Abstract
Evolution of genetic mechanisms of sex determination led to two processes causing sex differences in somatic phenotypes: gonadal differentiation and sex chromosome dosage inequality. In species with heteromorphic sex chromosomes, the sex of the individual is established at the time of formation of the zygote, leading to inherent sex differences in expression of sex chromosome genes beginning as soon as the embryonic transcriptome is activated. The inequality of sex chromosome gene expression causes sexual differentiation of the gonads and of non-gonadal tissues. The difference in gonad type in turn causes lifelong differences in gonadal hormones, which interact with unequal effects of X and Y genes acting within cells. Separating the effects of gonadal hormones and sex chromosomes has been possible using mouse models in which gonadal determination is separated from the sex chromosomes, allowing comparison of XX and XY mice with the same type of gonad. Sex differences caused by gonadal hormones and sex chromosomes affect basic physiology and disease mechanisms in most or all tissues.
Keywords: Sexual differentiation, sex determination, sex chromosomes, X chromosome, Y chromosome, dosage compensation, cell-autonomous, sex difference
1. Introduction
The evolution of sex chromosomes in mammals involved two major events affecting separate classes of molecular pathways causing sex differences in tissues. One event was the emergence of sex-chromosome-encoded genetic mechanisms determining the differentiation of a testis or ovary from the early gonad, leading to sex differences caused by different levels of gonadal hormones in the two sexes. The second event was the evolution of heteromorphic sex chromosomes resulting in an inherent XX vs. XY difference in expressed doses of X and Y genes, which are incompletely compensated by X inactivation and other balancing mechanisms. In particular, the evolution of sexual disparity of X chromosome number led to major sex-specific adjustments in gene expression and in crosstalk between sex chromosomes and autosomes. Theories of sexual differentiation have long emphasized the first, gonad-specifying attribute of sex chromosomes, and underestimated the second mechanism based on difference in X and Y gene dosage. Moreover, although concepts of sexual differentiation focus on the inherent sexual inequality, the sex chromosomes also balance the effects of each other, making XX and XY cells more similar than they might otherwise be. The relative importance of the sex-differentiating roles of sex chromosomes, and of the sex-balancing roles, is not yet well understood, but bears significantly on the causes of sex differences in disease.
2. The classic vs. revised theories of sex determination and sexual differentiation
A widely-held tenet of sex determination is that “in most mammals, sex determination is initiated by transient expression of Sry …”, the Y gene initiating testis differentiation (Miura et al., 2018; Zhao et al., 2010). Here, I challenge the thinking behind this narrative, because (1) ubiquitous sex differences in cell phenotypes emerge well before the expression of Sry, and (2) after differentiation of gonads, significant sex differences in cells are not the result of the this “sex determination” process. The dominant narrative is flawed because “sex determination” excludes some components of sexual differentiation. The error can be dismissed as merely semantic, but if so, it is important to define terms to optimize communication among investigators. In my view, however, the issue is deeper, so that the classic narrative has limited the conceptualization of the evolution and processes of sexual differentiation, and therefore has undermined proper design and interpretation of studies of sexual differentiation.
The dominant classic tenet appears to derive from a definition of sex based exclusively on type of gonad: females have ovaries and make eggs, and males have testes and make sperm. This definition has the advantage of being simple, long-standing, and can be consistently applied across all sexually reproducing plants and animals. In animals with genetic sex determination (e.g., mammals and birds), however, sex determination has been taken over by genetic mechanisms common to all cells, so that the onset of sexual differentiation of the gonads no longer defines sex because it does not encompass all sexual differentiation processes.
The central question of this chapter is: “What are the biological forces that make females and males different?” The focus is on sex as a set of diverse forces, not as an identity or simply defined condition. The discussion is about biological forces, not social forces, even though social forces have profound sex-biasing effects on phenotypes, which are nearly impossible to disentangle from the effects of biological factors with which they are strongly correlated, especially for human phenotypes that are difficult to model in animals. To answer the central question, a definition of sex based on gonadal type is not fundamental enough to encompass all forces that differentiate the two sexes. We explore the more expansive idea that all biological sex differences stem originally from the inherent imbalance of sex chromosome factors, present in the zygote, and acting in the embryo and throughout life.
In this sex-chromosome-centric view, the expression of Sry is not the sex determinant, but the testis-determining factor, one of several genes on the Y chromosome that make males different from females in diverse tissues (Figure 1)(Arnold, 2011; Arnold, 2017a). Sry is arguably the most important of sex-differentiating factors, because its expression leads to lifelong secretion of testicular hormones, rather than ovarian hormones. The different effects of gonadal hormones account for many sex differences in non-gonadal phenotypes, including the external and internal genitalia that are fundamental to classifying humans as male or female. XX and XY non-gonadal cells also experience the inherently sex-biased effects of X or Y genes, stemming from the inequality of representation of these genes within the male and female genome, which we call “sex chromosome effects” causing sex differences. Importantly, many sex-biased phenotypes are impacted by both gonadal hormones and sex chromosome effects, although the interaction of these different molecular mechanisms to date is poorly studied.
Figure 1. The sex-chromosome-centric view of sexual differentiation.
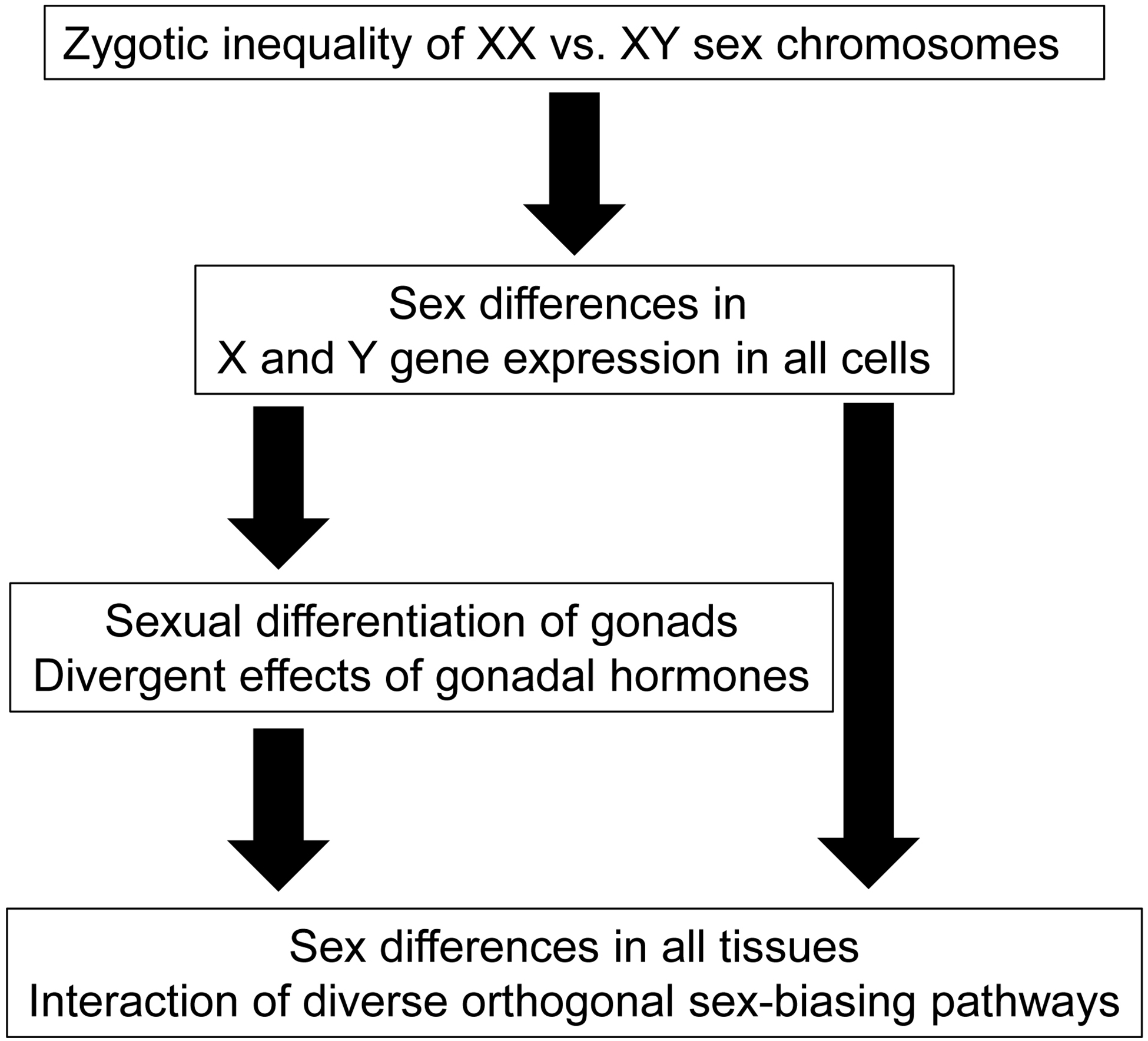
All phenotypic sex differences stem from the difference in number and type of sex chromosomes present in the zygote. In the undifferentiated gonadal ridge, Sry expression in XY cells induces differentiation of testes. Ovaries differentiate in XX animals in the absence of Sry, because of the expression of autosomal or X-linked genes. Differences in gonadal hormone levels contribute to sex differences in tissue function. In parallel, sex differences are also caused in all non-gonadal tissues by differences in effects of X and Y genes. Hormonal and sex chromosome factors interact to promote or reduce sex differences in phenotypes.
