Abstract
Omiganan is an indolicidin analog with antimicrobial properties that could be beneficial for patients with atopic dermatitis. In this randomized, double‐blind, placebo‐controlled, phase II trial we explored the efficacy, pharmacodynamics, and safety of topical omiganan once daily in 36 patients with mild to moderate atomic dermatitis. Patients were randomized to apply topical omiganan 1%, omiganan 2.5%, or vehicle gel to one target lesion once daily for 28 consecutive days. Small but significant improvements in local objective SCORing Atopic Dematitis index and morning itch were observed in the omiganan 2.5% group compared with the vehicle gel group (−18.5%; 95% confidence interval, −32.9 to −1.0; P = 0.04; and −8.2; 95% confidence interval, −16.3 to −0.2; P = 0.05, respectively). A shift from lesional to nonlesional skin microbiota was observed in both omiganan treatment groups, in contrast to the vehicle group. Thus, treatment with topical omiganan improved dysbiosis in patients with mild to moderate atopic dermatitis, and small but statistically significant improvements in clinical scores were detected. Our findings warrant further exploration in future clinical trials.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ One of the hypothesized key factors involved in the pathogenesis of atopic dermatitis (AD), an inflammatory chronic skin disease, is colonization and infection of the skin with Staphylococcus aureus. Omiganan has demonstrated antibacterial and anti‐inflammatory properties and thus represents a promising new treatment for AD patients. Skin microbiome‒associated outcomes have the potential to function as biomarker in early‐phase clinical AD trials.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ What are the effects on clinical efficacy, patient‐reported outcomes, pharmacodynamics, safety, and tolerability of topical omiganan in patients with mild to moderate AD?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ Omiganan improved dysbiosis in patients with atopic dermatitis. Small but statistically significant clinical effects on the target lesion objective SCORing Atopic Dematitis index and morning itch were observed, rendering the clinical relevance debatable. Clinical improvement did not correlate with the improvement in dysbiosis.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The results of this trial indicate that the role of S. aureus in the pathophysiology of AD appears to be minor in patients with mild to moderate disease severity. Although a clear proof of concept was not achieved, we demonstrated proof of mechanism for the drug, i.e., improvement of dysbiosis, a clear sign of target engagement.
Atopic dermatitis (AD) is a common skin disorder with a prevalence up to 3% in adults and up to 20% in children of the Western world. 1 Patients with AD have xerotic erythematous skin with oozing and crusting that typically occurs on the flexor sites of the body. Severe pruritus is the main and most bothersome symptom for most patients, and can lead to reduced quality of life and reduced quality of sleep. 2 Current topical therapies for AD include bland emollients in combination with anti‐inflammatory agents, such as corticosteroids and calcineurin inhibitors. Side effects can be serious, including hypothalmic‒pituitary‒adrenal axis suppression with extensive topical corticosteroid therapy. Recently dupilumab, the first monoclonal anti‒interleukin‐4 antibody treatment for patients with moderate to severe AD, was registered, and it is likely that others will follow. However, for patients with mild disease, novel therapeutic agents with a favorable side effect profile are needed.
The pathophysiology of atopic AD is multifactorial and only partially understood. One of the factors involved in the pathogenesis is colonization and infection of the skin with Staphylococcus aureus, which can produce and secrete toxins and act as superantigens leading to inflammation. 3 Colonization rates in a study of Park and colleagues showed that up to 90% of AD patients were colonized with this pathogen, compared with only 5–30% of the healthy subjects. 4 A deficiency in antimicrobial peptides (AMPs), which are important in the host defense system of the skin, accounts for the susceptibility to this bacterium in AD patients. 5 A dysregulated type 2 T‐helper response in the skin leading to production of pro‐inflammatory cytokines, such as interleukin (IL)‐4 and IL‐13, is most likely responsible for this AMP deficiency. 6 , 7 Therefore, topical AMPs represent a promising new therapeutic option for patients with AD.
Omiganan is a novel, synthetic, cationic peptide and an analog of indolicidin, a short AMP of the cathelicidin family. It has demonstrated antimicrobial activity against a wide variety of gram‐positive and ‐negative bacteria and fungi. 8 , 9 , 10 Cationic peptides are also suggested to have immunomodulatory roles in both pro‐ and anti‐inflammatory pathways, depending on the context. 11 , 12 The combination of antibacterial and anti‐inflammatory properties make omiganan a promising compound for the topical treatment of AD. Although omiganan has been investigated before in several other indications, e.g., acne vulgaris and rosacea, the current trial is the to elucidate its effects in patients with AD.
In this study we aimed to explore the clinical efficacy, patient‐reported outcomes, pharmacodynamics, safety, and tolerability of topical omiganan on one target lesion in patients with mild to moderate AD.
MATERIALS AND METHODS
Study design, randomization, and treatments
We conducted a randomized, double‐blind, placebo‐controlled, single‐center, phase 2 study to explore the clinical efficacy, patient‐reported outcomes (PROs), pharmacodynamics (PD), safety, and tolerability of omiganan in 36 patients with mild to moderate AD. The Declaration of Helsinki was the guiding principle for trial execution, and the study was approved by the independent medical ethics committee, Medisch Ethische Toetsingscommissie van de Stichting Beoordeling Ethiek Biomedisch Onderzoek (Assen, The Netherlands), before any trial procedure. All patients provided written informed consent before participation. The study was conducted from June 2015 to November 2015 at the Centre for Human Drug Research, Leiden, The Netherlands. Eligible patients were randomized to apply either topical omiganan 1%, omiganan 2.5%, or vehicle gel (which served as placebo with an identical appearance), once daily (q.d.) for 28 consecutive days to one target lesion. This was done 1:1:1 in blocks of three according to a randomization list with codes generated by an unblinded statistician. In addition to the active ingredient, omiganan pentahydrochloride, excipients of the compound are glycerine, hydroxyethyl cellulose, benzoic acid, sodium benzoate, and water. For safety reasons, only the predefined target lesion (one antecubital fossa) was treated in this first‐in‐human study with drug/placebo but not with emollient. Study drug application was recorded using a validated mobile‐phone e‐diary application. Subjects, study personnel, and investigators were blinded for the allocated treatment throughout the study. Emollients (unguentum leniens) were distributed for use once daily, but not to the target lesion. Patient visits were scheduled at days −14 (run‐in period), 0, 1, 3, 7, 14, 21, 28 (end of treatment), 35, and 42 (end of study). During the run‐in period, patients applied emollients to the skin (not to the target lesion) and triamcinolone 0.1% ointment to the eczema lesions if needed, but not the target lesion. All study details are provided in the protocol and assessment schedule (refer to Supplementary Material [Link] , [Link] ).
