Staphylococcus aureus and Streptococcus pneumoniae infections cause significant morbidity and mortality in humans. For both, nasal colonization is a risk factor for infection. Studies of nasal microbiota identify Dolosigranulum pigrum as a benign bacterium present when adults are free of S. aureus or when children are free of S. pneumoniae. Here, we validated these in vivo associations with functional assays. We found that D. pigrum inhibited S. aureus in vitro and, together with a specific nasal Corynebacterium species, also inhibited S. pneumoniae. Furthermore, genomic analysis of D. pigrum indicated that it must obtain key nutrients from other nasal bacteria or from humans. These phenotypic interactions support the idea of a role for microbe-microbe interactions in shaping the composition of human nasal microbiota and implicate D. pigrum as a mutualist of humans. These findings support the feasibility of future development of microbe-targeted interventions to reshape nasal microbiota composition to exclude S. aureus and/or S. pneumoniae.
KEYWORDS: Dolosigranulum pigrum, Corynebacterium, Staphylococcus aureus, Streptococcus pneumoniae, microbe-microbe interactions, interspecies interactions, upper respiratory tract, nasal, microbiota, comparative genomics
ABSTRACT
Multiple epidemiological studies identify Dolosigranulum pigrum as a candidate beneficial bacterium based on its positive association with health, including negative associations with nasal/nasopharyngeal colonization by the pathogenic species Staphylococcus aureus and Streptococcus pneumoniae. Using a multipronged approach to gain new insights into D. pigrum function, we observed phenotypic interactions and predictions of genomic capacity that support the idea of a role for microbe-microbe interactions involving D. pigrum in shaping the composition of human nasal microbiota. We identified in vivo community-level and in vitro phenotypic cooperation by specific nasal Corynebacterium species. Also, D. pigrum inhibited S. aureus growth in vitro, whereas robust inhibition of S. pneumoniae required both D. pigrum and a nasal Corynebacterium together. D. pigrum l-lactic acid production was insufficient to account for these inhibitions. Genomic analysis of 11 strains revealed that D. pigrum has a small genome (average 1.86 Mb) and multiple predicted auxotrophies consistent with D. pigrum relying on its human host and on cocolonizing bacteria for key nutrients. Further, the accessory genome of D. pigrum harbored a diverse repertoire of biosynthetic gene clusters, some of which may have a role in microbe-microbe interactions. These new insights into D. pigrum’s functions advance the field from compositional analysis to genomic and phenotypic experimentation on a potentially beneficial bacterial resident of the human upper respiratory tract and lay the foundation for future animal and clinical experiments.
IMPORTANCE Staphylococcus aureus and Streptococcus pneumoniae infections cause significant morbidity and mortality in humans. For both, nasal colonization is a risk factor for infection. Studies of nasal microbiota identify Dolosigranulum pigrum as a benign bacterium present when adults are free of S. aureus or when children are free of S. pneumoniae. Here, we validated these in vivo associations with functional assays. We found that D. pigrum inhibited S. aureus in vitro and, together with a specific nasal Corynebacterium species, also inhibited S. pneumoniae. Furthermore, genomic analysis of D. pigrum indicated that it must obtain key nutrients from other nasal bacteria or from humans. These phenotypic interactions support the idea of a role for microbe-microbe interactions in shaping the composition of human nasal microbiota and implicate D. pigrum as a mutualist of humans. These findings support the feasibility of future development of microbe-targeted interventions to reshape nasal microbiota composition to exclude S. aureus and/or S. pneumoniae.
INTRODUCTION
Colonization of the human nasal passages by Staphylococcus aureus or Streptococcus pneumoniae is a major risk factor for infection by the colonizing bacterium at a distant body site (1–5). Interventions that reduce the prevalence of colonization also reduce the risk of infection and transmission (6, 7). S. aureus and S. pneumoniae are major human pathogens that cause significant morbidity and mortality worldwide (8–11). There are also concerns regarding rising rates of antimicrobial resistance (12) and the potential for long-term effects of antibiotics early in life (13). Thus, efforts have recently focused on the identification of candidate bacteria that confer colonization resistance against S. aureus (14–21) and S. pneumoniae (22–25), with particular urgency for S. aureus in the absence of an effective vaccine.
Dolosigranulum pigrum has emerged in multiple studies of the human upper respiratory tract (URT) microbiota, colonizing with or without Corynebacterium species, as potentially beneficial and/or protective against colonization by S. aureus and S. pneumoniae (26–53) (reviewed in references 14, 54, 55, 56, and 57). However, little is known about this Gram-positive, catalase-negative, Firmicute bacterium, first described in 1993 (58). Microbiota studies sampling either nostrils or nasopharynx have shown very similar results; therefore, for simplicity, we use “nasal” or “nasal passages” to denote the area inclusive of the nostrils through the nasopharynx. D. pigrum and S. aureus are inversely correlated in adult nasal microbiota (30, 41, 59), whereas, in pediatric nasal microbiota, D. pigrum and members of the genus Corynebacterium are overrepresented when S. pneumoniae is absent (26, 33). Moreover, children with D. pigrum colonization of the nasal passages are less likely to have acute otitis media (27, 40) and it has been speculated that D. pigrum-dominated microbiota profiles might be more resistant to invasive pneumococcal disease (46). Furthermore, D. pigrum abundance in the nasal passages is inversely associated with wheezing and respiratory tract infections in infants (28) and an abundance of D. pigrum with Corynebacterium in adults provides greater community stability in the face of pneumococcal exposure (51). The intriguing inference from these studies that D. pigrum plays a beneficial role in human nasal microbiota deserves further investigation.
In contrast to the data mentioned above, there are very few reports of D. pigrum in association with human disease (60–68). Its frequent identification in human nasal microbiota (26–53, 69–81) and its rare association with infection are consistent with D. pigrum functioning as a commensal and, possibly, as a mutualist of humans––characteristics that support the idea of its potential for future use as a therapeutic. However, its metabolism and its interplay with other nasal bacteria remain uncharted territory. Using a multipronged approach, we have made significant advances in these areas. First, we identified specific species of candidate bacterial interactors with D. pigrum by analyzing nasal microbiota data sets from adults and children. Second, we used in vitro phenotypic assays to show that D. pigrum exhibits distinct interaction phenotypes with nasal Corynebacterium species, S. aureus, and S. pneumoniae. Third, on the basis of the genomes of 11 distinct D. pigrum strains, we identify key predicted functions and auxotrophies in its core genome plus a diversity of predicted biosynthetic gene clusters (BCGs) in its accessory genome. This critical shift to phenotypic and genomic experimentation marks a significant advance in understanding D. pigrum, a potentially beneficial member of human nasal microbiota.
RESULTS
Individual bacterial species are associated with D. pigrum in the nasal microbiota of both adults and children.
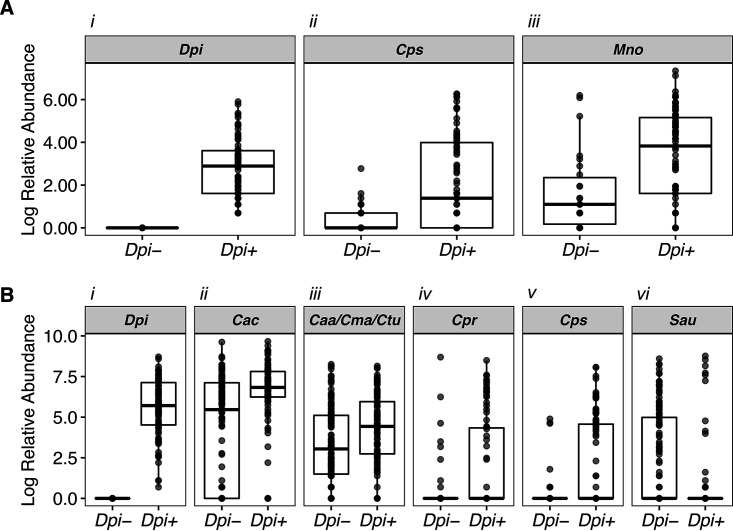
D. pigrum is the only member of its genus, and multiple 16S rRNA gene-based nasal microbiota studies have identified associations between Dolosigranulum and other genera, such as Corynebacterium (see, e.g., references 28, 29, 31, 36, 38, 40, 41, 43, and 82). In most cases, the taxonomic resolution in the aforementioned studies was limited to the genus or higher taxonomic levels. Thus, we sought to achieve finer taxonomic resolution and to determine which species are associated with D. pigrum. We identified two nostril data sets with V1-V2/V1-V3 16S rRNA gene sequences, regions that contain sufficient information for species-level taxonomic assignment of most nose-associated bacteria (26, 41). After parsing sequences into species-level phylotypes, we interrogated each data set using analysis of composition of microbiomes (ANCOM) (83) to identify bacterial species that display differential relative abundances in the absence or presence of D. pigrum sequences (Fig. 1; see also Table S1 in the supplemental material). In the nostrils of 99 children ages 6 to 78 months (26), Corynebacterium pseudodiphtheriticum exhibited increased differential relative abundance in the presence of D. pigrum, i.e., was positively associated with D. pigrum, as was Moraxella nonliquefaciens (Fig. 1A). In the nostrils of 210 adults from the Human Microbiome Project (HMP), three Corynebacterium species––Corynebacteriu accolens, C. propinquum, and C. pseudodiphtheriticum––and an unresolved supraspecies of C. accolens-macginleyi-tuberculostearicum were positively associated with D. pigrum (Fig. 1B, panels ii to v), whereas S. aureus was negatively associated with D. pigrum (Fig. 1B, panel vi). The associations identified in compositional microbiota data observed here and in prior studies (28, 29, 31, 36, 38, 40, 41, 43, 82) led to testable hypotheses about possible direct microbe-microbe interactions between D. pigrum and the specific nasal Corynebacterium species, as well as between D. pigrum and S. aureus. Therefore, we used in vitro phenotypic assays to test our hypotheses about direct microbe-microbe interactions.
FIG 1.
Individual nasal Corynebacterium species exhibit increased differential relative abundances in the presence of D. pigrum in human nostril microbiota. We used ANCOM to compare the species/supraspecies-level compositions of 16S rRNA gene nostril data sets from (A) 99 children ages 6 to 78 months and (B) 210 adults where D. pigrum was either absent (Dpi−) or present (Dpi+) on the basis of 16S rRNA gene sequencing data. Plots show only the taxa identified as statistically significant (sig = 0.05) after correction for multiple testing within ANCOM. The dark bar represents the median; lower and upper hinges correspond to the first and third quartiles. Each gray dot represents the value for a sample, and multiple overlapping dots appear black. Dpi = Dolosigranulum pigrum, Cac = Corynebacterium accolens, Caa/Cma/Ctu = supraspecies Corynebacterium accolens_macginleyi_tuberculostearicum, Cpr = Corynebacterium propinquum, Cps = Corynebacterium pseudodiphtheriticum, Mno = Moraxella nonliquefaciens. Only three species and one supraspecies of Corynebacterium from among the larger number of Corynebacterium supraspecies/species present in each data set met the significance threshold. Specifically, in the adult nostril data set, there were 21 species and 5 supraspecies groupings of Corynebacterium in addition to the reads of Corynebacterium that were nonassigned (NA) at the species level. These data were previously published (see Table S7 in reference 41). In the pediatric data set, there were 16 species of Corynebacterium in addition to those that were nonassigned among the species-level Corynebacterium reads (see Table S2). The Log relative abundance numerical data represented in this figure are available in Table S1.