3. Evolution of sex chromosomes: balance and inequality of X and Y
The inherent imbalance in XX vs. XY genomes can best be understood based on sex chromosome evolution. Early in the therian radiation, an autosomal mutation led to the emergence of the dominant testis-determining gene, Sry, which is expressed in the bipotential gonad to commit that tissue to a testicular fate (Sutton et al., 2011). The evolution of Sry led to several important consequences. (1) The genomic region near Sry was inherited only by gonadal males and it acquired genes that were good for males (Rice, 1992; Burgoyne and Mitchell, 2007). The near-Sry region lost ability to recombine with its partner proto-X chromosome (Bachtrog, 2013; Cortez et al., 2014; Hughes and Page, 2015). (2) Recombination of the X and Y chromosome was further reduced because of subsequent major inversions on the Y chromosome (Lahn and Page, 1999), leading to expansion of the male-only region to encompass nearly the entire Y chromosome, except for a small pseudoautosomal region (PAR) shared between the X and Y. The asexual inheritance (father to son, with no recombination) led to loss of most Y genes (Rice, 1994; Vallender and Lahn, 2004; Graves, 2016; Charlesworth, 1996). (3) The progressive loss of Y genes left X genes expressed at a lower dose in XY than XX cells, causing a sex difference in stoichiometric balance of X and autosomal genes that cooperate in gene networks (Veitia et al., 2015). For some X genes, the reduced balance with autosomal genes was critical, and favored evolution of dosage compensation mechanisms to adjust the X to autosomal expression ratios to within an acceptable range in both sexes. In XX cells, one X chromosome is transcriptionally silenced, so that both XX and XY cells express genes mostly from one X chromosome (Disteche, 2016). In addition, some evidence suggests that the expression of many X genes may be upregulated in both XX and XY cells to match the general level of expression of autosomal genes, with which they interact in gene networks (Nguyen and Disteche, 2006; Disteche, 2016; Sangrithi et al., 2017).
These dosage compensation mechanisms, X-inactivation and upregulation of X gene expression to better match autosomal expression, have the general consequence of making XX and XY cells phenotypically more similar to each other because they offset the effect of the difference in X chromosomal dose. The compensation is incomplete however, so that XX and XY genomes have inherent functional differences caused by difference in X and Y gene dose. Some X genes escape inactivation and are expressed from all X chromosomes, and therefore at constitutively higher levels in XX cells than XY cells in many tissues and developmental stages (Tukiainen et al., 2017; Carrel et al., 1999). In humans, about 23% of non-PAR X genes are expressed higher in XX than XY tissues (Figure 2). Genes appear be left out of, or protected from, the inactivation mechanism either because they are recently added to the X chromosome (Graves, 2016) or because they occupy positions in gene networks that are insensitive to dose, where one copy is as effective as two (Naqvi et al., 2018).
Figure 2. Inherent bias in human X chromosome gene expression across tissues.
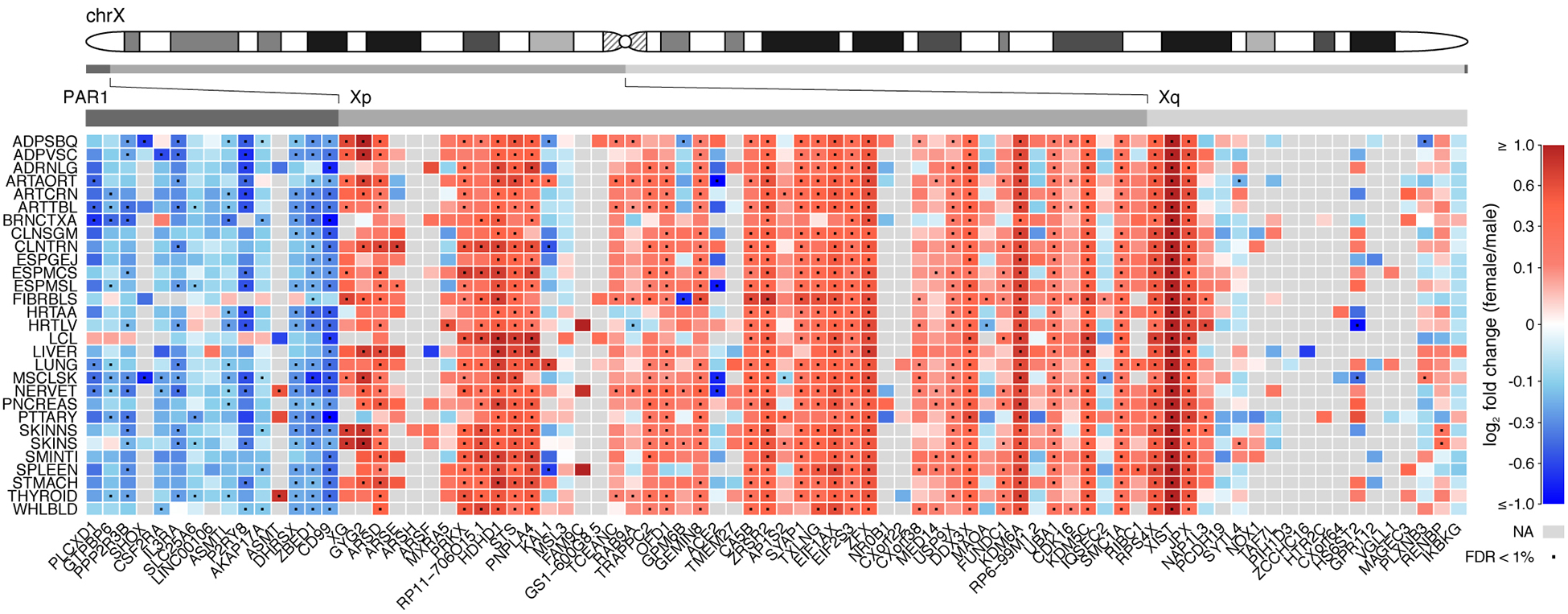
Analysis of over 5500 transcriptomes from 29 human tissues from GTEx consortium data shows the level of sex bias in expression of X genes reported to escape inactivation. Rows indicate different tissues, and columns represent X genes reported to escape X inactivation, mapped by chromosomal position. Red indicates expression higher in females, and blue indicates expression higher in males. Grey indicates expression too low to be analyzed. In the non-pseudoautosomal (non-PAR) region, about 23% of X genes overall showed higher expression in females, and in PAR1 region most genes were expressed higher in males. This general pattern can be attributed to escape from X inactivation in the non-PAR region in XX cells, and from some inactivation of PAR genes in XX cells but not XY cells. However, other sex-biasing factors (gonadal hormones, parental imprint) could contribute to sex differences as well, in either direction, and may account for colors that deviate from the pattern of adjacent genes (e.g., red squares in the PAR, and blue squares in the non-PAR). Reprinted from (Tukiainen et al., 2017) under terms of Creative Commons Attribution 4.0 International license.
The dosage sensitivity of genes was apparently a major factor influencing evolution of sex chromosome gene content. Dosage sensitivity determined, in part, which Y genes were lost or retained during evolution, and affected which X genes were subject to inactivation (Naqvi et al., 2018; Veitia et al., 2015; Bellott et al., 2014; Cortez et al., 2014). One example are X-Y gene pairs, which have similar DNA sequence to each other, and evolved from a common autosomal ancestor gene. X-Y gene pairs are involved in basic cellular functions such transcription factors and regulators of RNA splicing and translation, and are thus expressed widely across tissues (Bellott et al., 2014). The Y partner genes have survived on the Y chromosome despite hundreds of millions of years of degrading influences that eliminated most other Y genes, probably because of the particularly high dosage sensitivity of the X-Y pair that made deletion of the Y gene lethal or highly disadvantageous. The X and Y genes often share high sequence similarity, attesting to their common origin and shared functions. The X partner gene typically escapes X inactivation and is expressed higher in XX cells than XY cells (Disteche, 2016). The Y partner gene likely compensates for the lower expression of the X partner gene in XY cells, if two X copies in XX cells have similar function compared to that of one X copy plus one Y copy in XY cells.
