Abstract
Blocking interactions between PD-1 and PD-L1 opens a new era of cancer treatment involving immunity modulation. Although most immunotherapies use monoclonal antibodies, small-molecule inhibitors offer advantages. To facilitate development of small-molecule therapeutics, we implemented a rapid approach to characterize the binding interfaces of small molecule inhibitors with PD-L1. We determined its interaction with a synthetic macrocyclic peptide by using two mass-spectrometry-based approaches, hydrogen-deuterium exchange and fast photochemical oxidation of proteins or FPOP and corroborated the findings with our X-ray structure of the PD-L1-macrocycle complex. Although all three approaches show that macrocycle binds directly to PD-L1 over regions 46-87 and 114-125, the two protein footprinting approaches show additional binding at the N-terminus of PD-L1, and FPOP reveals some critical binding residues. The outcomes not only show the binding regions but also demonstrate the utility of MS-based footprinting to probe of protein-ligand inhibitory interactions in cancer immunotherapy.
Keywords: Cancer immunotherapy, PD-L1 inhibitor, macrocyclic peptide, X-ray crystallography, protein footprinting, fast photochemical oxidation of proteins (FPOP), hydrogen deuterium exchange, mass spectrometry
INTRODUCTION
Cancer immunotherapy, which harnesses the immune system to attack tumors, has seen major breakthroughs in the treatment of cancers resistant to conventional radiation and chemotherapy1-6. Combinations of immunotherapy with established genotoxic, targeted, or anti-angiogenic therapies have further enhanced responses in melanoma, non-small-cell lung carcinoma, and renal- cell carcinoma 7-9. An area of recent promise involves blocking immune checkpoint proteins that either up- or down-regulate immune cell molecular signaling 10 to promote antitumor immunity11-12.
PD-1 protein (Programmed Death-1, or CD279), which is a member of CD28 family, represents one of the inhibitory receptors that function as an immune checkpoint13-14. Following immune activation, PD-1 expression is increased on the surfaces of T lymphocytes, B lymphocytes, dendritic cells, and natural killer cells15. One of the ligands of PD-1 is PD-L1, a type I, transmembrane protein16 responsible for co-inhibitory signals in activated T-cells. PD-L1 promotes the inactivity, anergy, and even apoptosis of T-cells by binding to the immune cell surface17. Many cancer cells over-express PD-L1, preventing activated, tumor-infiltrating T-cells from mounting a sustained response against the tumor. Therefore, blocking the PD-1/PD-L1 immune checkpoint is an appealing approach to antitumor immunity that has achieved unprecedented results in cancer treatment in recent years (Figure 1A)18-19. For example, encouraging results were obtained recently for therapeutic monoclonal antibodies (mAbs) including pembrolizumab, nivolumab, and durvalumab that target either PD-1 or PD-L1 and restore T-cell activity, proliferation, and cytokine production 5, 18, 20-21.
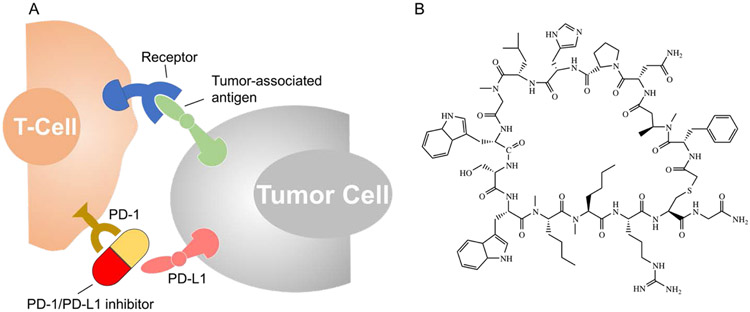
Figure 1.
(A) Cartoon showing inhibitors of programmed death-ligand 1 (PD-L1) or programmed cell death protein 1 (PD-1) can block the interaction between the two proteins, reactivating the immune response; (B) Structure of macrocyclic peptide (macrocycle), an inhibitor of PD-1/PD-L1 protein-protein interaction.
Small-molecule PD-1/PD-L1 inhibitors, which are mainly based on macrocyclic peptides22, peptidomimetics23, and nonpeptidic molecules24, are emerging and may have advantages over mAbs because they diffuse more easily and are taken up more readily in tumor tissue25. Their development has been impeded by the flat, non-contiguous PPI interface that challenges drug development and by the limited number of methods for determining protein-drug complex structures that can guide rationale design. The subject was recently reviewed26.
The usual approach to structure is X-ray crystallography, but thus far, only a few cocrystal structures of small molecules targeting PD-1 and PD-L1 were reported. Holak and co-workers20 found that some macrocyclic peptides can block the PD-1/PD-L1 interaction via direct binding to PD-L1 through the PD-1 interacting surface. Other nonpeptidic molecules, however, can achieve inhibitory interactions by inducing a dimeric interaction of PD-L1, which occludes the PD-1 interaction surface of PD-L121.
Another approach takes advantage of recent advances in mass spectrometry (MS)-based protein footprinting, which enables elucidation of protein conformations and interactions with high sensitivity, fast turnaround, and low sample consumption27-29. The large number of possible inhibitors to be explored demands structural tools that have fast turnaround26. Approaches include hydrogen-deuterium exchange (HDX)30-35, and hydroxyl radical labeling36-39, the latter further elaborated as fast photochemical oxidation of proteins (FPOP) by Hambly and Gross20, 40-44; for reviews, see45-46. Herein, we describe the independent use of HDX and FPOP for characterization of the interaction of PD-L1 and a macrocyclic peptidic inhibitor (see structure in Figure 1B, referred to as the “macrocycle” in this paper)22. We employed time-dependent measurements for both footprinting methods and demonstrated that PD-L1 interacts with the macrocycle on the N-lobe at three discontinuous regions.
In parallel, we obtained the X-ray crystal structure of the PD-L1-macrocycle complex to show that the contact sites mapped by footprinting agree well with those observed by in the solid-state structure. The contact sites approximately include those interfaces of PD-L1 that involve PD-1 binding to PD-L147. We further show, consistent with results from both footprinting approaches, that the binding induces protection on the N-terminal region of PD-L1, an interaction that was not observed in the crystal structure or reported in a similar crystal structure of another macrocyclic inhibitor and PD-L1 complex 20. Importantly, our results not only demonstrate that small macrocycles can inhibit PD-1/PD-L1 interactions but also help establish MS-based footprinting as an effective probe of protein-ligand inhibitory interactions and as a map of small-molecule drug interactions.
MATERIALS AND METHODS
PD-L1 and Macrocycle Preparation.