Patients
Patients were included if: (i) AD was mild to moderate and (intermittently) present for >1 year; (ii) between the age of 18 and 65 years; and (iii) willing to give written informed consent. Inclusion criteria were an objective SCORing Atopic Dematitis (oSCORAD) index of 8–40, and an affected body surface area (BSA) of 5–15%. For PD assessments throughout the study, all patients were required to have at least one target lesion (one antecubital fossa) affected by AD, a BSA of ≥0.5%, and a pruritus numeric rating scale (NRS) score of ≥30 on a 0–100 scale. Washout periods for any type of AD medication were as follows: for cyclosporine, mycophenolate mofetil, and other systemic immunosuppressive drugs, 4 weeks; phototherapy, 3 weeks; biologics, five half‐lives of the drug; topical calcineurin inhibitors, 10 days; topical corticosteroids, 2 weeks; and any other topical medication (prescription or over the counter), 2 weeks. Patients with other clinically‐significant (skin) conditions in the treatment area were excluded. Health status of included patients was verified by a detailed medical history, physical examination, vital signs, 12‐lead electrocardiogram, and laboratory testing (including hepatic and renal panels, complete blood count, chemistry panel, virology, and urinalysis). Patients were evaluated for the four most prevalent fillagrin mutations in Europe at screening (2282del4, R501x, S3247x, and R2447x).
Clinical efficacy, pharmacodynamic assessments, and PROs
One target lesion was assigned for treatment and PD assessments in the trial. Another nontreated eczema lesion served as a control lesion for part of the PD measurements (off‐target side). Clinical efficacy was assessed according to local oSCORAD index (percent BSA, erythema, edema/papulation, oozing/crusting, excoriation, lichenification, and dryness) of the target lesion. Standardized images were obtained using a three‐dimensional stereo‐camera system (LifeViz QuantifiCare, Valbonne, France) for assessment of the target lesion size and roughness analysis. The skin barrier status of lesional and nonlesional skin was assessed by transepidermal water loss assessment (AquaFlux AF200 System, Biox, London, UK). This was done under standard environmental conditions (temperature 22 ± 2°C, relative humidity <60%) and patients were acclimatized under relaxed conditions for at least 15 minutes before measurement. All measurements were performed at each study visit. PROs consisted of NRS itch (0–100) and the 5‐D itch scale. 13 , 14 The itch scores were divided into morning and evening itch to link the application time with efficacy outcomes.
Swab collection for microbiology
Patients were instructed not to wash or apply the study drug and keep the target lesion dry for at least 24 hours before each study visit. Skin swab samples were collected with sterile swabs that were dipped in NaCl Tween solution (REF 25‐806‐1PD, Puritan Sterile Polyester Tipped Applicator; Medtronic, Minneapolis, MN), before rubbing the tip of the swab firmly over 4 cm2 of the target lesion five times. Hereafter, the swab material was placed in a microtube (REF 72.694.105, Sarstedt, Numbrecht, Germany) containing 0.9% NaCl and 0.1% Tween 20. Analysis was performed as described by van den Munckhof et al. 15
Microbiology analysis―S. aureus quantification
A singleplex quantitative polymerase chain reaction (qPCR), adapted from literature, 16 targeting the nuc gene, was applied to quantify the S. aureus bacterial load. Quantification of bacterial load was done by comparing the results of the samples with the results of a standard curve with known concentrations. This standard curve was tested in parallel to the samples in each experiment. Samples that were below the limit of quantification (LOQ) were used in the analysis as half the lower LOQ.
Microbiome analysis
DNA extraction was performed automatically (MagNA Pure 96 DNA and Viral NA Large Volume Kit and Pathogen Universal 500, Roche Diagnostics, Basel, Switzerland). We used an input volume of 500‐μL sample and an elution volume of 100 µL. After DNA extraction, variable regions V3 and V4 of the 16S rRNA gene were amplified using primers described by Klindworth et al. 17 : Bakt_341F (5′ CCTACGGGNGGCWGCAG 3′) and Bakt_805R (5′ GACTACHVGGGTATCTAATCC 3′) with Illumina overhang adapter sequences added. The generated amplicons of around 460 base pairs were analyzed on a capillary system using a standard protocol, to confirm successful amplification of a PCR fragment of the expected size. As a next step, PCR products were cleaned using Ampure XP beads (Beckman Coulter, Brea, CA) to remove primer dimers and small aspecific PCR products and the purified PCR products were quantified (Quant‐iT PicoGreen dsDNA kit, Life Technologies, Carlsbad, CA), followed by serial dilution steps to reach the correct amount of input DNA. Index primers (Nextera XT Index kit, Illumina, San Diego, CA) were added by limited cycle PCR to the diluted PCR products. Before pooling, samples were normalized by using beads with maximum binding capacity and a sample preparation kit (Nextera XT).
