Abstract
Precise dosing of warfarin is important to achieve therapeutic benefit without adverse effects. Pharmacogenomics explains some interindividual variability in warfarin response, but less attention has been paid to drug‐drug interactions in the context of genetic factors. We investigated retrospectively the combined effects of cytochrome P450 (CYP)2C9 and vitamin K epoxide reductase complex (VKORC)1 genotypes and concurrent exposure to CYP2C9‐interacting drugs on long‐term measures of warfarin anticoagulation. Study participants predicted to be sensitive responders to warfarin based on CYP2C9 and VKORC1 genotypes, had significantly greater international normalized ratio (INR) variability over time. Participants who were concurrently taking CYP2C9‐interacting drugs were found to have greater INR variability and lesser time in therapeutic range. The associations of INR variability with genotype were driven by the subgroup not exposed to interacting drugs, whereas the effect of interacting drug exposure was driven by the subgroup categorized as normal responders. Our findings emphasize the importance of considering drug interactions in pharmacogenomic studies.
Despite the introduction of direct oral anticoagulants in 2010, warfarin remains the most widely prescribed anticoagulant 1 , 2 as well as one of the drugs responsible for the largest number of adverse drug reactions requiring an emergency department visit. 3 The combination of a narrow therapeutic index and the potential to precipitate life‐threatening adverse events made warfarin an attractive candidate for precision dosing algorithms, such as those based on pharmacogenomics.
The clinical utility of pharmacogenomics‐based dosing algorithms for the initiation of warfarin has been a controversial topic. Most notably, the results of the Clarification of Optimal Anticoagulation through Genetics (COAG) and European Pharmacogenetics of Anticoagulation Therapy (EU‐PACT) trials in 2013 were conflicting with the former reporting that a genotype‐guided dosing algorithm was not associated with greater time in therapeutic range (TTR) over the first 4 weeks of therapy, whereas the latter reported that a similar genotype‐guided dosing algorithm did improve TTR during the first 12 weeks of therapy. 4 , 5 These studies were randomized‐controlled trials with large cohorts, and both trials included extensive dose adjustment measures that were partially based on the most recent prothrombin time international normalized ratio (INR), which could dilute an underlying effect associated with cytochrome P450 (CYP)2C9 and vitamin K epoxide reductase complex (VKORC)1 genotype present in more typical clinical settings. Importantly, effects on long‐term measures of warfarin anticoagulation quality were not assessed.
An important factor that can affect pharmacogenomics‐based dosing algorithms of warfarin and long‐term measures of warfarin anticoagulation quality is the concurrent use of CYP2C9‐interacting drugs. However, the three most commonly cited pharmacogenomics‐based warfarin dosing algorithms 6 , 7 , 8 variably consider CYP2C9‐interacting drugs with amiodarone being the only drug in common. Hence, we considered the effect of CYP2C9‐interacting drugs to be an underexplored area and sought to study potential drug‐mediated effects with an expanded list of CYP2C9‐interacting drugs.
In this study, we investigated the association of long‐term measures of warfarin anticoagulation efficacy and stability with CYP2C9/VKORC1 genotype and concurrent use of CYP2C9‐interacting drugs in a retrospective clinical setting.
METHODS
Study cohort
The study cohort was a subset of participants in the NUgene biobank, a hospital‐based biobank at Northwestern Medicine. Participants in this study were identified using the inclusion and exclusion criteria outlined in Figure 1 . We excluded participants in the Electronic Medical Records and Genomics Network (eMERGE II) Pharmacogenomics study 9 because some providers received clinical decision support that may have altered prescribing behavior. At the time of the study, NUgene had enrolled 12,722 participants with 82% (11,416) described as white Americans. Of those 11,416 participants, 401 met all inclusion criteria for our study. Ninety‐six were excluded because they had fewer than 14 INR measurements during the study period, and 3 were excluded because they were not genotyped successfully for CYP2C9 or VKORC1. We had initially selected a cutoff of 6 INR measurements in our exclusion criteria, but quickly observed that many participants within the cohort who never had a recorded therapeutic INR also had fewer than 14 INR measurements. Because the goal of this study was to evaluate the effect of genotype and concurrent medication exposures on long‐term anticoagulation, we decided to omit participants with < 14 INR measurements to reduce the probability of a type II error.
Figure 1.
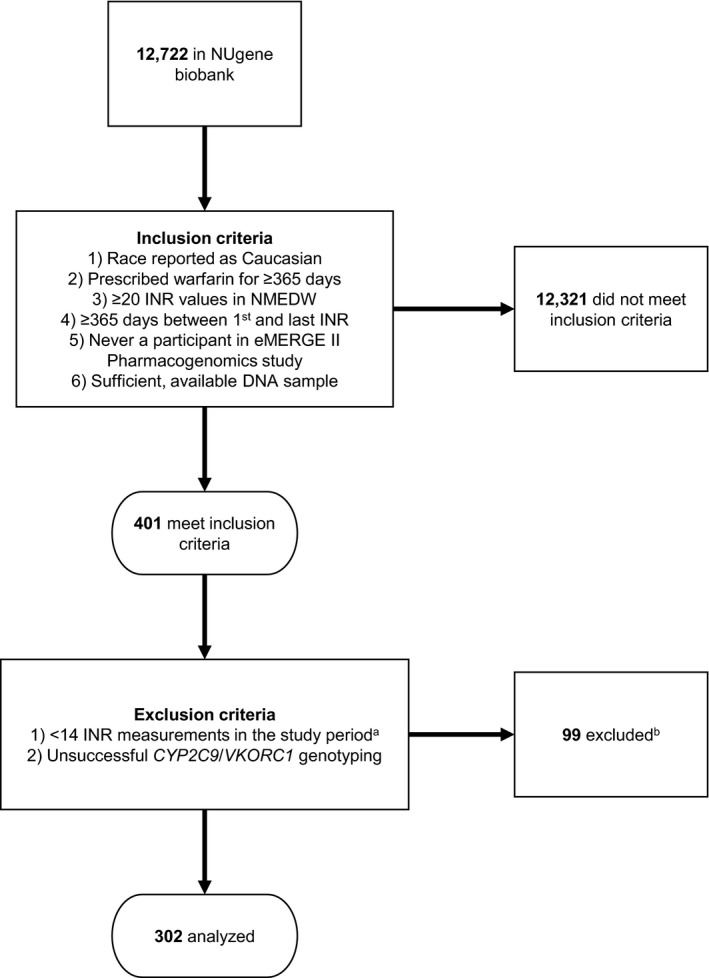
Flow diagram illustrating inclusion and exclusion criteria. aThe study period is the interval between 30 days after the first warfarin prescription start date and the last warfarin prescription end date. Ninety‐six participants were excluded because there were < 14 international normalized ratio (INR) measurements recorded. Three participants were excluded because genotyping for cytochrome P450 (CYP)2C9 and vitamin K epoxide reductase complex (VKORC)1 was unsuccessful. eMERGE, Electronic Medical Records and Genomics Network; NMEDW, Northwestern Medicine Enterprise Data Warehouse.
