Abstract
Osteogenesis imperfecta (OI) is a rare genetic disorder also known as a “brittle bone disease.” Around 90% of patients with OI harbor loss‐of‐function or dominant negative pathogenic variants in the COL1A1 and COL1A2 genes, which code for collagen type I α1 and α2 chains. Collagen‐related forms of the disorder are classified as Sillence OI types I–IV. OI phenotype expression ranges from mild to lethal. The current study aims to evaluate associations between interfamilial and intrafamilial phenotypic variability and genotype characteristics of patients with collagen‐related OI. The study was based on a systematic review of collagen‐related OI cases from the University of Tartu OI database (n = 137 individuals from 81 families) and the Dalgleish database (n = 479 individuals). Interfamilial variability analysis has shown that 17.74% of all studied OI‐related variants were associated with the same phenotype. The remaining 82.26% of pathogenic variants were associated with variable phenotypes. Additionally, higher interfamilial variability correlated with the COL1A1 gene (P value = 0.001) and dominant‐negative variants (P value = 0.0007). Within intrafamilial variability, 32.81% families had increasing or decreasing OI phenotype severity across generations. Higher intrafamilial variability of phenotypes correlated with the collagen I dominant negative variants (P value = 0.0246). The current study shows that, in line with other phenotype modification factors, OI interfamilial and intrafamilial diversity potential is associated with the genotype characteristics of the OI‐causing pathogenic variants. The results of the current study may advance knowledge of OI phenotype modification as well as assist family planning and the evaluation of disease progression in subsequent generations.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Osteogenesis imperfecta (OI) is a hereditary bone fragility disorder. The majority of patients with OI harbor pathogenic variants in the collagen type I genes (COL1A1 and COL1A2). OI is a genetically and phenotypically heterogeneous disorder. Previously, single cases of phenotype variability between carriers of the same pathogenic variant were described.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ The study aimed to perform analysis of phenotypic variability of collagen‐related OI in a large cohort of OI families. We analyzed cases with interfamilial and intrafamilial variability in order to evaluate associations between phenotypic variability and genotype characteristics of patients with OI with COL1A1 and COL1A2 pathogenic variants.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ Our study shows that families with loss‐of‐function collagen I pathogenic variants have lower phenotypic variability, whereas families with structural collagen I defects are more prone to show higher phenotypic variability.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ Our study adds to our understanding of OI phenotype modification and advances strategies for collagen‐related OI family planning and OI diagnosis.
Osteogenesis imperfecta (OI) is a heterogenous spectrum of rare, congenital bone fragility disorders. 1 The general prevalence of OI is 1/10–20,000. 2 , 3 A majority of patients with OI (90%) harbor autosomal dominant pathogenic variants in the COL1A1 and COL1A2 genes, which encode α1 and α2 chains of type I collagen. 4 The loss‐of‐function (LOF) pathogenic variants in type I collagen genes lead to reduced production of collagen, but the structure of the molecules is not altered. Missense pathogenic variants lead to a dominant‐negative (DN) effect and abnormal structure of collagen type I. 5
OI is characterized by frequent fractures, skeletal deformities, blue sclera, and hearing loss. In addition, patients with OI might have low bone mass, dentinogenesis imperfecta (DI), joint hypermobility, short stature, and pulmonary complications. 2 Based on phenotype and clinical and radiological characteristics, individuals with OI are divided into five clinical OI types: OI type I – mild, nondeforming OI with blue sclera; OI type II – perinatally lethal OI; OI type III – progressively deforming OI; OI type IV – common variable OI with normal sclera; and OI type V – OI with calcification in interosseous membranes. 2 , 6 OI genotype‐phenotype correlations exist to a certain extent. In general, DN collagen type I pathogenic variants are associated with more severe OI (types II, III, and IV), whereas LOF pathogenic variants mainly cause mild OI forms (type I). 5 , 7 , 8 However, interconnections between phenotype and genotype are much more complicated than presented above. The severity of the phenotype depends on the location of the pathogenic variant, changes in amino acid properties, and currently unknown modifying factors. 9 Therefore, OI symptoms develop individually, and there is still a lack of understanding as to the causes of phenotypic variability. 6 , 9
Phenotypic variability is common for many autosomal dominant monogenic disorders. 10 , 11 , 12 Comprehending the large spectrum of phenotype expressivity is challenging, as it induces misdiagnosis and underdiagnosis of the disease in asymptomatic family members. 13 A clinical phenotype may be hidden in one generation, but the genetic cause of the disease could still be transmitted to the offspring. 12 In the case of OI, affected family members may not only develop bone fragility phenotypes of differing expressivity, represented with different fracture numbers, but they may also differ in the presence of skeletal deformities, hearing loss, DI, sclera hue, and joint hypermobility. 14 , 15 , 16
The current study aims to systematize the OI interfamilial and intrafamilial phenotypic variability experience in a cohort of families from the University of Tartu (UT) OI database. We have conducted an analysis of interfamilial and intrafamilial variability in a large cohort of individuals with COL1A1‐related and COL1A2‐related OI. Interfamilial variability was explored in terms of correlation with genotype characteristics. To support the findings from the University of Tartu Osteogenesis Imperfecta database of the Clinic of Traumatology and Orthopedics, University of Tartu, Estonia (UT OI database), open access data were included in the analysis from the Dalgleish OI variant database of the Dalgleish Laboratory, Department of Genetics, University of Leicester, UK (Dalgleish database). 17 Intrafamilial variability analysis was performed in kindred with familial OI to assess the dependence of the degree of variable expressivity on genotype. This research provides valuable knowledge to enhance strategies for OI diagnosis in family planning and provides additional knowledge about OI phenotype modification.
METHODS
UT OI database description
The UT OI database was established in 1995 when OI treatment and follow‐up were centralized to one location in Estonia. The UT OI database includes 238 OI families of Estonian (n = 30), Ukrainian (n = 94), and Vietnamese (n = 114) origin. The database consists of clinical, phenotypic, genealogical, and genotype information collected from patients with OI and their healthy relatives during interviews. Clinical examination, phenotype descriptions, and mutational analysis are performed by UT’s medical and research team. Patients are classified into OI clinical types I–V, in concordance with the updated Sillence classification. Mutational analysis of the collagen I genes was performed previously using Sanger sequencing. 18 , 19 , 20
Interfamilial variability analysis
Our analysis of interfamilial variability is based on the collagen‐related OI cases in the UT OI database evaluated from 1995 to 2018 and on data from the Dalgleish OI variant database evaluated in April 2019 (https://oi.gene.le.ac.uk). 17
The study cohort was comprised of UT OI database families that harbored a non‐novel COL1A1 or COL1A2 variant (i.e., variants reported at least twice in the UT OI or Dalgleish databases). The remaining families carried novel variants, which lack phenotype data about other carriers for comparison. The selection process of the patients for the study is represented in the flow diagram (Figure 1 ). Eighty‐one families (137 individuals) who harbored 62 non‐novel COL1A1/COL1A2 variants that were previously independently reported in the OI variant database in 479 individuals, fulfilled the inclusion criteria.
