Abstract
Introduction
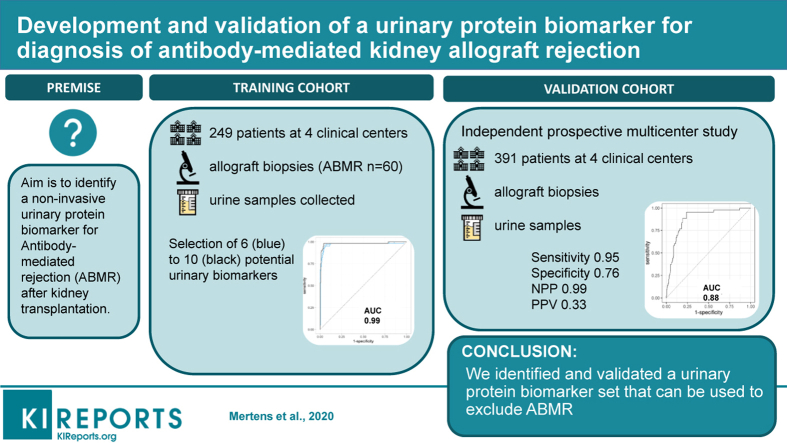
Antibody-mediated rejection (ABMR) impacts kidney allograft outcome. The diagnosis is made based on findings from invasive kidney transplant biopsy specimens. The aim of this study was to identify a noninvasive urinary protein biomarker for ABMR after kidney transplantation.
Methods
We performed a multicenter case-control study to identify a urinary biomarker for ABMR (training cohort, n = 249) and an independent, prospective multicenter cohort study for validation (n = 391). We used concomitant biopsies to classify the samples according to the Banff classification. After untargeted protein identification and quantification, we used a support vector machine to train the model in the training cohort. The primary endpoint was the diagnostic accuracy of the urinary biomarker for ABMR in the validation cohort.
Results
We identified a set of 10 urinary proteins that accurately discriminated patients with (n = 60) and without (n = 189) ABMR in the training cohort with an area under the curve (AUC) of 0.98 (95% confidence interval [CI], 0.96–1.00). The diagnostic accuracy was maintained in the validation cohort (AUC, 0.88; 95% CI, 0.8–0.93) for discriminating the presence (n = 43) from the absence (n = 348) of ABMR. The negative predictive value of the 10-protein marker set for exclusion of ABMR was 0.99, and the positive predictive value was 0.33. The diagnostic accuracy was independent of the reason for performing the biopsy, time after transplantation, and better than the accuracy of gross proteinuria (AUC, 0.76).
Conclusions
We identified and validated a urinary protein biomarker set that can be used to exclude ABMR.
Keywords: antibody-mediated rejection, noninvasive biomarker, renal transplantation, urinary proteomics
Graphical abstract
Long-term survival of kidney allografts has improved little during the past 2 decades. Rejection phenomena remain major determinants of late graft failure despite the use of powerful immunosuppressive agents. In particular, ABMR is known to impact graft outcome.1,2 Histologic examination of kidney allografts through invasive biopsies is currently the gold standard to detect rejection. The Banff classification was established to standardize the diagnosis of rejection phenotypes such as borderline lesions, T cell–mediated rejection (TCMR), and ABMR.3,4 Rejection can be suspected in clinical practice by a rise in the serum creatinine level or an increase of proteinuria, but it can also present subclinically with normal renal functional parameters. In addition, subclinical rejection, especially subclinical ABMR, is associated with impaired graft outcome.5
Noninvasive monitoring of renal allograft rejection is based on the measurement of serum creatinine levels, glomerular filtration rate, proteinuria, and the measurement of donor-specific human leukocyte antigen antibodies. These graft functional parameters lack sensitivity and specificity for rejection. Kidney transplant rejection that occurs with stable graft functional parameters remains currently undetected, unless systematic biopsies at predefined post-transplantation time points are performed. Such protocol biopsies are not performed in all transplant centers, leaving many cases of subclinical rejection undetected and thus not treated. Centers that do perform routine protocol biopsies will perhaps identify rejection at an earlier stage before chronic injury develops and when the disease process may be more responsive to treatment. In addition, kidney allograft biopsies remain expensive and invasive, with the inherent risk for postbiopsy complications.6
Noninvasive biomarkers with good sensitivity and specificity for kidney allograft disease processes are an unmet clinical need. Several biomarkers for acute rejection have been suggested,7 but none were developed specifically for ABMR. As ABMR is a main cause of kidney transplant failure but is often missed by current functional testing owing to the lack of validated and specific noninvasive markers, we aimed to develop and validate a novel urinary biomarker for ABMR after kidney transplantation.
Methods
Study Population
We included patients who received a kidney allograft in 4 European transplant centers (Necker Hospital Paris, France; University Hospitals Leuven, Belgium; Medical School Hannover, Germany; and University Hospital Centre, Limoges, France) after written informed consent was obtained. Samples were prospectively collected in the context of the BIOMArkers of Renal Graft INjuries (BIOMARGIN) study (www.biomargin.eu; P.I. Prof. P. Marquet). Protocol or indication renal allograft biopsies were performed, and urine samples were prospectively and consecutively collected at the time of the biopsies. In the 4 clinical centers, protocol biopsies were performed at 3 and 12 months, and sometimes at 24 months after transplantation according to local center practice, in addition to clinically indicated biopsies (biopsies at time of graft dysfunction). All adult patients who had received a single-kidney allograft at these institutions and who provided written informed consent were eligible. Recipients of combined transplantations were excluded. All transplantations were performed with negative complement-dependent cytotoxicity crossmatches. Institutional review boards and national regulatory agencies (when required) approved the study protocol at each clinical center.
The training cohort (grouping steps 1 and 2 of the initial BIOMARGIN program) consisted of 249 samples collected following a case-control study design. Patients and controls were selected based on sample availability and histology of the concomitant renal allograft biopsies, with exclusion of patients with diagnosis of glomerulonephritis or polyomavirus-associated nephropathy and those with unclear diagnosis. Based on local biopsy results, a preselection was made, which was then further refined by central reading by a group of 3 expert pathologists, independent from the local reading.