The PD-L1 ectodomain protein (see Supporting Information for materials and PD-L1 protein cloning, expression, purification, and in vitro characterization assays) was dissolved in formulation buffer at 9.6 mg/mL and stored in a −80 °C freezer until the time of footprinting. The macrocyclic peptide was dissolved in dry DMSO at 10 mM prior to storage in −20 °C freezer. At the time of footprinting, an aliquot of the PD-L1 stock solution (9.6 mg/mL) was diluted with 10 mM PBS buffer to 25 μM, to form the macrocycle-unbound samples. To prepare macrocycle-bound samples, an aliquot of the PD-L1 stock solution was diluted to 50 μM with 10 mM PBS, and mixed at 1:1 molar ratio with the macrocycle at gentle vortex for 1 h at room temperature. The final concentration of macrocycle-bound PD-L1 samples was 25 μM, with approximately 35 mM DMSO.
Continuous Hydrogen-Deuterium Exchange (HDX).
The design of the HDX was based on tight binding of the macrocycle PD-L1 (low nM Kd)22. The continuous labeling with deuterium was conducted as described previously20, 31, 41, 48, and detailed in Supporting Information. In brief, PD-L1 samples (bound or unbound) of 3 μL were mixed with 17 μL D2O buffer to initiate HDX labeling. Samples were then incubated on ice for different labeling times ranging from 30 sec to 4 h, followed by chemical quenching with 30 μL 3 M urea and 250 mM TCEP at pH 2.5. Pepsin digestion of PD-L1, chromatographic separation, MS detection, and subsequent data processing were conducted as described in Supporting Information. All analyses were done in triplicate. To minimize back exchange, all accessories including columns, injector, solvent lines were immersed in ice-water slush.
FPOP Footprinting.
FPOP, except as amended by adding a reporter peptide49, was performed as previously described and detailed in Supporting Information19. An estimate of the laser fluence was made as described in SI. The pulsed oxidative labeling experiments were initiated in a flow cell (at 25 μL/min) by submitting 50 μL of mixed solution containing protein (PD-L1 bound or unbound), LeuEnk (reporter peptide), H2O2, and histidine to approximately 1000 laser shots (operated at 7.4 Hz). The lifetime of hydroxyl radicals, which determines the extent of oxidative modifications given the same protein conformation, was adjusted by using different histidine concentrations ranging from 0.5 to 30 mM as described in Supporting Information. Each sample condition was prepared and studied in triplicate. Directly following FPOP, samples were submitted to trypsin digestion, LC-MS/MS separation and detection, data analysis and processing as detailed in SI.
X-ray Co-crystallization and Structure Determination.
Purified human PD-L1 ectodomain (7.5 mg/mL in 25 mM Tris, pH 7.5, 150 mM NaCl) was mixed at 1:1 molar ratio with the macrocycle and allowed to incubate on ice for at least 1 h prior to crystallization. Diffraction-quality crystals were obtained at room temperature by using the sitting-drop vapor diffusion technique. Drops (2 μL) containing 1 μL of the PD-L1/macrocycle solution and 1 μL of reservoir solution (20% peg 3000, 0.1 M sodium citrate, pH 5.5) were equilibrated against 1 mL of 100% reservoir solution. Prior to data collection, crystals were soaked briefly in a solution containing 20% peg 3000, 0.1 M sodium citrate at pH 5.5, and 15% glycerol and then flash frozen in liquid nitrogen. Data were collected on beamline 5.0.2 at the Advanced Light Source (Berkeley, CA) at cryogenic temperatures with a Quantum-315 CCD detector using 0.5° oscillations until greater than 90% overall completeness was observed. Data were processed with the programs DENZO and SCALEPACK 50.
The structure of the human PD-L1 ectodomain bound to macrocycle was determined by molecular replacement by using data collected to 2.0 Å resolution, a previously determined apo structure of human PD-L1 (PDB ID: 4Z1851) as the search model, and the PHENIX software suite52. The molecular replacement solution was subjected to several iterative cycles of torsion angle annealing in PHENIX and manual refitting in COOT53. Atoms for the macrocycle and water molecules were added to the model during the later rounds of refinement. The PD-L1-macrocycle structure PDB ID is 6PV9. DEPOSITION ID: D_1000243159. The data collection and refinement statistics for the PD-L1/macrocycle complex are detailed in SI Table S1.
RESULTS AND DISCUSSION
Macrocycle Binds Specifically to PD-L1 and Blocks the PD-1/PD-L1 Interaction in vivo and in vitro.
To characterize the PD-L1 macrocycle in biochemical binding, blocking and cellular assays, we used a PD-L1 specific antibody, Atezolizumab, as a positive control in all assays. For the biochemical binding and blocking assays, we used mammalian recombinant ectodomains for the proteins. Direct binding studies using the label-free Biacore assay revealed the PD-L1 macrocycle binds specifically to PD-L1 coated surface (Kd = 2.1 nM), and no binding was detected at concentrations up to 10 μM on surfaces coated with PD-1 (Supporting Information Table S2). For the biochemical PD-1/PD-L1 and CTA4-CD80 protein-protein interaction assays, the PD-L1 macrocycle specifically only inhibited the PD-1/PD-L1 interaction (IC50 = 1.6 nM; SI Table S2).
More importantly, the binding and blocking activity observed in the biochemical assays translates to functional cellular activity in a reporter assay that indirectly measures T-cell activation using a NFAT-luciferase reporter. This assay uses two cells lines: a CHO cell line that stably expresses the native (full-length) form of PD-L1 and a Jurkat cell line that stably expresses native (full-length) PD-1 and the NFAT-luciferase reporter. Co-cultivation of the two cell-lines results in activation of the T-cell receptor leading to NFAT-promoter-driven luciferase activity, which is inhibited by the interaction between PD-1 and PD-L1 on the cell surface. Blocking the interaction between PD-1 and PD-L1 would promote T-cell activation and re-activate the NFAT-promoter driven luciferase activity. In this assay the PD-L1 macrocycle inhibits the native PD-1/PD-L1 interaction resulting in re-activation NFAT-luciferase reporter (EC50 = 476 nM; SI Table S2). In summary, the PD-L1 macrocycle binds specifically to PD-L1 and blocks the PD-1/PD-L1 interaction both biochemically and in cells with a profile that is similar, although less potent, to the PD-L1 antibody.
HDX Kinetics Locates Discontinuous Binding Interfaces.
To determine the binding interfaces between PD-L1 and the macrocycle (structure is shown in Figure 1B), we compared comprehensive differential HDX analysis of the macrocycle-bound and unbound PD-L1. We identified 96 peptic peptides that are in common in the macrocycle-bound and unbound PD-L1 (the centroid of the isotopic profile of each peptide, as monitored by MS, was taken to determine the extent of HDX).