Figure 1.
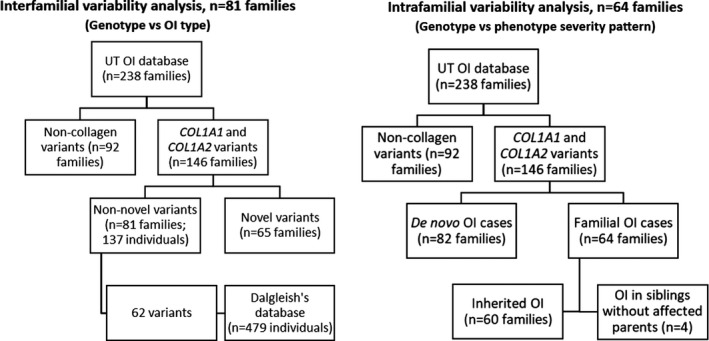
Flow diagram of the selection of patients with osteogenesis imperfecta (OI) from the University of Tartu Osteogenesis Imperfecta database of the Clinic of Traumatology and Orthopedics, University of Tartu, Estonia (UT OI database) for the interfamilial and intrafamilial analysis of phenotypic variability.
An open‐ended strategy was used to sort pathogenic variants included in the study according to their OI clinical types in the UT OI and Dalgleish databases. A systematic review of patients’ genotypes, clinical types, and the severity of phenotypes was used to form genotype‐phenotype severity groups. Groups were based on the following criteria: first, those that harbored OI mutations; and second, all clinical OI types reported in both databases. We analyzed associations between genotype (i.e., COL1A1 and COL1A2 gene; LOF and DN variant) and OI type in different groups (monophenotype and polyphenotype) using Fisher’s χ2 test for categorical variables. The threshold of statistical significance had a P value < 0.05. Statistical analysis of the data was completed with R version 3.3.2. software (R Team, Vienna, Austria). 21
Intrafamilial variability analysis
Intrafamilial analysis was performed in families with familial OI (i.e., > 1 affected individual). Of 146 collagen‐related OI families, 68 families fulfilled the criteria. In 64 families, there was a history of OI, and in 4 more families, OI was identified in a few siblings from the same generation without previous OI history (Figure 1 ). Affected family members from families with high intrafamilial variability (i.e., types I, III, and IV inside one family) underwent whole exome sequencing with Illumina HiSeq2000, following the previously described procedure. 22
The severity patterns of OI phenotypes were evaluated via typical OI signs: the number of fractures; age at the first fracture; limb, rib cage, and spine deformity degree; stature; and mobility status. Families were grouped according to severity pattern changes as follows: “no changes in severity,” “progressive severity,” and “decreasing severity.” Changes in phenotype severity between siblings and cousins were classified as “same severity” (i.e., identical symptoms) and “different severity” (i.e., symptoms differed between the siblings or cousins). Correlations between genotype (i.e., COL1A1 and COL1A2 gene; LOF and DN variant) and degree of intrafamilial diversity (i.e., the above‐mentioned groups of severity patterns) were checked with Fisher’s χ2 test for categorical variables. P values < 0.05 were considered to be statistically significant A statistical analysis of the data was completed with R version 3.3.2. software (R Team). 21
Ethical compliance
The study was conducted in accordance with the Helsinki Declaration and authorized by the Ethical Review Committee on Human Research of the University of Tartu (Permit no. 221/M‐34), the Ethical Review Board of Hue University Hospital (approval No. 75/CN‐BVYD), and the Sytenko Institute of Spine and Joint Pathology of the Ukrainian Academy of Medical Sciences. Informed written consent was collected from all subjects and controls or their legal representatives prior to collection of their clinical information and biological samples.
The data used to support the findings of this study are present in Table 1 and Table 2 of the current paper, as well as in the open access Dalgleish OI variant database.
Table 1.
Phenotypic diversity in COL1A1 a and COL1A2 b pathogenic variants discovered in patients with OI from the UT OI biobank
| No. | Pathogenic variant | Gene | Mutation type | Collagen defect | Protein alteration | Sillence OI type reported in the UT OI database studied cohort c | Sillence OI type reported in the Dalgleish OI variant database c |
|---|---|---|---|---|---|---|---|
| 1. Monophenotypes | |||||||
| 1.1 Mild monophenotypes | |||||||
| 1 | c.590G>A | COL1A1 | Missense | DN | p.Gly197Asp | I (1) | I (2) |
| 2 | c.3766G>A | COL1A1 | Missense | DN | p.Ala1256Thr | I (1) | I (1) |
| 3 | c.1354‐2A>G | COL1A1 | Splice site | LOF | ‐ | I (1) | I (1) |
| 4 | c.904‐9G>A | COL1A1 | Splice site | LOF | ‐ | I (2) | I (1) |
| 5 | c.579_delT | COL1A1 | Frameshift | LOF | p.Gly194Valfs*71 | I (1) | I (22) |
| 6 | c.459_delT | COL1A1 | Frameshift | LOF | p.Gly154Alafs*111 | I (1) | I (1) |
| 7 | c.3807G>A | COL1A1 | Nonsense | LOF | p.Trp1269* | I (1) | I (2) |
| 8 | c.3076C>T | COL1A1 | Nonsense | LOF | p.Arg1026* | I (1) | I (12) |
| 1.2 Moderate monophenotype | |||||||
| 1 | c.1630G>A | COL1A2 | Missense | DN | p.Gly544Ser | IV (1) | IV (1) |
| 1.3 Severe monophenotype | |||||||