For all participants included in the study, we extracted de‐identified data regarding warfarin prescription, INR measurements, age at first warfarin prescription, warfarin indication, and all other medication exposures with start and stop dates from the Northwestern Medicine Enterprise Data Warehouse. Additional demographic information (including weight, height, sex, and detailed race information) was obtained from the NUgene Study Questionnaire, a self‐reported measure collected upon enrollment into the NUgene project biobank. All information was de‐identified prior to receipt by two of the investigators (J.A.P. and M.H.). The NUgene project is institutional review board (IRB) approved and is compliant with Health Insurance Portability and Accountability Act (HIPAA). No additional IRB approval was required for this study. NUgene recruitment procedures and participant information have been described elsewhere. 10 Other information regarding the data obtained from the Northwestern Medicine Enterprise Data Warehouse can be found in Table S6 .
Genotyping
Genotyping methods for CYP2C9 and VKORC1 variants are described in the Supporting Information and Table S7 . All participants who met the inclusion criteria were assigned a genotype‐predicted warfarin response status (normal responder, sensitive responder, or highly sensitive responder) based on their CYP2C9 and VKORC1 genotypes that was adapted from the US Food and Drug Administration (FDA) prescribing information as used in previous studies and Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines. 11 , 12 , 13 , 14 Notably, no participants were homozygous for CYP2C9*3, and no participants carried the CYP2C9*4 variant, which can interfere with detection of CYP2C9*3.
CYP2C9 inhibitors and inducers
We identified drugs with potential CYP2C9‐mediated interactions with warfarin from two sources: the FDA label for warfarin, and the Flockhart Table of Drug Interactions. 15 This yielded 34 candidate CYP2C9‐interacting drugs (Tables S8 and S9 ). We omitted 14 because they were not prescribed in this cohort. A literature search was done for the remaining 20 drugs looking for evidence that the drug was a CYP2C9 inhibitor or inducer, and evidence of a clinically significant interaction with warfarin. This analysis, summarized in the Supporting Information, identified 10 CYP2C9 inhibitors and 3 CYP2C9 inducers as the interacting drugs considered in this study.
We examined exposure to CYP2C9‐interacting drugs by counting the number of unique medications taken during warfarin therapy. Participants were then stratified into three categories based on this number: 0 interacting drugs, 1 interacting drug, or 2 or more (2+) interacting drugs. We did not differentiate between CYP2C9 inhibitors and inducers when assigning these categories. Notably, exposures to multiple interacting drugs did not have to coincide as long as each exposure independently overlapped with the warfarin course of a given participant.
INR variability and TTR
We utilized four measures of anticoagulation quality in this study, denoted as INR variability (INRvarA), INRvarB, TTRa, and TTRb (Table S10 ). INRvarA and INRvarB are measures of INR variability without regard for distance from the therapeutic range, whereas TTRa and TTRb rely only on whether INR values are within the therapeutic range. A complete description of these variables is found in the Supporting Information.
Impact of concurrent CYP2C9‐interacting drugs on INR
The impact of exposure to CYP2C9‐interacting drugs on anticoagulation was evaluated by examining INR values immediately surrounding the start date of each concurrently prescribed medication. Importantly, each separate exposure was included in this analysis, even when there were two unique courses of the same drug. INR values obtained on the same day were not averaged for this analysis to lower the risk of masking short‐lived peaks and troughs. The study period from which INR values were sampled was defined by a minimum date (30 days before the start of the medication), and a maximum date defined as the earlier date between 30 days after the start date and 7 days after the end date. More detailed definitions of the study period are found in the Supporting Information.
Data analysis
All data cleaning and calculation of the outcome measures was done using Python 2.7.15 within the Spyder integrated development environment. All statistical analyses were conducted using SPSS Statistics (version 25.0.0.0) and specific tests are described in the Results. All tables and figures, except Figure S2 , were generated using SPSS Statistics. Figure S2 was generated using MATLAB R2018b.
One‐way analysis of variance (ANOVA) and χ2 tests were utilized to compare descriptive variables, such as age and sex, across combined genotype categories (normal responder, sensitive responder, and highly sensitive responder), and medication exposure categories (0, 1, 2+). One‐way ANOVA was utilized to compare combined genotype categories and medication exposure categories with the outcome variables of Ln(INRvarA), Ln(INRvarB), TTRa, and TTRb. Post hoc pairwise comparisons were made with Tukey’s HSD. Two‐way ANOVA was utilized with each outcome variable to determine if the genotype/medication exposure interaction term significantly contributed to the outcome variable. One‐way ANOVA was then done with subgroups defined by combined genotype and medication exposure. Comparisons of absolute INR changes before and after starting a given CYP2C9‐interacting medication were performed using t‐tests.
