Abstract
Synchrotron radiolysis generates hydroxyl radicals (•OH) that are successful footprinting reagents. Here, we describe a new reagent for the synchrotron platform, the trifluoromethyl radical (•CF3). The radical is produced by •OH displacement of •CF3 from sodium triflinate (Langlois reagent). Upon X-ray beam exposure, the reagent labels proteins extensively without any additional chemicals on the millisecond or shorter timescale. The •CF3 is comparably reactive to •OH and produces footprinting information that complements that of •OH alone. This reagent in combination with •OH should enable novel chemistry for protein footprinting on the synchrotron platform.
Graphical Abstract
“Protein footprinting” characterizes protein structure by determining solvent accessibility of backbone and side chains of proteins. Compared to NMR spectroscopy, X-ray crystallography, and cryo-EM, protein footprinting is not restricted by protein size or biomolecular media Moreover, protein structure and dynamics can be interrogated in physiologically relevant media (including live cells and animals)1. The method gives medium to high-resolution readouts of protein structure and solvent accessibility of side chains and often can provide useful information when other methods fail.2, 3. Footprinting is enabled by increasingly effective liquid chromatography with tandem mass spectrometry (LC/MS/MS), which has been the enabling tool in primary-structure proteomics.
One approach to protein footprinting is fast irreversible labeling of protein side chains. The prototype is •OH, introduced first by Chance and co-workers for protein footprinting.4 There are several methods to generate •OH including synchrotron radiolysis of water,5 laser photolysis of hydrogen peroxide,6–9 and others.10, 11 The advantages of •OH compared to other irreversible labeling reagents are its high reactivity, covering over one-half the common amino acids (AA), its size, comparable to that of water, and its polar nature compatible with soluble proteins. Other reactive reagents for protein footprinting are carbenes,12–15 iodine radical,16 sulfate radical anion.17 Slower reacting reagents complement •OH and are generally AA specific (e.g., NEM,18 GEE,19 BHD20) but can have relatively broad reactivity (e.g., DEPC21). These latter reagents require no special equipment, but have longer “exposure” times and the possibility sequential reactions label an already modified protein that has undergone conformational change.
The •CF3 radical may be an appropriate complement to •OH owing to its pyramidal geometry, high electrophilicity, and high hydrophobicity.22 Dolbier23 and Minisci24 demonstrated that perfluoroalkyl radicals (PFs) undergo reactions with alkenes and aromatic rings, and even abstract •H from alkanes. PFs are more reactive than analogous alkyl radicals,22 and their reaction rate constants are larger in aqueous media than in nonpolar organic solvents,25 providing motivation for exploring it in protein footprinting where multiple positions on a protein surface are available for modification.
Our original work with •CF3, carried out on the fast-photochemical oxidation of proteins (FPOP) platform, showed that trifluoromethylation can be induced by •OH radical activation of the Langlois reagent (sodium triflinate).26 We presume that once •OH forms, it rapidly reacts with the reagent at C=S, possibly without a transition-state barrier, to displace •CF3, which can be formed in any context that utilizes •OH or possibly other strong oxidizing agents.27
Synchrotron footprinting employs highly focused X-ray beams to generate •OH directly from water without the need for addition of peroxide.28 Thus, we hypothesized that radical trifluoromethylation can be initiated by water radiolysis with a synchrotron beam (Fig. 1). In this note, we describe a test of feasibility and utility of CF3 footprinting as a complement to that of •OH on the synchrotron platform and explore whether modified residue coverage can be increased while maintaining fast timescales of footprinting.
Figure 1.
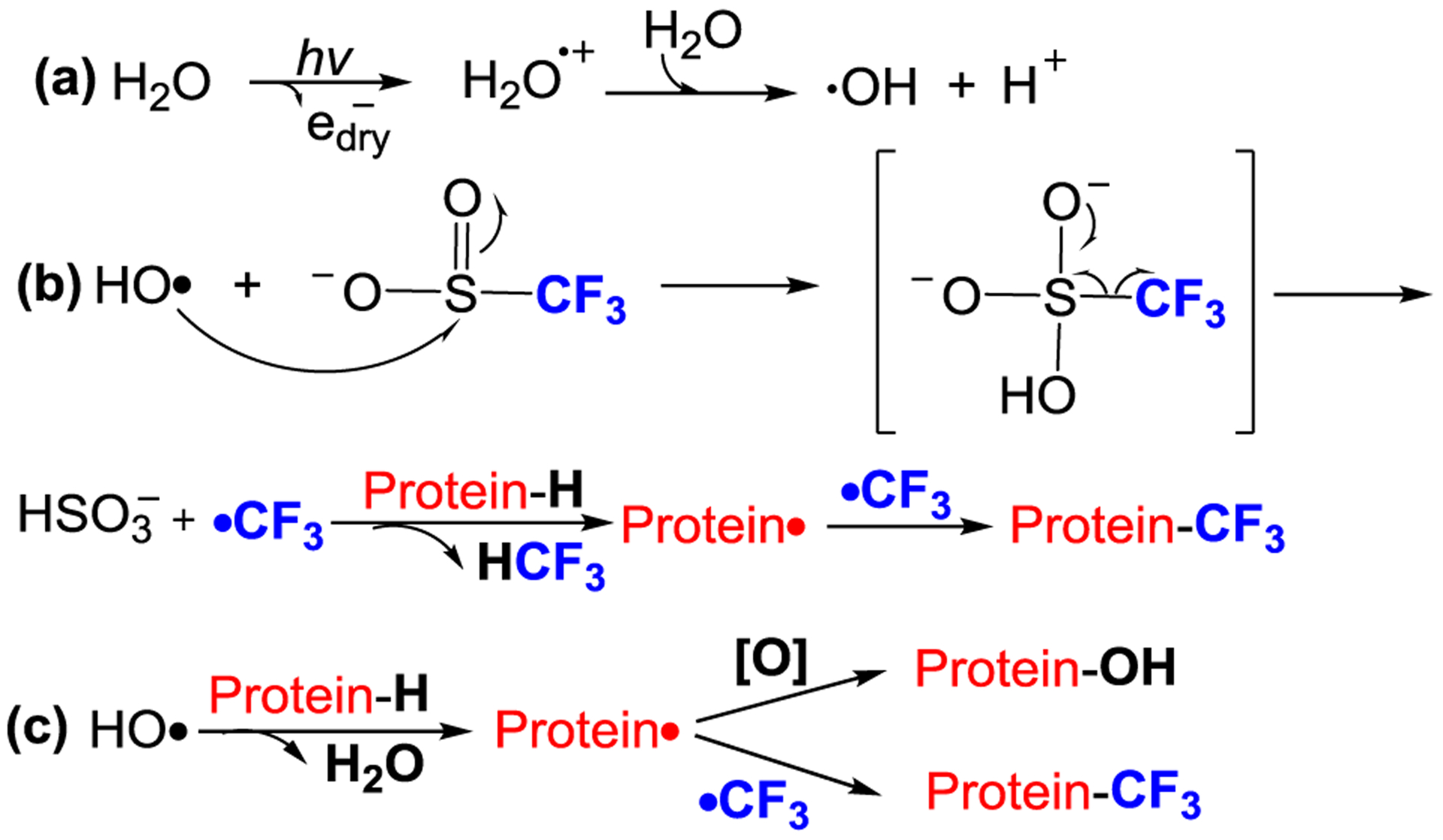
Proposed pathway for X-ray mediated radical trifluoromethylation for protein footprinting.