| 1 | c.1165G>A | COL1A1 | Missense | DN | p.Gly389Ser | III (1) | III (3) |
| 2 | c.742G>A | COL1A1 | Missense | DN | p.Gly284Arg | III (1) | III (1) |
| 2. Polyphenotypes | |||||||
| 2.1 Less variable phenotypes | |||||||
| 2.1.1 Mild‐to‐moderate OI phenotypes | |||||||
| 1 | c.2560G>A‡ | COL1A1 | Missense | DN | p.Gly854Ser | I, IV (1) | I/IV (2) |
| 2 | c.3235G>A | COL1A1 | Missense | DN | p.Gly1079Ser | I (1) | I, IV (20) |
| 3 | c.653G>A | COL1A1 | Missense | DN | p.Gly218Asp | I (2) | IV (1) |
| 4 | c.959G>A | COL1A1 | Missense | DN | p.Gly320Asp | IV (1) | I (1) |
| 5 | c.1451G>A | COL1A2 | Missense | DN | p.Gly484Glu | IV (1) | I (2) |
| 6 | c.3305G>T | COL1A2 | Missense | DN | p.Gly1102Val | I (1) | IV (1) |
| 7 | c.750+2T>A‡ | COL1A1 | Splice site | LOF | ‐ | I, IV (1) | I (1) |
| 8 | c.3045+1G>A | COL1A1 | Splice site | LOF | ‐ | IV (1) | I (1) |
| 9 | c.299‐1G>C‡ | COL1A1 | Splice site | LOF | ‐ | IV (1) | I (3) |
| 10 | c.1614+1G>A | COL1A1 | Splice site | LOF | ‐ | IV (1) | I (3) |
| 11 | c.858+1G>A‡ | COL1A1 | Splice site | LOF | ‐ | I, IV (2) | I (2) |
| 12 | c.804+1G>A | COL1A1 | Splice site | LOF | ‐ | IV (1) | I (1) |
| 13 | c.1002+2T>C | COL1A1 | Splice site | LOF | ‐ | IV (1) | I (1) |
| 14 | c.1299+1G>C | COL1A1 | Splice site | LOF | ‐ | IV (1) | I, IV (4) |
| 15 | c.2614‐1G>A | COL1A1 | Splice site | LOF | ‐ | IV (1) | I (1) |
| 16 | c.2523_delT | COL1A1 | Frameshift | LOF | p.Gly842Alafs*266 | I (1) | I, IV (4) |
| 17 | c.658C>T‡ | COL1A1 | Nonsense | LOF | p.Arg220* | I, IV (1) | I (11) |
| 18 | c.2089C>T | COL1A1 | Nonsense | LOF | p.Arg697* | IV (1) | I, IV (8) |
| 19 | c.1789G>T | COL1A1 | Nonsense | LOF | p.Glu597* | I (1) | IV (1) |
| 20 | c.1081C>T | COL1A1 | Nonsense | LOF | p.Arg361* | IV (2) | I (10) |
| 2.1.2 Severe‐to‐moderate phenotypes | |||||||
| 1 | c.2101G>T | COL1A1 | Missense | DN | p.Gly701Cys | III (1) | IV (1) |
| 2 | c.3226G>A | COL1A1 | Missense | DN | p.Gly1076Ser | IV (2) | III, IV (13) |
| 3 | c.1090G>A | COL1A2 | Missense | DN | p.Gly364Ser | III (1) | IV (1) |
| 4 | c.2288G>T | COL1A2 | Missense | DN | p.Gly763Val | III (1) | III/IV (2) |
| 5 | c.2324G>A | COL1A2 | Missense | DN | p.Gly775Glu | III (2) | III, IV (1) |
| 6 | c.3034G>A | COL1A2 | Missense | DN | p.Gly1012Ser | III (2) | III/IV (24) |
| 7 | c.865G>A | COL1A2 | Missense | DN | p.Gly289Ser | III (1) | III/IV, IV (2) |
| 8 | c.792+1G>A | COL1A2 | Splice site | LOF | ‐ | III (1) | IV (1) |
| 9 | c.2613+6T>C | COL1A1 | Splice site | LOF | ‐ | IV (1) | III (1) |
| 2.2 More variable phenotypes | |||||||
| 2.2.1 Opposite OI phenotypes | |||||||
| 1 | c.1072G>T‡ | COL1A2 | Missense | DN | p.Gly358Ser | I, III (1) | III (1) |
| 2 | c.2233G>C | COL1A2 | Missense | DN | p.Gly745Arg | III (1) | I (1) |
| 3 | c.874G>A | COL1A2 | Missense | DN | p.Gly292Ser | III (1) | I (5) |
| 2.2.2 Non‐lethal phenotypes | |||||||
| 1 | c.2362G>A | COL1A1 | Missense | DN | p.Gly788Ser | IV (2) | I, III, IV (8) |
| 2 | c.2461G>A‡ | COL1A1 | Missense | DN | p.Gly821Ser | I, IV (5) | I, III, IV (26) |
| 3 | c.757C>T | COL1A1 | Missense | DN | p.Arg253Phe | IV (1) | I, III, IV (11) |
| 4 | c.769G>A‡ | COL1A1 | Missense | DN | p.Gly257Arg | I, IV (1) | I, III, IV (37) |
| 5 | c.977G>A | COL1A1 | Missense | DN | p.Gly326Asp | I (1) | I, III, IV (3) |
| 6 | c.1057G>T‡ | COL1A1 | Missense | DN | p.Gly353Cys | I, IV (1) | III/IV (2) |
| 7 | c.1165G>T | COL1A1 | Missense | DN | p.Gly389Cys | I (2) | III/ IV (2) |
| 8 | c.2314G>A‡ | COL1A2 | Missense | DN | p.Gly772Ser | I, IV (1) | I, III, IV (18) |
| 9 | c.2503G>A | COL1A2 | Missense | DN | p.Gly835Ser | III (1) | I, III, IV (4) |
| 10 | c.982G>A‡ | COL1A2 | Missense | DN | p.Gly328Ser | IV, III (2) | I, III, IV (45) |
| 11 | c.1009G>A | COL1A2 | Missense | DN | p.Gly337Ser | III, IV (3) | I, III, IV (29) |
| 12 | c.1821+1G>A‡ | COL1A1 | Splice site | LOF | ‐ | III, IV, I (2) | I, IV (17) |
| 13 | c.1243C>T | COL1A1 | Nonsense | LOF | p.Arg415* | I (1) | I, III/IV (16) |
| 14 | c.1128_delT‡ | COL1A1 | Frameshift | LOF | p.Gly377Alafs*164 | I, IV, III (1) | I (9) |
| 2.2.3 Mild‐to‐lethal phenotypes | |||||||
| 1 | c.1102G>A | COL1A1 | Missense | DN | p.Gly368Ser | IV (1) | II/III, III (2) |
| 2 | c.1588G>A | COL1A1 | Missense | DN | p.Gly530Ser | III (1) | II, III, IV (12) |
| 3 | c.2299G>A | COL1A1 | Missense | DN | p.Gly767Ser | I, III, IV (3) | I, II, III, IV (34) |
| 4 | c.2596G>A | COL1A1 | Missense | DN | p.Gly866Ser | III (1) | II, III (10) |
| 5 | c.1378G>A | COL1A2 | Missense | DN | p.Gly460Ser | IV (1) | II/III, III, IV (16) |
OI types reported in the OI variant database were obtained from http://www.le.ac.uk/ge/collagen/.