The sequencing was performed on the Illumina MiSeq platform using an MiSeq v2 sequencing kit with 500 cycles (Illumina). De‐multiplexed FASTQ files were generated as output and the sequences of the FASTQ files were analyzed using Metagenomics workflow of the MiSeq Reporter software (Illumina), resulting in a taxonomy percentage summary of the sequenced bacterial sample. To calculate the relative abundance of microorganisms at the genus level, the unclassified DNA was excluded, hence the sum of the percentages of the DNA of all microorganisms found was set at 100%. Furthermore, the microorganisms comprising <10% were excluded from the analysis.
Biopsy biomarkers
Skin punch biopsies (4 mm) were collected from lesional skin at day 0 (predose) and day 28 and from nonlesional skin at day 0. Biopsies were placed in RNA later medium directly after harvest of the biopsy and stored and −80°C. The biopsies were analyzed at the DDL Laboratory (Rijswijk, The Netherlands). RNA extraction and real‐time quantitative PCR analysis was performed relative to the housekeeping gene glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). Because of limited material, a selection of protocol markers was made. The final set of markers was chosen based on disease involvement (IL‐31, eotaxin, interferon‐gamma (IFN‐γ)) and expected investigational drug effects (IFN‐α, IFN‐γ, and IL‐6). 18
Safety and tolerability
Safety and tolerability end points were assessed by frequency of treatment‐emergent adverse events (TEAEs), serious adverse events (SAEs), discontinuations due to AEs or deaths, laboratory values (hematology, chemistry, coagulation, and urinalysis), vital signs, electrocardiographic parameters, and physical examination.
Treatment compliance and exposure
Compliance of study drug application was recorded using a mobile e‐diary application, which entailed a notification and photo‐capture function enabling the date and time documentation of each gel application.
Statistical methods
All calculations were performed using SAS for Windows version 9.4 (SAS Institute, Cary, NC). No formal power calculation was performed given the exploratory character of this first‐in‐human study. Clinical efficacy and pharmacodynamic end points were analyzed with a mixed‐model analysis of variance using treatment, time, and treatment × time as fixed factors, and subject as random factor. Analyses were conducted in the clinical‐evaluable population. This population consisted of all subjects who applied the study medication for ≥21 days and completed the end‐of‐treatment visit. The results per variable are estimates of the difference of the various contrasts and back‐transformed estimates of the difference in percentage for log‐transformed parameters, 95% confidence intervals (in percent for log‐transformed parameters) and least‐square means (back transformed for log‐transformed parameters), and P‐values for contrast. The analyses of the mRNA expressions in the biopsies incorporated normalization for the housekeeping gene GAPDH. Moreover, it incorporated the values from nonlesional skin to correct for the high variability. Python version 3.5.2 software (Python Software Foundation, Wilmington, DE) was used for organization and visualization of the microbiome data.
RESULTS
Patients’ characteristics
Fifty‐nine patients were screened, of whom 36 were enrolled in the study. All enrolled patients completed the study ( Figure 1 ). Baseline characteristics were comparable between the treatment groups (Table 1 ). The overall mean BSA of the target lesion was 1.4% (±0.9).
Figure 1.
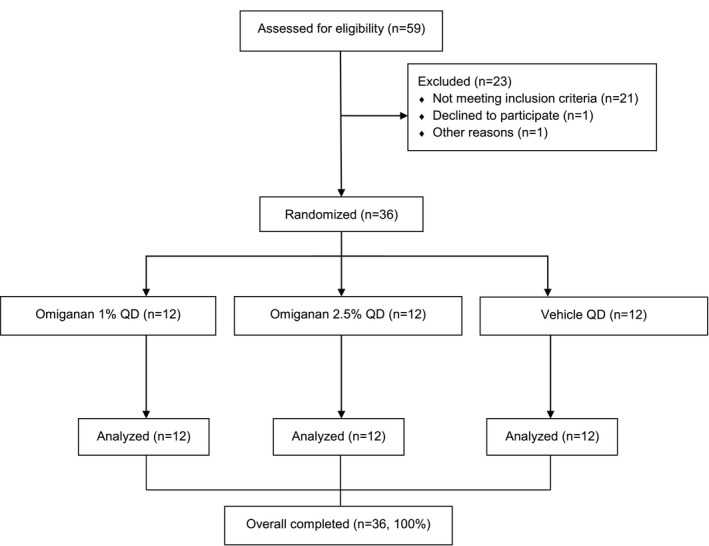
Flowchart of the study. QD = qualified dose (once daily).
Table 1.
Baseline characteristics of the study
|
Omiganan 1% N = 12 |
Omiganan 2.5% N = 12 |
Vehicle gel N = 12 |
Total N = 36 |
|
|---|---|---|---|---|
| Sex, n (%) | ||||
| Female | 9 (75) | 10 (83) | 8 (67) | 27 (75) |
| Male | 3 (25) | 2 (17) | 4 (33) | 9 (25) |
| Age, years (SD) | 25.0 (5.2) | 25.1 (7.1) | 24.7 (10.9) | 24.9 (7.8) |
| Fitzpatrick skin type, n (%) | ||||
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| II | 4 (33) | 5 (42) | 5 (42) | 15 (42) |
| III | 4 (33) | 5 (42) | 3 (25) | 12 (33) |
| IV | 1 (8) | 1 (8) | 2 (17) | 4 (11) |
| V | 2 (17) | 0 (0) | 1 (8) | 3 (8) |
| VI | 1 (8) | 0 (0) | 1 (8) | 2 (6) |
| Years since diagnosis, mean (SD) | 20.5 (9.4) | 19.2 (10.9) | 21.1 (12.2) | 20.3 (10.6) |
| Exacerbations per year, mean (SD) | 7.3 (6.9) | 9.1 (6.4) | 11.2 (9.8) | 9.2 (7.8) |
| Subjects with filaggrin mutation, n (%) | 0 (0%) | 3 (25%) | 4 (33%) | 7 (19%) |
| BSA (%)―target lesion, mean (SD) | 1.5 (0.9) | 1.0 (0.5) | 1.6 (1.1) | 1.4 (0.9) |
| oSCORAD index―target lesion, mean (SD) | 17.6 (7.5) | 16.3 (4.5) | 18.1 (8.4) | 17.3 (6.8) |
| BSA (%)―all lesions, mean (SD) | 9.1 (5.7) | 7.0 (7.2) | 8.9 (3.4) | 8.3 (5.6) |
| oSCORAD index―total body, mean (SD) | 18.4 (8.4) | 18.9 (6.4) | 17.8 (5.4) | 18.4 (6.7) |
| Previous treatment with corticosteroids (USA classification), n (%) | ||||
| Class IV corticosteroid | 6 (50) | 8 (67) | 6 (50) | 20 (56) |
| Class III corticosteroid | 6 (50) | 7 (58) | 7 (58) | 20 (56) |
| Class II corticosteroid | 5 (42) | 7 (58) | 6 (50) | 17 (47) |
| Class I corticosteroid | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Calcineurin inhibitor | 2 (17) | 7 (58) | 2 (17) | 11 (31) |
BSA, body surface area; oSCORAD, objective SCORing Atopic Dematitis; SD, standard deviation.