The inner markings of the boxes in Figures 2 and 3 are defined by the median, first quartile (q1), and third quartile (q3). The outer boundaries are defined by the outer most values within boundaries of q1 – 1.5 × interquartile range and q3 + 1.5 × interquartile range.
Figure 2.
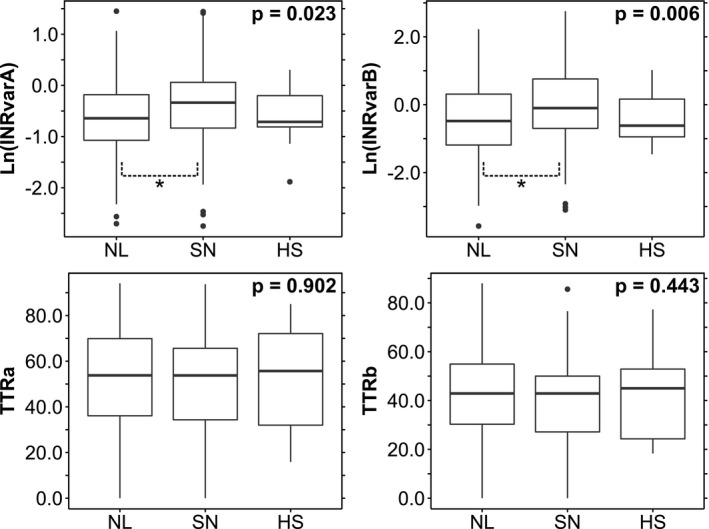
Associations of outcome measures with genotype‐predicted warfarin response. Bounds of the box‐and‐whisker plots are discussed in the Methods section. Each P value corresponds to a one‐way analysis of variance test for an international normalized ratio (INR) outcome measure against the three combined cytochrome P450 (CYP)2C9/vitamin K epoxide reductase complex (VKORC)1 genotype‐predicted responder categories (Table 2 ). Post hoc pairwise comparisons were made using Tukey’s HSD. *Indicates a significant pairwise comparison at the P < 0.05 level. HS, highly sensitive; NL, normal; SN, sensitive; TTR, time in therapeutic range.
Figure 3.
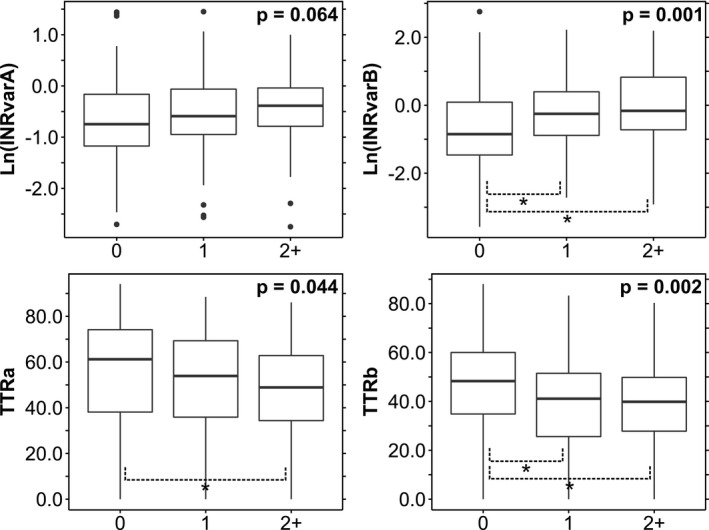
Associations of outcome measures with cytochrome P450 (CYP)2C9‐interacting drugs. Box‐and‐whisker plots are shown. Each P value corresponds to a one‐way analysis of variance test for an international normalized ratio (INR) outcome measure against the three categories of CYP2C9‐interacting drugs (Table 3 ). Post hoc pairwise comparisons were made using Tukey’s HSD. *Indicates a significant pairwise comparison at the P < 0.05 level. TTR, time in therapeutic range.
RESULTS
Study cohort
Inclusion and exclusion criteria for our study are presented in Figure 1 , and Table 1 presents characteristics of the 302 study participants who remained after application of these criteria. The mean age at first warfarin prescription was 62.6 years and the most common indication was atrial fibrillation (41.7%).
Table 1.