DN, dominant negative; LOF, loss‐of‐function; OI, osteogenesis imperfecta; UT, University of Tartu.
COL1A1 GenBank reference sequence (gDNA NG_007400.1, cDNA NM_000088.3). b COL1A2 GenBank reference sequence (gDNA NG_007405.1, cDNA NM_000089.3). cThe number of reported families in the UT OI database and the number of reported cases in the Dalgleish variant database are represented in parentheses.
“Stop codon” in mutation nomenclature.
Variants which caused variety of phenotypes intrafamilially are marked with a diesis (‡).
Table 2.
COL1A1 a and COL1A2 b pathogenic variants discovered in patients with OI with intrafamilial diversity
| No. | Pathogenic variant | Gene | Mutation type | Collagen defect | Protein alteration |
|---|---|---|---|---|---|
| Pathogenic variants discovered in patients with OI with decreasing OI severity within families | |||||
| 1 | c.1821+1G>A | COL1A1 | Splice site | LOF | ‐ |
| 2 | c.750+2T>A | COL1A1 | Splice site | LOF | ‐ |
| 3 | c.858+1G>A | COL1A1 | Splice site | LOF | ‐ |
| 4 | c.1128_delT | COL1A1 | Frameshift | LOF | p.Gly377Alafs*164 |
| 5 | c.769G>A | COL1A1 | Missense | DN | p.Gly257Arg |
| 6 |
c.370‐1G>A /c.2642A>C |
COL1A1 /COL1A2 |
Splice site/Missense | LOF/DN | ./p.Glu881Ala |
| 7 | c.3305G>T | COL1A2 | Missense | DN | p.Gly1102>Val |
| 8 | c.1072G>T | COL1A2 | Missense | DN | p.Gly358Ser |
| Pathogenic variants discovered in patients with OI with progressing OI severity within families | |||||
| 1 | c.3217G>A | COL1A1 | Missense | DN | p.Gly1073Ser |
| 2 | c.2362G>A | COL1A1 | Missense | DN | p.Gly788Ser |
| 3 | c.3652G>A | COL1A1 | Missense | DN | p.Ala1218Thr |
| 4 | c.1057G>T | COL1A1 | Missense | DN | p.353G>G/C |
| 5 | c.2560G>A | COL1A1 | Missense | DN | p.Gly854Ser |
| 6 | c.2461G>A | COL1A1 | Missense | DN | p.Gly821Ser |
| 7 | c.2461G>A | COL1A1 | Missense | DN | p.Gly821Ser |
| 8 | c.2314G>A | COL1A2 | Missense | DN | p.Gly772Ser |
| 9 | c.982G>A | COL1A2 | Missense | DN | p.Gly328Ser |
| 10 | c.658C>T | COL1A1 | Nonsense | LOF | p.Arg220* |
| 11 | c.495T>A | COL1A1 | Nonsense | LOF | p.Tyr165* |
| 12 | c.1816_delG | COL1A1 | Frameshift | LOF | p.Ala606Leufs*160 |
| 13 | c.103+2T>C | COL1A1 | Splice site | LOF | ‐ |
| Pathogenic variants discovered in patients with OI without changes in OI severity within families | |||||
| 1 | c.505G>A | COL1A1 | Missense | DN | p.Glu169Lys |
| 2 | c.1A>C | COL1A1 | Missense | DN | p.Met1Leu |
| 3 | c.3223G>A | COL1A1 | Missense | DN | p.Ala1075Thr |
| 4 | c.959G>A | COL1A1 | Missense | DN | p.Gly320Asp |
| 5 | c.1102G>A | COL1A1 | Missense | DN | p.Gly368Ser |
| 6 | c.1165G>T | COL1A1 | Missense | DN | p.Gly389Cys |
| 7 | c.977G>A | COL1A1 | Missense | DN | p.Gly326Asp |
| 8 | c.2005G>A | COL1A1 | Missense | DN | p.Ala669Thr |
| 9 | c.2362G>A | COL1A1 | Missense | DN | p.Gly788Ser |
| 10 | c.2299G>A | COL1A1 | Missense | DN | p.Gly767Ser |
| 11 | c.3766G>A | COL1A1 | Missense | DN | p.Ala1256Thr |
| 12 | c.1009G>A | COL1A2 | Missense | DN | p.Gly337Ser |
| 13 | c.1009G>A | COL1A2 | Missense | DN | p.Gly337Ser |
| 14 | c.1630G>A | COL1A2 | Missense | DN | p.Gly544Ser |
| 15 | c.1009G>A | COL1A2 | Missense | DN | p.Gly337Ser |
| 16 | c.1964G>T | COL1A2 | Missense | DN | p.Gly655Val |
| 17 | c.579_delT | COL1A1 | Frameshift | LOF | p.Gly194Valfs*71 |
| 18 | c.2821_delG | COL1A1 | Frameshift | LOF | p.Gly941Valfs*167 |
| 19 | c.2393_dupC | COL1A1 | Frameshift | LOF | p.Gly800Argfs*5 |
| 20 | c.2523_delT | COL1A1 | Frameshift | LOF | p.Gly842Alafs*266 |
| 21 | c.630_delG | COL1A1 | Frameshift | LOF | p.Glu210Aspfs*3 |
| 22 | c.2523_delT, | COL1A1 | Frameshift | LOF | p.Gly842Alafs*266 |
| 23 | 2093_2110_dup | COL1A2 | Frameshift | LOF | p.Leu699_Leu704dup |
| 24 | c.1897G>T | COL1A1 | Nonsense | LOF | p.Glu633* |
| 25 | c.2089C>T | COL1A1 | Nonsense | LOF | p.Arg697* |
| 26 | c.3262G>T | COL1A1 | Nonsense | LOF | p.Gly1088* |
| 27 | c.3262G>T | COL1A1 | Nonsense | LOF | p.Gly1088* |
| 28 | c.1081C>T | COL1A1 | Nonsense | LOF | p.Arg361* |
| 29 | c.3076C>T | COL1A1 | Nonsense | LOF | p.Arg1026* |
| 30 | c.1243C>T | COL1A1 | Nonsense | LOF | p.Arg415* |
| 31 | c.3807G>A | COL1A1 | Nonsense | LOF | p.Trp1269* |
| 32 | c.2179C>T | COL1A1 | Nonsense | LOF | p.Gln727* |
| 33 | c.1821+1G>A | COL1A1 | Splice site | LOF | ‐ |
| 34 | c.1155+2T>G | COL1A1 | Splice site | LOF | ‐ |
| 35 | c.299‐1G>C | COL1A1 | Splice site | LOF | ‐ |
| 36 | c.1767+5G>A | COL1A1 | Splice site | LOF | ‐ |
| 37 | c.1354‐2A>G | COL1A1 | Splice site | LOF | ‐ |
| 38 | c.904‐9G>A | COL1A1 | Splice site | LOF | ‐ |
| 39 | c.2613+6T>C | COL1A1 | Splice site | LOF | ‐ |
| 40 | c.804+1G>A | COL1A1 | Splice site | LOF | ‐ |
| 41 | c.103+2T>C | COL1A1 | Splice site | LOF | ‐ |
| 42 | c.2614‐1G>A | COL1A1 | Splice site | LOF | ‐ |
| 43 | c.2026‐1_2031het dup | COL1A2 | Splice site | LOF | ‐ |
DN, dominant negative, LOF, loss‐of‐function; OI, osteogenesis imperfecta.
