 2008, 1778, 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tristram-Nagle S, Chan R, Kooijman E, Uppamoochikkal P, Qiang W, Weliky DP and Nagle JF, J. Mol. Biol, 2010, 402, 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan J, Tristram-Nagle S. and Nagle JF, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys, 2009, 80, 021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kučerka N, Perlmutter JD, Pan J, Tristram-Nagle S, Katsaras J. and Sachs JN, Biophys. J, 2008, 95, 2792–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan JJ, Mills TT, Tristram-Nagle S. and Nagle JF, Phys. Rev. Lett, 2008, 100, 198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagle JF, Jablin MS and Tristram-Nagle S, Chem. Phys. Lipids, 2016, 196, 76–80. [DOI] [PubMed] [Google Scholar]
- 57.Peralta MF, Smith H, Moody D, Tristram-Nagle S. and Carrer DC, J. Phys. Chem. B, 2018, 122, 7332–7339. [DOI] [PubMed] [Google Scholar]
- 58.Raghunathan M, Zubovski Y, Venable RM, Pastor RW, Nagle JF and Tristram-Nagle S, J. Phys. Chem. B, 2012, 116, 3918–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mills TT, Toombes GE, Tristram-Nagle S, Smilgies DM, Feigenson GW and Nagle JF, Biophys. J, 2008, 95, 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mills TT, Tristram-Nagle S, Heberle FA, Morales NF, Zhao J, Wu J, Toombes GE, Nagle JF and Feigenson GW, Biophys. J, 2008, 95, 682–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sader HS, Ferraro MJ, Reller LB, Schreckenberger PC, Swenson JM and Jones RN, J. Clin. Microbiol, 2007, 45, 1640–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.López Cascales JJ, Zenak S, de la Torre JG, Lezama OG, Garro A. and Enriz RD, ACS Omega, 2018, 3, 5390–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arouri A, Dathe M. and Blume A, Biochim. Biophys. Acta, Bioenerg, 2009, 1788, 650–659. [DOI] [PubMed] [Google Scholar]
- 64.Lombardi L, Stellato MI, Oliva R, Falanga A, Galdiero M, Petraccone L, D’Errico G, De Santis A, Galdiero S. and Del Vecchio P, Sci. Rep, 2017, 7, 44425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Epand RM and Epand RF, J. Pept. Sci, 2011,17, 298–305. [DOI] [PubMed] [Google Scholar]
- 66.Gottfried EL, J. Lipid Res, 1967, 8, 321–327. [PubMed] [Google Scholar]
- 67.Cooper RA, J. Supramol. Struct. Cell. Biochem, 1978, 8, 413–430. [DOI] [PubMed] [Google Scholar]
- 68.Shai Y, Biochim. Biophys. Acta, Bioenerg, 1999, 1462, 55–70. [DOI] [PubMed] [Google Scholar]
- 69.Bechinger B, Biochim. Biophys. Acta, Bioenerg, 1999, 1462, 157–183. [DOI] [PubMed] [Google Scholar]
- 70.Golbek TW, Franz J, Fowler JE, Schilke KF, Weidner T. and Baio JE, Biointerphases, 2017, 12, 02D406. [DOI] [PubMed] [Google Scholar]
- 71.López Cascles JJ, Garro A, Porasso RD and Enriz RD, Phys. Chem. Chem. Phys, 2014, 16, 21694–21705. [DOI] [PubMed] [Google Scholar]