The independent validation cohort (step 3 of initial BIOMARGIN program) consisted of 391 consecutive samples collected according to the study protocol in the 4 centers between June 24 2014 and July 2 2015. In the validation cohort, no samples were excluded. There was no overlap in samples between the training and the validation cohort. All biopsies included in this study were reviewed and graded in a blinded fashion by the expert pathologists independent from the local reading. All biopsies were rescored semiquantitatively according to the updated Banff 2017 classification.3
Urine Proteome Analysis
We collected fresh urine samples in the morning before the biopsies were performed. No protease inhibitors were added to the samples, and the pH of the urine samples was measured but not adjusted. Urinary creatinine, hemoglobin, leukocytes, glucose, and protein content levels were checked locally using dipstick tests. Upon receipt of samples, urine creatinine and protein content were measured centrally at CHU Limoges using enzymatic assays on an Architect c8000 clinical chemistry analyzer (Abbott, Abbott Park, IL) No freeze-thaw cycles were allowed. In both the training and the validation cohorts, we performed untargeted proteomics using nano–reversed-phase liquid chromatography and shotgun mass spectrometry (Nano Acquity Ultra Performance LC system, Waters, Milford, MA, coupled to an LTQ Orbitrap Velos mass spectrometer, Thermo Scientific, Waltham, MA) (see Supplementary Methods). For protein identification, we searched against the human uniprot database using Proteome Discoverer software version 2.1(Thermo Scientific), including 2 search engines, Mascot and SEQUEST. A precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.5 Da were used. Trypsin was chosen as the cleavage enzyme and 2 missed cleavages were allowed. Carbamidomethylation was set as a fixed modification on cysteine and methionine oxidation, and serine, tyrosine, and threonine phosphorylations were set as variable modifications. We filtered the resulting peptide identification results using a false discovery rate <5% based on the target-decoy approach. The first ranked peptides were included. Protein quantification was based on the unique, fully digested and unmodified peptides, except for cysteine carbamidomethylation, which was allowed as a modification. Precursor peak areas of all peptides identified in the liquid chromatography–mass spectrometry runs were exported using Proteome Discoverer version 2.1 (Thermo Scientific). To avoid missing data from data-dependent acquisition of mass spectrometry–based proteomics,8 we extracted time-intensity chromatograms from high-resolution MS1 (parent ion spectra) data to quantify all peptides identified with high confidence.
Statistical Analysis
We normalized the proteome data by quantile normalization, starting from the peak intensities extracted from the MS1 data. We used the training cohort to build the model to discriminate patients with, versus those without, ABMR using analysis of variance on the unique peptides. The corresponding proteins were selected by a false discovery rate–corrected P value of 0.05 and at least a log2–fold difference of 0.6. We selected proteins based on their relative abundance in urine samples and subsequently 2 unique peptides per protein. This selection was based on uniqueness for targeted quantitative analysis and the quality of the chromatographic peaks (Gaussian shape, absence of fronting or tailing, peak width). The selected peptides were then used to build a linear support vector machine on the training cohort, with 100 times resampling internal cross-validation. A model was considered valid when adding an extra protein to the model did not increase the percentage of correctly classified samples. Finally, the support vector machines modeled on the training cohort were applied on the independent validation cohort. Receiver operating characteristic (ROC) curves were then generated to evaluate the diagnostic accuracy of the models in the validation cohort. Sensitivity, specificity, negative predictive value, and positive predictive value were determinted in both the training and validation cohorts. All analyses were performed using R Project software version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria).9,10
Results
Demographics
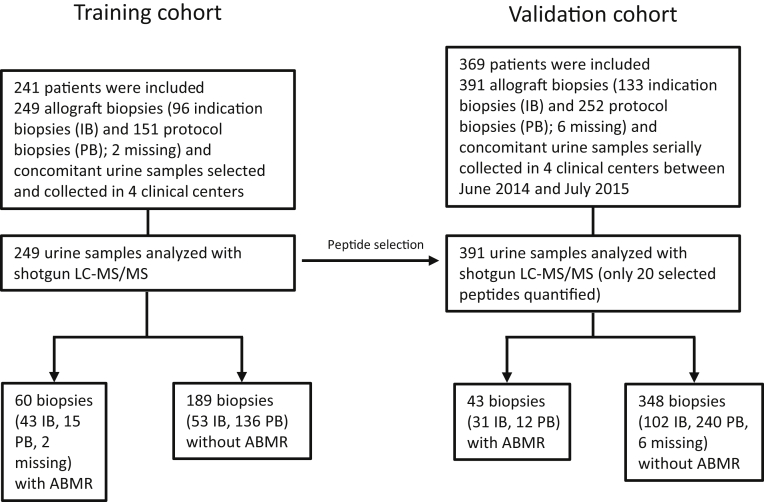
The clinical and histologic characteristics of the patients and samples included in the training and the validation cohorts are provided in Table 1. In the training cohort, 60 of 249 (24.1%) patients had ABMR compared with 43 of 391 (11.0%) patients in the validation cohort (Figure 1). There were differences in the baseline characteristics of patients with versus without ABMR. Patients with ABMR were younger at the time of transplantation, were more often female, and were more often diagnosed based on an indication biopsy than patients without ABMR, both in the training and in the validation cohorts (Table 1). Supplementary Table S1 provides the histologic characteristics of the biopsies included in the validation cohort (N = 391) according to rejection subtypes.
Table 1.