COL1A1 GenBank reference sequence (gDNA NG_007400.1, cDNA NM_000088.3). b COL1A2 GenBank reference sequence (gDNA NG_007405.1, cDNA NM_000089.3).
“Stop codon” in mutation nomenclature.
RESULTS
Interfamilial diversity
All studied variants were divided into two main groups: monophenotypes (variants that caused the same OI type in all affected individuals), and polyphenotypes (variants that caused variable OI types; Figure 2 ). Only 17.74% of the variants (11/62) were monophenotypic, whereas a majority of variants, 82.26% (51/62), were in the polyphenotype group (Table 1 ).
Figure 2.
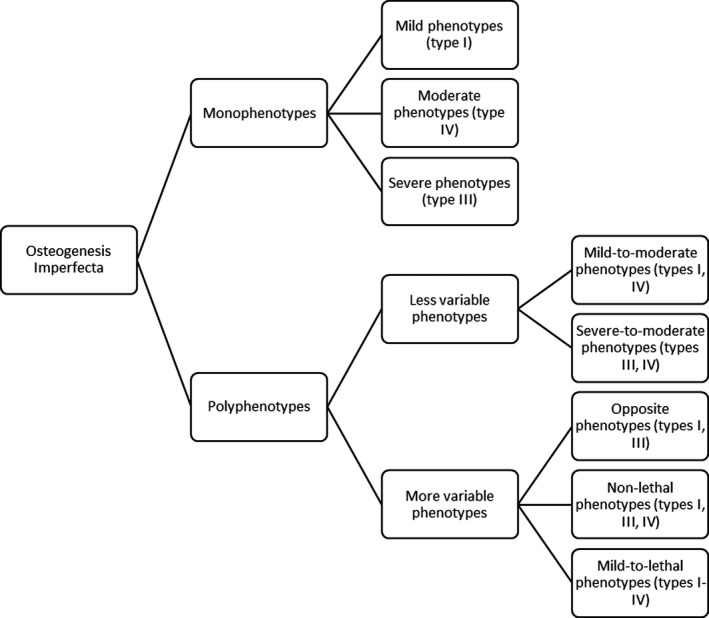
Clustering of the osteogenesis imperfecta (OI) phenotypic variability groups, formed on the basis of the genotype‐phenotype variability analysis and the severity scale of clinical symptoms. Mild phenotype ‐ OI type I, moderate phenotype ‐ OI type IV, severe phenotype ‐ OI type III, and lethal phenotype ‐ OI type II.
OI type I alone was caused by eight variants, all of which altered the COL1A1 gene. Two of them were DN variants—c.590G>A, p.(Gly197Asp) and c.3766G>A, p.(Ala1256Thr)—and six were LOF variants. A frameshift COL1A1 c.579delT, p.(Gly194Valfs)* variant, present in one of our patients, was one of the most common variants in the Dalgleish database and was identified 22 times in patients with OI type I only. OI type IV was caused by a single monophenotypic DN COL1A2 variant: c.1630G>A, p.(Gly544Ser). OI type III was caused by two DN variants in the COL1A1 gene: c.1165G>A, p.(Gly389Ser) and c.742G>A, p.(Gly284Arg).
We divided the polyphenotype group into variants, which comprised the less variable borderline mild‐to‐moderate and severe‐to‐moderate OI types (I, IV and III, and IV) and more variable types. The latter were represented with three classes: the opposite phenotypes (types I and III); nonlethal OI (types I, III, and IV), and mild‐to‐lethal OI (types I–IV).
The mild‐to‐moderate OI group included 20 pathogenic variants (14 LOF and 6 DN Gly substitutions). Severe‐to‐moderate OI was caused by nine variants (seven variants were DN variants and two LOF). Among the patients from the UT OI database was a pair of monozygotic twin girls (age 13, lock time May 2016), who carried the COL1A2, c.3034G>A, p.(Gly1012Ser) pathogenic variant. Both individuals were identically affected with OI type III. The twin girls had the identical, severe skeletal deformities (severe lower and upper limb deformities, severe kypho‐scoliosis, and chest deformity), a moderate number of fractures (10–15), had DI and white eye sclera, and no signs of hearing loss. Twenty‐five other individuals with the same variant from the Dalgleish database were described to have OI types III and IV.
The opposite phenotypes or extreme ends of the phenotype spectrum were caused by three DN Gly substitutions in the collagen I α2 chain: c.1072G>T, p.(Gly358Ser); c.2233G>C, p.(Gly745Arg); and c.874G>A, p.(Gly292Ser).
The nonlethal OI group included 14 variants. Eleven variants were DN (seven in the COL1A1 and four in the COL1A2 genes), and three were LOF variants (c.1821+1G>A, c.1243C>T, p.(Arg415*) and c.1128_delT, all in the COL1A1 gene).