Although the introduction of CF3 into large biomolecules (e.g., proteins) remains rare, it is attracting interest. Baran and coworkers29 reported that direct protein (beta-lactamase) trifluoromethylation in Tris buffer retained protein functional activity after isolation. Recently, the Davis30 and Krska31 groups independently developed methods for radical trifluoromethylation of proteins and peptides. The potential applications of trifluoromethylation chemistry in protein footprinting, protein engineering, protein F-NMR, and positron emission tomography (PET) represent considerable opportunities for its extension to problems in biophysics and biomedicine.32
EXPERIMENTAL SECTION
Materials.
The proteins, peptides, and reagents used in this study are described in Supporting Information (P.S2).
X-ray protein footprinting:
X-ray protein footprinting experiments were performed on beamline 3.2.1 at the Advanced Light Source (ALS) at Lawrence National Berkeley Laboratory (LBNL). Protein samples for •OH footprinting experiments (XF-OH) were prepared by reconstituting lyophilized apomyoglobin or holomyoglobin in PBS (pH 7.4) to yield a final concentration of 5 μM, and samples for XF-CF3 experiments were prepared the same way except sodium triflinate was added to a final concentration of 20 mM. All samples were prepared fresh and stored on ice until X-ray exposure and were exposed using a capillary flow setup as previously described.4
Enzymatic digestions.
After labeling, triplicate samples were precipitated with acetone (−20 °C). The resulting protein pellet was collected and denatured with 10 μL of 8 M urea, followed by dilution in 50 mM Tris-HCl buffer (pH 8.0) until the urea concentration was less than 1 M. Trypsin was added for digestion to a protease-to-protein ratio of 1:20 (w/w) at 37 °C for 12 h, followed by quench with 1 μL of formic acid.
Mass Analysis.
For intact-protein analysis, mass spectra of the modified protein were taken with a Bruker Maxis Q-TOF mass spectrometer. For bottom-up analysis, an aliquot of 5 μL tryptic peptides was loaded onto a custom-built silica capillary column (0.075 × 150 mm) packed with C18 reversed-phase material and analyzed on Q Exactive Plus hybrid quadrupole orbitrap mass spectrometer coupled with a Nanospray Flex ion source (Thermo Fisher, Santa Clara, CA). Descriptions of the HPLC gradient and MS instrument parameters are in the Supporting Information (P.S2).
Data Analysis.
The data processing was with PMI software (Protein Metrics, San Carlos, CA, USA) coupled with an in-house database. Experimental details are described in Supporting Information.
RESULTS AND DISCUSSION
Our objective is to test whether radical trifluoromethylation is compatible with the X-ray synchrotron platform. We mixed the protein only with Langlois reagent in PBS buffer (pH 7.4) without any additional reagents (e.g., transition metals, peroxide) and fixed the concentration of the Langlois reagent at 20 mM because this concentration worked efficiently in the original development.26 The synchrotron platform permits the X-ray exposure to be adjusted over a range of times (exposure times of 0, 25, 50, and 75 ms).
There is concern that the presence of the Langlois reagent affects protein HOS in the absence of forming •CF3, although that seems unlikely when the required amounts of denaturants are at the molar level. Nevertheless, we performed circular dichroism (CD) spectroscopy to test the biocompatibility of Langlois’ reagent with both holo and apo myoglobin incubated with the reagent (Figure S5 and Figure S6). No significant secondary structural change was seen as evidenced by the nearly identical CD spectra of apo/holo myoglobin with Langlois reagent, showing helical structure. Our perturbation analysis is consistent with previous bioactivity assay in which CF3-modified enzyme retained its functional activity after isolation.30 A more stringent test of effects on tertiary structure could be done by NMR or HDX, and we recommend this for studies on new proteins. Such studies are beyond the scope of this announcement.
The key to generating •CF3 is the •OH, formed by radiolysis of water (Fig. 1a). The electron-deficient •OH likely reacts with the sulfur of CF3SO2− to displace •CF3 accompanied by an entropy-driven loss of SO2. A 4,000-fold excess of the Langlois reagent relative to the protein molecule (and the •OH concentration) was chosen to favor •CF3 vs. •OH modification (Fig. 1b). In a “sea” of functionality and structural context of the protein environment, however, •OH may be more reactive to certain residues than with •CF3. Our design is appropriate because the CF3-modified protein is the dominant product. In a minor pathway (Fig. 1c), •OH abstracts •H from the protein to produce a protein radical, which either couples with •CF3 to provide CF3- substituted products or reacts with •OH or dissolved oxygen to generate oxygen-incorporated products (See Table S1 in Supporting Information). The small fraction of oxidized product can also be attributed to rapid oxidation of methionine by •OH. From our bottom-up analysis at the residue level, we found methionine is sensitive to •OH but is silent to •CF3 labeling.