We were able to cover more than 95% of the PD-L1 sequence, with some regions covered by multiple overlapping peptides that arose by cleavage at multiple pepsin sites and appeared in the mass spectrum with various charge states. Although the maximal deuterium uptake level should be 85%, which is the %D2O in the buffer, we observed that the highest deuterium uptake for some peptides was approximately 80%, suggested there is a small extent (5%) of back exchange.
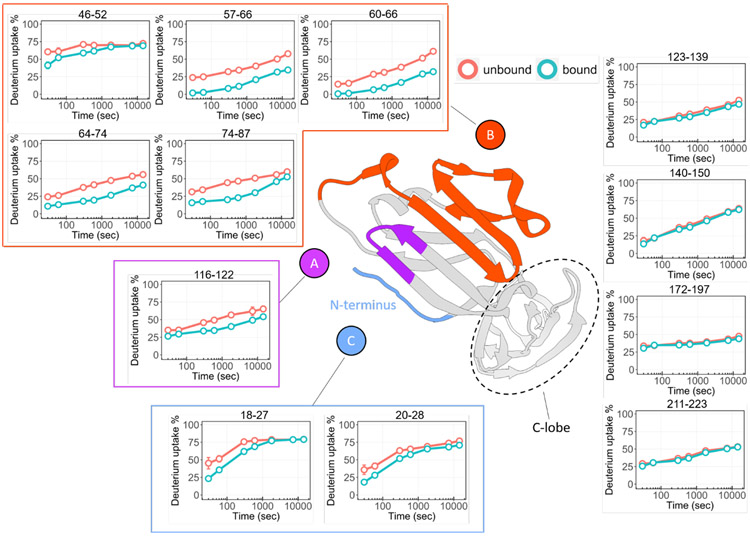
Because the HDX rates of protein backbone amides are highly dependent on the local hydrogen-bonding environment and solvent accessibility 32, we expected regions of PD-L1 associated with macrocycle binding to exchange more slowly and consequently show a larger difference in deuterium uptake compared to the unbound. For convenience of comparing the bound-versus-unbound states, we computed the average differential deuterium uptake for the triplicate analyses across the seven labeling times for each peptide (SI,Table S3). By requiring a threshold of 5% to assign with confidence significant differences that report on binding, we identified three discontinuous regions of PD-L1 that are involved in binding (represented by peptides N-terminal to 28, 46-87, and 116-122). We selected 12 peptides (from SI, Table S3) to represent the full PD-L1 protein and measured the time-dependent HDX of the bound and unbound states (Figure 2). The entire region, starting from residue 123 to the C-terminus showed consistently low differential deuterium uptake (i.e., below 4%), indicating that the C-lobe region of PD-L1 is not the macrocycle binding interface.
Figure 2. Peptide-level HDX kinetics analysis of PD-L1.
The comparison between macrocycle-bound (teal) and unbound (orange) states shows significant changes of HDX for mainly three regions, region A is represented by peptide 116-122 (denoted in purple), region B includes peptides 46-52, 57-66, 60-66, 64-74, 74-87 (denoted in orange), and region C that contains N-terminal peptides 18-27, 20-28 (denoted in light blue). The HDX results mapped onto the crystal structure of PD-L1 (PDB 4Z18) show all three regions are discontinuously located on the N-lobe. No deuterium uptake differences observed for peptides on C-lobe. Error bars correspond to ± SD from triplicate measurements and are nearly always smaller than the diameters of the open circles representing data points. For binding regions, the differences between the curves representing bound and unbound are at least 5 times the SD.
The kinetic curves of the first identified binding region, as represented by two peptides 18-27 and 20-28, featured an obvious deuterium uptake difference in the beginning (first 10 min), revealing protection afforded by binding of the macrocycle to PD-L1; however, the differences became less at longer labeling times—the two curves converge at a plateau of ~80% exchange (Figure 2). The high HDX extent is consistent with an expected highly dynamic and flexible N terminus (and C terminus)54, and the convergence suggests a significant off rate of the macrocycle that allows the HDX extent of the bound state to become ultimately equal to that of the unbound55-56.
The second putative macrocycle binding interface pertains to amino acids 46 to 87, exemplified by peptides 46-52, 57-66, 60-66, 64-74, and 74-87 (Figure 2), which all showed pronounced difference in HDX between the bound and unbound states (>17%, Supporting Information Table S3). The bound PD-L1 exhibited significantly less HDX than did the unbound (Figure 2). The results suggest that the region of the protein represented by peptides 57-66 and 60-66 is more strongly involved in binding, whereas regions represented by peptides 64-74 and 74-87 show smaller and converging changes, indicating some reduced flexibility induced by a nearby binding site. Furthermore, for regions 57-66 and 60-66, the HDX of the bound state is low (<2%) at the start of the exchange (Figure 2), indicating that this region becomes more rigid upon macrocycle binding.
Although regions 57-66, 60-66, 64-74, and 74-87 showed increases in differential HDX for the first 5 min of exchange (SI, Figure S1), the differences then decreased at longer times for regions 64-87, corroborating that region 57-66 is most directly involved in binding32, 57-58. Region 116-122 shows statistically significant differences at multiple HDX time points, affirming interactions of PD-L1 with the macrocycle (Figure 2). In summary, discontinuous regions 18-28, 57-87, and 116-122 are all involved in the interactions of the protein with the macrocycle.
Binding Interfaces Determined by Fast Photochemical Oxidation of Proteins (FPOP).
To confirm the PD-L1/macrocycle binding interfaces, we employed a second approach of targeting the protein backbones by footprinting with OH radicals, which impart fast and irreversible mapping of the protein’s surface solvent accessibility, as described previously 20, 42, 59-60. Hydroxyl radicals are remarkably reactive and short-lived 61, and the approach can identify some key binding residues. To enable the acquisition of “FPOP hydroxyl radical dose-response curves”, we incorporated a reporter peptide (e.g., Leu-enkephalin) in the FPOP protocol, the FPOP yields of which serves as good approximate of reaction time. These dose-response curves resemble those of HDX, and although the time axis is on the microsecond frame, it is not converted to actual time but reported as %modification of the reporter (see below) 49.
To vary the time of footprinting, we adjusted the concentrations of the scavenger histidine (lower histidine means longer radical lifetime). To normalize the differential scavenging and compare the FPOP outcomes between macrocycle-bound and unbound PD-L1 at the peptide level, we plotted the modification extents of Leu-enkephalin at each histidine concentration versus the corresponding modification extents of each tryptic peptide of the protein (y-axis). The modification extents of reporter peptide can be considered as a “ruler” of •OH lifetime (i.e., longer times yield more modifications)62. For each peptide, the curves for bound and unbound should overlap if their solvent-accessibilities are the same (Figure 3), whereas for bound regions, the more protected state should show lower oxidative modification49.