Descriptive variables for study cohort
| Variable |
Total n = 302 |
NL n = 183 |
SN n = 110 |
HS n = 9 |
a P value |
0 drugs n = 91 |
1 drug n = 121 |
2 + drugs n = 90 |
P value |
|---|---|---|---|---|---|---|---|---|---|
| Age in 2017 b ; mean, SD | 70.0, 13.8 | 69.8, 13.8 | 70.2, 13.9 | 71.2, 12.7 | 0.941 | 69.6, 14.5 | 71.4, 12.6 | 68.5, 14.4 | 0.288 |
| Age at first warfarin Rx | 62.6, 15.8 | 62.6, 16.3 | 62.5, 15.0 | 62.8, 15.0 | 0.997 | 62.5, 17.8 | 63.2, 14.7 | 61.8, 15.1 | 0.833 |
| Weight, lbs | 196.3, 57.7 | 196.9, 56.0 | 192.0, 57.5 | 237.9, 81.7 | 0.07 | 189.9, 50.4 | 198.8, 59.1 | 199.4, 62.6 | 0.453 |
| BMI, kg/m2 | 29.6, 7.6 | 29.9, 7.4 | 28.8, 7.8 | 32.7, 8.0 | 0.236 | 28.7, 6.6 | 29.9, 7.9 | 30.0, 8.3 | 0.433 |
| Sex; number, % | 0.303 | 0.451 | |||||||
| Male | 171 (56.6) | 99 (54.1) | 65 (59.1) | 7 (77.8) | 47 (51.6) | 73 (60.3) | 51 (56.7) | ||
| Female | 131 (43.3) | 84 (45.9) | 45 (40.9) | 2 (22.2) | 44 (48.4) | 48 (39.7) | 39 (43.3) | ||
| Warfarin indication c | 0.533 | 0.74 | |||||||
| Atrial fibrillation | 126 (41.7) | 74 (40.4) | 48 (43.6) | 4 (44.4) | 33 (36.3) | 56 (50.9) | 37 (41.1) | ||
| Thrombosis | 69 (22.8) | 38 (20.8) | 28 (25.5) | 3 (33.3) | 24 (26.3) | 25 (22.7) | 20 (22.2) | ||
| Stroke | 8 (2.6) | 6 (3.3) | 2 (1.8) | 0 (0) | 3 (3.3) | 3 (2.7) | 2 (2.2) | ||
| Orthopedic | 8 (2.6) | 4 (2.2) | 3 (2.7) | 1 (11.1) | 4 (4.4) | 3 (2.7) | 1 (1.1) | ||
| Other/unknown | 109 (36.1) | 70 (38.3) | 37 (33.6) | 2 (22.2) | 33 (36.3) | 39 (35.5) | 37 (41.1) | ||
| CYP2C9 | 0.826 | ||||||||
| *1/*1 | 197 (65.2) | 166 (90.7) | 31 (28.2) | 0 (0) | 57 (62.6) | 81 (66.9) | 59 (65.6) | ||
| *1/*2 | 61 (20.2) | 17 (9.3) | 44 (40.0) | 0 (0) | 18 (19.8) | 25 (20.7) | 18 (20.0) | ||
| *1/*3 | 29 (9.6) | 0 (0) | 27 (24.5) | 2 (22.2) | 11 (12.1) | 8 (6.6) | 10 (11.1) | ||
| *2/*2 | 10 (3.3) | 0 (0) | 8 (7.3) | 2 (22.2) | 4 (4.4) | 5 (4.1) | 1 (1.1) | ||
| *2/*3 | 5 (1.7) | 0 (0) | 0 (0) | 5 (55.6) | 1 (1.1) | 2 (1.7) | 2 (2.2) | ||
| *3/*3 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| VKORC1 rs9923231 | 0.436 | ||||||||
| G/G | 105 (34.8) | 92 (50.3) | 13 (11.8) | 0 (0) | 33 (36.3) | 38 (31.4) | 34 (37.8) | ||
| A/G | 146 (48.3) | 91 (49.7) | 50 (45.5) | 5 (55.6) | 40 (44.0) | 60 (49.6) | 46 (51.1) | ||
| A/A | 51 (16.9) | 0 (0) | 47 (42.7) | 4 (44.4) | 18 (19.8) | 23 (19.0) | 10 (11.1) | ||
| CYP2C9‐interacting drugs | 0.657 | ||||||||
| 0 | 91 (30.1) | 52 (28.4) | 36 (32.7) | 3 (33.3) | |||||
| 1 | 121 (40.1) | 71 (38.8) | 47 (42.7) | 3 (33.3) | |||||
| 2+ | 90 (29.8) | 60 (32.8) | 27 (24.5) | 3 (33.3) |
Mean values are in bold.
BMI, body mass index; CYP, cytochrome P450; HS, highly sensitive; NL, normal; SN, sensitive; VKORC, vitamin K epoxide reductase complex.
Analysis of variance tests were used to compare groups for continuous variables (age in 2017, age at first warfarin dose, weight, and BMI).
Chi‐squared tests were used to compare all other variables or age at death. All participants with age > 90 were reported as 90.
A given participant may have up to two indications.
All variants tested in the cohort were in Hardy‐Weinberg equilibrium and exhibited minor allele frequencies (Table S1 ) consistent with those reported in the literature. 16 , 17 Based on combined CYP2C9/VKORC1 genotypes, we categorized 183 study participants as normal responders, 110 as sensitive responders, and 9 as highly sensitive responders (defined in Table 2 ) in accordance with the FDA drug label for warfarin. There were no significant differences in clinical characteristics including age, sex, and body mass index among the three groups (Table 1 ).
Table 2.
Genotype‐predicted warfarin responder categories
| CYP2C9 genotype | |||||||
|---|---|---|---|---|---|---|---|
| *1/*1 | *1/*2 | *1/*3 | *2/*2 | *2/*3 | *3/*3 | ||
| VKORC1 genotype | G/G | Normal | Normal | Sensitive | Sensitive | Sensitive | Highly sensitive |
| G/A | Normal | Sensitive | Sensitive | Sensitive | Highly sensitive | Highly sensitive | |
| A/A | Sensitive | Sensitive | Highly sensitive | Highly sensitive | Highly sensitive | Highly sensitive | |
Genotype‐predicted warfarin responder groups were determined by cytochrome P450 (CYP)2C9 *1, *2, and *3 and VKORC1 c. ‐1639 G>A genotype. Our study cohort did not contain any participants who were genotyped as *3/*3 for CYP2C9. VKORC, vitamin K epoxide reductase complex.
Exposures to CYP2C9‐interacting drugs
Table 3 lists the CYP2C9‐interacting drugs that were considered in this study along with the number of study participants who were exposed concurrently to each during warfarin therapy. The three most frequent CYP2C9‐interacting drugs were amiodarone (91 participants), sulfamethoxazole (80 participants), and metronidazole (73 participants). A total of 344 instances of concurrent warfarin/CYP2C9‐interacting drug exposures were recorded. Of the 302 study participants, 91 had no exposure to any of the drugs listed in Table 3 , whereas 121 were exposed to one, 61 to two, 17 to three, 10 to four, and 2 to five CYP2C9‐interacting drugs. Study participants were stratified by 0, 1, and 2+ CYP2C9‐interacting drug exposures, and there were no significant differences in clinical characteristics among the three groups (Table 1 ).