Demographics of the patients and biopsies included in the training and the validation phase
| Variable | Training phase (N = 249) |
Validation phase (N = 391) |
||||
|---|---|---|---|---|---|---|
| No antibody-mediated rejection (N = 189) | Antibody-mediated rejection (N =60) | P value | No antibody-mediated rejection (N = 348) | Antibody-mediated rejection (N = 43) | P value | |
| Transplant characteristics | ||||||
| Recipient age at transplantation, yr | 531 (50.07) ± 14.5 (14.8 – 78.8) | 48.9 (46.2) ± 15.1 (16.6 – 72.4) | 0.042 | 52.1 (50.9) ± 14.7 (2.7 – 78.4) | 45.5 (44.1) ± 17.8 (7.4 – 71.8) | 0027 |
| Recipient age at time of biopsy, yr | 55.0 (53.1) ± 14.0 (19.8 – 79.9) | 52.5 (50.9) ± 14.8 (23.2 – 76.0) | 0.31 | 54.1 (52.8) ± 14,3 (19.0 – 78.7) | 54.1 (51.5) ± 14.6 (19.9 – 79.6) | 0.61 |
| Recipient gender, male/female | 119/69 (63.3%/36.7%) | 30/27 (52.6%/47.4%) | 0.023 | 228/120 (65.5%/34.5%) | 17/26 (39.5%/60.5%) | 0.0013 |
| Repeat transplantation, yes/no | 24/165 (12.7%/87.3%,) | 18/42 (30%/70%) | 0.0029 | 63/285 (18.1%/81.9%) | 10/33 (23.3%/76.7%) | 0.41 |
| Recipient ethnicity | 0.28 | 1.00 | ||||
| European | 163 (87.2%) | 46 (70%) | 305 (88.4%) | 40 (93%) | ||
| Asian | 6 (3.2% | 2 (3.4%) | 3 (0.9%) | 0 (0%) | ||
| African | 6 (3.2%) | 3 (5.25) | 7 (2.0%) | 0 (9%) | ||
| Other | 12 (6.4%) | 7 (12.1%) | 30 (8.7%) | 3 (7.0%) | ||
| Donor age, yr | 52.5 (51.7) ± 16.0 (8–89) | 49 (47.4) ± 16.3 (14–75) | 0.13 | 53 (51.4) ± 14.9 (5–91) | 44 (41.9) ± 17.4 (7–75) | 0.0019 |
| Donor gender, male/female | 85/94 (47.5%/52.5%) | 18/35 (34%/66%) | 0.050 | 167/176 (48.7%/51.3%) | 24/16 (60%/40%) | 0.028 |
| Deceased/living donor | 151/38 (79.9%/20.1%) | 49/10 (83%/17%) | 0.27 | 263/83 (76%/24%) | 36/5 (87.8%/12.2%) | 0.018 |
| Heart-beating/non–heart-beating donor | 137/14 (90.7%/9.3%) | 48/1 (98%/2%) | 0.12 | 233/30 (88.6%/11.4%) | 34/2 (94.4%/5.6%) | 0.39 |
| Cold ischemia time, hr | 13.1 (13,0) ± 7.9 (0.3–37.2) | 14.4 (15.1) ± 8.6 (1.5–38.2) | 0.13 | 12.6 (12) ± 8.0 (0.3–35.8) | 13.1 (13.4) ± 7.0 (0.4–29) | 0.36 |
| Biopsy characteristics | ||||||
| Indication/protocol biopsy | 53/136 (28%/72%) | 43/15 (74.1%/25.9%) | <0.0001 | 102/240 (29.8%/70.2%) | 31/12 (72.1%/27.9%) | <0.0001 |
| Time after transplantation, d | 371 (866.0) ± 1,387.3 (5 – 10,063) | 992.5 (1771.7) ± 2258.4 (6 - 9435) | 0.018 | 335 (672.1) ± 1,265.9 (12–10,023) | 1132 (2693.3) ± 3198.7 (6–12,564) | <0.0001 |
| Biopsy time after transplantation | 0.65 | 0.001 | ||||
| <1 yr | 88 (46.6%) | 33 (56.9%) | 189 (55.3%) | 12 (27.9%) | ||
| >1 yr | 101 (53.4%) | 25 (43.1%) | 153 (44,7%) | 31 (72.1%) | ||
| Proteinuria, g/g creatinine | 0.1 (0.3) ± 0.6 (0.03–4.6) | 0.5 (1.2) ± 1.4 (0.04–6.6) | <0.0001 | 0.1 (0.3) ± 0.7 (0.02–7.3) | 0.7 (1.4) ± 1.8 (0.03–8.1) | <0.0001 |
| MDRD eGFR, ml/min per 1.73 m2a | 45.6 (47.8) ± 22.1 (5.4–140.8) | 34.5 (37.0) ± 19.3 (5.6–97.1) | 0.00062 | 44.4 (46.5) ± 18.7 (5.4–119.3) | 29.9 (37.8) ± 24.5 (7.7–110.8) | 0.00055 |
| Immunosuppression at time of biopsy | ||||||
| Cyclosporine, yes/no | 22/167 (11.6%/88.4%) | 8/50 (13.8%/86.2%) | 0.65 | 32/310 (9.4%/90.6%) | 6/37 (13.9%/86.1%) | 0.41 |
| Tacrolimus, yes/no | 152/37 (80.4%/19.6%) | 45/13 (77.6%/22.4%) | 0.71 | 297/45 (86.8%/13.2%) | 34/9 (79.1%/20.9%) | 0.17 |
| Mycophenolate, yes/no | 162/27 (85.7%/14.3%) | 53/5 (91.4%/8.6%) | 0.37 | 282/60 (82.5%/17.5%) | 38/5 (88.4%/11.6%) | 0.39 |
| Azathioprine, yes/no | 10/179 (5.3%/94.7%) | 0/58 (0%/100%) | 0.12 | 8/334 (2.3%/97.7%) | 1/42 (2.3%/97.7%) | 1.00 |
| mTOR inhibitor, yes/no | 7/182 (3.7%/96.3%) | 6/52 (10.3%/89.7%) | 0.084 | 44/298 (12.9%/87.1%) | 3/40 (7%/93%) | 0.33 |
| Corticosteroids, yes/no | 162/27 (85.7%/14.3%) | 55/3 (94.8%/5.2%) | 0.069 | 311/31 (91%/9%) | 38/5 (88.4%/11.6%) | 0.58 |
| Histological diagnosis | ||||||
| No rejection | 146 (77.2%) | 0 (0%) | NA | 332 (95.4%) | 0 (0%) | NA |
| T cell–mediated rejection | ||||||
| No | 146 (77.2%) | 35 (58.3%) | NA | 332 (95.4%) | 40 (93%) | NA |
| Borderline changes | 31 (16.4%) | 18 (12.1%) | NA | 13 (3.7%) | 2 (4.7%) | NA |
| Grade 1 or 2 | 12 (6.3%) | 7 (11.7%) | NA | 3 (0.9%) | 1 (2.3%) | NA |
| Antibody-mediated rejection | 0 (0%) | 60 (100%) | NA | 0 (0%) | 43 (100%) | NA |
| Mixed rejection | 0 (0%) | 25 (41.7%) | NA | 0 (0%) | 3 (7%) | NA |
| Interstitial fibrosis/tubular atrophy | ||||||
| Grade 0 | 92 (48.7%) | 31 (51.7%) | NA | 167 (50%) | 17 (39.5%) | NA |
| Grade 1 | 25 (132%) | 17 (28.3%) | NA | 87 (25%) | 6 (14%) | NA |
| Grade 2–3 | 71 (37.6%) | 12 (20%) | NA | 94 (27%) | 19 (44.2%) | NA |
| Polyomavirus-associated nephropathyb | 0 (0%) | 0 (0%) | NA | 14 (4.0%) | 0 (0%) | NA |
| De novo/recurrent glomerulonephritisb | 0 (0%) | 0 (0%) | NA | 20 (5.7%) | 6 (14%) | NA |
eGFR, estimated glomerular filtration rate; IFTA, interstitial fibrosis and tubular atrophy; MDRD, modification of diet in renal disease; NA, not applicable; mTOR, mammalian target of rapamycin;
Values are depicted as follows: median (mean) ± SD (minimum – maximum) or as absolute numbers (percentages). P values were calculated using the Mann-Whitney-Wilcoxon test (nonparametric comparisons) for continuous variables and Fisher exact test with 2-tailed P value for categorical variables.