ANCOM Log abundance values. (A) Adult nostril ANCOM data (plotted in Fig. 1A). (B) Pediatric nostril ANCOM data (plotted in Fig. 1B). Download Table S1, XLSX file, 0.02 MB (25.7KB, xlsx) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nasal Corynebacterium species can enhance the growth of D. pigrum in vitro.
We hypothesized that the strong positive association between D. pigrum and the nasal passage-associated Corynebacterium species might be due to these Corynebacterium species releasing metabolites that enhance the growth of D. pigrum. To test this, we quantified D. pigrum growth yields on unconditioned agar medium compared to the yields seen on cell-free agar medium conditioned by growth of C. pseudodiphtheriticum, C. propinquum, or C. accolens (Fig. 2). Conditioning agar medium by prior growth of any of these three nasal Corynebacterium species increased the yield (measured as CFU) of two D. pigrum strains (CDC4709-98 and KPL1914) by 1 to 2 orders of magnitude compared to growth on unconditioned agar medium (Fig. 2A and B). Additionally, one strain of C. pseudodiphtheriticum (Fig. 2A) and the C. accolens strain (Fig. 2B) increased the growth yield of D. pigrum CDC2949-98, a strain with a higher baseline growth yield. The increases in D. pigrum growth yield on Corynebacterium cell-free conditioned agar medium (CFCAM) might have resulted from increased growth rate or increased viability or both and might be consistent with the nasal Corynebacterium species either removing a toxin from the medium or releasing a metabolite that enhances growth and/or survival of D. pigrum.
FIG 2.
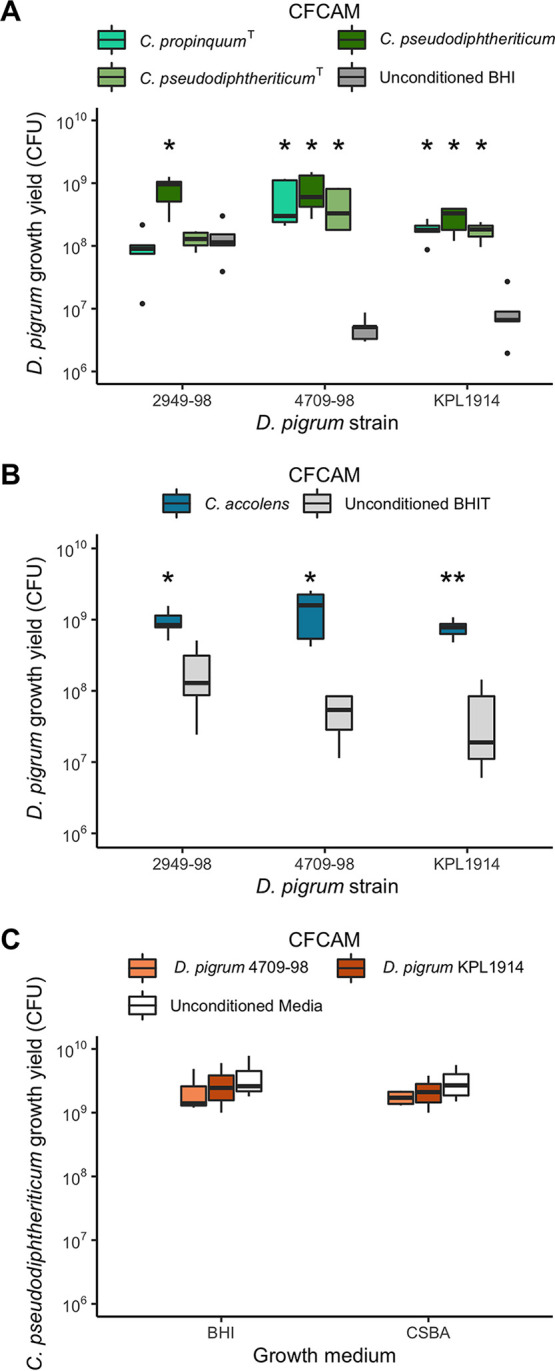
D. pigrum growth yields increase on cell-free conditioned agar medium (CFCAM) from nasal Corynebacterium species but not in reverse. (A and B) Growth yield of D. pigrum strains CDC2949-98, CDC4709-98, and KPL1914 was quantified as the number of CFU grown on a polycarbonate membrane placed on (A) cell-free conditioned BHI agar from C. propinquum (aqua green) or C. pseudodiphtheriticum (dark and light green) or (B) cell-free conditioned BHI-triolein (BHIT) agar from C. accolens (blue) and compared to growth on unconditioned BHI agar (dark gray) or unconditioned BHIT agar (light gray), respectively. (C) Growth yield of C. pseudodiphtheriticum KPL1989 on CFCAM from D. pigrum strains (orange) compared to unconditioned medium (white) was assessed similarly. BHIT was used for growth of C. accolens since it is a fatty acid auxotroph and releases needed oleic acid from triolein. Preconditioning strains were grown on a 0.2-μm-pore-size, 47-mm-diameter polycarbonate membrane for 2 days to generate CFCAM. After removal, we then placed a new membrane on the CFCAM onto which we spread 100 μl of target bacterial cells that had been resuspended to an OD600 of 0.50 in 1× PBS. After 2 days of growth, CFU were enumerated as described in Materials and Methods. CFU counts were compared independently for each individual strain (A and B, n = 5) or medium (C, n = 4) using a Wilcoxon rank sum test with Bonferroni correction for multiple comparisons to the unconditioned medium. Dark bars represent medians, lower and upper hinges correspond to the first and third quartiles, and outlier points are displayed individually. *, P ≤ 0.05; **, P ≤ 0.001. CSBA, citrated sheep blood agar.
In contrast to the increase in D. pigrum growth yield on C. pseudodiphtheriticum CFCAM (Fig. 2A), there was no increase in C. pseudodiphtheriticum strain KPL1989 growth yield on D. pigrum CFCAM (Fig. 2C). Thus, this growth enhancement goes in one direction from nasal Corynebacterium species to D. pigrum. This is consistent with unilateral cooperation of nasal Corynebacterium species––C. pseudodiphtheriticum, C. propinquum or C. accolens––with D. pigrum in the nostril microbiota and supports the observed positive in vivo community-level relationships (Fig. 1).
The positive association between C. accolens and D. pigrum in adult nostril microbiota data sets indicates that in vivo positive interactions between C. accolens and D. pigrum prevail (Fig. 1B, panel ii). However, in vitro, we observed either a positive or a negative interaction between C. accolens and D. pigrum depending on the assay conditions. C. accolens, unlike C. propinquum and C. pseudodiphtheriticum, is a fatty acid auxotroph, and triolein, a model host epithelial-surface triacylglycerol, served as a source of needed oleic acid in our assays. We observed increased D. pigrum growth yield on a semipermeable membrane atop C. accolens CFCAM consisting of brain heart infusion (BHI) agar supplemented with triolein (BHIT) (Fig. 2B). In contrast, D. pigrum was inhibited when placed directly onto this same C. accolens CFCAM (Table 1). This inhibition is reminiscent of our previous finding that the C. accolens triacylglycerol lipase LipS1 hydrolyzes triacylglycerols, releasing free fatty acids that inhibit S. pneumoniae (33), and S. pneumoniae served as a positive control for the effect of C. accolens in this assay (Table 1). Both D. pigrum and S. pneumoniae belong to the order Lactobacillales, and we hypothesized that D. pigrum might be similarly susceptible to free fatty acids such as the oleic acid that C. accolens releases from triolein. Indeed, we observed that oleic acid inhibited D. pigrum when we challenged D. pigrum with oleic acid using a disk diffusion assay with S. pneumoniae as a positive control (Table 2). We also challenged D. pigrum with various concentrations of oleic acid spread onto plates of BHI agar medium. Similarly to the membrane-mediated effect seen in the C. accolens CFCAM experiment described above, we observed D. pigrum growth at higher concentrations of oleic acid when it was placed on a semipermeable membrane atop the oleic acid-coated medium than when it was placed directly on the oleic acid-coated medium (Table 3). This indicates that the membrane provided some protection from inhibition by oleic acid. Overall, these in vitro data indicate that C. accolens can both inhibit the growth of D. pigrum by releasing antibacterial-free fatty acids from host triacylglycerols, such as oleic acid from triolein (Tables 1 and 2), and enhance the growth of D. pigrum by releasing an as-yet-unidentified factor(s) (Fig. 2B). Collectively, these results point to a complex set of molecular interactions between these two species.
TABLE 1.
In contrast to growth on a semipermeable membrane, D. pigrum is inhibited when grown directly on cell-free C. accolens-conditioned BHI agar supplemented with triolein as a source of oleic acid
| Conditioning strain | Growth of target straina
|
||
|---|---|---|---|
|
S. pneumoniae
603 (6B) |
D. pigrum
CDC4709-98 |
D. pigrum KPL1914 |
|
| C. accolens KPL1818 | 0 | 0 | 0 |
| C. propinquumT DSM44285 | 0 | + | + |
| C. pseudodiphtheriticum KPL1989 | + | + | + |
0, no growth; +, growth detected, n ≥ 3.
TABLE 2.
Oleic acid inhibits D. pigrum growth
| Oleic acid concn (μg/disc) |
ZOI (mm)a
|
||
|---|---|---|---|
|
S. pneumoniae 603 (6B) |
D. pigrum CDC4709-98 |
D. pigrum KPL1914 |
|
| 20 | 10.3 ± 4.7 | 12.0 ± 2.9 | 17.0 ± 2.1 |
| 50 | 22.0 ± 5.4 | 26.8 ± 4.4 | 28.4 ± 7.0 |
| 100 | 26.3 ± 6.7 | 35.8 ± 4.5 | 39.4 ± 5.0 |
Mean zone of inhibition (ZOI) ± standard deviation (SD) produced in a disc diffusion assay. ZOIs were measured as the smallest diameter of inhibited growth, and measurements included disc diameter (6 mm). Results from biological replicates (n = 4 for S. pneumoniae, n = 5 for D. pigrum) were averaged.
TABLE 3.
A 0.2-μm-pore-size, 47-mm-diameter polycarbonate membrane provides D. pigrum with some protection against inhibition by oleic acid in vitroa
| Plated vol of oleic acid (μg) |
Result for indicated D. pigrum strain |
|||
|---|---|---|---|---|
| Growth directly on agar |
Growth on membrane |
|||
| KPL1914 | CDC4709-98 | KPL1914 | CDC4709-98 | |
| 500 | 0 | 0 | 0 | + |
| 50 | 0 | 0 | + | + |
| 5 | + | + | + | + |
| 0.5 | + | + | + | + |
| 0.05 | + | + | + | + |
| 0 (BHI agar) | + | + | + | + |
| 0 (CSBA) | + | + | + | + |
0, no growth; +, growth detected; n = 3. CSBA, citrated sheep blood agar.