Paradoxically, although the evolutionary survival of the Y partner genes of X-Y gene pairs is explained by a balance of effects of the X and Y partners, X-Y gene pairs are considered a prime source of sexual inequality of XX and XY cells because their balance is variable across tissues, ages, or environmental conditions such as disease. For example, the Y genes are typically expressed at lower levels than the X partner in many tissues (Deng et al., 2014). Moreover, the partner genes sometimes show different patterns of expression across tissues (Xu et al., 2008). These inequalities can be explained if the balance of effects of the two genes might be critical in only some tissues or at specific developmental periods. Elsewhere, the Y gene was free to evolve male-specific functions, different from that of the X partner. An interesting case is the X-Y gene pair Utx-Uty, which is found in sex chromosomes of many mammalian species. Utx (a.k.a. Kdm6a) is a histone demethylase. Knock out of Utx is lethal in XX mouse embryos, but the presence of a functional copy of Uty prevents some of the mortality, illustrating the overlapping function of Uty with Utx (Shpargel et al., 2012). Nevertheless, the histone demethylase activity of Utx is not found in Uty, illustrating divergence of function of the two orthologous genes. In the brain, the pattern of expression of Utx and Uty are somewhat different (Xu et al., 2008). Thus, functional differences in XX and XY cells likely result from different levels of expression of both Utx and Uty.
Another illustrative example of X-Y gene pairs involves mouse Sly and its X partners, Slx and Slxl1. These genes participate in meiotic drive, an example of intergenomic conflict. Factors (drivers) arise on the Y chromosome that promote its inheritance in preference to the X chromosome. That process sets up counteracting pressures to select for suppressor sequences on the X chromosomes that favor its inheritance. Drivers and suppressors were duplicated on each chromosome, in successive waves that offset and overcame each other in evolutionary time. In mice, 95% of the Y chromosome represents massive duplications of only three gene families, and the X chromosome also contains extensive duplicated regions (Soh et al., 2014). The duplicated X-Y gene pair Sly-Slx participates in meiotic drive, because deletions of Sly undermine sperm viability, and deletions of Slx rescue these sperm differentiation defects. Sly deficiency also corrects the enhanced production of male offspring produced by Slx deficiency (Cocquet et al., 2009; Cocquet et al., 2012).
Further evidence supports the idea that the balance of X and Y gene effects is tenuous and varies across tissues, genotypes, age, and evolutionary time. XY male mice, of many strains, have greater body weight than XX female mice. In C57BL/6 mice, the number of X chromosomes causes sex differences in body weight and fat, but the Y chromosome has little effect (Chen et al., 2012) (Figure 3). In MF1 strain mice, however, XO mice weigh less than XX or XY mice, so that the addition of a second sex chromosome of either type to the XO genotype has the same effect (Chen et al., 2013). In MF1 mice, the X and Y chromosomes have balanced effects on body weight and fat, but the two chromosomes have unequal effects in C57BL/6, illustrating that the balance the two chromosomes can come and go depending on factors that interact with X and Y genes.
Figure 3. The number of X chromosomes influences body weight in FCG and XY* mice.
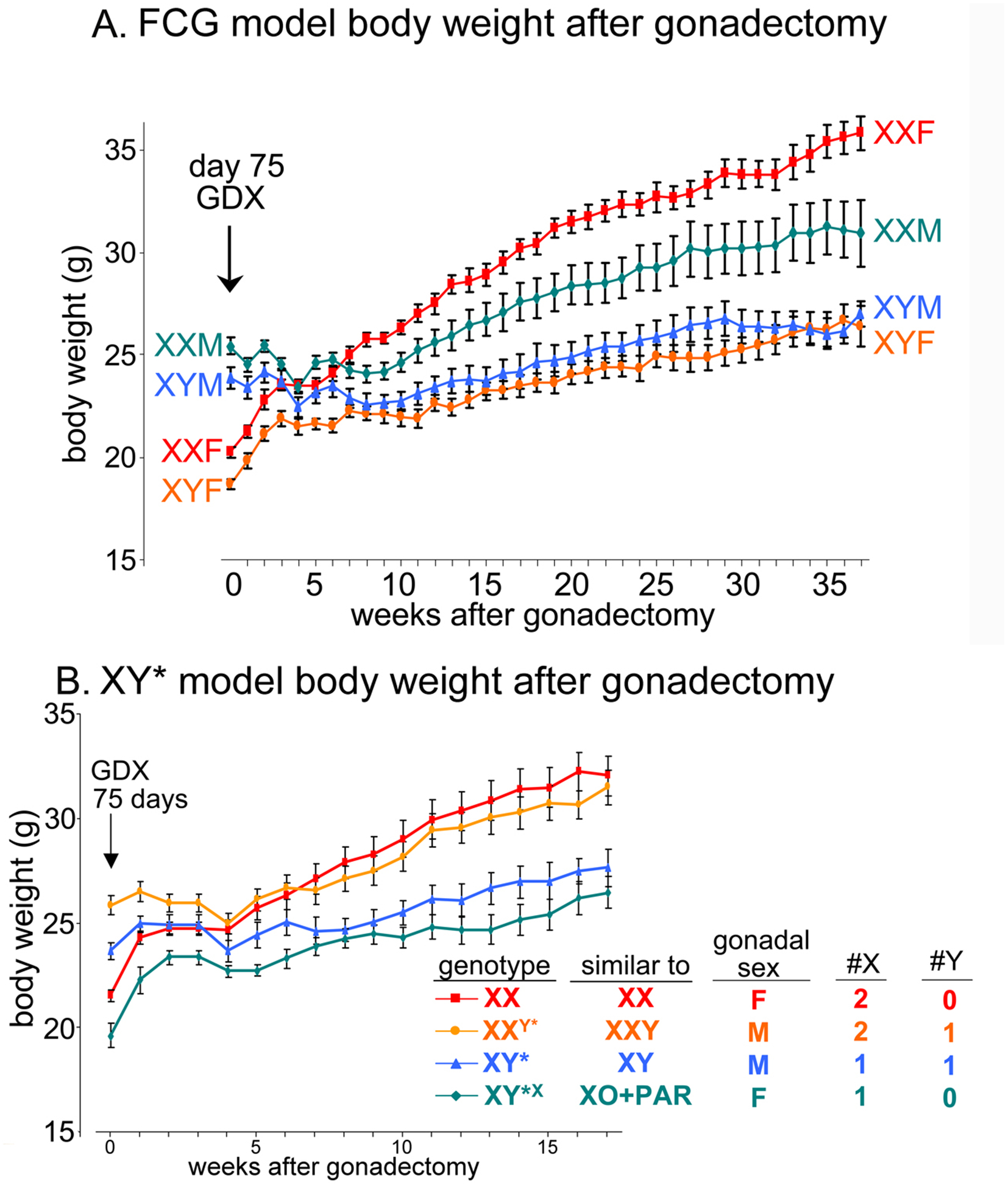
A. In FCG mice, gonadal males at postnatal day 75 (week 0) weighed about 25% more than gonadal females. When gonads were removed on that day, the sex differences disappeared by 4 weeks after gonadectomy (GDX). After that, XX mice slowly gained much more body weight than XY mice, irrespective of previous gonadal type. B. In the XY* model, the same experiment replicates the larger body size of two groups with testes, relative to two groups with ovaries. After gonadectomy, the groups with two X chromosomes gained more weight than those with one X chromosome, illustrating that the body weight is affected by the number of X chromosomes. M = gonadal male, F = gonadal female. PAR = pseudoautosomal region. Reprinted from (Chen et al., 2012) under terms of the Creative Commons Attribution License.
The tension between the gonad-centric and sex-chromosome-centric views of sexual determination uncovers a fundamental difference in thinking about evolutionary forces that lead to sex differences in phenotype. Within the gonad-centric view, sex differences are related predominantly to the evolution of sexual reproduction. Animals with testes or ovaries have evolved functional differences throughout the body that support the two reproductive roles. The evolution of sex differences in internal and external genitalia are obvious examples, because the different anatomical structures support the different roles. Sex differences in other tissues, such as the brain, have also long been rationalized as the result of separate selection pressures on males and females to make the brain support sex-biased reproductive functions such as ovulation, sex behavior, and aggression. In this view, sex differences are often seen as adaptive, rather than side-products of other evolutionary processes. The sex-chromosome-centric view emphasizes a different set of selection pressures. The inherent sexual inequality in genomic dose and effect of X and Y genes is the result of evolutionary factors generally unrelated to sexual function. As discussed above, sex differences caused by imbalance of X and Y genes occur because of loss of recombination of the X and Y chromosomes, loss of genomic regions on the Y chromosome, dosage compensation on the X and Y chromosomes, intergenomic conflict, etc. Thus, gonadal females get stuck with having two X chromosomes, and gonadal males with one X and one Y, for evolutionary reasons that do not primarily enhance reproduction. Moreover, the inequality of chromosome size affects many genes lacking a direct influence on reproduction. Of course, the different effects of X and Y genes in the two sexes must be compatible with their reproductive roles, so that both types of evolutionary pressure operate simultaneously within the constraints of tissue requirements related to sex roles. The evolution of different reproductive roles of cells and tissues, controlled dominantly by hormonal effects of gonadal secretions, occurs on a background of inherent sex differences in the genome.
