Abstract
In silico toxicology (IST) approaches to rapidly assess chemical hazard, and usage of such methods is increasing in all applications but especially for regulatory submissions, such as for assessing chemicals under REACH as well as the ICH M7 guideline for drug impurities. There are a number of obstacles to performing an IST assessment, including uncertainty in how such an assessment and associated expert review should be performed or what is fit for purpose, as well as a lack of confidence that the results will be accepted by colleagues, collaborators and regulatory authorities. To address this, a project to develop a series of IST protocols for different hazard endpoints has been initiated and this paper describes the genetic toxicity in silico (GIST) protocol. The protocol outlines a hazard assessment framework including key effects/mechanisms and their relationships to endpoints such as gene mutation and clastogenicity. IST models and data are reviewed that support the assessment of these effects/mechanisms along with defined approaches for combining the information and evaluating the confidence in the assessment. This protocol has been developed through a consortium of toxicologists, computational scientists, and regulatory scientists across several industries to support the implementation and acceptance of in silico approaches.
Keywords: In silico, in silico toxicology, computational toxicology protocols, (Q)SAR, expert alerts, expert review, genetic toxicology
Graphical Abstract
1. Introduction
The use of computational methods to assess the biological properties of chemicals is well established in many different industry sectors including the pharmaceutical, cosmetic, food, plant protection, biocides, and general chemical industries. (Marchant, 2012; Hasselgren et al., 2013) Computational methods are used during different stages of product development for purposes such as optimizing potency towards a protein target, determining the reactivity of a chemical, predicting the rate of transmembrane permeability, or predicting toxicological endpoints. In the field of toxicology, computational (in silico) methods are widely used to predict toxicological effects directly relevant to human health, as well as to support hazard and risk assessment activities or to prioritize chemicals for in vitro or in vivo testing. The first regulation to formally include the use of in silico approaches to address information requirements for the purposes of hazard identification and risk assessment was REACH (Registration, Evaluation, Authorisation and Restriction of Chemicals) (REACH, 2006). This regulation, which applies to chemicals manufactured or imported into the European Union where their import or use is not covered by other specified legislation. In addition, since 2014, with the implementation of the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) M7 guideline (ICH, 2014; ICH, 2017), regulatory authorities, such as the US Food and Drug Administration (US FDA), the Japanese Pharmaceutical and Medical Devices Agency (PMDA), and the European Medicines Agency (EMA) accept in silico assessments of the mutagenic potential of drug impurities. The ICH M7 guideline represented a milestone for the regulatory acceptance of computational methods for hazard assessment in pharmaceuticals and the implementation of the guideline has influenced the use of in silico assessments for other applications, such as the risk assessment of extractables and leachables, both for pharmaceuticals and for other industries. Other examples include the revision of the Toxic Substances Control Act (TSCA) to include predictive models and expert review as part of an overall assessment as well as the US FDA Center for Devices and Radiological Health (CDRH) issuing a guidance for industry on the use of International Standard ISO 10993–1 for biological evaluation of medical devices and indicating that in the absence of experimentally derived carcinogenicity information, structure-activity relationship modeling for these materials may be used (CDRH, 2016). These efforts and advancements in the adoption of in silico methods help to support the replacement, reduction and refinement (3Rs) of animal testing and are well aligned with the rapid screening approach that is common practice in the early development of chemical products (Ford, 2016; Stanton and Kruszewski, 2016).
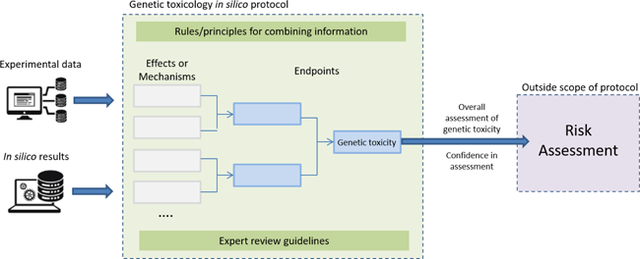
In a previous publication (Myatt et al., 2018), the use of in silico toxicology was discussed in more detail and highlighted that the field is, to some extent, hampered by lack of clarity concerning appropriate procedures related to the application, interpretation, and utilization of in silico approaches. To improve this situation, and to provide guidance, an in silico toxicology (IST) protocol template has been designed. The general IST protocol, as well as the protocol for genetic toxicology (the GIST protocol) described here, have been developed by an international consortium comprising over 50 organizations including industry, academia and government agencies, utilizing the extensive experience of its members and hence representing the state of the art of in silico toxicology application.
1.1. Genetic Toxicology In Silico Protocol Overview
Genetic toxicology (genotoxicity) concerns the effects induced by genetic alterations that may occur in somatic and/or germ cells following exposure to a chemical agent. Chemical agents can induce changes in DNA through direct or indirect interactions and the consequences of the genetic alterations may manifest as death and/or mutations in exposed cell populations. If somatic cells are affected, this might, for example, result in the development of cancer or neurodegenerative diseases (OECD, 2015). Alternatively, if the damage occurs in germ cells, it might manifest as reproductive defects or heritable changes that could eventually result in genetic diseases (OECD, 2015).
Genotoxicity testing for hazard identification and risk assessment is designed to characterize the ability of a chemical agent to induce genetic alterations (OECD, 2016a). A comprehensive assessment of genotoxicity incorporates a battery of tests that evaluate for:
Gene mutation (mutagenicity): Permanent, transmissible changes in the DNA that result from the induction of DNA adducts, insertions, inversions, and small deletions.
Clastogenicity: Structural chromosomal damage leading to sections of a chromosome being duplicated, deleted, or rearranged.
Aneugenicity: Numerical chromosomal abnormalities (aneuploidy) where an abnormal number of chromosomes is generated, often by disruption of the microtubule apparatus necessary for the orderly segregation of chromosomes during nuclear division.
In addition to test methods that evaluate these endpoints, tests directly detecting the presence of DNA damage (e.g., sister chromatid exchange (SCE) assay, alkaline comet assay), or the repair of certain types of DNA damage (e.g., unscheduled DNA synthesis (UDS) test) have been used. In addition, the upregulation of DNA repair enzymes and related stress response pathways that focus on individual genes (e.g., Gadd45a (Gentronix, 2018), p53 (Witt et al., 2017), ATAD5 (Fox et al., 2012)) or panels of genes (Li et al., 2015), or differential cytotoxicity using isogenic cell lines that have been knocked out for different DNA repair enzymes (Yamamoto et al., 2011) have also been utilized. Later in the section, these types of methods are referred to as “primary DNA damage”. Some of these test methods are no longer commonly used due to limitations in sensitivity (UDS) or the lack of a mechanistic underpinning (SCE), while others are generally used as screens to generate complementary information to provide a weight-of-evidence for mechanistic understanding.
Depending on the industry sector, slightly different combinations of tests may be required as outlined in published guidance documents to support regulatory data requirements, such as the ICH S2(R1) guidance for drugs (ICH, 2012), European Food Safety Authority (EFSA) guidance (EFSA, 2011) for food and feed safety assessment, the REACH guidance (ECHA, 2011) for registration of chemicals or ISO 10993–1 (CDRH, 2016) for evaluation of medical devices. In this publication, the intention is not to adhere to any specific guidance, but rather to base the assessments on a decision scheme (simple version shown in Figure 1), outlining a strategy for assessing genotoxicity based on coverage of the three major endpoints of genotoxicity as well as the generic term “primary DNA damage”, mentioned earlier in this section. Implicitly, this leaves room for alternatives in terms of specific study types. The commonly used genetic toxicology studies and the respective mechanisms/effects they identify are shown in Figure 2.
Figure 1.
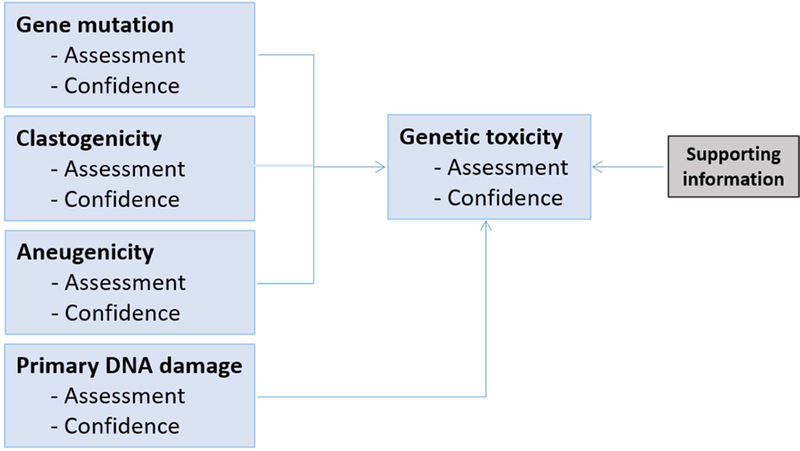
Basic decision scheme for genetic toxicity.
Figure 2.
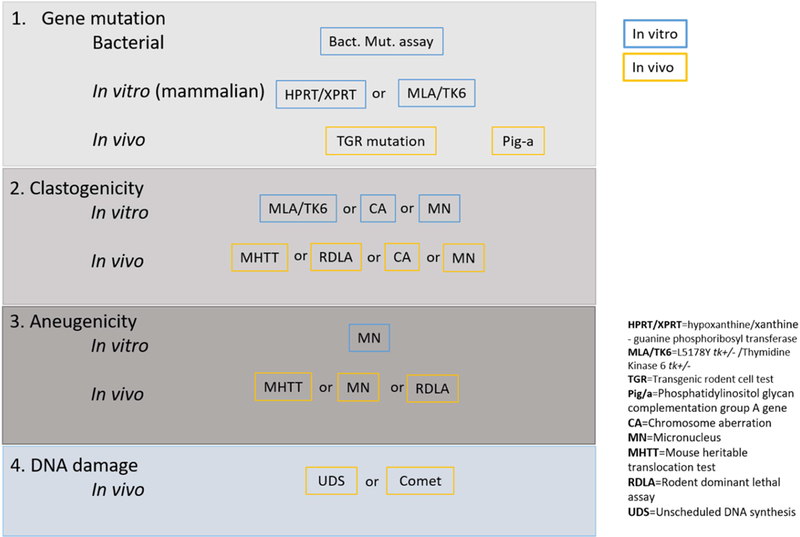
Common genetic toxicology mechanism/effects and corresponding studies
The purpose of this GIST protocol is to outline the process for determining whether a chemical agent is genotoxic or not, as well as the level of confidence related to the assessment. The process allows for the potential inclusion of additional information based on the results of other test methods or other supporting information, such as a history of safe use in food (Constable et al., 2007). The process of performing a risk assessment of a chemical agent will depend on many factors, such as the exposure conditions and in what context the agent is being investigated. Defined risk assessment is considered out of scope for the GIST protocol and should be performed in a situation dependent context although the GIST protocol can be used to support this activity.
2. In silico methodologies
2.1. Data availability for in silico models
The general protocol paper outlined some of the in silico methodologies that can be used to generate predictions (Myatt et al., 2018). These include i) rule-based (or “expert”) systems that identify the presence of a structural moiety, also referred to as a structural alert, that may indicate genotoxic potential, and ii) statistical (quantitative structure-activity relationship ((Q)SAR)) models that use a variety of molecular descriptors such as structural fragments or physicochemical properties to predict activity. Here, these two types are collectively referred to as “(Q)SAR” models. In addition, “read-across” (OECD, 2014) is a methodology that utilizes experimental or computed properties, such as physicochemical properties, together with structural similarity and experimental data for structural analogs to extrapolate from source chemical(s) to a target (query) chemical(s) (OECD, 2014).
The types of in silico tools that can be developed for a specific endpoint are, to a great extent, driven by the availability (amount and quality) of experimental data for model development, as well as the degree to which the chemicals of interest exert their toxicity via a common mechanism. In silico tools are most easily developed for endpoints with a well understood and similar mode of action for which a large number of data points are available. Bacterial mutagenicity is a relevant example of such an endpoint and consequently, in this area of genetic toxicology, the development of (Q)SAR models is the most established, largely due to the realization of an electrophilic mode of action for many genotoxic agents (Miller and Miller, 1981; Ashby and Tennant, 1988) and the availability of a large data set (> 2,000 compounds). Conversely, endpoints with less data or studies where the response can be due to several different mechanistic pathways (e.g., chromosome damage) are more challenging for (Q)SAR modeling.
Table 1 lists an estimate of the number of compounds in the public domain with associated genetic toxicology data, as published in the Leadscope toxicity database (Leadscope, 2018). In many cases, multiple results for the same assay and chemical will be available, sometimes with conflicting results and/or conclusions. Private or commercial organizations may have access to additional compounds with experimental data (e.g. from product development), which can be combined with publicly available data. There are some factors to consider, with respect to using experimental data for modeling or readacross: (1) data conflicts need to be resolved (this is not always possible) and experimental protocols need to be examined to ensure that only data measured and interpreted under similar conditions are merged, and (2) chemical structures need to be accurate. The general protocol provides more specific details regarding considerations when using data for modeling or read-across (Myatt et al., 2018). In the case of genetic toxicology, it may be relevant to look at modeling certain subsets of assay results. For example, data generated using the Escherichia coli (E. coli) WP2 uvrA pKM101 and Salmonella typhimurium TA102 (TA102) strains have sometimes been modeled separately (Stavitskaya, 2013) from data generated using the Salmonella strains TA98, TA100, TA1535, and TA1537, because the mechanistic basis of mutation induction is different for these two groups. The second group have GC base pairs at the primary reversion site, which are not as sensitive to detecting certain oxidizing mutagens, crosslinking agents and hydrazines which are instead better detected using the TA102 or E.coli strains which have an AT base pair at the primary reversion site (OECD, 1997a). An additional consideration is the distribution of the biological response. It is not unusual for the available data sets to be skewed so that one assay result classification (e.g., negative/positive) occurs much more frequently. Usually, inactive compounds are more abundant but regardless, the resulting imbalance will require specific strategies to be applied during the modeling procedure to avoid unbalanced predictions due to the prior probability resulting from the training set distribution.
Table 1.
Estimated number of compounds with genetic toxicology data in the public domain.*
| Assay/study type | Number of compounds |
|---|---|
| Bacterial mutagenicity | 10,440 |
| Chromosome aberration (in vitro) | 1,690 |
| Chromosome aberration (in vivo) | 360 |
| Mammalian cell mutagenicity (in vitro) | 2,390 |
| Micronucleus (in vitro) | 290 |
| Micronucleus (in vivo) | 1,850 |
Number of compounds with at least one experimental result listed as part of the Leadscope toxicity database (Leadscope, 2018).
2.2. In silico tools
2.2.1. Mutagenicity:
2.2.1.1. Bacterial mutagenicity:
The majority of in silico tools developed for this endpoint have been built using data generated in the bacterial reverse mutation (Ames) assay that relies primarily on Salmonella typhimurium tester strains. Historically, this assay has been viewed as the “gold standard” of mutagenicity testing and the first SARs relating chemical structure and bacterial mutagenicity using data generated using the Ames assay were published in 1988 by Ashby and Tennant (Ashby and Tennant, 1988). Several tools are available for modeling this endpoint, including expert rule-based systems and statistical models. The application of two complementary models, one rule-based and one statistical-based model, is described and recommended in the ICH M7 guideline for the evaluation of potential mutagenic impurities in pharmaceuticals (ICH, 2017) and by EFSA for dietary risk assessment (EFSA 2016).
2.2.1.2. Mammalian cell mutagenicity:
Both statistical models and rule-based systems utilizing mouse lymphoma assay (MLA) (L5178Y cells) data are available. Historically, application of these models often resulted in many false positive predictions, which was in part due to some of the experimental data from which the models were derived being liberally interpreted as evidence of mutagenicity. The criteria for interpretation of the experimental data were re-evaluated by Moore et al. (Moore et al., 2003; Moore et al., 2006), resulting in more stringent criteria, which led to changes to some of the experimental conclusions. In this context, and to ensure the best possible predictive power, it is important for the compilation of training sets to take the most contemporary data evaluation criteria into account. It should be noted that the currently available in silico MLA models do not provide information differentiating mutagenicity versus clastogenicity, and either or both endpoints may be implicated in a positive response in this assay.