MS analysis of the intact protein identifies the expected mass shift (+68 Da) corresponding to CF3 substitution of one hydrogen (Fig. 2a and Figures S1 and S2) and demonstrates extensive labeling. The •OH is produced uniformly throughout the solution and produces •CF3 with access to the protein surface. With only 25 ms X-ray exposure, apo-Mb was modified by •CF3 up to at least three times (the resulting peaks showing modification are resolved by MS, Fig. 2a, middle and Figures S1 and S2). Notably, the deconvoluted mass spectra indicate a different footprinting for apo and holo Mb. For holo (Fig. 2a, front), the most abundant modified protein contains one CF3 modification, whereas for apo, we see more extensive oxidation including mono, di, and trifuoromethylation. This result is expected, because the apo is less structured and has more surface area. In addition to CF3 modification, we also observe a mass shift (+16 Da) corresponding to •OH substitution. This signal is due in part to basal oxidation that was seen in the sample without X-ray irradiation.
Figure 2.
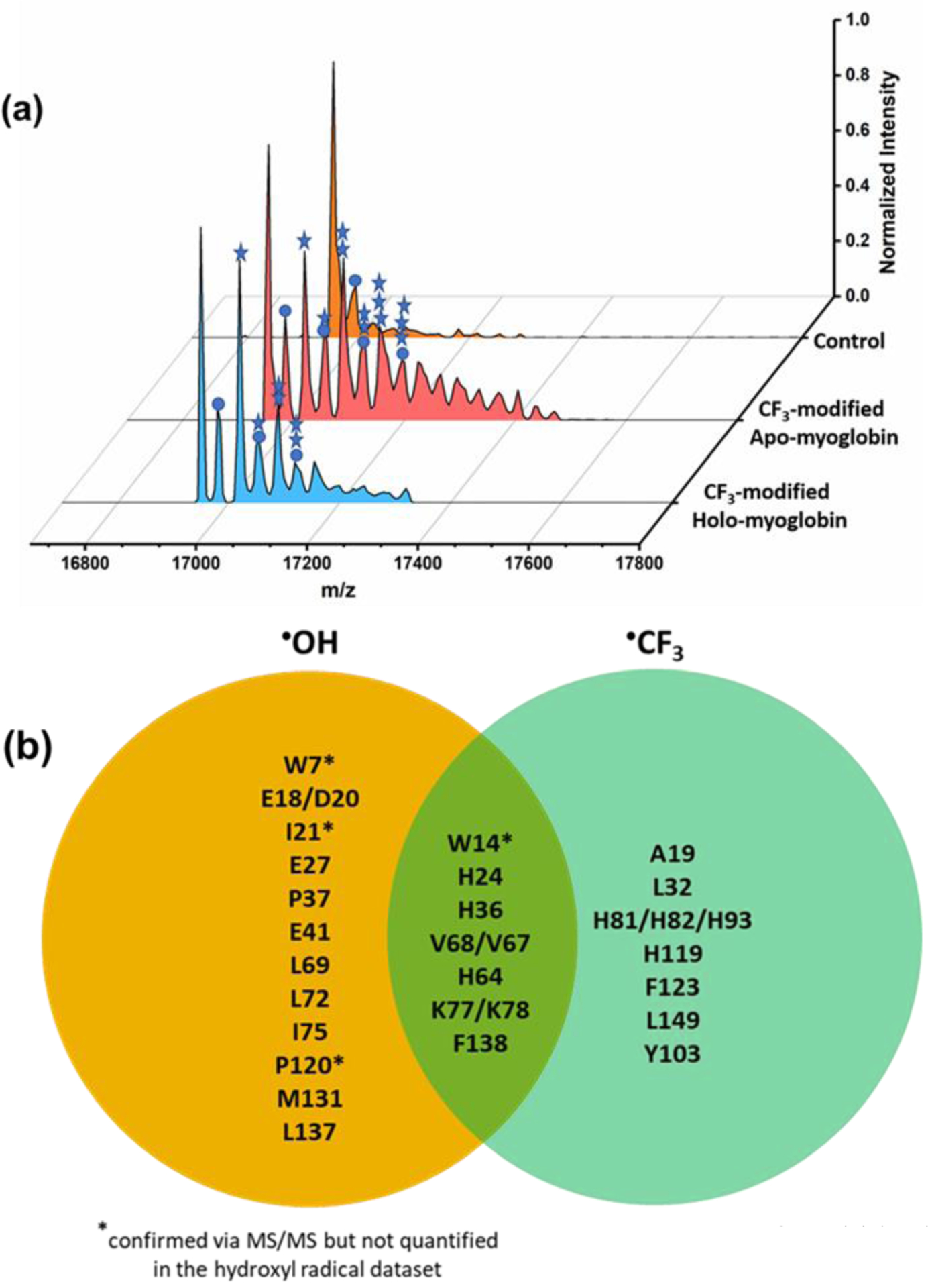
(a) Deconvoluted mass spectra. CF3-modified holo-Mb (front) and apo-Mb (middle) at 25 ms of X-ray irradiation. Apo-Mb (back) without x-ray irradiation shows no CF3-modifications. (b) Stars represent number of trifluoromethylations (+68 Da), and circles represent H substitution by •OH. This figure adapted from Figure S1. Comparison of chemical reactivities for •OH and •CF3 provided by a Venn diagram comparing residues reactive with •OH (in orange) and with •CF3 (in light green); residues in the overlap region are reactive with both •OH and •CF3.