4. Categories of X and Y genes causing phenotypic differences between XX and XY cells
It is useful to list all of the types of sexual inequality that are currently known or suspected in XX vs. XY cells because of the inherent imbalance of representation of sex chromosome genes (Arnold, 2017a). (1) The male-specific expression of Y genes causes XY cells to differ from XX cells (Arnold, 2017b; Case and Teuscher, 2015). (2) The constitutive difference in number of X chromosomes leads to four classes of X genes that underlie sex differences in phenotype. (2A) X genes escaping X inactivation are expressed higher in XX than XY cells (Carrel et al., 1999; Tukiainen et al., 2017). Sex differences in trophoblast expression of the putative X escapee gene Ogt is reported to cause sex differences in resilience to prenatal insults (Nugent et al., 2018). (2B) Genes in the pseudoautosomal regions (PAR) of the sex chromosomes are often expressed at higher levels in XY cells, relative to XX cells, because some of them are subject to X inactivation in XX cells (Tukiainen et al., 2017)(Figure 2). (2C) X genes that experience a parental imprint can be expressed at a higher or lower level in XX vs. XY cells because a paternal imprint affects only XX cells (Arnold, 2017a). (2D) The difference in epigenetic regulation of the X and Y chromosomes could have ramifications for epigenetic status of the autosomes. In Drosophila, for example, the large heterochromatic Y chromosome has indirect epigenetic effects on autosomal gene expression, which are not mediated by expression of Y genes (Silkaitis and Lemos, 2014). In mammals, it is conceivable that Xist, which initiates X inactivation in cis in XX cells, also has trans effects on autosomes. For example, silencing of Xist in adult XX mice leads to hematologic cancer (Yildirim et al., 2013), although this could be the result of reactivation of the inactive X chromosome instead of direct effects of Xist on autosomes. Moreover, a large heterochromatic X chromosome, present only in XX cells, is postulated to sequester heterochromatizing factors and reduce their availability to regulate autosomal heterchromatin, affecting autosomal gene expression (Wijchers and Festenstein, 2011).
The molecular differences between XX and XY cells, just listed, operate during ontogeny of all individuals, and thus can often be measured in inbred laboratory strains of animals, especially mice. Other forces cause sex differences in populations of humans and other animals, because of natural genetic heterogeneity not modeled in inbred strains (reviewed by (Arnold, 2017a)). These factors cause average differences between groups of males and females. (1) XY males have hemizygous exposure of X alleles, so that they express X allelic variations more prominently than XX females (Midgeon, 2007). XY individuals with X-linked lethal alleles are removed from the population, and shift the mean phenotype of males vs. females. (2) XX individuals are mosaic for X alleles and X gene imprints, because the parental origin of the active X varies among XX cells but not among XY cells. The mosaicism itself can have a protective effect for XX but not XY tissues. For example, in XX mice with a null mutation in one allele of the X-linked Hccs gene, cells expressing the null allele are selectively removed from the heart during embryonic development (Drenckhahn et al., 2008). (3) Some autosomal alleles are likely to be better adapted to one sex than the other, and thus will be found in greater frequency in one sex, creating an average genetic difference in autosomes of males and females. (4) The different modes of inheritance of the mitochondrial and nuclear genomes lead to sex-biasing effects. Because the mitochondrial genome is inherited in the female lineage, mitochondrial alleles might arise that are beneficial to females but harmful to males. Such a mismatch of mitochondrial genomes of the mother and nuclear genomes of her sons can have differential harmful effects in males relative to females (Camus et al., 2012; Frank, 2012; Innocenti et al., 2011).
5. Mouse models and methods that separate effects of sex chromosomes vs. gonadal hormones
For well over a century, investigators have been able manipulate gonadal hormone levels effectively to test their roles in causing sex differences in physiological and behavioral traits. The classic method is to reduce hormone levels by gonadectomy, and increase them by treating with hormones. Hormone effects are also manipulated by pharmacologic or genetic manipulations of steroid receptors or synthetic enzymes. In contrast, manipulating the number of sex chromosomes has been much more difficult. Thus, the first reports of sex chromosome effects causing sex difference relied on naturalistic observations rather than experimental manipulation. In the marsupial tammar wallaby, sex differences in the differentiation of the pouch or scrotum precede the differentiation of gonads (Renfree and Short, 1988). In mice and other mammalian embryos, the size of XX and XY embryos differs long before differentiation of gonads (Burgoyne et al., 1995). The embryonic mouse transcriptome differs in XX and XY cells as soon as the embryonic transcriptome is activated at the 8-cell stage or earlier (Lowe et al., 2015; Werner et al., 2017; Bramble et al., 2016), and sex differences in the transcriptome are found at later embryonic stages prior to gonadal differentiation (Dewing et al., 2003; Nef et al., 2005). In other cases, sex differences in phenotype are found in adult intersex animals such as avian gynandromorphs in which the sex difference is attributable to sex chromosome effects rather than hormonal effects (Agate et al., 2003; Witschi, 1939; Zhao et al., 2010).
The investigation of sex chromosome effects has been advanced by the production of mouse models in which whole mice with the same gonadal sex are engineered to have different complements of sex chromosomes (Burgoyne and Arnold, 2016; Arnold, 2014; Cox et al., 2014). In these models, the hormones are held relatively “constant” across groups, while sex chromosome complement is varied. In the Four Core Genotypes (FCG) model, the Sry gene is deleted from the Y chromosome, producing the Y¯ chromosome and XY¯ gonadal females that are compared to XX gonadal females. In addition, an Sry transgene is introduced onto chromosome 3, so that testis-determination is specified independently of the sex chromosome complement. In the FCG model, XY¯(Sry+) fathers mate with XX females to produce four types of offspring: XX and XY gonadal males, and XX and XY gonadal females (Figure 3). The model separates sex-biasing factors into two groups: those that differ in XX vs. XY mice (comparing XX and XY mice with the same type of gonad), and those that differ in gonadal males and females (comparing gonadal males and females with the same sex chromosome complement) (De Vries et al., 2002). The model is widely applicable to assess the causes of sex differences in virtually any somatic trait. As expected, many phenotypes differ in gonadal males and females, confirming the long-standing concept that gonadal hormones cause many or most sex differences in somatic phenotypes throughout the body. However, an increasing number of XX vs. XY differences have also been discovered (Cox et al., 2014; Arnold et al., 2017; Arnold et al., 2016; Arnold et al., 2013; Arnold and Chen, 2009).
The discovery of a phenotypic difference in XX vs. XY FCG mice is the first step in discovering the mechanisms of action of X or Y genes causing sex differences in phenotype. A second mouse model, the XY* model, is useful for figuring out whether the XX vs. XY phenotypic difference is attributable to X or Y genes. The XY* model produces genotypes very similar to XO, XX, XY, and XXY (Burgoyne and Arnold, 2016) (Figure 3). The XY* model allows measurement of the effects of X chromosome number (XO vs. XX gonadal females, and XY vs. XXY gonadal males). It also allows measurement of effects of presence / absence of the Y chromosome: XO vs. XY, and XX vs. XXY. Because Sry is present on the Y chromosome in this model, the comparisons of mice with and without the Y chromosome are confounded by differences in the type of gonad and levels of gonadal hormones. However, if such gonadal effects are found not to cause the sex difference in phenotype in FCG mice, the effects of the Y chromosome in the XY* model can be attributed to non-hormonal (direct) effects of Y genes. Using the XY* model, sex differences have been attributed more often to the effects of X genes, but Y chromosome effects have been found as well (Umar et al., 2018; Arnold et al., 2016).
6. Sex differences caused by sex chromosome effects
The FCG and XY* mouse models have been used to discover sex differences in various mouse phenotypes that are caused by sex chromosome complement, as previously reviewed (Arnold et al., 2016; Arnold, 2014; Cox et al., 2014; Arnold et al., 2017). A few examples are selected for discussion here.
In the aggregate, the studies indicate that XX vs. XY differences participate in causing sex differences in a wide variety of tissues and traits, and affect multiple mouse models of disease. The XX vs. XY differences affect autoimmune disease and susceptibility to infection, brain and behavior, metabolic traits including adiposity and fat metabolism, and numerous cardiovascular traits. Although sex chromosome effects are now widely recognized, numerous questions remain. For most traits, it is not known which X or Y genes (or non-genic regions of the sex chromosomes) account for the sex chromosome effect. This question is particularly of translational importance. The identification of specific X or Y genes causing a sex chromosome effect will greatly facilitate research about the role of such genes in sex differences in human disease. For example, one can study effects of variation of that gene in human populations, and cellular pathways influenced by the gene in human cell systems. The FCG and XY* models can be used to discover effects of X or Y genes contributing to sex differences in mouse models, after which manipulation of specific candidate genes leads to identification of the genes responsible [reviewed by (Burgoyne and Arnold, 2016)].
A second major outstanding question is how the sex differences caused by sex chromosome complement interact with sex differences caused by gonadal hormonal effects. Typically, both of these factors contribute to sex differences of the same phenotypes in mice. In what molecular pathways do diverse sex-biasing factors interact to affect the same disease process?