For assays using other mammalian mutagenicity cell lines, such as those detecting mutations at hypoxanthine-guanine phosphoribosyl transferase (HPRT), and at a transgene of xanthineguanine phosphoribosyl transferase (XPRT) which are treated as equivalent by some industry sectors and regulatory agencies, there are currently not enough data available to generate useful models, although these assays may be referenced in expert systems. This also applies to in vivo mutagenicity studies. In addition to referencing them in expert systems, such data can be used for read-across to support a weight-of-evidence scenario, if they are available. It is expected that there will eventually be enough data available in the public domain to support model development.
2.2.2. Clastogenicity:
Both statistical and rule-based tools for in vitro and in vivo clastogenicity are available as commercial and free tools. Clastogenicity can result from numerous and diverse mechanisms of action (Bender et al., 1974; Snyder, 2000; Kaina, 2004; Snyder, 2010). Furthermore, cytotoxicity can confound the results of in vitro clastogenicity assessments (Kirkland et al., 2007b; Parry et al., 2010b; Galloway et al., 2011; Honda et al., 2018). Consequently, it is challenging to build highly predictive in silico models for this endpoint. In addition, the supporting datasets are mostly quite small and therefore the applicability domain of these models is often limited from a chemical space perspective. In general, the in silico models are better at identifying reactive compounds that damage DNA directly, thereby leading to clastogenicity, than they are at correctly predicting compounds involved in indirect, non-DNA-reactive effects leading to clastogenicity (e.g., off-target interactions disturbing cellular homeostasis or non-covalent intercalation between DNA base pairs). For better prediction of indirect, non-DNA-reactive effects, supplemental structural similarity searching or the use of specific models for the prediction of off-targets known to be involved in clastogenic effects can provide additional important information (Olaharski et al., 2009; Hsu et al., 2018).
2.2.2.1. In vitro chromosomal aberration:
The majority of available data in the public domain has been generated using Chinese hamster ovary (CHO) or Chinese hamster lung (CHL) cell lines. Initially, in the 1980’s, it appeared that the two cell lines differed significantly in their sensitivity with respect to identifying genotoxic compounds but subsequent in-depth comparisons demonstrated that the apparent differences were due simply to when cells were sampled after the start of exposure (e.g., (Sofuni et al., 1990; Galloway et al., 2011). Currently, from a regulatory context data generated using these two cell lines are interchangeable, as well as those using other mammalian cell lines such as human peripheral blood lymphocytes, as long as the same protocol is followed (OECD, 2016b).
2.2.2.2. In vitro micronucleus:
Few data following a standardized protocol are available in the public domain for this endpoint, due to the relatively recent adoption of an OECD test guideline (number 487) for this assay (OECD, 2010; OECD, 2016i), and as a consequence, statistical modeling would be limited with a narrow applicability domain. The derivation of expert/structural alerts is therefore the most promising in silico approach until more data are published; however, the in vitro micronucleus (MN) assay is becoming more widely used than the in vitro chromosomal aberration (CA) assay and it is assumed that the body of data will grow in the near future. It is also likely that some larger organizations have proprietary models for this endpoint. For the currently available public data, the majority of positive data have not been differentiated between clastogenicity and aneugenicity with regard to mechanism of action. In individual cases, read-across may be possible if suitable chemical analogs, e.g. as defined in the ReadAcross Assessment Framework (ECHA, 2008; ECHA, 2017), are available.
2.2.2.3. In vivo chromosomal aberration:
The number of available data points is small (mainly bone marrow studies performed in rats), since this assay is often reserved for mechanistic investigations rather than as a core genotoxicity assay, limiting the use of statistical models beyond specific compound classes. The derivation of expert alerts and the application of read-across, when experimental data for analogs are available, may be the most relevant methodologies for this endpoint.
2.2.2.4. In vivo micronucleus:
Most publicly available data have been generated using bone marrow and/or peripheral blood studies performed in mice or bone marrow studies performed in rats. The difference in experimental procedure is a result of the fact that micronucleated erythrocytes are removed from the blood by the spleen in rats but not mice (Dertinger et al., 2011b; Hayashi, 2016). The ability to use flow cytometry to measure the frequency of micronucleated erythrocytes has greatly increased test chemical throughput while making the data collected more robust (Hayashi, 2016). Furthermore, this approach has been used to evaluate micronuclei in immature erythrocytes in the blood of rats (MacGregor et al., 2006). Data are available in sufficient amounts to build a statistical model although it will have a limited applicability domain. Rat and mouse MN data should be analyzed separately as the different species have differences in responses (positive/negative) to some chemical agents. Different strains, sexes, or administration routes are usually not separated as there is not enough data to support this and the individual datasets would be too small. Read-across or rule-based systems may better address differences in response where such factors are thought to be important and if they can be related to certain chemical classes in a systematic manner. Historically, the increases in MN formation in vivo have not been evaluated to determine if the response is due to clastogenicity or aneugenicity. Any in silico model built using these data will therefore not be specific as to the nature of the type of chromosomal changes, but rather the endpoint of the assay, MN formation per se. In some cases, the mechanism of MN formation can be inferred by combined interpretation with other assay results or specific staining techniques (e.g., kinetochore staining) (Hennig et al., 1988) or more recently based on size distribution using flow cytometric methods (Torous et al., 1998b). A specific example of a combined interpretation would be a negative prediction for in vitro or in vivo CA but a positive in vitro or in vivo MN prediction, leading to an overall prediction that clastogenic effects are unlikely but that aneugenic effects are possible. It is clear that interpretation of experimental data could lead to such a conclusion and although more uncertain, one could in theory interpret the in silico results in a similar manner. It would also be possible to look at the most similar examples in the training set and in a read-across approach determine what the mechanism might be. Since the availability of data is scarce and because the underlying mechanism has rarely been determined in historical data, this is not usually feasible with the current body of data available for modeling.
2.2.3. Aneugenicity:
Historically, data generated using the in vitro and in vivo MN assays were not routinely evaluated in such a way that the mechanism of MN formation could be determined. Consequently, it is often not possible to differentiate an aneugen from a clastogen when evaluating the majority of data published. The number of data points available for modeling where the mechanism has been unambiguously determined is small and would therefore not support statistical modeling. The limited data for this endpoint could be suitable for read-across if analogs with mechanistic information could be found, or for deriving expert alerts. Although not regularly reported, an increase in the number of mononucleated cells with micronuclei can indicate an aneugenic mode of action (Rosefort et al., 2004) and could be used to differentiate between the two modes of action. With new automated methods (Torous et al., 1998a; Dertinger et al., 2011a) for identifying and scoring micronuclei becoming more widely used and special methods for differentiation between chromosome fragments (clastogenicity) and whole chromosomes (aneugenicity) like kinetochore staining or analysis of the micronuclei size, it is anticipated that this situation will improve in the next few years
2.2.4. Other endpoints:
Other methods relevant for experimental genotoxicity testing have not yet been generally accepted for making regulatory decisions and/or the data generated by these test methods are not available in sufficient amounts to build reliable in silico models (see (Mahadevan et al., 2011; Zeiger et al., 2015; Dearfield et al., 2017)). These test methods include those that evaluate the upregulation of specific DNA damage response elements such as GADD45A (Knight et al., 2009; Hughes et al., 2012), H2AX (Kim et al., 2011; Mishima, 2017), ATAD5 (Fox et al., 2012), and TP53 (Clewell et al., 2014; Witt et al., 2017) using reporter genes, or multiple DNA damage response elements evaluated using targeted transcriptomic platforms (Aubrecht and Caba, 2005; Sakai et al., 2014; Li et al., 2015; Corvi et al., 2016; Li et al., 2017); those that evaluate the differential responses in wild-type and isogenic DNA repair deficient DT-40 cells (Yamamoto et al., 2011; Nishihara et al., 2016) or TK6 cells (Saha et al., 2018); those that integrate multiple endpoints into an assessment of genotoxicity (Wilde et al., 2017; Bryce et al., 2018); and those that integrate DNA damage response into an overall assessment of toxicity using high throughput transcriptomic profiling to derive points of departure for risk assessment (Farmahin et al., 2017; Mav et al., 2018). Any potential tools built using any of these test methods will not be further discussed in the GIST protocol as they tend to be less extensively validated, even though they may be useful in some cases. Once such test methods are accepted by the wider community and their use is justified through validation exercises, in silico methods built from their data should be formally incorporated within this framework.
2.3. Applying in silico tools:
The practical aspect of applying in silico tools was discussed in detail in the general protocol (Myatt et al., 2018) including how to select models based on their performance, applicability domain, and model complementarity as well as the factors to consider when running chemicals through the models such as ensuring chemical drawing conventions are adhered to that follow any requirement s of the model developer. Hence, this will not be further discussed here. Criteria for the selection of suitable in silico methodologies, as well as reporting strategies were also detailed in the general strategy paper (Myatt et al., 2018).
2.4. Expert review of in silico tools
The application of in silico tools for hazard identification may involve an expert review of both the models and the predictions. It is important to determine that the models were built according to accepted criteria (Myatt, 2016) and using relevant training datasets. The endpoint training data used will dictate what can be predicted. For example, if only compounds tested in E. coli uvrA pKM101 and S. typhimurium TA102 are used to build a bacterial mutagenicity model, then the output is only relevant for these strains and may not be extrapolated to predict the outcome of a full OECD guideline compliant bacterial reverse mutation assay which requires at a minimum the inclusion of five bacterial strains. Ideally, to ensure that the data originates from comparable protocols, only experimental data generated using guideline-compliant conditions should be used. However, in practice, pragmatic approaches may need to be considered to ensure that the models cover a wide chemical space without any unnecessary compromise to data quality. As an illustration, experimental data from an assay involving a limited number of bacterial strains are often included in model building if the compound is shown to be mutagenic in at least one strain and as long as the other experimental conditions adhere to established guidelines. The justification for this is that the test guidelines only require one strain to be positive for the test article to be considered mutagenic. For compounds to be considered negative, it is preferable to have negative data from all recommended strains (OECD, 1997a). This is often not available and a certain degree of compromise in both the number of strains and data quality is usually accepted. For the purpose of this protocol, assessing the underlying data used in the model building is an important component of assigning a reliability score to the prediction, which will be discussed further in section 3.1.
3. Laboratory data
3.1. Experimental assays and studies
Tables 2 and 3 list in vitro and in vivo assays, respectively, that are frequently used to assess genotoxicity, as well as annotation of the mechanism(s) each assay may identify. For detailed descriptions of the experimental protocols, the OECD test guidelines may be consulted. Test methods that are no longer supported by the OECD are listed in the tables but are not discussed further. Data that were generated in the past using such assays may be considered appropriate for use only if no additional or higher relevance data are available. In general, it is also important when using historical data to evaluate if the relevant regulatory guidelines or data requirements have changed since the data were generated, so that they can be assigned a contemporary quality score. For example, changes to some of the in vitro protocols used in the pharmaceutical sector were made after a 2006 EURL ECVAM workshop (Kirkland et al., 2007a) on assessing false positive rates of mammalian in vitro tests. The new protocols introduced requirements for p53 competent cell lines and lowering of maximum tested concentrations, amongst other things, to reduce the number of unnecessary follow-up in vivo studies (Kirkland and Fowler, 2010; Parry et al., 2010a; Fowler et al., 2012a; Fowler et al., 2012b). A number of these recommendations were adopted under the OECD test guideline revisions performed in 2014–2016.
Table 2.
In vitro genetic toxicology assays.
| OECD Test Guideline | Name | Endpoint | Comments |
|---|---|---|---|
| 471 (OECD, 1997a) | Bacterial reverse mutation test (Ames) | Gene mutation | |
| 473 (OECD, 2016c) | In vitro mammalian chromosomal aberration test | Clastogenicity | |
| 476 (OECD, 2016f) | In vitro mammalian cell gene mutation test (HPRT/XPRT) | Gene mutation | |
| 487 (OECD, 2016i) | In vitro mammalian cell micronucleus test | Clastogenicity / Aneugenicity | Kinetochore staining or MN sizing required to differentiate between clastogenicity and aneugenicity |
| 490 (OECD, 2016k) | In vitro mammalian cell gene mutation tests using thymidine kinase gene (MLA/TK6) | Gene mutation / Clastogenicity | Mutant colony sizing may differentiate clastogenic and mutagenic events |
| 472 (OECD, 2015) | Genetic toxicology: Escherichia coli, reverse assay | Gene mutation | Deleted by OECD (integrated into OECD 471) |
| 479 (OECD, 1986a) | Genetic toxicology: In vitro sister chromatid exchange assay in mammalian cells | Chromosome aberrations | Deleted by OECD |
| 480 (OECD, 1986a) | Genetic toxicology: Saccharomyces cerevisiae, gene mutation assay | Gene mutation | Deleted by OECD |
| 481 (OECD, 1986b) | Genetic toxicology: Saccharomyces cerevisiae, mitotic recombination assay | Mitotic recombination | Deleted by OECD |
| 482 (OECD, 1986c) | Genetic toxicology: DNA damage and repair, unscheduled DNA synthesis in mammalian cells in vitro | Unscheduled DNA synthesis | Deleted by OECD |
| In vitro comet assay | DNA damage | Not listed by the OECD but commonly used in the pharmaceutical sector |
Table 3.
In vivo genetic toxicology assays.
| OECD Test Guideline | Name | Endpoint | Comments |
|---|---|---|---|
| 474 (OECD, 2016d) | Mammalian erythrocyte micronucleus test | Clastogenicity / aneugenicity | Kinetochore staining or MN sizing required to differentiate clastogens and aneugens |
| 475 (OECD, 2016e) | Mammalian bone marrow chromosome aberration test | Clastogenicity | |
| 478 (OECD, 2016g) | Genetic toxicology: Rodent dominant lethal test | Chromosome aberration by clastogenicity/aneugenicity (gene mutations) | Germ cell assay |
| 483 (OECD, 2016h) | Mammalian spermatagonial chromosome aberration test | Clastogenicity | Germ cell assay |
| 485 (OECD, 1986e) | Genetic toxicology: Mouse heritable translocation assay | Clastogenicity / aneugenicity | Not updated in 2014–16 revisions |
| 486 (OECD, 1997b) | Unscheduled DNA synthesis (UDS) test with mammalian liver cells in vivo | Unscheduled DNA synthesis | Not updated in 2014–16 revisions |
| 488 (OECD, 2013) | Transgenic rodent somatic and germ cell gene mutation assays | Gene mutation | Somatic and male germ cell assays. Many data reported before adoption of the OECD test guideline so critical data review essential |
| 489 (OECD, 2016j) | In vivo mammalian alkaline comet assay | DNA damage | Many data reported before adoption of the OECD test guideline so critical data review essential |
| 484 (OECD, 1986d) | Genetic toxicology: Mouse spot test | Gene mutation | Deleted by OECD |
| Pig-a | Gene mutation | No guideline adopted by the OECD but is under review for inclusion (Gollapudi et al., 2015). This assay may be considered as having high relevance in the pharmaceutical sector due to the inclusion in the ICH M7 Note 3 as a follow-up assay to a positive bacterial mutagenicity result |
3.2. Expert review of experimental data
An important step in any hazard identification process is a search for existing experimental data from endpoint-relevant in vitro and in vivo assays. In this context, it is pertinent to assess the quality of any identified data as well as its relevance to any of the mechanistic assessments related to the major genotoxicity endpoints. In the general protocol publication (Myatt et al., 2018), we proposed to assess data quality using Klimisch scores (Klimisch et al., 1997) as this is a widely accepted methodology used by ECHA, for example, in the Read-Across Assessment Framework (ECHA, 2017) and can readily be generated using the ToxRTool (European Commission, 2018b). Klimisch scores rank data from 1 to 4, depending on how the experiment was conducted (and reported), taking into consideration for example, whether the experiment was compliant with Good Laboratory Practice (GLP) and whether details of the experiment are available for review. These scores provide a consistent and reproducible way to classify the reliability of the test results.