Clinical efficacy
A reduction of the target lesion oSCORAD index was observed in both active treatment groups compared with vehicle gel, mainly due to a reduction in percent BSA. This reduction was statistically significant for omiganan 2.5% (−18.5%; 95% confidence interval (CI), −32.9% to −1.0%), but not for omiganan 1% (−13.4%; 95% CI, −28.4% to 4.6%) (Figure 2a ), which may indicate a dose‐dependency. The reduction in the oSCORAD index in the omiganan 2.5% group was accompanied with a trend in reduction of percent BSA of the target lesion (−0.31; 95% CI, −0.64 to 0.03) (Figure 2b ).
Figure 2.
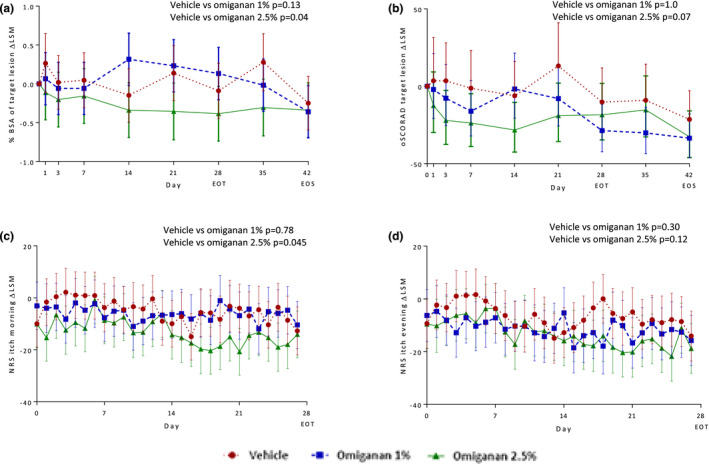
Change from baseline in body surface area, objective SCORing Atopic Dematitis (oSCORAD) index and morning and evening itch in the omiganan 1% and 2.5% treatment groups compared with the vehicle gel group. Change from baseline graphs: delta least‐squares mean (LSM) data over time of clinical assessments for (a) body surface area (BSA) and oSCORAD (b) of the target lesion. In the lower panels, patient‐reported outcomes are depicted, i.e., itch in the (c) morning and (d) evening.
PROs
A significant decrease in score on the 0‒100 NRS itch scale was observed during the morning in the omiganan 2.5% group compared with the vehicle gel group (−8.2; 95% CI, −16.3 to −0.2), but not in the omiganan 1% group (−1.1; 95% CI, −9.5 to 7.4). Itch during the evening slightly decreased in both active treatment groups. However, these decreases were not statistically significant (Figure 2c,d ).
Exploratory pharmacodynamics
Skin barrier function as measured by transepidermal water loss assessment improved in all treatment groups (−4.5, −8.8, and −12 in the vehicle gel group, omiganan 1% group, and omiganan 2.5% group, respectively) (Figure 3a ). The three‐dimensional photo analysis revealed no significant changes in roughness of the skin surface (data not shown). Biomarkers demonstrated a high degree of variability in general. At baseline, a significant difference between lesional and nonlesional mRNA expression in skin relative to GAPDH was observed for the markers eotaxin, IFN‐γ, and IL‐31 (Figure 3b–d ). There was no difference between lesional and nonlesional skin for the markers IFN‐α and IL‐6. No significant reductions in biomarkers were observed for any of the treatment groups. An example of the relative mRNA expression of eotaxin before and after treatment is shown in Figure 3e .
Figure 3.