The eGFR is calculated using the MDRD formula.
In the training cohort, N = 1 had missing data on IFTA grade. In the validation cohort, N = 1 had missing data on IFTA grade, N = 14 on polyomavirus-associated nephropathy, and N = 13 on glomerulonephritis.
Figure 1.
Study design. ABMR, antibody-mediated rejection; LC-MS/MS, liquid chromatography and shotgun mass spectrometry.
Biomarker Development in the Training Cohort
After extraction of the MS1 data (intact peptide precursor data) from the training cohort, we were able to quantify 1658 peptides, corresponding to 531 individual proteins. Ten proteins are Alpha-1 B glycoprotein (A1BG); afamin (AFM); apolipoprotein A1 (APOA1); apolipoprotein A4 (APOA4); Ig heavy constant α1 (IGHA1); Ig heavy constant γ4 (IGHA4); leucine rich α2 glycoprotein 1 (LRG1); alpha-1 antitrypsin (SERPINA1); antithrombin (SERPINC1); and transferrin (TF).
differed significantly between patients with versus without ABMR (Table 2). Two unique peptides per protein were then selected based on peptide peak intensity and shape. In addition, peptides were selected based on their use for targeted proteomics experiments. The final set of differentially expressed peptides used in the statistical model thus contained 10 proteins, represented by 20 unique peptides (Supplementary Table S2).
Table 2.
List of significantly upregulated proteins that segregated presence from absence of antibody-mediated rejection in the training cohort (N = 249)
| Gene identification | UniProt protein accession number | Total number of peptides identified | Total number of unique peptides identified | Median log2-fold change | FDR-corrected P value |
|---|---|---|---|---|---|
| A1BG | P04217 | 12 | 4 | 1.13 | 0.011 |
| AFM | P43652 | 14 | 14 | 1.00 | 0.0001 |
| APOA1 | P02647 | 16 | 3 | 0.61 | 0.045 |
| APOA4 | P06727 | 21 | 21 | 0.60 | 0.0001 |
| IGHA1 | P01876 | 12 | 4 | 0.87 | 0.00030 |
| IGHG4 | P01861 | 2 | 2 | 0.78 | 0.0076 |
| LRG1 | P02750 | 9 | 9 | 0.68 | <0.0001 |
| SERPINA1 | P01009 | 24 | 18 | 1.29 | <0.0001 |
| SERPINC1 | P01008 | 9 | 7 | 0,86 | 0.00022 |
| TF | P02787 | 53 | 31 | 1,37 | <0.0001 |
FDR, false discovery rate.
Training of the Statistical Model
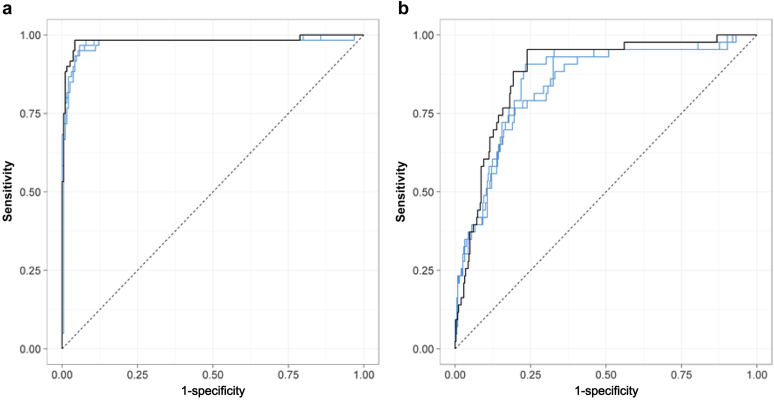
We next determined the minimal number of proteins needed in the model, using 100-times resampling cross-validation on the training cohort. This indicated that a minimum of 6 proteins was needed to obtain an accurate model for diagnosis of ABMR (Supplementary Figure S1). Next, a support vector machine was built on the 10 proteins (model 10) with ABMR as dependent variable. This model reached an AUC ROC of 0.98% (95% CI, 0.96– 1.00) (Figure 2). After fixing the cutoff point for optimal sensitivity and specificity, this model reached a sensitivity of 95% and a specificity of 96% for diagnosis of ABMR (Table 3). Depending on the cutoff point, the diagnostic performance of the model changes (Supplementary Table S3), so the treshold can be chosen in relation to the intended use of the diagnostic test. To make a selection between the many possible combinations of minimal sets of 6 proteins, 3 additional models were constructed, containining the most abundant proteins based on either sequence coverage (model 6A), number of peptide spectral matches (model 6B), or peptide peak intensity (model 6C) (Supplementary Table S4). These 3 models were then fixed and applied unaltered to the validation cohort.
Figure 2.
Diagnostic accuracy of the protein biomarker in (a) the training cohort (N = 249) and (b) the independent validation cohort (N = 391). Receiver operating characteristic curves are shown for the full model with 10 proteins (black line) and the 3 models with 6 proteins (blue line). The full model with 10 proteins reached an area under the curve of 0.98 (95% confidence interval [CI], 0.96—1.00) and 0.88 (95% CI, 0.83—0.93) in the training and validation cohorts, respectively.
Table 3.
Diagnostic accuracy of all 4 models for diagnosis of antibody-mediated rejection in the training (N = 249) and the validation cohorts (N = 391)
| Model name | AUC | TP | TN | FP | FN | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|---|
| Training cohort | |||||||||
| Model 10 | 0.98 | 57 | 182 | 7 | 3 | 0.95 | 0.96 | 0.89 | 0.98 |
| Model 6A | 0.98 | 57 | 179 | 10 | 3 | 0.95 | 0.95 | 0.85 | 0.98 |
| Model 6B | 0.98 | 57 | 178 | 11 | 3 | 0.95 | 0.94 | 0.84 | 0.98 |
| Model 6C | 0,97 | 57 | 178 | 11 | 3 | 0.95 | 0.94 | 0.84 | 0.98 |
| Validation cohort | |||||||||
| Model 10 | 0.88 | 41 | 263 | 85 | 2 | 0.95 | 0.76 | 0.33 | 0.99 |
| Model 6A | 0.84 | 36 | 243 | 105 | 7 | 0.84 | 0.70 | 0.26 | 0.97 |
| Model6B | 0.84 | 36 | 241 | 107 | 7 | 0.84 | 0.69 | 0.25 | 0.97 |
| Model 6C | 0.86 | 40 | 243 | 105 | 3 | 0.93 | 0.70 | 0.28 | 0.99 |
FN, false negatives; FP, false positives; NPV, negative predictive value; PPV, positive predictive value; TN, true negatives; TP, true positives; UC: area under the curve.