D. pigrum inhibits S. aureus growth.
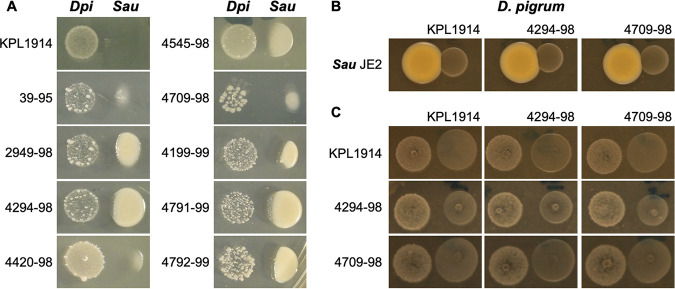
The ANCOM of the adult nostril microbiota data set revealed a negative association between S. aureus and D. pigrum (Fig. 1B, panel vi). Direct antagonism would be the simplest mechanism underpinning this observation. Therefore, we assayed for the effect of 10 different strains of D. pigrum on S. aureus. We gave D. pigrum a head start to compensate for its lower growth rate in vitro. S. aureus growth was inhibited when it was inoculated adjacent to a pregrown inoculum of each of these 10 D. pigrum strains on agar medium (Fig. 3A). In contrast, a pregrown inoculum of S. aureus did not inhibit D. pigrum (Fig. 3B). Furthermore, comparing three representative D. pigrum strains, none of the D. pigrum strains, when pregrown, inhibited the other D. pigrum strains (Fig. 3C).
FIG 3.
Ten different strains of D. pigrum inhibit methicillin-resistant S. aureus USA300 strain JE2 whereas S. aureus does not inhibit D. pigrum. (A) Ten pregrown D. pigrum isolates produced a diffusible activity that inhibited the growth of S. aureus strain JE2. (B) When S. aureus was pregrown in this assay, there was no visible inhibition of subsequently inoculated D. pigrum. (C) Similarly, we did not observe any inhibition in a pairwise comparison of three representative strains of D. pigrum in this assay. All growth was on BHI agar in the independent experiments (n ≥ 3) represented in panels A, B, and C. Representative images are shown for each strain. The respective pregrown strain (D. pigrum or S. aureus) was resuspended in PBS, and then a 5-μl drop was placed on BHI agar and pregrown for 48 h (D. pigrum) or 24 h (S. aureus). After that, the indicator strain was inoculated at a location adjacent to the pregrown strain. Inhibition was assessed after 24 and 48 h (48-h results are shown here). For panels A and B, similar results were observed using S. aureus Newman.
D. pigrum production of lactic acid is unlikely to be the primary mechanism for negative associations with S. pneumoniae or S. aureus.
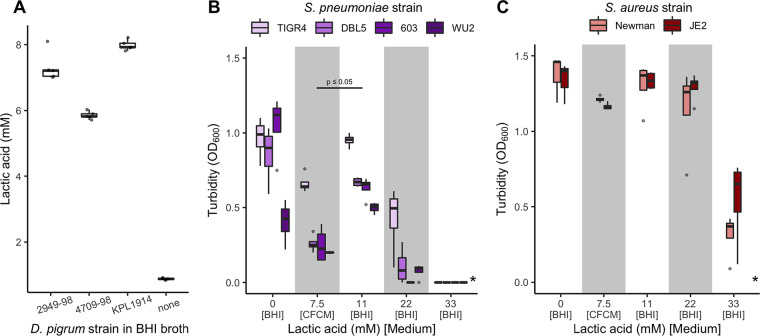
D. pigrum lactic acid production has been proposed as a mechanism to explain epidemiologic observations of negative associations between D. pigrum and S. pneumoniae (82). Under nutrient-rich conditions in vitro, three tested strains of D. pigrum produced from 5.7 to 8.2 mM l-lactic acid, with strain KPL1914 producing the highest concentration (Fig. 4A). Therefore, we assayed for growth of S. pneumoniae in D. pigrum KPL1914 cell-free conditioned medium (CFCM) and in BHI broth supplemented with various concentrations of l-lactic acid. Three of the four S. pneumoniae strains tested showed some growth in 22 mM lactic acid (Fig. 4B), and all strains displayed more growth in BHI medium supplemented with 11 mM l-lactic acid than in the D. pigrum KPL1914 CFCM, which had 7.5 mM D. pigrum-produced l-lactic acid (Fig. 4B). Furthermore, growth of S. pneumoniae alone under these conditions resulted in a higher concentration of l-lactic acid than did D. pigrum growth (see Fig. S1 in the supplemental material). Thus, the restriction of S. pneumoniae growth in D. pigrum CFCM is unlikely to have been due to production of lactic acid by D. pigrum. More likely, it reflected competition for nutrients since fresh medium was not added to the CFCM, which, therefore, would have a lower concentration of sugars than BHI broth. However, the possibility of D. pigrum production of a toxin and/or an antipneumococcal compound in BHI broth cannot be excluded. We also tested the in vitro effect of l-lactic acid on two strains of S. aureus. Both showed some growth in 33 mM lactic acid (Fig. 4C). Thus, D. pigrum did not produce enough l-lactic acid to restrict S. aureus growth under the tested conditions. Furthermore, growth of S. aureus alone resulted in a concentration of l-lactic acid similar to that seen with D. pigrum growth (Fig. S1). In contrast to the S. pneumoniae results, we would not expect depletion of sugars to have a large effect on S. aureus growth in D. pigrum CFCM given its ability to utilize a broader repertoire of energy sources, e.g., amino acids. Indeed, both S. aureus strains showed only a minimal decrease in growth in D. pigrum CFCM. This also revealed differences in D. pigrum production of the anti-S. aureus activity during growth on BHI agar medium (Fig. 3) versus growth in BHI broth (Fig. 4C). Levels of excretion of metabolites may differ during growth in liquid versus on agar medium, and the mechanism of the D. pigrum anti-S. aureus activity is yet to be identified.
FIG 4.
Lactate production by D. pigrum is insufficient to inhibit pathobiont growth. Strains of S. pneumoniae and S. aureus grew in the presence of higher levels of l-lactic acid than those produced by D. pigrum in vitro. (A) The concentration of l-lactic acid (mM) produced by three D. pigrum strains was measured after 24 h of gentle shaken aerobic growth in BHI broth at 37°C (n = 5) compared to the basal concentration of l-lactic acid in BHI medium alone (none). (B) The average growth (OD600) of 4 S. pneumoniae strains in D. pigrum KPL1914 CFCM or in unconditioned BHI broth supplemented with different concentrations of l-lactic acid measured after 19 to 20 h of static aerobic growth at 37°C (n = 4). (C) The average growth (OD600) of 2 S. aureus strains in D. pigrum KPL1914 CFCM or in unconditioned BHI broth supplemented with different concentrations of l-lactic acid measured after 19 to 20 h of shaken aerobic growth at 37°C (n = 4). In panels A and B, the average pH of the D. pigrum CFCM was 6.40 (±0.06). The pH of BHI without lactic acid (0 mM) was adjusted to match the pH of the CFCM, to control for any effect of pH alone. Average levels of growth of S. pneumoniae in CFCM and 11 mM l-lactic acid were analyzed independently for each individual strain using a Wilcoxon rank sum test. Dark bars represent medians, lower and upper hinges correspond to the first and third quartiles, and outlier points are displayed individually, except in panel A, where dots for all individual sample values are represented. *, none of the S. pneumoniae or S. aureus strains displayed growth in 55 mM l-lactate.
S. pneumoniae produces higher levels of l-lactic acid than S. aureus, D. pigrum, and C. pseudodiphtheriticum. The concentration (mM) of l-lactic acid produced by the assayed strains was measured after 24 h of aerobic growth with gentle shaking (∼50 to ∼60 rpm) in BHI broth at 37°C (n ≥ 4) compared to the basal concentration of l-lactic acid alone in BHI broth (none). Strains were grown and lactic acid concentrations determined as described in Materials and Methods for Fig. 4A with the following differences: (i) optical density was measured in a 96-well format for the lactate assay using a plate reader and (ii) we generated a standard curve of lactate concentrations using measurements from the plate reader to calculate the concentrations of lactate in the CFCM for each strain. Spn, S. pneumoniae 603; Sau, S. aureus JE2; Dpi, D. pigrum KPL1914; Cpsd, C. pseudodiphtheriticum KPL1989; none, BHI medium alone. Download FIG S1, PDF file, 0.04 MB (37KB, pdf) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
D. pigrum and C. pseudodiphtheriticum inhibit S. pneumoniae growth together but not alone.
Since the effects seen with C. pseudodiphtheriticum were positively associated with the presence of D. pigrum (Fig. 1), we investigated their combined effects on S. pneumoniae growth. Agar medium conditioned with a coculture of C. pseudodiphtheriticum strain KPL1989 and D. pigrum strain CDC4709-98 inhibited S. pneumoniae growth, whereas agar medium conditioned with a monoculture of either C. pseudodiphtheriticum or D. pigrum alone did not (Fig. 5; see also Fig. S2). This might have been due to cocultivation resulting either in a greater level of nutrient competition than monoculture of either commensal alone or in the production of a diffusible compound(s) toxic/inhibitory to S. pneumoniae by either D. pigrum or C. pseudodiphtheriticum or both D. pigrum and C. pseudodiphtheriticum when grown together. (Of note, cocultivation of D. pigrum and C. pseudodiphtheriticum in liquid BHI medium did not increase the level of l-lactic acid compared to growth of D. pigrum alone [Fig. S1].) Along with the Corynebacterium species enhancement of D. pigrum growth yield (Fig. 2) and the D. pigrum inhibition of S. aureus growth (Fig. 3), these data indicate that the negative associations of D. pigrum with S. aureus and S. pneumoniae are likely mediated by different molecular mechanisms.
FIG 5.