Having demonstrated trifluoromethylation on the synchrotron platform, we next examined for the synchrotron, how the •CF3 chemistry complements the •OH footprinting by modifying the protein with •OH or •CF3 in independent experiments. The modified sites were identified using a canonical proteomics strategy, and the modification sites are displayed on a Venn diagram that reveals unique and common modifications in each dataset (Fig. 2b). The product-ion spectra combined with extracted ion chromatograms (EIC) show modifications of 31 of 153 total residues in Mb by •OH and •CF3, giving 20% (31/153) reactivity coverage. Notably, •CF3 modifies nine residues (in blue) that are silent to •OH. On the other hand, 13 AAs (in yellow) are modified by •OH, but they are unreactive with •CF3. Both •CF3 and •OH can modify AAs containing aromatic side chains (e.g., His, Trp) and aliphatic side chains (e.g., Leu, Val). The % •OH modifications of apo myoglobin with 25 ms X-ray irradiation (Table S1) compared to the •CF3modifications are less abundant than CF3-modification except for M131 and V72, L76, V68. We also found that W14 and M131 are mainly •OH modified in apo myoglobin with 25 ms X-ray irradiation.
Noteworthy is that •CF3 modifies normally inert Ala19, as confirmed by EIC and MS/MS (see SI Figure S8). From energetics, •CF3 can abstract •H from methane that contains the most inert aliphatic C-H bonds (Figure S4), and C-H abstraction by perfluoroalkyl radicals are demonstrated in the literature.24,25 Ala19, as labeled, has the third largest solvent-accessible surface area (SASA) among the 15 Ala in myoglobin (Table S2) and is the only Ala on a loop region (the others are located in structured helices (Figure S9)). This modification is absent in •OH labeling and was not seen in previous radical trifluoromethylation.26 We attribute this lack of reactivity to a depletion of a local concentration of H2O2 around Ala, discriminating against its modification on the FPOP platform. Clearly, trifluoromethylation is a complement to hydroxylation.
We then examined the AA residue reactivity by quantifying the modifications in isomers resolvable by LC. One representative EIC for a CF3-modified peptide 119–133 (75 ms X-ray exposure) (Fig. 3a) reveals a CF3-modifed species as the major product. MS/MS assigns the major CF3-modifications to residues His119 and Phe123 and a minor modification to Met 131. Given that •OH is extremely reactive with methionine, footprinting with •OH is often dominated by reactions at this residue, muting the reactivity of other proximal residues. In this case, trifluoromethylation occurs with different specificity, allowing other residues (His 119 and Phe123) to be labeled. For another peptide 64–77 containing no methionine, the reactivity pattern for the two radicals is also different (Fig. 3b). The •CF3 reacts with His 64 whereas •OH with Val 68 and Leu 69, Leu72, and Leu76. Trifluoromethylation of Val 68, Leu72 and 76 also occurs but less abundantly compared to that of His 64. From their product-ion (MS/MS) spectra and the EIC, •OH and •CF3 modifications occur concurrently for CF3-radical footprinting, suggesting that a titration with •CF3 can show the effects of both •CF3 and •OH modifications.
Figure 3.
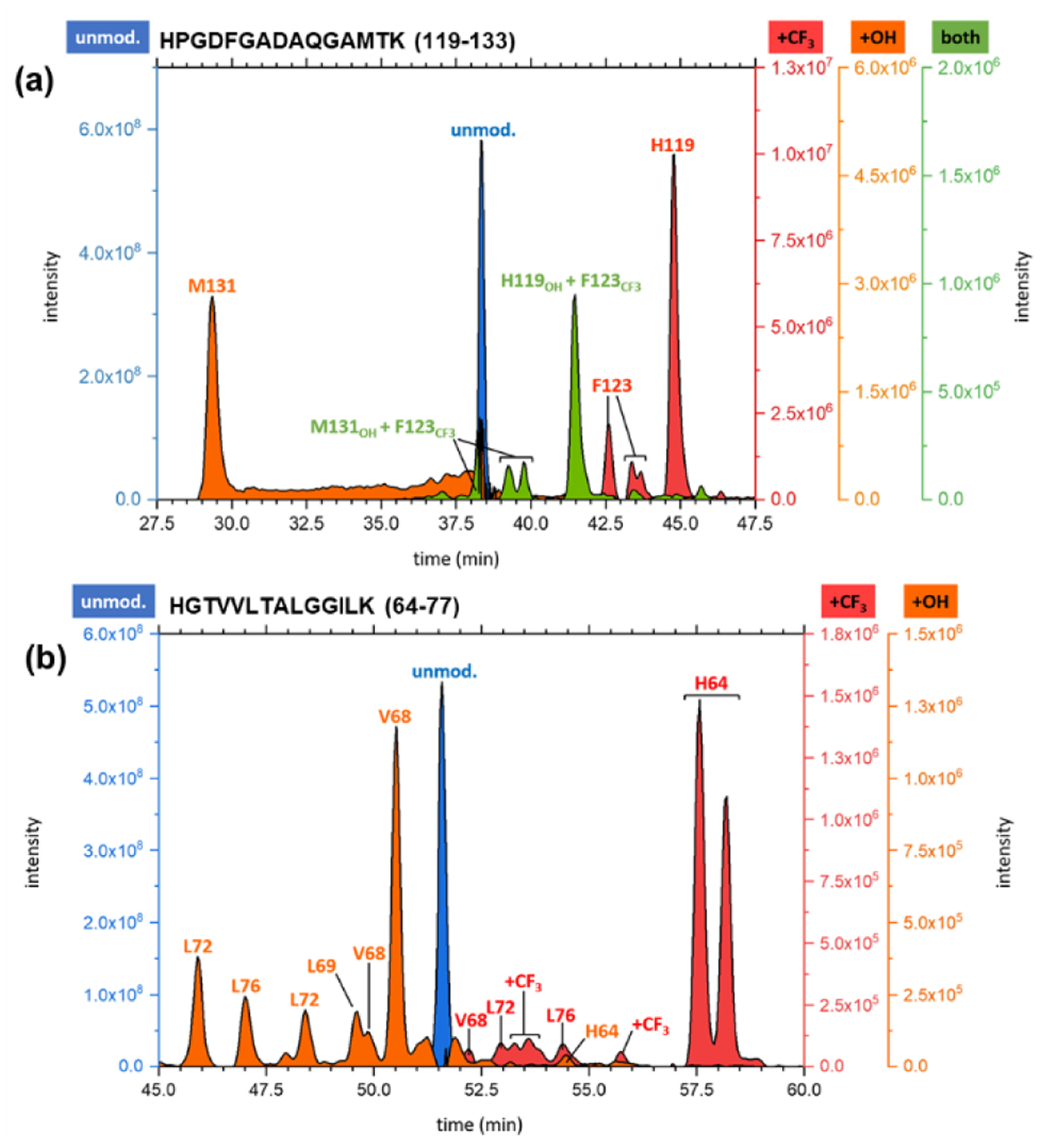
(a) EIC for peptide 119–133 from a 75 ms exposure of holo-Mb for CF3 footprinting on the synchrotron platform. (b) EIC for peptide 64–77 taken from the 50 ms exposure of holo-Mb. EICs for the unmodified (blue), hydroxyl-radical modified (orange), CF3-radical labeled (red), and di-modified (green) peptides confirmed via MS/MS are shown.