An expert review of any identified experimental dataset may be performed to assign the appropriate Klimisch score. A detailed description of how this can be performed has been published by ECHA (ECHA, 2011). For assays relating to genotoxicity that are mentioned in this GIST protocol, the experimental conditions can be examined in relation to the relevant OECD test guideline. For test methods that no longer have a current OECD test guideline, such as the in vitro SCE assay in mammalian cells, a historical version of the guideline can be used to determine whether the experimental conditions were relevant at the time of data generation. Although these data are considered of lower relevance as use of the assay was discontinued for being scientifically questioned, there are situations where no other experimental data are available and they may be used in a weight-of-evidence scenario.
In addition, historical data that were generated under conditions described in a previous version of a test guideline can be used if the data were generated and reported in such a way that they can be reevaluated in accordance with the current guideline version and best practice (e.g., as described by the International Workshop on Genotoxicity Testing (Kirkland, 1994; Kirkland, 2000; Kirkland, 2003; Kirkland et al., 2007c; Kirkland et al., 2007d; Kirkland et al., 2011; Martus et al., 2015)). This may not always be possible, and an expert review will determine what reliability can be assigned on a case-by-case basis considering the particular chemical class, as well as the experimental details.
In situations where multiple experimental results are available for a test substance, different scenarios can be envisaged. If several experiments are found for the same assay that were performed in different laboratories or under slightly different (guideline compliant) experimental conditions, the data with the best Klimisch score may be given stronger weight. In cases where there are multiple conflicting results with the same Klimisch score and it cannot be determined through the expert review if one result is more reliable than the other(s), the results can either be considered unusable for hazard identification if they are of low quality, or a conservative approach might be taken where the occurrence of a positive result takes precedence. It would be critical in this situation to scrutinize the experimental conditions in detail, taking into account factors such as compound purity, potential cytotoxicity, solvent effects, etc. Alternatively, a weight-of-evidence approach could be taken with the final call being dependent on the judgement of a subject matter expert.
There are particular elements, or fields, relating to the experiment that are essential to review and document when assessing data. This practice supports an efficient and thorough review of the final assessments and ensures that the review is conducted in a consistent way. Table 4 lists the relevant fields for any in vitro test and shows an example of an Ames assay result for fluorobenzene. Table 5 lists the required fields for an in vivo MN assay with bosutinib as an example.
Table 4.
Relevant fields to document for an in vitro assay. Example: bacterial mutation assay
| Compound identifier: | CASRN: 462–06-6; Fluorobenzene; Benzene, fluoro- |
| Compound purity: | Not reported |
| Compound solubility: | Not reported |
| Solvent | DMSO |
| Study call: | Positive |
| Title: | Genetic Toxicity Evaluation of Fluorobenzene in Salmonella/E.coli Mutagenicity Test or Ames Test. Study 639736 |
| Reference: | https://tools.niehs.nih.gov/cebs3/ntpViews/?activeTab=summary&studyNumber=639736 |
| Study type: | Bacterial mutagenicity (Ames) |
| Source: | National Toxicology Program |
| Species: | Salmonella typhimurium |
| Strain/Cell type (Number of tests): | TA98 (N=6); TA100 (N=7); TA1535 (N=2) |
| Metabolic activation (Number of tests): | Absent (N=4); Present (N=11) |
| Metabolic activation system: | Aroclor 1254 treated rat liver S-9 fraction (30%), Aroclor 1254 induced hamster liver S-9 fraction (30%) |
| Dose summary: | 0 – 1666 µg/plate; 0 – 750 µg/plate |
| Toxicity: | No cytotoxicity reported |
| Method: | Pre-incubation; Plate test – vapor from liquid |
| Controls used: Strain (wo S9/w S9): | Positive controls: TA98 (4-Nitro-o-phenylenediamine/2-aminoanthracene), TA100 (Sodium azide/2-aminoanthracene), TA1535 (Sodium azide/2-aminoanthracene) |
| Control values within historical ranges: | Yes |
| OECD test guideline: | 471 |
| Current guideline compliance | No, insufficient bacterial tester strains used |
| Study Report: | https://tools.niehs.nih.gov/cebs3/ntpViews/?activeTab=detail&studyNumber=639736&reportFormat=XLS |
| GLP compliance: | No |
| Year conducted | 1991 |
| Klimisch score: | 3 |
| Rationale for reliability incl. deficiencies: | Not tested up to guideline recommended concentrations and insufficient number of strains and not tested according to GLP |
Table 5.
Relevant fields to document for an in vivo assay: example in vivo micronucleus
| Compound identifier: | CASRN: 918639–08-4; Bosutinib |
| Compound purity: | 99.49% |
| Compound solubility: | Not reported |
| Study call: | Negative |
| Title: | SKI-606: Single dose oral (gavage) bone marrow micronucleus study in male mice |
| Reference: | http://www.accessdata.fda.gov/drugsatfda_docs/nda/2012/203341Orig1s000PharmR.pdf#page=151 |
| Study type: | In vivo clastogenicity assay in rodent (micronucleus assay) |
| Source: | FDA CDER |
| Species (Number of subjects): | CD mouse (N=6) |
| Target cell/organ: | Bone marrow |
| Sex: | Male |
| TK parameters | Dose 2000 mg/kg, Cmax = 9811 ± 3998 ng/mL, tmax = 2.0 hr, AUC0–24 = 172495 ± 26050 ng*hr/mL |
| No observed adverse event level: | 2000 µg/kg (from single dose toxicology study) (unit as stated in original report); it is assumed that this is an error and the correct level is 2000 mg/kg |
| Dose summary: | 0, 500, 1000, and 2000 mg/kg as 10 ml/kg, single oral dose |
| Number of days of treatment: | 1 |
| Timepoints for tissue harvesting: | 24h; 48h |
| Controls used: | Positive control: Cyclophosphamide (50 mg/kg) |
| Current guideline compliance | Yes (OECD 474) |
| Study Report: | http://www.accessdata.fda.gov/drugsatfda_docs/nda/2012/203341Orig1s000PharmR.pdf |
| GLP compliance: | Yes |
| Year conducted | 2003 |
| Klimisch score: | 1 |
| Rationale for reliability incl. deficiencies: |
In addition to understanding the quality of the data, which relates to the technical aspect of the information, the scientific relevance to the toxicological endpoint result needs to be determined. “Relevance” was defined in the general protocol (Myatt et al., 2018) and relates to the predictivity of a specific toxicological effect or mechanism (gene mutation, clastogenicity, aneugenicity) to the toxicological endpoint (genotoxicity). As an example, the bacterial mutagenicity assay is considered highly relevant with respect to genotoxicity, whereas an in vitro CA test may be considered to have lower relevance (Custer, 2015). The rationale is related to how these tests are managed in a practical setting, where a bacterial mutagenicity assay is often not followed up with in vivo testing and, in many industries, a positive result in this assay is often considered sufficient to stop the development of a candidate active substance. Other industry sectors may adopt a different level of concern and a manufacturing chemical intermediate might, for example, be subject to further testing or used under strictly controlled conditions. In contrast, a positive result in an in vitro CA test can be de-risked or confirmed by performing an in vivo CA study. From a 3Rs point of view, it is desirable to perform in vivo testing as a last resort and to incorporate genotoxicity testing into general toxicology testing that may be required for other purposes. Generally, all in vivo studies are considered to have high relevance with respect to an overall assessment of genotoxicity. Conversely, tests where the OECD test guideline has been deleted (OECD, 2016a) have been assigned a low relevance. The relevance score is to some extent more subjective than the reliability score as different organizations and industry sectors, as well as regulatory agencies working to the data requirements of differing regulations, may apply different criteria in this respect. Even within an organization, different toxicologists may have individual preferences and experiences influencing their choice of assays. Furthermore, the chemical agent (with its physicochemical properties and structural aspects) may dictate which assays are relevant. This protocol reflects a general view of assay relevance, but it is recognized that there could be situations where an expert review may justify a different interpretation. The suitability of different assays in terms of follow-up actions, mechanisms identified by each assay and many other aspects have been reviewed and discussed in a publication by the Health and Environmental Sciences Institute (HESI) In Vitro Genetic Toxicity Testing Review Subgroup (Dearfield et al., 2011).
3.3. Sources of genetic toxicology data
Table 6 provides a non-exhaustive list of available sources of genetic toxicology data. There are also databases that comprise several sources. Individual databases will support different types of queries such as various identifiers (Chemical Abstracts Service registration number (CASRN), synonym or chemical name) and/or chemical structure. If possible, it is desirable to know the batch of compound that was tested, as well as the associated characterization data, as it is relevant to know the purity of the tested chemical. The presence of potential impurities is important; even small quantities of a mutagenic impurity may result in a false positive result. This type of information is not always available in public databases but can often be found in corporate databases. Structure searches should be performed with care, considering factors such as stereochemistry, tautomerism, salt form, and counter ions, for example. It might be necessary to search for both the parent compound and alternative forms when searching for a particular chemical if it is not known how the structures have been reported. It may be helpful to perform a substructure search, which looks for compounds with open substitution patterns, or a “family search” that will retrieve different salt forms and also analogs with different chirality. Some databases additionally provide regulatory authority classification with respect to mutagenicity and carcinogenicity; the International Agency for Research on Cancer (IARC), will for example, provide carcinogenicity classification and ECHA provides carcinogenicity, mutagenicity or reproductive toxicity (CMR) classifications.
Table 6.
Some sources of genetic toxicology data (non-exhaustive list).*
| Database | Description |
|---|---|
| ATSDR | Open access database from the Agency for Toxic Substances and Disease Registry (ATSDR) includes toxicological profiles for the hazardous substances including genotoxicity (ATSDR, 2018) |
| CCRIS | Chemical Carcinogenesis Research Information System (CCRIS), open access database covering chemical carcinogens and genotoxicants, including structures and experimental data, covering the period 1985 – 2011 (CCRIS, 2011) |
| Drugs@FDA | Open access database from US FDA CDER product approval reviews (FDA, 2018) |
| EPA Comptox Dashboard | Open access Distributed Structure-Searchable Toxicity (DSSTox) Database Network from the United States Environmental Protection Agency (EPA) including content from other sources (e.g., CPDB, ISSCAN, Tox21 and ToxCast) |
| ECHA | Open access European Chemicals Agency (ECHA) database containing experimental data and read across results for chemicals manufactured and imported in Europe as regulated by the REACH guidance (ECHA, 2018) |
| ELSIE | The Extractables and Leachables Safety Information Exchange (ELSIE) database is a collection of experimental data shared and accessed by consortium members (ELSIE, 2018) |
| EURL ECVAM | Open access Genotoxicity & Carcinogenicity consolidated database containing available genotoxicity and carcinogenicity data for Ames positive compounds (European Commission, 2018a) |
| GENE-TOX | GENE-TOX provides genetic toxicology (mutagenicity) test data from expert peer review of open scientific literature for more than 3,000 chemicals from the EPA (GENE-TOX, 1998). GENETOX covers the years 1991 – 1998. |
| IPS INCHEM | Open access International Program on Chemical Safety search for variety of summary documents (INCHEM, 2015) |
| IRIS | Open access data from the EPA in support of human health risk assessment, focusing on hazard identification and dose-response assessment (IRIS, 2015) |
| ISSCAN | Open access database on chemical carcinogens, including structures and experimental data from Istituto Superiore di Sanità (Benigni et al., 2008) |
| ISSMIC | Open access database on in vivo micronucleus mutagenicity results from Istituto Superiore di Sanità (Benigni and Bossa, 2008; Benigni et al., 2012) |
| JECDB | Open access Japanese Existing Chemical Data Base (JECDB) containing high production volume chemicals (JECDB, 2018) |
| Leadscope | Commercial genetic toxicity databases from numerous sources (including US FDA CDER product approval reviews, FDA CFSAN, National Toxicology Program (NTP), CCRIS) as well as ongoing data harvesting from the literature (Leadscope, 2018) |
| NTP – CEBS | Chemical Effects in Biological Systems (CEBS). Open access database of NTP results (NTP, 2018) |
| PAN | Open access Pesticide Action Network (PAN) Pesticide Database (PAN, 2018) |
| PharmaPendium | Commercial preclinical toxicity and clinical safety data from FDA and EMA approval documents (PharmaPendium, 2018) |
| RTECS | Registry of Toxic Effects of Chemicals (RTECS). Commercial database available through third parties (e.g., Leadscope) (Sweet et al., 1999; RTECS, 2018) |
| TOXNET/ChemIDPlus | Open access on-line toxicity search system from the US National Library of Medicine with access to archived versions of CCRIS and GENE-TOX (Wexler, 2001; TOXNET, 2018) |
| OECD QSAR Toolbox | Open access to database of genotoxicity as well as other toxicology data. (OECD, 2019) |
| VITIC | Commercial database from Lhasa Limited, including data from published and unpublished sources (VITIC, 2018) |
Modified from Amberg et al. (Amberg et al., 2016)
3.3. Other data references
In addition to the above listed databases, other sources of data, such as model training sets or other compilations of experimental data, can be searched for supporting information. In some cases, substances may have already undergone a risk assessment by a regulatory committee, and this output can be useful either directly or in a modified format in the hazard identification process. For instance, for the evaluation of bacterial mutagenicity the ICH M7(R1) addendum (ICH, 2017) provides detailed information on risk assessment of a number of chemicals and is applicable in the pharmaceutical sector. The addendum discusses acceptable intakes of certain chemical residues or impurities that are mutagens and/or carcinogens and that are common in pharmaceutical manufacturing. Another source is the “European Commission, Joint Research Centre (JRC) Genotoxicity & Carcinogenicity Consolidated Database of Ames Positive Chemicals” (European Commission, 2018a) (also listed in Table 6), which contains >700 unique chemical compounds that are bacterial mutagens and have a variety of additionally reported in vitro and in vivo genotoxicity and carcinogenicity data. This database contains an “overall call” based on a set of defined criteria for the reliability and quality of the data when results from more than one source are available.
The Joint FAO/WHO Expert Committee on Food Additives (JECFA) maintains a database of flavors, food additives, contaminants, toxicants, and veterinary drugs that have been reviewed with respect to human safety (JECFA, 2018). Similarly, there are databases with food and flavor substances that are Generally Recognized as Safe (GRAS). The FDA maintains the Select Committee on GRAS Substances (SCOGS)(GRAS, 2018) database containing reports of the opinion and conclusions on food substances while The Flavor and Extract Manufacturers Association of the United States (FEMA) (FEMA, 2018b) maintains the FEMA GRAS lists (FEMA, 2018a) of GRAS flavors. In addition, organizations like the National Institute for Occupational Safety and Health (NIOSH) (NIOSH, 2018), the Occupational and Health Administration (OSHA) (OSHA, 2018), the OECD (OECD, 2018) and various regulatory agencies have searchable data repositories that can be accessed.
For many commercial organizations, it may be difficult to find structural analogs for proprietary compounds in public databases due to differences in chemical space. In these cases it is relevant to search proprietary databases, as these often contain high quality sources of information. From the documentation and reporting point of view, as well as for any regulatory submission, this may be an issue as a thorough expert review and final assessment needs to be documented and disclosed to reviewers to enable their independent evaluation. Any analogs or other relevant structures should preferably be included in the final report for full transparency of the assessment.
4. Combined assessment of in silico predictions and experimental data
4.1. Reliability score
The general protocol (Myatt et al., 2018) provides detailed information on how to combine in silico predictions with experimental data, where these are available. The process will not be outlined in this publication, but involves expert review of the model(s), the prediction(s) as well as a review on the quality of the experimental data. In general, it is preferable that experimental data be of Klimisch score 1 or 2, depending on the situation, to be considered of high enough quality to support decision making. It is recognized that this is not always possible. However, depending on the use case, there could be situations where expert review and data quality assessment is not feasible and a lower level of confidence is acceptable, such as screening.