The individual proteins included in models 6A, 6B, and 6C are provided in Supplementary Table S2. Model 6A is based on sequence coverage, model 6B on the number of peptide spectral matches, and model 6C on peptide peak intensity.
Validation of the Statistical Model
The data acquisition for the assessment of the selected peptides was the same as for the untargeted discovery approach. After acquisition of the data, only the relevant data of the selected peptides were retrieved and analyzed. Thus, in the independent validation cohort (N = 391), only the 20 peptides from the 10 proteins selected in the training cohort were quantified to validate the statistical model developed on the training cohort.
The diagnostic accuracy of the full 10-protein model reached a diagnostic accuracy of 0.88 (95% CI, 0.83–0.93) (Figure 2), with a sensitivity of 95% and a specificity 76% for diagnosis of ABMR at the cutoff value defined in the training cohort (Table 3). Only 2 patients with ABMR were misclassified, which translated into a negative predictive value of 99%. There was no apparent trend in the diagnoses of the misclassified cases; misclassification occurred in all diagnostic categories (Supplementary Tables S5 and S6). The model also captured almost half of the patients with TCMR in the validation cohort (16/391) as ABMR, indicating that the model is not entirely reflecting processes that are specific for ABMR (Supplementary Table S6). This is also reflected by the fact that the model also picks up polyomavirus nephropathy and glomerulonephritis. The results for the 3 models containing 6 proteins yielded similar diagnostic performance (Table 3).
Sensitivity Analysis
The full 10-protein model retained good diagnostic accuracy for ABMR both at time of graft dysfunction (in indication biopsies) and at time of stable graft function (protocol biopsies), and both early (before 1 year) and late (after 1 year) after transplantation. When we adjust for time after transplantation (as a log10-transformed variable) in a multivariate model, the urinary marker remains significantly associated with diagnosis of ABMR (P < 0.0001). The 10-protein model also retained accuracy in subgroups of proteinuria (Table 4). The 10-protein model had better diagnostic accuracy than gross proteinuria as a marker of ABMR (Supplementary Figure S2). In a sensitivity analysis in patients with human leukocyte antigen donor-specific antibodies, the biomarker demonstrates an ROC AUC of 0.92 (95% CI, 0.84–1.00) for diagnosis of ABMR. In addition, in patients without human leukocyte antigen donor-specific antibodies, there was good diagnostic performance of the urinary marker for ABMR with an ROC AUC of 0.88 (95% CI, 0.83–0.93) (Supplementary Figure S3). Supplementary Table S7 provides an overview of the sensitivity analyses of the diagnostic accuracy of the urinary marker in the presence or absence of hematuria, leukocyturia, and bacteriuria. The urinary marker also has significant diagnostic performance in all 4 centers; the AUC for all 4 clinical centers was calculated, and all of them yielded good results with AUC of the ROC curves ranging from 85.1% to 94.6% (KU Leuven AUC, 85.1%; 95% CI, 72.2–94.1; Paris-Necker AUC, 91.1%; 95% CI, 84.1%–96.1; Limoges AUC, 94.6%; 95% CI, 83–100; Hannover 89.7%, 95% CI, 82.4–95.1).
Table 4.
Diagnostic accuracy of the 10-protein model (model 10) for noninvasive diagnosis of antibody-mediated rejection in the validation cohort (N = 391) in different subgroups, according to biopsy type, time after transplantation, and different levels of proteinuria
| Characteristic | Total | TP | TN | FP | FN | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|---|
| Biopsy type | |||||||||
| Protocol biopsy | 252 | 11 | 200 | 40 | 1 | 91.7 | 83.3 | 21.6 | 99.5 |
| Indication biopsy | 133 | 30 | 57 | 45 | 1 | 96.8 | 55.9 | 40 | 98.3 |
| Missing | 6 | 0 | 6 | 0 | 0 | NA | 100 | NA | 100 |
| Biopsy timing | |||||||||
| Early (<1 yr post-transplant) | 201 | 12 | 144 | 45 | 0 | 100 | 76.2 | 21.1 | 100 |
| Late (>1 yr post-transplant) | 184 | 29 | 113 | 40 | 2 | 93.5 | 73.9 | 42.0 | 98.3 |
| Missing | 6 | 0 | 6 | 0 | 0 | NA | 100 | NA | 100 |
| Proteinuria | |||||||||
| <0.3 g/g creatinine | 295 | 14 | 238 | 41 | 2 | 87.5 | 85.1 | 20.0 | 99 |
| 0.3-1 g/g creatinine | 49 | 9 | 14 | 26 | 0 | 100 | 35 | 25.7 | 100 |
| 1-3 g/g creatinine | 25 | 11 | 2 | 12 | 0 | 100 | 14.3 | 47.8 | 100 |
| >3 g/g creatinine | 10 | 6 | 0 | 4 | 0 | 100 | 0 | 60 | NA |
| Missing | 12 | 1 | 9 | 2 | 0 | 100 | 81.8 | 33.3 | 100 |
NA, not applicable; NPV, negative predictive value; PPV, positive predictive value; TN, true negatives; TP, true positives.
Correlation of the Biomarker Model With Histologic and Clinical Variables
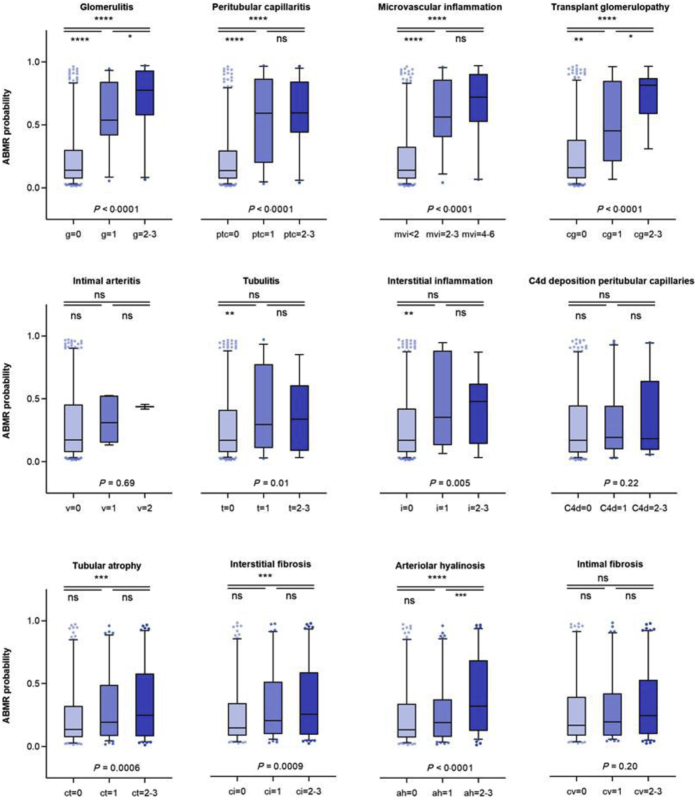
Figure 3 shows the probability of ABMR, as assessed by the 10-protein model, per histologic lesion grade in the validation cohort. The marker is significantly associated with lesions of ABMR (glomerulitis, peritubular capillaritis, microvascular inflammation score, transplant glomerulopathy, and intimal arteritis). Less significant associations were observed with lesions of TCMR (tubulitis, interstitial inflammation) and nonspecific chronic damage (arteriolar hyalinosis, interstitial fibrosis, and tubular atrophy). The model cannot be used to distinguish TCMR from no rejection samples; when comparing the urinary protein scores of the TCMR cases versus no rejection cases, no significant difference was noted (P = 0.11). The 3 ABMR-TCMR mixed cases seem to behave as ABMR cases (Supplementary Figure S4).