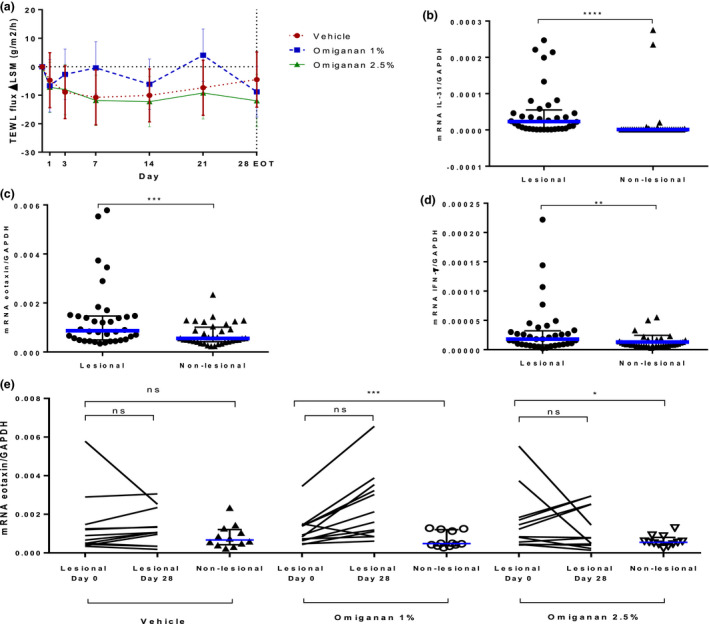
Pharmacodynamic effects of topical omiganan in the omiganan 1%, omiganan 2.5%, and vehicle gel groups. (a) Transepidermal water loss (TEWL) over time is depicted, showing improvement in the treatment groups and the vehicle group. Relative mRNA expressions in skin punch biopsy of markers (b) interleukin‐31, (c) eotaxin, and (d) interferon‐γ in lesional vs. nonlesional skin in mild to moderate atopic dermatitis patients at baseline. Data presented as median with interquartile range. Statistical significance is as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, based on a paired t‐test on log‐transformed data. (e) Relative mRNA expression of eotaxin per treatment group before treatment of lesional skin (day 0) and after end of treatment of lesional skin (day 28) and nonlesional (NL) skin. No treatment effect could be seen with this marker.
Microbiology and microbiome
No significant reductions in total bacterial load were observed in the two active treatment groups, i.e., omiganan 1% and omiganan 2.5%, compared with the vehicle gel group (data not shown). For the bacterial load information of S. aureus data by qPCR, a large proportion of samples showed results below the limit of quantification. There were no differences between treatment groups for the quantified samples. Microbiome data demonstrate a high degree of variability between patients in the presence and abundance of the genera. However, in general, the presence of the Staphylococcus genus dominated lesional skin compared with nonlesional skin (Figure 4 ). Individual data can be found in Supplementary Material S3 . After both active treatments, Staphylococcus abundance decreased significantly from 64% to 37% for omiganan 1% (P = 0.05) and from 70% to 42% (P = 0.01) for omiganan 2.5% compared with vehicle (Figures 4 , 5 ), whereas abundance in the vehicle group remained stable. With the decrease of Staphylococcus abundance, the summary of the microbiome diversity (Shannon index) increased significantly up to a total change of 0.11 for omiganan 1% (P = 0.03) and 0.08 for omiganan 2.5% (P = 0.03 vs. vehicle). At baseline, a moderate correlation was found between the target lesion oSCORAD index and Staphylococcus abundance (r = 0.409) and between the target lesion oSCORAD index and diversity index (r = 0.333). However, the decrease in the target lesion oSCORAD index as seen in both treatment groups did not correlate with the reduction in Staphylococcus abundance and increase in diversity (r = 0.182 and r = −0.096, respectively) (Figure 6 )
Figure 4.
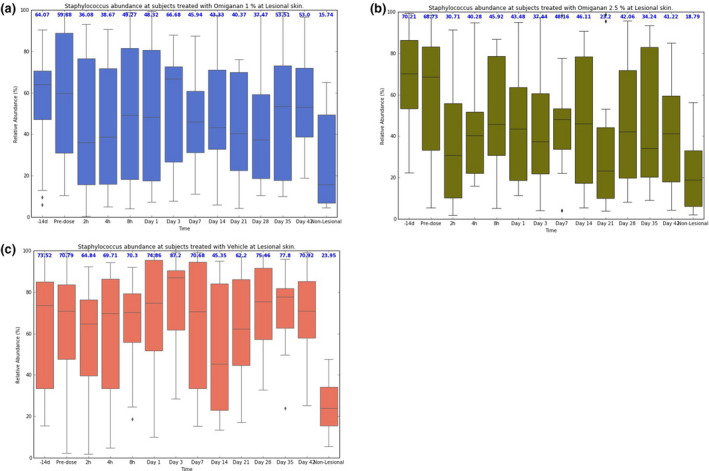
Course of cutaneous microbiome after omiganan treatment over time. Relative Staphylococcus abundance over time in the (a) omiganan 1%, (b) omiganan 2.5%, and (c) vehicle groups. A reduction is seen in both active treatment groups, but not in the vehicle group. Nonlesional boxes are presented as control on the right side. Median values are shown in blue above the boxplots.
Figure 5.
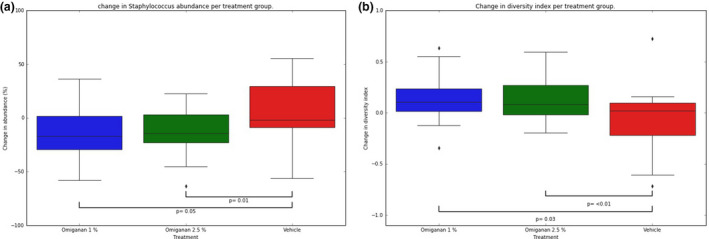
Omiganan improves dysbiosis of the target lesion. (a) Staphylococcus abundance and (b) diversity index from baseline to day 28 (end of treatment) per treatment group, with P‐values of the differences calculated using a mixed model for data over time.
Figure 6.
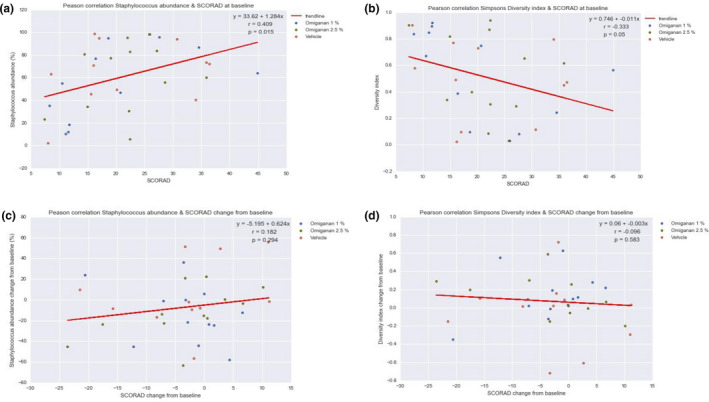
Correlation analysis of target lesion objective SCORing Atopic Dematitis (oSCORAD) index and Staphylococcus abundance. Correlations are shown between the (a) local target lesion oSCORAD index and Staphylococcus abundance in the microbiome and for the (b) target lesion oSCORAD and diversity index. (c,d) Delta correlations.