To enable a standardized method of performing an assessment of experimental results and in silico results together, an extension to the Klimisch score (Table 7) has been introduced to allow scoring of in silico components alongside experimental results using a Reliability Score (RS) (Myatt et al., 2018). Experimental data of Klimisch score 1 and 2 are essentially unchanged in their original Klimisch description but are referred to as RS1 and RS2. Furthermore, the lower quality Klimisch categories 3 and 4 have been placed in the lowest RS category of 5. This accommodates the use of in silico results of high quality in categories RS3 and RS4, illustrating their higher acceptability in certain regulatory contexts, compared to low quality experimental data or single, lower quality in silico result (RS5). For genetic toxicology, this is of particular importance for both REACH and ICH M7 applications, for example.
Table 7.
Reliability of toxicity assessments based on computational models and experimental data. (Myatt et al., 2018)
| Reliability Score | Klimisch Score | Description | Summary |
|---|---|---|---|
| 1 | 1 | Data reliable without restriction | • Well documented and accepted study or data from the literature • Performed according to valid and/or accepted test guidelines (e.g., OECD) • Preferably performed according to good laboratory practices (GLP) |
| 2 | 2 | Data reliable with restriction | • Well documented and sufficient • Primarily not performed according to GLP • Partially complies with test guideline |
| 3 | - | Expert Review | • Read-across • Expert review of in silico result(s) and/or Klimisch 3 or 4 data |
| 4 | - | Multiple concurring prediction results | |
| 5 | - | Single acceptable in silico result | |
| 5 | 3 | Data not reliable | • Inferences between the measuring system and test substance • Test system not relevant to exposure • Method not acceptable for the endpoint • Not sufficiently documented for an expert review |
| 5 | 4 | Data not assignable | • Lack of experimental details • Referenced from short abstract or secondary literature |
4.2. Toxicological effect or mechanism assessment
Toxicological effects are defined as observations derived from the experimental tests considered relevant for genetic toxicology (i.e., the in vitro and in vivo tests listed in Tables 2 and 3). An assessment will take into account all of the experimental and in silico information available for the query compound for each effect separately, in a weight-of-evidence scenario. A simple hypothetical example is shown in Figure 3.
Figure 3.
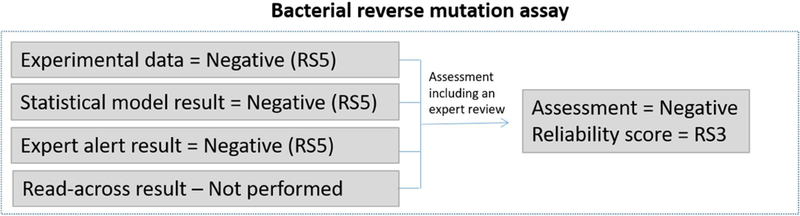
Effect assessment of the reverse bacterial mutation assay.
In this case, experimental data were found for the compound and it was reported to be inactive in a limited (too few strains) bacterial reverse mutation test. After expert review, it was concluded that the assay was run under appropriate conditions, but only in strains TA98 and TA100. The result is hence not sufficient to support a full assessment of bacterial mutagenicity and a Klimisch score of 3 is assigned to the data, which results in a reliability score of RS5. Two complementary in silico models for bacterial mutagenicity (incorporating E. coli/S. typhimurium TA102 and additional Salmonella data into both models) were applied and the compound was predicted to be negative in both models. The individual models have initial reliability scores of RS5 but since they concur, the combined score would be RS4. Further expert review showed that the predictions were of good quality and there were, for example, no reactive features identified. The in silico predictions are therefore assigned a reliability score of RS3. The weight-of-evidence for this compound supports the assessment that the compound is not a bacterial mutagen and the overall reliability score for bacterial gene mutation is set to RS3. For comparison, if the experimental results had been reported as positive in one of the two strains, the Klimisch score would still have been 3 and the initial reliability score would have been RS5. However, during expert review of the experimental data, it would have been appropriate to consider the result sufficient for an assessment of bacterial mutagenicity and to change the reliability score to RS3 as one positive strain is considered enough to make a positive call for the compound.
4.3. Toxicological endpoint assessments
Combining the genotoxic effect assessments that relate to a specific genotoxic endpoint is required to generate an overall endpoint call. Figure 4 shows a continuation of the hypothetical example from Figure 3 and illustrates the inclusion of a mammalian gene mutation result.
Figure 4.
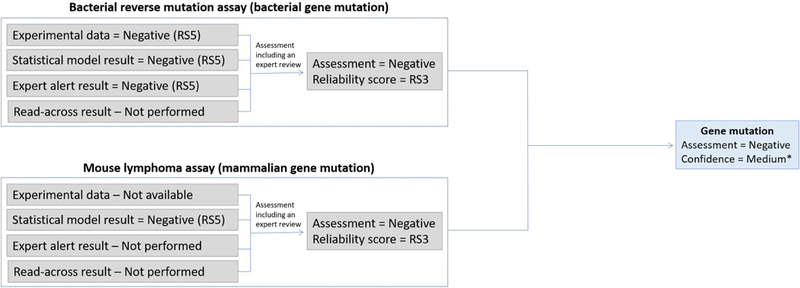
Combining information to assess the “gene mutation” endpoint. *The assignment of the “Confidence” is discussed in the following sections.
To perform this summary assessment, the concept of “Confidence” was introduced. Where “Reliability” relates to the quality of the experimental data or the in silico prediction and “Relevance” relates the assay to the mechanism or toxicological effect, “Confidence” combines the two parameters in addition to assessing the completeness (or coverage) of the information. It provides a method for merging information on the technical reliability of a result with the relevance of the assay from which it was derived, for predicting the toxicological endpoint being assessed. The determined confidence for each endpoint (gene mutation, clastogenicity, aneugenicity) will eventually propagate to the confidence for the overall call as to genotoxicity. As was discussed earlier with respect to relevance, the assigned confidence is somewhat subjective. To provide a starting point for how to combine terms, a set of rules has been devised for combining results, based on a conservative approach for combining relevance and reliability for the most commonly occurring components of genotoxicity hazard identification. This rule set is available in the supplementary material of this publication and can be adapted to accommodate organizational preferences or other needs. There may be times when it is not desirable to perform a full evaluation of all the genotoxicity endpoints. For example, only bacterial mutagenicity is required for an ICH M7 assessment. Subsets of the components can be used as appropriate in a situation dependent manner. A scheme including the genotoxic effects and endpoints that are amenable to the generation of in silico tools, using data currently available in the public domain, is shown in Figure 5. It is possible that private organizations have additional types of data that could also be used to generate in silico tools.
Figure 5.
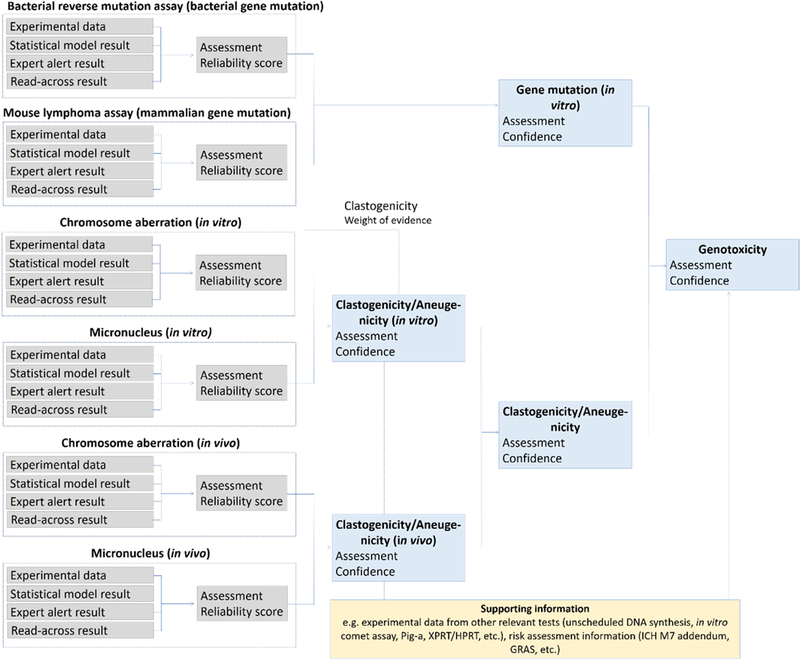
Current in silico components most relevant to genotoxicity.
An expert review of all the endpoint evaluations (described in section 4.4) may be performed to balance the relevance of each assay call to the overall genetic toxicology assessment. Depending on the use case, the confidence required may vary. For situations where false negatives may be acceptable and not be associated with health consequences, such as prioritization for more in-depth experimental testing, a lower level of confidence may be acceptable. However, in a human health hazard identification and risk assessment situation, a more conservative view is taken and higher confidence is required. In the general protocol (Myatt et al., 2018), we outlined the general principles around the influence that a particular level of confidence has.
4.4. Expert review of combined endpoint assessments
The expert review of genotoxic effects may include review of the in silico predictions and experimental data, as outlined earlier. The assessments might involve an expert review to weigh the individual assay results and in silico predictions, as well as any other information, such as experimental data for structural analogs or details that would influence the interpretation or translatability of a result. For example, a compound with antibacterial properties may be difficult to assay in a bacterial reverse mutation assay, due to the expected high cytotoxicity in a bacterial reverse mutation assay, and therefore, mammalian cell systems are usually recommended in these cases. Along similar lines, if such a compound is predicted with in silico tools to be negative in a bacterial mutation test, even with high reliability, but predicted by an in silico model to be a mammalian mutagen, the expert review may consider that the bacterial reverse mutation result may be misleading in the context of a combined “gene mutation” review. Even though the bacterial reverse mutation result would normally be considered to be of higher relevance due to the availability of more chemically diverse and abundant data for this endpoint, the mechanistic expert review could in this case rank the mammalian in silico prediction higher. It may at this point also be important to include information from primary DNA damage experiments (or models) to determine the mechanism of action. Table 8 includes some points to consider during an endpoint assessment. The expert review will also determine the level of confidence that can be placed in the endpoint summary.
Table 8.
Some elements of a mechanism expert review.
| Expert review elements | Considerations |
|---|---|
| 1. Chemical class assay response | Information such as if the compound belongs to a chemical class that may not be suited to particular assays, for example antibiotics in the bacterial reverse mutation assay, or interaction of the test substance with selected vehicle. In such cases, the bacterial mutagenicity result might be considered inappropriate and results from mammalian cell assays should be used. |
| 2. Mode of action | Combinations of assay results within a particular mechanistic class may provide information on the mode of action of a compound (e.g., different bacterial strains are specific for different types of mutations). |
| 3. Alerts that predict a particular mechanism | Some alerts may provide information on the mechanism through which a compound acts. |
4.5. Worked examples
A number of case studies that have been contributed by co-authors are discussed in the following section. For some of these, it is not possible to disclose the chemical structures as they are proprietary compounds. However, the included examples have been selected to show various aspects of the GIST protocol; emphasizing how the various model outputs and experimental data components can be fitted into this framework without judging the validity of the generated components.
4.5.1. Toxicological effect or mechanism examples
4.5.1.1. Acid chloride (bacterial gene mutation)
Figure 6 shows a case study of an acid chloride impurity which is being assessed for bacterial gene mutation potential for ICH M7 risk assessment. No experimental data could be found for the compound and two in silico tools, one statistical- and one rule-based, were applied. The prediction from the statistical model indicates that the compound may be a bacterial mutagen due to the presence of the acid halide functionality. The rule-based model gives an “Indeterminate” prediction and also highlights the acid halide functionality. Acid halides are a structural alert class for bacterial mutagenicity that were discussed recently (Amberg et al., 2015) and it was shown that with the exception of dimethylcarbamic chloride, the compounds tested and available for model building were active in the bacterial reverse mutation assay due to a reaction between the DMSO solvent and the test agent. When retested in other solvents, the majority of compounds show no mutagenic activity. Despite the positive and indeterminate in silico predictions, each with a reliability of RS5, an expert review revealed that the underlying data for the statistical model supporting the prediction are with high certainty false positives and the prediction was refuted. Expert review of the supporting text for the alert supports this outcome. The overall assessment of bacterial mutagenicity concludes that the compound is predicted to be inactive (negative) and the reliability score is set to RS3. The approach to this assessment is aligned with current ICH M7 guidance.
Figure 6.
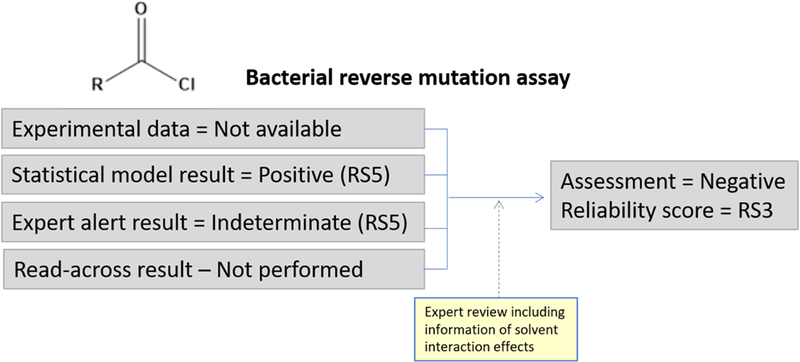
Assessment of an acid chloride compound
4.5.1.2. Drug impurity - API X (bacterial gene mutation)
There may be situations when an expert review can give an indication that the experimental results might not be correct. This is illustrated in the following example using Active Pharmaceutical Ingredient (API) X. API X was initially tested in the bacterial reverse mutation assay and found to have mutagenic activity. In contrast, as shown in Figure 7, the in silico predictions from both the statistical and the expert alert models predict API X to be inactive in the bacterial reverse mutation assay. An expert review of the information indicates that the models as well as the predictions appear robust and the reliability score which initially is set to RS4 due to two concurring models, is raised to RS3 after the expert review. In cases where experimental data are positive and in silico predictions are negative, the conservative approach would be to accept the positive experimental data, in which case the assessment would be positive with a reliability score of RS1, RS2, or RS5, depending on the quality of the experimental data. However, if the scientific review suggests that there is a valid reason to question the experimental result, the initial assessment for the compound could be Indeterminate, given the conflicting results from the experimental and in silico outputs, although this outcome would not be acceptable as a final conclusion from a drug regulatory standpoint. A reliability score is not assigned if the assessment is considered indeterminate. Given that the structure of API X is not predicted to be DNA reactive, it could be relevant to consider if there are other reasons for the observation of mutagenic activity related to the experimental procedures and/or the test article. One of the more frequently occurring reasons for an unexpected positive response in bacterial mutagenicity assays is the presence of a potent mutagenic impurity in the test article. In this particular case, an aldehyde was identified as a degradation product in API X and shown to be mutagenic. Follow-up testing of purified API X found it to be non-mutagenic and the bacterial gene mutation assessment would at this point be updated from the “Indeterminate” to “Negative” with a reliability score of RS2. Since the formation of the degradant could be avoided by modification of the synthetic route, it had no direct bearing on the classification of API X.
Figure 7.
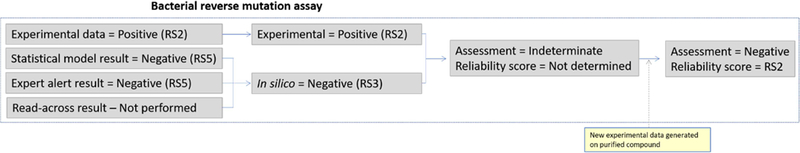
The conflicting in silico and experimental results of API X feeding into the overall bacterial gene mutation assessment.