Figure 3.
Distribution of the probability of antibody-mediated rejection (ABMR) as assessed by the urinary protein marker per histologic lesion grade in the validation cohort (N = 385). The urinary protein marker score was significantly associated with lesions of antibody-mediated rejection (glomerulitis, peritubular capillaritis, microvascular inflammation score, transplant glomerulopathy, and intimal arteritis). Less significant associations were seen with lesions of T cell–mediated rejection (tubulitis, interstitial inflammation) and nonspecific chronic damage (arteriolar hyalinosis, interstitial fibrosis, and tubular atrophy). Significance was assessed with nonparametric 1-way analysis of variance and pairwise comparisons with t test. ah, arteriolar hyalinosis; C4d, C4d deposition in peritubular capillaries; cg, transplant glomerulopathy; ci, interstitial fibrosis; ct, tubular atrophy; cv, intimal fibrosis; g, glomerulitis; ptc, peritubular capillaritis; i, nterstitial inflammation; mvi, microvascular inflammation; ns, not significant; peritubular capillaritis; t, tubulitis; v, intimal arteritis. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
The fact that TCMR cannot be classified from any TCMR samples using this model is also shown in the ROC curve with an AUC value of 64.4% (95% CI, 51.3–77.3) (Supplementary Figure S5). When we compare only the pure cases of ABMR and TCMR, the model does seem to have some potential to discriminate the 2 groups (Supplementary Figure S4), with an ROC AUC of 71% (95% CI, 54.5–85.5).
Comparison With Traditional Clinical Biomarkers and Added Value of the Urinary Protein Biomarker Panel
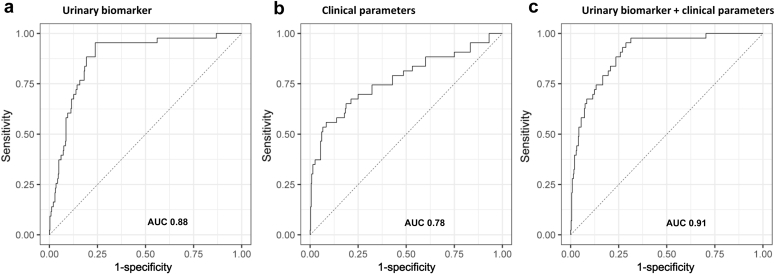
In another study of the BIOMARGIN group, a clinical model for diagnosis of ABMR was built consisting of 8 parameters (donor-specific antibodies, proteinuria, estimated glomerular filtration rate, time after transplantation, recipient age at time of transplantation, donor age, recipient sex, and protocol vs. indication biopsy).11 When this model is applied to the proteomics validation dataset and compared with the diagnostic accuracy of our urinary marker, the urinary biomarker model clearly outperforms the clinical model (AUC 0.88 vs. AUC 0.78). Figure 4 shows the ROC curves of the different models. After adding the urinary marker to the clinical model, the diagnostic accuracy increases significantly to an ROC AUC of 0.91. Also, the clinical model does not perform well for diagnosis of subclinical ABMR.
Figure 4.
(a) Receiver operating curve (ROC) curve of the urinary biomarker model including all 10 protein biomarkers. The area under the curve (AUC) value is 0.88 (AUC, 0.88; 95% confidence interval [CI], 0.83–0.93). (b) ROC curve of the clinical model including 8 clinical parameters. The AUC value is 0.78 (AUC, 0.78; 95% CI, 0.70–0.86). (c) ROC curve of the combined model including both the urinary biomarker and the clinical parameters. The AUC value is 0.91 (AUC, 0.91; 95% CI, 0.86–0.94).
Discussion
In this study, we propose a novel biomarker based on urinary proteomics for diagnosis of ABMR with high diagnostic performance and clinically useful test parameters. The urinary biomarker consists of 10 proteins, most of which have already been suggested as biomarkers in (native) kidney diseases, supporting their relevance. The model including these 10 proteins reaches a very high negative predictive value (99%), making it a clinically useful marker for excluding ABMR. The presented urinary protein biomarker performs well independent of gross proteinuria. In patients with undetectable gross proteinuria, this urinary marker still has good discriminative value for ABMR, whereas in patients with high proteinuria, reflecting a higher pretest probability of ABMR, the positive predictive value increases. In clinical practice, the combination of our urinary protein marker with other noninvasive markers for TCMR, as previously proposed, would be interesting to assess different graft injury processes simultaneously.
A major strength of this study is the validation of the biomarker in an independent cross-sectional cohort with actual disease prevalence. This independent validation is often lacking in biomarker discovery. Other urinary protein biomarkers for kidney allograft rejection have been reported.12,13 However, an important difference with these previous studies is that we analyzed all individual patient samples separately, eliminating major drawbacks inherent to the analysis of pooled samples, where results of the samples cannot be linked back to the individually identified pure phenotypes.