Safety and tolerability
Eighteen of 36 patients (50%) had at least 1 TEAE. All TEAEs were of mild (n = 24) or moderate severity (n = 3), The most frequent TEAEs were headache, upper respiratory tract infection, and influenza‐like illness. They were all self‐limiting and were considered unlikely related (n = 28) to treatment. No local AEs were reported. No discontinuations due to adverse events occurred. Application did not result in any clinically significant changes in safety laboratory parameters or vital signs. No SAEs, discontinuations due to AEs, or deaths occurred.
Treatment compliance and exposure
Treatment compliance was high and comparable in all treatment groups and ranged from 89% to 100%. 19 The mean usage of study drug per day ranged from 1.2 to 1.3 mg/cm2 in all treatment groups.
DISCUSSION
This is the first randomized, controlled, double‐blind clinical trial exploring the clinical efficacy, pharmacodynamics, and safety of omiganan in patients with AD. Treatment with omiganan 2.5% resulted in a clinically small but statistically significant reduction in the target lesion oSCORAD index and morning itch after 28 days of treatment compared with treatment with vehicle gel. This reduction was mainly caused by a reduction in BSA. As proof of pharmacology, a shift from lesional to a nonlesional microbiome profile in terms of a reduction of Staphylococcus genus and increase in diversity was seen in both active treatment groups compared with the vehicle gel group.
The shift in microbiome profile observed was predominantly driven by a reduction in Staphylococcus genus. This can be explained by the previously reported activity of omiganan against this genus. 8 The abundance of Staphylococcus is known to increase in skin affected by AD. Our study did show a moderate correlation between the clinical score (oSCORAD index) and the disturbed microbiome profile at baseline, which is in concordance with correlations of ≈0.50 described in other studies. 20 , 21 , 22 The correlation of improvement of the oSCORAD index and the degree of dysbiosis would suggest a major role of the microbiome in the pathogenesis of AD. However, a decrease in abundance of Staphylococcus, or an increase in microbial diversity, did not correlate with oSCORAD index improvement in our study. In some patients, this relationship was even reversed. In contrast, in previous studies with other treatments, such as topical corticosteroids and emollients, a correlation of clinical improvement and Staphylococcus reduction and/or increase in diversity index was reported. 20 , 22 A delay in clinical improvement after a recovery of the microbiome or variable individual responsiveness of the microbiome in the small treatment group in our study may represent underlying mechanisms prohibiting clear insight in the relation between the microbiome and clinical efficacy. It is also known that some individuals, by nature, have a greater abundance of Staphylococcus of up to 30% and/or a greater microbial diversity on the skin compared with others, although this does not appear to contribute to disease activity. 23 , 24 An alternative explanation could be that recovering the microbiome in mild to moderate AD patients does not lead to an improvement of clinical symptoms, but the effect is the other way around, wherein reduction of inflammation leads to normalization of the microbiome. This notion is supported by the fact that there is no evidence for a beneficial effect of antimicrobial interventions in noninfected AD, and the fact that the microbiome can recover with topical corticosteroids alone, which evidently lack antimicrobial properties. 25 More studies are needed to provide full insight in the relationship between the microbiome and inflammation in AD, as the outcomes of our study are not fully conclusive.
The clinical effects on the target lesion oSCORAD and affected BSA may be explained by the immunomodulatory effects of omiganan. Although there are no data available on omiganan, comprehensive data of immunomodulatory potential are available on another member of the cathelicidin family, namely LL‐37, which is the only endogenous human cathelicidin. LL‐37 has anti‐inflammatory properties, including inhibition of AIM2 inflammasome formation and suppression of IFN‐γ, tumor necrosis factor‐α, IL‐4, and IL‐12. 11 , 26 , 27 IFN‐γ expression remained unchanged after treatment (data not shown), but the other pro‐inflammatory markers were not included in this study. Moreover, LL‐37 is involved in skin barrier homeostasis and presumably suppresses itch. 28 On the contrary, LL‐37 is also involved in several pro‐inflammatory pathways when present in excess, such as in the downregulation of IL‐10 and mast cell release of inflammatory mediators. 29 , 30 In summary, there is evidence from AMP family members that the clinical effects of omiganan can rely on immunomodulatory properties rather than antimicrobial properties, but more in vitro and in vivo studies with omiganan are needed to draw conclusions.
Microbiome as biomarker
In a previous literature review we described the potential of skin microbiome‒associated outcomes as a biomarker in early‐phase clinical AD trials. 31 Although the exact relation between the skin microbiome and AD pathophysiology remains debatable based on this review, the implementation looks promising with respect to antimicrobial therapies. 31 In an earlier longitudinal observation study of AD patients, the authors found no major dissimilarity and robust microbiome profiles over time for both lesion and nonlesion skin. 15 In the current study we observed that the clear improvements in microbiome correlated only weakly with clinical response. Therefore, the microbiome may be considered a disease biomarker in AD to a lesser extent. For this part, because only a single AD lesion was treated and because high interindividual microbiome variability was observed, a larger study with total body application is needed to explore the full potential. However, the microbiome in this study provided proof of pharmacology of topical omiganan in patients with mild to moderate atopic dermatitis by serving as a drug mechanistic biomarker when assessing target engagement.