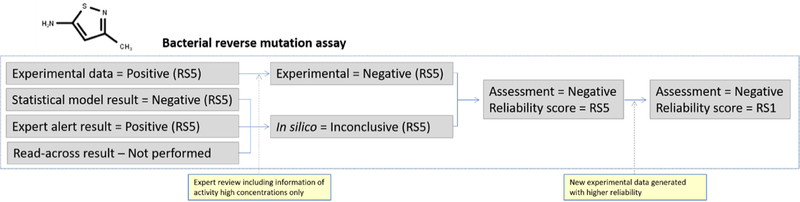
4.5.1.3. 3-Methyl-5-isothiazolamine (bacterial gene mutation)
Figure 8 shows the assessment components for 3-methyl-5-isothiazolamine related to bacterial gene mutation. Experimental data were available in the public domain for this compound where it was reported to have been tested in TA98, TA100, TA1535, TA1537 and TA1538 with and without metabolic activation using induced rat liver S9 and hamster liver S9 (Cameron et al., 1985). Further examination of the data revealed that the experiments were conducted under acceptable conditions and that the tested concentration range went to higher levels than normally required by OECD TG 471, the test guideline for the bacterial reverse mutation assay, but the compound was not tested in an E. coli or S. typhimurium TA102 strain, which is required to fulfill the current OECD test guideline. The standard maximum concentration is usually set to 5 mg/plate and this study reported maximum concentrations of 7.43 mg/plate. The data were initially assigned as positive with a Klimisch score of 3, indicating that the experiment was partially compliant with guidelines. However, when assessing the individual bacterial strain concentration responses, the biological relevance of the data was further questioned as the compound was only active in TA1538 (a strain not required by the OECD test guideline) at concentrations higher than the guideline recommended 5 mg/plate and only with hamster S9 metabolic activation. With rat S9 and at concentrations up to 5mg/plate, the compound was found to be inactive. At this point, an expert review of the data indicated that as the compound was negative at concentrations up to regulatory requirement of 5 mg/plate, the compound could potentially be viewed as negative with a reliability score of RS5 as this cannot be increased, considering that the compound was not tested in E. coli or S. typhimurium TA102 strains. Additionally, there is discrepancy seen with the two metabolic activation systems. In silico methods were applied to further refine the hazard identification. When reviewing these results, the statistical model output from a Salmonella model classified the compound as out of domain and the E. coli model predicted it to be negative. Review of the E. coli model results indicated that the prediction was not supported by many analogs or structural descriptors and is mainly driven by physicochemical properties. The expert alert model predicts 3methyl-5-isothiazolamine to be positive for bacterial gene mutation. In this case, however, there are compounds in the reference set that contain the thiazolamine functionality that the alert is based on, but they are not necessarily isothiazolamines. Additionally, further review shows that the majority of the reference structures also have other alerts such as aromatic nitro groups. At this point, there is contradictory information to consider: the low reliability (RS5) experimental result indicating the compound is negative up to 5 mg/plate but active at higher concentrations, and the inconclusive in silico results. By formally following the proposed scheme, it would be acceptable to view the compound as negative, but with a reliability score of RS5, as the expert review did not reveal evidence supporting a higher score. In a conservative scenario, if this compound, for example, is an impurity that has consequences for human safety, retesting the compound in a guideline acceptable study would be preferred. Indeed, when 3-methyl-5-isothiazolamine was retested according to the OECD guideline in a full 5-strain bacterial reverse mutation assay, with and without induced rat liver metabolic activation, the compound was found to be non-mutagenic (Ahlberg et al., 2016). At this point, the assessment could be updated with a “Negative” result with a reliability score of RS1 assigned. It should be noted that this assessment refers specifically to bacterial gene mutation and that any other available experimental data, such as MLA data, would be used to support the corresponding endpoint they relate to, which may or may not differ from the bacterial mutagenicity assessment. For a more comprehensive analysis of potential genotoxicity, such data may need to be considered and follow-up testing may need to be performed.
Figure 8.
Bacterial gene mutation assessment of 3-Methyl-5-isothiazolamine
4.5.2. Toxicological endpoint examples
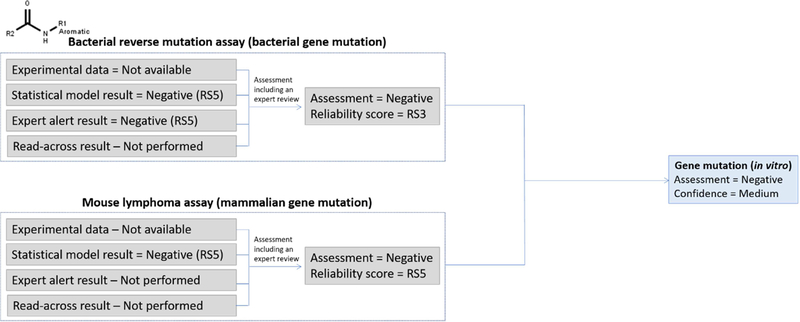
4.5.2.1. Aromatic amide (gene mutation)
Figure 9 shows the assessment of a compound containing an aromatic amide functionality. Bacterial gene mutation and mammalian gene mutation effects/mechanisms were identified as relevant to the assessment of the gene mutation endpoint. Two independent and concurring in silico models were run to predict bacterial gene mutation, one expert rule-based and the second statistical-based, and both model predictions were negative. An expert review was performed on the in silico model results and the review concluded that the predictions were well supported and there was sufficient evidence to increase the reliability to RS3 from the individual models’ scores of RS5. A single statistical model predicted the compound as negative for mammalian gene mutations (built using MLA training set data). An expert review was performed but the evidence concerning the prediction was not considered sufficient to raise the reliability score higher than RS5. The results from the bacterial and mammalian gene mutation endpoints were used as part of the assessment of the overall gene mutation potential. The confidence was assigned as “Medium” as outlined in the suggested set of rules in the supplementary information. It should be noted that this prediction itself refers to the in vitro gene mutation response. In a scenario where this result would feed into a framework supporting overall genotoxic potential, it would be pertinent to consider that certain aromatic amides and sulfonamides do not show activity in the bacterial assay due to the amide bond not being metabolized by S9, but may be active in an in vivo experiment.
Figure 9.
Gene mutation assessment of an aromatic amide compound
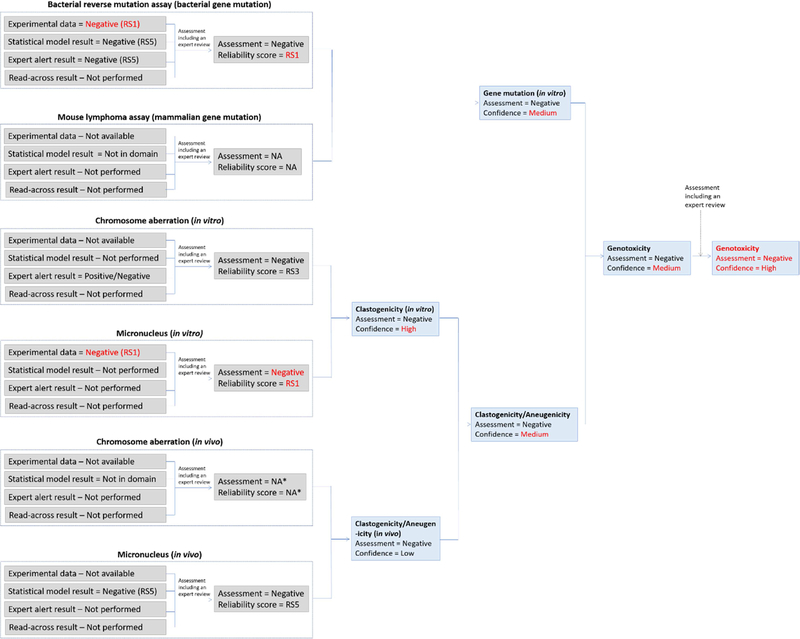
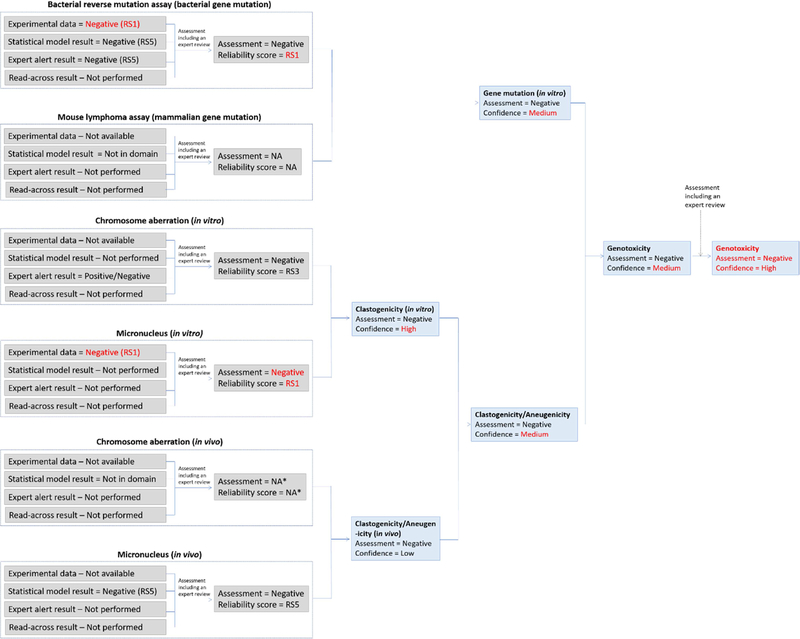
4.5.2.2. Plant protection product active ingredient metabolite assessment (genetic toxicology)
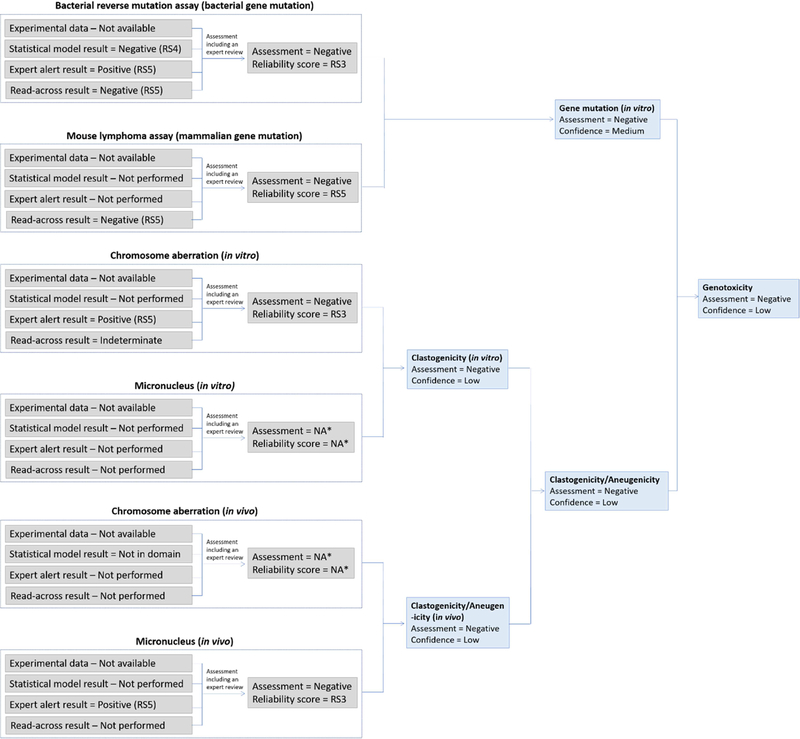
A herbicide metabolite was assessed using in silico methods for genotoxicity. Experimental data generated on the active ingredient (AI) was available and the data confirmed that the AI has no genotoxic potential based on negative bacterial gene mutation, in vitro mammalian gene mutation and in vitro CA assay results, as well as a negative in vivo CA study. The metabolite was noted to have high structural similarity to the AI. Figure 10 shows the initial in silico genotoxicity assessment of the metabolite. The metabolite was predicted by two methodologies to be inactive in the bacterial reverse mutation assay. It was out of domain for the mammalian gene mutation model as well as the in vivo CA model (however, the related endpoint “in vivo MN prediction” was in domain). Two expert alert systems for in vitro CA induction were applied, one indicating that the compound has clastogenic potential due to the presence of a carboxylic acid related alert, and the other that it does not. Expert review of the in silico results was performed by looking at specific details of the alert and the surrounding SAR. Sufficient experimental data for analogs matching the alert convinced the assessor that the alert could be dismissed, and the in vitro CA endpoint was set to negative with a reliability score of RS3, after the expert review. Expert review was also performed on the gene mutation endpoints as well as the predicted in vivo MN results to confirm that these were of sufficient quality. In the case of gene mutation, the reliability score could be increased to RS3, but this was not the case for the in vivo MN assessment and it remained at RS5.
Figure 10.
The initial in silico genetic toxicology assessment for the plant protection product active ingredient metabolite. Note the change in assessment outcome for in vitro CA before and after expert review. *NA refers to “Not available” since these results were not possible to generate.
Following the suggested conservative scheme included in the supplementary material, for combining toxicological effect outputs, the gene mutation endpoint was considered as negative with a low confidence due to the lack of information on the mammalian gene mutation endpoint. Similarly, the in vitro and in vivo clastogenicity/aneugenicity endpoints were considered to have low confidence related to the negative assessments as there was limited information available. The combination of these assessments resulted in the metabolite being considered of low genotoxic potential but with a low confidence. It should be noted that this assessment did not take aneugenicity into account at all with the exception of a predicted negative in vivo micronucleus result. This is an additional reason to consider this assessment of being of low confidence.
Following the in silico assessment exercise, the metabolite was tested in experimental assays for confirmation. The compound was tested in an OECD and GLP compliant bacterial reverse mutation assay and found to be inactive. It was also tested in an OECD and GLP compliant in vitro micronucleus assay and again, no activity was detected. Figure 11 shows how these experimental data would influence the assessment if the protocol framework was applied. The increased reliability scores from the bacterial gene mutation and the in vitro micronucleus tests would result in high confidence in the individual endpoints as well as in the overall genotoxicity assessment, which now would result in a negative outcome with medium confidence. It may appear surprising that the confidence is only set at medium, despite highly reliable experimental results demonstrating no genotoxic activity. However, to distinguish from a situation where in vivo studies were also performed, the confidence cannot, in a general sense, be higher as there needs to be room to increase the weight-of-evidence by the inclusion of in vivo results or expert review. The addition of an in vivo negative outcome would have brought the confidence up to “high”. However, in this particular case, an expert opinion was included in the final outcome, which raised the confidence to high. Sufficient experimental data were available for the parent AI in a full regulatory battery of in vitro and in vivo studies, showing that the AI had no genotoxic potential. The structural similarity between the metabolite and the AI was high and the available in vitro data for the metabolite showed similar responses, therefore no further concern was raised about the in vivo activity of the metabolite. Furthermore, it is recognized that there are different regulatory guidelines with respect to in vivo studies and that in some industries, an in vivo test would not be required for a high confidence assessment.
Figure 11.
Influence of including the experimental results in genetic toxicology assessment for the plant protection AI metabolite. Differences compared to Figure 10 are indicated in red text. *NA refers to “Not available” since these results were not possible to generate.
4.5.2.3. Plant protection product groundwater metabolite assessment (genetic toxicology)
Figure 12 illustrates the in silico genotoxicity assessment of a plant protection product AI metabolite with the potential to leach into the groundwater. The AI is categorized as an IARC Class 2 carcinogen and hence may bear risk to humans, and control strategies are required. It has, however, been shown experimentally to be non-genotoxic and it is hypothesized that the carcinogenicity is mediated through an endocrine disruption mechanism. The metabolite is a polar molecule containing functional groups in a similar environment to the parent molecule. The bacterial gene mutation assessment was performed by read-across and the application of statistical models and expert alerts. The read-across exercise concluded that the metabolite is likely to be negative but the analysis was not considered robust due to the lipophilicity of the metabolite being outside the range of the analogs. Therefore, the result was set at RS5 even though read-across could technically be considered an expert reviewed method and could therefore have been set to RS3 directly with a more robust analysis. Two independent statistical models for bacterial mutagenicity were applied, both indicating that the metabolite was negative, and the reliability score was set to RS4 as there were two concurring and independent models. The rule-based method highlighted an alert (positive, RS5), but after an expert review, the alert was dismissed, as the chemical environment of the alerting moiety was dissimilar between the training set examples and the metabolite and the alert was therefore considered not relevant (the result from this model is considered negative with a reliability score of RS3). No in silico assessment was made for mammalian gene mutation using computational models but comparison (read-across) with the predicted genotoxicity profile of the parent molecule indicated that there should be no concern for mammalian mutagenicity. The call for the in vitro gene mutation endpoint was set to negative with medium confidence.