Table 3.
List of cytochrome P450 (CYP)2C9‐interacting drugs selected for this study (number of participants exposed given in parentheses)
| CYP2C9 inhibitors | CYP2C9 inducers |
|---|---|
| Amiodarone (91) | Bosentan (5) |
| Capecitabine (1) | Carbamazepine (5) |
| Fenofibrate (29) | Rifampin (9) |
| Fluconazole (34) | |
| Fluvastatin (4) | |
| Metronidazole (73) | |
| Miconazole (9) | |
| Sulfamethoxazole (80) | |
| Voriconazole (3) | |
| Zafirlukast (1) |
Assessment of anticoagulation variability
Descriptive statistics regarding INR measurements in the study cohort and the corresponding outcome measures are presented in Table S2 . On average for each participant, 78 INR measurements were collected within the study period with a range of 14 to 1,004 INR values. Overall, the study cohort was under‐anticoagulated with 39% of INR values being in the subtherapeutic range (INR < 2.0), whereas 45.3% were within the therapeutic range (2.0 ≤ INR ≤ 3.0), and 15.6% were supratherapeutic (INR > 3.0).
Association of INR variability and TTR with CYP2C9/VKORC1 genotype
We examined the association of CYP2C9/VKORC1 genotype parsed into normal, sensitive, and highly sensitive responders with measures of warfarin anticoagulation stability and efficacy. Figure 2 shows the results of one‐way ANOVA analyses testing the associations of the three genotype‐predicted responder groups with the four outcome variables. Both measures of INR variability, Ln(INRvarA) and Ln(INRvarB), were significantly associated with CYP2C9/VKORC1 genotype (P < 0.05), but post hoc analyses (Tukey’s HSD) revealed that only the difference between the normal and sensitive groups was statistically significant for both outcome variables (Table S3 ). The highly sensitive group exhibits wide 95% confidence intervals across both analyses probably because of the small sample size (n = 9). Neither TTRa nor TTRb was significantly associated with CYP2C9/VKORC1 genotype. Taken together, these results suggest that CYP2C9/VKORC1 genotype influences INR variability but not TTR.
Association of CYP2C9‐interacting drug exposure with TTR
Drug interactions influence warfarin metabolism by impeding or enhancing CYP2C9 activity. We examined the impact of CYP2C9‐interacting drug exposure on INR variability and TTR. Figure 3 shows the results of four separate one‐way ANOVA analyses testing the association of interacting drug exposure group (0 interacting drugs exposures, 1 interacting drug exposures, and 2+ interacting drug exposures) with the four outcome variables. Ln(INRvarB), TTRa, and TTRb were significantly associated with interacting drug exposure (P < 0.05), and post hoc analyses (Tukey’s HSD) revealed that the differences between the 0 and 1 as well as the 0 and 2+ interacting drug exposure groups were significantly associated with Ln(INRvarB) and TTRb (Table S3 ). On the other hand, only the difference between the 0 and 2+ interacting drug exposure groups was significantly associated with TTRa. Although the corresponding analysis for Ln(INRvarA) was not statistically significant, it approached threshold (P = 0.064) and followed the same trend as Ln(INRvarB). These findings suggest that exposure to CYP2C9‐interacting drugs can influence both INR variability and TTR.
Interaction between CYP2C9/VKORC1 genotype and CYP2C9‐interacting drugs
We investigated whether CYP2C9/VKORC1 genotype and concurrent exposure to CYP2C9‐interacting drugs interact with each other to influence the quality and stability of warfarin anticoagulation. We conducted two‐way ANOVA analyses, including both genotype and interacting drug exposure, for each outcome variable and found that the interaction term in the Ln(INRvarA) analysis was significant (P < 0.05; Table S4 ). The interaction term for the Ln(INRvarB) analysis also approached significance (P = 0.093). To further probe this interaction, we conducted subgroup analyses of all four outcome variables. For each outcome variable, we subdivided participants by either their CYP2C9/VKORC1 genotype or their interacting drug exposure. Then, one‐way ANOVA analyses were run on each subgroup (Table S4 ).
In the absence of interacting drug exposures, CYP2C9/VKORC1 genotype‐predicted warfarin responder phenotype was significantly associated with Ln(INRvarA), Ln(INRvarB), and TTRb. This finding reflects the influence of genotype without the confounding effect of drug interactions. By contrast, there was no significant association of genotype with any outcome measures in participants exposed to at least one interacting drug suggesting that the impact of CYP2C9/VKORC1 genotype was masked by the effect of drug interactions. In the converse analyses, interacting drug exposure was significantly associated with all outcome measures in the normal responder subgroup, but was not associated with any outcome variable in sensitive and highly sensitive responders.
We also investigated the influence of interacting drugs in the subgroup without VKORC1 variants (n = 105). Among this subgroup, there were participants with CYP2C9 genotypes *1/*1 (n = 75), *1/*2 (n = 17), *1/*3 (n = 10), and *2/*2 (n = 3). A two‐way ANOVA considering independent variables of interacting drugs and CYP2C9 genotype (*1/*1 vs. any variant genotype) demonstrated an association for just Ln(INRvarA) but only in the group with genotype *1/*1 (P = 0.003). This finding suggests that drug‐drug interactions in the context of warfarin therapy are CYP2C9 genotype‐dependent. Furthermore, the association of CYP2C9‐interacting drugs on INR variability in participants with the CYP2C9 *1/*1 genotype likely represents drug‐induced phenoconversion. 18
Impact of CYP2C9‐interacting drugs on INR
To quantify the impact of starting a CYP2C9‐interacting drug on INR, we examined INR values determined within a time period around the start date of a concurrent medication and then characterized the trend of INRs before and after starting the drug (see Methods for definition of the time period and parameters that were calculated). Figure 4 illustrates (1) the maximum change in INR and (2) the change in average INR after starting each category of CYP2C9‐interacting drug (Table S5 ). Only metronidazole displayed a statistically significant change in average INR with a magnitude of −0.30. Amiodarone, metronidazole, and sulfamethoxazole were associated with significant differences in maximum change in INR with effect sizes ranging from +0.50 (metronidazole) to +1.04 (amiodarone). CYP2C9 inducers displayed significant differences in maximum change in INR as well with an effect size of −0.93, although there were only 7 instances in this category. Individual CYP2C9 inhibitors other than amiodarone, metronidazole, and sulfamethoxazole were not significantly associated with any of the measured differences.