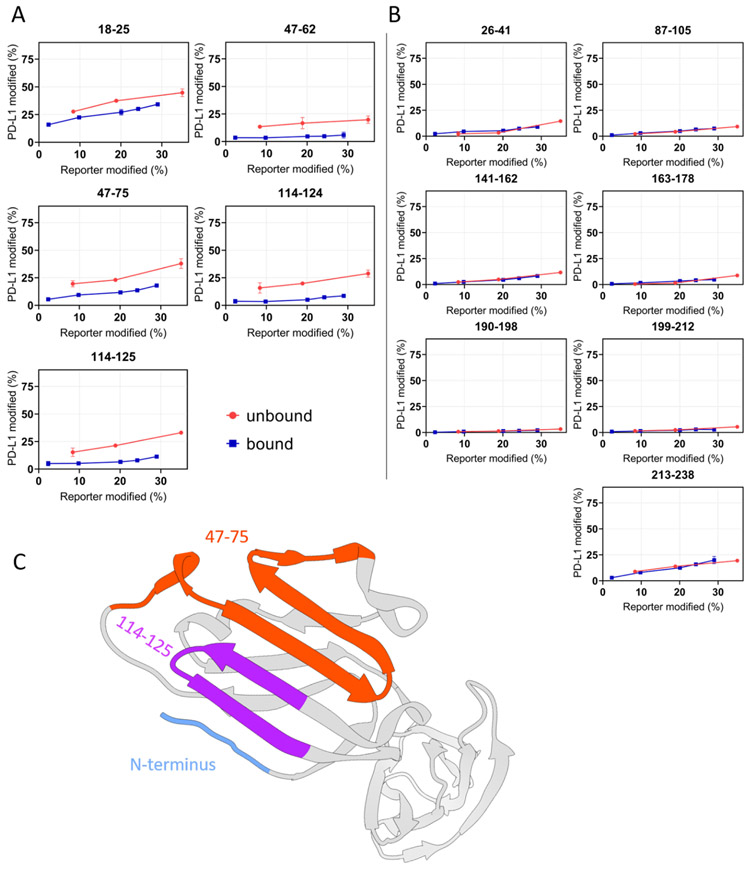
Figure 3. “Time-dependent” FPOP response curves of each peptide of PD-L1.
(A) Differential yields of FPOP observed between macrocycle-bound and unbound PD-L1 (represented by 5 tryptic peptides) suggest three discontinuous binding interfaces on PD-L1, including N-terminus, 47-75, and 114-125. (B) The overlapping of FPOP response curves indicate comparable surface solvent accessibility of these regions between bound and unbound states. These regions are not involved in the macrocycle binding. (C) FPOP results mapped onto the PD-L1 structure (PDB 4Z18) shows comparable outcome as that of HDX. The error bars correspond to ± SD from triplicate measurements and show that for binding sites, the differences between the accumulated differences for three points representing bound and unbound are at least 3 times the propagated errors (sq root of the sum of the squares of the SDs). This corresponds to 99.7% confidence in the assignment.
Decreases in time-dependent FPOP yields (attributed to decreases in solvent accessibility) reveal that three regions on the N-lobe of PD-L1 form the macrocycle binding interfaces, as represented by five peptides: 18-25 (N-terminus), 47-62, 47-75, 114-124, and 114-125 (Figure 3). These assignments agree well with our HDX results that also identify three discontinuous regions as the critical binding interfaces between the protein and the macrocycle. The FPOP kinetics also reveal that many regions do not change solvent-accessibility in going from unbound to macrocycle-bound state (e.g., regions represented by tryptic peptides 87-105, 141-162, 190-198, etc.). These overlapping curves serve as negative controls, and convincingly indicate that these regions are not involved in association with macrocycle whereas those that do are associated with binding.
Residue-Specific Binding Sites of PD-L1.
With MS/MS, we can also identify some site-specific amino-acid modifications (SI, Figure S2). Most of these identified residues, not surprisingly, have side-chains that are either sulfur-containing (Met18, Met36, Met59, Met115) or aromatic (Phe19, Trp57, Tyr160, Phe211, His220, etc.), which are highly reactive towards hydroxyl radicals36. Other less reactive residues including Leu, Val, Arg, and Lys reacted to smaller extents with the free radicals. The residue-specific FPOP modifications of all identified residues are detailed comprehensively (SI, Table S4).
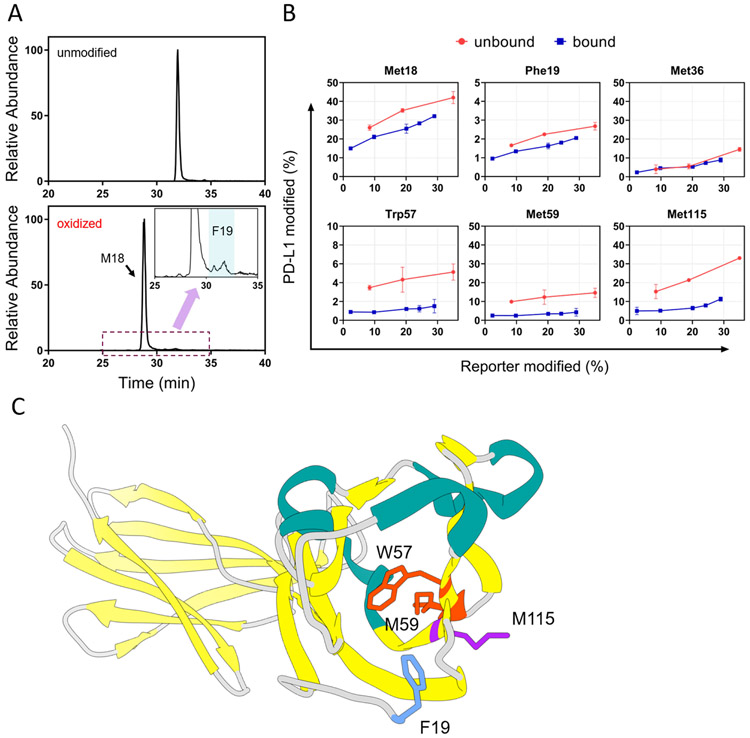
The site-specific FPOP modifications provide a “magnified” view of the PD-L1-macrocycle interacting region and pinpoint some of the critical residues or short regions responsible for ligand binding. For instance, we detected significant differences in the FPOP yields on residues Met18, Trp57, Met59, and Met115 (Figure 4B). It is noteworthy that, although the oxidation of Phe19 was not high (Figure 4A), the time-dependent quantitation at the residue level showed noticeable decreases of Phe19 modification for the bound state (Figure 4B and Figure S2). Met36, on the other hand, showed similar FPOP yields for the two states (Figure 4B). These residues are typically highly reactive in •OH-mediated modifications, even under the high-scavenging environment where identification and quantitation of other FPOP-labeled residues can be challenging. At this point, we can only suggest that those residues that show differences are critical binding residues (or adjoin critical binding adjacent residues). A more conclusive picture will arise with the development of comprehensive footprinting and/or residue-specific HDX. A productive and efficient strategy would be to use the HDX and FPOP results to guide site-specific metagenesis.