Figure 12.
In silico assessment of a plant protection product metabolite.
In vitro CA was also investigated using read-across. The weight of the evidence did not give a clear indication of potential for CA induction and was considered Indeterminate. Rule-based methods predicted the metabolite to be positive in the in vivo MN test and in the in vitro CA assay. Both of these predictions were given reliability scores of RS5. Expert review of the examples related to the in vitro CA prediction questioned the relevance as they did not bear strong structural similarity to the metabolite. The alert triggered in the in vitro CA model was also generic in nature and hence not specific to the structural environment in the metabolite (or the AI, for that matter). Furthermore, the parent AI triggered the same in silico response but had been confirmed to show no clastogenic effects in vivo. For the predicted positive outcome in the in vivo MN test, expert review suggested that the predicted activity would be due to carbamate and simple substituted acrylamide compounds, formed as downstream metabolites of the metabolite, rather than to the metabolite under review. For the analogs investigated with experimental data, only the carbamates appeared to truly flag as being related to any activity. Due to the physicochemical properties of the metabolite, it was considered highly unlikely that these would form in vivo and hence, the in vivo alert was overruled. The summary assessment for the metabolite concluded that there was medium confidence that there was no gene mutation potential and low confidence for the lack of clastogenic potential. Aneugenic effects have not been covered. The overall genetic toxicology assessment was therefore set to negative with low confidence. After review of the submission, the regulatory authority also concluded that some experimental testing be conducted to specifically ascertain the predicted lack of genotoxic potential.
5. Reporting
“Good in silico practice” requires a reproducible, transparent, and standardized procedure and it is important to document the entire process of performing the genetic toxicology assessment. This is comparable to Good Laboratory Practice (GLP) documentation of in vitro or in vivo studies and will enable the results to be reviewed rapidly and thoroughly by, for example, regulatory agencies. The general protocol (Myatt et al., 2018) lists relevant types of information that should be included in the report to ensure that the information is complete. Specifically, chemical structures (including analogs in case of read-across) and the models used need to be well documented.
Discussion
The GIST protocol should be applied in a context-dependent manner and in accordance with relevant guidelines. For example, if the application is for an ICH M7 assessment, then in addition to the recommendations provided in the guidance document, there are publications that provide more detailed procedures as well as case examples to illustrate best practices (Barber et al., 2015; Amberg et al., 2016). Similarly, there are, for example, guidelines for chemical registration through the REACH regulation (REACH, 2006; ECHA, 2008; ECHA, 2017) and Canada’s Chemicals Management Program (Canada, 2016), the EFSA definition of residue guidance (EFSA, 2016) and the Toxic Substances Control Act (TSCA) (TSCA, 2016).
The protocol presented in this publication represents the current state-of-the-art in in silico genetic toxicology. As new methods, both experimental and computational, are developed and as new data become available, the recommendations presented herein will need to be revised and updated. Additionally, it is important that the protocol reflects current regulatory standards, data requirements, and changes as these are revised. For example, as more in vitro micronucleus data are generated with differentiation of clastogenicity from aneugenicity mechanisms, statistical modeling may become an option for the separate mechanistic endpoints. Since aneugenicity is generally considered to be a thresholded endpoint, this would involve an important change in the current GIST protocol. Similarly, new assays will be accepted in a regulatory context. Also, there are considerable efforts underway to develop and evaluate the impact that methods such as toxicogenomics, flow cytometric biomarker assays, and other mechanistic platforms (see Section 2.2.4) can have on genotoxicity testing. As with any other protocol, it is therefore important to regularly revise the GIST protocol to include new developments and remove outdated sections as appropriate.
Conclusion
Applying a standardized format for performing and reporting in silico assessments for hazard identification will enable a transparent and consistent review of the results. This is beneficial both for organizations and individuals performing such analyses as well as review boards and regulatory agencies that consider such analyses. Along the same principles of standard practice for in vitro or in vivo data generation, the aim is to foster the use of good in silico practices to promote the use of these methodologies to their full potential.
Supplementary Material
Details a framework for assessment of genetic toxicity based on available data and in silico models
Outlines key effects/mechanisms and their relationship to genetic toxicity
Defines principles and procedures for combining the information
Outlines a methodology to assess the confidence any assessment
Acknowledgements
Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number R43ES026909. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: FDA CDER Disclaimer: This article reflects the views of the authors and should not be construed to represent FDA’s views or policies. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
Publisher's Disclaimer: NIEHS Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the National Institutes of Health (NIH), NIEHS. The mention of commercial products, their sources, or their use should not be construed as either an actual or implied endorsement of such products by the NIH/NIEHS.
Publisher's Disclaimer: MHRA Disclaimer: Any opinions expressed in this document are the author’s and are not necessarily shared by other assessors at the Medicines and Healthcare products Regulatory Agency (MHRA). As such, they cannot be considered to be UK policy. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the UK’s MHRA.
Publisher's Disclaimer: Health Canada Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of Health Canada. The mention of commercial products, their sources, or their use should not be construed as either an actual or implied endorsement of such products by Health Canada.
European Commission Disclaimer: The views expressed are solely those of the authors and the content does not necessarily represent the views or position of the European Commission.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- CHEMOINFORMATICS AND BEYOND. Chemoinformatics for Drug Discovery.
- Ahlberg E, Amberg A, Beilke LD, Bower D, Cross KP, Custer L, Ford KA, Van Gompel J, Harvey J, Honma M, Jolly R, Joossens E, Kemper RA, Kenyon M, Kruhlak N, Kuhnke L, Leavitt P, Naven R, Neilan C, Quigley DP, Shuey D, Spirkl HP, Stavitskaya L, Teasdale A, White A, Wichard J, Zwickl C, Myatt GJ, 2016. Extending (Q)SARs to incorporate proprietary knowledge for regulatory purposes: A case study using aromatic amine mutagenicity. Regul Toxicol Pharmacol 77, 1–12. 10.1016/j.yrtph.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Amberg A, Beilke L, Bercu J, Bower D, Brigo A, Cross KP, Custer L, Dobo K, Dowdy E, Ford KA, Glowienke S, Van Gompel J, Harvey J, Hasselgren C, Honma M, Jolly R, Kemper R, Kenyon M, Kruhlak N, Leavitt P, Miller S, Muster W, Nicolette J, Plaper A, Powley M, Quigley DP, Reddy MV, Spirkl HP, Stavitskaya L, Teasdale A, Weiner S, Welch DS, White A, Wichard J, Myatt GJ, 2016. Principles and procedures for implementation of ICH M7 recommended (Q)SAR analyses. Regul Toxicol Pharmacol 77, 13–24. 10.1016/j.yrtph.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Amberg A, Harvey JS, Czich A, Spirkl H-P, Robinson S, White A, Elder DP, 2015. Do Carboxylic/Sulfonic Acid Halides Really Present a Mutagenic and Carcinogenic Risk as Impurities in Final Drug Products? Organic Process Research & Development 19, 1495–1506. 10.1021/acs.oprd.5b00106 [DOI] [Google Scholar]
- Ashby J, Tennant RW, 1988. Chemical structure, Salmonella mutagenicity and extent of carcinogenicity as indicators of genotoxic carcinogenesis among 222 chemicals tested in rodents by the U.S. NCI/NTP. Mutat Res 204, 17–115. [DOI] [PubMed] [Google Scholar]
- ATSDR, 2018, ATSDR Toxic Substances Portal, https://www.atsdr.cdc.gov/substances/index.asp
- Aubrecht J, Caba E, 2005. Gene expression profile analysis: an emerging approach to investigate mechanisms of genotoxicity. Pharmacogenomics 6, 419–28. 10.1517/14622416.6.4.419 [DOI] [PubMed] [Google Scholar]
- Barber C, Amberg A, Custer L, Dobo KL, Glowienke S, Van Gompel J, Gutsell S, Harvey J, Honma M, Kenyon MO, Kruhlak N, Muster W, Stavitskaya L, Teasdale A, Vessey J, Wichard J, 2015. Establishing best practise in the application of expert review of mutagenicity under ICH M7. Regul Toxicol Pharmacol 73, 367–77. 10.1016/j.yrtph.2015.07.018 [DOI] [PubMed] [Google Scholar]
- Bender MA, Griggs HG, Bedford JS, 1974. Mechanisms of chromosomal aberration production III. Chemicals and ionizing radiation. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 23, 197–212. 10.1016/0027-5107(74)90140-7 [DOI] [PubMed] [Google Scholar]
- Benigni R, Bossa C, 2008. Structure alerts for carcinogenicity, and the Salmonella assay system: a novel insight through the chemical relational databases technology. Mutat Res 659, 248–61. 10.1016/j.mrrev.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Benigni R, Bossa C, Richard AM, Yang C, 2008. A novel approach: chemical relational databases, and the role of the ISSCAN database on assessing chemical carcinogenicity. Ann Ist Super Sanita 44, 48–56. [PubMed] [Google Scholar]
- Benigni R, Bossa C, Tcheremenskaia O, Battistelli CL, Crettaz P, 2012. The new ISSMIC database on in vivo micronucleus and its role in assessing genotoxicity testing strategies. Mutagenesis 27, 87–92. 10.1093/mutage/ger064 [DOI] [PubMed] [Google Scholar]
- Bryce SM, Bernacki DT, Smith-Roe SL, Witt KL, Bemis JC, Dertinger SD, 2018. Investigating the Generalizability of the MultiFlow (R) DNA Damage Assay and Several Companion Machine Learning Models With a Set of 103 Diverse Test Chemicals. Toxicol Sci 162, 146–166. 10.1093/toxsci/kfx235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron TP, Hughes TJ, Kirby PE, Palmer KA, Fung VA, Dunkel VC, 1985. Mutagenic activity of 5 thiazole compounds in the Salmonella/microsome and mouse lymphoma TK+/− assays. Mutation Research/Genetic Toxicology 155, 17–25. 10.1016/0165-1218(85)90020-5 [DOI] [PubMed] [Google Scholar]
- Canada, 2016, Canada’s Chemicals Management Program, https://www.canada.ca/en/healthcanada/services/chemical-substances/chemicals-management-plan.html
- CCRIS, 2011, Chemical Carcinogenesis Research Information System (CCRIS), https://www.nlm.nih.gov/pubs/factsheets/ccrisfs.html
- CDRH, 2016, Use of International Standard ISO 10993–1. Biological Evaluation of Medical Devices - Part 1: Evaluation and Testing within a Risk Management Process Guidance for Industry and Food and Drug Administration Staff (2016), https://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm348890.pdf
- Clewell RA, Sun B, Adeleye Y, Carmichael P, Efremenko A, McMullen PD, Pendse S, Trask OJ, White A, Andersen ME, 2014. Profiling dose-dependent activation of p53-mediated signaling pathways by chemicals with distinct mechanisms of DNA damage. Toxicol Sci 142, 56–73. 10.1093/toxsci/kfu153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constable A, Jonas D, Cockburn A, Davi A, Edwards G, Hepburn P, Herouet-Guicheney C, Knowles M, Moseley B, Oberdörfer R, Samuels F, 2007. History of safe use as applied to the safety assessment of novel foods and foods derived from genetically modified organisms. Food and Chemical Toxicology 45, 2513–2525. 10.1016/j.fct.2007.05.028 [DOI] [PubMed] [Google Scholar]
- Corvi R, Vilardell M, Aubrecht J, Piersma A, 2016. Validation of Transcriptomics-Based In Vitro Methods. Adv Exp Med Biol 856, 243–257. 10.1007/978-3-319-33826-2_10 [DOI] [PubMed] [Google Scholar]
- Custer LL, Powley MW, 2015. Genotoxicity Testing of API. in: Graziano MJ, Jacobson-Kram D, (Ed.), Genotoxicity and Carcinogenicity Testing of Pharmaceuticals Springer International Publishing, pp. 35–54. [Google Scholar]
- Dearfield KL, Gollapudi BB, Bemis JC, Benz RD, Douglas GR, Elespuru RK, Johnson GE, Kirkland DJ, LeBaron MJ, Li AP, Marchetti F, Pottenger LH, Rorije E, Tanir JY, Thybaud V, van Benthem J, Yauk CL, Zeiger E, Luijten M, 2017. Next generation testing strategy for assessment of genomic damage: A conceptual framework and considerations. Environ Mol Mutagen 58, 264–283. 10.1002/em.22045 [DOI] [PubMed] [Google Scholar]
- Dearfield KL, Thybaud V, Cimino MC, Custer L, Czich A, Harvey JS, Hester S, Kim JH, Kirkland D, Levy DD, Lorge E, Moore MM, Ouedraogo-Arras G, Schuler M, Suter W, Sweder K, Tarlo K, van Benthem J, van Goethem F, Witt KL, 2011. Follow-up actions from positive results of in vitro genetic toxicity testing. Environ Mol Mutagen 52, 177–204. 10.1002/em.20617 [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Torous DK, Hayashi M, MacGregor JT, 2011a. Flow cytometric scoring of micronucleated erythrocytes: an efficient platform for assessing in vivo cytogenetic damage. Mutagenesis 26, 139–145. 10.1093/mutage/geq055 [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Torous DK, Hayashi M, MacGregor JT, 2011b. Flow cytometric scoring of micronucleated erythrocytes: an efficient platform for assessing in vivo cytogenetic damage. Mutagenesis 26, 139–45. 10.1093/mutage/geq055 [DOI] [PubMed] [Google Scholar]
- DSSTox, 2018, Distributed Structure-Searchable Toxicity (DSSTox) Database, https://www.epa.gov/chemical-research/distributed-structure-searchable-toxicity-dsstoxdatabase [DOI] [PubMed]
- ECHA, 2008, Guidance on information requirements and chemical safety assessment Chapter R.6: QSARs and grouping of chemicals , https://echa.europa.eu/documents/10162/13632/information_requirements_r6_en.pdf/77f49f81-b76d-40ab-8513-4f3a533b6ac9
- ECHA, 2011, Guidance on information requirements and chemical safety assessment Chapter R.4: Evaluation of available information, https://echa.europa.eu/documents/10162/13643/information_requirements_r4_en.pdf/d6395ad2-1596-4708-ba86-0136686d205e
- ECHA, 2017, Read-Across Assessment Framework, https://echa.europa.eu/documents/10162/13628/raaf_en.pdf