6. 1. Independent sex biasing factors interact to cause sex differences in physiology and disease
6.1.1. Metabolism and Body Weight
The greater body weight of male mice, relative to females, is reduced by gonadectomy of adult mice, indicating that gonadal hormones largely are responsible for this difference. However, after gonadectomy of FCG mice, the body weight of XX mice increases slowly over weeks until it is much greater than that of XY mice, irrespective of their type of gonad (Figure 3A)(Chen et al., 2012). The sex chromosome effect is eventually as large as the effect of gonadal hormones in young mice at the outset of the experiment. As discussed above, use of the XY* model allows us to test mice with different numbers of X or Y chromosomes. These studies show that the XX-XY difference is caused by X genes, not Y genes (Figure 3B). The greater body weight is mostly the result of an X chromosome effect on the amount of body fat. In mice eating a high fat diet, sex chromosomes cause sex differences in the level of plasma cholesterol, and accumulation of liver trigylcerides (Link et al., 2015; Chen et al., 2012). In the case of body weight, the gonadal hormonal effect counteracts the sex chromosome effect: male hormones are associated with greater body weight, but male sex chromosomes (XY) with lower body weight (Figure 3). To date, little is known about the molecular site of interaction of the two effects, and whether they influence the same molecular pathways or distinct pathways that both impinge on the same emergent phenotype.
6.1.2. Sex differences in volumes of brain regions
Concepts of sexual differentiation in brain and behavior have stemmed disproportionately from study of a few phenotypes with large sex differences. In animals, a few brain regions are 3–6 times larger in one sex than the other (Forger, 2009; Arnold and Gorski, 1984), where it has been easiest to measure the effects of variables that cause the sex differences. These large sex differences are related to sexual function, such as sexual behavior, ovulation, aggression, and courtship. The principles learned for these brain regions have been thought to represent general principles by which sex variables (e.g., gonadal hormones) influence brain development and function (Breedlove et al., 1999). The advent of high-resolution brain-wide structural MRI has allowed investigators to ask if these principles also apply more generally across all brain regions. Importantly, study of FCG mice shows an unexpected incidence of sex chromosome effects not previously found in the most sexually dimorphic brain regions (Corre et al., 2016). Of 62 brain regions examined, 30 were significantly different among FCG groups, with 20 showing effects of gonadal hormones and 14 effects of sex chromosomes (Figure 4). The brain regions affected by gonadal hormones or sex chromosomes tended to be distinct. When gonadal hormones were removed before puberty and brain anatomy was measured in adulthood, most differences between XX and XY brain regions persisted, although on average they were modestly attenuated (Vousden et al., 2018). In some brain regions, the effects of gonadal hormones and sex chromosomes were synergistic, such that male-female hormonal effects where in the same direction as XY-XX sex chromosome effects. In other regions, the two sets of variables were antagonistic or compensatory, such that removal of hormones caused an increase in effect of sex chromosome complement. These complex interactions of hormonal and sex chromosome factors illustrate the orthogonal actions of the two sets of factors in a brain-region dependent fashion (Vousden et al., 2018).
Figure 4. Sex chromosome and hormonal effects on brain volumes from FCG mice.
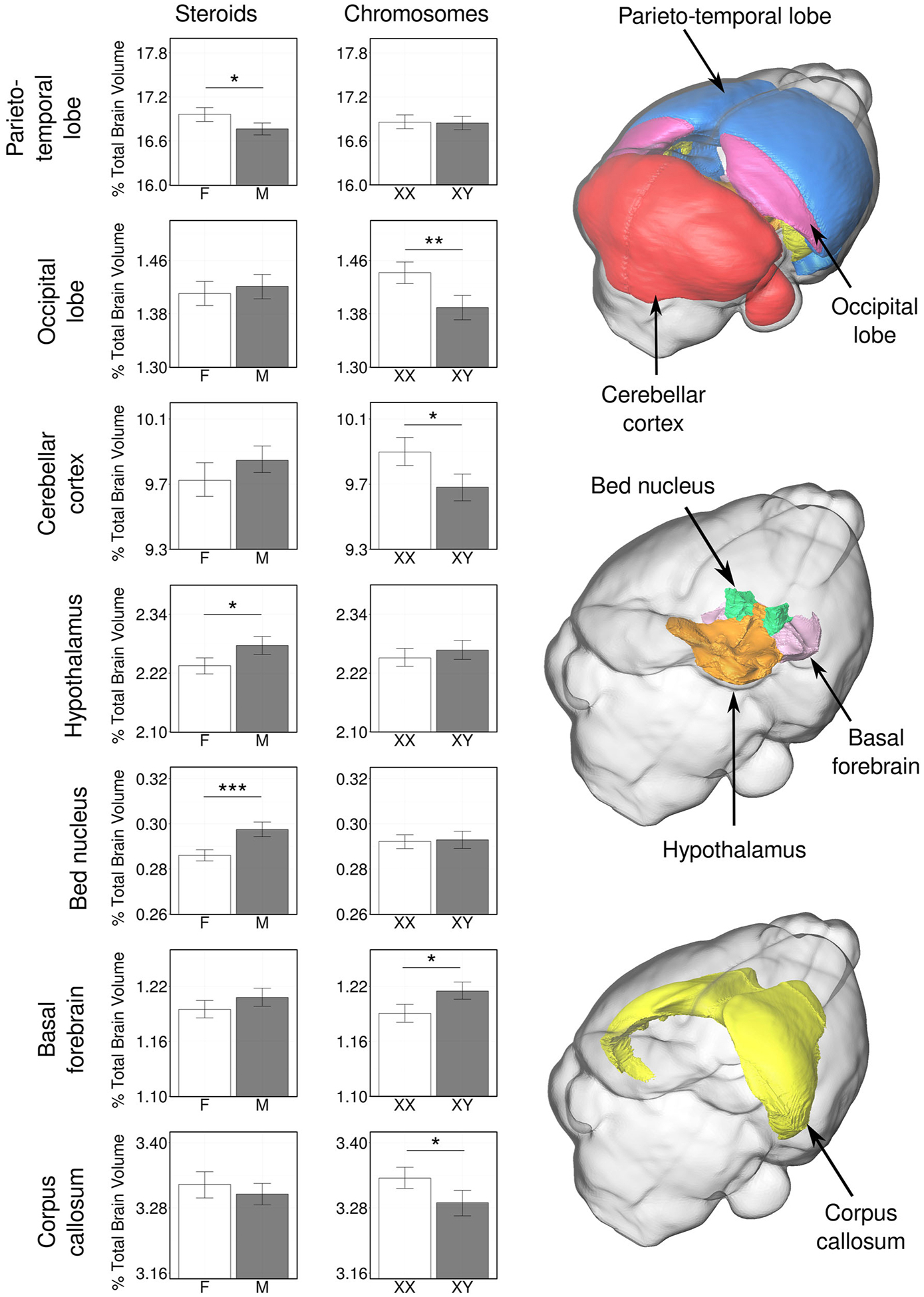
Whole-brain MRI was used to measure volumetric variation in gonad-intact FCG mice. Among seven brain regions illustrated, some showed significant effects of gonadal hormones (parieto-temporal lobe of the cerebral cortex, the hypothalamus, and the bed nucleus of the stria terminalis). Others showed effects of sex chromosomes (basal forebrain, occipital lobe of the cerebral cortex, cerebellar cortex, corpus callosum). Asterisks indicate changes were significant at false discovery rate of 5% or less. Error bars represent 95% confidence intervals. Reprinted by permission of Springer Nature from (Corre et al., 2016).
6.1.3. Sex chromosome regulation of brain sensitivity to gonadal hormones
The potential interaction of sex chromosome and sex hormone effects is also implied by the recent discovery in FCG mice that sex chromosomes influence the metabolism of gonadal hormones in the brain, and regulate the sensitivity of specific brain regions to sex steroid hormones. The amygdala has long been recognized as a site of action of gonadal steroids, to cause sex differences in function. In the anterior amygdala of E16 mouse embryos, XY mice, relative to XX, have greater expression of aromatase, the enzyme converting androgens to estrogens, irrespective of gonadal type (Cisternas et al., 2015). The same sex chromosome effect on aromatase is found in cultures of embryonic amygdala neurons (Cisternas et al., 2017; Cisternas et al., 2015). In vitro, estrogen receptor beta (Esr2) is expressed higher in XY than XX cells (Cisternas et al., 2017). Treatment of neurons with estradiol or dihydrotestosterone increases expression of aromatase and Esr2 in XX cells only, and abolishes the difference in Esr2 expression caused by sex chromosomes. Esr1 expression in vitro is also regulated by both sex chromosome complement and by gonadal hormones. This example is particularly interesting because it demonstrates a regulation by sex chromosomes both of local synthesis of estradiol, and of the sensitivity to estradiol, to regulate amygdalar phenotypes. The regulation of aromatase by sex chromosomes could shift the balance of action of estrogens and their androgenic precursors, contributing to hormonally driven sex differences in amygdala development. It will be exciting to discover the molecular pathways by which X or Y genes regulate steroid sensitivity of this and other brain regions (Cisternas et al., 2018; Cambiasso et al., 2017).