Figure 4.
(A) XICs corresponding to modified and unmodified peptide 18-25 (amino acid sequence MFTVTVPK) showed Met18 is the major modification site, whereas modification on Phe19 is less abundant. The insert represents a zoomed-in region where peptides with Phe19 oxidation elutes. (B) Residue-level time-dependent curves of macrocycle-bound and unbound PD-L1. Residues identified that show significant differential FPOP yields between bound and unbound states of PD-L1 include M18, F19, W57, M59, and M115. Residues were identified by manual interpretation of the product-ion (MS/MS) spectra. Error bars correspond to ± SD from triplicate measurements. The error bars correspond to ± SD from triplicate measurements and show that for binding sites, the differences between the accumulated differences for three points representing bound and unbound are at least 3 times the propagated errors (sq root of the sum of the squares of the SDs). This corresponds to 99.7% confidence in the assignment. (C) Close-up view of these residues show they are spatially localized on the N-lobe of PD-L1.
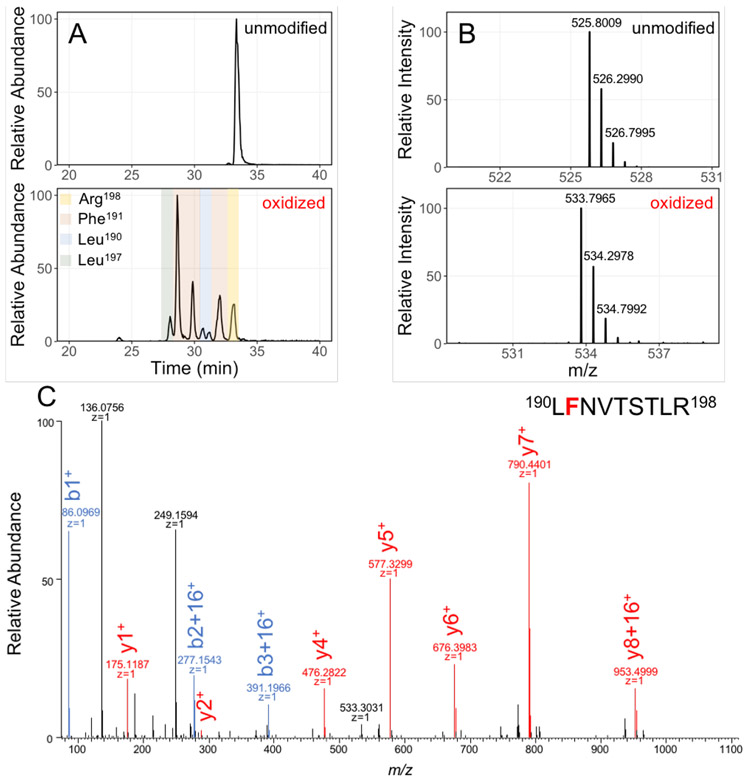
Nevertheless, we can demonstrate that the incomplete data we have are reliable. We show the extracted ion chromatograms (XICs) and precursor-ion spectra of the unmodified and singly oxidized species of peptide 190-198 (Figure 5A and 5B). Although the peptide-level extent of modification for this peptide was below 2%, we could see multiple chromatographic peaks for the oxidized species (+15.9949 Da), corresponding to isomeric peptides eluting at different retention times. Manual interpretation of product-ion spectra of these modified peptides locate modifications on spatially proximate residues Leu190, Phe191, Leu197, and Arg198 (for an example, see Figure 5C).
Figure 5.
Example of LC-MS/MS separation, detection, and identification of unmodified and oxidatively modified species of peptide 190-198. (A) XICs of unmodified and singly oxidized peptide 190-198; (B) mass spectra of precursor ions (+2 charge) show a 16 Da shift from unmodified to modified peptide 190-198; (C) the product-ion (MS/MS) spectra of the singly oxidized peptide 190-198 locates modification on Phe191.
X-ray Crystallography of PD-L1/Macrocycle Complex Reveals Two Binding Pockets.
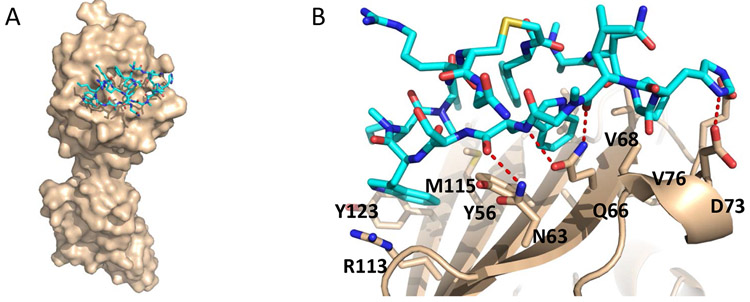
A parallel crystallography effort to characterize the interactions between human PD-L1 and the macrocycle afforded a high-resolution crystal structure of the protein-inhibitor complex. Two major hydrophobic binding pockets comprised of residues Ile54, Tyr56, Gln66, Arg113, Met115, and Tyr123 form on the surface of PD-L1 to accommodate the two tryptophan side chains of the macrocycle (Figure 6B). In addition, Asn63, Gln66, Glu71, and Asp73 form specific interactions with polar and charged atoms of the macrocycle (Figure 6B). The macrocycle used in the current study binds to the N-lobe of the PD-L1 ectodomain (Figure 6A) in a similar fashion to the two macrocyclic peptide inhibitors described previously by Holak and co-workers63. The N-terminal peptide of PD-L1, however, was not found to interact with the macrocycle according to the X-ray crystal structure.
Figure 6.
(A) Overall structure of the macrocycle (cyan) bound to human PD-L1 (beige). The structure confirms only the N-terminal lobe is involved in binding; (B) Residue-specific binding interface between macrocycle and human PD-L1. The macrocycle carbon atoms are colored cyan while the PD-L1 carbon atoms are colored beige. Oxygen atoms (red), nitrogen atoms (blue), and sulfur atoms (yellow) are colored accordingly.