- ECHA, 2018, https://echa.europa.eu/
- EFSA, 2011. Scientific opinion on genotoxicity testing strategies applicable to food and feed safety assessment. EFSA Journal 9, 2379. [Google Scholar]
- EFSA, 2016. EFSA Panel on Plant Protection products and their Residues. Guidance on the establishment of the residue definition for dietary risk assessment. EFSA Journal 14, e04549–n/a. 10.2903/j.efsa.2016.4549 [DOI] [Google Scholar]
- ELSIE, 2018, http://www.elsiedata.org/elsie-database/
- EPA, 2018. Strategic Plan to Promote the Development and Implementation of Alternative Test Methods Within the TSCA Program
- EPA, 2019, https://comptox.epa.gov/dashboard
- European Commission, J. R. C. J., 2018a, EURL ECVAM Genotoxicity & Carcinogenicity Consolidated Database of Ames Positive Chemicals, https://eurlecvam.jrc.ec.europa.eu/databases/genotoxicity-carcinogenicity-db
- European Commission, J. R. C. J., 2018b, ToxRTool - Toxicological data Reliability Assessment Tool, https://eurl-ecvam.jrc.ec.europa.eu/about-ecvam/archive-publications/toxrtool
- Farmahin R, Williams A, Kuo B, Chepelev NL, Thomas RS, Barton-Maclaren TS, Curran IH, Nong A, Wade MG, Yauk CL, 2017. Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch Toxicol 91, 2045–2065. 10.1007/s00204-016-1886-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA, 2018, https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm
- FEMA, 2018a, FEMA GRAS Flavoring Substance List, https://www.femaflavor.org/femagras%E2%84%A2-flavoring-substance-list
- FEMA, 2018b, The Flavor and Extract Manufacturers Association of the United States (FEMA), https://www.femaflavor.org/
- Ford KA, 2016. Refinement, Reduction, and Replacement of Animal Toxicity Tests by Computational Methods. ILAR Journal 57, 226–233. 10.1093/ilar/ilw031 [DOI] [PubMed] [Google Scholar]
- Fowler P, Smith K, Young J, Jeffrey L, Kirkland D, Pfuhler S, Carmichael P, 2012a. Reduction of misleading (“false”) positive results in mammalian cell genotoxicity assays. I. Choice of cell type. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 742, 11–25. 10.1016/j.mrgentox.2011.10.014 [DOI] [PubMed] [Google Scholar]
- Fowler P, Smith R, Smith K, Young J, Jeffrey L, Kirkland D, Pfuhler S, Carmichael P, 2012b. Reduction of misleading (“false”) positive results in mammalian cell genotoxicity assays. II. Importance of accurate toxicity measurement. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 747, 104–117. 10.1016/j.mrgentox.2012.04.013 [DOI] [PubMed] [Google Scholar]
- Fox JT, Sakamuru S, Huang R, Teneva N, Simmons SO, Xia M, Tice RR, Austin CP, Myung K, 2012. High-throughput genotoxicity assay identifies antioxidants as inducers of DNA damage response and cell death. Proc Natl Acad Sci U S A 109, 5423–8. 10.1073/pnas.1114278109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S, Lorge E, Aardema MJ, Eastmond D, Fellows M, Heflich R, Kirkland D, Levy DD, Lynch AM, Marzin D, Morita T, Schuler M, Speit G, 2011. Workshop summary: Top concentration for in vitro mammalian cell genotoxicity assays; and report from working group on toxicity measures and top concentration for in vitro cytogenetics assays (chromosome aberrations and micronucleus). Mutat Res 723, 77–83. 10.1016/j.mrgentox.2011.01.003 [DOI] [PubMed] [Google Scholar]
- GENE-TOX, 1998, https://toxnet.nlm.nih.gov/newtoxnet/genetox.htm
- Gentronix, 2018, Gentronix, http://www.gentronix.co.uk/product/greenscreen/
- Gollapudi BB, Lynch AM, Heflich RH, Dertinger SD, Dobrovolsky VN, Froetschl R, Horibata K, Kenyon MO, Kimoto T, Lovell DP, Stankowski LF Jr., White PA, Witt KL, Tanir JY, 2015. The in vivo Pig-a assay: A report of the International Workshop On Genotoxicity Testing (IWGT) Workgroup. Mutat Res Genet Toxicol Environ Mutagen 783, 23–35. 10.1016/j.mrgentox.2014.09.007 [DOI] [PubMed] [Google Scholar]
- GRAS, 2018, Food Substances Generally Recognized as Safe, https://www.fda.gov/Food/IngredientsPackagingLabeling/GRAS/SCOGS/default.htm
- Hasselgren C, Muthas D, Ahlberg E, Andersson S, Carlsson L, Noeske T, Stålring J, Boyer S, 2013. CHEMOINFORMATICS AND BEYOND. Chemoinformatics for Drug Discovery John Wiley & Sons, Inc, pp. 267–290. [Google Scholar]
- Hayashi M, 2016. The micronucleus test-most widely used in vivo genotoxicity test. Genes Environ 38, 18 10.1186/s41021-016-0044-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig UG, Rudd NL, Hoar DI, 1988. Kinetochore immunofluorescence in micronuclei: a rapid method for the in situ detection of aneuploidy and chromosome breakage in human fibroblasts. Mutat Res 203, 405–14. [DOI] [PubMed] [Google Scholar]
- Honda H, Fujita Y, Kasamatsu T, Fuchs A, Fautz R, Morita O, 2018. Necessity for retrospective evaluation of past-positive chemicals in in vitro chromosomal aberration tests using recommended cytotoxicity indices. Genes and Environment 40, 2 10.1186/s41021-017-0091-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C-W, Hewes KP, Stavitskaya L, Kruhlak NL, 2018. Construction and application of (Q)SAR models to predict chemical-induced in vitro chromosome aberrations. Regulatory Toxicology and Pharmacology 99, 274–288. 10.1016/j.yrtph.2018.09.026 [DOI] [PubMed] [Google Scholar]
- Hughes C, Rabinowitz A, Tate M, Birrell L, Allsup J, Billinton N, Walmsley RM, 2012. Development of a high-throughput Gaussia luciferase reporter assay for the activation of the GADD45a gene by mutagens, promutagens, clastogens, and aneugens. J Biomol Screen 17, 1302–15. 10.1177/1087057112453312 [DOI] [PubMed] [Google Scholar]
- ICH, S2(R1) Genotoxicity Testing and Data Interpretation for Pharmaceuticals Intended for Human Use 2012. [PubMed]
- ICH, 2014. M7 Assessment and control of DNA reactive (mutagenic) impurities in pharmaceuticals to limit potential carcinogenic risk
- ICH, 2017. M7 Assessment and control of DNA reactive (mutagenic) impurities in pharmaceuticals to limit potential carcinogenic risk (R1)
- INCHEM, 2015, http://www.inchem.org/
- IRIS, 2015, https://www.epa.gov/iris
- JECDB, 2018, https://www.echemportal.org/echemportal/index.action
- JECFA, 2018, Joint FAO/WHO Expert Committee on Food Additives (JECFA), http://apps.who.int/foodadditives-contaminants-jecfa-database/search.aspx
- Kaina B, 2004. Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: a long road traveled and still a far way to go. Cytogenet Genome Res 104, 77–86. 10.1159/000077469 [DOI] [PubMed] [Google Scholar]
- Kim S, Jun DH, Kim HJ, Jeong KC, Lee CH, 2011. Development of a high-content screening method for chemicals modulating DNA damage response. J Biomol Screen 16, 259–65. 10.1177/1087057110392993 [DOI] [PubMed] [Google Scholar]
- Kirkland D, Fowler P, 2010. Further analysis of Ames-negative rodent carcinogens that are only genotoxic in mammalian cells in vitro at concentrations exceeding 1 mM, including retesting of compounds of concern. Mutagenesis 25, 539–53. 10.1093/mutage/geq041 [DOI] [PubMed] [Google Scholar]
- Kirkland D, Pfuhler S, Tweats D, Aardema M, Corvi R, Darroudi F, Elhajouji A, Glatt H, Hastwell P, Hayashi M, Kasper P, Kirchner S, Lynch A, Marzin D, Maurici D, Meunier J-R, Müller L, Nohynek G, Parry J, Parry E, Thybaud V, Tice R, van Benthem J, Vanparys P, White P, 2007a. How to reduce false positive results when undertaking in vitro genotoxicity testing and thus avoid unnecessary follow-up animal tests: Report of an ECVAM Workshop. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 628, 31–55. 10.1016/j.mrgentox.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Kirkland D, Pfuhler S, Tweats D, Aardema M, Corvi R, Darroudi F, Elhajouji A, Glatt H, Hastwell P, Hayashi M, Kasper P, Kirchner S, Lynch A, Marzin D, Maurici D, Meunier JR, Muller L, Nohynek G, Parry J, Parry E, Thybaud V, Tice R, van Benthem J, Vanparys P, White P, 2007b. How to reduce false positive results when undertaking in vitro genotoxicity testing and thus avoid unnecessary follow-up animal tests: Report of an ECVAM Workshop. Mutat Res 628, 31–55. 10.1016/j.mrgentox.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Kirkland DJ, 1994. Preface. Mutation Research/Environmental Mutagenesis and Related Subjects 312, 195–199. 10.1016/0165-1161(94)00006-9 [DOI] [Google Scholar]
- Kirkland DJ, 2000. Preface. Environmental and Molecular Mutagenesis 35, 159–161. [DOI] [PubMed] [Google Scholar]
- Kirkland DJ, 2003. Preface. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 540, 119–121. 10.1016/j.mrgentox.2003.07.001 [DOI] [Google Scholar]
- Kirkland DJ, Hayashi M, Jacobson-Kram D, Kasper P, Gollapudi B, Müller L, Uno Y, 2011. Summary of major conclusions from the 5th IWGT, Basel, Switzerland, 17–19 August 2009. [DOI] [PubMed] [Google Scholar]
- Mutation Research/Genetic Toxicology and Environmental Mutagenesis 723, 73–76. 10.1016/j.mrgentox.2011.03.003 [DOI] [Google Scholar]
- Kirkland DJ, Hayashi M, Jacobson-Kram D, Kasper P, MacGregor JT, Müller L, Uno Y, 2007c. The International Workshops on Genotoxicity Testing (IWGT): History and achievements. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 627, 1–4. 10.1016/j.mrgentox.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Kirkland DJ, Hayashi M, Jacobson-Kram D, Kasper P, MacGregor JT, Müller L, Uno Y, 2007d. Summary of major conclusions from the 4th IWGT, San Francisco, 9–10 September, 2005. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 627, 5–9. 10.1016/j.mrgentox.2006.08.009 [DOI] [PubMed] [Google Scholar]
- Klimisch HJ, Andreae M, Tillmann U, 1997. A systematic approach for evaluating the quality of experimental toxicological and ecotoxicological data. Regul Toxicol Pharmacol 25, 1–5. 10.1006/rtph.1996.1076 [DOI] [PubMed] [Google Scholar]
- Knight AW, Birrell L, Walmsley RM, 2009. Development and validation of a higher throughput screening approach to genotoxicity testing using the GADD45a-GFP GreenScreen HC assay. J Biomol Screen 14, 16–30. 10.1177/1087057108327065 [DOI] [PubMed] [Google Scholar]
- Leadscope, 2018, http://www.leadscope.com/toxicity_databases/
- Li HH, Chen R, Hyduke DR, Williams A, Frotschl R, Ellinger-Ziegelbauer H, O’Lone R, Yauk CL, Aubrecht J, Fornace AJ Jr., 2017. Development and validation of a high-throughput transcriptomic biomarker to address 21st century genetic toxicology needs. Proc Natl Acad Sci U S A 114, E10881–e10889. 10.1073/pnas.1714109114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Hyduke DR, Chen R, Heard P, Yauk CL, Aubrecht J, Fornace AJ Jr., 2015. Development of a toxicogenomics signature for genotoxicity using a dose-optimization and informatics strategy in human cells. Environ Mol Mutagen 56, 505–19. 10.1002/em.21941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor JT, Bishop ME, McNamee JP, Hayashi M, Asano N, Wakata A, Nakajima M, Saito J, Aidoo A, Moore MM, Dertinger SD, 2006. Flow cytometric analysis of micronuclei in peripheral blood reticulocytes: II. An efficient method of monitoring chromosomal damage in the rat. Toxicol Sci 94, 92–107. 10.1093/toxsci/kfl076 [DOI] [PubMed] [Google Scholar]
- Mahadevan B, Snyder RD, Waters MD, Benz RD, Kemper RA, Tice RR, Richard AM, 2011. Genetic toxicology in the 21st century: reflections and future directions. Environ Mol Mutagen 52, 339–54. 10.1002/em.20653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant CA, 2012. Computational toxicology: a tool for all industries. Wiley Interdisciplinary Reviews: Computational Molecular Science 2, 424–434. 10.1002/wcms.100 [DOI] [Google Scholar]
- Martus HJ, Hayashi M, Honma M, Kasper P, Gollapudi B, Mueller L, Schoeny R, Uno Y, Kirkland DJ, 2015. Summary of major conclusions from the 6th International Workshop on Genotoxicity Testing (IWGT), Foz do Iguacu, Brazil. Mutat Res Genet Toxicol Environ Mutagen 783, 1–5. 10.1016/j.mrgentox.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Mav D, Shah RR, Howard BE, Auerbach SS, Bushel PR, Collins JB, Gerhold DL, Judson RS, Karmaus AL, Maull EA, Mendrick DL, Merrick BA, Sipes NS, Svoboda D, Paules RS, 2018. A hybrid gene selection approach to create the S1500+ targeted gene sets for use in high-throughput transcriptomics. PLOS ONE 13, e0191105 10.1371/journal.pone.0191105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EC, Miller JA, 1981. Searches for ultimate chemical carcinogens and their reactions with cellular macromolecules. Cancer 47, 2327–45. [DOI] [PubMed] [Google Scholar]
- Mishima M, 2017. Chromosomal aberrations, clastogens vs aneugens. Front Biosci (Schol Ed) 9, 1–16. [DOI] [PubMed] [Google Scholar]
- Moore MM, Honma M, Clements J, Bolcsfoldi G, Burlinson B, Cifone M, Clarke J, Delongchamp R, Durward R, Fellows M, Gollapudi B, Hou S, Jenkinson P, Lloyd M, Majeska J, Myhr B, O’Donovan M, Omori T, Riach C, San R, Stankowski LF Jr., Thakur AK, Van Goethem F, Wakuri S, Yoshimura I, 2006. Mouse lymphoma thymidine kinase gene mutation assay: follow-up meeting of the International Workshop on Genotoxicity Testing--Aberdeen, Scotland, 2003--Assay acceptance criteria, positive controls, and data evaluation. Environ Mol Mutagen 47, 1–5. 10.1002/em.20159 [DOI] [PubMed] [Google Scholar]
- Moore MM, Honma M, Clements J, Bolcsfoldi G, Cifone M, Delongchamp R, Fellows M, Gollapudi B, Jenkinson P, Kirby P, Kirchner S, Muster W, Myhr B, O’Donovan M, Oliver J, Omori T, Ouldelhkim M-C, Pant K, Preston R, Riach C, San R, Stankowski LF Jr, Thakur A, Wakuri S, Yoshimura I, 2003. Mouse Lymphoma Thymidine Kinase Gene Mutation Assay: International Workshop on Genotoxicity Tests Workgroup Report—Plymouth, UK 2002. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 540, 127–140. 10.1016/j.mrgentox.2003.07.003 [DOI] [PubMed] [Google Scholar]
- Myatt GJ, Ahlberg E, Akahori Y, Allen D, Amberg A, Anger LT, Aptula A, Auerbach S, Beilke L, Bellion P, Benigni R, Bercu J, Booth ED, Bower D, Brigo A, Burden N, Cammerer Z, Cronin MTD, Cross KP, Custer L, Dettwiler M, Dobo K, Ford KA, Fortin MC, GadMcDonald SE, Gellatly N, Gervais V, Glover KP, Glowienke S, Van Gompel J, Gutsell S, Hardy B, Harvey JS, Hillegass J, Honma M, Hsieh JH, Hsu CW, Hughes K, Johnson C, Jolly R, Jones D, Kemper R, Kenyon MO, Kim MT, Kruhlak NL, Kulkarni SA, Kummerer K, Leavitt P, Majer B, Masten S, Miller S, Moser J, Mumtaz M, Muster W, Neilson L, Oprea TI, Patlewicz G, Paulino A, Lo Piparo E, Powley M, Quigley DP, Reddy MV, Richarz AN, Ruiz P, Schilter B, Serafimova R, Simpson W, Stavitskaya L, Stidl R, Suarez-Rodriguez D, Szabo DT, Teasdale A, Trejo-Martin A, Valentin JP, Vuorinen A, Wall BA, Watts P, White AT, Wichard J, Witt KL, Woolley A, Woolley D, Zwickl C, Hasselgren C, 2018. In silico toxicology protocols. Regul Toxicol Pharmacol 96, 1–17. 10.1016/j.yrtph.2018.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt GJ, Beilke LD, Cross TKP, 2016. In Silico Tools and their Application. Reference Module in Chemistry, Molecular Sciences and Chemical Engineering 2016.