6.1.4. Cardiovascular and pulmonary disease
Women and men differ in the susceptibility and progression of numerous cardiovascular diseases, including atherosclerosis, aneurysms, ischemia/reperfusion injury, and systemic and pulmonary hypertension (Arnold et al., 2017). In animal models, the sex differences appear to be caused by a combination of sex chromosome and hormonal effects. These diseases have quite diverse molecular mechanisms, indicating that a wide range of cellular processes are influenced by sex chromosome complement. In models of abdominal aortic aneurysms, which affect men much more than women, testosterone exacerbates disease, and XY sex chromosome complement is worse than XX (Alsiraj et al., 2016). Thus, both male hormones and male sex chromosomes appear to promote disease. In a model of ischemia / reperfusion injury, estrogens are generally protective, but XX sex chromosome complement is worse that XY, indicating compensatory or counteracting effects of estrogens and sex chromosomes (Li et al., 2014). Hypertension affects men more than women. Ovarian hormones appear to be protective, but XX mice are more affected in mouse models than XY mice, again suggesting counteracting hormonal / sex chromosomal effects (Ji et al., 2010). The sex differences in humans can often be complex, depending on changes in age, hormonal status (e.g., pre- vs. postmenopausal), which might imply an age-related change in balance of hormonal and sex chromosome effects.
Pulmonary arterial hypertension affects women more than men. In a mouse model of experimental pulmonary hypertension using FCG mice, XY mice were less affected by hypoxia than XX mice. Further study of XY* mice indicated that the sex chromosome effects were caused by the presence of the Y chromosome, rather than the number of X chromosomes. Four Y genes, members X-Y gene pairs, are expressed in mouse lung, making them interesting candidates for the protective effect of the Y chromosome in this disease model (Umar et al., 2018).
6.1.5. Immune and Autoimmune Function
In Experimental Autoimmune Encephalomyelitis (EAE), a mouse model of multiple sclerosis (MS), (EAE), different sex chromosome factors interact to produce sex differences in the disease. Mice are immunized with autoantigens that trigger an immune response resembling MS. Disease progression and neuropathology is worse in XX than XY mice (Smith-Bouvier et al., 2008; Palaszynski et al., 2005). However, when bone marrow chimeric mice are produced in which the sex chromosome complement (XX vs. XY) in the immune system is independent from that of the brain, XY brains are found to respond worse in EAE than XX brains (Du et al., 2014). The different direction of the sex chromosome effects may be related to the complex sex difference in humans, in which incidence of MS is greater in women than men, but disease progression is worse in men than women. Conceivably different sex chromosome genes influence the immune system and the response of brain tissue to the immunological attack. The sex chromosome effect is found also in models of other autoimmune diseases such as lupus erythematosus (Smith-Bouvier et al., 2008; Moore et al., 2013). Sex chromosome effects also affect immunological response to some viruses (Robinson et al., 2011).
6.1.6. Interaction of sex chromosome effects and age
An experimental model of stroke involves temporary occlusion of the middle cerebral artery followed by reperfusion of the brain. Estrogens are known to protect from stroke damage in this model, causing a sex difference. In young FCG mice, no sex chromosome effect was discovered (Manwani et al., 2015). In aged mice, however, sex chromosome complement is the primary contributor to sex differences in this model. XX mice had greater lesions than XY mice, irrespective of gonadal sex, as well as greater activation of microglia and secretion of pro-inflammatory cytokines (McCullough et al., 2016). Among the aging processes that could contribute to an increase in sex chromosome effects is an age-related increase in escape from X inactivation (Disteche, 2016).
6. 2. Sex differences in the germline
Germ cells have long been known to show sex differences in behavior, including onset of meiosis during embryonic life. Although some evidence pointed to the possibility that XX and XY cells are sexually differentiated before they migrate into the undifferentiated gonadal ridge (Chuva de Sousa Lopes et al., 2008), most evidence supported the idea that germline sex differences were imposed by the testicular vs. ovarian environment in which the cells differentiate (Spiller et al., 2017). Thus, the sex differences were perceived to be downstream of the processes causing differentiation of gonadal somatic cells, induced by Sry in males vs. ovary-determining genes in females. Recently studies demonstrate, however, that the phenotypic differentiation of XX and XY germ cells depends also on sex chromosome complement. During the specification of germ cells in the epiblast and migration into the gonads, the active X chromosome of XX and XY germ cells is upregulated to be on par with autosomes. At early stages of differentiation of the gonads (E12.5 to 14.5), however, XX cells show X to autosome expression ratios above one, and XY cells have ratios below one [(Sangrithi et al., 2017); see also (Chuva de Sousa Lopes et al., 2008)]. This sex difference depends on the number of X chromosomes, not gonadal sex, and is another illustration that sex differences in development are not only dependent on gonadal type.
6. 3. Y chromosome-mediated sex differences in physiology and disease
One method for testing the involvement of the Y chromosome in traits is the use of Y chromosome consomic mice, in which Y chromosomes from different mouse strains are crossed onto the same genetic background. Phenotypic differences among strains reveal the effects of variation of Y genes. The use of Y consomic strains has been expanded impressively recently in studies of EAE in which males differing only in their Y chromosomes show quite substantially different disease progression (Case et al., 2015; Case and Teuscher, 2015; Case et al., 2013; Spach et al., 2009). Consomic Y chromosome strains also differ in their response to influenza A infection (Krementsov et al., 2017).The strain differences do not appear to be caused by different levels of testosterone. These studies allow the confident conclusion that genetic elements on the Y chromosome modulate disease, but may not necessarily imply that Y genes normally contribute to sex differences in disease mechanisms (Arnold, 2017b). On the one hand, Y genetic elements may have a “masculinizing” function in autoimmune disease in the sense that they make males different from females. But because the Y chromosome also balances the effects of the X chromosome, the variation in Y consomic strains may represent different mismatches of Y elements with X and autosomal genes with which they are artificially paired during breeding of consomic strains. The balancing role of the Y chromosome does, however, suggest a contribution of Y genes to sex differences more generally in human populations. In populations with different Y chromosome haplotypes, specific Y variants may be more or less compatible with certain X chromosomes or autosomes. In the same way that mismatches of the mitochondrial and nuclear genomes have a different effect on the two sexes, discussed above, the Y chromosome mismatches with the rest of the genome may induce male-specific variation in natural populations.
7. Comparison to birds
The study of sex differences in birds has contributed substantially to concepts of sex determination and sexual differentiation, and evolution of sex chromosomes. Males are homogametic (ZZ) and females are heterogametic (ZW). In the absence of easy transgenic methods in birds, two methods have historically been used to investigate a causal role for sex chromosomes in sex differences. One method is to transplant tissues from one sex to the other, to determine if the sexual phenotype of the donor is maintained or modified in the hormonal and tissue environment of the other sex. Using this approach, several studies found that some tissues retained their sex-specific character when transplanted to the other sex (Kozelka, 1932; Mueller, 1977). Although the suggestion was that genetic factors within the transplanted tissue might cause the sex differences, there remained the possibility that hormones might have caused permanent differences in the tissue before transplantation (Lillie, 1939). The second line of evidence came from study of lateral gynandromorphs, which are intersex birds with male plumage and a testis on one side of the body and female plumage and an ovary on the other. Early investigators realized that the presence of tissues typical of both sexes, within the same body, was unlikely to have an endocrine explanation, because hormones are expected to affect both sides (Macklin, 1923; Witschi, 1939).
Avian gynandromorphs have been analyzed since the advent of molecular tools that allow assessment of genotype and Z or W gene expression (Agate et al., 2003; Zhao et al., 2010; Morris et al., 2018). In a gynandromorphic zebra finch, brain regions controlling male song were larger on the genetically male side than on the genetically female side, implying sex chromosome effects on sexual development of the brain (Agate et al., 2003). In chickens, sexually dimorphic traits such as comb, wattles, leg spurs, and body size were male-like on the genetically male side, and more female-like on the genetically female side. Although the genotype of cells on the two sides is mixed ZZ-ZW (Zhao et al., 2010), the male side had more ZZ cells than the female side, and the female side had more ZW cells (Agate et al., 2003; Zhao et al., 2010; Morris et al., 2018). Interestingly, cells of one sex, introduced into chick embryos of the opposite sex, took up residence in the gonads and expressed markers typical of the donor sex, confirming that sex-specific development of gonadal somatic tissue is specified by sex chromosome complement (Zhao et al., 2010).
Zhao et al (2010) suggested a distinction between birds and mammals, in which sexual phenotype is specified cell-autonomously in birds, in contrast to hormonal specification in mammals. This distinction is unwarranted, because of extensive evidence that gonadal hormones cause sex differences in avian tissues (Balthazart et al., 2009; Arnold and Itoh, 2011; Witschi, 1961), and because of extensive evidence reviewed here that sex chromosomes have cell-autonomous sex-differentiating effects in mammals. However, the lack of a Z chromosome-wide mechanism of dosage compensation in birds (Itoh et al., 2007; Mank, 2009) means that most Z genes are expressed higher in every ZZ cell relative to ZW cells, making these genes available as drivers of sex differences. In mammals, the more effective X dosage compensation means that fewer X genes may be inherently sex-biased in their expression, so they may be less likely to acquire the ability to induce sex differences in cell function (Arnold et al., 2008).