Another strength of this study is the supporting literature on the proteins involved in our biomarker. Many of these proteins have already been reported as potential biomarkers for renal dysfunction, mostly in native kidney diseases. α1-B glycoprotein,14 afamin15, apolipoprotein A1 and A4,16, 17, 18, 19, 20 leucine-rich α2-glycoprotein 1,21, 22, 23 α1-antitrypsin,24,25 antithrombin,26,27 transferrin,28,29 and Ig heavy chain α1 and γ4 are not specific for disease processes or histologic lesions and have been associated with a wide range of kidney diseases or kidney dysfunction. It could be that some of the proteins in the panel might represent general injury or injury mechanisms, rather than being specific for antibody-mediated injury. This notion is also suggested by the association of our 10-protein marker with polyoma-associated nephropathy and glomerulonephritis. Moreover, major injury patterns in TCMR and viral nephropathy are interstitial inflammation and tubulitis, also often present in ABMR. The same is true with glomerulonephritis. One key feature of ABMR is glomerulitis, and conversely, glomerulonephritis can also show glomerulitis. Nevertheless, despite the nonspecificity of the individual molecules, the combination of these proteins seems to be highly sensitive in diagnosing ABMR. Moreover, the proteins were not selected because of their biologic function, but solely on their statistical relevance. Thus, the statistical relevance of the individual proteins is most probably the result of the resulting phenotype, not the cause.
The high negative predictive value of our 10-protein marker for detecting ABMR could be of importance in clinical practice—for example, to decide in which patients invasive biopsies can be omitted. The relative low positive predictive value should be put in perspective of the low prevalence of ABMR, where a 20% to 30% risk of having ABMR based on a positive result with this noninvasive, inexpensive urinary protein marker warrants a more invasive approach for histologic proof through biopsy. As the false-positive cases with our biomarker consist mostly of other inflammatory intrarenal diseases such as polyomavirus nephropathy and glomerulonephritis, performing a biopsy in such cases still contributes to clinical decision-making. Depending on biopsy practice by center, a different threshold may be chosen. On the one hand, a high threshold leads to high specificity for ABMR and a high positive predictive value, which could be important in centers performing only indication biopsies for reason of graft dysfunction that want to avoid too many false-positive results. On the other hand, centers performing protocol biopsies could apply a lower threshold, resulting in a higher negative predictive value and the ability to rule out ABMR and avoid performing too many biopsies yielding negative results.
Our study has several limitations. First, using mass spectrometry–based proteomics, the minimum set of proteins to make a model seems to consist of at least 6 proteins. However, more accessible quantification of these proteins could be considered–for example, by translating the 10-protein test into a test based on enzyme-linked immunoassay. If the platform for the biomarker is changed to such a targeted technique, additional validation will be necessary. Second, the nonspecificity of the separate proteins in this urinary biomarker indicates that further invasive histopathologic evaluation is necessary in case of positive test findings. Third, based on previous literature, not all individual proteins in our panel are specific for ABMR. Testing this panel in cohorts at higher risk of, for example, recurrent glomerular disease, is warranted. Also, our validation cohort with real-life disease prevalence had very low incidence of TCMR, with a majority of our TCMR samples meeting only criteria for borderline changes. Although this reflects the natural disease prevalence in our clinical centers, this could differ from other clinical centers with different clinical practice and perhaps overestimate the discriminative performance of our marker for ABMR versus TCMR. Finally, the temporal evolution of the 10-protein marker and the response to treatment needs further evaluation in larger prospective studies with repeated sampling.
In conclusion, our study shows that (i) a protein marker set including 10 proteins is able to predict ABMR with high accuracy and (ii) this marker panel can be used in clinical practice. Because of its very high negative predictive value the test could be used to rule out many biopsies in clinical centers that perform protocol biopsies at regular time points. Clinical validation of a test based on enzyme-linked immunoassay will be needed to guarantee easy implementation of the marker in daily clinical practice.
Disclosure
All the authors declared no competing interests.
Acknowledgments
The BIOMARGIN study is funded by the Seventh Framework Programme (FP7) of the European Commission, in the HEALTH.2012.1.4-1 theme for innovative approaches to solid organ transplantation, with grant agreement 305499. MN is senior clinical investigator of The Research Foundation Flanders (FWO) (1844019N). EVL holds a fellowship grant (1143919N) from The Research Foundation Flanders (FWO). We thank the clinical centers of the BIOMARGIN consortium, the clinicians and surgeons, nursing staff ,and the patients. We thank Aline Schindelé and the team at Venn Life Sciences SAS (France) for their invaluable help with sample collection, annotation, shipment, and the eCRF (Electronic Case Report Form) database.
Footnotes
Supplementary Methods. Urinary sample collection and preparation and liquid chromatography and shotgun mass spectrometry proteomics.
Figure S1. Number of proteins needed in a model based on cross-validation in training cohort to obtain a good classification of phenotypes.
Figure S2. The unmodeled proteinuria data (in g/g creatinine) reached an area under the curve of 0.75 (95% CI, 0.67 to 0.83) for the training cohort (A) and 0.76 (95% CI, 0.66 to 0.85) for the validation cohort (B).
Figure S3. Receiver operating characteristic (ROC) curves are shown for the full model with 10 proteins for DSA-positive (left panel) and DSA-negative patients (right panel) using the validation dataset. For the-DSA positive ABMRs, the AUC value is 92.4% (95% CI, 84–100). For the DSA-negative ABMRs, the AUC is 88% (95% CI, 83–93).
Figure S4. Probability scores using the 10-protein model for ABMR, TCMR, no rejection (NR), and mixed ABMR-TCMR cases.
Figure S5. Receiver operating characteristic (ROC) curves are shown for the full model with 10 proteins for TCMR versus no TCMR (left panel) and pure ABMR versus pure TCMR cases (right panel) using the validation dataset. For TCMR versus no TCMR, the AUC value is 64.4% (95% CI, 51.3–77.3). For the comparison of pure cases of ABMR and TCMR, the AUC is 71% (95% CI, 54.5–85.5).
Table S1. Histologic characteristics of the biopsies included in the BIOMARGIN study in the validation phase (N = 391) according to rejection subtypes
Table S2. Selected list of peptides used for training the support vector machine model in the training cohort.
Table S3. Impact of the chosen treshold on the diagnostic performance of the model including all 10 proteins (model 10).
Table S4. Proteins included in the different statistical models. The models including 6 proteins were chosen based on their abundance in urine samples.
Table S5. Subclassification of the number of biopsies that were classified using the model compared to the biopsy result for the training cohort.
Table S6. Subclassification of the number of biopsies that were classified using the model compared to the biopsy result for the validation cohort.
Table S7. Overview of the sensitivity analyses of the diagnostic accuracy of the urinary marker in the presence or absence of hematuria, leukocyturia, and bacteriuria.