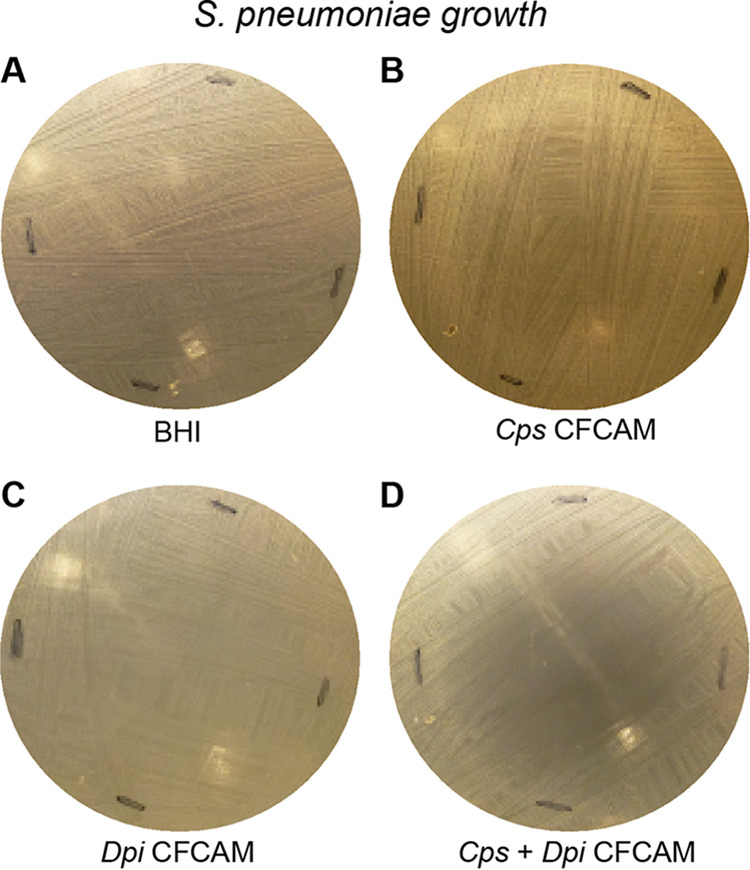
D. pigrum and C. pseudodiphtheriticum grown together but not D. pigrum alone inhibited S. pneumoniae in an in vitro agar medium-based assay. Representative images are shown of S. pneumoniae 603 growth on (A) BHI medium alone or on CFCAM from (B) C. pseudodiphtheriticum KPL1989, (C) D. pigrum KPL1914, or (D) both D. pigrum and C. pseudodiphtheriticum grown in a mixed inoculum (n = 4). To condition the medium, we cultivated D. pigrum and/or C. pseudodiphtheriticum on a membrane, which was then removed prior to spreading a lawn of S. pneumoniae. For monoculture, 100 μl of either D. pigrum or C. pseudodiphtheriticum, resuspended to an OD600 = 0.50, were inoculated onto the membrane. For mixed coculture, 50 μl of D. pigrum (OD600 = 0.50) were mixed with 50 μl of C. pseudodiphtheriticum (OD600 = 0.50) to yield a final volume of 100 μl for the inoculum, such that each bacterial species was present in the coculture inoculum at half the amount used for the corresponding monoculture inoculum. Images were cropped. Black marks indicate edges where the membrane had been.
Grown together, D. pigrum and C. pseudodiphtheriticum inhibit S. pneumoniae. A second set of representative images of S. pneumoniae 603 growth on (A) BHI medium alone or on CFCAM in the presence of (B) C. pseudodiphtheriticum KPL1989, (C) D. pigrum KPL1914, or (D) both D. pigrum and C. pseudodiphtheriticum in a mixed inoculum (n = 4) is shown. To condition the medium, we cultivated D. pigrum and/or C. pseudodiphtheriticum on a membrane, which was then removed prior to spreading a lawn of S. pneumoniae. Images were cropped. Black marks indicate edges of regions where the membrane had been. Download FIG S2, PDF file, 0.1 MB (90.1KB, pdf) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To learn more about the functional capacity and genomic structure of D. pigrum, we next turned to genomic analysis, which provided insights into some of the epidemiologic and phenotypic observations presented above.
The genomes of 11 D. pigrum strains reveal a small genome consistent with a highly host-adapted bacterium.
We analyzed one publicly available genome of D. pigrum (ATCC 51524) and sequenced 10 additional strains (see Text S1 in the supplemental material), which were selected to ensure representation of distinct strains (see Materials and Methods). To start, we focused on basic genomic characteristics. The 11 D. pigrum strain genomes had an average size of 1.86 Mb (median 1.88 Mb) with 1,693 predicted coding sequences (CDS) (see Tables A and B in Text S1). Approximately 1,200 CDS were core (see Fig. A and B and Table C in Text S1) and exhibited a high degree of nucleotide and amino acid sequence conservation (see Fig. C in Text S1). As shown in Text S1, we further analyzed synteny of two closed genomes (see Fig. D in Text S1) and BLAST ring comparisons (see Fig. E in Text S1) and constructed a core-gene-based phylogeny (see Fig. F in Text S1).
Supplemental genomic structure analysis. Download Text S1, PDF file, 2.5 MB (2.5MB, pdf) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
D. pigrum is a predicted auxotroph for amino acids, polyamines, and enzymatic cofactors.
The nasal environment is low in and/or lacking in key nutrients such as methionine (84), and the small genome size (1.86 Mb) of D. pigrum is consistent with reduced biosynthetic capacity. To gain insight into how D. pigrum functions in the nasal environment, we examined all 11 genomes, finding evidence of auxotrophy for some amino acids (e.g., methionine), polyamines (e.g., putrescine and spermidine), and enzymatic cofactors (e.g., biotin) across all strains. In turn, we identified putative degradation pathways (e.g., methionine), transporter pathways (e.g., polyamines and biotin), and salvage pathways (e.g., folate), suggesting that D. pigrum acquires some required nutrients exogenously. Section I in Text S2 contains additional details plus predictions of acquisition of metal cofactors. The auxotrophy predictions may be incomplete, since we were unable to grow D. pigrum in a chemically defined medium with all 20 amino acids that was putatively replete on the basis of these predictions. Apparent auxotrophy for a number of required nutrients indicates that these must be available either from the host or from neighboring microbes in human nasal passages, e.g., possibly from nasal Corynebacterium species.
Supplemental genomic functional predictions. Download Text S2, PDF file, 0.8 MB (865.4KB, pdf) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Whole-genome sequencing indicates that D. pigrum metabolizes carbohydrates to lactic acid via homofermentation.
D. pigrum produced lactate during in vitro cultivation (Fig. 4A). Lactic acid bacteria mainly perform either homofermentation or heterofermentation of carbohydrates (85). Therefore, we examined the genomic capacity of D. pigrum for carbohydrate metabolism (see section II in Text S2). D. pigrum genomes lacked genes required for a complete tricarboxylic acid cycle, which is consistent with fermentation. Moreover, we identified genes encoding a complete glycolytic pathway in all 11 strains that are consistent with homofermentation. All 11 strains harbored a predicted l-lactate dehydrogenase (EC 1.1.1.27) which catalyzes the reduction of pyruvate to lactate, regenerating NAD+ for glycolysis (GAPDH [glyceraldehyde-3-phosphate dehydrogenase] step), consistent with homofermentation to l-lactate as the main product of glycolysis.
The accessory genome of 11 D. pigrum strains contains a diversity of biosynthetic gene clusters predicted to encode antibiotics.
Lactic acid production alone appears insufficient to account for the negative in vitro associations of D. pigrum with S. aureus and with S. pneumoniae (Fig. 4). To delve further into the genetic capacity of D. pigrum for possible mechanisms of inhibition, we explored the accessory genome of the 11 sequenced strains. Consistent with a prior report (60), D. pigrum appears to be broadly susceptible to antibiotics (see section III in Text S2). What emerged in our analysis was a diversity of biosynthetic gene clusters (BGCs) (see Table A and Fig. A in Text S2), including a diversity of BGCs predicted to encode candidate antibiotics. Strikingly, although 10 of 10 strains tested displayed inhibition of S. aureus growth in vitro (Fig. 3), there was no single BGC common to all 10 strains that might encode a compound with antibiotic activity. On the basis of these data, we hypothesize that D. pigrum uses a diverse repertoire of BGCs to produce bioactive molecules that play key roles in interspecies interactions with its microbial neighbors, e.g., for niche competition, and potentially with its host. This points to a new direction for future research on the functions that underlie the positive associations of D. pigrum in human nasal microbiota with health and highlights the need to develop a system for genetic engineering of D. pigrum.
DISCUSSION
D. pigrum was shown to be associated with health in multiple genus-level compositional studies of human URT/nasal passage microbiota. In nasal passage microbiota data sets, we identified positive associations of D. pigrum with specific species of Corynebacterium in adults and children and a negative association of D. pigrum with S. aureus in adults (Fig. 1). We observed phenotypic support for these associations. First, unilateral cooperation from three common nasal Corynebacterium species enhanced D. pigrum growth yields (Fig. 2). Second, D. pigrum inhibited S. aureus (Fig. 3). Our genomic analysis revealed auxotrophies consistent with D. pigrum reliance on cocolonizing microbes and/or the human host for key nutrients. Genomic analysis also showed an aerotolerant anaerobe that performs homofermentation to lactate. However, D. pigrum lactate production (Fig. 4A) was insufficient to inhibit either S. pneumoniae (Fig. 4B) or S. aureus (Fig. 4C) and therefore is not the sole contributor to negative associations with S. pneumoniae and S. aureus in vivo. Consistent with the reports of a negative association between D. pigrum (usually in conjunction with Corynebacterium) and S. pneumoniae, we observed that cocultivation of D. pigrum and C. pseudodiphtheriticum produced a diffusible activity that robustly inhibited S. pneumoniae (Fig. 5; see also Fig. S2 in the supplemental material) whereas monoculture of either did not. Finally, we uncovered a surprisingly diverse repertoire of BGCs in 11 D. pigrum strains, revealing potential mechanisms for niche competition that were previously unrecognized. These data mark a significant advance in the study of D. pigrum and set the stage for future research on molecular mechanisms.
The in vitro interactions of D. pigrum with S. aureus and with S. pneumoniae support inferences from composition-level microbiota data of competition between D. pigrum and each pathobiont. However, these interactions differed in vitro. D. pigrum alone inhibited S. aureus, but D. pigrum plus C. pseudodiphtheriticum, together, robustly inhibited S. pneumoniae. These results point to a more complex set of interactions among these specific bacterial members of the human nasal microbiota which likely exists in the context of a network of both microbe-microbe and microbe-host interactions. To date, mechanisms for only a few such interactions have been described. For example, a C. accolens triacylglycerol lipase (LipS1) releases antipneumococcal free fatty acids from model host surface triacyclglycerols in vitro, pointing to habitat modification as a possible contributor to S. pneumoniae colonization resistance (33).
Multiple mechanisms could result in D. pigrum inhibition of S. aureus in vitro, including nutrient competition and excretion of a toxic primary metabolite or of an anti-S. aureus secondary metabolite (i.e., an antibiotic). Initial bioassay-guided fractionation approaches failed to identify such a mechanism. However, the data showing the existence of diverse repertoires of BGCs among the 11 D. pigrum strains are intriguing because they include predicted bacteriocins, including lanthipeptides. For example, 4 of the 11 strains harbored putative type II lanthipeptide biosynthetic gene clusters. These clusters are characterized by the presence of the LanM enzyme, containing both dehydration and cyclization domains needed for lanthipeptide biosynthesis (86). Alignment of these enzymes with the enterococcal cytolysin LanM revealed conserved catalytic residues in both domains (87). Cleavage of the leader portion of the lanthipeptide is necessary to produce an active compound, and the presence of peptidases and transporters within these BGCs suggests that these D. pigrum strains might secrete an active lanthipeptide, which could play a role in niche competition with other microbes. Additionally, 8 of the 11 D. pigrum genomes examined contain putative bacteriocins, or bactericidal proteins and peptides. Intriguingly, the D. pigrum strains exhibiting the strongest inhibition of S. aureus (CDC4709-98, CDC39-95, and KPL1914) (Fig. 3) were the only strains that contained both a lanthipeptide BGC and a bacteriocin, further indicating that D. pigrum may employ multiple mechanisms to inhibit S. aureus growth. Also, if both are required for the in vitro inhibition, this might explain the negative results from bioassay-guided fractionation. Mechanisms are coming to light that account for how other nasal bacteria interact with S. aureus. For example, commensal Corynebacterium species excrete a yet-to-be-identified substance that inhibits S. aureus autoinducing peptides blocking agr quorum sensing (QS), and shifting S. aureus toward a commensal phenotype (88). Also, the yet-to-be-identified mechanism of C. pseudodiphtheriticum contact-dependent inhibition of S. aureus is mediated through phenol-soluble modulins, the expression of which increases during activation of agr QS (89). Within broader Staphylococcus-Corynebacterium interactions, C. propinquum outcompetes coagulase-negative Staphylococcus, but not S. aureus, for iron in vitro using the siderophore dehydroxynocardamine, the genes for which are transcribed in vivo in human nostrils (90). Interphylum Actinobacteria-Firmicutes interactions also occur between Cutibacterium acnes and Staphylococcus species (reviewed in reference 14). For example, some strains of C. acnes produce an antistaphylococcus thiopeptide, cutimycin, in vivo and the presence of the cutimycin BGC is correlated with microbiota composition at the level of the individual human hair follicle (91). Of note, Actinobacteria competition with coagulase-negative Staphylococcus species could also have network-mediated (indirect) effects on S. aureus via the well-known competition among Staphylococcus species (reviewed in reference 92), which can be mediated by, e.g., antibiotic production (15–17, 19), interference with S. aureus agr QS (18, 20, 93, 94), or extracellular protease activity (95), among other means (14). Further rounding out the emerging complexity of microbe-microbe interactions in nasal microbiota, multiple strains of Staphylococcus, particularly S. epidermidis, inhibit the in vitro growth of other nasal and skin bacteria, including D. pigrum, via yet-to-be-identified mechanisms (16). The evidence described above points to a wealth of opportunity to use human nasal microbiota as a model system to learn how bacteria use competition to shape their community.