Both HDX and FPOP outcome reveal that the N-terminus of PD-L1 is associated with binding of macrocycle (Figure 2 and Figure 3C). However, the X-ray crystal structure does not see it (Figure 6B). This difference may be attributed to the different N-terminal solvent accessibility of PD-L1 obtained under solution environment and solid state64-65. That the N-terminal of PD-L1 interacts with its binding targets (small molecules or proteins) is not without precedent. Crystal structures of hPD-L1/PD-1 complex have seen hydrogen bond formation between Phe19 of PD-L1 and side-chain amine of Lys78 of PD-147. Nonpeptidic small molecules, including BMS-200 and BMS-202, interact with the N-terminus of PD-L1 through hydrogen bond with Thr2066, and a water-mediated interaction with the backbone carbonyl of Phe1921, respectively. Moreover, the PD-L1 N-terminal region participates in the interaction with durvalumab, through an ionic interaction and van der Waals contacts, involving N-terminal residues Thr20, Val23, Asp2667. Nevertheless, there is good agreement between interacting regions of PD-L1 identified by HDX and FPOP and the molecular interactions observed in the crystal structure of the protein-macrocycle complex.
To compare the outcomes of HDX, FPOP, and X-ray crystallography, we show in Table 1 the macrocycle binding regions identified by HDX, FPOP, and X-ray, respectively. Overall, we obtained good qualitative agreement between the HDX and FPOP results, despite several distinct differences between the two approaches. First, FPOP modifies side chains of protein with a reactive species, whereas HDX reports on the protein backbone. Second, the shortest D2O incubation time in our HDX experiment is 30 s, providing an averaged ensemble of conformations over time, whereas FPOP affords a “snapshot” on microsecond time scale for reactions of primary radicals.
Table 1.
Regions identified by HDX and FPOP show good agreement with those identified by the crystal structure of the complex.
| Contact Region (residue #) |
Observed in X-ray* | Identified by HDX | Identified by FPOP |
|---|---|---|---|
| 18(N-terminal)-28 | None | 18-28 | 18-25 |
| 46-87 | 54-56, 63, 66-68, 71, 73, 76 | 57-87 | 47-75 |
| 113-125 | 113, 115, 121, 123 | 116-122 | 114-125 |
A heavy-atom to heavy-atom distance cut off of less than or equal to 4 Å was used to select residues in close contact to the macrocycle.
Nevertheless, the FPOP and HDX results show conclusively the regions where the macrocycle interacts with the PD-L1 surface via discontinuous regions. Both approaches reveal decreased solvent accessibility on the N-terminal region and the region spanning 114-125, upon macrocycle binding (Figure 2 and Figure 3A). Further, FPOP showed protection for peptide 47-75 (Figure 3A). Region 76 to 86, however, was not covered in the analysis owing to multiple trypsin cleavage sites that led to many small peptides that are not retained by HPLC. HDX filled in the coverage of this region, by showing peptide 74-87 also exhibited protection upon binding (Figure 2). Both approaches show that the entire C-lobe of PD-L1 is not affected by macrocycle binding (Figure 2 and Figure 3C).
Additionally, FPOP provided some site-specific binding information that is largely consistent with the X-ray crystallography structure. For example, residue Met115, which showed FPOP protection upon macrocycle binding (Figure 4B and 4C), constitutes one of the two hydrophobic binding pockets identified from X-ray structure, comprising amino acids Arg113, Met115, and Tyr123 (Figure 6B). Interestingly, the amino acids Trp57 and Met59 also reported protection in FPOP (Figure 4B,C); these two are adjacent to the amino acids that form the other X-ray identified hydrophobic binding pocket (i.e., Ile54, Tyr56, and Gln66; Figure 6B). Side-chains of tryptophan and methionine are 10-20 fold more reactive towards hydroxyl radical compared to Ile and Gln 36-37, 68, this may justify the preferential FPOP modifications in this binding pocket. By and large, the FPOP site-specific information effectively narrows the localization of critical binding regions and affords a coarse-grained guide to potential “hot spots” on PD-L1.
CONCLUSIONS
In summary, our approaches of time dependent HDX and FPOP measurements not only provide assurance that the modification reactions occur normally, but also add statistical weight to the bound and unbound states comparison. The footprinting results are consistent with the X-ray crystal structure and substantiate that the structure is largely relevant in vitro. Moreover, the MS footprinting will usually be a faster approach, gaining binding information especially in the early stages of drug development or when crystallization is difficult. Those results can also guide site-directed mutagenesis studies if more detail is needed. Taken together, the data presented here provide an in-depth understanding of small-molecule PD-L1 interaction and suggest that modulation of the immune system with small-molecules may complement those efforts with large biologics.
Supplementary Material
Acknowledgements
This research was supported by the National Institutes of Health, National Institute of General Medical Sciences, Grant 2P41GM103422 and by a grant from Gilead to the WU MS laboratory.
Footnotes
Accession Number for Protein:
The PD-L1-macrocycle structure PDB ID is 6PV9. DEPOSITION ID: D_1000243159
Supporting Information. Details about Materials and Methods of cloning, expression, purification of PD-L1 ectodomain. Experimental descriptions of bioassays including direct binding Biacore assay, PD-1/PD-L1 interaction assay, CD80/CTLA4 interaction assay, PD-1/PD-L1 NFAT-Luciferase cell-cell assay, details on HDX and FPOP, and summary of X-ray crystallography parameters, summary of bioassay results, a summary of HDX averaged differential deuterium uptake of each peptic peptide, summary of differential deuterium uptake between bound and unbound states plotted as function of exchange time, selected product-ion spectra (MS/MS) of tryptic peptides with FPOP oxidative modification, list of residue-specific FPOP modification extents.
Competing Interests
The authors declare no competing interests.