Itch reduction not clinically meaningful
Although there was a small, statistically significant reduction in the morning itch with omiganan 2.5% treatment compared with vehicle, the minimal clinically important difference (MCID), which is the smallest patient‐reported outcome change considered clinically meaningful, 32 was not achieved. The mean reduction in NRS morning itch in the 2.5% treatment group was 8.2 (on a scale of 0–100), whereas the MCID for itch was represented by a 20‒30‐point reduction. 33 The evening itch did not decrease significantly. When looking at the time of dose administration, most patients applied the gel in the morning (60%), which may explain why the effects on itch in the evening were less apparent. Itch reduction did not correlate with a reduction in IL‐31 mRNA expression, a biomarker for itch, in the skin biopsies at end of treatment, which highlights the questionable clinical relevance of the observed reduction. 34
Limitations
For safety reasons, only one target lesion was treated with the study drug, so the efficacy remains unknown with regard to all lesions. When treating and trying to recover only one lesion in terms of inflammation and Staphylococcus reduction, auto‐contamination of other lesions may occur, which may be relevant, assuming Staphylococcus plays a major role in the pathogenesis. However, our results indicate a clear shift in microbiome profile with a reduction in Staphylococcus genus. It remains unclear whether this includes S. aureus or other Staphylococcus species, because analysis of species level was not feasible with our NGS determination method. Unfortunately, the qPCR analysis was not able to perform quantitation in many samples, which precluded statistical analysis. Cultures for comparison with our NGS method were not performed in this study. Another limitation concerns the scoring of itch of the target lesion only. It may be difficult for patients to discern the itch from the target lesion from the itch related to other AD lesions and this may have influenced the scoring. Furthermore, study groups were relatively small for making definite conclusions about efficacy, and the clinical relevance of the decrease in target lesion oSCORAD and morning itch is therefore debatable.
In conclusion, in patients with mild to moderate AD, the topical administration of omiganan q.d. to a limited treatment area for up to 28 days is safe and well tolerated. Pharmacologic activity in terms of a significant reduction in the oSCORAD of the predefined target lesion and the patient‐reported itch was observed with the highest dose of 2.5%. However, because these reductions were small, the clinical relevance of both is debatable. The microbiome showed a significant shift from a lesional to nonlesional skin profile with both active treatments, indication low correlations with clinical improvement. Future studies with optimization of the treatment regimen, i.e., dose (exploration of more concentrations), frequency (b.i.d. instead of q.d.), and other subindications (such as infected AD), are needed to determine the true potential of omiganan in patients with AD.
Funding
Cutanea Life Sciences, Wayne, Pennsylvania, USA.
Conflict of Interest
G.L.F. was employed by Cutanea Life Sciences. The final draft was approved by G.L.F. on behalf of the sponsor, but no major comments were made. The authors declared no competing interests for this work.
Author Contributions
T.N.v.d.K., H.v.d.W., R.R., D.C.J.G.v.A., E.H.A.v.d.M., M.L.d.K., G.L.F., E.P.P., J.B., R.R., and M.B.A.v.D. wrote the manuscript. T.N.v.d.K, H.v.d.W., G.K.H., M.L.d.K., G.L.F., E.P.P., J.B., R.R., and M.B.A.v.D. designed the research. T.N.v.d.K., H.v.d.W., G.K.H., R.R., S.L., E.P.P., J.B., R.R., and M.B.A.v.D. performed the research. T.N.v.d.K, H.v.d.W., R.R., S.L., D.C.J.G.v.A., E.H.A.v.d.M., M.L.d.K., G.L.F, E.P.P., J.B., R.R., and M.B.A.v.D. analyzed the data.
Supporting information
Supinfo S1
Supinfo S2
Supinfo S3
Acknowledgments
We are grateful for the careful review of the manuscript by Dr Karen Broekhuizen.
Trial identifiers: EudraCT: 2014‐003689‐26, ClinicalTrials.gov Identifier: NCT02456480
References
- 1. DaVeiga, S.P. Epidemiology of atopic dermatitis: a review. Allergy Asthma Proc. 33, 227–234 (2012). [DOI] [PubMed] [Google Scholar]
- 2. Bieber, T. Atopic dermatitis. N. Engl. J. Med. 358, 1483–1494 (2008). [DOI] [PubMed] [Google Scholar]
- 3. Tuffs, S.W. , Haeryfar, S.M.M. & McCormick, J.K. Manipulation of innate and adaptive immunity by staphylococcal superantigens. Pathogens. 7, 53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park, H.Y. et al. Staphylococcus aureus colonization in acute and chronic skin lesions of patients with atopic dermatitis. Ann. Dermatol. 25, 410–416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ong, P.Y. et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 347, 1151–1160 (2002). [DOI] [PubMed] [Google Scholar]
- 6. Nomura, I. et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J. Immunol. 171, 3262–3269 (2003). [DOI] [PubMed] [Google Scholar]
- 7. Kisich, K.O. , Carspecken, C.W. , Fieve, S. , Boguniewicz, M. & Leung, D.Y. Defective killing of Staphylococcus aureus in atopic dermatitis is associated with reduced mobilization of human beta‐defensin‐3. J. Allergy Clin. Immunol. 122, 62–68 (2008). [DOI] [PubMed] [Google Scholar]
- 8. Fritsche, T.R. , Rhomberg, P.R. , Sader, H.S. & Jones, R.N. In vitro activity of omiganan pentahydrochloride tested against vancomycin‐tolerant, ‐intermediate, and ‐resistant Staphylococcus aureus . Diagn. Microbiol. Infect. Dis. 60, 399–403 (2008). [DOI] [PubMed] [Google Scholar]