Direct cooperation could contribute to the observed positive associations between bacterial species in epidemiological microbiome studies. Conditioning medium with any of the three nasal Corynebacterium species positively associated with D. pigrum in vivo in human nasal microbiota (Fig. 1) enhanced the growth yield of some D. pigrum strains (Fig. 2). This is possibly accounted for by excretion of a limiting nutrient or by removal of a toxic medium component. The genomic predictions of auxotrophy might favor nasal Corynebacterium species providing cooperation to D. pigrum by excretion of a limiting nutrient. Indeed, mass spectrometry indicated that a number of nutrients are limiting in the nose (84).
There were several limitations of our study. First, we analyzed the genomes of 11 strains that were primarily isolated in the setting of disease. It is unclear whether these strains were contaminants or pathogenic contributors (60). However, D. pigrum strains are infrequently associated with disease (61–68). These 11 D. pigrum strains did not appear to encode potential virulence factors (see section III in Text S2 in the supplemental material), which is consistent with D. pigrum acting primarily as a mutualistic species of humans. Second, the ongoing search for a fully defined chemical medium permissive for D. pigrum growth precluded experimental verification of predicted auxotrophies and further investigation of how the presence of nasal Corynebacterium enhances D. pigrum growth yields. Third, the D. pigrum anti-S. aureus factor has eluded purification and identification efforts with standard chemistry approaches and D. pigrum is not yet genetically tractable, limiting the use of genetic approaches to identify it. Fourth, to date, there has been no animal model for nasal colonization with D. pigrum and Corynebacterium species, which stymies direct in vivo testing of the hypothesis of pathobiont inhibition and points to another area of need within the nasal microbiome field.
In summary, we validated in vivo associations from human bacterial microbiota studies with functional assays that support the hypothesis that D. pigrum is a mutualist with respect to its human host, rather than a purely commensal bacterium. Further, the data from these phenotypic interactions support the idea of a role for microbe-microbe interactions in shaping the composition of human nasal microbiota and, thus, the possibility of developing microbe-targeted interventions to reshape community composition. The next step will be to identify the molecular mechanisms of those interactions and to assess their role in the human host. Such work could establish the premise for future studies to investigate the therapeutic potential of D. pigrum as a topical nasal probiotic for use in patients with recurrent infections with S. pneumoniae, possibly in conjunction with a nasal Corynebacterium species, or with S. aureus, in conjunction with established S. aureus decolonization techniques (96).
MATERIALS AND METHODS
Species-level reanalysis of a pediatric nostril microbiota data set.
Laufer et al. analyzed nostril swabs collected from 108 children ages 6 to 78 months (26). Of these, 44% were culture positive for S. pneumoniae and 23% were diagnosed with otitis media. 16S rRNA gene V1-V2 sequences were generated using Roche/454 with primers 27F and 338R. We obtained 184,685 sequences from the authors, of which 94% included sequence matching primer 338R and 1% included sequence matching primer 27F. We performed demultiplexing in QIIME (97) (split_libraries.py), filtering reads for those ≥250 bp in length, those with a quality score of ≥30, and those with barcode type hamming_8. Then, we eliminated sequences from samples for which there were no metadata (n = 108 for metadata), leaving 120,963 sequences on which we performed de novo chimera removal in QIIME (USEARCH 6.1) (98, 99), yielding 120,274 16S rRNA V1-V2 sequences. We then aligned the 120,274 chimera-cleaned reads in QIIIME (PyNAST) (100), using eHOMDv15.04 (41) as a reference database, and trimmed the reads using “o-trim-uninformative-columns-from-alignment” and “o-smart-trim” scripts (101). A total of 116,620 reads (97% of the chimera-cleaned reads) were recovered after the alignment and trimming steps. After these initial cleaning steps, we retained only the 99 samples with more than 250 reads. We analyzed these data set of 99 samples with a total of 114,909 reads using MED (101) with minimum substantive abundance of an oligotype (-M) equal to 4 and maximum variation allowed in each node (-V) equal to 6 nucleotides (nt), which equals 1.6% of the 379-nucleotide length of the trimmed alignment. Of the 114,909 sequences, 82.8% (95,164) passed the -M and -V filtering and are represented in the MED output. Oligotypes were assigned taxonomy in R with the dada2::assignTaxonomy() function (an implementation of the naive Bayesian RDP classifier algorithm with a kmer size of 8 and a bootstrap value of 100) (102, 103) using the eHOMDv15.1 V1-V3 Training Set (version 1) (41) and a bootstrap value of 70. We then collapsed oligotypes within the same species/supraspecies, yielding the data shown in Table S2 in the supplemental material.
Species-level reanalysis of a pediatric nostril microbiota dataset (related to Fig. 1B). Download Table S2, XLSX file, 0.1 MB (92.7KB, xlsx) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbiota community comparison (Fig. 1).
The pediatric 16S rRNA gene V1-V2 data set analyzed at the species level (Table S2) and the HMP adult 16S rRNA gene V1-V3 data set previously analyzed at the species level (see Table S7 in reference 41) were used as input for the ANCOM, including all identified taxa (i.e., we did not remove taxa with low relative abundance). ANCOM (version 1.1.3) was performed using the presence or absence of D. pigrum, on the basis of the 16S rRNA gene sequencing data, as group definer. ANCOM default parameters were used (sig = 0.05, tau = 0.02, theta = 0.1, repeated = FALSE [i.e., Kruskal-Wallis test]), except that we performed a correction for multiple comparisons (multcorr = 2) instead of using the default no correction (multcorr = 3) (83). The Log relative abundance values for the taxa identified as statistically significant (sig = 0.05) are represented in Fig. 1 and are also available in Table S1.
Cultivation from frozen stocks.
Bacterial strains (see Table A in Text S1 in the supplemental material; see also Table S3) were cultivated as described here unless stated otherwise. Across the various methods, strains were grown at 37°C with 5% CO2 unless otherwise noted. D. pigrum strains were cultivated from frozen stocks on BBL Columbia colistin-nalidixic acid (CNA) agar with 5% sheep blood (BD Diagnostics) for 2 days. Corynebacterium species were cultivated from frozen stocks on BHI agar (C. pseudodiphtheriticum and C. propinquum) or on BHI agar supplemented with 1% Tween 80 (C. accolens) for 1 day. The resuspensions described below were made by harvesting colonies from agar medium and resuspending in 1× phosphate-buffered saline (PBS). Of note, we primarily use agar medium because in our experience D. pigrum exhibits more consistent growth on agar medium than in liquid medium. Likewise, growth on a semisolid surface is likely to better represent growth on nasal surfaces than would growth under the well-mixed conditions of shaking liquid medium.
Non-D. pigrum bacterial strains used in this study. Download Table S3, DOCX file, 0.03 MB (35.8KB, docx) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Preconditioning growth yield assays (Fig. 2).
To assess the growth yield of D. pigrum on a polycarbonate membrane atop media conditioned by Corynebacterium spp., each Corynebacterium strain was resuspended from growth on agar medium to an optical density at 600 nm (OD600) of 0.50 in 1× PBS. Then, 100 μl volumes of each resuspension were individually spread onto a 0.2-μm-pore-size, 47-mm-diameter polycarbonate membrane (EMD Millipore, Billerica, MA) atop 20 ml of either BHI agar for C. pseudodiphtheriticum and C. propinquum or BHI agar supplemented with triolein (BHIT) (CAS catalog no. 122-32-7, Acros) spread atop the agar medium, as previously described (33), for C. accolens. After 2 days of growth, the membranes with Corynebacterium cells were removed, leaving CFCAM. On each plate of CFCAM, we placed a new membrane onto which we spread 100 μl of D. pigrum cells that had been resuspended to an OD600 of 0.50 in 1× PBS. After 2 days, the membranes with D. pigrum were removed, placed in 3 ml 1× PBS, and subjected to vortex mixing for 1 min to resuspend cells. Resuspensions were diluted 1:10 six times, dilutions were inoculated onto BBL CNA agar with 5% sheep blood, and CFU were enumerated after 2 to 3 days of growth. To assess the growth yield of C. pseudodiphtheriticum on a polycarbonate membrane atop media conditioned by D. pigrum, strains KPL1914 and CDC4709-98 were grown for 2 days as described above. C. pseudodiphtheriticum KPL1989 growth yield was then measured as described above.
Growth of D. pigrum directly on BHI agar medium supplemented with triolein and conditioned by growth of nasal Corynebacterium species (Table 1).
Onto BHI agar supplemented with 200 U/ml of bovine liver catalase (C40-500MG, Sigma) (BHIC), we spread 50 μl of 100 mg/ml of triolein (BHICT). We then spread 50 μl of a resuspension (OD600 of 0.50) of each Corynebacterium strain onto a 0.2-μm-pore-size, 47-mm-diameter polycarbonate membrane placed atop 10 ml of BHICT agar in a 100-mm-by-15-mm petri dish. After 2 days, we removed each membrane with Corynebacterium cells, leaving CFCAM. Using a sterile cotton swab, we then spread a lawn of either D. pigrum (from cells resuspended to an OD600 of 0.50 in 1× PBS) or S. pneumoniae (taken directly from agar medium) onto the CFCAM. Each lawn was then grown for 1 to 2 days before documentation of growth or inhibition of growth was performed with digital photography.
Oleic acid disc diffusion assay (Table 2).
A lawn of D. pigrum or S. pneumoniae was spread onto 10 ml of BHIC agar using a sterile cotton swab as described above. Oleic acid (Sigma-Aldrich) was dissolved to reach final concentrations of 2 mg/ml, 5 mg/ml, and 10 mg/ml in ethanol, and then we added 10 μl of each to separate, sterile 0.2-μm-pore-size, 6-mm-diameter filter discs (Whatman), with 10 μl of ethanol alone added to a disc as a control. After the solvent was allowed to evaporate, filter discs were placed on the bacterial lawns, which were then allowed to grow for 1 day before measurement of zones of inhibition and photography were performed.