Noteworthy is that specific •CF3 modification on protein and peptides were recently exploited by several groups.30, 31 The reactions to modify certain amino acids were proposed and timescales of labeling were min to h. We show that •CF3 labeling of proteins can be realized on the ms timescale of the synchrotron. The •CF3 undergoes reactions with aromatic rings, and even abstracts •H from aliphatic carbon.24–26 Our goal of a complementary approach to high coverage footprinting with a broadly reactive reagent is closer to realization with this synchrotron platform.
Lastly, we evaluated whether •CF3 footprinting affords a protein footprint that is responsive to a change in protein structure. The modification pattern responding to heme binding in Mb is reflected in both •CF3 and •OH footprinting (Fig. 4). For AAs labeled by both •CF3 and •OH, H24, H36, H64, and F138 show similar trends of increased modifications for holo-Mb, whereas K77 maintains a constant modification rate for holo and apo Mb. On the other hand, V18 shows an opposite trend, where •CF3 labeling is greater for holo-Mb and •OH labeling is less. The divergence at V18 may be due to the low reactivity of V18 (< 3%), decreasing reliability owing to S/N issues. Overall, the outcome indicates the two reagents respond similarly to changes in structure although they have different reactivities. This similarity is due in part to the water solubility of the Langlois reagent, allowing a homogeneous production of the reactive species in places where bulk water penetrates.
Figure 4.
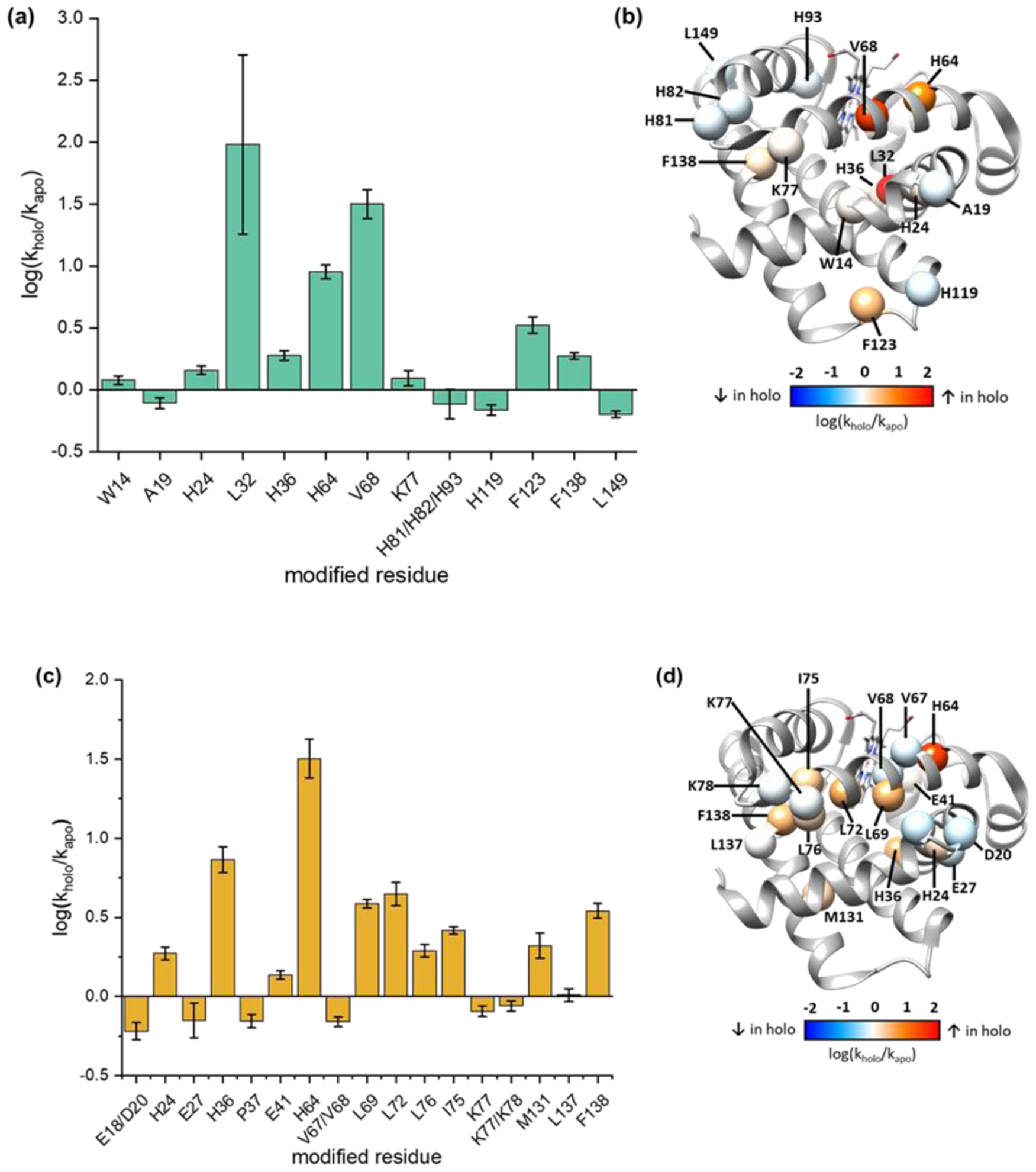
Comparison of X-ray-induced •CF3 and •OH at the AA residue level. Changes in the modification rates respond to heme binding, for the rates of •CF3 modification (a) and of •OH modification (c). Changes mapped on to the structure of holo-Mb (PDB: 1WLA) to identify regions with increased (red), decreased (blue), or unchanged (white) rates of modification upon heme binding, for •CF3 modification (b) and •OH modification (d).