Figure 4.
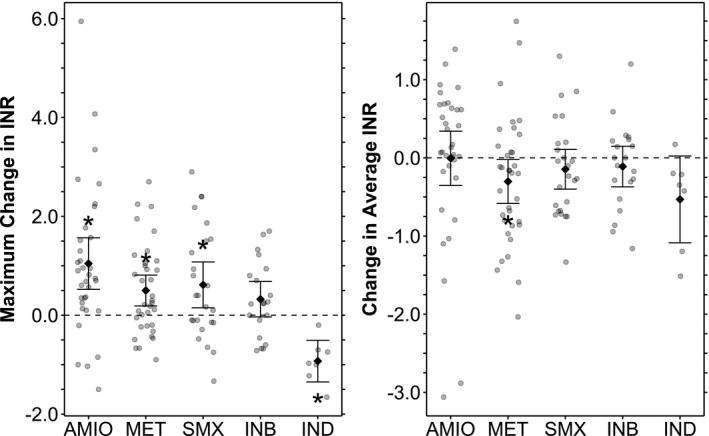
International normalized ratio (INR) differences before and after cytochrome P450 (CYP)2C9‐interacting drug exposure. Mean values with 95% confidence intervals are shown superimposed on the individual data. Maximum change in INR refers to the magnitude of maximum perturbation from the baseline INR after initiating an interacting drug. The baseline INR is defined by average INR in a study period prior to exposure to an interacting drug. The maximum perturbation is defined by peak INR after initiation of a CYP2C9 inhibitor, and by trough INR after initiation of a CYP2C9 inducer. Change in average INR refers to the difference between baseline INR as described above and the average INR in a time period shortly after initiation of an interacting drug. Tabulated data are presented in Table S11 . For each listed mean INR difference, the P value corresponds to a t‐test comparing the two sets of INRs included in the difference. *Indicates statistical significance at the P < 0.05 level. AMIO, amiodarone; INB, CYP2C9‐inhibitors; IND, CYP2C9‐inducers; MET, metronidozole; SMX, suflamethoxazole.
DISCUSSION
In this study, we investigated how CYP2C9/VKORC1 genotype impacts long‐term warfarin anticoagulation efficacy and stability by assessing TTR and INR variability, respectively. Our analysis (Figure 2 ) was consistent with CYP2C9/VKORC1 genotype being a significant predictor of INR variability, although this effect was driven only by the difference between the normal and sensitive warfarin responder groups. Importantly, INR variability is only a function of how noisy the INR data are without regard for how close the INR values are to the therapeutic range. Hence, although CYP2C9/VKORC1 genotype was found to be a significant predictor of INR variability, overall it did not predict how often INR values were in the therapeutic range. By contrast, exposure to CYP2C9‐interacting drugs was significantly associated with both INR variability and TTR.
Our findings on the association of genotype with INR variability is inconsistent with that reported by Iwuchukwu et al. 12 in which no association of CYP2C9 and/or VKORC1 genotype with INR variability was observed. Differences in study design and potential sources of ascertainment bias may help explain this discordance. Specifically, we excluded participants with fewer than 14 INR measurements during the study period, but included those who never achieved a therapeutic INR. By contrast, Iwuchukwu et al. excluded all participants with fewer than six INR measurements and excluded those who never recorded an INR measurement in the therapeutic range. Our study was designed to analyze participants in whom long‐term warfarin anticoagulation could be assessed without sacrificing the power of our study, and an exclusion threshold of 14 INR measurements satisfied this goal. It is possible that participants in the Iwuchukwu et al. study who were excluded because of failure to achieve a therapeutic INR were enriched in specific CYP2C9/VKORC1 genotypes. 12
Furthermore, our study cohort consisted exclusively of participants described as white who were presumed to have predominantly western European ancestry, whereas the study population reported by Iwuchukwu et al. included 13% African Americans. 12 The possibility of population stratification confounding results of warfarin pharmacogenomics was evident in the COAG trial, in which 27% of the study population was categorized as African American. Although genetic variants in VKORC1 may explain warfarin dose variability between white and black populations, 19 CYP2C9*2 and CYP2C9*3 variants make insignificant contributions to a dosing algorithm in African Americans. 20 Additionally, the COAG trial used a genotype‐guided dosing algorithm 6 that does not separately examine African Americans for relevant genetic variants. 2 For example, there are four novel single nucleotide variants on chromosome 6 associated with greater risk of major bleeding in persons of African descent who were taking warfarin. 21 Importantly, CYP2C9*2 and CYP2C9*3 did not have a significant association with warfarin‐related bleeding events in this African ancestry population. Hence, examining the effects of CYP2C9 on warfarin anticoagulation quality may be more useful in a study population consisting of persons of western European descent, a population that has been shown to have a sizable prevalence of CYP2C9 and VKORC1 variants 22 , 23 , 24 that explain a significant amount of variance in stable warfarin dose. 6 , 7 , 11 , 19 , 25
Regarding our finding that exposure to CYP2C9‐interacting drugs is predictive of both INR variability and TTR, our study is unique in analyzing CYP2C9‐interacting drugs together rather than analyzing each drug separately. We specifically chose a grouped analysis because our outcome variables reflect how often and with what magnitude a participant’s anticoagulation quality is perturbed from a stable baseline rather than reflecting directionality of that perturbation. The data in Figure 3 support the conclusion that any concurrent CYP2C9‐interacting drug exposure adversely affects a participant’s anticoagulation quality. Interestingly, our search for mechanistically predictable INR effects (Figure 4 ) found that although the maximum change in INR shortly after starting each CYP2C9‐interacting drug was consistent with that drug’s identity as an inhibitor or inducer, the difference in average INR shortly before and after starting that drug was not. This suggests that the clinical relevance of drug‐drug interactions on warfarin dosing may extend beyond immediate, mechanistically predictable effects.