8. Conclusions
It is now widely recognized that the sex chromosomes of mammals are a major source of inherent sex bias in the genome, because of the unavoidable sex difference in copy number of the two chromosomes. Nevertheless, much work is needed to identify the specific genes and mechanisms by which the two chromosomes cause sex differences in tissue function. Moreover, it will be imperative to understand how diverse forces, from different sex chromosome genes and from gonadal hormones, interact with each other to cause sex differences. Discovering these sex-biasing pathways is important for better understanding of sex differences in a wide variety of diseases.
Reference List
- Agate RJ, Grisham W, Wade J, Mann S, Wingfield J, Schanen C, Palotie A, Arnold AP (2003) Neural not gonadal origin of brain sex differences in a gynandromorphic finch. Proc Natl Acad Sci U S A 100:4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsiraj Y, Thatcher SE, Charnigo R, Kuey C, Blalock E, Daugherty A, Cassis LA (2016) Female Mice with an XY Sex Chromosome Complement Develop Severe Angiotensin II-Induced Abdominal Aortic Aneurysms. Circulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP (2011) The end of gonad-centric sex determination in mammals. Trends Genet 28:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP (2014) Conceptual frameworks and mouse models for studying sex differences in physiology and disease: Why compensation changes the game. Exp Neurol 259:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP (2017a) A general theory of sexual differentiation. J Neurosci Res 95:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP (2017b) Y chromosome’s roles in sex differences in disease. Proc Natl Acad Sci U S A 114:3787–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Cassis LA, Eghbali M, Reue K, Sandberg K (2017) Sex Hormones and Sex Chromosomes Cause Sex Differences in the Development of Cardiovascular Diseases. Arterioscler Thromb Vasc Biol 37:746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Chen X (2009) What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol 30:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Chen X, Link JC, Itoh Y, Reue K (2013) Cell-autonomous sex determination outside of the gonad. Dev Dyn 242:371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Gorski RA (1984) Gonadal steroid induction of structural sex differences in the CNS. Annu Rev Neurosci 7:413–442. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Itoh Y (2011) Factors causing sex differences in birds. Avian Biology Research 4:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Itoh Y, Melamed E (2008) A bird’s-eye view of sex chromosome dosage compensation. Annu Rev Genomics Hum Genet 9:109–127. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Reue K, Eghbali M, Vilain E, Chen X, Ghahramani N, Itoh Y, Li J, Link JC, Ngun T, Williams-Burris SM (2016) The importance of having two X chromosomes. Philos Trans R Soc Lond B Biol Sci 371:20150113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D (2013) Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balthazart J, Arnxold AP, Adkins-Regan E (2009) Sexual differentiation of brain and behavior in birds In: Hormones, Brain, and Behavior (Pfaff DW, Arnold AP, Etgen AM, Fahrbach SE, Rubin RT, eds), pp 1745–1788. San Diego: Elsevier. [Google Scholar]
- Bellott DW, et al. (2014) Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508:494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramble MS, Roach L, Lipson A, Vashist N, Eskin A, Ngun T, Gosschalk JE, Klein S, Barseghyan H, Arboleda VA, Vilain E (2016) Sex-Specific Effects of Testosterone on the Sexually Dimorphic Transcriptome and Epigenome of Embryonic Neural Stem/Progenitor Cells. Sci Rep 6:36916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breedlove SM, Cooke BM, Jordan CL (1999) The orthodox view of brain sexual differentiation. Brain Behav Evol 54:8–14. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Arnold AP (2016) A primer on the use of mouse models for identifying direct sex chromosome effects that cause sex differences in non-gonadal tissues. Biol Sex Differ 7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS, Mitchell MJ (2007) The role of mouse Y chromosome genes in spermatogenesis In: Y Chromosome and Male Germ Cell Biology (Lau YFC, Chan WY, eds), pp 27–45. Hackensack NJ: World Scientific Publishers. [Google Scholar]
- Burgoyne PS, Thornhill AR, Boudrean SK, Darling SM, Bishop CE, Evans EP (1995) The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos Trans R Soc Lond B Biol Sci 350:253–260. [DOI] [PubMed] [Google Scholar]
- Cambiasso MJ, Cisternas CD, Ruiz-Palmero I, Scerbo MJ, Arevalo MA, Azcoitia I, Garcia-Segura LM (2017) Interaction of sex chromosome complement, gonadal hormones and neuronal steroid synthesis on the sexual differentiation of mammalian neurons. J Neurogenet 31:300–306. [DOI] [PubMed] [Google Scholar]
- Camus MF, Clancy DJ, Dowling DK (2012) Mitochondria, maternal inheritance, and male aging. Curr Biol 22:1717–1721. [DOI] [PubMed] [Google Scholar]
- Carrel L, Cottle AA, Goglin KC, Willard HF (1999) A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci U S A 96:14440–14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LK, Teuscher C (2015) Y genetic variation and phenotypic diversity in health and disease. Biol Sex Differ 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LK, Wall EH, Dragon JA, Saligrama N, Krementsov DN, Moussawi M, Zachary JF, Huber SA, Blankenhorn EP, Teuscher C (2013) The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Res 23:1474–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LK, Wall EH, Osmanski EE, Dragon JA, Saligrama N, Zachary JF, Lemos B, Blankenhorn EP, Teuscher C (2015) Copy number variation in Y chromosome multicopy genes is linked to a paternal parent-of-origin effect on CNS autoimmune disease in female offspring. Genome Biol 16:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B (1996) The evolution of chromosomal sex determination and dosage compensation. Curr Biol 6:149–162. [DOI] [PubMed] [Google Scholar]
- Chen X, McClusky R, Chen J, Beaven SW, Tontonoz P, Arnold AP, Reue K (2012) The number of X chromosomes causes sex differences in adiposity in mice. PLoS Genet 8:e1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, McClusky R, Itoh Y, Reue K, Arnold AP (2013) X and Y chromosome complement influence adiposity and metabolism in mice. Endocrinol 154:1092–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes SM, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A (2008) X chromosome activity in mouse XX primordial germ cells. PLoS Genet 4:e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisternas CD, Cabrera Zapata LE, Arevalo MA, Garcia-Segura LM, Cambiasso MJ (2017) Regulation of aromatase expression in the anterior amygdala of the developing mouse brain depends on ERbeta and sex chromosome complement. Sci Rep 7:5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisternas CD, Garcia-Segura LM, Cambiasso MJ (2018) Hormonal and genetic factors interact to control aromatase expression in the developing brain. J Neuroendocrinol 30. [DOI] [PubMed] [Google Scholar]
- Cisternas CD, Tome K, Caeiro XE, Dadam FM, Garcia-Segura LM, Cambiasso MJ (2015) Sex chromosome complement determines sex differences in aromatase expression and regulation in the stria terminalis and anterior amygdala of the developing mouse brain. Mol Cell Endocrinol 414:99–110. [DOI] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS (2012) A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet 8:e1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, Burgoyne PS (2009) The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol 7:e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre C, Friedel M, Vousden DA, Metcalf A, Spring S, Qiu LR, Lerch JP, Palmert MR (2016) Separate effects of sex hormones and sex chromosomes on brain structure and function revealed by high-resolution magnetic resonance imaging and spatial navigation assessment of the Four Core Genotype mouse model. Brain Struct Funct 221:997–1016. [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grutzner F, Kaessmann H (2014) Origins and functional evolution of Y chromosomes across mammals. Nature 508:488–493. [DOI] [PubMed] [Google Scholar]
- Cox KH, Bonthuis PJ, Rissman EF (2014) Mouse model systems to study sex chromosome genes and behavior: Relevance to humans. Front Neuroendocrinol 35:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries GJ, Rissman EF, Simerly RB, Yang LY, Scordalakes EM, Auger CJ, Swain A, Lovell-Badge R, Burgoyne PS, Arnold AP (2002) A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci 22:9005–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Nguyen DK, Disteche CM (2014) X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet 15:367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewing P, Shi T, Horvath S, Vilain E (2003) Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Res Mol Brain Res 118:82–90. [DOI] [PubMed] [Google Scholar]
- Disteche CM (2016) Dosage compensation of the sex chromosomes and autosomes. Semin Cell Dev Biol 56:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn JD, Schwarz QP, Gray S, Laskowski A, Kiriazis H, Ming Z, Harvey RP, Du XJ, Thorburn DR, Cox TC (2008) Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev Cell 15:521–533. [DOI] [PubMed] [Google Scholar]