The modification extent increases, however, upon binding to the heme possibly because heme may increases the local concentration of Langlois’ reagent or changes the residue microenvironment.33 We previously showed that in radiolysis, internal waters (in the heme pocket) can be activated, suggesting that sulfinate may get into that pocket.34 Previous •CF3 and •OH footprinting26 and radical iodination16 show a single peptide region (peptide 80–96) undergoes significantly different modification between apo and holo. A plot of the fraction of unmodified peptide versus exposure time (Fig. 5a), shows a similar trend. The results reveal that H64, V68 and F138 surrounding the heme pocket undergo significantly different modification extents in the apo vs. holo. Although H63 reacts comparably with the two radicals, we speculate that •CF3 is more reactive with the hydrophobic V68 and F138 than is •OH because it too is hydrophobic.
Figure 5.
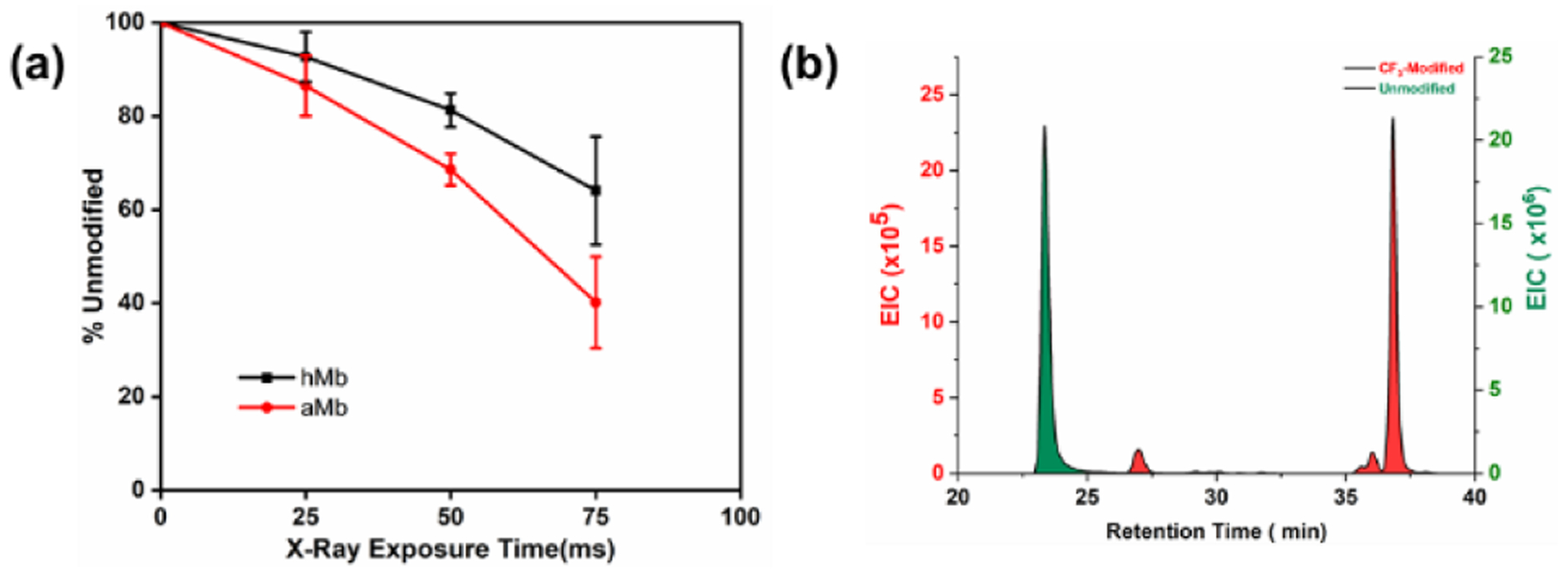
(a) The fraction of unmodified peptide versus exposure time at peptide 80–96. (b)The EIC of peptide 80–96.
Peptide 80–96 contains the ordered F-Helix in holo-Mb, but this helix does not form for apo, as shown by NMR.35 Top-down HDX/MS reveals this region exchanges faster for apo than holo.36 Although the red color disappeared for apo, the F-helix may not completely revert to a random coil after heme loss. The relatively higher CF3 modification extent for peptide 80–96 from apo is consistent with this and with the report that the F-helix is partially retained37 Although the divergence requires further investigation as different platforms were used for these experiments, the •CF3 reaction extent clearly shows increased side-chain solvent accessibility in the region of aMb that becomes the F helix for holo.