REFERENCES
- 1.Atkins MB; Sznol M, Cancer Immunotherapy: Past Progress and Future Directions. Semin Oncol 2015, 42 (4), 518–22. [DOI] [PubMed] [Google Scholar]
- 2.Gubin MM; Schreiber RD, CANCER. The odds of immunotherapy success. Science 2015, 350 (6257), 158–9. [DOI] [PubMed] [Google Scholar]
- 3.June CH; O'Connor RS; Kawalekar OU; Ghassemi S; Milone MC, CAR T cell immunotherapy for human cancer. Science 2018, 359 (6382), 1361–1365. [DOI] [PubMed] [Google Scholar]
- 4.Page DB; Postow MA; Callahan MK; Allison JP; Wolchok JD, Immune modulation in cancer with antibodies. Annu Rev Med 2014, 65, 185–202. [DOI] [PubMed] [Google Scholar]
- 5.Ribas A; Wolchok JD, Cancer immunotherapy using checkpoint blockade. Science 2018, 359 (6382), 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg SA, Raising the bar: the curative potential of human cancer immunotherapy. Sci Transl Med 2012, 4 (127), 127ps8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazzari C; Karachaliou N; Bulotta A; Vigano M; Mirabile A; Brioschi E; Santarpia M; Gianni L; Rosell R; Gregorc V, Combination of immunotherapy with chemotherapy and radiotherapy in lung cancer: is this the beginning of the end for cancer? Ther Adv Med Oncol 2018, 10, 1758835918762094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer RJ; Powles T; Atkins MB; Escudier B; McDermott DF; Suarez C; Bracarda S; Stadler WM; Donskov F; Lee J-L; Hawkins RE; Ravaud A; Alekseev BY; Staehler MD; Uemura M; Donaldson F; Li S; Huseni MA; Schiff C; Rini BI, IMmotion151: A Randomized Phase III Study of Atezolizumab Plus Bevacizumab vs Sunitinib in Untreated Metastatic Renal Cell Carcinoma (mRCC). Journal of Clinical Oncology 2018, 36 (6_suppl), 578–578. [Google Scholar]
- 9.Sui H; Ma N; Wang Y; Li H; Liu X; Su Y; Yang J, Anti-PD-1/PD-L1 Therapy for Non-Small-Cell Lung Cancer: Toward Personalized Medicine and Combination Strategies. J Immunol Res 2018, 2018, 6984948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L; Flies DB, Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 2013, 13 (4), 227–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riley JL, PD-1 signaling in primary T cells. Immunol Rev 2009, 229 (1), 114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leach DR; Krummel MF; Allison JP, Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996, 271 (5256), 1734–6. [DOI] [PubMed] [Google Scholar]
- 13.Francisco LM; Sage PT; Sharpe AH, The PD-1 pathway in tolerance and autoimmunity. Immunol Rev 2010, 236, 219–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fife BT; Pauken KE, The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann N Y Acad Sci 2011, 1217, 45–59. [DOI] [PubMed] [Google Scholar]
- 15.Keir ME; Butte MJ; Freeman GJ; Sharpe AH, PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 2008, 26, 677–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okazaki T; Honjo T, PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol 2007, 19 (7), 813–24. [DOI] [PubMed] [Google Scholar]
- 17.Flies DB; Sandler BJ; Sznol M; Chen L, Blockade of the B7-H1/PD-1 pathway for cancer immunotherapy. Yale J Biol Med 2011, 84 (4), 409–21. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhan MM; Hu XQ; Liu XX; Ruan BF; Xu J; Liao C, From monoclonal antibodies to small molecules: the development of inhibitors targeting the PD-1/PD-L1 pathway. Drug Discov Today 2016, 21 (6), 1027–36. [DOI] [PubMed] [Google Scholar]
- 19.Zarganes-Tzitzikas T; Konstantinidou M; Gao Y; Krzemien D; Zak K; Dubin G; Holak TA; Domling A, Inhibitors of programmed cell death 1 (PD-1): a patent review (2010-2015). Expert Opin Ther Pat 2016, 26 (9), 973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magiera-Mularz K; Skalniak L; Zak KM; Musielak B; Rudzinska-Szostak E; Berlicki L; Kocik J; Grudnik P; Sala D; Zarganes-Tzitzikas T; Shaabani S; Domling A; Dubin G; Holak TA, Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint. Angew Chem Int Ed Engl 2017, 56 (44), 13732–13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zak KM; Grudnik P; Guzik K; Zieba BJ; Musielak B; Domling A; Dubin G; Holak TA, Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1). Oncotarget 2016, 7 (21), 30323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller MM, M. C, Allen MP, Bowshe MS, Boy KM, Gillis, Langley DR, Mull E, Poirier MA, Sanghvi N, Sun L-Q, Tenney DJ, Yeung K-S, Zhu J, Reid PC, Scola PM Macrocyclic inhibitors of the pd-1/pd-l1 and cd80(b7-1)/pd-l1 protein/protein interactions. 20140294898 A1, 2014.
- 23.Sasikumar PG, Ramachandra M, Vadlamani SK, Vemula RK, Satyam LK, Subbarao K, Shrimali RK, Kandepu S Immunosuppression modulating compounds. EP2585099 A2, 2013.
- 24.Chupak LS, Zheng X Compounds useful as immunomodulators. WO2015034820 A1, 2015.
- 25.Chen T; Li Q; Liu Z; Chen Y; Feng F; Sun H, Peptide-based and small synthetic molecule inhibitors on PD-1/PD-L1 pathway: A new choice for immunotherapy? Eur J Med Chem 2019, 161, 378–398. [DOI] [PubMed] [Google Scholar]
- 26.Guzik K; Tomala M; Muszak D; Konieczny M; Hec A; Błaszkiewicz U; Pustuła M; Butera R; Dömling A; Holak TA, Development of the inhibitors that target the PD-1/PD-L1 interaction—a brief look at progress on small molecules, peptides and macrocycles. Molecules 2019, 24 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jemal M, High-throughput quantitative bioanalysis by LC/MS/MS. Biomed Chromatogr 2000, 14 (6), 422–9. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths J, A brief history of mass spectrometry. Anal Chem 2008, 80 (15), 5678–83. [DOI] [PubMed] [Google Scholar]
- 29.Domon B; Aebersold R, Mass spectrometry and protein analysis. Science 2006, 312 (5771), 212–7. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z; Smith DL, Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Science 1993, 2 (4), 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y; Rempel DL; Zhang J; Sharma AK; Mirica LM; Gross ML, Pulsed hydrogen-deuterium exchange mass spectrometry probes conformational changes in amyloid beta (Abeta) peptide aggregation. Proc Natl Acad Sci U S A 2013, 110 (36), 14604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wales TE; Engen JR, Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev 2006, 25 (1), 158–70. [DOI] [PubMed] [Google Scholar]
- 33.Katta V; Chait BT, Hydrogen/deuterium exchange electrospray ionization mass spectrometry: a method for probing protein conformational changes in solution. Journal of the American Chemical Society 1993, 115 (14), 6317–6321. [Google Scholar]
- 34.Kan ZY; Walters BT; Mayne L; Englander SW, Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis. Proc Natl Acad Sci U S A 2013, 110 (41), 16438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engen JR, Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem 2009, 81 (19), 7870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu G; Chance MR, Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chemical reviews 2007, 107 (8), 3514–43. [DOI] [PubMed] [Google Scholar]