- NIOSH, 2018, https://www.cdc.gov/niosh/index.htm
- Nishihara K, Huang R, Zhao J, Shahane SA, Witt KL, Smith-Roe SL, Tice RR, Takeda S, Xia M, 2016. Identification of genotoxic compounds using isogenic DNA repair deficient DT40 cell lines on a quantitative high throughput screening platform. Mutagenesis 31, 69–81. 10.1093/mutage/gev055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP, 2018, https://tools.niehs.nih.gov/cebs3/ui/
- OECD, 1986a. Test No. 479: Genetic Toxicology: In vitro Sister Chromatid Exchange Assay in Mammalian Cells Section 4, OECD Publishing, Paris: 10.1787/9789264071384-en. [DOI] [Google Scholar]
- OECD, 1986b. Test No. 481: Genetic Toxicology: Saacharomyces cerevisiae, Miotic Recombination Assay Section 4, OECD Publishing, Paris: 10.1787/9789264071421-en. [DOI] [Google Scholar]
- OECD, 1986c. Test No. 482: Genetic Toxicology: DNA Damage and Repair, Unscheduled DNA Synthesis in Mammalian Cells in vitro Section 4, OECD Publishing, Paris: 10.1787/9789264071445-en. [DOI] [Google Scholar]
- OECD, 1986d. Test No. 484: Genetic Toxicology: Mouse Spot Test, OECD Guidelines for the Testing of Chemicals Section 4, OECD Publishing, Paris: 10.1787/9789264071483-en. [DOI] [Google Scholar]
- OECD, 1986e. Test No. 485: Genetic toxicology, Mouse Heritable Translocation Assay Section 4, OECD Publishing, Paris: 10.1787/9789264071506-en. [DOI] [Google Scholar]
- OECD, 1997a. Test No. 471: Bacterial Reverse Mutation Test Section 4, OECD Publishing, Paris: 10.1787/9789264071247-en. [DOI] [Google Scholar]
- OECD, 1997b. Test No. 486: Unscheduled DNA Synthesis (UDS) Test with Mammalian Liver Cells in vivo Section 4, OECD Publishing, Paris: 10.1787/9789264071520-en. [DOI] [Google Scholar]
- OECD, 2010. Test No. 487: In Vitro Mammalian Cell Micronucleus Test doi: 10.1787/9789264091016-en. [DOI]
- OECD, 2013. Test No. 488: Transgenic Rodent Somatic and Germ Cell Gene Mutation Assays Section 4, OECD Publishing, Paris: 10.1787/9789264203907-en. [DOI] [Google Scholar]
- OECD, 2014. Guidance on Grouping of Chemicals, Second Edition Series on Testing & Assessment No. 194 http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono(2014)4&doclanguage=en.
- OECD, 2015. Guidance Document on Revisions to OECD Genetic Toxicology Test Guidelines https://www.oecd.org/env/ehs/testing/Draft%20Guidance%20Document%20on%20OECD%20Genetic%20Toxicology%20Test%20Guidelines.pdf.
- OECD, 2016a. Overview of the set of OECD Genetic Toxicology Test Guidelines and updates performed in 2014–2015, Series on Testing and Assessment nr 238. Paris. [Google Scholar]
- OECD, 2016b. Test No. 473: In Vitro Mammalian Chromosomal Aberration Test
- OECD, 2016c. Test No. 473: In Vitro Mammalian Chromosomal Aberration Test doi: 10.1787/9789264264649-en. [DOI] [Google Scholar]
- OECD, 2016d. Test No. 474: Mammalian Erythrocyte Micronucleus Test Section 4, OECD Publishing, Paris: 10.1787/9789264264762-en. [DOI] [Google Scholar]
- OECD, 2016e. Test No. 475: Mammalian Bone Marrow Chromosomal Aberration Test Section 4, OECD Publishing, Paris: 10.1787/9789264264786-en. [DOI] [Google Scholar]
- OECD, 2016f. Test No. 476: In Vitro Mammalian Cell Gene Mutation Tests using the Hprt and xprt genes Section 4, OECD Publishing, Paris: 10.1787/9789264264809-en. [DOI] [Google Scholar]
- OECD, 2016g. Test No. 478: Rodent Dominant Lethal Test Section 4, OECD Publishing, Paris: 10.1787/9789264264823-en. [DOI] [Google Scholar]
- OECD, 2016h. Test No. 483: Mammalian Spermatogonial Chromosomal Aberration Test Section 4, OECD Publishing, Paris: 10.1787/9789264264847-en. [DOI] [Google Scholar]
- OECD, 2016i. Test No. 487: In Vitro Mammalian Cell Micronucleus Test Section 4, OECD Publishing, Paris: 10.1787/9789264264861-en. [DOI] [Google Scholar]
- OECD, 2016j. Test No. 489: In Vivo Mammalian Alkaline Comet Assay Section 4, OECD Publishing, Paris: 10.1787/9789264264885-en. [DOI] [Google Scholar]
- OECD, 2016k. Test No. 490: In Vitro Mammalian Cell Gene Mutation Tests Using the Thymidine Kinase Gene Section 4, OECD Publishing, Paris: 10.1787/9789264264908-en. [DOI] [Google Scholar]
- OECD, 2018, Existing Chemicals Database, https://hpvchemicals.oecd.org/UI/Default.aspx
- OECD, 2019, OECD QSAR Toolbox, https://qsartoolbox.org/
- Olaharski AJ, Gonzaludo N, Bitter H, Goldstein D, Kirchner S, Uppal H, Kolaja K, 2009. Identification of a Kinase Profile that Predicts Chromosome Damage Induced by Small Molecule Kinase Inhibitors. PLoS Computational Biology 5, e1000446 10.1371/journal.pcbi.1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OSHA, 2018, https://www.osha.gov/
- PAN, 2018, http://www.pesticideinfo.org/
- Parry JM, Parry E, Phrakonkham P, Corvi R, 2010a. Analysis of published data for top concentration considerations in mammalian cell genotoxicity testing. Mutagenesis 25, 531–8. 10.1093/mutage/geq046 [DOI] [PubMed] [Google Scholar]
- Parry JM, Parry E, Phrakonkham P, Corvi R, 2010b. Analysis of published data for top concentration considerations in mammalian cell genotoxicity testing. Mutagenesis 25, 531–538. 10.1093/mutage/geq046 [DOI] [PubMed] [Google Scholar]
- PharmaPendium, 2018, http://www.pharmapendium.com/
- REACH, 2006, http://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:02006R1907-20161011&from=EN
- Rosefort C, Fauth E, Zankl H, 2004. Micronuclei induced by aneugens and clastogens in mononucleate and binucleate cells using the cytokinesis block assay. Mutagenesis 19, 277–284. 10.1093/mutage/geh028 [DOI] [PubMed] [Google Scholar]
- RTECS, 2018, https://www.cdc.gov/niosh/rtecs/default.html
- Saha LK, Kim S, Kang H, Akter S, Choi K, Sakuma T, Yamamoto T, Sasanuma H, Hirota K, Nakamura J, Honma M, Takeda S, Dertinger S, 2018. Differential micronucleus frequency in isogenic human cells deficient in DNA repair pathways is a valuable indicator for evaluating genotoxic agents and their genotoxic mechanisms. Environ Mol Mutagen 59, 529–538. 10.1002/em.22201 [DOI] [PubMed] [Google Scholar]
- Sakai R, Kondo C, Oka H, Miyajima H, Kubo K, Uehara T, 2014. Utilization of CDKN1A/p21 gene for class discrimination of DNA damage-induced clastogenicity. Toxicology 315, 8–16. 10.1016/j.tox.2013.10.009 [DOI] [PubMed] [Google Scholar]
- Snyder RD, 2000. Use of catalytic topoisomerase II inhibitors to probe mechanisms of chemicalinduced clastogenicity in Chinese hamster V79 cells. Environ Mol Mutagen 35, 13–21. [PubMed] [Google Scholar]
- Snyder RD, 2010. Possible structural and functional determinants contributing to the clastogenicity of pharmaceuticals. Environ Mol Mutagen 51, 800–14. 10.1002/em.20626 [DOI] [PubMed] [Google Scholar]
- Sofuni T, Matsuoka A, Sawada M, Ishidate M Jr., Zeiger E, Shelby MD, 1990. A comparison of chromosome aberration induction by 25 compounds tested by two Chinese hamster cell (CHL and CHO) systems in culture. Mutat Res 241, 175–213. [DOI] [PubMed] [Google Scholar]
- Stanton K, Kruszewski FH, 2016. Quantifying the benefits of using read-across and in silico techniques to fulfill hazard data requirements for chemical categories. Regul Toxicol Pharmacol 81, 250–259. 10.1016/j.yrtph.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Stavitskaya L, Minnier BL, Benz RD, Kruhlak NL, Development of Improved QSAR Models for Predicting A-T Base Pair Mutations Poster presented at the Genetic Toxicity Association, Newark, Delaware, 2013. [Google Scholar]
- Sweet DV, Anderson VP, Fang JCF, 1999. An overview of the Registry of Toxic Effects of Chemical Substances (RTECS): Critical information on chemical hazards. Chemical Health and Safety 6, 12–16. 10.1016/S1074-9098(99)00058-1 [DOI] [Google Scholar]
- Torous DK, Dertinger SD, Hall NE, Tometsko CR, 1998a. An automated method for discriminating aneugen- vs. clastogen-induced micronuclei. Environmental and Molecular Mutagenesis 31, 340–344. [DOI] [PubMed] [Google Scholar]
- Torous DK, Dertinger SD, Hall NE, Tometsko CR, 1998b. An automated method for discriminating aneugen- vs. clastogen-induced micronuclei. Environ Mol Mutagen 31, 340–4. [PubMed] [Google Scholar]
- TOXNET, 2018, https://toxnet.nlm.nih.gov/
- TSCA, 2016. Toxic Substances Control Act (TSCA) https://www.congress.gov/bill/114thcongress/senate-bill/697/all-info.
- VITIC, 2018, https://www.lhasalimited.org/products/vitic-nexus.htm
- Wexler P, 2001. TOXNET: An evolving web resource for toxicology and environmental health information. Toxicology 157, 3–10. 10.1016/S0300-483X(00)00337-1 [DOI] [PubMed] [Google Scholar]
- Wilde S, Dambowsky M, Hempt C, Sutter A, Queisser N, 2017. Classification of in vitro genotoxicants using a novel multiplexed biomarker assay compared to the flow cytometric micronucleus test. Environ Mol Mutagen 58, 662–677. 10.1002/em.22130 [DOI] [PubMed] [Google Scholar]
- Witt KL, Hsieh JH, Smith-Roe SL, Xia M, Huang R, Zhao J, Auerbach SS, Hur J, Tice RR, 2017. Assessment of the DNA damaging potential of environmental chemicals using a quantitative high-throughput screening approach to measure p53 activation. Environ Mol Mutagen 58, 494–507. 10.1002/em.22112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KN, Hirota K, Kono K, Takeda S, Sakamuru S, Xia M, Huang R, Austin CP, Witt KL, Tice RR, 2011. Characterization of environmental chemicals with potential or DNA damage using isogenic DNA repair-deficient chicken DT40 cell lines. Environ Mol Mutagen 52, 547–61. 10.1002/em.20656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger E, Gollapudi B, Aardema MJ, Auerbach S, Boverhof D, Custer L, Dedon P, Honma M, Ishida S, Kasinski AL, Kim JH, Manjanatha MG, Marlowe J, Pfuhler S, Pogribny I, Slikker W, Stankowski LF Jr., Tanir JY, Tice R, van Benthem J, White P, Witt KL, Thybaud V, 2015. Opportunities to integrate new approaches in genetic toxicology: an ILSI-HESI workshop report. Environ Mol Mutagen 56, 277–85. 10.1002/em.21923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The general protocol paper outlined some of the in silico methodologies that can be used to generate predictions (Myatt et al., 2018). These include i) rule-based (or “expert”) systems that identify the presence of a structural moiety, also referred to as a structural alert, that may indicate genotoxic potential, and ii) statistical (quantitative structure-activity relationship ((Q)SAR)) models that use a variety of molecular descriptors such as structural fragments or physicochemical properties to predict activity. Here, these two types are collectively referred to as “(Q)SAR” models. In addition, “read-across” (OECD, 2014) is a methodology that utilizes experimental or computed properties, such as physicochemical properties, together with structural similarity and experimental data for structural analogs to extrapolate from source chemical(s) to a target (query) chemical(s) (OECD, 2014).
The types of in silico tools that can be developed for a specific endpoint are, to a great extent, driven by the availability (amount and quality) of experimental data for model development, as well as the degree to which the chemicals of interest exert their toxicity via a common mechanism. In silico tools are most easily developed for endpoints with a well understood and similar mode of action for which a large number of data points are available. Bacterial mutagenicity is a relevant example of such an endpoint and consequently, in this area of genetic toxicology, the development of (Q)SAR models is the most established, largely due to the realization of an electrophilic mode of action for many genotoxic agents (Miller and Miller, 1981; Ashby and Tennant, 1988) and the availability of a large data set (> 2,000 compounds). Conversely, endpoints with less data or studies where the response can be due to several different mechanistic pathways (e.g., chromosome damage) are more challenging for (Q)SAR modeling.
Table 1 lists an estimate of the number of compounds in the public domain with associated genetic toxicology data, as published in the Leadscope toxicity database (Leadscope, 2018). In many cases, multiple results for the same assay and chemical will be available, sometimes with conflicting results and/or conclusions. Private or commercial organizations may have access to additional compounds with experimental data (e.g. from product development), which can be combined with publicly available data. There are some factors to consider, with respect to using experimental data for modeling or readacross: (1) data conflicts need to be resolved (this is not always possible) and experimental protocols need to be examined to ensure that only data measured and interpreted under similar conditions are merged, and (2) chemical structures need to be accurate. The general protocol provides more specific details regarding considerations when using data for modeling or read-across (Myatt et al., 2018). In the case of genetic toxicology, it may be relevant to look at modeling certain subsets of assay results. For example, data generated using the Escherichia coli (E. coli) WP2 uvrA pKM101 and Salmonella typhimurium TA102 (TA102) strains have sometimes been modeled separately (Stavitskaya, 2013) from data generated using the Salmonella strains TA98, TA100, TA1535, and TA1537, because the mechanistic basis of mutation induction is different for these two groups. The second group have GC base pairs at the primary reversion site, which are not as sensitive to detecting certain oxidizing mutagens, crosslinking agents and hydrazines which are instead better detected using the TA102 or E.coli strains which have an AT base pair at the primary reversion site (OECD, 1997a). An additional consideration is the distribution of the biological response. It is not unusual for the available data sets to be skewed so that one assay result classification (e.g., negative/positive) occurs much more frequently. Usually, inactive compounds are more abundant but regardless, the resulting imbalance will require specific strategies to be applied during the modeling procedure to avoid unbalanced predictions due to the prior probability resulting from the training set distribution.
Table 1.
Estimated number of compounds with genetic toxicology data in the public domain.*
| Assay/study type | Number of compounds |
|---|---|
| Bacterial mutagenicity | 10,440 |
| Chromosome aberration (in vitro) | 1,690 |
| Chromosome aberration (in vivo) | 360 |
| Mammalian cell mutagenicity (in vitro) | 2,390 |
| Micronucleus (in vitro) | 290 |
| Micronucleus (in vivo) | 1,850 |
Number of compounds with at least one experimental result listed as part of the Leadscope toxicity database (Leadscope, 2018).