We also sought evidence of drug‐drug‐gene interactions in this study population. Drug‐drug‐gene interactions with CYP2C9, which we define as drug‐drug interactions that vary based on CYP2C9 genotype, have been previously suggested based on in vitro 26 , 27 , 28 and in vivo 7 , 29 , 30 evidence. Two‐way ANOVA analyses, including CYP2C9/VKORC1 genotype and exposure to CYP2C9‐interacting drugs during warfarin therapy, revealed a significant interaction term for Ln(INRvarA; Table S4 ). To explore our hypothesis further, we conducted subgroup analyses and found that for all outcome variables there was a significant association with CYP2C9‐interacting drugs only in the genotype‐predicted normal response group (Table S4 ). This observation raises the possibility that variant CYP2C9 enzyme alleles may respond differently to interacting drugs.
Another interesting finding from our subgroup analyses was that CYP2C9/VKORC1 genotype was only a significant predictor of INR variability and TTR in the subgroup with no CYP2C9‐interacting drug exposures. This is an important observation because it could explain negative results regarding the relationship of INR variability and TTR with CYP2C9 and VKORC1 variants if concurrent medication exposures were not considered. 12 Furthermore, our results raise concern that exposure to CYP2C9‐interacting drugs could confound efforts to discern the effects of genotype alone in previous studies of warfarin pharmacogenomics if such exposures were not comprehensively considered.
This study highlights a limitation of the three most commonly cited pharmacogenomics‐based dosing algorithms, 6 , 7 , 8 which variably consider CYP2C9‐interacting drugs other than amiodarone. The Gage algorithm considers no drugs other than amiodarone. The International Warfarin Pharmacogenetics Consortium algorithm 7 further includes concurrent use of the CYP2C9 inducers rifampin, carbamazepine, and phenytoin, whereas the Lenzini algorithm 8 includes concurrent fluvastatin use. The 2017 CPIC update on warfarin dosing 11 references the International Warfarin Pharmacogenetics Consortium and Gage algorithms, but does not comment further about CYP2C9‐interacting drugs. Our study considered a more comprehensive list of clinically significant CYP2C9‐interacting drugs, some of which might be considered in future pharmacogenomics‐based dosing algorithms.
Study limitations
There are limitations to our study. First, our sample size lacked power to discern associations with less common genotype groups, such as highly sensitive responders. Second, we group many CYP2C9‐interacting drugs together when evaluating effects on INR variability and TTR. Conclusions derived from this approach are informative at the population level but may be less useful when determining the impact of a single drug on an individual’s warfarin regimen. Third, we have limited data about each of the participants’ risk of thrombosis and bleeding that may have influenced dosing patterns and INR measurements. We were also unable to assess adherence to warfarin regimen, which may be a source of INR variability. Fourth, we excluded participants with < 14 INR measurements to better assess long‐term INR trends, but this could introduce bias.
Despite limitations, we have shown that long‐term measures of warfarin anticoagulation are significantly associated with CYP2C9/VKORC1 genotype and concurrent exposure to CYP2C9‐interacting drugs. Future studies should examine INR variability over longer study periods, and control for a broader set of CYP2C9‐interacting drugs when examining the pharmacogenomics of warfarin as well as other medications dependent upon this metabolic pathway. This type of study should also be done in other populations, such as African Americans using genes that have been found to be related to warfarin metabolism in those groups. 21 , 31
In conclusion, we observed that exposure to CYP2C9‐interacting drugs was more predictive of long‐term warfarin anticoagulation efficacy and stability than CYP2C9/VKORC1 genotype in a self‐identified white population. This illustrates that pharmacogenomics phenomena related to warfarin can be confounded by concurrent medication exposures, and perhaps a similar interaction occurs with other drugs metabolized by CYP2C9. Our findings further suggest that genotype‐based dosing algorithms that consider an expanded list of CYP2C9‐interacting drugs beyond amiodarone may be valuable.
Funding
This study was supported by the Northwestern Medicine Catalyst Fund and National Institutes of Health (NIH) grants HG008673 (M.E.S.), MD009217 (M.A.P.), and MD010723 (M.A.P.).
Conflict of Interest
Dr. George serves on Scientific Advisory Boards for Amgen, Inc. and Otsuka Pharmaceuticals, and received grant funding from Praxis Precision Medicines, Inc. All other authors declared no competing interests for this work.
Author Contributions
S.A., M.S.H., L.J.R.‐T, M.A.P., and A.L.G. designed the research. S.A., M.S.H., R.B.F., T.V.A., L.J.R.‐T., M.E.S., and J.A.P. performed the research and analyzed the data. S.A., M.S.H., and A.L.G. wrote the manuscript.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Pharmacogenomics explains some of the interindividual variability in response to warfarin, but less attention has been paid to drug‐drug interactions in the context of genetic factors.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ What is the impact of cytochrome P450 (CYP)2C9‐interacting drug exposure on long‐term measures of warfarin anticoagulation when CYP2C9 and vitamin K epoxide reductase complex (VKORC)1 genotypes are taken into account?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ Exposure to CYP2C9‐interacting drugs was more predictive of long‐term warfarin anticoagulation measured by international normalized ratio variability and time in therapeutic range than CYP2C9/VKORC1 genotype in a self‐identified white population.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ Our findings emphasize the importance of considering an expanded range of drug interactions in pharmacogenomic studies of warfarin and potentially other medications.