To illustrate further footprinting in this region, we obtained an EIC for peptide 80–96. Well resolved chromatograms and MS/MS show three His residues are labeled by •CF3 (Fig. 5b). As expected, the CF3-modified peptide elutes at longer time than the unmodified, consistent with strong absorption of the fluorinated compound on the adsorbent surface. There is even good separation for CF3-modified isomers that contain identical numbers of fluorine atoms. This property of fluorine-containing reagents may be exploited in the future to improve LC resolution or enrich fluorine-containing peptides in more complex biological samples.38
In conclusion, we successfully implemented a new reagent for synchrotron-based protein footprinting. The reactivity of •CF3 complements that of •OH and gives higher coverage. Even for the same peptide regions, •CF3 exhibits different reactivities with AAs side chains. The extension enriches the synchrotron platform by taking advantage of the complementary reactivities of •CF3 and •OH and shows that the synchrotron platform is adaptable to other novel chemistries that may enhance footprinting coverage. Further, the footprinting does not require metal catalysis or peroxide as in other experiments. Although access to synchrotrons is limited, there are now multiple beamlines around the world (Advanced Light Source, National Synchrotron Light Source-II, SOLEIL) that are primarily devoted to footprinting. Thus, this technology can be immediately adopted at the other facilities.
In another application, trifluoromethylation may be useful for protein engineering because trifluoromethylated proteins seem to maintain their enzymatic ability; furthermore, specific protein trifluoromethylation is tunable with appropriate dosimetry and adjustable labeling timescales. In the future, we will investigate how to tune •CF3 reactivity and extend this novel chemistry to other protein systems to enhance protein footprinting in structural biology.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH 2P41GM103422 and R24GM136766 (to MLG) and P30EB09998(to MRC). The XFMS was supported in part by NIH Grant 1R01GM126218 and conducted at the Advanced Light Source and Joint BioEnergy Institute, supported by the Office of Science and Office of Biological and Environmental Research, of the United States DOE under contract DE-AC02-05CH11231. We acknowledge the support from Advanced Light Source (ALS) fellowship for this work. We are also grateful to Protein Metrics (PMI) for providing software for data processing and to Chunyang Guo for helping design TOC graphic.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website. Contents: Materials; global mass analysis; LC/MS/MS analysis of proteolytic peptides; calculation of modification ratio; examples LC-MS chromatograms for global analysis, protein mass spectra for +20 charge state, sequence coverage, representative LC-MS results of CF3 modification of Trp, representative LC-MS results of CF3 modification at Ala; circular dichroism analysis (PDF).
REFERENCES
- 1.Rinas A; Mali VS; Espino JA; Jones LM, Development of a Microflow System for In-Cell Footprinting Coupled with Mass Spectrometry. Anal. Chem 2016, 88, 10052–10058. [DOI] [PubMed] [Google Scholar]
- 2.Huang W; Ravikumar KM; Chance MR; Yang S, Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: a protection factor analysis. Biophys J 2015, 108, 107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaur P; Kiselar J; Yang S; Chance MR, Quantitative Protein Topography Analysis and High-Resolution Structure Prediction Using Hydroxyl Radical Labeling and Tandem-Ion Mass Spectrometry (MS). Mol. Cell. Proteom 2015, 14, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maleknia SD; Brenowitz M; Chance MR, Millisecond Radiolytic Modification of Peptides by Synchrotron X-rays Identified by Mass Spectrometry. Anal. Chem 1999, 71, 3965. [DOI] [PubMed] [Google Scholar]
- 5.Kiselar JG; Maleknia SD; Sullivan M; Downard KM; Chance MR, Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. Int. J. Radiat. Biol 2002, 78, 101. [DOI] [PubMed] [Google Scholar]
- 6.Aye TT; Low TY; Sze SK, Nanosecond Laser-Induced Photochemical Oxidation Method for Protein Surface Mapping with Mass Spectrometry. Anal. Chem 2005, 77, 5814–5822. [DOI] [PubMed] [Google Scholar]
- 7.Hambly DM; Gross ML, Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J. Am. Soc. Mass. Spectrom 2005, 16, 2057–2063. [DOI] [PubMed] [Google Scholar]
- 8.Watson C; Janik I; Zhuang T; Charvátová O; Woods RJ; Sharp JS, Pulsed electron beam water radiolysis for submicrosecond hydroxyl radical protein footprinting. Anal. Chem 2009, 81, 2496–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie B; Sood A; Woods RJ; Sharp JS, Quantitative Protein Topography Measurements by High Resolution Hydroxyl Radical Protein Footprinting Enable Accurate Molecular Model Selection. Sci. Rep 2017, 7, 4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minkoff BB; Blatz JM; Choudhury FA; Benjamin D; Shohet JL; Sussman MR, Plasma-Generated OH Radical Production for Analyzing Three-Dimensional Structure in Protein Therapeutics. Sci. Rep 2017, 7, 12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu G; Chance MR, Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem. Rev 2007, 107, 3514. [DOI] [PubMed] [Google Scholar]
- 12.Richards FM; Lamed R; Wynn R; Patel D; Olack G, Methylene as a possible universal footprinting reagent that will include hydrophobic surface areas: overview and feasibility: properties of diazirine as a precursor. Protein Sci 2000, 9, 2506–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B; Rempel DL; Gross ML, Protein Footprinting by Carbenes on a Fast Photochemical Oxidation of Proteins (FPOP) Platform. J. Am. Soc. Mass. Spectrom 2016, 27, 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jumper CC; Schriemer DC, Mass Spectrometry of Laser-Initiated Carbene Reactions for Protein Topographic Analysis. Anal. Chem 2011, 83, 2913–2920. [DOI] [PubMed] [Google Scholar]