The mild‐to‐lethal OI group consisted of five DN variants. All of them were Gly substitutions with Ser (four variants in the COL1A1 gene: c.1102G>A, p.(Gly368Ser); c.1588G>A, p.(Gly530Ser); c.2299G>A, p.(Gly767Ser); c.2596G>A, p.(Gly866Ser); and one variant c.1378G>A, p.(Gly460Ser) in the COL1A2 gene). The COL1A1 c.2299G>A, p.(Gly767Ser) variant was present in three patients from the UT OI database with OI types I, IV, and III (Figure 3 ). The current variant is present in 34 individuals reported in the Dalgleish database. This was the only variant in the current study that was responsible for a whole spectrum of OI phenotypes—from mild to lethal.
Figure 3.
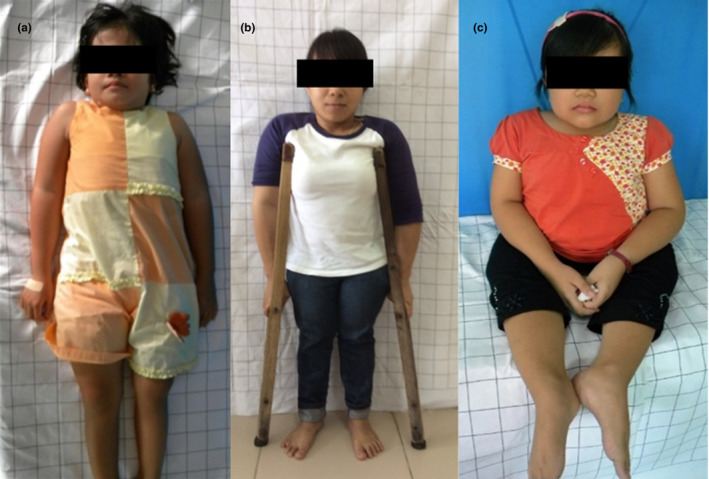
Three unrelated female patients with different clinical types of osteogenesis imperfecta (OI) from the Clinic of Traumatology and Orthopedics, University of Tartu, Estonia (UT OI database) harboring the same pathogenic dominant negative variant c.2299G>A, p.Gly767Ser in the COL1A1 gene. Patients developed (a) type I, (b) type IV, and (c) type III of OI, supporting high interfamilial variability of this pathogenic variant.
We have identified an interfamilial diversity pattern that depends on the mutated gene and the suspected consequence defect of the variant (Figure 4 ). Patients lacking phenotypic diversity harbored mutations in the COL1A1 gene (P value 0.001), whereas patients with variable phenotypes were more likely to harbor the COL1A2 pathogenic variants. Interfamilial diversity also correlated with a collagen I defect type (P value 0.0007). LOF pathogenic variants lacked diversity, whereas DN variants were associated with higher degrees of phenotype variability.
Figure 4.
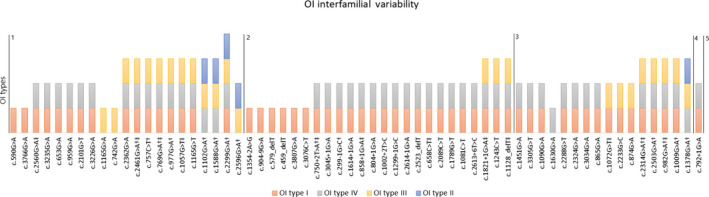
Diagram of osteogenesis imperfecta (OI) clinical types identified in carriers of the studied COL1A1 and COL1A2 variants. Data taken from the Clinic of Traumatology and Orthopedics, University of Tartu, Estonia (UT OI database) and the Dalgleish variant database. Area 1‐2 COL1A1 dominant negative; area 2‐3 COL1A1 loss‐of‐function; area 3‐4 COL1A2 dominant negative; area 4‐5 COL1A2 loss‐of‐function pathogenic variants. Red color ‐ mild OI (OI type I); grey color ‐ moderate OI (OI type IV); yellow color ‐ severe OI (OI type III); and blue color ‐ lethal OI (OI type II).
Intrafamilial diversity
Although all affected family members were carriers of the same pathogenic variant, in some families, phenotypes tended to vary. Of 64 families with a history of OI, 21 families (32.81%) had variability of OI between different generations, and 43 families (67.19%) lacked phenotype variability (Table 2 ). The number of families lacking signs of OI phenotypic variability was as follows in the three studied populations: Estonian (n = 13), Ukrainian (n = 15), and Vietnamese (n = 15) families. Of them, 27 (62.79%) harbored LOF variants. Variability between siblings and cousins within the same generation was less common and was registered in 7/68 families (10.29%).
Follow‐ups conducted with families comprising three and four generations has shown that general changes in OI severity occur gradually in one direction, either decreasing or increasing through the generations. In addition, most families with intrafamilial diversity presented cases of mild‐to‐moderate OI within the single family. Only two families included all nonlethal OI types: I, IV, and III. Both of these Estonian OI families from the UT OI database had reduced OI severity in the younger generations. whole exome sequencing analysis of these families has shown that all affected family members, regardless of their phenotype, harbor the same pathogenic variants (COL1A1 c.1821+1G>GA; COL1A1, c.1128_delT, p.(Gly337Alafs*164)). Totally decreasing severity in younger generations was discovered in 8 families (36.37%), and half of them harbored LOF mutations. The families identified were of Ukrainian (n = 3), Estonian (n = 3), and Vietnamese (n = 2) origin.
Progressing severity was registered in 13 families (61.90%; data registered from two (n = 8), three (n = 3), and four (n = 2) generations). This was identified in eight Ukrainian (n = 8) families, four Vietnamese (n = 4) families, and one Estonian (n = 1) OI family. Patients with DN pathogenic variants were more common among families with progressing OI severity.
Intrafamilial variability of phenotypes correlated with the type of collagen variant. Families lacking variability were associated with harboring an LOF pathogenic variant; however, the majority of families with progressing OI severity mostly had DN mutations (P value = 0.0231). There were no significant correlations between changes in severity and whether the COL1A1 or COL1A2 gene was altered (P value = 0.2376).