Growth of D. pigrum directly on versus atop a membrane on oleic acid-coated agar medium (Table 3).
Oleic acid was dissolved in 100% ethanol to a concentration of 5 mg/ml and then further diluted 10-fold 5 times in ethanol. For each dilution, 100 μl was spread on a separate plate of BHI agar medium. Next, 10 μl of D. pigrum KPL1914 and CDC4709-98, each resuspended to an OD600 = 0.3, were inoculated both directly onto the oleic acid-coated agar medium and atop of a 0.2-μm-pore-size, 47-mm-diameter polycarbonate membrane (EMD Millipore, Billerica, MA) on the same plate. After the plates had been maintained for 2 days at 37°C, we assessed and photographed the growth. In addition, for each dilution and strain, one spot on the membrane was resuspended in PBS to assess CFU counts after serial dilutions and plating on blood agar plates (see above).
D. pigrum-S. aureus side-by-side coculture assays (Fig. 3).
D. pigrum cells were grown from frozen stocks as described above. S. aureus JE2 was grown overnight on BBL Columbia CNA agar with 5% sheep blood. Bacterial cells were harvested with sterile cotton swabs and resuspended in sterile 1× PBS to a minimal OD600 of 0.3 for D. pigrum and 0.1 for S. aureus. Next, 5-μl drops were individually inoculated on BHI agar medium and incubated for 2 days for D. pigrum and 1 day for S. aureus. Then, 5-μl drops of resuspended bacteria to be screened for inhibition were inoculated at different distances from the pregrown bacteria. Inhibition was assessed daily and photographically documented.
Measurement of l-lactic acid concentration (Fig. 4A).
S. pneumoniae, S. aureus, C. pseudodiphtheriticum, and D. pigrum cells were grown from frozen stocks as described above. Cells were then harvested with a sterile cotton swab, resuspended to an OD600 of 0.50 in 1× PBS, and inoculated at 1:25 into BHI broth for overnight growth with gently shaking (∼50 to ∼60 rpm) at 37°C under atmospheric conditions. For mixed subcultures, i.e., C. pseudodiphtheriticum and D. pigrum, a 1:1 mixture was inoculated at 1:25 into fresh BHI broth. The overnight culture was then inoculated at 1:25 into fresh BHI broth and grown for 24 h at 37°C prior to measurement of the lactic acid concentration (mmol/liter) using a d-lactic acid/l-lactic acid kit (catalog no. 11112821035, R-Biopharm AG) per the instructions of the manufacturer and BHI as a negative control.
Growth of S. aureus and S. pneumoniae in D. pigrum cell-free conditioned liquid medium (CFCM in Fig. 4B and C).
After growth in BHI, as described for l-lactic acid measurement, the D. pigrum KPL1914 cells were removed with a 0.22-μm-pore-size sterile filter, yielding cell-free conditioned medium (CFCM). Each of S. aureus strains Newman and JE2 and S. pneumoniae strains TIGR4, DBL5, 603, and WU2 was grown on BBL Columbia CNA agar with 5% sheep blood for 1 day, harvested with a sterile cotton swab, resuspended to an OD600 of 0.30 in 1× PBS, inoculated at 1:100 into both D. pigrum CFCM and BHI broth, and grown for 19 to 20 h at 37°C in shaking (S. aureus; 50 rpm) or static (S. pneumoniae) culture under atmospheric conditions. Growth yield was quantified as OD600 absorbance.
Growth of S. aureus and S. pneumoniae in BHI broth supplemented with l-lactic acid (see lactic acid data in Fig. 4B and C).
Strains of S. aureus and S. pneumoniae were grown and harvested as described above for inoculation. BHI broth, supplemented with l-lactic acid (CAS no. 79-33-4; Fisher BioReagents) at various concentrations from 11 mM to 55 mM, was sterilized through a 0.22-μm-pore-size filter. After inoculation of each strain separately into BHI broth with l-lactic acid, cultures were grown as described above for growth in CFCM. Growth yield was quantified as OD600 absorbance.
Growth assay for S. pneumoniae on BHI agar medium conditioned by monoculture versus coculture of D. pigrum and/or C. pseudodiphtheriticum (Fig. 5; see also Fig. S2 in the supplemental material).
D. pigrum and C. pseudodiphtheriticum strains were grown from freezer stocks as described above. Cells were harvested with sterile cotton swabs and resuspended in sterile PBS to an OD600 of 0.5. We then spotted 100 μl of 1:1 mixed resuspension on a polycarbonate membrane (see above) on BHI agar medium containing 400 U/ml bovine liver catalase. After 2 days of growth, the polycarbonate membrane with D. pigrum and/or C. pseudodiphtheriticum was removed from each plate, leaving CFCAM. S. pneumoniae 603 (104) was grown overnight on BBL Columbia CNA agar with 5% sheep blood as described above, and, using a sterile cotton swab, a lawn was streaked onto the CFCAM and allowed to grow for 24 h. Growth/inhibition was assessed daily and photographically recorded. Imaging was difficult due to the transparency of S. pneumoniae lawns.
Statistical analyses.
R version 3.6.2 was used for statistical analysis and data visualization. The Wilcoxon rank sum test (equivalent to the Mann-Whitney test) was performed using wilcox.test() with paired = FALSE, alternative = “two.sided”.
Ethics approval and consent to participate.
We isolated D. pigrum KPL1914 and C. pseudodiphtheriticum KPL1989 from the nostril of an adult as part of a protocol to study the bacterial microbiota of the nostrils of healthy adults that was initially approved by the Harvard Medical School Committee on Human Studies (105) and subsequently approved by the Forsyth Institute Institutional Review Board.
Data availability.
We declare that all data that support the findings of this study are available within the paper (and its supplemental material); from publicly available repositories, i.e., GenBank (BioProject accession numbers PRJNA379818 and PRJNA379966); or from the corresponding authors upon reasonable request. All computer code for published tools used in this work is referenced in Materials and Methods; custom-made code (i.e., loop code) is described in Materials and Methods. Further details are available from the corresponding authors on reasonable request.
ACKNOWLEDGMENTS
We thank Richard R. Facklam and Lynn Shewmaker for providing strains; Joshua Metlay for providing data; Markus Hilty and Stephany Flores Ramos for manuscript edits; and Markus Hilty, Lindsey Bomar, Srikanth Mairpady Shambat, Annelies Zinkernagel, and members of the Lemon lab for helpful input and discussions.
This work was supported by the National Institutes of Health through the National Institute of General Medical Sciences (grant R01 GM117174 to K.P.L.) and the National Institute of Deafness and other Communication Disorders (grant R01 DC013554 to M.M.P.); by the Swiss National Science Foundation and Swiss Foundation for Grants in Biology and Medicine (grant P3SMP3_155315 to S.D.B.); by the Novartis Foundation for Medical-Biological Research (grant 16B065 to S.D.B.); and by the Promedica Foundation (grant 1449/M to S.D.B.). The funders had no role in the preparation of the manuscript or the decision to publish.
Our contributions were as follows: conceptualization, S.D.B., M.M.P., and K.P.L.; methodology, S.D.B., S.M.E., and M.T.H.; investigation, S.D.B., S.M.E., I.F.E., and Y.K.; interpretation of data, S.D.B., S.M.E., M.M.P., I.F.E., M.T.H., Y.K., and K.P.L.; visualization, S.D.B., S.M.E., and I.F.E.; writing of the original draft, S.D.B., S.M.E., and K.P.L.; editing and review, S.D.B., M.M.P., S.M.E., I.F.E., and K.P.L.; supervision, S.D.B. and K.P.L.; funding acquisition, S.D.B., M.M.P., and K.P.L.
Consent for publication is not applicable.
We declare no competing interests.
REFERENCES
- 1.Bogaert D, De Groot R, Hermans PW. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 2.von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 3.Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 4.Kluytmans J, van Belkum A, Verbrugh H. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10:505–520. doi: 10.1128/CMR.10.3.505-520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young BC, Wu CH, Gordon NC, Cole K, Price JR, Liu E, Sheppard AE, Perera S, Charlesworth J, Golubchik T, Iqbal Z, Bowden R, Massey RC, Paul J, Crook DW, Peto TE, Walker AS, Llewelyn MJ, Wyllie DH, Wilson DJ. 2017. Severe infections emerge from commensal bacteria by adaptive evolution. Elife 6:e30637. doi: 10.7554/eLife.30637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bode LG, Kluytmans JA, Wertheim HF, Bogaers D, Vandenbroucke-Grauls CM, Roosendaal R, Troelstra A, Box AT, Voss A, van der Tweel I, van Belkum A, Verbrugh HA, Vos MC. 2010. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med 362:9–17. doi: 10.1056/NEJMoa0808939. [DOI] [PubMed] [Google Scholar]
- 7.Lexau CA, Lynfield R, Danila R, Pilishvili T, Facklam R, Farley MM, Harrison LH, Schaffner W, Reingold A, Bennett NM, Hadler J, Cieslak PR, Whitney CG, Active Bacterial Core Surveillance Team. 2005. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. JAMA 294:2043–2051. doi: 10.1001/jama.294.16.2043. [DOI] [PubMed] [Google Scholar]
- 8.Wahl B, O'Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, Luksic I, Nair H, McAllister DA, Campbell H, Rudan I, Black R, Knoll MD. 2018. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000-15. Lancet Glob Health 6:e744–e757. doi: 10.1016/S2214-109X(18)30247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.GBD 2016 Lower Respiratory Infections Collaborators. 2018. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 18:1191–1210. doi: 10.1016/S1473-3099(18)30310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG, Jr.. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, Fowler VG, Jr.. 2019. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 17:203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO. 2012. Pneumococcal vaccines WHO position paper - 2012 - recommendations. Vaccine 30:4717–4718. doi: 10.1016/j.vaccine.2012.04.093. [DOI] [PubMed] [Google Scholar]
- 13.Schulfer A, Blaser MJ. 2015. Risks of antibiotic exposures early in life on the developing microbiome. PLoS Pathog 11:e1004903. doi: 10.1371/journal.ppat.1004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brugger SD, Bomar L, Lemon KP. 2016. Commensal-pathogen interactions along the human nasal passages. PLoS Pathog 12:e1005633. doi: 10.1371/journal.ppat.1005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brotz-Oesterhelt H, Grond S, Peschel A, Krismer B. 2016. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 16.Janek D, Zipperer A, Kulik A, Krismer B, Peschel A. 2016. High frequency and diversity of antimicrobial activities produced by nasal staphylococcus strains against bacterial competitors. PLoS Pathog 12:e1005812. doi: 10.1371/journal.ppat.1005812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, Latif H, Kim JN, Lockhart A, Artis K, David G, Taylor P, Streib J, Dorrestein PC, Grier A, Gill SR, Zengler K, Hata TR, Leung DY, Gallo RL. 2017. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 9:eaah4680. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paharik AE, Parlet CP, Chung N, Todd DA, Rodriguez EI, Van Dyke MJ, Cech NB, Horswill AR. 2017. Coagulase-negative staphylococcal strain prevents staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22:746–756.e5. doi: 10.1016/j.chom.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Sullivan JN, Rea MC, O'Connor PM, Hill C, Ross RP. 2019. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol Ecol 95:fiy241. doi: 10.1093/femsec/fiy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, Winter HL, Sanford JA, O'Neill AM, Liggins MC, Nakatsuji T, Cech NB, Cheung AL, Zengler K, Horswill AR, Gallo RL. 2019. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med 11:eaat8329. doi: 10.1126/scitranslmed.aat8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, Glose KA, Fisher EL, Hunt RL, Li B, Chiou J, Pharkjaksu S, Khongthong S, Cheung GYC, Kiratisin P, Otto M. 2018. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562:532–537. doi: 10.1038/s41586-018-0616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tano K, Hakansson EG, Holm SE, Hellstrom S. 2002. Bacterial interference between pathogens in otitis media and alpha-haemolytic Streptococci analysed in an in vitro model. Acta Otolaryngol 122:78–85. doi: 10.1080/00016480252775788. [DOI] [PubMed] [Google Scholar]