- Du S, Itoh N, Askarinam S, Hill H, Arnold AP, Voskuhl RR (2014) XY sex chromosome complement, compared with XX, in the CNS confers greater neurodegeneration during experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 111:2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger NG (2009) Control of cell number in the sexually dimorphic brain and spinal cord. J Neuroendocrinol 21:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA (2012) Evolution: mitochondrial burden on male health. Curr Biol 22:R797–R799. [DOI] [PubMed] [Google Scholar]
- Graves JA (2016) Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet 17:33–46. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Page DC (2015) The Biology and Evolution of Mammalian Y Chromosomes. Annu Rev Genet 49:507–527. [DOI] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH, Dowling DK (2011) Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science 332:845–848. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP. (2007) Dosage compensation is less effective in birds than in mammals. J BIol 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Zheng W, Wu X, Liu J, Ecelbarger CM, Watkins R, Arnold AP, Sandberg K (2010) Sex chromosome effects unmasked in angiotensin II-induced hypertension. Hypertension 55:1275–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozelka AW (1932) Integumental grafting as a means of analyzing the factors determining the secondary sexual characteristics of leghorn fowl. J Exp Zool 61:431–495. [Google Scholar]
- Krementsov DN, Case LK, Dienz O, Raza A, Fang Q, Ather JL, Poynter ME, Boyson JE, Bunn JY, Teuscher C (2017) Genetic variation in chromosome Y regulates susceptibility to influenza A inflection. PNAS in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC (1999) Four evolutionary strata on the human X chromosome. Science 286:964–967. [DOI] [PubMed] [Google Scholar]
- Li J, Chen X, McClusky R, Ruiz-Sundstrom M, Itoh Y, Umar S, Arnold AP, Eghbali M (2014) The number of X chromosomes influences protection from cardiac ischaemia/reperfusion injury in mice: one X is better than two. Cardiovasc Res 102:375–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie FR (1939) General biological introduction In: Sex and Internal Secretions (Allen E, Danforth CH, Doisy EA, eds), pp 3–14. Baltimore: Williams and Wilkins Co. [Google Scholar]
- Link JC, Chen X, Prien C, Borja MS, Hammerson B, Oda MN, Arnold AP, Reue K (2015) Increased High-Density Lipoprotein Cholesterol Levels in Mice With XX Versus XY Sex Chromosomes. Arterioscler Thromb Vasc Biol 35:1778–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe R, Gemma C, Rakyan VK, Holland ML (2015) Sexually dimorphic gene expression emerges with embryonic genome activation and is dynamic throughout development. BMC Genomics 16:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklin MT (1923) A description of material from a gynandromorph fowl. J Exp Zool 38:355–375. [Google Scholar]
- Mank JE (2009) The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet 25:226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manwani B, Bentivegna K, Benashski SE, Venna VR, Xu Y, Arnold AP, McCullough LD (2015) Sex differences in ischemic stroke sensitivity are influenced by gonadal hormones, not by sex chromosome complement. J Cereb Blood Flow Metab 35:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough LD, Mirza MA, Xu Y, Bentivegna K, Steffens EB, Ritzel R, Liu F (2016) Stroke sensitivity in the aged: sex chromosome complement vs. gonadal hormones. Aging (Albany NY ) 8:1432–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midgeon BR (2007) Females are Mosaics. Oxford: Oxford University Press. [Google Scholar]
- Miura K, Tomita A, Kanai Y (2018) Sex determination and differentiation in mammals In: Reproductive and developmental strategies: The continuity of life (Kobayashi K, Kitano T, Iwao Y, Kondo M, eds), pp 407–432. Springer; Japan. [Google Scholar]
- Moore S, Patel R, Hannsun G, Yang J, Tiwari-Woodruff SK (2013) Sex chromosome complement influences functional callosal myelination. Neurosci 245:166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KR, Hirst CE, Major AT, Ezaz T, Ford M, Bibby S, Doran TJ, Smith CA (2018) Gonadal and endocrine analysis of a gynandromorphic chicken. Endocrinol in press. [DOI] [PubMed] [Google Scholar]
- Mueller N (1977) Control of sex difference in the plumage of the house sparrow, Passer domesticus. J Exp Zool 202:45–48. [Google Scholar]
- Naqvi S, Bellott DW, Lin KS, Page DC (2018) Conserved microRNA targeting reveals preexisting gene dosage sensitivities that shaped amniote sex chromosome evolution. Genome Res 28:474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, Parker KL, Vassalli JD (2005) Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol 287:361–377. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM (2006) Dosage compensation of the active X chromosome in mammals. Nat Genet 38:47–53. [DOI] [PubMed] [Google Scholar]
- Nugent BM, O’Donnell CM, Epperson CN, Bale TL (2018) Placental H3K27me3 establishes female resilience to prenatal insults. Nat Commun 9:2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaszynski KM, Smith DL, Kamrava S, Burgoyne PS, Arnold AP, Voskuhl RR (2005) A Yin-Yang effect between sex chromosome complement and sex hormones on the immune response. Endocrinol 146:3280–3285. [DOI] [PubMed] [Google Scholar]
- Renfree MB, Short RV (1988) Sex determination in marsupials: evidence for a marsupial-eutherian dichotomy. Philosophical Transactions of the Royal Society of London B:Biological Sciences 322:41–53. [DOI] [PubMed] [Google Scholar]
- Rice WR (1992) Sexually antagonistic genes - experimental evidence. Science 256:1436–1439. [DOI] [PubMed] [Google Scholar]
- Rice WR (1994) Degeneration of a nonrecombining chromosome. Science 263:230–232. [DOI] [PubMed] [Google Scholar]
- Robinson DP, Huber SA, Moussawi M, Roberts B, Teuscher C, Watkins R, Arnold AP, Klein SL (2011) Sex chromosome complement contributes to sex differences in Coxsackievirus B3 but not Influenza A virus pathogenesis. Biol Sex Differ 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Royo H, Mahadevaiah SK, Ojarikre O, Bhaw L, Sesay A, Peters AH, Stadler M, Turner JM (2017) Non-Canonical and Sexually Dimorphic X Dosage Compensation States in the Mouse and Human Germline. Dev Cell 40:289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Sengoku T, Yokoyama S, Magnuson T (2012) UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 8:e1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkaitis K, Lemos B (2014) Sex-biased chromatin and regulatory cross-talk between sex chromosomes, autosomes, and mitochondria. Biol Sex Differ 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Bouvier DL, Divekar AA, Sasidhar M, Du S, Tiwari-Woodruff SK, King JK, Arnold AP, Singh RR, Voskuhl RR (2008) A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med 205:1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh YQ, et al. (2014) Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159:800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spach KM, Blake M, Bunn JY, McElvany B, Noubade R, Blankenhorn EP, Teuscher C (2009) Cutting edge: the Y chromosome controls the age-dependent experimental allergic encephalomyelitis sexual dimorphism in SJL/J mice. J Immunol 182:1789–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller C, Koopman P, Bowles J (2017) Sex Determination in the Mammalian Germline. Annu Rev Genet 51:265–285. [DOI] [PubMed] [Google Scholar]
- Sutton E, et al. (2011) Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Invest 121:328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A, Lappalainen T, Regev A, Ardlie KG, Hacohen N, MacArthur DG (2017) Landscape of X chromosome inactivation across human tissues. Nature 550:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar S, Cunningham CM, Itoh Y, Moazeni S, Vaillancourt M, Sarji S, Centala A, Arnold AP, Eghbali M (2018) The Y Chromosome Plays a Protective Role in Experimental Hypoxic Pulmonary Hypertension. Am J Respir Crit Care Med 197:952–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender EJ, Lahn BT (2004) How mammalian sex chromosomes acquired their peculiar gene content. BioEssays 26:159–169. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Veyrunes F, Bottani S, Birchler JA (2015) X chromosome inactivation and active X upregulation in therian mammals: facts, questions, and hypotheses. J Mol Cell Biol 7:2–11. [DOI] [PubMed] [Google Scholar]
- Vousden DA, Corre C, Spring S, Qiu LR, Metcalf A, Cox E, Lerch JP, Palmert MR (2018) Impact of X/Y genes and sex hormones on mouse neuroanatomy. Neuroimage 173:551–563. [DOI] [PubMed] [Google Scholar]
- Werner RJ, Schultz BM, Huhn JM, Jelinek J, Madzo J, Engel N (2017) Sex chromosomes drive gene expression and regulatory dimorphisms in mouse embryonic stem cells. Biol Sex Differ 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers PJ, Festenstein RJ (2011) Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet 27:132–140. [DOI] [PubMed] [Google Scholar]
- Witschi E (1939) Modification of the development of sex in lower vertebrates and in mammals In: Sex and Internal Secretions (Allen E, Danforth CH, Doisy EA, eds), pp 145–226. Baltimore: Williams & Wilkins. [Google Scholar]
- Witschi E (1961) Sex and secondary characteristics In: Biology and Comparative Physiology of Birds (Marshall AJ, ed), pp 115–168. New York: Academic Press. [Google Scholar]
- Xu J, Deng X, Watkins R, Disteche CM (2008) Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci 28:4521–4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT (2013) Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152:727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, McBride D, Nandi S, McQueen HA, McGrew MJ, Hocking PM, Lewis PD, Sang HM, Clinton M (2010) Somatic sex identity is cell autonomous in the chicken. Nature 464:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]