DISCUSSION
Interfamilial variability
Our study has shown that 82.26% of OI COL1A1/2 pathogenic variants are associated with interfamilial phenotypic variability, which is higher than the clinical variability found by Maioli et al. 8 This difference could be partly explained by the fact that our study included individuals from different populations, whereas the study by Maioli et al. did not. In addition, the utilization of data from the Dagleish database means that diagnoses were made by different medical professionals, and, thus, classification bias might be present. 17
It can be summarized that monophenotypic variants caused a clear phenotype picture, especially in cases of mild OI and when all reported patients were classified with the same OI type. However, in cases of polyphenotypic variants, the clinical picture differed from patient to patient, which caused a challenge for the classification procedure due to medical treatment, subjective classification, the limited number of cases, and maturation of the patients. 23 However, none of the mentioned factors can affect causation of the lethal and nonlethal OI forms by a single pathogenic variant.
Similarly to the previous report, our results show that the degree of variability depends on the affected gene and the type of collagen defect. 8 The correlation between the degree of variability and the affected gene might be explained by a lack of LOF variants in the COL1A2 gene, as compared with the COL1A1 gene, and a dominating number of Gly substitutions. COL1A1/2 LOF pathogenic variants are associated with less variable phenotypes compared with DN pathogenic variants. Our results also find support from the data of the Dalgleish OI variant database. 17
LOF mutations led to haploinsufficiency phenotypes via degradation of the abnormal collagen type I, inefficient transport of the mRNA to cytoplasm or premature termination codons, and nonsense‐mediated mRNA decay. 24 The null allele is followed by a reduction in the amount of protein and a lower penetration of the phenotype (i.e., mild OI). However, cases with mutations in the last exons of the COL1A1/2 genes might escape nonsense‐mediated mRNA decay, cause truncated transcripts, and result in structural abnormalities of collagen, similarly to the missense mutations. 25 Additionally, some splice site mutations might not fully escape translation and result in a truncated protein. Thus, we suppose that these cases might explain the higher phenotypic variability, similarly to DN pathogenic variants in some LOF cases. Missense mutations disrupt the triple helical domain of the collagen molecule, altering protein stability, and causing qualitative collagen defects and higher penetrations with severe OI. These observations give insights into the probable modification capacity of OI phenotypes. It could be speculated that modification factors (gene transcription, expression levels, or the presence of a genetic modifier) might particularly affect collagen protein properties, thus altering its structure and function rather than quantity, as individuals harboring LOF variants lack phenotypic variability.
Intrafamilial variability
The degree of intrafamilial phenotype variability was lower (32.81%) compared with interfamilial variability (80%). This might be explained by the lower degree of genetic variations between related patients. In contrast to interfamilial phenotype variability, intrafamilial variability depended only on the type of collagen defect, not on the gene affected.
It is known that, due to recombination, siblings might have higher genetic variance compared with parent‐child variance. However, siblings had less variable OI phenotypes compared with family members from different generations. 26 One possible explanation could be newly arising de novo mutations in the gametes, which might become a source of additional parent‐child genetic variance. 27 , 28
The progression of OI symptoms in the following generations was proposed earlier; however, our data show that both progressing and decreasing phenotype severity can be observed in OI families. 29 For example, among Estonian families, the majority of cases demonstrated the reduction of OI symptoms in subsequent generations. The Clinic of Traumatology and Orthopedics of Tartu University Hospital followed up with Estonian OI families for 25 years. We cannot exclude the notion that OI phenotypes changing to milder forms might be partly explained by supportive medical care administered to Estonian patients with OI from the first month after birth. Estonian patients with OI got early treatment with medication and corrections of deformities, which might prevent fractures and complex medical management. All of the aforementioned procedures and regular systems of management for patients with OI were absent in Vietnam and in Ukraine until recently. Variability of the phenotypes cannot be explained with differences in treatment on their own, as mildly affected and untreated parents might have moderately affected children who may develop a need for pharmacological treatment and orthopedic interventions, in contrast to their parents. The higher range of progressing OI severity in Ukrainian and Vietnamese patients with OI might also be explained by the higher proportion of DN mutations in their cohorts, as compared with the Estonian OI cohort. 18 , 19 Further studies of phenotype variability and treatment associations are needed.
Previous studies have shown that some cases of intrafamilial diversity in collagen‐related OI can be explained by mosaic parents who are mildly affected or asymptomatic. 30 In addition, heterozygous family members might be mildly affected compared with severely affected homozygous individuals. 31 , 32 Rarely, cases of compound heterozygous mutations or double mutations in different OI genes might also affect phenotype variability inside a single family. Additional studies should be conducted to identify the presence of compound heterozygosity in different OI genes and to identify cases of gonadal mosaicism among patients in the study cohort.
Beneficial diagnostic outcomes from the results of the current study might be provided to inform family planning in those families with a history of OI. It may be predicted that families with LOF mutations have a lower chance of developing disease progression in the following generations in the case of mutation transmission, as LOF variants lack phenotypic variability.
Further collagen type I functional studies might shed light on the reasons for OI diversity among patients from the same family. Not only genetics, but also epigenetics, environmental factors, and complex genetic and environmental interactions might contribute to the development of specific OI phenotypes. This leads to many questions and the need for future research on OI variability.
CONCLUSIONS
The existing OI paradigm is rapidly evolving and filling the gaps in our understanding of this genetic disorder. In our study, we have analyzed interfamilial and intrafamilial phenotypic diversity in a large cohort of patients with collagen‐related OI from different populations. We formed phenotypic groups of collagen‐related OI based on a genotype‐phenotype severity scale. We have also identified a potential correlation between the collagen defect type, gene, and interfamilial phenotypic variability. Intrafamilial OI phenotypic variability correlated only with the type of collagen defect. The current results advance our understanding of OI variability and contribute toward family planning. Moreover, the results of the current study give additional information regarding the variability of OI phenotype inside a family depending on the genotype, as families harboring LOF pathogenic variants have lower variability of OI severity compared with patients with DN variants.
We believe that the current data help to illuminate the modification of the OI phenotype and advances strategies for consultation and family planning with patients with collagen‐related OI connected to risks of disease progression in subsequent generations.
Funding
This work was supported by an institutional research grant (IUT20–46) from the Estonian Ministry of Education and Research, by the projects DIOXMED and EVMED from the Estonian Ministry of Education and Research, and by the H2020 ERA‐chair grant (agreement 668989, project Transgeno).
Conflict of Interest
All authors declared no competing interests for this work.
Author Contributions
L.Z., K.M., and T.R. wrote the manuscript. L.Z., K.M., T.R., S.K., and A.M. designed the research. L.Z., B.H.D., K.M., and T.R. performed the research. L.Z., K.M., T.R., S.K., and A.M. analyzed the data.