- 15.Manzi L; Barrow AS; Scott D; Layfield R; Wright TG; Moses JE; Oldham NJ, Carbene footprinting accurately maps binding sites in protein–ligand and protein–protein interactions. Nat. Commun 2016, 7, 13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J; Cui W; Giblin D; Gross ML, New Protein Footprinting: Fast Photochemical Iodination Combined with Top-Down and Bottom-Up Mass Spectrometry. J. Am. Soc. Mass. Spectrom 2012, 23, 1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gau BC; Chen H; Zhang Y; Gross ML, Sulfate radical anion as a new reagent for fast photochemical oxidation of proteins. Anal. Chem 2010, 82, 7821–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramakrishnan D; Xing W; Beran RK; Chemuru S; Rohrs H; Niedziela-Majka A; Marchand B; Mehra U; Zábranský A; Doležal M; Hubálek M; Pichová I; Gross ML; Kwon HJ; Fletcher SP, Hepatitis B Virus X Protein Function Requires Zinc Binding. J. Virol 2019, 93, e00250–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen J; Zhang H; Gross ML; Blankenship RE, Membrane orientation of the FMO antenna protein from Chlorobaculum tepidum as determined by mass spectrometry-based footprinting. Proc. Natl. Acad. Sci. U S A 2009, 106, 6134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo C; Cheng M; Gross ML, Protein-Metal-Ion Interactions Studied by Mass Spectrometry-Based Footprinting with Isotope-Encoded Benzhydrazide. Anal. Chem 2019, 91, 1416–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limpikirati P; Pan X; Vachet RW, Covalent Labeling with Diethylpyrocarbonate: Sensitive to the Residue Microenvironment, Providing Improved Analysis of Protein Higher Order Structure by Mass Spectrometry. Anal. Chem 2019, 91, 8516–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Studer A, A “Renaissance” in Radical Trifluoromethylation. Angew. Chem. Int. Ed 2012, 51, 8950–8958. [DOI] [PubMed] [Google Scholar]
- 23.Shtarev AB; Tian F; Dolbier WR; Smart BE, Absolute Rates of Intermolecular Carbon-Hydrogen Abstraction Reactions by Fluorinated Radicals. J. Am. Chem. Soc 1999, 121 (32), 7335–7341. [Google Scholar]
- 24.Liguori L; Bjørsvik H-R; Bravo A; Fontana F; Minisci F, A new direct homolytic iodination reaction of alkanes by perfluoroalkyl iodides. Chem. Commun 1997, (16), 1501–1502. [Google Scholar]
- 25.Zhang L; Cradlebaugh J; Litwinienko G; Smart BE; Ingold KU; Dolbier JWR, Absolute rate constants for some hydrogen atom abstraction reactions by a primary fluoroalkyl radical in water. Org. Biomol. Chem 2004, 2, 689–694. [DOI] [PubMed] [Google Scholar]
- 26.Cheng M; Zhang B; Cui W; Gross ML, Laser-Initiated Radical Trifluoromethylation of Peptides and Proteins: Application to Mass-Spectrometry-Based Protein Footprinting. Angew. Chem. Int. Ed. Engl 2017, 56, 14007–14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Application of Langlois’ Reagent in Trifluoromethylation Reactions. Adv. Synth. Catal 2014, 356, 2895–2906. [Google Scholar]
- 28.Kiselar J; Chance MR, High-Resolution Hydroxyl Radical Protein Footprinting: Biophysics Tool for Drug Discovery. Annu. Rev. Biophys 2018, 47, 315–333. [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara Y; Dixon JA; O’Hara F; Funder ED; Dixon DD; Rodriguez RA; Baxter RD; Herlé B; Sach N; Collins MR; Ishihara Y; Baran PS, Practical and innate carbon–hydrogen functionalization of heterocycles. Nature 2012, 492, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imiołek M; Karunanithy G; Ng W-L; Baldwin AJ; Gouverneur V; Davis BG, Selective Radical Trifluoromethylation of Native Residues in Proteins. J. Am. Chem. Soc 2018, 140, 1568–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichiishi N; Caldwell JP; Lin M; Zhong W; Zhu X; Streckfuss E; Kim H-Y; Parish CA; Krska SW, Protecting group free radical C–H trifluoromethylation of peptides. Chem. Sci 2018, 9, 4168–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verhoog S; Kee CW; Wang Y; Khotavivattana T; Wilson TC; Kersemans V; Smart S; Tredwell M; Davis BG; Gouverneur V, 18F-Trifluoromethylation of Unmodified Peptides with 5–18F-(Trifluoromethyl)dibenzothiophenium Trifluoromethanesulfonate. J. Am. Chem. Soc 2018, 140 (5), 1572–1575. [DOI] [PubMed] [Google Scholar]
- 33.Liu T; Marcinko TM; Kiefer PA; Vachet RW, Using Covalent Labeling and Mass Spectrometry To Study Protein Binding Sites of Amyloid Inhibiting Molecules. Anal. Chem 2017, 89, 11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta S; D’Mello R; Chance MR, Structure and dynamics of protein waters revealed by radiolysis and mass spectrometry. Proc. Natl. Acad. Sci. U S A 2012, 109, 14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eliezer D; Wright PE, Is Apomyoglobin a Molten Globule? Structural Characterization by NMR. J. Mol. Biol 1996, 263, 531. [DOI] [PubMed] [Google Scholar]
- 36.Pan J; Han J; Borchers CH; Konermann L, Hydrogen/Deuterium Exchange Mass Spectrometry with Top-Down Electron Capture Dissociation for Characterizing Structural Transitions of a 17 kDa Protein. J. Am. Chem. Soc 2009, 131, 12801–12808. [DOI] [PubMed] [Google Scholar]
- 37.Vahidi S; Stocks BB; Liaghati-Mobarhan Y; Konermann L, Mapping pH-Induced Protein Structural Changes Under Equilibrium Conditions by Pulsed Oxidative Labeling and Mass Spectrometry. Anal. Chem 2012, 84 (21), 9124–9130. [DOI] [PubMed] [Google Scholar]
- 38.Cheng M; Guo C; Gross ML, The Application of Fluorine-Containing Reagents in Structural Proteomics. Angew. Chem., Int. Ed. Engl [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