- 37.Xu G; Chance MR, Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Anal Chem 2005, 77 (14), 4549–55. [DOI] [PubMed] [Google Scholar]
- 38.Tullius TD; Dombroski BA; Churchill ME; Kam L, Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol 1987, 155, 537–58. [DOI] [PubMed] [Google Scholar]
- 39.Takamoto K; Chance MR, Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct 2006, 35, 251–276. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y; Wecksler AT; Molina P; Deperalta G; Gross ML, Mapping the Binding Interface of VEGF and a Monoclonal Antibody Fab-1 Fragment with Fast Photochemical Oxidation of Proteins (FPOP) and Mass Spectrometry. J Am Soc Mass Spectrom 2017, 28 (5), 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan Y; Chen G; Wei H; Huang RY; Mo J; Rempel DL; Tymiak AA; Gross ML, Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. J Am Soc Mass Spectrom 2014, 25 (12), 2084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li KS; Chen G; Mo J; Huang RY; Deyanova EG; Beno BR; O'Neil SR; Tymiak AA; Gross ML, Orthogonal Mass Spectrometry-Based Footprinting for Epitope Mapping and Structural Characterization: The IL-6 Receptor upon Binding of Protein Therapeutics. Anal Chem 2017, 89 (14), 7742–7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hambly DM; Gross ML, Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom 2005, 16 (12), 2057–63. [DOI] [PubMed] [Google Scholar]
- 44.Gau BC; Sharp JS; Rempel DL; Gross ML, Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal Chem 2009, 81 (16), 6563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li KS; Shi L; Gross ML, Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization. Accounts of Chemical Research 2018, 51 (3), 736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang B; Cheng M; Rempel D; Gross ML, Implementing fast photochemical oxidation of proteins (FPOP) as a footprinting approach to solve diverse problems in structural biology. Methods 2018, 144, 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zak KM; Kitel R; Przetocka S; Golik P; Guzik K; Musielak B; Domling A; Dubin G; Holak TA, Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1. Structure 2015, 23 (12), 2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen E; Salinas ND; Huang Y; Ntumngia F; Plasencia MD; Gross ML; Adams JH; Tolia NH, Broadly neutralizing epitopes in the Plasmodium vivax vaccine candidate Duffy Binding Protein. Proc Natl Acad Sci U S A 2016, 113 (22), 6277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niu B; Mackness BC; Rempel DL; Zhang H; Cui W; Matthews CR; Zitzewitz JA; Gross ML, Incorporation of a Reporter Peptide in FPOP Compensates for Adventitious Scavengers and Permits Time-Dependent Measurements. J Am Soc Mass Spectrom 2017, 28 (2), 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z; Minor W, Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997, 276, 307–26. [DOI] [PubMed] [Google Scholar]
- 51.Fedorov AA, Fedorov EV, Samantha D, Hillerich B, Seidel RD, Almo SC, To be published.
- 52.Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung LW; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010, 66 (Pt 2), 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P; Lohkamp B; Scott WG; Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010, 66 (Pt 4), 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H; Shu Q; Rempel DL; Frieden C; Gross ML, Continuous and Pulsed Hydrogen-Deuterium Exchange and Mass Spectrometry Characterize CsgE Oligomerization. Biochemistry 2015, 54 (42), 6475–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z; Vachet RW, Kinetics of Protein Complex Dissociation Studied by Hydrogen/Deuterium Exchange and Mass Spectrometry. Anal Chem 2015, 87 (23), 11777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Mol NJ; Catalina MI; Fischer MJ; Broutin I; Maier CS; Heck AJ, Changes in structural dynamics of the Grb2 adaptor protein upon binding of phosphotyrosine ligand to its SH2 domain. Biochim Biophys Acta 2004, 1700 (1), 53–64. [DOI] [PubMed] [Google Scholar]
- 57.Hulme EC; Trevethick MA, Ligand binding assays at equilibrium: validation and interpretation. Br J Pharmacol 2010, 161 (6), 1219–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferraro DM; Lazo N; Robertson AD, EX1 hydrogen exchange and protein folding. Biochemistry 2004, 43 (3), 587–94. [DOI] [PubMed] [Google Scholar]
- 59.Jones LM; J BS; J AC; Gross ML, Fast photochemical oxidation of proteins for epitope mapping. Anal Chem 2011, 83 (20), 7657–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gau B; Garai K; Frieden C; Gross ML, Mass spectrometry-based protein footprinting characterizes the structures of oligomeric apolipoprotein E2, E3, and E4. Biochemistry 2011, 50 (38), 8117–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pryor WA, Oxy-radicals and related species: their formation, lifetimes, and reactions. Annu Rev Physiol 1986, 48, 657–67. [DOI] [PubMed] [Google Scholar]
- 62.Niu B; Gross ML, MS-Based Hydroxyl Radical Footprinting: Methodology and Application of Fast Photochemical Oxidation of Proteins (FPOP) In Mass Spectrometry-Based Chemical Proteomics, Tao WAZ, Y., Ed. Wiley: 2019; pp 363–416. [Google Scholar]
- 63.Magiera-Mularz K; Skalniak L; Zak KM; Musielak B; Rudzinska-Szostak E; Berlicki L; Kocik J; Grudnik P; Sala D; Zizigas-Zarganis T; Shaabani S; Domling A; Dubin G; Holak TA, Bioactive macrocyclic inhibitors of the PD-1/PD-L1 immune checkpoint. Angew Chem Int Ed Engl 2017. [DOI] [PMC free article] [PubMed]
- 64.Kozak M; Jurga S, A comparison between the crystal and solution structures of Escherichia coli asparaginase II. Acta Biochim Pol 2002, 49 (2), 509–13. [PubMed] [Google Scholar]
- 65.Sikic K; Tomic S; Carugo O, Systematic comparison of crystal and NMR protein structures deposited in the protein data bank. Open Biochem J 2010, 4, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guzik K; Zak KM; Grudnik P; Magiera K; Musielak B; Torner R; Skalniak L; Domling A; Dubin G; Holak TA, Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1. J Med Chem 2017, 60 (13), 5857–5867. [DOI] [PubMed] [Google Scholar]
- 67.Lee HT; Lee JY; Lim H; Lee SH; Moon YJ; Pyo HJ; Ryu SE; Shin W; Heo YS, Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab. Sci Rep 2017, 7 (1), 5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niu B; Zhang H; Giblin D; Rempel DL; Gross ML, Dosimetry determines the initial OH radical concentration in fast photochemical oxidation of proteins (FPOP). J Am Soc Mass Spectrom 2015, 26 (5), 843–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.