Acknowledgments
The authors thank all of the patients and their relatives who participated in the study. We also extend our appreciation to the following people and organizations for their help and support with data collection: the Ukrainian Association of Crystal People, OI Club in Vietnam, workers of the Department of Traumatology and Orthopedics and Department of Pathophysiology, the University of Tartu, and Hue University of Medicine and Pharmacy. This work is generated within the European Reference Network for rare BONe Diseases.
References
- 1. Byers, P.H. & Steiner, R.D. Osteogenesis imperfecta. Annu. Rev. Med. 43, 269–282 (1992). [DOI] [PubMed] [Google Scholar]
- 2. Sillence, D.O. , Rimoin, D.L. & Danks, D.M. Clinical variability in osteogenesis imperfecta‐variable expressivity or genetic heterogeneity. Birth Defects Orig. Artic. Ser. 15, 113–129 (1979). [PubMed] [Google Scholar]
- 3. Lindahl, K. et al Genetic epidemiology, prevalence, and genotype–phenotype correlations in the Swedish population with osteogenesis imperfecta. Eur. J. Hum. Genet. 23, 1042–1050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forlino, A. , Cabral, W.A. , Barnes, A.M. & Marini, J.C. New perspectives on osteogenesis imperfecta. Nat. Rev. Endocrinol. 7, 540–557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marini, J.C. et al Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum. Mutat. 28, 209–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van Dijk, F.S. & Sillence, D.O. Osteogenesis imperfecta: clinical diagnosis, nomenclature and severity assessment. Am. J. Med. Genet. A 164A, 1470–1481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li, L. et al Genotypic and phenotypic characterization of Chinese patients with osteogenesis imperfecta. Hum. Mutat. 40, 588–600 (2019). [DOI] [PubMed] [Google Scholar]
- 8. Maioli, M. et al Genotype–phenotype correlation study in 364 osteogenesis imperfecta Italian patients. Eur. J. Hum. Genet. 27, 1090–1100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marini, J.C. et al Osteogenesis imperfecta. Nat. Rev. Dis. Prim. 3, 17052 (2017). [DOI] [PubMed] [Google Scholar]
- 10. McGettrick, A.J. , Knott, V. , Willis, A. & Handford, P.A. Molecular effects of calcium binding mutations in Marfan syndrome depend on domain context. Hum. Mol. Genet. 9, 1987–1994 (2000). [DOI] [PubMed] [Google Scholar]
- 11. Castori, M. Ehlers‐Danlos syndrome, hypermobility type: an underdiagnosed hereditary connective tissue disorder with mucocutaneous, articular, and systemic manifestations. ISRN Dermatol. 2012, 1–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cooper, D.N. , Krawczak, M. , Polychronakos, C. , Tyler‐Smith, C. & Kehrer‐Sawatzki, H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 132, 1077–1130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deodhar, A.A. & Woolf, A.D. Fragile without fractures. Ann. Rheum. Dis. 59, 166–171 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pillion, J.P. , Vernick, D. & Shapiro, J. Hearing loss in osteogenesis imperfecta: characteristics and treatment considerations. Genet. Res. Int. 2011, 983942 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dalgleish, R. The human type I collagen mutation database. Nucleic Acids Res. 25, 181–187 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swinnen, F.K.R. et al Osteogenesis Imperfecta: the audiological phenotype lacks correlation with the genotype. Orphanet J. Rare Dis. 6, 88 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dalgleish, R. The human collagen mutation database 1998. Nucleic Acids Res. 26, 253–255 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho Duy, B. et al Mutation analysis of the COL1A1 and COL1A2 genes in Vietnamese patients with osteogenesis imperfecta. Hum. Genomics 10, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhytnik, L. et al Mutational analysis of COL1A1 and COL1A2 genes among Estonian osteogenesis imperfecta patients. Hum. Genomics 11, 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Binh, H.D. et al The clinical features of osteogenesis imperfecta in Vietnam. Int. Orthop. 41, 21–29 (2017). [DOI] [PubMed] [Google Scholar]
- 21. Chen, R. et al Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maasalu, K. et al Whole‐exome sequencing identifies de novo mutation in the COL1A1 gene to underlie the severe osteogenesis imperfecta. Hum. Genomics 9, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lim, J.Y. et al A novel Ser40Trp variant in IFITM5 in a family with osteogenesis imperfecta and review of the literature. Clin. Dysmorphol. 28, 118–123 (2019). [DOI] [PubMed] [Google Scholar]
- 24. Garnero, P. , Schott, A.‐M. , Prockop, D. & Chevrel, G. Bone turnover and type I collagen C‐telopeptide isomerization in adult osteogenesis imperfecta: associations with collagen gene mutations. Bone 44, 461–466 (2009). [DOI] [PubMed] [Google Scholar]
- 25. Slayton, R.L. , Deschenes, S.P. & Willing, M.C. Nonsense mutations in the COL1A1 gene preferentially reduce nuclear levels of mRNA but not hnRNA in osteogenesis imperfecta type I cell strains. Matrix Biol. 19, 1–9 (2000). [DOI] [PubMed] [Google Scholar]
- 26. Visscher, P.M. , Hill, W.G. & Wray, N.R. Heritability in the genomics era — concepts and misconceptions. Nat. Rev. Genet. 9, 255–266 (2008). [DOI] [PubMed] [Google Scholar]
- 27. Acuna‐Hidalgo, R. , Veltman, J.A. & Hoischen, A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 17, 241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Veltman, J.A. & Brunner, H.G. De novo mutations in human genetic disease. Nat. Rev. Genet. 13, 565–575 (2012). [DOI] [PubMed] [Google Scholar]
- 29. Moraes, M. et al Variable expressivity of osteogenesis imperfecta in a Brazilian family due to p. G1079S mutation in the COL1A1 gene. Genet. Mol. Res. 11, 3246–3255 (2012). [DOI] [PubMed] [Google Scholar]
- 30. Frederiksen, A.L. , Duno, M. , Johnsen, I.B.G. , Nielsen, M.F. & Krøigård, A.B. Asymptomatic parental mosaicism for osteogenesis imperfecta associated with a new splice site mutation in COL1A2. Clin. Case Rep. 4, 972–978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laine, C.M. et al WNT1 Mutations in early‐onset osteoporosis and osteogenesis imperfecta. N. Engl. J. Med. 368, 1809–1816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Symoens, S. et al Deficiency for the ER‐stress transducer OASIS causes severe recessive osteogenesis imperfecta in humans. Orphanet J. Rare Dis. 8, 154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]