- 23.Coleman A, Cervin A. 2019. Probiotics in the treatment of otitis media. The past, the present and the future. Int J Pediatr Otorhinolaryngol 116:135–140. doi: 10.1016/j.ijporl.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Cardenas N, Martin V, Arroyo R, Lopez M, Carrera M, Badiola C, Jimenez E, Rodriguez JM. 2019. Prevention of recurrent acute otitis media in children through the use of Lactobacillus salivarius PS7, a target-specific probiotic strain. Nutrients 11:376. doi: 10.3390/nu11020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning J, Dunne EM, Wescombe PA, Hale JD, Mulholland EK, Tagg JR, Robins-Browne RM, Satzke C. 2016. Investigation of Streptococcus salivarius-mediated inhibition of pneumococcal adherence to pharyngeal epithelial cells. BMC Microbiol 16:225. doi: 10.1186/s12866-016-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. 2011. Microbial communities of the upper respiratory tract and otitis media in children. mBio 2:e00245-10. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. 2012. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, Bogaert D. 2014. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med 190:298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 29.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D. 2014. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 30.Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, Stegger M, Skov R, Andersen PS. 2015. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv 1:e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, Bochkov YA, Grindle K, Johnston SL, Gern JE, Sly PD, Holt PG, Holt KE, Inouye M. 2015. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosch A, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, de Steenhuijsen Piters WAA, de Groot PCM, Pernet P, Keijser BJF, Sanders EAM, Bogaert D. 2016. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 9:336–345. doi: 10.1016/j.ebiom.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. 2016. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7:e01725-15. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M, Wang R, Liao Y, Buijs MJ, Li J. 2016. Profiling of oral and nasal microbiome in children with cleft palate. Cleft Palate Craniofac J 53:332–338. doi: 10.1597/14-162. [DOI] [PubMed] [Google Scholar]
- 35.Salter SJ, Turner C, Watthanaworawit W, de Goffau MC, Wagner J, Parkhill J, Bentley SD, Goldblatt D, Nosten F, Turner P. 2017. A longitudinal study of the infant nasopharyngeal microbiota: the effects of age, illness and antibiotic use in a cohort of South East Asian children. PLoS Negl Trop Dis 11:e0005975. doi: 10.1371/journal.pntd.0005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosch A, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Biesbroek G, Kool J, Pernet P, de Groot PCM, Eijkemans MJC, Keijser BJF, Sanders EAM, Bogaert D. 2017. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am J Respir Crit Care Med 196:1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 37.Kelly MS, Surette MG, Smieja M, Pernica JM, Rossi L, Luinstra K, Steenhoff AP, Feemster KA, Goldfarb DM, Arscott-Mills T, Boiditswe S, Rulaganyang I, Muthoga C, Gaofiwe L, Mazhani T, Rawls JF, Cunningham CK, Shah SS, Seed PC. 2017. The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr Infect Dis J 36:e211–e218. doi: 10.1097/INF.0000000000001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasegawa K, Linnemann RW, Mansbach JM, Ajami NJ, Espinola JA, Petrosino JF, Piedra PA, Stevenson MD, Sullivan AF, Thompson AD, Camargo CA, Jr.. 2017. Nasal airway microbiota profile and severe bronchiolitis in infants: a case-control study. Pediatr Infect Dis J 36:1044–1051. doi: 10.1097/INF.0000000000001500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langevin S, Pichon M, Smith E, Morrison J, Bent Z, Green R, Barker K, Solberg O, Gillet Y, Javouhey E, Lina B, Katze MG, Josset L. 2017. Early nasopharyngeal microbial signature associated with severe influenza in children: a retrospective pilot study. J Gen Virol 98:2425–2437. doi: 10.1099/jgv.0.000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lappan R, Imbrogno K, Sikazwe C, Anderson D, Mok D, Coates H, Vijayasekaran S, Bumbak P, Blyth CC, Jamieson SE, Peacock CS. 2018. A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol 18:13. doi: 10.1186/s12866-018-1154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. 2018. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems 3:e00178-18. doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen Z, Xie G, Zhou Q, Qiu C, Li J, Hu Q, Dai W, Li D, Zheng Y, Wen F. 2018. Distinct nasopharyngeal and oropharyngeal microbiota of children with influenza A virus compared with healthy children. Biomed Res Int 2018:6362716. doi: 10.1155/2018/6362716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Copeland E, Leonard K, Carney R, Kong J, Forer M, Naidoo Y, Oliver BGG, Seymour JR, Woodcock S, Burke CM, Stow NW. 2018. Chronic rhinosinusitis: potential role of microbial dysbiosis and recommendations for sampling sites. Front Cell Infect Microbiol 8:57. doi: 10.3389/fcimb.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boelsen LK, Dunne EM, Mika M, Eggers S, Nguyen CD, Ratu FT, Russell FM, Mulholland EK, Hilty M, Satzke C. 2019. The association between pneumococcal vaccination, ethnicity, and the nasopharyngeal microbiota of children in Fiji. Microbiome 7:106. doi: 10.1186/s40168-019-0716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toivonen L, Hasegawa K, Waris M, Ajami NJ, Petrosino JF, Camargo CA, Jr, Peltola V. 2019. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax 74:592–599. doi: 10.1136/thoraxjnl-2018-212629. [DOI] [PubMed] [Google Scholar]
- 46.Camelo-Castillo A, Henares D, Brotons P, Galiana A, Rodriguez JC, Mira A, Munoz-Almagro C. 2019. Nasopharyngeal microbiota in children with invasive pneumococcal disease: identification of bacteria with potential disease-promoting and protective effects. Front Microbiol 10:11. doi: 10.3389/fmicb.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Man WH, Clerc M, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Kool J, Keijser BJF, Sanders EAM, Bogaert D. 2019. Loss of microbial topography between oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am J Respir Crit Care Med 200:760–770. doi: 10.1183/13993003.congress-2019.PA4995. [DOI] [PubMed] [Google Scholar]
- 48.Man WH, van Houten MA, Merelle ME, Vlieger AM, Chu M, Jansen NJG, Sanders EAM, Bogaert D. 2019. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med 7:417–426. doi: 10.1016/S2213-2600(18)30449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Man WH, van Dongen TMA, Venekamp RP, Pluimakers VG, Chu M, van Houten MA, Sanders EAM, Schilder AGM, Bogaert D. 2019. Respiratory microbiota predicts clinical disease course of acute otorrhea in children with tympanostomy tubes. Pediatr Infect Dis J 38:e116–e125. doi: 10.1097/INF.0000000000002215. [DOI] [PubMed] [Google Scholar]
- 50.Gan W, Yang F, Tang Y, Zhou D, Qing D, Hu J, Liu S, Liu F, Meng J. 2019. The difference in nasal bacterial microbiome diversity between chronic rhinosinusitis patients with polyps and a control population. Int Forum Allergy Rhinol 9:582–592. doi: 10.1002/alr.22297. [DOI] [PubMed] [Google Scholar]
- 51.de Steenhuijsen Piters WAA, Jochems SP, Mitsi E, Rylance J, Pojar S, Nikolaou E, German EL, Holloway M, Carniel BF, Chu M, Arp K, Sanders EAM, Ferreira DM, Bogaert D. 2019. Interaction between the nasal microbiota and S. pneumoniae in the context of live-attenuated influenza vaccine. Nat Commun 10:2981. doi: 10.1038/s41467-019-10814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Boeck I, Wittouck S, Martens K, Claes J, Jorissen M, Steelant B, van den Broek MFL, Seys SF, Hellings PW, Vanderveken OM, Lebeer S. 2019. Anterior nares diversity and pathobionts represent sinus microbiome in chronic rhinosinusitis. mSphere 4:e00532-19. doi: 10.1128/mSphere.00532-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Man WH, Scheltema NM, Clerc M, van Houten MA, Nibbelke EE, Achten NB, Arp K, Sanders EAM, Bont LJ, Bogaert D. 2020. Infant respiratory syncytial virus prophylaxis and nasopharyngeal microbiota until 6 years of life: a subanalysis of the MAKI randomised controlled trial. Lancet Respir Med S2216–S2600:30170–30479. doi: 10.1016/S2213-2600(19)30470-9. [DOI] [PubMed] [Google Scholar]
- 54.Krismer B, Weidenmaier C, Zipperer A, Peschel A. 2017. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol 15:675–687. doi: 10.1038/nrmicro.2017.104. [DOI] [PubMed] [Google Scholar]
- 55.Man WH, de Steenhuijsen Piters WA, Bogaert D. 2017. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bomar L, Brugger SD, Lemon KP. 2018. Bacterial microbiota of the nasal passages across the span of human life. Curr Opin Microbiol 41:8–14. doi: 10.1016/j.mib.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Esposito S, Principi N. 2018. Impact of nasopharyngeal microbiota on the development of respiratory tract diseases. Eur J Clin Microbiol Infect Dis 37:1–7. doi: 10.1007/s10096-017-3076-7. [DOI] [PubMed] [Google Scholar]
- 58.Aguirre M, Morrison D, Cookson BD, Gay FW, Collins MD. 1993. Phenotypic and phylogenetic characterization of some Gemella-like organisms from human infections: description of Dolosigranulum pigrum gen. nov., sp. nov. J Appl Bacteriol 75:608–612. doi: 10.1111/j.1365-2672.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 59.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, Relman DA. 2013. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laclaire L, Facklam R. 2000. Antimicrobial susceptibility and clinical sources of Dolosigranulum pigrum cultures. Antimicrob Agents Chemother 44:2001–2003. doi: 10.1128/aac.44.7.2001-2003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall GS, Gordon S, Schroeder S, Smith K, Anthony K, Procop GW. 2001. Case of synovitis potentially caused by Dolosigranulum pigrum. J Clin Microbiol 39:1202–1203. doi: 10.1128/JCM.39.3.1202-1203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoedemaekers A, Schulin T, Tonk B, Melchers WJ, Sturm PD. 2006. Ventilator-associated pneumonia caused by Dolosigranulum pigrum. J Clin Microbiol 44:3461–3462. doi: 10.1128/JCM.01050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin JC, Hou SJ, Huang LU, Sun JR, Chang WK, Lu JJ. 2006. Acute cholecystitis accompanied by acute pancreatitis potentially caused by Dolosigranulum pigrum. J Clin Microbiol 44:2298–2299. doi: 10.1128/JCM.02520-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lecuyer H, Audibert J, Bobigny A, Eckert C, Janniere-Nartey C, Buu-Hoi A, Mainardi JL, Podglajen I. 2007. Dolosigranulum pigrum causing nosocomial pneumonia and septicemia. J Clin Microbiol 45:3474–3475. doi: 10.1128/JCM.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnsen BO, Ronning EJ, Onken A, Figved W, Jenum PA. 2011. Dolosigranulum pigrum causing biomaterial-associated arthritis. APMIS 119:85–87. doi: 10.1111/j.1600-0463.2010.02697.x. [DOI] [PubMed] [Google Scholar]