- 9. Fritsche, T.R. , Rhomberg, P.R. , Sader, H.S. & Jones, R.N. Antimicrobial activity of omiganan pentahydrochloride against contemporary fungal pathogens responsible for catheter‐associated infections. Antimicrob. Agents Chemother. 52, 1187–1189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fritsche, T.R. , Rhomberg, P.R. , Sader, H.S. & Jones, R.N. Antimicrobial activity of omiganan pentahydrochloride tested against contemporary bacterial pathogens commonly responsible for catheter‐associated infections. J. Antimicrob. Chemother. 61, 1092–1098 (2008). [DOI] [PubMed] [Google Scholar]
- 11. Niyonsaba, F. , Kiatsurayanon, C. , Chieosilapatham, P. & Ogawa, H. Friends or foes? Host defense (antimicrobial) peptides and proteins in human skin diseases. Exp. Dermatol. 26, 989–998 (2017). [DOI] [PubMed] [Google Scholar]
- 12. van der Kolk, T. et al. Comprehensive, multimodal characterization of an imiquimod‐induced human skin inflammation model for drug development. Clin. Transl. Sci. 11, 607–615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen, M.P. , Karoly, P. & Braver, S. The measurement of clinical pain intensity: a comparison of six methods. Pain 27, 117–126 (1986). [DOI] [PubMed] [Google Scholar]
- 14. Elman, S. , Hynan, L.S. , Gabriel, V. & Mayo, M.J. The 5‐D itch scale: a new measure of pruritus. Br. J. Dermatol. 162, 587–593 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van den Munckhof, E.H.A. et al. Inter‐ and intra‐patient variability over time of lesional skin microbiota in adult patients with atopic dermatitis. Acta Derm. Venereol. 100, adv00018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pichon, B. et al. Development of a real‐time quadruplex PCR assay for simultaneous detection of nuc, Panton‐Valentine leucocidin (PVL), mecA and homologue mecALGA251. J. Antimicrob. Chemother. 67, 2338–2341 (2012). [DOI] [PubMed] [Google Scholar]
- 17. Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next‐generation sequencing‐based diversity studies. Nucleic Acids Res. 41, e1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niemeyer‐van der Kolk, T.P. et al. Omiganan enhances imiquimod‐induced inflammatory responses in a human skin challenge model. 2020;Feb 10. doi: 10.1111/cts.12741. [DOI] [PMC free article] [PubMed]
- 19. Rijsbergen, M. et al. Mobile e‐diary application facilitates the monitoring of patient‐reported outcomes and a high treatment adherence for clinical trials in dermatology. J. Eur. Acad. Dermatol. Venereol. 34, 633–639 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kong, H.H. et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim, M.H. et al. A metagenomic analysis provides a culture‐independent pathogen detection for atopic dermatitis. Allergy Asthma Immunol. Res. 9, 453–461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonzalez, M.E. et al. Cutaneous microbiome effects of fluticasone propionate cream and adjunctive bleach baths in childhood atopic dermatitis. J. Am. Acad. Dermatol. 75, 481–493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Human Microbiome Project Consortium . A framework for human microbiome research. Nature 486, 215–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grice, E.A. & Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 9, 244–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bath‐Hextall, F.J. , Birnie, A.J. , Ravenscroft, J.C. & Williams, H.C. Interventions to reduce Staphylococcus aureus in the management of atopic eczema: an updated Cochrane review. Br. J. Dermatol. 163, 12–26 (2010). [DOI] [PubMed] [Google Scholar]
- 26. Dombrowski, Y. et al Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Translat. Med. 3, 82ra38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kahlenberg, J.M. & Kaplan, M.J. Little peptide, big effects: the role of LL‐37 in inflammation and autoimmune disease. J. Immunol. 191, 4895–4901 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Umehara, Y. , Kamata, Y. , Tominaga, M. , Niyonsaba, F. , Ogawa, H. & Takamori, K. Cathelicidin LL‐37 induces semaphorin 3A expression in human epidermal keratinocytes: implications for possible application to pruritus. J. Invest. Dermatol. 135, 2887–2890 (2015). [DOI] [PubMed] [Google Scholar]
- 29. Niyonsaba, F. et al. Antimicrobial peptides human beta‐defensins and cathelicidin LL‐37 induce the secretion of a pruritogenic cytokine IL‐31 by human mast cells. J. Immunol. 184, 3526–3534 (2010). [DOI] [PubMed] [Google Scholar]
- 30. Niyonsaba, F. , Someya, A. , Hirata, M. , Ogawa, H. & Nagaoka, I. Evaluation of the effects of peptide antibiotics human beta‐defensins‐1/‐2 and LL‐37 on histamine release and prostaglandin D(2) production from mast cells. Eur. J. Immunol. 31, 1066–1075 (2001). [DOI] [PubMed] [Google Scholar]
- 31. Niemeyer‐van der Kolk, T. , van der Wall, H.E.C. , Balmforth, C. , Van Doorn, M.B.A. & Rissmann, R. A systematic literature review of the human skin microbiome as biomarker for dermatological drug development. Br. J. Clin. Pharmacol. 84, 2178–2193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Copay, A.G. , Subach, B.R. , Glassman, S.D. , Polly, D.W. Jr. & Schuler, T.C. Understanding the minimum clinically important difference: a review of concepts and methods. Spine J. 7, 541–546 (2007). [DOI] [PubMed] [Google Scholar]
- 33. Reich, A. et al. Itch assessment with visual analogue scale and numerical rating scale: determination of minimal clinically important difference in chronic itch. Acta Dermato‐Venereologica. 96, 978–980 (2016). [DOI] [PubMed] [Google Scholar]
- 34. Lee, C.H. & Yu, H.S. Biomarkers for itch and disease severity in atopic dermatitis. Curr. Probl. Dermatol. 41, 136–148 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supinfo S1
Supinfo S2
Supinfo S3