Supporting information
Supplementary Material
References
- 1. Barnes, G.D. , Lucas, E. , Alexander, G.C. & Goldberger, Z.D. National trends in ambulatory oral anticoagulant use. Am. J. Med. 128, 1300–1305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arwood, M.J. et al Anticoagulation endpoints with clinical implementation of warfarin pharmacogenetic dosing in a real‐world setting: A proposal for a new pharmacogenetic dosing approach. Clin. Pharmacol. Ther. 101, 675–683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang, L. , McLeod, H.L. & Weinshilboum, R.M. Genomics and drug response. N. Engl. J. Med. 364, 1144–1153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kimmel, S.E. et al A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 369, 2283–2293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pirmohamed, M. et al A randomized trial of genotype‐guided dosing of warfarin. N. Engl. J. Med. 369, 2294–2303 (2013). [DOI] [PubMed] [Google Scholar]
- 6. Gage, B.F. et al Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin. Pharmacol. Ther. 84, 326–331 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klein, T.E. et al Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med. 360, 753–764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lenzini, P. et al Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin. Pharmacol. Ther. 87, 572–578 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bush, W.S. et al Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin. Pharmacol. Ther. 100, 160–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ormond, K.E. , Cirino, A.L. , Helenowski, I.B. , Chisholm, R.L. & Wolf, W.A. Assessing the understanding of biobank participants. Am. J. Med. Genet. A 149A, 188–198 (2009). [DOI] [PubMed] [Google Scholar]
- 11. Johnson, J.A. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics‐guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 102, 397–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iwuchukwu, O.F. et al Genetic determinants of variability in warfarin response after the dose‐titration phase. Pharmacogenet. Genomics 26, 510–516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mega, J.L. et al Genetics and the clinical response to warfarin and edoxaban: findings from the randomised, double‐blind ENGAGE AF‐TIMI 48 trial. Lancet 385, 2280–2287 (2015). [DOI] [PubMed] [Google Scholar]
- 14. Vandell, A.G. et al Genetics and clinical response to warfarin and edoxaban in patients with venous thromboembolism. Heart 103, 1800–1805 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flockhart Table of Drug Interactions . <https://drug‐interactions.medicine.iu.edu/MainTable.aspx> (2019).
- 16. Yasuda, S.U. , Zhang, L. & Huang, S.M. The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin. Pharmacol. Ther. 84, 417–423 (2008). [DOI] [PubMed] [Google Scholar]
- 17. Scott, S.A. , Khasawneh, R. , Peter, I. , Kornreich, R. & Desnick, R.J. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics 11, 781–791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shah, R.R. & Smith, R.L. Addressing phenoconversion: the Achilles' heel of personalized medicine. Br. J. Clin. Pharmacol. 79, 222–240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Limdi, N.A. et al Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood 115, 3827–3834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schelleman, H. et al Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin. Pharmacol. Ther. 84, 332–339 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De, T. et al Association of genetic variants with warfarin‐associated bleeding among patients of African descent. JAMA 320, 1670–1677 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hill, C.E. & Duncan, A. Overview of pharmacogenetics in anticoagulation therapy. Clin. Lab. Med. 28, 513–524 (2008). [DOI] [PubMed] [Google Scholar]
- 23. Dean, L. Warfarin therapy and VKORC1 and CYP genotype In: Medical Genetics Summaries (eds. Pratt V., McLeod H., Dean L., Kattman B. & Malheiro A.). (National Center for Biotechnology Information (US), Bethesda, MD, 2012). <https://www.ncbi.nlm.nih.gov/pubmed/28520347>. [PubMed] [Google Scholar]
- 24. Takahashi, H. & Echizen, H. Pharmacogenetics of warfarin elimination and its clinical implications. Clin. Pharmacokinet. 40, 587–603 (2001). [DOI] [PubMed] [Google Scholar]
- 25. Wadelius, M. et al The largest prospective warfarin‐treated cohort supports genetic forecasting. Blood 113, 784–792 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hummel, M.A. , Dickmann, L.J. , Rettie, A.E. , Haining, R.L. & Tracy, T.S. Differential activation of CYP2C9 variants by dapsone. Biochem. Pharmacol. 67, 1831–1841 (2004). [DOI] [PubMed] [Google Scholar]
- 27. Hanatani, T. et al CYP2C9*3 influences the metabolism and the drug‐interaction of candesartan in vitro. Pharmacogenomics J. 1, 288–292 (2001). [DOI] [PubMed] [Google Scholar]
- 28. Zhang, N. , Seguin, R.P. , Kunze, K.L. , Zhang, Y.Y. & Jeong, H. Characterization of inhibition kinetics of (S)‐warfarin hydroxylation by noscapine: implications in warfarin therapy. Drug Metab. Dispos. 41, 2114–2123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar, V. , Locuson, C.W. , Sham, Y.Y. & Tracy, T.S. Amiodarone analog‐dependent effects on CYP2C9‐mediated metabolism and kinetic profiles. Drug Metab. Dispos. 34, 1688–1696 (2006). [DOI] [PubMed] [Google Scholar]
- 30. Kumar, V. , Brundage, R.C. , Oetting, W.S. , Leppik, I.E. & Tracy, T.S. Differential genotype dependent inhibition of CYP2C9 in humans. Drug Metab. Dispos. 36, 1242–1248 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perera, M.A. et al Genetic variants associated with warfarin dose in African‐American individuals: a genome‐wide association study. Lancet 382, 790–796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