- 66.Haas W, Gearinger LS, Hesje CK, Sanfilippo CM, Morris TW. 2012. Microbiological etiology and susceptibility of bacterial conjunctivitis isolates from clinical trials with ophthalmic, twice-daily besifloxacin. Adv Ther 29:442–455. doi: 10.1007/s12325-012-0023-y. [DOI] [PubMed] [Google Scholar]
- 67.Sampo M, Ghazouani O, Cadiou D, Trichet E, Hoffart L, Drancourt M. 2013. Dolosigranulum pigrum keratitis: a three-case series. BMC Ophthalmol 13:31. doi: 10.1186/1471-2415-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Venkateswaran N, Kalsow CM, Hindman HB. 2014. Phlyctenular keratoconjunctivitis associated with Dolosigranulum pigrum. Ocul Immunol Inflamm 22:242–245. doi: 10.3109/09273948.2013.841484. [DOI] [PubMed] [Google Scholar]
- 69.Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, Pieper DH. 2010. A poke into the diversity and associations within human anterior nare microbial communities. ISME J 4:839–851. doi: 10.1038/ismej.2010.15. [DOI] [PubMed] [Google Scholar]
- 70.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M, Sanders E. 2011. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Camarinha-Silva A, Jauregui R, Pieper DH, Wos-Oxley ML. 2012. The temporal dynamics of bacterial communities across human anterior nares. Environ Microbiol Rep 4:126–132. doi: 10.1111/j.1758-2229.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 72.Camarinha-Silva A, Wos-Oxley ML, Jauregui R, Becker K, Pieper DH. 2012. Validating T-RFLP as a sensitive and high-throughput approach to assess bacterial diversity patterns in human anterior nares. FEMS Microbiol Ecol 79:98–108. doi: 10.1111/j.1574-6941.2011.01197.x. [DOI] [PubMed] [Google Scholar]
- 73.Sakwinska O, Bastic Schmid V, Berger B, Bruttin A, Keitel K, Lepage M, Moine D, Ngom Bru C, Brussow H, Gervaix A. 2014. Nasopharyngeal microbiota in healthy children and pneumonia patients. J Clin Microbiol 52:1590–1594. doi: 10.1128/JCM.03280-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson SW, Knox NC, Golding GR, Tyler SD, Tyler AD, Mabon P, Embree JE, Fleming F, Fanella S, Van Domselaar G, Mulvey MR, Graham MR. 2016. A study of the infant nasal microbiome development over the first year of life and in relation to their primary adult caregivers using cpn60 universal target (UT) as a phylogenetic marker. PLoS One 11:e0152493. doi: 10.1371/journal.pone.0152493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez-Losada M, Alamri L, Crandall KA, Freishtat RJ. 2017. Nasopharyngeal microbiome diversity changes over time in children with asthma. PLoS One 12:e0170543. doi: 10.1371/journal.pone.0170543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chonmaitree T, Jennings K, Golovko G, Khanipov K, Pimenova M, Patel JA, McCormick DP, Loeffelholz MJ, Fofanov Y. 2017. Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One 12:e0180630. doi: 10.1371/journal.pone.0180630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luna PN, Hasegawa K, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, Piedra PA, Sullivan AF, Camargo CA, Jr, Shaw CA, Mansbach JM. 2018. The association between anterior nares and nasopharyngeal microbiota in infants hospitalized for bronchiolitis. Microbiome 6:2. doi: 10.1186/s40168-017-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caputo M, Zoch-Lesniak B, Karch A, Vital M, Meyer F, Klawonn F, Baillot A, Pieper DH, Mikolajczyk RT. 2019. Bacterial community structure and effects of picornavirus infection on the anterior nares microbiome in early childhood. BMC Microbiol 19:1. doi: 10.1186/s12866-018-1372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song C, Chorath J, Pak Y, Redjal N. 2019. Use of dipstick assay and rapid PCR-DNA analysis of nasal secretions for diagnosis of bacterial sinusitis in children with chronic cough. Allergy Rhinol (Providence) 10:2152656718821281. doi: 10.1177/2152656718821281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walker RE, Walker CG, Camargo CA, Jr, Bartley J, Flint D, Thompson JMD, Mitchell EA. 2019. Nasal microbial composition and chronic otitis media with effusion: a case-control study. PLoS One 14:e0212473. doi: 10.1371/journal.pone.0212473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Boeck I, Wittouck S, Wuyts S, Oerlemans EFM, van den Broek MFL, Vandenheuvel D, Vanderveken O, Lebeer S. 2017. Comparing the healthy nose and nasopharynx microbiota reveals continuity as well as niche-specificity. Front Microbiol 8:2372. doi: 10.3389/fmicb.2017.02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Steenhuijsen Piters WA, Bogaert D. 2016. Unraveling the molecular mechanisms underlying the nasopharyngeal bacterial community structure. mBio 7:e00009-16. doi: 10.1128/mBio.00009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mandal S, Van Treuren W, White RA, Eggesbo M, Knight R, Peddada SD. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26:27663. https://www.tandfonline.com/doi/full/10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krismer B, Liebeke M, Janek D, Nega M, Rautenberg M, Hornig G, Unger C, Weidenmaier C, Lalk M, Peschel A. 2014. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog 10:e1003862. doi: 10.1371/journal.ppat.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kandler O. 1983. Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 49:209–224. doi: 10.1007/BF00399499. [DOI] [PubMed] [Google Scholar]
- 86.Repka LM, Chekan JR, Nair SK, van der Donk WA. 2017. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem Rev 117:5457–5520. doi: 10.1021/acs.chemrev.6b00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dong SH, Tang W, Lukk T, Yu Y, Nair SK, van der Donk WA. 2015. The enterococcal cytolysin synthetase has an unanticipated lipid kinase fold. Elife 4:e07607. doi: 10.7554/eLife.07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. 2016. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol 7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hardy BL, Dickey SW, Plaut RD, Riggins DP, Stibitz S, Otto M, Merrell DS. 2019. Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. mBio 10:e02491-18. doi: 10.1128/mBio.02491-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stubbendieck RM, May DS, Chevrette MG, Temkin MI, Wendt-Pienkowski E, Cagnazzo J, Carlson CM, Gern JE, Currie CR. 2018. Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl Environ Microbiol 85:e02406-18. doi: 10.1128/AEM.02406-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Claesen J, Spagnolo J, Flores Ramos S, Kurita K, Byrd A, Aksenov lA, Melnik A, Wong W, Wang S, Hernandez R, Donia M, Dorrestein P, Kong H, Segre J, Linington R, Fischbach M, Lemon K. 2019. Cutibacterium acnes antibiotic production shapes niche competition in the human skin microbiome. bioRxiv doi: 10.1101/594010. [DOI]
- 92.Parlet CP, Brown MM, Horswill AR. 2019. Commensal staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol 27:497–507. doi: 10.1016/j.tim.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Otto M, Sussmuth R, Vuong C, Jung G, Gotz F. 1999. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett 450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 94.Otto M, Echner H, Voelter W, Gotz F. 2001. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun 69:1957–1960. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 96.Fritz SA, Hogan PG, Hayek G, Eisenstein KA, Rodriguez M, Epplin EK, Garbutt J, Fraser VJ. 2012. Household versus individual approaches to eradication of community-associated Staphylococcus aureus in children: a randomized trial. Clin Infect Dis 54:743–751. doi: 10.1093/cid/cir919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 100.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. 2015. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Malley R, Lipsitch M, Stack A, Saladino R, Fleisher G, Pelton S, Thompson C, Briles D, Anderson P. 2001. Intranasal immunization with killed unencapsulated whole cells prevents colonization and invasive disease by capsulated pneumococci. Infect Immun 69:4870–4873. doi: 10.1128/IAI.69.8.4870-4873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. 2010. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 1:e00129-10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ANCOM Log abundance values. (A) Adult nostril ANCOM data (plotted in Fig. 1A). (B) Pediatric nostril ANCOM data (plotted in Fig. 1B). Download Table S1, XLSX file, 0.02 MB (25.7KB, xlsx) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. pneumoniae produces higher levels of l-lactic acid than S. aureus, D. pigrum, and C. pseudodiphtheriticum. The concentration (mM) of l-lactic acid produced by the assayed strains was measured after 24 h of aerobic growth with gentle shaking (∼50 to ∼60 rpm) in BHI broth at 37°C (n ≥ 4) compared to the basal concentration of l-lactic acid alone in BHI broth (none). Strains were grown and lactic acid concentrations determined as described in Materials and Methods for Fig. 4A with the following differences: (i) optical density was measured in a 96-well format for the lactate assay using a plate reader and (ii) we generated a standard curve of lactate concentrations using measurements from the plate reader to calculate the concentrations of lactate in the CFCM for each strain. Spn, S. pneumoniae 603; Sau, S. aureus JE2; Dpi, D. pigrum KPL1914; Cpsd, C. pseudodiphtheriticum KPL1989; none, BHI medium alone. Download FIG S1, PDF file, 0.04 MB (37KB, pdf) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Grown together, D. pigrum and C. pseudodiphtheriticum inhibit S. pneumoniae. A second set of representative images of S. pneumoniae 603 growth on (A) BHI medium alone or on CFCAM in the presence of (B) C. pseudodiphtheriticum KPL1989, (C) D. pigrum KPL1914, or (D) both D. pigrum and C. pseudodiphtheriticum in a mixed inoculum (n = 4) is shown. To condition the medium, we cultivated D. pigrum and/or C. pseudodiphtheriticum on a membrane, which was then removed prior to spreading a lawn of S. pneumoniae. Images were cropped. Black marks indicate edges of regions where the membrane had been. Download FIG S2, PDF file, 0.1 MB (90.1KB, pdf) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental genomic structure analysis. Download Text S1, PDF file, 2.5 MB (2.5MB, pdf) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental genomic functional predictions. Download Text S2, PDF file, 0.8 MB (865.4KB, pdf) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Species-level reanalysis of a pediatric nostril microbiota dataset (related to Fig. 1B). Download Table S2, XLSX file, 0.1 MB (92.7KB, xlsx) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Non-D. pigrum bacterial strains used in this study. Download Table S3, DOCX file, 0.03 MB (35.8KB, docx) .
Copyright © 2020 Brugger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
We declare that all data that support the findings of this study are available within the paper (and its supplemental material); from publicly available repositories, i.e., GenBank (BioProject accession numbers PRJNA379818 and PRJNA379966); or from the corresponding authors upon reasonable request. All computer code for published tools used in this work is referenced in Materials and Methods; custom-made code (i.e., loop code) is described in Materials and Methods. Further details are available from the corresponding authors on reasonable request.