Supplementary Material
References
- 1.Nankivell B.J., Alexander S.I. Rejection of the kidney allograft. N Engl J Med. 2010;363:1451–1462. doi: 10.1056/NEJMra0902927. [DOI] [PubMed] [Google Scholar]
- 2.Loupy A., Lefaucheur C. Antibody-mediated rejection of solid-organ allografts. N Engl J Med. 2018;379:1150–1160. doi: 10.1056/NEJMra1802677. [DOI] [PubMed] [Google Scholar]
- 3.Haas M., Loupy A., Lefaucheur C. The BANFF 2017 kidney meeting report: revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18:293–307. doi: 10.1111/ajt.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solez K., Axelsen R.A., Benediktsson H. International standardization of criteria for the histologic diagnosis of renal allograft rejection: the Banff working classification of kidney transplant pathology. Kidney Int. 1993;44:411–422. doi: 10.1038/ki.1993.259. [DOI] [PubMed] [Google Scholar]
- 5.Loupy A., Vernerey D., Tinel C. Subclinical rejection phenotypes at 1 year post-transplant and outcome of kidney allografts. J Am Soc Nephrol. 2015;26:1721–1731. doi: 10.1681/ASN.2014040399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lees J.S., McQuarrie E.P., Mordi N. Risk factors for bleeding complications after nephrologist-performed native renal biopsy. Clin Kidney J. 2017;10:573–577. doi: 10.1093/ckj/sfx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anglicheau D., Naesens M., Essig M. Establishing biomarkers in transplant medicine: a critical review of current approaches. Transplantation. 2016;100:2024–2038. doi: 10.1097/TP.0000000000001321. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B., Kall L., Zubarev R.A. DeMix-Q: quantification-centered data processing workflow. Mol Cell Proteomics. 2016;15:1467–1478. doi: 10.1074/mcp.O115.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robin X., Turck N., Hainard A. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R Core Team . Foundation for Statistical Computing; Vienna: 2013. R: a language and environment for statistical computing. [Google Scholar]
- 11.Van Loon E., Gazut S., Yazdani S. Development and validation of a peripheral blood mRNA assay for the assessment of antibody-mediated kidney allograft rejection: a multicentre, prospective study. EBioMedicine. 2019;46:463–472. doi: 10.1016/j.ebiom.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sigdel T.K., Salomonis N., Nicora C.D. The identification of novel potential injury mechanisms and candidate biomarkers in renal allograft rejection by quantitative proteomics. Mol Cell Proteomics. 2014;13:621–631. doi: 10.1074/mcp.M113.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigdel T.K., Gao Y., He J. Mining the human urine proteome for monitoring renal transplant injury. Kidney Int. 2016;89:1244–1252. doi: 10.1016/j.kint.2015.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piyaphanee N., Ma Q., Kremen O. Discovery and initial validation of α1-B glycoprotein fragmentation as a differential urinary biomarker in pediatric steroid-resistant nephrotic syndrome. Proteomics Clin Appl. 2011;5:334–342. doi: 10.1002/prca.201000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang L., Duan N., Xu D. Urine afamin and afamin-creatinine ratio as biomarkers for kidney injury. Biomark Med. 2018;12:1241–1249. doi: 10.2217/bmm-2018-0126. [DOI] [PubMed] [Google Scholar]
- 16.Prikryl P., Vojtova L., Maixnerova D. Proteomic approach for identification of IgA nephropathy-related biomarkers in urine. Physiol Res. 2017;66:621–632. doi: 10.33549/physiolres.933380. [DOI] [PubMed] [Google Scholar]
- 17.Puig-Gay N., Jacobs-Cacha C., Sellares J. Apolipoprotein A-Ib as a biomarker of focal segmental glomerulosclerosis recurrence after kidney transplantation: diagnostic performance and assessment of its prognostic value—a multi-centre cohort study. Transpl Int. 2019;32:313–322. doi: 10.1111/tri.13372. [DOI] [PubMed] [Google Scholar]
- 18.Kronenberg F. Apolipoprotein L1 and apolipoprotein A-IV and their association with kidney function. Curr Opin Lipidol. 2017;28:39–45. doi: 10.1097/MOL.0000000000000371. [DOI] [PubMed] [Google Scholar]
- 19.Goek O.N., Kottgen A., Hoogeveen R.C. Association of apolipoprotein A1 and B with kidney function and chronic kidney disease in two multiethnic population samples. Nephrol Dial Transplant. 2012;27:2839–2847. doi: 10.1093/ndt/gfr795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stangl S., Kollerits B., Lamina C. Association between apolipoprotein A-IV concentrations and chronic kidney disease in two large population-based cohorts: results from the KORA studies. J Intern Med. 2015;278:410–423. doi: 10.1111/joim.12380. [DOI] [PubMed] [Google Scholar]
- 21.Lee H., Fujimoto M., Ohkawara T. Leucine rich α-2 glycoprotein is a potential urinary biomarker for renal tubular injury. Biochem Biophys Res Commun. 2018;498:1045–1051. doi: 10.1016/j.bbrc.2018.03.111. [DOI] [PubMed] [Google Scholar]
- 22.Liu J.J., Pek S.L.T., Ang K. Plasma leucine-rich α-2-glycoprotein 1 predicts rapid eGFR decline and albuminuria progression in type 2 diabetes mellitus. J Clin Endocrinol. Metab. 2017;102:3683–3691. doi: 10.1210/jc.2017-00930. [DOI] [PubMed] [Google Scholar]
- 23.Fu J., Wei C., Zhang W. Gene expression profiles of glomerular endothelial cells support their role in the glomerulopathy of diabetic mice. Kidney Int. 2018;94:326–345. doi: 10.1016/j.kint.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzger J., Kirsch T., Schiffer E. Urinary excretion of twenty peptides forms an early and accurate diagnostic pattern of acute kidney injury. Kidney Int. 2010;78:1252–1262. doi: 10.1038/ki.2010.322. [DOI] [PubMed] [Google Scholar]
- 25.Choi Y.W., Kim Y.G., Song M.Y. Potential urine proteomics biomarkers for primary nephrotic syndrome. Clin Proteomics. 2017;14:18. doi: 10.1186/s12014-017-9153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Z., Wang F., Liang M. SerpinC1/antithrombin iii in kidney-related diseases. Clin Sci (Lond) 2017;131:823–831. doi: 10.1042/CS20160669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F., Zhang G., Lu Z. Antithrombin III/SerpinC1 insufficiency exacerbates renal ischemia/reperfusion injury. Kidney Int. 2015;88:796–803. doi: 10.1038/ki.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunner H.I., Gulati G., Klein-Gitelman M.S. Urine biomarkers of chronic kidney damage and renal functional decline in childhood-onset systemic lupus erythematosus. Pediatr Nephrol. 2019;34:117–128. doi: 10.1007/s00467-018-4049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Źyłka A., Dumnicka P., Kuśnierz-Cabala B. Markers of glomerular and tubular damage in the early stage of kidney disease in type 2 diabetic patients. Mediators Inflamm. 2018:7659243. doi: 10.1155/2018/7659243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.